Abstract
Modification of the Schizosaccharomyces pombe genome is often laborious, time consuming due to the lower efficiency of homologous recombination. Here, we constructed metabolically engineered S. pombe strains using a CRISPR-Cas9 system and also demonstrated D-lactic acid (D-LA) production from glucose and cellobiose. Genes encoding two separate pyruvate decarboxylases (PDCs), an L-lactic acid dehydrogenase (L-LDH), and a minor alcohol dehydrogenase (SPBC337.11) were disrupted, thereby attenuating ethanol production. To increase the cellular supply of acetyl-CoA, an important metabolite for growth, we introduced genes encoding bacterial acetylating acetaldehyde dehydrogenase enzymes (Escherichia coli MhpF and EutE). D-LA production by the resulting strain was achieved by expressing a Lactobacillus plantarum gene encoding D-lactate dehydrogenase. The engineered strain efficiently consumed glucose and produced D-LA at 25.2 g/L from 35.5 g/L of consumed glucose with a yield of 0.71 g D-LA / g glucose. We further modified this strain by expressing beta-glucosidase by cell surface display; the resulting strain produced D-LA at 24.4 g/L from 30 g/L of cellobiose in minimal medium, with a yield of 0.68 g D-LA / g glucose. To our knowledge, this study represents the first report of a S. pombe strain that was metabolically engineered using a CRISPR-Cas9 system, and demonstrates the possibility of engineering S. pombe for the production of value-added chemicals.
Keywords: Schizosaccharomyces pombe, Genome editing, Lactic acid, CRISPR-Cas9 system
Highlights
-
•
Schizosaccharomyces pombe were metabolically engineered using a CRISPR-Cas9 system.
-
•
D-lactic acid (D-LA) producing Schizosaccharomyces pombe strains were constructed.
-
•
25.2 g/L of D-LA was produced with a yield of 0.71 g-D-LA / g-glucose.
-
•
Beta-glucosidase was expressed on this engineered S. pombe strain.
-
•
D-LA was produced at 24.4 g/L from 30 g/L of cellobiose directly.
1. Introduction
Production of chemicals by microbial fermentation is the focus of increasing interest in the chemical industry (Choi et al., 2015). Here, we describe the metabolic engineering of the fission yeast Schizosaccharomyces pombe for the biosynthesis of D-lactic acid (D-LA). Lactic acid (LA) has broad industrial applications (Choi et al., 2015). One appealing use is as a monomer for the production of polylactic acid (PLA), a raw material for the generation of biodegradable plastics. Classically, LA has been obtained using lactic acid bacteria, the natural hosts for LA fermentation (Okano et al., 2010). There have been various trials to develop bacterial strains that overproduce LA, along with attempts to improve the economics of LA synthesis. Polymerization of L-LA and D-LA leads to poly (L-lactic acid) (PLLA) and poly (D-lactic acid) PDLA, respectively, and PLLA has been the major PLA produced to date. However, the thermochemical properties, physical properties, and biodegradation rate of PLA can be adjustable by making stereocomplex PLA incorporating different ratios of PLLA and PDLA (Auras et al., 2011). Therefore, there is an increasing demand for D-LA production as well as for that of L-LA.
The fission yeast S. pombe has proven to be a useful model organism (Forsburg, 2005) because of the higher degree of similarity of its genome to those of higher eukaryotes (compared to the classic model Saccharomyces cerevisiae). S. pombe, which is taxonomically and evolutionarily distant from budding yeast, has been extensively characterized both genetically and physiologically (Takegawa et al., 2009). In contrast to budding yeast, S. pombe has not been widely used in the manufacture of fermented beverages and foods such as wine, beer, and bread. Given that fission yeast shares many genetic and biochemical characteristics with higher eukaryotes (Zhao and Lieberman, 1995), this species also has been used extensively for high-level heterologous protein production (Giga-Hama, 1997). Although many types of recombinant proteins have been successfully expressed in fission yeast (Celik and Calık, 2012, Delic et al., 2013), to our knowledge, only one study about the production of value-added chemicals using fission yeast has been reported (Suyama et al., in press). Metabolic pathway engineering in yeasts (especially buddying yeast) has proven to be tremendously important for improving basic understanding of metabolism and for production of chemicals (Nielsen and Keasling, 2016). However, S. pombe remains less well studied than S. cerevisiae, and lags behind budding yeast in the availability of molecular tools.
Modification of the S. pombe genome is often laborious, time consuming, and requires screening of large number of candidate colonies to obtain the desired modification (Storici et al., 2001). Furthermore, the integration of exogenous DNA via homologous recombination (HR) occurs at lower efficiency in S. pombe than in S. cerevisiae (Gao et al., 2013, Mudge et al., 2011). As a result, targeted integration and engineering of the fission yeast genome requires extended homology, necessitating additional cloning steps. Single gene disruption in S. pombe is typically achieved by substituting the target gene with a selectable marker through HR, which provides site-specific mutagenesis at low efficiency. Recently, a CRISPR-Cas9 system applicable for use in S. pombe was developed (Jacobs et al., 2014, Jakočiūnas et al., 2016, Stovicek et al., 2017). Using double-stranded oligonucleotides centered around the target protospacer adjacent motif (PAM) sequence for homology-directed DNA double-strand break repair, target locus-specific point mutation was achieved. This method is expected to be a powerful tool for metabolic engineering of S. pombe, as well as S. cerevisiae.
S. pombe, which exhibits the potential of the tolerance to low pH conditions, has emerged as a promising alternative for the production of LA (Rupes et al., 1999). S. pombe, like S. cerevisiae, has a strong tendency for ethanol production; hence decreasing or inactivating the ethanol synthesis pathway is essential to obtaining high-yield production of LA. In S. cerevisiae, disruption of the genes encoding pyruvate decarboxylases (PDCs), enzymes that catalyze the first step of ethanol fermentation from pyruvate, is an effective metabolic approach to inhibiting ethanol production (Kim and Hahn, 2015; van Rossum et al., 2016). However, disruption of PDC-encoding genes in budding yeast results in a severe growth defect for growth on glucose-containing medium; semi-rational approaches to improving the viability of pdc-negative S. cerevisiae strains remain limited (van Maris et al., 2004). Elimination of alcohol dehydrogenase (ADH), which converts acetaldehyde to ethanol, is an alternative approach to decreasing ethanol production (Kim and Hahn, 2015). However, the inhibition of ethanol production by this mechanism impairs NAD+ regeneration by ADH, resulting in an increase in glycerol production via the compensatory activation of glycerol-3-phosphate dehydrogenase (GPD) to maintain redox balance (Kim and Hahn, 2015). Therefore, additional deletion of the GPD-encoding genes has been employed to decrease glycerol production. Although these approaches can provide improved yield of the desired product, these disruptions cause significant impairment of growth. Thus, ethanol pathway engineering to reduce by-product synthesis is one of the most critical issues in the engineering of industrial yeast strains. However, to our knowledge, only a limited number of reports have described the engineering of S. pombe via deletion of PDC- or ADH-encoding genes.
In the present study, we constructed D-LA-producing S. pombe strains by using metabolic pathway engineering based on CRISPR-Cas9-mediated genome editing. PDC-, ADH-, and GPD-encoding genes were deleted, and the Lactobacillus plantarum D-lactate dehydrogenase (D-LDH) -encoding gene then was introduced into the fission yeast genome. Elimination of by-product-forming pathways and introduction of two copies of the D-LDH-encoding gene permitted us to achieve a large increase in LA productivity. In addition, beta-glucosidase was expressed on the cell surface (Tanaka et al., 2013), facilitating D-LA production from cellobiose at high yield. Since there are great potentials of S. pombe as a microbial cell factory to produce value-added chemicals and fuels from renewable lignocellulosic biomass, efficient utilization of cellulosic sugars is an inevitable goal. Thus, our study demonstrates the utility of S. pombe as a candidate host for the production of value-added chemicals from glucose and cellobiose.
2. Material and methods
2.1. Strains and media
All strains used in this study are listed in Table 1. S. pombe FY12804 (NBRP) was used as a parental strain. Yeast strains were cultivated in YM medium (BD Diagnostic Systems, Sparks, MD, USA) or EMM medium (ForMedium, Norfolk, United Kingdom).
Table 1.
Strains and plasmids used in this study.
| Schizosaccharomyces pombe strain | Relevant phenotype, description | Source |
|---|---|---|
| FY12804 | h90 ura4-D18 leu1-32 | YGRC/NBRP |
| ade6Δ-1 | ade6Δ Insertion of 1 bp | This study |
| ade6Δ-2 | ade6 Δ Deletion of 1 bp and insertion of a stop codon | This study |
| ade6Δ-3 | ade6 Δ Deletion of 8 bp | This study |
| ade6Δ-4 | ade6 Δ Insertion of repeated stop codons | This study |
| ade6Δ::gfp | ade6 Δ::Pcam1-GFP | This study |
| ATR5 | ku70Δ, pdc1Δ::Ptif51-mhpF, pdc2Δ::Ptif51-eutE, l-ldhΔ, adh SPBC337.11Δ | This study |
| ATR5-LA1 |
ku70Δ, pdc1Δ::Ptif51-mhpF, pdc2Δ::Ptif51-eutE, l-ldhΔ, adh SPBC337.11Δ, gut2Δ::Pef1a-c-d-ldh, |
This study |
| ATR5-LA2 |
ku70Δ, pdc1 Δ::Ptif51-mhpF, pdc2Δ::Ptif51-eutE, l-ldhΔ, adh SPBC337.11Δ, gut2Δ::Pef1a-c-d-ldh, adh8Δ::Pef1a-c-d-ldh |
This study |
| ATR5-LA2-BGL |
ku70Δ, pdc1Δ::Ptif51-mhpF, pdc2Δ::Ptif51-eutE, l-ldhΔ, adh SPBC337.11Δ, gut2 Δ::Pef1a-c-d-ldh, adh8Δ::Pef1a-c-d-ldh, gut2Δ::Phsp-BGL |
This study |
| Plasmid | ||
| pDUAL-FFH61 | Vector under ef1a-c promoter control | RIKEN BRC |
| pDUAL-FFH61-dLDH | Vector for expression of d-LDH from Lactobacillus plantarum | This study |
| pDUAL-hsp-SPBC359.04c_BGL | Vector for cell surface display expression of protein fusion of BGL from Aspergillus aculeatus with SPBC359.04c anchor protein | This study |
| pMZ374 | Vector for expressing adh1: cas9/rrk1: sgRNA in fission yeast: Empty sgRNA target | Addgene (#59896) |
| pMZ374-ade6–1450 | pMZ374 derivative, ade6 sgRNA target | This study |
| pMZ374-pdc1–412 | pMZ374 derivative, pdc1(412) sgRNA target | This study |
| pMZ374-pdc2–423 | pMZ374 derivative, sgRNA target | This study |
| pMZ374-ku70–41 | pMZ374 derivative, ku70(41) sgRNA target | This study |
| pMZ374-adh8–434 | pMZ374 derivative, adh8(434) sgRNA target | This study |
| pMZ374-adhBC-468 | pMZ374 derivative, adh SPBC337.11(468) sgRNA target | This study |
| pMZ374-gpd2–76 | pMZ374 derivative, gpd2(76) sgRNA target | This study |
| pMZ374-gut2–220 | pMZ374 derivative, gut2(220) sgRNA target | This study |
| pMZ374-gut2–1749 | pMZ374 derivative, gut2(1749) sgRNA target | This study |
| pMZ374-ldh-556 | pMZ374 derivative, ldh(556) sgRNA target | This study |
2.2. CRISPR-Cas9 system target site selection and donor design
A 20-bp seed sequence together with the NGG PAM sequence (N20NGG) in the S. pombe genome was selected using CRISPR direct (http://crispr.dbcls.jp/) (Naito et al., 2015). HR donor sequences as editing templates were designed to have about 500-bp homology arms to either side (upstream and downstream) flanking the Cas9 cutting site and the sites of the intended mutations, deletions, or insertions.
2.3. Plasmid construction and HR donor construction
Plasmids used in this study are listed in Table 1, and primers are listed in Supplementary Table S1. Cas9 and gRNA expression plasmid pMZ374 was purchased from Addgene. A cas9 expression plasmid targeting the ade6-1450 region was constructed using the KOD-Plus-mutagenesis Kit (TOYOBO, Co., Ltd., Osaka, Japan) according to the manufacturer's instructions. pMZ374 was used as a template with the primer pair F-ade6-1450 + R-ade6-1450. The resultant plasmid was named pMZ374-ade6. HR donor DNA for editing the ade6 region was constructed as follows. The upstream and downstream regions were amplified by PCR with the primer pairs up_F-ade6 + up_R-ade6-1 and down_F-ade6-1 + down_R-ade6, respectively. The amplified fragments were conjugated by overlap extension PCR with the primer pair up_F-ade6 + down_R-ade6. Other plasmids for Cas9 targeting and HR donor DNA as editing templates were constructed similarly and are summarized in Table 1.
The gfp expression cassette (Pcam1-GFP-TADH1) was amplified by PCR with the primer pair cassette-F-ade6-gfp + cassette-R-ade6-gfp using pDUAL-HFG31 (RIKEN BRC) as the template (Matsuyama et al., 2006). Upstream and downstream regions were amplified with the primer pairs up_F-ade6-4 + up_R-ade6-gfp and down_F-ade6-4 + down_R-ade6-gfp, respectively. The three amplified fragments were conjugated by overlap extension PCR with up_F-ade6-4 + down_R-ade6-gfp primers, yielding HR donor DNA for introduction into the chromosomal ade6 locus of a cassette directing the expression of gfp under control of the cam1 promoter (Matsuyama et al., 2004).
To construct the d-ldh expression cassette, the D-LDH-encoding gene from L. plantarum NCIMB 8826 was amplified by PCR using bacterial genomic DNA. The PCR product was cloned into the NheI and SalI sites of pDUAL-FFH61 (RIKEN BRC), and the resultant plasmid was named pDUAL-FFH61-dLDH. Then the d-ldh expression cassette (Pef1a-c-D-LDH -TADH1) was amplified by PCR with the primer pair Cassette_F-gut2-220 + Cassette_R-gut2-220. Upstream and downstream regions were amplified with the primer pairs of up_F-gut-220 + up_R-gut2-220 and down_F-gut2-220 + down_R-gut2-220, respectively. The three amplified fragments were conjugated by overlap extension PCR with up_F-gut2-220 + down_R-gut2-220 primers, resulting in HR donor DNA for introduction into the chromosomal gut2 region of the d-ldh expression cassette. HR donor DNA for introduction into the chromosomal adh8 region of the d-ldh expression cassette was constructed similarly using primer pairs, Cassette_F-adh8 + Cassette_R-adh8, up_F-adh8 + up_R-adh8 and down_F- adh8 + down_R-gut2.
To construct the bgl (beta-glucosidase) expression cassette (Phsp90-SPBC359.04c_BGL-Tnmt1), the hsp90 promoter from S. pombe was amplified with the primer pair SpeI_hsp90_for + hsp90_Sac1_re. The amplified fragments were cloned between the SpeI and SacI sites of pDUAL-FFH61. Next, the nmt1 terminator from S. pombe was amplified with the primer pair Xho_tnmt1_for + tnmt1_EcoR1_re. The amplified fragment was cloned between the XhoI and NotI sites of the plasmid mentioned above, and the resultant plasmid was named pDUAL-hsp. Then, a gene encoding the Aspergillus aculaetus beta-glucosidase fused to the anchor protein SPBC359.04c was amplified with the primers hsp_XhoI_SPBC359.04c_for + BGL_Not1_re using pDUAL-SPBC359.04c_BGL (Tanaka et al., 2013) as a template. The amplified fragment was cloned between the XhoI and NotI sites of pDUAL-hsp; the resultant plasmid was named pDUAL-hsp-SPBC359.04c_BGL. Using this plasmid as a template, three fragments were generated by PCR using the primer pairs Cassette_F-gut2-1749 + Cassette_R-gut2-1749, up_F-gut2-1749 + uP_R-gut2-1749, and down_F-gut2-1749 + down_R-gut2-1749. The amplified fragments were conjugated by overlap extension PCR with the up_F-gut2-1749 + down_R-gut2-1749 primers, yielding HR donor DNA for introduction into the chromosomal gut2 region of the bgl expression cassette.
2.4. Genome editing
All mutagenesis experiments were carried out by co-transformation of 10 µL of the pMZ374 vector series with 20 µL of the respective PCR product being used as the HR donor DNA for the target locus. Strains were grown in 5 mL of YM medium at 30 °C with shaking at 180 rpm until the culture reached an optical density (OD) at 600 nm of 0.5; cells then were transformed by electroporation using a Gene Pulser Xcell II (Bio-Rad) using standard methodologies. Transformants were spread onto EMM plates supplemented with 225 mg/L leucine and grown at 30 °C for approximately 7 days. Transformants were screened by colony PCR and DNA sequencing. For the curing of the pMZ374-based vector, the edited colony harboring the pMZ374 derivative was inoculated into YM medium, cultured for 2–3 days at 30 °C with shaking, diluted, and spread onto YM plates containing 5-fluoroorotic acid (4 mg/L). The resulting uracil auxotrophs (i.e., strains cured of the pMZ374-series plasmids) were then used in the next round of genome editing.
2.5. Culturing
For D-LA fermentation from glucose or cellobiose, metabolically engineered strains were pre-cultured in 5 mL YM medium for 3 days at 30 °C with shaking in a 15-mL test tube, and then washed twice with 1% NaCl and diluted to an initial OD of 3.0 in EMM medium supplemented with 225 mg/L each of uracil, adenine, and leucine and containing glucose (50 or 30 g/L) or cellobiose (30 g/L). The resulting cultures (final volume is 5 mL) were incubated at 30 °C with shaking anaerobically with sampling at the indicated time points.
2.6. Analytical methods
Cell growth was determined by measuring the OD at 600 nm using a UVmini-1240 spectrophotometer (Shimadzu Corporation, Kyoto, Japan). Extracellular metabolites, including glucose and cellobiose, were analyzed using a Prominence HPLC System (Shimadzu) equipped with a Shodex SUGAR KS-801 column (6 µm, 300 mm × 8.0 mm; Shodex). Water was used as the mobile phase at a flow rate of 0.8 mL/min, and the column was maintained at 50 °C. The peak elution profile was monitored using a refractive index detector. D-LA was analyzed using an HPLC equipped with a SCR-102H column (7 µm, 8.0 mm ID × 300 mm.; Shimadzu). p-Toluenesulfonic acid (5 mM) was used as the mobile phase at a flow rate of 1.5 mL/min, and the column was maintained at 50 °C. The peak elution profile was monitored using a conductivity detector. The enantiomeric purity of D-/L-LA was determined using a D-/L-Lactic Acid Assay Kit (detection limit; 0.21 mg/L) (Megazyme, Wicklow, Ireland).
3. Results
3.1. Genome editing in S. pombe using CRISPR-Cas9 system
A CRISPR-Cas9 system was tested for efficacy in mutating the S. pombe ade6 gene, which when mutated causes accumulation of a red-colored precursor in medium containing low concentrations of adenine. We generated sgRNA constructs targeting ade6 and prepared four different HR donor fragments: ade6Δ-1 (1-bp mutation), ade6Δ-2 (stop codon insertion), ade6Δ-3 (8-bp deletion), and ade6Δ-4 (insertion of repeated stop codon) (Fig. 1A). Each of these mutations were located near a protospacer adjacent motif (PAM) necessary for Cas9/sgRNA recognition. The lengths of the upstream and downstream regions in each HR donor were about 500 bp in this study due to high recombination efficiency. We introduced each donor fragment, along with an expression vector providing production of Cas9 and the respective sgRNA, into the parental strain FY12804, and transformants were spread onto EMM plates supplemented with a low concentration of adenine. After 3–4 days of growth, several white colonies were observed; sequencing confirmed that these colonies harbored intact ade6 loci. After another 5–10 days, a number of red colonies were observed (Fig. 1B); screening by PCR and sequencing confirmed that these colonies harbored the targeted ade6 mutations. For plates allowed to grow for up to 14 days, red colonies represented approximately 80% of the transformants. In this and following experiments, we observed that transformation during logarithmic growth phase was critical for obtaining the desired Cas9-mediated gene-editing events.
Fig. 1.
A) Homologous recombination (HR) donors for mutation, insertion, and deletion of the S. pombe ade6 gene. PAM sequence (AGG) is shown in red, and mutated sequences are shown in green. B) Representative plate (10 days post-transformation of ade6Δ-4) from the ade6 mutagenesis experiment. C) Colony PCR of the wild-type (parental) strain (lane 1, 2) and 4 clones transformed with the Cas9/sgRNA expression vector and ade6-targeting HR donor template containing gfp expression cassette (lane 3–6). Lane M indicates DNA marker. D) Fluorescent images of GFP-expressing strain constructed by CRISPR-Cas9 system. A gfp expression cassette was introduced into ade6 locus. No fluorescence was observed on wild type strains under the same conditions (data not shown). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We next attempted the introduction of a gene expression cassette into a targeted locus by the CRISPR-Cas9 system. The gene encoding green fluorescent protein (GFP) was chosen as a model for targeting insertion into the ade6 locus. We prepared HR donor for expression of GFP under control of the cam1 promoter (20, 21), such that the gfp gene was flanked by 500-bp arms corresponding to sequences upstream and downstream of the chromosomal target. Thus, CRISPR-Cas9-mediated introduction of the gfp expression cassette was expected to result in ade6 gene disruption. By 7–10 days of post-transformation, we obtained red colonies, indicative of ade6 gene disruption; screening of these transformants by colony PCR confirmed introduction of gfp gene cassettes into ade6 (Fig. 1C). Specifically, flanking primers amplified a 1500-bp band from wild-type strains (lane 1, 2); in contrast, three of four red colonies yielded 3000-bp bands, consistent with size expected for the ade6::gfp integration (lanes 3–5). One red colony yielded a wild-type like band (lane 6), suggesting that the ade6 locus in this transformant was damaged by repair of a Cas9 cleavage event without integration of the HR donor. As shown in Fig. 1D, bright fluorescence was observed in one such transformant, showing successful GFP expression. In addition, the color of this colony was red and adenine-limiting medium, confirming that the gfp expression cassette inserted into ade6 as targeted.
3.2. Construction of D-LA biosynthetic pathway and disruption of competing pathways
Encouraged by these findings, we generated D-LA producing S. pombe by CRISPR-Cas9-mediated genome editing. To improve recombination efficiency, Ku domain protein Pku70 was disrupted (Baumann and Cech, 2000). Disruption was confirmed by colony PCR and sequencing of the resulting product. The homologous recombination efficiency was evaluated by ade6 gene disruption. From Fig. S1, several numbers of white colonies were observed in the plate of wild type strain, and almost all colonies were red in the plate of ku70 deleted strain. This result shows ku70 deletion were beneficial to homologous recombination.
To channel the metabolic fluxes toward LA formation and impair by-product formation, we tried to delete the gene encoding ADH1, which is known to play a major role in ethanol production (Tsai et al., 1992). However, adh1 deletion mutants exhibited a severe growth defect (OD600 was less than 0.5 over 2-week cultivation and we could not complete next round of genome editing). Hence, we employed another metabolic engineering strategy, that of increasing the acetyl-CoA supply. We sought to increase acetyl-CoA levels not for use in the synthesis of pyruvate-derived products, but rather to enhance cell growth or carbon catabolic rates, as is seen in S. cerevisiae (van Rossum et al., 2016). S. pombe encodes four distinct pyruvate dehydrogenases; we chose to delete pdc101 and pdc202 while introducing two kinds of bacterial acetylating acetaldehyde dehydrogenases, which was expected to facilitate the direct conversion of acetaldehyde to acetyl-CoA without additional energy consumption (Song et al., 2016). Previous studies have reported the successful functional expression of the E. coli MhpF protein in S. cerevisiae (Guadalupe Medina et al., 2010), although E. coli EutE appeared to yield superior performance in budding yeast (Kozak et al., 2014). We chose to introduce both mhpF and eutE (each under the control of the tif promoter) by integration into the pdc101 and pdc202 loci, respectively. In addition, we deleted our strain for endogenous loci encoding L-LDH and a minor ADH (SPBC337.11). The resultant strain, designated ATR-5, showed growth comparable to that of the wild-type parent under aerobic conditions (Fig. S1). Using ATR-5 as an acetyl-CoA-overproducing and PDC-deficient chassis strain, we then introduced an expression cassette encoding the Lactobacillus plantarum D-LDH under the control of the ef1a promoter; specifically, this cassette was integrated at (and disrupted) the S. pombe locus encoding GPD2 (a glycerol-3-phosphate dehydrogenase) (Fig. 2). The resultant strain, designated ATR5-LA1, was used for further fermentation experiments.
Fig. 2.
Engineered metabolic pathway for D-LA production by S. pombe. Six genes (gut2, pdc101, pdc202, l-ldh, adh8, adh SPBC337.11, shown in blue) were disrupted and three genes (d-ldh, eutE, mhpF, shown in red) were introduced. gut2, glycerol-3-phosphate dehydrogenase; l-ldh, l-lactate dehydrogenase; pdc101, pyruvate decarboxylase (SPAC1F8.07c); pdc202, pyruvate decarboxylase (SPAC13A11.06); adh8, alcohol dehydrogenase (SPBC1773.06c); eutE, ethanolamine utilization protein from E. coli; mhpF, acetaldehyde dehydrogenase from E. coli; d-ldh, d-lactate dehydrogenase from Lactobacillus plantarum NCIMB 8826; BGL, beta-glucosidase from Aspergillus aculeatus. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. D-LA production from glucose using genome-edited S. pombe strains ATR5-LA1 and ATR5-LA2
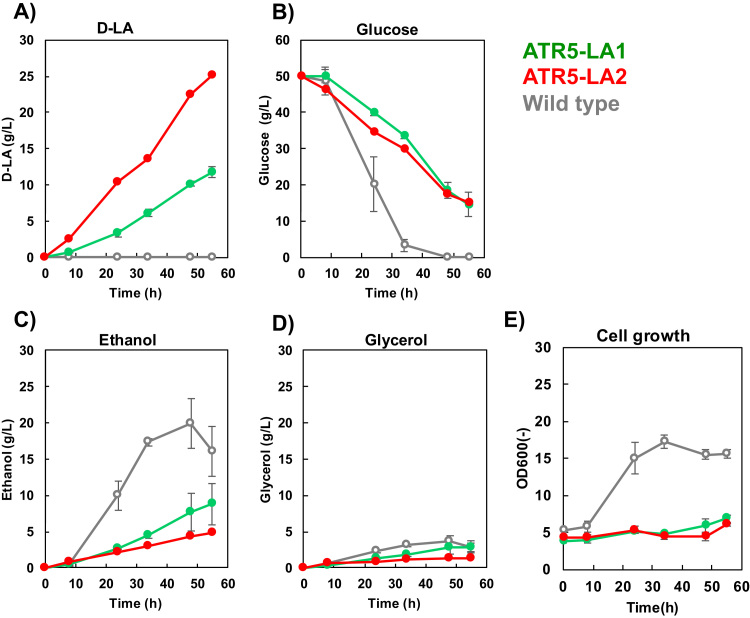
To evaluate the potential of the ATR5-LA1 strain for D-LA acid production, test tube-scale fermentation from glucose was performed under non-neutralizing conditions starting from EMM medium containing 50 g/L glucose. After 55 h of cultivation, ATR5-LA1 consumed glucose at 35.5 g/L and produced D-LA at 11.8 g/L, corresponding to a yield of 0.33 g D-LA / g glucose (Fig. 3A). The by-products ethanol and glycerol were produced at 8.7 g/L of and 2.9 g/L, respectively (Fig. 3C, D). In comparison, a 50-h cultivation of the wild-type strain consumed glucose at 50 g/L and produced ethanol and glycerol at 20 g/L and 3.6 g/L of glycerol, respectively, instead of D-LA. Cell growth of ATR5-LA1 was clearly suppressed during cultivation (OD600 is 6.8 for 55 h), although OD600 of wild-type strain was increased up to 15.5 (Fig. 3E). To improve D-LA production and reduce by-product formation, we further modified ATR5-LA1 by introducing a second copy of the d-ldh expression cassette by targeted disruption of the endogenous adh8 gene, yielding strain ATR5-LA2. In a 55-h fermentation, this strain consumed glucose at 35.5 g/L and produced D-LA at 25.2 g/L (0.71 g D-LA / g glucose), ethanol at 4.8 g/L, and glycerol at 1.3 g/L (Fig. 3; red symbols). Thus, compared to ATR5-LA1, ATR5-LA2 exhibited an approximate doubling of D-LA production and an approximate halving of by-product synthesis. Notably, although low levels of L-LA were produced by the wild type strain, ATR5-LA2 produced 99.9% optically pure D-LA, consistent with disruption of the endogenous gene encoding L-LDH.
Fig. 3.
Lactic acid (LA) production from EMM medium containing 50 g/L glucose using strains ATR5-LA1 and ATR5-LA2. Time courses of D-LA (A), glucose (B), ethanol (C), glycerol (D) concentrations and cell growth (E) are shown. ATR5-LA1 is represented by green symbols, ATR5-LA2 is represented by red symbols, and the wild-type parental strain is represented by gray symbols. There experiments were started three separate colonies and data are presented as the mean ± standard deviation (SD) from triplicate experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. D-LA production from cellobiose using beta-glucosidase-displaying strain ATR5-LA2-BGL
Metabolic engineering to broaden the substrate scope of S. pombe with respect to access to various carbon sources is first step for efficient utilization of cellulose. Cell surface display is a powerful technology for endowing microorganisms with an enhanced ability to degrade biomass. We previously developed a beta-glucosidase-displaying strain of S. pombe capable of cellobiose assimilation (Tanaka et al., 2013). In the present work, we introduced a gene encoding a cell surface-displayed beta-glucosidase into the ATR-LA2 strain. Specifically, an Aspergillus aculeatus beta-glucosidase-encoding gene was fused to sequences encoding anchor protein SPBC359.04c, and the resulting fusion protein was expressed under the control of the hsp promoter. An anchor protein SPBC359.04c was most suitable anchor for cellobiose degradation (Tanaka et al., 2013), and the strong promoter hsp90 (Suyama et al., in press) was utilized to increase protein expression level. In a 55-h fermentation starting from medium containing cellobiose at 30 g/L (Fig. 4), the resulting strain (designated ATR-LA2-BGL) completely consumed the cellobiose, producing D-LA at 22.8 g/L, corresponding to a yield of 0.68 g D-LA / g-glucose (1 g of cellobiose corresponds to 1.11 g of glucose). Starting from medium containing 30 g/L of glucose as the carbon source, ATR-LA2-BGL produced D-LA at 24.4 g/L, corresponding to a yield of 0.81 g-D-LA / g-glucose. For fermentations from either cellobiose or glucose, ATR-LA2-BGL produced ethanol at less than 2.5 g/L (Fig. 4). As a negative control, ATR5 could neither produce LA nor consume cellobiose (data not shown).
Fig. 4.
Lactic acid (LA) production from EMM medium containing 30 g/L glucose (A) or cellobiose (B) using strain ATR5-LA2-BGL. Glucose is represented by brown symbols, cellobiose by green symbols, ethanol by blue symbols, glycerol by purple symbols, and D-LA by orange symbols. There experiments were started three separate colonies and data are presented as the mean ± standard deviation (SD) from triplicate experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
In this study, we constructed D-LA producing S. pombe strains by genome editing via a CRISPR-Cas9 system. All strains used were generated by CRISPR-Cas9. Targeted gene deletions and insertion of gene expression cassettes were carried out efficiently and repeatedly. As a result, D-LA production from glucose and cellobiose were successfully achieved (Fig. 4). Although lactic acid bacteria produced LA with high yield (95%>) and titer (100 g/L>) (Sauer et al., 2017), S. pombe exhibit high tolerance to low extracellular pH, which was the advantage of using S. pombe for the organic acid production (Kildegaard et al., 2014, Suyama et al., 2017). In the absence of pH control, ATR5-LA2 produced 25.2 g/L of D-LA from 35.5 g/L of consumed glucose, and ATR-LA2-BGL produced 24.4 g/L of D-LA from 30 g/L of cellobiose in minimal medium. These fermentations corresponded to yields of 0.71 and 0.68 g-D-LA / g-glucose, respectively. In previous studies in S. cerevisiae, L-LA was produced with yields of 0.3–0.93 g-L-LA / g-glucose (Dato et al., 2014, Lee et al., 2015, Valli et al., 2006), and D-LA was produced with yields of 0.5–0.82 g-D-LA / g-glucose (Baek et al., 2016, Baek et al., 2017, Kim et al., 2015; Ishida et al., 2005, Yamada et al., 2017). However, the volumetric productivity (g/L/h) and titer (g/L) of D-LA was relatively lower (0.44 g/L/h in this study) than those in previous reports (0.39–0.79 g/L/h) We anticipate that D-LA production by our strains could be further improved by enhancement of glucose uptake (Kim et al., 2015), given that we detected free glucose in cultures that used cellobiose as a carbon source (Fig. 4B). In addition, the enhancement of glucose uptake would decrease initial cell density and may contribute to achieving industrial application.
CRISPR-Cas9 systems have been employed to edit yeast genomes, including that of S. cerevisiae. The genome editing efficiency has been high and has been employed for a variety of metabolic engineering experiments. CRISPR-Cas9's applicability in S. pombe was recently reported (Jacobs et al., 2014), but we are not aware of other reports on the use of this system for metabolic engineering of S. pombe. We demonstrated the application of CRISPR-Cas9 for the generation of several kinds of mutations, including point mutations, deletions, and gene expression cassette insertion (Fig. 1). The insertion of a gfp expression cassette was confirmed by fluorescence of one such engineered strain. ku70 disruption by CRISPR-Cas9 was also achieved, and the resulting strain (expected to be impaired for the non-homologous end joining pathway) might exhibited improved efficiency for subsequent genome editing.
To improve the carbon flux toward LA formation, we employed two approaches: disruption of a competing pathway (ethanol production from pyruvate) and increasing the D-LDH-catalyzed reaction by introducing an additional d-ldh expression cassette. In S. cerevisiae, genes encoding PDCs and ADHs have been deleted in various combinations to increase LA production (Ishida et al., 2006, Kim et al., 2015, Lee et al., 2015, van Rossum et al., 2016). Those experiments have shown that combinatorial deletion of three PDC-encoding genes or of the two major PDC-encoding genes (PDC1 and PDC5) are sufficient to impair pyruvate accumulation in budding yeast, although the resulting strains exhibit severe growth defects on glucose. On the other hand, the S. pombe genome encodes five PDC isotypes and four ADH isotypes (Wood et al., 2002). In the present study, we deleted two pdc genes and two adh genes in combination (Fig. 2); the resulting quadruple-mutant (ATR-5) did not exhibit defects in growth (Fig. S1). In contrast, deletion of the gene encoding ADH1, the major enzyme converting acetaldehyde to ethanol, caused significant growth defects on glucose (OD600 was less than 0.5 over 2-week cultivation), suggesting ADH1-dependent NAD+ regeneration might be indispensable for S. pombe growth. Introduction of a gene encoding a bacterial D-LDH via disruption of the endogenous gut2 (GPD-encoding) gene provided D-LA production with a concomitant decrease in glycerol production (Fig. 3D), presumably because high D-LDH activity compensated for the NAD+ regeneration lost upon mutation of gut2. Ethanol production also was decreased upon introduction of the d-ldh gene (Fig. 3C); we infer that redox balance is maintained via D-LDH activity, despite the absence of the ADH enzymes. In a further step, an additional copy of the D-LDH-encoding gene was introduced by targeted disruption of adh8; in the resulting strain (ATR5-LA2), ethanol production (Fig. 3D) was reduced and D-LA production was improved (Fig. 3A). Cell growth of ATR5-LA1 and ATR5-LA2 was quite slower than that of wild-type strain (Fig. 3E). These results suggested that carbon flux had been redirected from cell growth to D-LA production by D-LDH introduction. Subsequent attempts to disrupt gpd2 or adh1 in this strain yielded strains with significant growth defects (OD600 of both strains were less than 0.5 over 2-week cultivation), suggesting that complete elimination of the GPD- or ADH-encoding genes was not suitable for S. pombe growth on glucose.
Cell surface display is a powerful tool for endowing novel functions on the host cell by displaying functional proteins on the cell surface (Tanaka et al., 2012; Tanaka and Kondo, 2015). Amylase-displaying yeasts have amylolytic activity, and cellulase-displaying yeast and bacteria have cellulose-depolymerizing activity. In cell surface display, a target protein is genetically fused to an anchor protein, and the fusion protein is expressed in the desired host cells. Previously, we developed beta-glucosidase-displaying S. pombe using SPBC359.04c as an anchor protein (Tanaka et al., 2013). Here, we introduced a BGL-expressing cassette into ATR5-LA2 and demonstrated that the resulting strain (ATR5-LA2-BGL) was able to produce D-LA from cellobiose with a yield similar to that obtained when using glucose as the carbon source (Fig. 4). In samples from intermediate time points, free glucose was detected in the cellobiose-containing culture medium during fermentation by ATR5-LA2-BGL, suggesting that glucose uptake is the rate-limiting step in this fermentation. Improving glucose uptake may increase the LA productivity of this strain.
Conflict of interest
We declare no conflicts of interest.
Acknowledgments
This work was supported primarily by a Grant-in-Aid for Young Scientists (A) (grant number 16H06132 to T.T.), and partially supported by Special Coordination Funds for Promoting Science and Technology, Creation of Innovation Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe), MEXT, Japan.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.meteno.2017.08.002.
Appendix A. Supplementary material
Supplementary material
References
- Auras R.A., Lim L., Selke S.E., Tsuji H. John Wiley & Sons; Hoboken, NJ: 2011. Poly (lactic acid): Synthesis, Structures, Properties, Processing, and Applications. [Google Scholar]
- Baek S.H., Kwon E.Y., Kim Y.H., Hahn J.S. Metabolic engineering and adaptive evolution for efficient production of D-lactic acid in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2016;100 doi: 10.1007/s00253-015-7174-0. (2737-274) [DOI] [PubMed] [Google Scholar]
- Baek S.H., Kwon E.Y., Bae S.J., Cho B.R., Kim S.Y., Hahn J.S. Improvement of d-lactic acid production in Saccharomyces cerevisiae under acidic conditions by Evolutionary and rational metabolic engineering. Biotechnol. J. 2017 doi: 10.1002/biot.201700015. (in press) [DOI] [PubMed] [Google Scholar]
- Baumann P., Cech T.R. Protection of telomeres by the Ku protein in fission yeast. Mol. Biol. Cell. 2000;11:3265–3275. doi: 10.1091/mbc.11.10.3265. (31. 8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik E., Calık P. Production of recombinant proteins by yeast cells. Biotechnol. Adv. 2012;30:1108–1118. doi: 10.1016/j.biotechadv.2011.09.011. [DOI] [PubMed] [Google Scholar]
- Choi S., Song C.W., Shin J.H., Lee S.Y. Biorefineries for the production of top building block chemicals and their derivatives. Metab. Eng. 2015;28:223–239. doi: 10.1016/j.ymben.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Dato L., Berterame N., Ricci M., Paganoni P., Palmieri L., Porro D., Branduardi P. Changes in SAM2 expression affect lactic acid tolerance and lactic acid production in Saccharomyces cerevisiae. Microb. Cell Factor. 2014;13:147. doi: 10.1186/s12934-014-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delic M., Valli M., Graf A.B., Pfeffer M., Mattanovich D., Gasser B. The secretory pathway: exploring yeast diversity. FEMS Microbiol. Rev. 2013;37:872–914. doi: 10.1111/1574-6976.12020. [DOI] [PubMed] [Google Scholar]
- Forsburg S.L. The yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe: models for cell biology research. Gravit. Space Biol. Bull. 2005;18:3–9. [PubMed] [Google Scholar]
- Gao J., Kan F., Wagnon J.L., Storey A.J., Protacio R.U., Davidson M.K., Wahls W.P. Rapid, efficient and precise allele replacement in the fission yeast Schizosaccharomyces pombe. Curr. Genet. 2013;60:109–119. doi: 10.1007/s00294-013-0406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giga-Hama Y. Springer-Verlag; Berlin: 1997. Fission Yeast Schizosaccharomyces pombe: An Attractive Host for Heterologous Protein Production. [Google Scholar]
- Guadalupe Medina V., Almering M.J., van Maris A.J., Pronk J.T. Elimination of glycerol production in anaerobic cultures of a Saccharomyces cerevisiae strain engineered to use acetic acid as an electron acceptor. Appl. Environ. Microbiol. 2010;76:190–195. doi: 10.1128/AEM.01772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida N., Suzuki T., Tokuhiro K., Nagamori E., Onishi T., Saitoh S., Kitamoto K., Takahashi H. D-lactic acid production by metabolically engineered Saccharomyces cerevisiae. J. Biosci. Bioeng. 2006;101:172–177. doi: 10.1263/jbb.101.172. [DOI] [PubMed] [Google Scholar]
- Jacobs J.Z., Ciccaglione K.M., Tournier V., Zaratiegui M. Implementation of the CRISPR-Cas9 system in fission yeast. Nat. Commun. 2014;5:5334. doi: 10.1038/ncomms6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakočiūnas T., Jensen M.K., Keasling J.D. CRISPR/Cas9 advances engineering of microbial cell factories. Metab. Eng. 2016;34:44–59. doi: 10.1016/j.ymben.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Kildegaard K.R., Hallström B.M., Blicher T.H., Sonnenschein N., Jensen N.B., Sherstyk S., Harrison S.J., Maury J., Herrgård M.J., Juncker A.S., Forster J., Nielsen J., Borodina I. Evolution reveals a glutathione-dependent mechanism of 3-hydroxypropionic acid tolerance. Metab. Eng. 2014;26:57–66. doi: 10.1016/j.ymben.2014.09.004. [DOI] [PubMed] [Google Scholar]
- Kim D., Song J.Y., Hahn J.S. Improvement of glucose uptake rate and production of target chemicals by overexpressing hexose transporters and transcriptional activator Gcr1 in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2015;81:8392–8401. doi: 10.1128/AEM.02056-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Hahn J.S. Efficient production of 2,3-butanediol in Saccharomyces cerevisiae by eliminating ethanol and glycerol production and redox rebalancing. Metab. Eng. 2015;31:94–101. doi: 10.1016/j.ymben.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Kozak B.U., van Rossum H.M., Benjamin K.R., Wu L., Daran J.M., Pronk J.T., van Maris A.J. Replacement of the Saccharomyces cerevisiae acetyl-CoA synthetases by alternative pathways for cytosolic acetyl-CoA synthesis. Metab. Eng. 2014;21:46–59. doi: 10.1016/j.ymben.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Kang C.D., Lee S.H., Park Y.K., Cho K.M. Engineering cellular redox balance in Saccharomyces cerevisiae for improved production of L-lactic acid. Biotechnol. Bioeng. 2015;112:751–758. doi: 10.1002/bit.25488. [DOI] [PubMed] [Google Scholar]
- Matsuyama A., Shirai A., Yashiroda Y., Kamata A., Horinouchi S., Yoshida M. pDUAL, a multipurpose, multicopy vector capable of chromosomal integration in fission yeast. Yeast. 2004;21:1289–1305. doi: 10.1002/yea.1181. [DOI] [PubMed] [Google Scholar]
- Matsuyama A., Arai R., Yashiroda Y., Shirai A., Kamata A., Sekido S., Kobayashi Y., Hashimoto A., Hamamoto M., Hiraoka Y., Horinouchi S., Yoshida M. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- Mudge D.K., Hoffman C.A., Lubinski T.J., Hoffman C.S. Use of a ura5p-lys7p cassette to construct unmarked gene knock-ins in Schizosaccharomyces pombe. Curr. Genet. 2011;58:59–64. doi: 10.1007/s00294-011-0360-4. [DOI] [PubMed] [Google Scholar]
- Naito Y., Hino K., Bono H., Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31:1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J., Keasling J.D. Eng. Cell. Metab. 2016;164:1185–1197. doi: 10.1016/j.cell.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Okano K., Tanaka T., Ogino C., Fukuda H., Kondo A. Biotechnological production of enantiomeric pure lactic acid from renewable resources: recent achievements, perspectives, and limits. Appl. Microbiol. Biotechnol. 2010;85:413–423. doi: 10.1007/s00253-009-2280-5. [DOI] [PubMed] [Google Scholar]
- Rupes I., Jia Z., Young P.G. Ssp1 promotes actin depolymerization and is involved in stress response and new end take-off control in fission yeast. Mol. Biol. Cell. 1999;10(5):1495–1510. doi: 10.1091/mbc.10.5.1495. (1999 May) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M., Russmayer H., Grabherr R., Peterbauer C.K., Marx H. The efficient clade: lactic acid bacteria for industrial chemical production. Trends Biotechnol. 2017;35(8):756–769. doi: 10.1016/j.tibtech.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Song J.Y., Park J.S., Kang C.D., Cho H.Y., Yang D., Lee S., Cho K.M. Introduction of a bacterial acetyl-CoA synthesis pathway improves lactic acid production in Saccharomyces cerevisiae. Metab. Eng. 2016;35:38–45. doi: 10.1016/j.ymben.2015.09.006. [DOI] [PubMed] [Google Scholar]
- Storici F., Lewis L.K., Resnick M.A. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 2001;19:773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- Stovicek V., Holkenbrink C., Borodina I. CRISPR/Cas system for yeast genome engineering: advances and applications. FEMS Yeast Res. 2017 doi: 10.1093/femsyr/fox030. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama A., Higuchi Y., Urushihara M., Maeda Y., Takegawa K. Production of 3-hydroxypropionic acid via the malonyl-CoA pathway using recombinant fission yeast strains. J. Biosci. Bioeng. 2017 doi: 10.1016/j.jbiosc.2017.04.015. (in press) [DOI] [PubMed] [Google Scholar]
- Takegawa K., Tohda H., Sasaki M., Idiris A., Ohashi T., Mukaiyama H., Giga-Hama Y., Kumagai H. Production of heterologous proteins using the fission-yeast (Schizosaccharomyces pombe) expression system. Biotechnol. Appl. Biochem. 2009;53:227–235. doi: 10.1042/BA20090048. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Kondo A. Cell surface engineering of industrial microorganisms for biorefining applications. Biotechnol. Adv. 2015;33:1403–1411. doi: 10.1016/j.biotechadv.2015.06.002. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Yamada R., Ogino C., Kondo A. Recent developments in yeast cell surface display toward extended applications in biotechnology. Appl. Microbiol. Biotechnol. 2012;95:577–591. doi: 10.1007/s00253-012-4175-0. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Matsumoto S., Yamada M., Yamada R., Matsuda F., Kondo A. Display of active beta-glucosidase on the surface of Schizosaccharomyces pombe cells using novel anchor proteins. Appl. Microbiol. Biotechnol. 2013;97:4343–4352. doi: 10.1007/s00253-013-4733-0. [DOI] [PubMed] [Google Scholar]
- Tsai C.S., Shi J.L., Beehler B.W., Beck B. Enzyme activities of D-glucose metabolism in the fission yeast Schizosaccharomyces pombe. Can. J. Microbiol. 1992;38:1313–1319. doi: 10.1139/m92-216. [DOI] [PubMed] [Google Scholar]
- Valli M., Sauer M., Branduardi P., Borth N., Porro D., Mattanovich D. Improvement of lactic acid production in Saccharomyces cerevisiae by cell sorting for high intracellular pH. Appl. Environ. Microbiol. 2006;72:5492–5499. doi: 10.1128/AEM.00683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Maris A.J., Geertman J.M., Vermeulen A., Groothuizen M.K., Winkler A.A., Piper M.D., van Dijken J.P., Pronk J.T. Directed evolution of pyruvate decarboxylase-negative Saccharomyces cerevisiae, yielding a C2-independent, glucose-tolerant, and pyruvate-hyperproducing yeast. Appl. Environ. Microbiol. 2004;70:159–166. doi: 10.1128/AEM.70.1.159-166.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rossum H.M., Kozak B.U., Pronk J.T., van Maris A.J. Engineering cytosolic acetyl-coenzyme A supply in Saccharomyces cerevisiae: pathway stoichiometry, free-energy conservation and redox-cofactor balancing. Metab. Eng. 2016;36:9. doi: 10.1016/j.ymben.2016.03.006. [DOI] [PubMed] [Google Scholar]
- Wood V., Gwilliam R., Rajandream M.A., Lyne M., Lyne R., Stewart A., Sgouros J., Peat N., Hayles J., Baker S., Basham D., Bowman S., Brooks K., Brown D., Brown S., Chillingworth T., Churcher C., Collins M., Connor R., Cronin A., Davis P., Feltwell T., Fraser A., Gentles S., Goble A., Hamlin N., Harris D., Hidalgo J., Hodgson G., Holroyd S., Hornsby T., Howarth S., Huckle E.J., Hunt S., Jagels K., James K., Jones L., Jones M., Leather S., McDonald S., McLean J., Mooney P., Moule S., Mungall K., Murphy L., Niblett D., Odell C., Oliver K., O'Neil S., Pearson D., Quail M.A., Rabbinowitsch E., Rutherford K., Rutter S., Saunders D., Seeger K., Sharp S., Skelton J., Simmonds M., Squares R., Squares S., Stevens K., Taylor K., Taylor R.G., Tivey A., Walsh S., Warren T., Whitehead S., Woodward M., Volckaert G., Aert R., Robben J., Grymonprez B., Weltjens I., Vanstreels E., Rieger M., Schäfer M., Müller-Auer S., Gabel C., Fuchs M., Düsterhöft A., Fritzc C., Holzer E., Moestl D., Hilbert H., Borzym K., Langer I., Beck A., Lehrach H., Reinhardt R., Pohl T.M., Eger P., Zimmermann W., Wedler H., Wambutt R., Purnelle B., Goffeau A., Cadieu E., Dréano S., Gloux S., Lelaure V., Mottier S., Galibert F., Aves S.J., Xiang Z., Hunt C., Moore K., Hurst S.M., Lucas M., Bowman M., Gaillardin C., Tallada V.A., Garzon A., Thode G., Daga R.R., Cruzado L., Jimenez J., Sánchez M., del Rey F., Benito J., Domínguez A., Revuelta J.L., Moreno S., Armstrong J., Forsburg S.L., Cerutti L., Lowe T., McCombie W.R., Paulsen I., Potashkin J., Shpakovski G.V., Ussery D., Barrell B.G., Nurse P. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Yamada R., Wakita K., Mitsui R., Ogino H. Enhanced d-lactic acid production by recombinant Saccharomyces cerevisiae following optimization of the global metabolic pathway. Biotechnol. Bioeng. 2017;114(9):2075–2084. doi: 10.1002/bit.26330. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Lieberman H.B. Schizosaccharomyces pombe: a model for molecular studies of eukaryotic genes. DNA Cell Biol. 1995;14:359–371. doi: 10.1089/dna.1995.14.359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material