Abstract
Humankind depends on the sustainability of soils for its survival and wellbeing. Threatened by a rapidly changing world, our soils suffer from degradation and biodiversity loss, making it increasingly important to understand the role of soil biodiversity in soil aggregation, a key parameter for soil sustainability. We here provide evidence of the contribution of soil biota to soil aggregation on macro- and microaggregate scales, and evaluate how specific traits, soil biota groups and species interactions contribute to this. We conducted a global meta-analysis comprising 279 soil biota species. Our study shows a clear positive effect of soil biota on soil aggregation, with bacteria and fungi generally appearing more important for soil aggregation than soil animals. Bacteria contribute strongly to both macro- and microaggregates while fungi strongly affect macro-aggregation. Motility, body size and population density were important traits modulating effect sizes. Investigating species interactions across major taxonomic groups revealed their beneficial impact on soil aggregation. At the broadest level our results highlight the need to consider biodiversity as a causal factor in soil aggregation. This will require a shift from the current management and physicochemical perspective to an approach that fully embraces the significance of soil organisms, their diversity and interactions.
Keywords: soil aggregation, biodiversity, meta-analysis, fungi, bacteria, animalia
The world’s soils provide many critical functions and services. For example, 95% of our food is produced by plants growing in soil 1, soil is the largest terrestrial carbon sink 2, and soil properties contribute to human health 3. In fact, past human civilizations may have failed in part due to insufficient soil stewardship 4. An essential feature of soil that allows it to provide these important functions is its structure: the arrangement of soil particles into aggregates and associated pore networks. Soil structure provides the stage for soil’s immense biodiversity 5, exerts key controls over all soil-borne biogeochemical processes 6, and is a central aspect of soil sustainability in agroecosystems.
While soil aggregation has always been viewed as a biota-driven process 7,8, it has been examined overwhelmingly in an agricultural context where the focus has been on management practices (e.g., tillage) and the physiochemical factors influencing soil aggregation (e.g., texture, soil carbon). This has, in part, resulted in the current situation where there has been comparatively limited quantification of the role of soil biodiversity in soil aggregation, despite the fact that functional roles of soil biodiversity in general are coming into focus 9,10.
Soil biota potentially contribute to soil aggregation in a number of ways (summarized in Supplementary Table 1 and 11,12). For example, bacteria can exude biopolymers that act as binding agents for aggregates on micrometer scale 13,14, fungal hyphae can entangle particles to hold them together (micrometer to millimeter scale15), and geophagous animals, such as earthworms, grind and remold ingested particles into new aggregates and create biopores (millimeter to centimeter scale; 16). Due to these various contributions of soil biota to soil aggregation, there is also a clear potential for complementarity among soil aggregation mechanisms, as has been shown in isolated studies 17,18. Overall, net effects of soil biota on soil aggregation are expected to be positive 6,12, although this has never been shown in a quantitative synthesis.
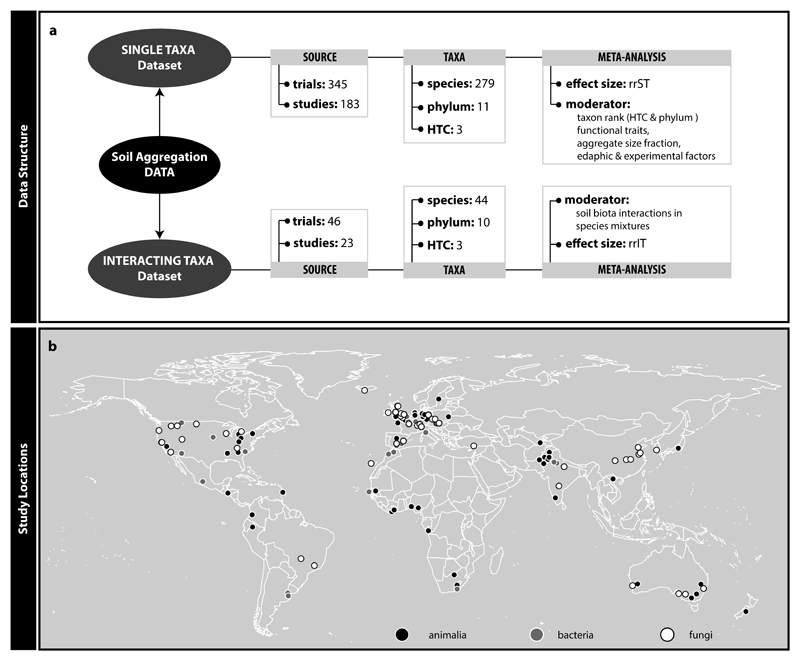
A quantitative understanding of the contribution of various soil biota groups to soil aggregation is required to predict possible consequences of soil biodiversity loss. However, efforts in this respect are at present limited to only one quantitative data synthesis, addressing the effect of arbuscular mycorrhizal fungi on soil aggregation (Glomeromycota; 19). This paper revealed significantly positive but also context-dependent effects on soil aggregation. For other soil biota groups, we currently lack such a quantitative synthesis. To fill this gap, we here conducted a global meta-analysis on published articles reporting any experimentally based soil biota effects on soil aggregation. We included effects of single species and/or species mixtures. We considered soil animals, bacteria and fungi and also pertinent organismal traits. We compiled data from 183 studies comprising 279 different soil biota species (members of 11 phyla) into a global dataset including study sites on six continents (Fig. 1, Supplementary Table 2). The data derived from field and experimental studies (52 and 131 of 183 studies, respectively), manipulating already existing soil biodiversity or introducing new soil biota, and with agricultural or ecological focus (78 and 106 of 183 studies, respectively). Using these data, we generated two datasets, one for testing effects of single taxa, the other for assessing effects of taxa interactions. We used these to address several questions, including: (i) How do major taxonomic groups differ in their effects on soil aggregation? (ii) Do functional traits of soil organisms affect their soil aggregation capability? (iii) Does the interaction of species influence soil aggregation, within or across taxa? Additionally, we evaluate how abiotic variables (experimental and edaphic factors) modulate the observed soil biota effects.
Figure 1.
Information on data structure (a) and study locations (b). We constructed two datasets (Single Taxa and Interacting Taxa dataset) from the data we collected following the literature search. These datasets differ in data volume, comprised taxonomic groups (higher taxonomic category, phylum, and species), effect size calculated and moderator variables tested. The map marks locations of studies included in this meta-analysis. There were a total of 129 different locations of which 59 investigated the impact of animals, 34 of bacteria and 55 of fungi on soil aggregation (note that in some sites more than one higher-level taxon was investigated). The map was drawn with the maps() function in R 50.
Results & Discussion
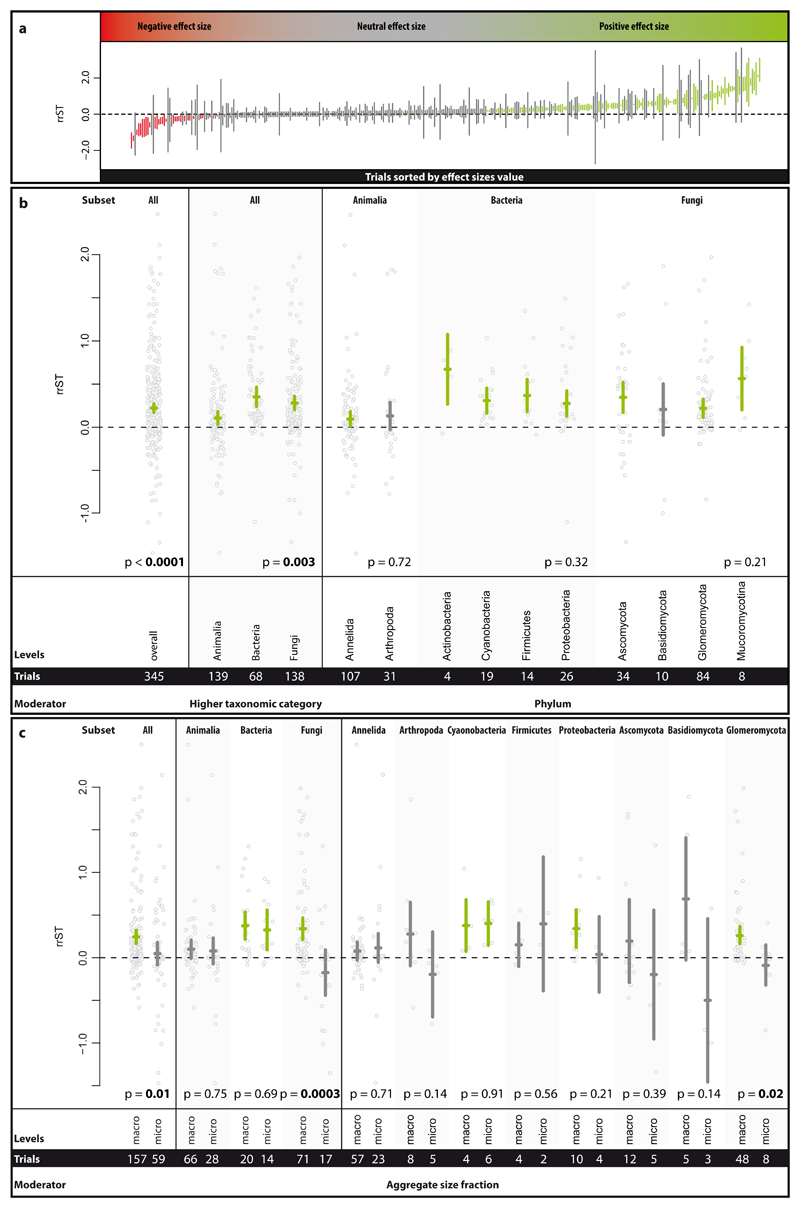
We here present a global-scale quantitative synthesis of the published evidence for soil biota contributions to soil aggregation, providing unprecedented coverage of soil biota and study settings (345 trials provided by 183 studies). Our analysis provides evidence that soil biota, taken together, have a significant positive effect (increase of 24% [CI: 18-31%]) on soil aggregation at parity of all other given study factors, including site and soil conditions (Fig. 2a, b). This provides strong support for the notion that soil biota, by virtue of their activity and products, contribute to soil aggregation (e.g. 6,12). However, our analysis also clearly reveals that this effect is not uniform in either size or direction (Fig. 2a). For example, looking at the distribution of effects across all trials, most soil biota trials had no detectable effect (60.6%), while about a third (30.4%) of the trials reported positive effects (Supplementary Figure 2). It is also noteworthy that 9.0% of trials resulted in negative effects on soil aggregation. Among the 30 negative effect sizes values, 23 trials belonged to Animalia and 7 to Fungi, and none to Bacteria. Effect sizes overall ranged from 77% decreases to over 10-fold increases in soil aggregation following soil biota additions. To verify soil biota effects, we carried out sensitivity analyses (see supplementary material), which revealed that the magnitude and direction of the effect were robust to the exclusion of trials that had the potential to introduce a bias (Supplementary Figure 5).
Figure 2.
Effect size range in the Single Taxa dataset (a) and effect of higher taxonomic category (HTC), phylum and aggregate size fraction on the effect size rrST (b and c). In panel a, the effect size values of each trial (not study) sorted by increasing effect size value are depicted, comprising 30 negative, 210 neutral and 105 positive effect size values. Effect sizes of which the 95% confidence intervals (CI) overlap the zero line (dashed line) have no clear positive or negative effect. In panel b, the summary effect over all studies included in the analysis of the Single Taxa dataset is presented. The influence of the phylum on the effect size was tested in respective subsets (soil animals, bacteria and fungi). In panel c, the impact of the aggregate size fractions (macro- vs. microaggregates) on the effect size was tested over all trials (of the Single Taxa dataset) and the respective HTC and phylum subsets. Effects are displayed as means and 95% CIs; below the p-values, the moderator levels and number of trials are shown. Significance test for between-level differences of moderators were based on a permutation test (random effects design); significant (p < 0.05) effects are marked in bold. Data points (grey) represent original data distribution of all corresponding trials.
Impact of major taxonomic groups on soil aggregation
Having established this overall pattern, we next proceeded to test in more detail for potential differences among taxonomic groups, using the Single Taxa dataset. At the highest level of taxonomic hierarchy all groups (i.e. animals, bacteria, fungi) had a positive effect size (Fig. 2a); however, bacteria and fungi had a significantly higher soil aggregation capability than soil animals. On the lowest examined rank, the phylum, we detected no significant differences among phyla within soil animals, bacteria and fungi (Fig. 2b). Arthropoda and Basidiomycota were the only taxa for which we found no clear positive, but rather a neutral effect.
Soil aggregate measurements are routinely divided into macro- and microaggregates (defined as larger and smaller than 250 µm, respectively) since many aspects, including dominant binding agents, typically differ between these two fractions 20,21. We found that the overall impact of soil biota on macro- and microaggregates differed significantly, with stronger positive effects for macroaggregates (Fig. 2c). This pattern appears to be driven by the pronounced effect for fungi, with animals and bacteria not showing this pattern. Within the fungi, this trend was consistent for all three phyla for which we had sufficient numbers of studies; however, the pattern was only statistically significant for the Glomeromycota, one of the best examined groups within the fungi 19.
These analyses are the first quantitative support from a broad data synthesis for the widely held claim that fungi, owing to their hyphal growth, contribute to stabilizing and forming macroaggregates 8. Fungal hyphae can cross-link and enmesh aggregates 15, and also produce exobiopolymers (e.g. polysaccharides 13) that act as temporary binding agents. Even though microaggregates can also form within macroaggregates, fungal effects for microaggregation are evidently much weaker. By contrast, our analysis showed that bacteria make strong contributions to both micro- and macroaggregates, even though they are generally thought to be more important as contributors of persistent binding agents for microaggregates 22,23. At the microaggregate scale, bacterial filaments, as found in some Cyanobacteria and Proteobacteria, likely function in the same way as fungal filaments for larger aggregates 24,25. Our data show that bacteria are equally important for macroaggregates, likely by contributing exobiopolymers serving as transient binding agents for macroaggregates 26.
Impact of functional traits on soil aggregation
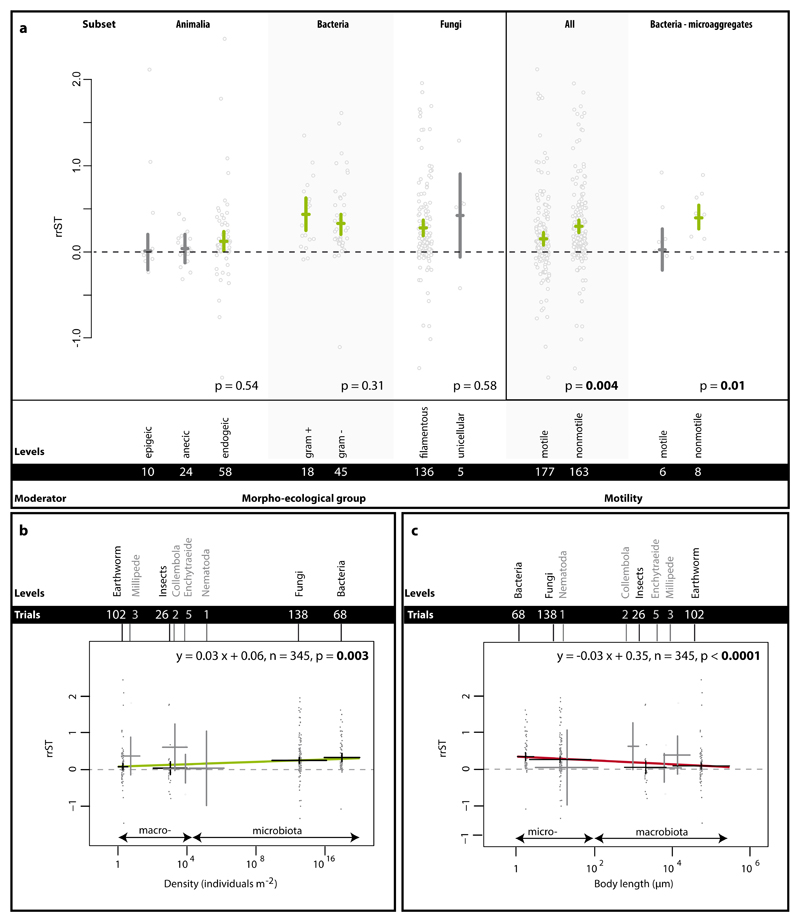
To further investigate these patterns, we examined morpho-ecological, functional and morphometric traits of the species included in the Single Taxa dataset. Despite the diversity of organisms included, we selected traits that could be universally compared, namely size and motility. For both of these we found significant patterns. Conversely, for morpho-ecological groups, which by necessity could only be defined for particular groups of biota, we found no effects (Fig. 3).
Figure 3.
Impact of functional traits on the effect size rrST in the Single Taxa dataset. In panel a, the traits motility and morpho-ecological group (presented specifically for animals (here comprising only earthworms), bacteria and fungi) are shown. Effects are represented as means and 95% CIs; below the p-values, moderator levels and number of trials included in the analysis can be found. Significance test for between-level differences of moderators were based on a permutation test (random effects design); significant (p < 0.05) effects are marked in bold. In panels b and c, the relationship between soil aggregation and organism density and body length are shown. Regression equation, number of trials and p-value are from meta-regression. Data points (grey) represent original data distribution of all corresponding trials.
Morpho-ecological group
Morphological characteristics such as growth form (filamentous vs. unicellular in fungi) or cell wall composition (gram-negative vs. gram-positive in bacteria) determine the way organisms can interact with their environment; this is exceptionally important for bacteria and fungi since they largely interact with their environment via their body surface. In fungi, both filamentous and single celled (yeast-like) forms positively influence soil aggregation (Fig. 3), but there are very limited data available for yeasts and the effect was not significant.
In bacteria, cell wall composition did not influence soil aggregation capabilities. Despite the architectural and molecular differences between gram positive and gram negative bacteria 27, the overall cell wall charge is negative for both bacterial groups and this may enhance their surface adhering capability e.g. towards clay particles and soil aggregates.
The morpho-ecological groups (epigeic, endogeic and anecic) in earthworms reflect habitat and feeding strategies. Epigeic species live and feed mainly in the upper organic layer, endogeic species in the upper mineral layer producing horizontal burrows, while anecic species inhabit vertical burrows ranging from the epigeic to endogeic habitat 28. Although all earthworms contribute to soil aggregation by basically the same mechanisms (Supplementary Table 1), we found that only endogeic earthworms significantly and positively affected soil aggregation (Fig. 3). These findings support the suggested role of endogeic earthworms as major players 12 but do not corroborate similar ideas about anecic species; the latter have been thought to improve soil aggregation by intensive burrowing and incorporation of organic material into the mineral layers 29.
Motility
To reach new areas and thus resources (e.g. nutrient patches), soil biota utilize a variety of motion types, e.g. locomotion by use of muscles or flagella, passive motion via growth (e.g. hyphal growth or colony growth), dispersion by vectors or water flow 30–32. However, how this trait affects soil aggregation has never before been examined. Here, we investigated the impact of motility on soil aggregation and found that across all biota there was a clear effect, but it was confounded by taxonomic group (Supplementary Figure 6). Thus we re-analyzed the data exclusively for bacteria, comprising both motile and nonmotile species, and found the same pattern: nonmotile bacteria had a larger effect on soil aggregation than motile bacteria. Differentiating between macro- and microaggregates, we found a significant effect at the microaggregate scale (Fig. 3a). This can probably be attributed to the localized impact of bacterial exobiopolymers and formation of biofilms produced by nonmotile species.
Body size
The size of organisms determines their interaction with biotic and abiotic environmental factors and their metabolic rate. Here we grouped body size into two broad categories across all soil biota and found clear evidence that soil microbiota has a more positive influence on soil aggregation than soil macrobiota (microbiota (body width < 100µm, n = 207): 0.30 [CI: 0.23 - 0.36]; macrobiota (body width > 100µm, n = 138): 0.10 [CI: 0.03 - 0.18]; between group differences: p >0.0001). These results correspond with our findings that fungi and bacteria have a strong positive impact on soil aggregation (Fig. 2). To further investigate these findings, we examined the related features of population density (Fig. 3b) and body length (Fig. 3c). Here, it was evident that with smaller body length and concomitantly the increased population density, soil aggregation was enhanced. Thus, size matters but population density is as important. This means that bacteria and fungi are important biotic soil aggregators likely due to their sheer abundance, smaller body size, and lower motility (for bacteria only), all of which can (Supplementary Table 1) contribute to soil structure.
Impact of within and across taxa diversity on soil aggregation
In soil, organisms are not operating in isolation but intensely interact with other soil biota as part of the soil food web that also involves symbiosis, parasitism, and competition. To investigate the possible consequences of taxa interactions on soil aggregation, we analyzed the Interacting Taxa dataset.
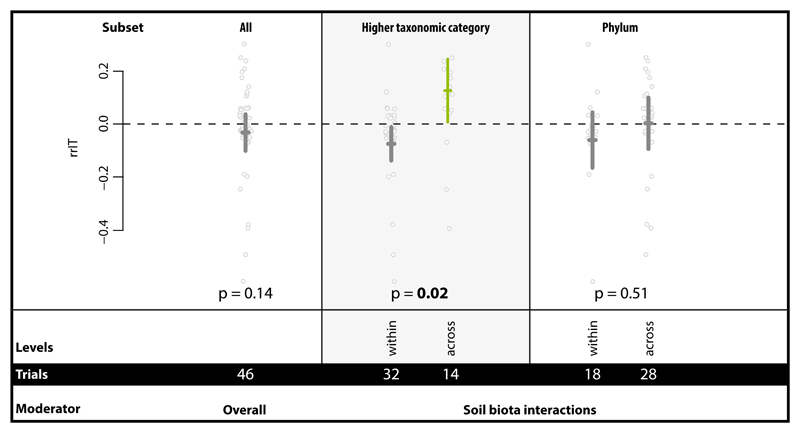
Overall, we found no enhanced soil aggregation capability in species mixtures compared to the best performing monoculture in the same study. However, soil biota interactions across higher taxonomic category (HTC) levels (animals, fungi, bacteria) had a remarkable positive effect which was not present when looking just at within-HTC combinations of species (Fig. 4). Interactions within or across phyla also did not influence the effect size. This finding represents the first quantitative evidence that diversity across different taxa more positively affects soil aggregation than diversity within same taxon; this is at least true for higher taxonomic categories. Soil is a biodiversity hotspot. There are estimates that 109 bacterial cells can be found in just 1g of soil along with 200m of fungal hyphae, 105 invertebrates and a myriad of protozoa and micro-algae 5. Hence it is inherent in the nature of soil that different higher taxonomic levels co-occur and potentially interact in the same aggregate or pore space. In other words, this large effect contributed by across-HTC pairs of species very much represents the reality of soils.
Figure 4.
Impact of soil biota interactions in mixtures across and within taxonomic groups (HTC and phylum level, respectively) on soil aggregation. This was tested with the effect size rrIT in the Interacting Taxa dataset for which additionally the overall summary effect is presented. More detailed information on taxonomic group combinations both on HTC and phylum level can be found in Supplementary Figure 7. Effects are represented as means and 95% CIs; below the p-values, moderator levels and number of trials included in the analysis are given. Significance test for between-level differences of moderators were based on a permutation test (random effects design); significant (p < 0.05) effects are marked in bold. Data points (grey) represent original data distribution of all corresponding trials.
In our dataset, the across-HTC diversity was mainly represented by bacterial:fungal interactions with animal:bacteria and animal:fungi interactions being strongly underrepresented (Supplementary Figure 7). While the data reported (see Supplementary Table 3) did not allow us to exclude that mixtures received more initial biomass than monocultures, typically organisms are added as inoculum with the assumption of growth during the experiment, suggesting that this density effect may be secondary to any complementarity. The overall positive influence of fungi and bacteria as shown for single species (Fig. 2) is also detectable when tested in mixtures. These findings support the hypothesis that there is functional complementarity contributing to soil aggregation 33, and the results highlight that this functional complementarity mainly resides at the level of HTC. The presence of pronounced organismal interaction effects highlights the opportunity to use soil biota mixtures tailored for enhancing soil aggregation (e.g. inoculation for use in restoration). This result also emphasizes the need to manage for overall high levels of soil biodiversity, especially across HTC, in agroecosystems, which would facilitate the development of such interactions. Our study also clearly suggests that future experiments would profit from being aimed at understanding the mechanistic basis of such interactions and their complementarity. Despite this generally emerging picture of functional importance of soil biodiversity levels, still relatively few studies directly address this feature in experiments (e.g. 34), most likely due to limitations in terms of experimentally generating and maintaining complex soil biodiversity gradients. Renewed efforts are necessary in this regard, and, given the importance of soil aggregation as an ecosystem process, future work should experimentally examine the role of varying levels of soil biodiversity on soil aggregation. Our results point to the need to include a broad taxonomic range of organisms in such experiments, which presents additional challenges, but is absolutely required in order not to miss essential complementary contributions.
Conclusion
In our dataset we looked at effects of experimentally selected soil biota, not the summary effect of all of a given soil’s biodiversity, for which no studies are available. However, it seems likely that, given our data, such overall biota effects would also be positive, despite the presence of negative effects and the numerical dominance of observations of neutral effects in our dataset. This is in particular the case because we observed strongly positive effect sizes resulting from pairwise interaction among different broad groups of soil organisms, a situation common in the soil environment.
This study collated all available experimental soil biodiversity data, comparing soil aggregation effects among very different organism groups, operating at different temporal and spatial scales, and representing different trait spectra. At the broadest level our results highlight the need to consider biodiversity when thinking about soil aggregation.
Materials & Methods
The meta-analysis follows the PRISMA guidelines35 (for further information see Supplementary Figure 1)
Literature search
To generate the data for our analyses, we conducted a two-step literature search in March 2016. In the first step, we used the search string “acar* OR actinomycet* OR alga* OR amoeb* OR animal* OR ant* OR archaea* OR arthropod* OR ascomycet* OR bacteri* OR basidiomycet* OR collembola* OR dinoflagellat* OR enchytrae* OR (filamentous fung*) OR fung* OR larva* OR mite* OR mycorrhiza* OR nematod* OR oligochaet* OR protist* OR (sapro* fung*) OR springtail* OR termite* OR worm*OR yeast* OR (mean diameter) OR water-stab* AND (soil aggregat* or “soil structur*”)” in the databases Web of Knowledge™ by Thomson Reuters and Scopus® excluding languages other than English, German, Spanish, Portuguese and French. The literature search results were uploaded to EndNote™, a reference management software facilitating article screening. After removing duplicates, the retrieved approximately 5000 articles were screened by title and abstract to identify potential candidate articles for our dataset. For the second step, we traced back citations from these articles and relevant reviews which yielded 60 additional articles. The resulting 731 articles were then screened according to the following inclusion/ exclusion criteria: (i) studies needed to report soil aggregation measurements when a test organism (i.e. single species) or a defined organism group (e.g. natural, soil-extracted Glomeromycota mixture) was present or not, including organism-free or organism-reduced controls. In the case of field studies usually involving earthworms, ants and termites, adjacent soil samples differing in biota composition had been measured for soil aggregation. (ii) The growth substrate had to be soil or a soil-sand mixture and usually involved sterilized substrates (e.g. autoclaved), and (iii) the effects of added stress factors (e.g. reduced irrigation, salt or heavy metal application, tillage or fertilization) were excluded and only data from the last harvest were included. (iv) In cases where a paper presented several experimental treatment combinations, we chose those in which, no additional organic matter application and no plant as additional factor in the test system were used. Identified but not available articles, were provided by the literature archive of the Botanical Museum of Berlin and by contact with study authors. A total of 183 articles met our inclusion criteria and were used in our analyses to evaluate the contribution of soil biodiversity to soil aggregation. We use the term “soil biodiversity” here in its broad sense to encompass taxa diversity, functional (trait) diversity, and interactions among soil biota36.
Effect size
For the analyses, we used the natural log response ratio (rr) of metrics representing aggregate stability (Supplementary Table 4) for treatment and control groups following the function rrX = log(XT/XC). We calculated two different effect sizes for our two datasets (Fig. 1). For the Single Taxa dataset, soil aggregation data for the organism treatment (XT) and its corresponding organism-free control (XC) were incorporated in the effect size, hereafter called rrST. For the Interacting Taxa dataset, the soil aggregation data of species mixtures (XT) and the corresponding best performing monoculture (XC) were used to calculate the effect size, hereafter called rrIT (see also Supplementary Figure 8). The effect sizes were calculated in R (package ‘metafor’) by incorporating the control and treatment means, their standard deviations and the sample sizes (N). If only standard error (SE) was presented, standard deviation (SD) was calculated as follows: SD = SE * sqrt(N). Where neither SD nor SE were reported, the median of the calculated effect size variances was used as surrogate for the missing data 37.
Moderator variables
For Single Taxa dataset, we collected data for species’ taxonomic identity, functional traits, soil aggregate size fractions and soil and experiment related factors. To characterize species’ taxonomic identity, we used (i) the higher taxonomic categories (HTC) with three levels - namely: Animalia, Bacteria and Fungi, and (ii) phylum with overall 10 levels assigned to Animalia, Bacteria or Fungi, respectively. The phyla Nematoda and Bacteriodites were strongly underrepresented with one and two species, respectively, and were not included as moderator levels. Within Animalia, we distinguished annelids and arthropods. Bacteria had four levels (Actinobacteria, Cyanobacteria, Firmicutes and Proteobacteria) and fungi had also four levels (Ascomycota, Basidiomycota, Glomeromycota and Mucoromycotina).
For the functional traits, we compiled data from the peer-reviewed literature. Traits had to be (i) relevant for the soil aggregation process and (ii) applicable to a broad range of organisms ranging from macro- to microbiota, and common to cryptic species. Motility had two levels (motile and nonmotile). As motile we defined active, spontaneous and target-oriented types of locomotion including all modes of movements of animals and flagellated microbes. Fungal growth or any movement caused by Brownian motion and the corresponding organisms were grouped as nonmotile. Body size was grouped in the two levels micro- and macrobiota 38; in macrobiota, we grouped all species with a body width >100µm. Morphometrical and density data were derived from Veresoglou et al. 39. Morpho-ecological groups were formed separately for levels of higher taxonomic categories. For animals only earthworms were included, and they had three levels (epigeic, anecic and endogeic) representing feeding and casting strategies. For bacteria, two levels were included (gram+ and gram-) representing cell wall characteristics. In fungi, the moderator had two levels (filamentous and unicellular) representing growth characteristics.
Aggregate size fraction (ASF) had two levels (micro and macro) according to the common definition of micro- (< 250 µm) and macroaggregates (250-4000 µm). If studies reported data on multiple aggregate size classes (e.g. <53µm, 53-100µm) then these data were combined into a single index following the calculation of the mean weight diameter 40:
where x̄i is the mean diameter of the measured aggregate size fraction and wi is the ratio of the weight collected in the corresponding size fraction and the total sample weight.
Additionally, we included data describing soil (soil pH, sand content, soil organic matter content, bulk density) and experiment related factors (experimental setting, experimental duration, substrate sterilization, organic matter application, plant) to evaluate their importance in modulating soil aggregation effects of soil biota. Further information and the corresponding results can be found in the Supplementary Information.
For the Interacting Taxa dataset, the moderator variable soil biota interactions in species mixtures was tested on HTC and phylum level, respectively. This moderator had the two levels within and across. Soil biota interactions within taxonomic groups included in our dataset was present when species of the same HTC or phylum, respectively, were combined in the species interaction treatments. For example, combinations of two different earthworm species were scored as within-phylum level (annelida:annelida) and within-HTC level (Animalia:Animalia) interactions. For across taxonomic group interactions of soil biota, we used data from studies where interacting species belonged to different phyla or HTCs, respectively. For example, a collembolan combined with an arbuscular mycorrhizal fungus resulted in a phylum level interaction of Arthropoda:Glomeromycota and in HTC level interactions of Animalia:Fungi. This moderator variable was only tested against the effect size rrIT in the Interacting Taxa dataset (Fig. 1).
Statistics
Following construction of the two datasets (Single Taxa and Interacting Taxa dataset; Fig. 1A), we obtained multiple effect size values per study, which violates the assumption of independence for meta-analysis data. Thus to reduce the number of effect size values per study when possible we used three data merging approaches: (i) phylogenetic corrected 41, (ii) common control corrected and (iii) multiple trials merging. Phylogenetic corrected merging was accomplished in R v.3.3.1 42 via the rma.mv() function in the ‘metafor’ package 43 by building models with species as random factor and an implemented phylogenetic correlation matrix (derived from incorporated maximum likelihood trees) constructed by the ‘phytools’ package 44; for detailed information see Supplementary Information. Common controls correction was implemented using the ‘metagear’ package 45, which aligns the prior constructed variance-covariance matrix with the corresponding effect size dataset. Multiple trial merging was performed by calculating random-effects models by the function rma.uni() in the ‘metafor’ package resulting in a combined effect size value and corresponding variance.
The resulting datasets were analyzed by random-effects meta-analyses (‘metafor’ package) incorporating study weighing by the inverse of the effect size variance and the restricted maximum likelihood method. Since data were not normally distributed, we applied permutation and bootstrapping approaches for estimation of p-values and CIs, using functions implemented in the ‘metafor’ and ‘boot’ packages (3999 iterations; 46,47). Finally, we applied different types of sensitivity analyses to verify (i) presence of potential confounding factors via subset analyses (Supplementary Figure 5; 48), (ii) presence of publication bias (Supplementary Figure 9; 49) and (iii) robustness of results by the ‘disproportional impact of studies’ approach (Supplementary Figure 10-21; only robust results are presented; 37). Analysis outcomes of sensitivity analyses and further information can be found in the according section of the Supplementary Information.
Supplementary Material
Acknowledgement
We thank J. Antonovics for input on this manuscript. AL and MCR acknowledge funding from Deutsche Forschungsgemeinschaft (DFG, grant no: RI 1815/16-1); MCR additionally acknowledges funding from an ERC Advanced Grant (694368) and the Federal Ministry for Education and Research (BMBF)-funded project INPLAMINT.
Footnotes
Data availability. All data generated or analyzed during this study are included in this published article (and its Supplementary Information and Supplementary Data files).
Author contribution
A.L. designed and performed the research; W.Z. contributed analytical tools; all authors contributed to writing the paper.
Competing interests
The authors declare no competing financial interests.
References
- 1.FAO. Healthy soils are the basis for healthy food production. 2015:1–4. [Google Scholar]
- 2.Lal R. Soil carbon sequestration impacts on global climate change and food security. Science. 2004;304:1623–1627. doi: 10.1126/science.1097396. [DOI] [PubMed] [Google Scholar]
- 3.Wall DH, Nielsen UN, Six J. Soil biodiversity and human health. Nature. 2015;528:69–76. doi: 10.1038/nature15744. [DOI] [PubMed] [Google Scholar]
- 4.Diamond J. Collapse: How Societies Choose to Fail or Succeed. Pinguin Group; 2011. [Google Scholar]
- 5.Dindal DL. Soil Biology Guide. Wiley-Interscience: 1990. [Google Scholar]
- 6.Bronick CJ, Lal R. Soil structure and management: a review. Geoderma. 2005;124:3–22. [Google Scholar]
- 7.Rillig MC, Mummey DL. Mycorrhizas and soil structure. New Phytol. 2006;171:41–53. doi: 10.1111/j.1469-8137.2006.01750.x. [DOI] [PubMed] [Google Scholar]
- 8.Tisdall JM, Oades JM. Organic-matter and water-stable aggregates in soils. J Soil Sci. 1982;33:141–163. [Google Scholar]
- 9.Wagg C, Bender SF, Widmer F, van der Heijden MGA. Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc Natl Acad Sci USA. 2014;111:5266–5270. doi: 10.1073/pnas.1320054111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delgado-Baquerizo M, et al. Circular linkages between soil biodiversity, fertility and plant productivity are limited to topsoil at the continental scale. New Phytol. 2017;215:1186–1196. doi: 10.1111/nph.14634. [DOI] [PubMed] [Google Scholar]
- 11.Lehmann A, Leifheit EF, Rillig MC. In: Mycorrhizal Mediation of Soil - Fertility, Structure, and Carbon Storage. Johnson N, Gehring C, Jansa J, editors. Elsevier; 2016. [Google Scholar]
- 12.Six J, Bossuyt H, Degryze S, Denef K. A history of research on the link between (micro)aggregates, soil biota, and soil organic matter dynamics. Soil Till Res. 2004;79:7–31. [Google Scholar]
- 13.Chenu C. Influence of a fungal polysaccharide, scleroglucan, on clay microstructure. Soil Biol Biochem. 1989;21:299–305. [Google Scholar]
- 14.Deng JZ, et al. Synergistic effects of soil microstructure and bacterial EPS on drying rate in emulated soil micromodels. Soil Biol Biochem. 2015;83:116–124. [Google Scholar]
- 15.Degens BP. Macro-aggregation of soils by biological bonding and binding mechanisms and the factors affecting these: A review. Aust J Soil Res. 1997;35:431–459. [Google Scholar]
- 16.Blanchart E, et al. SWORM: an agent-based model to simulate the effect of earthworms on soil structure. Eur J Soil Sci. 2009;60:13–21. [Google Scholar]
- 17.Leifheit EF, Verbruggen E, Rillig MC. Arbuscular mycorrhizal fungi reduce decomposition of woody plant litter while increasing soil aggregation. Soil Biol Biochem. 2015;81:323–328. [Google Scholar]
- 18.Siddiky MRK, Schaller J, Caruso T, Rillig MC. Arbuscular mycorrhizal fungi and collembola non-additively increase soil aggregation. Soil Biol Biochem. 2012;47:93–99. [Google Scholar]
- 19.Leifheit EF, Veresoglou SD, Lehmann A, Morris EK, Rillig MC. Multiple factors influence the role of arbuscular mycorrhizal fungi in soil aggregation-a meta-analysis. Plant Soil. 2014;374:523–537. [Google Scholar]
- 20.Miller RM, Jastrow JD. In: Mycorrhizae in Sustainable Agriculture. Bethlenfalvayand CF, Linderman RG, editors. Soil Science Society of America; Madison, Wi: 1992. [Google Scholar]
- 21.Tisdall JM, Smith SE, Rengasamy P. Aggregation of soil by fungal hyphae. Aust J Soil Res. 1997;35:55–60. [Google Scholar]
- 22.Marshall KC. Interfaces in Microbial Ecology. Harvard University Press; 1976. [Google Scholar]
- 23.Hattori T. Microbial Life in Soil. Marcel Dekker; 1973. [Google Scholar]
- 24.Belnap J, Gardner JS. Soil microstructure in soils of the Colorado Plateau - the role of the cyanobacterium Microcoleus vaginatus. Great Basin Nat. 1993;53:40–47. [Google Scholar]
- 25.Smith SM, Abed RMM, Garcia-Pichel F. Biological soil crusts of sand dunes in Cape Cod National Seashore, Massachusetts, USA. Microb Ecol. 2004;48:200–208. doi: 10.1007/s00248-004-0254-9. [DOI] [PubMed] [Google Scholar]
- 26.Santaella C, Schue M, Berge O, Heulin T, Achouak W. The exopolysaccharide of Rhizobium sp. YAS34 is not necessary for biofilm formation on Arabidopsis thaliana and Brassica napus roots but contributes to root colonization. Environ Microbiol. 2008;10:2150–2163. doi: 10.1111/j.1462-2920.2008.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown L, Wolf JM, Prados-Rosales R, Casadevall A. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol. 2015;13:620–630. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisenhauer N. The action of an animal ecosystem engineer: Identification of the main mechanisms of earthworm impacts on soil microarthropods. Pedobiologia. 2010;53:343–352. [Google Scholar]
- 29.Lee KE. Earthworms: their ecology and relationships with soils and land use. Academic Press; 1985. [Google Scholar]
- 30.Pijper A. In: Ergebnisse der Mikrobiologie, Immunitätsforschung und experimentellen Therapie. Kikuth W, et al., editors. Springer Verlag; Heidelberg: 1957. [DOI] [PubMed] [Google Scholar]
- 31.Quillin KJ. Ontogenetic scaling of hydrostatic skeletons: Geometric, static stress and dynamic stress scaling of the earthworm Lumbricus terrestris. J Exp Biol. 1998;201:1871–1883. doi: 10.1242/jeb.201.12.1871. [DOI] [PubMed] [Google Scholar]
- 32.Tan Y, Gannon JT, Baveye P, Alexander M. Transport of bacteria in an aquifer sand - experiments and model simulations. Water Resour Res. 1994;30:3243–3252. [Google Scholar]
- 33.Lehmann A, Rillig MC. Understanding mechanisms of soil biota involvement in soil aggregation: A way forward with saprobic fungi? Soil Biol Biochem. 2015;88:298–302. [Google Scholar]
- 34.de Graaff MA, Adkins J, Kardol P, Throop HL. A meta-analysis of soil biodiversity impacts on the carbon cycle. Soil. 2015;1:257–271. [Google Scholar]
- 35.Moher D, Liberati A, Tetzlaff J, Altman DG, Group TP Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009;6 [PMC free article] [PubMed] [Google Scholar]
- 36.Bender SF, Wagg C, van der Heijden MGA. An Underground Revolution: Biodiversity and Soil Ecological Engineering for Agricultural Sustainability. Trends Ecol Evol. 2016;31:440–452. doi: 10.1016/j.tree.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 37.Copas J, Shi JQ. Meta-analysis, funnel plots and sensitivity analysis. Biostatistics. 2000;1:247–262. doi: 10.1093/biostatistics/1.3.247. [DOI] [PubMed] [Google Scholar]
- 38.Bardgett RD. The Biology of Soil: A Community and Ecosystem Approach. 1st edn. Oxford University Press; 2005. [Google Scholar]
- 39.Veresoglou SD, Halley JM, Rillig MC. Extinction risk of soil biota. Nat Commun. 2015;6 doi: 10.1038/ncomms9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kemper WD, Rosenau RC. Aggregate stability and size distribution. American Society of Agronomy - Soil Science Society of America; 1986. [Google Scholar]
- 41.Anderson JT. Plant fitness in a rapidly changing world. New Phytol. 2016;210:81–87. doi: 10.1111/nph.13693. [DOI] [PubMed] [Google Scholar]
- 42.R: A Language and Environment for Statistical Computing. Vienna, Austria: 2014. [Google Scholar]
- 43.Viechtbauer W. Conducting Meta-Analyses in R with the metafor Package. J Stat Softw. 2010;36:1–48. [Google Scholar]
- 44.Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3:217–223. [Google Scholar]
- 45.Lajeunesse MJ. Facilitating systematic reviews, data extraction, and meta-analysis with the metagear package for R. Methods Ecol Evol. 2016;7:323–330. [Google Scholar]
- 46.Canty A, Ripley B. Boot: Bootstrap R (S-Plus) Functions. R package version 1.3-18. 2015 [Google Scholar]
- 47.Knapp G, Hartung J. Improved tests for a random effects meta-regression with a single covariate. Stat Med. 2003;22:2693–2710. doi: 10.1002/sim.1482. [DOI] [PubMed] [Google Scholar]
- 48.Borenstein M, Higgins JPT. Meta-Analysis and Subgroups. Prev Sci. 2013;14:134–143. doi: 10.1007/s11121-013-0377-7. [DOI] [PubMed] [Google Scholar]
- 49.Sterne JAC, Egger M. Funnel plots for detecting bias in meta-analysis: Guidelines on choice of axis. J Clin Epidemiol. 2001;54:1046–1055. doi: 10.1016/s0895-4356(01)00377-8. [DOI] [PubMed] [Google Scholar]
- 50.Becker RA, Wilks AR, Brownrigg R, Minka TP, Deckmyn A. maps: Draw Geographical Maps. 2016 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.