Abstract
Earlier studies have shown that the d120 mutant of herpes simplex virus 1, which lacks both copies of the α4 gene, induces apoptosis in all cell lines tested. In some cell lines d120-induced apoptosis, manifested by the release of cytochrome c, activation of caspase 3, and fragmentation of cellular DNA, is blocked by the overexpression of Bcl-2. In these cells viral protein kinase US3 delivered in trans blocks apoptosis induced by the mutant virus at a premitochondrial stage. We report that the US3 protein kinase targets the pro-apoptotic BAD member of the Bcl-2 family. Specifically, the US3 protein kinase mediates a posttranslational modification of BAD and blocks its cleavage, which is reported to activate apoptosis. Thus, US3 protein kinase is the sole viral protein required to block activation of caspase 3, prevent cleavage of poly(ADP-ribose) polymerase, and block fragmentation of cellular DNA induced by BAD.
In earlier publications this and other laboratories reported that the interaction of herpes simplex virus 1 (HSV-1) with cells results in programmed cell death and that the virus has evolved mechanisms that block apoptosis, whether it is induced by viral gene products or by exogenous agents. Specifically, wild-type HSV does not induce apoptosis, and infection with wild-type virus blocks apoptosis induced by osmotic or thermal shock or by Fas ligand (1–7). A number of HSV-1 mutants have been reported to induce apoptosis. These include mutants lacking the infected cell protein no.4 or infected cell protein no. 27 (4, 8, 9), two regulatory proteins expressed immediately after infection, a mutant lacking glycoprotein D (10), and a mutant carrying a temperature-sensitive mutation that blocks the release of viral DNA from capsids at nuclear pores in cells infected and maintained at nonpermissive temperatures (3, 11). Detailed analyses of the mutant d120 from which both copies of the gene encoding infected cell protein no. 4 had been deleted revealed that the virus induces apoptosis in all of the cell lines tested but that the mechanisms by which the virus induces apoptosis is cell type dependent (5). In HEp-2 cells the d120 mutant caused the translocation of cytochrome c from mitochondria, activation of caspase 3, and fragmentation of cellular DNA (5, 12). Apoptosis was blocked in a HEp-2-derived cell line that overexpressed Bcl-2 (12).
Earlier studies have also reported that d120 rescuants in which the deleted gene encoding infected cell protein no. 4 was repaired continued to induce apoptosis but that DNA fragments sharing the US3 gene blocked apoptosis (13). Other laboratories have since confirmed that the US3 protein kinase contributes to HSV-mediated protection from a variety of exogenous apoptotic inducers (7, 14–16). In a recent study we have shown that the US3 protein kinase blocked d120-induced apoptosis at a premitochondrial stage and that activation of caspase 3 could be blocked by the US3 protein kinase expressed as late as 6 to 9 h after infection of HEp-2 cells with the d120 mutant (17). Because overexpression of US3 blocked d120-induced apoptosis at a premitochondrial stage, these studies suggested the possibility that the US3 protein kinase targets a member of the Bcl-2 protein family.
Members of the Bcl-2 family of proteins regulate the execution of programmed cell death. The members of this family can be functionally separated into apoptotic antagonists, including Bcl-2, Bcl-XL, and Bcl-w, and apoptotic agonists, such as BAD, BID, and BAX. These key apoptotic regulators mediate their pro- or antiapoptotic signals through their relative abundance, subcellular localization, and posttranslational modifications.
Pro- and antiapoptotic family members are capable of dimerizing through the three Bcl-2 homology domains (BH1, BH2, and BH3), apparently titering out each other's functions (18–20). Specifically, BH1, BH2, and BH3 domains form a hydrophobic cleft to which the BH3 domain can bind (21). Some proapoptotic Bcl-2 family members, such as BAD, contain only the BH3 domain, which is essential for binding to antiapoptotic family members, such as Bcl-2 and Bcl-XL, and for their proapoptotic function (19).
Cell survival signals block BAD from inducing apoptosis by phosphorylation (22). Some of these signals activate phosphatidylinositol 3-kinase with subsequent activation of Akt, which phosphorylates BAD at Ser-136 (23, 24). Survival signals also promote the activation of 90-kDA ribosomal S6 kinase (RSK) and protein kinase A (PKA), which have both been shown to phosphorylate BAD at Ser-112 (25, 26). Phosphorylation of BAD at Ser-112 and Ser-136 has been demonstrated to abrogate its proapoptotic activity by promoting its association with 14-3-3 proteins, which sequester phosphorylated BAD, thereby preventing its localization to the mitochondria and association with Bcl-XL (26–29). Furthermore, phosphorylation of BAD at Ser-155 disrupts its interaction with Bcl-XL (30–32). Conversely, activation of BAD appears to be carried out by phosphatases. Thus two different phosphatases, calcineurin and PP1α, dephosphorylate BAD at Ser-112 and Ser-136, thereby releasing BAD from 14-3-3 proteins, stimulating its binding to Bcl-2/Bcl-XL and ultimately leading to cytochrome c release, caspase activation, and apoptosis (33, 34). Recently it has been reported that death receptor engagement induces the caspase-mediated cleavage of BAD, yielding an Mr 15,000 truncated protein that is a more potent inducer of apoptosis than the full-length BAD (35). This cleavage could serve as a general mechanism to maintain and enhance the proapoptotic functions of BAD.
In this report, we show that the US3 protein kinase, in the absence of other HSV proteins, posttranslationally modified the BAD protein and blocked BAD-induced caspase 3 activation, caspase-dependent cleavage of poly(ADP-ribose) polymerase (PARP), and fragmentation of cellular DNA.
Materials and Methods
Cells.
Rabbit skin cells were originally obtained by J. McLaren and grown in DMEM supplemented with 5% newborn calf serum.
Plasmids.
An EcoRI/NotI fragment from pEBG-mBAD (Cell Signaling Technology, Beverly, MA) containing the mouse BAD ORF fused to glutathione S-transferase was cloned into the baculovirus (Bac) transfer vector MTS-1. MTS-1 contains a cytomegalovirus promoter inserted into the XhoI/EcoRI sites of pAcSG2 (PharMingen). An EcoRI/BamHI fragment encoding glutathione S-transferase was removed, creating BAD-MTS-1 that contains the BAD ORF under the control of the cytomegalovirus promoter.
Baculoviruses.
A BAD-expressing Bac was constructed by cotransfecting the BAD-MTS-1 transfer plasmid with Baculogold DNA (PharMingen) per the manufacturer's instructions. Bac-US3 has been described (17). Efficient Bac gene expression in mammalian cells requires treatment with sodium butyrate, a histone deacetylase inhibitor (36). In all experiments in which rabbit skin cells were infected with Bac, all infected or treated cultures were exposed to medium containing 5 mM sodium butyrate after 2 h of viral infection at 37°C.
Immunoblot Assays.
Cells were harvested as follows. The cells were scraped into their medium, pelleted by low-speed centrifugation, rinsed twice with PBS A (0.14 M NaCl/3 mM KCl/10 mM Na2HPO4/1.5 mM KH2PO4), lysed in RIPA buffer (1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS in PBS A), and stored on wet ice for 10 min before centrifugation at 14,000 × g for 10 min. The protein concentration of the supernatant fluids was determined with the aid of the Bio-Rad protein assay (Bio-Rad) according to directions provided by the manufacturer. Protein samples, denatured in disruption buffer (50 mM Tris, pH 7.0/2.75% sucrose/5% 2-mercaptoethanol/2% SDS) were electrophoretically separated in a 12% denaturing polyacrylamide gel (100 μg of protein per lane), electrically transferred to a nitrocellulose sheet, blocked, and reacted with a rabbit polyclonal antibody specific for PARP (Santa Cruz Biotechnology), BAD, BAD–Ser-136P, BAD–Ser-155P (Cell Signaling Technology), or mouse monoclonal antibody specific for BAD–Ser-112P. Protein bands were visualized with either alkaline phosphatase or through enhanced chemiluminescent detection according to the instructions of the manufacturer (Amersham Pharmacia).
Measurement of DEVDase Activity.
Cellular extracts were assayed for caspase-3 activity with a tetrapeptide (Asp-Glu-Val-Asp) conjugated to phenylnitraniline (DEVD-pNA). Rabbit skin cells grown in 25 cm2 flask cultures were either mock infected or infected with recombinant Bac as indicated. At 18 h after infection, the cells were scraped in their medium, rinsed twice with PBS A, resuspended in 150 μl of lysis buffer (50 mM Hepes, pH 7.4/0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate/1 mM DTT/0.1 mM EDTA), and left on ice for 5 min. The lysates were then centrifuged at 14,000 × g for 10 min at 4°C, and the supernatant was collected and tested for DEVDase activity as recommended by the manufacturer (Biomol, Plymouth Meeting, PA). The released chromophore was measured in a spectrophotometer at 405 nm after 2 h.
DNA Fragmentation Assay.
Infected or uninfected cells were collected; washed in PBS; lysed in a solution containing 10 mM Tris⋅HCl (pH 8.0), 10 mM EDTA, and 0.5% Triton X-100; and centrifuged at 14,000 rpm for 25 min in an Eppendorf microcentrifuge to pellet chromosomal DNA. The supernatant was digested with RNase A (0.1 mg/ml) at 37°C for 1 h, digested with proteinase K (1 mg/ml) at 50°C for 2 h in the presence of 1% SDS, precipitated in cold ethanol, and subjected to electrophoresis in 1.5% agarose gels containing 0.5 mg of ethidium bromide/ml. Oligonucleosomal DNA fragments were visualized by UV light transillumination. Photographs were taken with the aid of Eagle Eye II (Stratagene).
Results
Induction of Apoptosis by a Bac Expressing the BAD Protein.
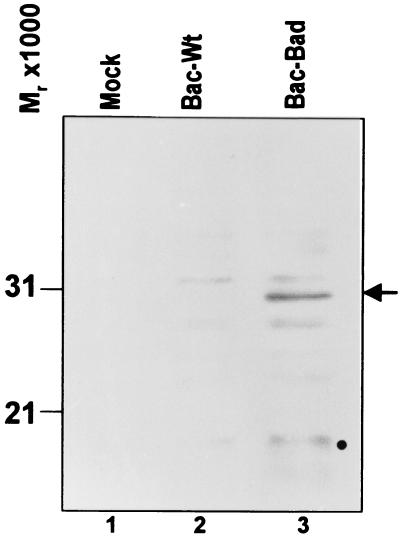
To facilitate the studies on the role of the US3 protein kinase in blocking apoptosis, it became critical to express the BAD protein to all cells in a dose-dependent manner. We chose to deliver BAD by exposing cells to a recombinant Bac, designated Bac-BAD, that expresses the BAD ORF driven by the human cytomegalovirus immediate early promoter as described in Materials and Methods. Rabbit skin cells were either mock infected or exposed to 5 plaque-forming units (pfu) of Bac-WT or Bac-BAD per cell. The cells were harvested 18 h after the addition of Bac-BAD, solubilized, electrophoretically separated in a denaturing polyacrylamide gel, transferred to a nitrocellulose sheet, and reacted with the anti-BAD antibody. The results of the immunoblot assay for the expression of BAD protein are shown in Fig. 1. The BAD protein was detected in cells infected with Bac-BAD but not in mock-infected or Bac-WT-infected cells. In none of the experiments described in this study were we able to detect endogenous BAD. In addition, a potential BAD cleavage product of Mr 15,000 was also detected.
Figure 1.
Photograph of electrophoretically separated cell lysates reacted with polyclonal rabbit antibody to the BAD protein. Replicate 25 cm2 flask cultures of rabbit skin cells were mock-infected or infected with 5 pfu of Bac-WT or Bac-Bad per cell and maintained at 37°C. The cells were harvested and, 18 h after infection, solubilized, electrophoretically separated on denaturing gels, electrically transferred to a nitrocellulose sheet, and reacted with rabbit antibody to the BAD protein. The arrow points to the intact BAD protein. Note the presence of a fainter BAD-related band with an Mr of 15,000, possibly representing the active cleavage product of BAD.
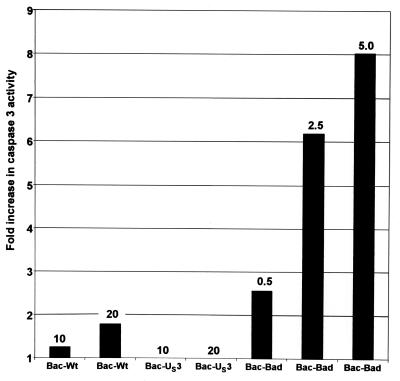
To determine whether Bac-mediated overexpression of BAD is sufficient to induce apoptosis, rabbit skin cells were mock-infected or exposed to 10 or 20 pfu of Bac-WT or Bac-Us3, or to 0.5, 2.5, or 5.0 pfu of Bac-BAD per cell. Cells were harvested at 18 h after infection, lysed, and tested for DEVDase activity. The results, normalized with respect to the level of caspase activity in mock-infected cells, are shown in Fig. 2 and were as follows. Cells infected with either 10 or 20 pfu of Bac-WT per cell exhibited less than a 2-fold increase in DEVDase activity. In cells infected with either 10 or 20 pfu of Bac-US3 per cell, the DEVDase activity was identical to that of mock-infected cells. The DEVDase activity of cells infected with 0.5, 2.5, or 5.0 pfu of Bac-BAD per cell exhibited a 2.5-, 6-, or 8-fold increase in DEVDase activity, respectively, relative to that of mock-infected cells. These results indicated that whereas Bac-WT and Bac-US3 did not induce significant amounts of caspase activity, Bac-BAD induced significant caspase activity in a dose-dependent manner.
Figure 2.
The effect of Bac-mediated gene expression on DEVDase activity in cells. Replicate 25 cm2 flask cultures of rabbit skin cells were either mock-infected or infected with 10 or 20 pfu of Bac-WT or Bac-US3 or 0.5, 2.5, or 5.0 pfu of Bac-BAD per cell. The cells were harvested at 18 h after Bac-BAD infection and assayed for DEVDase activity colorimetrically at 405 nm as described in Materials and Methods. The results are expressed as the fold increase in activity over that of mock-infected cells.
US3 Protein Kinase Blocks DEVDase Activity Induced by BAD Expression.
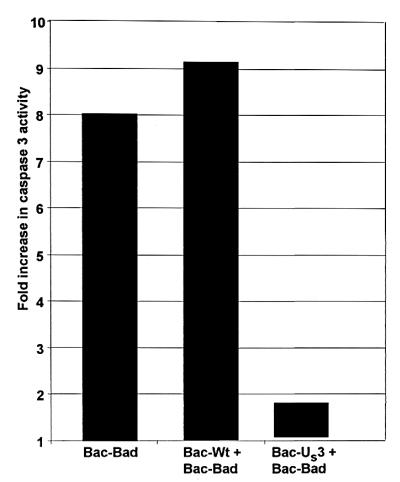
The objective of this series of experiments was to determine whether the US3 protein kinase is sufficient to prevent BAD-induced apoptosis. Rabbit skin cells were either mock-infected or infected with 20 pfu of Bac-WT or Bac-US3 per cell 6.5 h before exposure of the cells to 5.0 pfu of Bad-BAD per cell. The cells were harvested 18 h after Bac-BAD infection and tested for DEVDase activity. As shown in Fig. 3, rabbit skin cells that were either mock-infected or infected with Bac-WT before exposure to Bac-BAD exhibited a 8- or 9-fold increase in DEVDase activity, respectively, relative to that of mock-infected cells that were not exposed to Bac-BAD. In contrast, cells infected with Bac-US3 before Bac-BAD addition exhibited significantly less DEVDase activity (less than 2-fold increase). These results indicate that US3 is sufficient to prevent BAD-induced caspase activation.
Figure 3.
The effect of US3 protein kinase on DEVDase activity induced by BAD. Replicate rabbit skin cell cultures were exposed to 5 pfu of Bac-BAD per cell 6.5 h after infection of the cells with 20 pfu of either Bac-WT or Bac-US3 per cell. The cells were harvested and processed as described in the legend to Fig. 2. The results are expressed as the fold increase in DEVDase activity over that of mock-infected cells.
US3 Protein Kinase Blocks the Cleavage of PARP Induced by BAD Expression.
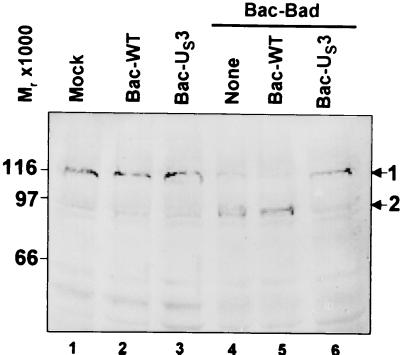
To further verify that Bac-BAD induced caspase activity and that Bac-US3 blocked Bac-BAD-induced caspase activity, we examined the status of PARP, a substrate of caspase 3. Active caspase 3 cleaves the Mr 110,000 PARP to yield a truncated polypeptide with a Mr of 85,000. To determine whether US3 could prevent BAD-induced PARP cleavage, rabbit skin cells were either mock infected or exposed to 20 pfu of Bac-WT or Bac-US3 per cell 6.5 h before exposure to 5 pfu of Bac-Bad per cell. The cells were harvested at 18 h after the addition of Bac-Bad, solubilized, electrophoretically separated in a denaturing polyacrylamide gel, transferred to a nitrocellulose sheet, and reacted with the anti-PARP antibody. The results (Fig. 4) were as follows. The Mr 85,000 PARP cleavage product was not detected in mock-infected, Bac-WT-infected, or Bac-US3-infected cells that had not been treated with Bac-Bad (lanes 1–3). The Mr 85,000 PARP cleavage product was readily detected in mock-infected or in Bac-WT-infected cells that had been exposed to Bac-Bad (lanes 4 and 5), but not in Bac-US3-infected cells that had been exposed to Bac-Bad (lane 6).
Figure 4.
Photograph of electrophoretically separated cell lysates reacted with polyclonal rabbit antibody to the PARP protein. Replicate 25 cm2 flask cultures of rabbit skin cells were mock infected or exposed to 5 pfu of Bac-BAD per cell 6.5 h after exposure of the cells to pfu of either Bac-WT or Bac-US3 per cell. The cells were harvested 18 h after Bac-BAD infection, solubilized, electrophoretically separated on denaturing gels, electrically transferred to a nitrocellulose sheet, and reacted with rabbit antibody to the PARP protein. 1, Full-length PARP; 2, Mr 85,000 PARP cleavage product.
We conclude that, consistent with the results described above, the US3 protein kinase prevented the cleavage of PARP by caspases induced by BAD.
US3 Protein Kinase Blocks DNA Fragmentation Induced by BAD Expression.
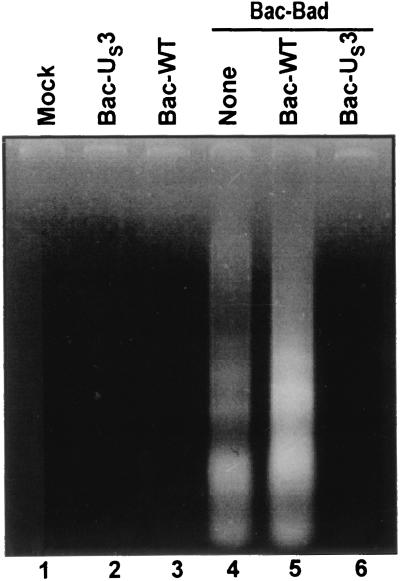
The objective of this series of experiments was to further verify that US3 blocks BAD-induced apoptosis by determining whether US3 prevents the fragmentation of cellular DNA induced by BAD. Rabbit skin cells were either mock-infected or were infected with 20 pfu of Bac-WT or Bac-US3 per cell 6.5 h before exposure to 5 pfu Bac-Bad per cell. The cells were harvested 18 h after exposure to Bac-Bad and processed as described in Materials and Methods. As shown in Fig. 5, lanes 4 and 5, cellular DNA was fragmented in cells that were mock-infected or infected with Bac-WT before exposure to Bac-Bad, but not in mock-infected cells or cells infected with Bac-WT or Bac-US3 (Fig. 5, lanes 1–3). In contrast, DNA fragmentation was not observed in extracts of Bac-Bad-treated cells that had been exposed to Bac-US3 (lane 6). We conclude that, consistent with the evidence presented above, US3 protein kinase is sufficient to block BAD-induced apoptosis.
Figure 5.
Photograph of agarose gel containing electrophoretically separated low-molecular-weight DNA fragments. Replicate 25 cm2 flask cultures of rabbit skin cells were either mock-infected or were infected with 5 pfu of Bac-BAD per cell 6.5 h after exposure to 20 pfu of either Bac-WT or Bac-US3 per cell. The cells were harvested at 18 h after Bac-Bad infection and processed as described in Materials and Methods.
Bac-US3 Mediates Posttranslational Modification of BAD.
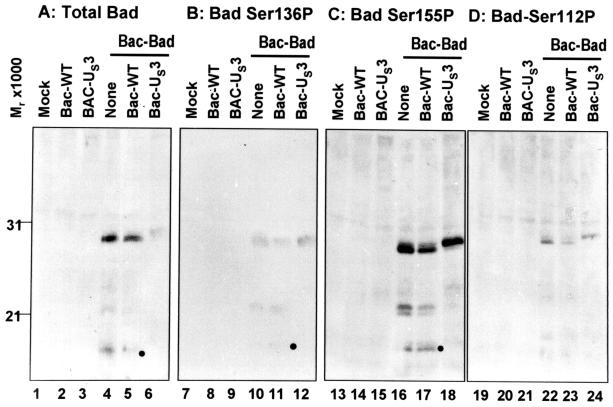
BAD phosphorylated at Ser-112 and Ser-136 is sequestered by the 14-3-3 proteins. In turn, this step blocks the translocation of BAD to the mitochondria and its proapoptotic activity (26–29). In addition, phosphorylation of BAD at Ser-155 appears to be necessary to release the protein from its association with the Bcl-2 family member Bcl-XL (30–32). It was of interest to determine whether US3 prevents BAD-induced apoptosis by posttranslational modification of BAD. To resolve this question, rabbit skin cells were mock-infected or infected with 20 pfu of Bac-WT or Bac-US3 per cell 6.5 h before the exposure of the cells to Bac-Bad (5 pfu per cell). The cells were harvested 18 h after exposure to Bac-BAD, solubilized, electrophoretically separated in denaturing polyacrylamide gels, transferred to a nitrocellulose sheet, and reacted with antibody specific for total BAD protein or to BAD–Ser-136P, BAD–Ser-155P, or BAD–Ser-112P. The results (Fig. 6) were as follows:
Figure 6.
Photograph of electrophoretically separated cell lysates reacted with antibody specific for either BAD or various BAD phosphoforms. Replicate 25 cm2 flask cultures of rabbit skin cells were either mock-infected or infected with 5 pfu of Bac-Bad per cell 6.5 h after infection with 20 pfu of either Bac-WT or Bac-Us3 per cell. Cultures were harvested 18 h after Bac-Bad infection, solubilized, electrophoretically separated on denaturing gels, electrically transferred to a nitrocellulose sheet, and reacted with antibody specific to total BAD (A), phosphoserine136 BAD (B), phosphoserine155 BAD (C), or phosphoserine112 BAD (D).
(i) The total amount of BAD protein detected by anti-BAD antibody in lysates of cells infected with Bac-US3 before exposure to Bac-BAD migrated more slowly and was significantly reduced relative to those amounts detected in lysates of cells exposed to BAC-WT or mock-infected before exposure to Bac-BAD (compare lane 6 with lanes 4 and 5). In this series of experiments we detected BAD only in cells exposed to Bac-BAD (compare lanes 1–3, 7–9, 13–15, 19–21). It is worth noting that Bac-US3 did not affect the accumulation of an irrelevant viral protein expressed by a recombinant Bac (data not shown). These results indicate that US3 mediates both a posttranslational processing of BAD and a reduction in the amount of total BAD protein.
(ii) The levels of BAD phosphorylated at Ser-136, Ser-155, and Ser-112 present in mock-infected cells were similar to those present in cells infected with Bac-WT or Bac-US3. The significant reduction of BAD in cells exposed to Bac-US3 suggests that although the total amount of Bad protein decreased in cells treated with Bac-US3, the amount of BAD protein phosphorylated at specific serines remained the same as in cells exposed to Bac-Bad in the absence of Bac-US3. Thr results suggest therefore that US3 down-regulated primarily if not exclusively active, proapoptotic, nonphosphorylated BAD and concomitantly mediated posttranslational modification of all phosphorylated forms of the protein.
(iii) BAD cleavage products were present in cells that had been mock-infected or infected with Bac-WT before the addition of Bac-BAD, but were absent in cells infected with Bac-US3 before Bac-Bad addition. Recently it has been reported that caspase-mediated cleavage of BAD results in a Mr 15,000 truncated product that stimulates the apoptotic activity of BAD (35). That Bac-US3 prevents the cleavage of BAD is consistent with the evidence that US3 protein kinase prevents BAD from inducing apoptosis.
Taken together, these results indicate that US3 (i) mediated a posttranslational modification of full-length Bad, (ii) mediated a decrease in the total amount of BAD, and (iii) prevented the accumulation of a Mr15,000 cleavage product of BAD, all of which could serve to block the proapoptotic activity of BAD.
Discussion
Earlier reports have shown that the d120 mutant of HSV-1 induced apoptosis in a cell-type-independent manner, but that the pathway to apoptosis is cell-type dependent. Specifically, in some cell lines, the d120 mutant caused the release of cytochrome c, the activation of caspase 3, and fragmentation of cellular DNA (3, 5). In at least one cell line tested, overexpression of Bcl-2 blocked all of these events (12). In more recent studies we have shown that the US3 viral protein kinase blocks apoptosis induced by the d120 mutant at a premitochondrial stage (17). These results suggested the possibility that the US3 protein kinase acts on a proapoptotic member of the Bcl-2 family. In this paper we report that a Bac expressing the US3 protein kinase gene under a promoter active in mammalian cells blocks apoptosis induced by BAD. We also show that this function of the US3 protein does not require the participation of other HSV proteins. The following observations and measurements are relevant to our results:
(i) HSV-1 replication, defined by the accumulation of infectious virus, lasts ≈18–24 h, although viral protein synthesis can last much longer. The replication of HSV is accompanied by the development of cytopathic effects. Earlier, this laboratory reported that in cells overexpressing Bcl-2, the development of cytopathic effects is delayed without significant effect on viral replication (12). These results suggest that cytopathic effects at the end of the replicative cycle reflect injuries to the cell resulting from proapoptotic events. These proapoptotic events most likely are not specifically targeted by the virus to be blocked, inasmuch as they are not detrimental to viral replication, as they occur very late during infection. Alternatively, some proapoptotic events may benefit viral replication. If such were the case, the virus would dictate the cellular apoptotic environment by blocking apoptotic events that are detrimental to viral replication and by allowing apoptotic events that benefit viral replication. The available data suggest that the virus has evolved a number of inducers of apoptosis, and each is very likely blocked by specific gene products. A number of different HSV-1 gene products have been reported to inhibit apoptosis, among them US3, gD, and gJ (4, 7, 10). It must be stressed, however, that in the normal course of wild-type virus replication (i.e., at 18–24 h), late apoptotic manifestations such as caspase 3 activation and DNA fragmentation are not detected, and, as noted in the introduction, wild-type virus protects cells from these apoptotic manifestations induced by exogenous agents.
(ii) HSV-1 encodes two well-characterized protein kinases. The UL13 protein kinase has been shown to mediate the phosphorylation of a large number of viral and cellular proteins. In contrast, the US3 protein kinase appears to have a narrow, less defined range of substrates. Its functions in the course of viral infection have always been somewhat of a puzzle: although a virus deleted for the US3 ORF is growth impaired in mice (37), the gene is not essential for viral replication in tissue culture, although the cytopathic effects of the deletion mutant are very different from those of wild-type virus (38). By light microscopy the cells have a crenated appearance but exhibit no evidence of classical apoptosis resulting in activation of caspases or degradation of cellular DNA. These observations lead us to conclude that the stimulus for the activation of apoptosis that is blocked by the US3 protein kinase either is not activated in wild-type infected cells or is blocked by another HSV gene function. The nature of this stimulus is under investigation.
(iii) In the studies described in this report, we measured both the total BAD protein accumulating in cells infected with Bac-BAD and the accumulation of posttranslationally processed Bac-BAD. Several interesting and potentially significant observations emerged from these studies. First, BAD accumulating in cells infected with Bac-US3 before exposure to Bac-BAD decreased significantly relative to that present in cells infected with Bac-BAD alone or in the presence of Bac-WT. These results indicate that a fraction of Bac-BAD in cells exposed to Bac-US3 was targeted for degradation. Another interesting feature is the presence of BAD cleavage products reported to activate apoptosis in cells infected with Bac-BAD alone or in the presence of Bac-WT but not in cells infected with Bac-US3 only. Second, some of the BAD accumulating in cells infected with Bac-BAD alone was phosphorylated at Ser-136, Ser-155, and/or Ser-112. An additional posttranslational modification of BAD manifested by a slower electrophoretic mobility in denaturing polyacrylamide gels was observed in cells infected with Bac-US3 before exposure to Bac-BAD. The observation of interest is that the amounts of intact BAD that was phosphorylated at Ser-155 and Ser-112 in cells infected only with Bac-Bad and in cells infected with Bac-US3 were relatively similar. Taken together, these results suggest the following scenario: In cells infected with Bac-BAD, at least a fraction of BAD was phosphorylated at Ser-112, Ser-136, and/or Ser-155. In addition, a fraction of BAD was cleaved either before or after phosphorylation of the serines. The US3 protein kinase precluded the accumulation of cleaved BAD and posttranslationally processed virtually all of the full-length BAD. In light of the relatively equivalent amounts of BAD phosphorylated at Ser-112, Ser-136, or Ser-155, the data can be interpreted to signify that BAD targeted for destruction by US3 protein kinase was BAD molecules lacking one or more of the phosphorylated serines, that is, active BAD.
Acknowledgments
These studies were aided by grants from the National Cancer Institute (CA87761, CA83939, CA71933, and CA78766).
Abbreviations
- HSV
herpes simplex virus
- PARP
poly(ADP-ribose) polymerase
- Bac
baculovirus
- pfu
plaque-forming units
References
- 1.Koyama A H, Miwa Y. J Virol. 1997;71:2567–2571. doi: 10.1128/jvi.71.3.2567-2571.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sieg S, Yildrim Z, Smith D, Kayagaki N, Yagata H, Huang Y, Kaplan D. J Virol. 1996;70:8747–8751. doi: 10.1128/jvi.70.12.8747-8751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galvan V, Roizman B. Proc Natl Acad Sci USA. 1998;95:3931–3936. doi: 10.1073/pnas.95.7.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leopardi R, Roizman B. Proc Natl Acad Sci USA. 1996;93:9583–9587. doi: 10.1073/pnas.93.18.9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galvan V, Brandimarti R, Roizman B. J Virol. 1999;73:3219–3226. doi: 10.1128/jvi.73.4.3219-3226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aubert M, O'Toole J, Blaho J A. J Virol. 1999;73:10359–10370. doi: 10.1128/jvi.73.12.10359-10370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jerome K R, Fox R, Chen Z, Sears A E, Lee Hy, Corey L. J Virol. 1999;73:8950–8957. doi: 10.1128/jvi.73.11.8950-8957.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLuca N A, McCarth A, Schaffer P A. J Virol. 1985;56:558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auber M, Blaho J A. J Virol. 1999;73:2803–2813. doi: 10.1128/jvi.73.4.2803-2813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou G, Galvan V, Campadelli-Fiume G, Roizman B. J Virol. 2000;74:11782–11791. doi: 10.1128/jvi.74.24.11782-11791.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batterson W, Furlong D, Roizman B. J Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galvan V, Brandimarti R, Munger J, Roizman B. J Virol. 2000;74:1931–1938. doi: 10.1128/jvi.74.4.1931-1938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leopardi R, Van Sant C, Roizman B. Proc Natl Acad Sci USA. 1997;94:7891–7896. doi: 10.1073/pnas.94.15.7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asano S, Honda T, Goshima F, Watanabe D, Miyake Y, Sugiura Y, Nishiyama Y. J Gen Virol. 1999;80:51–56. doi: 10.1099/0022-1317-80-1-51. [DOI] [PubMed] [Google Scholar]
- 15.Asano S, Honda T, Goshima F, Nishiyama Y, Sugiura Y. Neurosci Lett. 2000;294:105–108. doi: 10.1016/s0304-3940(00)01554-8. [DOI] [PubMed] [Google Scholar]
- 16.Hata S, Koyama A H, Shiota H, Adachi A, Goshima F, Nishiyama Y. Microbes Infect. 1999;1:601–607. doi: 10.1016/s1286-4579(99)80059-8. [DOI] [PubMed] [Google Scholar]
- 17.Munger J, Chee A V, Roizman B. J Virol. 2001;75:5491–5497. doi: 10.1128/JVI.75.12.5491-5497.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oltvai Z N, Milliman C L, Korsmeyer S J. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 19.Chittenden T, Flemington C, Houghton A B, Ebb R G, Gallo G J, Elangovan B, Chinnadurai G, Lutz RJ. EMBO J. 1995;14:5589–5596. doi: 10.1002/j.1460-2075.1995.tb00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin X M, Oltvai Z N, Korsmeyer S J. Nature (London) 1994;369:321–323. doi: 10.1038/369321a0. [DOI] [PubMed] [Google Scholar]
- 21.Sattler M, Liang H, Nettesheim D, Meadows R P, Harlan J E, Eberstadt M, Yoon H S, Shuker S B, Chang B S, Minn A J, et al. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 22.Franke T F, Cantley L C. Cell. 1997;91:231–241. [Google Scholar]
- 23.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 24.Scheid M P, Duronio V. Proc Natl Acad Sci USA. 1998;95:7439–7444. doi: 10.1073/pnas.95.13.7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan Y, Ruan H, Demeter M R, Comb M J. J Biol Chem. 1999;274:34859–34867. doi: 10.1074/jbc.274.49.34859. [DOI] [PubMed] [Google Scholar]
- 26.Harada H, Becknell B, Wilm M, Mann M, Huang L J, Taylor S S, Scott J D, Korsmeyer S J. Mol Cell. 1999;3:413–422. doi: 10.1016/s1097-2765(00)80469-4. [DOI] [PubMed] [Google Scholar]
- 27.Pastorino J G, Tafani M, Farber J L. J Biol Chem. 1998;274:19411–19416. doi: 10.1074/jbc.274.27.19411. [DOI] [PubMed] [Google Scholar]
- 28.Zha J, Harada H, Yang E, Jockel J, Korsmeyer S J. Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 29.Hsu S Y, Karpia A, Zhu L, Hsueh A J. Mol Endocrinol. 1997;12:1858–1867. doi: 10.1210/mend.11.12.0023. [DOI] [PubMed] [Google Scholar]
- 30.Zhou X M, Lui Y, Payne G, Lutz R J, Chittenden T. J Biol Chem. 2000;275:25046–25051. doi: 10.1074/jbc.M002526200. [DOI] [PubMed] [Google Scholar]
- 31.Tan Y, Demeter M R, Ruan H, Comb M J. J Biol Chem. 2000;275:25865–25869. doi: 10.1074/jbc.M004199200. [DOI] [PubMed] [Google Scholar]
- 32.Datta S R, Katsov A, Hu L, Petros A, Fesik S W, Yaffe M B, Greenberg M E. Mol Cell. 2000;6:41–51. [PubMed] [Google Scholar]
- 33.Salomoni P, Condorelli F, Sweeney S M, Calabretta B. Blood. 2000;96:676–684. [PubMed] [Google Scholar]
- 34.Wang H-G, Pathan N, Ethel I M, Krajewski S, Yamaguchi Y, Shibasaki F, McKeon F, Bobo T, Franke T F, Reed J C. Science. 1999;284:339–343. doi: 10.1126/science.284.5412.339. [DOI] [PubMed] [Google Scholar]
- 35.Condorelli F, Salomoni P, Cotteret S, Cesi V, Srinivasula S M, Alnemri E S, Calabretta B. Mol Cell Biol. 2001;21:3025–3036. doi: 10.1128/MCB.21.9.3025-3036.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Condreay J P, Witherspoon S M, Clay W C, Kost T A. Proc Natl Acad Sci USA. 1999;96:127–132. doi: 10.1073/pnas.96.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meignier B, Longnecker R, Mavromara-Nazos P, Sears A E, Roizman B. Virology. 1988;162:251–254. doi: 10.1016/0042-6822(88)90417-5. [DOI] [PubMed] [Google Scholar]
- 38.Purves F C, Longnecker RM, Leader D P, Roizman B. J Virol. 1987;61:2896–2901. doi: 10.1128/jvi.61.9.2896-2901.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]