ABSTRACT
Bacterial β-lactamases readily inactivate most penicillins and cephalosporins by hydrolyzing and “opening” their signature β-lactam ring. In contrast, carbapenems resist hydrolysis by many serine-based class A, C, and D β-lactamases due to their unique stereochemical features. To improve the resistance profile of penicillins, we synthesized a modified penicillin molecule, MPC-1, by “grafting” carbapenem-like stereochemistry onto the penicillin core. Chemical modifications include the trans conformation of hydrogen atoms at C-5 and C-6 instead of cis, and a 6-α hydroxyethyl moiety to replace the original 6-β aminoacyl group. MPC-1 selectively inhibits class C β-lactamases, such as P99, by forming a nonhydrolyzable acyl adduct, and its inhibitory potency is ∼2 to 5 times higher than that for clinically used β-lactamase inhibitors clavulanate and sulbactam. The crystal structure of MPC-1 forming the acyl adduct with P99 reveals a novel binding mode for MPC-1 that resembles carbapenem bound in the active site of class A β-lactamases. Furthermore, in this novel binding mode, the carboxyl group of MPC-1 blocks the deacylation reaction by occluding the critical catalytic water molecule and renders the acyl adduct nonhydrolyzable. Our results suggest that by incorporating carbapenem-like stereochemistry, the current collection of over 100 penicillins and cephalosporins can be modified into candidate compounds for development of novel β-lactamase inhibitors.
KEYWORDS: β-lactamase inhibitor, carbapenem-like, class C β-lactamase
INTRODUCTION
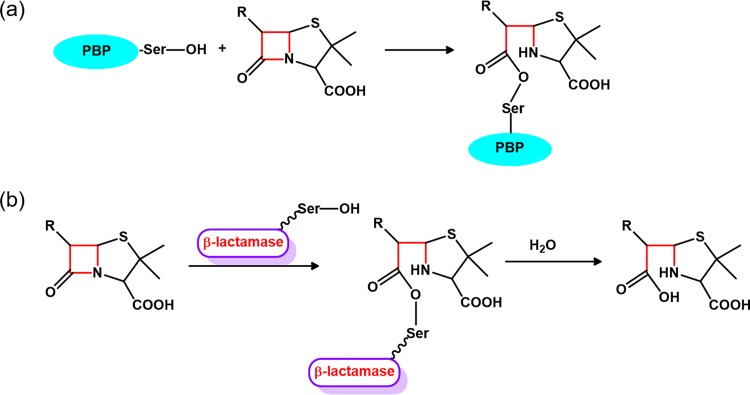
Since the discovery of penicillin G as the first β-lactam antibiotic nearly a century ago, this family of antibacterial drugs has expanded to include over 50 clinically used drugs and serve as the most effective treatment for bacterial infection. All β-lactam antibiotics target the penicillin binding proteins (PBPs), a group of dd-transpeptidases that play essential roles in bacterial cell wall synthesis (1). With the signature four-membered β-lactam ring that is spatially strained and thus reactive to hydrolysis, this family of compounds can readily form an acyl adduct with the catalytic serine residue within the penicillin-binding (PB) domain of PBPs (2) (Fig. 1a). As a result, the transpeptidase activity of PBPs is irreversibly inhibited, leading to disruption in cell wall synthesis and bacterial lysis.
FIG 1.
Mechanism of β-lactam antibiotics. (a) β-Lactam antibiotics irreversibly inhibit penicillin binding proteins (PBPs) by forming a covalent acyl adduct with the catalytic serine (Ser) residue. (b) Serine-based class A and C β-lactamases inactivate β-lactam antibiotics by hydrolyzing and “opening” the β-lactam ring in a two-step process.
However, after widespread usage of β-lactam antibiotics in the past decades, resistance toward this group of compounds has emerged on a global scale as bacteria rapidly have evolved to adapt to the selection pressure imposed by these drugs. Among the multiple resistance mechanisms reported in the literature, inactivation by bacterial β-lactamases is the most prominent one (3). So far over 1,000 β-lactamases have been reported, and these enzymes are grouped into four major classes (A to D) based on sequence similarity (4). While the exact catalytic machineries for the four classes are quite different, all β-lactamases can hydrolyze and “open” the signature β-lactam ring to render the antibiotics ineffective against PBPs (Fig. 1b). By expressing one or a few of these enzymes, bacteria readily acquire resistance against β-lactam antibiotics. Alarmingly, some strains, such as carbapenem-resistant Enterobacteriaceae and NDM-1-producing Klebsiella pneumoniae, have become resistant to all clinically available β-lactam antibiotics, with no effective treatment available.
Whether a β-lactam antibiotic can defy the hydrolytic prowess of β-lactamases depends heavily on its chemical structure and how it fits into the active sites of these enzymes. This is particularly true for the clinically ubiquitous class A and C β-lactamases because they share similar catalytic mechanisms but have distinctively shaped active sites. Both class A and C β-lactamases are serine-based enzymes that hydrolyze β-lactam antibiotics in a two-step process (Fig. 1b). During the first step, acylation, the catalytic serine residue attacks the reactive β-lactam ring to form an acyl adduct. In the subsequent step, deacylation, the acyl adduct is hydrolyzed by a water molecule and released from the active site (3). However, class A β-lactamases have a narrowly shaped active site, with its closely packed Ω-loop serving as a steric boundary. In class C β-lactamases the corresponding Ω-loop is significantly longer and folds into a more extended conformation toward the solvent, leading to an “enlarged” active site (5, 6).
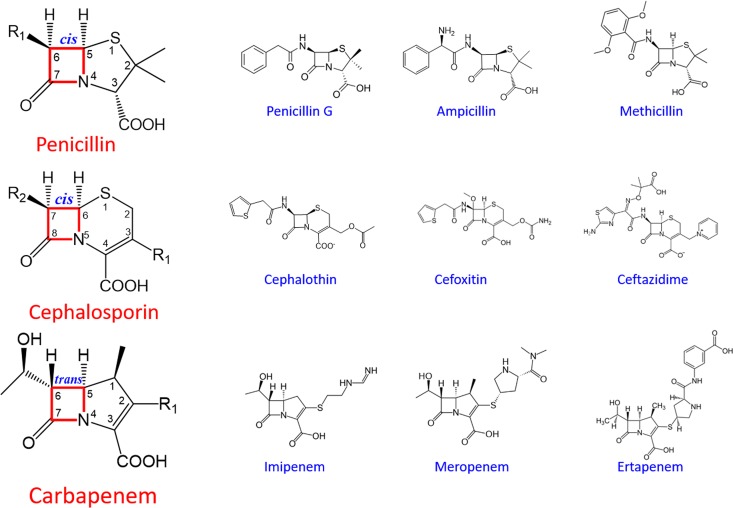
For the penicillin type of β-lactam antibiotics that have a penicillin nucleus with the β-lactam ring fused to five-membered thiazolidine ring, the acylamino side chain at C-6 is critical for conferring resistance against β-lactamases (Fig. 2). Those first- and second-generation penicillins, like penicillin G and methicillin, usually contain a simple side chain linking to an aromatic ring terminus and can be readily hydrolyzed by most β-lactamases. The third- and fourth-generation penicillins, like carbenicillin and piperacillin, contain longer and branched side chains with carboxyl or bulky heterocyclic amide groups. These antibiotics usually are resistant to class A β-lactamases due to steric clash between the “narrow” active site and the bulky branched acylamino side chains (7). Such a mechanism also applies to the cephalosporin type of β-lactam antibiotics that contain a cephem nucleus with the β-lactam ring fused to the six-membered dihydrothiazine ring (Fig. 2). Newer-generation cephalosporins with a branched acylamino side chain at C-7 are usually resistant against class A β-lactamases but still susceptible to class C β-lactamases due to their “enlarged” active site that can accommodate the bulky side chains (6).
FIG 2.
Structures of the three classes of β-lactam antibiotics.
Interestingly, the carbapenem type of β-lactam antibiotics show resistance to both class A and C β-lactamases (8). Carbapenems contain a five-membered pyrrolidine ring fused to the β-lactam core and a short hydroxyethyl side chain at C-6 in α conformation (Fig. 2). This side chain is a significant departure from the long acylamino side chain in penicillins and cephalosporins with β conformation and is particularly important for the broad-spectrum resistance of carbapenems. Structural studies have shown that this side chain interferes with the binding of carbapenems to serine-based β-lactamases prior to the acylation step and significantly slows down the kinetics of hydrolysis at the deacylation step (9). By acting as “slow substrates,” carbapenems become resistant against these inactivating enzymes and can even serve as de facto inhibitors of β-lactamases to further potentiate their antibacterial efficacy (10).
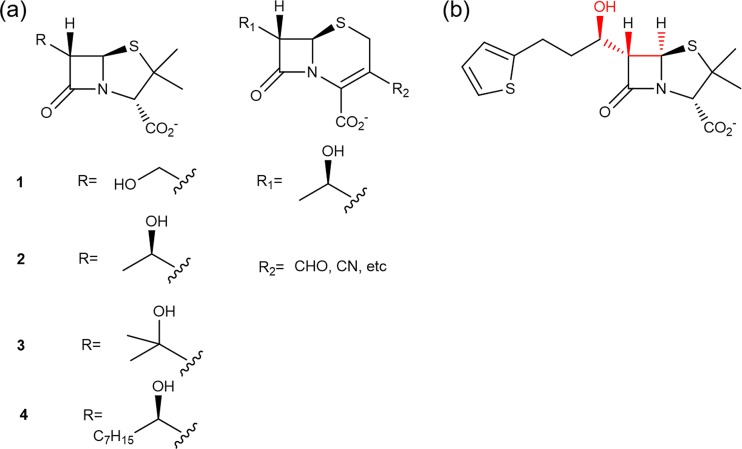
Given the extended resistance profile of carbapenems conferred by their hydroxyethyl side chain, it is reasonable to expect that adding this feature to penicillins and cephalosporins would broaden their resistance spectra as well. Previous studies have shown that replacing the acylamino side chain with the hydroxyethyl moiety converts penicillins and cephalosporins into inhibitors for class A β-lactamases (Fig. 3a) (11–13). However, these modified compounds cannot inhibit class C β-lactamases. In fact, such inactivity toward class C β-lactamases has also been observed among clinically used β-lactamase inhibitors like clavulanate, sulbactam, and tazobactam (3). With class C β-lactamases being one of the most frequently encountered resistance mechanisms in clinically significant bacterial pathogens, there is an urgent need for novel and potent class C β-lactamase inhibitors.
FIG 3.
Modification of penicillins and cephalosporins with carbapenem-like stereochemical features. (a) Previously reported compounds with carbapenem-like hydroxyethyl side chain “grafted” onto a penem or cephem nucleus. (b) Structure of our designed molecule, MPC-1. The carbapenem-like features are highlighted in red.
We reason that penicillins and cephalosporins can be modified with carbapenem-like stereochemistry to become class C β-lactamase inhibitors. Our plan is to retain the extended structure of the acylamino side chain and the associated aromatic groups found on penicillins and cephalosporins because these features have been shown to facilitate the binding of these drugs to class C β-lactamases (14). The 6-α hydroxyethyl group will be added to replace only the corresponding acylamino moiety within the side chain to adopt carbapenem-like stereochemistry (Fig. 3b).
To test our hypothesis, we synthesized such a “hybrid” molecule and characterized its in vitro efficacy. Our designed molecule exerts a potent and specific inhibitory effect against class C β-lactamases but is ineffective against class A β-lactamases. We also determined the crystal structure of this molecule in complex with class C β-lactamase and uncovered a novel binding mode that underlies its inhibitory mechanism. Our results suggest that indeed modifying penicillins and cephalosporins by adding carbapenem-like stereochemistry may lead to potential specific inhibitors for class C β-lactamases.
RESULTS
Design and synthesis of penicillin derivative MPC-1 with carbapenem-like stereochemistry.
We decided to synthesize a penicillin derivative (here named MPC-1) that contains a penicillin nucleus and a hydroxyalkyl side chain supplemented with a thiophene ring at the tail (Fig. 3b). The stereochemistry at C-5 and C-6 of the penicillin nucleus has been modified with the two H atoms in trans conformation to resemble carbapenem. Furthermore, a hydroxylalkyl group is introduced at C-6 with R conformation to mimic the hydroxyethyl moiety in carbapenem and to replace the original amide group in penicillins and cephalosporins. Additionally, a thiophene ring is added to the tail end of the alkyl side chain because such a nonbranched aromatic side chain is commonly found in first- or second-generation antibiotics that can readily bind to class C β-lactamases (7, 14).
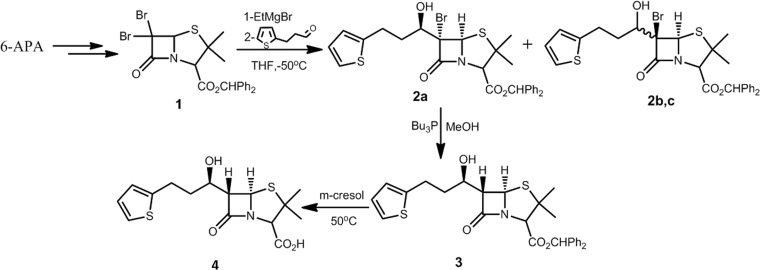
The synthesis route (Fig. 4) for MPC-1 is designed with consideration for efficiency and reproducibility (15). The synthesis started with 6-aminopenicillanic acid (6-APA), which was converted to 6,6-dibromopenicillanic acid according to a procedure previously reported (16). Then the C-2 carboxyl acid was protected with diphenyldiazomethane to give the key intermediate, compound 1. Treatment of compound 1 with Grignard reagent and 2-thiophenepropanal in anhydrous tetrahydrofuran (THF) produces the desired compound 2a, along with a mixture of two isomers, 2b and 2c. The stereochemistry of C-6 and the side chain stereogenic carbon of 2a and 2b and 2c can be assigned by nuclear magnetic resonance (NMR) according to a previous analysis (17). The reduction of 2a by Bu3P gave stereochemistry inversion product 3, with its stereochemical assignment at C-6 determined based on the coupling constant JH5-H6, with a value of 1.48 (15). Finally, the protective benzhydryl group was removed through reaction with m-cresol (18). All products reported showed 1H and 13C NMR spectra in agreement with the assigned structures; details of these spectra are in the supplemental material. The stereochemistry of the target compound, MPC-1, was further confirmed by the crystal structure of P99 β-lactamase in complex with MPC-1.
FIG 4.
Synthetic route to MPC-1 (compound 4).
MPC-1 shows no antibacterial activity as evaluated by in vitro assays.
Previous studies have shown that replacing the acylamino side chain with the hydroxyethyl moiety leads to loss of antibiotic activity in modified penicillins and cephalosporins (12). It is not clear if this would apply to our designed molecule, MPC-1. To test its antibacterial efficacy, we used a MIC assay and found that the MIC of MPC-1 for non-β-lactamase-producing Escherichia coli strain BL21(DE3) is over 512 μg/ml (data not shown), significantly higher than that of penicillin G. This result suggests that indeed MPC-1 has little antimicrobial activity, probably due to the carbapenem-like stereochemistry at C-6 and the C-8 hydroxyl side chain in R conformation.
MPC-1 specifically inhibits class C β-lactamase P99 but does not inhibit the class A β-lactamase PenP.
After confirming that MPC-1 does not have antibiotic activity, we proceeded to characterize its efficacy as a β-lactamase inhibitor. We used two β-lactamases, PenP and P99, for this study. PenP is a class A β-lactamase from Bacillus licheniformis with over 80% sequence similarity to the clinically significant TEM-1. Our previous studies have also confirmed that the kinetic profile of PenP is nearly identical to that for TEM-1, showing a narrow substrate profile with more effective hydrolysis of only first- and second-generation penicillins and cephalosporins (19, 20). P99, on the other hand, is an AmpC β-lactamase (also termed class C β-lactamase) from Enterobacter cloacae. P99 is an extended-spectrum β-lactamase with a broader substrate profile than PenP, probably because its enlarged active site can accommodate newer-generation penicillins and cephalosporins with branched side chains (21).
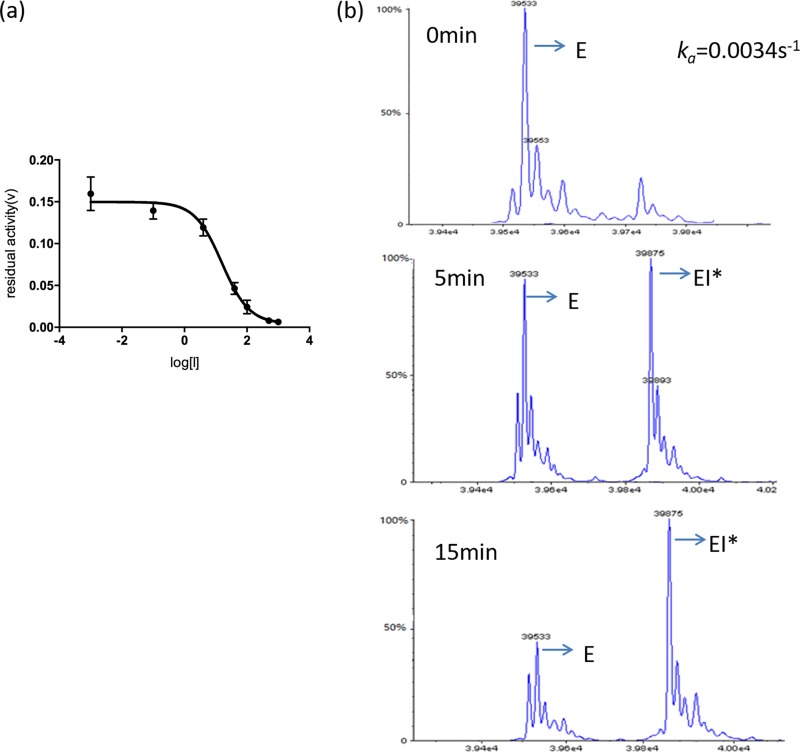
The inhibitory effect of MPC-1 on PenP and P99 was assessed by measuring its 50% inhibitory concentration (IC50) in the nitrocefin assay (Table 1). Two clinically available β-lactamase inhibitors, i.e., clavulanate and sulbactam, were used as references. Our results show that MPC-1 is not an effective inhibitor of the class A β-lactamase PenP because its IC50 is higher than 1 mM. In comparison, clavulanate and sulbactam can inhibit PenP with high potency, as their IC50s are less than 2 μM. However, MPC-1 is a potent and specific inhibitor of the class C β-lactamase P99, with an IC50 of 15 μM (Fig. 5a). This inhibitory potency against P99 is particularly exciting, as it is ∼2 to 5 times stronger than that for the clinically used clavulanate or sulbactam, for which the IC50s are 77.8 μM and 29.88 μM, respectively.
TABLE 1.
β-Lactamase-inhibitory activity of MPC-1
| β-Lactamase | IC50 (μM) |
||
|---|---|---|---|
| MPC-1 | Clavulanate | Sulbactam | |
| PenP | >1,000 | 1.26 ± 0.85 | 1.99 ± 1.22 |
| P99 | 15.8 ± 1.26 | 77.8 ± 8.74 | 29.9 ± 2.53 |
FIG 5.
MPC-1 inhibits class C β-lactamase through slow acylation and defective deacylation. (a) Determination of IC50 of MPC-1 against class C β-lactamase P99 with nitrocefin assay. (b) Time-dependent mass spectrum profile of P99–MPC-1 acyl adduct formation when the MPC-1 concentration is 10 μM. ka is derived by fitting the ratio of [EI*]/[E + EI*] versus time to equation 2.
MPC-1 binds to class C β-lactamase and forms an irreversible acyl adduct.
To further understand the interaction between MPC-1 and P99, we added MPC-1 to purified P99 and, after incubation, subjected the mixture to electrospray ionization mass spectroscopy (ESI-MS). While the mass spectra revealed intact P99 at the start of the experiment, a new species with a 342-Da increase in mass emerged after 5 min (Fig. 5b). The mass of this species matched exactly the acyl adduct between P99 and MPC-1, suggesting that MPC-1 probably acted like a substrate for P99 and initiated the acylation reaction to form an enzyme inhibitor (EI*) acyl adduct at the active site of P99. We then measured the kinetic parameters of MPC-1 as a P99 inhibitor by ESI-MS. Our data show that the acylation efficiency (k2/Kd) (where Kd is dissociation constant for the formation of the reversible noncovalent EI complex, and k2 is the rate constant for the formation of the covalent acyl adduct EI*) of MPC-1 is only (4.0 ± 0.1) × 102 M−1 s−1, significantly lower than that for natural substrates like penicillins and cephalosporins (∼106 M−1 s−1) (22). This low acylation rate suggests that the modified structures of MPC-1 drastically slowed down the acylation. Furthermore, the deacylation rate (k3) is also very low, at (1.2 ± 0.7) × 10−3 s−1, almost comparable to that of avibactam (7 × 10−4 s−1), a recently approved inhibitor for class A, C, and D β-lactamases (23). The extremely low k3 value explains why MPC-1 can serve as a P99 inhibitor. The Ki was 7.9 ± 0.6 μM.
MPC-1 potentiates the efficacies of penicillins and cephalosporins in P99-expressing resistant E. coli.
In the clinical setting, β-lactamase inhibitors such as clavulanate and sulbactam are used in combination with β-lactam antibiotics to potentiate their efficacies by reducing the hydrolytic activity of β-lactamases. To test whether MPC-1 can achieve a similar effect, we compared the antibacterial efficacies of several antibiotics in the presence and absence of MPC-1 in vivo. For this study, we first transformed a plasmid carrying the P99 gene into E. coli BL21(DE3) to confer resistance toward penicillins and cephalosporins. Then the MICs of various antibiotics, in both the presence and absence of MPC-1, were measured (Table 2). For penicillin G, the MIC was reduced by MPC-1 in a concentration-dependent manner, from 512 μg/ml in the absence of MPC-1 to 16 μg/ml in the presence of 512 μg/ml of MPC-1, a 32-fold reduction. For ampicillin, the MIC was 32 μg/ml in the absence of MPC-1 and only 4 μg/ml in the presence of 16 μg/ml of MPC-1, an 8-fold reduction. A similar effect was also observed for cephalothin and cefoxitin, with 4- to 8-fold reductions in MIC. These data clearly demonstrate that MPC-1 can potentiate the activity of penicillins and cephalosporins against P99-positive E. coli by inhibiting the hydrolytic activity of P99. The efficacy of such potentiation appears to be most prominent for antibiotics with mid-range MICs, like ampicillin, when the antibiotic and MPC-1 have similarly low acylation rates.
TABLE 2.
MPC-1 potentiates the efficacy of penicillins and cephalosporins in P99-expressing E. coli strain BL21(DE3)
| Drug | MIC (μg/ml) of drug at indicated concn of MPC-1 (μg/ml) |
|||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 4 | 16 | 32 | 64 | 128 | 256 | 512 | |
| Penicillin G | 512 | 512 | 256 | 256 | 128 | 128 | 32 | 16 |
| Ampicillin | 32 | 4 | ||||||
| Cephalothin | 512 | 128 | ||||||
| Cefoxitin | 8 | 4 | ||||||
Structure of MPC-1 in complex with P99 reveals a novel binding mode that blocks deacylation.
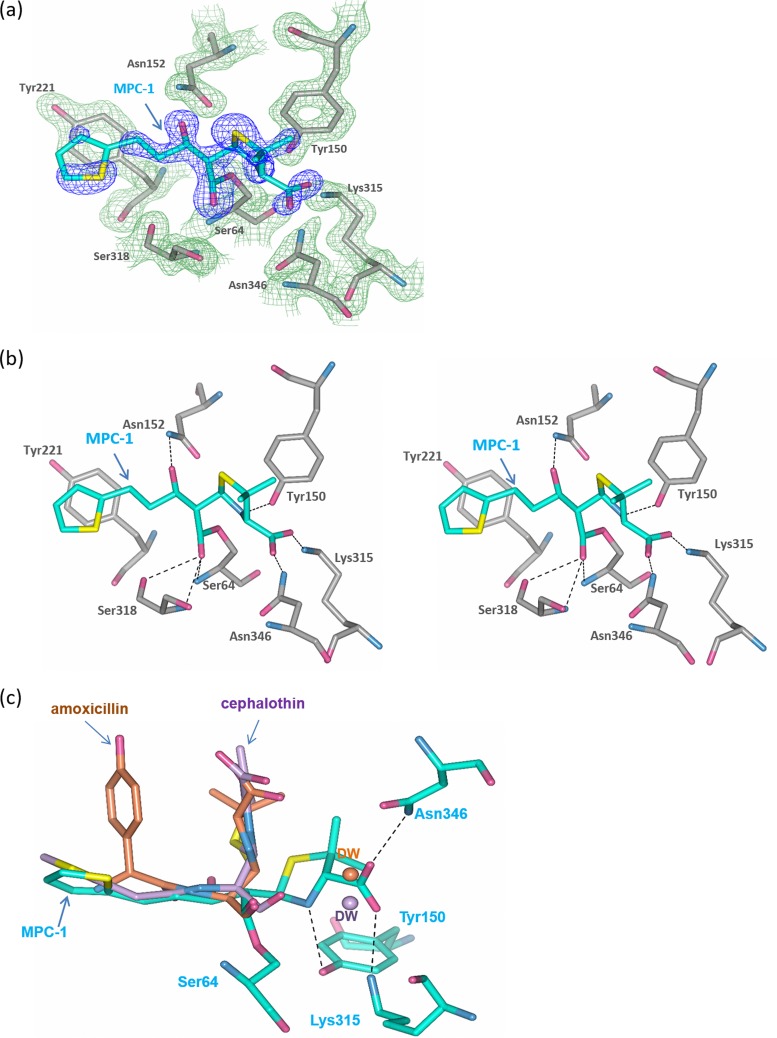
To determine the molecular mechanism of how MPC-1 forms nonhydrolyzable EI* acyl adduct with P99, we determined the structure of P99 in complex with MPC-1 at 1.8 Å (Table 3). The overall structure is nearly identical to the P99 apo structure (PDB code 1XX2), with a root mean square deviation (RMSD) of only 0.35 Å. Key residues at the active site, including the catalytic residue Ser64 and the general base residues Lys67 and Tyr150, are in exactly the same conformation as in the apo structure (Fig. 6a). Furthermore, both the difference map (Fo-Fc) and the composite map (2Fo-Fc) clearly show the presence of MPC-1 with its β-lactam ring opened and the carbonyl carbon covalently linked to Oγ of the catalytic Ser64 in the active site (Fig. 6a).
TABLE 3.
Statistics of X-ray crystallography data collection and structure refinement for P99–MPC-1
| Parameter | Value |
|---|---|
| Data collection statistics | |
| Space group | P22121 |
| Unit cell parameters | |
| a, b, c (Å) | 61.160, 69.330, 76.850 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| Resolution range (Å) | 69.33–1.80 (1.90–1.80) |
| No. of total reflections | 209,165 |
| No. of unique reflections | 30,924 |
| I/σ | 16.1 (6.2) |
| Completeness (%) | 99.8 (95.6) |
| Rmerge (%) | 8.7 (29.2) |
| Refinement statistics | |
| Resolution (Å) | 76.85–1.8 |
| Rcryst/Rfree (%) | 21.3/23.3 |
| RMSD bonds (Å)/angles(°) | 0.008/1.211 |
| No. of reflections | |
| Working set | 29,328 |
| Test set | 1,559 |
| No. of: | |
| Protein atoms | 2,828 |
| Ligand/ion atoms | 22 |
| Water molecules | 142 |
| Average B-factor (Å2) | |
| Main chain | 9.994 |
| Side chain | 13.692 |
FIG 6.
MPC-1 binds to the active site of AmpC in a distinct flat mode. (a) Active-site structure of P99–MPC-1 complex. The compound MPC-1 is in cyan, while the active-site residues are in gray. The Fo-Fc omit map (light yellow for active site residues and blue for MPC-1) is drawn in mesh format and contoured at 2.0 σ. (b) Interactions between compound MPC-1 and the active-site residues. The dashed lines indicate hydrogen bonds. (c) Superposition of P99–MPC-1 structure with the AmpC structures in complex with amoxicillin (PDB code 1LL9) and cephalothin (PDB code 1KVM), respectively. Amoxicillin and cephalothin are colored in brown and purple, respectively. The corresponding deacylation water (DW) in these two structures is shown as a sphere within the active site.
Close inspection of the P99 structure reveals several important features of its interaction with MPC-1. First of all, the covalent linkage between MPC-1 and Ser64 of P99 confines to the classical acyl adduct configuration found in the active site of class C β-lactamases, with the carbonyl oxygen of the β-lactam moiety pointing to the oxyanion hole formed by the amide of Ser64 and Ser318 (14, 21) (Fig. 6b). Second, the thiophene ring at the end of the alkyl side chain in MPC-1 forms π-π stacking interaction with the side chain of Tyr221 in P99. This interaction is similar to that seen in AmpC–cephalothin structure and reaffirms the importance of this moiety in mediating favorable binding to P99 (14). Additionally, the introduced hydroxyl group at C-7 of MPC-1 forms a hydrogen bond with Asn152. This interaction is equivalent to the hydrogen bond between Asn152 and the amide group in the side chain of penicillin or cephalosporin (Fig. 6b). In summary, the structure reveals that our designed molecule, MPC-1, retains nearly all the interactions that a natural substrate can form with the P99 active site. As a result, MPC-1 can specifically bind to class C β-lactamases like P99.
Interestingly, the EI* acyl adduct in our structure adopts a novel “flat” conformation that has not been reported for penicillins or cephalosporins. In previously reported structures of P99 in complex with amoxicillin and cephalothin, the acyl adducts adopt a “bent” conformation with their thiazolidine or dihydrothiazine ring pointing toward the solvent and at roughly 90° relative to the main chain of the adduct along the β-lactam moiety (Fig. 6c) (14, 21). In our structure, MPC-1 exhibits a flat conformation with its thiazolidine ring extending along the main chain of the acyl adduct and fully embedded in the active site. With this conformation, the carboxyl side chain on the thiazolidine ring forms multiple H-bond interactions with nearby residues Lys315, Ser316, and Asn346 (Fig. 6c). Most importantly, in this flat conformation, the carboxyl group occupies the position of the critical deacylation water—WAT 402—as shown in other two complex structures (Fig. 6c). As a result, the deacylation reaction cannot proceed and the acyl adduct EI* becomes irreversible.
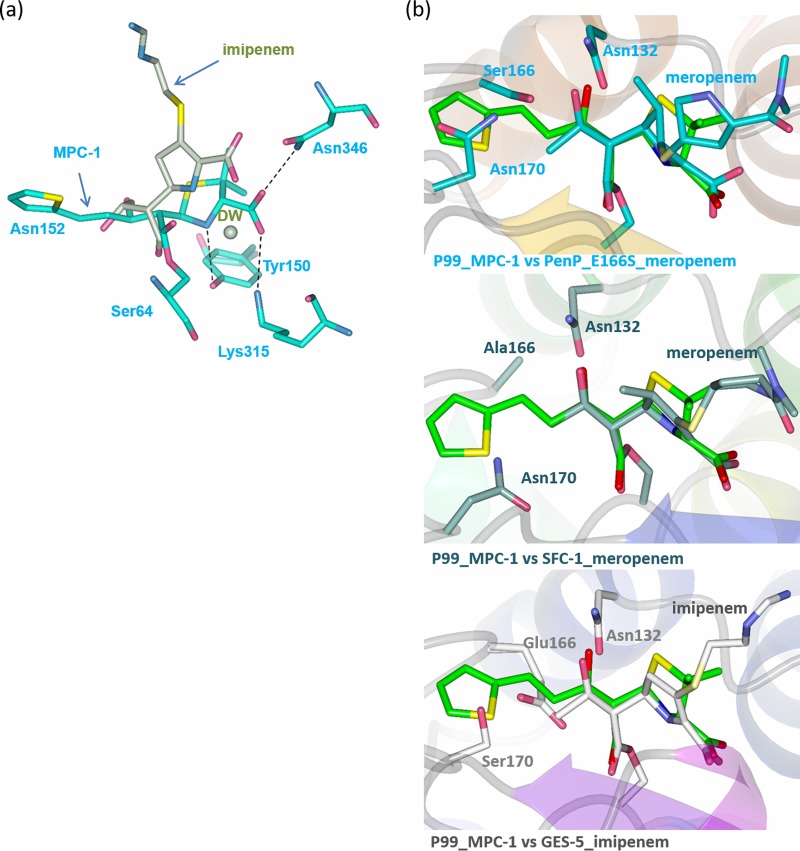
To investigate whether the novel binding mode of MPC-1 acyl adduct is due to its carbapenem-like stereochemistry, we first compared our structure to that of the class C β-lactamase AmpC with covalently bound imipenem (24). An unusual feature for the AmpC-imipenem structure is the catalytically incompetent conformation of the acyl adduct with its carbonyl oxygen “flipped” out of the oxyanion hole (Fig. 7a). The impact of such a flipped conformation on the structural configuration for the rest of imipenem is not clear. Nonetheless, the pyrrole ring of imipenem adopts a “bent” conformation similar to that of amoxicillin and cephalothin in the active site of P99 but in distinct contrast to the flat conformation of MPC-1 (Fig. 7a).
FIG 7.
MPC-1 adopts a configuration similar to that of carbapenems while binding to the active sites of β-lactamases. (a) Superimposition of P99–MPC-1 structure with the structure of AmpC in complex with imipenem (PDB code 1LL5). Imipenem and the conserved deacylation water (DW) are in light green. (b) Superposition of the P99–MPC-1 structure with three class A β-lactamases, including PenP, in complex with meropenem (PDB code 1BT5), SFC-1 in complex with imipenem (PDB code 4EV4), and GES-5 in complex with imipenem (PDB code 4H8R).
To further understand the molecular mechanism of the novel flat conformation seen in MPC-1, we decided to compare our structure to that of class A β-lactamases with covalently bound carbapenems. In fact, a series of such structures has been reported, including TEM-1 with imipenem, BlaC with tebipenem, SFC-1 with meropenem, and GES-5 with imipenem (9, 25–27). In particular, SFC-1 and GES-5 are two class A β-lactamases that have acquired carbapenemase activity due to mutations at Ala69Cys/Ala238Cys and Gly170Ser. Thus, the carbapenem acyl adducts formed in the active sites of SFC-1 and GES-5 are regarded as catalytically competent and would be used for our comparison. Structural superposition reveals that the flat conformation of MPC-1 overlies almost identically the meropenem and imipenem structures seen in SFC-1 and GES-5 (26, 27) (Fig. 7b). Specifically, the covalent linkage between MPC-1 and the catalytic residue Ser64 in P99 is identical to the corresponding part that connects meropenem or imipenem to the catalytic residue Ser70 in SFC-1 or GES-5. Additionally, the thiazolidine ring of MPC-1 overlies very well the pyrroline ring of the acylated meropenem and imipenem. Furthermore, the hydroxyl moiety at C-7 position of MPC-1 superimposes exactly the hydroxyethyl side chain of meropenem or imipenem. Thus, the flat conformation of MPC-1 indeed results from its carbapenem-like stereochemistry after it forms the catalytically competent acyl adduct.
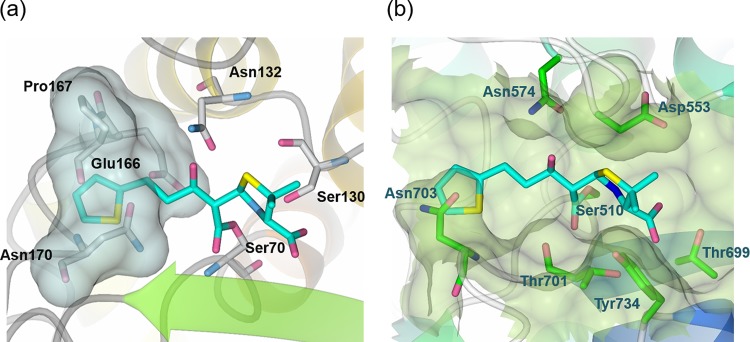
Lastly, to confirm that the flat conformation of MPC-1 is responsible for its specificity for class C β-lactamases, we generated a model of MPC-1 in complex with PenP, a class A β-lactamase, after superimposing PenP (PDB code 4BLM) onto our P99 structure (28). The model shows that MPC-1, in its flat conformation, cannot fit into the active site of PenP because the extended thiophene group would clash with Glu166 and Asn170 (Fig. 8a). We also generated a model of MPC-1 in complex with the penicillin-binding (PB) domain of E. coli PBP1b (PDB code 5HL9) after superimposing the PB domain onto our P99 structure (29). Although the large number of PBPs identified in nature have functionally and structurally distinct N-terminal domains, their C-terminal PB domain is highly conserved (30). This MPC-1–PB model reveals a situation similar to that revealed by the MPC-1–PenP model: i.e., the flat MPC-1 molecule cannot fit into the active site of PB domain because the extended side chain would clash with residue Asn703 (Fig. 8b). In summary, the novel flat conformation of MPC-1 renders it incompatible with the active site of class A β-lactamases or PBPs. Thus, MPC-1 is a specific inhibitor only for class C β-lactamases and is not a class A inhibitor or an antibiotic.
FIG 8.
MPC-1 causes steric hindrance with the active site of class A β-lactamase and PBP. Model structures of MPC-1 in complex with PenP β-lactamase (a) and E. coli PBP1b (b) were generated by docking MPC-1 onto relevant structures (PDB codes 4BLM [PenP β-lactamase] and 5HL9 [E. coli PBP1b]).
DISCUSSION
β-Lactamase inhibitors such as clavulanate, sulbactam, and tazobactam have played significant roles in clinical treatment of bacterial infection. These molecules are structurally similar to β-lactam but lack any side chain at C-6. They can effectively inhibit class A β-lactamases by forming slowly hydrolyzed acyl adducts in the active site to block substrate turnover (3). Furthermore, some of these inhibitor acyl adducts can undergo complex secondary reactions within the active site that lead to irreversible modification of the catalytic serine residue and permanent loss of β-lactamase activity. Although they show little antimicrobial activity when used alone, these inhibitors greatly enhance the therapeutic efficacy of β-lactam antibiotics when used in combination with them. For example, the highly successful antibiotic amoxicillin-clavulanic acid (Augmentin) is indeed a combination of amoxicillin and clavulanic acid (31).
However, resistance to these widely used inhibitors are on the rise. Class A β-lactamases like the TEM and SHV series acquire mutations at certain active-site residues, like Met69, Ser130, and Lys234, to either impede inhibitor acylation or speed up acyl adduct hydrolysis (32). Furthermore, these inhibitors are not effective against the class B metallo-β-lactamases or class C extended-spectrum β-lactamases that are frequently found in multidrug-resistant Gram-negative bacteria, like Enterobacter spp. and Pseudomonas aeruginosa. Therefore, there is an urgent need to develop inhibitors for class B and class C β-lactamases. In 2015, the FDA approved avibactam, a non-β-lactam inhibitor for class A, C, and D β-lactamases. This is the first inhibitor approval in 22 years and rekindled interest in β-lactamase inhibitor development.
In this study, we have synthesized a penicillin derivative, MPC-1, that can serve as a specific inhibitor for class C β-lactamases. MPC-1 contains a penicillin nucleus modified with two carbapenem-like stereochemical features, i.e., the C-5–C-6 trans conformation and the C-8 hydroxyl group. Additionally, MPC-1 contains an alkyl side chain with terminal thiophene ring at C-8 to mimic penicillins and cephalosporins that bind favorably to class C β-lactamases. Such a “hybrid” molecule is expected to bind to class C β-lactamases like a good substrate and then form a slowly hydrolyzable acyl adduct to act as a specific inhibitor.
Our biochemical and ESI-MS studies confirm that indeed MPC-1 can bind specifically to the class C β-lactamase P99 and form a slowly hydrolyzed acyl adduct. The structure of P99 in complex with MPC-1 reveals that the EI* adduct adopts a novel flat conformation, with its thiazolidine ring fully extended along the active site. This flat conformation is in clear contrast to the bent conformation adopted by amoxicillin and cephalothin in the active sites of class C β-lactamases but largely similar to the orientation of imipenem and meropenem when covalently bound to the class A β-lactamases SFC-1 and GES-5. Furthermore, the flat conformation of MPC-1 in the active site of P99 positions the carboxyl group on its thiazolidine ring to a site overlapping the critical deacylation water molecule. As a result, the deacylation reaction is abolished and the acyl adduct becomes an inhibitor of P99. Given the structural and kinetic similarities shared by different class C β-lactamases (3, 5), we believe that the mechanistic findings from our study on how MPC-1 specifically inhibits P99 can be applied to other class C β-lactamases, like AmpC and CMY varieties, as well. In summary, the carbapenem-like stereochemical features of MPC-1 allows it to adopt a novel flat conformation in the active sites of class C β-lactamases that leads to blocked deacylation and irreversible inhibition. This mode of inhibition is totally different from that observed for clavulanate, sulbactam, avibactam, and other investigational β-lactamase inhibitors (3, 33) and offers a novel approach to design class C β-lactamase inhibitors.
Our designed hybrid molecule, MPC-1, shows modestly higher potency, 2- to 5-fold, against class C β-lactamases than do commercially available β-lactamase inhibitors, like clavulanate and sulbactam, but it shows little activity against class A β-lactamases. This difference is likely due to the extended alkyl side chain with terminal thiophene ring in the C-6 position of MPC-1. Such a moiety has been shown to be beneficial for binding to class C β-lactamases because it can form favorable interactions with active-site residues. In contrast, clavulanate and sulbactam contain no side chains at the C-6 position and as a result cannot fit tightly into the enlarged active sites of class C β-lactamases.
One obvious limitation of MPC-1 in its current form is the slow acylation kinetics. At a k2/Kd of only 102 M−1 s−1, MPC-1 cannot readily compete with natural substrates like penicillins and cephalosporins to effectively inhibit class C β-lactamases in vivo. To improve the binding affinity of MPC-1 to class C β-lactamases, different side chains at the C-6 position should be tried with the aim to form more favorable interactions, like hydrogen bonds. Hopefully, applying our hybrid design strategy and adding carbapenem-like stereochemical features to existing penicillins and cephalosporins will yield a large library of lead compounds to serve as potential class C β-lactamase inhibitors.
MATERIALS AND METHODS
Synthesis of MPC-1.
The synthesis route for the target compound MPC-1 is shown in Fig. 4. All reaction products were routinely visualized with UV lighting (254 and 365 nm) and purified by column chromatography. 1H and 13C NMR spectra were determined in CDCl3 solutions with a Bruker spectrometer (400 MHz). Peak positions are given in parts per million (δ) downfield from tetramethylsilane as the internal standard, and J values are given in hertz.
Compound 1 was synthesized as described previously.
Diphenylmethyl 6α-bromo-6β-(1R-hydroxy-3-(thiophen-2-yl)propyl) penicillanate (compound 2a).
A solution of ethylmagnesium bromide in ether (3 M, 0.61 ml, and 1.82 mmol) was added dropwise to a solution of diphenylmethyl 6,6-dibromopenicillanate 1 (2 g and 3.8 mmol) in anhydrous THF (40 ml) at −50°C, and the reaction mixture was stirred for an additional 2 h at the same temperature. 3-(Thiophen-2-yl)propanal (1 g and 7.6 mmol) was added dropwise to the solution and the mixture was stirred for 2 h at −50°C. The reaction was then quenched by the addition of a saturated solution of ammonium chloride, the mixture was warmed to room temperature, and the solvent was evaporated in vacuo. Both water and ethyl acetate were added to the residue, and the organic layer was washed with 5% NaHCO3 and brine, dried over MgSO4, and concentrated to dryness in vacuo. The product mixture was purified by flash column chromatography to give the desired compound, compound 2a (1.1 g; 49%), along with the mixture of two isomers, 2b and 2c (0.5g, 22%). Compound 2a: 1H NMR (CDCl3, 400 MHz) δ 1.25 (s, 3H), 1.53 (s, 3H), 1.92 to 2.10 (m, 2H), 2.97 to 3.18 (m, 2H), 4.04 to 4.07 (m, 1H), 4.58 (s, 1H), 5.63 (s, 1H), 6.87 (d, 1H), 6.93 to 6.97 (m, 2H), 7.14 to 7.15 (m, 1H), 7.31 to 7.40 (m, 10H); 13C NMR (CDCl3, 400 MHz) δ 25.3, 25.6, 33.4, 34.3, 64.7, 68.1, 69.7, 70.8, 74.8, 78.6, 123.3, 125.0, 126.7, 127.1, 127.4, 128.3, 128.4, 128.6, 138.9, 139.0, 143.6, 165.9, 168.9; ESI-MS calculated for C28H28BrNO4S2 ([M+Na]+), 609.5; found, 609.5. Mixture of 2b and 2c: 1H NMR (CDCl3, 400 MHz) δ 1.28 (s, 3H), 1.67 (s, 3H), 1.96 to 2.27 (m, 2H), 2.97 to 3.18 (m, 2H), 4.01 to 4.15 (m, 1H), 4.61 (s, 1H), 5.49 (s, 0.65H), 5.52 (s, 0.35H), 6.84 to 6.86 (m, 1H), 6.92 to 6.96 (m, 2H), 7.13 to 7.16 (m, 1H), 7.28 to 7.39 (m, 10H); 13C NMR (CDCl3, 400 MHz) δ 25.8, 26.0, 26.1, 32.8, 33.0, 34.9, 35.9, 64.8, 70.0, 70.1, 71.0, 72.3, 72.6, 73.0, 77.2, 78.5, 123.5, 123.6, 124.8, 124.9, 126.9, 127.0, 127.0, 127.5, 128.2, 128.4, 139.0, 139.1, 143.3, 166.3, 168.4.
Diphenylmethyl 6α-(1R-hydroxy-3-(thiophen-2-yl)propyl) penicillanate (compound 3).
Tri-n-butylphosphine (0.59 ml, 2.35 mmol) was added to a solution of compound 2 (0.92 g, 1.57 mmol) in methanol (40 ml) at 0 ∼ 5°C and the reaction mixture was stirred for 1 h at the same temperature. The reaction mixture was then concentrated under reduced temperature, and the residual oil was purified by silica gel column chromatography to afford the desired product (0.47 g; 59%).1H NMR (CDCl3, 400 MHz) δ 1.26 (s, 3H), 1.63 (s, 3H), 1.93 to 2.02 (m, 2H), 3.00 to 3.07 (m, 2H), 3.40 to 3.42 (m, 1H), 4.12 to 4.17 (m, 1H), 4.58 (s, 1H), 5.34 (d, J = 1.48 Hz, 1H), 6.84 to 6.85 (d, 1H), 6.93 to 6.96 (m, 2H), 7.15 to 7.16 (m, 1H), 7.30 to 7.38 (m, 10H); 13C NMR (CDCl3, 400 MHz) δ 25.7, 26.1, 32.8, 37.1, 63.5, 65.6, 67.5, 67.8, 69.8, 78.3, 123.4, 124.7, 126.9, 127.0, 127.5, 128.1, 128.3, 128.5, 128.6, 139.2, 139.3, 143.7, 166.9, 172.0; ESI-MS calculated for C28H29NO4S2 ([M+Na]+), 530.6; found, 530.5.
6α-(1R-Hydroxy-3-(thiophen-2-yl)propyl) penicillanate (compound 4).
A solution of compound 3 (200 mg, 0.39 mmol) in m-cresol (3 ml) was heated at 50°C for 3 h under N2. The mixture was then cooled to room temperature and concentrated in vacuo, and the residue oil was purified by silica gel column chromatography to afford the desired product (60 mg, 46%).1H NMR (CDCl3, 400 MHz) δ 1.55 (s, 3H), 1.68 (s, 3H), 1.86 to 1.98 (m, 2H), 2.91 to 3.05 (m, 2H), 3.33 (d, 1H), 4.15 to 4.18 (m, 1H), 4.39 (s, 1H), 5.32 (s, 1H), 6.83 (d, 1H), 6.91 to 6.94 (m, 1H), 7.12 to 7.14 (d, 1H); 13C NMR (CDCl3, 400 MHz) δ 25.9, 26.8, 30.6, 37.1, 61.5, 65.5, 65.6, 66.5, 70.9, 123.4, 124.6, 126.9, 143.8, 172.4, 174.0; ESI-MS calculated for C15H19NO4S2 ([M-H]−), 340.5; found, 340.3.
All materials, data, and associated protocols will be made promptly available to readers after publication, without undue qualifications in material transfer agreements.
Protein expression and purification.
The DNA encoding P99 β-lactamase was subcloned into a modified pET 30a vector containing an N-terminal His6 tag and the human rhinovirus (HRV) 3C protease cleavage site. Protein expression for the wild-type and substitution constructs was done using E. coli strain BL21(DE3) following a standard procedure. Briefly, inoculated bacterial culture was grown at 37°C until the optical density at 600 nm (OD600) reached 0.6 to 0.8; then protein expression was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 500 μM, and the bacteria were grown at 30°C for an additional 5 h and collected by centrifugation. The His6-tagged P99 proteins were purified by HisTrap affinity column (GE Healthcare) and then protease 3C was used to cleave the tag. The target proteins were further purified by gel filtration chromatography (Superdex 75; GE Healthcare) in a buffer of 20 mM Tris (pH 7.5) and 50 mM NaCl. The desired fractions were collected and concentrated by Amicon Ultra-15 centrifugal filter devices (Millipore; nominal molecular weight limit [NMWL] = 10,000).
Determination of MIC.
MPC-1 was dissolved in dimethyl sulfoxide (DMSO) first and then diluted in water to prepare a stock solution at a concentration of 2.048 mg/ml. MPC-1 stock was then treated with a series of 2-fold dilutions in lysogeny broth (LB) to prepare MPC-1-containing media at concentrations ranging from 0.5 to 512 μg/ml. Simultaneously, the bacterial strain E. coli BL21(DE3) was inoculated in LB medium and grown overnight; then the overnight culture was diluted to an OD600 of 0.08 to 0.1, 5 μl was added into previously prepared MPC-1-containing media, and the resulting solution was incubated at 37°C overnight. MIC was determined by observation to evaluate the potency at which MPC-1 enhanced the activity of antibiotics against β-lactamase-producing bacteria. The MICs for antibiotics alone or combined with MPC-1 were also determined. A stock solution was prepared by mixing different ratios of MPC-1 and each antibiotic, and E. coli BL21(DE3) was replaced with P99-transformed BL21(DE3) as constructed before. The corresponding MIC was then measured using the method described above.
Measurement of inhibitory activity.
To evaluate the inhibitory effect of MPC-1, the enzymatic activities of P99 and PenP were determined with nitrocefin as the reporter substrate. First of all, the Km of nitrocefin was measured by following the standard spectrophotometric method. Specifically, different concentrations ([S]) of nitrocefin were hydrolyzed by optimal concentration of β-lactamases within 200 s, the hydrolytic velocity was calculated based on the absorbance change readout at 500 nm, values for v (see below) versus [S] values were fitted into the Michaelis-Menten equation, and the Km of nitrocefin was determined.
Next, various concentrations ([I]) of MPC-1 were mixed with PenP or P99 in the assay buffer (50 mM potassium phosphate, pH 7.0) and the mixture was incubated at 25°C for 10 min. Nitrocefin at a concentration equal to the value of Km was added to each enzyme-inhibitor mixture, the absorbance change was monitored at 500 nm over 100 s by UV-visible spectrophotometer to determine the initial velocity, and each measurement was repeated three times to calculate standard deviation. By fitting v versus log[I] into equation 1, IC50 can be determined. Two clinically available β-lactamase inhibitors—clavulanate and sulbactam—were also tested as references.
| (1) |
Ki was then determined as IC50/2 based on the competitive-inhibition model (34).
Determination of ka.
The kinetics studies of P99 with MPC-1 as an inhibitor are based on a simple model (Fig. 9). The ESI-MS method as previously described (35, 36) was adopted to determine the kinetic parameters ka, k2/Kd, and k3.
FIG 9.
Model used for kinetics studies of P99 with MPC-1 as an inhibitor.
Specifically, the enzyme-inhibitor binding interaction was initiated by mixing 35 μl of 5 μM enzyme in 20 mM ammonium acetate (pH 7.0) with 35 μl of seven different MPC-1 concentrations ranging from 10 μM to 400 μM in the same buffer. At the desired intervals, the reactions were quenched by addition of 70 μl of 1% (vol/vol) formic acid in acetonitrile. The resulting solutions were characterized by electrospray ionization mass spectrometry (ESI-MS). Normally, there are two major peaks in the mass spectrum, one for the enzyme itself (E) and the other for the acyl adduct (EI*). The relative concentration of free enzyme ([E]) and the acyl adduct ([EI*]) can be determined by the integration of the area under the measurement of the intensity of these two peaks, while the total amount of enzyme ([Etotal]) can be calculated from the sum of [E] and [EI*]. The obtained values for [EI*]/[Etotal] were plotted as a function of the duration time (t) with equation 2 to give the apparent first-order constant ka:
| (2) |
In this equation, B represents the relative concentration of EI* at steady state.
Determination of k2/Kd and k3.
According to the model in Fig. 9, ka is related to k2 and Kd according to equation 3:
| (3) |
A linear variation of ka plotted versus [I] indicated that Kd is ≫[I]; thus, k2/Kd could be obtained from the slope of the line, while k3 is determined by extrapolation to [I] = 0.
Crystallization and structure determination.
The crystals of P99 were grown at 16°C by the hanging-drop vapor diffusion method. One microliter of protein solution in a buffer of 20 mM Tris–50 mM NaCl (pH 7.5) was mixed with 1 μl of reservoir buffer, which was composed of 0.1 M bis-Tris (pH 6.5) and 25% polyethylene glycol 3350 (PEG 3350). Crystals appeared in 14 days and were harvested after they had grown to ∼100 μm. MPC-1 was soaked into the crystals by incubating P99 crystals in reservoir buffer containing saturated MPC-1 for 1 h. Initial crystals were screened with an in-house Rigaku X-ray diffraction system using both a RAXIS image plate and PILATUS 200K hybrid pixel array detector. Crystals with MPC-1 soaked into active site were then taken to Shanghai Synchrotron Beamline 17U1 for data collection. Diffraction data were collected at 100 K, integrated by Mosflm (37), and scaled by the SCALA module (38) in CCP4. All the structures were solved by molecular replacement using the Phaser module in the CCP4i suite of programs with the P99 wild-type structure (PDB code 1XX2) as a search model (39). The subsequent structural refinement was conducted using the REFMAC module in CCP4 (40). Manual structure rebuilding was done using WINCOOT (41). The structure figures were prepared using the CCP4 mg package (42) in CCP4.
Accession number(s).
The structure determined in this study has been deposited in the Protein Data Bank with PDB code 5XHR.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Research Grants Council GRF grants PolyU 5640/11M, PolyU 151043/14M, and 151052/16M; Research Grants Council CRF grant C5030-14E; Research Grants Council AoE grant AoE/M-09/12; Health and Medical Research Fund grant 14130502; the Shenzhen Basic Research Program of China (JCYJ20160531184305919); and the Research Committee of the Hong Kong Polytechnic University.
We thank Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U for help with data collection.
X.P. synthesized the compound MPC-1, conducted in vitro biophysical and biochemical assays, and solved the X-ray structures. Y.H. assisted with biochemistry and X-ray crystallography. K.-F.C. supervised chemical synthesis. Y.Z. designed the experiments, analyzed the data, supervised the project, and wrote the manuscript. All authors have given approval for the final version of the manuscript.
We declare no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01288-17.
REFERENCES
- 1.Fisher JF, Meroueh SO, Mobashery S. 2005. Bacterial resistance to beta-lactam antibiotics: compelling opportunism, compelling opportunity. Chem Rev 105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 2.Frère JM, Duez C, Ghuysen JM, Vandekerkhove J. 1976. Occurrence of a serine residue in the penicillin-binding site of the exocellular DD-carboxy-peptidase-transpeptidase from Streptomyces R61. FEBS Lett 70:257–260. doi: 10.1016/0014-5793(76)80770-3. [DOI] [PubMed] [Google Scholar]
- 3.Drawz SM, Bonomo RA. 2010. Three decades of beta-lactamase inhibitors. Clin Microbiol Rev 23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helfand MS, Bonomo RA. 2003. Beta-lactamases: a survey of protein diversity. Curr Drug Targets Infect Disord 3:9–23. doi: 10.2174/1568005033342181. [DOI] [PubMed] [Google Scholar]
- 5.Jacoby GA. 2009. AmpC β-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenollar-Ferrer C, Frau J, Donoso J, Muñoz F. 2008. Evolution of class C β-lactamases: factors influencing their hydrolysis and recognition mechanisms. Theor Chem Acc 121:209–218. doi: 10.1007/s00214-008-0463-2. [DOI] [Google Scholar]
- 7.Goo KS, Sim TS. 2011. Designing new beta-lactams: implications from their targets, resistance factors and synthesizing enzymes. Curr Comput Aided Drug Des 7:53–80. doi: 10.2174/157340911793743538. [DOI] [PubMed] [Google Scholar]
- 8.Nicolau DP. 2008. Carbapenems: a potent class of antibiotics. Expert Opin Pharmacother 9:23–37. doi: 10.1517/14656566.9.1.23. [DOI] [PubMed] [Google Scholar]
- 9.Pan X, Wong WT, He Y, Jiang Y, Zhao Y. 2014. Perturbing the general base residue Glu166 in the active site of class A beta-lactamase leads to enhanced carbapenem binding and acylation. Biochemistry 53:5414–5423. doi: 10.1021/bi401609h. [DOI] [PubMed] [Google Scholar]
- 10.Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. 2011. Carbapenems: past, present, and future. Antimicrob Agents Chemother 55:4943–4960. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishimura S, Yasuda N, Sasaki H, Matsumoto Y, Kamimura T, Sakane K, Takaya T. 1990. Synthesis and beta-lactamase inhibitory activity of 7-alpha-hydroxyethyl cephem sulfone and sulfoxide derivatives. J Antibiot 43:114–117. doi: 10.7164/antibiotics.43.114. [DOI] [PubMed] [Google Scholar]
- 12.Mourey L, Miyashita K, Swaren P, Bulychev A, Samama JP, Mobashery S. 1998. Inhibition of the NMC-A beta-lactamase by a penicillanic acid derivative and the structural bases for the increase in substrate profile of this antibiotic resistance enzyme. J Am Chem Soc 120:9382–9383. doi: 10.1021/ja9817996. [DOI] [Google Scholar]
- 13.Miyashita K, Massova I, Taibi P, Mobashery S. 1995. Design, synthesis, and evaluation of a potent mechanism-based inhibitor for the TEM β-lactamase with implications for the enzyme mechanism. J Am Chem Soc 117:11055–11059. doi: 10.1021/ja00150a003. [DOI] [Google Scholar]
- 14.Beadle BM, Trehan I, Focia PJ, Shoichet BK. 2002. Structural milestones in the reaction pathway of an amide hydrolase: substrate, acyl, and product complexes of cephalothin with AmpC beta-lactamase. Structure 10:413–424. doi: 10.1016/S0969-2126(02)00725-6. [DOI] [PubMed] [Google Scholar]
- 15.Testero SA, O'Daniel PI, Shi Q, Lee M, Hesek D, Ishiwata A, Noll BC, Mobashery S. 2009. Regiospecific syntheses of 6alpha-(1R-hydroxyoctyl)penicillanic acid and 6beta-(1R-hydroxyoctyl)penicillanic acid as mechanistic probes of class D beta-lactamases. Org Lett 11:2515–2518. doi: 10.1021/ol900668k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volkmann RA, Carroll RD, Drolet RB, Elliott ML, Moore BS. 1982. Efficient preparation of 6,6-dihalopenicillanic acids. Synthesis of penicillanic acid S,S-dioxide (sulbactam). J Org Chem 47:3344–3345. [Google Scholar]
- 17.DiNinno F, Beattie TR, Christensen BG. 1977. Aldol condensations of regiospecific penicillanate and cephalosporanate enolates. Hydroxyethylation at C-6 and C-7. J Org Chem 42:2960–2965. [DOI] [PubMed] [Google Scholar]
- 18.Padayatti PS, Sheri A, Totir MA, Helfand MS, Carey MP, Anderson VE, Carey PR, Bethel CR, Bonomo RA, Buynak JD, van den Akker F. 2006. Rational design of a beta-lactamase inhibitor achieved via stabilization of the trans-enamine intermediate: 1.28 A crystal structure of wt SHV-1 complex with a penam sulfone. J Am Chem Soc 128:13235–13242. doi: 10.1021/ja063715w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong WT, Chan KC, So PK, Yap HK, Chung WH, Leung YC, Wong KY, Zhao Y. 2011. Increased structural flexibility at the active site of a fluorophore-conjugated beta-lactamase distinctively impacts its binding toward diverse cephalosporin antibiotics. J Biol Chem 286:31771–31780. doi: 10.1074/jbc.M110.198895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan X, He Y, Lei J, Huang X, Zhao Y. 2017. Crystallographic snapshots of class A beta-lactamase catalysis reveal structural changes that facilitate beta-lactam hydrolysis. J Biol Chem 292:4022–4033. doi: 10.1074/jbc.M116.764340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trehan I, Morandi F, Blaszczak LC, Shoichet BK. 2002. Using steric hindrance to design new inhibitors of class C beta-lactamases. Chem Biol 9:971–980. doi: 10.1016/S1074-5521(02)00211-9. [DOI] [PubMed] [Google Scholar]
- 22.Galleni M, Amicosante G, Frère JM. 1988. A survey of the kinetic parameters of class C beta-lactamases. Cephalosporins and other beta-lactam compounds. Biochem J 255:123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Kern G, Walkup GK, Fisher SL. 2012. Avibactam is a covalent, reversible, non-beta-lactam beta-lactamase inhibitor. Proc Natl Acad Sci U S A 109:11663–11668. doi: 10.1073/pnas.1205073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beadle BM, Shoichet BK. 2002. Structural basis for imipenem inhibition of class C beta-lactamases. Antimicrob Agents Chemother 46:3978–3980. doi: 10.1128/AAC.46.12.3978-3980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tremblay LW, Fan F, Blanchard JS. 2010. Biochemical and structural characterization of Mycobacterium tuberculosis beta-lactamase with the carbapenems ertapenem and doripenem. Biochemistry 49:3766–3773. doi: 10.1021/bi100232q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fonseca F, Chudyk EI, van der Kamp MW, Correia A, Mulholland AJ, Spencer J. 2012. The basis for carbapenem hydrolysis by class A beta-lactamases: a combined investigation using crystallography and simulations. J Am Chem Soc 134:18275–18285. doi: 10.1021/ja304460j. [DOI] [PubMed] [Google Scholar]
- 27.Smith CA, Frase H, Toth M, Kumarasiri M, Wiafe K, Munoz J, Mobashery S, Vakulenko SB. 2012. Structural basis for progression toward the carbapenemase activity in the GES family of beta-lactamases. J Am Chem Soc 134:19512–19515. doi: 10.1021/ja308197j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knox JR, Moews PC. 1991. Beta-lactamase of Bacillus licheniformis 749/C. Refinement at 2 A resolution and analysis of hydration. J Mol Biol 220:435–455. [DOI] [PubMed] [Google Scholar]
- 29.King DT, Wasney GA, Nosella M, Fong A, Strynadka NC. 2017. Structural insights into inhibition of Escherichia coli penicillin-binding protein 1B. J Biol Chem 292:979–993. doi: 10.1074/jbc.M116.718403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 31.Shlaes DM. 2013. New beta-lactam-beta-lactamase inhibitor combinations in clinical development. Ann N Y Acad Sci 1277:105–114. doi: 10.1111/nyas.12010. [DOI] [PubMed] [Google Scholar]
- 32.Salverda ML, De Visser JA, Barlow M. 2010. Natural evolution of TEM-1 beta-lactamase: experimental reconstruction and clinical relevance. FEMS Microbiol Rev 34:1015–1036. doi: 10.1111/j.1574-6976.2010.00222.x. [DOI] [PubMed] [Google Scholar]
- 33.Lahiri SD, Johnstone MR, Ross PL, McLaughlin RE, Olivier NB, Alm RA. 2014. Avibactam and class C β-lactamases: mechanism of inhibition, conservation of the binding pocket, and implications for resistance. Antimicrob Agents Chemother 58:5704–5713. doi: 10.1128/AAC.03057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cer RZ, Mudunuri U, Stephens R, Lebeda FJ. 2009. IC(50)-to-K (i): a web-based tool for converting IC(50) to K (i) values for inhibitors of enzyme activity and ligand binding. Nucleic Acids Res 37:W441–W445. doi: 10.1093/nar/gkp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu WP, Kincaid E, Sun Y, Bauer MD. 2001. Kinetics of beta-lactam interactions with penicillin-susceptible and -resistant penicillin-binding protein 2x proteins from Streptococcus pneumoniae. Involvement of acylation and deacylation in beta-lactam resistance. J Biol Chem 276:31494–31501. [DOI] [PubMed] [Google Scholar]
- 36.Chan PH, So PK, Ma DL, Zhao Y, Lai TS, Chung WH, Chan KC, Yiu KF, Chan HW, Siu FM, Tsang CW, Leung YC, Wong KY. 2008. Fluorophore-labeled beta-lactamase as a biosensor for beta-lactam antibiotics: a study of the biosensing process. J Am Chem Soc 130:6351–6361. doi: 10.1021/ja076111g. [DOI] [PubMed] [Google Scholar]
- 37.Leslie AG. 2006. The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr 62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 38.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 39.McCoy AJ. 2007. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. 2004. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr 60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.