Abstract
Resistance gene Cf-9 of cultivated tomato (Lycopersicon esculentum) confers recognition of the AVR9 elicitor protein of the fungal pathogen Cladosporium fulvum. The Cf-9 locus, containing Cf-9 and four homologs (Hcr9s), originates from Lycopersicon pimpinellifolium (Lp). We examined naturally occurring polymorphism in Hcr9s that confer AVR9 recognition in the Lp population. AVR9 recognition occurs frequently throughout this population. In addition to Cf-9, we discovered a second gene in Lp, designated 9DC, which also confers AVR9 recognition. Compared with Cf-9, 9DC is more polymorphic, occurs more frequently, and is more widely spread throughout the Lp population, suggesting that 9DC is older than Cf-9. The sequences of Cf-9 and 9DC suggest that Cf-9 evolved from 9DC by intragenic recombination between 9DC and another Hcr9. The fact that the 9DC and Cf-9 proteins differ in 61 aa residues, and both mediate recognition of AVR9, shows that in nature Hcr9 proteins with the same recognitional specificity can vary significantly.
Recognition of a diverse range of pathogens, followed by an adequate defense response, is crucial for the survival of plants. Resistance (R) genes, which mediate recognition of products of matching avirulence (Avr) genes, play a key role in the recognition of pathogens (1). Most R gene products contain a leucine-rich repeat (LRR) domain with putative solvent-exposed amino acid residues that decorate the surface of the protein, where specific interactions with other proteins are thought to occur (2). R proteins with different specificities differ predominantly at putative solvent-exposed positions, which are often thought to result from adaptive evolution (3).
Plants need to generate R genes with new specificities because pathogens continuously try to circumvent recognition by the host plant. New R genes are thought to evolve by sequence exchange between homologous genes and by accumulation of random point mutations in codons that encode amino acids located at putative solvent-exposed positions (3, 4).
The continuous generation of new recognitional specificities by the host, followed by subsequent adaptation of the pathogen to circumvent this recognition, can be seen as an “arms race” between plants and pathogens (5). Recent observations suggest that in nature, this arms race is a slow process and that the battle between plants and pathogens is more likely to be similar to “trench warfare.” In this model, frequencies of R genes in the plant population fluctuate in time, following the frequency of the matching Avr gene in the pathogen population (6). Consistent with this model, gene-for-gene pairs like AvrRpm1-RPM1 and AvrPto-Pto are ancient (6, 7), and plants carrying or lacking the RPM1 gene coexist in the plant population (6).
The tomato R genes Cf-9 and Cf-4 mediate recognition of strains of the leaf mold fungus Cladosporium fulvum carrying the Avr9 or Avr4 gene, respectively (8). Recognition by resistant plants results in the activation of multiple defense responses that limit further fungal growth. The hypersensitive response is a macroscopically visible phenomenon in which plant cells surrounding the infection site quickly die. The Avr9 and Avr4 genes both encode proteins that are secreted by the fungus into the extracellular space of tomato leaves during infection of susceptible plants. Injection of these elicitor proteins into the extracellular space of tomato leaves carrying the matching Cf gene is sufficient to trigger a hypersensitive response. The Cf genes encode receptor-like proteins with extracellular LRRs and are predicted to be anchored in the plasma membrane (2). Cf-4 differs from Cf-9 in 67 aa residues and contains three deletions when compared with Cf-9 (9). By exchanging domains between Cf-4 and Cf-9, we showed that specificity in Cf-4 resides in the N-terminal domain, the number of LRRs, and three Cf-4-specific amino acid residues at putative solvent-exposed positions (10). In Cf-9, specificity is likely scattered throughout the LRR domain (10, 11).
The Cf-9 gene is a member of a gene family called Hcr9s (homologs of Cladosporium fulvum resistance gene Cf-9), present on the short arm of chromosome 1 of tomato. Thus far, 18 Hcr9s have been reported (12, 13). The Cf-9 gene is the third Hcr9 (Hcr9-9C) of a cluster of five homologs, named Hcr9-9A to -9E. The Cf-9 locus has been introgressed into cultivated tomato (Lycopersicon esculentum) from its wild relative Lycopersicon pimpinellifolium (Lp) (14). Lp contains many different recognitional specificities for proteins of C. fulvum and was used as a rich germplasm for Cf R genes (15). The natural habitat of Lp is a narrow, 2,500-km-long coastal area of Ecuador and Peru, bordered by the Pacific Ocean and the Andes Mountains (16). Lp is predominantly self-fertilizing, and previous studies on the genetic variation in this species showed that allele frequencies can differ significantly between regions of the Lp habitat (17).
The large genetic variation in the Lp population prompted us to investigate whether this population contains Hcr9s that are polymorphic but still mediate recognition of the same elicitor protein of C. fulvum. If this is the case, we might get better insight into how existing recognitional specificities are maintained in nature and how new specificities evolve. Here we show that AVR9 recognition occurs frequently throughout the Lp population, suggesting that this trait did not evolve recently. In addition to Cf-9, we discovered a second gene, designated 9DC, which also mediates AVR9 recognition. Cf-9 likely evolved by intragenic recombination between 9DC and another Hcr9. It appears that in nature, Hcr9 proteins that have the same recognitional specificity can be highly polymorphic.
Materials and Methods
Accessions of Lp were donated by the C. M. Rick Tomato Genetics Resource Center of the University of California, Davis (http://tgrc.ucdavis.edu/). Transgenic tomato plants (L. esculentum cv. MoneyMaker) carrying Cf-9 or Hcr9-9D were a gift from J. Jones (Sainsbury Laboratory, Norwich, U.K.). Plants were grown under standard greenhouse conditions. To select for AVR9-responsive plants, leaflets were injected with 10 μg/ml AVR9. The hypersensitive response was visible within 2 days after injection. The wild-type and mutant AVR9 proteins F21A, F10A, and R08K used for injections have been described (18).
DNA manipulations were performed by standard protocols (19). PCRs were performed with AmpliTaq (Perkin–Elmer), Pfu (Stratagene), or the Expand High Fidelity PCR System (Roche Diagnostics), according to the manufacturer's instructions. Restriction enzymes, T4 ligase, and Escherichia coli DH5α cells were from Life Technologies (Breda, The Netherlands). Primers were synthesized by Amersham Pharmacia. Primer sequences were as follows (given in the 5′-to-3′ direction, followed by the position of the 5′ end of the primer, relative to the ATG of Cf-9; restriction sites are underlined): CF1: ggcatcgattgtgacgagacg, 245; BR1: attattggatcccaagtctaacaatatc, 1485; CS1: gccgttcaagttgggtgtt, 1093; CS3: tctgaaagataatgatcaagtg, 2639; CS5: tttccaacttacaatcccttc, 713; CS11:cccccctgcagtcactaatatcttttcttgtgc, 2606; CS12: tcttctctatcaacataacaag, −44; DS1: gagagctcaacctttacgaa, 587; DS3: ctatgtgaggtagctagtag, −124; DS9: tttttccatgggttgtgtaaaacttgtg, −7.
For the construction of chimeric Hcr9s, genomic DNA was isolated (20) from Lp plants and used as a template for PCR. Fragments of Hcr9s encoding LRRs 1–17 were amplified with primers CF1 and BR1 and cloned into the ClaI and BamHI restriction sites, thereby constructing chimeric Hcr9s (Fig. 1A).
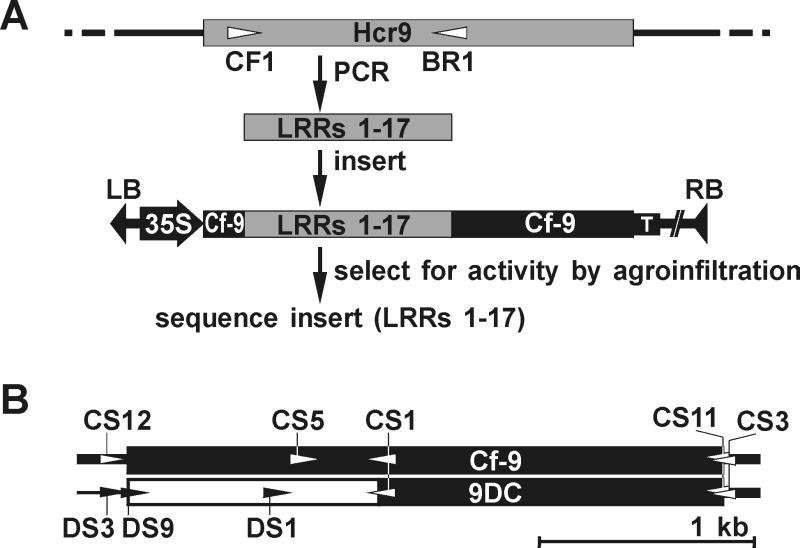
Figure 1.
Selection procedure for chimeric Hcr9 constructs that confer AVR9 recognition and position of the various specific primers. (A) CF1 and BR1 are Hcr9-specific primers (triangles) located at positions corresponding to the B-domain and LRR 17, respectively, of the encoded Hcr9 protein. Amplified fragments encoding LRRs 1–17 were inserted into a binary expression vector that contains the Cf-9 ORF lacking the fragment encoding LRRs 1–17. 35S, cauliflower mosaic virus 35S constitutive promoter; T, nos terminator; RB and LB, right and left borders of the T-DNA, respectively. For further details, see Materials and Methods. (B) Position of specific primers in and around Cf-9 and 9DC ORFs. Triangles indicate the annealing position and direction of primers relative to the ORF of Cf-9 and 9DC. For further details see Materials and Methods.
To detect 9DC or Cf-9 in Lp plants, genomic DNA was used as a template for PCR with specific primers (Fig. 1B). Primers CS5 and CS1 were used to amplify a 378-bp Cf-9-specific fragment, whereas primers DS1 and CS1 were used to amplify a 507-bp 9DC-specific fragment. In the same reaction mix, primers 5′-ggttatcttatggctactctg-3′ and 5′-gcgccatccgaatgtagag-3′ were included to amplify a 778-bp fragment of the aspartate carbamoyl transferase (Act) gene, which served as a positive control for the amplification reaction (21).
The complete 9DC and Cf-9 ORFs were amplified from genomic DNA by PCR with primers DS3 and CS3 or CS12 and CS3, respectively (Fig. 1B). Amplified fragments were cloned into pGEM-T Easy (Promega) and sequenced. The presence of polymorphic sites in the sequences was determined unambiguously by direct sequencing of the PCR products or by sequencing of independent clones.
To clone the 9DC ORF into a binary expression vector, primers DS9 and CS11 were designed (Fig. 1B). Amplified fragments were inserted into the NcoI and PstI restriction sites of pRH80 (22), between the 35S promoter and the terminator (T). The 35S-9DC-T cassette subsequently was inserted into the binary plasmid pMOG800 (22), with the use of the XbaI and KpnI restriction sites.
Agroinfiltration of tobacco plants (Nicotiana tabacum cv. Petite Havana SR1) was performed as described (22). To compare the activity of 9DC with Cf-9, dilution series of Agrobacterium cultures were infiltrated, and necrotic responses were quantified as described (10).
Results
Identification of an Additional Hcr9 That Mediates AVR9 Recognition in Lp.
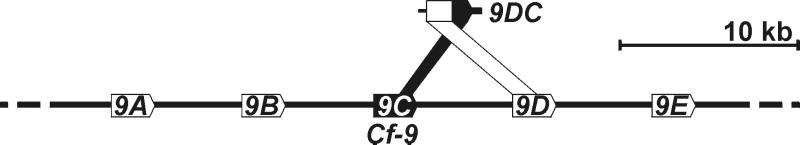
In a previous study, six accessions of Lp were identified as AVR9-responsive (15). We chose one AVR9-responsive plant of accession LA1301 to characterize the Hcr9 mediating AVR9 recognition. Hcr9s are highly similar, and the encoded proteins differ predominantly in LRRs 1–17 (12). We found that this region determines specificity in Cf-9 (10). To identify the Hcr9 that confers AVR9 recognition in this Lp accession, a library of chimeric Cf-9 genes was generated in a binary vector, with fragments encoding LRRs 1–17 amplified from genomic DNA of the AVR9-responsive plant (Fig. 1A; see Materials and Methods). Transient coexpression of the chimeric Cf-9 genes and Avr9 in tobacco by agroinfiltration (22) was used to select for fragments that complemented Cf-9 function. Of the 13 chimeric constructs tested, three conferred AVR9 recognition (data not shown). The DNA sequence of the inserts encoding LRRs 1–17 revealed that the 3′ part (0.4 kb, encoding LRRs 12–17) was identical to Cf-9 (Hcr9-9C), whereas the 5′ part (0.8 kb, encoding LRRs 1–11) was nearly identical to Hcr9-9D, which is located directly downstream of Cf-9 at the Cf-9 locus (Fig. 2). Therefore, the gene was designated 9DC.
Figure 2.
Organization of the Hcr9s at the Cf-9 locus and their homology with 9DC. The five Hcr9s (9A to 9E) present at the Cf-9 locus are indicated (Lower), as is the area of homology of Hcr9-9C and Hcr9-9D with 9DC (Upper). Arrows indicate ORFs with transcriptional direction. Note that no mechanism or direction in time is implied.
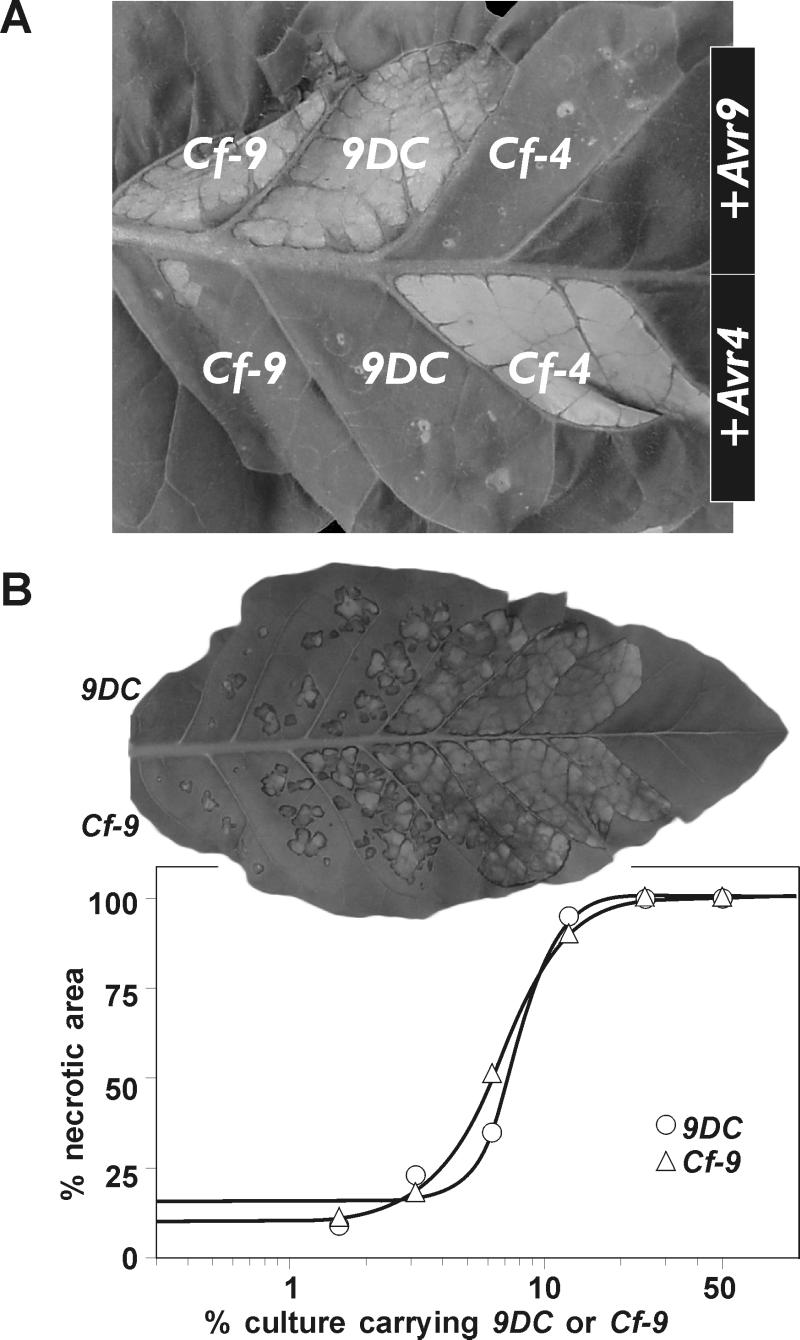
Based on sequence information on the fragment encoding LRR1–17, we expected that the 5′ untranslated region (UTR) of 9DC would be identical to Hcr9-9D, and the 3′ UTR would be identical to Cf-9. With an Hcr9-9D-specific primer in the 5′ UTR and a Cf-9-specific primer in the 3′ UTR (primers DS3 and CS3, respectively, Fig. 1B), we could indeed amplify the entire 9DC ORF from genomic DNA of the AVR9-responsive plant. To test the encoded 9DC protein for mediation of AVR9 recognition, the 9DC ORF was inserted into a binary expression vector and coexpressed with Avr9. This procedure demonstrated that 9DC indeed confers AVR9 recognition (Fig. 3A), whereas dilution experiments showed that its activity with respect to AVR9 recognition is similar to that of Cf-9 (Fig. 3B). Furthermore, injection of 9DC-agroinfiltrated leaves with mutant AVR9 peptides that are either inactive (F21A), less active (F10A), or more active (R08K), as compared with wild-type AVR9 (18), demonstrated that 9DC confers AVR9 recognition with the same specificity as Cf-9 (data not shown). Thus, 9DC functions similarly to Cf-9 in these assays.
Figure 3.
Comparative transient expression studies of 9DC and Cf-9. (A) 9DC confers AVR9 recognition. An Agrobacterium culture carrying the 9DC ORF in a binary expression vector was mixed with Agrobacterium carrying Avr9 and infiltrated into a tobacco leaf sector. As controls, the ORFs of Cf-9 and Cf-4 were included and Avr4 was used for coexpression. The photograph was taken 7 days after infiltration. (B) Activity of 9DC and Cf-9 in AVR9 recognition. Agrobacterium cultures carrying 9DC or Cf-9 were diluted in a culture carrying Avr9 and infiltrated into neighboring tobacco leaf sectors. The photograph was taken 7 days after infiltration. The percentage of the infiltrated area that had become necrotic was measured and plotted against the percentage of culture containing 9DC or Cf-9. Note that the dose–response curves for 9DC and Cf-9 are similar.
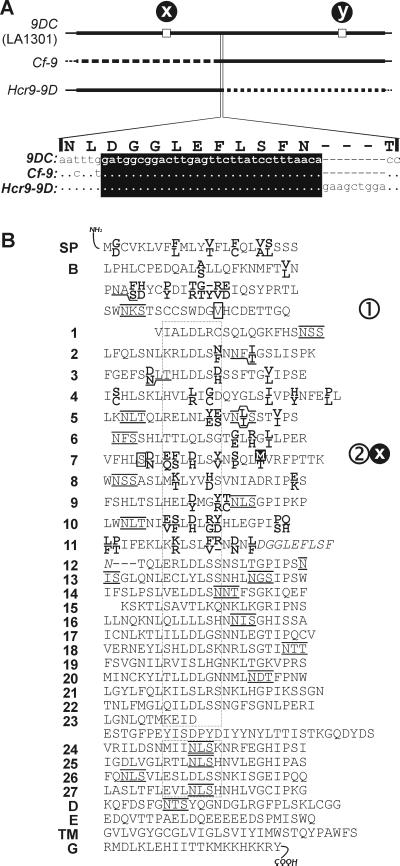
The 5′ half of the 9DC ORF (1,966 bp) and 104 bp of the 5′ UTR differ in only 1 nt from Hcr9-9D (site x, Fig. 4A), resulting in a difference in amino acid sequence as compared with the Hcr9-9D protein (site x, Fig. 4B). The DNA sequence of the 3′ half of the 9DC ORF (1,550 bp) and 26 bp of the 3′ UTR also differ from Cf-9 in only 1 nt (site y, Fig. 4A), which does not result in a difference in amino acid sequence. The 5′ end of the 31-bp recombination region (Fig. 4A, Lower) is bordered by a Cf-9-specific nucleotide (t), whereas the 3′ end is bordered by an Hcr9-9D-specific insertion of three codons.
Figure 4.
Comparison of nucleotide and protein sequences of 9DC, Cf-9, and Hcr9-9D. (A) Schematic representation of the DNA sequences of 9DC present in Lp accession LA1301, and Cf-9 and Hcr9-9D. Thick lines indicate ORFs. Squares at positions x and y indicate nucleotides (C755T and T2160A, respectively) that are different from the DNA sequence of Hcr9-9D and Cf-9, respectively. The amino acid sequence of the 9DC protein encoded by the area of recombination is enlarged. The DNA sequence with the recombination region (boxed in black) is indicated. Dots indicate nucleotides that are identical to 9DC; minuses indicate nucleotides that are lacking. (B) Alignment of 9DC and Cf-9 proteins. Amino acid residues of 9DC and Cf-9 that are identical are shown in the central line. 9DC- and Cf-9-specific residues are shown in bold at the top and bottom lines, respectively. Potential N-glycosylation sites (NXS/T) in 9DC and Cf-9 are overlined and underlined, respectively. The amino acid sequence encoded by the recombination region (A) is shown in italics. The dotted box indicates the various β-sheets (consensus XXLXLXX), each of which contains five putative solvent-exposed amino acid residues (X). The amino acid residue in the black box (site x) indicates the difference (Met-Thr) between the 9DC protein of LA1301 and the N-terminal half of the Hcr9-9D protein. Residues in white boxes are polymorphic in different 9DC alleles: site 1 (Val-Ile) and site 2 (Ser-Phe) (see Fig. 6). Amino acid residues encoded at SNPs y and 3 (Figs. 4A and 6) are not indicated because these do not result in a polymorphic amino acid sequence. SP, signal peptide; B, B domain; 1–27, LRR domain; D, D domain; E, acidic domain; TM, transmembrane domain; G, cytoplasmic tail.
Most strikingly, the 9DC protein encoded by the 9DC gene of Lp accession LA1301 differs in 61 aa residues from Cf-9 (Fig. 4B). Of these residues, 45 are located in the first 11 LRRs, of which 22 are present at putative solvent-exposed positions. The 9DC protein also lacks three potential glycosylation sites. The amino acids that are polymorphic between 9DC and Cf-9 are similar in extent and position to those observed between Cf-4 and Cf-9 (9). Nineteen amino acid residues of 9DC that differ from Cf-9 are identical to those occurring in Cf-4, of which five are located at putative solvent-exposed positions.
Molecular Basis of AVR9 Recognition in the Lp Population.
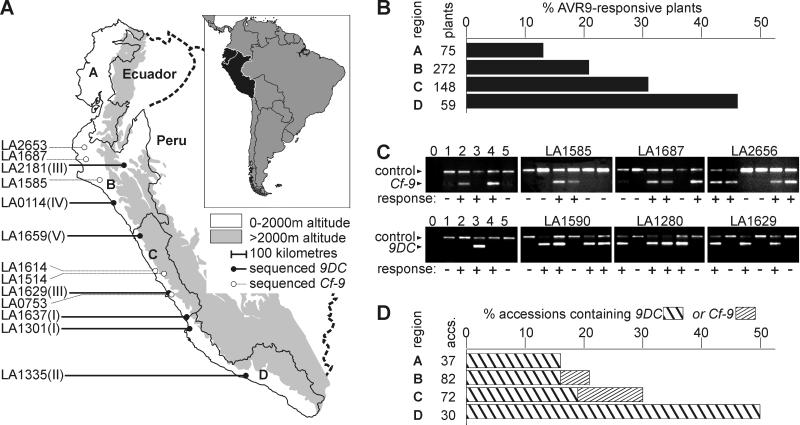
Having identified the gene that mediates AVR9 recognition in accession LA1301, we set out to examine the Hcr9 mediating AVR9 recognition in other accessions of Lp. We took advantage of the collection of Lp accessions maintained at the University of California, Davis. Multiple plants of 231 accessions were injected with the AVR9 elicitor. Of 570 injected plants, 143 developed a specific hypersensitive response. Responsive plants belong to 72 accessions, of which 27 contained both responsive and nonresponsive plants. It appeared that AVR9-recognizing plants are present throughout the geographical distribution range of Lp (data not shown). To calculate frequencies of AVR9 recognition, the distribution range of Lp was divided arbitrarily into four regions (regions A–D, Fig. 5A). Interestingly, this procedure revealed that the frequency of AVR9 recognition gradually increases in the southern direction, up to an almost 3-fold higher level in the south when compared with the northern part of the Lp distribution range (Fig. 5B).
Figure 5.
Frequency of AVR9 recognition and occurrence of 9DC and Cf-9 in the Lp population. (A) The natural distribution range of Lp. The Lp distribution range is bordered by the Pacific Ocean in the west and the 2,000-m elevation line of the Andes Mountains in the east. This area is divided into four regions: Ecuador (A) and northern (B), central (C), and southern (D) Peru. Accessions from which entire 9DC or Cf-9 ORFs have been sequenced are indicated on the left, with allelic classes I–V between brackets (see Fig. 6). (B) Frequency of AVR9-responsive plants per region. For each region, the number of AVR9-responsive plants was divided by the total number of AVR9-injected plants originating from that region. Not all plants could be mapped to regions. (C) Amplification of fragments of Cf-9 and 9DC. Specific primers were tested (lanes 0–5) and used to detect the presence of 9DC and Cf-9 genes in Lp accessions that contain both AVR9-responsive and nonresponsive plants (panels marked with LA numbers). Specific amplification products of Cf-9, 9DC, and Act (control) genes were obtained as explained in Materials and Methods. Templates were genomic DNA isolated from the following sources: 1, MoneyMaker (MM)-Cf0 tomato; 2, MM-Cf9 tomato; 3, AVR9-responsive Lp plant of accession LA1301; 4, Cf-9-transgenic MM-Cf0 tomato; and 5, Hcr9-9D-transgenic MM-Cf0 tomato. Water (lane 0) was used as a negative control. AVR9 responsiveness is indicated by − or + below the panels. (D) Frequency of 9DC and Cf-9 genes per region. One AVR9-responsive plant of each accession was analyzed for the presence of 9DC or Cf-9 genes. The number of accessions that contained 9DC or Cf-9 was divided by the total number of accessions from that region. The frequency has been adjusted for the number of plants tested per accession. Not all accessions could be mapped to regions.
From each accession containing AVR9-responsive plants, one responsive plant was randomly selected for genomic DNA isolation and subsequent PCR analysis. To detect Cf-9 or 9DC, primers were developed to specifically amplify fragments from Cf-9 or 9DC, but not from any other known Hcr9 (Fig. 5C, lanes 0–5). The identity of the amplified fragments was confirmed by sequencing. All AVR9-responsive plants contained either Cf-9 or 9DC, indicating that these are the only two genes that confer AVR9 recognition in the Lp population. None of the AVR9-responsive plants tested contained both Cf-9 and 9DC. Significantly, in accessions with both responsive and nonresponsive plants, the Cf-9 or 9DC fragments were detected only in AVR9-responsive plants (Fig. 5C). A 9DC fragment was amplified from 56 of the AVR9-responsive plants, whereas from the remaining 16 a Cf-9 fragment was amplified. Thus, 9DC occurs more frequently in the Lp population as compared to Cf-9. Accessions with 9DC are present throughout the entire distribution range of Lp, whereas Cf-9 is found only in accessions of Lp collected from northern and central Peru (Fig. 5D).
Sequence Polymorphism in Cf-9 and 9DC.
To determine whether polymorphism occurs in Cf-9 and 9DC, we cloned and sequenced six Cf-9 ORFs and six additional 9DC ORFs of different accessions, representing separated geographical collection sites (Fig. 5A). All six Cf-9 sequences were identical to the Cf-9 ORF that was introgressed into L. esculentum (12). In contrast, DNA sequences of the six additional 9DC ORFs showed three single nucleotide polymorphisms (SNPs) when compared with 9DC of accession LA1301 (Fig. 6). Two of these (sites 1 and 2) result in a polymorphic amino acid sequence (Fig. 4B).
Figure 6.
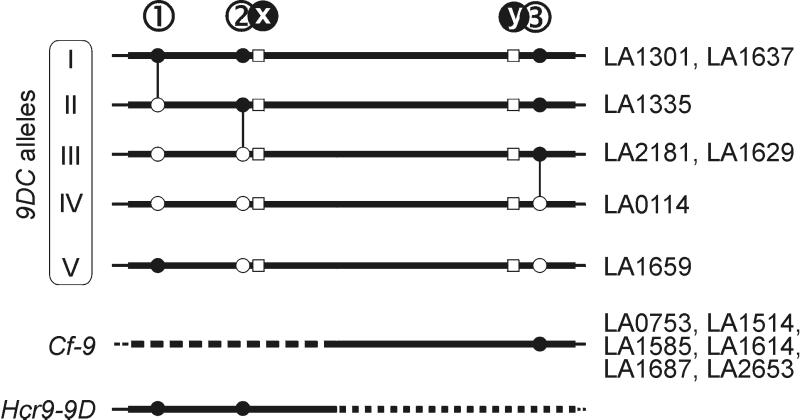
Polymorphism among 9DC alleles and their relation with Cf-9 and Hcr9-9D. Accessions from which 9DC or Cf-9 have been sequenced are indicated on the right. Solid lines represent DNA sequences of the 9DC alleles. Thick lines indicate ORFs. Open squares at positions x and y indicate the nucleotides conserved among 9DC genes (see also Fig. 4A). Circles at positions 1, 2, and 3 indicate SNPs (G244A, C713T, and T2304C, respectively) between 9DC ORFs, which differentiate these ORFs into five distinct allelic classes. Alleles I–IV are related to each other by the accumulation of point mutations (vertical connections). Allele V likely resulted from recombination between two different 9DC alleles. Cf-9 contains a 3′ half that is identical to that of alleles I–III, except for site y. Hcr9-9D contains a 5′ half that is identical to that of allele I, except for site x.
The SNPs differentiate the 9DC genes into five different allelic classes (I–V, Fig. 6). Alleles I and III each were identified in two separate accessions. Alleles I–IV may have evolved from each other by consecutive accumulation of point mutations (Fig. 6). However, the combination of SNPs in allele V suggests that this allele has resulted from recombination between different 9DC alleles. The geographical distribution of the 9DC alleles does not reveal any direction of genetic drift (Fig. 5A).
Discussion
Tremendous efforts in the past decade have resulted in the cloning of many R genes that confer recognition of very different pathogens. However, how R genes are generated and maintained in nature is still poorly understood. Most knowledge of the evolution of R genes comes from the comparison of R genes with different recognitional specificities. In this report, we examined naturally occurring polymorphism between R genes with the same recognitional specificity. The two genes that confer AVR9 recognition in the Lp population encode highly polymorphic proteins, but one likely evolved from the other by a single intragenic recombination event. Maintenance of both Cf genes in the Lp population is likely a result of trench warfare, where the frequency of Avr9 in the pathogen population is counterbalanced by the frequency of the matching Cf gene in the plant population.
Role of Amino Acid Polymorphism in Cf Proteins.
Cf proteins differ predominantly in amino acid residues at putative solvent-exposed positions, which may be a result of adaptive evolution to mediate recognition of a particular avirulence factor (3). However, we have shown that from the 67 aa residues that vary between Cf-4 and Cf-9, only three Cf-4-specific residues present at putative solvent-exposed positions are essential to confer AVR4 recognition (10). A comparison between Cf-9 and 9DC proteins described in this report reveals that significant variation in amino acid residues is also allowed for AVR9 recognition in nature. These results suggest that the variation observed between Cf proteins that mediate recognition of different avirulence factors is not a result of adaptive evolution. Variation may rather serve as a reservoir of diversity that facilitates the generation of R proteins with new specificities resulting from recombination and additional point mutations. These events can result in the sudden appearance (and disappearance) of a functional R gene according to the “birth-and-death” model of evolution as postulated by Michelmore and Meyers (4).
The Origin of Cf-9.
The sequences of 9DC and Cf-9 are nearly identical in their 3′ halves, indicating that they are evolutionarily related by an intragenic recombination event. This similarity in sequences also suggests that 9DC maps at the same position as Cf-9 in the tomato genome. Indeed, a test cross between accessions PI126915 (containing Cf-9) and PI126946 (containing 9DC) indicated that Cf-9 and 9DC map at the same chromosomal position (M.K., unpublished results).‡ The observation that 9DC occurs more frequently, is more dispersed in the Lp population, and contains more sequence polymorphism when compared with Cf-9 suggests that the 9DC gene is older than Cf-9 and that 9DC is an ancestor of Cf-9.
Thus, intragenic recombination between 9DC and another Hcr9 has likely resulted in Cf-9, which contains only the 3′ half of 9DC, but still mediates AVR9 recognition. The 5′ half of 9DC apparently ended up at the same locus as part of Hcr9-9D. Hcr9-9D does not confer AVR9 recognition (Fig. 5C), probably because of the absence of specific amino acids that are required for AVR9 recognition, as identified by Cf-4 and Cf-9 domain-swap analysis (10, 11).
Introgression of Cf-9, instead of 9DC, into cultivated tomato has been a matter of chance. Accessions that contain 9DC, like PI126946, have been used in breeding programs (23).‡ Indeed, one AVR9-responsive commercial tomato cultivar was found to contain 9DC instead of Cf-9 (R. Luderer, M. de Kock, and M.K., unpublished results).
Intragenic Recombination Between R Gene Homologs.
Intragenic recombination has been reported for many R gene families and is thought to be an important evolutionary force that generates new specificities. However, intragenic recombination resulting in new recognitional specificities has been reported only for L genes of flax (24). Most intragenic recombination events described in the literature were identified during screens for loss-of-function mutants of R genes. For example, intragenic recombination between Cf-2 and Hcr2–5B resulted in a homolog that was not functional in AVR2 recognition (25). Moreover, intragenic recombination between the functional RPP8 gene and its adjacent homolog RPP8A probably resulted in an inactive rpp8 homolog (26). By searching for polymorphism in Hcr9s conferring AVR9 recognition in the Lp population, we have shown that intragenic recombination also occurs in nature, without having an effect on the recognitional specificity of the newly generated Hcr9.
AVR9 Recognition in the Lp Population.
The high frequency of AVR9-responsive plants in the Lp population suggests that the locus mediating AVR9 recognition provides a selective advantage in nature. This advantage also has been observed in modern resistance breeding, where the Cf-9 locus, which originates from Lp, has not yet been overcome by a fit, virulent isolate (8). The selective advantage of the Cf-9 or 9DC locus can be due to conferring AVR9 recognition itself, or can be the result of the presence of additional linked R genes with recognitional specificities for yet unidentified Avr gene products of C. fulvum (12, 27).
An interesting observation is that AVR9 recognition occurs almost 3-fold more frequently in the southern than in the northern regions of the Lp distribution range. Perhaps this difference in frequency of occurrence reflects differences in pathogen pressure in these regions, which may be a result of climatic differences, favoring the incidence of C. fulvum. Coastal temperatures in southern Peru range from 15°C to 22°C compared with 18°C to 25°C in Ecuador (16). A more moderate temperature is known to favor the occurrence of tomato leaf mold (28).
AVR9 Recognition in the Lp Population Complies with the Trench Warfare Model.
Previous studies on the presence of multiple genetic markers in the Lp population revealed that the largest genetic variation exists in northern Peru (17). In this area, Lp is a facultative outcrosser, which correlates with the presence of large flowers to attract bees and long stamens that prevent self-pollination. In Ecuador and central and southern Peru, Lp is genetically more uniform and mainly self-fertilizing. These observations led to the hypothesis that northern Peru is the center of origin of Lp, from which the species has spread in both northern and southern directions, giving self-fertilizing plants a selective advantage as pioneers. The study of Rick and coworkers (17) also revealed that certain alleles only occur in certain regions of the Lp distribution range.
In contrast, we have shown that AVR9 recognition occurs throughout the entire Lp distribution range. The predominantly self-fertilizing nature of Lp may be reflected in the accumulation of point mutations in 9DC alleles I–IV. However, recombination between 9DC alleles, resulting in allele V, probably has occurred in an outcrossing population. A previous study showed that unequal crossing over at the Cf-9 locus occurs more frequently in heterozygous plants than in homozygous plants (12). This difference in the rate of unequal crossing over may suggest that intragenic recombination leading to Cf-9 and Hcr9-9D occurred in a heterozygous background of an outcrossing population. Taken together, these observations suggest that AVR9 recognition was present in the center of origin of Lp before the species started to spread. This early presence of AVR9 recognition implies that it is a trait that did not evolve recently. In addition, we observed that AVR9-recognizing and nonrecognizing plants coexist in the same area (Fig. 5 B and C). These observations fully comply with the trench warfare model of gene-for-gene interactions between plants and pathogens (6). According to this model, R genes are maintained in the plant population with a frequency that fluctuates with time, following the frequency of the matching Avr gene in the pathogen population. This model also implicates that R gene frequencies differ significantly between different areas. However, we observed a gradual decline in the frequency of AVR9 recognition in the northern direction of the Lp distribution range. We believe that the regional R gene frequency is an average of fluctuating R gene frequencies of local populations. The R gene frequency at the regional level may represent an equilibrium that does not fluctuate significantly in time. In either case, it is conceivable that trench warfare between plants and pathogens maintains R genes with a particular recognitional specificity in a natural plant population over a long period.
Acknowledgments
We thank Roger Chetelat (Tomato Genetucs Resource Center, Davis, CA) for critically reading the manuscript and providing seeds of the Lp collection; Ietje Boukema (Centre for Genetic Resources, Plant Research International, Wageningen, The Netherlands) for providing seeds and for helpful discussions; Jonathan Jones (Sainsbury Laboratory, Norwich, U.K.) for providing Cf-9- and Hcr9-9D-transgenic tomato; and Bert Essenstam, Mart Berns, and Henk Smid (Unifarm, Wageningen, The Netherlands) for excellent plant care. This study was supported by the Dutch Foundation for Earth and Life Sciences, the Dutch Foundation for Applied Sciences, the European Molecular Biology Organization, and the European Community.
Abbreviations
- R gene
resistance gene
- Avr gene
avirulence gene
- Lp, Lycopersicon pimpinellifolium
Hcr9, homolog of Cladosporium fulvum resistance gene Cf-9
- LRR
leucine-rich repeat
- UTR
untranslated region
- SNP
single nucleotide polymorphism
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF401036).
Boukema, I. W., Meeting on Cladosporium fulvum Cke in Tomato, February 26–27, 1980, Wageningen, The Netherlands.
References
- 1.Flor H H. J Agric Res. 1946;73:335–357. [Google Scholar]
- 2.Jones D A, Jones J D G. Adv Bot Res. 1997;24:91–167. [Google Scholar]
- 3.Richter T E, Ronald P C. Plant Mol Biol. 2000;42:195–204. [PubMed] [Google Scholar]
- 4.Michelmore R W, Meyers B C. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- 5.Dawkins R, Krebs J R. Proc R Soc London. 1979;205:489–511. doi: 10.1098/rspb.1979.0081. [DOI] [PubMed] [Google Scholar]
- 6.Stahl E A, Dwyer G, Mauricio R, Kreitman M, Bergelson J. Nature (London) 1999;400:667–671. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- 7.Riely B K, Martin G B. Proc Natl Acad Sci USA. 2001;98:2059–2064. doi: 10.1073/pnas.98.4.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joosten M H A J, De Wit P J G M. Annu Rev Phytopathol. 1999;37:335–367. doi: 10.1146/annurev.phyto.37.1.335. [DOI] [PubMed] [Google Scholar]
- 9.Thomas C M, Jones D A, Parniske M, Harrison K, Balint-Kurti P J, Hatzixanthis K, Jones J D G. Plant Cell. 1997;9:2209–2224. doi: 10.1105/tpc.9.12.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van der Hoorn R A L, Roth R, De Wit P J G M. Plant Cell. 2001;13:273–285. doi: 10.1105/tpc.13.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wulff B B H, Thomas C M, Smoker M, Grant M, Jones J D G. Plant Cell. 2001;13:255–272. doi: 10.1105/tpc.13.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parniske M, Hammond-Kosack, Golstein C, Thomas C M, Jones D A, Harrison K, Wulff B B H, Jones J D G. Cell. 1997;91:821–832. doi: 10.1016/s0092-8674(00)80470-5. [DOI] [PubMed] [Google Scholar]
- 13.Parniske M, Jones J D G. Proc Natl Acad Sci USA. 1999;96:5850–5855. doi: 10.1073/pnas.96.10.5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tichelaar E C. Rep Tomato Genet Coop. 1984;34:55–57. [Google Scholar]
- 15.Laugé R, Goodwin P, De Wit P J G M, Joosten M H A J. Plant J. 2000;23:735–745. doi: 10.1046/j.1365-313x.2000.00843.x. [DOI] [PubMed] [Google Scholar]
- 16.Warnock S J. Hortic Sci. 1991;26:466–471. [Google Scholar]
- 17.Rick C M, Fobes J F, Holle M. Plant Syst Evol. 1977;127:139–170. [Google Scholar]
- 18.Kooman-Gersmann M, Vogelsang R, Vossen P, Van den Hooven H W, Mahé E, Honée G, De Wit P J G M. Plant Physiol. 1998;117:609–618. doi: 10.1104/pp.117.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Van der Beek J G, Verkerk R, Zabel P, Lindhout P. Theor Appl Genet. 1992;84:106–112. doi: 10.1007/BF00223988. [DOI] [PubMed] [Google Scholar]
- 21.Overduin B, Hogenhout S A, Van der Biezen E A, Haring M A, Nijkamp H J, Hille J. Mol Gen Genet. 1993;240:43–48. doi: 10.1007/BF00276882. [DOI] [PubMed] [Google Scholar]
- 22.Van der Hoorn R A L, Laurent F, Roth R, De Wit P J G M. Mol Plant–Microbe Interact. 2000;13:439–446. doi: 10.1094/MPMI.2000.13.4.439. [DOI] [PubMed] [Google Scholar]
- 23.Laugé R, Joosten M H A J, Haanstra J P W, Goodwin P H, Lindhout P, De Wit P J G M. Proc Natl Acad Sci USA. 1998;95:9014–9018. doi: 10.1073/pnas.95.15.9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellis J G, Lawrence G J, Luck J E, Dodds P N. Plant Cell. 1999;11:495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dixon M S, Hatzixanthis K, Jones D A, Harrison K, Jones J D G. Plant Cell. 1998;10:1915–1925. doi: 10.1105/tpc.10.11.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDowell J M, Dhandaydham M, Long T A, Aarts M G M, Goff S, Holub E B, Dangl J L. Plant Cell. 1998;10:1861–1874. doi: 10.1105/tpc.10.11.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laugé R, Dmitriev A P, Joosten M H A J, De Wit P J G M. Mol Plant–Microbe Interact. 1998;11:301–308. [Google Scholar]
- 28.Small T. Ann Appl Biol. 1930;17:71–80. [Google Scholar]