ABSTRACT
Genetics-based drug susceptibility testing has improved the diagnosis of drug-resistant tuberculosis but is limited by our lack of knowledge of all resistance mechanisms. Next-generation sequencing has assisted in identifying the principal genetic mechanisms of resistance for many drugs, but a significant proportion of phenotypic drug resistance is unexplained genetically. Few studies have formally compared the transcriptomes of susceptible and resistant Mycobacterium tuberculosis strains. We carried out comparative whole-genome transcriptomics of extensively drug-resistant (XDR) clinical isolates using RNA sequencing (RNA-seq) to find novel transcription-mediated mechanisms of resistance. We identified a promoter mutation (t to c) at position −11 (t−11c) relative to the start codon of ethA that reduces the expression of a monooxygenase (EthA) that activates ethionamide. (In this article, nucleotide changes are lowercase and amino acid substitutions are uppercase.) Using a flow cytometry-based reporter assay, we show that the reduced transcription of ethA is not due to transcriptional repression by ethR. Clinical strains harboring this mutation were resistant to ethionamide. Other ethA promoter mutations were identified in a global genomic survey of resistant M. tuberculosis strains. These results demonstrate a new mechanism of ethionamide resistance that can cause high-level resistance when it is combined with other ethionamide resistance-conferring mutations. Our study revealed many other genes which were highly up- or downregulated in XDR strains, including a toxin-antitoxin module (mazF5 mazE5) and tRNAs (leuX and thrU). This suggests that global transcriptional modifications could contribute to resistance or the maintenance of bacterial fitness have also occurred in XDR strains.
KEYWORDS: promoter, RNA sequencing, RNA-seq, drug resistance, drug susceptibility testing, flow cytometry, fluorescent reporter, mymA, tuberculosis
INTRODUCTION
Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), has progressively developed resistance to the most effective first- and second-line antituberculosis drugs (1). Patients infected with extensively drug-resistant (XDR) strains (strains resistant to the fluoroquinolones and aminoglycosides, in addition to rifampin [RIF] and isoniazid, in which resistance to the last two drugs defines multidrug resistance [MDR]) have extremely high rates of mortality, despite the use of long and intensive treatment regimens (2, 3). The ultimate control of drug resistance will require multiple interventions, one of which will be individualized therapy based on rapid comprehensive drug susceptibility testing (DST).
Current molecular genetics-based tests, such as the GeneXpert MTB/RIF and GenoType MTBDRplus assays, have accelerated the clinical detection of known mutations causing RIF and/or isoniazid resistance (4, 5). These and other genetic tests detect only MDR-TB and a limited number of mutations associated with resistance to second-line drugs (6). Whole-genome sequencing (WGS) has the potential to rapidly detect all possible drug resistance-conferring mutations (7). However, recent studies have demonstrated that genotypic DST using WGS lacks sensitivity for the detection of resistance to many second-line drugs, including fluoroquinolones (8–11). Improving the sensitivity of genetic susceptibility testing will be possible only with a more comprehensive understanding of the genetic determinants of drug resistance.
Our current understanding of drug resistance in M. tuberculosis has developed through studying resistant mutants isolated in vitro and the accumulation of mutations in resistant clinical isolates (12). These studies have identified various genetic mechanisms of resistance, including target modification, loss of the enzymatic function required to activate prodrugs, and altered drug efflux (13, 14).
In addition to intragenic mutations, there is increasing evidence that alterations to gene transcription are an important mechanism of conferring drug resistance. Promoter mutations which result in the upregulation of inhA, which encodes the target for isoniazid, were the first to be described (15). Pyrazinamide (PZA) resistance has been associated with mutations in the regulatory region upstream of pncA, the enzyme responsible for activating PZA (16–18). Aminoglycoside cross-resistance in M. tuberculosis can arise due to mutations in the regulatory region of whiB7 (which encodes a transcriptional activator), which results in increased expression of eis (which acetylates and inactivates kanamycin), as well as tap (which encodes an efflux pump that extrudes streptomycin) (19). eis promoter mutations have also been described. Recently, cross-resistance between clofazimine (CFZ) and bedaquiline (BDQ) was shown to be due to mutations within Rv0678 (20, 21), a transcriptional repressor, which results in derepression and upregulation of the multisubstrate efflux pump mmpL5.
Despite the discovery of these varied transcriptionally driven mechanisms of resistance, there have been few systematic whole-genome transcriptional comparisons of suitably matched susceptible and resistant M. tuberculosis strains, and none to date has used RNA-sequencing (RNA-seq). In this study, we therefore selected phylogenetically closely related susceptible and resistant clinical strains and subjected them to comparative transcriptomics using RNA-seq to identify novel mechanisms of resistance.
RESULTS
Comparative transcriptomics.
In order to identify novel mechanisms of resistance mediated at the level of transcription, we subjected drug-resistant and drug-susceptible strains of M. tuberculosis to comparative transcriptomics using RNA sequencing (Table 1). We reasoned that strains with highly complex resistance profiles were most likely to have acquired mutations resulting in transcriptional changes. Using a whole-genome-based phylogenetic analysis, we identified 3 XDR clinical isolates from a well-documented outbreak in KwaZulu-Natal, South Africa, and a closely related drug-susceptible strain to act as a control (1). All strains were from the LAM4 branch of lineage 4. In pairwise comparisons, the 3 XDR strains differed from each other by 7 single nucleotide polymorphisms (SNPs) or less (Fig. 1A). The maximum difference between the drug-susceptible strain and an XDR strain was 76 SNPs, of which 6 occurred in known drug resistance-conferring genes.
TABLE 1.
Strain details, including resistance mutations and RNA sequencing coverage
| Strain | Spoligotype | Resistance mutation for each druga |
RNA-seq coverage (fold) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| INH | RIF | STR | EMB | KAN | ETH | OFL | |||
| TKK-01-0084 | LAM4 | 288.63 | |||||||
| TKK-01-0025 | LAM4 | inhA t−8a | rpoB L452P | gidB L16R | embB M306V | rrs A1401G | inhA t−8a | gyrA A90V | 214.34 |
| katG S315T | rpoB D435G | gidB del | |||||||
| TKK-01-0033 | LAM4 | inhA t−8a | rpoB L452P | gidB L16R | embB M306V | rrs A1401G | inhA t−8a | gyrA A90V | 239.13 |
| katG S315T | rpoB D435G | gidB del | |||||||
| TKK-01-0040 | LAM4 | inhA t−8a | rpoB L452P | gidB L16R | embB M306V | rrs A1401G | inhA t−8a | gyrA A90V | 269.24 |
| katG S315T | rpoB D435G | gidB del | |||||||
INH, isoniazid; RIF, rifampin; STR, streptomycin; EMB, ethambutol; KAN, kanamycin; ETH, ethionamide; OFL, ofloxacin; del, deletion.
FIG 1.
(A) Phylogenetic tree representing the distribution of the 4 strains selected for RNA-seq (shown in red and boxed). (B) Hierarchical gene clustering of the 4 strains selected for RNA-seq on the basis of their relative gene expression shows that the drug-susceptible strain clusters separately from the others. (C) Venn diagram representing the numbers of genes differentially expressed at levels 7-fold or greater relative to their levels of expression by the susceptible control strain. The blue, red, and green circles represent pairwise comparisons with TKK-01-0033, TKK-01-0025, and TKK 01-0040, respectively.
To determine if there were global transcriptional differences between our strains, we first carried out hierarchical clustering of their transcriptional profiles. This separated the expression profiles of the three drug-resistant strains from the expression profile of the susceptible control (Fig. 1B). To identify genes either up- or downregulated in the XDR strains, we performed pairwise comparisons for each resistant strain with the drug-susceptible control. In the resistant strains, up to 40 genes were significantly over- or underexpressed at the 95% confidence level (P ≤ 0.05) and up to 10 genes were significantly over- or underexpressed at the 99% level (P ≤ 0.01) relative to their expression in the susceptible control (see Table S1 in the supplemental material). Importantly, in all three pairwise comparisons, the inhA gene showed a greater than 8-fold upregulation of expression in the resistant strains at the 99% confidence interval. All three resistant strains harbored a t-to-a mutation at position 8 (t−8a) in the promoter region of fabG1, which is known to cause the upregulation of inhA. The detection of this transcriptional change therefore acted as an internal validation of our approach. Apart from inhA, no other genes were significantly upregulated in all three comparisons. Two genes, fabG1 (also in the inhA operon) and Rv1761c (a gene of unknown function), had expression levels in two strains (TKK-01-0040 and TKK-01-0033) significantly different from that in the susceptible control.
After defining differential gene expression at the statistically significant levels (95% and 99% confidence intervals), we extended our analysis to all genes that had a high mean fold change in transcript levels (≥7-fold up or down) relative to the susceptible control (Fig. 1C and Table S2). In addition to fabG1 and inhA, we found that 5 other genes fell into this classification: mazF5; mazE5, encoding a toxin-antitoxin module; two tRNAs (leuX and thrU); and ethA. ethA was of particular interest, as it encodes a monooxygenase required for the activation of the prodrug ethionamide (ETH) (22, 23), a key component of treatment of infections cause by MDR strains. Loss-of-function mutations in ethA result in ethionamide resistance (23, 24). Following Benjamini-Hochberg correction, ethA was found to be significantly downregulated in one of our pairwise comparisons described above.
Comparative genome-transcriptome analysis.
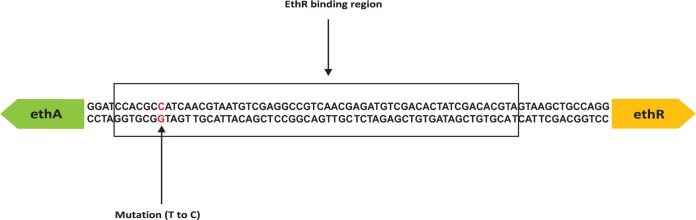
In order to understand the genetic basis of the transcriptional changes defined by our RNA-seq experiments, we used comparative genomics to identify mutations located in intergenic regions associated with genes that were highly over- or underexpressed in our resistant strains relative to our susceptible control strain. This analysis identified an intergenic region mutation (t to c) at position −11 (t−11c) relative to the start codon of ethA. The detected mutation was located within the promoter region of ethA as well as within the binding domain of the divergently expressed transcriptional regulator ethR, which is known to repress ethA (25) (Fig. 2). The location of the mutation suggested that it could lead to the downregulation of EthA by (i) directly reducing ethA transcription independently of ethR regulation, (ii) increasing ethR transcription, leading to the repression of ethA, or (iii) affecting the binding of ethR, leading to the increased repression of ethA transcription.
FIG 2.
Representation of the intergenic region between ethA and ethR. The location of the single nucleotide polymorphism (SNP) is found 11 bp upstream of ethA and is indicated in red. The ethR binding region is indicated by the black box (25).
Functional characterization of ethA and ethR promoters.
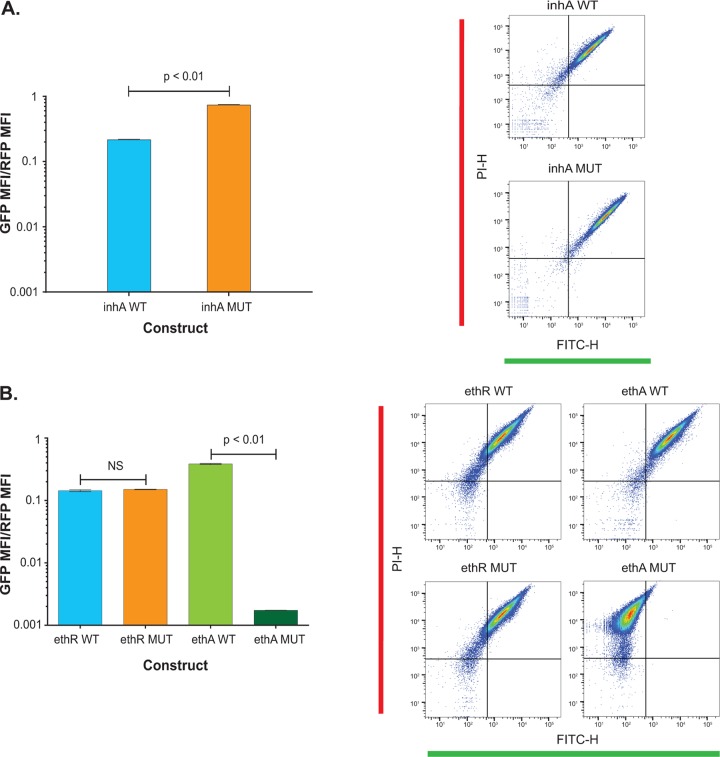
To determine if the t−11c mutation functionally influenced either ethA or ethR transcription, we used a dual-color fluorescent protein promoter assay. The episomal construct pLDW-DC* has a constitutively expressed red fluorescent protein (RFP), TagRFP, and a promoterless Emerald green fluorescent protein (GFP), in front of which promoters with or without mutations can be cloned. Promoter activity is expressed as the ratio of green to red fluorescence, normalizing for any variability in plasmid number. To validate our approach, we used the fabG1-inhA promoter with and without the mutant promoter sequence of inhA with a g−17t mutation. The construct harboring the g−17t mutant promoter sequence of inhA resulted in a 3.4-fold increase in the ratio of the median fluorescent intensity (MFI) of green fluorescence to the MFI of red fluorescence (the MFI ratio) relative to the MFI ratio for the wild type (Fig. 3A).
FIG 3.
Analysis of promoter activity between the wild-type (WT) and mutant (MUT) constructs. (Left) Ratios of the median fluorescent intensity (MFI) of the green fluorescent protein (GFP) to the MFI of the red fluorescent protein (RFP), as well as statistical differences between the wild-type and mutant constructs for the inhA promoter (A) and ethA and ethR promoters (B). P values are indicated on the bar charts. (Right) Single cell counts from flow cytometry. RFP expression is represented on the y axis as the PI-H (height) channel, and GFP expression is represented on the x axis as the FITC-H (height) channel.
We then assayed constructs with the wild-type 250-bp upstream region of ethA or ethR and 2 matched mutant constructs with either the t−11c mutation (relative to ethA) or the corresponding t−65c mutation in the ethR construct (Fig. 3B). We observed no significant change in the MFI ratio between the two ethR promoter constructs. In contrast, the t−11c mutant promoter resulted in an MFI ratio that was significantly lower than that obtained with the wild-type control. These results suggest that the t−11c intergenic region mutation does not affect the transcription of ethR but does diminish the expression of ethA to levels that could result in ethionamide resistance.
ethA expression in clinical isolates of M. tuberculosis.
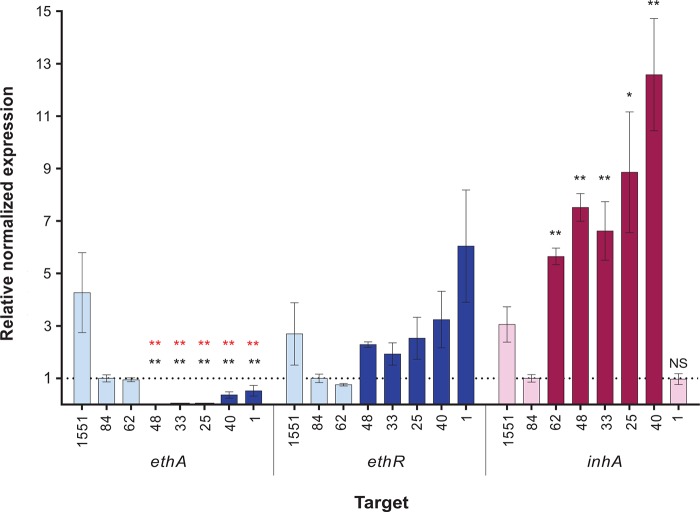
To confirm the transcriptional changes identified by RNA-seq in strains harboring the t−11c mutation, we used quantitative reverse transcription-PCR (RT-qPCR) to measure the expression levels of ethA in clinical isolates (Table S3). In the 5 strains with an ethA t−11c promoter mutation tested, the relative normalized expression levels of the monooxygenase were significantly lower or close to zero compared to those in the control strains. All tested strains with the inhA promoter mutations had increased relative normalized levels of expression of inhA compared to those in strains without the mutation (Fig. 4).
FIG 4.
Relative gene expression of ethA, ethR, or inhA in clinical strains of M. tuberculosis (see Table S5 in the supplemental material). Gene expression levels were normalized to the sigA expression level for each strain. Relative normalized expression represents the fold change in the normalized expression by each strain compared to that by drug-susceptible clinical strain TKK-01-0084. Light blue bars, strains that do not contain t−11c ethA promoter mutations; dark blue bars, strains that have the t−11c ethA promoter mutation; light pink bars, strains without inhA promoter mutations; dark pink bars, strains with inhA promoter mutations. The TKK strain numbers are abbreviated to their last two digits; e.g., 62 represents strain TKK-01-0062. The statistical significance of the relative normalized expression for ethA and inhA was derived using unpaired t tests between each strain and clinical drug-susceptible strain TKK-01-0084 (asterisks in black). In addition, the statistical significance of the relative normalized expression for ethA was derived using unpaired t tests between each strain and strain TKK-01-0062, which does not harbor a t−11c ethA promoter mutation (asterisks in red). 1551 corresponds to the laboratory strain CDC1551, which was excluded from this analysis. **, P ≤ 0.01; *, P ≤ 0.05; NS, not significant.
ethA promoter mutations and ethionamide resistance in clinical isolates.
To determine if ethA promoter mutations were associated with clinical resistance, we tested a panel of clinical isolates which, on the basis of genome analyses, harbored putative ethionamide resistance-conferring mutations for quantitative ethionamide susceptibility (Table 2). The panel included three strains that had the t−11c intergenic region mutation but no other mutations previously associated with clinical ethionamide resistance (inhA promoter mutations and intragenic region mutations in ethA, ethR, ndh, and mshA) (26). Recently, loss-of-function mutations in another M. tuberculosis monooxygenase, mymA (Rv3083), have been proposed to be an additional resistance mechanism (27). Interestingly, during our selection of strains, we were able to identify a group of isolates with a deletion spanning mymA (Fig. S1). Sequence confirmation in five of these strains showed an identical deletion of 2,891 bp, indicating a unique polymorphism event suggestive of clonal expansion. Strains with this mutation were included in our analysis.
TABLE 2.
ETH MICs for clinical strainsa
| Strain | DST | Spoligotype | Putative ETH resistance-conferring mutations |
MIC (mg/liter) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ethA promoter | inhA promoter | inhA intragenic region | ethA intragenic region | ethR intragenic region | mymA | mshA | ndh | ||||
| TKK-01-0001 | MDR | KZN | t−11c | 20 | |||||||
| TKK-01-0035 | MDR | KZN | t−11c | 10 | |||||||
| TKK-01-0075 | MDR | KZN | t−11c | 5 | |||||||
| TKK-01-0025 | XDR | KZN | t−11c | t−8a | 80 | ||||||
| TKK-01-0040 | XDR | KZN | t−11c | t−8a | >80 | ||||||
| TKK-01-0048 | MDR | KZN | t−11c | t−8a | 20 | ||||||
| TKK-01-0033 | XDR | KZN | t−11c | t−8a | 80 | ||||||
| TKK-02-0001 | XDR | Beijing | c−15t | I194V | P94P | A189T | 40 | ||||
| TKK-02-0046 | Poly (P/N + RIF) | Beijing | c−15t | I194V | P94P | A189T | 20 | ||||
| TKK-01-0005 | Poly (STR + ETH) | Beijing | c−15t | A189T | 80 | ||||||
| TKK-01-0062 | PXDR | Beijing | g−17t | A381P | 20 | ||||||
| TKK-01-0032 | MDR | S | t−8g | V18A | 5 | ||||||
| TKK-01-0013 | XDR | Beijing | Y276H | A189T | 5 | ||||||
| TKK-02-0018 | MDR | T3 | V202L | del | 15 | ||||||
| TKK-02-0019 | PXDR | V202L | del | 20 | |||||||
| TKK-01-0026 | MDR | T3 | del | 2.5 | |||||||
| TKK-02-0069 | PXDR | del | 5 | ||||||||
| TKK-01-0081 | DS | KZN | 2.5 | ||||||||
| TKK-01-0084 | DS | KZN | 2.5 | ||||||||
| TKK-01-0047 | DS | Beijing | 2.5 | ||||||||
| TKK-01-0027 | DS | Beijing | 2.5 | ||||||||
| H37Rv | DS | NA | 2.5 | ||||||||
| CDC 1551 | DS | NA | 1.25 | ||||||||
del, deletion; Poly, polydrug-resistant (resistance to one or more drugs but not MDR/XDR); P/N, para-aminosalicylic acid–nicotinamide; RIF, rifampin; STR, streptomycin; ETH, ethionamide; PXDR, preextensively drug resistant (MDR and resistance to either a fluoroquinolone or an injectable); DS, drug susceptible; NA, not applicable.
The ETH MICs for strains that had only the ethA t−11c promoter mutation ranged from 5 to 20 mg/liter, showing low-level resistance to ETH (Table 2). However, in combination with the t−8a inhA promoter mutation, we observed higher levels of resistance, suggesting that the phenotypic consequences of the two promoter mutations are additive. The two strains with only mymA deletions had ETH MICs of 2.5 and 5 mg/liter, which were only marginally elevated relative to the MICs of strains without any ethionamide drug resistance-conferring mutations (1.25 to 2.5 mg/liter).
Global distribution of ethA promoter mutations.
In order to determine how widespread ethA promoter mutations are among clinical M. tuberculosis isolates, we exploited a recent genome analysis of globally isolated drug-resistant and -susceptible strains (28). From a total of 5,310 strains, we identified 402 with a mutation within the ethA-ethR intergenic region relative to the sequence of H37Rv (Table S4). One hundred thirty-nine of these were the t−11c mutation, and all were identified in South Africa-derived lineage 4 isolates. The most common mutation was a−7g, found in 212 strains, nearly all of which (205 strains) were from Eastern Europe. Eleven other infrequently occurring mutations were identified, but one of these also mapped to the −11 site (t−11g). Using parsimony to define independent mutational events across the phylogeny (29), we found that the a−7g mutation had independently evolved at least 32 times, suggesting that this mutation was under selective pressure and supporting the possibility that it has a role in conferring drug resistance. In contrast, the t−11c mutation was predicted to have evolved only once, which is compatible with the ongoing transmission and clonal expansion of XDR strains from South Africa in which the mutation was found.
DISCUSSION
The aim of our study was to use a comparative whole-genome transcriptomic approach to identify novel mechanisms of resistance mediated at the transcriptional level. We were able to identify a promoter mutation upstream of ethA. We confirmed by our dual-color promoter assay and quantitative RT-PCR that this mutation leads to the reduced transcription of ethA, which encodes a monooxygenase that activates the prodrug ethionamide (22, 23). Strains harboring only this t−11c mutation and no other ethionamide resistance-determining genotypes were resistant to ethionamide (MIC range, 5 to 20 mg/liter), indicating that this mutation should be included in genetics-based diagnostic tests. In support of this, a recent genomewide association analysis also reported an association between the t−11c mutation and ethionamide resistance (8).
In our analysis of the global distribution of ethA-ethR intergenic region mutations, we identified the t−11c mutation solely in South African strains. This data set, however, consisted of isolates from only 43 countries and notably lacked representation from several regions of the world where TB is epidemic, such as South America. Nonetheless, a previous study using direct sequencing of drug resistance loci detected five different variants in the promoter region of ethA, one of which was a t−11c mutation found in an isolate from Peru (30). There were, however, no clinical data available to rule out whether the patient in question had any travel history to South Africa. We therefore cannot confirm whether this mutation is geographically restricted. The a−7g mutation was more dispersed, but the majority of strains harboring this mutation were from Eastern Europe. A previous study reported the phenotype of 172 strains with the a−7g mutation, and only 56 of these were resistant to ethionamide (31). This could be due to inconsistencies in drug susceptibility testing or because the level of resistance conferred lay close to the breakpoint used in susceptibility testing but suggests that not all mutations within the ethA-ethR intergenic region result in levels of resistance similar to those that we observed for strains with the t−11c mutation (30).
Few studies have used quantitative drug susceptibility testing to correlate the ethionamide MIC with the genotype (32), so it is unclear to what extent individual mutations contribute to resistance and how they might interact. Although there may be additional mechanisms of ethionamide resistance that have yet to be identified, our results suggest that the t−11c mutation causes a modest increase in the ethionamide MIC but in combination with a mutation in the inhA promoter (considered to cause low-level ethionamide resistance [24]) leads to high-level resistance. Among the other ethionamide-resistant strains assayed, most had more than one mutation potentially contributing to their increased MIC. The pathway to clinical ethionamide resistance may therefore be the stepwise accumulation of multiple mutations rather than the selection of a single high-level-resistance-conferring mutation, as seen with some other antituberculosis drugs.
In the panel of clinical isolates that we selected to evaluate the phenotype associated with the t−11c mutation, we identified polymorphisms in other genes implicated in ethionamide resistance. We found four mutations at three positions in the inhA promoter region, all of which have been previously described (33). One of these strains had a t−8g inhA promoter mutation in combination with a nonsynonymous mutation (V18A) in ndh, which encodes a type II NADH dehydrogenase. Mutations in ndh can result in increased levels of NADH and reduce the level of binding of the isoniazid and ethionamide NAD adducts to their target, InhA (33). However, this strain had a low MIC, suggesting that neither of these mutations causes high-level resistance.
We cannot rule out the possibility of the existence of other ethionamide resistance-conferring mechanisms in our strains. EthA is 1 of 30 other monooxygenases within the M. tuberculosis genome (24), and a recently characterized monooxygenase, mymA (Rv3083) (27), was proposed to be an additional enzyme responsible for the activation of ethionamide. We identified strains with a 2,891-bp deletion spanning mymA, lipR, and half of Rv3085 (Fig. S1). Two of these strains had no other known mutations associated with ethionamide resistance but were susceptible to ethionamide when a standard MIC cutoff was employed (Table 2), suggesting that mymA is not important for drug resistance in clinical isolates.
Our initial comparative transcriptional analysis identified only a limited number of genes whose level of expression was statistically significantly different from that in the control. This may have been due to the increased variability associated with the propagation of clinical isolates in culture media. We therefore looked at genes whose expression was highly divergent in the resistant strains in all pairwise comparisons. In addition to ethA, this identified mazF5 and mazE5, which encode a toxin-antitoxin system, one of nine MazEF homologues in M. tuberculosis. A mazF3, mazF6, and mazF9 triple-null mutant was less able to survive exposure to antituberculosis drugs (34), so these systems could potentially be involved in mediating resistance, although it is unclear how the downregulation of mazE5 would influence drug susceptibility. Two tRNAs, leuX and thrU, were among the genes most highly upregulated in the resistant strains. Beyond a fundamental role in translation, tRNAs and their degradation products have been shown to regulate stress responses and adaptive changes in translation (35). It is therefore conceivable that the upregulation of these two tRNAs may be a manifestation of more global regulatory changes that have occurred during the evolution of drug resistance. Future studies comprising strains from different outbreaks and lineages are, however, needed to determine whether these transcriptional changes are limited to the XDR outbreak from KwaZulu-Natal in 2005 (1).
The treatment of MDR-TB is currently undergoing a revolution, with the introduction of new drugs and regimens (36). WHO has recently approved the use of a 9-month short course of therapy, and the 4-month intensive phase of this regimen includes ethionamide (or its analogue, prothionamide). Although the contribution of individual drugs to treatment efficacy is unclear, it is recommended that short-course treatment be withheld from MDR-TB patients with preexisting resistance to any individual drug. Pretreatment screening for ethionamide resistance is therefore critical for the implementation of short-course MDR treatment. However, testing for phenotypic susceptibility to ethionamide is notoriously difficult (37). Our results contribute to the development of a genetics-based resistance test, but further studies are required to define the interaction of diverse mutations and drug resistance-conferring loci as well as establish a clinically relevant critical concentration for ethionamide.
MATERIALS AND METHODS
Strains and growth conditions.
Three XDR isolates and 1 fully drug-susceptible clinical isolate from the LAM4 (KZN) spoligotype of M. tuberculosis (Table 1) were obtained from archived cultures from single colonies whose genomes had previously been sequenced (1). Cultures were grown in triplicate at 37°C in BD Difco Middlebrook 7H9 broth supplemented with BBL Middlebrook oleic acid-albumin-dextrose-catalase enrichment medium, 0.5% glycerol, and 0.01% Tween 80 with continuous shaking at 200 rpm. Additional strains were selected from the same collection on the basis of specific ethA, ethR, inhA, and mymA genotypes (Table 2) (1).
RNA extraction and quality control. (i) RNA extraction.
RNA was harvested from 25-ml cultures grown to an optical density at 600 nm (OD600) of between 0.5 and 0.8, using a modified TRIzol method (38). Briefly, the cultures were centrifuged at 4,000 rpm for 20 min at 25°C and the pellet was resuspended in 1 ml of TRIzol reagent (Invitrogen, USA). Thereafter, approximately 100 μl of 0.1-mm zirconia/silica glass beads (BioSpec Products, USA) was added and the cultures were subjected to four pulses of bead beating, using a Roche MagNA Lyser instrument, at 7,000 rpm for 60 s with 2-min intermittent incubations on ice. Immediately after bead beating, 200 μl of chloroform was added, followed by centrifugation at 15,000 rpm for 15 min at 4°C and separation of the aqueous phase. The RNA was precipitated with 500 μl of 100% isopropanol and incubated at −20°C for 1 h. After centrifugation at 15,000 rpm for 10 min at 4°C, the RNA pellet was washed with 1 ml 75% ethanol, centrifuged at 10,000 rpm for 5 min at 4°C, and air dried. The RNA pellet was then dissolved in 30 μl of RNase-free water.
(ii) DNase treatment and purification.
The RNA was subjected to DNase treatment using a DNase I RNase-free kit (Thermo Scientific, USA) per the manufacturer's instructions. The RNA was then purified using an RNeasy minikit (Qiagen, Germany), during which a second round of DNase digestion utilizing the RNase-free DNase set (Qiagen, Germany) took place. The integrity of the RNA samples was confirmed using a 23S rRNA/16S rRNA ratio (≥1.2) determined by an Experion StdSens analysis kit (Bio-Rad, USA).
RNA sequencing and bioinformatics analysis. (i) RNA-seq library preparation.
A Qubit RNA assay kit (Invitrogen, USA) was used with a Qubit (version 2.0) fluorometer to quantify the RNA. Following RNA quantification, the rRNA was depleted using a Ribo-Zero magnetic kit (Illumina, USA). Enriched mRNA was analyzed on an RNA-specific E-gel EX 2% agarose gel (Invitrogen, USA) to confirm rRNA removal. After purification of the mRNA using an RNeasy minikit (Qiagen, Germany), RNA sequencing libraries were constructed using a NEBNext Ultra directional RNA library preparation kit for Illumina (New England BioLabs Inc., USA). The prepared libraries were indexed with NEBNext multiplex oligonucleotides for Illumina (New England BioLabs Inc. USA) and sequenced with 50-bp single-end reads on an Illumina HiSeq 2000 platform at the Norwegian Sequencing Centre, Oslo, Norway.
(ii) Bioinformatics.
The sequence reads (Bioproject PRJNA414397, SRA SRP120003) were aligned to the M. tuberculosis H37Rv genome (NCBI accession number NC_000962.2) using the SeqMan NGen program from DNAStar Lasergene (version 11) software. Transcripts for each sample were quantified and normalized as the number of reads per kilobase per million reads (RPKM). The three replicate RPKM values for each sample were standardized on the basis of their mean transcript values and were used to assess gene expression and fold change differences in expression between isolates using the ArrayStar program (DNAStar). Pairwise comparisons between strains were conducted, with confidence intervals and statistics being determined using Student's t test and with multiple-testing corrections being made using the Benjamini-Hochberg correction to reduce the false discovery rate (FDR). Intergenic SNPs present only in the three XDR strains and not in the drug-susceptible strain were identified from whole-genome sequencing data from a previous study (1). A transcriptomic-genomic analysis was then conducted to identify promoter SNPs associated with at least a 4-fold up- or downregulation of the downstream gene.
Whole-genome phylogeny.
Sequence reads for the four KZN strains were downloaded from the Sequence Read Archive (run accession numbers SRR832991, SRR833024, SRR833121, and SRR924700). The reads were aligned to the H37Rv genome (NCBI accession number NC_000962.3) using the SeqMan NGen program (DNAStar), resulting in median alignment depths ranging from 184 to 330 times for individual isolates. SNPs were called and filtered as previously described (39). The concatenated SNPs were used to create a distance-based neighbor-joining tree.
RT-qPCR.
RNA was reverse transcribed into cDNA using an iScript cDNA synthesis kit (Bio-Rad, USA). Quantitative real-time PCR was conducted using iTaq Universal SYBR green supermix (Bio-Rad, USA) and forward and reverse primers for selected genes of interest. Primers were designed for ethA, inhA, ethR, and a housekeeping gene, sigA (see Table S5 in the supplemental material). Expression levels were normalized to the expression level of the reference gene, sigA.
Flow cytometry promoter reporter assay.
To create a dual-color reporter, the Multisite Gateway three-fragment vector construction method (Invitrogen, USA) was used. The ethA-ethR intergenic region, mycobacterial codon-optimized Emerald GFP, and mycobacterial codon-optimized TagRFP constitutively expressed by the promoter pUV15 were individually cloned into entry vectors. These were combined with a destination vector based on an episomal mycobacterial vector containing a kanamycin resistance cassette (aph), the mycobacterial origin of replication, and the Escherichia coli origin of replication. Four separate ethA-ethR intergenic regions corresponding to the wild-type and mutant sequences (generated by PCR using genomic DNA from resistant clinical isolates) upstream of ethA and the same pair in the reverse orientation corresponding to the sequences upstream of ethR were used (Table S6). Additional plasmids with the inhA promoter with and without a g−17t mutation and a nonpromoter region (intragenic katG sequence) cloned in front of the GFP were constructed (Fig. S2). The promoter sequences for each construct were confirmed. The respective plasmids were transformed into H37Rv using standard protocols (40).
Strains harboring the dual-color reporters were grown to mid-log phase (OD600, 0.5 to 0.8) in 7H9 medium containing 25 mg/liter kanamycin. One milliliter of each strain was then filtered through a 10-μm-pore-size filter and acquired on a BD FACSAria III flow cytometer using BD DIVA software. A total of 100,000 events were recorded, with single-cell acquisition set at a threshold rate of ∼5,000 to 7,000 events per second. Green and red fluorescence were detected using the fluorescein isothiocyanate (FITC) and propidium iodide (PI) filters, respectively. The gating strategy employed during acquisition and software analysis, in which FlowJo (version 10) software was used, differentiated single cells/events on the basis of the relationship between cell size (forward scatter [FSC]) and granularity (side scatter [SSC]). Secondary gating on events with a red fluorescent signal was done using FlowJo software to ensure that only cells containing expression vectors were included in our analysis. The median fluorescent intensity (MFI) of the red and green fluorescent signals was extracted. The MFI of green fluorescence was normalized to the MFI of red fluorescence for each replicate before calculation of the median and standard deviation. A two-sided t test was used to determine statistical significance.
Drug susceptibility testing.
One hundred microliters of three dilutions of each strain including 1 ×106, 1 ×104, and 1 ×103 cells was plated onto quadrant plates containing BD Difco Middlebrook 7H10 agar with various concentrations of ethionamide (1.25, 2.5, 5, 10, 20, 40, and 80 mg/liter), and the number of CFU was counted after 3 weeks of incubation at 37°C.
Global distribution of ethA promoter mutations.
A global data set of 5,310 M. tuberculosis strains from five continents (28) was searched for all instances of ethA promoter mutations. To identify individual mutation events arising across the phylogeny, we performed parsimony-based analysis using PAUP software, version 4.0b10 (29), as described by Manson et al. (28).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge and thank M. R. Farhat and M. Murray for providing us with additional information on previous work that they had conducted surrounding the ethA-ethR locus, A. R. Baulard for his helpful comments and suggestions, K. A. Cohen for her insights and information regarding the clinical strains used in this study, and V. Munsamy-Govender for her assistance in locating and propagating some of these strains.
This project received funding from the Africa Health Research Institute (AHRI) and has also been funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, U.S. Department of Health and Human Services, under grant no. U19AI110818 and contract no. HHSN272200900018C to the Broad Institute.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01461-17.
REFERENCES
- 1.Cohen KA, Abeel T, Manson McGuire A, Desjardins CA, Munsamy V, Shea TP, Walker BJ, Bantubani N, Almeida DV, Alvarado L, Chapman SB, Mvelase NR, Duffy EY, Fitzgerald MG, Govender P, Gujja S, Hamilton S, Howarth C, Larimer JD, Maharaj K, Pearson MD, Priest ME, Zeng Q, Padayatchi N, Grosset J, Young SK, Wortman J, Mlisana KP, O'Donnell MR, Birren BW, Bishai WR, Pym AS, Earl AM. 2015. Evolution of extensively drug-resistant tuberculosis over four decades: whole genome sequencing and dating analysis of Mycobacterium tuberculosis isolates from KwaZulu-Natal. PLoS Med 12:e1001880. doi: 10.1371/journal.pmed.1001880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Donnell MR, Padayatchi N, Kvasnovsky C, Werner L, Master I, Horsburgh CR. 2013. Treatment outcomes for extensively drug-resistant tuberculosis and HIV co-infection. Emerg Infect Dis 19:416–424. doi: 10.3201/eid1903.120998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pietersen E, Ignatius E, Streicher EM, Mastrapa B, Padanilam X, Pooran A, Badri M, Lesosky M, van Helden P, Sirgel FA, Warren R, Dheda K. 2014. Long-term outcomes of patients with extensively drug-resistant tuberculosis in South Africa: a cohort study. Lancet (London, England) 383:1230–1239. doi: 10.1016/S0140-6736(13)62675-6. [DOI] [PubMed] [Google Scholar]
- 4.Boehme CC, Nicol MP, Nabeta P, Michael JS, Gotuzzo E, Tahirli R, Gler MT, Blakemore R, Worodria W, Gray C, Huang L, Caceres T, Mehdiyev R, Raymond L, Whitelaw A, Sagadevan K, Alexander H, Albert H, Cobelens F, Cox H, Alland D, Perkins MD. 2011. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet (London, England) 377:1495–1505. doi: 10.1016/S0140-6736(11)60438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hillemann D, Rusch-Gerdes S, Richter E. 2007. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol 45:2635–2640. doi: 10.1128/JCM.00521-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hillemann D, Rusch-Gerdes S, Richter E. 2009. Feasibility of the GenoType MTBDRsl assay for fluoroquinolone, amikacin-capreomycin, and ethambutol resistance testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol 47:1767–1772. doi: 10.1128/JCM.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown AC, Bryant JM, Einer-Jensen K, Holdstock J, Houniet DT, Chan JZM, Depledge DP, Nikolayevskyy V, Broda A, Stone MJ, Christiansen MT, Williams R, McAndrew MB, Tutill H, Brown J, Melzer M, Rosmarin C, McHugh TD, Shorten RJ, Drobniewski F, Speight G, Breuer J. 2015. Rapid whole-genome sequencing of Mycobacterium tuberculosis isolates directly from clinical samples. J Clin Microbiol 53:2230–2237. doi: 10.1128/JCM.00486-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desjardins CA, Cohen KA, Munsamy V, Abeel T, Maharaj K, Walker BJ, Shea TP, Almeida DV, Manson AL, Salazar A, Padayatchi N, O'Donnell MR, Mlisana KP, Wortman J, Birren BW, Grosset J, Earl AM, Pym AS. 2016. Genomic and functional analyses of Mycobacterium tuberculosis strains implicate ald in d-cycloserine resistance. Nat Genet 48:544–551. doi: 10.1038/ng.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker TM, Kohl TA, Omar SV, Hedge J, Del Ojo Elias C, Bradley P, Iqbal Z, Feuerriegel S, Niehaus KE, Wilson DJ, Clifton DA, Kapatai G, Ip CLC, Bowden R, Drobniewski FA, Allix-Béguec C, Gaudin C, Parkhill J, Diel R, Supply P, Crook DW, Smith EG, Walker AS, Ismail N, Niemann S, Peto TEA, Davies J, Crichton C, Acharya M, Madrid-Marquez L, Eyre D, Wyllie D, Golubchik T, Munang M. 2015. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis 15:1193–1202. doi: 10.1016/S1473-3099(15)00062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coll F, McNerney R, Preston MD, Guerra-Assunção JA, Warry A, Hill-Cawthorne G, Mallard K, Nair M, Miranda A, Alves A, Perdigão J, Viveiros M, Portugal I, Hasan Z, Hasan R, Glynn JR, Martin N, Pain A, Clark TG. 2015. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med 7:51. doi: 10.1186/s13073-015-0164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Li D, Zhao L, Fleming J, Lin N, Wang T, Liu Z, Li C, Galwey N, Deng J, Zhou Y, Zhu Y, Gao Y, Wang T, Wang S, Huang Y, Wang M, Zhong Q, Zhou L, Chen T, Zhou J, Yang R, Zhu G, Hang H, Zhang J, Li F, Wan K, Wang J, Zhang X-E, Bi L. 2013. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat Genet 45:1255–1260. doi: 10.1038/ng.2735. [DOI] [PubMed] [Google Scholar]
- 12.Cohen KA, Bishai WR, Pym AS. 2014. Molecular basis of drug resistance in Mycobacterium tuberculosis. Microbiol Spectr 2(3):MGM2-0036-2013. doi: 10.1128/microbiolspec.MGM2-0036-2013. [DOI] [PubMed] [Google Scholar]
- 13.Siddiqi N, Das R, Pathak N, Banerjee S, Ahmed N, Katoch VM, Hasnain SE. 2004. Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a tap-like efflux pump. Infection 32:109–111. doi: 10.1007/s15010-004-3097-x. [DOI] [PubMed] [Google Scholar]
- 14.Louw GE, Warren RM, van Pittius NC, Leon R, Jimenez A, Hernandez-Pando R, McEvoy CRE, Grobbelaar M, Murray M, van Helden PD, Victor TC. 2011. Rifampicin reduces susceptibility to ofloxacin in rifampicin-resistant Mycobacterium tuberculosis through efflux. Am J Respir Crit Care Med 184:269–276. doi: 10.1164/rccm.201011-1924OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mdluli K, Sherman DR, Hickey MJ, Kreiswirth BN, Morris S, Stover CK, Barry CE. 1996. Biochemical and genetic data suggest that InhA is not the primary target for activated isoniazid in Mycobacterium tuberculosis. J Infect Dis 174:1085–1090. doi: 10.1093/infdis/174.5.1085. [DOI] [PubMed] [Google Scholar]
- 16.Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M, Zhang Y. 1997. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother 41:540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juréen P, Werngren J, Toro JC, Hoffner S. 2008. Pyrazinamide resistance and pncA gene mutations in Mycobacterium tuberculosis. Antimicrob Agents Chemother 52:1852–1854. doi: 10.1128/AAC.00110-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yüksel P, Tansel Ö. 2009. Characterization of pncA mutations of pyrazinamide-resistant Mycobacterium tuberculosis in Turkey. New Microbiol 32:153–158. [PubMed] [Google Scholar]
- 19.Reeves AZ, Campbell PJ, Sultana R, Malik S, Murray M, Plikaytis BB, Shinnick TM, Posey JE. 2013. Aminoglycoside cross-resistance in Mycobacterium tuberculosis due to mutations in the 5′ untranslated region of whiB7. Antimicrob Agents Chemother 57:1857–1865. doi: 10.1128/AAC.02191-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartkoorn RC, Uplekar S, Cole ST. 2014. Cross-resistance between clofazimine and bedaquiline through upregulation of mmpL5 in Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:2979–2981. doi: 10.1128/AAC.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andries K, Villellas C, Coeck N, Thys K, Gevers T, Vranckx L, Lounis N, De Jong BC, Koul A. 2014. Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS One 9:e102135. doi: 10.1371/journal.pone.0102135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baulard AR, Betts JC, Engohang-Ndong J, Quan S, McAdam RA, Brennan PJ, Locht C, Besra GS. 2000. Activation of the pro-drug ethionamide is regulated in mycobacteria. J Biol Chem 275:28326–28331. [DOI] [PubMed] [Google Scholar]
- 23.DeBarber AE, Mdluli K, Bosman M, Bekker LG, Barry CE. 2000. Ethionamide activation and sensitivity in multidrug-resistant Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 97:9677–9682. doi: 10.1073/pnas.97.17.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morlock GP, Metchock B, Sikes D, Crawford JT, Cooksey RC. 2003. ethA, inhA, and katG loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother 47:3799–3805. doi: 10.1128/AAC.47.12.3799-3805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engohang-Ndong J, Baillat D, Aumercier M, Bellefontaine F, Besra GS, Locht C, Baulard AR. 2004. EthR, a repressor of the TetR/CamR family implicated in ethionamide resistance in mycobacteria, octamerizes cooperatively on its operator. Mol Microbiol 51:175–188. [DOI] [PubMed] [Google Scholar]
- 26.Vilchèze C, Jacobs WR Jr. 2014. Resistance to isoniazid and ethionamide in Mycobacterium tuberculosis: genes, mutations, and causalities. Microbiol Spectr 2(4):MGM2-0014-2013. doi: 10.1128/microbiolspec.MGM2-0014-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant SS, Wellington S, Kawate T, Desjardins CA, Silvis MR, Wivagg C, Thompson M, Gordon K, Kazyanskaya E, Nietupski R, Haseley N, Iwase N, Earl AM, Fitzgerald M, Hung DT. 2016. Baeyer-Villiger monooxygenases EthA and MymA are required for activation of replicating and non-replicating Mycobacterium tuberculosis inhibitors. Cell Chem Biol 23:666–677. doi: 10.1016/j.chembiol.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manson AL, Cohen KA, Abeel T, Desjardins CA, Armstrong DT, Barry CE III, Brand J, Chapman SB, Cho S-N, Gabrielian A, Gomez J, Jodals AM, Joloba M, Jureen P, Lee JS, Malinga L, Maiga M, Nordenberg D, Noroc E, Romancenco E, Salazar A, Ssengooba W, Velayati AA, Winglee K, Zalutskaya A, Via LE, Cassell GH, Dorman SE, Ellner J, Farnia P, Galagan JE, Rosenthal A, Crudu V, Homorodean D, Hsueh P-R, Narayanan S, Pym AS, Skrahina A, Swaminathan S, Van der Walt M, Alland D, Bishai WR, Cohen T, Hoffner S, Birren BW, Earl AM. 2017. Genomic analysis of globally diverse Mycobacterium tuberculosis strains provides insights into the emergence and spread of multidrug resistance. Nat Genet 49:395–402. doi: 10.1038/ng.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swofford D. 1991. PAUP: phylogenetic analysis using parsimony, version 3.1. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 30.Farhat MR, Sultana R, Iartchouk O, Bozeman S, Galagan J, Sisk P, Stolte C, Nebenzahl-Guimaraes H, Jacobson K, Sloutsky A, Kaur D, Posey J, Kreiswirth BN, Kurepina N, Rigouts L, Streicher EM, Victor TC, Warren RM, van Soolingen D, Murray M. 2016. Genetic determinants of drug resistance in Mycobacterium tuberculosis and their diagnostic value. Am J Respir Crit Care Med 194:621–630. doi: 10.1164/rccm.201510-2091OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casali N, Nikolayevskyy V, Balabanova Y, Harris SR, Ignatyeva O, Kontsevaya I, Corander J, Bryant J, Parkhill J, Nejentsev S, Horstmann RD, Brown T, Drobniewski F. 2014. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet 46:279–286. doi: 10.1038/ng.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rueda J, Realpe T, Mejia GI, Zapata E, Rozo JC, Ferro BE, Robledo J. 2015. Genotypic analysis of genes associated with independent resistance and cross-resistance to isoniazid and ethionamide in Mycobacterium tuberculosis clinical isolates. Antimicrob Agents Chemother 59:7805–7810. doi: 10.1128/AAC.01028-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vilcheze C, Weisbrod TR, Chen B, Kremer L, Hazbon MH, Wang F, Alland D, Sacchettini JC, Jacobs WRJ.. 2005. Altered NADH/NAD+ ratio mediates coresistance to isoniazid and ethionamide in mycobacteria. Antimicrob Agents Chemother 49:708–720. doi: 10.1128/AAC.49.2.708-720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiwari P, Arora G, Singh M, Kidwai S, Narayan OP, Singh R. 2015. MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat Commun 6:6059. doi: 10.1038/ncomms7059. [DOI] [PubMed] [Google Scholar]
- 35.Kirchner S, Ignatova Z. 2015. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet 16:98–112. doi: 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- 36.World Health Organization. 2016. WHO treatment guidelines for drug-resistant tuberculosis 2016. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 37.Kaniga K, Cirillo DM, Hoffner S, Ismail NA, Kaur D, Lounis N, Metchock B, Pfyffer GE, Venter A. 2016. A multilaboratory, multicountry study to determine MIC quality control ranges for phenotypic drug susceptibility testing of selected first-line antituberculosis drugs, second-line injectables, fluoroquinolones, clofazimine, and linezolid. J Clin Microbiol 54:2963–2968. doi: 10.1128/JCM.01138-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alli OAT, Mangan, Butcher JA, Alli O. 2009. Optimization of RNA extraction in Mycobacterium tuberculosis for studying intracellular gene expression. Afr J Clin Exp Microbiol 10:64–79. [Google Scholar]
- 39.Eldholm V, Pettersson JH-O, Brynildsrud OB, Kitchen A, Rasmussen EM, Lillebaek T, Rønning JO, Crudu V, Mengshoel AT, Debech N, Alfsnes K, Bohlin J, Pepperell CS, Balloux F. 2016. Armed conflict and population displacement as drivers of the evolution and dispersal of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 113:13881–13886. doi: 10.1073/pnas.1611283113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Kessel JC, Hatfull GF. 2007. Recombineering in Mycobacterium tuberculosis. Nat Methods 4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.