SUMMARY
The mechanisms by which clock neurons in the Drosophila brain confer an ~24-hr rhythm onto locomotor activity are unclear, but involve the neuropeptide diuretic hormone 44 (DH44), an ortholog of corticotropin-releasing factor. Here we identified DH44 receptor 1 as the relevant receptor for rest:activity rhythms and mapped its site of action to hugin-expressing neurons in the subesophageal zone (SEZ). We traced a circuit that extends from Dh44-expressing neurons in the pars intercerebralis (PI) through hugin+ SEZ neurons to the ventral nerve cord. Hugin neuropeptide, a neuromedin U ortholog, also regulates behavioral rhythms. The DH44 PI-Hugin SEZ circuit controls circadian locomotor activity in a daily cycle but has minimal effect on feeding rhythms, suggesting that the circadian drive to feed can be separated from circadian locomotion. These findings define a linear peptidergic circuit that links the clock to motor outputs to modulate circadian control of locomotor activity.
In Brief
Circadian clocks allow organisms to coordinate their behavior with time of day. King et al. demonstrate that a specific circuit comprising peptidergic neurons in Drosophila connects the central pacemaker neurons to motor outputs. Peptides released by neurons in this circuit, DH44 and Hugin, regulate rest:activity rhythms.
INTRODUCTION
Drosophila melanogaster has been instrumental for understanding the molecular and cellular basis of circadian clocks. At the molecular level, a transcription-translation feedback loop keeps the circadian clock running at an ~24-hr pace. At the cellular level, ~150 clock-expressing neurons in the Drosophila brain synchronize as a network to coordinate behavioral rhythms [1, 2]. Of these clock neurons, the ventrolateral neurons (LNvs) are the most important for driving locomotor activity rhythms in free-running conditions of constant darkness (DD) [3, 4]. In addition, the LNvs maintain the phase and amplitude of molecular oscillations among different clock neurons through neuropeptide pigment-dispersing factor (PDF) signaling [2, 5, 6]. Although we have some understanding of the signaling mechanisms within the central clock network that generate circadian rhythms, the mechanisms for relaying circadian timing information from the clock to neural circuits controlling behavior are poorly understood.
A screen for circadian output neurons in Drosophila identified Dh44-expressing neurons in the pars intercerebralis (PI), a functional homolog of the mammalian hypothalamus [7], as relevant for rest:activity rhythms [8]. Dh44+ PI neurons lack clocks themselves but are indirectly connected to the small LNvs (sLNvs), the central pacemaker neurons, through a subset of dorsal clock neurons (DN1s). The diuretic hormone 44 (DH44) neuropeptide is the fly ortholog of corticotropin-releasing factor (CRF) and modulates rest:activity rhythms [8]. To identify signals downstream of DH44 that regulate rest:activity rhythms, we sought to identify the relevant receptor and its site of action. Here we find that a null mutation in Dh44 receptor 1 (Dh44-R1) disrupts the amplitude of free-running rest:activity rhythms. We find that DH44-R1 acts in neurons expressing hugin, a neuropeptide ortholog of neuromedin U (NMU) [9], which also regulates rest:activity rhythms. Dh44+ PI neurons are anatomically and functionally connected to hugin+ neurons in the subesophageal zone (SEZ), a sensorimotor control center in flies [10]. hugin+ neurons display cyclic neuropeptide release that is controlled by the clock and have descending projections into the ventral nerve cord (VNC), where they potentially regulate motor circuits driving locomotion. Although Dh44-R1 and hugin modulate circadian locomotor activity, manipulations of the Dh44 PI-Hugin SEZ circuit have little to no effect on feeding rhythms. We propose that the sLNv→DN1→DH44 PI→Hugin SEZ→VNC pathway defines a linear circuit that modulates rest:activity rhythms.
RESULTS
DH44-R1 Is the Predominant DH44 Receptor Regulating Circadian Rhythms of Rest:Activity
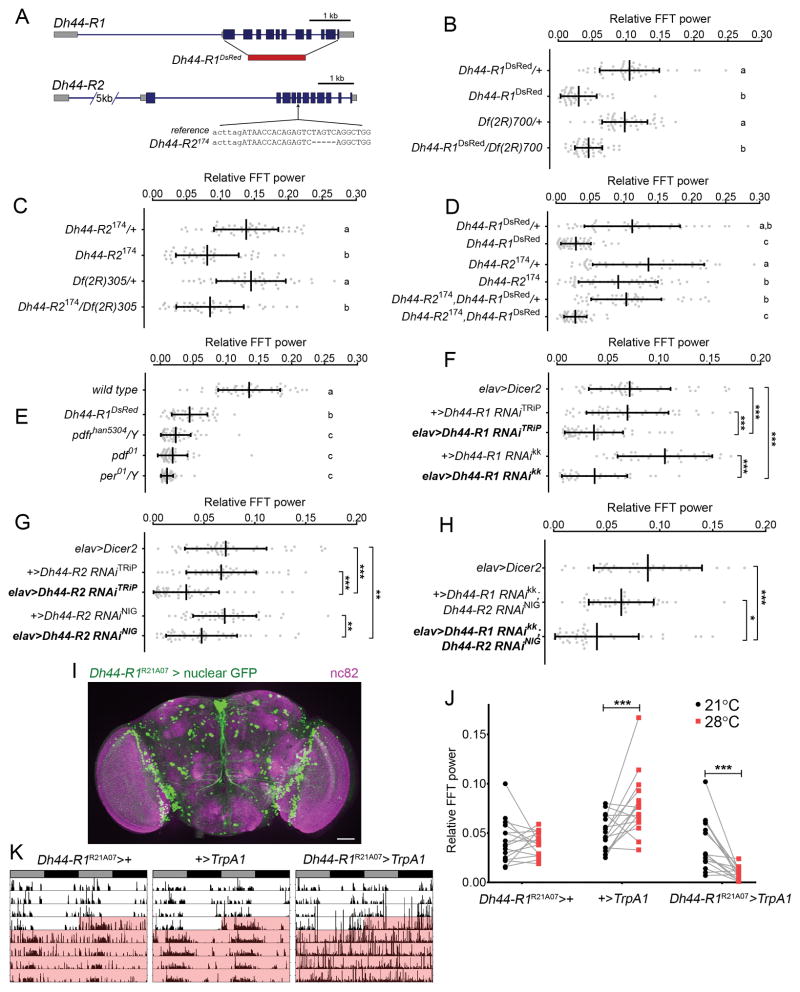
DH44 neurons as well as the peptide itself are required for normal rest:activity rhythms in DD [8]. Because DH44 can signal through two G protein-coupled receptors, DH44-R1 and DH44-R2, we asked which receptor was necessary for rhythmic behavior [11, 12]. Using CRISPR/CAS9-mediated genome editing, we generated mutant alleles of both Dh44 receptor genes, Dh44-R1DsRed and Dh44-R2174. The Dh44-R1DsRed allele is a deletion of the entire protein-coding region and replaces exons 2–11 with a DsRed selection marker, which decreases mRNA levels of Dh44-R1 (Figures 1A and S1A). Dh44-R2174 allele is a 5-bp deletion in exon 6 of the gene (Figure 1A). Dh44-R2174 mutants have normal levels of Dh44-R2 mRNA (Figure S1A); however, the frameshift mutation is predicted to result in a non-functional truncated protein with only two transmembrane domains (Figure S1B).
Figure 1. DH44 Receptors Regulate Rest:Activity Rhythms.
(A) Sequence alterations to Dh44-R1 and Dh44-R2 loci. Blue denotes the coding DNA sequence, and red denotes the replacement of Dh44-R1 with the DsRed sequence.
(B–D) Amplitude of circadian rest:activity rhythms under constant darkness (DD) represented by FFT power (mean ± SD) for Dh44-R1DsRed mutants (B), Dh44-R2174 mutants (C), Dh44-R2174,Dh44-R1DsRed double mutants (D), and their heterozygous controls.
(E) Amplitude of rest:activity rhythms in Dh44-R1DsRed mutants, clock output mutants (Pdf01 and Pdfrhan5304), and clock mutant (per01).
(F–H) Amplitude of rest:activity rhythms under DD conditions represented by FFT power (mean ± SD) for flies with RNAi-mediated knockdown of Dh44-R1 (F), Dh44-R2 (G), or both Dh44-R1 and Dh44-R2 (H) in all neurons. *p < 0.05, **p < 0.01, ***p < 0.001 by Sidak’s test following one-way ANOVA.
(I) Brain with Dh44-R1R21A07-GAL4+ neurons labeled with nuclear GFP (green) and counterstained with nc82 (anti-bruchpilot; magenta). Scale bar, 50 μm.
(J) FFT power for rest:activity rhythms at 21°C (black) and 28°C (red). ***p < 0.00093 by Sidak’s test following two-way repeated-measure ANOVA.
(K) Representative records of individual fly activity in DD for 4 days at 21°C and then for 4 days at 28°C for TrpA1 activation (red).
In (B)–(E), groups with the same letter are not significantly different from each other (p > 0.05 by Tukey’s test following one-way ANOVA). See also Figures S1 and S2 and Tables S1 and S2.
We assessed circadian rhythms of locomotor activity in Dh44-R1DsRed and Dh44-R2174 mutants under DD. Both Dh44-R1DsRed and Dh44-R2174 mutants displayed rest:activity rhythms with wild-type period length (Figures S1C–S1E; Table S1). However, the amplitude of the behavioral rhythm was affected in Dh44-R1DsRed mutants (Figure 1B), as assayed by fast Fourier transform (FFT) [8]. Dh44-R2174 mutants were largely normal, although FFT analysis shows that they had modestly weaker rest:activity rhythms compared to control heterozygotes (Figure 1C). Both Dh44-R1DsRed and Dh44-R2174 failed to complement large chromosomal deficiencies that remove the respective genes, consistent with Dh44-R1DsRed and Dh44-R2174 being null alleles (Figures 1B and 1C). To investigate the relationship between the two DH44 receptors, we tested the behavior of flies mutant for both receptors. Double heterozygotes, Dh44-R1DsRed,Dh44-R2174/+, had strong rest:activity rhythms similar to those of single heterozygous mutants. In contrast, flies homozygous for both mutations (Dh44-R1DsRed,Dh44-R2174) exhibited weak rest:activity rhythms like those seen in Dh44-R1DsRed mutants (Figures 1D and S1F). Because loss of Dh44-R2 does not modify the phenotype of the Dh44-R1DsRed mutant, we conclude that DH44-R1 is the primary DH44 receptor regulating rest: activity rhythms.
The phenotype of Dh44-R1DsRed mutants suggests a modulatory role of DH44 signaling, which is generally the case for peptide signaling. Indeed, whereas flies lacking core clock genes, such as period (per), are completely arrhythmic, this is not the case for mutants of PDF, the major neuropeptide in the clock circuit, or PDF receptor [3, 13–17]. We found that 54% of Pdf01 mutants, but none of the per0 flies, were rhythmic (Table S1). Nevertheless, rest:activity rhythms of Dh44-R1DsRed flies were stronger than those of Pdf01 and Pdfrhan5304 mutants (Figure 1E), suggesting that DH44 is not the only signal downstream of PDF relevant for rest:activity rhythms. We examined expression levels of the DH44 receptors across the day, but did not see any evidence for cycling of Dh44-R1 or Dh44-R2 mRNA (Figure S1F).
To verify a role for Dh44-R1 in neurons, we pan-neuronally knocked it down using RNAi. elav-GAL4-driven expression of two different RNAi lines reduced mRNA levels of Dh44-R1 to approximately 50% of the levels in controls (Figures S2A and S2B). Compared to control flies, elav>Dh44-R1 RNAi flies showed lower amplitude of rest:activity rhythms (Figure 1F). Interestingly, knockdown of Dh44-R2 also dampened rest: activity rhythms (Figure 1G), more so than the genetic mutant, perhaps because of compensation with the global knockout. Nevertheless, simultaneous knockdown of both receptors in neurons resulted in the same rhythm phenotype as knockdown of a single DH44 receptor (Figure 1H). These data are consistent with the results from genetic mutant analysis and suggest that effects of Dh44-R1 and Dh44-R2 on circadian rhythms are not additive or synergistic; thus, any role of DH44-R2 is not independent of DH44-R1. Because of the stronger phenotype of the Dh44-R1 mutant, we conclude that DH44-R1 is the more relevant receptor for rest:activity rhythms.
Dh44-R1-Expressing Neurons Regulate Rest:Activity Rhythms
To identify the site of DH44-R1 action relevant for rest:activity rhythms, we first examined expression of a Dh44-R1R21A07-GAL4 driver (which includes 3.65 kb from the Dh44-R1 promoter). Dh44-R1R21A07-GAL4 is expressed broadly in the brain and in a pattern similar to an in situ characterization of Dh44-R1 mRNA expression (Figure 1I) [18]. RNAi-mediated knockdown of Dh44-R1 in Dh44-R1R21A07-GAL4+ neurons reduced the strength of rest:activity rhythms (Figures S2C and S2D), supporting the idea that the driver targets neurons that mediate effects of DH44-R1.
We next determined whether activating Dh44-R1-expressing neurons is sufficient to degrade rest:activity rhythms. We expressed the Drosophila temperature-activated cation channel, TrpA1 [19], in Dh44-R1-expressing neurons and tested rest: activity rhythms of individual flies at 21°C and then at 28°C. At 21°C, 93.5% of the Dh44-R1R21A07>TrpA1 flies were rhythmic. However, after transitioning the flies to 28°C to activate TrpA1, only 31% of the flies were rhythmic. FFT power for Dh44-R1R21A07>TrpA1 flies also decreased after transitioning to 28°C (Figures 1J and 1K). Sustained activation of Dh44-R1-expressing neurons is sufficient to disrupt rest:activity rhythms, indicating that these neurons have a role in regulating rest:activity rhythms.
Effects of Dh44-R1 on Rest:Activity Rhythms Are Mediated by hugin+ Neurons in the SEZ
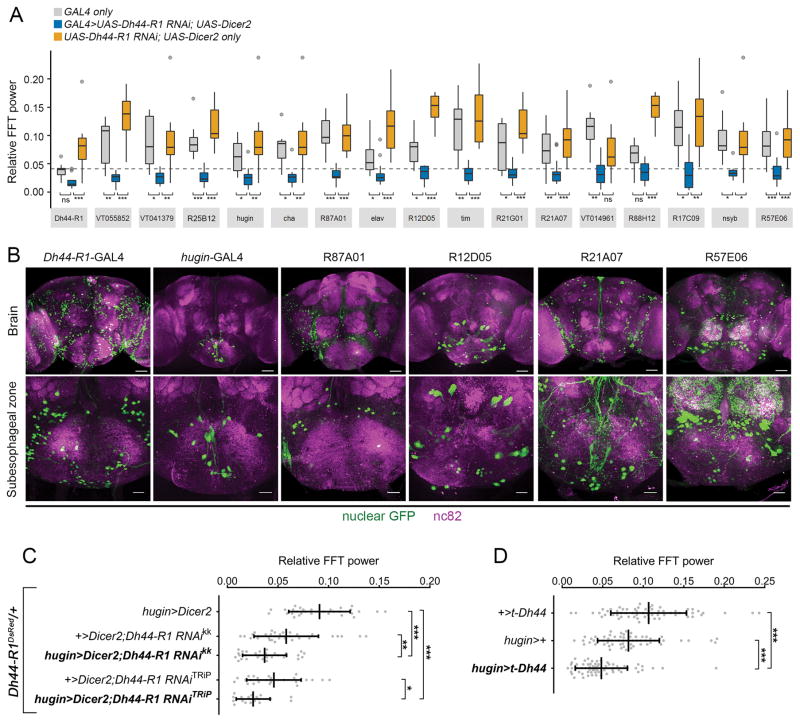
To identify the specific neurons requiring Dh44-R1 for rest: activity rhythms, we targeted RNAi knockdown of Dh44-R1 to random subsets of brain cells using 168 independent GAL4s (Figure S3) [20, 21]. We found that 15 GAL4s driving Dh44-R1 RNAi weakened rest:activity rhythms comparable to the phenotype observed with pan-neuronal nsyb-GAL4- or elav-GAL4-targeted knockdown (Figures 2A and S3). Of the GAL4 hits, three are regulated by Dh44-R1 genomic sequences: Dh44-R1-GAL4 [18], R21A07-GAL4, and R57E06-GAL4. We examined the expression of GAL4 hits in the brain and found that the SEZ stood out as a region of overlap, labeled by five GAL4 drivers (Figure 2B). Interestingly, axons of Dh44+ PI neurons terminate in the SEZ.
Figure 2. Dh44-R1 in hugin+ Neurons Regulates Rest:Activity Rhythms.
(A) Amplitude (FFT values) of circadian rest:activity rhythms in flies with different GAL4s driving Dh44-R1 RNAi knockdown (blue) and GAL4 genetic controls (gray and orange). 17 GAL4 lines that yielded the weakest rhythms by FFT analysis are shown from the GAL4 screen. Data are summarized with Tukey’s boxplots. The gray dashed line denotes 1 SD below the average FFT value of the RNAi knockdown phenotype from all 168 GAL4 lines screened.
(B) Images of SEZ-localized and -proximal GAL4 hits expressing nuclear GFP (green) in the brain (scale bars, 50 μm) and SEZ (scale bars, 20 μm). Brains were counterstained with nc82 (magenta).
(C) Amplitude of rest:activity rhythms with Dh44-R1 knocked down in hugin+ neurons in a Dh44-R1DsRed heterozygous background and genetic control flies under DD conditions.
(D) Amplitude of rest:activity rhythms under DD conditions in flies expressing a transgenic tethered DH44 peptide in hugin+ neurons (hugin>t-Dh44).
In (C) and (D), data are shown as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 by Sidak’s test following one-way ANOVA; ns, not significant. See also Figure S3 and Tables S1 and S2.
We focused on hugin-GAL4, because its expression is restricted to about 20 hugin+ neurons in the SEZ [22]. hugin (hug) encodes a prepropeptide that produces two neuropeptides, pyrokinin-2 and Hugin-γ, one of which (pyrokinin-2) is homologous to mammalian NMU [9]. We followed up on the initial phenotype and found that knockdown of Dh44-R1 in hugin+ neurons weakens rest:activity rhythms. Although only one of the two Dh44-R1 RNAi transgenes significantly reduced circadian rhythmicity in a wild-type background, both yielded a consistent weak rhythm phenotype in a sensitized Dh44-R1DsRed/+ heterozygous background, suggesting incomplete knockdown in wild-type flies (Figure 2C).
To verify the circadian relevance of DH44 expression in hugin cells, we expressed a membrane-tethered form of DH44 (t-DH44) in hugin+ neurons. Membrane-tethered peptides cell autonomously and constitutively activate their cognate receptors, and were used previously to study PDF signaling in the circadian network [23, 24]. Expression of t-DH44 in hugin+ neurons weakened rest:activity rhythms (Figure 2D), supporting the idea that Dh44-R1 functions in hugin+ SEZ neurons to modulate rest:activity rhythms.
hugin+ Neurons in the SEZ Receive Inputs from Dh44+ Neurons in the PI
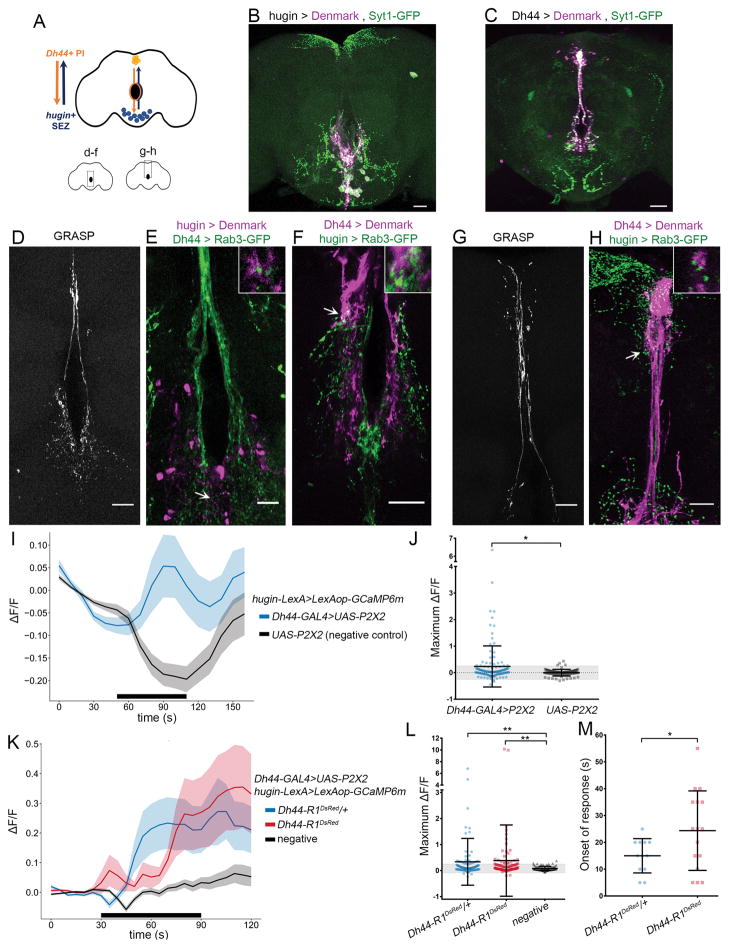
Because the function of DH44-R1 partially maps to hugin+ neurons, we sought to determine whether hugin+ neurons receive synaptic inputs from Dh44+ neurons (Figure 3A). To analyze the circuitry, we labeled the projections of each neuronal subset—hugin+ and Dh44+—with fluorescent markers: syt1-GFP to identify presynaptic membranes [25], and Denmark to identify postsynaptic membranes [26]. hugin+ neurons have both pre-synaptic and postsynaptic components within the SEZ and near the esophagus. Interestingly, hugin+ axon terminals also project to the PI (Figure 3B). Conversely, axons from Dh44+ PI neurons terminate within the SEZ, and Dh44+ dendritic compartments are located both in the PI and near the esophagus (Figure 3C).
Figure 3. hugin+ Neurons in the SEZ Receive Inputs from Dh44+ PI Neurons.
(A) Schematic of a circuit between Dh44+ neurons in the pars intercerebralis (PI) and hugin+ neurons in the subesophageal zone (SEZ).
(B and C) hugin-GAL4 (B) or Dh44-GAL4 (C) expressing presynaptic (syt1-GFP; green) and postsynaptic markers (Denmark; magenta) in the brain.
(D) Neurexin-GRASP signal near the esophagus in a brain expressing Dh44-GAL4>UAS-neurexin-spGFP1-10; hugin-LexA>LexAop-CD4-spGFP11.
(E) Dh44+ axon terminals (green) and hugin+ dendrites (magenta) near the esophagus in the brain.
(F) Hugin+ axon terminals (green) and Dh44+ dendrites (magenta) near the esophagus in the brain.
(G) Neurexin-GRASP signal in the PI of a brain expressing Dh44-GAL4>UAS-neurexin-spGFP1-10; hugin-LexA>LexAop-CD4-spGFP11.
(H) hugin+ axon terminals (green) and Dh44+ dendrites (magenta) in the PI.
(I) GCaMP signal over time in hugin+ neurons with activation of Dh44+ cells (blue; n = 129 cells, 11 brains) or no activation (black; n = 83 cells, 8 brains). The black bar denotes duration of ATP application. Data are represented as mean ± SEM.
(J) Maximum GCaMP change (ΔF/F) in individual cells. Mean ± SD. The shaded gray region indicates within 2 SD of the mean value for the UAS-P2X2 group. *p = 0.0119 by Mann-Whitney test. U = 4259, Z = ~2.51.
(K) GCaMP signal over time in hugin+ neurons upon activation of Dh44+ cells in Dh44-R1DsRed/+ heterozygotes (blue; n = 99 cells, 9 brains) or Dh44-R1DsRed mutants (red; n = 108 cells, 11 brains). Negative control is UAS-P2X2; hugin-LexA>LexAop-GCaMP6m in Dh44-R1DsRed/+ heterozygotes (black; n = 82 cells, 7 brains). The black bar denotes duration of ATP application. Data are represented as mean ± SEM.
(L) Maximum GCaMP change (ΔF/F) in individual cells. Mean ± SD. The shaded gray region indicates within 2 SD of the mean for the negative control group. **p < 0.005 by Kruskal-Wallis test followed by Dunn’s test.
(M) Onsets of response in Dh44-R1DsRed mutants and heterozygotes. Mean ± SD. *p = 0.0339 by two-tailed Welch’s t test.
Insets in (E), (F), and (H) show 3× magnification of a single confocal section from the regions indicated by arrows. Scale bars, 35 μm (B and C), 20 μm (D and F–H), and 10 μm (E). See also Table S2.
To test for synaptic connections between Dh44+ and hugin+ neurons, we used a GFP reconstitution across synaptic partners (GRASP) method that labels synaptic sites [27, 28]. We used Dh44-GAL4 to express neurexin-bound GFP fragments 1–10 and hugin-LexA to express CD4 membrane-bound GFP fragment 11. In these flies, GRASP signal was observed in the SEZ, near the esophagus, and along the midline of the brain (Figure 3D). To determine the polarity of the connectivity detected with GRASP and to confirm that Dh44+ and hugin+ projections overlap in the same region, we simultaneously labeled the axons of one group with Rab3-GFP [29] and the somatodendritic membrane of the other group using Denmark. We found that axons from Dh44+ PI neurons intersect with hugin+ dendrites near the esophagus and in the SEZ (Figure 3E). Intriguingly, hugin+ axon terminals also contact Dh44+ dendrites near the esophagus (Figure 3F). In addition, we detected a GRASP signal between Dh44+ and hugin+ neurons in the PI (Figure 3G), where hugin+ axon terminals contact Dh44+ neurons (Figure 3H). GRASP and polarity analysis indicate that Dh44+ PI and hugin+ SEZ neurons make extensive synaptic contacts through reciprocal projections.
To test whether Dh44+ and hugin+ neurons are functionally connected, we expressed and activated ATP-gated P2X2 receptors [30] in Dh44+ neurons while imaging Ca2+ in hugin+ neurons with GCaMP6m [31]. Addition of ATP to activate Dh44+ neurons increased GCaMP signal in a subset of hugin+ neurons (Figures 3I and 3J). Some neurons showed a decreased GCaMP signal upon ATP application; however, this is most likely an experimental artifact, because we also observed decreases in the negative control group. We estimated the number of hugin+ neurons that responded to Dh44+ PI activation as the number of neurons with a GCaMP signal increase greater than 2 SDs from the mean response in the negative control. We found that approximately 15% of the ~20 hugin+ neurons responded and increased GCaMP signal upon activation of Dh44+ PI neurons (Figure 3J), suggesting that hugin+ neurons are a heterogeneous group.
We next asked to what extent DH44 signaling is required for the Ca2+ response in hugin+ neurons following Dh44+ PI activation. Thus, we performed the P2X2 and GCaMP6 imaging experiments in Dh44-R1DsRed mutants. We did not observe significant differences in the amplitude of the responses in hugin+ neurons between mutants and heterozygotes (Figures 3K and 3L), but the onset of response to Dh44+ PI activation was delayed in Dh44-R1DsRed mutants (Figure 3M). Thus, the functional connection between Dh44+ PI and hugin+ SEZ is partly dependent on DH44 signaling. It is likely that, in addition to DH44, Dh44+ neurons express other neurotransmitters that may signal in the circadian output circuit. Taken together, the functional and anatomical data are consistent with hugin+ SEZ neurons receiving inputs from Dh44+ PI neurons.
hugin+ Neurons Are Circadian Output Neurons with Descending Projections into the VNC
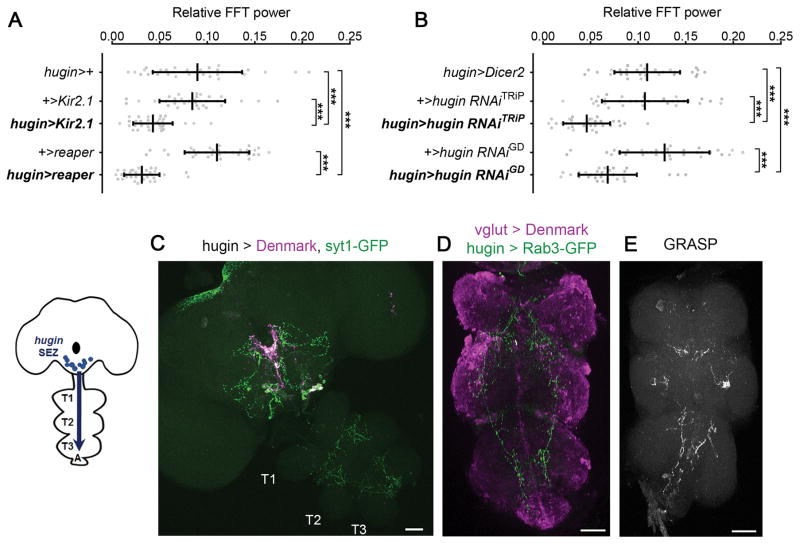
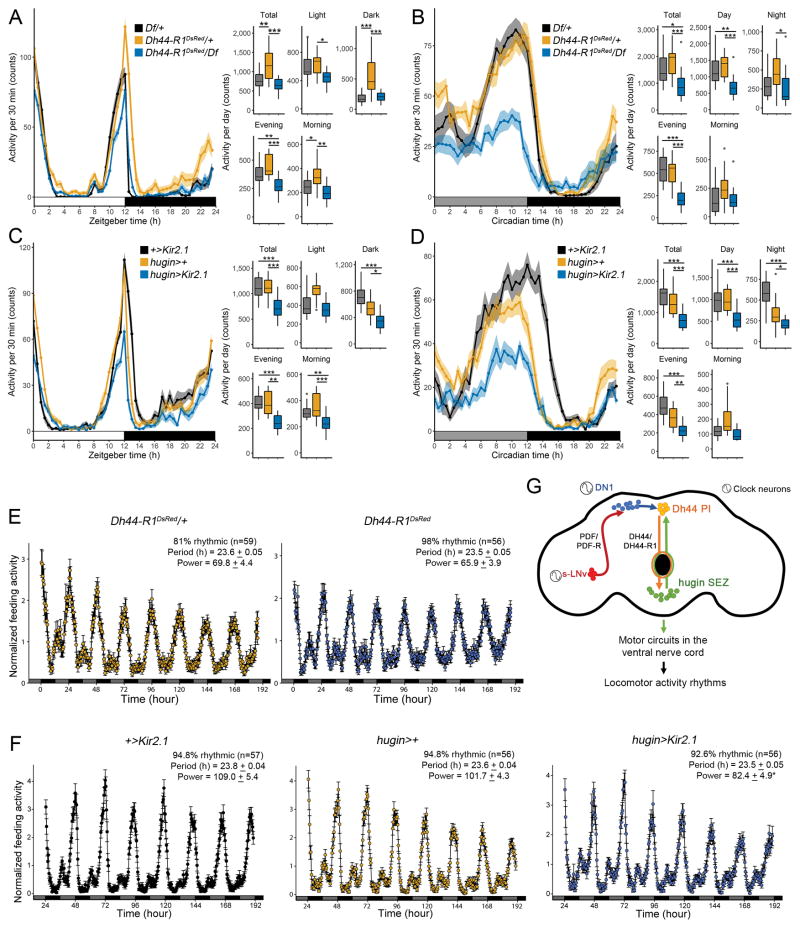
The findings reported above suggested that hugin+ neurons regulate rest:activity rhythms. To test this idea, we expressed Kir2.1, an inwardly rectifying potassium channel, with the hugin-GAL4 driver to hyperpolarize and silence hugin-expressing neurons [32]. Flies expressing hugin>Kir2.1 showed weaker rhythms (Figure 4A), and ablating hugin+ neurons using the pro-apoptotic gene reaper [33] resulted in an even stronger phenotype (Figure 4A).
Figure 4. hugin+ Neurons Are Circadian Output Neurons that Project to the Ventral Nerve Cord.
(A) Amplitude of rest:activity rhythms in control flies and flies with hugin+ neurons silenced (hugin>Kir2.1) or ablated (hugin>reaper) under DD conditions.
(B) Amplitude of rest:activity rhythms in control flies and flies with RNAi-mediated knockdown of hugin in hugin+ neurons.
(C) hugin-GAL4 expressing postsynaptic Denmark (magenta) and presynaptic syt1-GFP (green) markers in the central brain and VNC. The VNC is formed of first (T1), second (T2), and third (T3) thoracic and abdominal ganglia (A).
(D) hugin-LexA expressing presynaptic Rab3-GFP (green) and vglut-GAL4 expressing Denmark (magenta) markers in the VNC. (E) GRASP signal in the VNC of flies expressing vglut-GAL4>UAS-CD4-spGFP1-10; hugin-LexA>LexAop-CD4-spGFP11.
In (A) and (B), data are shown as mean ± SD. ***p < 0.001 by Sidak’s test following one-way ANOVA. Scale bars, 50 μm (C–E). See also Figure S4 and Tables S1 and S2.
Next, we tested whether Hugin neuropeptide is the signal from hugin+ neurons that controls behavioral rhythms by knocking down hugin expression and assaying behavior. To test for efficacy of knockdown with two different RNAi transgenes against hugin, we drove their expression pan-neuronally with elav-GAL4, and saw >90% reduction in hugin mRNA levels (Figure S4A). Expression of the RNAi transgenes in hugin+ neurons resulted in weaker rest:activity rhythms (Figure 4B). These data show that hugin+ neurons and Hugin neuropeptide modulate rest:activity rhythms.
We also examined the projections of hugin+ neurons to identify their targets. A subset of the hugin+ neurons are descending neurons [22], which have cell bodies in the central brain and project to the VNC, a region containing motor circuits responsible for locomotion [34]. We confirmed that hugin+ neurons in the central brain send axonal projections to the VNC using the presynaptic marker syt1-GFP (Figure 4C). Double labeling experiments revealed that hugin+ SEZ neurons are negative for vglut (vesicular glutamate transporter)-GAL4, a marker for motor neurons (data not shown) [35]. To determine whether hugin+ neurons contact vglut+ neurons in the VNC, we used the presynaptic marker Rab3-GFP and postsynaptic marker Denmark. hugin+ presynaptic terminals localize with vglut+ dendritic projections in thoracic segments T2 and T3 and the abdominal (A) segment of the VNC (Figure 4D). GRASP also revealed contacts between hugin+ and vglut+ neurons in the thoracic and abdominal ganglia (Figure 4E). We hypothesize that descending projections from the hugin+ neurons to the VNC signal to motor circuits.
Neuropeptide Release from hugin+ Neurons Is Clock Regulated
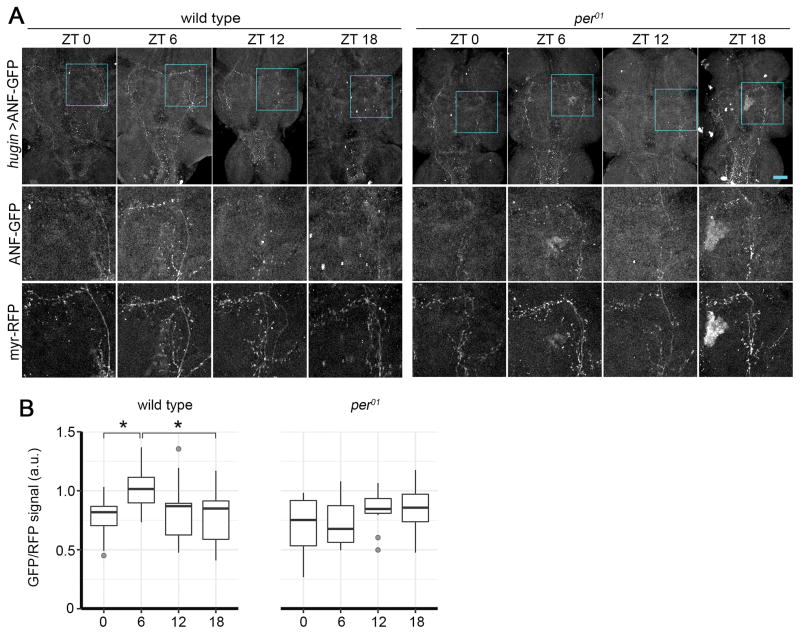
LNvs and DN1s show rhythmic electrical activity with peak spontaneous firing rates around the early morning [36–38]. Dh44+ circadian output neurons also show rhythms of intracellular Ca2+ [39], which is most likely indicative of rhythmic neural activity and peptide release [40]. To determine whether peptide release is rhythmic in hugin+ neurons, we used ANF-GFP, a transgenic neuropeptide reporter [41]. We expressed UAS-ANF-GFP and UAS-myr-RFP, used to normalize the ANF-GFP signal, in hugin+ neurons and detected the ANF-GFP signal in cell bodies, the projections to the PI, and the descending projections in the VNC. Because the ANF-GFP signal in the hugin+ projections to the VNC is most likely to reflect neuropeptide release that affects motor circuits, we measured ANF-GFP in these projections. We found that ANF-GFP was rhythmic in the descending projections, with ~33% reduction in levels from the peak at midday (zeitgeber time 6; ZT6) to the trough in the middle of the night (ZT18) (Figure 5). We also measured ANF-GFP levels in hugin+ descending projections in per01 mutants, which do not have a molecular clock, and found rhythms were lost, confirming that the rhythms of neuropeptide release from hugin+ neurons are clock controlled. However, mRNA levels of hugin do not appear to cycle (Figure S4B).
Figure 5. Neuropeptide Levels in Projections of hugin+ Neurons Are Regulated by the Circadian Clock.
(A) ANF-GFP signal in VNCs from wild-type or per01 flies expressing hugin>ANF-GFP. Scale bar, 50 μm. Close-ups of the boxed regions (top) are shown in the middle and bottom rows to highlight the ANF-GFP and corresponding myr-RFP (red fluorescent protein) signals, respectively.
(B) Tukey’s boxplots of ANF-GFP fluorescence levels in the entire VNC. n = 10–15 flies/time point and genotype. *p < 0.0359 by two-way ANOVA and Tukey’s test for comparison within genotype.
The DH44 PI-Hugin SEZ Circuit Controls Locomotor Rhythms without Affecting Feeding Rhythms
All the data described above assayed the strength of rhythms in DD, which is the paradigm typically used to assess internal clock function. However, clocks also modulate the daily distribution of activity, which is particularly evident in a light:dark (LD) cycle. In LD cycles, flies display morning and evening peaks of locomotor activity separated by an afternoon siesta, all of which are controlled by different clock neurons [4, 42, 43]. However, little to nothing is known about the output circuits controlling diurnal behavior. To determine the contribution of DH44 signaling to the timing of diurnal behavior, we analyzed behavior of Dh44-R1 and Dh44-R2 mutants under standard 12:12 LD conditions. Compared to heterozygous flies, Dh44-R1-deficient mutants (Dh44-R1DsRed/Df) had a reduced evening peak of locomotor activity (Figure 6A). However, Dh44-R2174 mutants displayed a normal pattern of activity in LD (Figures S5A and S5B). In DD, where the pattern typically consists of a single broad evening peak of activity, Dh44-R1DsRed/Df mutants showed a strong reduction of this peak (Figure 6B).
Figure 6. The DH44-Hugin Circuit Alters Locomotor Activity without Affecting Feeding.
(A and B) Locomotor activity profiles of Dh44-R1DsRed/Df mutants averaged over 3 days in LD (A) or DD (B).
(C and D) Locomotor activity profiles of hugin>Kir2.1 flies averaged over 3 days in LD (C) or DD (D).
(A–D) Traces (left) show activity counts/30 min (mean ± SEM). Tukey’s boxplots (right) summarize the distribution of activity counts per day during a total 24-hr day, day (ZT or CT 0–12), night (ZT or CT 12–24), evening (ZT or CT 9–13), and morning (ZT or CT 21–1). n = 15–16/genotype. CT, circadian time.
(E and F) Normalized feeding activity in Dh44-R1DsRed/+ and Dh44-R1DsRed flies (E) and hugin>Kir2.1 and genetic control flies (F) in DD conditions. Period and power data are summarized as mean ± SEM. (G) Model of a circadian output circuit for locomotor activity rhythms in Drosophila. The circuit extends from the master pacemaker sLNvs (red), through DN1 clock neurons (blue), and to Dh44+ PI neurons (orange). This circadian output circuit continues through hugin+ SEZ neurons (green) to the VNC.
*p < 0.05, **p < 0.01, ***p < 0.001 by one-way ANOVA and Tukey’s test. See also Figure S5 and Table S2.
Neuronal inactivation of hugin+ neurons with Kir2.1 expression attenuated the evening peak of activity in both LD and DD conditions (Figures 6C and 6D), recapitulating the phenotype of Dh44-R1-deficient mutants. We hypothesize that Dh44-R1 and hugin>Kir2.1 mutants have dampened clock output signals, which attenuates the evening peak in particular. Together, the data suggest that DH44-R1 acting in hugin+ neurons modulates circadian locomotor activity in LD and DD conditions.
The role of Hugin/NMU [22, 44] and DH44/CRF [45–47] in feeding-related behaviors raised the possibility that the DH44 PI-Hugin SEZ circuit affects locomotor activity rhythms indirectly by driving feeding. We performed continuous, long-term monitoring of fly feeding behavior using the fly liquid-food interaction counter (FLIC) system [48] to directly assess whether manipulations of the DH44 PI-Hugin SEZ circuit alter fly feeding rhythms. Dh44-R1DsRed mutants exhibited strong feeding rhythms that were indistinguishable from those of controls (Figures 6E and S5C). Feeding rhythms in hugin>Kir2.1 flies were also strong, although slightly reduced in strength compared to corresponding controls (Figures 6F and S5C), perhaps indicating that the hugin+ cells are functionally heterogeneous and the subset unresponsive to DH44 makes a minor contribution to the modulation of feeding behavior. Overall, these results show that the degradation of rest:activity rhythms in these flies was not secondary to alterations in feeding behavior. They also suggest that distinct output circuits mediate control of feeding and rest:activity rhythms.
DISCUSSION
The neural circuits that transmit information from clock neurons to motor outputs to control rest:activity rhythms are poorly understood. We showed previously that Dh44+ PI cells are circadian output neurons indirectly connected to sLNvs, the central pacemaker neurons [8]. Here we identified hugin+ neurons as downstream circadian neurons that modulate rest:activity rhythms. Our data suggest that information flows from the clock network to Dh44+ PI neurons, to hugin+ SEZ neurons, and finally to the VNC, which contains motor circuitry for locomotor activity (Figure 6G).
Although both Dh44-R1 and Dh44-R2 mutants showed some defects in their rest:activity rhythms, the amplitude of behavioral rhythms was significantly weaker in Dh44-R1 mutants than in Dh44-R2 mutants. In addition, the Dh44-R2 mutation did not modify the Dh44-R1 mutant phenotype, suggesting that Dh44-R1 is the predominant DH44 receptor that regulates rest:activity rhythms. Dh44-R1 may function both independently as well as together with Dh44-R2, which could explain the small circadian deficiency in Dh44-R2 mutants. To localize the neurons where Dh44-R1 functions to regulate rest:activity rhythms, we tested 168 GAL4 drivers and identified 15 that weaken rest:activity rhythms when used to drive RNAi targeted to Dh44-R1 (Figure 2A). Although no obvious area of expression was common to all GAL4 lines, several GAL4s target expression to the SEZ, specifically hugin+ neurons, suggesting that the SEZ is a major neuroanatomical region receiving DH44 signals. However, Dh44-R1 may be required in multiple groups of neurons for robust rest:activity rhythms, similar to how the collective network of clock neurons is required for sustaining molecular oscillations and behavioral rhythms [1, 2].
We also asked whether Dh44-R1 and hugin+ neurons regulate the output of morning and evening peaks of activity under LD conditions. Dh44-R1 mutants have a normal morning peak, suggesting that the timing signal from morning oscillators in sLNvs is propagated to motor outputs through alternative circuits. However, the evening peak of activity is reduced. An effect of DH44 on the evening peak of activity, which is the peak that persists in free-running conditions [4], is actually consistent with disrupted free-running rhythms in Dh44-R1 mutants, but it would require a link between Dh44+ cells and evening oscillators in dorsolateral neurons (LNds) [49]. This may occur through direct connections, because LNds project to the region of the PI [50]. Dh44+ cells may also receive evening signals from DN1s, which control the evening peak of activity in addition to the morning peak [51–53].
Several points about this study are worth noting. First, the pathway reported here does not necessarily function as a linear feedforward circuit. hugin+ SEZ neurons not only project to the VNC but may also project back to Dh44+ PI neurons. Indeed, GRASP revealed membrane contacts between hugin+ and Dh44+ neurons in both the PI and SEZ. Reciprocal connections between Dh44+ and hugin+ neurons may comprise a feedback circuit mechanism for propagating rhythmic signals in the output circuit. As discussed below, zebrafish orthologs of DH44 and Hugin are also linked in a circuit that regulates arousal but, in that case, Hugin acts upstream of DH44 [54]. It is possible hugin+ neurons signal to Dh44+ neurons, as they do in the Drosophila larval brain [55]. Second, although we describe one discrete circuit, this circuit almost certainly integrates with other circuits involved in circadian rhythms of locomotor activity. Leucokinin (LK)-expressing neurons also regulate rest:activity rhythms, and the LK receptor is expressed in Dh44+ neurons [39, 56]. Thus, LK and DH44 may comprise another interconnected circuit for rest:activity rhythms. Third, it is clear that circadian output circuits are modulatory rather than strictly essential, because loss of one neuropeptide (i.e., DH44, Hugin, or LK) does not cause complete arrhythmicity but reduces the amplitude of rest:activity rhythms. Even PDF, a neuropeptide expressed in the LNvs, is not completely essential. Consistent with previous reports of rhythms in the absence of PDF signaling [16, 17], we found that 54.3% of the Pdf-null flies are still rhythmic. However, behavioral rhythms are weaker in Pdf mutants than in Dh44-R1 and hugin loss-of-function mutants.
Hugin is an ortholog of mammalian NMU [9]. We find that Drosophila Hugin regulates circadian rhythms of locomotor activity, in particular by promoting activity at specific times of day, which is consistent with behavioral effects of NMU-related peptides in vertebrates. Although not associated with changes in rhythms, nmu overexpression in zebrafish larvae promotes hyperactivity and inhibits sleep during both the day and night [54]. In addition, consistent with our fly data, nmu mutant larval and adult zebrafish are less active during the daytime [54]. However, Hugin/NMU may even have a conserved role in circadian rhythms, because NMU injections into the rat brain can shift the phase of locomotor activity rhythms [57]. Moreover, cells expressing a different neuromedin, neuromedin S, are important for rest:activity rhythms controlled by the mammalian suprachiasmatic nucleus, although the peptide itself does not appear to be relevant [58].
In addition to the circadian clock, locomotor activity is regulated by various internal states, such as hunger and arousal, as well as environmental cues, such as light and temperature. These other states and inputs could modify locomotor activity through alternate circuits that access motor command centers in parallel to circadian output circuits. Alternatively, they could directly modulate circadian locomotor circuits. For example, the DH44 PI-Hugin SEZ circuit is located close to the esophagus in the brain and may be receptive to feeding signals. Indeed, Dh44+ PI neurons are proposed to function as a postingestive nutrient sensor [45], and the SEZ contains gustatory cells activated by tastants [59]. We addressed whether manipulations of Dh44-R1 or hugin+ SEZ neurons affect the flies’ overt feeding rhythms and found that these were largely unaltered, suggesting that effects of the DH44 PI-Hugin SEZ circuit on locomotor rhythms are not mediated by an increase in hunger or food-seeking behavior. Thus, although it is possible that the drive to eat contributes to rest:activity rhythms, the cellular basis of locomotor rhythms can be distinguished from that of feeding rhythms. Indeed, locomotor activity rhythms are also more robust than feeding rhythms [60], most likely because activity restricted to specific times of day serves many functions other than feeding.
STAR+METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-GFP | Thermo Fisher Scientific | Cat# A-11122; RRID: AB_221569 |
| Mouse monoclonal anti-bruchpilot | A. Hofbauer, University of Regensburg; Bavaria; Germany | Cat# nc82; RRID: AB_2314865 |
| Rat monoclonal anti-RFP | ChromoTek | Cat# 5f8-100, RRID: AB_2336064 |
| FITC-AffiniPure Donkey polyclonal anti-Rabbit IgG (H+L) | Jackson ImmunoResearch Labs | Cat# 711-095-152, RRID: AB_2315776 |
| Cy3-AffiniPure Donkey Anti-Rat IgG (H+L) | Jackson ImmunoResearch Labs | Cat# 712-165-153, RRID: AB_2340667 |
| Cy5-AffiniPure Donkey Anti-Mouse IgG (H+L) | Jackson ImmunoResearch Labs | Cat# 715-175-151, RRID: AB_2340820 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: w1118 | Bloomington | RRID: BDSC_5905 |
| D. melanogaster: Df(2R)BSC305 | Bloomington | RRID: BDSC_23688 |
| D. melanogaster: Df(2R)BSC700, P+PBac{w[+mC] = XP3.RB5}BSC700 | Bloomington | RRID: BDSC_26552 |
| D. melanogaster: Dh44-R1 RNAi TRiP.JF03208 (attP2) | Bloomington | RRID: BDSC_28780 |
| D. melanogaster: Dh44-R2 RNAi TRiP.JF03289 (attP2) | Bloomington | RRID: BDSC_29610 |
| D. melanogaster: elav[C155]; UAS-Dicer2 | Bloomington | RRID: BDSC_25750 |
| D. melanogaster: UAS-Dicer2 | Bloomington | RRID: BDSC_24650 |
| D. melanogaster: UAS-Dicer2 | Bloomington | RRID: BDSC_24651 |
| D. melanogaster: Dh44-R1 R21A07-GAL4 (attP2) | Bloomington | RRID: BDSC_49855 |
| D. melanogaster: hugin-GAL4 | Bloomington | RRID: BDSC_58769 |
| D. melanogaster: hugin-LexA (GMR61H10-lexA (attP40)) | Bloomington | RRID: BDSC_52715 |
| D. melanogaster: hugin RNAi TRiP.JF03122 (attP2) | Bloomington | RRID: BDSC_28705 |
| D. melanogaster: UAS-reaper | Bloomington | RRID: BDSC_5823 |
| D. melanogaster: UAS-nls.GFP | Bloomington | RRID: BDSC_7032 |
| D. melanogaster: UAS-Denmark, UAS-syt.eGFP | Bloomington | RRID: BDSC_33064 |
| D. melanogaster: UAS-Denmark | Bloomington | RRID: BDSC_33062 |
| D. melanogaster: 13XLexAop2-GFP-Rab3 (VK00027) | Bloomington | RRID: BDSC_52239 |
| D. melanogaster: 13XLexAop2-IVS-GCaMP6m-p10 (attp1) | Bloomington | RRID: BDSC_44275 |
| D. melanogaster: vglut-GAL4 (ok371) | Bloomington | RRID: BDSC_26160 |
| D. melanogaster: Dh44-GAL4 (attp2) | Vienna Drosophila Resource Center | VDRC: 207474 Flybase: FBti0169412 |
| D. melanogaster: Dh44-R1 RNAi kk | Vienna Drosophila Resource Center | VDRC: 110708 Flybase: FBtp0067029 |
| D. melanogaster: Hugin RNAi GD | Vienna Drosophila Resource Center | VDRC: 26766 Flybase: FBtp0025825 |
| D. melanogaster: Dh44-R2 RNAi NIG | NIG-Fly (National Institute of Genetics Mishima, Japan) | NIG: 12370R-4 Flybase: FBtp0074111 |
| D. melanogaster: per01 | [61] | Flybase: FBal0013649 |
| D. melanogaster: pdf01 | [3] | Flybase: FBal0102946 |
| D. melanogaster: pdfrhan5304 | [14] | Flybase: FBal0218040 |
| D. melanogaster: Dh44-R1-GAL4 | [18] | Flybase: FBtp0112068 |
| D. melanogaster: neurexin-GRASP (LexAop-CD4-spGFP11; UAS-nrx-spGFP1-10) | [28] | N/A |
| D. melanogaster: CD4-GRASP (LexAop-CD4-spGFP11; UAS-CD4-spGFP1-10) | [62] | N/A |
| D. melanogaster: UAS-P2X2 | [30] | Flybase: FBtp0021869 |
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| D. melanogaster: UAS-Kir2.1 | [32] | Flybase: FBtp0014166 |
| D. melanogaster: UAS-TrpA1 | [19] | Flybase: FBtp0040248 |
| D. melanogaster: UAS-ANF-GFP | [41] | Flybase: FBtp0016739 |
| D. melanogaster: vasa-Cas9 | Bloomington | RRID: BDSC_51323 |
| D. melanogaster: Dh44-R1DsRed | This paper | N/A |
| D. melanogaster: Dh44-R2174 | This paper | N/A |
| D. melanogaster: Dh44-LexA (attp40) | This paper | N/A |
| D. melanogaster: UAS-t-Dh44 (attp40) | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pBPLexA::p65Uw | [63] | Addgene plasmid# 26231 |
| Plasmid: pCFD4-U6:1_U6:3tandemgRNAs | [64] | Addgene plasmid# 49411 |
| Plasmid: pDsRed-attP | Melissa Harrison & Kate O’Connor-Giles & Jill Wildonger | Addgene plasmid# 51019 |
| Plasmid: pJFRC7-20XUAS-IVS | [63] | N/A |
| Software and Algorithms | ||
| Fiji | [65] | https://fiji.sc/ |
| ClockLab | Actimetrics | http://actimetrics.com/products/clocklab/ |
| Graphpad Prism (v 7.03) | Graphpad Software | https://www.graphpad.com/ |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Amita Sehgal (amita@mail.med.upenn.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila lines
Flies were maintained on cornmeal-molasses medium at 25°C. The w1118 iso31 strain was used as wild-type. When tested as controls, UAS and GAL4 fly lines were tested as heterozygotes after crossing to iso31. Most of the GAL4 lines used in the screen were selected for their restricted expression in the brain from the Janelia Fly Light collection [20] at the Bloomington Drosophila Stock Center (BDSC) and the Vienna Tiles collection [21] at the Vienna Drosophila Resource Center (VDRC). See Table S2 for a list of the complete genotype for the animals used in each experiment.
METHOD DETAILS
Generating Dh44-LexA driver
Dh44-LexA was generating using the same ~2.2 kb Dh44 enhancer fragment (chr3R:9639799-9641976 from dm6) in Dh44-GAL4 (VT039046 from VDRC). The Dh44 fragment was directionally cloned into a pBPLexA::p65Uw plasmid (Addgene 26231) between two attR sites using the Gateway TOPO cloning kit (Thermo Fisher Scientific). Flies were generated by site-specific PhiC31 integration at an attP40 site [63]. Despite using the same enhancer fragment as Dh44-GAL4, we observed that Dh44-LexA was expressed in other neurons, in addition to the six Dh44+ pars intercerebralis neurons. Therefore, Dh44-LexA was only used in experiments where anatomical analysis could exclude the spurious expression pattern. Transgenic fly injections were done by Rainbow Transgenic Flies (Camarillo, CA).
Generating Dh44-R1 and Dh44-R2 mutants
Dh44-R1DsRed and Dh44-R2174 mutants were generated with the CRISPR/CAS9 system. Guide RNA sequences to target Dh44-R1 and Dh44-R2 were determined using a target finder (http://flycrispr.molbio.wisc.edu/tools). Guide RNAs were cloned into the pCFD4 plasmid [64]. For the Dh44-R1 mutation, a homology directed repair template (HDR) was also used. 5′ and 3′ homology arms spanning 1 Kb upstream and downstream of the desired deletion were cloned into the pHD-DsRed-attP plasmid [66]. Guide RNAs and HDR template were injected into vasa-Cas9 flies at Rainbow Transgenic Flies. Mutations were identified with PCR screening and sequencing. To PCR identify mutations at the CRISPR target site in Dh44-R2, two forward primers and one reverse primer were used. One forward primer primes outside the CRISPR target site, referred as Primer outside (Po), and another forward primer overlaps the CRISPR target site, referred as Primer indel (Pi). Thus, Po amplifies from both wild-type and mutant alleles. Pi can only amplify from the wild-type allele, and any mutation will disrupt the binding of Pi. To PCR verify HDR insertion at Dh44-R1, one primer was targeted against a genomic region outside of the HDR template and the other primer was targeted against a region within the HDR template. Thus, a PCR product can only be produced when the HDR template has been integrated into the genomic Dh44-R1 locus. See also Table S3 for guide RNA and primer sequences.
Generating UAS-t-Dh44
t-Dh44 cDNA was chemically synthesized using optimal Drosophila codon usage and with an optimal Drosophila Kozak translation initiation site upstream of the start methionine (CAAA) as described in [23]. t-Dh44 cDNA and encoded peptide sequence are as follows:
cDNA: GAATT CCAAA ATGTC CGCCC TGCTC ATCTT GGCTT TGGTC GGTGC TGCAG TTGCC AACAA ACCCT CCCTG AGCAT CGTGA ATCCG CTAGA TGTCC TGCGT CAACG CCTGC TACTT GAGAT AGCCC GTCGC CAGAT GAAGG AGAAT AGCCG ACAGG TGGAG CTGAA TCGAG CCATC CTGAA GAACG TGGGC AACGA GCAGA AGCTC ATCAG TGAGG AGGAT CTGGG AAACG GAGCT GGCTT TGCTA CTCCA GTGAC ACTAG CCCTT GTGCC TGCAC TGTTG GCAAC CTTCT GGTCG CTCCT GTAAT CTAGA
Peptide: MSALLILALVGAAVANKPSLSIVNPLDVLRQRLLLEIARRQMKENSRQVELNRAILKNVGNEQKLISEEDLGNGAGFATPVTL ALVPALLATFWSLL
The cDNA was cloned into pJFRC7-20XUAS-IVS plasmids using NotI and NheI and cloned vectors were injected into fly strains carrying the attP40 landing site to obtain transgenic flies [63].
Behavior experiment: circadian rest:activity rhythm
Rest:activity rhythm assays were performed with the Drosophila Activity Monitoring System (Trikinetics, Waltham MA) as described previously [8, 67]. Flies were entrained to a 12 hr light: 12 hr dark (LD) cycle for > 3 days at 25°C. ~7 d old male flies were individually placed into glass tubes with 5% sucrose/2% agar food and monitored in constant darkness (DD) for 7 d at 25°C. For TrpA1 experiments, flies were raised at 18°C. ~7 d old male flies were entrained to an LD cycle for 3 days at 21°C, then transferred to DD for 5 days at 21°C, followed by 5 days DD at 28°C. The GAL4 screen was initially performed with 8–16 flies. All other behavioral experiments were performed at least 2 independent times with at least 16 flies/genotype each.
Immunohistochemistry, GRASP, and microscopy
Fly brains from ~4–7 d old males were dissected in phosphate-buffered saline with 0.1% Triton-X (PBST) and fixed in 4% formaldehyde for 20 min at room temperature. Brains were rinsed 3 × 10 min with PBST, blocked for 60 min in 5% Normal Donkey Serum in PBST (NDST), and incubated in primary antibody diluted in NDST for > 16 hr at 4°C. Brains were rinsed 3 × 10 min in PBST, incubated 2 hr in secondary antibody diluted in NDST, rinsed 3 × 10 min in PBST, and mounted with Vectashield (Vector Laboratories). Primary antibodies used are rabbit anti-GFP at 2μg/mL (Thermo Fisher Scientific A-11122), rat anti-RFP at 1μg/mL (ChromoTek 5F8), and mouse anti-brp at 1:100 (Developmental Studies Hybridoma Bank nc82). Secondary antibodies used are FITC donkey anti-rabbit (Jackson ImmunoResearch 711-095-152), Cy3 donkey anti-rat (712-165-153), and Cy5 donkey anti-mouse (715-175-151) at 1:500. For GRASP experiments, endogenous signal without antibody labeling was imaged. Eight-bit images were acquired using a Leica TCS SP5 laser scanning confocal microscope with a 40x/1.3 NA or 20x/0.7 NA objective and a 1-μm z-step size. Maximum intensity z-projection images were generated in Fiji, a distribution of ImageJ software [65].
P2X2 activation and calcium imaging
Adult male flies ~7–9 d old were anesthetized on ice and dissected in hemolymph-like saline (HL3) consisting of (in mM): 70 NaCl, 5 KCl, 1.5 CaCl2, 20 MgCl2, 10 NaHCO3, 5 trehalose, 115 sucrose, 5 HEPES, pH 7.1 [68]. Imaging experiments were performed using a naked brain preparation in a small bath of HL3 in a perfusion chamber (AutoMate Scientific, Berkeley CA). The brain was stabilized under nylon fibers attached to a platinum wire frame. Solutions were perfused over the brain at a rate of ~5 mL/min with a gravity-fed ValveLink perfusion system (Automate Scientific). After 1 min of baseline GCaMP6s imaging, ATP was delivered to the chamber by switching perfusion flow from the channel containing HL3 to another channel containing 5 mM ATP (Sigma–Aldrich, St Louis, MO) in HL3, pH 7.1. ATP was perfused for 1 min. GCaMP6 calcium imaging was performed on a Leica TCS SP5 confocal microscope. Twelve-bit images were acquired with a 40 × /0.8 water immersion objective at 256 × 256cpixel resolution. Z stacks were acquired every 5 or 10 s.
ANF-GFP
Adult males were entrained to a LD cycle. For each time point, ventral nerve cords were dissected in PBST and fixed in 4% PFA/PBS for 20 m at room temperature. Tissues were washed 3 times in PBST, mounted in Vectashield media, and imaged on a Leica TCS SP5 confocal microscope using identical laser power and scan settings for all samples. Eight-bit images were acquired using a Leica TCS SP5 laser scanning confocal microscope with a 20x/0.7 NA objective and a 1-μm z-step size.
Behavior experiment: locomotor activity
The number of beam crossings (activity counts) per 30 min was measured using the Drosophila Activity Monitoring System. ~7 d old individual male flies were monitored for 3 d in LD and then 3 d in DD. Locomotor activity analysis was performed 2 or 3 independent times with 16 flies per genotype with similar results.
Feeding behavior experiment: FLIC
Feeding rhythm analysis was performed using the Fly Liquid-food Interaction Counter (FLIC) [48]. Liquid food for the Dh44-R1DsRed experiments was prepared as a 10% sucrose (w/v) solution. Liquid food for the hugin > Kir2.1 experiments was a 10% sucrose solution plus 45 mg/L MgCl2 · 6H2O as an additional source of ions for a more robust signal. ~2–3 d old male flies were entrained in a LD cycle for > 3 d at 25ºC, then transferred to DD 25ºC for 8 d. Feeding events, measured as constant food contact for a minimum of 1 s, were monitored for 8 d in DD.
Quantitative reverse transcription PCR (qPCR)
Total RNA was extracted from 3–7 d old male flies (30 heads or 5 whole bodies) using TRIzol reagent (Thermo Fisher Scientific). RNA was reverse transcribed to generate cDNA using a High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific). qPCR was performed on a ViiA 7 Real-Time PCR System (Applied Biosystems) using SYBR Green PCR master mix (Thermo Fisher Scientific). Primers (5′ to 3′) for qPCR used in the study are: actin-F: GCGCGGTTACTCTTTCACCA; actin-R: ATGTCACGGACGATTT CACG; Dh44-F: GCAGGCAAATGAAGGAGAAC; Dh44-R: CCACGTTCTTCAGGATGG; Dh44-R1-F: CAGCACCCCCGAAAAGTACG; Dh44-R1-R: ATTAGCACCGCACAGACAGG; Dh44-R2-F: CCGGAACAGGGTATCAGTCG; Dh44-R2-R: AGAAGCCCTGCGTGCT TATG; hugin-F: ATGTGTGGTCCTAGTTATTGCAC; hugin-R: TCCCAAATCCAGTTTGCTCGT. Because the region targeted by the Dh44-R1 primers above was deleted in the Dh44-R1DsRed mutant, the following primers were used to measure mRNA levels of Dh44-R1: Dh44-R1-CRISPR-F: CCTGATGAGGCAAGGACTCG and Dh44-R1-CRISPR-R: AGATCTGCGACACGGAAGTG.
QUANTIFICATION AND STATISTICAL ANALYSIS
The statistical details of experiments can be found in the figure legends. All statistical tests were performed in GraphPad Prism (version 7.03). Tukey’s boxplots were generated in R (version 3.3.1) using ggplot2 package. In the boxplots, the line inside the box indicates the median, and the bottom and top lines represent the 1st and 3rd quartiles (the 25th and 75th percentiles). The upper whisker extends to the highest value that is within 1.5 * IQR above the 3rd quartile, where IQR is the inter-quartile range (the distance between the 25th and 75th percentiles). The lower whisker extends to the lowest value within 1.5 * IQR below the 1st quartile. Data beyond the end of the whiskers are outliers and plotted as points.
Behavior experiment: circadian rest:activity rhythm
Circadian rhythms was analyzed with ClockLab software (Actimetrics, Wilmette IL). Period and rhythm strength were determined for each individual fly using activity data collected from days 2–7 of DD. Period length was determined using χ2 periodogram analysis, and relative power (or amplitude) of circadian rhythm was determined using fast Fourier transform (FFT). Fly activity was considered rhythmic if the χ2 periodogram showed a peak above the 95% confidence interval and the FFT value was > 0.01 [8]. Data from flies that survived the duration of the experiments were pooled and analyzed. Behavioral data were analyzed with one-way analysis of variance (ANOVA). Tukey’s test was used as the post hoc test in Figures 1B–1E. Sidak’s test was used as the post hoc test in all other experiments to compare means between the two control genotypes (flies containing GAL4 or UAS only) and experimental genotype (flies containing both GAL4 and UAS). Differences between groups were considered significant if p < 0.05 by the post hoc test. TrpA1 data were analyzed with two-way repeated-measures ANOVA followed by a Sidak’s test. Differences in FFT power between temperatures and within a genotype were considered significant if p < 0.05 by Sidak’s test.
Calcium imaging
Image processing and fluorescence intensity measurement was performed in Fiji. A summed intensity Z-projection at each time point was used for analysis. StackReg plugin for Fiji was used to correct for xy movements over time in the projected image [69]. Regions of interest (ROIs) were manually drawn to encompass individual GCaMP-positive cell bodies, and mean fluorescence intensities was measured from a ROI at each time point. For each individual cell, fluorescence traces over time were normalized using this equation: ΔF/F = (Fn–F0)/F0, where Fn is the fluorescence intensity recorded at time point n, and F0 is the average fluorescence value during the 30 s-baseline preceding ATP application. Maximum ΔF/F was calculated by subtracting the average ΔF/F in the 30 s preceding ATP delivery from the largest ΔF/F value during the 60 s of ATP application. Brains with cells that have unstable baselines were discarded from quantification. The sample sizes, including the total number of cell bodies and number of brains, quantified are indicated in legend. We used two-tailed Mann-Whitney U test (for 2 group comparison) or Kruskal-Wallis test followed by Dunn’s multiple comparison test (for 3 group comparison) to compare differences in maximum ΔF/F between groups. A responding cell was defined as a cell with a maximum ΔF/F greater than 2*SD(ΔF/F of the negative control group). The onset of response for a cell was defined as the time where ΔF/F cross a threshold corresponding to 2*SD(ΔF/F during the 30 s baseline preceding ATP).
ANF-GFP experiments
FIJI software was used to measure fluorescent signal in axon terminal. Background subtraction was performed using the ‘‘rolling ball’’ method, then a max intensity Z-projection was generated. To create a selection mask of the axons, a 1.5 pixel range Gaussian blur was applied to a Z projected image of the myr-RFP signal, and the threshold was adjusted to select for the brightest myr-RFP signal. Fluorescent artifacts, such as autofluorescent puncta in the T3 and abdominal segments, were removed from the mask and not measured. The mask was transferred to the max Z-projected images and used to measure the mean pixel intensity of the ANF-GFP and myr-RFP signals. We also took a background signal for each ventral nerve cord. Normalized GFP/RFP signal was determined as (mean ANFGFP - mean background GFP) ÷ (mean myrRFP - mean background RFP) for each ventral nerve cord. ANF-GFP data were analyzed with two-way ANOVA. After determining the interaction effect between time and genotype variables was significant (p < 0.05), Tukey’s post hoc test was used to compare the means between time points within a genotype.
Behavior experiment: locomotor activity
Each fly’s 24-h activity profile was determined from the average of 3 d of data. Locomotor activity profiles for each genotype were then generated from the average of 15–16 flies’ activity profiles. We defined light or day activity as cumulative activity counts occurring between ZT or CT 0-11.5 (inclusive of start and end times), dark or night activity between ZT/CT 12-23.5, evening activity between ZT/CT 9.5–12.5, and morning activity between ZT/CT 21.5–23.5 and 0–0.5. Statistical tests were done with one-way ANOVA followed by a Tukey post hoc test. Differences between groups were considered significant if p < 0.05 by the Tukey test. Tukey’s boxplots were generated in R.
Feeding behavior
Period and rhythm strength of feeding behavior were determined from feeding events during days 2–7 of darkness (DD) using ClockLab software. Only flies that survived the duration of the experiment were included in the data analysis. Period length was determined using χ2 periodogram analysis, and ~24 hr rhythm strength was determined by subtracting the corresponding p = 0.01 χ2 significance value from the amplitude of the maximum period. Flies were categorized as rhythmic (power > 10) or arrhythmic (power < 10). Normalized feeding activity was calculated within each fly for comparison across flies and experiments. Feeding activity of a fly for a given 30 min period was divided by the average behavioral count of 30 min over the duration of the experiment. Plots of normalized feeding activity begin at day 2 of the experiment after flies have acclimated to the FLIC monitor enclosure.
Quantitative reverse transcription PCR (qPCR)
Two-tailed Welch’s t test was used to compare differences in gene expression between experimental and control groups. We used one-way ANOVA test and JTK_CYCLE algorithm (version 3) [70] to determine if there was cycling in gene expression.
Supplementary Material
Highlights.
DH44-R1 functions in hugin+ neurons to regulate rest:activity rhythms
A neuromedin U ortholog, Hugin, is a circadian output molecule
An LNv→DN1→DH44 PI→Hugin SEZ→VNC circuit links the clock to motor output
The DH44 PI-Hugin SEZ circuit regulates locomotor activity, but not feeding rhythms
Acknowledgments
We thank Drs. Monica Dus, Paula Haynes, Young-Joon Kim, and Kyunghee Koh for sharing fly lines. Stocks from the Bloomington Drosophila Stock Center (NIH P40OD018537) and Vienna Drosophila Resource Center were used in this study. The work was supported by NIH R37NS048471 (to A.S.). A.N.K. was supported in part by NIH grants T32GM008216 and F31NS100395.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, A.N.K., A.F.B., D.J.C., and A.S.; Methodology, A.N.K., A.F.B., A.E.S., A.P.D., D.J.C., and A.S.; Investigation, A.N.K., A.E.S., A.P.D., and D.J.C.; Formal Analysis, A.N.K., A.E.S., and A.P.D.; Resources, A.F.B., D.S., and M.N.N.; Writing – Original Draft, A.N.K. and A.S.; Writing – Review & Editing, A.N.K., A.F.B., A.E.S., A.P.D., D.S., M.N.N., D.J.C., and A.S.; Visualization, A.N.K., A.E.S., and A.P.D.; Supervision, A.S.
Supplemental Information includes five figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.cub.2017.05.089.
References
- 1.Yao Z, Shafer OT. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science. 2014;343:1516–1520. doi: 10.1126/science.1251285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M. Drosophila free-running rhythms require intercellular communication. PLoS Biol. 2003;1:E13. doi: 10.1371/journal.pbio.0000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- 4.Grima B, Chelot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- 5.Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci. 2004;24:7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshii T, Wülbeck C, Sehadova H, Veleri S, Bichler D, Stanewsky R, Helfrich-Förster C. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila’s clock. J Neurosci. 2009;29:2597–2610. doi: 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Velasco B, Erclik T, Shy D, Sclafani J, Lipshitz H, McInnes R, Hartenstein V. Specification and development of the pars in-tercerebralis and pars lateralis, neuroendocrine command centers in the Drosophila brain. Dev Biol. 2007;302:309–323. doi: 10.1016/j.ydbio.2006.09.035. [DOI] [PubMed] [Google Scholar]
- 8.Cavanaugh DJ, Geratowski JD, Wooltorton JR, Spaethling JM, Hector CE, Zheng X, Johnson EC, Eberwine JH, Sehgal A. Identification of a circadian output circuit for rest:activity rhythms in Drosophila. Cell. 2014;157:689–701. doi: 10.1016/j.cell.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melcher C, Bader R, Walther S, Simakov O, Pankratz MJ. Neuromedin U and its putative Drosophila homolog hugin. PLoS Biol. 2006;4:e68. doi: 10.1371/journal.pbio.0040068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKellar CE. Motor control of fly feeding. J Neurogenet. 2016;30:101–111. doi: 10.1080/01677063.2016.1177047. [DOI] [PubMed] [Google Scholar]
- 11.Johnson EC, Bohn LM, Taghert PH. Drosophila CG8422 encodes a functional diuretic hormone receptor. J Exp Biol. 2004;207:743–748. doi: 10.1242/jeb.00818. [DOI] [PubMed] [Google Scholar]
- 12.Hector CE, Bretz CA, Zhao Y, Johnson EC. Functional differences between two CRF-related diuretic hormone receptors in Drosophila. J Exp Biol. 2009;212:3142–3147. doi: 10.1242/jeb.033175. [DOI] [PubMed] [Google Scholar]
- 13.Lear BC, Merrill CE, Lin JM, Schroeder A, Zhang L, Allada R. A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron. 2005;48:221–227. doi: 10.1016/j.neuron.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Hyun S, Lee Y, Hong ST, Bang S, Paik D, Kang J, Shin J, Lee J, Jeon K, Hwang S, et al. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron. 2005;48:267–278. doi: 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 15.Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 16.Wülbeck C, Grieshaber E, Helfrich-Förster C. Pigment-dispersing factor (PDF) has different effects on Drosophila’s circadian clocks in the accessory medulla and in the dorsal brain. J. Biol. Rhythms. 2008;23:409–424. doi: 10.1177/0748730408322699. [DOI] [PubMed] [Google Scholar]
- 17.Shafer OT, Taghert PH. RNA-interference knockdown of Drosophila pigment dispersing factor in neuronal subsets: the anatomical basis of a neuropeptide’s circadian functions. PLoS ONE. 2009;4:e8298. doi: 10.1371/journal.pone.0008298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee K-M, Daubnerová I, Isaac RE, Zhang C, Choi S, Chung J, Kim Y-J. A neuronal pathway that controls sperm ejection and storage in female Drosophila. Curr Biol. 2015;25:790–797. doi: 10.1016/j.cub.2015.01.050. [DOI] [PubMed] [Google Scholar]
- 19.Pulver SR, Pashkovski SL, Hornstein NJ, Garrity PA, Griffith LC. Temporal dynamics of neuronal activation by Channelrhodopsin-2 and TRPA1 determine behavioral output in Drosophila larvae. J Neurophysiol. 2009;101:3075–3088. doi: 10.1152/jn.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenett A, Rubin GM, Ngo T-TB, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kvon EZ, Kazmar T, Stampfel G, Yáñez-Cuna JO, Pagani M, Schernhuber K, Dickson BJ, Stark A. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature. 2014;512:91–95. doi: 10.1038/nature13395. [DOI] [PubMed] [Google Scholar]
- 22.Melcher C, Pankratz MJ. Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PLoS Biol. 2005;3:e305. doi: 10.1371/journal.pbio.0030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi C, Fortin J-P, McCarthy Ev, Oksman L, Kopin AS, Nitabach MN. Cellular dissection of circadian peptide signals with genetically encoded membrane-tethered ligands. Curr Biol. 2009;19:1167–1175. doi: 10.1016/j.cub.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi C, Cao G, Tanenhaus AK, McCarthy Ev, Jung M, Schleyer W, Shang Y, Rosbash M, Yin JCP, Nitabach MN. Autoreceptor control of peptide/neurotransmitter corelease from PDF neurons determines allocation of circadian activity in Drosophila. Cell Rep. 2012;2:332–344. doi: 10.1016/j.celrep.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang YQ, Rodesch CK, Broadie K. Living synaptic vesicle marker: synaptotagmin-GFP. Genesis. 2002;34:142–145. doi: 10.1002/gene.10144. [DOI] [PubMed] [Google Scholar]
- 26.Nicolaï LJJ, Ramaekers A, Raemaekers T, Drozdzecki A, Mauss AS, Yan J, Landgraf M, Annaert W, Hassan BA. Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proc. Natl. Acad. Sci. USA. 2010;107:20553–20558. doi: 10.1073/pnas.1010198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, Bargmann CI. GFP reconstitution across synaptic partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 28.Fan P, Manoli DS, Ahmed OM, Chen Y, Agarwal N, Kwong S, Cai AG, Neitz J, Renslo A, Baker BS, et al. Genetic and neural mechanisms that inhibit Drosophila from mating with other species. Cell. 2013;154:89–102. doi: 10.1016/j.cell.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shearin HK, Dvarishkis AR, Kozeluh CD, Stowers RS. Expansion of the Gateway MultiSite recombination cloning toolkit. PLoS ONE. 2013;8:e77724. doi: 10.1371/journal.pone.0077724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lima SQ, Miesenböck G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121:141–152. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 31.Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baines RA, Uhler JP, Thompson A, Sweeney ST, Bate M. Altered electrical properties in Drosophila neurons developing without synaptic transmission. J Neurosci. 2001;21:1523–1531. doi: 10.1523/JNEUROSCI.21-05-01523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White K, Tahaoglu E, Steller H. Cell killing by the Drosophila gene reaper. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- 34.Enriquez J, Venkatasubramanian L, Baek M, Peterson M, Aghayeva U, Mann RS. Specification of individual adult motor neuron morphologies by combinatorial transcription factor codes. Neuron. 2015;86:955–970. doi: 10.1016/j.neuron.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahr A, Aberle H. The expression pattern of the Drosophila vesicular glutamate transporter: a marker protein for motoneurons and glutamatergic centers in the brain. Gene Expr. Patterns. 2006;6:299–309. doi: 10.1016/j.modgep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 36.Sheeba V, Gu H, Sharma VK, O’Dowd DK, Holmes TC. Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophysiol. 2008;99:976–988. doi: 10.1152/jn.00930.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao G, Nitabach MN. Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neurosci. 2008;28:6493–6501. doi: 10.1523/JNEUROSCI.1503-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flourakis M, Kula-Eversole E, Hutchison AL, Han TH, Aranda K, Moose DL, White KP, Dinner AR, Lear BC, Ren D, et al. A conserved bicycle model for circadian clock control of membrane excitability. Cell. 2015;162:836–848. doi: 10.1016/j.cell.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cavey M, Collins B, Bertet C, Blau J. Circadian rhythms in neuronal activity propagate through output circuits. Nat Neurosci. 2016;19:587–595. doi: 10.1038/nn.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shakiryanova D, Tully A, Hewes RS, Deitcher DL, Levitan ES. Activity-dependent liberation of synaptic neuropeptide vesicles. Nat Neurosci. 2005;8:173–178. doi: 10.1038/nn1377. [DOI] [PubMed] [Google Scholar]
- 41.Rao S, Lang C, Levitan ES, Deitcher DL. Visualization of neuropeptide expression, transport, and exocytosis in Drosophila mela-nogaster. J Neurobiol. 2001;49:159–172. doi: 10.1002/neu.1072. [DOI] [PubMed] [Google Scholar]
- 42.Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- 43.Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- 44.Howard AD, Wang R, Pong SS, Mellin TN, Strack A, Guan XM, Zeng Z, Williams DL, Jr, Feighner SD, Nunes CN, et al. Identification of receptors for neuromedin U and its role in feeding. Nature. 2000;406:70–74. doi: 10.1038/35017610. [DOI] [PubMed] [Google Scholar]
- 45.Dus M, Lai JSY, Gunapala KM, Min S, Tayler TD, Hergarden AC, Geraud E, Joseph CM, Suh GSB. Nutrient sensor in the brain directs the action of the brain-gut axis in Drosophila. Neuron. 2015;87:139–151. doi: 10.1016/j.neuron.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spina M, Merlo-Pich E, Chan RK, Basso AM, Rivier J, Vale W, Koob GF. Appetite-suppressing effects of urocortin, a CRF-related neuropeptide. Science. 1996;273:1561–1564. doi: 10.1126/science.273.5281.1561. [DOI] [PubMed] [Google Scholar]
- 47.Stengel A, Tache Y. CRF and urocortin peptides as modulators of energy balance and feeding behavior during stress. Front Neurosci. 2014;8:52. doi: 10.3389/fnins.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ro J, Harvanek ZM, Pletcher SD. FLIC: high-throughput, continuous analysis of feeding behaviors in Drosophila. PLoS ONE. 2014;9:e101107. doi: 10.1371/journal.pone.0101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo F, Cerullo I, Chen X, Rosbash M. PDF neuron firing phase-shifts key circadian activity neurons in Drosophila. eLife. 2014;3:1–21. doi: 10.7554/eLife.02780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 51.Guo F, Yu J, Jung HJ, Abruzzi KC, Luo W, Griffith LC, Rosbash M. Circadian neuron feedback controls the Drosophila sleep–activity profile. Nature. 2016;536:292–297. doi: 10.1038/nature19097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang L, Chung BY, Lear BC, Kilman VL, Liu Y, Mahesh G, Meissner R-AA, Hardin PE, Allada R. DN1(p) circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr Biol. 2010;20:591–599. doi: 10.1016/j.cub.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Liu Y, Bilodeau-Wentworth D, Hardin PE, Emery P. Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr Biol. 2010;20:600–605. doi: 10.1016/j.cub.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiu CN, Rihel J, Lee DA, Singh C, Mosser EA, Chen S, Sapin V, Pham U, Engle J, Niles BJ, et al. A zebrafish genetic screen identifies neuromedin U as a regulator of sleep/wake states. Neuron. 2016;89:842–856. doi: 10.1016/j.neuron.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schlegel P, Texada MJ, Miroschnikow A, Schoofs A, Hückesfeld S, Peters M, Schneider-Mizell CM, Lacin H, Li F, Fetter RD, et al. Synaptic transmission parallels neuromodulation in a central food-intake circuit. eLife. 2016;5:462–465. doi: 10.7554/eLife.16799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cannell E, Dornan AJ, Halberg KA, Terhzaz S, Dow JAT, Davies SA. The corticotropin-releasing factor-like diuretic hormone 44 (DH44) and kinin neuropeptides modulate desiccation and starvation tolerance in Drosophila melanogaster. Peptides. 2016;80:96–107. doi: 10.1016/j.peptides.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakahara K, Hanada R, Murakami N, Teranishi H, Ohgusu H, Fukushima N, Moriyama M, Ida T, Kangawa K, Kojima M. The gut-brain peptide neuromedin U is involved in the mammalian circadian oscillator system. Biochem Biophys Res Commun. 2004;318:156–161. doi: 10.1016/j.bbrc.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 58.Lee IT, Chang AS, Manandhar M, Shan Y, Fan J, Izumo M, Ikeda Y, Motoike T, Dixon S, Seinfeld JE, et al. Neuromedin S-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron. 2015;85:1086–1102. doi: 10.1016/j.neuron.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harris DT, Kallman BR, Mullaney BC, Scott K. Representations of taste modality in the Drosophila brain. Neuron. 2015;86:1449–1460. doi: 10.1016/j.neuron.2015.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu K, Zheng X, Sehgal A. Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell Metab. 2008;8:289–300. doi: 10.1016/j.cmet.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gordon MD, Scott K. Motor control in a Drosophila taste circuit. Neuron. 2009;61:373–384. doi: 10.1016/j.neuron.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pfeiffer BD, Ngo TTB, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Port F, Chen HM, Lee T, Bullock SL. Optimized CRISPR/ Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA. 2014;111:E2967–E2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’Connor-Giles KM. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196:961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams JA, Su HS, Bernards A, Field J, Sehgal A. A circadian output in Drosophila mediated by neurofibromatosis-1 and Ras/MAPK. Science. 2001;293:2251–2256. doi: 10.1126/science.1063097. [DOI] [PubMed] [Google Scholar]
- 68.Yao Z, Macara AM, Lelito KR, Minosyan TY, Shafer OT. Analysis of functional neuronal connectivity in the Drosophila brain. J Neurophysiol. 2012;108:684–696. doi: 10.1152/jn.00110.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thevenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 70.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.