Abstract
Comparative analysis of metagenomes can be used to detect sub-metagenomes (species or gene sets) that are associated with specific phenotypes (e.g., host status). The typical workflow is to assemble and annotate metagenomic datasets individually or as a whole, followed by statistical tests to identify differentially abundant species/genes. We previously developed subtractive assembly (SA), a de novo assembly approach for comparative metagenomics that first detects differential reads that distinguish between two groups of metagenomes and then only assembles these reads. Application of SA to type 2 diabetes (T2D) microbiomes revealed new microbial genes associated with T2D. Here we further developed a Concurrent Subtractive Assembly (CoSA) approach, which uses a Wilcoxon rank-sum (WRS) test to detect k-mers that are differentially abundant between two groups of microbiomes (by contrast, SA only checks ratios of k-mer counts in one pooled sample versus the other). It then uses identified differential k-mers to extract reads that are likely sequenced from the sub-metagenome with consistent abundance differences between the groups of microbiomes. Further, CoSA attempts to reduce the redundancy of reads (from abundant common species) by excluding reads containing abundant k-mers. Using simulated microbiome datasets and T2D datasets, we show that CoSA achieves strikingly better performance in detecting consistent changes than SA does, and it enables the detection and assembly of genomes and genes with minor abundance difference. A SVM classifier built upon the microbial genes detected by CoSA from the T2D datasets can accurately discriminates patients from healthy controls, with an AUC of 0.94 (10-fold cross-validation), and therefore these differential genes (207 genes) may serve as potential microbial marker genes for T2D.
Keywords: metagenome, concurrent subtractive assembly, Wilcoxon rank-sum test, comparative metagenomics
1 Introduction
The human body is host to trillions of bacteria cells, outnumbering human cells by 1.3 to 1 (in contrast to the widely cited 10:1 ratio), according to a recent estimate [40]. Moreover, the genes encoded by human microbiome are hundreds of times more than the human complement [47]. It has been reported that those microorganisms are involved in ~20% of human malignancies [6]. The gut microbiota has been linked to a variety of conditions including inflammatory bowel disease [23], cardiovascular disease [19], rheumatoid arthritis [38], Parkinson’s disease [37], autism spectrum disorder [16], colon cancer [9, 39], and liver cirrhosis [34], among others. However, only 10 microbes are designated to be carcinogenic to human beings by the International Agency for Cancer Research (IACR) [6]. Therefore, it is intriguing to explore microbes that are directly related to the development of human diseases.
The development of next generation sequencing has pushed the advancement of metagenomics, which presents us a great opportunity to identify microorganisms that are enriched or depleted during disease and explore possible mechanisms behind the association. The human microbiome project has shown the association between the shifts in our microbiota and diseases such as obesity [17] and periodontitis [15]. Although the change in identify of the species (or abundance) does not ensure a causal role for the microbes, we can narrow down the set of candidate genomes or genes by such studies. One new trend of microbiome research is microbiome-wide association studies (MWAS), which are analogous to genome-wide association studies (GWAS) [20]. MWAS may take a case-control approach, revealing the association between microbiomes and human diseases. However, the limitation of this approach is that it cannot distinguish whether the microbiome drives the disease, the disease drives the microbiome, or both are modified by confounding factors. On the other hand, longitudinal studies may allow researchers to test whether changes in the microbiome precede or follow the development of disease [5, 11].
In the seeking of disease-associated microbes, we should note the significant compositional variations of microbiota from individual to individual [10]. Regarding this interpatient variability, the correct strategy is to identify conserved microbial community behaviors in microbiota-associated diseases [15]. Microbial marker gene surveys have been used extensively to reveal the association of microbiota with diseases such as diabetes and Crohn’s disease [31]. For instance, Qin et al. identified 15 optimal marker genes from the gut microbiome in liver cirrhosis by comparing 98 patients and 83 healthy control individuals [34]. Based on only the 15 biomarkers, they were able to construct a classifier that can discriminate patients with a decent accuracy [34]. Similarly, gut microbiota was explored to detect colorectal cancer and a metagenomic classifier was trained using the taxonomic abundances of 22 marker species [45]. The typical workflow of these marker-gene surveys is to assemble the metagenomes and then predict the genes, potential marker genes can then be identified by detecting significant differences in their distribution across healthy and disease populations. The analysis of differential abundance is critical for these surveys and computational tools have been developed for the analysis, including a recently developed approach that relies on a novel normalization technique and a statistical model accounting for undersampling [31].
Due to the complexity of microbial communities, the de novo partition of metagenomic space into specific biological entities remains to be difficult. To address this problem, researchers have utilized various features, including compositional features such as tetra-nucleotide statistics [13] and coverage signals of genetic sequences [1, 44]. However, the assumptions of those methods are not universally true. For example, the methods relying on abundances of genetic sequences are admittedly weak in segregating taxonomically related organisms [1]. In the process of exploring other features, it has been realized that utilizing co-abundance across multiple samples improves the resolution of genome segregation from metagenomic data sets [29, 2, 43]. Similarly, we should also utilize information from multiple samples for the sake of identifying conserved differential patterns.
We have previously introduced a method called subtractive assembly (SA) [42], which is a de novo method to compare metagenomes by identifying and assembling the differential reads. We have demonstrated that SA can recover the differential genomes by effectively extracting the differential reads based on sequence signatures (frequencies of k-mers). Also, SA can improve the quality of metagenomic assembly when only a subset of closely-related genomes change in their abundances between the groups of samples in comparison. Application of SA to gut metagenomes from women with type 2 diabetes (T2D) [17] reveals compositional features and a large collection of unique or abundant genes in T2D gut metagenomes (some of the genes identified by SA were otherwise missed by direct assembly of the original datasets). SA utilizes both the compositional and coverage features through the composition and frequency of k-mers, contributing to its superior performance. However, the SA method pools the samples for each group before comparison and therefore loses power in detecting minor but consistent changes without using information from individual samples. In addition, SA picks up genes in species which only appear in a few samples but with high abundances, as a result, many of the “differential” genes assembled are not actually consistently abundant across samples in the same group. Therefore additional profiling of gene abundance is required in order to search for genes consistently more abundant in one group versus the other.
In this paper, we further developed the subtractive assembly approach for the detection of consistently differential genomes or genes by using k-mer frequencies in individual samples (co-abundance). We adopted KMC 2 [7] for k-mer counting in our implementation, since KMC 2 is one of the fastest k-mer counting approaches, which was claimed to be twice faster than the strongest competitors such as Jellyfish 2 [26]. Differential reads extracted from individual samples were then pooled for assembly. We call our new method Concurrent Subtractive Assembly approach (CoSA). We observed that some reads are extremely redundant (those sampled from abundant common species across samples). We further developed a strategy to remove redundant reads based on k-mer counts: only some of the reads that contain highly abundant k-mers are retained for assembly. Using simulated datasets, we showed that CoSA achieves much better performance in detecting consistent changes than the original subtractive assembly (SA) approach. Moreover, we applied it to analyzing T2D gut metagenomes to identify microbial marker genes, based on which we built a classifier that accurately discriminates patients from healthy controls.
2 Materials and Methods
2.1 Overview
Concurrent Subtractive Assembly (CoSA) is designed to identify the short reads that make up the conserved/consistent compositional differences across multiple samples based on sequence signatures (k-mer frequencies), and then to only assemble the differential reads, aiming to reveal the consistent differences between two groups of metagenomic samples (e.g., metagenomes from cancer patients vs. metagenomes from healthy controls).
2.2 k-mer counting
CoSA is a k-mer-based method, and therefore the first step is the counting of all k-mers in metagenomic samples. For comparative metagenomic studies, the sheer size of the datasets is a fundamental challenge. We employed KMC 2 for k-mer counting. We specified the maximal value of a count (the cs flag) as 65,536 instead of 255 by default. On one hand this helps identify the more frequently observed differential k-mers by using a larger cut-off value; on the other hand we can store each count using a 16-bit unsigned integer, which demands a reasonable amount of memory or disk space when dealing with billions of k-mers.Meanwhile, we exclude k-mers occurring less than two times by the ci option based on the fact that a large number of singletons are products from sequencing errors, as previously employed by both BFCounter [28] and khmer [46].
After k-mer counting with KMC 2, CoSA goes through the outputs of KMC by using the KMC API and stores all observed k-mers in a hash table, implemented using the libcuckoo library (downloaded from https://github.com/efficient/libcuckoo). Libcuckoo [25] provides a high-performance concurrent hash table, by which we can efficiently update the hash table using multiple threads. With the k-mers in the hash table, CoSA accesses the outputs of KMC again and writes to disk the counts of the k-mers based on their orders in the hash table for every sample. By storing the counts on the disk, we can load the counts of k-mers in batches and therefore significantly reduce the memory requirement for recording the counts of all k-mers in every sample.
2.3 Identification of differential k-mers using Wilcoxon rank-sum test
CoSA by default loads 107 k-mers into a two-dimensional array each time and iteratively tests if the frequencies of each k-mer are differential between the two groups of samples. To compare k-mers in different metagenomic samples, we calculate the frequency of each k-mer in each metagenomic sample. In case the frequency of a rare k-mer is extremely small, we compute the frequency of a k-mer as the number of occurrences per million k-mers. Then the normalized frequencies are used for WRS test (a nonparametric test), for which we employ the “mannwhitneyutest” function from ALGLIB (http://www.alglib.net). The WRS test is used to detect k-mers that have different frequencies in one group of the samples (e.g., the patient group) than the other group of samples (e.g., the healthy control) with statistical significance. The k-mers that pass the test (p-value cut-off is set to 0.05 by default) are identified as differential k-mers.
We tested different k-mer sizes empirically. Bigger k-mer size increases the memory assumption by CoSA, but has very little impact on the results of extracted reads and downstream application of the reads. We therefore set the default k-mer size to 23.
2.4 Identification of differential reads based on differential k-mers
Reads that are composed of differential k-mers tend to be from differential genomes. Thus, we extract differential reads in each sample based on the differential k-mers using a voting strategy. With the voting threshold as 0.5, for example, a read is considered to be differential if 50% of its k-mers belong to differential k-mers. We empirically tested the voting threshold and found a value in the range of 0.3 ~ 0.8 gives a good balance between the number of extracted reads and efficiency of the differential gene assembly. However, users may change this parameter (−v) in their own applications of CoSA.
2.5 Reduction of reads redundancy
We noticed that some k-mers are extremely abundant in the extracted reads file (these k-mers are likely from the reads sampled from abundant species that are common across many samples). When the differential reads contain these k-mers, the distribution of k-mers is skewed and this can challenge the assembly algorithm. To address this issue, we reduced the reads redundancy by excluding reads that contain highly abundant k-mers. The reads redundancy removal relies on a list of highly abundant k-mers prepared based on k-mer counts. A read is determined to be redundant if it contains many k-mers on the abundant k-mer list. Specifically, for each read, the fraction of abundant k-mer (over all k-mers) is computed and used for determing the fate of the read: if the fraction is smaller than a random number between 0 and 1 generated by the program, the read is retained; otherwise, it is discarded. In this way, a read that has a higher ratio of abundant k-mers will have a higher chance to be discarded.
2.6 Assembly of extracted reads and downstream annotations
Following the read extraction, any metagenomic assembler can be employed in subtractive assembly. Here, we used MegaHIT (with meta-large presets option)[ 24](version 1.0.2) to assemble the differential reads, to illustrate the usage of CoSA. For each group (e.g., T2D patients, or healthy controls), differential reads extracted from individual samples were pooled and assembled together by MegaHIT. We note that we only pooled reads from multiple samples in the same group for assembly. We used MegaHIT as it is one of the recently developed assemblers that are memory efficient and fast. But in principle, other assemblers such as IDBA-UD [33] and metaSPAdes [3] can be used as well. In order to identify differential genes, protein coding genes were predicted from the contigs using FragGeneScan [35] (version 1.30).
To estimate the abundance of the genes, all the reads from each sample were aligned against the gene set by using Bowtie 2 [22](version 2.2.6). We counted a gene’s abundance based on the counts of both uniquely and multiplely mapped reads. The contribution of multiplely mapped reads to a gene was computed according to the proportion of the multiplely mapped read counts divided by the gene’s unique abundance [34]. The read counts were then normalized per kilobase of gene per million of reads in each sample.
2.7 Building classifiers
After the gene abundance profile was built, we attempted to build a classifier that can discriminate patients from healthy controls. We first used L1-based feature selection method in the “scikit learn” python package [32] to select genes. After the feature selection, we built classifiers using Random Forest (RF) and Support Vector Machine (SVM). We used RF as it has been shown to be a suitable model for exploiting non-normal and dependent data such as metagenomic data [18] and it was used for prediction of T2D in [17]. On the other hand, SVMs are widely used in computational biology due to their high accuracy and their ability to deal with high-dimensional and large datasets [4]. We used the SVM (linear kernal) and RF (10 trees) in the “scikit learn” python package. We evaluated the predictive power of a model as the Area Under Curve (AUC) using a tenfold cross-validation method.
We tested different p-value cut-offs and voting thresholds used in CoSA for evaluating their impact on the accuracy of the classifiers built from genes derived by CoSA.
2.8 Simulated and real metagenome datasets
To test the performance of CoSA in detecting minor effects, we first generated two groups of metagenomic datasets using five bacterial genomes from the FAMeS dataset [27] by MetaSim [36], with each group representing a unique population structure; and for each group, we simulated 10 samples.
As a showcase for CoSA, we further applied our method to the T2D cohort. The T2D cohort was derived from two groups of 70-year-old European women, one group of 50 with T2D and the other a matched group of healthy controls (NGT group; 43 participants). We did not use 3 samples of T2D datasets that were outliers based on neighbor-joining clustering using a dissimilarity measure for k = 9 [14]. We tested our original SA approach using the T2D cohort, and in this study, we focused on the comparison of CoSA with SA using the T2D datasets. Table 1 summarizes the simulated datasets and the T2D microbiome datasets we used for testing.
Table 1.
Summary of the simulated and T2D datasets.
| Simulated | T2D and healthy | |
|---|---|---|
|
| ||
| Number of datasets | 20 | 93 |
| Total bps | 2.29Gbp | 225.30Gbp |
| Number of k-mers | 9,112,554 | 4,121,225,700 |
2.9 Availability of CoSA
We implemented CoSA in C++. Because CoSA employs k-mer frequencies from individual samples, it introduces a new dimension for different samples and therefore increases the requirement of computational resources, especially for large cohort of datasets such as the T2D datasets. To reduce the running time and memory usage, we implemented CoSA with multiple threading. Also, counts of k-mers are written to disk and then loaded back in batches for the detection of differential k-mers (since it is impossible to load all k-mer counts into the memory at the same time). The software is available for download at sourceforge (https://sourceforge.net/projects/concurrentsa/).
3 Results
We first report the results of CoSA using simulated datasets. We then report the comparison of CoSA with our original SA method using the T2D cohort. Finally we report the results of using CoSA for extracting and charactering disease associated sub-microbiome using the T2D datasets.
3.1 Evaluation of CoSA using simulated datasets
Instead of using fold change of k-mers, CoSA detects differential genomes by testing k-mer frequencies with Wilcoxon rank-sum test. Also, it employs k-mer frequencies concurrently from multiple samples for each group in comparison. In theory CoSA has the capability of detecting minor but consistent changes between groups of samples. To test the performance of CoSA in such case, we simulated metagenomic samples using two population (community) structures (Table 2). The Streptococcus thermophilus LMD-9 genome is two times more in population one (P1) than in population two (P2) in terms of relative abundance. Similarly, Prochlorococcus marinus NATL2A is the differential genome that is two times more abundant in P2 than in P1. Since there is only a fold change of two for the differential genomes, it is hard to detect the minor effects through fold change of k-mers (as a result SA performed poorly on this simulated dataset; see below).
Table 2.
Evaluation of CoSA using simulated datasets: community structure and reads extraction.
| Population | Reads extracted/simulated | |||
|---|---|---|---|---|
| P1a | P2 | P1 | P2 | |
| Ferroplasma acidarmanus fer1 | 1b | 1 | 0/38,568 c | 19/38,569 |
| Lactobacillus gasseri ATCC 33323 | 2 | 2 | 122/75,528 | 77/76,152 |
| Pediococcus pentosaceus ATCC 25745 | 4 | 4 | 178/146,787 | 25/147,199 |
| Prochlorococcus marinus NATL2A | 8 | 16 | 8/295,230 | 587,980/588,579 |
| Streptococcus thermophilus LMD-9 | 16 | 8 | 590,820/593,858 | 0/297,227 |
simulated population 1;
relative abundance of the F. acidarmanus genome in population 1;
0 reads were extracted out of 38,568 reads from the F. acidarmanus genome in P1.
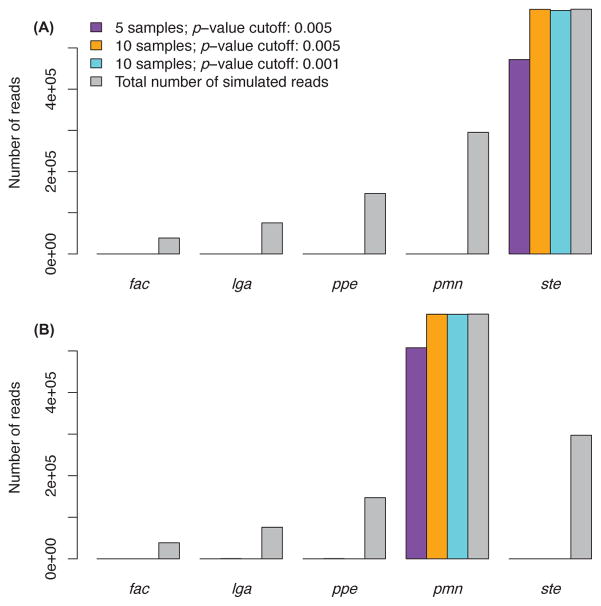
We evaluated CoSA with different parameters, including p-value cut-off and number of samples for each group in comparison. First, we compared the efficacy of read extraction using either 5 or 10 samples for each population. The results show that CoSA extracted more reads from the differential genomes by using more samples (Figure 1). For example, using a p-value cut-off of 0.005, CoSA extracted 593,739 (99.98%) out of 593,858 short reads (expected) for the S. thermophilus LMD-9 genome when 10 samples were used (see Table 2). When using only 5 samples for each population, CoSA could only extract 471,786 (79.44%) reads. Meanwhile, CoSA extracted very few reads from the non-differential genomes in both cases. Using a lower p-value cut-off of 0.001 (see Table 2 for the results) reduced the number of extracted reads from both differential and non-differential genomes. But CoSA still extracted most of the reads from the differential genomes. In conclusion, CoSA effectively extracted reads from differential genomes with a minor fold change whereas a minimal number of reads were extracted from non-differential genomes. We note that a very stringent p-value cut-off (e.g., 0.001) works well for this simulated case; however, for real microbiome datasets that have more complex population structure, a less stringent p-value cut-off might be needed for differential reads extraction (because of the sharing of k-mers among species) as shown in the application of CoSA to the T2D microbiomes (see below).
Fig. 1.
CoSA effectively extracted reads from differential genomes. The upper and lower subfigures refer to read extraction for one of the samples of population 1 and 2, respectively. The x-axis shows the 5 different species; fac: Ferroplasma acidarmanus fer1, lga: Lactobacillus gasseri ATCC 33323, ppe: Pediococcus pentosaceus ATCC 25745, pmn: Prochlorococcus marinus NATL2A, ste: Streptococcus thermophilus LMD-9. Bars of different colours (purple, yellow, cyan) indicate separate runs of CoSA using different parameters or different number of samples while the grey bars indicate simulated reads for each genome. The y-axis shows the number of reads extracted (or expected shown in gray bars).
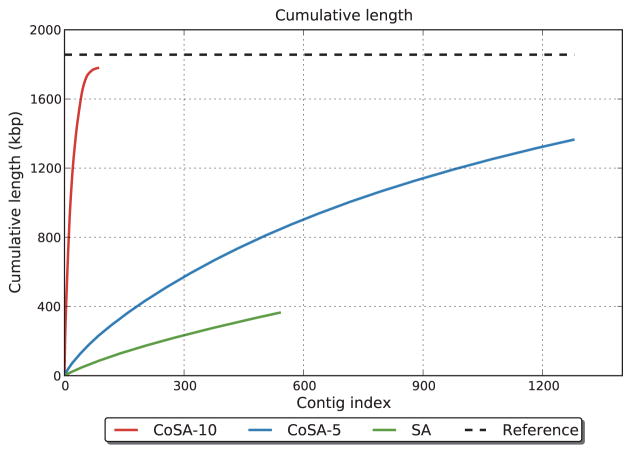
We further compared the assembly quality for the differential genomes with different number of samples, with the help of QUAST [12] and MUMer [21]. For the S. thermophilus LMD-9 genome in the same sample as above, we recovered 95.76% of the reference genome when 10 samples per population were used; but only 73.32% of the genome were assembled when we used 5 samples for each group (see Figure 2 for the comparison). Not only we assembled a higher fraction of the genome for the differential genomes, but also we obtained fewer but longer contigs. We produced 84 contigs with N50 of 51,061 using 10 samples and 1,280 contigs with N50 of 1,180 using 5 samples. With more samples, CoSA is capable of better assembling the differential genomes. By contrast, our original SA approach relies on ratios of k-mers to detect differential reads and only a small fraction (19.64%) of the genome can be assembled using the reads it extracted.
Fig. 2.
Evaluation of the assembly quality of the differential genomes. The results indicate that CoSA outperforms SA for detecting minor but consistent effect when multiple samples are used, and that using more samples by CoSA results in better assembly of the differential genomes (CoSA-10, 10 samples were used; CoSA-5, 5 samples were used).
3.2 Evaluation of CoSA using the T2D microbiomes
As shown in the above, CoSA was able to detect minor, but conserved differential genomes using the simulated datasets. Here we applied CoSA to the T2D microbiome cohort. As shown in Table 3, CoSA has resulted in a greater reduction of the sequencing data (retaining 8.99% of the total bases) than the original SA reads (which retained 17.59% of the original sequencing data). Extracted reads were then used for assembly and gene annotation. Although reads extraction by CoSA resulted in a smaller collection of microbial genes than the SA approach (since CoSA retained much fewer reads than SA), genes from CoSA tend to be more consistently differential across the samples between the groups. We pooled the genes derived from CoSA (1,008,068 genes) and SA (1,648,016 genes), resulting a collection of 2,656,084 genes, and futher quantified the abundances of the genes in this collection. The gene abundance profile was then used for WRS test between the patient and the healthy control groups, with correcting for multiple testing using false discovery rate (q-value) computed by the tail area-based method of the R fdrtool package [41]. Table 3 summaries the test results, indicating that CoSA produced more significantly differential genes than SA. We note that none of the genes derived by SA had q-value less than 0.05. Sequences and annotations of the 357,591 genes assembled by CoSA (with q-value ≤ 0.05) are available for download at the CoSA sourceforge project page.
Table 3.
Summary of subtractive assembly results of the T2D datasets.
| CoSA* | SA | |
|---|---|---|
|
| ||
| Total base pair in extracted reads | 11.59 Gbp (8.99%) | 22.68 Gbp (17.59%) |
| # of predicted genes a | 1,008,068 | 1,648,016 |
| # of significant genes (q-value ≤ 0.07) | 563,743 | 285,666 |
| # of significant genes (q-value ≤ 0.05) | 357,591 | 0 |
p-value=0.2 and voting threshold=0.8 were used for reads extraction;
only counted genes assembled from extracted reads from patients (but not healthy individuals).
3.3 Prediction of T2D using microbial genes
It has been shown that metagenomes can be used for classification and prediction of diabetes status [17]. Karlssons and colleagues trained a Random Forest (RF) model based on a training set of the NGT and T2D subjects using the profiles of species and MGCs (megenomic gene clusters), and evaluated its performance using a tenfold cross-validation approach and calculated the predictive power as the area under the ROC curve (AUC). Their results showed that T2D was identified more accurately with MGCs (highest AUC = 0.83) than with microbial species (highest AUC = 0.71), suggesting that the functional composition of the microbiota determined by MGCs correlates better with diabetes than the species composition. We applied CoSA to T2D datasets (including datasets from patients and healthy individuals) using different settings of parameters and compared the performance of classifiers built from the assembled microbial genes (from both T2D patients and healthy-controls). Table 4 summarizes the results. We used two different classify algorithms, one is SVM with linear kernel and the other is RF whose forest includes 10 trees.
Table 4.
Comparison of the accuracy of T2D prediction using microbial genes derived by CoSA and SA.
| CoSA | |||||
|---|---|---|---|---|---|
| Strict | Normal | Loose | SA | ||
| Reads extraction | P-value cut-off | 0.001 | 0.05 | 0.2 | -a |
| Voting threshold | 0.5 | 0.3 | 0.8 | 0.5 | |
| Total base pair | 19.13Mbp | 6.08Gbp | 19.23Gbp | 36.26Gbp | |
|
| |||||
| Classification | # of genes | 249 | 296,979 | 1,741,472 | 2,098,590 |
| # of genes selected b | 249c | 207 | 230 | 210 | |
| Classifier | RF | SVM | SVM | SVM | |
| AUC d | 0.79 | 0.94 | 0.89 | 0.85 | |
SA uses ratios of k-mer counts to determine differential k-mers;
genes were selected using L1-based feature selection method;
no feature selection was applied for this case;
average accuracy using 10-fold cross-validation.
Using p-value of 0.05 and voting threshold of 0.3 (called Normal in Table 4) for reads extraction in CoSA followed by assembly and abundance quantification, we derived 296,979 genes. Our collection of genes resulted in a SVM that achieved a prediction accuracy of 0.94 (AUC), a significant improvement in the prediction accuracy as compared to the AUC reported in [17] (AUC=0.83).
We also tested CoSA using more stringent parameters for reads extraction (p-value = 0.001 and voting threshold = 0.5). The reads extraction only resulted in a small reads file with 19.13 Mbp in total. Not surprisingly we were only able to assemble and predict 249 genes from this small collection of sequencing reads. Interestingly, a RF model (without using feature selection) built from this small set of microbial genes achieved an AUC of 0.79. This accuracy is worse than our best model (AUC=0.94), and Karlsson’s RF model based on MGC (AUC=0.83), but it is much better than Karlsson’s RF model based on bacterial species (AUC=0.71). The advantage of using this setting (we called it Strict) is that only a small number of reads were extracted and only a small number of genes need to be quantified and used for building classifiers, and it still achieves reasonable prediction accuracy.
On the other hand, a much larger collection of microbial genes may make feature selection a more serious problem for building predictors, and therefore compromise the accuracy of predictors trained using these microbial genes. For example, we applied CoSA using a looser setting (p-value=0.2 and voting-threshold= 0.8; called Loose in Table 4), which resulted in the extraction of many more reads. Not surprisingly, many more genes can be assembled. However, more genes to start with doesn’t necessarily result in a better classifier for prediction. The best classier built using this larger collection of genes achieved an AUC of only 0.89. Similarly, using our original subtractive assembly approach (SA), an even greater collection of microbial genes can be assembled. However, the best predictor built using this larger collection of genes only achieved an AUC of 0.85.
Sequences and annotations (by myRAST [30] and hmmscan [8]) of the 207 differential genes that resulted in the highest prediction accuracy (AUC=0.94) are available for download at the CoSA sourceforge project website. Some of the functions and associated pathways are consistent with what we observed based on SA [42], including murein hydrolases (protein ID: k87 534 1 134 +) and multidrug resistance efflux pumps (protein ID: k87 34893 1 275 −).
4 Discussion
We developed a pipeline based on CoSA, which efficiently extracts reads that are likely sequenced from differential genes across samples for the identification of conserved microbial marker genes. Considering the heterogeneity nature of the microbiomes across human subjects, it is important to have a method that can detect disease-associated features that are consistent across samples. Tests of our approach using both simulated and real microbiomes show the importance of using multiple samples for such purposes.
The time and space complexity of CoSA is related to the number of datasets and the size of each dataset. The running time and memory cost is small for small datasets such as the simulated microbiome datasets. However, the computational time and memory usage can be substantial for large cohorts of datasets such as the T2D datasets. The total running time of CoSA for the simulated datasets was 44 mins (38 mins for k-mer counting and 6 mins for the detection of differential k-mers and therefore differential reads), and the peak memory usage was 2G. However, for the large T2D cohort, the running time for k-mer counting was 6.9 hours and the next step of detecting differential k-mers and reads took 27.5 hours. The peak memory usage for the T2D datasets was also substantial, which was 229Gb. Considering the increasing capacity of sequencing technologies, we will further investigate other strategies to reduce the memory usage and running time of CoSA.
In the current implemention of CoSA, WRS test is applied to k-mer counts normalized by the total k-mers (which is equivalent to the total reads) in each sample, for the detection of k-mers with differential abundances across healthy-controls and patients. This choice is mostly driven by the practical conveniency. Our results showed that this simple strategy of normalization worked well in practice. However, it has been shown that such a normalization approach may have limitations for applications in detecting metagenome-wise marker-gene surveys [31]. We will explore the possibility of using other normalization techniques such as the cumulative-sum scaling approach in CoSA.
Acknowledgments
This work was supported by the NIH grant 1R01AI108888 to Ye.
References
- 1.Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 2013 Jun;31(6):533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 2.Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, Lahti L, Loman NJ, Andersson AF, Quince C. Binning metagenomic contigs by coverage and composition. Nat Methods. 2014 Nov;11(11):1144–1146. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 3.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012 May;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Hur A, Ong CS, Sonnenburg S, Scholkopf B, Ratsch G. Support vector machines and kernels for computational biology. PLoS Comput Biol. 2008 Oct;4(10):e1000173. doi: 10.1371/journal.pcbi.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012 Mar;13(4):260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012 Jun;13(6):607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 7.Deorowicz S, Kokot M, Grabowski S, Debudaj-Grabysz A. KMC 2: fast and resource-frugal k-mer counting. Bioinformatics. 2015 May;31(10):1569–1576. doi: 10.1093/bioinformatics/btv022. [DOI] [PubMed] [Google Scholar]
- 8.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011 Jul;39(Web Server issue):29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrett WS. Cancer and the microbiota. Science. 2015 Apr;348(6230):80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge X, Rodriguez R, Trinh M, Gunsolley J, Xu P. Oral microbiome of deep and shallow dental pockets in chronic periodontitis. PLoS ONE. 2013;8(6):e65520. doi: 10.1371/journal.pone.0065520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, Jansson JK, Dorrestein PC, Knight R. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature. 2016 Jul;535(7610):94–103. doi: 10.1038/nature18850. [DOI] [PubMed] [Google Scholar]
- 12.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013 Apr;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV. Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science. 2012 Feb;335(6068):587–590. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- 14.Jiang B, Song K, Ren J, Deng M, Sun F, Zhang X. Comparison of metagenomic samples using sequence signatures. BMC Genomics. 2012 Dec;13:730. doi: 10.1186/1471-2164-13-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jorth P, Turner KH, Gumus P, Nizam N, Buduneli N, Whiteley M. Metatranscriptomics of the human oral microbiome during health and disease. MBio. 2014 Apr;5(2):e01012–01014. doi: 10.1128/mBio.01012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang DW, Park JG, Ilhan ZE, Wallstrom G, Labaer J, Adams JB, Krajmalnik-Brown R. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS ONE. 2013;8(7):e68322. doi: 10.1371/journal.pone.0068322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, Nielsen J, Backhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013 Jun;498(7452):99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 18.Knights D, Costello EK, Knight R. Supervised classification of human microbiota. FEMS Microbiol Rev. 2011 Mar;35(2):343–359. doi: 10.1111/j.1574-6976.2010.00251.x. [DOI] [PubMed] [Google Scholar]
- 19.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013 May;19(5):576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kostic AD, Howitt MR, Garrett WS. Exploring host-microbiota interactions in animal models and humans. Genes Dev. 2013 Apr;27(7):701–718. doi: 10.1101/gad.212522.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012 Mar;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis JD, Chen EZ, Baldassano RN, Otley AR, Griffiths AM, Lee D, Bittinger K, Bailey A, Friedman ES, Hoffmann C, Albenberg L, Sinha R, Compher C, Gilroy E, Nessel L, Grant A, Chehoud C, Li H, Wu GD, Bushman FD. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe. 2015 Oct;18(4):489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li D, Luo R, Liu CM, Leung CM, Ting HF, Sadakane K, Yamashita H, Lam TW. Megahit v1. 0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Andersen DG, Kaminsky M, Freedman MJ. Algorithmic improvements for fast concurrent cuckoo hashing. Proc. 9th ACM European Conference on Computer Systems (EuroSys); April 2014. [Google Scholar]
- 26.Marcais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011 Mar;27(6):764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mavromatis K, Ivanova N, Barry K, Shapiro H, Goltsman E, McHardy AC, Rigoutsos I, Salamov A, Korzeniewski F, Land M, Lapidus A, Grigoriev I, Richardson P, Hugenholtz P, Kyrpides NC. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat Methods. 2007 Jun;4(6):495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- 28.Melsted P, Pritchard JK. Efficient counting of k-mers in DNA sequences using a bloom filter. BMC Bioinformatics. 2011 Aug;12:333. doi: 10.1186/1471-2105-12-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen HB, Almeida M, Juncker AS, Rasmussen S, Li J, Sunagawa S, Plichta DR, Gautier L, Pedersen AG, Le Chatelier E, et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol. 2014 Aug;32(8):822–828. doi: 10.1038/nbt.2939. [DOI] [PubMed] [Google Scholar]
- 30.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014 Jan;42(Database issue):D206–214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013 Dec;10(12):1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. Scikit-learn: Machine learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- 33.Peng Y, Leung HC, Yiu SM, Chin FY. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012 Jun;28(11):1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 34.Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, Zhou J, Ni S, Liu L, Pons N, Batto JM, Kennedy SP, Leonard P, Yuan C, Ding W, Chen Y, Hu X, Zheng B, Qian G, Xu W, Ehrlich SD, Zheng S, Li L. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014 Sep;513(7516):59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 35.Rho M, Tang H, Ye Y. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 2010 Nov;38(20):e191. doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richter DC, Ott F, Auch AF, Schmid R, Huson DH. MetaSim: a sequencing simulator for genomics and metagenomics. PLoS ONE. 2008 Oct;3(10):e3373. doi: 10.1371/journal.pone.0003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheperjans F, Aho V, Pereira PA, Koskinen K, Paulin L, Pekkonen E, Haapaniemi E, Kaakkola S, Eerola-Rautio J, Pohja M, Kinnunen E, Murros K, Auvinen P. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov Disord. 2015 Mar;30(3):350–358. doi: 10.1002/mds.26069. [DOI] [PubMed] [Google Scholar]
- 38.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, Huttenhower C, Littman DR. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013 Nov;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014 Mar;15(3):317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016 Aug;14(8):e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008 Jun;24(12):1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- 42.Wang M, Doak TG, Ye Y. Subtractive assembly for comparative metagenomics, and its application to type 2 diabetes metagenomes. Genome Biol. 2015 Nov;16:243. doi: 10.1186/s13059-015-0804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu YW, Simmons BA, Singer SW. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016 Feb;32(4):605–607. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 44.Wu YW, Ye Y. A novel abundance-based algorithm for binning metagenomic sequences using l-tuples. J Comput Biol. 2011 Mar;18(3):523–534. doi: 10.1089/cmb.2010.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, Amiot A, Bohm J, Brunetti F, Habermann N, Hercog R, Koch M, Luciani A, Mende DR, Schneider MA, Schrotz-King P, Tournigand C, Tran Van Nhieu J, Yamada T, Zimmermann J, Benes V, Kloor M, Ulrich CM, von Knebel Doeberitz M, Sobhani I, Bork P. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014 Nov;10:766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Q, Pell J, Canino-Koning R, Howe AC, Brown CT. These are not the k-mers you are looking for: Efficient online k-mer counting using a probabilistic data structure. PLoS ONE. 2014;9(7):e101271. doi: 10.1371/journal.pone.0101271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu B, Wang X, Li L. Human gut microbiome: the second genome of human body. Protein Cell. 2010 Aug;1(8):718–725. doi: 10.1007/s13238-010-0093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]