Significance
Cancer cells derived from mesenchymal tissues or induced to adopt a mesenchymal state have been demonstrated to be largely resistant to standard chemotherapies, necessitating the identification of new effective treatment strategies. From a high throughput screen, we have discovered FiVe1, a compound capable of irreversibly inhibiting the growth of mesenchymally transformed cancer cells by binding to and interfering with the organization and phosphorylation of vimentin (VIM) during mitosis. In contrast to the many naturally and synthetically derived compounds targeting microtubules, we report that chemically targeting an intermediate filament protein, VIM, promotes mitotic catastrophe. As VIM expression is restricted to mesenchymal cells, these results provide a mechanistic basis toward developing genotype-selective chemotherapeutics for the treatment of mesenchymal cancers.
Keywords: vimentin, epithelial-to-mesenchymal transition, cancer stem cell, mitosis, drug discovery
Abstract
Expression of the transcription factor FOXC2 is induced and necessary for successful epithelial–mesenchymal transition, a developmental program that when activated in cancer endows cells with metastatic potential and the properties of stem cells. As such, identifying agents that inhibit the growth of FOXC2-transformed cells represents an attractive approach to inhibit chemotherapy resistance and metastatic dissemination. From a high throughput synthetic lethal screen, we identified a small molecule, FiVe1, which selectively and irreversibly inhibits the growth of mesenchymally transformed breast cancer cells and soft tissue sarcomas of diverse histological subtypes. FiVe1 targets the intermediate filament and mesenchymal marker vimentin (VIM) in a mode which promotes VIM disorganization and phosphorylation during metaphase, ultimately leading to mitotic catastrophe, multinucleation, and the loss of stemness. These findings illustrate a previously undescribed mechanism for interrupting faithful mitotic progression and may ultimately inform the design of therapies for a broad range of mesenchymal cancers.
For many tumor types, resistance to conventional chemotherapy and subsequent tumor relapse have been attributed to the presence of a slower cycling, drug-resistant population of cells termed cancer stem cells (CSCs) or tumor initiating cells. We and others have demonstrated that activation of a latent embryonic program, called the epithelial–mesenchymal transition (EMT), endows epithelial-derived cancer cells with the properties of stem cells as well as migratory and metastatic potential (1). Cells undergoing EMT exhibit the loss of epithelial cell–cell contacts (e.g., E-cadherin), the induction of matrix degrading proteases (e.g., matrix metalloproteinases), and the acquisition of motility-inducing intermediate filaments [e.g., vimentin (VIM)], features which promote metastatic progression by allowing dissemination from the local tumor niche (2). A number of transcription factors (e.g., Snail, Twist, ZEB1) and microenvironment-derived extracellular signaling molecules (e.g., TGF-β1) are capable of inducing the EMT transcriptional program. Among these factors, we have demonstrated that the transcription factor Forkhead Box C2 (FOXC2) is a central regulator of EMT in breast cancer (3, 4). FOXC2 expression is both up-regulated and required for the induction of CSC properties by the classical EMT-inducing factors Twist, Snail, and TGF-β1 (4). Knockdown of FOXC2 in this context sensitizes these cells to traditional DNA-damaging chemotherapies and inhibits their tumor initiating potential (4). Additionally, exogenous FOXC2 expression is sufficient to endow metastasis-incompetent cells with the potential to form secondary tumors in liver, lung, brain, and bone (4). Further, FOXC2 expression is frequently correlated with poor survival rates in breast cancer patients and is often up-regulated in triple negative breast cancers (3–5).
Consequently, an attractive approach to combat chemotherapy resistance and metastatic dissemination in breast cancer is, therefore, to specifically target mechanisms necessary for the survival of EMT-derived CSCs. Given FOXC2’s role as a central mediator of the EMT-CSC phenotype, small molecules that are selectively toxic to this genotype should, in principle, eliminate EMT-derived CSCs from multiple genetic backgrounds and render non-CSC populations sensitive to the effects of standard chemotherapies. Additionally, the characterization of these compounds should help unravel mechanisms governing cancer stemness. Synthetic lethal chemical screens have previously provided a rich source of new insight into cell death mechanisms (6–8). Herein, we undertook a synthetic lethal screen to identify small molecules selectively cytotoxic to FOXC2-expressing mesenchymally transformed breast cancer cells. We identified a compound that binds the filamentous protein vimentin, which is expressed in all mesenchymal cells, and by so doing, disrupts mitotic progression. Finally, we characterize the activity of this compound in other mesenchymally derived cancers.
Results
A High Throughput Screen Identifies Selective Inhibitors of FOXC2-Expressing Cells.
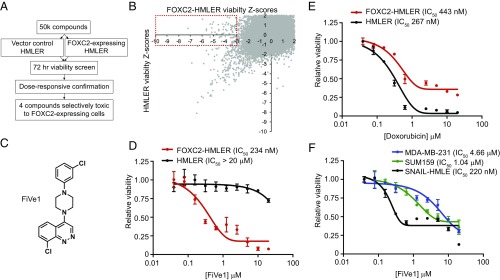
To identify molecules selectively toxic to the FOXC2-positive genotype, we employed the use of the engineered cancer cell line HMLER as an isogenic screening system. HMLER cells are derived from human mammary epithelial cells (HMECs), but are virally transformed with the K-Ras oncogene (G12V), human telomerase reverse transcriptase (hTERT), and SV40 large-T antigen (9). Expression of these genes endows HMLER cells with functional immortality and tumorigenic potential at high seeding number (>106 cells) (4). HMLER cells retain epithelial characteristics and sensitivity to various EMT-inducing stimuli (e.g., TGF-β1) and have historically been used to investigate the effects of single EMT-related genes on cancer stemness (1). We previously demonstrated that retroviral expression of FOXC2 endows these cells with metastatic potential and the properties of stem cells, including the ability to form mammospheres, resist conventional chemotherapeutic agents, and form tumors at limiting dilutions (103 cells) (4). We therefore generated FOXC2-expressing HMLER cells (designated FOXC2-HMLER throughout) using retroviral transgene delivery, with the isogenic HMLER cell line as a control (4).
We established a high throughput screening assay in which each cell line was plated at 1,000 cells per well in 384-well plates, treated for 72 h with 2 μM compound and viability of each line determined in parallel by Cell Titer Glo (Promega) luminance measurements (Fig. 1A). We then screened a library of ∼50,000 diverse heterocyclic compounds and biologically active small molecules for cytotoxic activity against both cell lines. A total of 71 compounds were identified, which decreased the viability of FOXC2-HMLER cells at least three Z scores from plate mean but did not alter the luminance signal of control HMLER cells one Z score from plate mean (Fig. 1B). Screening hits which passed this initial selectivity filter were then retested in eight-point dose–response format (8 nM to 20 μM) to determine cytotoxic indices. This led to the identification of four compounds that selectively inhibit the growth of FOXC2-HMLER cells with submicromolar half maximal inhibitory (IC50) concentrations and are generally nontoxic to HMLER cells at concentrations up to 20 μM (Fig. 1 C and D and SI Appendix, Fig. S1). For comparison, the commonly used chemotherapy drug doxorubicin (Adriamycin) displays modest preferential toxicity to control HMLER cells (IC50 267 nM) relative to FOXC2-HMLER cells (IC50 447 nM, Fig. 1E). One cinnoline-containing compound, which we termed FiVe1 for FOXC2-inhibiting Vimentin effector 1, was chosen for further study due to its selective cytotoxicity (FOXC2-HMLER IC50 234 nM; HMLER IC50 > 20 μM) as well as its perceived chemical tractability and novelty (Fig. 1 C and D). Importantly, FiVe1 was found to inhibit the growth of three additional mesenchymal breast cancer cell lines, SNAIL-HMLE, MDA-MB-231, and SUM159, which express FOXC2 from its endogenous locus, suggesting FiVe1’s antiproliferative effects are not restricted to the HMLER system (4) (Fig. 1F).
Fig. 1.
A high throughput screen identifies FiVe1 as a selective inhibitor of FOXC2-expressing breast cancer cells. (A) Schematic representation of the high throughput screening platform used to identify compounds selectively toxic to FOXC2-HMLER cells. (B) Scatterplot of primary screening data displaying plate normalized Z scores of viability for FOXC2-HMLER cells versus control HMLER cells. Contents of red box indicate primary screening hits culled for further analysis. (C) Relative viability measurements of FOXC2 or control HMLER cells exposed to the indicated doses of doxorubicin for 72 h (n = 3, mean and SEM). (D) Structure of FiVe1. (E) Relative viability measurements of FOXC2 and control HMLER cells exposed to the indicated doses of FiVe1 for 72 h (n = 3, mean and SEM). (F) Relative viability measurements of the indicated FOXC2-expressing breast cancer cells after 72-h treatment with FiVe1 (n = 3, mean and SEM).
FiVe1 Inhibits the Morphological Changes Associated with EMT.
One of the distinguishing characteristics separating mesenchymally transformed cells from their epithelial counterparts is a spindle-shaped, fusiform morphology (10). These morphological differences are due, in part, to the loss of the adherens junctions as a result of suppressed E-cadherin transcription and to the acquisition of the intermediate filament protein vimentin, which affords structural integrity and motility to the transformed cell (10). We observed that submicromolar doses of FiVe1 were capable of converting most FOXC2-expressing breast cancer cell types to a more epithelial morphology within 6 h (SI Appendix, Fig. S2). Similarly, treatment with 500 nM FiVe1 was also found to block the morphological changes associated with TGF-β1–induced EMT in MCF10A and EPh4Ras cells (SI Appendix, Fig. S3A). Immunofluorescent staining of MCF10A cells treated with FiVe1 and TGF-β1 indicated that FiVe1 did not modulate the levels of E-cadherin or FOXC2, although the staining pattern of the mesenchymal marker vimentin was altered (SI Appendix, Fig. S3 B and C). Additionally, Western blot analyses of SNAIL-HMLE and FOXC2-HMLER cells indicated that FiVe1 treatment did not alter the expression levels of FOXC2, vimentin, or E-cadherin in these cell types (SI Appendix, Fig. S3D). Together, these results suggested that FiVe1 likely targets a pathway responsible for the maintenance of mesenchymal morphology but does not induce growth arrest by altering the EMT transcriptional program.
FiVe1 Irreversibly Inhibits Properties Associated with Cancer Stemness.
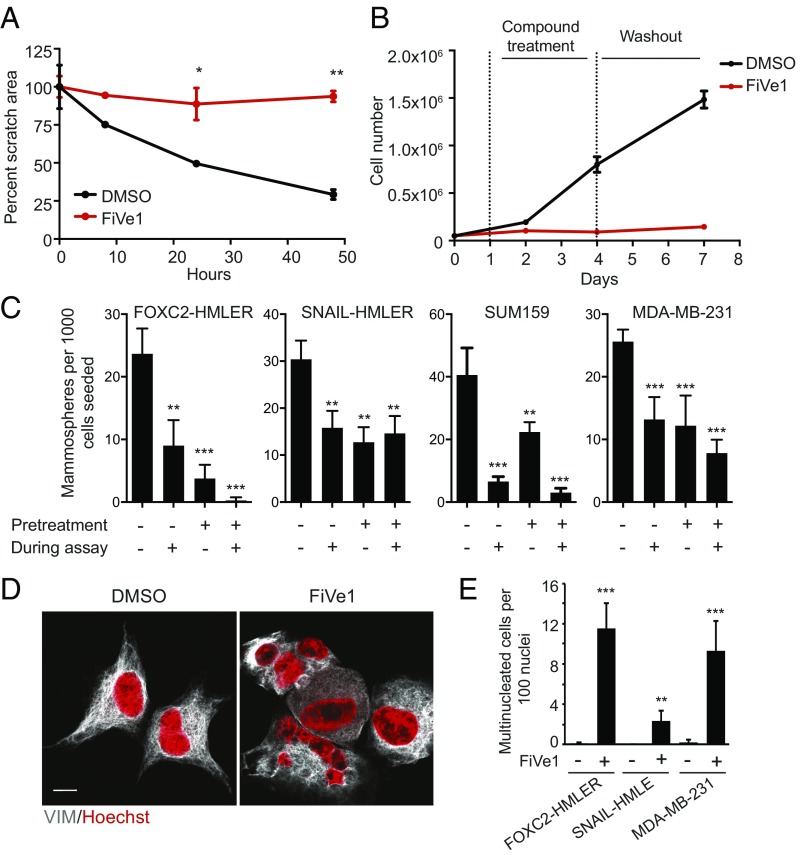
Passage of cancer cells through EMT endows them with migratory potential, a property which is thought to contribute to their ability to disseminate from the primary tumor and form metastases (11). To determine whether FiVe1 could inhibit the migration of mesenchymally transformed cells we employed the use of a quantitative scratch assay, which has been used as an in vitro surrogate to monitor wound healing and the migratory potential of cells (12, 13). Monitoring relative scratch area from a monolayer of FOXC2-HMLER cells indicated that 1-μM FiVe1 treatment fully inhibited wound closure over a period of 48 h (Fig. 2A).
Fig. 2.
FiVe1 irreversibly inhibits stemness-associated properties in mesenchymally transformed breast cancer cells. (A) Quantification of wound closure in an in vitro scratch assay with FOXC2-HMLER cells (n = 3, mean and SD, FiVe1, 1 μM). (B) Growth curves of FOXC2-HMLER cells treated with 1 μM FiVe1 during days 1–4 as noted by dashed vertical lines (n = 3, mean and SD). (C) Quantification of mammospheres from an in vitro mammosphere formation assay with the indicated cell types. Pretreatment indicates a 72-h treatment with 1 μM FiVe1 before plating in mammosphere formation conditions. During assay indicates 1 μM compound treatment during the mammosphere assay (14 d) (n = 3, mean and SD). (D) Representative images of FOXC2-HMLER cells treated for 24 h with DMSO or 500 nM FiVe1 and then stained with DAPI and anti-VIM antibodies. (Scale bar, 10 μm.) (E) Quantification of multinucleation (n > 3 nuclei per cell) in the indicated cell types treated with 500 nM FiVe1 for 24 h (n = 3, mean and SD; *P < 0.05, **P < 0.005, ***P < 0.0005; t test).
We then performed cell growth experiments to determine whether the cytotoxic effects of FiVe1 were reversible. Treating FOXC2-HMLER cells seeded at low density (5,000 cells/cm2) with 1 μM FiVe1 was sufficient to fully repress cell growth over a period of 72 h (Fig. 2B). Interestingly, after compound washout, these cells were unable to proliferate past the initial seeding density, indicating that the effects of FiVe1 were irreversible (Fig. 2B).
EMT induction endows breast cancer cells with the ability to grow as mammospheres in culture (1). This assay principle is based on studies by Dontu et al., demonstrating that only undifferentiated mammary stem cells can survive in suspension culture, whereas differentiated mammary epithelia die as a consequence of anoikis (14). FiVe1 treatment inhibited the ability of the FOXC2-expressing lines FOXC2-HMLER, SNAIL-HMLER, SUM159, and MDA-MB-231 cells to form mammospheres in culture (Fig. 2C). We additionally tested whether pretreating cells for 72 h with 1 μM FiVe1 was sufficient to inhibit their ability to form mammospheres. Indeed, pretreating these cell lines before plating resulted in similar levels of growth inhibition compared with conditions in which FiVe1 was present throughout the duration of the assay (14 d), further confirming that FiVe1 irreversibly inhibits the stemness potential of these cell types (Fig. 2C).
FiVe1 Targets the Intermediate Filament Protein Vimentin.
We next determined whether the mechanism by which FiVe1 inhibits cell growth is through programmed apoptotic cell death. Although prolonged treatment of FOXC2-HMLER cells with FiVe1 induced low levels of cleaved caspase 3 staining as monitored by immunofluorescent staining, FiVe1 treatment was not found to significantly induce caspase 3/7 activity (Caspase-Glo, Promega) at concentrations up to 40 μM (SI Appendix, Fig. S4 A and B). Additionally, FiVe1 treatment did not induce an observable increase in the cleaved protein content of the downstream caspase substrate poly ADP ribose polymerase (PARP) in FOXC2-HMLER cells as monitored by Western blotting (SI Appendix, Fig. S4C). In contrast, the pan-kinase inhibitor staurosporine was found to induce both caspase 3/7 activity and PARP cleavage at submicromolar doses in FOXC2-HMLER cells (SI Appendix, Fig. S4 B and C). Lastly, we found that the pan-caspase inhibitor Z-VAD(OMe)-FMK was unable to rescue FiVe1-induced cell growth arrest, indicating the compound’s primary mechanism of action does not involve induction of apoptosis (SI Appendix, Fig. S4D).
To understand how FiVe1 might irreversibly inhibit cell growth, we performed imaging experiments to examine whether the nuclear architecture is altered in response to compound treatment. We found that 24-h treatment of FOXC2-HMLER cells with FiVe1 (250 nM) resulted in the appearance of irregularly shaped nuclei and multinucleation, with some cells containing up to seven observable nuclei (Fig. 2 D and E). The FOXC2-expressing, mesenchymally transformed cell lines SNAIL-HMLE and MDA-MB-231 were also found to exhibit multinucleation phenotypes in response to FiVe1 over this treatment period (Fig. 2E). Multinucleation and mitotic failure have been reported as morphological outcomes from inhibiting essential mitosis-related kinases (e.g., Aurora kinases) (13). To determine whether FiVe1 inhibited any cell cycle or cytokinesis-related kinases, we profiled FiVe1 against a panel of ∼100 kinases at 1 and 10 μM (SI Appendix, Table S1). FiVe1 inhibited none of the kinases tested.
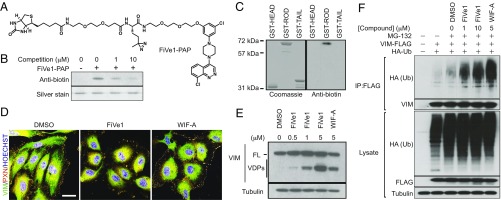
We next generated a photo-activatable affinity probe (PAP) molecule for target identification experiments. A structure activity relationship study of FiVe1 suggested that the cinnoline heterocycle is intolerant to modification and crucial for potency and selectivity. In contrast, the 3-chloro-phenyl moiety was found to be tolerant of bulky substitutions at the five position. Based on this information, we synthesized a diazirine and biotin-containing photoactivatable affinity probe compound, termed FiVe1-PAP, which retained selective cytotoxic activity to FOXC2-HMLER cells (Fig. 3A and SI Appendix, Fig. S5A). We found that a synthetic intermediate to FiVe1-PAP, ADP-2341, displayed enhanced solubility with respect to FiVe1 and retained potent and selective cellular activity in proliferation assays (SI Appendix, Fig. S5 B and C). To identify the relevant cellular target of FiVe1, we incubated live FOXC2-HMLER cells with 2.5 μM FiVe1-PAP for 15 min and subjected these cells to UV irradiation. A sample containing both FiVe1-PAP and a 20-fold molar excess of free FiVe1 was used to distinguish specific labeling events. Surprisingly, ammonium sulfate fractionation of the labeled lysate followed by Western blotting for biotin revealed an abundant band in the 20% fraction whose intensity was dramatically increased in the presence of FiVe1 competition (SI Appendix, Fig. S5D). To ensure this observation was not due to compound aggregation and nonspecific labeling, we performed the same experiment using the soluble analog ADP-2341 as a competitor and observed a similar increase in labeling of an ∼50-kDa protein (SI Appendix, Fig. S5D). One possible explanation for this observation is that the large molar excess of free competitor alters the structure of a protein complex to make it amenable to labeling by the lower concentrations of the affinity probe. LC-MS/MS analysis of this band indicated that the protein present was the intermediate filament protein VIM. We confirmed that VIM was present in the 20% ammonium sulfate fraction by Western blotting and that this band overlaid with anti-biotin Western blots and the Coomassie-stained band that was sent for MS-based analysis (SI Appendix, Fig. S5E).
Fig. 3.
Vimentin is the relevant cellular target of FiVe1. (A) Structure of the photoactivatable affinity probe FiVe1-PAP. (B) Anti-biotin Western blot analysis of recombinant VIM protein exposed to in vitro crosslinking conditions. (FiVe1-PAP, 50 nM; competition, ADP-2341.) (C) Anti-biotin Western blot analysis of the indicated GST-VIM fusion proteins exposed to in vitro crosslinking conditions (FiVe1-PAP, 20 nM). (D) Maximum intensity projections from confocal imaging of vimentin (VIM) and paxillin (PXN) immunostained HUVECs after treatment with 1 μM of the indicated compounds for 1 h. (Scale bar, 20 μm.) (E) Western blot analysis of VIM degradation in FOXC2-HMLER cells after 4 h of treatment with the indicated compounds (FL, full-length VIM; VDPs, VIM degradation products; WIF-A, withaferin A). (F) Western blot analysis of anti-FLAG immunoprecipitated protein content from HEK293T cells transfected with VIM-FLAG and HA-Ub constructs after treatment with the indicated compounds for 4 h.
We then showed that FiVe1-PAP labels recombinant full-length VIM in vitro and that this binding could be competed with a molar excess of ADP-2341 (Fig. 3B). VIM consists of a long rod domain, which forms a coiled coil in the presence of other VIM molecules, as well as head and tail domains, which are thought to be unstructured (15, 16) (SI Appendix, Fig. S5F). To identify which portion of the protein was the relevant site of FiVe1-PAP labeling, we expressed these domains as GST fusion proteins. In vitro labeling experiments suggested that FiVe1-PAP binds specifically and potently to the rod domain of VIM (Fig. 3C). VIM is one of four type III intermediate filament proteins, whose members additionally include peripherin, glial fibrillary acidic protein (GFAP), and desmin, which share 59%, 61%, and 62% sequence similarity to VIM, respectively (SIM alignment, ExPASy) (17). In vitro labeling experiments with recombinant preparations of these proteins revealed FiVe1-PAP labeled VIM exclusively, indicating the specificity of this interaction relative to closely related filamentous proteins (SI Appendix, Fig. S5G).
Withaferin A (WIF-A), a steroidal lactone natural product derived from Withania somnifera, has been previously reported as a potent inhibitor of cancer growth (18, 19) (SI Appendix, Fig. S5H). Early mechanism of action studies suggested that WIF-A inhibits the activities of VIM through covalent modification of the single cysteine residue present on the rod domain of the protein (20). WIF-A treatment was found to be selectively cytotoxic to FOXC2-HMLER cells with an ∼10-fold cytotoxic index relative to control cells, confirming the notion that targeting VIM is lethal to mesenchymally transformed cells (SI Appendix, Fig. S5I). We reasoned that toxicity to both cell lines at doses above 200 nM might be explained by WIF-A’s reported off-target inhibitory activities, which include the covalent modification of GFAP, β-tubulin, NF-κB, and Sp1 (21–23). Although these off-target interactions indicate WIF-A is unsuitable as a selective chemical probe of VIM, we nevertheless used it as a positive control in certain assays for which WIF-A has been previously reported to inhibit VIM function.
WIF-A is thought to induce cell death of VIM-expressing cancer cells, at least in part, by inducing filamentous network collapse and degradation of VIM (18, 19). To monitor the assembly status of the VIM filamentous architecture, we performed immunofluorescent analyses of FiVe1- and WIF-A–treated human umbilical endothelial cells (HUVECs), which have been shown to display a high elaborated VIM architecture sensitive to WIF-A treatment (20). These studies revealed that 1-h treatment with FiVe1 or WIF-A promoted the destruction of fine VIM-containing filamentous structures and the contraction of the VIM apparatus relative to the boundary of the cell (Fig. 3D). Additionally, we observed that WIF-A and FiVe1 induced rapid morphological changes in the appearance of FOXC2-HMLER cells as determined by immunofluorescent analysis for VIM staining, a result consistent with the idea that the previously described morphological changes are due to VIM reorganization (SI Appendix, Fig. S5J). Further, treating FOXC2-HMLER cells with FiVe1 dramatically reduced the appearance of dibromobimane-crosslinked dimeric VIM protein content by Western blotting, an assay which has previously been used to monitor the filamentous status of VIM in cells and in solution (SI Appendix, Fig. S5K) (24). Additionally, treatment of FOXC2-HMLER cells with FiVe1 or WIF-A induced the dose-dependent accumulation of lower molecular weight VIM degradation products as visualized by Western blotting (Fig. 3E). WIF-A has also been reported to induce the degradation of VIM protein by a ubiquitin proteasome-mediated mechanism (20). Analyzing the protein content of FLAG-immunoprecipitated VIM from HEK293T cells, we observed a clear accumulation of high molecular weight ubiquitinated VIM species consistent with the idea that both compounds trigger the degradation of VIM through an ubiquitin-mediated mechanism (Fig. 3F).
Specific VIM Targeting Results in Mitotic Catastrophe.
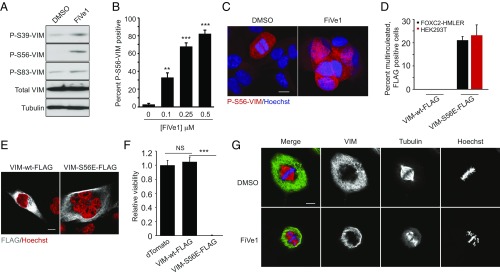
The VIM filamentous network is anchored to the nucleus and forms a cage-like structure around dividing nuclei and the mitotic spindle (25–27). For the appropriate segregation of nuclei to daughter cells during mitosis, VIM undergoes a series of sequential phosphorylation events by cell cycle- and cytokinesis-related kinases which promote its filamentous disassembly (28–31). Studies by Inagaki and colleagues have demonstrated that expression of certain phosphorylation-incompetent (Ser to Ala) VIM mutants results in multinucleation and aneuploidy in vitro and in vivo (29, 32). We hypothesized that FiVe1 might similarly interfere with this sequence of VIM phosphorylation and reorganization events during mitosis. We therefore performed Western blotting analysis of FOXC2-HMLER cells treated for 24 h with FiVe1 using commercially available antibodies targeting specific phosphorylated VIM species (P-S39, P-S56, and P-S83). We found that FiVe1 treatment induced a modest increase in P-S39– and P-S83–VIM protein content but led to a marked increase in the steady-state levels of P-S56–VIM (Fig. 4A). Immunofluorescent staining experiments for P-S56–VIM corroborated this observation, with FiVe1 treatment resulting in a concentration-dependent increase in P-S56–VIM positive FOXC2-HMLER cells (Fig. 4B). In contrast, only a small fraction of cells undergoing mitosis stained positive for P-S56–VIM in DMSO-treated controls, consistent with previous reports identifying phosphorylation at S56 as a modification catalyzed by cyclin-dependent kinase 1 (CDK1) (Fig. 4C) (31). To determine whether this increase in P-S56–VIM protein content might be responsible for FiVe1’s ability to induce multinucleation, we performed transient overexpression experiments with vectors encoding FLAG-tagged wild-type VIM (VIM-WT-FLAG) or a phosphomimetic mutant of VIM at S56 (VIM-S56E-FLAG) in FOXC2-HMLER and HEK293T cells. Seventy-two-hour expression of the VIM-S56E-FLAG transgene was found to induce multinucleation in both cell lines (Fig. 4 D and E). We additionally overexpressed these transgenes by stable lentiviral delivery in FOXC2-HMLER cells. Whereas VIM-WT-FLAG–expressing cells grew similarly to dTomato-expressing control cells over 7 d, VIM-S56E-FLAG cells were found to be replication incompetent over this period (Fig. 4F). Together, these results suggest that a specific, sustained phosphorylation modification on VIM is sufficient to recapitulate the activity of FiVe1.
Fig. 4.
Specific engagement of VIM by FiVe1 induces defects in chromosomal alignment during metaphase. (A) Western blotting analysis of phosphorylated VIM protein content from FOXC2-HMLER cells treated for 24 h with FiVe1 (500 nM). Quantification (B) and representative images (C) of immunofluorescent staining for P-S56-VIM from FOXC2-HMLER cells treated with the indicated doses of FiVe1 (n = 3, mean and SD). (D) Quantification of multinucleated, FLAG positive FOXC2-HMLER or HEK293T cells 72 h after transfection with the indicated VIM expression constructs (n = 3, mean and SD). (E) Representative images of FOXC2-HMLER cell immunostained for FLAG protein content 72 h after transfection with the indicated VIM expression constructs. (Scale bar, 10 μm.) (F) Relative viability measurements of FOXC2-HMLER cells 7 d after transduction with lentiviruses encoding the indicated overexpressed transgene products (n = 8, mean and SD). (G) Representative confocal images of thymidine-synced FOXC2-HMLER cells at metaphase immunostained for VIM and β-tubulin (TUB) (FiVe1, 250 nM). (Scale bar, 10 μm.) (**P < 0.005, ***P < 0.0005; NS, not significant; t test).
Previously reported, phosphorylation-incompetent VIM mutants have been demonstrated to induce multinucleation by physically interfering with the process of cytokinesis as evidenced by VIM positive, intermediate filament bridges connecting daughter cells (29). Given FiVe1 induces a hyperphosphorylated VIM phenotype, we sought to determine whether FiVe1’s engagement of VIM might interfere at an alternative stage of mitosis. We therefore performed time course confocal imaging studies of FiVe1-treated FOXC2-HMLER cells after their release from thymidine blocking-based cell cycle synchronization. While FiVe1-treated cells were found to condense their chromosomes during anaphase normally, FiVe1-treated cells exhibited a number of altered phenotypes during metaphase. Consistent with our previous observations, FiVe1 treatment resulted in a collapsed VIM filamentous structure, which appeared more closely associated to mitotic spindle poles compared with DMSO-treated controls (Fig. 4G). Additionally, we observed that FiVe1 treatment inhibited the ability of chromosomes to align to the metaphase plate, frequently occupying regions distal to the spindle pole (Fig. 5G). We further observed that FiVe1 treatment disrupted the ability of β-tubulin to faithfully form the fine spindle microtubules of the mitotic spindle, instead resulting in a compressed phenotype with radiating projections connecting unaligned chromosomes (Fig. 5G). To assess the uniqueness of this phenotype, we next evaluated the metaphase phenotypes of other chemical inhibitors which have been shown to alter mitotic progression. These included inhibitors of centromere-associated protein E (CENP-E, also called kinesin-7, GSK92395), kinesin spindle protein (KSP, also called Eg5, Ispinesib), polo-like kinases (PLK1/2/3, BI2536), and Aurora kinases (AURKA/B/C, VX-680). Treating thymidine-synced FOXC2-HMLER cells with these inhibitors revealed that the majority of these compounds did not induce phenotypes similar to FiVe1, with the exception of GSK92395, which also demonstrated an unaligned chromosome phenotype in line with CENP-E’s reported function of aligning chromosomes to the metaphase plate (33) (SI Appendix, Fig. S6). However, GSK92395 treatment did not induce VIM filamentous collapse or alter the morphology of spindle tubulin. Taken together, these results suggest that FiVe1 induces mitotic failure and eventual multinucleation through a mechanism which involves interfering with the metaphase organization of chromosomes and the spindle apparatus.
Fig. 5.
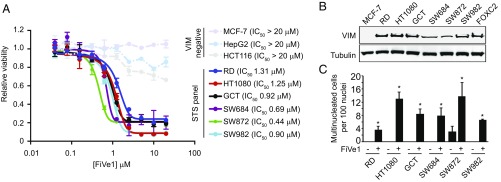
FiVe1 inhibits the growth of soft tissue sarcoma (STS) cell lines. (A) Relative proliferation measurements of the indicated VIM negative and STS cell lines treated with a concentration response of FiVe1 (n = 3, mean and SEM). (B) Western blotting analysis for VIM protein levels across the STS panel (FOXC2 = FOXC2-HMLER). (C) Multinucleation analysis of the indicated STS lines treated for 24 h with 1 μM FiVe1 (n = 3, mean and SD; *P < 0.05, t test).
We next evaluated whether targeting VIM by WIF-A might promote similar defects in mitotic progression. WIF-A induced a significant increase in multinucleated FOXC2-HMLER cells at concentrations (250–500 nM) below those at which it induced an apoptotic, nonadherent phenotype during this treatment period (24 h, SI Appendix, Fig. S7A). Additionally, treating thymidine synched FOXC2-HMLER cells with WIF-A resulted in the characteristic appearance of unaligned chromosomes during metaphase, although not all metaphase plates displayed an unaligned phenotype as observed with a maximally efficacious dose of FiVe1 (SI Appendix, Fig. S7 B and C). While differences in target engagement or off-target modification might explain this difference, together these data indicate that chemical targeting of VIM likely represents a generalizable path to interfere with mitotic progression.
FiVe1 Target and Phenotype Specificity.
To confirm that the multinucleation and VIM reorganization phenotypes of FiVe1-treated cells were responsible for the cytotoxic activity of this compound series, we investigated the activity of three closely related FiVe1 analogs which did not inhibit FOXC2-HMLER growth at concentrations up to 20 μM (SI Appendix, Fig. S8 A and B). None of these compounds were found to induce morphological changes in FOXC2-HMLER cells and were not observed to induce VIM reorganization in HUVECs at concentrations 10-fold higher than used for FiVe1 (SI Appendix, Fig. S8 C and D). Additionally, these compounds were not found to promote the accumulation of VIM degradation products or induce multinucleation in FOXC2-HMLER cells (SI Appendix, Fig. S8 E and F). Consistent with the hypothesis that FiVe1 functions solely through a VIM-dependent mechanism, cancer cell lines (HCT116, HepG2, and MCF-7), which do not express VIM (34) (SI Appendix, Fig. S9 A and B), were found to be largely insensitive to FiVe1 at concentrations up to 20 μM (Fig. 5A). Conversely, VIM overexpression sensitized MCF-7 cells to the antiproliferative effects of FiVe1 (SI Appendix, Fig. S9 C and D). Additionally, in contrast to the CENP-E inhibitor GSK92395, treating HCT116, HepG2, and MCF-7 cells with FiVe1 did not result in the characteristic appearance of unaligned chromosomes during metaphase in these VIM-negative cell lines (SI Appendix, Fig. S9E).
FiVe1 Inhibits the Growth of Soft Tissue Sarcoma Cell Lines.
We next determined whether FiVe1 might similarly inhibit the in vitro growth of other VIM-expressing cancer types in addition to mesenchymally transformed breast cancer cells. We therefore assessed the antiproliferative activity of FiVe1 against a selection of soft tissue sarcomas (STSs), which collectively refers to a broad grouping of over 50 subtypes of connective tissue-derived cancers for which targeted therapies are lacking (35). STS tumors arise from tissues of mesenchymal origin and, by default, express VIM irrespective of subtype (18). We evaluated a panel of six STS cell lines, which included the histological subtypes of fibrosarcoma (HT1080 and SW684), rhabdomyosarcoma (RD), fibrous histiocytoma (GCT), liposarcoma (SW872), and synovial sarcoma (SW982). We first confirmed that all STS cell lines expressed VIM at similar levels to FOXC2-HMLER cells by Western blotting analysis (Fig. 5B). FiVe1 was found to effectively inhibit the growth of all cell lines with similar IC50 values (0.44–1.31 μM, Fig. 5A). FiVe1 additionally promoted a robust multinucleation phenotype in all STS cell lines over a 24-h treatment period (Fig. 5C). Interestingly, we found that SW872 cells, a liposarcoma cell line that displayed a significant degree of basal multinucleation, were most sensitive to FiVe1 treatment (IC50 0.44 μM), indicating that existing mutations promoting genomic instability might serve as predictive markers of FiVe1 sensitivity. Together, these results indicate that FiVe1’s mechanism involving VIM-dependent mitotic catastrophe is broadly relevant to other cancers of mesenchymal origin. Unfortunately, the short half-life of FiVe1 due to P450-mediated clearance precluded xenograft rodent studies.
VIM is expressed broadly in mesenchymal tissues and has been demonstrated to be a contributing factor in normal physiological processes such as wound healing and neovascularization (19, 36). To determine whether FiVe1 treatment was toxic to noncancerous, primary VIM-expressing cells, we performed cell growth experiments with HUVECs and human lung fibroblasts (HLFs) treated with FiVe1. FiVe1 was found to inhibit the growth of HUVECs (IC50 1.70 μM) and HLFs (IC50 2.32 μM) with low micromolar potency in 72-h proliferation assays (SI Appendix, Fig. S10A). We additionally found that FiVe1 could fully inhibit cell growth in low cell density conditions, although a 10-fold increase in concentration was necessary to observe full growth inhibition (10 μM vs. 1 μM for FOXC2-HMLER, SI Appendix, Fig. S10 B and C). In contrast to our studies with FOXC2-HMLER cells, washout experiments indicated the cytotoxic effects of FiVe1 were reversible in HUVEC and HLFs (SI Appendix, Fig. S10 B and C). Interestingly, FiVe1 treatment was not found to induce multinucleation (n ≥ 3) in HUVECs, but did induce multinucleation in HLFs to a lesser degree (∼2%) compared with our results with other cancer cell lines (SI Appendix, Fig. S10D). We observed the majority of HUVECs and HLFs treated with FiVe1 to be arrested in a binucleated state, consistent with the idea that normal cells might halt cell division in response to mitotic perturbation. Future experiments with additional normal, mesenchymal cell types will be needed to substantiate these observations and to elucidate the molecular basis for the selectivity of this phenotype.
Discussion
Using a high throughput synthetic lethal screen, we have identified a series of compounds which selectively inhibit the growth of FOXC2-expressing HMLER cells. The study of one such molecule, FiVe1, demonstrated that it selectively and irreversibly inhibited the growth of multiple FOXC2-expressing breast cancer cell lines. Target identification studies with FiVe1 indicated that it targets the type III intermediate filament protein VIM, which is broadly expressed in mesenchymal tissues and is frequently used as a marker for cells having undergone an EMT (35). VIM expression in post-EMT cells endows these populations with their characteristic fusiform morphology as well as structural integrity after loss of epithelial cell–cell junctions (35). We demonstrated that FiVe1 treatment promotes the collapse of VIM architecture and promotes VIM degradation via a ubiquitin–proteasome-dependent mechanism, rapidly promoting morphological rearrangement to a more epithelium-like state in mesenchymally transformed cells. VIM filaments undergo constant dynamic remodeling, which endows VIM-expressing cells with motility, contributing to their metastatic capacity (37, 38). We demonstrated that FiVe1 fully inhibited VIM-dependent cellular motility in scratch assays. VIM knockdown in post-EMT cells has been demonstrated to reorganize cellular morphology and inhibit migratory capacity, underscoring the notion that targeting VIM can antagonize EMT-induced CSC traits (39).
In addition to its role as a structural protein, others have demonstrated that VIM serves as a communication hub for multiple signaling pathways (37, 38). VIM directly binds to a number of cell signaling-associated kinases, including phosphorylated ERK and AKT1, an event which facilitates their enzymatic activity while shielding from degradative enzymes (38). Additionally, VIM makes key contacts with focal adhesions through vimentin matrix adhesions, which are thought to modulate the interaction between certain integrins and their ligands (37). Further, VIM associates with a number of cell stress-related proteins including HSP90, 14–3-3, and p53 (37, 38). We cannot exclude the possibility that FiVe1 may alter VIM’s interactions with these molecules in a mode which contributes to the observed antiproliferative efficacy of the compound. Clearly, future investigation with FiVe1 will be necessary to fully understand the potentially pleiotropic consequences of targeting VIM. Nonetheless, our data suggest that FiVe1 will serve as a valuable chemical probe to assess the consequences of VIM assembly in these contexts.
We found that FiVe1 induced irreversible cell cycle arrest in FOXC2-HMLER cells as a result of inducing multinucleation, an observation in line with previous reports, demonstrating that multinucleation-inducing compounds can elicit irreversible effects on cell proliferation (40). While a loss of mesenchymal morphology and migratory capacity could be explained by inducing collapse of the VIM filamentous structure, multinucleation had not been observed for the previously reported VIM targeting compound WIF-A. Like other intermediate filaments, VIM undergoes a series of carefully orchestrated phosphorylation events, which promote its filamentous disassembly for the segregation of nuclei to daughter cells during cell division (41). These phosphorylation steps are catalyzed sequentially throughout mitosis, starting in prometaphase by the activity of CDK1 and ending in anaphase with Aurora-B– and Rho-kinase–catalyzed phosphorylation (41). Expression of specific or combined point mutations (Ser to Ala) of VIM on these phosphorylation sites induces multinucleation and the accumulation of VIM filamentous bridges connecting daughter cells, suggesting that the inability of VIM filaments to disassemble physically blocks the segregation of nuclei during cytokinesis (41). In contrast, FiVe1 treatment resulted in the steady-state hyperphosphorylation at S56. Additionally, the overexpression of phosphomimetic mutants of VIM at this position (S56E) were found to recapitulate the phenotype induced by FiVe1, resulting in multinucleation and cell cycle arrest, together indicating that inadequate or excessive phosphorylation of VIM is sufficient to induce mitotic defects. Whether FiVe1 induces additional phosphoryl modifications on VIM on positions unavailable to probe with commercial antibodies will be the work of future studies. Our imaging experiments additionally suggested FiVe1 induced unique metaphase abnormalities, including defects in chromosomal alignment and tubulin rearrangement. VIM filaments are transported on microtubules through the activities of conventional kinesin and cytoplasmic dynein (42, 43). Given the similarity of FiVe1’s metaphase phenotype to that of the kinesin inhibitor GSK92395, we speculate that FiVe1 may alter previously undescribed interactions between VIM and the mitotic kinesins responsible for appropriate spindle assembly and chromosomal alignment; however, further studies will be necessary to determine which of VIM’s interactors are disrupted to result in these phenotypes. Our studies indicate that FiVe1 intercedes at an earlier stage of the cell cycle (i.e., metaphase) than reported VIM mutants, indicating that appropriate phosphorylation of VIM is necessary throughout mitosis for fidelitous cell division, not only at cytokinesis as earlier studies would suggest. In contrast to the various compounds targeting microtubule polymerization (e.g., taxol, colchicine, nocodazole), to our knowledge, no small molecule binding an intermediate filament protein has been shown to induce mitotic failure, indicating that targeting VIM represents a previously undescribed mechanism for inhibiting cell growth.
STSs are a rare and heterogeneous class of tumors, grouped solely by their mesenchymal origin (36). Current standards of care for STS patients are currently limited to surgical resection and treatment with typical DNA-damaging chemotherapies (e.g., doxorubicin, ifosfamide) or radiation, interventions which yield 5-y median survival rates of only 50% for late-stage STS patients (44). Additionally, only a handful of targeted therapies have been approved or are under investigation for STS indications (45). Clearly, the identification of novel drugs targeting mechanisms conserved across multiple sarcoma subtypes would have a significant clinical impact in these populations. We found that FiVe1 treatment inhibited the growth of six STS cell lines of diverse histological origins, suggesting that targeting VIM not only inhibits the growth of post-EMT CSCs but also broadly inhibits the growth of mesenchymal cancers. Unlike other mitosis-inhibiting drugs (e.g., taxol) which target filamentous proteins present in all cells, FiVe1 induces mitotic defects only in VIM-expressing cells. Given’s VIM’s restriction to mesenchymal tissues, we speculate that targeting VIM would likely induce fewer side effects than conventional chemotherapies, which have the potential to induce off-target toxicity to rapidly dividing epithelial tissues. Our current efforts are aimed at screening a broader panel of STS cell lines to identify whether certain subtypes and/or genotypes display enhanced sensitivity to FiVe1. Additionally, we are undertaking a medicinal chemistry campaign to improve the potency and pharmacological properties of the FiVe1 scaffold to assess the translational potential of targeting VIM as therapy for mesenchymal cancers.
Methods
Chemicals.
Doxorubicin was from Thermo Fisher Scientific. Withaferin A and staurosporine were from Cayman Chemical. FiVe1 (initially E906-0461), F727-0225, F646-0707, and E896-1088 were purchased as powders from ChemDiv. After initial confirmation of cellular activity, FiVe1 was resynthesized in house and this material was used for all in vitro experiments. The syntheses of FiVe1, FiVe1-PAP, and ADP-2341 are described in SI Appendix. All commercially obtained chemicals were dissolved in DMSO and used without further purification.
Cell Lines.
The propagation conditions and generation of FOXC2-HMLER, HMLER, SNAIL-HMLER, SNAIL-HMLE, and SUM159 cells have been described previously (4). MCF-7, HepG2, HCT116, and MDA-MB-231 cells were from ATCC and maintained in DMEM (Corning) supplemented with 10% FBS (Corning) with Anti-anti (Gibco). The soft tissue sarcoma cell line panel was purchased from ATCC (TCP-1019) and maintained in the recommended medium for each cell type. HUVEC cells (pooled donor) were maintained in EBM-2 medium (both from Lonza Walkersville, Inc.). Primary human lung fibroblasts were maintained in fibroblast medium (both from Sciencell Research Laboratories). For cell growth experiments, HLFs were grown in DMEM supplemented with 2% FBS and Anti-anti. For all experiments, primary human cells were used at a passage no greater than 4.
High Throughput Screening and Miniaturized Cell Viability Experiments.
For high throughput screening, FOXC2-HMLER and HMLER cells were plated at 103 cells per well in white 384-well plates in 50 μL of MEGM medium (Lonza). Cells were allowed to attach for 1 h before compound was transferred to each well as a DMSO solution using a 100 nL pintool head affixed to a PerkinElmer FX instrument. After 72-h incubation, 30 μL of a Cell Titer Glo solution (diluted 1:6 in water; Promega) was dispensed into each well and luminance values were recorded using an Envision plate reader. Compounds which decreased viability of FOXC2-HMLER cells three Z scores below plate mean but did not decrease control HMLER viability more than one Z score below plate mean were deemed primary screening hits. Selected hit compounds were reordered and tested for selective toxicity in 10-point response assays as above. All other miniaturized, selective viability experiments were performed as above using the indicated growth medium.
Quantification of Multinucleation.
Cells were plated at 5 × 104 cells per well in 12-well dishes in their respective growth medium. After 24-h incubation, cells were treated with compound for an additional 24 h and then fixed in a 4% paraformaldehyde solution for 10 min. Cells were washed three times in DPBS and then stained with an anti-VIM antibody as above. Cells were costained with Hoechst 33342 (2 μg/mL) in DPBS for 1 h. Immunofluorescent images were recorded on a Nikon Eclipse Ti microscope at 100× magnification. Two images of separate imaging fields were recorded for three biological replicates of each condition reported. Cells containing three or more nuclei were counted manually and were considered multinucleated. Total nuclei per imaging field measurements were determined by a custom ImageJ (NIH) macro which utilized thresholding and counting functions within the software. Multinucleation counts are reported as total number of multinucleated cells (n > 3) per 100 nuclei. A total of 500–1,700 nuclei were typically analyzed per treatment condition.
Experiments with VIM Mutants.
The VIM-WT-FLAG transient overexpression plasmid was from Origene [pCMV6-Entry, National Center for Biotechnology Information (NCBI) ref. seq. NM_003380.2, RC201546]. The VIM-S56E-FLAG transient overexpression plasmid was generated from the above Origene vector with a Q5 Site-Directed Mutagenesis Kit (NEB) following manufacturer’s instructions. The VIM-WT-FLAG lentiviral expression vector was from Origene (pLenti, NCBI ref. seq. NM_003380.2, RC201546L1). The S56E lentiviral mutant plasmid was generated by subcloning the mutagenized ORF from the VIM-S56E-FLAG construct to pLENTI. Transient expression of transgenes was performed by transfecting 1 μg per 24-well plate using FuGeneHD (4:1, microliters of transfection reagent:micrograms of plasmid). Lentiviruses were generated in HEK293T cells using pSPAX2 and pMD2.G packaging vectors (Addgene plasmids 12260 and 12259). Lentiviruses were harvested from the supernatant from a 15-cm dish of confluent HEK293T, concentrated with a 30-kDa cutoff concentrator (Amicon), and then added to 105 FOXC2-HMLER cells per well in a 6-well plate. Cells were transduced for 48 h before trypsinization and replating in 96-well plates (2,000 cells per well) to analyze growth (Cell Titer Glo) over 7 d.
Washout Experiments.
For monolayer experiments with compound washout, the indicated cell type was plated at 5 × 104 cells per well in six-well dishes. Medium containing the indicated dose of FiVe1 was present days 1–4. Cells were washed three times with DPBS before replenishment with fresh medium without compound on day 4. At the specified time intervals, cells were exposed to a trypsin solution and then counted using a Countess automated cell counter (Invitrogen).
Confocal Microscopy.
For visualization of VIM reorganization, HUVECs were seeded at 5 × 104 cells per well on glass coverslips in 12-well plates and incubated for 24 h. Cells were treated for 1 h before fixation. For pS56-VIM staining, FOXC2-HMLER cells were seeded at 2.5 × 104 per well on laminin-coated coverslips in 12-well plates and incubated for 24 h. Cells were treated for an additional 24 h with the indicated dose of FiVe1. Cells were fixed with a 4% paraformaldehyde solution (Electron Microscopy Sciences) for 10 min. After three DPBS washes, cells were blocked for 30 min in 5% FBS, 0.3% Triton X-100 in DPBS. Cells were then stained with primary antibodies overnight at 4 °C in 1% BSA, 0.1% Triton X-100 in DPBS according to the dilutions in SI Appendix, Table S2. After three DPBS washes, coverslips were exposed to Alexa Fluor-conjugated secondary antibodies (1:500; Invitrogen) and Hoechst 33342 (2 μg/mL; Invitrogen) for 2 h at room temperature in the dark. After three DPBS washes, coverslips were mounted to glass slides using ProLong Gold Antifade mounting medium (Invitrogen). Z-stack images were acquired using a Zeiss confocal microscope with a 63× oil objective in the Scripps Core Microscopy Facility. Maximum intensity projections across multiple imaging planes were generated using ZEN Black software (Zeiss).
Mammosphere Formation Assays.
Mammosphere formation assays were performed as described previously (4). Mammospheres were allowed to form for 14 d. Mammospheres over 70 μm in diameter were quantified.
Western Blotting.
Western blotting was performed essentially as described previously (46). Cells were collected by brief trypsinization and centrifugation at 500 × g for 5 min. Cells were lysed by the addition of RIPA buffer with protease and phosphatase inhibitors (Roche). Lysates were incubated on ice for 30 min before eliminating insoluble protein content by centrifugation at 12,000 × g for 5 min at 4 °C. Protein concentrations were determined from absorbance values obtained by a NanoDrop instrument. Equal amounts of protein were then mixed with 2× loading buffer (100 mM Tris⋅HCl, 1% SDS, 10% glycerol, 0.1% bromophenol blue 10% mercaptoethanol) and exposed to 95 °C for 5 min. Protein was separated by SDS/PAGE using 4–12% Bis-Tris gels (Invitrogen) and then transferred to PVDF membranes (Invitrogen) using semidry transfer. Membranes were blocked for 1 h at room temperature in 5% nonfat dry milk in TBST (Tris buffered saline with 0.1% Tween 20). Membranes were incubated overnight with primary antibodies in blocking buffer overnight at the dilutions indicated in SI Appendix, Table S2. After three washes with TBST, membranes were then exposed to HRP-conjugated secondary antibodies (1:5,000 in blocking buffer; Sigma) for 1 h followed by 1 h of additional washing with TBST. Relative protein content was visualized using film and SuperSignal West Dura Substrate. For VIM degradation blots, 20–60 μg of protein content was loaded per gel lane and the anti-VIM antibody V9 (ab8069) was used to blot for VIM protein content. Phosphoprotein blots were performed as above with the exception that primary antibodies were incubated in 5% BSA in TBST instead of milk.
FiVe1 Target Identification.
Ten-centimeter dishes of confluent FOXC2-HMLER cells were exposed to 2.5 μM of FiVe1-PAP with the addition of either DMSO or 50 μM FiVe1 or 50 μM ADP-2341 as competition for 15 min at 37 °C. Plates were then UV crosslinked using a Stratalinker 2400 instrument for 10 min. RIPA lysates from UV-irradiated cells were then fractionated with ammonium sulfate in percent increments of 20. Individual fractions were separated via SDS/PAGE and relevant probe labeling was assessed by anti-biotin Western blotting performed as above. Anti-biotin Western blots were compared with a Coomassie blue-stained SDS/PAGE gel run in parallel. A relevant gel slice was excised and VIM identity was identified by LC-MS/MS performed by the Scripps Center for Metabolomics and Mass Spectrometry.
In Vitro Crosslinking Experiments.
Full-length, recombinant VIM, GFAP, desmin, and peripherin were purchased from Origene. For VIM fusion proteins, DNA inserts corresponding to the indicated VIM domains were ordered as codon optimized gBlocks from IDT and subcloned via Gibson assembly into pGEX-6P-3 (GE Life Sciences) for expression in BL21 cells. Expression was carried out for 16 h at 30 °C and proteins were purified using GST agarose (Sigma) in batch format according to manufacturer’s recommendations. Binding assays were performed by adding 1 μg of recombinant protein to 100 μL of 20 mM Tris-buffered water without NaCl. Compounds were delivered as 100× DMSO stocks such that the final concentration of DMSO was 1% for all samples. Reactions were incubated for 30 min at 37 °C and then UV irradiated for 10 min in a Stratalinker 2400 instrument. Reactions were mixed with an equal volume of 4× loading buffer before 35–50 μL of this solution was separated by SDS/PAGE and labeling determined by anti-biotin Western blotting as above. Silver staining was performed using the Pierce Silver Stain Kit according to supplied protocols.
Kinase Profiling.
Kinase profiling was performed at Nanosyn according to established in-house methods.
Supplementary Material
Acknowledgments
We thank Dr. Shoutian Zhu for insightful advice regarding target identification experiments. This work was supported by the Skaggs Institute for Chemical Biology and the National Cancer Institute of the NIH (Grant R01CA200970).
Footnotes
Conflict of interest statement: M.J.B., S.A.M., P.G.S., and L.L.L. are listed as inventors on a patent application related to the small molecules described in this manuscript.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716009114/-/DCSupplemental.
References
- 1.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.May CD, et al. Epithelial-mesenchymal transition and cancer stem cells: A dangerously dynamic duo in breast cancer progression. Breast Cancer Res. 2011;13:202. doi: 10.1186/bcr2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mani SA, et al. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci USA. 2007;104:10069–10074. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hollier BG, et al. FOXC2 expression links epithelial-mesenchymal transition and stem cell properties in breast cancer. Cancer Res. 2013;73:1981–1992. doi: 10.1158/0008-5472.CAN-12-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taube JH, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA. 2010;107:15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta PB, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 8.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 9.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye X, Weinberg RA. Epithelial-mesenchymal plasticity: A central regulator of cancer progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 12.Liang CC, Park AY, Guan JL. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 13.Tsuno T, et al. Inhibition of Aurora-B function increases formation of multinucleated cells in p53 gene deficient cells and enhances anti-tumor effect of temozolomide in human glioma cells. J Neurooncol. 2007;83:249–258. doi: 10.1007/s11060-007-9335-1. [DOI] [PubMed] [Google Scholar]
- 14.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrmann H, Bär H, Kreplak L, Strelkov SV, Aebi U. Intermediate filaments: From cell architecture to nanomechanics. Nat Rev Mol Cell Biol. 2007;8:562–573. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- 16.Meier M, et al. Vimentin coil 1A-A molecular switch involved in the initiation of filament elongation. J Mol Biol. 2009;390:245–261. doi: 10.1016/j.jmb.2009.04.067. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs E, Weber K. Intermediate filaments: Structure, dynamics, function, and disease. Annu Rev Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 18.Lahat G, et al. Vimentin is a novel anti-cancer therapeutic target; insights from in vitro and in vivo mice xenograft studies. PLoS One. 2010;5:e10105. doi: 10.1371/journal.pone.0010105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Grin B, et al. Withaferin a alters intermediate filament organization, cell shape and behavior. PLoS One. 2012;7:e39065. doi: 10.1371/journal.pone.0039065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bargagna-Mohan P, et al. The tumor inhibitor and antiangiogenic agent withaferin A targets the intermediate filament protein vimentin. Chem Biol. 2007;14:623–634. doi: 10.1016/j.chembiol.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bargagna-Mohan P, et al. Withaferin A targets intermediate filaments glial fibrillary acidic protein and vimentin in a model of retinal gliosis. J Biol Chem. 2010;285:7657–7669. doi: 10.1074/jbc.M109.093765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antony ML, et al. Growth arrest by the antitumor steroidal lactone withaferin A in human breast cancer cells is associated with down-regulation and covalent binding at cysteine 303 of β-tubulin. J Biol Chem. 2014;289:1852–1865. doi: 10.1074/jbc.M113.496844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heyninck K, Lahtela-Kakkonen M, Van der Veken P, Haegeman G, Vanden Berghe W. Withaferin A inhibits NF-kappaB activation by targeting cysteine 179 in IKKβ. Biochem Pharmacol. 2014;91:501–509. doi: 10.1016/j.bcp.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Pérez-Sala D, et al. Vimentin filament organization and stress sensing depend on its single cysteine residue and zinc binding. Nat Commun. 2015;6:7287. doi: 10.1038/ncomms8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maison C, Horstmann H, Georgatos SD. Regulated docking of nuclear membrane vesicles to vimentin filaments during mitosis. J Cell Biol. 1993;123:1491–1505. doi: 10.1083/jcb.123.6.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehto VP, Virtanen I, Kurki P. Intermediate filaments anchor the nuclei in nuclear monolayers of cultured human fibroblasts. Nature. 1978;272:175–177. doi: 10.1038/272175a0. [DOI] [PubMed] [Google Scholar]
- 27.Martys JL, Ho CL, Liem RK, Gundersen GG. Intermediate filaments in motion: Observations of intermediate filaments in cells using green fluorescent protein-vimentin. Mol Biol Cell. 1999;10:1289–1295. doi: 10.1091/mbc.10.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goto H, et al. Phosphorylation of vimentin by Rho-associated kinase at a unique amino-terminal site that is specifically phosphorylated during cytokinesis. J Biol Chem. 1998;273:11728–11736. doi: 10.1074/jbc.273.19.11728. [DOI] [PubMed] [Google Scholar]
- 29.Yasui Y, et al. Protein kinases required for segregation of vimentin filaments in mitotic process. Oncogene. 2001;20:2868–2876. doi: 10.1038/sj.onc.1204407. [DOI] [PubMed] [Google Scholar]
- 30.Goto H, et al. Aurora-B regulates the cleavage furrow-specific vimentin phosphorylation in the cytokinetic process. J Biol Chem. 2003;278:8526–8530. doi: 10.1074/jbc.M210892200. [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi T, et al. Phosphorylation by Cdk1 induces Plk1-mediated vimentin phosphorylation during mitosis. J Cell Biol. 2005;171:431–436. doi: 10.1083/jcb.200504091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka H, et al. Cytokinetic failure-induced tetraploidy develops into aneuploidy, triggering skin aging in phosphovimentin-deficient mice. J Biol Chem. 2015;290:12984–12998. doi: 10.1074/jbc.M114.633891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood KW, et al. Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proc Natl Acad Sci USA. 2010;107:5839–5844. doi: 10.1073/pnas.0915068107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu C, et al. BioGPS: An extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zambo I, Veselý K. WHO classification of tumours of soft tissue and bone 2013: The main changes compared to the 3rd edition. Cesk Patol. 2014;50:64–70. [PubMed] [Google Scholar]
- 36.Rogel MR, et al. Vimentin is sufficient and required for wound repair and remodeling in alveolar epithelial cells. FASEB J. 2011;25:3873–3883. doi: 10.1096/fj.10-170795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivaska J, Pallari HM, Nevo J, Eriksson JE. Novel functions of vimentin in cell adhesion, migration, and signaling. Exp Cell Res. 2007;313:2050–2062. doi: 10.1016/j.yexcr.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 38.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68:3033–3046. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Virtakoivu R, et al. Vimentin-ERK signaling uncouples slug gene regulatory function. Cancer Res. 2015;75:2349–2362. doi: 10.1158/0008-5472.CAN-14-2842. [DOI] [PubMed] [Google Scholar]
- 40.Scaife RM. Selective and irreversible cell cycle inhibition by diphenyleneiodonium. Mol Cancer Ther. 2005;4:876–884. doi: 10.1158/1535-7163.MCT-05-0009. [DOI] [PubMed] [Google Scholar]
- 41.Izawa I, Inagaki M. Regulatory mechanisms and functions of intermediate filaments: A study using site- and phosphorylation state-specific antibodies. Cancer Sci. 2006;97:167–174. doi: 10.1111/j.1349-7006.2006.00161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prahlad V, Yoon M, Moir RD, Vale RD, Goldman RD. Rapid movements of vimentin on microtubule tracks: Kinesin-dependent assembly of intermediate filament networks. J Cell Biol. 1998;143:159–170. doi: 10.1083/jcb.143.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon M, Moir RD, Prahlad V, Goldman RD. Motile properties of vimentin intermediate filament networks in living cells. J Cell Biol. 1998;143:147–157. doi: 10.1083/jcb.143.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nathenson MJ, Sausville E. Looking for answers: The current status of neoadjuvant treatment in localized soft tissue sarcomas. Cancer Chemother Pharmacol. 2016;78:895–919. doi: 10.1007/s00280-016-3055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dancsok AR, Asleh-Aburaya K, Nielsen TO. Advances in sarcoma diagnostics and treatment. Oncotarget. 2016;8:7068–7093. doi: 10.18632/oncotarget.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bollong MJ, et al. A small molecule inhibits deregulated NRF2 transcriptional activity in cancer. ACS Chem Biol. 2015;10:2193–2198. doi: 10.1021/acschembio.5b00448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.