Abstract
Paraquat (PQ; a widely used herbicide) exerts its harmful effect to human, mammals and microorganisms upon intracellular conversion to superoxide radical. Cellular responses against toxic paraquat remain not fully understood, especially on the adaptive metabolic changes as a consequence of oxidative burden. In this study, alterations of metabolic processes of Escherichia coli (E. coli) by paraquat were systematically investigated by two-dimensional gel electrophoresis (2-DE) in conjunction with peptide mass fingerprinting (PMF). In host cells, the first line mechanism was scrutinized by a remarkable induction of endogenous superoxide dismutase (E. coli SOD). The second line involved in the metabolic adaptation and compensation for energy production by up- or down-regulation of the enzymes implicated in glycolysis and tricarboxylic acid cycle. Notably, down-regulation of aconitase enzyme and changes of enzyme isoform from the acidic (pI~5.29) to the higher basidic form (pI~5.59) were detected. Meanwhile, up-regulation of fumarase approximately 4-5 folds were observed. Importantly, overexpression of human manganese superoxide dismutase (human Mn-SOD) in E. coli cells could in turn down-regulate the expression of fumarase enzyme. This observation was not found when the cells expressing human catalase were tested. Other mechanisms such as changes of purine nucleoside phosphorylase and protein transporters (D-ribose-binding protein and oligopeptide binding protein) were also accounted. However, among all the differentially expressed proteins, the fumarase enzyme is evidenced to be a major target responsible for superoxide-generating paraquat, which may further be applied as a potential biomarker for paraquat toxicity in the future.
Keywords: Escherichia coli, proteomic, oxidative stress, two-dimensional gel electrophoresis, superoxide dismutase
Introduction
Paraquat (1,1′-dimethyl-4,4′-bipyridi-nium dichloride; PQ) has long been used as a potent herbicide worldwide (Sagar, 1987[19]). Its killing effect on green plant tissue is quick and non-selective by inhibition of photosynthesis (Dodge and Harris, 1970[2]). Accidental or voluntary uptake of pure paraquat causes a noxious effect to human, potentially leading to acute respiratory distress syndrome, neurotoxicity, lung damage, kidney failure and skin cancer (Li et al., 2004[7]; Yoon, 2009[27]). In bacteria, inhibition of growth by the reaction of paraquat with unidentifed cellular moieties or biochemical processes has been revealed (Carr et al., 1986[1]).
In E. coli, the majority of genes that are responsible for paraquat toxicity belong to the soxRS regulon, coding for a sensor-transcriptional activator. It has previously been shown that several genes including sodA (encoding Mn-SOD), zwf (glucose-6-phosphate dehydrogenase), micF (antisense RNA, repressor of porin OmpF translation), nfo (endonuclease IV, a DNA repair enzyme for oxidative damage), fpr (ferredoxin reductase), acrAB (drug efflux pumps), and fumC (redox-resistant fumarase) are regulated by this regulon (Greenberg et al., 1990[3]). Induction of nitroreductase A, another member of soxRS regulon which contributes to the defenses against oxidative stress by minimizing the redox cycling attendant upon the univalent reduction of nitro compounds, quinones, and dyes, was reported (Liochev et al., 1999[10]). Activation of the membrane component of glucose transporting system was also potentiated. Such induction of glucose transport was also dependent on the soxRS regulon (Rungrassamee et al., 2008[18]). Stimulation of polyamine synthesizing enzymes e.g. ornithine and lysine decarboxylases and their metabolized products were also scrutinized (Tkachenko, 2004[23]). Meanwhile, down-regulation of Mut Y gene (coding for adenine DNA glycosylase) was disclosed (Yoon et al., 2003[28]). Using the genome-wide transcriptional analysis, a total of 112 genes was modulated in response to paraquat. These genes served for several functions e.g. regulation of iron transport and storage; and participation in sugar and amino acid transport, detoxification, protein modification, osmotic protection, and peptidoglycan synthesis (Pomposiello et al., 2001[15]). However, notification has to be made that a large number of paraquat-responsive genes have no known function in which many adaptive metabolic changes as a consequence of oxidative burden remain uncharacterized.
In the present study, effect of paraquat on cellular alterations of E. coli host has been investigated using proteomic in conjunction with peptide mass fingerprinting. In order to confirm the responsive mechanisms against paraquat, cells expressing either human manganese superoxide dismutase (Mn-SOD) or human catalase have been included for comparison. The interrelation between differentially expressed proteins as well as the feasibility to search for novel biomarkers for paraquat toxicity has been discussed.
Material and Methods
Bacterial strain and plasmids
E. coli strain BL21(DE3) [F-, ompT, hsdSB(rB-, mB-), dcm, gal, λ(DE3)] was used throughout the study. Plasmids pET46MnSOD (Yainoy et al., 2007[26]) and pET46CAT (encoding human Mn-superoxide dismutase and human catalase) were individually transformed into the E. coli host. Plasmid pET46 was used as a control plasmid.
Preparation of protein samples for proteomic analysis
Cells carrying pET46MnSOD, pET46CAT or pET46 (except for E. coli host) were grown in 5 ml 37ºC for overnight. Cells were adjusted to O.D. of 1.0. One millilitre of cells was subsequently inoculated in 50 ml LB/Amp and incubated at 37ºC for 2 hours. Cells were supplemented with 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside) and 200 ppm MnCl2. Paraquat solution was then added to the cultures to yield the final concentration of 0.8 mM and the cells were incubated for additional 14 hours. Preparation of crude protein extracts was performed as previously described (Isarankura-Na-Ayudhya et al., 2008[4], 2009[6]) with minor modification. Briefly, cells were collected by centrifugation at 6,000 rpm for 10 min at 4 ºC, washed three times, and resuspended in Tris buffer. The suspension was then mixed with 350 μl of lysis solution (7 M urea, 2 M thiourea, 4 % CHAPS; freshly prepared by supplementation with 10 mg/ml dithiothreitol (DTT) and 10 μl/ml protease inhibitor cocktail). Sonic disintegration using Branson sonifier (model 450) equipped with a microtip was carried out to disrupt the cells. Whole cell lysates was collected by centrifugation at 15,000 rpm for 15 min at room temperature. The protein amount was quanitfied by Bradford's method and a standard curve was prepared by varying bovine serum albumin concentrations.
Two-dimensional gel electrophoresis (2-DE)
Two-dimensional gel electrophoresis was performed using 2-D Electrophoresis System (GE Health care, USA) as described previously (Isarankura-Na-Ayudhya et al., 2009[6]; Panpumthong and Vattanaviboon, 2006[11]) with some modification. Three hundred micrograms of bacterial protein extract was mixed with 250 μl of rehydration buffer (8 M urea, 4 % CHAPS, 2 mM TBP, 0.001 % bromphenol blue, 2.8 mg/ml dithiothreitol and 12 μl/ml destreak) containing 1 % 3-10 IPG buffer. The mixture was stand for 10 min at room temperature and the insoluble material was removed by centrifugation at 13,000 rpm for 10 min at 20°C. The 13-cm IPG strips with pH range of 3-10 of an isoelectric focusing system (IPGphoreTM) was placed in the IPGphor strip holder gel-side-down in rehydration solution that contained the sample proteins, then covering them with mineral oil. Samples were run through steps of strip rehydration (30 volts, 12 h) and isoelectric focusing (500 volts for 1 h, 1,000 volts for 1 h, and 8,000 volts to reach 16,000 volt·h). The maximum current was maintained at 50 mA per one strip. Once complete, the strip was equilibrated twice times (15 min each) in equilibration buffer (50 mM Tris pH 8.8, 6 M urea, 30 % glycerol, 2 % SDS, 0.03 % bromphenol blue) supplemented with 65 mM DTT and 135 mM iodoacetamide to allow the cysteine residues to be reduced and then carbamidomethylated. The second dimensional separation was carried out using HoeferTM DALT on 12.5 % SDS-polyacrylamide gels. Separation of protein was executed under the applied voltage of 250 volt, 10 mA per strip at 20 °C for 30 min following by 250 volt, 20 mA per strip at 20°C for 2.5 hours until the bromphenol blue dye front reached 0.5 cm from the bottom of the gel. The gels were further stained with colloidal Coomassie brilliant blue G for overnight. Excess dye was removed by rinsing several times with deionized distilled water. Gels were scanned with the Canoscan LiDE20 scanner (Canon, USA). Differential analysis was partly performed by Delta2D 4.0 evaluation version (Decodon, GmbH, FRG) software tool.
MALDI-TOF Mass spectrometry and peptide mass fingerprinting (PMF) analysis
Mass spectrometry and peptide mass fingerprinting (PMF) analysis were carried out as described (Isarankura-Na-Ayudhya et al., 2009[6]). Initially, protein spots were manually excised from the gels, transferred to 96-well microtitre plate, and then soaked in 50 % methanol and 5 % acetic acid for overnight. Tryptic digestion was performed using sequencing grade of modified trypsin (Promega, UK) The in-gel-digested peptide fragments were extracted from the gel segments on a Spot Handling Workstation (GE Health Care, USA) using the preset protocols from the manufacture.
Protein identification was performed using MALDI-TOF mass spectrometer (Model ReflexIV, Bruker Daltonics, Germany) based on peptide fingerprint map. Briefly, the peptide solution was prepared by mixing the extracted peptides with solution of 10 mg/ml α-cyano-4-hydroxycinnamic acid (LaserBio Labs, France) in 66 % acetonitrile and 0.1 % trifluoroacetic acid (TFA) and further spotted onto a 96-well target plate. The mass spectra were acquired in the positive ion reflector delayed extraction mode using approximately 200 laser shots. Peak lists were created using the XMASS software (Bruker Daltonics). The BioTool 2.0 software (Bruker Daltonics) integrated with the MASCOT 2.2 search engine (MatrixScience, http://www.matrixscience.com/) was applied for spot proteins identification by querying the trypsin-digested peptide fragment data. The reference database used in this study was MSDB 20060831 with 3239079 sequences and 1079594700 residues. The searching criteria were set up as follows: complete carbamidomethylation of cysteine and partial methionine oxidation were exploited; an initial mass tolerance of ± 200 ppm was used in all searches; the number of missed cleavage sites was allowed up to 1. Search result scores which is greater than 58 was considered to be of significant difference (p<0.05). The accuracy of the experimental to theoretical pI and molecular weight of proteins were carefully taken into consideration.
Results
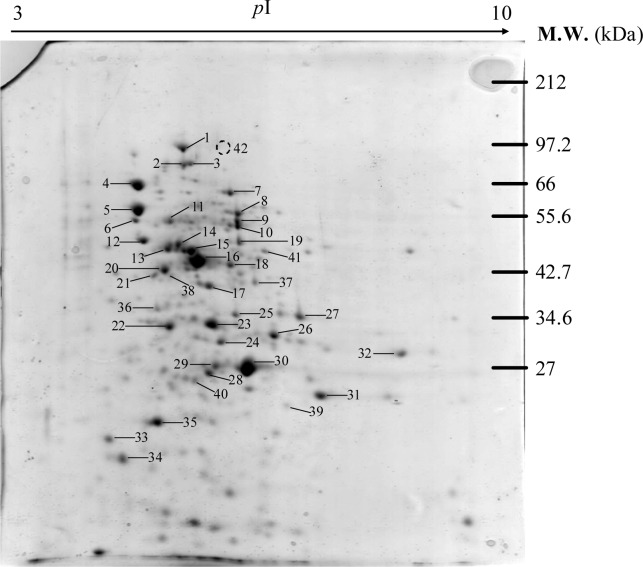
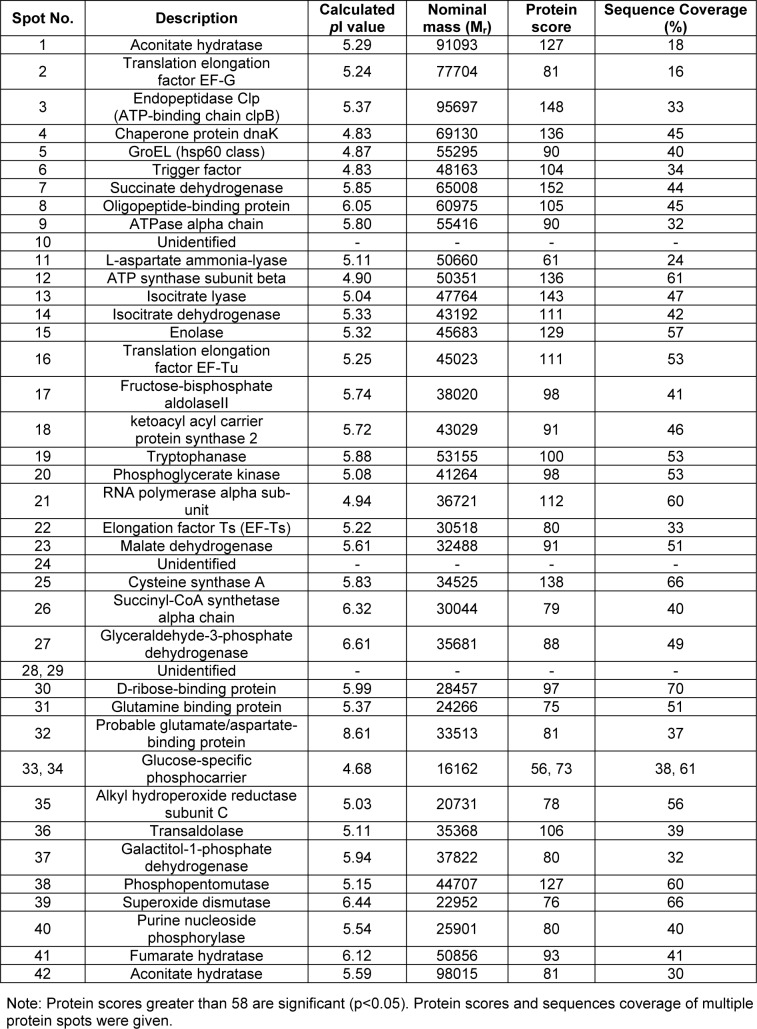
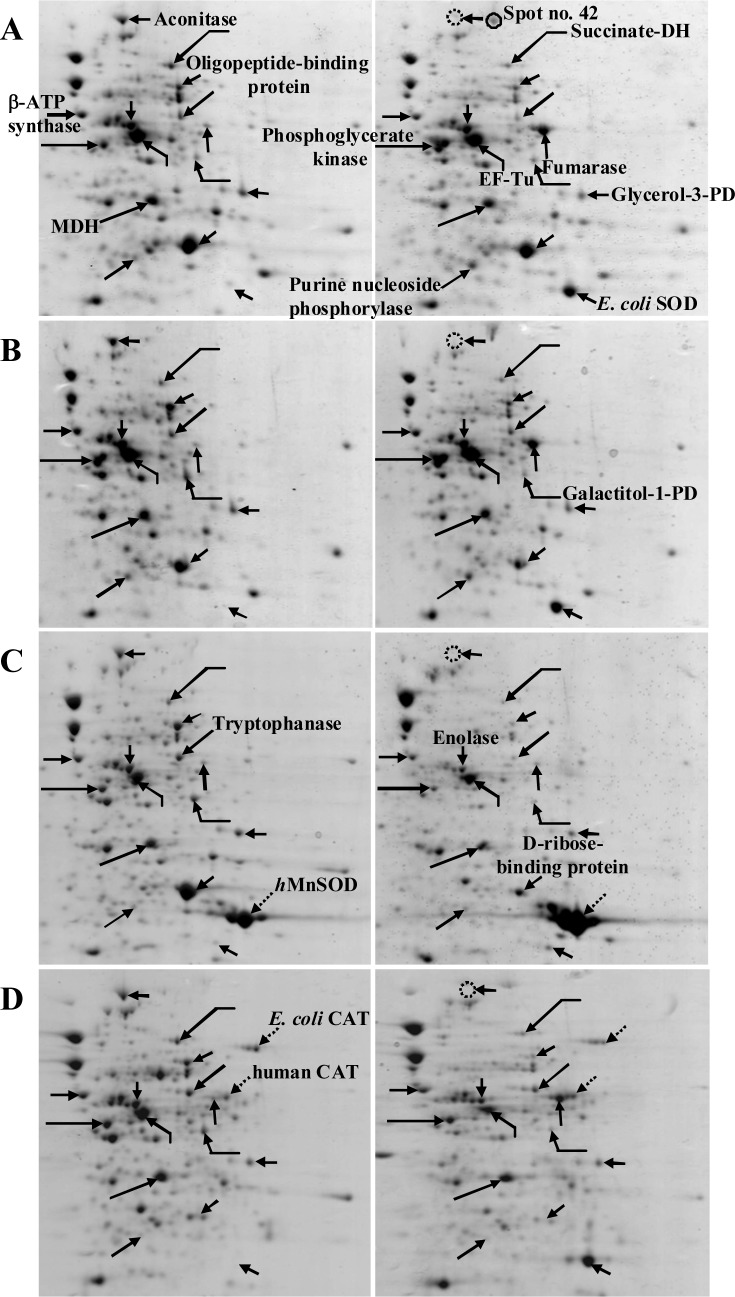
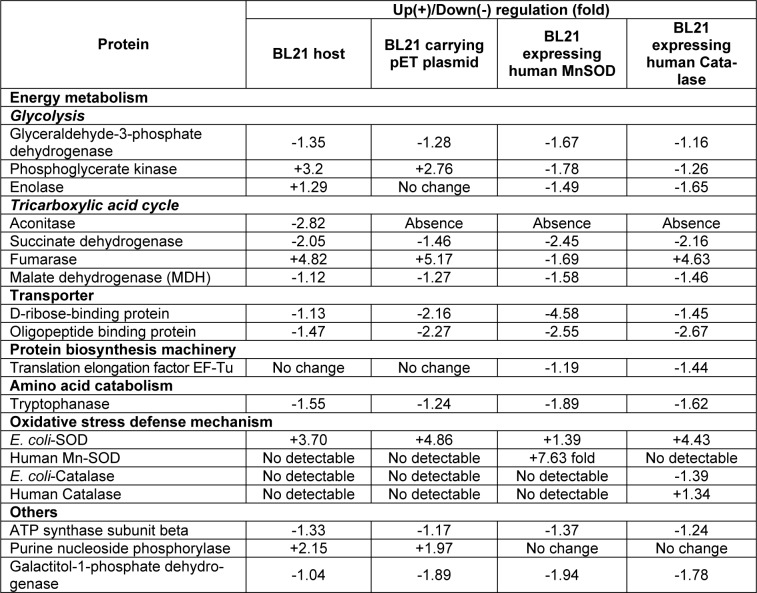
Figure 1(Fig. 1) demonstrates a master map of protein expression profile of E. coli BL21 host. Approximately 250 protein spots were revealed using the pI range of 3 to 10. Forty two spots were identified and the protein names were summarized in Table 1(Tab. 1). Exposure of E. coli cells to 0.8 mM paraquat led to remarkable changes of protein, especially on the energy metabolism and oxidative stress defense mechanism (Figure 2(Fig. 2) and Table 2(Tab. 2)). In the case of host cells, five genes were up-regulated while nine genes were found in the opposite manner. For examples, enzymes involved in glycolysis metabolism such as glyceraldehyde-3-phosphate dehydrogenase and phospho-glycerate kinase were found to be down-regulated and up-regulated, respectively (Figure 2A(Fig. 2)).
Figure 1. Master map representing protein profiles of E. coli BL21 grown in LB broth.
Table 1. Proteins of E. coli BL21 identified by mass spectrometry and peptide mass fingerprinting (PMF) analysis.
Figure 2. Comparison of protein profiles of E. coli BL21 host (A), E. coli BL21 carrying pET46 control plasmid (B), E. coli BL21 expressing human Mn-SOD (C) and E. coli BL21 expressing human CAT (D). Left and right panels represent protein profiles of cells in the absence and presence of 0.8 mM paraquat, respectively.
Table 2. Changes of differentially expressed proteins of E. coli BL21 following exposure to paraquat.
For the tricarboxylic acid (TCA) cycle, aconitase was down-regulated (2.82 fold) by paraquat while fumarase was found to be stimulated upto 4.82 fold. Importantly, another form of aconitase (shown as spot no. 42) was induced. Increased level of purine nucleoside phosphorylase (2.15 fold) together with the reduction of ATP synthase (1.33 fold) was also taken into consideration. Moreover, the E. coli SOD, which is responsible for the dismutation of superoxide anion to hydrogen peroxide and oxygen, was up-regulated upto 3.7 fold. It is worth to note that similar fashion of responses was accounted for the cells carrying a control plasmid (Figure 2B(Fig. 2)).In contrast to those findings, overexpression of human Mn-SOD in E. coli abolished the stimulatory level of fumarase (Figure 2C(Fig. 2)). The presence of human Mn-SOD could also compensate the expression of host SOD. A marked decrease of D-ribose-binding protein (4.58 fold) as compared to those of the others was detected. In addition, it is interesting to note that the purine nucleoside phosphorylase remained unchanged as compared to the aforementioned observations.
Next question was addressed of whether overexpression of other oxidative scavenging enzymes (e.g. human catalase) rendered any significant changes on the protein profile upon exposure to paraquat. As represented in Figure 2D(Fig. 2), expression of E. coli SOD was still required in order to detoxify the hazadous effect of paraquat. Disappearance of aconitase together with the overproduction of fumarase was in good agreement with those of the host cells and cells carrying control plasmid (Figures 2A and B(Fig. 2)). Contrarily, down-regulation of phosphoglycerate kinase as well as no alteration of purine nucleoside phosphorylase resembled those of the cells expressing human Mn-SOD (Figure 2C(Fig. 2)).
Discussion
In the present study, responses of E. coli against detrimental effect of paraquat have been systematically investigated by proteomic approach in combination with genetic manipulation of oxidative scavenging enzymes. Paraquat has widely been established to exert intracellular production of superoxide anion (O2˙-), which consequently leading to the inhibition of bacterial cell growth. Experimentation has been initiated by determining changes of protein expression profiles of host cells in the presence of paraquat. Our results revealed that the first line of defense mechanism was the up-regulation of endogenous superoxide dismutase upto 3-5 fold (Figure 2A(Fig. 2) and Table 2(Tab. 2)). In paralelle, metabolic adaptation and compensation for energy production have also been accounted. Down-regulation and up-regulation of two major enzymes involved in tricarboxylic acid (TCA) cycle (namely aconitase and fumarase, respectively) were affected by the existence of paraquat. Both enzymes are known to be controlled by SoxRS (superoxide response) regulon (Liochev and Fridovich, 1992[9]). In E. coli, there are two types of aconitase: a stationary-phase enzyme (AcnA), which is induced by iron and oxidative stress; and a major citric acid cycle but less stable enzyme (AcnB), produced during exponential phase (Tang et al., 2002[22]). As shown in Figure 2A(Fig. 2), the major form of aconitase was reduced while elevation of another form (spot no. 42) was potentiated as a consequence of paraquat exposure. It can be presumed that this form of aconitase was adapted for maintaining citric acid cycle activity because impairment of the TCA cycle would render a decrease in respiration by limiting the electrons flow from substrates to molecular oxygen. Moreover, reduction of the AcnB coincided with the decrease of oligopeptide binding protein (Table 2(Tab. 2)) since there was some interconnection between protein folding and the absence of a fully functional citric acid cycle (Tang et al., 2002[22]). For the fumarase, E. coli contains three biochemically distinct fumarases (FumA-C). The FumA and FumB are heat-labile and iron-containing fumarases, the FumC represents as a heat-stable fumarase with no iron as cofactor (Park and Gunsalus, 1995[12]). Notification has to be made that superoxide radicals can cause increased fumC gene expression while fumA and fumB expression are unaffected. Therefore, dramatic induction of fumaraseC was revealed once the paraquat was added (Figures 2A and B(Fig. 2)). It has been shown previously that induction of FumC can be mediated either by accumulation of superoxide stress or by deletion of SodA and SodB (genes encoding superoxide dismutases) (Liochev et al., 1999[8]). In our case, overproduction of human Mn-SOD could in turn block the induction of fumarase (Figure 2C(Fig. 2)). However, none of such effect was further observed in the case of E. coli expressing human catalase (Figure 2D(Fig. 2)). Additionally, the purine nucleoside phosphorylase was up-regulated approximately two fold in the case of host cells and cells carrying plasmid control while those of the cells expressing oxidative scavenging enzymes (human Mn-SOD and catalase) remained unchanged (Table 2(Tab. 2)). Our findings coincide with the evidence that the polynucleotide phosphorylase played imperative role in protecting E. coli cells against oxidative stress, possibly by reduction of oxidized RNA (Wu et al., 2009[25]). Taken altogether, such intrinsic responses particularly on metabolic adaptation in combination with overexpression of human MnSOD in engineered E. coli explain the underlying mechanisms on how the cells conferred efficient toleration against paraquat toxicity (Yainoy et al., 2007[26]). Moreover, investigations of proteomic profiling of cells expressing chimeric enzymes e.g. SOD-bacterial hemoglobin (Isarankura-Na-Ayudhya et al., 2010[5]) or SOD-CAT in response to paraquat in the absence or presence of antioxidant compounds (Piacham et al., 2003[14], 2006[13]; Prachayasittikul et al., 2008[16][17]; Suksrichavalit et al., 2008[21], 2009[20]; Wongsawatkul et al., 2008[24]) are underway conducted as ongoing research in our laboratory.
In conclusion, changes of protein expression profiles of E. coli cells as a consequence of paraquat exposure have successfully been discovered using proteomic approach. Among all the differentially expressed proteins, fumarase enzyme is evidenced to be the most significant target responsible for superoxide anion derived from paraquat. Up-regulation of the fumarase can be triggered by superoxide anion while dismutation of such radical by the overexpressed MnSOD down-regulates the level of enzyme production. This interrelation is quite specific since overexpression of human catalase gives rise to the same phenomenon as those observed in the host cells. Therefore, our findings not only explore the underlying mechanism of cellular responses against paraquat but also open up a high feasibility to further apply the fumarase enzyme as a potential biomarker for paraquat toxicity in the future.
Acknowledgements
W.S. and W.P. are undergraduate students under supervision of C.I. C.T. is a Ph.D. student financially supported by the Royal Golden Jubilee Ph.D. scholarship from the Thailand Research Fund under the supervision of V.P. The authors would like to thank Ms. Janthima Jaresitthikunchai for technical assistance. This research is partially supported by the annual governmental budget of Mahidol University (B.E.2551-2555).
References
- 1.Carr RJ, Bilton RF, Atkinson T. Toxicity of paraquat to microorganisms. Appl Environ Microbiol. 1986;52:1112–1116. doi: 10.1128/aem.52.5.1112-1116.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dodge AD, Harris N. The mode of action of paraquat and diquat. Biochem J. 1970;118:43P–44P. doi: 10.1042/bj1180043p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greenberg JT, Monach P, Chou JH, Josephy PD, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci U S A. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isarankura-Na-Ayudhya C, Panpumthong P, Tangkosakul T, Boonpangrak S, Prachayasittikul V. Shedding light on the role of Vitreoscilla hemoglobin on cellular catabolic regulation by proteomic analysis. Int J Biol Sci. 2008;4:71–80. doi: 10.7150/ijbs.4.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isarankura-Na-Ayudhya C, Yainoy S, Tantimongcolwat T, Bulow L, Prachayasittikul V. Engineering of a novel chimera of superoxide dismutase and Vitreoscilla hemoglobin for rapid detoxification of reactive oxygen species. J Biosci Bioeng. 2010;in press doi: 10.1016/j.jbiosc.2010.07.001. Available from: doi10.1016/j.jbiosc.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Isarankura-Na-Ayudhya P, Isarankura-Na-Ayudhya C, Treeratanapaiboon L, Kasikun K, Thipkeaw K, Prachayasittikul V. Proteomic profiling of Escherichia coli in response to heavy metals stress. Eur J Sci Res. 2009;25:679–688. [Google Scholar]
- 7.Li S, Crooks PA, Wei X, de Leon J. Toxicity of dipyridyl compounds and related compounds. Crit Rev Toxicol. 2004;34:447–460. doi: 10.1080/10408440490503143. [DOI] [PubMed] [Google Scholar]
- 8.Liochev SI, Benov L, Touati D, Fridovich I. Induction of the soxRS regulon of Escherichia coli by superoxide. J Biol Chem. 1999;274:9479–9481. doi: 10.1074/jbc.274.14.9479. [DOI] [PubMed] [Google Scholar]
- 9.Liochev SI, Fridovich I. Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc Natl Acad Sci U S A. 1992;89:5892–5896. doi: 10.1073/pnas.89.13.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liochev SI, Hausladen A, Fridovich I. Nitroreductase A is regulated as a member of the soxRS regulon of Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:3537–3539. doi: 10.1073/pnas.96.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panpumthong P, Vattanaviboon P. Improvement of proteomic profile of Plasmodium falciparum by two-step protein extraction in two-dimensional gel electrophoresis. Thammasat Int J Sc Tech. 2006;11:61–68. [Google Scholar]
- 12.Park SJ, Gunsalus RP. Oxygen, iron, carbon, and superoxide control of the fumarase fumA and fumC genes of Escherichia coli: role of the arcA, fnr, and soxR gene products. J Bacteriol. 1995;177:6255–6262. doi: 10.1128/jb.177.21.6255-6262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piacham T, Isarankura-Na-Ayudhya C, Nantasenamat C, Yainoy S, Ye L, Bulow L, Prachayasittikul V. Metalloantibiotic Mn(II)-bacitracin complex mimicking manganese superoxide dismutase. Biochem Biophys Res Comm. 2006;341:925–930. doi: 10.1016/j.bbrc.2006.01.045. [DOI] [PubMed] [Google Scholar]
- 14.Piacham T, Isarankura-Na-Ayudhya C, Prachayasittikul V, Bulow L, Ye L. A polymer supported manganese catalyst useful as a superoxide dismutase mimic. Chem Comm. 2003:1254–1255. doi: 10.1039/b301683h. [DOI] [PubMed] [Google Scholar]
- 15.Pomposiello PJ, Bennik MH, Demple B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J Bacteriol. 2001;183:3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prachayasittikul S, Buraparuangsang P, Worachartcheewan A, Isarankura-Na-Ayudhya C, Ruchirawat S, Prachayasittikul V. Antimicrobial and antioxidative activities of bioactive constituents from Hydnophytum formicarum Jack. Molecules. 2008;13:904–921. doi: 10.3390/molecules13040904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prachayasittikul S, Suksrichavalit T, Isarankura-Na-Ayudhya C, Ruchirawat S, Prachayasittikul V. Antimicrobial and antioxidative activities of 1-adamantylthio derivative of 3-substituted pyridines. EXCLI J. 2008;7:63–70. [Google Scholar]
- 18.Rungrassamee W, Liu X, Pomposiello PJ. Activation of glucose transport under oxidative stress in Escherichia coli. Arch Microbiol. 2008;190:41–49. doi: 10.1007/s00203-008-0361-y. [DOI] [PubMed] [Google Scholar]
- 19.Sagar GR. Uses and usefulness of paraquat. Hum Toxicol. 1987;6:7–11. doi: 10.1177/096032718700600102. [DOI] [PubMed] [Google Scholar]
- 20.Suksrichavalit T, Prachayasittikul S, Nantasenamat C, Isarankura-Na-Ayudhya C, Prachayasittikul V. Copper complexes of pyridine derivatives with superoxide scavenging and antimicrobial activities. Eur J Med Chem. 2009;44:3259–3265. doi: 10.1016/j.ejmech.2009.03.033. [DOI] [PubMed] [Google Scholar]
- 21.Suksrichavalit T, Prachayasittikul S, Piacham T, Isarankura-Na-Ayudhya C, Nantasenamat C, Prachayasittikul V. Copper complexes of nicotinic-aromatic carboxylic acids as superoxide dismutase mimetics. Molecules. 2008;13:3040–3056. doi: 10.3390/molecules13123040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Y, Quail MA, Artymiuk PJ, Guest JR, Green J. Escherichia coli aconitases and oxidative stress: post-transcriptional regulation of sodA expression. Microbiology. 2002;148:1027–1037. doi: 10.1099/00221287-148-4-1027. [DOI] [PubMed] [Google Scholar]
- 23.Tkachenko AG. Mechanisms of protective functions of Escherichia coli polyamines against toxic effect of paraquat, which causes superoxide stress. Biochemistry (Mosc) 2004;69:188–194. doi: 10.1023/b:biry.0000018950.30452.53. [DOI] [PubMed] [Google Scholar]
- 24.Wongsawatkul O, Prachayasittikul S, Isarankura-Na-Ayudhya C, Satayavivad J, Ruchirawat S, Prachayasittikul V. Vasorelaxant and antioxidant activities of Spilanthes acmella Murr. Int J Mol Sci. 2008;9:2724–2744. doi: 10.3390/ijms9122724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J, Jiang Z, Liu M, Gong X, Wu S, Burns CM, Li Z. Polynucleotide phosphorylase protects Escherichia coli against oxidative stress. Biochemistry. 2009;48:2012–2020. doi: 10.1021/bi801752p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yainoy S, Isarankura-Na-Ayudhya C, Tantimongcolwat T, Prachayasittikul V. Cloning of active human manganese superoxide dismutase and its oxidative protection in Escherichia coli. Pak J Biol Sci. 2007;10:3541–3548. doi: 10.3923/pjbs.2007.3541.3548. [DOI] [PubMed] [Google Scholar]
- 27.Yoon SC. Clinical outcome of paraquat poisoning. Korean J Intern Med. 2009;24:93–94. doi: 10.3904/kjim.2009.24.2.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon SH, Lee HS, Choi JY, Kang HK, Lee JJ, Hyun JW, Choi J, Ye SK, Chung MH. MutY is down-regulated by oxidative stress in E. coli. Free Radic Res. 2003;37:873–879. doi: 10.1080/1071576031000150760. [DOI] [PubMed] [Google Scholar]