Abstract
Despite global eradication efforts over the past century, malaria remains a devastating public health burden, causing almost half a million deaths annually (WHO, 2016). A detailed understanding of the mechanisms that control malaria infection has been hindered by technical challenges of studying a complex parasite life cycle in multiple hosts. While many interventions targeting the parasite have been implemented, the complex biology of Plasmodium poses a major challenge, and must be addressed to enable eradication. New approaches for elucidating key host-parasite interactions, and predicting how the parasite will respond in a variety of biological settings, could dramatically enhance the efficacy and longevity of intervention strategies. The field of systems biology has developed methodologies and principles that are well poised to meet these challenges. In this review, we focus our attention on the Liver Stage of the Plasmodium lifecycle and issue a “call to arms” for using systems biology approaches to forge a new era in malaria research. These approaches will reveal insights into the complex interplay between host and pathogen, and could ultimately lead to novel intervention strategies that contribute to malaria eradication.
Keywords: malaria, plasmodium, liver, systems biology, computational modeling, omics-technologies
Introduction
Parasitic diseases infect over half a billion people worldwide, and are a tremendous public health burden. Malaria is the most lethal, causing infection and death primarily in young children in sub Saharan Africa (WHO, 2016). In humans, five Plasmodium species are known to cause disease, with the greatest burden arising from infection with P. falciparum and P. vivax. Despite multifaceted control efforts, the adaptive nature of the Plasmodium parasite has confounded vaccine development (Neafsey et al., 2015; Schats et al., 2015), and has contributed to the emergence of widespread drug resistance (reviewed in Blasco et al., 2017).
The life cycle of Plasmodium is complex. The parasite cycles between mosquito and mammalian hosts, with elaborate developmental and differentiation processes within each. Every transition represents an opportunity to arrest the parasite, and to stop subsequent life cycle progression. A systematic approach that identifies key components required by the parasite at each stage of its life cycle could ultimately elucidate fundamental pathogenesis strategies, which will aid the development of cohesive intervention approaches. By contrast, any approach that reduces the biology of the parasite to a single antigen or drug target leaves open the possibility of parasite adaptation and, ultimately, intervention failure. Here, we propose a systems biology approach to interrogate the Plasmodium parasite that, although not without its challenges, will result in a global view of the host-parasite interactions during key transition states in the life cycle. This view could inform interventions that are not easily circumvented by the parasite and therefore contribute to malaria eradication.
Plasmodium parasites have a complex life cycle that engages multiple host environments
Plasmodium infection of mammals begins with injection of the sporozoite into the skin of the vertebrate host during the bite of a female Anopheles mosquito. After migration through the skin and entrance into a capillary, sporozoites travel through the blood stream to the liver. The parasite then traverses through the sinusoidal barrier to gain access to hepatocytes (Mota et al., 2001; Ishino et al., 2004; Tavares et al., 2013; Cha et al., 2016; Yang et al., 2017). Once within the liver parenchyma, sporozoites infect a host hepatocyte within which they will reside for the next 2–10 days (reviewed in Kaushansky and Kappe, 2015b; Vaughan and Kappe, 2017). Following liver stage development, parasites exit the liver, re-enter the blood stream and infect erythrocytes. During asexual blood stage infection, parasites undergo cycles of replication, followed by destruction of the host cell. It is this cycle that causes disease symptoms.
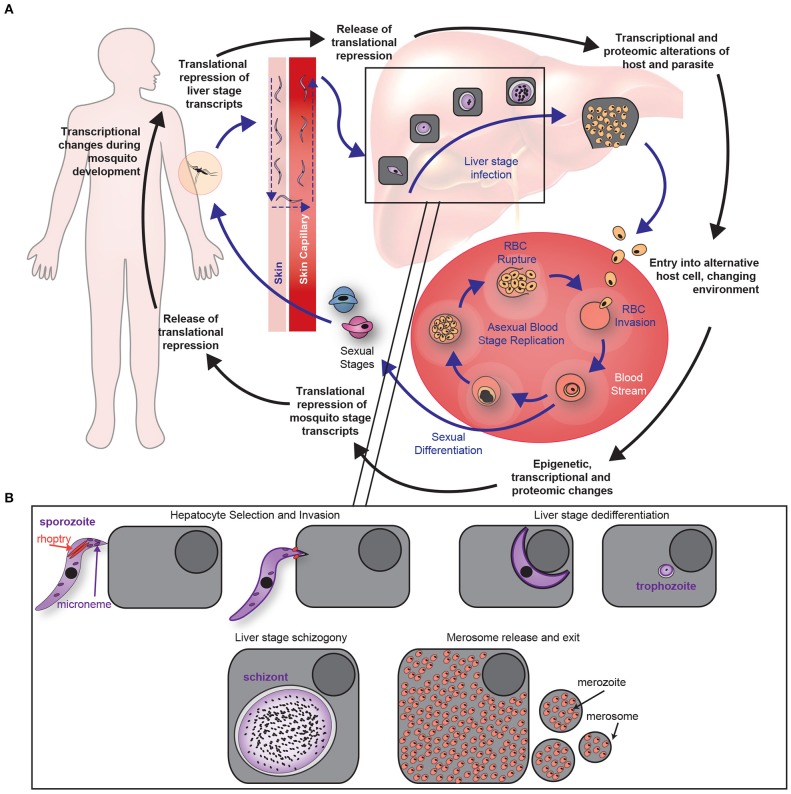
During the blood stage, a portion of parasites commit to sexual development (Coleman et al., 2014; Kafsack et al., 2014; Sinha et al., 2014; Poran et al., 2017) and initiate a differentiation process that occurs largely in the bone marrow (Joice et al., 2014). Once female and male forms have nearly completed maturation, they re-enter the blood stream and are transmitted to mosquitoes. In the mosquito midgut, fertilization occurs, generating a motile diploid (ookinete), which then replicates its DNA and develops into a stationary oocyst. Sporozoites then form within the midgut oocyst, become motile, and travel to the salivary glands. Once within the salivary glands, the parasite is transmitted to the next mammalian host during a blood meal. Each of these stage transitions is initiated by, and induces, broad, systematic changes that alter cellular behaviors (Table 1, Figure 1). Yet, these changes cannot be fully represented by any single transcript or individual cellular measurement. Rather, comprehensive changes within interconnected networks occur on multiple scales. This includes changes in gene regulatory networks, protein interactions with other biomolecules, and morphological variation of host and parasite subcellular structures. Together, these changes drive stage transitions. The goal must therefore be to establish a comprehensive picture of the host and parasite effector molecules and networks that are required to facilitate life cycle transitions.
Table 1.
Stage transitions in the Plasmodium life cycle.
| Life cycle stage transition | System-level alteration reported | References |
|---|---|---|
| Development from midgut sporozoite to salivary gland sporozoite | Transcriptome changes | Matuschewski et al., 2002; Mikolajczak et al., 2008 |
| Transmission between mosquito and mammalian host | Translational repression | Zhang et al., 2010; Gomes-Santos et al., 2011; Muller et al., 2011; Lindner et al., 2013; Silvie et al., 2014; Silva et al., 2016 |
| Development through Liver Stage | Transcriptome and proteome changes | Tarun et al., 2008; Albuquerque et al., 2009; Vaughan et al., 2009 |
| Exit from Liver Stage and Entry into Blood Stage | Transcriptome changes | Tarun et al., 2008 |
| Differentiation into sexual forms | Epigenetic and Transcriptome changes | Coleman et al., 2014; Kafsack et al., 2014; Sinha et al., 2014; Poran et al., 2017 |
| Transmission from mammalian to mosquito host | Translational repression | Mair et al., 2006; Guerreiro et al., 2014; Lasonder et al., 2016 |
| Gametocyte to gamete transformation | Proteome changes | Khan et al., 2005 |
Figure 1.
Plasmodium life cycle. (A) Each stage of the malaria life cycle is accompanied by unique transcriptional or translational changes, which ultimately allow for successful transition to each stage of the life cycle. Red Blood Cell is abbreviated “RBC.” (B) Liver stage infection of a hepatocyte is a unique microenvironment that allows the parasite to invad and differentiate into several forms to ensure growth, replication, and eventual egress from the hepatocyte. These key transitions occur in specific subcellular locations during liver stage infection.
Plasmodium parasites significantly alter the biology of their hosts
To illustrate the need to comprehensively evaluate changes during the Plasmodium life cycle, we will consider one stage of the complex life cycle of the parasite in detail—the Liver Stage of infection. Once within the liver sinusoid, the parasite traverses through phagocytic Kupffer cells, liver-resident macrophages, and liver endothelial sinusoidal cells, to access hepatocytes, while avoiding phagocytosis (Mota et al., 2001; Ishino et al., 2004; Usynin et al., 2007; Tavares et al., 2013; Cha et al., 2016; Yang et al., 2017). Once in the liver parenchyma, the parasite continues to traverse through several hepatocytes before selecting a suitable host for invasion. While the precise properties that make one hepatocyte more hospitable than another remain unknown, altered levels of specific hepatocyte receptors dramatically alter infection rates (Silvie et al., 2003; Ishino et al., 2004; Rodrigues et al., 2008; Yalaoui et al., 2008a; Kaushansky et al., 2015).
Following establishment of an intracellular niche within the hepatocyte, Plasmodium replicates extensively, stretching the hepatocyte to 50–100 times its normal volume (Shortt and Garnham, 1948; Vaughan et al., 2012). This rapid expansion is surprising, given the cell's strict cell size regulations under normal conditions (Sinturel et al., 2017). This observation suggests that Plasmodium effectively overwrites the hepatocyte's hardwiring to exert massive influence over the host cell. Plasmodium likely disrupts a multitude of classical signaling pathways during infection, only a small fraction of which have been described (Kaushansky et al., 2013a,b; Ruivo et al., 2016). Interrogating single proteins in a pathway to determine functionality is limiting in this context, and ignores secondary effects within the complex cell system. Instead, it is critical to comprehensively and quantitatively evaluate changes that occur during infection to illuminate mechanisms of control employed by the parasite.
What can systems biology do for malaria?
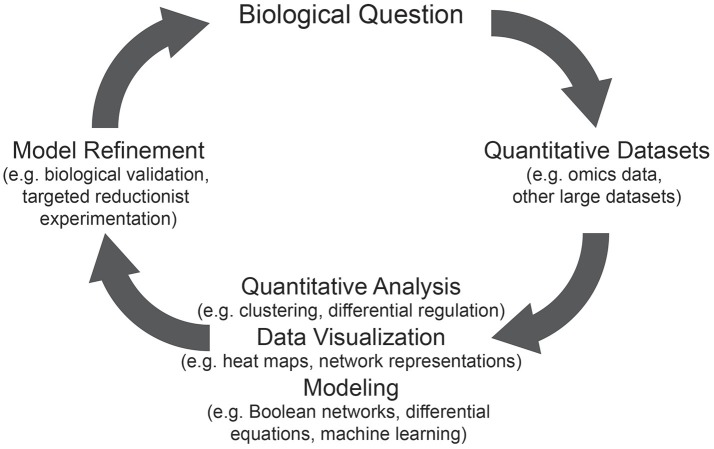
Understanding biology is a systems-level problem. Interactions between components of a system lead to the emergence of properties that cannot be understood from the study of the components individually. The study of systems biology is predicated on two basic assumptions. First, that the whole is far greater than the sum of its parts; and second, that a more comprehensive understanding of the components and their relationships within a system will allow for more accurate predictions of the system's behavior. It is through this lens that systems biology aims to determine the relationships and interactions of the components of a system. In practice, systems biology is a set of principles and processes by which we take complex systems apart and put them back together, with the aim of understanding the properties of the entire biological system. The approach generally starts with the systematic and comprehensive identification and quantification of molecules, called omics datasets, as a biological system transitions from one state to another. Initially, these data are evaluated using standard statistical tools, resulting in ordered lists, with significance values for each observed difference. These data provide the basis for the deployment of simple tools such as pathway analysis and clustering to interpret the data, or more complex analysis, such as regression or inference methods, to suggest causal or correlative relationships between components. These approaches can, and have, identified major molecular players at each stage, but fall short of a detailed and comprehensive understanding. Visualizing the data is also important for generating insights and predictive models that describe key determinants of the stage transition being interrogated (Figure 2). Predictions that are generated are then tested, often using “classic” or “reductionist” approaches. This process results in the refinement of both the model, and of our biological understanding. Molecular details are important, and systems biology must not ignore them. Examining the individual components of a system allows us to understand their molecular and physicochemical properties, as well as the function of the components in context of the entire system (Van Regenmortel, 2004). As the data that informs a model becomes more detailed, the predictions generated become more mechanistic. This level of insight is critical for rational intervention.
Figure 2.
The processes involved in generating systems biology-informed models. To inform the biological question, quantitative datasets are generated, which are then used in quantitative analysis, data visualization, and modeling to describe how the system behaves. These tools can be used interchangeably and/or in succession before further refining the model. Refinement of the model can then provide new biological insights.
Modeling is not unique to systems biology. Indeed, all scientists generate “models,” sometimes in the form of cartoons to aid in the design of the next line of inquiry. Systems biology models are often in the form of networks composed of balls and sticks, where balls (also called nodes) represent genes or proteins, and the sticks (or lines, also called edges) between them represent a relationship between molecular players. These simple visualizations can themselves facilitate the development of novel hypotheses. Many representations allow scientists to superimpose multiple types of data onto these networks (for example, molecule types, confidence of the interactions or subcellular organization). There are many popular and facile tools for these network visualizations (reviewed in Gehlenborg et al., 2010; Pavlopoulos et al., 2017). Once established, these networks can be mined to design subsequent experiments, and also used as a foundation for more complex models of the dynamics of molecular interactions and information flow. Depending on the complexity and the question to be addressed, models can take many forms. Some widely used approaches include Boolean networks, ordinary differential equations, and stochastic simulations. Different model classes involve different approximations, assumptions and levels of granularity. All models are best informed by quantitative, high quality, and biochemical data. While omics approaches can contribute to a more comprehensive view than possible with classical biochemical approaches, many models also incorporate data from rigorous reductionist approaches. Regardless of model class, the power of modeling lies in its capacity to capture insights that are difficult to reach through intuition alone.
Many of the technological and computational tools of systems biology are modular, and the resulting data can be integrated in different ways to inform the biological question (Danziger et al., 2014). Indeed, modeling biological data increasingly aims to incorporate a range of types of information, which monitor changes at different scales. This allows the researcher to determine what types of data are most informative when predicting a biological outcome of interest (Hwang et al., 2005a,b; Janes et al., 2005; Bonneau et al., 2007; AlQuraishi et al., 2014) and design subsequent experiments accordingly. Nevertheless, in most applications, applying computational analysis to quantitative datasets enables predictions (or new hypotheses) about how a perturbation, such as a gene deletion, drug treatment, or new environment, will influence the system as a whole.
The role of quantitative and comprehensive datasets in malaria research
In the case of malaria, numerous studies have generated omics data during life cycle transitions (Table 1). These include the cataloging of genes (genomics), mRNA transcripts (transcriptomics), translated protein (proteomics), metabolites (metabolomics), and translational repression/de-repression of transcripts as the parasite transitions through its life cycle stages. The genome of the Plasmodium parasite was initially published in 2002 (Gardner et al., 2002), and has been refined since. Initial transcriptomes and proteomes of P. yoelii and P. berghei Liver Stages have been generated, which have provided lists of the components involved in liver stage development, and further revealed the requirement of fatty acid synthesis from both the parasite and host during liver infection (Tarun et al., 2008; Albuquerque et al., 2009; Vaughan et al., 2009).
Insights that originate from transcriptomic analysis of Liver Stage infection reveal that Type I interferons and ER stress are systematically upregulated during liver stage infection and can modulate the level of liver stage infection (Liehl et al., 2014; Miller et al., 2014; Inacio et al., 2015; Kaushansky and Kappe, 2015a). Additional information can be obtained by monitoring changes in the parasite and host during infection under different environmental conditions. These, and related datasets can inform models that predict causality and cellular outcomes. The goal of this effort would be to identify networks of parasite and/or host factors that facilitate the development or demise of the parasite during its infection of the liver.
Protein-protein interactions
A major goal of host-pathogen studies is to elucidate specific interactions that dictate success or failure of the pathogen. While transcriptomics and proteomics can catalog changes that occur in the infected cell, a list of alterations alone does not provide mechanistic insight, and is unsatisfying to most cell biologists. Databases of protein interactions in many organisms, including humans, are becoming highly populated (Hein et al., 2015; Huttlin et al., 2015). Yet, many immunopurification—mass-spectrometry (IP-MS) based approaches to study protein-protein interactions do not meet the standard of quantitative and comprehensive. As datasets become larger, statistical tools can be used to predict which interactions are more likely to be specific, compared to commonly identified (abundant or promiscuous) proteins (Mellacheruvu et al., 2013). The pitfalls of qualitative and low throughput data have been partially overcome in model organisms such as yeast, where whole-genome GFP tag libraries have been generated and used in IP-MS experiments, although even these datasets remain incomplete, and are error prone (Ghaemmaghami et al., 2003; Huh et al., 2003; Mellacheruvu et al., 2013).
More sophisticated approaches designed to distinguish between bona fide and spurious interactions are being developed and applied. For example, Isotopic Differentiation of Interactions as Random or Targeted (I-DIRT) and variants (Tackett et al., 2005; Selbach and Mann, 2006; Trinkle-Mulcahy et al., 2008; Byrum et al., 2012; Trinkle-Mulcahy, 2012) exploit isotopic labeling and immunopurification to distinguish between interactions that occur before cell lysis, from those interactions that are introduced during the purification process. While these approaches improve confidence in interactions, they are neither widely adopted, nor have they been applied in genome scale studies.
The role of imaging in defining quantitative stage transitions
Since the initial discovery of liver stage parasites by microscopy in 1948 (Shortt and Garnham, 1948), imaging has been an invaluable tool of malaria research. However, most common imaging methods are neither quantitative nor comprehensive, limiting their capacity to inform modeling approaches. This is particularly troubling for applying a systems biology approach, as cellular outcomes are what we aim to predict, but are often poorly defined. Traditional imaging also falls short of reaching the temporal resolution necessary to elucidate the dynamic cellular processes during invasion and throughout liver stage infection.
A number of new imaging modalities enhance our ability to increase resolution, quantification and throughput. A comprehensive review of the advances made in increasing throughput, quantification and resolution in the imaging field is outside the scope of this review, we will highlight some examples that are particularly relevant to malaria research. One example, correlated light microscopy and electron microscopy (CLEM) combines fluorescence microscopy with electron microscopy, thereby increasing the throughput of monitoring rare events like liver stage infection at EM-level resolution (van Rijnsoever et al., 2008), and has already been applied to monitor liver stage development (Grutzke et al., 2014). Intravital imaging (IVM) has been adapted for malaria research and facilitates analysis of live tissue with microscopic resolution to reveal cellular responses that closely mimic in vivo infection, both spatially and temporally (Pittet and Weissleder, 2011; De Niz et al., 2017). Additional instrumentation, such as the Lattice Light Sheet Microscope (Betzig et al., 2006), enhances temporal and spatial resolution, with applications in both in vivo and in vitro systems, which could enable a more quantitative assessment of cellular outcomes.
The power of new genetic tools and screens in determining function
An essential component of systems biology is the experimental testing of predictions made by modeling efforts. This testing is greatly assisted by the capacity to perform genetic perturbations. Indeed, one of the major shortfalls of employing systems biology is that testing predictions is largely performed by single candidate-based approaches, and thus often fails to recapitulate the complexity of the system. In many cases, it remains difficult to determine if the model is incorrect, or if reductionist approaches cannot fully capture the emergent properties associated with a complex system. New genome-editing approaches, like CRISPR/Cas, can assess multiple perturbations in combination, in both mammalian and parasite genomes, which will facilitate testing more complex models (Cong et al., 2013; Mali et al., 2013; Ghorbal et al., 2014; Wagner et al., 2014; Lu et al., 2016).
In addition to evaluating individual or groups of gene candidates for function, new genome-editing approaches also have the ability to globally evaluate both host and parasite genes. Whole genome CRISPR/Cas9 knockout screens are now common in mammalian cells (Cong et al., 2013; Mali et al., 2013) and can be adapted to the Plasmodium genome. The Plasmodium Genetic Modification Project (PlasmoGEM), a new community resource from the Wellcome Trust Sanger Institute, aims to produce new tools for the genetic modification of malaria parasites at genome scale. This resource has already demonstrated that two-thirds of P. berghei genes contribute to normal blood stage development (Bushell et al., 2017). Subsequent studies should not only focus on the role of parasite genes in other life cycle stages, but also interrogate the role of host genes during each stage of parasite development.
Key questions and findings in malaria liver stage biology
The existing literature provides a basis upon which global experiments can be designed and modeled, and also highlights the most critical questions that remain. Given the potential and increasing power of systems biology, the challenge lies in how to use this approach to bolster the rich collection of findings that have been amassed by the Plasmodium research community, and address hurdles that have been unattainable by more traditional approaches. In this next section, we focus on some of the key findings on liver stage malaria with an emphasis on questions that remain.
Hepatocyte invasion
During hepatocyte invasion, the parasite attaches to the host cell, at least partially through circumsporozoite protein (CSP), which interacts directly with highly sulfated proteoglycans (HSPGs) on the cell surface to trigger CSP cleavage, inducing the sporozoite to switch to an invasive state (Table 2A) (Coppi et al., 2007, 2011). Thrombospondin-related anonymous protein (TRAP) is also involved in this process (Kappe et al., 1999; Matuschewski et al., 2002; Morahan et al., 2009). Additionally, Plasmodium proteins P36 and P52 play a role in invasion, parasitophorous vacuole membrane (PVM) formation, and protecting the host against apoptosis (Ishino et al., 2005; van Dijk et al., 2005; Ploemen et al., 2012). How each of these factors works in concert to facilitate productive invasion of the hepatocyte remains unknown.
Table 2.
Determinants of hepatocyte liver stage infection: (A) Plasmodium determinants of infection and (B) Host determinants of infection.
| Host/Parasite factor | Stage of infection | Main findings | References |
|---|---|---|---|
| (A) | |||
| SPECT | Traversal | Essential for cell traversal | Ishino et al., 2004, 2005 |
| PLP1 (SPECT2) | Essential for cell traversal | Ishino et al., 2004, 2005 | |
| CelTOS | Hypothesized to play a role in the exit step of traversal | Kariu et al., 2006 | |
| TRAP-like protein (TLP) | TLP-deficient sporozoites show a diminished ability to traverse | Moreira et al., 2008 | |
| PL (UIS10) | Hepatocyte Invasion | PL-deficient sporozoites show reduction in Liver Stage burden | Bhanot et al., 2005 |
| Circumsporozoite protein (CSP) | Multiple roles in motility and invasion, including transition from traversing state to invasive state | Coppi et al., 2007 | |
| P36 | Contributes to PVM formation | Ishino et al., 2005; Labaied et al., 2007 | |
| P52/P36p | Contributes to PVM formation | Ishino et al., 2005; Labaied et al., 2007 | |
| Cysteine proteases | Inhibition of sporozoite cysteine proteases completely inhibits infectivity | Coppi et al., 2005 | |
| Calcium Dependent Protein Kinase-6 (CDPK-6) | Sporozoites from CDPK-6-deficient parasites show decrease in invasion and CSP cleavage | Coppi et al., 2007 | |
| TRAP | Direct role in invasion through attachment with cytoplasmic tail | Kappe et al., 1999; Matuschewski et al., 2002; Morahan et al., 2009 | |
| Upregulated in Sporozoite 4 (UIS4) | Liver stage Development | UIS4-deficient P. berghei parasites severely impaired in Liver Stage development | Mueller et al., 2005 |
| Upregulated in Sporozoite (UIS3) | UIS3-deficient parasites severely impaired in Liver Stage development. UIS3 has been hypothesized to play a role in fatty acid uptake | Mikolajczak et al., 2007 | |
| EXP1 | Interacts with host Apolipoprotein H to promote liver stage development | Sa et al., 2017 | |
| LISP2 | Hypothesized to be involved in merozoite formation and exported to host cytosol | Orito et al., 2013 | |
| B9 | P9 mutants show liver stage growth arrest | Annoura et al., 2014 | |
| Sequestrin | Mutants lacking sequestrin show a reduction in liver stage development | Annoura et al., 2014 | |
| MSP1 | Conditional mutagenesis of MSP1 in sporozoites impaired merozoite formation | Combe et al., 2009 | |
| LISP1 | Hepatocyte Exit | In P. berghei, LISP1 is required for lysis of the PVM prior to egress | Ishino et al., 2009 |
| SUB1 | SUB1-deficient P. berghei parasites fail to rupture the PVM prior to egress | Tawk et al., 2013 | |
| (B) | |||
| CD68 | Traversal | Putative receptor of Kupffer cells, gateway for liver stage infection | Cha et al., 2016 |
| Hepatocyte Growth Factor | Hepatocyte Invasion | Secretion of HGF renders P. berghei host hepatocytes susceptible to infection | Carrolo et al., 2003 |
| CD81 | Required on hepatocytes for P. yoelii invasion with PVM formation | Silvie et al., 2003 | |
| Cholesterol | Involved in assembly of CD81 microdomains on the cell surface | Silvie et al., 2003, 2006, 2007 | |
| HSPGs | Binds CSP, increased sulfation on HSPGs triggers invasion of migrating sporozoite | Frevert et al., 1993; Coppi et al., 2007 | |
| EphA2 | Engages parasite protein P36 to facilitate hepatocyte invasion | Kaushansky et al., 2015 | |
| Scavenger Receptor B1 | Required for CD81 microdomain formation, additional roles independent of CD81 for P. berghei and P. vivax | Rodrigues et al., 2008; Yalaoui et al., 2008a; Manzoni et al., 2017 | |
| HGF/MET signaling | Liver Stage Development | Prevents the apoptosis of P. berghei infected cells, promoting successful infection | Leiriao et al., 2005 |
| Endosomes and lysosomes | Endosomes and lysosomes are localized around the PVM during development | Lopes da Silva et al., 2012; Grutzke et al., 2014 | |
| Phosphatidylcholine | Required for correct localization of proteins within the PVM; important for parasite survival | Itoe et al., 2014 | |
| P53 | Decreased levels of P53 are important for successful Liver Stage infection. | Kaushansky et al., 2013b | |
| Apolipoprotein H | Interacts with parasite protein EXP1 to promote successful Liver Stage infection | Sa et al., 2017 | |
| ALK4 | Knockdown reduces Liver Stage infection | Arang et al., 2017 | |
| CAMKK2 | Knockdown reduces Liver Stage infection | Arang et al., 2017 | |
| CSK | Knockdown reduces Liver Stage infection | Arang et al., 2017 | |
| FGFR4 | Knockdown reduces Liver Stage infection | Arang et al., 2017 | |
| FLT1 | Knockdown reduces Liver Stage infection | Arang et al., 2017 | |
| FLT3 | Knockdown reduces Liver Stage infection | Arang et al., 2017 | |
| IKBKB | Knockdown reduces Liver Stage infection | Arang et al., 2017 | |
| IRAK1 | Knockdown reduces Liver Stage infection | Arang et al., 2017 | |
| MAPK1 | Knockdown reduces Liver Stage infection | Arang et al., 2017 | |
| MAPKAPK2 | Knockdown reduces Liver Stage infection | Arang et al., 2017 | |
| MARK2 | Knockdown reduces Liver Stage infection | Prudencio et al., 2008; Arang et al., 2017 | |
| MARK4 | Knockdown reduces Liver Stage infection | Arang et al., 2017 | |
| MET | Knockdown reduces Liver Stage infection | Prudencio et al., 2008; Arang et al., 2017 | |
| PKCζ | Knockdown reduces Liver Stage infection | Prudencio et al., 2008; Arang et al., 2017 | |
| PRKWNK1 | Knockdown reduces Liver Stage infection | Prudencio et al., 2008 | |
| SGK2 | Knockdown reduces Liver Stage infection | Prudencio et al., 2008 | |
| STK35 | Knockdown reduces Liver Stage infection | Prudencio et al., 2008 | |
| TGFBR1 | Knockdown reduces Liver Stage infection | Arang et al., 2017 | |
| TYRO3 | Knockdown reduces Liver Stage infection | Arang et al., 2017 | |
| ULK1 | Knockdown reduces Liver Stage infection | Arang et al., 2017 | |
| WEE1 | Knockdown reduces Liver Stage infection | Arang et al., 2017 | |
A collection of host factors have also been described to impact parasite infection (Table 2B). Scavenger Receptor B1 (SRB1) and the tetraspanin CD81 both play roles in cholesterol-rich microdomain formation and are critical for hepatocyte invasion (Silvie et al., 2003; Rodrigues et al., 2008; Yalaoui et al., 2008a; Valacchi et al., 2011). More recently, it has been described that CD81 and SRB1 are involved in invasion in different species; CD81 is required for P. yoelii and P. falciparum infection, but appears to be dispensable for P. berghei and P. vivax infection. SRB1 plays a more substantial role in P. vivax and P. berghei infections (Silvie et al., 2003; Manzoni et al., 2017). It remains unknown if either protein makes contact with the sporozoite, although it has been suggested that SRB1 might directly engage the parasite, whereas CD81 indirectly impacts infection (Yalaoui et al., 2008b; Manzoni et al., 2017). The receptor tyrosine kinase EphA2 is also critical for hepatocyte infection, at least in part by engaging the parasite protein P36 (Kaushansky et al., 2015). While each of these factors contributes to the infection process, how they work in concert, and how changes in one invasion factor impacts another remain unknown. New approaches that integrate biochemical information and omics datasets are well-suited to merge with existing candidate-based research to create a more comprehensive view of the molecular components required for hepatocyte invasion (AlQuraishi et al., 2014; Gujral et al., 2014). The capacity to integrate biochemical data into a more global framework also paves the way for the identification of molecules or networks that could be targeted for intervention.
Liver stage development
Once the parasite has taken up residence in the hepatocyte, the sporozoite dedifferentiates over the course of 12 h in rodents, or 2–3 days in humans. This process results in a rounded trophozoite, which is characterized by dramatic changes in the parasite including the disassembly of molecular and cellular structures and the expulsion of invasion machinery (Bano et al., 2007) (reviewed in Kaushansky and Kappe, 2015b; Vaughan and Kappe, 2017). Following dedifferentiation, schizogony begins, which involves the massive replication the genome, and takes place over the course of 2–10 days, depending on the Plasmodium species. During this time, cellular structures including lysosomes and late endosomes sequester around the parasitophorous vacuole membrane and associate with the tubovesicular network (Lopes da Silva et al., 2012; Grutzke et al., 2014). The unfolded protein response is triggered, which promotes endoplasmic reticulum stress and the survival of the Liver Stage parasite (Inacio et al., 2015; Kaushansky and Kappe, 2015a). The most dramatic change, however, is the replication of the liver stage schizont, which produces tens of thousands of merozoites (membrane-bound, haploid, red blood cell invasive forms) that eventually invade erythrocytes during blood stage development. During this process, some parasites survive, and re-wire their host cells to resist certain types of apoptotic stimuli, while others succumb to host cell apoptosis or alternative cell death stimuli (Leiriao et al., 2005; van de Sand et al., 2005; Kaushansky et al., 2013a,b; Douglass et al., 2015). While these dramatic cellular changes have been qualitatively observed, they are rarely monitored quantitatively.
Some molecular determinants have been linked to Liver Stage survival and development. For example, Plasmodium proteins Upregulated in Infectious Sporozoites (UIS) UIS3 and UIS4 have been hypothesized to play an active role in host nutrient acquisition, in part because of the demonstration that UIS3 associates with the host Liver Fatty Acid Binding Protein (L-FABP) (Mikolajczak et al., 2007; Blume et al., 2011; Slavic et al., 2011; Favretto et al., 2013) and localizes both proteins to the PVM (Mueller et al., 2005). Fatty acids of both host and parasite origin, including host phosphatidylcholine, have been demonstrated to be required for optimal liver stage development (Mazumdar et al., 2006; Vaughan et al., 2009; Itoe et al., 2014). How each of these components specifically contributes to the observed cellular changes, and how each factor co-opts host defenses remains unknown. A more quantitative assessment of the cellular changes that occur, matched to molecular information, will enable the development of models that describe networks of host-parasite interactions required for development of the liver stage parasite.
Liver stage exit
Intracellular pathogens must exit their host cell in order to propagate and survive. The precise strategies they use directly impact their ability to disseminate within a host, transmit to new hosts, and engage or avoid host immune responses. Despite these important functions of exit, detailed investigations into the mechanisms governing Plasmodium exit have been lacking. This process is important not only for our basic understanding of liver stage development, but also for immunity. This is illustrated by the finding that the most potent stimulus of the immune system is elicited by parasites that develop through the liver stage and exit, but cannot undergo replication within the blood stage (Bijker et al., 2013).
Egress from hepatocytes occurs through the rupture of the PVM, followed by destabilization of the actin cytoskeleton, to allow the budding of merozoites from the host cell through the formation of merozoite-filled vesicles (merosomes). These structures are surrounded by a membrane of host origin (Graewe et al., 2011), and have been hypothesized to shuttle merozoites into the bloodstream to begin blood stage infection (Burda et al., 2017). This process inhibits exposure of phosphatidylserine (PS) on the outer surface of the cell, thereby simultaneously ensuring migration of parasites to the bloodstream and protection from host immune responses (Sturm et al., 2006; Tarun et al., 2006; Baer et al., 2007). Recent methodological advances have developed a platform for quantifying exit events (Stanway et al., 2009). This quantification, and a global assessment of molecular changes that occur during exit, will drive the development of models that describe networks of host-parasite interactions that underlie the exit process. Importantly, these networks could then be used to predict host and parasite determinants of dissemination to the blood stream, and the ability to engage or avoid host immune responses.
Conclusions and new directions
Parasites must successfully navigate a wide variety of different environmental milieus, and each alternative setting presents challenges for the parasite, as well as opportunities for intervention. Here, we have described how the tools and approaches of systems biology can be deployed to more comprehensively characterize the complex interaction between parasite and host. This will inform our understanding of how the parasite and the host interact, and also facilitate future strategies to combat the parasite. Interventions that have been designed and employed without a comprehensive understanding of the complex dynamic between the Plasmodium parasite and its host have only partially controlled malaria in the field.
Despite the challenges, many influential leaders have called for malaria eradication in recent years (Gates, 2007; WHO, 2017). This goal is most likely to be realized if control strategies are deployed rationally with the capacity to predict how a given treatment will impact systematic changes in the parasite and host alike, to facilitate readiness for these changes. The integration of systems biology could evaluate the capacity of the parasite to circumvent new interventions, and in doing so, contribute to the success of eradication efforts. While references to the principles of systems biology first occurred decades ago, the field was established in earnest ~15 years ago with the completion of the human genome. Since then, most systems biology studies have steered clear of the complexity that is introduced when multiple genomes collide, as is the case during infection. Pathogens and their host cells have coevolved, introducing alterations to both genomes along the way (Miller et al., 1976; Zimmerman et al., 2013). What has resulted is the capacity of a pathogen to fundamentally alter the biology of its host, by changing the size, shape, composition and function of the cell. Intracellular pathogens thus are expert cell biologists, controlling the host cell to their own advantage. As such, the study of host-pathogen interactions presents an unmatched opportunity for the field of systems biology, just as the approach of systems biology presents an unmatched opportunity for the eradication of malaria.
Author contributions
MZ, LSA, SAD, JDA, and AK were involved in the conception of the article. MZ, JDA, and AK wrote the article with assistance from LSA and SAD.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge Zach Grossnickle and Marissa Vignali for assistance with figure design. We thank Jarrod Johnson and Thurston Herricks for critical review of the manuscript. Funding for this review was provided by NIH 1R01GM101183, R21AI124266, 1K99/R00AI111785, P50 GM076547, P41 GM109824, R01 GM112108, and Center for Infectious Disease Research Internal Funding.
References
- Albuquerque S. S., Carret C., Grosso A. R., Tarun A. S., Peng X., Kappe S. H., et al. (2009). Host cell transcriptional profiling during malaria liver stage infection reveals a coordinated and sequential set of biological events. BMC Genomics 10:270. 10.1186/1471-2164-10-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- AlQuraishi M., Koytiger G., Jenney A., MacBeath G., Sorger P. K. (2014). A multiscale statistical mechanical framework integrates biophysical and genomic data to assemble cancer networks. Nat. Genet. 46, 1363–1371. 10.1038/ng.3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annoura T., van Schaijk B. C., Ploemen I. H., Sajid M., Lin J. W., Vos M. W., et al. (2014). Two Plasmodium 6-Cys family-related proteins have distinct and critical roles in liver-stage development. FASEB J. 28, 2158–2170. 10.1096/fj.13-241570 [DOI] [PubMed] [Google Scholar]
- Arang N., Kain H. S., Glennon E. K., Bello T., Dudgen D. R., Walter E. N. F., et al. (2017). Identifying host regulators and inhibitors of liver stage malaria infection using kinase activity profiles. Nat. Commun. 8:1232. 10.1038/s41467-017-01345-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer K., Klotz C., Kappe S. H., Schnieder T., Frevert U. (2007). Release of hepatic Plasmodium yoelii merozoites into the pulmonary microvasculature. PLoS Pathog. 3:e171. 10.1371/journal.ppat.0030171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bano N., Romano J. D., Jayabalasingham B., Coppens I. (2007). Cellular interactions of Plasmodium liver stage with its host mammalian cell. Int. J. Parasitol. 37, 1329–1341. 10.1016/j.ijpara.2007.04.005 [DOI] [PubMed] [Google Scholar]
- Betzig E., Patterson G. H., Sougrat R., Lindwasser O. W., Olenych S., Bonifacino J. S., et al. (2006). Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645. 10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- Bhanot P., Schauer K., Coppens I., Nussenzweig V. (2005). A surface phospholipase is involved in the migration of plasmodium sporozoites through cells. J Biol Chem. 280, 6752–6760. 10.1074/jbc.M411465200 [DOI] [PubMed] [Google Scholar]
- Bijker E. M., Bastiaens G. J., Teirlinck A. C., van Gemert G. J., Graumans W. M., van de Vegte-Bolmer, et al. (2013). Protection against malaria after immunization by chloroquine prophylaxis and sporozoites is mediated by preerythrocytic immunity. Proc. Natl. Acad. Sci. U.S.A. 110, 7862–7867. 10.1073/pnas.1220360110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco B., Leroy D., Fidock D. A. (2017). Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nat. Med. 23, 917–928. 10.1038/nm.4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume M., Hliscs M., Rodriguez-Contreras D., Sanchez M., Landfear S., Lucius R., et al. (2011). A constitutive pan-hexose permease for the Plasmodium life cycle and transgenic models for screening of antimalarial sugar analogs. FASEB J. 25, 1218–1229. 10.1096/fj.10-173278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau R., Facciotti M. T., Reiss D. J., Schmid A. K., Pan M., Kaur A., et al. (2007). A predictive model for transcriptional control of physiology in a free living cell. Cell 131, 1354–1365. 10.1016/j.cell.2007.10.053 [DOI] [PubMed] [Google Scholar]
- Burda P. C., Caldelari R., Heussler V. T. (2017). Manipulation of the host cell membrane during plasmodium liver stage egress. MBio. 8:17. 10.1128/mBio.00139-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell E., Gomes A. R., Sanderson T., Anar B., Girling G., Herd C., et al. (2017). Functional profiling of a plasmodium genome reveals an abundance of essential genes. Cell 170, 260–272 e8. 10.1016/j.cell.2017.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrum S., Smart S. K., Larson S., Tackett A. J. (2012). Analysis of stable and transient protein-protein interactions. Methods Mol. Biol. 833, 143–152. 10.1007/978-1-61779-477-3_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrolo M., Giordano S., Cabrita-Santos L., Corso S., Vigario A. M., Silva S., et al. (2003). Hepatocyte growth factor and its receptor are required for malaria infection. Nat. Med. 9, 1363–1369. 10.1038/nm947 [DOI] [PubMed] [Google Scholar]
- Cha S. J., Kim M. S., Pandey A., Jacobs-Lorena M. (2016). Identification of GAPDH on the surface of Plasmodium sporozoites as a new candidate for targeting malaria liver invasion. J. Exp. Med. 213, 2099–2112. 10.1084/jem.20160059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman B. I., Skillman K. M., Jiang R. H. Y., Childs L. M., Altenhofen L. M., Ganter M., et al. (2014). A Plasmodium falciparum histone deacetylase regulates antigenic variation and gametocyte conversion. Cell Host Microbe 16, 177–186. 10.1016/j.chom.2014.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combe A., Giovannini D., Carvalho T. G., Spath S., Boisson B., Loussert C., et al. (2009). Clonal conditional mutagenesis in malaria parasites. Cell Host Microbe 5, 386–396. 10.1016/j.chom.2009.03.008 [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppi A., Natarajan R., Pradel G., Bennett B. L., James E. R., Roggero M. A., et al. (2011). The malaria circumsporozoite protein has two functional domains, each with distinct roles as sporozoites journey from mosquito to mammalian host. J. Exp. Med. 208, 341–356. 10.1084/jem.20101488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppi A., Pinzon-Ortiz C., Hutter C., Sinnis P. (2005). The Plasmodium circumsporozoite protein is proteolytically processed during cell invasion. J. Exp. Med. 201, 27–33. 10.1084/jem.20040989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppi A., Tewari R., Bishop J. R., Bennett B. L., Lawrence R., Esko J. D., et al. (2007). Heparan sulfate proteoglycans provide a signal to Plasmodium sporozoites to stop migrating and productively invade host cells. Cell Host Microbe 2, 316–327. 10.1016/j.chom.2007.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danziger S. A., Ratushny A. V., Smith J. J., Saleem R. A., Wan Y., Arens C. E., et al. (2014). Molecular mechanisms of system responses to novel stimuli are predictable from public data. Nucleic Acids Res. 42, 1442–1460. 10.1093/nar/gkt938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Niz M., Burda P. C., Kaiser G., Del Portillo H. A., Spielmann T., Frischknecht F., et al. (2017). Progress in imaging methods: insights gained into Plasmodium biology. Nat. Rev. Microbiol. 15, 37–54. 10.1038/nrmicro.2016.158 [DOI] [PubMed] [Google Scholar]
- Douglass A. N., Kain H. S., Abdullahi M., Arang N., Austin L. S., Mikolajczak S. A., et al. (2015). Host-based prophylaxis successfully targets liver stage malaria parasites. Mol. Ther. 23, 857–865. 10.1038/mt.2015.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favretto F., Assfalg M., Molinari H., D'Onofrio M. (2013). Evidence from NMR interaction studies challenges the hypothesis of direct lipid transfer from L-FABP to malaria sporozoite protein UIS3. Protein Sci. 22, 133–138. 10.1002/pro.2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frevert U., Sinnis P., Cerami C., Shreffler W., Takacs B., Nussenzweig V. (1993). Malaria circumsporozoite protein binds to heparan sulfate proteoglycans associated with the surface membrane of hepatocytes. J. Exp. Med. 177, 1287–1298. 10.1084/jem.177.5.1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M. J., Hall N., Fung E., White O., Berriman M., Hyman R. W. (2002). Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511. 10.1038/nature01097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates M. (2007). Malaria Forum Keynote Address. Available online at: https://www.gatesfoundation.org/media-center/speeches/2007/10/melinda-french-gates-malaria-forum
- Gehlenborg N., O'Donoghue S. I., Baliga N. S., Goesmann A., Hibbs M. A., Kitano H., et al. (2010). Visualization of omics data for systems biology. Nat. Methods 7, S56–S68. 10.1038/nmeth.1436 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., et al. (2003). Global analysis of protein expression in yeast. Nature 425, 737–741. 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- Ghorbal M., Gorman M., Macpherson C. R., Martins R. M., Scherf A., Lopez-Rubio J. J. (2014). Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat. Biotechnol. 32, 819–821. 10.1038/nbt.2925 [DOI] [PubMed] [Google Scholar]
- Gomes-Santos C. S., Braks J., Prudencio M., Carret C., Gomes A. R., Pain A., et al. (2011). Transition of Plasmodium sporozoites into liver stage-like forms is regulated by the RNA binding protein Pumilio. PLoS Pathog. 7:e1002046. 10.1371/journal.ppat.1002046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graewe S., Rankin K. E., Lehmann C., Deschermeier C., Hecht L., Froehlke U., et al. (2011). Hostile takeover by Plasmodium: reorganization of parasite and host cell membranes during liver stage egress. PLoS Pathog. 7:e1002224. 10.1371/journal.ppat.1002224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutzke J., Rindte K., Goosmann C., Silvie O., Rauch C., Heuer D., et al. (2014). The spatiotemporal dynamics and membranous features of the Plasmodium liver stage tubovesicular network. Traffic 15, 362–382. 10.1111/tra.12151 [DOI] [PubMed] [Google Scholar]
- Guerreiro A., Deligianni E., Santos J. M., Silva P. A., Louis C., Pain A., et al. (2014). Genome-wide RIP-Chip analysis of translational repressor-bound mRNAs in the Plasmodium gametocyte. Genome Biol. 15:493. 10.1186/s13059-014-0493-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujral T. S., Peshkin L., Kirschner M. W. (2014). Exploiting polypharmacology for drug target deconvolution. Proc. Natl. Acad. Sci. U.S.A. 111, 5048–5053. 10.1073/pnas.1403080111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein M. Y., Hubner N. C., Poser I., Cox J., Nagaraj N., Toyoda Y., et al. (2015). A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723. 10.1016/j.cell.2015.09.053 [DOI] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., et al. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686–691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Huttlin E. L., Ting L., Bruckner R. J., Gebreab F., Gygi M. P., Szpyt J., et al. (2015). The BioPlex network: a systematic exploration of the human interactome. Cell 162, 425–440. 10.1016/j.cell.2015.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D., Rust A. G., Ramsey S., Smith J. J., Leslie D. M., Weston A. D., et al. (2005a). A data integration methodology for systems biology. Proc. Natl. Acad. Sci. U.S.A. 102, 17296–17301. 10.1073/pnas.0508647102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D., Smith J. J., Leslie D. M., Weston A. D., Rust A. G., Ramsey S., et al. (2005b). A data integration methodology for systems biology: experimental verification. Proc. Natl. Acad. Sci. U.S.A. 102, 17302–17307. 10.1073/pnas.0508649102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inacio P., Zuzarte-Luis V., Ruivo M. T., Falkard B., Nagaraj N., Rooijers K., et al. (2015). Parasite-induced ER stress response in hepatocytes facilitates Plasmodium liver stage infection. EMBO Rep. 16, 955–964. 10.15252/embr.201439979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino T., Boisson B., Orito Y., Lacroix C., Bischoff E., Loussert C., et al. (2009). LISP1 is important for the egress of Plasmodium berghei parasites from liver cells. Cell. Microbiol. 11, 1329–1339. 10.1111/j.1462-5822.2009.01333.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino T., Chinzei Y., Yuda M. (2005). Two proteins with 6-cys motifs are required for malarial parasites to commit to infection of the hepatocyte. Mol. Microbiol. 58, 1264–1275. 10.1111/j.1365-2958.2005.04801.x [DOI] [PubMed] [Google Scholar]
- Ishino T., Yano K., Chinzei Y., Yuda M. (2004). Cell-passage activity is required for the malarial parasite to cross the liver sinusoidal cell layer. PLoS Biol. 2:E4. 10.1371/journal.pbio.0020004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoe M. A., Sampaio J. L., Cabal G. G., Real E., Zuzarte-Luis V., March S., et al. (2014). Host cell phosphatidylcholine is a key mediator of malaria parasite survival during liver stage infection. Cell Host Microbe 16, 778–786. 10.1016/j.chom.2014.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes K. A., Albeck J. G., Gaudet S., Sorger P. K., Lauffenburger D. A., Yaffe M. B. (2005). A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science 310, 1646–1653. 10.1126/science.1116598 [DOI] [PubMed] [Google Scholar]
- Joice R., Nilsson S. K., Montgomery J., Dankwa S., Egan E., Morahan B., et al. (2014). Plasmodium falciparum transmission stages accumulate in the human bone marrow. Sci. Transl. Med. 6:244re5. 10.1126/scitranslmed.3008882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafsack B. F., Rovira-Graells N., Clark T. G., Bancells C., Crowley V. M., Campino S. G., et al. (2014). A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature 507, 248–252. 10.1038/nature12920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappe S., Bruderer T., Gantt S., Fujioka H., Nussenzweig V., Menard R. (1999). Conservation of a gliding motility and cell invasion machinery in Apicomplexan parasites. J. Cell Biol. 147, 937–944. 10.1083/jcb.147.5.937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariu T., Ishino T., Yano K., Chinzei Y., Yuda M. (2006). CelTOS, a novel malarial protein that mediates transmission to mosquito and vertebrate hosts. Mol. Microbiol. 59, 1369–1379. 10.1111/j.1365-2958.2005.05024.x [DOI] [PubMed] [Google Scholar]
- Kaushansky A., Douglass A. N., Arang N., Vigdorovich V., Dambrauskas N., Kain H. S., et al. (2015). Malaria parasites target the hepatocyte receptor EphA2 for successful host infection. Science 350, 1089–1092. 10.1126/science.aad3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushansky A., Kappe S. H. (2015a). Host ER stress during malaria parasite infection. EMBO Rep. 16, 883–884. 10.15252/embr.201540792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushansky A., Kappe S. H. (2015b). Selection and refinement: the malaria parasite's infection and exploitation of host hepatocytes. Curr. Opin. Microbiol. 26, 71–78. 10.1016/j.mib.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushansky A., Metzger P. G., Douglass A. N., Mikolajczak S. A., Lakshmanan V., Kain H. S., et al. (2013a). Malaria parasite liver stages render host hepatocytes susceptible to mitochondria-initiated apoptosis. Cell Death Dis. 4:e762. 10.1038/cddis.2013.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushansky A., Ye A. S., Austin L. S., Mikolajczak S. A., Vaughan A. M., Camargo N., et al. (2013b). Suppression of host p53 is critical for Plasmodium liver-stage infection. Cell Rep. 3, 630–637. 10.1016/j.celrep.2013.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. M., Franke-Fayard B., Mair G. R., Lasonder E., Janse C. J., Mann M., et al. (2005). Proteome analysis of separated male and female gametocytes reveals novel sex-specific Plasmodium biology. Cell 121, 675–687. 10.1016/j.cell.2005.03.027 [DOI] [PubMed] [Google Scholar]
- Labaied M., Harupa A., Dumpit R. F., Coppens I., Mikolajczak S. A., Kappe S. H. (2007). Plasmodium yoelii sporozoites with simultaneous deletion of P52 and P36 are completely attenuated and confer sterile immunity against infection. Infect. Immun. 75, 3758–3768. 10.1128/IAI.00225-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasonder E., Rijpma S. R., van Schaijk B. C., Hoeijmakers W. A., Kensche P. R., Gresnigt M. S., et al. (2016). Integrated transcriptomic and proteomic analyses of P. falciparum gametocytes: molecular insight into sex-specific processes and translational repression. Nucleic Acids Res. 44, 6087–6101. 10.1093/nar/gkw536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiriao P., Albuquerque S. S., Corso S., van Gemert G. J., Sauerwein R. W., Rodriguez A., et al. (2005). HGF/MET signalling protects Plasmodium-infected host cells from apoptosis. Cell. Microbiol. 7, 603–609. 10.1111/j.1462-5822.2004.00490.x [DOI] [PubMed] [Google Scholar]
- Liehl P., Zuzarte-Luis V., Chan J., Zillinger T., Baptista F., Carapau D., et al. (2014). Host-cell sensors for Plasmodium activate innate immunity against liver-stage infection. Nat. Med. 20, 47–53. 10.1038/nm.3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner S. E., Mikolajczak S. A., Vaughan A. M., Moon W., Joyce B. R., Sullivan W. J., Jr., et al. (2013). Perturbations of Plasmodium Puf2 expression and RNA-seq of Puf2-deficient sporozoites reveal a critical role in maintaining RNA homeostasis and parasite transmissibility. Cell. Microbiol. 15, 1266–1283. 10.1111/cmi.12116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes da Silva M., Thieleke-Matos C., Cabrita-Santos L., Ramalho J. S., Wavre-Shapton S. T., Futter C. E., et al. (2012). The host endocytic pathway is essential for Plasmodium berghei late liver stage development. Traffic 13, 1351–1363. 10.1111/j.1600-0854.2012.01398.x [DOI] [PubMed] [Google Scholar]
- Lu J., Tong Y., Pan J., Yang Y., Liu Q., Tan X., et al. (2016). A redesigned CRISPR/Cas9 system for marker-free genome editing in Plasmodium falciparum. Parasit. Vectors 9:198 10.1186/s13071-016-1487-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair G. R., Braks J. A., Garver L. S., Wiegant J. C., Hall N., Dirks R. W., et al. (2006). Regulation of sexual development of Plasmodium by translational repression. Science 313, 667–669. 10.1126/science.1125129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoni G., Marinach C., Topcu S., Briquet S., Grand M., Tolle M., et al. (2017). Plasmodium P36 determines host cell receptor usage during sporozoite invasion. Elife 6:e25903. 10.7554/eLife.25903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuschewski K., Ross J., Brown S. M., Kaiser K., Nussenzweig V., Kappe S. H. (2002). Infectivity-associated changes in the transcriptional repertoire of the malaria parasite sporozoite stage. J. Biol. Chem. 277, 41948–41953. 10.1074/jbc.M207315200 [DOI] [PubMed] [Google Scholar]
- Mazumdar J., Emma H. W., Masek K., Hunter A. C., Striepen B. (2006). Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc. Natl. Acad. Sci. U.S.A. 103, 13192–13197. 10.1073/pnas.0603391103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N. A., Li T., et al. (2013). The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736. 10.1038/nmeth.2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolajczak S. A., Jacobs-Lorena V., MacKellar D. C., Camargo N., Kappe S. H. (2007). L-FABP is a critical host factor for successful malaria liver stage development. Int. J. Parasitol. 37, 483–489. 10.1016/j.ijpara.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Mikolajczak S. A., Silva-Rivera H., Peng X., Tarun A. S., Camargo N., Jacobs-Lorena V., et al. (2008). Distinct malaria parasite sporozoites reveal transcriptional changes that cause differential tissue infection competence in the mosquito vector and mammalian host. Mol. Cell. Biol. 28, 6196–6207. 10.1128/MCB.00553-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. L., Sack B. K., Baldwin M., Vaughan A. M., Kappe S. H. (2014). Interferon-mediated innate immune responses against malaria parasite liver stages. Cell Rep. 7, 436–447. 10.1016/j.celrep.2014.03.018 [DOI] [PubMed] [Google Scholar]
- Miller L. H., Mason S. J., Clyde D. F., McGinniss M. H. (1976). The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N. Engl. J. Med. 295, 302–304. [DOI] [PubMed] [Google Scholar]
- Morahan B. J., Wang L., Coppel R. L. (2009). No TRAP, no invasion. Trends Parasitol. 25, 77–84. 10.1016/j.pt.2008.11.004 [DOI] [PubMed] [Google Scholar]
- Moreira C. K., Templeton T. J., Lavazec C., Hayward R. E., Hobbs C. V., Kroeze H., et al. (2008). The Plasmodium TRAP/MIC2 family member, TRAP-Like Protein (TLP), is involved in tissue traversal by sporozoites. Cell. Microbiol. 10, 1505–1516. 10.1111/j.1462-5822.2008.01143.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota M. M., Pradel G., Vanderberg J. P., Hafalla J. C., Frevert U., Nussenzweig R. S., et al. (2001). Migration of Plasmodium sporozoites through cells before infection. Science 291, 141–144. 10.1126/science.291.5501.141 [DOI] [PubMed] [Google Scholar]
- Mueller A. K., Camargo N., Kaiser K., Andorfer C., Frevert U., Matuschewski K., et al. (2005). Plasmodium liver stage developmental arrest by depletion of a protein at the parasite-host interface. Proc. Natl. Acad. Sci. U.S.A. 102, 3022–3027. 10.1073/pnas.0408442102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller K., Matuschewski K., Silvie O. (2011). The Puf-family RNA-binding protein Puf2 controls sporozoite conversion to liver stages in the malaria parasite. PLoS ONE 6:e19860. 10.1371/journal.pone.0019860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey D. E., Juraska M., Bedford T., Benkeser D., Valim C., Griggs A., et al. (2015). Genetic diversity and protective efficacy of the RTS,S/AS01 malaria vaccine. N. Engl. J. Med. 373, 2025–2037. 10.1056/NEJMoa1505819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orito Y., Ishino T., Iwanaga S., Kaneko I., Kato T., Menard R., et al. (2013). Liver-specific protein 2: a Plasmodium protein exported to the hepatocyte cytoplasm and required for merozoite formation. Mol. Microbiol. 87, 66–79. 10.1111/mmi.12083 [DOI] [PubMed] [Google Scholar]
- Pavlopoulos G. A., Paez-Espino D., Kyrpides N. C., Iliopoulos I. (2017). Empirical comparison of visualization tools for larger-scale network analysis. Adv. Bioinformatics 2017:1278932. 10.1155/2017/1278932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittet M. J., Weissleder R. (2011). Intravital imaging. Cell 147, 983–991. 10.1016/j.cell.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploemen I. H., Croes H. J., van Gemert G. J., Wijers-Rouw M., Hermsen C. C., Sauerwein R. W. (2012). Plasmodium berghei Δp52&p36 parasites develop independent of a parasitophorous vacuole membrane in Huh-7 liver cells. PLoS ONE 7:e50772. 10.1371/journal.pone.0050772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poran A., Nötzel C., Aly O., Mencia-Trinchant N., Harris C. T., Guzman M. L., et al. (2017). Single-Cell RNA sequencing reveals a signature of sexual commitment in malaria parasites. Nature. 551, 95–99. 10.1038/nature24280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudencio M., Rodrigues C. D., Hannus M., Martin C., Real E., Goncalves L. A., et al. (2008). Kinome-wide RNAi screen implicates at least 5 host hepatocyte kinases in Plasmodium sporozoite infection. PLoS Pathog. 4:e1000201. 10.1371/journal.ppat.1000201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues C. D., Hannus M., Prudencio M., Martin C., Goncalves L. A., Portugal S., et al. (2008). Host scavenger receptor SR-BI plays a dual role in the establishment of malaria parasite liver infection. Cell Host Microbe 4, 271–282. 10.1016/j.chom.2008.07.012 [DOI] [PubMed] [Google Scholar]
- Ruivo M. T., Vera I. M., Sales-Dias J., Meireles P., Gural N., Bhatia S. N., et al. (2016). Host AMPK Is a modulator of plasmodium liver infection. Cell Rep. 16, 2539–2545. 10.1016/j.celrep.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa E. C. C., Nyboer B., Heiss K., Sanches-Vaz M., Fontinha D., Wiedtke E., et al. (2017). Plasmodium berghei EXP-1 interacts with host Apolipoprotein H during Plasmodium liver-stage development. Proc. Natl. Acad. Sci. U.S.A. 114, E1138–E1147. 10.1073/pnas.1606419114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schats R., Bijker E. M., van Gemert G. J., Graumans W., van de Vegte-Bolmer M., van Lieshout L., et al. (2015). Heterologous protection against malaria after immunization with Plasmodium falciparum sporozoites. PLoS ONE 10:e0124243. 10.1371/journal.pone.0124243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M., Mann M. (2006). Protein interaction screening by quantitative immunoprecipitation combined with knockdown (QUICK). Nat. Methods. 3, 981–983. 10.1038/nmeth972 [DOI] [PubMed] [Google Scholar]
- Shortt H. E., Garnham P. C. (1948). Pre-erythrocytic stage in mammalian malaria parasites. Nature 161:126. 10.1038/161126a0 [DOI] [PubMed] [Google Scholar]
- Silva P. A., Guerreiro A., Santos J. M., Braks J. A., Janse C. J., Mair G. R. (2016). Translational control of UIS4 protein of the host-parasite interface is mediated by the RNA binding protein Puf2 in Plasmodium berghei sporozoites. PLoS ONE 11:e0147940. 10.1371/journal.pone.0147940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvie O., Briquet S., Muller K., Manzoni G., Matuschewski K. (2014). Post-transcriptional silencing of UIS4 in Plasmodium berghei sporozoites is important for host switch. Mol. Microbiol. 91, 1200–1213. 10.1111/mmi.12528 [DOI] [PubMed] [Google Scholar]
- Silvie O., Charrin S., Billard M., Franetich J. F., Clark K. L., van Gemert G. J., et al. (2006). Cholesterol contributes to the organization of tetraspanin-enriched microdomains and to CD81-dependent infection by malaria sporozoites. J. Cell Sci. 119(Pt 10), 1992–2002. 10.1242/jcs.02911 [DOI] [PubMed] [Google Scholar]
- Silvie O., Franetich J. F., Boucheix C., Rubinstein E., Mazier D. (2007). Alternative invasion pathways for Plasmodium berghei sporozoites. Int. J. Parasitol. 37, 173–182. 10.1016/j.ijpara.2006.10.005 [DOI] [PubMed] [Google Scholar]
- Silvie O., Rubinstein E., Franetich J. F., Prenant M., Belnoue E., Renia L., et al. (2003). Hepatocyte CD81 is required for Plasmodium falciparum and Plasmodium yoelii sporozoite infectivity. Nat. Med. 9, 93–96. 10.1038/nm808 [DOI] [PubMed] [Google Scholar]
- Sinha A., Hughes K. R., Modrzynska K. K., Otto T. D., Pfander C., Dickens N. J., et al. (2014). A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature 507, 253–257. 10.1038/nature12970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinturel F., Gerber A., Mauvoisin D., Wang J., Gatfield D., Stubblefield J. J., et al. (2017). Diurnal oscillations in liver mass and cell size accompany ribosome assembly cycles. Cell 169, 651–663.e14. 10.1016/j.cell.2017.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavic K., Delves M. J., Prudencio M., Talman A. M., Straschil U., Derbyshire E. T., et al. (2011). Use of a selective inhibitor to define the chemotherapeutic potential of the plasmodial hexose transporter in different stages of the parasite's life cycle. Antimicrob. Agents Chemother. 55, 2824–2830. 10.1128/AAC.01739-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanway R. R., Graewe S., Rennenberg A., Helm S., Heussler V. T. (2009). Highly efficient subcloning of rodent malaria parasites by injection of single merosomes or detached cells. Nat. Protoc. 4, 1433–1439. 10.1038/nprot.2009.172 [DOI] [PubMed] [Google Scholar]
- Sturm A., Amino R. C., van de Sand, Regen T., Retzlaff S., Rennenberg A., et al. (2006). Manipulation of host hepatocytes by the malaria parasite for delivery into liver sinusoids. Science 313, 1287–1290. 10.1126/science.1129720 [DOI] [PubMed] [Google Scholar]
- Tackett A. J., DeGrasse J. A., Sekedat M. D., Oeffinger M., Rout M. P., Chait B. T. (2005). I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J. Proteome Res. 4, 1752–1756. 10.1021/pr050225e [DOI] [PubMed] [Google Scholar]
- Tarun A. S., Baer K., Dumpit R. F., Gray S., Lejarcegui N., Frevert U., et al. (2006). Quantitative isolation and in vivo imaging of malaria parasite liver stages. Int. J. Parasitol. 36, 1283–1293. 10.1016/j.ijpara.2006.06.009 [DOI] [PubMed] [Google Scholar]
- Tarun A. S., Peng X., Dumpit R. F., Ogata Y., Silva-Rivera H., Camargo N., et al. (2008). A combined transcriptome and proteome survey of malaria parasite liver stages. Proc. Natl. Acad. Sci. U.S.A. 105, 305–310. 10.1073/pnas.0710780104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares J., Formaglio P., Thiberge S., Mordelet E., Van Rooijen N., Medvinsky A., et al. (2013). Role of host cell traversal by the malaria sporozoite during liver infection. J. Exp. Med. 210, 905–915. 10.1084/jem.20121130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawk L., Lacroix C., Gueirard P., Kent R., Gorgette O., Thiberge S., et al. (2013). A key role for Plasmodium subtilisin-like SUB1 protease in egress of malaria parasites from host hepatocytes. J. Biol. Chem. 288, 33336–33346. 10.1074/jbc.M113.513234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L. (2012). Resolving protein interactions and complexes by affinity purification followed by label-based quantitative mass spectrometry. Proteomics 12, 1623–1638. 10.1002/pmic.201100438 [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Boulon S., Lam Y. W., Urcia R., Boisvert F. M., Vandermoere F., et al. (2008). Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 183, 223–239. 10.1083/jcb.200805092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usynin I., Klotz C., Frevert U. (2007). Malaria circumsporozoite protein inhibits the respiratory burst in Kupffer cells. Cell. Microbiol. 9, 2610–2628. 10.1111/j.1462-5822.2007.00982.x [DOI] [PubMed] [Google Scholar]
- Valacchi G., Sticozzi C., Lim Y., Pecorelli A. (2011). Scavenger receptor class B type I: a multifunctional receptor. Ann. N.Y. Acad. Sci. 1229, E1–E7. 10.1111/j.1749-6632.2011.06205.x [DOI] [PubMed] [Google Scholar]
- van de Sand C., Horstmann S., Schmidt A., Sturm A., Bolte S., Krueger A., et al. (2005). The liver stage of Plasmodium berghei inhibits host cell apoptosis. Mol. Microbiol. 58, 731–742. 10.1111/j.1365-2958.2005.04888.x [DOI] [PubMed] [Google Scholar]
- van Dijk M. R., Douradinha B., Franke-Fayard B., Heussler V., van Dooren M. W., van Schaijk B., et al. (2005). Genetically attenuated, P36p-deficient malarial sporozoites induce protective immunity and apoptosis of infected liver cells. Proc. Natl. Acad. Sci. U.S.A. 102, 12194–12199. 10.1073/pnas.0500925102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Regenmortel M. H. (2004). Reductionism and complexity in molecular biology. Scientists now have the tools to unravel biological and overcome the limitations of reductionism. EMBO Rep. 5, 1016–1020. 10.1038/sj.embor.7400284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijnsoever C., Oorschot V., Klumperman J. (2008). Correlative light-electron microscopy (CLEM) combining live-cell imaging and immunolabeling of ultrathin cryosections. Nat. Methods 5, 973–980. 10.1038/nmeth.1263 [DOI] [PubMed] [Google Scholar]
- Vaughan A. M., Mikolajczak S. A., Wilson E. M., Grompe M., Kaushansky A., Camargo N., et al. (2012). Complete Plasmodium falciparum liver-stage development in liver-chimeric mice. J. Clin. Invest. 122, 3618–3628. 10.1172/JCI62684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan A. M., O'Neill M. T., Tarun A. S., Camargo N., Phuong T. M., Aly A. S., et al. (2009). Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell. Microbiol. 11, 506–520. 10.1111/j.1462-5822.2008.01270.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan A. M., Kappe S. H. I. (2017). Malaria parasite liver infection and exoerythrocytic biology. Cold Spring Harb. Perspect. Med. 7:a025486. 10.1101/cshperspect.a025486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J. C., Platt R. J., Goldfless S. J., Zhang F., Niles J. C. (2014). Efficient CRISPR-Cas9-mediated genome editing in Plasmodium falciparum. Nat. Methods 11, 915–918. 10.1038/nmeth.3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2016). World Malaria Report. Geneva: World Health Organization. [Google Scholar]
- WHO (2017). Overview of Malaria Elimination. Geneva: World Health Organization. [Google Scholar]
- Yalaoui S., Huby T., Franetich J. F., Gego A., Rametti A., Moreau M., et al. (2008a). Scavenger receptor BI boosts hepatocyte permissiveness to Plasmodium infection. Cell Host Microbe 4, 283–292. 10.1016/j.chom.2008.07.013 [DOI] [PubMed] [Google Scholar]
- Yalaoui S., Zougbede S., Charrin S., Silvie O., Arduise C., Farhati K., et al. (2008b). Hepatocyte permissiveness to Plasmodium infection is conveyed by a short and structurally conserved region of the CD81 large extracellular domain. PLoS Pathog. 4:e1000010. 10.1371/journal.ppat.1000010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A. S., O'Neill M. T., Jennison C., Lopaticki S., Allison C. C., Armistead J. S., et al. (2017). Cell traversal activity is important for Plasmodium falciparum liver infection in humanized mice. Cell Rep. 18, 3105–3116. 10.1016/j.celrep.2017.03.017 [DOI] [PubMed] [Google Scholar]
- Zhang M., Fennell C., Ranford-Cartwright L., Sakthivel R., Gueirard P., Meister S., et al. (2010). The Plasmodium eukaryotic initiation factor-2alpha kinase IK2 controls the latency of sporozoites in the mosquito salivary glands. J. Exp. Med. 207, 1465–1474. 10.1084/jem.20091975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman P. A., Ferreira M. U., Howes R. E., Mercereau-Puijalon O. (2013). Red blood cell polymorphism and susceptibility to Plasmodium vivax. Adv Parasitol. 81, 27–76. 10.1016/B978-0-12-407826-0.00002-3 [DOI] [PMC free article] [PubMed] [Google Scholar]