Abstract
Regenerative medicine has emerged as an important discipline that aims to repair injury or replace damaged tissues or organs by introducing living cells or functioning tissues. Successful regenerative medicine strategies will likely depend upon a simultaneous optimization strategy for the design of biomaterials, cell-seeding methods, cell-biomaterial interactions and molecular signaling within the engineered tissues. It remains a challenge to image three-dimensional (3-D) structures and functions of the cell-seeded scaffold in mesoscopic scale (>2~3 mm). In this study, we utilized angled fluorescence laminar optical tomography (aFLOT), which allows depth-resolved molecular characterization of engineered tissues in 3-D to investigate cell viability, migration and bone mineralization within bone tissue engineering scaffolds in situ.
Keywords: Mesoscopic fluorescence tomography, Bone tissue scaffold, 3-D cell printing, Cell migration, Bone mineralization
1. Introduction
Bone tissue engineering scaffolds act as vehicles for the delivery of progenitor cell populations or support structures for surrounding tissue ingrowth. Scaffold properties often determine the success of an engineered tissue and scaffolds must be designed for a specific application. Many outcomes shown that key parameters can allow for improved cell migration, proliferation and vascularization (Karageorgiou and Kaplan 2005). To improve tissue regeneration and integration, a scaffold should mimic surrounding tissue morphology, structure and function. The scaffold environment should promote cellular interactions and signaling and, as a result, differentiation. One challenge is to comprehensively observe and quantify the distribution, migration and differentiation of seeded cells throughout the scaffold. Bone tissue engineering could benefit from a precise and noninvasive imaging method to follow cells’ activities. Such an imaging technology could provide useful information on the interplay of scaffold designs, cells and chemical/environmental cues to optimize the development of bone grafts.
Currently, fluorescent confocal microscopy (FCM) is the gold standard method for quantifying 3-D cell distribution. FCM can visualize cells and molecules via a wide variety of fluorescence probes and is a powerful technique for molecular imaging. However, FCM has a limited imaging depth of 100 μm (Tan et al. 2004). In addition, two-photon microscopy (TPM) has been used to image engineered tissues (Tan et al. 2004), but is also associated with a limited penetration depth of 500 μm (So et al. 1998; Tan et al. 2007; Vroom et al. 1999). The current state-of-the-art method for quantifying 3-D cell distribution, migration or differentiation involves FCM imaging of cryo-sectioned scaffolds and 3-D image recompiling (Thevenot et al. 2008). Although robust, this approach is destructive and time-consuming. To overcome the depth limitation of confocal microscopy, fluorescence laminar optical tomography (FLOT), or mesoscopic fluorescence molecular tomography (MFMT), was developed. It uses an array of photon detectors or a charge-coupled device (CCD) camera to collect photons emitted from locations at different distances away from the illumination position, and then reconstructs the depth-resolved images through mathematical deconvolution (Bjorn et al. 2010; Hillman et al. 2007; Ozturk et al. 2016; Ozturk et al. 2013; Ozturk et al. 2014; Tang et al. 2016c; Yuan et al. 2009a; Yuan et al. 2009b; Zhao et al. 2012). FLOT can provide an axial resolution of 100–200 μm with 2–3 mm penetration depths (Chen et al. 2010; Hillman et al. 2007; Hillman et al. 2004; Tang et al. 2016a; Tang et al. 2016b). FLOT recently was applied to 3-D imaging of tissue engineering scaffolds (Ozturk et al. 2013; Zhao et al. 2012), quantifying depth-resolved distribution of fluorescence-labelled tumors (Chen et al. 2010; Ozturk et al. 2014) and visualizing neural activities in mouse brains in vivo (Tang et al. 2016a; Tang et al. 2016b). Recent studies demonstrated that angled illumination or detection modification (termed aFLOT) can improve both resolution and depth sensitivity (Chen and Chen 2011; Tang et al. 2016a; Tang et al. 2016b).
Therefore, aFLOT represents a promising novel imaging platform to nondestructively quantify depth-resolved information in cell-scaffold interactions in situ. The proposed study aims at investigating different applications of aFLOT in bone tissue engineering. First, we demonstrated the depth-resolve capability of aFLOT and the ability to quantify cell density inside a scaffold. Then, 3-D-printed bone osteons were evaluated using aFLOT to show its capability in accurately differentiating distinct cell populations. The 3-D migration of cells was also visualized using aFLOT. Finally, aFLOT was used to image the mineralization of alginate scaffolds seeded with human mesenchymal stem cells (hMSCs) by measuring hydroxyapatite (HA) deposition.
2. Materials and methods
2.1. Mesoscopic fluorescence tomography
2.1.1. The aFLOT system
The aFLOT system in Fig. S1 is similar to that reported previously (Chen et al. 2012; Tang et al. 2017; Tang et al. 2016b). Pigtailed 637-nm and 520-nm laser diodes were used as light sources (LP637-SF70 and LP520-SF15, Thorlabs Inc, Newton NJ, USA) to illuminate the samples at two wavelengths in a time-shared manner. A cylindrical lens was used to shape the collimated light into a line-field illumination with a full line-width at the half maximum of ~26 μm at the focal plane (Tang et al. 2017; Tang et al. 2016b). An iris adjusted the length of the line illumination. The backscattered light and emitted fluorescent light were collected through an objective lens, a filter wheel (695-nm and 561-nm long-pass emission filters for fluorescence imaging and no filter for reflectance images), and finally onto a 12-bit CCD camera (EM-CCD, PCO-TECH Inc, Romulus MI, USA) (Tang et al. 2017; Tang et al. 2016b). A pair of polarizers was applied to eliminate the specular reflection from the sample surface. The illumination angle was set at 45°, rendering an ~30° transmission angle in engineered tissues (n~1.33) (Tang et al. 2017; Tang et al. 2016b). During imaging, the focal plane was set slightly below the sample surface to compensate for the defocusing effect. The sample was scanned by moving laterally in the scanning direction X (perpendicular to the line illumination direction Y) using a motorized stage (ESP301, Newport Corp, Irvine CA, USA).
2.1.2. Data acquisition and image reconstruction
Before aFLOT data acquisition, the excitation laser line was first focused on the border (X=0) of the sample field of view (FOV) (Tang et al. 2017; Tang et al. 2016b). At each scanning position, one 2-D XY image (512 × 512, with pixel dimension of 30.8 μm) was obtained with 100 ms exposure times. The sample was then moved by a motorized stage in X direction with a step size of 30.8 μm to the next illumination/collection position. Another 2-D XY image was acquired. This process was repeated until scanning was completed on the entire sample (Tang et al. 2017; Tang et al. 2016b). The raw measurement had the format of XYT. A typical 3-D dataset recorded 512 × 512 × 300 XYT voxels. To fit the experimental data to the theoretical model used for 3-D reconstruction, the sample surface needs to be found (Tang et al. 2016b). The same FOV was scanned at lower laser power to obtain the surface profile of the samples. The emission filter was removed by rotating the filter wheel during the acquisition of the reflectance images (Tang et al. 2017; Tang et al. 2016b). Without the emission filter, most of the acquired signal was from the reflection of the illumination light at the sample surface, which would then be used to indicate the location of the sample-air interface. The raw measurement of reflectance dataset had the same format of XYT (X=512, Y=512, T=300) (Tang et al. 2016b).
First, the raw measurements of the reflectance data were directly stacked together according to the geometrical relationship between the illumination plane and the detection FOV (Huisken et al. 2004; Tang et al. 2017; Tang et al. 2016b) to indicate the contour of the alginate bead scaffolds. To reconstruct the 3-D aFLOT fluorescence images, image reconstruction was applied using the same method as described previously (Tang et al. 2017; Tang et al. 2016b). Briefly, we utilized first-order Born approximation to obtain a linear relationship between the CCD measurement F and the fluorophore distribution C (i.e., for each FOVXZ, F=JC, with J the weight or sensitivity matrix) (Bjorn et al. 2010; Chen et al. 2012; Hillman et al. 2004; Ouakli et al. 2010; Tang et al. 2017; Tang et al. 2016b). Monte Carlo simulation and the reciprocity principle were used to constitute J. J was later decomposed by singular value decomposition (SVD) (Culver et al. 2001; Tang et al. 2017; Tang et al. 2016b). Lastly, the undetermined system was solved by least square fitting and Tikhonov regularization (Chen and Chen 2011; Hillman et al. 2004; Tang et al. 2016b), with the regularization parameter determined by the L-curve criterion (Hansen and O’Leary 1993; Intes et al. 2003; Zhao et al. 2012). In this experiment, for each FOVXZ, 200 source-detector pairs and 200 scanning positions starting from the illumination surface were chosen to constitute F. Each reconstructed FOVXZ was composed of 200 × 200 pixels with the same pixel size of ~30.8 μm. The 3-D FLOT image (FOVXYZ) was constituted by juxtaposing an individual FOVXZ along the Y direction (Tang et al. 2017).
2.2. Cell culture
Human MSCs (Lonza Walkersville, Inc, Walkersville, MD, USA) were cultured in control media consisting of Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco), 1.0% v/v penicillin/streptomycin (Gibco), 0.1 mM non-essential amino acids (Gibco), and 4 mM L-glutamine (Gibco, as per the manufacturer’s specifications). hMSCs were expanded on tissue culture polystyrene flasks with media changes every 3 days. Prior to the experiment, cells were passaged using trypsin/EDTA (Lonza). L929 cells (ATCC, Manassas, VA, USA) were cultured as per the manufacturer’s specifications with Minimum Essential Medium (Life Technologies, Thermo Fisher) and 10% horse serum (Life Technologies). Cells were plated and allowed to grow to 80% confluency before initiating the assays.
2.3. Sample fabrication and cell seeding
2.3.1. Gelatin methacrylate synthesis
Gelatin methacrylate was synthesized according to previously published methods (Kuo et al. 2016). Type A porcine skin gelatin (Millipore Sigma, St. Louis, MO, USA; 300 bloom) was mixed at 10% (w/v) into phosphate buffered saline (PBS; Thermo Fisher) at 50 °C for 20 min. Methacrylic anhydride (MA; Millipore Sigma) was added at the ratio of 0.6 g MA/g gelatin under rigorous stirring for an hour. The reactants were then diluted 2-fold with PBS to stop the reaction. After centrifugation, the pellet was discarded and the supernatant was dialyzed against deionized water using dialysis cassettes (10 kDa MWCO, Thermo Fisher) for at least 3 days at 50 °C to remove salts and excess MA. The dialyzed GelMA was then lyophilized and kept at −80 °C for long-term storage. To form samples, GelMA was rehydrated in PBS for 10 min at 50 °C. Photoinitiator (2-hydroxy-1-(4-hydroxyethoxy)phenyl)-2-methyl-1-propanone (BASF Corp, Southfield, MI, USA) was then added into the GelMA solution at a concentration of 0.5% (w/v). If needed, cells were added at this point, and the prepolymer solution was UV cured into the desired shape for 1 min (0.09 mW/cm2) using a UV box (VWR, Radnor, PA, USA).
2.3.2. GelMA layered and 3-D printed scaffolds fabrication
To demonstrate the feasibility of aFLOT imaging of cell distribution inside engineered tissue scaffolds, L929 fibroblasts were labelled with orange fluorescent dye (CellTracker™ Orange CMRA Dye, 548⁄576 nm, Thermo Fisher). The labelled cells were suspended into a solution of 10w/v% gelatin methacrylate (GelMA), and 3-D constructs were obtained by photocrosslinking of the solution. The constructs were seeded at concentrations of 0.015 × 106, 0.05 × 106, 0.1 × 106, 0.5 × 106, 1 × 106 and 2 × 106 cells/mL. The hydrogels with different cell concentrations were then imaged by the aFLOT system.
To demonstrate the depth-resolve capability of the aFLOT system, we fabricated a three-layer structure with an acellular GelMA layer in the middle and two layers with fluorescent labelled cells (1M cells/mL) as illustrated in Fig. 2(E). The L929 fibroblasts were labelled either with orange fluorescent dye (CellTracker™ Orange CMRA Dye, 548⁄576 nm, Thermo Fisher) or far red fluorescent dye (DiIC18(5)-DS, 650/670 nm, Thermo Fisher) to demonstrate the multiwavelength capability of the aFLOT system. We also constructed a three-layer GelMA cylinder with far red labelled cells (2M cells/mL) in the middle, orange labelled cells at the left and right sides (or the other way, orange labelled cells in the middle, far red labelled cells at the left and right sides) as shown in Fig. S2.
Fig. 2.
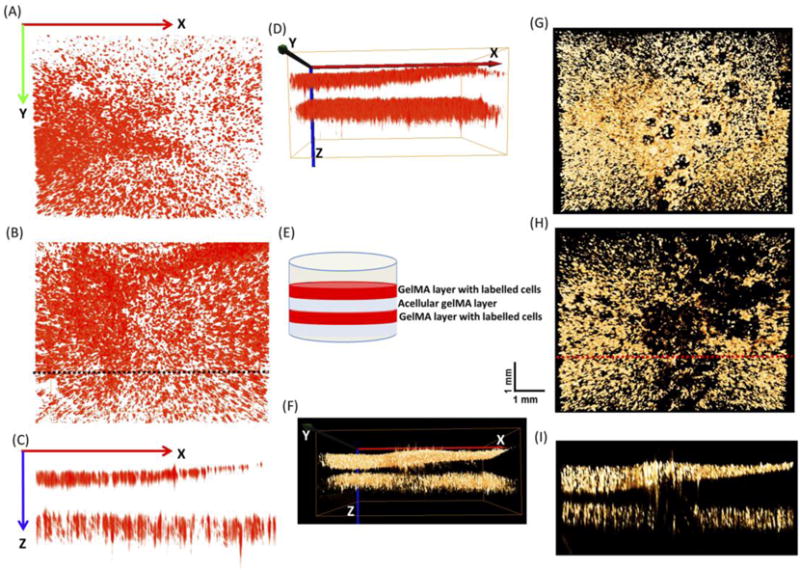
3-D aFLOT reconstructed three-layer structures (A) Upper layer of the 3-D aFLOT reconstructed three-layer structure labelled by far red fluorescence. (B) Lower layer of the 3-D aFLOT reconstructed three-layer structure labelled by far red fluorescence. (C) Cross section of the 3-D aFLOT reconstructed three-layer structure labelled by far red fluorescence as indicated by the black dashed line in Fig. 2(B). (D) Side view of the 3-D aFLOT reconstructed three-layer structure labelled by far red fluorescence. (E) The three-layer gel structure. (F) Side view of the 3-D aFLOT reconstructed three-layer gel structure labelled by orange fluorescence. (G) Upper layer of the 3-D aFLOT reconstructed three-layer structure labelled by orange fluorescence. (H) Lower layer of the 3-D aFLOT reconstructed three-layer structure labelled by orange fluorescence. (I) Cross section of the 3-D aFLOT reconstructed three-layer structure labelled by orange fluorescence, as indicated by the red dashed line in Fig. 2(H).
Furthermore, samples were 3-D printed using an extrusion-based bioprinter (3D-Bioplotter; EnvisionTEC Inc, Dearborn, MI, USA). The bioink containing orange- or far red-labelled cells was loaded into the low-temperature printer head and allowed to equilibrate for 30 min at 22 °C. Printed constructs were UV cured for 30 s (0.09 mW/cm2) using a UV box (VWR) (Trachtenberg et al. 2014). In this study, three letters, “UMD”, were printed using a labelled-cells suspension of GelMA, with far red- and orange-labelled cells, respectively. Another construct mimicking bone osteon was also printed, as shown in Fig. 3(B).
Fig. 3.
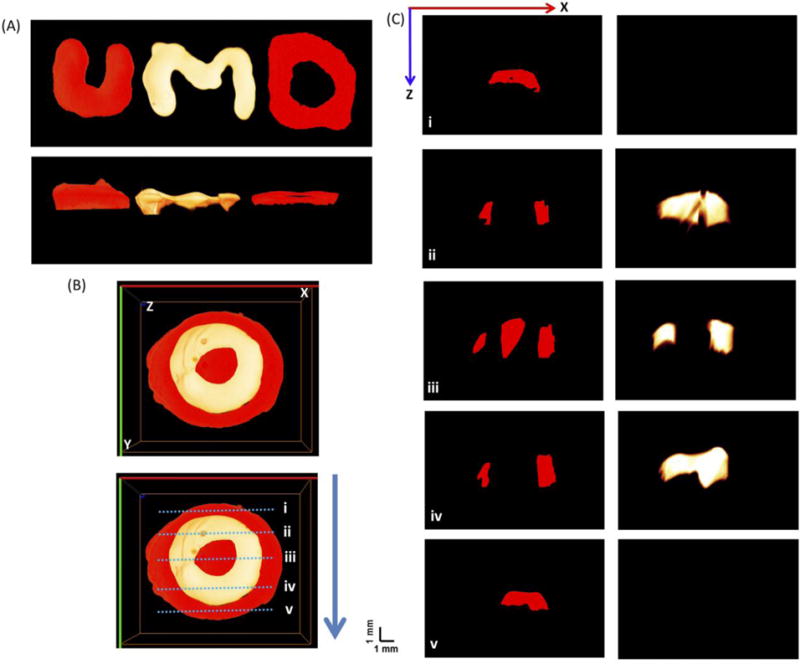
(A) Top view and side views of the 3-D aFLOT reconstructed 3-D printed U,M,D letters labelled by far red fluorescence and orange fluorescence, respectively. (B) Top view of the 3-D aFLOT reconstructed 3-D printed ‘osteon’ structure labelled by far red fluorescence and orange fluorescence, respectively. (C) Cross sections of the 3-D aFLOT reconstructed 3-D printed ‘osteon’ structure from the blue dashed lines labelled in (B). (3D renderings of the 3-D printed structures in Supplementary Video 1 and 2).
2.4. Cell migration model
To demonstrate the feasibility of aFLOT imaging to track cell migration inside a scaffold, orange-labelled (CellTracker™ Orange CMRA Dye, Thermo Fisher) hMSCs were encapsulated in 10w/v% GelMA to print cylindrical scaffolds at a concentration of 4 × 106 cells/mL. Acellular GelMA was printed along the periphery of the cylinder, while cells were printed in the center. Samples were cultured in growth media and imaged at different time points over the course of four days to study cell migration from the center toward the edge of the gel.
2.5. Cell differentiation study
To demonstrate the feasibility of aFLOT imaging to visualize cell differentiation, hMSCs were cultured in a tubular perfusion bioreactor under differentiation conditions (Yeatts et al. 2012). HA deposition, an indicator of hMSCs’ differentiation into osteoblasts, was assessed using both aFLOT and histology.
2.5.1. Alginate beads fabrication
Alginate beads were fabricated as previously described in the literature (Nguyen et al. 2015). Alginate solution (Millipore Sigma) was sterilized via sterile filtration then mixed with a cell pellet containing hMSCs. Beads were seeded with orange fluorescent dye-labelled cells (CellTracker™ Orange CMRA Dye, 548⁄576 nm, Thermo Fisher) at a concentration of 2 × 106 cells/mL. This solution was added dropwise by 20-gauge syringe to a 0.10 M calcium chloride solution, in which the alginate was ionically crosslinked into beads. The solution was stirred for 15 min. Beads were transferred to the tubular perfusion system, as described in (Yeatts et al. 2013; Yeatts and Fisher 2011), and cultured for 14 days in control media supplemented with 100 nM dexamethasone (Millipore Sigma), 10 mM β-glycerophosphate and 173 μM ascorbic acid.
2.5.2. HA deposition assay
For each time point during the 14-day study, alginate beads were removed from the bioreactor and washed for 15 min in PBS. The beads were stained for HA deposition in 300 μl of OsteoSense 680EX (Zaheer et al. 2001) (PerkinElmer, Waltham, MA, USA) per mL of PBS. After 15 min incubation, the beads were rinsed again and imaged using the aFLOT system. The labelled cells were imaged using the 520-nm laser diode, and the labelled HA was imaged by the 637-nm laser diode with 561-nm/695-nm long-pass emission filters, respectively.
2.5.3. Histology
At each time point, beads were collected and fixed in 4% paraformaldehyde (Millipore Sigma) for 12 h at 4 °C. Following fixation, the beads were dehydrated for histological processing by ethanol washes followed by two CitriSolv (Thermo Fisher) washes. The samples were then embedded in paraffin (Thermo Fisher) and sectioned into 5-μm-thick sections and placed on glass slides. Sections were oven dried at 64 °C for 2 h, deparaffinized in CitriSolv, and rehydrated in ethanol. Von Kossa staining was performed to visualize mineralization using a Nuclear Fast Red (Poly Scientific R&D Corp, Bay Shore, NY) counterstain following standard protocols.
3. Results and discussion
3.1. Imaging of cell distribution using aFLOT
We demonstrated here the feasibility of using aFLOT to image the cell distribution and quantify cell density inside a 3-D scaffold. At a cell density of 0.015 × 106 cells/mL, the fluorescent spots were sparsely distributed as shown in Fig. 1(A). With increasing cell density, the fluorescence from the 3-D reconstructed images became denser [Fig. 1(B–E)]. When the cell density was greater than 2 × 106 cells/mL, the fluorescence-labelled cell clusters became too dense to be resolved [i.e., the image became homogeneous, Fig. 1(F)]. To further quantify the correlation between aFLOT-detected cell number and the true cell density, the numbers of fluorescent pixels over a preset threshold value (Background + (Max-Background)/2) were counted and averaged from 10 random 3-D cubic voxels (each contained 30 × 30 × 30 pixels) from 3-D reconstructed images with different cell densities. The averaged pixel numbers were then normalized and plotted as a function of the true cell densities, as shown in Fig. 1(G). The pixel value of the 2 × 106 cells/mL group was set as 1, and was used to normalize the results from other groups. The normalized averaged pixel numbers show an excellent linear response to the true cell densities with R2 = 0.99. This demonstrates the capability of aFLOT to correlate fluorescence imaging results with cell density and quantify cell growth in 3-D samples in a nondestructive manner. Therefore, this technique presents the advantage of monitoring the same sample over time, unlike standard cell viability/proliferation assays, such as XTT (2,3-Bis[2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxyanilide tetrazolium salt), which is primarily an end point assay.
Fig. 1.
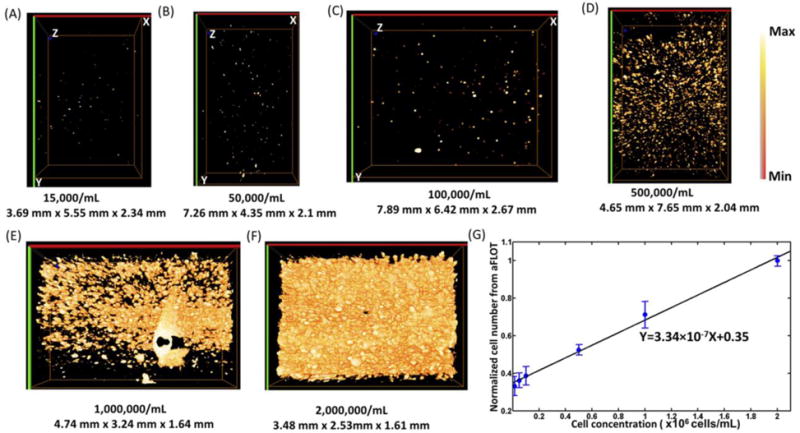
(A) 3-D aFLOT reconstruction images of orange-labelled fibroblast cells 15,000/mL (A), 50,000/mL (B), 100,000/mL (C), 500,000/mL (D), 1,000,000/mL(E), 2,000,000/mL (F), in the GelMA gel. (G) Normalized averaged cell numbers from aFLOT as a function of true cell concentration in the gel. The “Max” in the colormap is the maximum value in the image and “Min” represents the value: (Background + (Max-Background)/2).
3.2. Demonstrating the depth-resolve imaging capability of aFLOT
To demonstrate the depth-resolve imaging capability of our aFLOT system, different fluorescence-labelled cells were seeded in the three-layer GelMA scaffolds. Different views at different depths are shown in Fig. 2. The aFLOT system could image samples with greater than 3-mm thickness, for both wavelength channels (far red and orange). To further demonstrate the capability of the imaging system in performing multiwavelength imaging, other GelMA constructs seeded with both far red-labelled cells and orange-labelled cells were fabricated and imaged as shown in Fig. S2. Fig. 2 further indicates the feasibility of the aFLOT system to image the distribution of fluorescent-labelled cells in a 3-D scaffold with great in-depth precision and, therefore, enabling quantification of cell infiltration into hydrogel scaffolds.
In addition, to validate the capability of the aFLOT system to reconstruct more-complex structures or patterns, several samples were 3-D printed using a suspension of GelMA and far red- or orange-labelled L929 cells, as shown in Fig. 3. The aFLOT successfully reconstructed the printed patterns regardless of the spectral range, as shown in Fig. 3(A), demonstrating the ability of aFLOT to recover the patterns with accuracy.
An osteon-like structure was also 3-D printed [Fig. 3(B)] to illustrate the potential application of aFLOT in bone tissue engineering. Osteons are a repetitive unit found in cortical bones consisting of organized concentric calcified regions with interpenetrated vasculature. Osteons are represented here by two cell populations labelled with different fluorescent proteins. The biomimetic osteon-like scaffolds were also imaged, as shown in Fig. 3(B). The concentric layers, or lamellae, were successfully printed using cells labelled with different fluorescent proteins and reconstructed by the aFLOT system. Fig. 3(C) shows the cross sections of the reconstructed scaffolds from the blue dashed lines labelled in Fig. 3(B), demonstrating the capability of the aFLOT system to analyze precise areas of a sample and identify different cell populations.
3.3. Demonstrating the feasibility of aFLOT imaging to track cell migration
Cell migration is fundamental to many biological and pathological processes. Development, immune response and wound healing all rely on the directed migration of cells (Kirfel et al. 2004). Moreover, in the case of tissue engineering, understanding cell migration within the native organ is important for evaluating cellular interactions with biomaterials and, therefore, creating successful outcomes.
In this study, we tracked the migration of hMSCs within a 3-D printed GelMA scaffold from a high cell density area (4 × 106 cells/mL) to a low cell density area (no cells). As illustrated in Fig. 4(A–C), the scaffolds with 4 × 106 cells/mL have strong fluorescence, the pink color indicates the acellular GelMA reconstructed from stacking of the reflectance images. On the first day, no cells outside the high density zone can be seen. After two days, cells outside of the high density zone can be detected. Finally after four days, cells have migrated further into the no cell area. To quantify the migration distance, the cells numbers was plotted over migration distance at day 2 and day 4 as shown in Fig. 4(D). The average migrated distance was defined as ; X is the number of different migration distances, Ni is the pixel number of the migrated cells at the migration distance Di and T is the total number of the pixels that migrated. (The migrated cells are defined as the fluorescent pixels over a preset threshold value [Background + (Max-Background)/2].) The average migrated distance after two days is 650 ± 38 μm and it increased to 1208 ± 167 μm after four days. The average migration rate of 12.6 ± 2.33 μm/h is consistent with what previously has been reported in the literature (Herzmann et al. 2016; Lee et al. 2006).
Fig. 4.
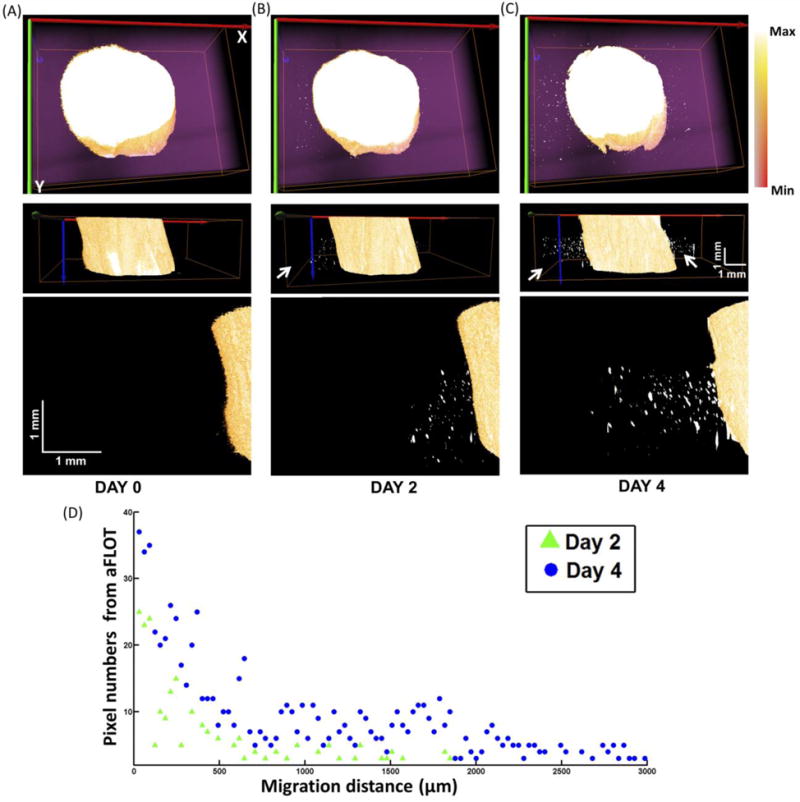
Top view, side views and zoomed in side views to the left of the 3-D aFLOT reconstructed migration model at day 0 (A), day 2 (B) and day 4 (C). The Max in the colormap is the maximum value in the image and Min represents the value: (Background + (Max-Background)/2). The migrated and proliferated cells are indicated by the white arrows. (D) Cells (Pixels) numbers was plotted over migration distance at day 2 and day 4.
The aFLOT system is a promising tool to accurately follow cell migration. It presents the advantage of accurately tracking cells in 3-D scaffolds when compared to conventional time-lapse microscopy. As migration of some cell types differs dramatically between 2-D and 3-D environments, it is critical to be able to evaluate the effect of the structure and/or biomaterial on 3-D cell migration (De Cock et al. 2012; Murphy et al. 2010).
3.4. Demonstrating the feasibility of aFLOT imaging to visualize markers of cell differentiation
The capability of aFLOT to indirectly image and quantify cell differentiation was demonstrated in this study by evaluating the mineralization of alginate scaffolds seeded with hMSCs and cultivated in a tubular perfusion system (TPS). Mechanical stimulation from a perfusion bioreactor was shown to induce osteoblastic differentiation of hMSCs (Yeatts and Fisher 2011; Yeatts et al. 2012), with a significant increase in mineralization of the scaffold.
In this study, mineralization of alginate scaffolds was evaluated using our aFLOT system along with the near-infrared (NIR) fluorescent bisphosphonate derivative (OsteoSense 680EX, PerkinElmer) that exhibits rapid and specific binding to HA in vitro and in vivo (Zaheer et al. 2001). Calibration of the aFLOT system in different HA concentrations was first carried out as shown in Fig. S3. The normalized pixel numbers from aFLOT show a linear relationship with HA percentage in the alginate beads, and the images of alginate beads doped with different HA concentrations are also shown in Fig. S3. However, due to the increasing scattering as the HA concentration increases, high mineralization results in a decrease in light penetration into the sample and, therefore, it is difficult to reconstruct deeper signals accurately. In the future, we can flip the sample to image samples from different views and thus increase the sample imaging area.
Fig. 5(A) and (B) show different views of the 3-D aFLOT reconstructed scaffolds seeded with hMSCs. Human MSC (orange-red) and HA (green-blue) distribution are superimposed with the alginate bead (pink). Three-dimensional cell distribution is clearly displayed and well coregistered with the bead contour. No HA deposition is seen at day 0. Cell distributions at different depths are further exhibited in Fig. 5(I). At day 7, HA deposition is still low; however, cell proliferation is shown by the appearance of larger ‘clusters’ [Fig. 5(E) and (J)]. At day 14, cells have continued to proliferate, as more large ‘clusters’ can be seen, and the HA deposition (green-blue) can be clearly seen in the alginate beads as shown in Fig. 5(B) and (F). Three-dimensional distribution of HA deposition and location with respect to the bead can be observed more clearly in Fig. 5(C) and (G). Most of the HA was deposited in the center of the bead, which can be clearly revealed at the depth of 1.5 mm in Fig. 5(M).
Fig. 5.
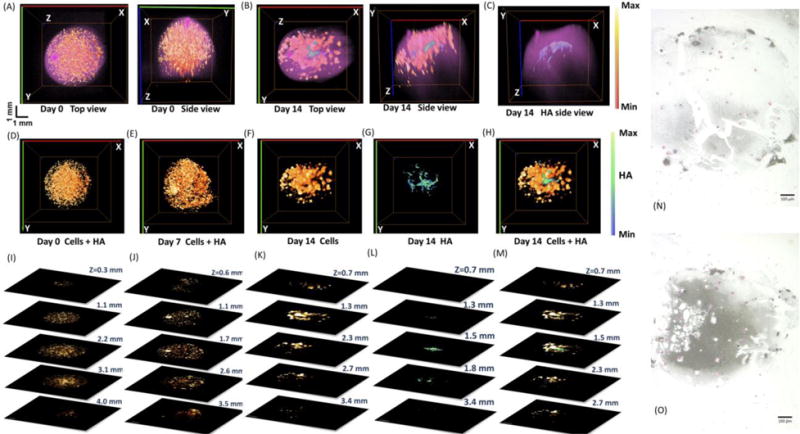
Top and side views of the 3-D aFLOT reconstructed hMSCs (orange-red) and HA (green-blue) distribution superimposed with the bead surface (pink) in the alginate beads at day 0 (A) and day 14 (B). (C) Side view of the 3-D aFLOT reconstructed HA distribution (green-blue) superimposed with the bead surface (pink) in the alginate beads at day 14. (D)–(F) Top views of the 3-D aFLOT reconstructed hMSCs (orange-red) at day 0 (D), day 7 (E) and day 14 (F). (G) Top view of the 3-D aFLOT reconstructed HA (green-blue) distribution at day 14. (H) Top views of the 3-D superimposed aFLOT reconstructed hMSCs (orange-red) and HA (green-blue) distribution in the alginate beads at day 14. (I)–(M) Corresponding XY cross sections of the 3-D aFLOT reconstructed alginate beads at different depths to (D)–(H). Scale bar: 1 mm. (N)–(O) Von Kossa staining of alginate scaffold at Day 7 (N) and Day 14 (O), respectively. Mineralization is stained in black, while cell nuclei appear in pink. (3D renderings of the reconstructed alginate beads in Supplementary Video 3–5).
To confirm mineralization of the scaffolds, Von Kossa staining was performed alongside aFLOT imaging. The Von Kossa stain uses silver nitrate to detect calcium salt deposits with a black or brownish-black precipitate, and nuclear fast red to stain cell nuclei red and the cell cytoplasm pink. Von Kossa staining confirmed increased mineralization in the alginate scaffolds over the 14-day study in Fig. 5(N–O). In addition, cell nuclei (shown in pink) are observed to increase in number from day 7 to day 14. This study demonstrates that aFLOT imaging is able to image osteoblastic activity and offers a practical, time-saving, nondestructive alternative to conventional histology.
4. Conclusions
In this paper, we demonstrated various potential applications of aFLOT in bone tissue engineering, and, more specifically, we nondestructively quantified the depth-resolved information of cell-scaffold interactions. We successfully quantified 3-D cell density, characterized cell distribution within a 3-D scaffold and visualized the 3-D cellular migration process. Finally, we could indirectly evaluate cell differentiation by quantifying the mineralization of alginate scaffolds seeded with hMSCs, by measuring HA deposition. The aFLOT is a promising tool in regenerative medicine, allowing the noninvasive investigation of structural, cellular and molecular information. Therefore, use of aFLOT enables both in vitro and in vivo longitudinal studies of cell viability, migration, proliferation, differentiation and signaling within 3-D scaffolds. This imaging technology could also provide useful information on the interplay of scaffold designs and materials, cell-seeding methods and chemical/environmental cues to optimize the development of tissue grafts.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the U.S. National Institutes of Health under Award No. R01 EB014946. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Author contributions
Y.C. and J.F. initiated this project. Q.T. and C.P. designed and performed experiments, processed data and wrote the manuscript. J.L., K.N. and T.G. helped with data analysis. Y.C., J.F. and J.C. supervised the project.
Competing financial interests
The authors declare no competing financial interests.
References
- Bjorn S, Ntziachristos V, Schulz R. Mesoscopic Epifluorescence Tomography: Reconstruction of superficial and deep fluorescence in highly-scattering media. Optics Express. 2010;18(8):8422–8429. doi: 10.1364/OE.18.008422. [DOI] [PubMed] [Google Scholar]
- Chen C-W, Yeatts AB, Coates EE, Fisher JP, Chen Y. OSA Technical Digest. Miami, Florida: Optical Society of America; 2012. Experimental Demonstration of Angled Fluorescent Laminar Optical Tomography for Tissue Engineering; p. BTu4A.5. 2012/04/28. [Google Scholar]
- Chen CW, Chen Y. Optimization of Design Parameters for Fluorescence Laminar Optical Tomography. Journal of Innovative Optical Health Sciences. 2011;4(3):309–323. [Google Scholar]
- Chen Y, Yuan S, Wierwille J, Naphas R, Li Q, Blackwell TR, Winnard PT, Raman V, Glunde K. Integrated Optical Coherence Tomography (OCT) and Fluorescence Laminar Optical Tomography (FLOT) IEEE J Selected Topics in Quantum Electronics. 2010;16(4):755–766. [Google Scholar]
- Culver JP, Ntziachristos V, Holboke MJ, Yodh AG. Optimization of optode arrangements for diffuse optical tomography: A singular-value analysis. Optics Letters. 2001;26(10):701–703. doi: 10.1364/ol.26.000701. [DOI] [PubMed] [Google Scholar]
- De Cock LJ, De Wever O, Hammad H, Lambrecht BN, Vanderleyden E, Dubruel P, De Vos F, Vervaet C, Remon JP, De Geest BG. Engineered 3D microporous gelatin scaffolds to study cell migration. Chemical Communications. 2012;48(29):3512–3514. doi: 10.1039/c2cc17006j. [DOI] [PubMed] [Google Scholar]
- Hansen PC, O’Leary DP. The use of the L-curve in the regularization of discrete ill-posed problems. SIAM Journal on Scientific Computing. 1993;14(6):1487–1503. [Google Scholar]
- Herzmann N, Salamon A, Fiedler T, Peters K. Analysis of migration rate and chemotaxis of human adipose-derived mesenchymal stem cells in response to LPS and LTA in vitro. Experimental cell research. 2016;342(2):95–103. doi: 10.1016/j.yexcr.2016.03.016. [DOI] [PubMed] [Google Scholar]
- Hillman EMC, Bernus O, Pease E, Bouchard MB, Pertsov A. Depth-resolved optical imaging of transmural electrical propagation in perfused heart. Optics Express. 2007;15(26):17827–17841. doi: 10.1364/oe.15.017827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman EMC, Boas DA, Dale AM, Dunn AK. Laminar optical tomography: demonstration of millimeter-scale depth-resolved imaging in turbid media. Optics Letters. 2004;29(14):1650–1652. doi: 10.1364/ol.29.001650. [DOI] [PubMed] [Google Scholar]
- Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EHK. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305(5686):1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- Intes X, Ripoll J, Chen Y, Nioka S, Yodh A, Chance B. In vivo continuous-wave optical breast imaging enhanced with Indocyanine Green. Medical physics. 2003;30(6):1039–1047. doi: 10.1118/1.1573791. [DOI] [PubMed] [Google Scholar]
- Karageorgiou V, Kaplan D. Porosity of 3D biomaterial scaffolds and osteogenesis. Biomaterials. 2005;26(27):5474–91. doi: 10.1016/j.biomaterials.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Kirfel G, Rigort A, Borm B, Herzog V. Cell migration: mechanisms of rear detachment and the formation of migration tracks. Eur J Cell Biol. 2004;83(11–12):717–24. doi: 10.1078/0171-9335-00421. [DOI] [PubMed] [Google Scholar]
- Kuo C-Y, Eranki A, Placone JK, Rhodes KR, Aranda-Espinoza H, Fernandes R, Fisher JP, Kim PCW. Development of a 3D Printed, Bioengineered Placenta Model to Evaluate the Role of Trophoblast Migration in Preeclampsia. ACS Biomaterials Science & Engineering. 2016;2(10):1817–1826. doi: 10.1021/acsbiomaterials.6b00031. [DOI] [PubMed] [Google Scholar]
- Lee DH, Park BJ, Lee M-S, Lee JW, Kim JK, Yang H-C, Park J-C. Chemotactic migration of human mesenchymal stem cells and MC3T3-E1 osteoblast-like cells induced by COS-7 cell line expressing rhBMP-7. Tissue engineering. 2006;12(6):1577–1586. doi: 10.1089/ten.2006.12.1577. [DOI] [PubMed] [Google Scholar]
- Murphy CM, Haugh MG, O’Brien FJ. The effect of mean pore size on cell attachment, proliferation and migration in collagen–glycosaminoglycan scaffolds for bone tissue engineering. Biomaterials. 2010;31(3):461–466. doi: 10.1016/j.biomaterials.2009.09.063. [DOI] [PubMed] [Google Scholar]
- Nguyen B-NB, Ko H, Moriarty RA, Etheridge JM, Fisher JP. Dynamic Bioreactor Culture of High Volume Engineered Bone Tissue. Tissue Engineering Part A. 2015;22(3–4):263–271. doi: 10.1089/ten.tea.2015.0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouakli N, Guevara E, Dubeau S, Beaumont E, Lesage F. Laminar optical tomography of the hemodynamic response in the lumbar spinal cord of rats. Optics Express. 2010;18(10):10068–10077. doi: 10.1364/OE.18.010068. [DOI] [PubMed] [Google Scholar]
- Ozturk MS, Chen CW, Ji R, Zhao LL, Nguyen BNB, Fisher JP, Chen Y, Intes X. Mesoscopic Fluorescence Molecular Tomography for Evaluating Engineered Tissues. Annals Of Biomedical Engineering. 2016;44(3):667–679. doi: 10.1007/s10439-015-1511-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk MS, Lee VK, Zhao LL, Dai GH, Intes X. Mesoscopic fluorescence molecular tomography of reporter genes in bioprinted thick tissue. Journal Of Biomedical Optics. 2013;18(10) doi: 10.1117/1.JBO.18.10.100501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk MS, Rohrbach D, Sunar U, Intes X. Mesoscopic Fluorescence Tomography of a Photosensitizer (HPPH) 3D Biodistribution in Skin Cancer. Academic Radiology. 2014;21(2):271–280. doi: 10.1016/j.acra.2013.11.009. [DOI] [PubMed] [Google Scholar]
- So PTC, Kim H, Kochevar IE. Two-photon deep tissue ex vivo imaging of mouse dermal and subcutaneous structures. Optics Express. 1998;3(9):339–350. doi: 10.1364/oe.3.000339. [DOI] [PubMed] [Google Scholar]
- Tan W, Sendemir-Urkmez A, Fahrner LJ, Jamison R, Leckband D, Boppart SA. Structural and functional optical imaging of three-dimensional engineered tissue development. Tissue Engineering. 2004;10(11–12):1747–1756. doi: 10.1089/ten.2004.10.1747. [DOI] [PubMed] [Google Scholar]
- Tan W, Vinegoni C, Norman JJ, Desai TA, Boppart SA. Imaging cellular responses to mechanical stimuli within three-dimensional tissue constructs. Microscopy Research And Technique. 2007;70(4):361–371. doi: 10.1002/jemt.20420. [DOI] [PubMed] [Google Scholar]
- Tang Q, Lin J, Tsytsarev V, Erzurumlu RS, Liu Y, Chen Y. Review of mesoscopic optical tomography for depth-resolved imaging of hemodynamic changes and neural activities. Neurophotonics. 2016a;4(1):011009–011009. doi: 10.1117/1.NPh.4.1.011009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Liu Y, Tsytsarev V, Lin J, Wang B, Kanniyappan U, Li Z, Chen Y. High-dynamic-range fluorescence laminar optical tomography (HDR-FLOT) Biomedical Optics Express. 2017;8(4):2124–2137. doi: 10.1364/BOE.8.002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Tsytsarev V, Frank A, Wu Y, Chen CW, Erzurumlu RS, Chen Y. In Vivo Mesoscopic Voltage-Sensitive Dye Imaging of Brain Activation. Sci Rep. 2016b;6:25269. doi: 10.1038/srep25269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Wang J, Frank A, Lin J, Li Z, Chen C-w, Jin L, Wu T, Greenwald BD, Mashimo H, et al. Depth-resolved imaging of colon tumor using optical coherence tomography and fluorescence laminar optical tomography. Biomedical Optics Express. 2016c;7(12):5218–5232. doi: 10.1364/BOE.7.005218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenot P, Nair A, Dey J, Yang J, Tang LP. Method to Analyze Three-Dimensional Cell Distribution and Infiltration in Degradable Scaffolds. Tissue Engineering Part C-Methods. 2008;14(4):319–331. doi: 10.1089/ten.tec.2008.0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtenberg JE, Mountziaris PM, Miller JS, Wettergreen M, Kasper FK, Mikos AG. Open‐source three‐dimensional printing of biodegradable polymer scaffolds for tissue engineering. Journal of Biomedical Materials Research Part A. 2014;102(12):4326–4335. doi: 10.1002/jbm.a.35108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroom JM, De Grauw KJ, Gerritsen HC, Bradshaw DJ, Marsh PD, Watson GK, Birmingham JJ, Allison C. Depth penetration and detection of pH gradients in biofilms by two-photon excitation microscopy. Applied And Environmental Microbiology. 1999;65(8):3502–3511. doi: 10.1128/aem.65.8.3502-3511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeatts AB, Both SK, Yang W, Alghamdi HS, Yang F, Fisher JP, Jansen JA. In vivo bone regeneration using tubular perfusion system bioreactor cultured nanofibrous scaffolds. Tissue Engineering Part A. 2013;20(1–2):139–146. doi: 10.1089/ten.tea.2013.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeatts AB, Fisher JP. Tubular perfusion system for the long-term dynamic culture of human mesenchymal stem cells. Tissue Eng Part C Methods. 2011;17(3):337–48. doi: 10.1089/ten.TEC.2010.0172. [DOI] [PubMed] [Google Scholar]
- Yeatts AB, Geibel EM, Fears FF, Fisher JP. Human mesenchymal stem cell position within scaffolds influences cell fate during dynamic culture. Biotechnology and Bioengineering. 2012;109(9):2381–2391. doi: 10.1002/bit.24497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan BH, Burgess SA, Iranmahboob A, Bouchard MB, Lehrer N, Bordier C, Hillman EMC. A system for high-resolution depth-resolved optical imaging of fluorescence and absorption contrast. Review Of Scientific Instruments. 2009a;80(4) doi: 10.1063/1.3117204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Li Q, Jiang J, Cable A, Chen Y. Three-dimensional coregistered optical coherence tomography and line-scanning fluorescence laminar optical tomography. Optics Letters. 2009b;34(11):1615–1617. doi: 10.1364/ol.34.001615. [DOI] [PubMed] [Google Scholar]
- Zaheer A, Lenkinski RE, Mahmood A, Jones AG, Cantley LC, Frangioni JV. In vivo near-infrared fluorescence imaging of osteoblastic activity. Nat Biotech. 2001;19(12):1148–1154. doi: 10.1038/nbt1201-1148. [DOI] [PubMed] [Google Scholar]
- Zhao LL, Lee VK, Yoo SS, Dai GH, Intes X. The integration of 3-D cell printing and mesoscopic fluorescence molecular tomography of vascular constructs within thick hydrogel scaffolds. Biomaterials. 2012;33(21):5325–5332. doi: 10.1016/j.biomaterials.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


