Abstract
Membrane lipid composition varies greatly within submembrane compartments, different organelle membranes, and also between cells of different cell stage, cell and tissue types, and organisms. Environmental factors (such as diet) also influence membrane composition. The membrane lipid composition is tightly regulated by the cell, maintaining a homeostasis that, if disrupted, can impair cell function and lead to disease. This is especially pronounced in the brain, where defects in lipid regulation are linked to various neurological diseases. The tightly regulated diversity raises questions on how complex changes in composition affect overall bilayer properties, dynamics, and lipid organization of cellular membranes. Here, we utilize recent advances in computational power and molecular dynamics force fields to develop and test a realistically complex human brain plasma membrane (PM) lipid model and extend previous work on an idealized, “average” mammalian PM. The PMs showed both striking similarities, despite significantly different lipid composition, and interesting differences. The main differences in composition (higher cholesterol concentration and increased tail unsaturation in brain PM) appear to have opposite, yet complementary, influences on many bilayer properties. Both mixtures exhibit a range of dynamic lipid lateral inhomogeneities (“domains”). The domains can be small and transient or larger and more persistent and can correlate between the leaflets depending on lipid mixture, Brain or Average, as well as on the extent of bilayer undulations.
Introduction
Cellular membranes are complex assemblies of lipids and proteins that separate cellular compartments, as well as the cell interior from the outside environment. A typical plasma membrane (PM) contains hundreds of different lipid species that are actively regulated by the cell (1, 2). The diverse set of lipids can regulate protein function through specific lipid-protein interactions and through general bilayer-protein interaction (i.e., changes in bilayer properties) (3, 4, 5). Additionally, lipids are non-uniformly distributed within the membrane plane (6, 7) and are thought to reside close to a critical point (8), where large fluctuations in regions (domains) of locally increased/depleted lipid content are to be expected. The lipid segregation can further modulate membrane proteins, affecting local concentrations, aggregation, and trafficking (9, 10). On one hand, lipid compositions vary significantly within a cell between the membranes of its different organelles and submembrane compartments (1, 11, 12, 13), and between different cells, PM lipid composition differs by organism, cell stage, environmental factors, and cell and tissue types (14, 15, 16, 17, 18). On the other hand, altered lipid composition is linked to many diseases, e.g., cancers, HIV, diabetes, atherosclerosis, cardiovascular disease, and Alzheimer’s disease (9, 19, 20).
The brain, in particular, appears to be especially susceptible to disease states that are enhanced or accelerated by lipid composition (21, 22, 23). For instance, specific phosphatidylinositolphosphate (PIP) lipids are involved in regulation of aspects of neuronal cell function, and PIP lipid imbalances have been linked to a number of major neurological diseases (23). Indeed, PIPs themselves can modulate ion flux through PM ion channels (24, 25) by direct interaction with the ion channels or by modulating membrane charge. Moreover, these neuronal membrane lipids can influence both the function and localization of proteins within the PM of the neuron and, in effect, regulate synaptic throughput (22).
These lipid differences raise questions as to how complex changes in composition affect overall bilayer properties, dynamics, and lipid organization of cellular membranes. Studying lipid structural heterogeneity is challenging because of the lack of experimental methods suitable for measuring nanoscale assemblies of soft bilayers and living cells in the required spatiotemporal resolution. Computational modeling has emerged as a powerful alternative method and has become indispensable for exploring dynamic biomembranes and lipids at the molecular level (26). The use of coarse-grained (CG) molecular dynamics simulations has increased the accessible length- and timescales (27) compared to all-atom simulations. At the CG resolution, a number of atoms are combined into functional groups, decreasing the number of particles in the system and smoothing the energy landscape. The smoother energy landscape allows for larger integration time steps and often leads to faster effective dynamics. CG methods neglect some of the atomistic degrees of freedom, losing accuracy, and are therefore not applicable to all problems. Currently at the CG resolution, multi-component membranes can be modeled that approach the complexity of realistic cell membranes (28, 29, 30, 31, 32, 33, 34, 35, 36).
Here, we developed a realistically complex model of a human “brain-like” PM and extend previous work on the idealized, “average” mammalian PM (28). Our results show both striking similarities and differences between the Brain and Average PM mixtures. Despite significant changes in lipid composition, the biggest contributors—increased cholesterol concentration and increased tail unsaturation in the brain—appear to act complementary to each other. The differences effectively influence the membrane in opposite directions, yet with similar magnitudes, leading to many overall bilayer properties being comparable. Both mixtures exhibit a range of lipid lateral in homogeneities, or domains. The domains are dynamic, and sizes fluctuate, and their size and correlation across the leaflets differ in the Average PM compared to the Brain and with the level of bilayer undulations.
Materials and Methods
Neuronal PM composition
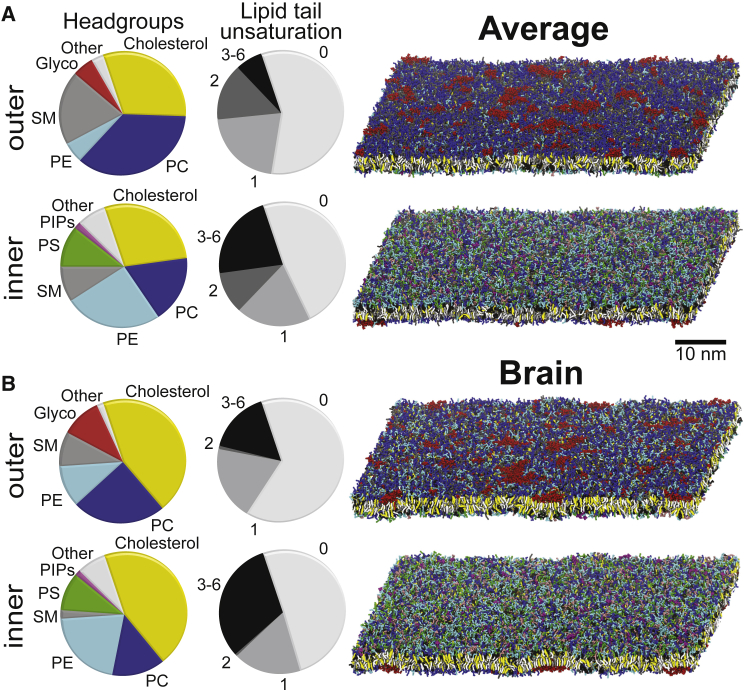
The Brain composition represents an idealized lipid composition of human brain tissue or, more specifically, a “typical” human neuronal PM mixture. Although several different membrane compositions exist for specific cells and cell regions within the brain (12, 13, 14), the relative dearth of available data made it more prudent to construct a model that possessed the general properties of membranes found within the brain. To capture a typical Brain PM composition, we derived a consensus from a number of studies that performed lipidomic measurements of neurons and brain tissue (37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49). Using the idealized mammalian PM mixture (28) as a reference, we adjusted percentages of lipid headgroup types and tail distributions based on the overall trends reported for brain tissue extractions and PM isolations that differed from the idealized mammalian PM (see Supporting Materials and Methods for details). An overview of the Brain and the Average compositions is given in Fig. 1, and the specific lipid types used, their ratio in the outer/inner leaflets, and the lipid counts in the simulations are listed in Table S1.
Figure 1.
PM lipid distributions. Pie charts with the overall distribution of the main lipid headgroups and level of tail unsaturation in the outer/inner leaflet, as well as snapshots of the outer/inner leaflet of the simulations after 80 μs, are shown for the Average (A) and Brain membranes (B). The lipids in the snapshots are colored as follows: PC, blue; PE, cyan; SM, gray; PS, green; glycolipids (Glyco), red; PI, pink; PA, white; PIPs, magenta; CER, ice blue; Lyso, orange; DAG, brown; and cholesterol, yellow.
Force field
The simulations were performed using the Martini CG model (50, 51). Most of the lipid force fields used were described in Ingólfsson et al. (28), including the PIP and ganglioside (GM) parameters, originally described in López et al. (52), and the improved cholesterol model (53). Control simulations using GMs with newly optimized clustering behavior (54) were also performed. Parameters for new Martini lipids were constructed according to the standard Martini 2.0 lipid building blocks and rules (50, 51), as previously described (28, 55). The details of the lipid force fields used are given in the Supporting Materials and Methods; all the lipid force fields, including the ones generated for this study, can be found at the Martini portal: http://www.cgmartini.nl.
Simulations
All the simulations were run using the GROMACS 4.6 simulation package (56), following the same setup described in Ingólfsson et al. (28). In short, a time step of 20 fs was used for all production runs with the standard Martini cutoffs, the same parameter set as denoted “common” in de Jong et al. (57). Each simulation contains ∼20,000 lipids (or ∼6000 for smaller control simulations) with >15 CG waters per lipid (one CG water representing four water molecules), counterions, and 150 mM NaCl; the exact lipid composition in each simulation is listed in Table S1. Membranes were constructed using the bilayer builder insane (55). For each bilayer mixture, the number of lipids in the inner/outer leaflet was adjusted based on an independent bilayer simulation with symmetrical composition of each leaflet (both leaflets being outer or inner). This process was iterated with changes in the cholesterol outer/inner leaflet distribution until the initial cholesterol distribution did not drift with time; i.e., the cholesterol distribution was allowed to adjust to its chemical potential in each leaflet, as detailed in the Supporting Materials and Methods and in Ingólfsson et al. (28). For the main simulations, large membrane undulations were restricted using weak position restraints on selected lipids in the outer leaflet (see Supporting Materials and Methods). Additional simulations with 10-fold weaker restraints and no restraints were also run. The temperature and pressure were controlled using the velocity rescaling thermostat (58) (at 310 K, with τT = 1.0 ps) and the Parrinello-Rahman barostat (59) (1 bar semi-isotropic pressure, with τp = 5.0 ps). Each membrane was simulated for 80 μs, corresponding to 320 μs of effective time, if accounting for the ∼4-fold faster diffusion at the Martini CG level (51). All analysis was done either with respect to time or averaging over the last 2–10 μs of each simulation, as indicated. The analysis was carried out partly using tools provided in the GROMACS package and partly by custom tools written in Python and C++, to perform bilayer surface construction and topological analysis (60, 61), as well as using the MDAnalysis package (62) and lipid-flow analysis methods (63), as described in the Supporting Materials and Methods.
Results and Discussion
An idealized neuronal PM mixture was constructed (Brain; see Materials and Methods) and compared to the average mammalian PM mixture from Ingólfsson et al. (28) (Average). To compare the physical properties of the different PM lipid mixtures, large lipid patches (∼20,000 lipids) of both lipid mixtures were simulated for 80 μs using the Martini CG force field (50, 51) and their properties were analyzed. Note that bilayer undulations were suppressed in these systems to facilitate the analysis and to be representative of real membranes that are constrained by both the cytoskeletal network and the presence of membrane proteins. Fig. 1 shows an overview of the main lipid headgroup and tail saturation distributions for both mixtures, as well as snapshots of the outer and inner leaflets after 80 μs. More detailed snapshots of the headgroups and tails are shown in Fig. S1 and a time-lapse sequence of the headgroups in Movie S1.
Global membrane properties; similar but different
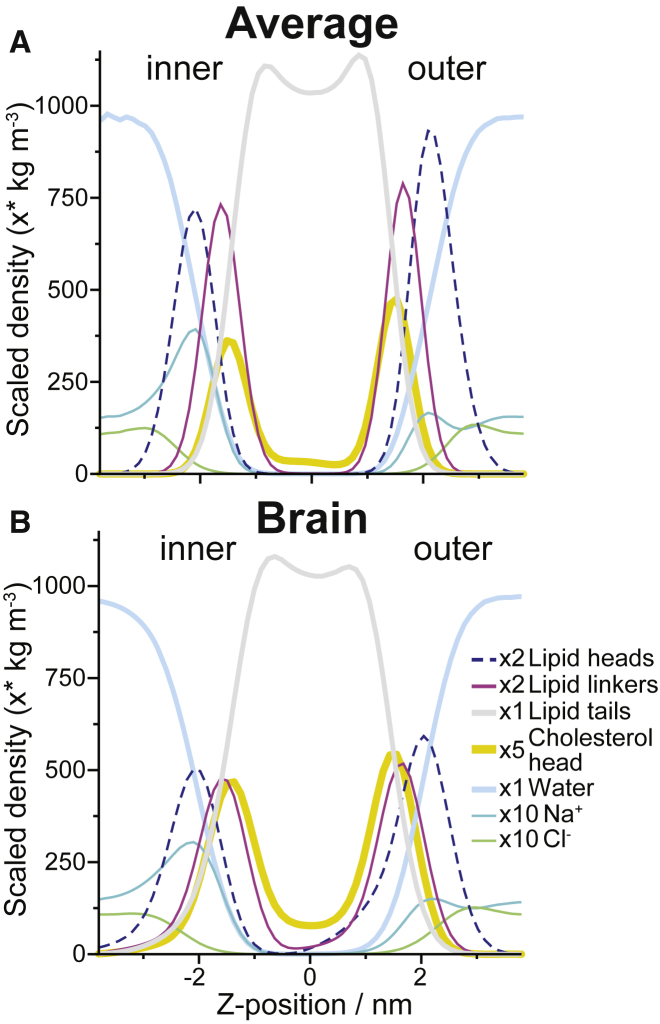
Common properties of the two mixtures are listed in Table 1. Comparing the lipid composition of the two PM mixtures (Fig. 1 A and Table S1), the biggest differences are the significantly higher cholesterol content in the Brain, 44.5% compared to 30% in the Average PM, and the increased amount of polyunsaturated tails in Brain (on average, each lipid tail in the Brain has 1.27 double bonds compared to 1.05 in the Average PM). Because cholesterol is known to flip-flop between the leaflets within the time frame of the simulations, the cholesterol in both mixtures was allowed to redistribute between the leaflets based on its chemical potential (see Supporting Material for details). In both mixtures, cholesterol preferentially localizes in the outer leaflet, but the emerging cholesterol asymmetry is much lower in the Brain (∼1%) than in the Average PM (∼5%). The bilayer average thickness (phosphate to phosphate distance) is comparable between the PMs (4.11 nm for the Average and 4.06 nm for the Brain, with SE <0.002). The density profiles along the box z-direction (Fig. 2) show similar peak locations. The Brain PM, despite having the same type of restraints on large-scale undulations, undulates more locally, leading to broader density distributions. Fitting the bilayer surfaces and measuring the average bilayer normal (all lipids over the last 2 μs) deviation from the z-axes, the Brain mixture deviates ∼80% more than the Average mixture (Table 1). Additionally, the Brain PM has more cholesterol in the middle of the bilayer (Fig. 2), consistent with previous simulations and experiments showing higher preference for cholesterol in the bilayer center in more polyunsaturated bilayers (64). How much more depends somewhat on how you define being in the bilayer middle. Considering cholesterols within 0.8 nm of the bilayer center as in the middle, the Brain PM has 13% of the cholesterols in the middle compared to 7% in the Average, or ∼75% more.
Table 1.
Membrane Properties
| Average |
Brain |
|||
|---|---|---|---|---|
| Outer | Inner | Outer | Inner | |
| Average number of unsaturations per tail | 0.77 | 1.32 | 0.90 | 1.63 |
| Cholesterol fraction | 0.54 | 0.46 | 0.51 | 0.49 |
| Average area per lipid (nm2)a | 0.513 | 0.553 | 0.460 | 0.485 |
| Average sn-1 tail orderb | 0.435 | 0.430 | 0.487 | 0.444 |
| Average sn-2 tail orderb | 0.374 | 0.301 | 0.391 | 0.224 |
| Average pos#3 tail orderb | 0.412 | 0.349 | 0.445 | 0.301 |
| Average diffusion ratesc (10−7 cm2/s) | 3.1 ± 0.3 | 4.3 ± 0.3 | 1.6 ± 0.2 | 2.8 ± 0.2 |
| Average bilayer normal deviationsd | 13.07 ± 0.01 | 23.36 ± 0.09 | ||
| Bilayer thickness (nm) | 4.109 ± 0.001 | 4.057 ± 0.002 | ||
| Flip-flop ratese (106 s−1) | ||||
| CHOL | 7.290 ± 0.018 | 4.820 ± 0.004 | ||
| DAG | 7.662 ± 0.049 | 2.800 ± 0.074 | ||
| CER | 0.027 ± 0.006 | 0.015 ± 0.005 | ||
The average area per lipid (Al) for the outer/inner leaflets was estimated in separate symmetrical simulations. SE values for Al are ∼0.001 nm2.
Lipid tail order was evaluated using the lipid tail order parameter (S). Flip-flopping lipids were excluded and averages weighted based on lipid counts in the respective leaflets. Either all bonds in the sn-1/sn-2 tails were averaged or the tail bond at position 3 was averaged between the tails. The weighted SE ∼0.002. Tail order parameters for each lipid class are reported in Table S3.
The weighted average of the lipid lateral diffusion coefficients (D) for all lipids that don’t flip-flop. Note that lipid diffusion coefficients are reported as is, and no correction is applied for overestimates due to the larger effective simulation speed of CG simulations (51) or underestimates due to the periodically bound finite system sizes (79). All diffusion values are reported in Table S2.
Average bilayer normal deviations are the average angle between the bilayer normal and the z-axis for each lipid (from the fitted bilayer surfaces) to the z-axes. Average over all lipids and the last 2 μs of the simulations (± SE, estimated using block averaging).
Flip-flop rates (± SE) were measured as described in (28). For details on all calculated properties, see Supporting Materials and Methods.
Figure 2.
Membrane density profile. The density profile of the two different PM mixtures Average (A) and Brain (B) is shown across the Z-dimension, averaged from 78 to 80 μs. The Z-dimension is a reasonable approximation of the bilayer normal for these membranes as their undulations have been restricted. The density of the smaller groups is scaled for clarity, as indicated on the figure key.
Tail order was evaluated for all lipid types (Table S2). The tail order varies considerably due to their different headgroup and tail characteristics, but also based on the lipid location in the outer/inner leaflet or in the Average or Brain mixtures. In combination with the higher cholesterol content, the outer leaflets of both PMs contain lipids with somewhat longer and more saturated tails (Table S1), leading to higher tail order in the outer than in the inner leaflets (Table 1). For the Brain PM, the higher cholesterol content acts to increase the overall tail order, whereas the higher level of tail unsaturation acts to decrease the tail order. These two effects mostly balance out, with the overall tail order nearly the same in the Brain and the Average PMs (with an average tail order of 0.385 in the Average mixture and 0.386 in the Brain). However, if we look at the tail order in the outer and inner leaflets separately, there is a significant difference (Table 1). The Brain outer leaflet is more ordered than the Average outer leaflet, whereas the inner leaflet of the Brain is less ordered than the inner leaflet in the Average mixture. The increased tail order in the outer leaflet with respect to the inner leaflet is 11% for the Average and 31% for the Brain. Note that cholesterols influence the packing of other lipids but are not included in the tail order calculations themselves, as they do not contain fatty acid tails; cholesterol is mostly rigid, and given its significantly higher concentration in the Brain PM, the hydrocarbon core of the Brain PM is more “ordered” than that of the Average PM.
Lipid diffusion was also evaluated for each lipid type (Table S3). Just as with the tail order, lipid diffusion rates vary between the different lipid species, as well as their locations in the outer/inner leaflet and membrane type. Lipids in the more ordered outer leaflets diffuse slower than in the less ordered inner leaflets (Table 1), and the difference between the outer/inner leaflet diffusion rates is higher in the Brain than in the Average, in line with the larger difference in tail order. The overall lipid diffusion is ∼40% slower in the Brain compared to the Average, even though the combined tail order is similar. This is presumably due to the higher cholesterol content in the Brain.
At the timescale of these simulations, cholesterol, diacylglycerol (DAG), and ceramide (CER) lipids flip-flop between the leaflets. The flip-flop rates are shown in Table 1, and similar to the lipid diffusion, lipid flip-flop is somewhat slower in the Brain. Previous simulation work showed a steep reduction in cholesterol flip-flop rate with increased cholesterol content and an increase in polyunsaturated bilayers (65). The effects of the ∼15% increase in cholesterol content between the Brain and Average appear to be mostly compensated with an increase in the level of tail unsaturation, resulting in only a modest reduction in flip-flop rates, ∼35% for cholesterol.
Lipid mixing and domain sizes
In both the Average and the Brain PMs, the different lipid species are not homogeneously mixed in the bilayer plane. Based on their mutual interactions, lipids preferentially associate with other lipid species. At the ends of simulation (80 μs) snapshots (Figs. 1 and S1), glycolipid domains (red) can be seen in both PM mixtures; otherwise at that resolution, the mixture appears random. The snapshots of the lipid tails in Fig. S1 show preferential co-localization of polyunsaturated tails in both membranes, with more clusters of polyunsaturated tails in the Brain mixture, though of a somewhat smaller size. This is consistent with both mixtures having a significant fraction of polyunsaturated tails, but with more unsaturation in the Brain than the Average (Fig. 1; Table S1). In both mixtures, most of the polyunsaturated tails are on lipids where the other tail is saturated. The Brain mixture has a fraction where the other tail is monounsaturated (∼5% of total lipids), but the Average mixture has a small fraction of lipids (∼1.5% of total lipids) with both tails polyunsaturated.
To quantify preferential lipid-lipid interactions, we calculated the enrichment/depletion of the different lipid headgroups and linker types in their immediate neighborhood (defined as <1.5 nm) (Fig. S2). The lipid-lipid interaction profile for the Brain is very similar to the Average mixture, which is described in Ingólfsson et al. (28); the main features are domains of glycolipids in the outer leaflet and increased self-association of PIPs in the inner leaflet. The glycolipid domains can also be clearly seen by looking at the local lipid mobility or variations in the bilayer thickness (Fig. S3).
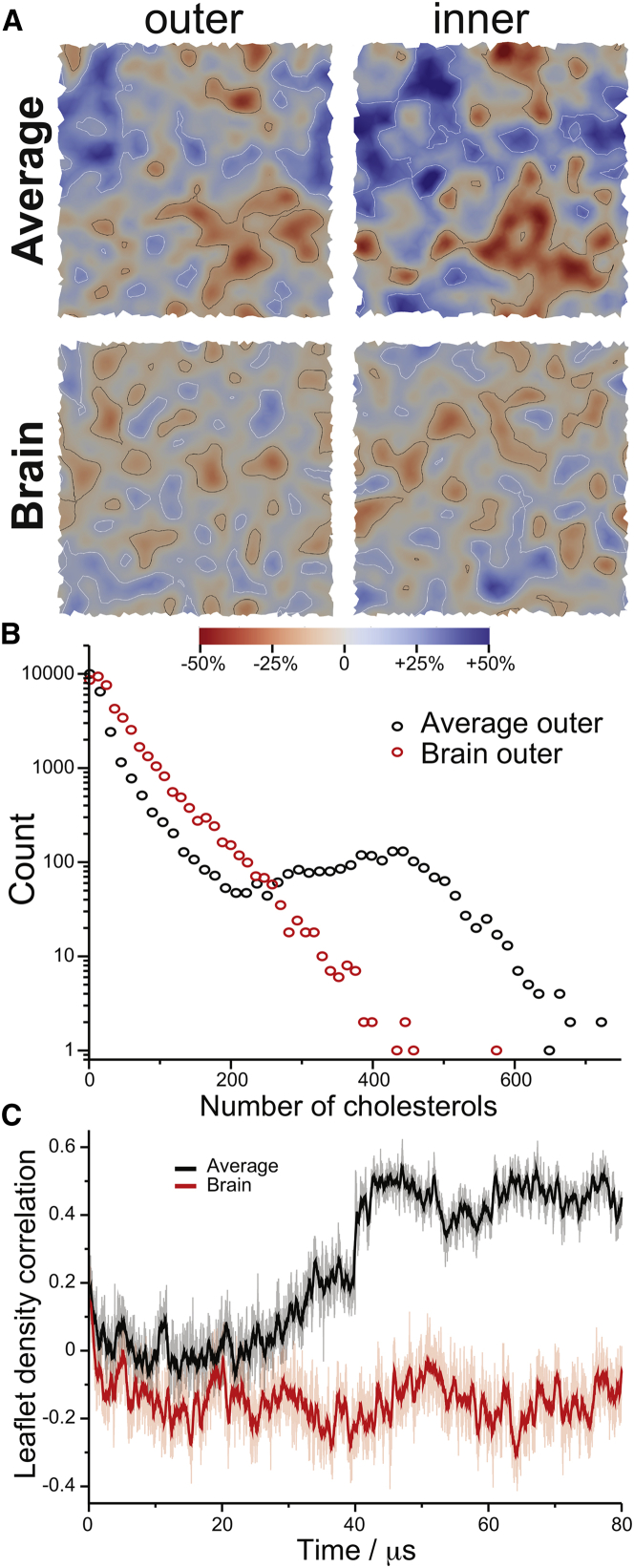
Cholesterol density is used to define bilayer domains that are enriched/depleted in cholesterol. Fig. 3 A shows the cholesterol density of the outer and inner leaflets of the last frame of the main Brain and Average PM simulations. Regions of high density (red) and low density (blue) are marked with contour lines (high density, black lines; low density, white lines). As the absolute cholesterol concentration varies between the PMs and their leaflets, we selected thresholds to define the high/low-density regions in each layer that maximize the number of domains in that layer (Fig. S4 A). Fig. S4 B shows the cumulative distribution function (CDF) of domain sizes with varying thresholds, demonstrating their sensitivity. Fig. 3 B shows domain size histograms of outer-leaflet high-density regions in the Brain and the Average mixtures. Histograms for the inner leaflets and low-density regions are shown in Fig. S4 C. Local cholesterol density fluctuates significantly in all layers. In main simulations (2 kJ mol−1 nm−2 undulation restraints), the Brain mixture has small and transient cholesterol domains and the Average mixture has larger, more persistent domains. Note that at this patch size (∼20,000 lipids), the buildup of larger domains in the Average mixtures takes tens of microseconds (Fig. S4 D). The size fluctuations are consistent with mixtures close to a critical point (66, 67), and the smaller domain sizes in the Brain mixture agree with the reduced phase separation observed with increased cholesterol content in giant PM vesicles (GPMVs) (68).
Figure 3.
Lipid domains. In-plane lateral redistribution of cholesterol was used to track lipid patches of increased/decreased order for the outer/inner leaflets in both the Average and Brain mixtures. (A) Cholesterol density was mapped for each snapshot using a Gaussian filter and colored based on regions of increased (red) or decreased (blue) average density. Thresholds for high-density regions (black contour lines) and low-density regions (white contour lines) were determined as the values that maximized the number of domains in that layer (Fig. S4, A and B). (B) Histograms of domain “size,” in number of cholesterols for the high-density regions of the outer leaflet; Fig. S4C shows the same histograms for all other regions. (C) Cross correlation between the cholesterol densities of the PM’s outer and inner leaflets, shown for every 5 ns (dimmer lines) and averaged over 500 ns (bold lines).
As seen in Fig. 3 A, the cholesterol density in the Average mixture is highly correlated between the leaflets, whereas the Brain mixture does not show such correlations. Fig. 3 C better depicts this, showing the cross correlation between the leaflets with time. In the Average mixture, the leaflet correlation builds up at about the same timescale as the larger domains form (Fig. S4 D), whereas in the Brain mixture, the leaflet correlation stays somewhat anti-correlated throughout the simulation. Averaging over the bilayer area and the last 10 μs of the simulations, the cross correlation is 0.45 ± 0.01 and −0.14 ± 0.01 in the Average and Brain mixtures, respectively. Numerous mechanisms have been proposed to drive leaflet coupling and domain registration, involving domain boundary line tension, inter-leaflet surface tension, cholesterol flip-flopping, bilayer undulation, local bilayer curvature, lipid curvature, and domain thickness mismatch (69, 70, 71, 72). Any speculation in complex lipid mixtures like these, where the local domain composition varies and the boundaries are ill defined, is therefore troublesome; but notably, more local undulations are observed in the Brain mixture, where the asymmetry between leaflets (e.g., order, diffusion, tail unsaturation) is higher.
To analyze the lateral velocities of lipid regions, we used the lipid Flows methodology (63) (Fig. S5). As is to be expected, with both the Brain and Average mixtures, the regions of slower lateral lipid movement corresponded to the higher-cholesterol-concentration domains, whereas the faster-moving lipids were found in areas of lower cholesterol concentration. The overall rates of lipid lateral displacement (Fig. S5 A) were slower in the Brain mixture than in the Average mixture. Again, this is consistent with the fact that the Brain mixture contains significantly more cholesterol. Furthermore, when calculating the leaflet correlation function (the degree to which the lipid motions are correlated between the leaflets, see Supporting Materials and Methods), the Average mixture displays a very strong correlation between both leaflets (Fig. S5 A, left images). In contrast, the Brain mixture does not indicate a high correlation of lipid motions between the leaflets. This is in agreement with the previous cholesterol-density cross correlation. Interestingly, when the smoothing of the trajectory is averaged over a shorter temporal range (<20 ns), smaller pockets of correlated lipid regions become apparent for the Brain mixture (Fig. S5 B). These short-term correlated lipid motions reiterate the presence of small, transient cholesterol domains in the undulation-controlled Brain mixture.
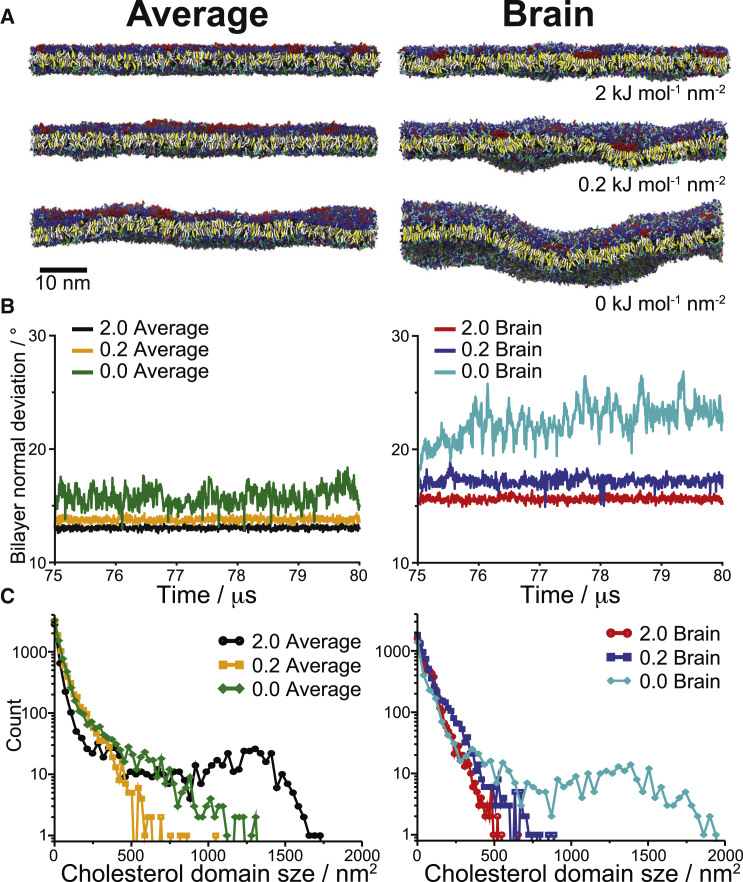
Although bilayer undulations in cells are restricted due to the presence of the underlying cytoskeleton network as well as the high fraction of membrane proteins, it is of interest to study how bilayer undulations couple to the lipid organization and domain formation. As an initial exploration of the effects of undulations, additional simulations with either weaker or no restraints on undulations (0.2 and 0 kJ mol−1 nm−2 compared to 2 kJ mol−1 nm−2 in the main simulations) were performed. These simulations were started from the main Brain and Average PM simulations at 75 μs and simulated for 5 μs. Fig. 4 A shows a side view of the last frame of each simulation, demonstrating the undulation amplitude. We quantified the undulations by plotting the bilayer normal angle deviations with respect to the membrane normal, averaged over the entire membrane surface (Fig. 4 B). As expected, with weaker or no undulation restraints the bilayer undulations increase. At each level of restraint, the Brain mixture undulates more than the Average, pointing toward a lower bending modulus for the Brain membrane. Overall, the average bulk bilayer properties are similar for the different levels of undulations, e.g., the number of neighboring lipids showed no obvious deviation in the weaker and no-restraints simulations (see Fig. S2 for results on the main simulations and Fig. S6 C for the undulating case). All lipid neighbor enrichments/depletions changed by no more than 5%, i.e., no trends were observed with increasing curvature. The longer-scale lipid domain behavior, however, does change with different levels of undulations. Fig. 4 C shows the size histogram of cholesterol-enriched domains in the outer leaflet for the last 2 μs of each simulation (see Fig. S6 B) for results on different parts of the simulations. (Note that in Fig. 4 C, we plotted the cholesterol domain size and not the number of cholesterols as in Fig. 3 B. The shape of both curves is similar, compare Figs. 3 B and 4 C, black and red curves. Remarkably, with increased undulations, the domain sizes decrease in the Average mixture while they increase in the Brain. The reason for this behavior is unclear and requires more simulations at extended timescales to fully sample the coupling between domain sizes and undulatory modes. A clear coupling can be appreciated in the case of the glycolipids, which prefer regions of high negative curvature of the outer membrane leaflet (Fig. S6), consistent with previous simulation results (29). Notably, the bilayers tend to bend at glycolipid domain boundaries (Fig. S6 A), which may explain the growth of the cholesterol domains observed in the Brain membrane given the preferential co-localization of glycolipids and cholesterol (Fig. S2).
Figure 4.
Effects of bilayer undulations. Starting from the main Brain and Average PM simulations at 75 μs, simulations with 10-fold weaker and no restraints on bilayer undulations (0.2 and 0 kJ mol−1 nm−2, respectively) were run for 5 μs. (A) Side-view snapshots of the final structure of each simulation. The lipids are colored according to the same scheme as in Fig. 1. (B) The average bilayer undulations with time are shown as the average angle between the bilayer normal of each lipid (from the fitted bilayer surfaces) and the z-axis. (C) Size histograms of cholesterol-enriched domains in the outer leaflet of each simulation.
Additionally, after the Average PM model was published (28), a few small updates to Martini lipid parameters were made, as well as alternative parameters for the GM1 and GM3 lipids (54) (see Supporting Materials and Methods), and the effects of these changes were explored in smaller control simulations for both mixtures (Fig. S7). The average bilayer properties of the smaller alternative-parameter systems are very similar to those of the larger main simulations. The biggest difference is in the reduced ganglioside clustering using the recently modified version of the Martini GM1 and GM3 ganglioside lipid parameters, optimized to better match the size of ganglioside clusters seen in atomistic simulations (54). As the smaller systems are not much larger than the largest cholesterol domains above, analyzing the cholesterol density using the same method was not very informative, but qualitative comparison of spatial two-dimensional cholesterol density maps averaged over the last 200 ns of the simulations (Fig. S7 B) show larger, more connected densities in Average PM than in the Brain at this undulation level (2 kJ mol−1 nm−2 restraints).
Conclusions
We assembled a realistically complex lipid model of a human neuronal PM (Brain), and despite significant differences in lipid composition (Fig. 1; Table S1), the overall bilayer properties show striking similarities to the recently published idealized mammalian plasma membrane (Average) (28). The higher cholesterol content of the Brain is balanced by more tail unsaturation, resulting in some average bilayer properties being comparable to those of the Average PM (see values for bilayer thickness and lipid tail order, diffusion, flip-flop, and average neighbors in Figs. 2 and S3; Tables 1, S2, and S3. Looking more closely, there are marked differences; the cholesterol asymmetry between the outer/inner leaflets is less pronounced in the Brain (Table 1), presumably due to saturation of preferred cholesterol lipid interactions; lipids in the Brain mixture diffuse and flip-flop more slowly (Tables 1 and S2), and the difference in properties between the outer and inner leaflets is greater in the Brain. Possible future work could involve exploring modulation of the cholesterol concentration or lipid tail unsaturation components independently.
Both mixtures are inhomogeneous and show significant fluctuation in local lipid concentrations. Defining domains as regions of high or low cholesterol density, we mapped the size and leaflet correlations of these domains. In the undulation-restrained simulations (2 kJ mol−1 nm−2 restraints), the Brain mixture has more cholesterol domains, but they are smaller and transient, whereas in the Average mixture, after considerable simulation time, larger persistent domains emerge (Figs. 3 and 4; Figs. S4 and S6). Interestingly, on the same timescale as the emergence of larger cholesterol domains in the Average PM, the registration between the leaflets goes up, whereas in the Brain, the leaflets rapidly become somewhat anti-registered (Fig. 3; Fig. S5). However, the domain size distribution turned out to be very sensitive to the level of bilayer undulation (Fig. 4; Fig. S6). In particular, for the Brain membrane, high-amplitude undulatory modes are easily accessed, leading to coalescence of domains.
There are many interesting questions raised by the marked differences and similarities between the PMs. What is the acceptable range of changes in bilayer properties before cellular function is impaired? Are PM proteins such as ion channels and neuroreceptors sensitive to the presence of smaller, more transient, deregistered membrane domains? Is the only function of high cholesterol content in neurons to make the bilayers less permeable to ions, or are there additional benefits? Despite huge leaps forward in the fields of computational membrane studies, it is clear that in terms of understanding the full complexity and adaptability of cell-specific PMs, we have barely scratched the (highly complicated!) surface.
Author Contributions
H.I.I., T.S.C., and F.C.L. designed the research. H.I.I. and T.S.C. performed the simulations. H.I.I., T.S.C., H.B., and P.T.B. performed the analysis; and all authors contributed to writing the paper.
Acknowledgments
This work was partially funded by Laboratory Directed Research and Development at the Lawrence Livermore National Laboratory (16-FS-007). This work has been supported in part by the Joint Design of Advanced Computing Solutions for Cancer (JDACS4C) program established by the U.S. Department of Energy (DOE) and the National Cancer Institute (NCI) of the National Institutes of Health. We thank the Livermore Institutional Grand Challenge for the computing time. This work was performed under the auspices of the U.S. DOE by the Lawrence Livermore National Laboratory under contract DE-AC52-07NA27344, Los Alamos National Laboratory under contract DE-AC5206NA25396, Oak Ridge National Laboratory under contract DE-AC05-00OR22725, and Frederick National Laboratory for Cancer Research under contracts HHSN261200800001E. Release LLNL-JRNL-733434.
Editor: D. Peter Tieleman.
Footnotes
Helgi I. Ingólfsson and Timothy S. Carpenter contributed equally to this work.
Supporting Materials and Methods, seven figures, three tables, and one movie are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31132-3.
Supporting Citations
References (73, 74, 75, 76, 77, 78) appear in the Supporting Material.
Supporting Material
References
- 1.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sampaio J.L., Gerl M.J., Shevchenko A. Membrane lipidome of an epithelial cell line. Proc. Natl. Acad. Sci. USA. 2011;108:1903–1907. doi: 10.1073/pnas.1019267108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee A.G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Andersen O.S., Koeppe R.E., II Bilayer thickness and membrane protein function: an energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 5.Lundbaek J.A., Collingwood S.A., Andersen O.S. Lipid bilayer regulation of membrane protein function: gramicidin channels as molecular force probes. J. R. Soc. Interface. 2010;7:373–395. doi: 10.1098/rsif.2009.0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelman D.M. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 7.Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 8.Veatch S.L., Cicuta P., Baird B. Critical fluctuations in plasma membrane vesicles. ACS Chem. Biol. 2008;3:287–293. doi: 10.1021/cb800012x. [DOI] [PubMed] [Google Scholar]
- 9.van Meer G. Cellular lipidomics. EMBO J. 2005;24:3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sezgin E., Levental I., Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017;18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klose C., Surma M.A., Simons K. Organellar lipidomics—background and perspectives. Curr. Opin. Cell Biol. 2013;25:406–413. doi: 10.1016/j.ceb.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 12.O’Brien J.S., Sampson E.L. Lipid composition of the normal human brain: gray matter, white matter, and myelin. J. Lipid Res. 1965;6:537–544. [PubMed] [Google Scholar]
- 13.Kishimoto Y., Agranoff B.W., Burton R.M. Comparison of the fatty acids of lipids of subcellular brain fractions. J. Neurochem. 1969;16:397–404. doi: 10.1111/j.1471-4159.1969.tb10380.x. [DOI] [PubMed] [Google Scholar]
- 14.Breckenridge W.C., Gombos G., Morgan I.G. The lipid composition of adult rat brain synaptosomal plasma membranes. Biochim. Biophys. Acta. 1972;266:695–707. doi: 10.1016/0006-3002(72)90012-1. [DOI] [PubMed] [Google Scholar]
- 15.Christie W.W. Rapid separation and quantification of lipid classes by high performance liquid chromatography and mass (light-scattering) detection. J. Lipid Res. 1985;26:507–512. [PubMed] [Google Scholar]
- 16.Atilla-Gokcumen G.E., Muro E., Eggert U.S. Dividing cells regulate their lipid composition and localization. Cell. 2014;156:428–439. doi: 10.1016/j.cell.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levental K.R., Lorent J.H., Levental I. Polyunsaturated lipids regulate membrane domain stability by tuning membrane order. Biophys. J. 2016;110:1800–1810. doi: 10.1016/j.bpj.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tulodziecka K., Diaz-Rohrer B.B., Levental I. Remodeling of the postsynaptic plasma membrane during neural development. Mol. Biol. Cell. 2016;27:3480–3489. doi: 10.1091/mbc.E16-06-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez C., Sandin M., Melander O. Plasma lipid composition and risk of developing cardiovascular disease. PLoS One. 2013;8:e71846. doi: 10.1371/journal.pone.0071846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holthuis J.C.M., Menon A.K. Lipid landscapes and pipelines in membrane homeostasis. Nature. 2014;510:48–57. doi: 10.1038/nature13474. [DOI] [PubMed] [Google Scholar]
- 21.Mattson M.P., Magnus T. Ageing and neuronal vulnerability. Nat. Rev. Neurosci. 2006;7:278–294. doi: 10.1038/nrn1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller C.P., Reichel M., Kornhuber J. Brain membrane lipids in major depression and anxiety disorders. Biochim. Biophys. Acta. 2015;1851:1052–1065. doi: 10.1016/j.bbalip.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 23.Waugh M.G. PIPs in neurological diseases. Biochim. Biophys. Acta. 2015;1851:1066–1082. doi: 10.1016/j.bbalip.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Zhou P., Yu H., Li M. Phosphatidylinositol 4,5-bisphosphate alters pharmacological selectivity for epilepsy-causing KCNQ potassium channels. Proc. Natl. Acad. Sci. USA. 2013;110:8726–8731. doi: 10.1073/pnas.1302167110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi N., Hamada-Nakahara S., Suetsugu S. TRPV4 channel activity is modulated by direct interaction of the ankyrin domain to PI(4,5)P2. Nat. Commun. 2014;5:4994. doi: 10.1038/ncomms5994. [DOI] [PubMed] [Google Scholar]
- 26.Ingólfsson H.I., Arnarez C., Marrink S.J. Computational ‘microscopy’ of cellular membranes. J. Cell Sci. 2016;129:257–268. doi: 10.1242/jcs.176040. [DOI] [PubMed] [Google Scholar]
- 27.Ingólfsson H.I., Lopez C.A., Marrink S.J. The power of coarse graining in biomolecular simulations. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2014;4:225–248. doi: 10.1002/wcms.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingólfsson H.I., Melo M.N., Marrink S.J. Lipid organization of the plasma membrane. J. Am. Chem. Soc. 2014;136:14554–14559. doi: 10.1021/ja507832e. [DOI] [PubMed] [Google Scholar]
- 29.Koldsø H., Shorthouse D., Sansom M.S.P. Lipid clustering correlates with membrane curvature as revealed by molecular simulations of complex lipid bilayers. PLOS Comput. Biol. 2014;10:e1003911. doi: 10.1371/journal.pcbi.1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koldsø H., Reddy T., Sansom M.S.P. Membrane compartmentalization reducing the mobility of lipids and proteins within a model plasma membrane. J. Phys. Chem. B. 2016;120:8873–8881. doi: 10.1021/acs.jpcb.6b05846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reddy T., Sansom M.S.P. The role of the membrane in the structure and biophysical robustness of the dengue virion envelope. Structure. 2016;24:375–382. doi: 10.1016/j.str.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnarez C., Marrink S.J., Periole X. Molecular mechanism of cardiolipin-mediated assembly of respiratory chain supercomplexes. Chem. Sci. (Camb.) 2016;7:4435–4443. doi: 10.1039/c5sc04664e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holdbrook D.A., Huber R.G., Khalid S. Dynamics of crowded vesicles: local and global responses to membrane composition. PLoS One. 2016;11:e0156963. doi: 10.1371/journal.pone.0156963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marino K.A., Prada-Gracia D., Filizola M. Impact of lipid composition and receptor conformation on the spatio-temporal organization of μ-opioid receptors in a multi-component plasma membrane model. PLOS Comput. Biol. 2016;12:e1005240. doi: 10.1371/journal.pcbi.1005240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeon J.-H., Javanainen M., Vattulainen I. Protein crowding in lipid bilayers gives rise to non-gaussian anomalous lateral diffusion of phospholipids and proteins. Phys. Rev. X. 2016;6:021006–021017. [Google Scholar]
- 36.van Eerden F.J., van den Berg T., Marrink S.J. Molecular dynamics of photosystem II embedded in the thylakoid membrane. J. Phys. Chem. B. 2017;121:3237–3249. doi: 10.1021/acs.jpcb.6b06865. [DOI] [PubMed] [Google Scholar]
- 37.Wells M.A., Dittmer J.C. A comprehensive study of the postnatal changes in the concentration of the lipids of developing rat brain. Biochemistry. 1967;6:3169–3175. doi: 10.1021/bi00862a026. [DOI] [PubMed] [Google Scholar]
- 38.Lutzke B.S., Braughler J.M. An improved method for the identification and quantitation of biological lipids by HPLC using laser light-scattering detection. J. Lipid Res. 1990;31:2127–2130. [PubMed] [Google Scholar]
- 39.Kracun I., Rosner H., Lauc G. Human brain gangliosides in development, aging and disease. Int. J. Dev. Biol. 1991;35:289–295. [PubMed] [Google Scholar]
- 40.Diagne A., Fauvel J., Douste-Blazy L. Studies on ether phospholipids. II. Comparative composition of various tissues from human, rat and guinea pig. Biochim. Biophys. Acta. 1984;793:221–231. doi: 10.1016/0005-2760(84)90324-2. [DOI] [PubMed] [Google Scholar]
- 41.Homan R., Anderson M.K. Rapid separation and quantitation of combined neutral and polar lipid classes by high-performance liquid chromatography and evaporative light-scattering mass detection. J. Chromatogr. B Biomed. Sci. Appl. 1998;708:21–26. doi: 10.1016/s0378-4347(97)00651-8. [DOI] [PubMed] [Google Scholar]
- 42.Han X., Holtzman D.M., McKeel D.W., Jr. Plasmalogen deficiency in early Alzheimer’s disease subjects and in animal models: molecular characterization using electrospray ionization mass spectrometry. J. Neurochem. 2001;77:1168–1180. doi: 10.1046/j.1471-4159.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- 43.Han X., Gross R.W. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 44.Sharon R., Bar-Joseph I., Selkoe D.J. Altered fatty acid composition of dopaminergic neurons expressing α-synuclein and human brains with α-synucleinopathies. J. Biol. Chem. 2003;278:49874–49881. doi: 10.1074/jbc.M309127200. [DOI] [PubMed] [Google Scholar]
- 45.Chan R.B., Oliveira T.G., Di Paolo G. Comparative lipidomic analysis of mouse and human brain with Alzheimer disease. J. Biol. Chem. 2012;287:2678–2688. doi: 10.1074/jbc.M111.274142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abbott S.K., Else P.L., Atkins T.A., Hulbert A.J. Fatty acid composition of membrane bilayers: importance of diet polyunsaturated fat balance. Biochim. Biophys. Acta. 2012;1818:1309–1317. doi: 10.1016/j.bbamem.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 47.Lam S.M., Wang Y., Shui G. Brain lipidomes of subcortical ischemic vascular dementia and mixed dementia. Neurobiol. Aging. 2014;35:2369–2381. doi: 10.1016/j.neurobiolaging.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arai Y., Sampaio J.L., Huttner W.B. Lipidome of midbody released from neural stem and progenitor cells during mammalian cortical neurogenesis. Front. Cell. Neurosci. 2015;9:325. doi: 10.3389/fncel.2015.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oliveira T.G., Chan R.B., Sousa N. The impact of chronic stress on the rat brain lipidome. Mol. Psychiatry. 2016;21:80–88. doi: 10.1038/mp.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marrink S.J., De Vries A.H., Mark A.E. Coarse grained model for semiquantitative lipid simulations. J. Phys. Chem. B. 2004;108:750–760. [Google Scholar]
- 51.Marrink S.J., Risselada H.J., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 52.López C.A., Sovova Z., Marrink S.J. Martini force field parameters for glycolipids. J. Chem. Theory Comput. 2013;9:1694–1708. doi: 10.1021/ct3009655. [DOI] [PubMed] [Google Scholar]
- 53.Melo M.N., Ingólfsson H.I., Marrink S.J. Parameters for Martini sterols and hopanoids based on a virtual-site description. J. Chem. Phys. 2015;143:243152. doi: 10.1063/1.4937783. [DOI] [PubMed] [Google Scholar]
- 54.Gu R.-X., Ingólfsson H.I., Tieleman D.P. Ganglioside-lipid and ganglioside-protein interactions revealed by coarse-grained and atomistic molecular dynamics simulations. J. Phys. Chem. B. 2017;121:3262–3275. doi: 10.1021/acs.jpcb.6b07142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wassenaar T.A., Ingólfsson H.I., Marrink S.J. Computational lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. J. Chem. Theory Comput. 2015;11:2144–2155. doi: 10.1021/acs.jctc.5b00209. [DOI] [PubMed] [Google Scholar]
- 56.Pronk S., Páll S., Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Jong D.H., Baoukina S., Marrink S.J. Martini straight: boosting performance using a shorter cutoff and GPUs. Comput. Phys. Commun. 2016;199:1–7. [Google Scholar]
- 58.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 59.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 60.Bremer P.-T., Weber G.H., Bell J.B. Analyzing and tracking burning structures in lean premixed hydrogen flames. IEEE Trans. Vis. Comput. Graph. 2010;16:248–260. doi: 10.1109/TVCG.2009.69. [DOI] [PubMed] [Google Scholar]
- 61.Bennett J.C., Krishnamoorthy V., Bremer P.-T. Feature-based statistical analysis of combustion simulation data. IEEE Trans. Vis. Comput. Graph. 2011;17:1822–1831. doi: 10.1109/TVCG.2011.199. [DOI] [PubMed] [Google Scholar]
- 62.Michaud-Agrawal N., Denning E.J., Beckstein O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011;32:2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chavent M., Reddy T., Sansom M.S.P. Methodologies for the analysis of instantaneous lipid diffusion in MD simulations of large membrane systems. Faraday Discuss. 2014;169:455–475. doi: 10.1039/c3fd00145h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marrink S.J., de Vries A.H., Wassall S.R. Cholesterol shows preference for the interior of polyunsaturated lipid membranes. J. Am. Chem. Soc. 2008;130:10–11. doi: 10.1021/ja076641c. [DOI] [PubMed] [Google Scholar]
- 65.Bennett W.F.D., MacCallum J.L., Tieleman D.P. Molecular view of cholesterol flip-flop and chemical potential in different membrane environments. J. Am. Chem. Soc. 2009;131:12714–12720. doi: 10.1021/ja903529f. [DOI] [PubMed] [Google Scholar]
- 66.Honerkamp-Smith A.R., Veatch S.L., Keller S.L. An introduction to critical points for biophysicists; observations of compositional heterogeneity in lipid membranes. Biochim. Biophys. Acta. 2009;1788:53–63. doi: 10.1016/j.bbamem.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Levental I., Veatch S. The continuing mystery of lipid rafts. J. Mol. Biol. 2016;428(24 Pt A):4749–4764. doi: 10.1016/j.jmb.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Levental I., Byfield F.J., Janmey P.A. Cholesterol-dependent phase separation in cell-derived giant plasma-membrane vesicles. Biochem. J. 2009;424:163–167. doi: 10.1042/BJ20091283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Risselada H.J., Marrink S.J. The molecular face of lipid rafts in model membranes. Proc. Natl. Acad. Sci. USA. 2008;105:17367–17372. doi: 10.1073/pnas.0807527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Putzel G.G., Uline M.J., Schick M. Interleaflet coupling and domain registry in phase-separated lipid bilayers. Biophys. J. 2011;100:996–1004. doi: 10.1016/j.bpj.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perlmutter J.D., Sachs J.N. Interleaflet interaction and asymmetry in phase separated lipid bilayers: molecular dynamics simulations. J. Am. Chem. Soc. 2011;133:6563–6577. doi: 10.1021/ja106626r. [DOI] [PubMed] [Google Scholar]
- 72.Fujimoto T., Parmryd I. Interleaflet coupling, pinning, and leaflet asymmetry-major players in plasma membrane nanodomain formation. Front. Cell Dev. Biol. 2017;4:155. doi: 10.3389/fcell.2016.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shevchenko A., Simons K. Lipidomics: coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 2010;11:593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- 74.Clark J., Anderson K.E., Hawkins P.T. Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat. Methods. 2011;8:267–272. doi: 10.1038/nmeth.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Castillo N., Monticelli L., Tieleman D.P. Free energy of WALP23 dimer association in DMPC, DPPC, and DOPC bilayers. Chem. Phys. Lipids. 2013;169:95–105. doi: 10.1016/j.chemphyslip.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 76.Xie H., McDonnell K.T., Qin H. Proceedings of the VIS ’04 Conference on Visualization. IEEE Computer Society; Washington, DC: 2004. Surface reconstruction of noisy and defective data sets; pp. 259–266. [Google Scholar]
- 77.Kazhdan M., Bolitho M., Hoppe H. Poisson Surface Reconstruction. In: Polthier K., Sheffer A., editors. Eurographics Symposium on Geometry Processing. Eurographics Association; 2006. pp. 61–70. [Google Scholar]
- 78.Floater M.S., Hormann K. Surface parameterization: a tutorial and survey. In: Dodgson N.A., Floater M.S., Sabin M.A., editors. Advances in Multiresolution for Geometric Modelling. Springer); 2005. pp. 157–186. [Google Scholar]
- 79.Venable R.M., Ingólfsson H.I., Pastor R.W. Lipid and peptide diffusion in bilayers: the Saffman-Delbrück model and periodic boundary conditions. J. Phys. Chem. B. 2017;121:3443–3457. doi: 10.1021/acs.jpcb.6b09111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.