Abstract
Background
The Aspergillus niger genome contains a large repertoire of genes encoding carbohydrate active enzymes (CAZymes) that are targeted to plant polysaccharide degradation enabling A. niger to grow on a wide range of plant biomass substrates. Which genes need to be activated in certain environmental conditions depends on the composition of the available substrate. Previous studies have demonstrated the involvement of a number of transcriptional regulators in plant biomass degradation and have identified sets of target genes for each regulator. In this study, a broad transcriptional analysis was performed of the A. niger genes encoding (putative) plant polysaccharide degrading enzymes. Microarray data focusing on the initial response of A. niger to the presence of plant biomass related carbon sources were analyzed of a wild-type strain N402 that was grown on a large range of carbon sources and of the regulatory mutant strains ΔxlnR, ΔaraR, ΔamyR, ΔrhaR and ΔgalX that were grown on their specific inducing compounds.
Results
The cluster analysis of the expression data revealed several groups of co-regulated genes, which goes beyond the traditionally described co-regulated gene sets. Additional putative target genes of the selected regulators were identified, based on their expression profile. Notably, in several cases the expression profile puts questions on the function assignment of uncharacterized genes that was based on homology searches, highlighting the need for more extensive biochemical studies into the substrate specificity of enzymes encoded by these non-characterized genes. The data also revealed sets of genes that were upregulated in the regulatory mutants, suggesting interaction between the regulatory systems and a therefore even more complex overall regulatory network than has been reported so far.
Conclusions
Expression profiling on a large number of substrates provides better insight in the complex regulatory systems that drive the conversion of plant biomass by fungi. In addition, the data provides additional evidence in favor of and against the similarity-based functions assigned to uncharacterized genes.
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4164-x) contains supplementary material, which is available to authorized users.
Keywords: Transcriptional regulators, Plant biomass degradation, CAZy genes, XlnR, AmyR, GalX, AraR, RhaR, Aspergillus niger
Background
Aspergillus niger is a saprobic fungus that degrades a broad range of plant polysaccharides. Its genome encodes a versatile set of polysaccharide degrading enzymes [1, 2], which can be classified into families of glycoside hydrolases (GHs), polysaccharide lyases (PLs), carbohydrate esterases (CEs) and auxiliary activities (AAs) according to the CAZy (Carbohydrate-Active Enzymes) database (www.cazy.org; [3]). The classification is based on amino acid sequence and structural similarity. Among the 176 genes of A. niger CBS513.88 [4] that are predicted to encode CAZymes involved in plant biomass degradation less than half have been biochemically characterized, while the others have been assigned to CAZy families merely based on homology to functionally characterized genes.
In addition to the production of a wide variety of CAZyme encoding genes, the efficient depolymerization of the polysaccharides present in plant biomass requires a fine-tuned regulatory system. The expression of fungal CAZy genes have been shown to be controlled by multiple transcriptional regulators, most of which belong to fungi specific Zn2Cys6zinc binuclear family of transcriptional factors [5]. In A. niger, several regulators related to plant polysaccharide degradation have been identified [6]. These include XlnR [7], AraR [1], AmyR [8], InuR [9], RhaR [10], ManR/ClrB [11, 12], ClrA [13], GalX [14] and GaaR [15] that have been reported as transcriptional activators of CAZymes (Table 1). These regulators respond to mono- and small oligosaccharides that act as inducers (Table 1) [16], but so far, a limited set of target genes of these regulators have been identified. While some genes can be controlled by a single regulator, co-regulation of several CAZyme encoding genes has been described in Aspergillus species.
Table 1.
Transcriptional activators involved in plant polysaccharide degradation and/or sugar catabolism in A. niger
| Regulatora | Inducer | Function | Reference |
|---|---|---|---|
| AmyR | D-glucose or maltose | Starch degradation | [8] |
| AraR | L-arabitol | Arabinan degradation, Pentose Catabolic Pathway | [22, 66] |
| ClrA | celluloseb | Degradation of cellulose | [13] |
| GaaR | 2-keto-3-deoxy-L-galactonate | Degradation of polygalacturonic acid and more complex pectins, transport of D-galacturonic acid, D-galacturonic acid catabolism | [29, 47] |
| GalX | D-galactose or derivative | D-galactose catabolism | [15] |
| InuR | sucrose | Inulin and sucrose degradation | [9, 43] |
| ManR/ClrB | Galactomannan and cellulose degradation | [11, 12] | |
| RhaR | L-rhamnose derivative | Rhamnogalacturonan I degradation, L-rhamnose catabolism | [10, 27] |
| XlnR | D-xylose | Xylan, xyloglucan and cellulose degradation, Pentose Catabolic Pathway | [7, 21, 22] |
aThe A. niger deletion strains of the underlined regulators were used in this study
bBased on data from Neurospora crassa [45]
AmyR, a transcriptional regulator that controls the genes involved in starch degradation, was the first well-studied regulator in several Aspergillus species [17, 18]. In Aspergilli, AmyR is induced by maltose and regulates genes encoding α-amylases, glucoamylase and α-glucosidases all of which are involved in depolymerization of starch, the major storage polysaccharide in plants [6]. In addition, AmyR has been shown to have a broader physiological role in A. niger by controlling some of the genes encoding D-glucose and D-galactose releasing enzymes, i.e. β-glucosidases, and α- and β-galactosidases [8]. Also, D-glucose or its metabolic product has been suggested to have a possible role as the inducer of the AmyR system in A. niger.
XlnR has an important role in biomass degradation by controlling the expression of genes encoding enzymes that degrade xylan, cellulose and xyloglucan, which are the most abundant polysaccharides in nature [19–21]. The xlnR gene has also been shown to be present in almost in all filamentous ascomycete fungi [22]. The range of genes regulated by XlnR include genes encoding endoxylanase, β-xylosidase, α-glucuronidase, acetylxylan esterase, arabinoxylan arabinofuranohydrolase, feruloyl esterase, α- and β-galactosidases, endoglucanase and cellobiohydrolase, as well as aglB and lacA genes that encode enzymes putatively involved in xyloglucan or galactomannan degradation [23].
A homolog of XlnR, AraR, is a transcriptional regulator induced by L-arabinose and its degradation product, L-arabitol [22]. These monomers are building blocks of arabinan present in side chains of arabinoxylan and pectin. Two arabinan hydrolyzing enzymes produced by A. niger, α-L-arabinofuranohydrolases A and B, are controlled by AraR [22]. In addition, AraR controls the expression of the genes involved in L-arabinose catabolism. AraR and XlnR also co-regulate genes from pentose catabolic pathway and pentose phosphate pathway [24].
The expression of the genes encoding inulinases and invertase, which hydrolyze plant storage polymer inulin, is controlled by the transcriptional regulator InuR in A. niger [9]. Inulinolytic enzyme encoding genes are also inducted by sucrose, and moreover, the repertoire of the genes regulated by InuR has been suggested to include other genes related to degradation of inulin and sucrose.
Several plant polysaccharides, e.g. xylan, pectin and glucogalactomannan, include D-galactose, which is released by fungal α- and β-galactosidases and endogalactanases. While a galactose-related regulator GalR was reported to be unique for Aspergillus nidulans [25], it has also been found in related species of Aspergillus section nidulantes [26]. In contrast, GalX is more generally present in Aspergillus species. In A. niger, GalX regulates the expression of the genes from oxido-reductive pathway for D-galactose catabolism [14].
In addition to AraR, the other pectinolytic regulators described from A. niger are RhaR [10] and GaaR [15]. RhaR, induced by a metabolic conversion product of L-rhamnose, influences the degradation of rhamnogalacturonan I part of pectin by controlling several genes involved in L-rhamnose release and catabolism [10, 27], as well as a L-rhamnose transporter [28]. The more recently described transcriptional regulator GaaR is induced by 2-keto-3-deoxy-L-galactonate, a metabolic conversion product of D-galacturonic acid, and involved in the release of galacturonic acid from polygalacturonic acid and more complex pectins, as well as transport of galacturonic acid and induction of the galacturonic acid catabolic genes [15, 29].
Other plant-biomass related transcriptional regulators described in Aspergillus species include the regulator of mannan degrading enzymes, ManR/ClrB, which was first described in Aspergillus oryzae, with a role in galactomannan and cellulose degradation [11, 12]. In A. oryzae, ManR/ClrB is induced by the disaccharide mannobiose, but not mannose [11, 12]. Furthermore, the genomes of Aspergilli possess various homologs of plant-polysaccharide related regulators from other fungal species, and the gene expression studies have also indicated the presence of several additional regulators involved in this process, including e.g. those responding to the presence of ferulic acid, glucuronic acid and galacturonic acid [6].
The aim of this study was to evaluate co-regulation/co-expression of characterized and putative CAZymes to gain more insight in the function of uncharacterized CAZyme encoding genes in plant biomass utilization and to identify new targets of transcriptional regulators. The focus of the study was on the initial response of A. niger to the presence of a carbon source. For this, microarray data were analyzed of A. niger N402 (wild type) that was grown on a set of 23 carbon sources (including eight monosaccharides, two oligosaccharides, 11 polysaccharides, a crude plant biomass substrate and ferulic acid), and of regulatory mutant strains (ΔxlnR, ΔaraR, ΔamyR, ΔrhaR and ΔgalX) that were grown on their specific inducing compounds. Hierarchical clustering of the expression data revealed several gene clusters that appear to be under control of the same regulators.
Results and discussion
Microarray data were analyzed of A. niger N402 that was grown on 23 carbon sources (Tables 2, 3) and of the regulatory mutants ΔxlnR, ΔaraR, ΔamyR, ΔrhaR and ΔgalX that were grown on their inducing compounds (Tables 1, 3). The mycelial samples were collected after 2 h or 4 h (for N402 and ΔamyR on D-maltose) exposure to the carbon source of interest. Thus, this study focused on the initial response to the presence of a carbon source to avoid changes due to carbon source limitation or changes in the polymeric substrates. Although we can therefore not exclude that genes that were not expressed may have been induced after longer incubation times or on substrates that were not included in this analysis, it provides a detailed understanding of the initial response of A. niger to the presence of plant-biomass related carbon sources.
Table 2.
A. niger strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| A. niger N402 | cspA1 | [67] |
| FP-304 (ΔrhaR) | cspA1, ΔkusA::amdS+, pyrA5, ΔrhaR::pyrA+ | [10] |
| UU-A101.1 (ΔamyR) | cspA1, ΔargB, pyrA6, leuA1, nicA1, ΔamyR::argB+ | [8] |
| FP-306 (ΔgalX) | cspA1, ΔkusA::amdS+, pyrA5, ΔgalX::pyrA+ | [14] |
| UU-A033.21 (ΔaraR) | cspA1, pyrA6, nicA1, leuA1, ΔargB::pIM2101 ΔaraR::argB+ | [22] |
| UU-A062.10 (ΔxlnR) | cspA1, pyrA6, ΔargB, nicA1, leuA, pyrA6, ΔxlnR::pyrA+ | [22] |
Table 3.
Composition, purity and concentration of the carbon sources used in this study
| Substrate | Conc. | Company | Purity | Composition |
|---|---|---|---|---|
| D-glucose | 25 mM | Sigma-Aldrich | ≥99.5% | – |
| D-fructose | 25 mM | Sigma-Aldrich | ≥99% | – |
| D-galactose | 25 mM | Sigma-Aldrich | ≥99% | – |
| L-arabinose | 25 mM | Sigma-Aldrich | ≥99% | – |
| D-xylose | 25 mM | Sigma-Aldrich | ≥99% | – |
| D-mannose | 25 mM | Sigma-Aldrich | ≥99% | – |
| D-galacturonic acid | 25 mM | Sigma-Aldrich | ≥99% | – |
| L-rhamnose | 25 mM | Sigma-Aldrich | ≥99% | – |
| Maltose | 25 mM | Sigma-Aldrich | ≥99% | – |
| Sucrose | 25 mM | Sigma-Aldrich | ≥99% | – |
| Ferulic acid | 25 mM | Sigma-Aldrich | ≥99% | – |
| Inulin from chicory | 1% | Sigma-Aldrich | ns | ns |
| Cellulose | 1% | Sigma-Aldrich | ns | ns |
| Guar gum | 1% | Sigma-Aldrich | ns | ns |
| Xyloglucan | 1% | Sigma-Aldrich | ns | ns |
| Xylan (from beechwood) | 1% | Sigma-Aldrich | >90% | >90% D-xylose residues |
| Polygalacturonic acid (from apples) | 1% | Sigma-Aldrich | ≥85% | ns |
| Apple pectin | 1% | Sigma-Aldrich | ns | ns |
| Galactan (from potato) | 1% | Megazyme | ns | Gal: Ara: Rha: GalA = 88: 2: 3: 7 |
| Debranched 1,5-α-L-arabinan (from sugar beet) | 1% | Megazyme | ~95% | Ara: Gal: Rha: GalA = 88: 4: 2: 6 |
| Rhamnogalacturonan I (from potato) | 1% | Megazyme | >97% | GalA: Rha: Ara: Xyl: Gal: Os = 62: 20: 3.3: 1: 12: 1.7 |
| Mannan (ivory nut) | 1% | Megazyme | >98% | 99% Mannan, Ara + Xyl traces |
| Citrus pulp | 1% | ns | ns | Glc: GalA: Ara: Gal: Xyl: Man: Rha = 39: 35: 11: 7: 4: 4: 1 |
| Sugar beet pulp | 1% | ns | ns | Glc: Ara: GalA: Gal: Xyl: Man: Rha = 33: 28: 26: 7: 2: 2: 1 |
| Soy bean hulls | 1% | ns | ns | Glc: GalA: Xyl: Ara: Man: Gal: Rha = 49: 16: 15: 8: 7: 4: 1 |
Gal D-galactose, Ara L-arabinose, Rha L-rhamnose, GalA D-galacturonic acid, Xyl D-xylose, Man D-mannose, Os other sugars. ns = not specified
Clustering the expression profiles of A. niger (putative) CAZyme encoding genes that are related to plant polysaccharide degradation by Pearson correlation resulted in nine clusters, A-I (Additional file 1). After the initial clustering analysis, genes with a signal value below 50 under all growth conditions were removed from the analysis. These genes were considered not to be significantly expressed (Additional files 2 and 3). The genes that were significantly expressed (signal value >50) at least under one condition are shown in Tables 4, 5, 6, 7, 8, 9, 10, 11. In addition, the fold-changes of the significantly expressed genes between N402 and the regulatory mutant strains were determined (Tables 4, 5, 6, 7, 8, 9, 10, 11). Negative fold-changes indicate genes for which the expression is lower in the mutant than in the wild type strain, while positive fold-changes indicate higher expression in the mutant than in the wild type strain. If the negative fold-change is larger than 2.5, we consider this gene under control of the respective regulator.
Table 4.
Significantly expressed A. niger N402 genes from cluster B in the expression profiling tree
| Up/Down-regulatedc | Regulated by | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genea | Enzymeb | CAZy family | ΔxlnR | ΔaraR | ΔrhaR | ΔgalX | ΔamyR | This study | Literature | Reference | |
| B-1 | An03g01050 | GLN | GH5 | 4.8 | −74.6 | 3.1 | 29.9 | 6.3 | AraR | nr | [38] |
| An14g01800 (aglD) | AGL | GH27 | −1.5 | −97.4 | – | – | – | AraR | nr | [38] | |
| An01g00330 (abfA) | ABF | GH51 | 1.0 | −113.9 | −1.7 | 4.6 | 38.3 | AraR | AraR, GaaR | [15, 22, 34, 68] | |
| An03g00940 (xlnC/xynA) | XLN | GH10 | −17.6 | −27.6 | – | – | – | AraR, XlnR | ClrA, XlnR | [13, 21, 38] | |
| An03g00960 (axhA) | AXH | GH62 | −17.3 | −28.5 | – | – | – | AraR, XlnR | AraR, XlnR | [21, 22, 30] | |
| An16g06800 (eglB) | EGL | GH5 | 4.1 | −6.2 | −1.3 | −1.1 | 1.0 | AraR | nr | [38] | |
| An15g04550 (xynA) | XLN | GH11 | – | −6.6 | – | – | – | AraR | nr | [38] | |
| B-2 | An12g05010 (axeA) | AXE | CE1 | −51.5 | 1.5 | – | – | – | XlnR | XlnR | [21, 38] |
| An01g09960 (xlnD/xynD) | BXL | GH3 | −502.6 | 1.6 | −1.8 | 2.1 | 5.9 | XlnR | XlnR | [21, 69] | |
| An08g01900 | BXL | GH43 | −4.0 | 1.2 | – | – | – | XlnR | nr | [38] | |
| An11g02100 | BGL | GH1 | −5.3 | 2.0 | 1.0 | 2.1 | 2.0 | XlnR | nr | [38] | |
| An14g05800 (aguA) | AGU | GH67 | −32.5 | 2.1 | −3.7 | 1.5 | 1.5 | RhaR, XlnR | XlnR | [21, 70] | |
| An09g00120 (faeA) | FAE | SF7d | −78.3 | −1.5 | – | – | – | XlnR | XlnR | [21, 36] | |
| An08g01760 | CBH | GH6 | −3.8 | – | – | – | – | XlnR | nr | [38] | |
| An09g03300 (axlA/xylS) | AXL | GH31 | −10.4 | −6.3 | 1.3 | 3.8 | 4.3 | AraR, XlnR | nr | [41] | |
| An01g00780 (xlnB/xynB) | XLN | GH11 | −40.7 | −5.6 | 1.7 | 1.1 | −2.2 | AraR, XlnR | XlnR | [21, 38] | |
aGenes with expression value of <50 in all studied A. niger N402 cultures are not included in the table
bEnzyme codes are provided in Additional file 2
cFold-change between A. niger N402 and the regulatory mutants grown on their relevant carbon source. - = expression value <50 in both N402 and regulatory mutant strain. Negative fold-changes >2.5 were considered as proof of regulator function and are depicted in bold
dSub-family (SF) classification of fungal FAEs according to Dilokpimol et al. [49]
nr not reported
Table 5.
Significantly expressed A. niger N402 genes from cluster C in the expression profiling tree
| Up/Down-regulatedc | Regulated by | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genea | Enzymeb | CAZy family | ΔxlnR | ΔaraR | ΔrhaR | ΔgalX | ΔamyR | This study | Literature | Reference | |
| C-1 | An01g13610 (amyD) | AMY | GH13 | 1.2 | – | – | 1.3 | – | nd | nr | [43] |
| An18g04810 (rgxC) | RGX | GH28 | – | – | −17.1 | – | – | RhaR | GaaR, RhaR | [15, 34, 40] | |
| An06g00290 (lacC) | LAC | GH35 | 1.3 | −1.7 | −20.2 | 1.6 | 1.5 | RhaR | AmyR, AraR, GaaR, RhaR | [15, 34, 40] | |
| C-2 | An11g04040 (pgxA) | PGX | GH28 | – | – | – | – | – | nd | GaaR | [15, 34, 40] |
| An03g06740 (pgxB) | PGX | GH28 | – | – | – | – | – | nd | GaaR | [15, 34, 40] | |
| An02g12450 (pgxC) | PGX | GH28 | −1.3 | −3.2 | – | – | – | RhaR | AraR, GaaR | [15, 34, 40] | |
| An12g07500 (pgaX) | PGX | GH28 | – | – | – | – | – | nd | GaaR | [15, 34, 40] | |
| An02g00140 (xynB) | BXL | GH43 | – | 2.3 | – | 1.6 | 1.9 | nd | GaaR | [34, 38] | |
| An03g06310 (pmeA) | PME | CE8 | – | – | – | – | – | nd | GaaR, AraR, RhaR | [15, 34, 71] | |
| An14g04370 (pelA) | PEL | PL1 | 1.2 | −1.2 | −1.5 | 1.0 | 1.0 | nd | GaaR | [15, 34, 72, 73] | |
| An14g01130 (rglA) | RGL | PL4 | – | – | – | – | – | nd | GaaR | [15, 34, 35] | |
| C-3 | An18g05620 (agdF) | AGD | GH31 | – | – | −3.4 | 2.7 | 3.2 | RhaR | nr | [41] |
| An09g02160 (rgaeA) | RGAE | CE12 | – | – | −22.1 | 2.3 | – | RhaR | GaaR, RhaR | [10, 34, 74] | |
| An04g09360 (rgaeB) | RGAE | CE12 | – | 1.5 | −38.1 | 2.0 | – | RhaR | GaaR, RhaR | [10, 34, 39] | |
| An01g14650 (rgxA) | RGX | GH28 | – | – | −26.4 | – | – | RhaR | RhaR | [10, 40] | |
| An01g06620 | RHA | GH78 | −1.2 | 1.3 | −90.2 | −1.1 | 2.8 | RhaR | RhaR | [10, 38] | |
| An12g05700 | RHA | GH78 | – | – | −132.0 | – | 4.8 | RhaR | GaaR, RhaR | [10, 34, 38] | |
| An14g02920 (urhgA) | URH | GH105 | – | 2.3 | −50.2 | 2.1 | 3.6 | RhaR | AraR, GaaR, RhaR | [10, 34, 39] | |
| An07g00240 | RHA | GH78 | 2.0 | – | −17.0 | – | – | RhaR | RhaR | [10, 38] | |
| An10g00290 | RHA | GH78 | – | – | −8.2 | – | – | RhaR | RhaR | [10, 38] | |
| An01g14600 | XLN | GH11 | – | – | −13.5 | – | – | RhaR | nr | [38] | |
| An03g02080 (rgxB) | RGX | GH28 | – | – | −137.7 | – | – | RhaR | RhaR | [40] | |
| An04g09070 | RHA | GH78 | −2.8 | 1.1 | −40.4 | 1.1 | 2.0 | RhaR, XlnR | RhaR | [10, 38] | |
aGenes with expression value of <50 in all studied A. niger N402 cultures are not included in the table
bEnzyme codes are provided in Additional file 2
cFold-change between A. niger N402 and the regulatory mutants grown on their relevant carbon source. - = expression value <50 in both N402 and regulatory mutant strain. Negative fold-changes >2.5 were considered as proof of regulator function and are depicted in bold
nd not detected, nr not reported
Table 6.
Significantly expressed A. niger N402 genes from cluster D in the expression profiling tree
| Up/Down-regulatedc | Regulated by | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genea | Enzymeb | CAZy family | ΔxlnR | ΔaraR | ΔrhaR | ΔgalX | ΔamyR | This study | Literature | Reference | |
| D-1 | An02g01400 (abnB) | ABN | GH43 | −1.6 | 1.4 | 1.6 | −1.1 | – | nd | nr | [39] |
| An11g07660 (exgD) | EXG | GH5 | −1.2 | 1.0 | 1.2 | −1.3 | −1.3 | nd | nr | [38] | |
| An04g09890 (agsA) | AGS | GH13 | 1.4 | – | – | −2.1 | −1.9 | nd | nr | [41, 75] | |
| An18g04800 | RHA | GH78 | – | – | 1.7 | 1.9 | – | nd | GaaR | [34, 38] | |
| An15g01890 | BGL | GH3 | 1.4 | 1.4 | 1.0 | −1.2 | 1.2 | nd | nr | [38] | |
| An13g02110 | AFC | GH29 | – | – | – | – | – | nd | nr | [38] | |
| An01g01340 | UGH | GH88 | – | – | – | – | – | nd | AraR | [34, 38] | |
| An14g05340 (urhgB) | URH | GH105 | – | – | – | – | – | nd | GaaR, RhaR | [34, 39] | |
| D-2 | An04g06930 (amyC) | AMY | GH13 | – | 1.0 | 1.2 | 6.2 | −3.4 | AmyR | AmyR | [38] |
| An03g06550 (glaA) | GLA | GH15 | −2.3 | −4.6 | 5.9 | 9.1 | −44.4 | AmyR, AraR | AmyR | [8, 38, 76, 77] | |
| An11g03340 (aamA) | AMY | GH13 | 4.3 | −9.2 | 12.8 | 38.4 | −35.8 | AmyR | AmyR | [41, 42] | |
| An04g06920 (agdA) | AGD | GH31 | −1.1 | −1.2 | 4.3 | 6.7 | −42.7 | AmyR | AmyR | [8, 38, 78] | |
| An12g02460 (agtB) | AGT | GH13 | – | – | – | – | −3.8 | AmyR | nr | [41] | |
| An03g05530 | XG-EGL | GH12 | – | – | −1.8 | −1.9 | −3.0 | AmyR | nr | [4] | |
aGenes with expression value of <50 in all studied A. niger N402 cultures are not included in the table
bEnzyme codes are provided in Additional file 2
cFold-change between A. niger N402 and the regulatory mutants grown on their relevant carbon source. - = expression value <50 in both N402 and regulatory mutant strain. Negative fold-changes >2.5 were considered proof of regulator function and are depicted in bold
nd = not detected; nr = not reported
Table 7.
Significantly expressed A. niger N402 genes from cluster E in the expression profiling tree
| Up/Down-regulatedc | Regulated by | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genea | Enzymeb | CAZy family | ΔxlnR | ΔaraR | ΔrhaR | ΔgalX | ΔamyR | This study | Literature | Reference | |
| E-1 | An15g04900 (eglD) | LPMO | AA9 | 2.1 | −1.2 | −1.7 | −1.6 | −1.8 | nd | nr | [38] |
| An03g00190 (pelB) | PEL | PL1 | – | – | – | 1.2 | 1.0 | nd | nr | [73] | |
| E-2 | An07g09760 | BGL | GH3 | – | – | – | – | – | nd | nr | [38] |
| An08g01100 | EXG | GH5 | 1.6 | 1.1 | 1.1 | – | 3.8 | nd | nr | [38] | |
| An11g03200 (inuA/inuB) | INU | GH32 | – | – | – | – | – | nd | InuR | [8, 79] | |
| An12g08280 (inuE/inu1) | INX | GH32 | 2.9 | 2.0 | 1.9 | 5.2 | 4.1 | nd | InuR | [8] | |
| An02g04900 (pgaB) | PGA | GH28 | 2.6 | −2.3 | – | 2.2 | −2.1 | nd | GaaR | [15, 34, 80] | |
| An08g11070 (sucA/suc1) | SUC | GH32 | – | – | – | – | – | nd | InuR | [9, 81, 82] | |
aGenes with expression value of <50 in all studied A. niger N402 cultures are not included in the table
bEnzyme codes are provided in Additional file 2
cFold-change between A. niger N402 and the regulatory mutants grown on their relevant carbon source. - = expression value <50 in both N402 and regulatory mutant strain
nd not detected, nr not reported
Table 8.
Significantly expressed A. niger N402 genes from cluster F in the expression profiling tree
| Up/Down-regulatedc | Regulated by | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genea | Enzymeb | CAZy family | ΔxlnR | ΔaraR | ΔrhaR | ΔgalX | ΔamyR | This study | Literature | Reference | |
| F-1 | An02g00610 | GUS | GH2 | 11.7 | – | – | – | – | nd | nr | [38] |
| An09g03070 (agsE) | AGS | GH13 | 1.6 | 1.1 | −2.0 | −11.0 | −5.8 | AmyR, GalX | nr | [43, 75] | |
| An16g02730 (abnD) | ABN | GH43 | 3.6 | – | – | – | −2.5 | AmyR | GaaR, RhaR | [15, 34, 39] | |
| An09g00260/An09g00270 (aglC) | AGL | GH36 | 1.3 | 1.0 | −3.5 | 1.3 | 1.3 | RhaR | AmyR | [8, 44] | |
| F-2 | An09g05350 | FAE | SF9d | 1.5 | – | – | – | −1.1 | nd | nr | [4] |
aGenes with expression value of <50 in all studied A. niger N402 cultures are not included in the table
bEnzyme codes are provided in Additional file 2
cFold-change between A. niger N402 and the regulatory mutants grown on their relevant carbon source. - = expression value <50 in both N402 and regulatory mutant strain. Negative fold-changes >2.5 were considered proof of regulator function and are depicted in bold
dSub-family (SF) classification of fungal FAEs according to Dilokpimol et al. [49]
nd not detected, nr not reported
Table 9.
Significantly expressed A. niger N402 genes from cluster G in the expression profiling tree
| Up/Down-regulatedc | Regulated by | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genea | Enzymeb | CAZy family | ΔxlnR | ΔaraR | ΔrhaR | ΔgalX | ΔamyR | This study | Literature | Reference | |
| G-1 | An17g00520 | BGL | GH3 | – | 1.6 | – | – | 1.5 | nd | nr | [38] |
| An11g06540 (mndA) | MND | GH2 | 4.1 | −3.0 | −2.8 | 1.2 | 2.6 | AraR, RhaR | nr | [4] | |
| An03g03740 (blg4) | BGL | GH1 | 4.1 | 1.0 | −1.2 | 5.8 | 9.6 | nd | nr | [38] | |
| An12g01850 (mndB) | MND | GH2 | −2.5 | −1.5 | 1.0 | 1.9 | 13.4 | XlnR | nr | [38] | |
| An02g07590 | na | GH3 | – | 1.6 | 1.0 | 1.3 | – | nd | nr | [38] | |
| An09g05880 (agdE) | AGD | GH31 | 1.0 | 1.2 | 1.6 | 1.3 | −1.1 | nd | nr | [41] | |
| G-2 | An09g01190 (abnA) | ABN | GH43 | 5.5 | −2.4 | 1.8 | 1.4 | 13.1 | nd | AraR, GaaR | [34, 83] |
| An01g10350 (lacB) | LAC | GH35 | 1.9 | −2.7 | 1.3 | 3.5 | 12.9 | AraR | AraR, GaaR | [15, 34, 38] | |
| An18g05940 (galA) | GAL | GH53 | – | – | – | – | – | nd | AraR, GaaR, RhaR | [34, 84] | |
| An01g12150 (lacA) | LAC | GH35 | −8.9 | −6.3 | 1.4 | 6.4 | 13.6 | AraR, XlnR | AraR, XlnR | [23, 34, 85] | |
| An08g10780 (gbgA) | BXL | GH43 | −1.6 | −29.7 | 2.8 | 11.7 | 5.2 | AraR | AraR | [34, 39] | |
| An08g01710 (abfC) | ABF | GH51 | 1.8 | −8.7 | 1.8 | 7.7 | 31.1 | AraR | AraR, GaaR | [15, 34, 39] | |
| An15g02300 (abfB) | ABF | GH54 | 2.2 | −6.2 | 1.3 | 18.8 | 159.4 | AraR | AraR | [34, 46] | |
| G-3 | An01g01320 | AGL | GH27 | – | −3.8 | – | – | – | AraR | nr | [38] |
| An17g00300 (xarB) | BXL/ABF | GH3 | −2.8 | −1.9 | 1.2 | 1.2 | 1.0 | XlnR | nr | [47] | |
| An01g04880 (axlB) | AXL | GH31 | −2.2 | 1.0 | 1.8 | 1.8 | 1.2 | nd | nr | [41] | |
| An16g02760 (afcA) | AFC | GH95 | −5.2 | 2.2 | −1.3 | 1.7 | 5.9 | XlnR | nr | [38] | |
| An01g03340 (xgeA) | XG-EGL | GH12 | −1.1 | −2.1 | – | – | – | nd | nr | [38] | |
| An04g09690 (pmeB) | PME | CE8 | – | – | – | – | – | nd | AraR, GaaR | [15, 34, 39] | |
| An04g09700 (xghA) | XGH | GH28 | – | −4.6 | – | – | – | AraR | AraR, GaaR | [34, 86] | |
| An01g11520 (pgaI) | PGA | GH28 | – | – | – | – | – | nd | GaaR | [15, 34, 50] | |
| An19g00270 (pelD) | PEL | PL1 | – | – | – | – | – | nd | AraR, GaaR, RhaR | [15, 34, 72, 87] | |
aGenes with expression value of <50 in all studied A. niger N402 cultures are not included in the table
bEnzyme codes are provided in Additional file 2
cFold-change between A. niger N402 and the regulatory mutants grown on their relevant carbon source. - = expression value <50 in both N402 and regulatory mutant strain. Negative fold-changes >2.5 were considered proof of regulator function and are depicted in bold
na no assigned or predicted function, nd not detected, nr not reported
Table 10.
Significantly expressed A. niger N402 genes from cluster H in the expression profiling tree
| Up/Down-regulatedc | Regulated by | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genea | Enzymeb | CAZy family | ΔxlnR | ΔaraR | ΔrhaR | ΔgalX | ΔamyR | This study | Literature | Reference | |
| H-1 | An04g03170 | BGL | GH1 | – | – | – | 2.3 | 3.3 | nd | nr | [38] |
| An14g01770 | BGL | GH3 | 1.0 | 1.5 | −1.2 | 1.0 | 1.5 | nd | AmyR | [8, 38] | |
| An11g00200 (bg1M) | BGL | GH3 | – | 3.7 | – | – | 2.2 | nd | nr | [38] | |
| An12g02550 (faeC) | FAE | CE1 | – | – | – | – | – | nd | nr | [39] | |
| An12g10390 (faeB) | FAE | SF1d | – | 2.4 | −2.8 | 1.1 | 6.5 | RhaR | AraR, GaaR, RhaR | [10, 34, 84] | |
| H-2 | An15g04570 | LPMO | AA9 | – | −2.7 | 1.3 | 1.3 | – | AraR | nr | [38] |
| An14g04200 (rhgB) | RHG | GH28 | – | – | – | – | – | nd | nr | [88] | |
| An16g09090 | na | GH3 | −1.1 | 1.2 | −1.1 | 1.2 | 2.0 | nd | nr | [38] | |
| An18g03570 (bglA/bgl1) | BGL | GH3 | −13.1 | 2.9 | −2.4 | 1.5 | 101.3 | XlnR | nr | [53] | |
| An16g02100 | EGL | GH5 | −1.4 | −1.1 | 1.3 | 1.3 | 1.7 | nd | nr | [38] | |
| An04g02700 | AGL | GH36 | – | – | 1.2 | 1.6 | – | nd | AmyR | [8, 38] | |
| An18g04100 (gp43) | EXG | GH5 | – | −5.7 | −1.5 | 6.0 | 1.8 | AraR | nr | [38] | |
| An01g06120 (gdbA) | GDB | GH13 | −1.2 | 1.0 | −2.1 | 1.9 | 6.3 | nd | nr | [41] | |
| An01g10930 (agdB) | AGD | GH31 | 2.2 | 3.4 | 1.0 | 1.9 | 1.3 | nd | AmyR | [8, 41] | |
| An11g03120 (xynD) | BXL | GH43 | −1.7 | 4.5 | 1.2 | 4.9 | 22.5 | nd | nr | [38] | |
| An06g00170 (aglA) | AGL | GH27 | – | −4.2 | −1.6 | 6.5 | – | AraR | AmyR | [8, 89, 90] | |
| An02g11150 (aglB) | AGL | GH27 | −1.5 | 1.1 | 2.5 | 9.6 | 5.5 | nd | XlnR | [23] | |
| An02g13240 (agdC) | AGD | GH13 | 1.0 | 1.1 | −1.2 | 2.8 | 2.4 | nd | nr | [41] | |
| An05g02410 | GUS | GH2 | – | 1.7 | 1.9 | 4.5 | 3.9 | nd | nr | [38] | |
| An14g04190 (gbeA) | GBE | GH13 | −1.7 | 1.3 | −1.6 | 2.9 | 6.1 | nd | nr | [41] | |
aGenes with expression value of <50 in all studied A. niger N402 cultures are not included in the table
bEnzyme codes are provided in Additional file 2
cFold-change between A. niger N402 and the regulatory mutants grown on their relevant carbon source. - = expression value <50 in both N402 and regulatory mutant strain. Negative fold-changes >2.5 were considered proof of regulator function and are depicted in bold
dSub-family (SF) classification of fungal FAEs according to Dilokpimol et al. [49]
na no assigned or predicted function, nd not detected, nr not reported
Table 11.
Significantly expressed A. niger N402 genes from clusters A and I in the expression profiling tree
| Up/Down-regulatedc | Regulated by | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genea | Enzymeb | CAZy family | ΔxlnR | ΔaraR | ΔrhaR | ΔgalX | ΔamyR | This study | Literature | Reference | |
| A | An15g00320 (sucB) | SUC | GH32 | 1.5 | −1.3 | −2.1 | – | 1.7 | nd | InuR | [9, 52] |
| An15g07160 (pelF) | PEL | PL1 | 1.5 | 1.4 | −2.1 | −1.1 | 1.7 | nd | GaaR | [15, 35, 72] | |
| I | An08g05230 | LPMO | AA9 | – | – | −2.9 | −1.2 | 1.3 | RhaR | nr | [38] |
| An14g02670 | LPMO | AA9 | −1.1 | −1.7 | −1.6 | 1.0 | – | nd | XlnR | [21, 38] | |
| An03g05380 | XG-EGL | GH12 | – | – | – | – | – | nd | nr | [38] | |
| An10g00870 (plyA) | PLY | PL1 | – | – | – | – | – | nd | nr | [91] | |
| An02g10550 (abnC) | ABN | GH43 | −27.8 | 15.2 | 1.4 | −1.8 | 49.2 | XlnR | nr | [39] | |
| An15g03550 | ABN | GH43 | – | 10.0 | – | – | 6.0 | nd | nr | [38] | |
| An07g07630 | BGL | GH3 | – | 2.9 | – | – | 3.1 | nd | nr | [4] | |
| An11g06080 | BGL | GH3 | – | 2.0 | 1.4 | 2.4 | 3.8 | nd | nr | [4] | |
aGenes with expression value of <50 in all studied A. niger N402 cultures are not included in the table
bEnzyme codes are provided in Additional file 2
cFold-change between A. niger N402 and the regulatory mutants grown on their relevant carbon source. - = expression value <50 in both N402 and regulatory mutant strain. Negative fold-changes >2.5 were considered proof of regulator function and are depicted in bold
nd not detected, nr not reported
AraR and XlnR regulated genes involved in cellulose, xyloglucan, xylan and arabinan degradation cluster together based on their expression profile
The genes of cluster B were specifically induced on L-arabinose, D-xylose and/or polygalacturonic acid. This cluster can be divided into sub-clusters B-1 and B-2 that contain seven and ten significantly expressed genes, respectively (Additional file 1, Table 4). The highest expression for genes of sub-cluster B-1 was detected on L-arabinose, except for axhA that was also induced on L-arabinose, but was higher expressed on polygalacturonic acid. The axhA gene encodes an arabinoxylan arabinofuranohydrolase and is specific for arabinoxylan degradation [30]. The high expression of this and other genes of cluster B on polygalacturonic acid may be due to impurity of the substrate (Table 3). The expression of XlnR-regulated genes has been shown to decrease with increasing concentrations of D-xylose due to carbon catabolite repression [31]. Small traces of D-xylose and L-arabinose in the polygalacturonic acid substrate may therefore lead to higher expression of the xylanolytic, arabinanolytic and cellulolytic genes than on 25 mM of D-xylose or L-arabinose used in our study. One gene of sub-cluster B-1 has been characterized as an endoglucanase (eglB), which has activity towards cellulose [21]. The other significantly expressed genes of this cluster encode an arabinofuranosidase (abfA), two putative endoxylanases (xlnC and xynA), a putative α-galactosidase (aglD) and a putative β-endogalactanase (An03g01050) (Table 4).
The highest expression level of these genes was found on L-arabinose (Additional file 1). Regulation of two of these genes, abfA and axhA, is controlled by the transcriptional activator AraR [23] that is induced by L-arabitol, a metabolic product of L-arabinose [32]. Co-regulation of AraR-regulated arabinanolytic genes (i.e. abfA, abfB and abnA) has been suggested previously [22, 33] and abfA has been shown to be controlled by GaaR [15, 34]. A previous principal component analysis (PCA) clustering of the pectinolytic genes has been shown to result in one cluster which contained abfA, abfB, abnA and lacA [35], which matches a more resent hierarchical clustering of the expression of pectinolytic genes in wild type and gaaR deletion mutant strains resulting in a cluster containing abfA, abfB, abfC, lacA, lacB and An03g01620 [15]. However, in this study, the abfB, abnA and lacA genes were separated from abfA, which indicates that abfA has a distinct expression profile from the other genes. This is likely due to the large set of carbon sources that were tested in our study, which provides a more detailed view of the expression of these genes than has been published previously, and also reveals the complexity of the expression of plant-biomass related genes. It should be noted that in nature, fungi are confronted with mixtures of carbon sources, and therefore likely activate a combination of the gene sets we observed in response to pure substrates.
Similar expression profiles for the other genes in this sub-cluster (eglB, xlnC, aglD, xynA and An03g01050) suggest that they are also regulated by AraR. This is supported by the reduced expression of these genes in the ΔaraR strain on L-arabinose compared to N402 (Table 4). The axhA and xlnC genes are also regulated by XlnR [21], which was confirmed in our analysis, as these genes were down-regulated in the ΔxlnR strain. In addition, xlnC has been reported to be ClrA-regulated [13]. Thus, our results indicate a broader role for AraR as some of the genes related to cellulose (eglB), galactomannan (aglD, mndA), pectin (lacA, lacB, xghA), xyloglucan (axlA) and xylan (gbgA, xlnB, xlnC, An01g01320) degradation were significantly down-regulated in the ΔaraR strain.
The genes of sub-cluster B-2 were significantly down-regulated in the ΔxlnR strain (Table 4), thus suggesting that they are controlled by XlnR. Indeed, five of these genes (axeA, xlnD, aguA, faeA and xlnB) have previously been shown to be regulated by XlnR [21, 36, 37]. The highest expression for most genes of this sub-cluster was detected on D-xylose, except for aguA and An11g02100 that were higher expressed on polygalacturonic acid, and axlA and An16g00540 that were higher expressed on L-arabinose (Additional file 1). High expression of axlA on D-xylose has previously been reported [9, 37]. This gene encodes a putative α-xylosidase that is suggested to have a role in xyloglucan degradation [38]. An16g00540 encodes an α-L-fucosidase, which also has a putative role in xyloglucan hydrolysis, indicating co-regulation of some of the genes involved in this process. An11g02100 and An08g01760 encode the cellulolytic enzymes β-glucosidase and cellobiohydrolase, respectively. This is in line with the previous finding that XlnR is a regulator of xylanolytic, xyloglucanolytic and cellulolytic genes [21]. The co-regulation of AraR- and XlnR-regulated genes in cluster B that are involved in cellulose, xyloglucan, xylan and arabinan degradation supports combined action of regulators. Co-regulation of these genes is an efficient strategy for polysaccharide degradation, since L-arabinose, D-xylose and D-glucose often co-occur in plant cell wall polysaccharides.
Expression of pectinolytic genes involved in degradation of the pectin main chains were clustered
Cluster C contains 28 significantly upregulated genes of which most are pectin backbone hydrolyzing genes, mainly from CAZy families GH28 (several types of pectin hydrolases) and GH78 (α-rhamnosidases) (Table 5). It can be divided into the sub-clusters C-1, C-2 and C-3 (Additional file 1). Sub-cluster C-3 contains 12 significantly expressed genes, of which 10 have been shown to be regulated by RhaR and are specifically induced on L-rhamnose [10]. The other two genes of this cluster, agdF and An01g14600, were also specifically induced on L-rhamnose and down-regulated in the ΔrhaR strain suggesting that they are also under control of this regulator (Table 5). However, our results suggest a broader role for RhaR, since in addition to its target genes of cluster C, some other genes were identified that were down-regulated in the ΔrhaR strain, such as aguA, aglC and mndA.
Notably, the agdF gene has previously been assigned to encode a putative enzyme of the starch degrading GH31 family [38]. Our data does not support a function in starch degradation as, in addition to induction on L-rhamnose, this gene was significantly up-regulated in the ΔamyR strain (Table 5), while the opposite would be expected for a starch-related gene. The expression profile of An01g14600, which encodes a putative enzyme of the GH11 endoxylanase family, is unexpected as no link between this family and rhamnogalacturonan degradation has been described. Therefore, our data suggests the involvement of agdF and An01g14600 in rhamnogalacturonan degradation, although their enzymatic function is unclear at this point. A high expression level on L-rhamnose has been previously reported for rgaeB, rgxA, rgxB, urhgA and rglB [39, 40]. In our analysis, rgaeB appears to have a slightly different expression profile from the other genes of sub-cluster C-3 as it is located in a separate branch of the hierarchal cluster (Additional file 1). The inclusion of the L-rhamnose and D-galacturonic acid mixture data enabled us to evaluate the co-operation of these two sugars as inducers by comparing them to the individual sugar cultivations. Interestingly, despite the dominant role for galacturonic acid and GaaR in regulation of pectinolytic genes [16, 34], the mixture of L-rhamnose and D-galacturonic acid clusters more closely with L-rhamnose than with D-galacturonic acid in our analysis. This may indicate that the induction by L-rhamnose is more discriminative than the induction by D-galacturonic acid in distinguishing genes by expression pattern.
Sub-cluster C-1 contains three significantly expressed genes, two of which are regulated by RhaR on L-rhamnose and by GaaR: lacC and rgxC (Table 5) [10, 15]. The lacC and rgxC genes were previously reported to be expressed on D-galacturonic acid, polygalacturonic acid and L-rhamnose, in contrast to the genes of sub-cluster C-3 that were specifically induced on L-rhamnose [39]. High expression of lacC and rgxC on galactan could be due to the small traces of D-galacturonic acid and L-rhamnose in the substrate (Table 3). The lacC has also been reported to be under control of AraR [34] and AmyR [8], but it was not observed to be down-regulated in the ΔamyR strain in our study (Table 5). The third gene of the sub-cluster C-1, amyD, has been classified as an α-amylase [38], but its expression was not detected on D-maltose in A. niger N402 [41]. In our study, the gene was expressed on D-galacturonic acid, polygalacturonic acid and the mixture of D-galacturonic acid and L-rhamnose (Additional file 1). A role for amyD in starch degradation is therefore doubtful.
The pectinolytic genes in sub-cluster C-2 are involved in the degradation of homogalacturonan (Table 5). These genes are not regulated by RhaR but were induced on D-galacturonic acid and polygalacturonic acid in this study (Additional file 1) and most of them are under control of GaaR [15, 34]. The significantly expressed genes of sub-cluster C-2 include four exopolygalacturonases (pgxA, pgxB, pgxC and pgaX), a pectin methyl esterase (pmeA), a pectin lyase (pelA), and rhamnogalacturonan lyase (rglA) (Table 5), all of which have been shown to be GaaR-regulated [15]. In addition, regulation by AraR has been reported for pgxC, and by AraR and RhaR for pmeA [34]. Also, gene An02g00140, which encodes a putative β-xylosidase, showed significant expression (Table 5). The expression profiles of pelA, pmeA and pgaX genes were previously shown to cluster and these genes were suggested to play a major role in the initial degradation of pectin [35]. This is also supported by the results reported from sugar beet pectin [15]. In line with our results, strong induction on D-galacturonic acid and polygalacturonic acid has been reported for pgxB, pgxC and pgaX, while lower expression has been observed for pgxA on these substrates [15, 40]. The pelA gene was well expressed on all tested substrates, but its highest expression was detected on polygalacturonic acid (Additional file 1). In agreement with the previous studies [15, 39], the rglA gene was expressed on D-galacturonic acid, polygalacturonic acid and galactan, but not on L-rhamnose. The GaaR-regulated pmeA gene [15] was slightly induced on D-galacturonic acid and polygalacturonic acid in our study and that of de Vries et al. [35]. In contrast to the results of Kowalczyk et al. [34], the regulation of pmeA by AraR or RhaR was not detected. The function of five out of eight putative α-rhamnosidase encoding genes (i.e. An01g06620, An12g05700, An07g00240, An10g00290 and An04g09070) in sub-cluster C-2 is supported by our analysis as they are specifically induced on L-rhamnose and are under control of RhaR [10, 38]. In addition, An12g05700 is controlled by GaaR and RhaR, and An18g04800 by GaaR [34].
The pectinolytic genes found in cluster C were expressed on L-rhamnose, D-galacturonic acid and/or polygalacturonic acid, suggesting that these genes encode initial pectin degrading enzymes. Pectinolytic genes that showed no significant, or constitutive expression, may be induced on pectin-related substrates after longer incubation times. Expression of plyA, pgaII, pgaB, pgaD, pgaE, pelB, pelC and pelF was low or not significant on all substrates in our study. However, expression of these genes on D-galacturonic acid, polygalacturonic acid and sugar beet pectin has been reported to increase in time [35] and pgaB, pgaE and pelF have been shown to be regulated by GaaR [15, 34].
Constitutively expressed genes clustered with genes involved in starch degradation
In cluster D, sub-cluster D-1 contains nine significantly expressed genes encoding enzymes from different GH families, while in sub-cluster D-2 six genes are present that mainly encode enzymes from GH families assigned to starch degradation (GH13, 15 and 31) (Table 6). The genes of sub-cluster D-1 were not down-regulated in any of the tested regulatory mutant strains, indicating that they are not regulated by these transcriptional activators (Table 6). They show a relatively distant separation from each other, and most showed low, but similar expression levels on all substrates (Additional file 1) indicating that the genes in sub-cluster D-1 are likely constitutively expressed. Indeed, the abnB gene, present in sub-cluster D-1, was previously reported to be constitutively expressed on D-fructose, D-xylose, sorbitol, L-rhamnose, D-galacturonic acid, polygalacturonic acid and sugar beet pectin [39].
The sub-cluster D-2 contains genes that are involved in starch degradation and are down-regulated in the ΔamyR strain. Two glaA and agdA genes, encoding a glucoamylase and an α-glucosidase, respectively [38, 41], showed high expression on all substrates, while the highest expression levels were detected in N402 on maltose (Additional file 1), in line with the previous study [41]. Gene aamA, which encodes an acid α-amylase [42], has also been reported to be highly expressed on maltose [41], but was expressed at a much lower level in our study. For this gene, significant expression was also detected on L-arabinose, polygalacturonic acid and sugar beet pulp (Additional file 1). The similar expression patterns and the down-regulation of glaA, agdA and aamA genes in the ΔamyR strain (Table 6) indicates their co-regulation by AmyR, as has been suggested by Yuan et al. [41]. All three genes were up-regulated in the ΔgalX mutant on D-galactose to a higher level than the expression on maltose in N402 (Additional file 1). The α-amylase gene amyC was also most highly expressed on D-galactose in the ΔgalX mutant. Like glaA, agdA and aamA, expression of this gene was reported to be reduced in the ΔamyR strain [41]. However, the expression profile of amyC in our study differs from the other three amylolytic genes, because a similar expression level of this gene was found on D-maltose, L-rhamnose and guar gum, making its induction on D-maltose less specific (Additional file 1). In a previous study, expression of amyC was similar on D-xylose and D-maltose after 2 h of incubation, but the gene was not expressed after 8 h on xylose, while its expression on maltose was still detected [41].
Low expression for agtB encoding a putative 4-α-glucanotransferase was detected on all substrates, with only significant expression levels and down-regulation in the ΔamyR strain (Additional file 1, Table 6). This data is in contrast with a previous study [41], where expression was only detected after 8 h on D-maltose and agtB was reported to be AmyR independent. Co-expression of agtB and agsC, encoding a putative α-glucan synthase, has previously been observed [41]. Even though agsC was not significantly expressed in our study (Additional file 2), it did cluster with agtB in our initial correlation analysis (Additional files 1 and 2).
An03g05530 is also found in sub-cluster D-2, even though its highest expression level was detected on L-rhamnose and D-galacturonic acid. However, this gene is significantly down-regulated in the ΔamyR strain, which may explain its presence in sub-cluster D-2.
InuR-regulated inulinolytic genes were co-expressed on sucrose and inulin
Cluster E contains eight significantly expressed genes that have relatively distant positions in the expression profile tree (Additional file 1). Sub-cluster E-1 consists of only eglD and pelB encoding a putative LPMO and a pectin lyase, respectively (Table 7), that showed a low overall expression. While this is in contrast to the reported lack of expression for pelB in A. niger cultures on sugar beet pectin, galacturonic acid, rhamnose and xylose [39], the low expression we observed may indicate that expression levels of pelB are always around the detection cut-off. Sub-cluster E-2 contains six genes that were expressed on guar gum, inulin, sugar beet pulp and/or sucrose (Table 7, Additional file 1). High expression on inulin and to a lesser extent on guar gum was observed for a putative exo-inulinase encoding gene inuE, which clustered with an endo-inulinase encoding inuA, but expression levels of the latter gene were much lower. In addition to inuE and inuA, sub-cluster E-2 contains the extracellular inulinolytic gene sucA. These genes were all regulated by InuR, and co-regulation and expression on sucrose and inulin was previously reported for these genes [43]. The more distant position of sucA in the expression profile tree can be explained by its relative expression levels on sucrose, inulin and sugar beet pulp, the latter resulting in the highest expression for sucA. An08g01100 and to a lesser extent An07g09760 were specifically induced on guar gum, but are located close to inuE and inuA in the expression profile tree (Additional file 1). The correlation analysis also demonstrated which substrates are most similar when the expression of all the tested genes was taken into account. Guar gum was most closely related to inulin, sucrose and sugar beet pulp. The sugar beet pulp used in this study contains significant amounts of sucrose (data not shown), which explains the clustering of this substrate with sucrose and inulin. Our results suggest that guar gum may also contain some traces of sucrose, even though this was not reported by the supplier.
Other inulinolytic genes described for A. niger, i.e. sucB, sucC and inuQ, were not present in cluster E. Absence of expression of the intracellular invertase encoding sucC gene, and inuQ, which was described to be a pseudogene, confirmed a previous study [44]. The other intracellular invertase encoding gene, sucB, was reported to have an overall low expression on other substrates than sucrose and inulin [44], which was also confirmed by our study.
Only five significantly expressed genes are positioned in cluster F (Additional file 1, Table 8), with only one gene, An09g05350, in sub-cluster F-2. It was expressed on D-glucose, D-fructose, D-maltose and rhamnogalacturonan. The four genes that form sub-cluster F-1 differ in their expression profile, and therefore the reason for the clustering of these genes may be that they did not fit into any of the other clusters. It should be noted that the genes of cluster F are distantly separated from each other within the expression profiling tree (Additional file 1). A putative α-glucan synthase encoding gene (agsE) showed high expression levels on all substrates in N402, which confirms a previous study [41]. However, expression of this gene was strongly reduced in the ΔamyR strain (Table 8), which was not observed in the study of Yuan et al. [41]. The opposite was found for α-galactosidase encoding aglC that has been reported to be under control of AmyR [41], while our study only detected significant down-regulation in the ΔrhaR strain. Expression of endoarabinanase encoding abnD was previously reported to be constitutive [39], but more recently it was shown to be GaaR-dependent on D-galacturonic acid and GaaR and RhaR-dependent on sugar beet pectin [15, 34]. However, we only detected significant expression levels of abnD on D-maltose in N402 and down-regulation in the ΔamyR strain, suggesting control by this regulator.
Genes related to degradation of pectin side chains cluster separately from those acting on the pectin main chain
Most of the significantly expressed genes of cluster G (Table 9) were highly expressed on D-galacturonic acid and polygalacturonic acid (Additional file 1). The difference between these genes and D-galacturonic and polygalacturonic acid induced genes of cluster C is that the cluster G genes are less specifically induced on D-galacturonic acid and polygalacturonic acid, as they also show high expression levels on other carbon sources. Cluster G, the largest cluster detected with 23 genes, can be divided into the sub-clusters G-1, G-2, and G-3 (Additional file 1).
Expression of some of the genes in cluster G has been previously analyzed on D-fructose, L-rhamnose, D-xylose, sorbitol, D-galacturonic acid, polygalacturonic acid and sugar beet pectin [15, 39, 40]. Specific induction has been observed for pmeB, xghA, pgaI, abfB, abfC, lacA, lacB, galA and abnA on D-galacturonic acid, polygalacturonic acid and sugar beet pectin [15, 45], and all these genes have been shown to be GaaR-controlled, except lacA and abfB [15, 34]. Furthermore, the abfB and abfC genes were also highly expressed on D-xylose [39, 46]. In our study, induction of these genes on D-galacturonic acid and polygalacturonic acid was also observed. In addition, abfB, abfC, lacA, lacB, galA and abnA, all members of sub-cluster G-2, were highly expressed on galactan (Additional file 1). Co-regulation of abfB, abnA and galA was suggested previously [33, 35], but in our study only abnA and galA fall in the same cluster, while the expression profile of abfA is different.
High expression for most of the sub-cluster G-2 genes, except lacA and galA, was observed on arabinan, while high expression on L-arabinose was observed for abfB, abfC and lacA, all of which were down-regulated in the ∆araR strain. The genes of this sub-cluster all encode enzymes that could be involved in the degradation of the pectinolytic side chains, suggesting a strong link between function and expression.
High expression levels of the genes of sub-cluster G-1 were detected on polygalacturonic acid, but to a much lower extent than for the sub-cluster G-2 genes. The highest expression for three genes of sub-cluster G-1, mndA, mndB and bgl4, was found on mannan. The mndA gene encodes a β-mannosidase [44], involved in mannan degradation, while mndB and bgl4 encode a putative β-mannosidase and β-glucosidase, respectively. Their co-expression with mndA supports these functions as both activities are needed for complete degradation of galactoglucomannan. However, these genes were not inducted by mannose. This is in line with the ManR/ClrB regulator from A. oryzae induced by mannobiose, but not by mannose [11, 12]. The highest expression for the other genes of this sub-cluster, agdE, An17g00520 and An02g07590, was detected on polygalacturonic acid.
The highest expression levels of all the genes of sub-cluster G-3 were found on polygalacturonic acid. The GaaR, AraR and RhaR-regulated pelD gene [15, 34] was specifically induced on polygalacturonic acid, in contrast to a previous study where this gene was reported to be non-expressed [39]. Expression of pgaI, which is under control of GaaR [15], and pmeB and xghA, which are under control of GaaR and AraR [15, 34], has previously been reported on D-galacturonic acid and polygalacturonic acid [39], which was confirmed in our study. Three genes of sub-cluster G-3, xarB, axlB and afcA, which encode a putative bi-functional xylosidase/arabinofuranosidase [47], an α-glucosidase and an α-fucosidase, respectively, were down-regulated in the ΔxlnR strain, suggesting control by XlnR. One gene of this sub-cluster, xghA, was down-regulated in the ΔaraR strain, suggesting regulation by AraR in line with Kowalczyk et al. [34]. As mentioned earlier, the polygalacturonic acid specific induction of arabinanolytic and xylanolytic genes may be due to impurity of the substrate with small traces of D-xylose and L-arabinose.
Cluster H contains a diverse set of genes that are expressed on a broad range of substrates
Six and 15 significantly expressed genes form sub-cluster H-1 and H-2, respectively (Additional file 1, Table 10). The faeB gene was expressed at a basal level on L-rhamnose, D-xylose, sorbitol, D-fructose D-galacturonic acid, polygalacturonic acid and sugar beet pectin, while faeC, which is also found in this sub-cluster, was not expressed on these substrates [39]. The genes of sub-cluster H-1 were all specifically induced on ferulic acid. While induction of faeB on ferulic acid has previously been reported [48], the faeC was also induced on this substrate, suggesting co-regulation of these two feruloyl esterase encoding genes, which was confirmed by a recent study [49]. Interestingly, the other genes of the sub-cluster H-1 specifically induced on ferulic acid encode putative β-glucosidases (An04g03170, An14g01770 and bgm1) and a putative LPMO (An15g04570).
The genes of sub-cluster H-2 were expressed at a constant level on most carbon sources tested, but showed low expression on D-glucose, D-fructose, sucrose and sugar beet pulp (Additional file 1). As mentioned before, the sugar beet pulp used in this analysis contains sucrose (data not shown). These genes may therefore be under strong carbon catabolite repression. Binding sites for CreA have been found in the promoter regions of all these genes [38], and low overall expression of a putative α-glucosidase encoding agdC has previously been described [41]. Another α-glucosidase encoding gene, agdB, has been reported to be strongly induced on D-maltose and down-regulated in the ΔamyR strain [41]. Our study, however, revealed that this gene was highly expressed on most carbon sources tested and no down-regulation in the ΔamyR strain was observed (Table 10).
Two clusters of putatively not co-expressed genes were detected
In clusters A and I, only a small number of genes (two and eight, respectively) were significantly expressed (Additional file 1, Table 11). Furthermore, the genes in clusters A and F share no specific trends in their expression profiles and are relatively distantly separated from each other within the expression profiling tree (Additional file 1), and are probably not co-expressed.
In cluster A, the significantly expressed genes, sucB and pelF, encode enzymes from CAZy families GH32 and PL1, respectively (Table 11, Additional file 1). The overall expression of these genes was very low on all substrates. Furthermore, the genes were not significantly down- or up-regulated in the studied regulatory mutant strains, indicating that these genes are not regulated by any of these transcriptional activators. The low overall expression of pelF, a gene encoding a putative pectin lyase, has been reported previously [39] and it has been shown to be regulated by GaaR [15]. Notably, pelF did not cluster with any of the other pectinolytic genes in our data. In contrast, pelF clustered distantly with the other pectinolytic genes in a previous study [35], which, however, included a smaller set of genes and a more focused set of growth conditions that may explain the differences with our study. In addition, the previous hierarchical clustering suggested induction of pelF during starvation or derepressed conditions [15]. Gene sucB encodes an intracellular invertase with transfructosylation activity [50, 51]. Its expression profile was distinct from other inulinolytic genes (Additional file 1). The sucB gene has been reported to be under control of the inulinolytic regulator InuR [52] and to be constitutively expressed at low level [43]. In our study, significant expression of sucB was found on inulin, which supports regulation by InuR. In addition, sucB expression was observed on D-maltose in the ΔamyR strain (Additional file 1). This suggests interaction between AmyR and InuR, similarly as was described for XlnR and AraR in A. niger [22, 53].
All genes in cluster I were expressed at low level on D-maltose and sugar beet pulp in the N402 strain. Some of these genes (i.e. An15g03550, abnC, An07g07630 and An11g06080) were up-regulated in the ΔamyR strain. The highest expressed gene of this cluster was a putative endoarabinanase encoding abnC, which was highly expressed on all the tested substrates except D-maltose and sugar beet pulp (Additional file 1). Expression levels of this gene have previously been reported to be elevated after 24 h on D-fructose, L-rhamnose, sorbitol, D-xylose and D-galacturonic acid [39]. The abnC gene was significantly down-regulated in the ΔxlnR strain on D-xylose, which indicates that this gene is regulated by XlnR (Table 11). The abnC gene and An15g03550, both encoding putative endoarabinanases from family GH43, were highly expressed on galactan, while An15g03550 was also highly expressed on mannan. The highest expression levels of An08g05230 and An14g02670 encoding putative LPMOs from family GH61, An03g05380 encoding putative xyloglucan-active endoglucanase and plyA encoding putative pectate lyase were detected on arabinan (Additional file 1).
Upregulation of genes in regulatory mutants suggests interaction between the different regulatory systems
While the down-regulation of gene expression in A. niger regulatory mutants compared to the wild type strain can be taken as evidence of control by this regulator, we surprisingly also found a significant number of genes for which the expression in a regulatory mutant was higher than in the wild type. While in most cases this was a moderate increase (less than 3-fold,), for 46 genes the difference was higher and 13 of these had fold-changes >10. The largest set of strongly upregulated genes was observed in the amyR mutant on maltose. Interestingly, this seems to especially affect L-arabinose related genes as the fold-change for abfA, abfB, abfC, abnA, abnC and An159g3550 (putative ABN) was 38, 160, 31, 13, 49 and 6, respectively. In addition, bglA was also 100-fold upregulated. Antagonistic interactions between regulators have been observed before, in particular for the two pentose-related regulators XlnR and AraR [54]. However, more recently, this was also observed for three pectinolytic regulators, GaaR, AraR and RhaR [34], suggesting that this is more common phenomenon has been so far considered. The nature of the antagonistic interaction and whether this is a direct or indirect is not clear at this point and requires further study.
Conclusions
This study aimed to reveal co-expression patterns of plant biomass polysaccharide degradation related genes from A. niger, using a more global approach than is usually performed by including a wide range of carbon sources, as well as five regulatory mutants, thus generating an unprecedented view of this system. The broader range of substrates revealed the highly complex expression patterns of these CAZy genes, and demonstrated that the focused analyses of the transcriptional regulators involved in this process that have been identified so far only revealed initial indications of the overall regulatory system. In fact, many of the genes tested in this study were shown to be under control of more than one regulator (Fig. 1a). Interestingly, the role of the regulators appears to be less linked to a specific polysaccharide when the genes encoding a certain enzyme activity and the regulators that act on them were combined (Fig. 1b). This could imply that the role of the enzymes may in fact be broader than currently assumed. E.g. the role of BXL in removing xylose from xylogalacturonan could explain the influence of GaaR on the expression of some BXL-encoding genes.
Fig. 1.
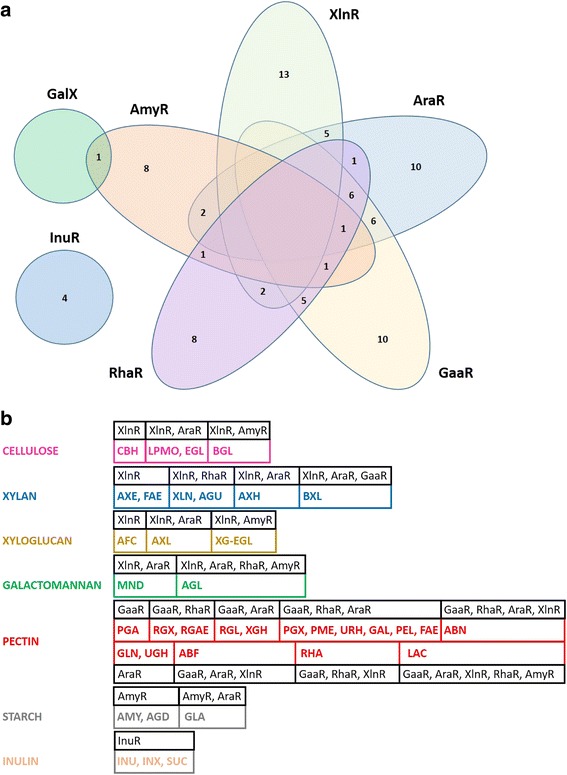
Global analysis of the expression profiles of CAZy genes related to plant polysaccharide degradation. a Number of genes under control of one or more regulators are indicated in a VENN diagram. b Comparison of the influence of regulators on enzyme activities linked to the polysaccharide they act on. Regulatory effects on individual genes encoding the same enzyme activity were combined in the boxes
Previous studies in T. reesei and N. crassa also addressed induction of CAZy genes under different conditions and in some cases by using deletion mutants of plant biomass related regulators [55–58]. However, these studies, similar to previous studies in Aspergillus, used a limited number of carbon sources and did not compare different regulatory mutants. It can therefore be expected that also in these studies the complexity of the regulatory network driving plant biomass degradation is underestimated. More detailed studies in A. niger as well as other fungi are needed to fully grasp the organization of the regulatory network and reveal the differences between fungal species.
Methods
Strains, media and culture conditions
The A. niger strains used in this study are listed in Table 2. Strains were grown at 30 °C on minimal medium (MM) or complete medium (CM) [51] either or not containing 1.5% agar. Liquid cultures were grown on a rotary shaker at 250 rpm. Pre-cultures for RNA isolation were grown for 16 h in 1 L Erlenmeyer flasks that contained 250 ml CM supplemented with 2% D-fructose. Mycelium was washed with MM and 1 g (wet weight) aliquots were transferred for 2 h to 250 ml Erlenmeyer flasks containing 50 ml MM supplemented with 25 mM mono- or disaccharide or ferulic acid, or mixture of 25 mM L-rhamnose and 25 mM D-galacturonic acid, or 1% polysaccharide or complex plant biomass (Table 3). The only exceptions were D-maltose cultures of N402 and ∆amyR strains that were incubated for 4 h and for which 1% maltose was used. These data originate from a different study [8], but were included to help with the grouping of the genes and assess the AmyR effect. Mycelium was harvested by vacuum filtration, dried between towels and frozen in liquid nitrogen. While N402 liquid cultures were performed on all carbon sources listed in Table 3 as well as on the mixture of L-rhamnose and D-galacturonic acid, the regulatory mutant strains ΔxlnR, ΔaraR, ΔamyR, ΔrhaR and ΔgalX were grown on D-xylose, L-arabinose, maltose, L-rhamnose and D-galactose, respectively, and L-rhamnose and D-galacturonic acid. All cultures were performed as biological duplicates.
Microarray processing
RNA isolation and microarray hybridization were performed as described previously [59]. In brief, RNA for microarray analysis was extracted using TRIzol reagent (Invitrogen) and purified using TRIzol® Plus RNA Purification Kit (Sigma-Aldrich) according to the instructions of the manufacturer. The concentration of RNA was calculated from the absorbance at 260 nm in a spectrophotometer (Biochrom Libra S22). The quality of the RNA was analyzed with an Agilent 2100 Bioanalyzer using a RNA6000 LabChip kit (Agilent Technology). Microarray hybridization using the Affymetrix GeneChips A. niger Genome Array was performed at GenomeScan (Leiden, The Netherlands).
Transcriptome analysis
Microarray data was analyzed using the Bioconductor tool package version 2.8 (http://www.bioconductor.org/) together with house-made Perl (version .5.0) and Python (version 3.0) scripts. Probe intensities were normalized for background by the robust multi-array average (RMA) method [60] using the R statistical language and environment [61]. This method makes use of only perfect match (PM) probes.
Normalization was processed by the quantiles algorithm. The median polish summary method [62] was used to calculate the gene expression values. Further statistical analyses were performed with the CyberT tool package using multiple testing (http://cybert.ics.uci.edu/). BayesAnova and paired BayesT-test tests were performed on each gene through pairing carbon sources, PPDE (Posterior Probability of Differential Expression) analysis and multiple hypothesis testing correction are performed on the p-values [63]. Adjusted cut off value of p < 0.05 was used to determine the statistical significance of gene expression difference. Reproducibility of the replicates was verified by PCA analysis (Additional file 4). Genome scale PCA analysis was performed with the gene expression values of the different samples. The PCA was generated using R (v3.40) statistical language and environment, the PCA function from FactoMineR package (v1.35) and plotted using ggplot2 package (v 2.2.1). Replicates are plotted using the same color. Due to the large amount of data, the calculation of the matrix was not possible.
Gene expression clustering, visualization and annotation
Hierarchical clusters were made using complete linkage with the normalized expression data from selected CAZyme encoding genes by calculating the Pearson correlation distances [64]. Clusters were set manually based on the branch-length differences of the gene-tree. The genes were selected based on the annotation of the CAZy families and their (putative) role in plant biomass degradation. Clusters and expression correlation profiles were visualized by Genesis [65]. Genes with an expression value <50 were colored dark blue, the ones >1000 were colored red and the values ≥50 and ≤1000 were colored by a gradient of these 2 colors.
Gene functional annotations were based on previous study [1]. When the data of this study suggested a different function, this was verified by performing phylogenetic analysis of the CAZy family this gene belongs to. The phylogeny analysis was performed using all the A. niger genes of the corresponding family together with all functionally characterized fungal members of that family, which allowed us to verify to which activity this gene clustered.
Additional files
Expression profiling tree containing 168 A. niger genes encoding putative CAZymes (www.cazy.org). Clusters A-I can be distinguished. (PDF 1780 kb)
Expression of selected CAZy genes. (XLSX 215 kb)
Significantly and not significantly expressed genes encoding CAZymes in A. niger CBS513.88 in this study. (XLSX 19 kb)
PCA analysis of the gene expression values of the biological duplicate samples revealing the reproducibility of the duplicates. (PDF 90 kb)
Acknowledgements
Not applicable.
Availability of data and materials
The data sets supporting the results of this article are available in the GEO repository under accession number GSE98572 (token: cbszoqmqddgndkf).
Abbreviations
- AA
Auxiliary activity
- CAZy
Carbohydrate-active enzyme
- CE
Carbohydrate esterase
- CM
Complete medium
- GH
Glycoside hydrolase
- MM
Minimal medium
- PL
Polysaccharide lyase
- RMA
Robust multi-array average
Authors’ contributions
BSG performed part of the experiments and participated in the analysis and writing of the manuscript. MRM participated in the analysis and writing of the manuscript. JK analyzed part of the data. MZ performed the bioinformatics analysis. IBG performed part of the experiments. RPdV designed the study and participated in the analysis and writing of the manuscript. All authors read and approved the final manuscript.
Funding
BS and IBG were supported by a grant of the Dutch Technology Foundation STW, Applied Science division of NWO and the Technology Program of the Ministry of Economic Affairs UGC 07938 to RPdV. MZ was supported by a grant from the Netherlands Organisation for Scientific Research (NWO) and the Netherlands Genomics Initiative 93,511,035 to RPdV.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4164-x) contains supplementary material, which is available to authorized users.
Contributor Information
Birgit S. Gruben, Email: birgit0500@hotmail.com
Miia R. Mäkelä, Email: miia.r.makela@helsinki.fi
Joanna E. Kowalczyk, Email: j.kowalczyk@westerdijkinstitute.nl
Miaomiao Zhou, Email: m.zhou1@avans.nl.
Isabelle Benoit-Gelber, Email: isabelle.benoit@concordia.ca.
Ronald P. De Vries, Email: r.devries@westerdijkinstitute.nl
References
- 1.Benoit I, Culleton H, Zhou M, DiFalco M, Aguilar-Osorio G, Battaglia E, et al. Closely related fungi employ diverse enzymatic strategies to degrade plant biomass. Biotechnol Biofuels. 2015;8. [DOI] [PMC free article] [PubMed]
- 2.de Vries RP, Visser J. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol Mol Biol Rev. 2001;65:497–522. doi: 10.1128/MMBR.65.4.497-522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotech. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 5.Todd R, Zhao M, Ohm RA, Leeggangers H, Visser L, de Vries R. Prevalence of transcription factors in ascomycete and basidiomycete fungi. BMC Genomics. 2014;15:214. doi: 10.1186/1471-2164-15-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kowalczyk JE, Benoit I, de Vries RP. Regulation of plant biomass utilization in Aspergillus. Adv Appl Microbiol. 2014;88:31–56. doi: 10.1016/B978-0-12-800260-5.00002-4. [DOI] [PubMed] [Google Scholar]
- 7.van Peij NNME, Visser J, de Graaff LH. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol Microbiol. 1998;27:131–142. doi: 10.1046/j.1365-2958.1998.00666.x. [DOI] [PubMed] [Google Scholar]
- 8.vanKuyk PA, Benen JAE, Wösten HAB, Visser J, de Vries RPA. Broader role for AmyR in Aspergillus niger: regulation of the utilisation of D-glucose or D-galactose containing oligo- and polysaccharides. Appl Microbiol Biotechnol. 2012;93:285–293. doi: 10.1007/s00253-011-3550-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan X-L, Roubos JA, van den Hondel CAMJJ, Ram AFJ. Identification of InuR, a new Zn(II)2Cys6 transcriptional activator involved in the regulation of inulinolytic genes in Aspergillus niger. Mol Gen Genomics. 2008;279:11–26. doi: 10.1007/s00438-007-0290-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruben BS, Zhou M, Wiebenga A, Ballering J, Overkamp KM, Punt PJ, et al. Aspergillus niger RhaR, a regulator involved in L-rhamnose release and catabolism. Appl Microbiol Biotechnol. 2014;98:5531–5540. doi: 10.1007/s00253-014-5607-9. [DOI] [PubMed] [Google Scholar]
- 11.Ogawa M, Kobayashi T, Koyama Y. ManR, a transcriptional regulator of the β-mannan utilization system, controls the cellulose utilization system in Aspergillus oryzae. Biosci Biotechnol Biochem. 2013;77:426–429. doi: 10.1271/bbb.120795. [DOI] [PubMed] [Google Scholar]
- 12.Ogawa M, Kobayashi T, Koyama Y. ManR, a novel Zn(II)2Cys6 transcriptional activator, controls the β-mannan utilization system in Aspergillus oryzae. Fungal Genet Biol. 2012;49:987–995. doi: 10.1016/j.fgb.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Raulo R, Kokolski M, Archer DB. The roles of the zinc finger transcription factors XlnR, ClrA and ClrB in the breakdown of lignocellulose by Aspergillus niger. AMB Express. 2016;6:5. doi: 10.1186/s13568-016-0177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruben BS, Zhou M, de Vries RP. GalX regulates the D-galactose oxido-reductive pathway in Aspergillus niger. FEBS Lett. 2012;586:3980–3985. doi: 10.1016/j.febslet.2012.09.029. [DOI] [PubMed] [Google Scholar]
- 15.Alazi E, Niu J, Kowalczyk JE, Peng M, Aguilar Pontes MV, van Kan JAL, et al. The transcriptional activator GaaR of Aspergillus niger is required for release and utilization of D-galacturonic acid from pectin. FEBS Lett. 2016;590:1804–1815. doi: 10.1002/1873-3468.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vries RP. Regulation of Aspergillus genes encoding plant cell wall polysaccharide-degrading enzymes; relevance for industrial production. Appl Microbiol Biotechnol. 2003;61:10–20. doi: 10.1007/s00253-002-1171-9. [DOI] [PubMed] [Google Scholar]
- 17.Gomi K, Akeno T, Minetoki T, Ozeki K, Kumagai C, Okazaki N, et al. Molecular cloning and characterization of a transcriptional activator gene, amyR, involved in the amylolytic gene expression in Aspergillus oryzae. Biosci Biotechnol Biochem. 2000;64:816–827. doi: 10.1271/bbb.64.816. [DOI] [PubMed] [Google Scholar]
- 18.Tani S, Katsuyama Y, Hayashi T, Suzuki H, Kato M, Gomi K, et al. Characterization of the amyR gene encoding a transcriptional activator for the amylase genes in Aspergillus nidulans. Curr Genet. 2001;39:10–15. doi: 10.1007/s002940000175. [DOI] [PubMed] [Google Scholar]
- 19.Gielkens MMC, Dekkers E, Visser J, de Graaff LH. Two cellobiohydrolase-encoding genes from Aspergillus niger require D-xylose and the xylanolytic transcriptional activator XlnR for their expression. Appl Environ Microbiol. 1999;65:4340–4345. doi: 10.1128/aem.65.10.4340-4345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasper AA, Dekkers E, van Mil M, van de Vondervoort PJI, de Graaff LH. EglC, a new endoglucanase from Aspergillus niger with major activity towards xyloglucan. Appl Environ Microbiol. 2002;68:1556–1560. doi: 10.1128/AEM.68.4.1556-1560.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Peij NNME, Gielkens MMC, de Vries RP, Visser J, de Graaff LH. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl Environ Microbiol. 1998;64:3615–3619. doi: 10.1128/aem.64.10.3615-3619.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Battaglia E, Visser L, Nijssen A, van Veluw GJ, Wösten HAB, de Vries RP. Analysis of regulation of pentose utilisation in Aspergillus niger reveals evolutionary adaptations in Eurotiales. Stud Mycol. 2011;69:31–38. doi: 10.3114/sim.2011.69.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Vries RP, van den Broeck HC, Dekkers E, Manzanares P, de Graaff LH, Visser J. Differential expression of three α-galactosidase genes and a single β-galactosidase gene from Aspergillus niger. Appl Environ Microbiol. 1999;65:2453–2460. doi: 10.1128/aem.65.6.2453-2460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battaglia E, Zhou M, de Vries RP. The transcriptional activators AraR and XlnR from Aspergillus niger regulate expression of pentose catabolic and pentose phosphate pathway genes. Res Microbiol. 2014;165:531–540. doi: 10.1016/j.resmic.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Christensen U, Gruben BS, Madrid S, Mulder H, Nikolaev I, de Vries RP. Unique regulatory mechanism for D-galactose utilization in Aspergillus nidulans. Appl Environ Microbiol. 2011;77. [DOI] [PMC free article] [PubMed]
- 26.de Vries R, Riley R, Wiebenga A, Aguilar-Osorio G, Amillis S, Akemi Uchima C, et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017;18:28. doi: 10.1186/s13059-017-1151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khosravi C, Kun RS, Visser J, Aguilar-Pontes MV, de Vries RP, Battaglia E. In vivo functional analysis of L-rhamnose metabolic pathway in Aspergillus niger: a tool to identify the potential inducer of RhaR. BMC Microbiol. 2017;17:214. doi: 10.1186/s12866-017-1118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sloothaak J, Odoni DI, dos Santos VAP M, Schaap PJ, Tamayo-Ramos JA. Identification of a novel L-rhamnose uptake transporter in the filamentous fungus Aspergillus niger. PLoS Genet. 2016;12:e1006468. doi: 10.1371/journal.pgen.1006468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alazi E, Khosravi C, Homan T, du Pré S, Arentshorst M, Di Falco M, et al.. The pathway intermediate 2-keto-3-deoxy-L-galactonate mediates the induction of genes involved in D-galacturonic acid release and catabolism. FEBS Lett. 2017;591:1408–18. [DOI] [PMC free article] [PubMed]
- 30.Gielkens M, Visser J, de Graaff L. Arabinoxylan degradation by fungi: characterization of the arabinoxylan-arabinofuranohydrolase encoding genes from Aspergillus niger and Aspergillus tubingensis. Curr Genet. 1997;31:22–29. doi: 10.1007/s002940050172. [DOI] [PubMed] [Google Scholar]
- 31.de Vries RP, Visser J, de Graaff LH. CreA modulates the XlnR-induced expression on xylose of Aspergillus niger genes involved in xylan degradation. Res Microbiol. 1999;150:281–285. doi: 10.1016/S0923-2508(99)80053-9. [DOI] [PubMed] [Google Scholar]
- 32.de Vries RP, Flipphi MJA, Witteveen CFB, Visser J. Characterization of an Aspergillus nidulans L-arabitol dehydrogenase mutant. FEMS Microbiol Lett. 1994;123:83–90. doi: 10.1111/j.1574-6968.1994.tb07205.x. [DOI] [PubMed] [Google Scholar]
- 33.Flipphi MJA, Visser J, van der Veen P, de Graaff LH. Arabinase gene expression in Aspergillus niger: indications for coordinated regulation. Microbiology. 1994;140:2673–2682. doi: 10.1099/00221287-140-10-2673. [DOI] [PubMed] [Google Scholar]
- 34.Kowalczyk J, Lubbers R, Peng M, Battaglia E, Visser J, de Vries R. Combinational control of gene expression in Aspergillus niger grown on complex pectins. Sci Rep. 2017;7:12356. [DOI] [PMC free article] [PubMed]
- 35.de Vries RP, Jansen J, Aguilar G, Pařenicová L, Joosten V, Wülfert F, et al. Expression profiling of pectinolytic genes from Aspergillus niger. FEBS Lett. 2002;530:41–47. doi: 10.1016/S0014-5793(02)03391-4. [DOI] [PubMed] [Google Scholar]
- 36.de Vries RP, Visser J. Regulation of the feruloyl esterase (faeA) gene from Aspergillus niger. Appl Environ Microbiol. 1999;65:5500–5503. doi: 10.1128/aem.65.12.5500-5503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jørgensen TR, Goosen T, van den Hondel CA, Ram AFJ, Iversen JJL. Transcriptomic comparison of Aspergillus niger growing on two different sugars reveals coordinated regulation of the secretory pathway. BMC Genomics. 2009;10:44. doi: 10.1186/1471-2164-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coutinho P, Andersen M, Kolenova K, VanKuyk P, Benoit I, Gruben B, et al. Post-genomic insights into the plant polysaccharide degradation potential of Aspergillus nidulans and comparison to Aspergillus niger and Aspergillus oryzae. Fungal Genet Biol. 2009;46:S161–S169. doi: 10.1016/j.fgb.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 39.Martens-Uzunova ES, Schaap PJ. Assessment of the pectin degrading enzyme network of Aspergillus niger by functional genomics. Fungal Genet Biol. 2009;46:S170–S179. doi: 10.1016/j.fgb.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 40.Martens-Uzunova ES, Zandleven JS, Benen JAE, Awad H, Kools HJ, Beldman G, et al. A new group of exo-acting family 28 glycoside hydrolases of Aspergillus niger that are involved in pectin degradation. Biochem J. 2006;400:43–52. doi: 10.1042/BJ20060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan X-L, van der Kaaij RM, van den Hondel CAMJJ, Punt PJ, van der Maarel MJEC, Dijkhuizen L, et al. Aspergillus niger genome-wide analysis reveals a large number of novel α-glucan acting enzymes with unexpected expression profiles. Mol Gen Genomics. 2008;279:545–561. doi: 10.1007/s00438-008-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korman D, Bayliss F, Barnett C, Carmona C, Kodama K, Royer T, et al. Cloning, characterization, and expression of two alpha-amylase genes from Aspergillus niger var. awamori. Curr Genet. 1990;17:203–212. doi: 10.1007/BF00312611. [DOI] [PubMed] [Google Scholar]
- 43.Yuan X-L, Goosen C, Kools H, van den Maarel MJEC, van den Hondel CAMJJ, Dijkhuizen L, et al. Database mining and transcriptional analysis of genes encoding inulin-modifying enzymes of Aspergillus niger. Microbiology. 2006;152:3061–3073. doi: 10.1099/mic.0.29051-0. [DOI] [PubMed] [Google Scholar]
- 44.Ademark P, de Vries RP, Hägglund P, Stålbrand H, Visser J. Cloning and characterization of Aspergillus niger genes encoding an α-galactosidase and a β-mannosidase involved in galactomannan degradation. Eur J Biochem. 2001;268:2982–2990. doi: 10.1046/j.1432-1327.2001.02188.x. [DOI] [PubMed] [Google Scholar]
- 45.Tian C, Beeson WT, Iavarone AT, Sun J, Marletta MA, Cate JHD, et al. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc Natl Acad Sci U S A. 2009;106:22157–22162. doi: 10.1073/pnas.0906810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flipphi MJA, van Heuvel M, van der Veen P, Visser J, de Graaff LH. Cloning and characterization of the abfB gene coding for the major α-L-arabinofuranosidase (ABF B) of Aspergillus niger. Curr Genet. 1993;24:525–532. doi: 10.1007/BF00351717. [DOI] [PubMed] [Google Scholar]
- 47.de Souza WR, de Gouvea PF, Savoldi M, Malavazi I, de Souza Bernardes LA, Goldman MHS, et al. Transcriptome analysis of Aspergillus niger grown on sugarcane bagasse. Biotechnol Biofuels. 2011;4:40. doi: 10.1186/1754-6834-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Vries RP, VanKuyk PA, Kester HCM, Visser J. The Aspergillus niger faeB gene encodes a second feruloyl esterase involved in pectin and xylan degradation and is specifically induced in the presence of aromatic compounds. Biochem J. 2002;363:377–386. doi: 10.1042/bj3630377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dilokpimol A, Mäkelä MR, Aguilar-Pontes MV, Benoit-Gelber I, Hildén KS, de Vries RP. Diversity of fungal feruloyl esterases: updated phylogenetic classification, properties, and industrial applications. Biotechnol Biofuels. 2016;9:231. doi: 10.1186/s13068-016-0651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bussink HJD, Brouwer KB, de Graaff LH, Kester HCM, Visser J. Identification and characterization of a second polygalacturonase gene of Aspergillus niger. Curr Genet. 1991;20:301–307. doi: 10.1007/BF00318519. [DOI] [PubMed] [Google Scholar]
- 51.de Vries RP, Frisvad JC, van de Vondervoort PJI, Burgers K, Kuijpers AFA, Samson RA, et al. Aspergillus vadensis, a new species of the group of black Aspergilli. Antonie Van Leeuwenhoek. 2005;87:195–203. doi: 10.1007/s10482-004-3194-y. [DOI] [PubMed] [Google Scholar]
- 52.Goosen C, Yuan X-L, van Munster JM, Ram AFJ, van der Maarel MJEC, Dijkhuizen L. Molecular and biochemical characterization of a novel intracellular invertase from Aspergillus niger with transfructosylating activity. Eukaryot Cell. 2007;6:674–681. doi: 10.1128/EC.00361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siegel D, Marton I, Dekel M, Bravdo B-A, He S, Withers SG, et al. Cloning, expression, characterization, and nucleophile identification of family 3, Aspergillus niger β-glucosidase. J Biol Chem. 2000;275:4973–4980. doi: 10.1074/jbc.275.7.4973. [DOI] [PubMed] [Google Scholar]
- 54.de Groot MJL, van de Vondervoort PJI, de Vries RP, VanKuyk PA, Ruijter GJG, Visser J. Isolation and characterization of two specific regulatory Aspergillus niger mutants shows antagonistic regulation of arabinan and xylan metabolism. Microbiology. 2003;149:1183–1191. doi: 10.1099/mic.0.25993-0. [DOI] [PubMed] [Google Scholar]
- 55.Li J, Lin L, Li H, Tian C. Transcriptional comparison of the filamentous fungus Neurospora crassa growing on three major monosaccharides D-glucose, D-xylose and L-arabinose. Biotechnol Biofuels. 2014;7:31. doi: 10.1186/1754-6834-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.dos Santos CL, de Paula RG, Antoniêto ACC, Persinoti GF, Silva-Rocha R, Silva RN. Understanding the role of the master regulator xyr1 in Trichoderma reesei by global transcriptional analysis. Front Microbiol. 2016;7:175. doi: 10.3389/fmicb.2016.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun J, Tian C, Diamond S, Glass NL. Deciphering transcriptional regulatory mechanisms associated with hemicellulose degradation in Neurospora crassa. Eukaryot Cell. 2012;11:482–493. doi: 10.1128/EC.05327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benz JP, Chau BH, Zheng D, Bauer S, Glass NL, Somerville CRA. Comparative systems analysis of polysaccharide-elicited responses in Neurospora crassa reveals carbon source-specific cellular adaptations. Mol Microbiol. 2014;91:275–299. doi: 10.1111/mmi.12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benoit I, Zhou M, Vivas Duarte A, Downes D, Todd RB, Kloezen W, Post H, Heck AJ, Maarten Altelaar AF, de Vries RP. Spatial differentiation of gene expression in Aspergillus niger colony grown for sugar beet pulp utilization. Sci Rep. 2015;5:13592. doi: 10.1038/srep13592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 61.Core Team R. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 62.Bolstad BM. Comparing the effects of background, normalization and summarization on gene expression estimates. 2002. http://bmbolstad.com/stuff/components.pdf. Accessed June 2017.
- 63.Baldi P, Long ADA. Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 64.gibbons FD, Roth FP. Judging the quality of gene expression-based clustering methods using gene annotation. Genome Res. 2002;12:1574–1581. doi: 10.1101/gr.397002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 66.de Groot MJL, van den Dool C, Wösten HAB, Levisson M, VanKuyk PA, Ruijter GJG, et al. Regulation of the pentose catabolic pathway of Aspergillus niger. Food Technol Biotechnol. 2007;45:134–138. [Google Scholar]
- 67.Bos CJ, Debets AJM, Swart K, Huybers A, Kobus G, Slakhorst SM. Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr Genet. 1988;14:437–443. doi: 10.1007/BF00521266. [DOI] [PubMed] [Google Scholar]
- 68.Flipphi MJA, Visser J, van der Veen P, de Graaff LH. Cloning of the Aspergillus niger gene encoding α-l-arabinofuranosidase a. Appl Microbiol Biotechnol. 1993;39:335–340. doi: 10.1007/BF00192088. [DOI] [PubMed] [Google Scholar]
- 69.van Peij NN, Brinkmann J, Vrsanská M, Visser J, de Graaff LH. β-Xylosidase activity, encoded by xlnD, is essential for complete hydrolysis of xylan by Aspergillus niger but not for induction of the xylanolytic enzyme spectrum. Eur J Biochem. 1997;245:164–173. doi: 10.1111/j.1432-1033.1997.00164.x. [DOI] [PubMed] [Google Scholar]
- 70.de Vries R, van de Vondervoort P, Hendriks L, van de Belt M, Visser J. Regulation of the α-glucuronidase-encoding gene (aguA) from Aspergillus niger. Mol Gen Genomics. 2002;268:96–102. doi: 10.1007/s00438-002-0729-7. [DOI] [PubMed] [Google Scholar]
- 71.Khanh NQ, Ruttkowski E, Leidinger K, Albrecht H, Gottschalk M. Characterization and expression of a genomic pectin methyl esterase-encoding gene in Aspergillus niger. Gene. 1991;106:71–77. doi: 10.1016/0378-1119(91)90567-U. [DOI] [PubMed] [Google Scholar]
- 72.Harmsen JAM, Kusters-van Someren MA, Visser J. Cloning and expression of a second Aspergillus niger pectin lyase gene (pelA): indications of a pectin lyase gene family in A. niger. Curr Genet. 1990;18:161–166. doi: 10.1007/BF00312604. [DOI] [PubMed] [Google Scholar]
- 73.Kusters-van Someren MA, Harmsen JAM, Kester HCM, Visser J. Structure of the Aspergillus niger pelA gene and its expression in Aspergillus niger and Aspergillus nidulans. Curr Genet. 1991;20:293–299. doi: 10.1007/BF00318518. [DOI] [PubMed] [Google Scholar]
- 74.de Vries RP, Kester HCM, Poulsen CH, Benen JAE, Visser J. Synergy between enzymes from Aspergillus involved in the degradation of plant cell wall polysaccharides. Carbohydr Res. 2000;327:401–410. doi: 10.1016/S0008-6215(00)00066-5. [DOI] [PubMed] [Google Scholar]
- 75.Damveld RA, VanKuyk PA, Arentshorst M, Klis FM, van den Hondel CAMJJ, Ram AFJ. Expression of agsA, one of five 1,3-α-D-glucan synthase-encoding genes in Aspergillus niger, is induced in response to cell wall stress. Fungal Genet Biol. 2005;42:165–177. doi: 10.1016/j.fgb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 76.Boel E, Hansen MT, Hjort I, Høegh I, Fiil NP. Two different types of intervening sequences in the glucoamylase gene from Aspergillus niger. EMBO J. 1984;3:1581–1585. doi: 10.1002/j.1460-2075.1984.tb02014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fowler T, Berka RM, Ward M. Regulation of the glaA gene of Aspergillus niger. Curr Genet. 1990;18:537–545. doi: 10.1007/BF00327025. [DOI] [PubMed] [Google Scholar]
- 78.Nakamura A, Nishimura I, Yokoyama A, Lee D-G, Hidaka M, Masaki H, et al. Cloning and sequencing of an α-glucosidase gene from Aspergillus niger and its expression in A. nidulans. J Biotechnol. 1997;53:75–84. doi: 10.1016/S0168-1656(97)01664-7. [DOI] [PubMed] [Google Scholar]
- 79.Ohta K, Akimoto H, Matsuda S, Toshimitsu D, Nakamura T. Molecular cloning and sequence analysis of two endoinulinase genes from Aspergillus niger. Biosci Biotechnol Biochem. 1998;62:1731–1738. doi: 10.1271/bbb.62.1731. [DOI] [PubMed] [Google Scholar]
- 80.Parenicová L, Benen JA, Kester HC, Visser J. pgaA and pgaB encode two constitutively expressed endopolygalacturonases of Aspergillus niger. Biochem J. 2000;345:637–44. [PMC free article] [PubMed]
- 81.Bergès T, Barreau C, Peberdy JF, Boddy LM. Cloning of an Aspergillus niger invertase gene by expression in Trichoderma reesei. Curr Genet. 1993;24:53–59. doi: 10.1007/BF00324665. [DOI] [PubMed] [Google Scholar]
- 82.Somiari RI, Brzeski H, Tate R, Bieleck S, Polak J. Cloning and sequencing of an Aspergillus niger gene coding for β-fructofuranosidase. Biotechnol Lett. 1997;19:1243–1247. doi: 10.1023/A:1018454424640. [DOI] [Google Scholar]
- 83.Flipphi M, Panneman H, van der Veen P, Visser J, de Graaff L. Molecular cloning, expression and structure of the endo-1,5-α-L-arabinase gene of Aspergillus niger. Appl Microbiol Biotechnol. 1993;40:318–326. doi: 10.1007/BF00170387. [DOI] [PubMed] [Google Scholar]
- 84.de Vries RP, Pařenicová L, Hinz SWA, Kester HCM, Beldman G, Benen JAE, et al. The β-1,4-endogalactanase a gene from Aspergillus niger is specifically induced on arabinose and galacturonic acid and plays an important role in the degradation of pectic hairy regions. Eur J Biochem. 2002;269:4985–4993. doi: 10.1046/j.1432-1033.2002.03199.x. [DOI] [PubMed] [Google Scholar]
- 85.Kumar V, Ramakrishnan S, Teeri T, Knowles J, Hartley B. Saccharomyces cerevisiae cells secreting an Aspergillus niger β-galactosidase grow on whey permeate. Biotechnology. 1992;10:82–85. doi: 10.1038/nbt0192-82. [DOI] [PubMed] [Google Scholar]
- 86.van der Vlugt-Bergmans CJB, Meeuwsen PJA, Voragen AGJ, van Ooyen AJJ. Endo-xylogalacturonan hydrolase, a novel pectinolytic enzyme. Appl Environ Microbiol. 2000;66:36–41. doi: 10.1128/AEM.66.1.36-41.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gysler C, Harmsen J, Kester H, Visser J, Heim J. Isolation and structure of the pectin lyase D-encoding gene from Aspergillus niger. Gene. 1990;30:101–108. doi: 10.1016/0378-1119(90)90211-9. [DOI] [PubMed] [Google Scholar]
- 88.Suykerbuyk ME, Kester HC, Schaap PJ, Stam H, Musters W, Visser J. Cloning and characterization of two rhamnogalacturonan hydrolase genes from Aspergillus niger. Appl Environ Microbiol. 1997;63:2507–2515. doi: 10.1128/aem.63.7.2507-2515.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.den Herder IF, Mateo Rosell AM, van Zuilen CM, Punt PJ, van den Hondel CAMJJ. Cloning and expression of a member of the Aspergillus niger gene family encoding α-galactosidase. Mol Gen Genet. 1992;233:404–410. doi: 10.1007/BF00265437. [DOI] [PubMed] [Google Scholar]
- 90.Kulik N, Weignerová L, Filipi T, Pompach P, Novák P, Mrázek H, et al. The α-galactosidase type a gene aglA from Aspergillus niger encodes a fully functional α-N-acetylgalactosaminidase. Glycobiology. 2010;20:1410–1419. doi: 10.1093/glycob/cwq105. [DOI] [PubMed] [Google Scholar]
- 91.Benen JAE, Kester HCM, Pařenicová L, Visser J. Characterization of Aspergillus niger pectate lyase a. Biochemistry. 2000;39:15563–15569. doi: 10.1021/bi000693w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression profiling tree containing 168 A. niger genes encoding putative CAZymes (www.cazy.org). Clusters A-I can be distinguished. (PDF 1780 kb)
Expression of selected CAZy genes. (XLSX 215 kb)
Significantly and not significantly expressed genes encoding CAZymes in A. niger CBS513.88 in this study. (XLSX 19 kb)
PCA analysis of the gene expression values of the biological duplicate samples revealing the reproducibility of the duplicates. (PDF 90 kb)
Data Availability Statement
The data sets supporting the results of this article are available in the GEO repository under accession number GSE98572 (token: cbszoqmqddgndkf).


