Significance
Despite the revolution in cancer therapy initiated by checkpoint inhibitors, durable clinical responses remain sporadic in many types of cancer, including ovarian cancer. Understanding which antigens are essentially presented by tumor cells and further able to be recognized by T cells provides a major step toward novel effective targeted immunotherapies. In this study, we comprehensively analyzed the immunopeptidomic landscape of ovarian carcinoma and compared it to variety of benign sources to identify antigens exclusively presented on tumor cells. With personalized therapies moving into the focus of clinical cancer therapy, we further present insights on how gene-expression analysis and immunohistochemistry can support antigen selection for individualized immunotherapy.
Keywords: immunopeptidome, ovarian cancer, immunotherapy, mucin 16, mesothelin
Abstract
Immunotherapies, particularly checkpoint inhibitors, have set off a revolution in cancer therapy by releasing the power of the immune system. However, only little is known about the antigens that are essentially presented on cancer cells, capable of exposing them to immune cells. Large-scale HLA ligandome analysis has enabled us to exhaustively characterize the immunopeptidomic landscape of epithelial ovarian cancers (EOCs). Additional comparative profiling with the immunopeptidome of a variety of benign sources has unveiled a multitude of ovarian cancer antigens (MUC16, MSLN, LGALS1, IDO1, KLK10) to be presented by HLA class I and class II molecules exclusively on ovarian cancer cells. Most strikingly, ligands derived from mucin 16 and mesothelin, a molecular axis of prognostic importance in EOC, are prominent in a majority of patients. Differential gene-expression analysis has allowed us to confirm the relevance of these targets for EOC and further provided important insights into the relationship between gene transcript levels and HLA ligand presentation.
Epithelial ovarian cancer (EOC) remains the leading cause of death from gynecologic malignancies and the fifth leading cause of cancer-related death in the Western world, causing an estimated 22,000 new diagnoses and 14,000 deaths in the United States in 2016 (1). Most patients (>70%) are diagnosed with stage III or IV disease caused by a lack of specific early symptoms. Despite progress in chemotherapy regimens and the approval of bevacizumab for first-line therapy, most patients relapse within months or years after initial treatment (2, 3). Maintenance therapy of recurrent disease has recently improved as a result of the approval of poly(ADP-ribose) polymerase inhibitors (4). However, the only available curative treatment option remains complete surgical tumor removal at an early nonmetastatic stage. Novel therapies, particularly immunotherapeutic approaches, are therefore urgently needed.
Over the last two decades, EOC has been recognized as a highly immunogenic tumor, based on diverse clinical findings (5). Showing frequent immune cell infiltration, EOC was among the first cancers with an established association of T cell infiltration and clinical prognosis (6). Within these infiltrating T cell populations, tumor-reactive (7) and antigen-specific T cells (8, 9) have been identified. In contrast, tumor-resident regulatory T cells are negatively correlated with clinical outcome (10). Furthermore, immune stimulatory cytokines have been shown to induce compelling tumor responses in individual patients (11).
The effectiveness of immunotherapeutic approaches for cancer therapy has been illustrated by the approval and success of immune checkpoint inhibitors, especially in melanoma and lung cancer (12–14). Moreover, a second wave of antigen-specific immunotherapies is on the rise, showing success in melanoma (15) or leukemias (16, 17). Personalized antigen-specific immunotherapy has curative potential (18) and stunning results have been presented for individual patients (19).
An array of different antigen-specific immunotherapies has been developed for EOC during the last decade. Most accordant clinical trials could show immune responses [i.e., the induction of T cell responses (both CD4 and CD8)], but reports of long-lasting clinical improvements remain rare (20, 21). A major problem constitutes the selection of suitable antigens. Virtually all clinical trials have so far relied on long-established tumor-associated antigens, such as Her2neu, WT1, NY-ESO-1, and p53. The major shortcoming of these antigens is that these have never been shown to be frequently presented on HLA molecules of ovarian carcinoma cells. Induced immune responses toward these antigens might therefore misguide immune cells and thus fail to attack tumor cells. Hence, an in-depth knowledge of the HLA-presented antigenic profile of ovarian tumors (referred to as the HLA ligandome or immunopeptidome) is critically needed.
In this study, we present one of the most comprehensive analyses of the immunopeptidome of any solid cancer and the most comprehensive analysis of EOC reported so far, to the best of our knowledge. Exhaustive immunopeptidome analysis of ovarian tumors and comparative profiling with the immunopeptidome of benign tissues revealed frequently and exclusively presented immunogenic EOC antigens as peerless targets for antigen-specific immunotherapeutic approaches.
Results
Ovarian Tumors Show Strong MHC Class I and Class II Expression.
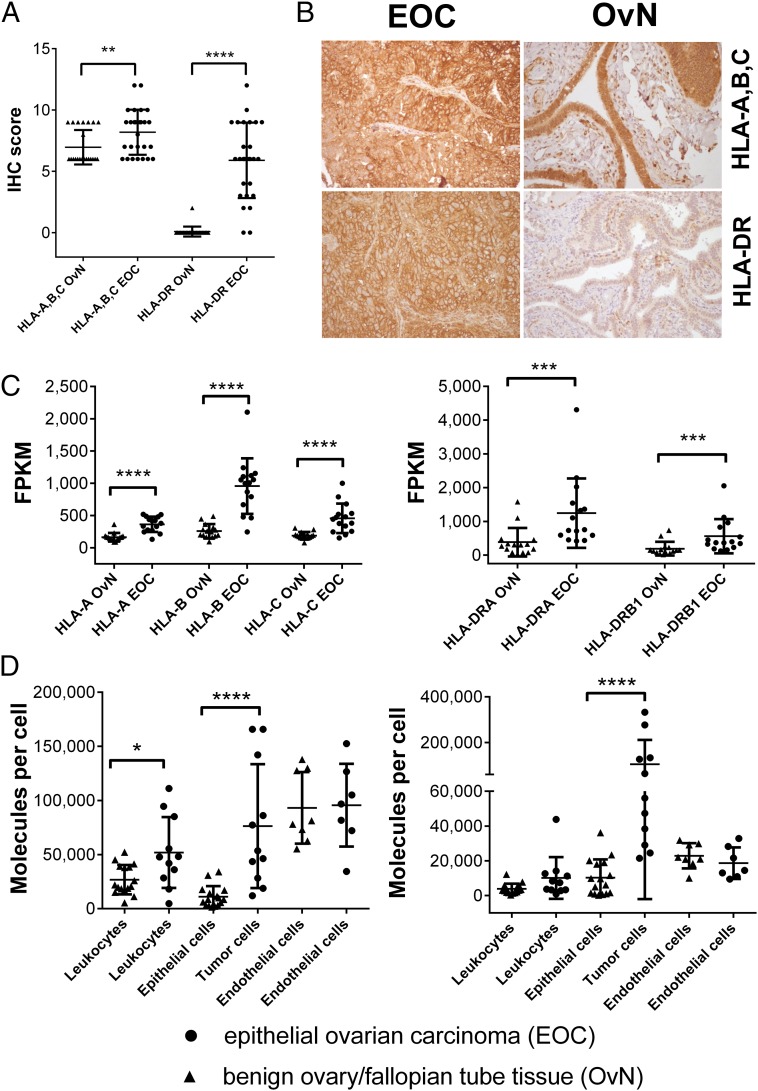
A major prerequisite for effective T cell-mediated killing is the expression of MHC molecules on the surface of tumor cells. We analyzed HLA-A, -B, -C, and HLA-DR expression on EOC and compared it to benign ovarian or fallopian tube tissue employing different techniques. Immunohistochemical (IHC) analyses of EOCs (n = 27) and fallopian tube tissues (n = 24) showed a significant overexpression of HLA-A, -B, and -C (P = 0.0057) on cancer tissue, and additionally revealed a strong yet heterogeneous expression of HLA-DR among EOC patients’ tumors, as opposed to fallopian tube tissue (P < 0.0001), which did not show staining for HLA-DR (Fig. 1 A and B). To confirm these results, we performed gene-expression analysis via RNA-Seq of 15 ovarian tumors, as well as 15 benign fallopian tube tissues (Fig. 1C). Although MHC class I and class II transcript levels (for HLA-DP and HLA-DQ; see Fig. S1) could be evidenced on both benign and cancerous tissue, gene-expression levels [fragments per kilobase of transcript per million mapped reads (FPKM values)] were significantly higher (all P < 0.01) within EOC throughout all MHC class I and class II alleles. Finally, we also quantified the number of HLA-A, -B, -C, and HLA-DR molecules by flow cytometry on different cell subsets of ovarian tumors (n = 11; n = 7 for endothelial cells) as well as benign tissues from ovary and fallopian tube (n = 16; n = 8 for endothelial cells) obtained by enzymatic dissociation. Our analysis aimed at the separate quantification of cell-type–specific HLA expression for leukocytes (CD45+), tumor/epithelial cells (EpCAM+) and endothelial cells (CD31+; the latter only in a subset of eight benign ovary/fallopian tube and seven EOC tissues) (for the complete gating strategy, see Fig. S2). The median number of HLA molecules per cell was heterogeneous both among different cell types and individual patients, ranging from ∼5,000–150,000 HLA class I and ∼500–330,000 HLA-DR molecules (Fig. 1D). The number of HLA-A, -B, and -C molecules was significantly higher on leukocytes (P = 0.03) isolated from tumor vs. benign tissue, potentially indicating an ongoing inflammatory reaction within the tumor. Differences in HLA class I expression were also visible when comparing tumor cells with epithelial cells derived from benign tissue. HLA class I molecule expression was significantly higher on tumor cells (∼75,000 molecules per cell; P < 0.0001) but remained in the range of other stromal cells, such as endothelial cells (∼95,000 molecules per cell). Furthermore, we evidenced a strong (∼105,000 molecules per cell) to some extent extraordinarily high (>300,000 molecules per cell) expression of HLA-DR on cancer cells, whereas benign epithelial cells were virtually negative for HLA-DR (P < 0.0001). Altogether, we could observe an increased MHC class I and class II expression within EOC.
Fig. 1.
EOCs show an increased MHC class I and II expression. (A) IHC assessment of HLA-A, -B, -C (Ab clone H2A) and HLA-DR (Ab clone L243) expression on EOC (n = 27), as well as fallopian tube samples OvN (n = 24). (B) Representative photomicorgraphs of EOC and benign fallopian tubes after IHC stainings at 200× magnification. (C) Gene-expression analysis of HLA class I and HLA-DR genes. RNA-Seq was used to characterize gene expression, determined as FPKM values. Expression levels for HLA-DP/HLA-DQ genes can be found in Fig. S1. (D) HLA class I (Left, Ab clone W6/32) and HLA-DR (Right, Ab clone L243) expression on different cell types within EOC and benign ovarian/fallopian tissue after enzymatic dissociation. Individual cell populations were characterized by distinct cell surface markers (leukocyte compartments: CD45+, tumor cells/epithelial cell compartments: CD45−EpCAM+, endothelial cell compartments: CD45−CD31+). Quantification of cell-type–specific surface expression was performed using a bead-based standard (QIFIKIT) (see SI Methods). Each data point represents the mean of triplicate experiments performed for each sample. For an overview on the gating strategy and the outlined process, see Fig. S2. Raw values for median fluorescence intensity and calculated copy numbers per cell can be found in Dataset S2. Throughout the figure, nonparametric Mann–Whitney test was used to test for significance (*P < 0.05; **P < 0.01, ***P < 0.001, ****P < 0.0001) due to rejected normality test (D’Agostino and Pearson). Data points represent individual samples unless stated otherwise. Horizontal lines indicate mean values ± SD.
HLA Ligandome Analysis and Comparative Profiling Reveal EOC-Specific Antigen Presentation.
To map the HLA ligand repertoire of EOC, we isolated HLA molecules from bulk tumor tissue and performed MS to characterize the HLA ligandome for a total of 42 EOCs (for patient characteristics and HLA typing, see Dataset S1). For MHC class I, we could identify 34,177 unique peptides (median 1,381 per sample) emanating from 10,677 different source proteins (median 1,334 per sample) reaching >95% of the estimated maximal attainable coverage in HLA ligand source proteins (Fig. S3A and Dataset S2).
Aiming to extract the most recurrent and specific HLA ligands for EOC from this vast catalog of data, we compared the HLA ligand source proteins to various histologically confirmed benign tissues from in-house datasets, including samples of liver (n = 15), colon (n = 20), ovary (n = 23), and kidney (n = 20), as well as peripheral blood mononuclear cells (PBMCs) from healthy donors (n = 30), all analyzed with the identical pipeline as used for EOCs. The total number of identified HLA class I ligand source proteins for respective benign sources varied between 3,667 and 7,233, achieving estimated maximal attainable coverages of 84–95%.
We used qualitative comparative analyses, as previously described (22, 23), to estimate the overlap in identification of HLA ligand source proteins from EOC and benign datasets. Differences in the depth of sample analyses (i.e., number of identified peptides per sample in EOC vs. benign tissues) were accounted for by ranking of the peptide identifications in EOC according to their abundance (i.e., area of the peptide precursor) and artificially truncating each dataset to the median number of peptide identifications of the respective benign tissues. The results from the comparative analyses of the EOC dataset with the individual benign tissue datasets, as well as the overlap in identified HLA ligand source proteins, is presented in Fig. 2A.
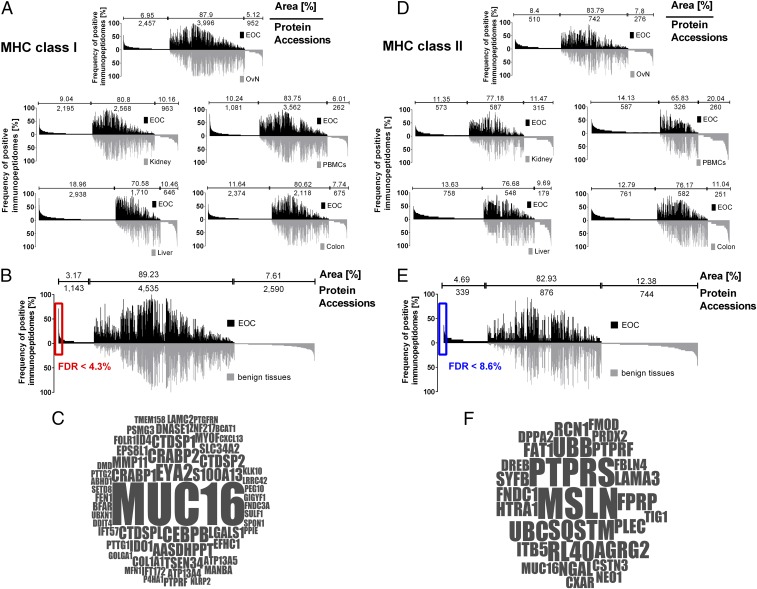
Fig. 2.
Immunopeptidome analysis of EOC and comparative profiling to various benign tissues. (A) Comparative profiling and overlap analyses of HLA class I ligand source proteins of EOC (n = 42) in comparison with benign ovary/fallopian tube tissue (n = 23), benign kidney tissue (n = 20), healthy donor PBMCs (n = 30), benign liver tissue (n = 15), and benign colorectal tissue (n = 20). Profiling is based on the frequency of positive immunopeptidomes (i.e., HLA-restricted representation of ligand source proteins within the different cohorts), as indicated on the y axis. The number of ligand source proteins that are unique to either EOC or benign tissues, as well as the number of shared source proteins, is indicated above each graph, together with the respective area under the curve in percentage of total area (i.e., number of ligand source proteins × frequency of presentation of each source protein). To cope with differences in the depth of sample analyses (i.e., number of identified peptides per sample in EOC vs. benign tissues) peptide identifications for EOC were ranked according to abundance (i.e., area of the peptide ID) and artificially truncated to the median number of peptide identifications of respective benign tissues. (B) Comparative profiling and overlap analysis of HLA class I ligand source proteins of EOC in comparison with the entirety of benign tissues, deliberately excluding benign ovarian/fallopian tube tissue during comparative profiling to avoid loss of differentiation antigens possibly shared among tumor and benign ovarian tissues. Differences in the depth of sample analysis were addressed by ranking and artificially truncating the EOC dataset to the median number of peptide identification of benign tissues. Number of protein accessions and area (%) are presented accordingly. The significance threshold for exclusively presented proteins (FDR) was calculated by comparing the number of EOC exclusively presented HLA ligand source proteins at different presentation frequencies in the investigated cohort to a virtual cohort generated by in silico-based random weighted sampling from the entirety of protein identifications of both original cohorts. The in silico-generated randomized immunopeptidomes are of the same size and number of proteins as the real cohorts, but their origin (i.e., whether they are presented on EOC or benign tissues) is ignored for this analysis. Randomization of HLA ligand source proteins, cohort assembly, and assessment of exclusively presented HLA ligand source proteins was repeated 1,000 times. The mean value of resulting exclusively presented antigens, which randomly associate with either cohort, is then compared with the number of exclusively presented antigens in the original cohort at different presentation frequencies. A minimal threshold for HLA class I ligand source proteins exclusively presented on EOC of >10% (≥5 samples) presentation frequency was chosen with a corresponding FDR of 0.043 (4.3%). Gene names of respective source proteins are presented in a word cloud in C. The size of the font correlates with the presentation frequency in the EOC cohort. (D) Comparative profiling and overlap analyses of HLA class II ligand source proteins of epithelial ovarian cancer (EOC, n = 30) in comparison with benign ovary/fallopian tube tissue (n = 19), benign kidney tissue (n = 15), healthy donor PBMCs (n = 15), benign liver tissue (n = 10), and benign colorectal tissue (n = 15). Differences in the depth of sample analysis are adjusted using the same approach as in A and B. (E) Comparative profiling and overlap analysis of HLA class II ligand source proteins of EOC in comparison with the entirety of benign tissues deliberately excluding benign ovarian tissue samples during comparative profiling to avoid losing differentiation antigens possibly shared among tumor and benign ovarian tissues. Differences in the depth of sample analysis were adjusted (A and B). A minimal threshold exclusively on EOC-presented HLA class II ligand source proteins of >10% (≥4 samples) presentation frequency was chosen with a corresponding FDR of 0.086 (8.6%). Gene names of respective source proteins are presented in a word cloud in F. The size of the font correlates with the presentation frequency in the EOC cohort.
To extract from this dataset the most promising HLA-presented tumor-specific antigens, we performed a comparative profiling of the EOC dataset to all different cohorts of benign tissues, with the exception of benign ovarian/fallopian tube tissue. The latter was analyzed in a separate comparative profiling analysis to avoid the loss of tissue-specific antigens, which are potentially shared by EOC and the tissue from where it originated. This analysis revealed 1,143 proteins to be exclusively presented on EOC tissue among the analyzed tissues (Fig. 2B). The false-discovery rate (FDR) for EOC-specific HLA ligand source proteins presented in ≥10% of patients was determined to be below 4.3%, using a random-weighted sampling approach, as previously described (23). The remaining exclusively presented TOP56 HLA ligand source proteins are presented according to their frequency of presentation in Fig. 2C. One source protein, MUC16, also known as cancer antigen 125 (CA-125) (24), was clearly remarkable. In total, 113 different MUC16-derived HLA ligands (Dataset S3) were presented in nearly 80% (33 of 42) of patients’ tumors. These data highlight the frequent processing and presentation of MUC16 by a multitude of different HLA allotypes unparalleled by any other EOC-specific antigen (Fig. S4) and mirrored only by frequently (>97%, 41 of 42) presented housekeeping proteins, such as β-actin (overall 181 different peptides identified). Among the other EOC-specific source proteins, we identified additional well-established tumor-associated antigens like CRABP1/2, FOLR1, or KLK10 (25), as well as antigens with well-documented immunoevasive functions, like Indoleamine-2,3-dioxygenase (IDO1) (26, 27) or Galectin 1 (LGALS1) (28, 29).
Because of the power of CD4 T cells in supporting or driving (19, 30, 31) an antitumor immune response, we used the same approach to further analyze MHC class II-presented peptides in EOC (n = 30), yielding 17,334 peptides (median 697 per sample) representing 3,544 source proteins (mean 337 per sample), reaching >85% of attainable coverage (Fig. S3B). The HLA benign ligand dataset for MHC class II consisted of ovary/fallopian tube tissue (n = 19), benign kidney tissue (n = 15), healthy donor PBMCs (n = 15), benign colorectal tissue (n = 15), and benign liver tissue (n = 10). The number of HLA ligand source proteins from benign datasets varied between 1,042 and 1,953 proteins, achieving estimated attainable coverages of 70–88%. We used the same comparative profiling approach as for MHC class I, leaving 339 exclusively presented HLA ligand source proteins and 28 proteins presented on ≥10% of EOC patients with an FDR of 8.6%. Analysis of these TOP28 MHC class II presented antigens revealed a more heterogeneous and complex picture (Fig. 2F). Notably, MHC presented peptides of mesothelin (MSLN) (32), an established ligand of MUC16 (33), could be identified on 50% of EOC patients in the entire dataset. MUC16 itself was also among the exclusively presented MHC class II antigens, with ligands being identified on 7 of 30 EOC patients. MUC16 and MSLN were also moderately correlated in their expression on a tissue microarray (TMA) (Spearman correlation coefficient r = 0.5237; 95% CI = 0.3159–0.6835, two-tailed significance P < 0.001) involving 71 optimally debulked high-grade serous ovarian cancer patients. However, when we analyzed the prognostic relevance of these two antigens, only MSLN but not MUC16 had a negative prognostic impact on overall survival (Fig. S5). To further evaluate whether protein expression analyzed by IHC (immunoreactivity score) could serve as a surrogate marker for HLA ligand presentation; we analyzed the expression of these two antigens by IHC and correlated the staining intensities to the presence or absence of HLA ligands on the respective tumors. For both, MUC16 and MSLN, staining scores were significantly higher on tumors (P = 0.0006 for MUC16 or 0.017 for MSLN), which presented HLA ligands of the respective source proteins. The same was true for CA-125 serum levels determined at the day of surgery (P = 0.0047), indicating that these parameters could indeed be used for a proper selection of candidate antigens for peptide vaccination.
In a separate comparative profiling approach, we compared the EOC dataset with the set of benign ovarian and fallopian tube samples. This approach revealed 2,457 MHC class I and 510 MHC class II ligand source proteins to be exclusively identified on EOC. Due to the lower number of benign samples in the analysis, the FDR was calculated for EOC-specific HLA ligand source proteins presented in ≥30% of ovarian cancer patients, resulting in an FDR below 6.3% for the 15 most-frequently presented EOC-specific class I antigens and an FDR below 12.3% for the 4 most-frequently presented class II antigens (Dataset S4). Notably, only one of the antigens, namely CRABP2, also showed exclusivity on EOC within the complete benign tissue dataset. Among the remaining EOC-specific antigens not presented on benign ovarian/fallopian tube tissue, but with presentation on other normal tissues, several proteins with established functions in cell adhesion (LAMB3, FNDC3B, FAB, THBS2), endo/phagocytosis (SIGLEC1, CLINT1), and cell proliferation (MKI67, TPX2) could be identified. However, due to their confirmed presentation on multiple normal tissues, these antigens should be considered with caution for any immunotherapeutic approaches.
Mining of the EOC Immunopeptidome for Established Vaccination as Well as Cancer Testis Antigens.
Besides the EOC-specific HLA ligand source proteins emanating from the comparative profiling approach, we further looked for established tumor-associated antigens that have been previously employed for clinical application (Her2neu, WT1, NY-ESO-1, and p53). Although we could identify HLA-presented peptides for all antigens except for NY-ESO-1, none of them were exclusively presented on EOC (Dataset S4). The only ligands showing EOC-specific presentation, albeit with low frequency, were HLA class I ligands (but not HLA class II) from Her2neu. Analysis for cancer-testis antigens from a published reference dataset (34) showed a similar picture, with many antigens also being presented on benign tissues. However, individual HLA ligands derived from cancer/testis antigens (Dataset S4) were uniquely presented on EOC and can further lend themselves as targets for a personalized immunotherapy.
Differential Gene-Expression Analysis of EOC and Benign Fallopian Tube Tissue.
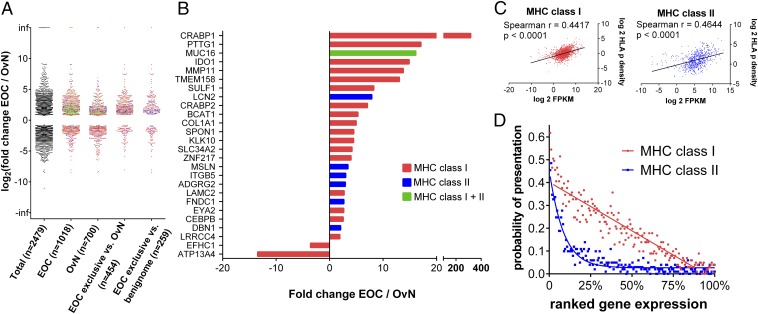
To analyze whether EOC exclusively presented antigens are also functionally associated with the tumor, we analyzed the gene-expression profile of 15 epithelial ovarian cancers and 15 benign fallopian tube tissues employing RNA-Seq (Dataset S5) and comparing the results to respective immunopeptidomic data. Differential analysis of healthy and cancer tissue revealed 2,479 genes to be significantly (FDR < 5%) differentially regulated between the two datasets. The distribution of these genes is presented together with additional distributions of immunopeptidomic subgroups in Fig. 3A. A closer look at the TOP EOC exclusively presented antigens that were identified by comparative profiling revealed 20 of 56 MHC class I and 7 of 28 MHC class II antigens to be significantly differentially expressed and all but two showed higher expression on EOC than on benign fallopian tube tissues (Fig. 3B), indicating a functional association with tumorigenesis. Moreover, many of these antigens also showed a low and restricted expression pattern on healthy tissues comparable to cancer/testis antigens according to public databases (for GTEX gene-expression profiles of TOP antigens and HLA ligands from cancer testis antigens, see Fig. S6). Of note, despite a lack of significant overexpression in the complete differential gene-expression dataset, some of the TOP antigens show very high expression values in individual tumors (for example NLRP2 in OvCa 84 and OvCa 105) (Dataset S5), which could be exploited for individualized approaches.
Fig. 3.
Differential analysis of gene expression of ovarian cancer (EOC) vs. benign fallopian tube samples (OvN) by RNA-Seq and correlation to the immunopeptidome. (A) Distribution of significantly differentially expressed genes (adjusted P < 0.05) from a gene-expression analysis of 15 ovarian cancer (EOC) and 15 benign fallopian tube samples (OvN). Among the differentially expressed genes, a total of 2,479 could be mapped to a single UniProtKB/Swiss-Prot accession of the human proteome, which has been previously used as a data source for immunopeptidome analysis and HLA ligand identification. Additional distributions show differentially expressed genes reflected in the immunopeptidome of ovarian cancer (EOC), benign tubal and ovarian tissue (OvN), and subsets of the immunopeptidome after comparative profiling (i.e., EOC exclusive genes after comparative profiling of the EOC dataset to tubo-ovarian tissues or a respective set of benign tissues). Differentially expressed genes that give rise only to MHC class I or MHC class II genes are colored in red (class I) or blue (class II), whereas genes that give rise to both classes of MHC ligands are colored in green. (B) Differentially expressed genes among TOP EOC exclusively presented antigens (i.e., TOP56 class I and TOP28 class II antigens of EOC immunopeptidome after comparative profiling to the set of benign tissues). (C) Correlation of gene expression (FPKM values) and immunopeptidomic presentation (expressed as HLA-peptide density; see SI Methods for further information) for OvCa 111. Nonparametric two-tailed Spearman rank correlation was employed to test for the degree of correlation. (D) Probability of MHC presentation in relation to gene expression. Gene-expression values (FPKM values) of OvCa 111 were ranked from high to low and divided into bins of 100 genes. For each bin, the number of genes that are presented on the immunopeptidomic level (MHC class I in red and MHC class II in blue) was determined and plotted accordingly as probability of presentation (i.e., a probability of 0.4 means that 40 of 100 genes were presented on the immunopeptidome level). Linear and sigmoidal trendlines have been added for visualization purposes.
We were further interested whether we could determine a direct correlation of gene expression and antigen presentation. For this purpose, we calculated the HLA-p density for each HLA ligand source protein, as previously described by Bassani-Sternberg et al. (35), and correlated it to respective FPKM values. Representative results for one patient are presented in Fig. 3C. In accordance with previous results (36), we could evidence a significant but overall rather moderate correlation of gene expression and HLA ligand presentation. Gene-expression analysis can nevertheless be of great importance for target evaluation, especially if antigens need to be prioritized for targeted immunotherapeutic approaches without access to MHC presentation data. Fig. 3D shows the probability of antigen presentation in dependence of gene expression for the same patient. Notably, whereas the HLA presentation probability of an antigen is decreasing in a linear fashion for HLA class I, a rather sigmoid relation is evident for HLA class II.
To also have a more detailed look into the biological pathways up-regulated in EOC, we performed gene-set enrichment analysis (37) using the Hallmark gene set hosted at the Broad Institute (MSigDB Collections). Notably, hallmarks highlighting an ongoing immune response were especially prominent among the enriched gene sets (Dataset S6).
Cellular Origin of EOC-Associated HLA-Presented Peptides.
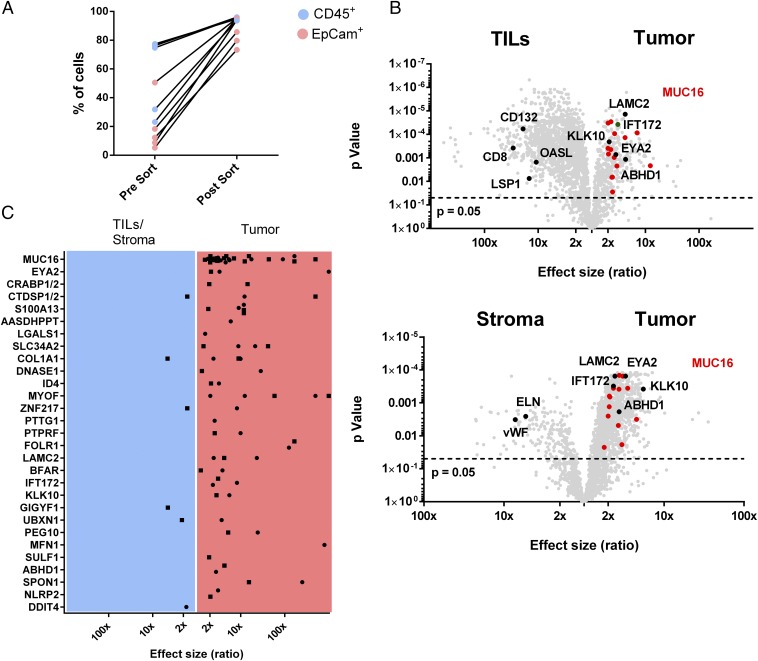
Because EOCs embody not only cancer cells but rather represent a heterogeneous mixture of different cell types, we asked whether exclusive MHC class I antigens were indeed originally presented by cancer cells. For this purpose, we enzymatically digested EOC tissue and separated CD45+ leukocytes, EpCAM+ tumor cells, and stroma cells, which are negative for both markers (enrichment efficiencies are presented in Fig. 4A and Dataset S2), subsequently performing HLA ligandomics individually for each of the subsets. We used label-free quantification to determine the source of each identified HLA ligand in a total of five EOCs (for a representative example see Fig. 4B). As expected, MUC16-derived HLA ligands, identified on four of five EOC samples, were always found to be overrepresented on enriched cancer cells with a median fivefold overrepresentation (range 1.8- to 135-fold) dependent on the enrichment efficiency. The same held true for the majority of exclusively presented antigens like CRABP1/2, EYA2, or KLK10, with few ambiguous exceptions, such as GIGYF1 or DDIT4 (Fig. 4C). Apart from tumor-associated antigens, we also recognized ligands from source proteins with cell-type–specific expression. For example, ligands derived from CD8, CD132, 2′-5′-oligoadenylate synthase-like protein (OASL) or lymphocyte specific protein 1 (LSP1) were highly overrepresented on CD45+ cells. On the other hand, van Willebrand factor (vWF) and elastin (ELN), most likely expressed by endothelial cells in the stroma, were found highly overrepresented within the stromal subset, emphasizing the strength of this cell-type–specific approach (Fig. 4B).
Fig. 4.
Cellular origin of EOC-presented antigens. (A) Percentage of CD45+ tumor infiltrating leukocytes (TILs) and CD45−EpCAM+ tumor cells before and after enrichment via MACS. (B) Volcano plots of the relative abundance of HLA ligands in the class I immunopeptidome of enriched cell populations of OvCa 84, analyzed by label-free quantitation. Panels show TIL (CD45+) vs. tumor cells (CD45−EpCAM+; Upper) and stroma cells (CD45−EpCAM−) vs. tumor cells (Lower). The horizontal dashed line indicates significance threshold after adjustment for multiple testing (P < 0.05). TOP56 EOC exclusive ligands MHC class I antigens [MUC16 (red), IFT172, EYA2, LAMC2, KLK10, and ABHD1] as well as ligands derived from established leukocyte-associated antigens (CD132, CD8, LSP1, OASL) and stroma cell-associated antigens (ELN, vWF) are highlighted. (C) Cumulative results from label free quantitation of five tumors (OvCa 84, OvCa 73, OvCa 70, OvCa 60, OvCa 57) after MACS enrichment. Dots show peptides significantly enriched on stroma or tumor cells whereas squares indicate peptides significantly enriched on TILs or tumor cells. Enrichment and P values for all EOC exclusive class I antigens identified in the five EOCs analyzed can be found in Dataset S2.
Immunogenicity Analysis.
For the applicability of vaccination approaches, immunogenicity is a major imperative. To evaluate the immunogenic potential of the identified HLA ligands, we performed in vitro priming of T cells from healthy blood donors, as previously published (38–40). Effective priming and expansion of antigen-specific T cells was confirmed by multimer staining (Fig. S7). The results for selected EOC-exclusive antigens are presented in Table 1. For the most-frequently presented EOC-specific antigen MUC16, 25 of the 28 peptides tested so far were shown to be immunogenic in at least one tested healthy donor. Similar frequencies could also be shown for other EOC-presented tumor-associated antigens (CRABP1/2, KLK10) or antigens with immunoevasive function (IDO1, LGALS1), but not for the control peptide YLLPAIVHI (DDX5) (41), which is frequently presented by HLA-A*02 on both tumor and normal benign tissues (peptide presentation >90% if only A*02-positive sources are considered). Immunogenicity could also not be demonstrated for peptides derived from antigens with broader expression profiles, such as MKI67, for which HLA presentation was shown to be absent on normal ovarian tissue but not on other benign tissues. Additional control experiments were performed to confirm that the same healthy donors did not show preexisting (memory) T cell responses ex vivo or after recall stimulation against the tested peptides (Fig. S7).
Table 1.
In vitro immunogenicity of TOP HLA-presented EOC antigens (MUC16, IDO1, LGALS1, KLK10, CRABP1/2) as well as antigens with additional HLA peptide presentation on normal tissues (MKI67, DDX5)
| Tissues with HLA peptide presentation of source antigen (%) | Tissues with HLA peptide presentation | ||||||||
| HLA | Sequence | Protein | Positive/tested donors | EOC | OvN | Benign sources | EOC | OvN | Benign sources |
| A*01 | STETSTVLY | MUC16 | 0/3 | 33/42 (79) | 5/23 (22) | 0/85 (0) | 3 | 0 | 0 |
| A*02 | IITEVITRL | MUC16 | 3/10 | 1 | 0 | 0 | |||
| A*02 | KMISAIPTL | MUC16 | 4/6 | 3 | 0 | 0 | |||
| A*03 | SVLADLVTTK | MUC16 | 0/1 | 1 | 0 | 0 | |||
| A*11 | STSQEIHSATK | MUC16 | 2/6 | 1 | 0 | 0 | |||
| A*11 | GTSGTPVSK | MUC16 | 0/5 | 1 | 0 | 0 | |||
| A*24 | TYSEKTTLF | MUC16 | 3/3 | 5 | 1 | 0 | |||
| A*24 | QFITSTNTF | MUC16 | 1/2 | 1 | 0 | 0 | |||
| A*24 | AVTNVRTSI | MUC16 | 1/3 | 2 | 0 | 0 | |||
| A*25 | EVITSSRTTI | MUC16 | 1/1 | 2 | 0 | 0 | |||
| A*25 | EVISSRGTSM | MUC16 | 1/3 | 3 | 0 | 0 | |||
| A*25 | EVTSSGRTSI | MUC16 | 2/3 | 3 | 0 | 0 | |||
| A*25 | ETILTFHAF | MUC16 | 2/2 | 5 | 0 | 0 | |||
| B*07 | SPHPVTALL | MUC16 | 1/1 | 4 | 0 | 0 | |||
| B*07 | SPQNLRNTL | MUC16 | 2/3 | 4 | 0 | 0 | |||
| B*07 | TPGNRAISL | MUC16 | 5/5 | 6 | 1 | 0 | |||
| B*07 | SPLFQRSSL | MUC16 | 1/3 | 1 | 0 | 0 | |||
| B*07 | SPHPVTALL | MUC16 | 1/2 | 4 | 0 | 0 | |||
| B*07 | LPHSEITTL | MUC16 | 1/3 | 2 | 0 | 0 | |||
| B*07 | TPGGTRQSL | MUC16 | 0/3 | 7 | 3 | 0 | |||
| B*07 | SPSKAFASL | MUC16 | 3/3 | 9 | 2 | 0 | |||
| B*07 | VPRSAATTL | MUC16 | 2/3 | 3 | 0 | 0 | |||
| B*15 | SQGFSHSQM | MUC16 | 4/5 | 1 | 0 | 0 | |||
| B*15 | FQRQGQTAL | MUC16 | 1/6 | 1 | 0 | 0 | |||
| B*18 | TETEAIHVF | MUC16 | 1/1 | 4 | 0 | 0 | |||
| B*27 | ERSPVIQTL | MUC16 | 1/2 | 1 | 0 | 0 | |||
| B*51 | DPYKATSAV | MUC16 | 3/3 | 7 | 1 | 0 | |||
| B*51 | DALVLKTV | MUC16 | 1/3 | 5 | 0 | 0 | |||
| A*03 | RSYHLQIVTK | IDO1 | 1/1 | 12/42 (29) | 4/23 (17) | 0/85 (0) | 5 | 2 | 0 |
| A*11 | RSYHLQIVTK | IDO1 | 1/1 | 5 | 2 | 0 | |||
| A*24 | RYMPPAHRNF | IDO1 | 4/4 | 2 | 0 | 0 | |||
| B*07 | NPKAFFSVL | IDO1 | 3/5 | 1 | 0 | 0 | |||
| A*25 | EVAPDAKSF | LGALS1 | 1/1 | 11/42 (26) | 1/23 (4) | 0/85 (0) | 5 | 1 | 0 |
| A*02 | RTTEINFKV | CRABP1/2 | 1/2 | 15/42 (36) | 0/23 (0) | 0/85 (0) | 9 | 0 | 0 |
| A*02 | RALAKLLPL | KLK10 | 2/2 | 9/42 (21) | 0/23 (0) | 0/85 (0) | 7 | 0 | 0 |
| A*11 | SSKSQTEVPK | MKI 67 | 0/5 | 21/42 (50) | 0/23 (0) | 14/85 (16) | 0 | 0 | 5 |
| B*08 | TPKEKAQAL | MKI 67 | 0/5 | 2 | 0 | 0 | |||
| A*02 | YLLPAIVHI | DDX 5 | 0/5 | 29/42 (69) | 18/23 (78) | 63/85 (74) | 23 | 13 | 36 |
Discussion
Tremendous progress in the field of cancer immunotherapy during the last years has led to its wide appreciation (42, 43) as a potentially curative addition (44) or alternative to standard chemotherapeutic approaches. Several papers demonstrate the importance of HLA-presented mutated (45–47) and wild-type tumor-associated antigens (48, 49) as valuable tumor rejection antigens. Therefore, large-scale identification of HLA-presented cancer-specific tumor antigens adds another important piece to the puzzle of our understanding how the immune system identifies and recognizes tumor cells.
In the present study, we focused on EOC, with the goal to comprehensively characterizing the immunopeptidome of EOC and evaluating the HLA-presented antigens for their usefulness in clinical applications. So far, only few HLA-presented antigens have been identified for EOC and most clinical studies have relied on predicted or established cancer testis antigens, not necessarily frequently presented by EOC, an assumption that could be confirmed by our analysis.
A major prerequisite for all immunotherapies that target MHC molecules is the persistent and strong expression of these molecules on their target cells. We were able to clearly demonstrate a high expression of HLA class I molecules on ovarian tumors on both the RNA and protein level without any evidence for HLA loss or down-regulation. Furthermore, we show on a single-cell level that EOC also strongly express HLA-DR molecules. We were further able to validate this strong expression by the identification of numerous MHC class II ligands, including peptides from established tumor-associated antigens. This high expression of MHC class II in EOC has to our knowledge not been appreciated in the literature, and clearly warrants further investigation to identify the mechanism responsible for this overexpression.
Profiling of the immunopeptidome of 42 ovarian tumors in comparison with more than 85 benign sources of different origins revealed several hundred EOC-associated antigens exclusively presented on EOC within all analyzed tissues. Among the HLA class I antigens not presented on any of the tissues in our benign dataset, MUC16 was clearly the most exceptional. Concerning both the number of HLA ligands identified (>80) and the frequency of presentation in the patient cohort (∼80%), this is unprecedented for any other tumor antigen and tumor entity we have investigated so far. Moreover, we could demonstrate that more than 85% of HLA ligands derived from MUC16 are immunogenic and able to prime T cells in healthy individuals, rendering MUC16 an unparalleled first-class antigen for EOC immunotherapy. Immunopeptidome profiling further provides a showcase for apparent mechanistic insights into EOC, which are reflected in the HLA ligandome of both HLA class I and class II ligands. HLA ligands from important kinases and phosphatases (EYA2), proteins associated with immunosuppression (IDO1, LGALS1), as well as established and suspected molecular markers for EOC (KLK10, FOLR1) are only a few to mention. Notably, for HLA class II, MSLN, an established ligand of MUC16, has been identified as the top exclusively presented antigen. Several studies have demonstrated the pivotal role of the MUC16/MSLN axis for cell invasion and metastasis in EOC, as well as in other tumors, such as pancreatic cancer (50) or mesothelioma (33), suggesting that T cell epitopes of these antigens should be further tested in other malignancies. We could show that MSLN staining is directly correlated with MUC16 staining, and it has been further reported that high MSLN expression is a negative prognostic factor in EOC (51).
In our study, several different benign tissues and cell types (PBMCs, liver, kidney, colon, ovary) have been used for this kind of selective immunopeptidome profiling. Due to restrictions in the number of different tissues available for investigation, we cannot completely exclude that individual antigens might also be presented by HLA molecules in other organs and tissues. The established functional relevance of those antigens for EOC, and particularly the immunogenicity of the respective peptides in healthy individuals, make a presentation in other tissues unlikely but clearly demand additional confirmation in a more extended set of tissues and primary cells. Further analysis specifically of the induced T cells and respective T cell receptors will be required to confirm the clinical usefulness and safety of these antigens. In particular, demonstration that the induced T cells show full functional capacity in vivo, especially with regard to tumor cell killing, is of essential importance for the clinical development of immunotherapies.
In the last 2 years, many articles have focused on the identification and utilization of HLA-presented mutated antigens and their value and importance for cancer immunotherapy has been undoubtedly presented in many individualized approaches. Because of the low mutational load of high-grade serous ovarian cancer, which is characterized by large-scale chromosomal alterations rather than mutations in individual genes, these approaches might, however, be only applicable for a small group of patients. We have previously shown that identification of mutated MHC ligands is indeed feasible by MS (47). Although identification of mutated HLA ligands in ovarian cancer patients has been beyond the scope of this article, we could demonstrate that the use of gene-expression data may increase the probability of selecting promising HLA-presented antigens when presentation data are not available.
In conclusion, our study provides deep insights into the immunopeptidome of EOC, highlighting new rational targets based on frequently naturally presented and immunogenic HLA ligands, which can be further developed for different immunotherapeutic approaches. We envisage that these findings will promote the emergence of novel immunotherapeutic approaches urgently needed in the management of EOC.
Methods
Detailed descriptions for each method can be found in SI Methods.
Patient Samples.
All tissue samples were collected at the University Hospital of Tübingen after obtaining patient informed consent, in accordance with the principles of the Declaration of Helsinki. The local institutional review board (Ethics Committee at the Medical Faculty and at the University Hospital of Tübingen) approved all study protocols.
Tissue Dissociation.
Tissues were minced into small pieces <2 mm3 and transferred into an enzymatic dissociation solution containing collagenase. Dissociation was performed on a rotating shaker (Infors HT) for 3 h at 37 °C. Remaining tissue fragments (typically <1% of initial weight) were removed using a 100-µm cell strainer (BD). Single-cell suspensions were washed twice with PBS and erythrocytes were lysed using ammonium chloride lysis buffer.
HLA Surface Molecule Quantification.
HLA surface expression was determined using QIFIKIT quantification flow cytometric assay (Dako) according to the manufacturer’s instructions. Cells were stained with either pan-HLA class I-specific monoclonal antibody W6/32, HLA-DR–specific L243 or respective isotype control, as well as additional fluorescently labeled antibodies to distinguish different cell populations.
Cell Separation.
Cell separation was performed using two consecutive magnetic-activated cell separation (MACS) protocols (depletion of CD45+ and enrichment of EpCAM+ cells), according to the manufacturer’s instructions (Miltenyi).
HLA Ligand Isolation.
HLA class I and II molecules were isolated by standard immunoaffinity purification as described previously (22). Between 0.5 and 3 g of tissue samples or 2.5 × 108 to 1 × 109 of enriched cell fractions were employed for lysis and HLA ligand isolation.
Immunopeptidome Analysis by LC-MS/MS and Data Processing.
Immunopeptidome analysis was performed on an linear trap quadrupole (LTQ) Orbitrap XL mass spectrometer (Thermo Fisher) equipped with a nanoelectron spray ion source and coupled to an Ultimate 3000 RSLC Nano UHPLC System (Dionex). MS data analysis was carried out using Proteome discoverer 1.3 (Thermo Fisher). Peak lists were searched against the human proteome as comprised in the UniProtKB/Swiss-Prot database (www.uniprot.org, released September 27, 2013; including 20,279 reviewed protein sequences) using Mascot search engine (Mascot 2.2.04, Matrix Science).
RNA-Seq Analysis.
RNA-Seq analysis was performed by an external service provider (CeGaT GmbH). In brief, RNA was isolated from cryopreserved tissue using the RNeasy Mini kit (Qiagen). One-hundred nanograms of isolated RNA was employed for library preparation using the TruSeq Stranded mRNA Kit (Illumina). Single-end sequencing was performed on a HiSEq. 2500 instrument with a target read length of 100 bp and a read depth of at least 50 million reads.
Immunogenicity Analysis of HLA Class I Ligands.
Priming of peptide-specific cytotoxic lymphocytes was conducted using an established protocol involving artificial antigen presenting cells (aAPCs) (38). One million T cells per well were cultured in 96-well plates (Corning) and stimulated with 200,000 loaded aAPCs in the presence of 5 ng/mL IL-12 (PromoCell). T cells were stimulated three times in total, with a weekly stimulation interval. Then, 40 U/mL IL-2 and 5 ng/mL IL-7 was added 2 d after each stimulation. The induction and expansion of peptide-specific T cells was assessed by MHC-multimer staining.
IHC.
Tissue sections were pretreated with EDTA-buffer solution (pH 8.6) at 95 °C for 36 min or citrate buffer solution (pH 6) at 100° (only L243, HCA2). IHC stainings were performed on (semi)automated immunostainer according to the manufacturer’s instructions. For immunoscoring, as well as employed antibodies and dilutions, please refer to SI Methods.
Data Availability.
BAM files of RNA-Seq data are available through National Center for Biotechnology Information Sequence Read Archive project PRJNA398141. MS raw data for epithelial ovarian cancer samples have been deposited to the ProteomeXchange Consortium via the PRIDE partner (52) repository with the dataset identifier PXD007635 or can be requested from the corresponding author.
Supplementary Material
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft, Collaborative Research Center 685, the German Cancer Consortium (DKTK), and the European Research Council Advanced Grant “Mutaediting.”
Footnotes
Conflict of interest statement: H.-G.R. is a shareholder of Immatics Biotechnologies, Tübingen, and CureVac GmbH, Tübingen. H.S. and D.J.K. are employees of Immatics Biotechnologies GmbH. The authors declare that Immatics did not provide neither financial nor scientific support in any direct relation to this manuscript or the underlying studies, and was not involved in data collection, analysis, or decision to publish.
This article is a PNAS Direct Submission.
Data deposition: RNA-Seq BAM Files have been uploaded to the National Center for Biotechnology Informaton Sequence Read archive (Bioproject ID PRJNA398141) and the ProteomeXchange Consortium via the PRIDE partner repository (ID PXD007635).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707658114/-/DCSupplemental.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Coleman RL, Monk BJ, Sood AK, Herzog TJ. Latest research and treatment of advanced-stage epithelial ovarian cancer. Nat Rev Clin Oncol. 2013;10:211–224. doi: 10.1038/nrclinonc.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herzog TJ, Pothuri B. Ovarian cancer: A focus on management of recurrent disease. Nat Clin Pract Oncol. 2006;3:604–611. doi: 10.1038/ncponc0637. [DOI] [PubMed] [Google Scholar]
- 4.Mirza MR, et al. ENGOT-OV16/NOVA Investigators Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med. 2016;375:2154–2164. doi: 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
- 5.Kandalaft LE, Powell DJ, Jr, Singh N, Coukos G. Immunotherapy for ovarian cancer: What’s next? J Clin Oncol. 2011;29:925–933. doi: 10.1200/JCO.2009.27.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348:203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 7.Schlienger K, et al. TRANCE- and CD40 ligand-matured dendritic cells reveal MHC class I-restricted T cells specific for autologous tumor in late-stage ovarian cancer patients. Clin Cancer Res. 2003;9:1517–1527. [PubMed] [Google Scholar]
- 8.Matsuzaki J, et al. Tumor-infiltrating NY-ESO-1-specific CD8+ T cells are negatively regulated by LAG-3 and PD-1 in human ovarian cancer. Proc Natl Acad Sci USA. 2010;107:7875–7880. doi: 10.1073/pnas.1003345107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisk B, Blevins TL, Wharton JT, Ioannides CG. Identification of an immunodominant peptide of HER-2/neu protooncogene recognized by ovarian tumor-specific cytotoxic T lymphocyte lines. J Exp Med. 1995;181:2109–2117. doi: 10.1084/jem.181.6.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curiel TJ, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 11.Vlad AM, et al. A phase II trial of intraperitoneal interleukin-2 in patients with platinum-resistant or platinum-refractory ovarian cancer. Cancer Immunol Immunother. 2010;59:293–301. doi: 10.1007/s00262-009-0750-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodi FS, et al. Immunologic and clinical effects of antibody blockade of cytotoxic T lymphocyte-associated antigen 4 in previously vaccinated cancer patients. Proc Natl Acad Sci USA. 2008;105:3005–3010. doi: 10.1073/pnas.0712237105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robert C, et al. Anti-programmed-death-receptor-1 treatment with pembrolizumab in ipilimumab-refractory advanced melanoma: A randomised dose-comparison cohort of a phase 1 trial. Lancet. 2014;384:1109–1117. doi: 10.1016/S0140-6736(14)60958-2. [DOI] [PubMed] [Google Scholar]
- 14.Wolchok JD, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenberg SA, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011;17:4550–4557. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porter DL, et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci Transl Med. 2015;7:303ra139. doi: 10.1126/scitranslmed.aac5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maude SL, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenberg SA. Raising the bar: The curative potential of human cancer immunotherapy. Sci Transl Med. 2012;4:127ps8. doi: 10.1126/scitranslmed.3003634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tran E, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mantia-Smaldone GM, Corr B, Chu CS. Immunotherapy in ovarian cancer. Hum Vaccin Immunother. 2012;8:1179–1191. doi: 10.4161/hv.20738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu B, et al. Ovarian cancer immunotherapy: Opportunities, progresses and challenges. J Hematol Oncol. 2010;3:7. doi: 10.1186/1756-8722-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berlin C, et al. Mapping the HLA ligandome landscape of acute myeloid leukemia: A targeted approach toward peptide-based immunotherapy. Leukemia. 2014;29:647–659, and erratum (2016) 30:1003–1004. doi: 10.1038/leu.2016.1. [DOI] [PubMed] [Google Scholar]
- 23.Kowalewski DJ, et al. HLA ligandome analysis identifies the underlying specificities of spontaneous antileukemia immune responses in chronic lymphocytic leukemia (CLL) Proc Natl Acad Sci USA. 2015;112:E166–E175. doi: 10.1073/pnas.1416389112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haridas D, et al. MUC16: Molecular analysis and its functional implications in benign and malignant conditions. FASEB J. 2014;28:4183–4199. doi: 10.1096/fj.14-257352. [DOI] [PubMed] [Google Scholar]
- 25.Luo LY, et al. The serum concentration of human kallikrein 10 represents a novel biomarker for ovarian cancer diagnosis and prognosis. Cancer Res. 2003;63:807–811. [PubMed] [Google Scholar]
- 26.Uyttenhove C, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 27.Sørensen RB, et al. The immune system strikes back: Cellular immune responses against indoleamine 2,3-dioxygenase. PLoS One. 2009;4:e6910. doi: 10.1371/journal.pone.0006910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van den Brûle F, et al. Galectin-1 accumulation in the ovary carcinoma peritumoral stroma is induced by ovary carcinoma cells and affects both cancer cell proliferation and adhesion to laminin-1 and fibronectin. Lab Invest. 2003;83:377–386. doi: 10.1097/01.lab.0000059949.01480.40. [DOI] [PubMed] [Google Scholar]
- 29.Rubinstein N, et al. Targeted inhibition of galectin-1 gene expression in tumor cells results in heightened T cell-mediated rejection; A potential mechanism of tumor-immune privilege. Cancer Cell. 2004;5:241–251. doi: 10.1016/s1535-6108(04)00024-8. [DOI] [PubMed] [Google Scholar]
- 30.Perez-Diez A, et al. CD4 cells can be more efficient at tumor rejection than CD8 cells. Blood. 2007;109:5346–5354. doi: 10.1182/blood-2006-10-051318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braumüller H, et al. T-helper-1-cell cytokines drive cancer into senescence. Nature. 2013;494:361–365. doi: 10.1038/nature11824. [DOI] [PubMed] [Google Scholar]
- 32.Pastan I, Hassan R. Discovery of mesothelin and exploiting it as a target for immunotherapy. Cancer Res. 2014;74:2907–2912. doi: 10.1158/0008-5472.CAN-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassan R, Ho M. Mesothelin targeted cancer immunotherapy. Eur J Cancer. 2008;44:46–53. doi: 10.1016/j.ejca.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Almeida LG, et al. CTdatabase: A knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 2009;37:D816–D819. doi: 10.1093/nar/gkn673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bassani-Sternberg M, Pletscher-Frankild S, Jensen LJ, Mann M. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol Cell Proteomics. 2015;14:658–673. doi: 10.1074/mcp.M114.042812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinzierl AO, et al. Distorted relation between mRNA copy number and corresponding major histocompatibility complex ligand density on the cell surface. Mol Cell Proteomics. 2007;6:102–113. doi: 10.1074/mcp.M600310-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter S, et al. Cutting edge: Predetermined avidity of human CD8 T cells expanded on calibrated MHC/anti-CD28-coated microspheres. J Immunol. 2003;171:4974–4978. doi: 10.4049/jimmunol.171.10.4974. [DOI] [PubMed] [Google Scholar]
- 39.Walter S, et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat Med. 2012;18:1254–1261. doi: 10.1038/nm.2883. [DOI] [PubMed] [Google Scholar]
- 40.Dutoit V, et al. Exploiting the glioblastoma peptidome to discover novel tumour-associated antigens for immunotherapy. Brain. 2012;135:1042–1054. doi: 10.1093/brain/aws042. [DOI] [PubMed] [Google Scholar]
- 41.Weinzierl AO, et al. Identification of HLA-A*01- and HLA-A*02-restricted CD8+ T-cell epitopes shared among group B enteroviruses. J Gen Virol. 2008;89:2090–2097. doi: 10.1099/vir.0.2008/000711-0. [DOI] [PubMed] [Google Scholar]
- 42.Couzin-Frankel J. Breakthrough of the year 2013. Cancer immunotherapy. Science. 2013;342:1432–1433. doi: 10.1126/science.342.6165.1432. [DOI] [PubMed] [Google Scholar]
- 43.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez SA, et al. A new era in anticancer peptide vaccines. Cancer. 2010;116:2071–2080. doi: 10.1002/cncr.24988. [DOI] [PubMed] [Google Scholar]
- 45.Matsushita H, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robbins PF, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gubin MM, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andersen RS, et al. Dissection of T-cell antigen specificity in human melanoma. Cancer Res. 2012;72:1642–1650. doi: 10.1158/0008-5472.CAN-11-2614. [DOI] [PubMed] [Google Scholar]
- 49.Lu YC, et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res. 2014;20:3401–3410. doi: 10.1158/1078-0432.CCR-14-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimizu A, et al. Coexpression of MUC16 and mesothelin is related to the invasion process in pancreatic ductal adenocarcinoma. Cancer Sci. 2012;103:739–746. doi: 10.1111/j.1349-7006.2012.02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng WF, et al. High mesothelin correlates with chemoresistance and poor survival in epithelial ovarian carcinoma. Br J Cancer. 2009;100:1144–1153. doi: 10.1038/sj.bjc.6604964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vizcaíno JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seidmann JD, Cho KR, Ronnett BM, Kurman RJ. Surface epithelial tumors of the ovary. In: Kurman RJ, Ellenson LH, Ronnett BM, editors. Blaustein's Pathology of the Female Genital Tract. 6th Ed Springer; New York: 2011. [Google Scholar]
- 54.Pham DL, et al. SOX2 expression and prognostic significance in ovarian carcinoma. Int J Gynecol Pathol. 2013;32:358–367. doi: 10.1097/PGP.0b013e31826a642b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
BAM files of RNA-Seq data are available through National Center for Biotechnology Information Sequence Read Archive project PRJNA398141. MS raw data for epithelial ovarian cancer samples have been deposited to the ProteomeXchange Consortium via the PRIDE partner (52) repository with the dataset identifier PXD007635 or can be requested from the corresponding author.