Abstract
While screening of small-molecular metabolites produced by most cultivatable microorganisms often results in rediscovery of known compounds, genome-mining programs allow to harness much greater chemical diversity and result in discovery of new molecular scaffolds. Here we report genome-guided identification of a new antibiotic klebsazolicin (KLB) from Klebsiella pneumoniae that inhibits growth of sensitive cells by targeting ribosome. A member of ribosomally-synthesized post-translationally modified peptides (RiPPs), KLB is characterized by the presence of unique N-terminal amidine ring essential for its activity. Biochemical in vitro studies indicate that KLB inhibits ribosome by interfering with translation elongation. Structural analysis of the ribosome-KLB complex reveals the compound bound in the peptide exit tunnel overlapping with the binding sites of macrolides or streptogramins-B. KLB adopts compact conformation and largely obstructs the tunnel. Engineered KLB fragments retain in vitro activity and can serve as a starting point for the development of new bioactive compounds.
INTRODUCTION
Since the discovery of the first antibiotics, natural products of microbial origin have been recognized as a highly valuable source of lead compounds for the development of new therapeutic agents1,2. Increasing instances of multidrug resistant bacteria and dissemination of plasmid-mediated resistance to the last-resort antibiotics stimulate development of new approaches to revive natural products discovery pipeline and enrich the arsenal of structural scaffolds suitable for optimization by medicinal chemists3–5.
Analysis of genomic data allows identification of gene clusters encoding biosynthetic pathways for potential drug candidates, which may otherwise escape attention due to their inactivity under laboratory growth conditions6. Human microbiome has been shown to contain a wealth of antibiotic biosynthesis genes and recently two new promising antibiotics, lactocillin and lugdunin, isolated from human commensals have been reported7,8. Ribosomally-synthesized post-translationally modified peptides (RiPPs) are among the most abundant antimicrobial agents synthesized by human microbiota7,9. Ribosomes are employed for the synthesis of RiPPs precursors, which next undergo post-translational modifications by dedicated enzymes encoded in compact gene clusters10. A frequently found feature of the modifying enzymes is their ability to sequentially modify multiple residues of cognate precursor peptide, which consists of a leader peptide and a core peptide. The leader peptide is usually removed prior to formation of the final biologically active product but it ensures recognition of the core peptide by the enzyme(s) during modification11. A prominent example is the YcaO-domain-containing enzymes12–14. YcaO is an ATP-dependent catalytic unit of cyclodehydratase of thiazole/oxazole-modified microcins (TOMMs)15. YcaO directly activates amide linkages adjacent to particular Cys/Ser/Thr residues across core peptide with subsequent events of cyclization leading to formation of thiazoline and (methyl)-oxazoline heterocycles. TOMM-cyclodehydratase is often accompanied by dehydrogenase that oxidizes all or some of the newly formed azolines to azoles16,17. Activation of amide carbonyl oxygen by YcaO-enzymes also might be involved in macrocyclization via formation of amidine bond, as it was proposed for the formation of the protein synthesis inhibitor bottromycin18. Comprehensive phylogenetic analysis of YcaO-containing gene clusters allowed cataloguing of nearly 1500 predicted TOMMs allocated in 13 families with at least one experimentally validated member and at least 9 additional putative families with no characterised representatives17. Unexplored TOMMs represent an untapped source of new molecular scaffolds.
In this study we report genome-guided identification and characterization of structurally unique TOMM from opportunistic human pathogen Klebsiellae pneumonia sp. ozaenae, klebsazolicin (KLB) that is a first member of a new class of protein synthesis inhibitors. We present biochemical, genetic and structural evidence that KLB targets the bacterial ribosome. KLB inhibits ribosome by interfering with the early stages of translation elongation. KLB binds to the bacterial ribosome in the nascent peptide exit tunnel in a site, which overlaps with that of macrolides and other inhibitors. However, unlike macrolides that obstruct the tunnel partially, the extended backbone of the KLB molecule adopts a compact conformation, crossing the tunnel several times and essentially plugging, making the passage of the nascent peptide unlikely. Engineered fragments of KLB retain in vitro activity and can serve as a starting point for the development of a new family of bioactive compounds.
RESULTS AND DISCUSSION
Identification of a new TOMM
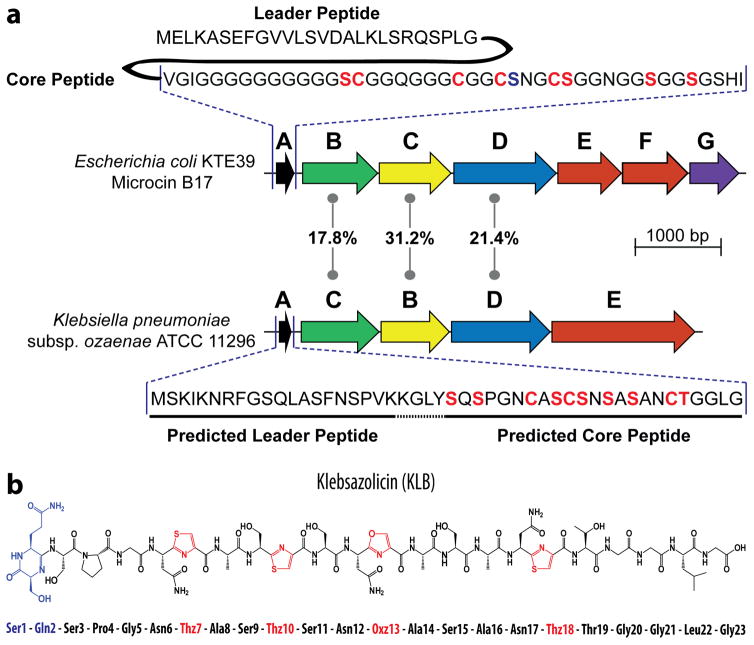
Microcin B17 (MccB17), a potent DNA-gyrase inhibitor encoded by a gene cluster mcbABCDEFG in some strains of Escherichia coli19, is an archetypal TOMM. Amino acid sequences of McbB, McbC, and McbD, the components of MccB17-synthetase complex, were used as queries in a PSI-BLASTP search against NCBI non-redundant protein database. A full set of homologous proteins was identified in the genome of Klebsiella pneumoniae subspecies ozaenae ATCC 11296. Analysis of the corresponding genomic region revealed that the genes encoding the identified homologs are organized in an operon-like gene cluster, which also includes genes for an ABC-transporter and a putative precursor peptide enriched with Ser and Cys residues (klpABCDE, Fig. 1a). The sequence of the putative precursor peptide KlpA does not match those of catalogued TOMMs, indicating that mature modified KlpA may have unusual structural and functional properties.
Figure 1. Organization of the biosynthetic gene cluster and production of klebsazolicin by E. coli host.
(a) Comparison of organization of biosynthetic gene clusters mcbABCDEFG for microcin B (top) and klpABCDE for KLB (bottom). Genes of the clusters are schematically depicted as coloured arrows. Homologous genes have the same colours. The extent of identity between the amino acid sequences of the homologous proteins forming BCD synthetase is indicated in the middle. The mcbEF genes encode the two-component ABC transporter that ensures export of mature MccB17. The mcbG gene encodes the immunity protein. The klpE gene is predicted to encode an ABC transporter. Gene cluster klpABCDE is annotated in agreement with the community consensus10. Amino acid sequence for the MccB17 precursor peptide (McbA) is shown above its gene cluster; residues converted to azoles are indicated in red, leader peptide is cleaved after maturation. Gene product of klpA is shown below its gene cluster with Ser, Cys, Thr residues of predicted core peptide labeled in red. (b) Chemical structure of KLB and its amino acid sequence.
Expression and structural characterization of a new TOMM
To investigate whether the expression of klpABCDE gene cluster leads to production of a TOMM, the entire cluster was expressed in a surrogate E. coli host using two-plasmid system that allows inducible separate expression of klpA and the klpBCDE genes. Supernatants of the bacterial cell cultures from induced and uninduced cells were analysed by reverse-phase HPLC and mass spectrometry (Supplementary Results, Supplementary Fig. 1a). A compound with a molecular mass of 1973.684 Da appeared after induction of the klp genes (Supplementary Fig. 1b). Its UV-absorbance spectrum was characteristic of azole-containing peptides (Supplementary Fig. 1c). We named the compound klebsazolicin (KLB, 1) and purified it from cultured medium by solid-phase extraction on a C18-cartridge followed by reverse-phase HPLC.
Mass-spectrometric and sequence analysis of the precursor peptide KlpA allowed identification of a core peptide that remains after the leader peptide is cleaved off – S1Q2S3P4G5N6C7A8S9C10S11N12S13A14S15A16N17C18T19G20G21L22G23 – and provided initial information on the residues involved in formation of azoles: Cys7, Cys10, Ser13 and Cys18 (Supplementary Fig. 2). Surprisingly, the first two N-terminal amino acids (Ser1 and Gln2) of the KLB were also involved in a modification leading to a loss of a water molecule (−18 Da). We were unable to map the exact position of this modification by mass spectrometry due to the lack of MS/MS fragmentation in this region. Therefore, the chemical structure of KLB was determined by two-dimensional NMR (Supplementary Table 1, Supplementary Fig. 3). We found that N-terminal amino acid residues Ser1 and Gln2 form a 6-membered heterocycle via amidine linkage (Fig. 1b). Interestingly, similar amidine linkage is found in bottromycin20 where its formation is seemingly catalyzed by a stand-alone YcaO protein18. In the case of KLB, the KlpBCD maturation complex seems to be responsible not only for the formation of the azole rings, but also for the activation of carbonyl of the Gln2 which ultimately leads to the linkage between the N-terminus of the core peptide (formed after the cleavage of the leader peptide) and the peptide backbone to yield the N-terminal amidine heterocycle.
KLB exhibits narrow spectrum of antibacterial activity
Antimicrobial activity of KLB was assessed by broth microdilution assay based on CSLI guidelines using a panel of Gram-positive and Gram-negative bacteria. KLB inhibits growth of E. coli, Klebsiella pneumoniae and Yersinia pseudotuberculosis in LB medium at 16–65 μM (Table 1). The growth of other Gram-negative bacteria, including Acinetobacter baumannii, Pseudomonas aeruginosa, Pseudomonas putida, Vibrio cholerae, as well as of all Gram-positive bacteria tested was not affected. KLB exhibited only bacteriostatic activity against sensitive Gram-negative species (Supplementary Fig. 4). The observed narrow spectrum of antibacterial activity of KLB is common to microcins, a class of antimicrobial peptides, which often require cellular membrane proteins, such as porins, receptors and ATP-dependent transporters, for penetration into cells. For example, in order to cross the outer membrane microcin J25 uses the FhuA ferrichrome receptor21; microcins MccB17 and C employ the OmpF porin22; microcins E492, M, H47 utilize catecholate siderophore receptors Fiu, Cir, FepA23. Microcins must also have a route across the inner membrane, such as SbmA24,25 or YejABEF26 inner-membrane proteins. Our results suggest, that similarly to MccB17, KLB is transported inside the sensitive E. coli cells via OmpF and SbmA transporters, since deletions of the corresponding genes lead to decreased sensitivity (ΔompF) or even complete resistance (ΔsbmA) to KLB (Table 1), while overexpression of these genes causes increased sensitivity (Table 1). It should be noted that SbmA is also used by proline-rich antimicrobial peptides targeting ribosome, such as Bac7 and oncocin27. Therefore, the narrow spectrum of antibacterial activity observed for KLB is likely due to the absence of the suitable transporters in the unsusceptible tested bacterial species.
Table 1.
Klebsazolicin MICs for various bacterial strains in rich LB medium and minimal M9+Glucose medium.
| Microorganism (Strain) | MIC, μg/ml | MIC, μM | |
|---|---|---|---|
| LB medium | Escherichia coli B | 32 | 16 |
| Escherichia coli K-12 MG1655 | 128 | 65 | |
| Escherichia coli MG1655 (ΔompF) | 256 | 130 | |
| Escherichia coli MG1655 (ΔsbmA) | >1024 | >519 | |
| Escherichia coli MG1655 (pBAD-sbmA-ompF) | 64 | 32 | |
| Escherichia coli MG1655 (pBAD-klpE) | 1024 | 519 | |
| Yersinia pseudotuberculosis | 64 | 32 | |
| Klebsiella pneumoniae | 128 | 65 | |
| Vibrio cholerae | >1024 | >519 | |
| Acinetobacter baumannii | >1024 | >519 | |
| Pseudomonas aeruginosa | >1024 | >519 | |
| Pseudomonas putida | >1024 | >519 | |
| Staphylococcus aureus | >1024 | >519 | |
| Bacillus subtilis | >1024 | >519 | |
| Bacillus cereus | >1024 | >519 | |
| Mycobacterium smegmatis | >1024 | >519 | |
| Listeria grayi | >1024 | >519 | |
| Streptomyces lividans | >1024 | >519 | |
| M9+Glucose | Escherichia coli B | 4 | 2 |
| Escherichia coli K-12 MG1655 | 16 | 8 | |
| Klebsiella pneumoniae | 64 | 32 |
Interestingly, KLB is more potent on the minimal M9 medium as compared to the LB medium with minimal inhibitory concentrations (MIC) for E. coli MG1655 strain being 8 μM and 65 μM, respectively (Table 1). This is likely due to competition for the access to the import systems between the KLB and multiple nutrients abundant in LB and depleted in minimal M9 medium. Competition for access to import systems was shown to play a critical role in apparent activity levels of the tetra-peptide antibiotic GE8111228 and microcin C29. Most likely, resistance to KLB in the producing host arises from the activity of the klpE gene product, which could be responsible for the active export of the compound from the cell, since expression of klpE in E. coli results in 8-fold increase in the MIC value (Table 1).
KLB inhibits protein synthesis by acting on the ribosome
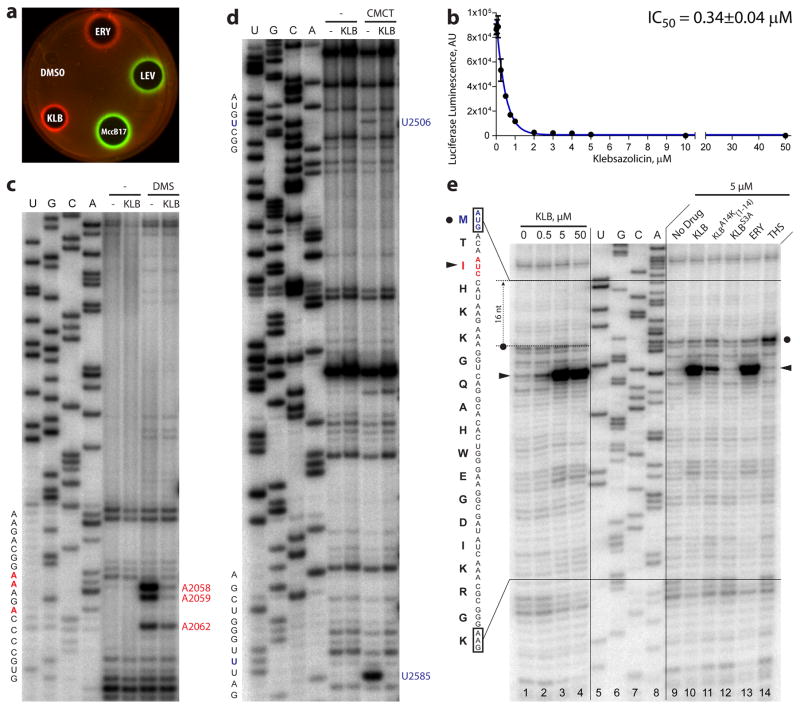
To gain insights into the mechanism of antibacterial action of KLB, we employed E. coli-based in vivo reporter system that is designed for screening of inhibitors targeting either DNA replication or protein synthesis30. In this assay, sub-inhibitory concentrations of antibacterial compounds that stall translation (e.g., erythromycin) induce expression of far-red fluorescent protein reporter Katushka2S (Fig. 2a, ERY, red pseudocolor ring). Compounds that trigger SOS response, such as inhibitors of DNA gyrase (e.g., levofloxacin), induce expression of Red Fluorescent Protein (RFP) reporter (Fig. 2a, LEV, green pseudocolor ring). As expected, MccB17, an inhibitor of DNA gyrase, induces expression of RFP (Fig. 2a, MccB17, green pseudocolor ring). KLB strongly induced expression of Katushka2S reporter, but not of RFP indicating that it inhibits protein synthesis and not the DNA replication (Fig. 2a, KLB, red pseudocolor ring).
Figure 2. Klebsazolicin is an inhibitor of protein synthesis both in vitro and in vivo.
(a) Induction of a two-color dual reporter system sensitive to inhibitors of the ribosome progression or inhibitors of DNA replication, respectively. Spots of erythromycin (ERY), levofloxacin (LEV), microcin B17 (MccB17), klebsazolicin (KLB) and solvent (DMSO) were placed on the surface of an agar plate containing E. coli ΔtolC cells transformed with the pDualrep2 plasmid30. Induction of expression of Katushka2S (red) is triggered by translation inhibitors, while RFP (green) is induced upon DNA damage. (b) Inhibition of protein synthesis by increasing concentrations of KLB in the in vitro cell-free translation in S30 extract. (c, d) KLB-dependent protection of the 23S rRNA nucleotides from chemical modification by DMS or CMCT, respectively. Sequencing lanes are shown on the left. Positions of the protected nucleotides are marked in red and blue. (e) Ribosome stalling by KLB on osmC mRNA as revealed by reverse transcription inhibition (toe-printing) in a recombinant cell-free translation system. Sequencing lanes for the osmC mRNA are shown in the middle. The sequence of the osmC mRNA and corresponding amino acid sequence are shown on the left. Band corresponding to the ribosome occupying the initiator codon is indicated by the black circle. Stalling of ribosomes at the third AUC (Ile) codon is shown by the black triangles. Note that due to the large size of the ribosome the reverse transcriptase stops at the nucleotide +16 relative to the codon located in the P site.
To confirm that KLB is a translation inhibitor we tested its ability to prevent in vitro synthesis of luciferase in E. coli S30 extract. We observed a dose-dependent inhibition of translation by KLB with IC50 = 0.34±0.04 μM (Fig. 2b, Supplementary Dataset 1). Importantly, the IC50 of KLB is within the range of values known for the other structurally unrelated ribosome targeting peptides, such as Onc112 (IC50 = 0.15 μM)31 and Bac7 (IC50 = 1 μM)32. For KLB and Onc112, but not for Bac7, the MIC values (65 μM and 3.5 μM31, respectively) are higher than IC50 values. This difference may reflect the fact that uptake is limiting antibacterial efficiency of Onc112 and, to a much greater degree, of KLB. The latter conjecture is supported by the observation that overexpression of sbmA and ompF transporter genes increases sensitivity to KLB (Table 1). Furthermore, there appears to be a correlation between the expression levels of the OmpF transporter in various E. coli strains and their sensitivity to KLB. For example, E. coli strain B, which has higher expression levels of OmpF compared to E. coli strain MG165533, also has higher sensitivity to KLB (Table 1).
A compound can inhibit protein synthesis by interfering with the activity of the ribosome or any of the various other molecules associated with translation process (translation factors, aminoacyl-tRNA synthetases, tRNAs, etc). In order to identify the target of KLB, we selected resistant mutants. An E. coli strain SQ110DTC34 that is particularly suited for identifying resistance mutations in the genes encoding rRNA was used. This strain lacks 6 out of 7 natural rrn alleles encoding rRNA. In addition, it is hypersensitive to many antibiotics due to the lack of the tolC gene, which encodes the major efflux pump in the outer membrane. To avoid accumulation of spontaneous mutations in sbmA and/or ompF genes that encode transporters for KLB uptake, the SQ110DTC strain was additionally transformed with the plasmid pBAD-sbmA-ompF overproducing these transporters. Applying ~109 SQ110DTC cells on an agar plate containing 200 μg/mL KLB (3-fold MIC) led to the appearance of several resistant colonies. Sequencing of the rRNA genes from four independent resistant clones revealed two mutations of the same 23S rRNA nucleotide: U2609G and U2609A (Supplementary Table 2). Nucleotide U2609 of the 23S rRNA is exposed at the surface of the peptide exit tunnel35,36 and its mutations were previously shown to confer resistance to ketolides37 pointing to the possibility of KLB binding in the same location.
To validate this hypothesis, we assessed KLB-dependent protection of nucleotides around the peptidyl transferase center (PTC) from chemical modification by dimethyl sulfate (DMS) or 1-cyclohexyl-3-(2-morpholinoethyl)-carbodiimide metho-p-toluenesulfonate (CMCT). KLB protected several nucleotides of the 23S rRNA around the PTC: A2058, A2059, U2506, U2585, and, to a lesser extent, A2062 (Fig. 2c, d). These results are also in accordance with the binding site of the KLB located in the peptide exit tunnel.
To elucidate the step of translation that is specifically inhibited by KLB, a primer extension inhibition (toe-printing) assay was used38. Toe-printing allows unambiguous identification of drug-induced ribosome stalling on mRNA with a single-nucleotide precision34. A large set of antibacterials targeting the ribosome has been already tested by toe-printing using osmC mRNA34. Therefore, in order to be able to directly compare our results for KLB with those for other antibiotics, we have chosen exactly the same experimental system. The addition of KLB to the PURExpress cell-free transcription-translation system programmed with the osmC mRNA results in a dose-dependent ribosome stalling at the third codon (Fig. 2e, lanes 1–4). We used thiostrepton and erythromycin, ribosome inhibitors with well-defined mechanisms of action, as controls for the inhibition of initiation and elongation steps, respectively. Addition of the thiostrepton leads to ribosome stalling at the 1st codon (Fig. 2e, lane 14), whereas erythromycin pauses translation at the third codon (Fig. 2e, lane 13), in agreement with previously published data on the mechanism of action of these antibiotics34. Thus, KLB emerges as an inhibitor of ribosome progression along mRNA causing stalling at a site similar to that observed upon the binding of chemically unrelated antibiotic erythromycin.
To further compare modes of action of KLB and erythromycin we analyzed the ability of these compounds to inhibit translation of hns mRNA that was reported previously to be partially erythromycin-resistant39. We observed strong inhibition of the full-length product accumulation on osmC mRNA by both erythromycin and KLB (Supplementary Fig. 5, lanes 4, 5, 6). However, the hns mRNA translation inhibition by either of the two compounds was only partial (Supplementary Fig. 5, lanes 1, 2, 3), again pointing to apparently similar modes of action by KLB and macrolides.
Altogether these results suggest that KLB exerts its antibacterial activity via inhibition of protein synthesis. It acts on the ribosome by binding to the peptide exit tunnel and inhibits the elongation of the polypeptide chain beyond more than three amino acid residues.
Extended KLB is curled in the peptide exit tunnel
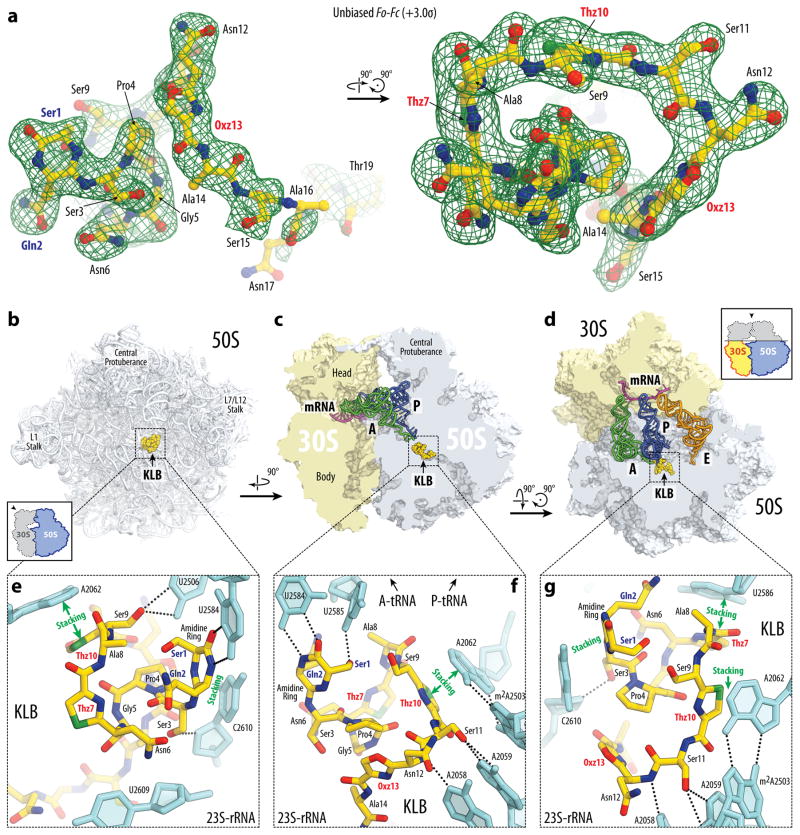
To unambiguously determine the mode of binding and action of KLB, we crystallized Thermus thermophilus 70S ribosomes in the presence of KLB, mRNA and A-, P-, and E-site tRNAs and determined the structure of the complex by X-ray crystallography at 2.7 Å resolution. The structure was solved by molecular replacement using atomic coordinates of the T. thermophilus 70S ribosome as a model. An unbiased difference Fourier map (Fig. 3a), which was calculated using the observed amplitudes from the crystal and the amplitudes and phases derived from a model of the ribosome without the bound antibiotic, revealed positive density peaks resembling characteristic features of the KLB chemical structure (Fig. 1b). A single binding site for KLB was observed within the peptide exit tunnel on the large ribosomal subunit (Fig. 3b–d; Movie S1).
Figure 3. Structure of KLB-70S ribosome complex with A- and P-tRNAs.
(a) Difference Fourier electron density map of KLB in complex with the T. thermophilus 70S ribosome (green mesh). The refined model of the compound is displayed in its respective electron density before refinement viewed from two different perspectives. The unbiased (Fobs – Fcalc) difference electron density map is contoured at 3.0σ. Carbon atoms are coloured yellow, nitrogens are blue, oxygens are red, and sulfurs are green. Note that only the first 19 residues of KLB are visible in the electron density. (b, c, d) Overview of the KLB binding site (yellow) on the T. thermophilus 70S ribosome viewed from three different perspectives. 30S subunit is shown in light yellow, 50S subunit is in light blue. mRNA is shown in magenta and tRNAs are displayed in green for the A-site, in dark blue for the P-site, and in orange for the E-site. In (b), the 50S subunit is viewed from the inter-subunit interface (30S subunit, mRNA and tRNAs are removed for clarity), as indicated by the inset. The view in (c) is from the cytoplasm onto the A site. The view in (d) is from the top after removing the head of the 30S subunit and protuberances of the 50S subunit, as indicated by the inset. (e, f, g) Close-up views of the KLB binding site shown in panels (b), (c), and (d), respectively. E. coli numbering of the nucleotides in the 23S rRNA is used.
The quality of the observed electron density and the resolution of the resulting structure allowed us to analyse interactions of KLB with the ribosome at atomic details. Importantly, the electron density confirms the presence of the amidine ring and azoles in the structure of ribosome-bound KLB (Fig. 3a, e–g). The N-terminal amidine cycle forms a number of contacts with the universally conserved nucleotides at the heart of the PTC (Fig. 3e–g; Movie S1). The amide linker connecting Ser1 and Gln2, that is enclosed into the 6-membered amidine ring, forms two H-bonds with U2584 (Fig. 3e, f). This interaction resembles a non-canonical U-U base-pair.
In agreement with the location of the resistance mutations, KLB forms extensive Van der Waals contacts with the U2609 nucleotide of the 23S rRNA (Fig. 3e). Also, in line with the observed KLB-dependent protection of the nucleotides U2506 and U2585 of the 23S rRNA against chemical modification by CMCT (Fig. 2d), hydroxyl group of the side chain of the Ser1 residue in KLB forms H-bond with the U2585 residue (Fig. 3f), while Ser9 interacts with the U2506 in a similar way (Fig. 3e). The observed protection of the nucleotides A2058 and A2059 of the 23S rRNA by KLB against DMS modification (Fig. 2c) can be explained by their direct interaction with the KLB (Fig. 3f, g). Several stacking interactions are formed between the heterocycles of the KLB and the nucleobases in the 23S rRNA (Fig. 3e–g). For example, the plane of the amidine ring stacks over the C2610 nucleotide of the 23S rRNA (Fig. 3e–g). The first and the second thiazole rings (Thz7 and Thz10) stack over the non-canonical C2586-C1782 and A2062-m2A2503 base-pairs, respectively (Fig. 3e–f, 4b). A2062 is the only nucleotide in the 23S rRNA whose conformation significantly changes upon binding of KLB (Supplementary Fig. 6a, b). This nucleotide rotates by more than ninety degrees to form Hoogsteen base-pair with the m2A2503 in the presence of KLB, similar to other inhibitors of the PTC (Fig. 4; Supplementary Fig. 7). Formation of the A2062-m2A2503 base-pair upon KLB binding is in agreement with the observed protection of the nucleotide A2062 of the 23S rRNA from DMS modification (Fig. 2c). Apparently, the last oxazole cycle of the KLB molecule guides its C-terminal part towards the exit of the peptide tunnel (Fig. 4f, g).
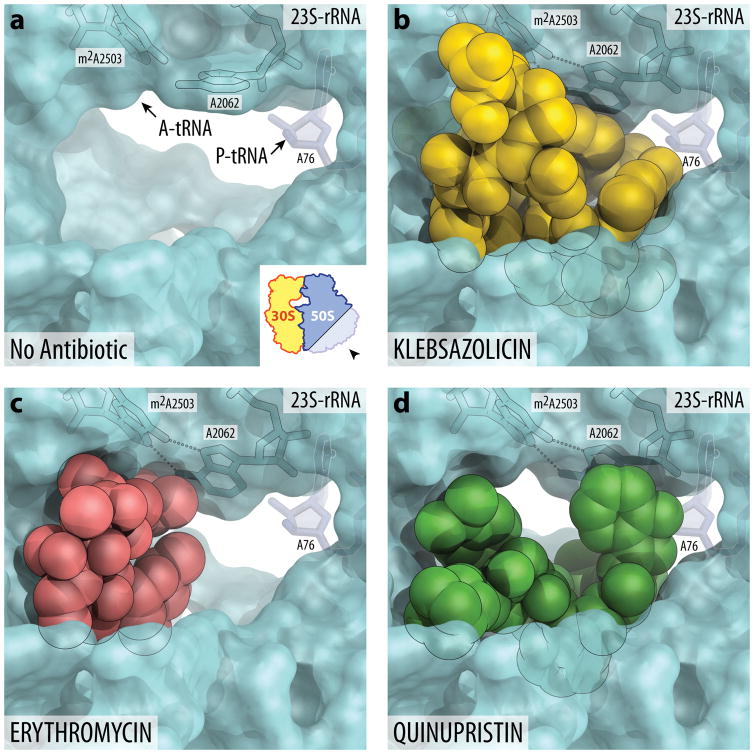
Figure 4. Occlusion of the nascent peptide exit tunnel by antibiotics.
(a) Lumen of the nascent peptide exit tunnel of the drug-free 70S ribosome (PDB entry 4Y4P36). The view is from the wide-open part of the tunnel onto the PTC as indicated by the inset. Nucleotide A76 of the P-site tRNA is shown in dark blue. A-site tRNA is not visible in this view, however, its location is indicate by the arrow. Note that nucleotide A2062 of the 23S rRNA is pointed toward the viewer and is not involved in Hoogsteen base pairing with A2503. (b, c, d) Occlusion of the nascent peptide exit tunnel by KLB (b), macrolide ERY (c), and type B streptogramin QIN (d). KLB structure is from the current work, ERY is from PDB entry 4V7X46, and QIN is from PDB entry 4U2642. Note that, unlike ERY or QIN, KLB almost completely occludes the lumen of the exit tunnel. Also note that binding of KLB, ERY, or QIN causes characteristic rotation of nucleotide A2062 by more than ninety degrees away from the viewer to form Hoogsteen base-pair with the A2503 of the 23S rRNA.
KLB binding site overlaps with other antibiotics
Several different classes of ribosome inhibitors bind and act upon the peptide exit tunnel. Superposition of the structure of KLB in complex with the 70S ribosome with the known structures of macrolide antibiotics shows that their binding sites largely overlap with each other (Fig. 4; Supplementary Fig. 7). However, macrolides do not completely obstruct the exit tunnel, but rather partially occlude it on one side still allowing for many of the experimentally tested nascent peptides to pass through (Fig. 4c; Supplementary Fig. 7c)39–41. In contrast to a small and compact erythromycin, extended KLB molecule adopts a folded conformation in the peptide exit tunnel thereby significantly occluding it (Fig. 4b; Supplementary Fig. 7b). Surprising is the ability of the ribosome to synthesize even a short peptide when a bulky KLB molecule is present that interacts with the key functional residues of the PTC, such as U2506 and U2585. Limited mobility of these nucleotides could result in decreased catalytic activity of the PTC, however, even in the presence of KLB all these nucleotides remain in their catalytically active states (Supplementary Fig. 6a, b). Moreover, PTC of the KLB-bound ribosome might retain some limited functionality due to the flexibility of the linear KLB molecule inside the ribosomal tunnel. Finally, the KLB binding site does not overlap with the aminoacyl moieties of the A- and P-site tRNAs (Supplementary Fig. 6c, d) and the main occlusion point occurs around the macrolide binding site leaving some space between the PTC active site and the bound KLB molecule, so that the 2–3 amino acid long nascent peptide can fit (Fig. 2e, lane 10).
The KLB binding site on the 70S ribosome also overlaps with that of the type B streptogramins42 (Fig. 4d; Supplementary Fig. 7d). The first and the second thiazole rings of the KLB stack over the same nucleotides as the amide and the 3-hydroxypicolinic acid groups of the quinupristin, a representative of the streptogramin B family (Supplementary Fig. 7b, d). The observed similarity in the binding modes of type B streptogramins and KLB is also reflected in the similar toe-print patterns obtained in the presence of KLB (Fig. 2e) and streptogramin B34. Therefore, regardless of the nature of the molecule that blocks the tunnel, the location of the binding site seems to determine the length of the peptide that can be synthesized in the presence of the inhibitor.
Binding sites on the ribosome for some antibacterial peptides, such as Onc112 or Bac7, also overlap with that of KLB32,43–45 (Supplementary Fig. 8). However, unlike these peptides, which are highly enriched in positively charged amino acids and interact with the negatively charged rRNA backbone, KLB interacts with the ribosome mainly via stacking of heterocycles over the rRNA bases (Fig. 4d–f; Supplementary Movie 1). Another striking difference between these peptides and KLB is that they bind to the ribosomal peptide exit tunnel in the elongated conformation, while KLB binds in a compact curled up globular conformation (Supplementary Fig. 8).
Altogether, structural comparisons of different chemically unrelated compounds that inhibit ribosome by binding to the peptide exit tunnel reveal that very similar structural elements of the ribosome are involved in all cases. This information can be utilized to design hybrid inhibitors. Moreover, the unique folded conformation of the KLB in the peptide exit tunnel provides a new structural scaffold for further improvement.
Significance of individual KLB-ribosome contacts
To assess the significance of individual contacts between the KLB and the ribosome and also to verify which single rRNA mutations could be responsible for the resistance three possible nucleotide substitutions at each of the positions A2058, A2059, U2609 and U2584C were introduced into the 23S rRNA gene of the rrnB operon carried by the pAM552 plasmid. An E. coli strain SQ171DTC was transformed with the mutated pAM552 plasmids, and, after curing off the wild-type pAM552 plasmid, all the 23S rRNA molecules in the cells were expected to be mutant (Supplementary Table 2). Among the mutations at positions A2058 and A2059 of the 23S rRNA, mutation A2059G caused the highest level of KLB-resistance (8-fold) most likely due to the steric clash of the N2-atom with the backbone oxygen of thiazole ring formed by Cys10 cyclization (Thz10) (Fig. 3e, f). Consistent with the results of the spontaneous-mutant selection experiments, the engineered mutations U2609A and U2609G, but not U2609C, resulted in resistance to KLB (Supplementary Table 2). Of a particular interest is the base-pair-like interaction via two H-bonds between the amidine heterocycle and the nucleotide U2584 of the 23S rRNA. Mutation U2584C increases KLB’s MIC by 8 fold, pointing to the extreme importance of this interaction for the activity of the inhibitor.
Next, we created a set of KLB mutants to further assess the effects of the individual contacts between the ribosome and KLB on its ability to inhibit translation (Table 2). The most unique feature that distinguishes KLB from the related TOMM peptides (e.g., MccB17) is its 6-membered amidine ring that interacts with the nucleotides U2584, U2585 and C2610 of the 23S rRNA. We engineered KLBS1A, KLBQ2N, and KLBS3A mutants (Table 2) and expressed them in E. coli. KLBS1A and KLBQ2N mutants contained the full set of azole rings as well as the amidine ring (Supplementary Fig. 9a, b). In contrast, the KLBS3A mutant lacked the amidine cycle (Supplementary Fig. 9c). Inability to form this amidine cycle renders KLBS3A completely inactive (Fig. 2e, Table 2). A contact of the Gln2 with the bridging phosphate oxygen between U2585 and U2586 also appeared to be essential, as the Q2N substitution resulted in a complete loss of activity both in vivo and in vitro (Table 2). Residue Ser1, whose side chain makes a single hydrogen bond with the universally conserved and essential nucleotide U2585, was also found to be crucial for the activity of the KLB, as evident from the lower activity of KLBS1A mutant (Table 2). Altogether, these data point to the key role of the N-terminal part of the KLB in its inhibitory activity.
Table 2.
Cell growth and protein synthesis inhibition by the mutant forms of klebsazolicin.
| KLB Mutant Name | KLB Mutant Sequence* | Luciferase synthesized in vitro, % | MIC, μg/ml (μM); E. coli MG1655 on M9+Glucose |
|---|---|---|---|
| No inhibitors | 100 | - | |
| KLB | SQSPGNCASCSNSASANCTGGLG | 2±1 | 16 (8) |
| KLBS1A | AQSPGNCASCSNSASANCTGGLG | 20±8 | 256 (130) |
| KLBQ2N | SNSPGNCASCSNSASANCTGGLG | 84±19 | >512 (>259) |
| KLBS3A | SQAPGNCASCSNSASANCTGGLG | 80±11 | >512 (>259) |
| KLBA14K | SQSPGNCASCSNSKSANCTGGLG | 1.9±1 | 32 (16) |
| KLBA14K(1-14) | SQSPGNCASCSNSK | 4.8±0.3 | >512 (>259) |
| KLBS11K | Low yield | - | - |
| KLBS13A/A14K | SQSPGNCASCSNAKSANCTGGLG | 43±4 | >512 (>259) |
| KLBS13A/A14K(1-14) | SQSPGNCASCSNAK | 51±5 | >512 (>259) |
| KLB(1-11) | SQSPGNCASCS | 73±6 | >512 (>259) |
Residues that are involved in cyclization resulting in the formation of the N-terminal amidine cycle are highlighted in blue, while residues involved in the formation of thiazole/oxazole cycles are highlighted in red.
To understand the role of the C-terminal part of the KLB in the inhibition of translation and to establish a minimal set of heterocycles that constitutes an active scaffold we created a set of KLB derivatives with mutated and/or truncated C-terminal parts (Table 2). Since original KLB sequence lacks protease recognition sites, we introduced a lysine residue between the azole rings at position 14. Full-length KLBA14K as well as truncated KLBA14K(1-14) produced after trypsin digestion appeared to be almost as active as the original KLB in the in vitro translation (Table 2) and in the toe-printing assay (Fig. 2e). However, unlike the full-length KLBA14K, its truncated version was unable to inhibit cell growth, suggesting that the nine C-terminal residues of KLB are required for the peptide uptake, but not for the interactions with the ribosome (Table 2). Further shortening of the KLB molecule as well as removal of the third azole ring in the context of the full-length peptide in the S13A-A14K mutant abolished both in vitro and in vivo inhibitory activities (Table 2; Supplementary Fig. 10). Thus, the N-terminal 14-amino-acid-long fragment of KLB including three out of four thiazole/oxazole cycles appears to be the minimal functional core capable of inhibiting the ribosome. Search for the klpABCDE-like gene clusters allowed identification of several clusters which encode predicted precursor peptides containing a conservative core peptide that likely constitutes a functional scaffold (Supplementary Fig. 11). We predict that the products of these clusters encode ribosome-targeting RiPPs.
KLB represents the first linear azole-containing peptide for which the mode of binding to its target was structurally characterized. KLB is a unique thiazole/oxazole containing peptide that inhibits protein synthesis by binding to the bacterial ribosome in the peptide exit tunnel. It binds in a tightly folded conformation and blocks the elongation after only three amino acids of the nascent peptide have been polymerized. A unique and essential amidine ring of KLB forms multiple interactions with the conserved core of the 23S rRNA. The structure of KLB appeared to be modular: its fourteen N-terminal residues are essential for the inhibition of the ribosome, while its nine C-terminal residues are likely to be important for the uptake of the molecule, but not essential for ribosome binding. It is highly likely that natural sensitivity/resistance to KLB is determined not by the differences in the ribosome target but is rather due to differences in proteins involved in its uptake. The modular structure of KLB should make it possible to rationally design hybrid ribosome-targeting antibiotics containing a common ribosome-targeting warhead and variable uptake parts that control specificity of antibacterial action.
ONLINE METHODS
Bacterial strains and growth conditions
The following bacterial strains were used in this study: E. coli BL21(DE3); E. coli BW25113 and isogenic BW25113 ΔtolC (JW5503 from Keio collection); E. coli MG1655 and isogenic E. coli MG1655 ΔompF, E. coli MG1655 ΔsbmA; E. coli SQ110DTC (ΔtolC, ΔrrnA, ΔrrnB, ΔrrnC, ΔrrnD, ΔrrnG, ΔrrnH)34, and SQ171DTC (ΔtolC, ΔrrnA, ΔrrnB, ΔrrnC, ΔrrnD, ΔrrnE, ΔrrnG, ΔrrnH, pAM552); E. coli B; Pseudomonas aeruginosa PA01; Pseudomonas putida KT2440; Acinetobacter baumannii ATCC 17978; Bacillus subtilis str. 168; Klebsiella pneumoniae 875 (isolated in St. Petersburg Research Institute of Children’s Infections of the Federal Medical and Biological Agency of Russia); Yersinia pseudotuberculosis IP 32953; Vibrio cholerae 0395 (ΔtoxT); Mycobacterium smegmatis B-24020; Bacillus cereus ATCC 14579; Listeria grayi; Streptomyces lividans B-16148; and Staphylococcus aureus ATCC 25923. Bacterial strains were grown in LB medium (1 liter contains 5 g of NaCl, 10 g of tryptone, 5 g of yeast extract) or M9 minimal medium (1 liter contains 12.8 g Na2HPO4·7H2O, 3 g KH2PO4, 0.5 g NaCl, 1 g NH4Cl, 1 ml of 2M MgSO4 solution, 100 μl of 1M CaCl2 solution, 10 μg/ml thiamine) supplemented with 1% glycerol or 0.4% glucose. When necessary, antibiotics were used at concentrations of 100 μg/ml for ampicillin, 50 μg/ml for kanamycin, and 60 μg/ml for spectinomycin.
Molecular cloning procedures
Genomic DNA of K. pneumoniae subsp. ozaenae ATCC 11296 was obtained from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (DSM-16358). Fragments containing klpBCDE genes and klpA gene were amplified using pairs of oligonucleotides klpcF-klpeR and klpaF-klpaR (Supplementary Table 3), respectively, and cloned into pBAD His/B and pET28a vectors using restriction enzymes SacI, PstI, NcoI, XhoI resulting in pBAD-klpBCDE and pET28-klpA plasmids. A fragment containing klpE gene was amplified using pair of oligonucleotides klpeF-klpeR2 (Supplementary Table 3) and cloned into pBAD His/B vector using restriction enzymes NcoI and SacI resulting in pBAD-klpE plasmid. Mutated klpA genes for the precursor peptide were generated by amplifying klpA using the following pairs of oligonucleotides: klpaF-klpaS3A, klpaF-klpaQ2N, klpaF-klpaS1A, klpaF-klpaA14K, klpaF-klpaA11K, klpaF-klpaS13AA14K (Supplementary Table 3), and cloned into pET28a vector using restriction enzymes NcoI and XhoI resulting in pET28a-klpASer3Ala, pET28a-klpAGln3Asn, pET28a-klpASer1Ala, pET28a-klpAAla14Lys, pET28a-klpAAla11Lys, pET28a-klpASer13Ala-Ala14Lys plasmids, respectively.
In order to prepare 13C/15N KLB for NMR studies, klpA gene was amplified using pair of oligonucleotides klpaF-klpaR2 (Supplementary Table 3) and cloned into the modified pET28a vector (with an additional site for PstI restriction enzyme within the multiple cloning site) using restriction enzymes NcoI and SacI resulting in pET28-klpA2 plasmid. Then, plasmid pBAD-klpBCDE was digested using restriction enzymes SacI and PstI and the fragment containing klpBCDE was cloned into the previously obtained pET28-klpA2 vector resulting in pET28-klpABCDE plasmid.
PCR-fragments containing sbmA and ompF genes were amplified using genomic DNA of E. coli BL21(DE3) and pairs of oligonucleotides sbmaF-sbmaR and ompfF-ompfR (Supplementary Table 3), respectively, and cloned consequently into pCOLADuet-1 vector using restriction sites NcoI, BamHI, NdeI, and XhoI resulting in pColA-sbmA-ompF plasmid. Plasmid pColA-sbmA-ompF was digested using restriction enzymes NcoI and XhoI and the fragment containing sbmA genes ompF was cloned into pBAD His/B yielding plasmid pBAD-sbmA-ompF.
Purification of klebsazolicin and its derivatives
E. coli BL21(DE3) strain transformed with plasmids pBAD-klpBCDE and pET28 encoding corresponding precursor peptide KlbA was cultured in 2 liters of LB medium supplemented with ampicillin and kanamycin until OD600 reached 0.6. Bacterial cells were harvested by centrifugation for 10 min at 4,000g, washed with PBS solution and used to inoculate 2 liters of M9 medium supplemented with 1% glycerol, 10 mM arabinose, 0.1 mM IPTG (isopropyl-β-D-thiogalactopyranoside), ampicillin, and kanamycin and were grown for 24 hours at 30°C. Bacterial cells were pelleted by centrifugation for 30 min at 12,000g, supernatant was loaded onto a Waters Sep-Pak 12 cc Vac C18 cartridge (55–105 μm particle size). The cartridge was extensively washed with 9% aqueous MeCN. KLB-containing fractions were eluted with 15 ml of 15% aqueous MeCN and subjected to a subsequent HPLC purification using semi-preparative Jupiter C18 HPLC column (300 Å; 250 × 4.6; 5 μm particle size). The column was equilibrated with 0.1% trifluoroacetic acid (TFA) and the bound material was eluted with a linear gradient of acetonitrile in 0.1% TFA (from 0 to 30% acetonitrile in 30 min). Fractions containing KLB-like compounds were lyophilized, dissolved in dimethyl sulfoxide (DMSO), and stored at −80 °C. Yields of KLB ranged from 15 to 40 mg per liter of medium, while yields of derivatives ranged from 2 to 20 mg per liter of medium.
To prepare 13C/15N KLB cultivation of E. coli BL21(DE3) containing plasmid pET28-klpABCDE was performed in modified M9 minimal medium (1 liter contains 12.8 g Na2HPO4·7H2O, 3 g KH2PO4, 0.5 g NaCl, 1 g (15NH4)2SO4 (15N2, 99%, Cambridge Isotope Laboratories), 1 ml of 2M MgSO4 solution, 100 μl of 1M CaCl2 solution, 10 μg/ml thiamine) supplemented with 0.1% D-glucose(U-13C6, 99%, Cambridge Isotope Laboratories), 0.1 mM IPTG and kanamycin.
MS analysis
High-resolution mass spectra were recorded on a Fourier Transform Ion Cyclotron Resonance Mass Spectrometer (Varian 902-MS) equipped with the 9.4 T magnet (FTMS) in positive MALDI mode. The instrument was calibrated using the ProteoMass Peptide MALDI-MS Calibration Kit (Sigma-Aldrich). The accuracy of the mass peak measurements was 2.5 ppm. Samples (0.5 μl) were spotted on a steel plate with 0.5 μl of a 2,5-Dihydroxybenzoic acid matrix (Sigma-Aldrich) and air-dried at room temperature.
Fragment ion spectra were recorded on an AB SCIEX TOF/TOF 5800 MALDI mass-spectrometer. The instrument was calibrated using the Mass Standards Kit for Calibration of AB Sciex TOF/TOF Instruments (AB Sciex). Samples (0.5 μl) were spotted on a steel plate with 0.5 μl of α-cyano-4-hydroxycinnamic acid matrix solution (AB Sciex) and air-dried at room temperature. Fragment ion spectra were generated by collision-induced dissociation (CID) in MS/MS mode. The accuracy of the mass peak measurements was 30 ppm for parent ions and 0.2 Da for daughter ions. Analysis of the MS and MS/MS data was carried out manually by GPMAW4.04 software (Lighthouse Data).
NMR spectroscopy
NMR spectra were recorded on a 5.0 mM 13C/15N KLB sample (enriched NMR active isotopes 13C and 15N) dissolved in DMSO-D6 and stored in Shigemi tube at 55°C that led to a well-resolved set of signals. All NMR spectra were recorded at the Varian DirectDrive NMR System 700 MHz spectrometer, equipped with the 5 mm-gradients salt tolerant H/C/N probe at the SpbSPU. The NMR data were processed and analyzed with the Mestrelab Mnova version 10. Full assignment of individual amino acids including one proline, four glycines, three asparagines, three alanines, one lysine, five serines and one non-canonical glutamine, as well as three thiazoles and one oxazole, was obtained from 1H, 13C, 1H-TOCSY, 1H-COSY, 13C-HSQC, 13C-HMBC, 13C-COSY, 13C-HSQCTOXY, 15N-HSQC and 15N-HSQCTOXY, HNCO experiments (Supplementary Table 1; Supplementary Fig. 3).
Alanines, glycines, asparagines, threonine and leucine were assigned (excluding carbonyl carbon) from 1H, 1H-TOCSY and 1H-COSY spectra of 1H signals (Supplementary Fig. 3a) and 13C (Supplementary Fig. 3b) and 13C-HSQC.The assignment of serines, proline and glutamine was performed from 13C-HSQCTOXY. Side chains of asparagines and a glutamine were assigned from 13C-HMBC. Thiazoles and oxazole were assigned from 13C-HSQC, 13C-HSQCTOXY, and 13C-HMBC. The C5-H5 correlations were assigned form 13C-HSQC (Supplementary Fig. 3c). The C4, C6 Oxz13 signals were assigned form 13C-COSY (Supplementary Fig. 3d). The signals of carbonyl carbons were assigned form 13C-HMBC, 13C-COSY and H-C projection of HNCO spectra. NH groups were assigned form 15N-HSQC (Supplementary Fig. 3e) and 15N-HSQCTOXY spectra. Sequence connectivity between different amino acids was established on the basis of the analysis of the observed 1H–13C HMBC correlations (Supplementary Fig. 3f). The connectivity of Pro4-Gly5 was evident from the HMBC correlations of Pro4-CO to Gly5-HN, and Pro4-CO to Gly5-Hα. The connectivity of Gly5-Asn6, Ala8-Ser9, Ser11-Asn12 and Thr19-Gly20 were assigned in a similar fashion. The connection between Ser15-Ala16 and Ala16-Asn17 was established through the HMBC correlations of Ser15-CO to Ala16-Hα and Ala16-CO to Asn17-Hα respectively. The connection between Gly20-Gly21 was established through the HMBC correlations of Gly20-CO to Gly21-HN. The connectivity of Gly21-Leu22 and Leu22-Gly23 were assigned in a similar fashion. The connectivity of Asn6-Thz7 was evident from the HMBC correlations of Thz7-C2 and Thz7-C4 to Asn6-Hα and Asn6-Hβ. The connectivity of Ser9-Thz10 and Asn17-Thz18 were assigned in a similar fashion. The connectivity of Oxz13-Ala14 was evident from the HMBC correlations of Oxz13-C6 to Ala14-Hα and Ala14-HN. The connectivity of Ala8-Thz7, Thz10-Ser11 and Thz18-Thr19 were assigned in a similar fashion. The connectivity of Ala12-Oxz13 was evident from the HMBC correlations of Oxz13-C2 to Ala12-Hα and Ala12-Hβ. The connectivity of Ser1′-Glu2 and Glu2-Ser3 was evident from the HMBC correlations of Glu2-CO to Ser1′-Hα, Ser3-Hα and Ser1′-HN, Ser3-HN (Supplementary Fig. 3g). We observe two groups of signals from Ser1, Gln2, Ser3 and Asn6, where population of states relates as 5/3, indicating that there is a tautomeric equilibrium between two protonated forms of amide moiety.
Protease treatment
Purified KLBA14K and KLBS13A-A14K (0.5 mg each) were treated with 10 μg of trypsin (from porcine pancreas, Sigma-Aldrich) for 12 hours at 37 °C in 100 mM NH4HCO3 (pH 8.5). The reaction mixtures were purified using Supelcosil LC-18 HPLC column (120 Å; 250 × 4.6; 5 μm particle size) operating at a flow rate of 1.0 ml/min and separated with a linear gradient of MeCN in 0.1% TFA (from 0 to 30% MeCN in 30 min). Absorbance was monitored at 254 nm. Fractions were collected manually and analyzed by MALDI-TOF MS. The fraction containing the mass ion of m/z 1291 [M+H]+ (Supplementary Fig. 10c) that corresponds to the N-terminal part KLBA14K (1-14) and the fraction containing the mass ion of m/z 1295 [M+H]+ (Supplementary Fig. 10d) that corresponds to the N-terminal part KLBS13A-A14K (1-14) were lyophilized and dissolved in DMSO. Purified KLBS13A-A14K (1-14) (0.2 mg) was digested with 10 μg of carboxypeptidase Y (from baker’s yeast; Sigma-Aldrich) for 6 hours at 37 °C in water, the reaction mixture was purified using HPLC as described above and the fraction containing the mass ion of m/z 982 [M+H]+ (Supplementary Fig. 10e) that corresponds to KLB (1-11) was lyophilized and dissolved in DMSO.
Evaluation of bioactivity
Minimum inhibitory concentrations (MIC) in LB and M9 medium were determined by the broth microdilution assay47. The test medium was supplemented with 3 mM L-arabinose for E. coli MG1655 carrying pBAD-sbmA-ompF or pBAD-klpE plasmids. Cell concentration was adjusted to approximately 5×105 cells/ml. Tested compounds were serially diluted 2-fold in a 96-well microplate (100 μl/well). Microplates were covered and incubated at 37°C (30°C for P. putida and S. lividans) with shaking. The OD600 of each well was measured and the lowest concentration of the tested compound that resulted in no growth after 16–20 hours (2 days for M. smegmatis and S. lividans) was reported as MIC.
Selection of spontaneous KLB-resistant mutants
Three independent overnight cultures of E. coli SQ110DTC carrying pBAD-sbmA-ompF (approx. 109 cells in 1 ml when tested) were spun off and washed 3 times in sterile LB medium before applying to the LB plates containing 200 μg/ml KLB (3x MIC). All selection plates also contained 1 mM arabinose, 50 μg/ml spectinomycin, 50 μg/ml kanamycin and 200 μg/ml carbenicillin to maintain the necessary plasmids. ~109 cells were applied to each plate. As a positive control for the mutant selection under these conditions, a separate plate containing 300 μg/ml erythromycin (but not KLB) was inoculated. In course of 24 h incubation at 37 °C 2–8 colonies became visible on each 3x MIC plates. Colonies were re-streaked onto fresh 200 μg/ml KLB (plus other antibiotics) plates to confirm resistance. rRNA genes of 4 mutants were sequenced and two rRNA mutations (U2609G and U2609A) were identified. From the control (erythromycin) plate, ~10 resistant colonies were isolated in similar manner. rRNA genes of 5 of them were sequenced and A2058G mutation was confirmed in all the cases.
Testing of KLB resistance mutations
Mutation U2584C (Table 2) was introduced by site directed mutagenesis in the 23S rRNA gene in plasmid pAM552 (a derivative of pLK3548) containing the entire E. coli rrnB operon under the control of the PL promoter. pAM552 plasmid was amplified by PCR using the primers listed in Supplementary Table 3. PCR product was self-ligated and transformed into E. coli JM109 strain for plasmid production and purification. Once mutant plasmids were verified by sequencing, they were transformed into the E. coli SQ171DTC strain and the cells were then cured of the wild type plasmid pCSacB by plating onto an LB/Amp plate supplemented with 5% sucrose49. Other pAM552 plasmids carrying mutations at positions corresponding to nucleotides A2058, A2059, U2609 of the 23S rRNA, which were used in this work, were obtained from Dr. Alexander Mankin. The complete replacement of wild type plasmid with the mutant one and expression of a homogeneous population of mutant ribosomes was verified by primer extension. MIC values of the engineered mutants were tested by microbroth dilution.
Testing of in vivo activity of KLB
Reporter strain BW25113-pDualrep2 was used as previously described30. Briefly, 2 μl of tested solutions of antibiotics were applied to agar plate that already contained lawn of the reporter strain. 10 mM KLB, MccB17, erythromycin and 20 μM levofloxacin solutions were used. After overnight incubation of the plate at 37 °C, it was scanned by ChemiDoc (Bio-Rad): “Cy3-blot” for RFP and “Cy5-blot” for Katushka2S.
In vitro translation analysis
The in vitro transcribed firefly luciferase mRNA, hns mRNA or osmC mRNA (final concentration 0.2 μM) were translated in the E. coli S30 cell-free system for linear templates (Promega). Reactions were carried out in 5 μL aliquots at 37°C for 30 minutes. Activity of in vitro synthesized luciferase was assessed using the Steady-Glo Luciferase Assay System (Promega). The hns and osmC mRNAs were translated in the presence of the FluoroTect™ GreenLys and products were analyzed by SDS-PAGE.
DMS and CMCT chemical protection assays
70S ribosomes were incubated in the presence of 50 μM of KLB for 10 minutes at 37°C. Chemical modification with DMS and CMCT, and subsequent primer extension reactions were performed as described previously50,51. Used primers were complementary to the nucleotides 2102–2119 and 2607–2624 of the 23S rRNA.
Toe-printing analysis
Toe-printing analysis was carried out essentially as previously described52 using osmC mRNA as a template for protein translation34. The concentrations of the tested compounds are shown above each lane in Figure 2e.
Crystallographic structure determination
Ribosome complexes with mRNA and tRNAs were formed by programming of 5 μM Tth 70S ribosomes with 10 μM mRNA and incubation at 55°C for 10 min, followed by addition of 20 μM P-site (fMet-tRNAiMet) and 20 μM A-site (Phe-tRNAPhe) substrates36. Each of the last two steps was allowed to reach equilibrium for 10 min at 37°C. The Tth 70S ribosome complexes were formed in the buffer containing 5 mM HEPES-KOH (pH 7.6), 50 mM KCl, 10 mM NH4Cl, and 10 mM Mg(CH3COO)2, and then crystallized in the buffer containing 100 mM Tris-HCl (pH 7.6), 2.9% (w/v) PEG-20K, 7–12% (v/v) MPD, 100–200 mM arginine, 0.5 mM β-mercaptoethanol. Crystals were grown by the vapor diffusion method in sitting drops at 19°C and stabilized as described previously36 with the KLB included in the stabilization buffers (200 μM KLB). Diffraction data were collected using beamline 24ID-C at the Advanced Photon Source. All crystals belonged to the primitive orthorhombic space group P212121 with approximate unit cell dimensions of 210Å x 450Å x 620Å and contained two copies of the 70S ribosome per asymmetric unit. Each structure was solved by molecular replacement using PHASER from the CCP4 program suite53. The search model was generated from the previously published structure of T. thermophilus 70S ribosome with bound mRNA and tRNAs (PDB entry 4Y4P from36). The initial molecular replacement solutions were refined by rigid body refinement with the ribosome split into multiple domains, followed by positional and individual B-factor refinement. The final models of the 70S ribosome in complex with KLB and mRNA/tRNAs was generated by multiple rounds of model building in COOT54, followed by refinement in PHENIX55. The statistics of data collection and refinement are compiled in Supplementary Table 4.
Supplementary Material
Acknowledgments
We thank Prof. Alexander Mankin and Tanja Florin (University of Illinois at Chicago) for providing pAM552 plasmids carrying various mutations, and for careful reading of the manuscript and valuable suggestions. We are thankful to the members of the K.V.S, P.V.S., and Y.S.P. laboratories for discussions and critical feedback. We thank Raktim Roy for critical reading of the manuscript. We also thank Ivan Kamyshko (MilliporeSigma) for material source consulting and for providing Supelco C5 HPLC column used for tRNA purification. We thank staff at NE-CAT beamline 24ID-C for help with data collection and freezing of the crystals, especially Drs. Kanagalakhatta Rajashankar, Malcolm Capel, Frank Murphy, Igor Kourinov, Anthony Lynch, Surajit Banerjee, David Neau, Jonathan Schuermann, Narayanasami Sukumar, James Withrow, Kay Perry, and Cyndi Salbego.
This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). The Pilatus 6M detector on 24ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
This work was supported by Illinois State startup funds (to Y.S.P.), US National Institutes of Health grant R01 AI117210 (to K.S.), Skoltech Institutional funds (to K.S.) and Russian Foundation for Basic Research grants 16-04-01100 (to P.V.S.) and 15-34-20139 (to I.A.O.). This work was also supported in part by the Ministry of Education and Science of the Russian Federation grant 14.B25.31.0004 (to K.S.), and the Russian Science Foundation grant 15-15-10017 (to M.M., used for the expression, purification, structure determination of KLB and testing its in vivo activity), the Russian Science Foundation grant 14-14-00072 (to P.V.S., used for the toe-printing assays, in vitro activity tests and chemical foot-printing), the Dynasty Foundation Fellowship (to M.M.) and FASIE grant №9186GU/2015 (to D.Y.T).
Footnotes
DATA AVAILABILITY STATEMENT
Structural data have been deposited in the RCSB Protein Data Bank (PDB) with coordinate accession number 5W4K. All other data generated or analyzed during this study are included in this published article (and its supplementary information files) or are available from the corresponding authors on reasonable request.
ACCESSION CODES
Coordinates and structure factors were deposited in the RCSB Protein Data Bank with accession code 5W4K for the Tth 70S ribosome in complex with klebsazolicin, mRNA, A-, P- and E-site tRNAs.
AUTHOR CONTRIBUTIONS
M.M. and I.U. assembled constructs for isolation of KLB and derivatives, M.M. I.U., D.G., D.Y.T. isolated and purified KLB and its derivatives; M.M., D.G, I.A.O evaluated bioactivity; D.G. and D.Y.T. selected resistant mutants; N.F.K and Y.S.P. designed and performed X-ray crystallography experiments; I.A.O., P.V.S., T.F. and A.S.M. designed biochemistry and genetic experiments; A.Y. and K.S. performed and analyzed the NMR experiments; M.M. performed and analyzed the MS experiments with critical input from T.A. and M.S. All authors interpreted the results. M.M., Y.S.P., P.V.S., and K.V.S. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES FOR MAIN TEXT
- 1.Lewis K. Platforms for antibiotic discovery. Nat Rev Drug Discov. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 2.Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 3.Laxminarayan R, et al. Antibiotic resistance-the need for global solutions. Lancet Infect Dis. 2013;13:1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 4.Walsh CT, Wencewicz TA. Prospects for new antibiotics: a molecule-centered perspective. J Antibiot (Tokyo) 2014;67:7–22. doi: 10.1038/ja.2013.49. [DOI] [PubMed] [Google Scholar]
- 5.Liu YY, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 6.Doroghazi JR, et al. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol. 2014;10:963–968. doi: 10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donia MS, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zipperer A, et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. 2016;535:511–516. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- 9.Donia MS, Fischbach MA. HUMAN MICROBIOTA. Small molecules from the human microbiota. Science. 2015;349:1254766. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oman TJ, van der Donk WA. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat Chem Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koehnke J, et al. The cyanobactin heterocyclase enzyme: a processive adenylase that operates with a defined order of reaction. Angew Chem Int Ed Engl. 2013;52:13991–13996. doi: 10.1002/anie.201306302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunbar KL, et al. Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis. Nat Chem Biol. 2014;10:823–829. doi: 10.1038/nchembio.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burkhart BJ, Schwalen CJ, Mann G, Naismith JH, Mitchell DA. YcaO-dependent posttranslational amide activation: biosynthesis, structure, and function. Chem Rev. 2017;117:5389–5456. doi: 10.1021/acs.chemrev.6b00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunbar KL, Melby JO, Mitchell DA. YcaO domains use ATP to activate amide backbones during peptide cyclodehydrations. Nat Chem Biol. 2012;8:569–575. doi: 10.1038/nchembio.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SW, et al. Discovery of a widely distributed toxin biosynthetic gene cluster. Proc Natl Acad Sci USA. 2008;105:5879–5884. doi: 10.1073/pnas.0801338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox CL, Doroghazi JR, Mitchell DA. The genomic landscape of ribosomal peptides containing thiazole and oxazole heterocycles. BMC Genomics. 2015;16:778. doi: 10.1186/s12864-015-2008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crone WJ, et al. Dissecting Bottromycin Biosynthesis Using Comparative Untargeted Metabolomics. Angew Chem Int Ed Engl. 2016;55:9639–9643. doi: 10.1002/anie.201604304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.San Millan JL, Kolter R, Moreno F. Plasmid genes required for microcin B17 production. J Bacteriol. 1985;163:1016–1020. doi: 10.1128/jb.163.3.1016-1020.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huo L, Rachid S, Stadler M, Wenzel SC, Muller R. Synthetic biotechnology to study and engineer ribosomal bottromycin biosynthesis. Chem Biol. 2012;19:1278–1287. doi: 10.1016/j.chembiol.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Salomon RA, Farias RN. The FhuA protein is involved in microcin 25 uptake. J Bacteriol. 1993;175:7741–7742. doi: 10.1128/jb.175.23.7741-7742.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavina M, Pugsley AP, Moreno F. Identification, mapping, cloning and characterization of a gene (sbmA) required for microcin B17 action on Escherichia coli K12. J Gen Microbiol. 1986;132:1685–1693. doi: 10.1099/00221287-132-6-1685. [DOI] [PubMed] [Google Scholar]
- 23.Patzer SI, Baquero MR, Bravo D, Moreno F, Hantke K. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology. 2003;149:2557–2570. doi: 10.1099/mic.0.26396-0. [DOI] [PubMed] [Google Scholar]
- 24.Runti G, et al. Functional characterization of SbmA, a bacterial inner membrane transporter required for importing the antimicrobial peptide Bac7(1–35) J Bacteriol. 2013;195:5343–5351. doi: 10.1128/JB.00818-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salomon RA, Farias RN. The peptide antibiotic microcin 25 is imported through the TonB pathway and the SbmA protein. J Bacteriol. 1995;177:3323–3325. doi: 10.1128/jb.177.11.3323-3325.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novikova M, et al. The Escherichia coli Yej transporter is required for the uptake of translation inhibitor microcin C. J Bacteriol. 2007;189:8361–8365. doi: 10.1128/JB.01028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattiuzzo M, et al. Role of the Escherichia coli SbmA in the antimicrobial activity of proline-rich peptides. Mol Microbiol. 2007;66:151–163. doi: 10.1111/j.1365-2958.2007.05903.x. [DOI] [PubMed] [Google Scholar]
- 28.Maio A, Brandi L, Donadio S, Gualerzi CO. The oligopeptide permease Opp mediates illicit transport of the bacterial P-site decoding inhibitor GE81112. Antibiotics (Basel) 2016;5 doi: 10.3390/antibiotics5020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vondenhoff GH, et al. Characterization of peptide chain length and constituency requirements for YejABEF-mediated uptake of microcin C analogues. J Bacteriol. 2011;193:3618–3623. doi: 10.1128/JB.00172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osterman IA, et al. Sorting out antibiotics’ mechanisms of action: a double fluorescent protein reporter for high throughput screening of ribosome and DNA biosynthesis inhibitors. Antimicrob Agents Chemother. 2016;60:7481–7489. doi: 10.1128/AAC.02117-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krizsan A, et al. Insect-derived proline-rich antimicrobial peptides kill bacteria by inhibiting bacterial protein translation at the 70S ribosome. Angew Chem Int Ed Engl. 2014;53:12236–12239. doi: 10.1002/anie.201407145. [DOI] [PubMed] [Google Scholar]
- 32.Seefeldt AC, et al. Structure of the mammalian antimicrobial peptide Bac7(1-16) bound within the exit tunnel of a bacterial ribosome. Nucleic Acids Research. 2016;44:2429–2438. doi: 10.1093/nar/gkv1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han MJ, Lee SY, Hong SH. Comparative analysis of envelope proteomes in Escherichia coli B and K-12 strains. J Microbiol Biotechnol. 2012;22:470–478. doi: 10.4014/jmb.1110.10080. [DOI] [PubMed] [Google Scholar]
- 34.Orelle C, et al. Tools for characterizing bacterial protein synthesis inhibitors. Antimicrob Agents Chemother. 2013;57:5994–6004. doi: 10.1128/AAC.01673-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 36.Polikanov YS, Melnikov SV, Soll D, Steitz TA. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat Struct Mol Biol. 2015;22:342–344. doi: 10.1038/nsmb.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garza-Ramos G, Xiong L, Zhong P, Mankin A. Binding site of macrolide antibiotics on the ribosome: new resistance mutation identifies a specific interaction of ketolides with rRNA. J Bacteriol. 2001;183:6898–6907. doi: 10.1128/JB.183.23.6898-6907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymology. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 39.Kannan K, Vazquez-Laslop N, Mankin AS. Selective protein synthesis by ribosomes with a drug-obstructed exit tunnel. Cell. 2012;151:508–520. doi: 10.1016/j.cell.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 40.Kannan K, et al. The general mode of translation inhibition by macrolide antibiotics. Proc Natl Acad Sci USA. 2014;111:15958–15963. doi: 10.1073/pnas.1417334111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis AR, Gohara DW, Yap MN. Sequence selectivity of macrolide-induced translational attenuation. Proc Natl Acad Sci USA. 2014;111:15379–15384. doi: 10.1073/pnas.1410356111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noeske J, et al. Synergy of streptogramin antibiotics occurs independently of their effects on translation. Antimicrob Agents Chemother. 2014;58:5269–5279. doi: 10.1128/AAC.03389-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy RN, Lomakin IB, Gagnon MG, Steitz TA. The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin. Nat Struct Mol Biol. 2015;22:466–469. doi: 10.1038/nsmb.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gagnon MG, et al. Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition. Nucleic Acids Research. 2016;44:2439–2450. doi: 10.1093/nar/gkw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seefeldt AC, et al. The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nat Struct Mol Biol. 2015;22:470–475. doi: 10.1038/nsmb.3034. [DOI] [PubMed] [Google Scholar]
- 46.Bulkley D, Innis CA, Blaha G, Steitz TA. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc Natl Acad Sci USA. 2010;107:17158–17163. doi: 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 48.Douthwaite S, Powers T, Lee JY, Noller HF. Defining the structural requirements for a helix in 23 S ribosomal RNA that confers erythromycin resistance. J Mol Biol. 1989;209:655–665. doi: 10.1016/0022-2836(89)93000-3. [DOI] [PubMed] [Google Scholar]
- 49.Zaporojets D, French S, Squires CL. Products transcribed from rearranged rrn genes of Escherichia coli can assemble to form functional ribosomes. J Bacteriol. 2003;185:6921–6927. doi: 10.1128/JB.185.23.6921-6927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moazed D, Noller HF. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 51.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 52.Orelle C, et al. Identifying the targets of aminoacyl-tRNA synthetase inhibitors by primer extension inhibition. Nucleic Acids Res. 2013;41:e144. doi: 10.1093/nar/gkt526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.