Abstract
RNA molecules have emerged as promising therapeutics. Like all other drugs, the safety profile and immune response are important criteria for drug evaluation. However, the literature on RNA immunogenicity has been controversial. Here, we used the approach of RNA nanotechnology to demonstrate that the immune response of RNA nanoparticles is size, shape, and sequence dependent. RNA triangle, square, pentagon, and tetrahedron with same shape but different sizes, or same size but different shapes were used as models to investigate the immune response. The levels of pro-inflammatory cytokines induced by these RNA nanoarchitectures were assessed in macrophage-like cells and animals. It was found that RNA polygons without extension at the vertexes were immune inert. However, when single-stranded RNA with a specific sequence was extended from the vertexes of RNA polygons, strong immune responses were detected. These immunostimulations are sequence specific, because some other extended sequences induced little or no immune response. Additionally, larger-size RNA square induced stronger cytokine secretion. 3D RNA tetrahedron showed stronger immunostimulation than planar RNA triangle. These results suggest that the immunogenicity of RNA nanoparticles is tunable to produce either a minimal immune response that can serve as safe therapeutic vectors, or a strong immune response for cancer immunotherapy or vaccine adjuvants.
Keywords: RNA nanotechnology, RNA nanoparticle, pRNA 3WJ, RNA polygon, immunogenicity, immunomodulation, immune response, nanobiotechnology
Introduction
When the first human genome was completely sequenced a decade ago, it was found that only 1.5% of the human genome codes for protein.1 A large portion of the remaining 98.5% was proposed to be “Junk DNA.”2 However, more and more evidence revealed that a substantial part of the so-called Junk DNA encodes small non-coding RNA.3, 4 Recently, many long non-coding RNAs were identified with the advancement of technology, making it possible to characterize long non-coding RNA molecules.5 It has been predicted that the third milestone in drug development will be RNA drugs, either RNA itself as drugs or chemicals/ligands that target RNA, following the first milestone of chemical drugs and the second milestone of protein drugs including antibodies, enzymes, hormones, or chemicals/ligands that target proteins.6
Many small RNA oligos and moieties, such as short interfering RNA (siRNA),7, 8 microRNA (miRNA),9, 10 ribozymes,11, 12 aptamers,13 and riboswitches,14 have been reported to be potential powerful therapeutics. Meanwhile, a variety of RNA species or analogs have also been reported to be immunogenic. These include viral single-stranded RNA (ssRNA),15, 16 double-stranded RNA (dsRNA),17, 18 specific mRNA,19, 20 specific siRNA,21, 22, 23 immune checkpoint RNA aptamers,24, 25, 26 and inosine incorporated synthetic RNA.27, 28 Previous studies have identified the immunological targets for these special RNA molecules, including cell surface or endosome Toll-like receptors (TLRs; TLR-3, -7, -8, and -9)15, 16, 17, 18, 29 and cytosolic sensors (protein kinase R [PKR], RIG-I, and MDA-5).30, 31, 32, 33 Recognition of these specific RNA sequences (SEQs) or structures by their corresponding immune receptors will trigger a series of immune events that are part of the host’s natural defense, including cytokine secretion, immune cell proliferation and survival, or activation of adaptive immunity.
The RNA nanotechnology concept was proposed in 1998 by showing that RNA dimers, trimers, tetramers, and hexamers can self-assemble from reengineered RNA monomers by a bottom-up assembly approach.34 Consequently, the field of RNA nanotechnology has emerged rapidly.35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45 This field is the study of RNA architectures in nanoscale with their major frame composed of RNA. In these nanoarchitectures, the scaffolds, targeting ligands, therapeutics, or regulators are mainly made up of RNA. The applications of RNA nanotechnology in cancer therapy have been extensively investigated.35, 36, 46, 47, 48, 49, 50, 51, 52, 53 RNA nanoparticles display distinct attributes including controllable size, shape, and stoichiometry, favorable thermodynamic stability in vitro and in vivo, specific targeting for diseased cells, and low accumulation in the liver and other vital organs.38, 39, 40, 41, 42, 43, 44, 45, 54, 55, 56, 57 Many RNA nanoparticles have been reported to be safe because they are negatively charged, and thus have a lower intention to bind to or enter normal cells that contain negatively charged cell membrane. Consequently, RNA nanoparticles have little impairment effect on normal cells or vital tissues46, 47, 48, 49, 53, 58 and do not induce cytokine or interferon.59, 60 The chemical properties of RNA nanoparticles are similar to siRNA, miRNA, ribozymes, aptamers, riboswitches, or other immunogenic RNA in that they are all composed of ribonucleotides. The main difference is that RNA nanoparticles are larger in size and are constructed by bottom-up self-assembly of multiple RNA oligos. This raises an interesting question of whether the size or shape of RNA plays a role in triggering potential immune response and toxicity, because RNA nanoparticles are similar in size to viruses. Additionally, it was reported that immunostimulatory nucleic acid molecules have been incorporated into phi29-based RNA polygons to enhance immunomodulation in vivo, while RNA polygons themselves display little to no immunoactivity.59 Other studies reported that the immunological properties of a hybrid RNA/DNA cube can be conditionally controlled by tuning the ratio between RNA/DNA strands.61, 62 These raise another question whether the RNA SEQ plays a role in triggering immune response and toxicity.
Toxicity, side effects, and immunogenicity are some of the most important criteria in drug evaluation, aside from drug efficacy and shelf life. A thorough investigation of the immune-compatibility of RNA nanoparticles is important for their translation to the clinic.63, 64, 65 To help explore the basic mechanism of the immunogenicity of RNA nanoparticles, here we use RNA polygons and tetrahedron (Th) that have the same size but different shapes or the same shape with different sizes, and variable SEQ extensions, as model systems. It was found that their immunomodulatory property is dependent on size, shape, and sequence. Our results suggest that RNA nanoparticles can be manually designed using their programmable property to exhibit no, low, or high immunogenicity, allowing for the development of potential RNA-based immunomodulators for cancer immunotherapy or as vaccine adjuvants.
Results
Design, Self-Assembly, and Physicochemical Characterization of RNA Nanoparticles with Extended Special SEQ
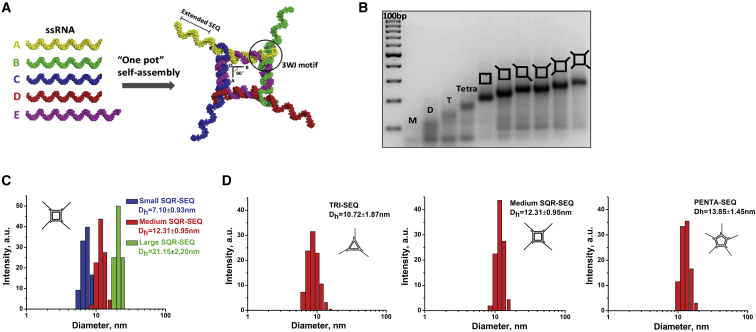
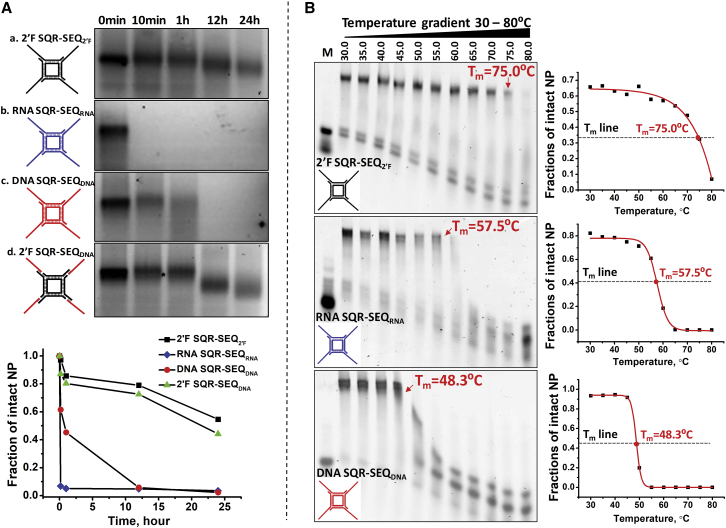
Different RNA SEQs were incorporated into RNA square (SQR) by direct extension of the four external sticky ends (Figure 1A). To examine the assembly, we evaluated the stepwise annealing of SQRs containing increasing copies of extended SEQs by gel electrophoresis. The observed homogeneous bands (Figure 1B) and size assessment by dynamic light scattering (DLS) (Figure 1C) confirm the expected formation of the designed nanoparticles and high assembly yield. By altering the length of each side of the SQR, smaller and larger SQR-SEQ nanoparticles were constructed using the same approach (Figure S1). The average hydrodynamic diameter (Dh) distribution was determined to be 7.10 ± 0.93, 12.31 ± 0.95, and 21.15 ± 2.20 nm for small, medium, and large SQR-SEQ nanoparticles, respectively (Figure 1C). By changing the number of external RNA strands of the medium SQR-SEQ nanoparticle, a triangle-SEQ (TRI-SEQ) and a pentagon-SEQ (PENTA-SEQ) were constructed with 10.72 ± 1.87 and 13.85 ± 1.45 nm of average Dh (Figure 1D). Next, a serum degradation assay was performed to evaluate the enzymatic stability of 2′-fluorine (2′F)-modified SQR-SEQ. Four SQR-SEQ nanoparticles with different compositions (2′F SQR-SEQ2′F, RNA SQR-SEQRNA, DNA SQR-SEQDNA, and 2′F SQR-SEQDNA) were studied for comparison. It was found that 2′F SQR-SEQ2′F remained most stable compared with the other counterparts (Figure 2A). DNA SQR-SEQDNA began to degrade within 10 min and was completely degraded by 12 hr. Substitution of the 2′F SQR core structure in place of the DNA SQR resulted in a remarkable improvement on nuclease resistance. A distinguishable increase in the migration rate of gel bands between 1 and 12 hr time points of 2′F SQR-SEQDNA suggests that the four external SEQDNA strands, which were hybridized to 2′F SQR, were degraded, whereas the 2′F SQR core structure remained stable. After 24 hr, 54.6% of 2′F SQR-SEQ2′F remained intact, compared with 3.5% for RNA SQR-SEQRNA, 2.3% for DNA SQR-SEQDNA, and 44.2% for 2′F SQR-SEQDNA. Furthermore, the melting temperature (Tm) of 2′F SQR-SEQ2′F, RNA SQR-SEQRNA, and DNA SQR-SEQDNA nanoparticles was determined to be 75.0°C, 57.5°C, and 48.3°C, respectively, by a perpendicular temperature gradient gel electrophoresis (TGGE) (Figure 2B). The serum degradation assay and Tm measurement indicate that the 2′F SQR-SEQ2′F nanoparticle exhibited the highest chemical and thermodynamic stability compared with other compositions.
Figure 1.
Assembly and Size Characterization of 2’F RNA Nanoparticles
(A) Assembly of 2′F SQR-SEQ with 3D modeled structures are shown. (B) Stepwise assembly of 2′F SQR-SEQ with different numbers of modules, evaluated by 3% agarose gel (ladder: 100 bp DNA). (C and D) Size distribution of small, medium, and large RNA nanoparticles measured by DLS (n = 3). D, dimer; M, monomer; T, trimer; Tetra, tetramer.
Figure 2.
Serum Stability Assay and Tm Measurement of Different SQR-SEQ Nanoparticles
(A) Serum stability of (a) 2′F SQR-SEQ2′F, (b) RNA SQR-SEQRNA, (c) DNA SQR-SEQDNA, and (d) 2′F SQR-SEQDNA assayed in 10% FBS supplemented cell culture medium. (B) Tm measurement of 2′F SQR-SEQ2′F, RNA SQR-SEQRNA, and DNA SQR-SEQDNA by temperature gradient gel electrophoresis (TGGE). Quantification analysis of intact nanoparticles % was performed by ImageJ.
Binding of 2′F SQR with Extended Special SEQ to Macrophage-like RAW 264.7 Cells
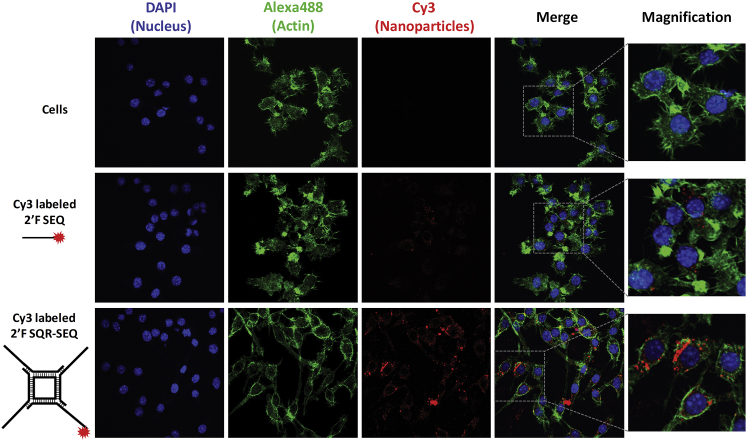
The effective delivery to targeted cells is important to elicit immunomodulatory activity. Unfortunately, many small nucleic acid therapeutics, such as short single-stranded antisense oligos, have shown difficulty in binding to or entering cells because of their negative charge and small size, and were either expelled by the negatively charged cell membrane or were excluded from macrophage engulfment.66, 67 To demonstrate increased cellular binding of RNA nanoparticles compared with small ssRNA, we incubated both Cy3-labeled 2′F SQR-SEQ (50 nM) and 2′F SEQ (200 nM) with macrophage-like RAW 264.7 cells at equimolar concentration of SEQ and evaluated them by confocal microscopy. Co-localization of the Cy3 signal (red) and cellular actin (green) in confocal imaging indicates that 2′F SQR-SEQ strongly bind to RAW 264.7 cells, whereas only weak Cy3 signal was observed in the 2′F SEQ group (Figure 3). This suggests that macrophage binding occurred in a size-dependent manner because larger RNA nanoparticles can be caught by the RAW 264.7 cells much easier than smaller ones. In addition, flow cytometry analysis showed that the cellular binding events rose with increasing copies of 2′F SEQ per nanoparticle (Figure S2).
Figure 3.
Cellular Binding Comparison of Medium 2’F SQR-SEQ and 2’F SEQ
Confocal images showing the cellular binding comparison of medium 2′F SQR-SEQ and 2′F SEQ to macrophage-like RAW 264.7 cells by co-localization of nucleus (blue), cytoplasm (green), and Cy3-labeled nanoparticles (red) signals.
Comparison of the Immunostimulation of RNA Nanoparticles with Different Sizes and Stoichiometries
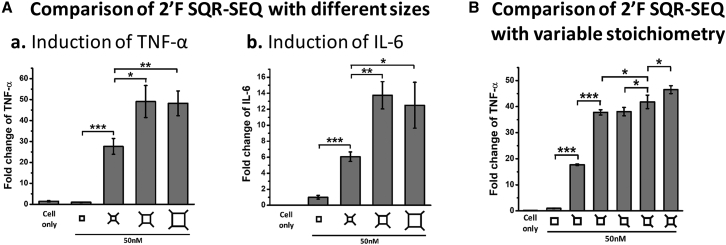
It has been reported that physicochemical properties of particles will affect their interaction with the body’s immune system, triggering different levels of immune responses.63, 65, 68, 69, 70, 71 In this study, 2′F small (7.10 nm), medium (12.31 nm), and large (21.15 nm) SQRs with SEQ extensions were constructed at the same concentration (50 nM) for the study of size-dependent immunomodulation. The immunostimulatory activity was evaluated by using ELISA to measure the stimulated cytokine levels of tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6). The small 2′F SQR-SEQ nanoparticle elevated the immunomodulation to some extent, while a more significant improvement was observed with medium and large 2′F SQR-SEQ nanoparticles (Figure 4A). These results indicate that RNA nanoparticles display immunoactivity in a size-dependent manner, and their size should be greater than 7 nm to exhibit significant immunomodulation. We also evaluated medium 2′F SQRs harboring different numbers of 2′F SEQ modules and observed that the responses increased in a stoichiometry-dependent manner. It can be found that incorporating more 2′F SEQ to 2′F SQR (50 nM) led to higher cytokine secretion levels (Figure 4B).
Figure 4.
Size- and Stoichiometry-Dependent Cytokines Induction in RAW 264.7 Cells by 2′F SQR-SEQ
(A) Cytokines (a) TNF-α and (b) IL-6 induction by small (S), medium (M), and large (L) 2′F SQR-SEQ. (B) Cytokine TNF-α induction by medium 2′F SQR-SEQ with different numbers of extensions (concentrations refer to nanoparticles; fold changes were determined by normalizing the cytokine level elicited by 2′F SQR without extension as 1; results are presented as mean ± SD; n = 3; *p < 0.05; **p < 0.01; ***p < 0.001, analyzed by Student’s t test).
Comparison of the Immunostimulation of RNA Nanoparticles with Different Shapes
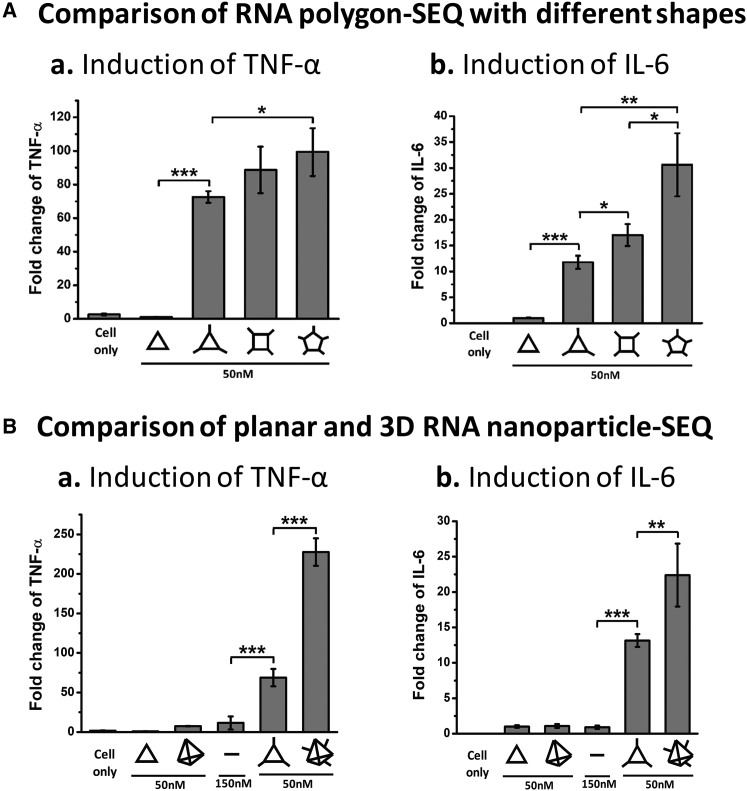
2′F RNA triangle, square, and pentagon with the extension of SEQ were designed to be close in size and constructed by stretching the inner AOB angle to 60°, 90°, or 108°.59 It was observed that higher cytokine secretion levels were induced with the transition from triangle, to square, and to pentagon at the same concentration (50 nM) (Figure 5A). This should be due to the different stoichiometry of the RNA polygons with different shapes (pentagon, square, and triangle can carry five, four, and three SEQ modules, respectively). As negative controls, nude 2′F RNA triangles without extended SEQ were assessed, but no obvious cytokine secretion was observed.
Figure 5.
Shape-Dependent Cytokines Induction in RAW 264.7 Cells by 2′F RNA Nanoparticles
(A) Cytokines (a) TNF-α and (b) IL-6 induction by 2′F TRI-SEQ, 2′F SQR-SEQ, and 2′F PENTA-SEQ. (B) Cytokines (a) TNF-α and (b) IL-6 induction by planar 2′F TRI-SEQ and 3D 2′F Th-SEQ (concentrations refer to nanoparticles; fold changes were determined by normalizing the cytokine level elicited by 2′F TRI without extension as 1; results are presented as mean ± SD; n = 3; *p < 0.05; **p < 0.01; ***p < 0.001, analyzed by Student’s t test).
To compare the effect of planar and three-dimensional RNA nanostructures on enhancing immunomodulation, we constructed a well-characterized 2′F RNA Th43 with three extended SEQ (Dh = 11.18 ± 2.28 nm) (Figures S1 and S3). 2′F Th-SEQ was used to compare with 2′F TRI-SEQ because both harbor three SEQ extensions. It was observed that the 2′F Th-SEQ (50 nM) elicited a much stronger immune response than the 2′F TRI-SEQ (50 nM) (Figure 5B). Taking the free 2′F SEQ control (150 nM) into consideration, it suggests a possible rule that the immunogenicity of RNA nanoparticles will increase with the transition from linear, to planar, and to three-dimensional structures. Our results demonstrate that the size and shape of RNA nanoparticles have an essential effect on the immune response elicited by the extended 2′F SEQ. It is suggested that it would be beneficial to control the immunoactivity of RNA nanoparticles through altering size or shape to reach a desired level.
Special RNA SEQs as Key Factors in the Immunogenicity of RNA Nanoparticles as Revealed in Cell Assay and Animal Trial
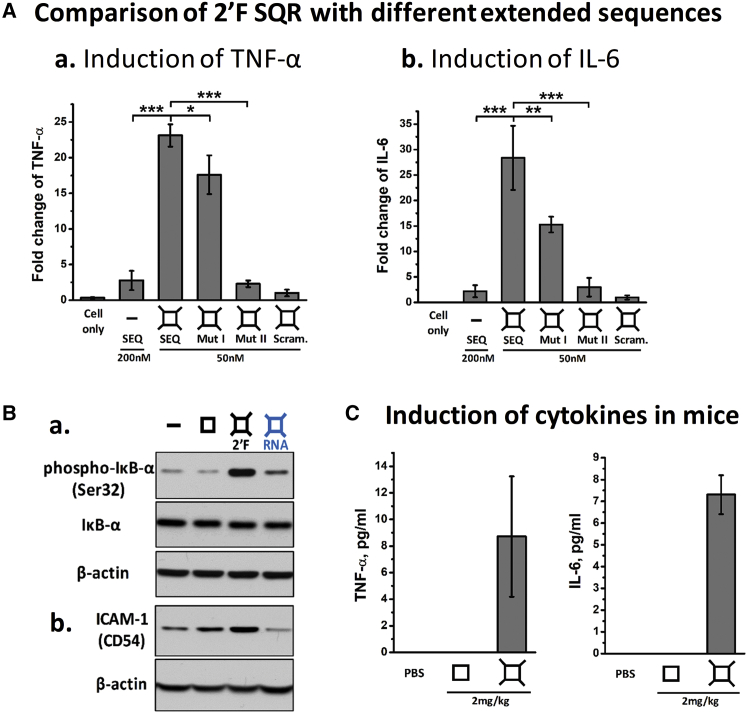
Some specific RNA SEQs were reported to trigger immune response because of their recognition and specific interaction with various TLRs (TLR-3, -7, and -8) or cytosolic sensors (PKR, RIG-1, and MDA-5) that are widely expressed on immune cells.15, 16, 17, 18, 29, 30, 31, 32, 33 To study the importance of SEQ specificity in immunomodulation of RNA nanoparticles, we employed four ssRNA SEQs (2′F SEQ, Mut I, Mut II, and a scramble SEQ control; see Table S1) as external extensions to the medium SQR for comparison. The free 2′F SEQ (200 nM) only induced low response (Figure 6A). However, extensions of SEQ from the vertexes of 2′F SQR (50 nM) resulted in strong immunostimulations of cytokines TNF-α and IL-6 (Figures 4 and 6). The results suggest that the specific RNA SEQs extensions play a central role, because the 2′F SQR-SEQ enhanced immune response, while the effects of 2′F SQR-Mut SEQ I (50 nM) and 2′F SQR-Mut SEQ II (50 nM) were reduced and almost abolished, respectively. Negligible immune response was detected for 2′F SQR with extended random SEQ (2′F SQR-scramble, 50 nM), our negative control. In addition, blocking of 2′F SEQ at the end of the vertexes by a complementary ssRNA strand resulted in decreased cytokine stimulation (Figure S4). More importantly, we have demonstrated the ability to tune their immunoactivity by simply varying the extended SEQ. To further confirm immunomodulation, we analyzed the activation of the cytokine signaling pathway in RAW 264.7 cells stimulated by the medium 2′F SQR-SEQ by immunoblotting assay. The significantly enhanced phosphorylation of IκB-α protein could subsequently activate NF-κB and inflammatory responses, and demonstrates that 2′F SQR-SEQ exhibited strong immunostimulatory activity (Figure 6Ba). ICAM-1 (Intercellular Adhesion Molecule 1), another protein marker involved in the pro-inflammatory pathways, was also upregulated in the 2′F SQR-SEQ-treated group (Figure 6Bb). As a control, the unmodified RNA SQR-SEQ only triggered low or negligible effects because of their instability in serum environment. Next, 2′F SQR-SEQ was systemically injected into CD-1 mice to evaluate the immunostimulation in vivo. After 3 hr post-injection, 2′F SQR-SEQ induced significant cytokine induction in mice, whereas the nude 2′F SQR control showed undetectable effect (Figure 6C). This result is consistent with the in vitro results, confirming that RNA nanoparticles are immune inert in vivo but can be manipulated to be immune active with the extensions of special RNA SEQs.
Figure 6.
Sequence-Dependent Cytokines Induction and Immune Markers Activation in RAW 264.7 Cells and Mice by 2′F SQR-SEQ
(A) Cytokines (a) TNF-α and (b) IL-6 induction by medium 2′F SQR with SEQ, mutated SEQ, and scramble RNA (concentrations refer to nanoparticles; fold changes were determined by normalizing the cytokine level elicited by 2′F SQR-Scramble as 1). (B) Activation of (a) phosphorylated-IκB-α (Ser32) and (b) ICAM-1 (CD54) stimulated by medium 2′F SQR-SEQ. (C) Cytokines TNF-α and IL-6 induction by medium 2′F SQR-SEQ in mice (concentrations refer to SEQ per body weight; results are presented as mean ± SD; n = 3; *p < 0.05; **p < 0.01; ***p < 0.001, analyzed by Student’s t test).
Discussion
Drug safety profiles including toxicity, side effects, and immunogenicity are essential for drug evaluation. Nanotechnology-based carriers have been widely used for the delivery of therapeutics because there are numerous advantages to using nanoparticles over traditional routes.41, 72, 73, 74 However, certain detrimental immune responses have also been reported, leading to some concerns.63, 64, 65 It has been pointed out that these side effects might be caused by a wide range of physicochemical properties such as surface charge, solubility, or hydrophobicity.63, 64, 65 Thus, controlling the immune response triggered by nanoparticles remains one of the challenges for many nano-delivery systems. Notably, the physicochemical properties of RNA nanoparticles are tunable, making them an attractive nanomaterial. Their size, shape, sequence, stoichiometry, and other properties can be controlled at ease, and the procedures and process for RNA nanoparticles construction is reproducible. Therefore, their immunomodulatory effect can be controlled precisely through rational design.
It has been previously shown that RNA nanoparticles themselves might not display intrinsic immunogenicity.59, 60 That premise is supported here by this work. Incorporation of a special SEQ to RNA nanoparticles endows them with strong immunostimulatory activity compared with RNA nanoparticles without extension controls (Figures 4, 5, and 6). It was found that size is one of the important determinants of immunostimulation. Previous studies reported that nanoparticles ranging from 10 to 100 nm are favorable for effective delivery to target tissues or cells, because they are large enough to avoid rapid renal excretion, but small enough for cell entry via receptor-mediated endocytosis.75, 76, 77 Larger particles will be recognized by the reticuloendothelial system (RES) more easily and prolong retention within the liver. In contrast, small nanoparticles less than 10 nm are rapidly eliminated from the body via renal filtration, though they can escape from trapping by the RES. In addition, the complement system is another key part of the immune system. It recognizes abnormal detrimental signals rapidly, coating intruders with opsonins and eliminating them by complement receptors bearing phagocytes. It was reported that some physicochemical parameters, especially surface charge and surface-projected polymers, affect complement sensing.78 For example, some cationic polymers such as polyethyleneimines are confirmed to activate complement pathways,79 whereas an anionic vesicle modified with carboxylic acid failed to induce activation.80 Thus, RNA nanoparticles benefit from their polyanionic nature that minimizes the non-specific interaction with negatively charged cell membranes. Moreover, the nanoparticle size also plays a role. It was reported that the bulky opsonins such as C3b molecules prefer to assemble and deposit onto the surface of larger nanoparticles or microparticles.81 For nanoparticles that are smaller than 30 nm, the opsonins tend to be released into the surrounding medium because of the limited accommodation, suggesting that a small RNA nanoparticle (10–20 nm) may be a poor activator of the complement system. In this study, the RNA polygons were designed to be smaller than 30 nm. RNA polygons with SEQ extensions in larger size (10–20 nm) significantly enhanced immunomodulation effects, compared with the small one (7.1 nm) (Figure 4A). As demonstrated by confocal microscopy imaging, the larger the RNA nanoparticle size, the more efficient it is in cell binding to RAW 264.7 cells (Figure 3). The mechanism might be attributed to the engulfing property of macrophages. Additionally, the levels of immunomodulation varied depending on RNA nanoparticle shapes (Figure 5). It is worth noting that the 3D 2′F Th-SEQ displayed a stronger cytokine induction compared with the planar 2′F TRI-SEQ, when both were similar in size and loaded with three 2′F SEQ modules (Figure 5B), demonstrating another dimension of immune response control. More importantly, cytokine induction by different SEQ extensions reveals that SEQ plays a dominant role in the immunogenicity of RNA nanoparticles, and only specific RNA SEQ would induce strong immunostimulation (Figure 6A). The SEQ that caused the strongest immune response is the one that holds strong homology to CpG-1826 oligodeoxynucleotide (ODN; 20-mer), a well-studied immunological adjuvant discovered from bacterial DNA that binds to the TLR-9.82
RNA nanotechnology is an emerging field, but the study of its immunogenicity is still at an early stage. More investigations on this aspect are of great significance for progressing toward clinical studies. Our studies have provided important implications for exploring the mechanism behind the immunogenicity of RNA nanoparticles. By tuning their properties, certain resulting RNA nanoparticles can be employed as safe therapeutic carriers without triggering immune response,46, 47, 48, 49, 58 or as potential immunomodulators for immunotherapy. Also, it should be noted that this study was performed in a murine macrophage model that might not fully represent the situations of human cells. There is a distinct difference between human cells and mouse cells regarding TLR-9 response. In humans, only B cells and plasmacytoid dendritic cells express TLR-9. In mice, however, most other dendritic cells and macrophages express TLR-9. Therefore, more studies are required in the future to perform on human cell models to explore a more comprehensive mechanism related to clinical applications.
Materials and Methods
RNA Nanoparticle Design, Synthesis, and Self-Assembly
The 3D computational model of RNA nanoparticle was generated in Swiss PDB Viewer and PyMOL Molecular Graphics System, using a pRNA-3WJ crystal structure (PDB: 4KZ2), as previously described.83 In brief, the ultra-stable pRNA-3WJ motif was placed at each vertex of the RNA core polygon structure, stretching the inner AOB angle to accommodate polygon geometry (Figure 1A).54, 59 ssRNAs were synthesized by in vitro T7 transcription, as previously described.84 For synthesizing 2′F-modified RNA, the Y693F mutant T7 polymerase and 2′F pyrimidine trinucleotides were used.85 DNA templates were produced by PCRs using DNA oligomers ordered from IDT (Integrated DNA Technologies). RNA transcripts were then purified using 8 M urea, 8% denaturing PAGE. All RNA nanoparticles were self-assembled in a one-pot manner by mixing individual RNA strands at equimolar concentrations (10 μM) in 1× TMS buffer (50 mM Tris [pH 8.0], 100 mM NaCl, and 10 mM MgCl2) and heated to 95°C, followed by slowly cooling down to 4°C. Stepwise self-assembly was verified by 3% agarose gel electrophoresis with ethidium bromide (EB) staining and imaged by Typhoon FLA 7000 (GE Healthcare).
DLS
RNA nanoparticles harboring SEQ extensions were preassembled at 0.5 μM in 50 μL 1× TMS buffer. The apparent hydrodynamic size of assembled RNA nanoparticle was measured by Zeta-sizer nano-ZS (Malvern Instruments) at 25°C with at least three independent measurements. The laser wavelength was 633 nm.
Serum Degradation Assay and TGGE Assay
Preassembled nanoparticles were incubated at the final concentration of 1 μM in cell culture medium that contained 10% fetal bovine serum (FBS) at 37°C for various time points and then were examined by 3% agarose gel at 90 V for 40 min. After being stained with EB and imaged by Typhoon FLA 7000, quantification analysis was performed using ImageJ software to calculate the percentage of intact nanoparticles over time. Quantified values of gel bands for each nanoparticle were divided by the value of first nanoparticles at 0 min and then plotted by OriginLab software.
TGGE analysis was performed on 6% native PAGE in 1× TBE buffer (89 mM Tris-borate [pH 8.0] and 2 mM EDTA) as reported before.86 In brief, a gradient temperature from 30°C to 80°C was applied perpendicular to electrical current in PAGE and run for 60 min at 20 W. Quantified values of bands for each nanoparticle were divided by the sum of the total values in corresponding lanes. Tm values were defined as the temperature at which 50% of loaded nanoparticle was dissociated.
Cell Culture
Murine macrophage-like RAW 264.7 cells were grown at 37°C in humidified air environment containing 5% CO2, cultured in DMEM containing both 10% (v/v) FBS and 100 U/mL penicillin.
Confocal Microscopy Imaging
RAW 264.7 cells were seeded on glass coverslips and cultured at 37°C overnight. RNA nanoparticles were labeled with Cy3 fluorophore at the 5′ end of one of the ssRNAs. 200 nM 2′F SEQ and 50 nM 2′F SQR-SEQ were incubated with cells for 4 hr at 37°C. After washing twice with 1× PBS, cells were fixed with 4% formaldehyde, followed by staining with ProLong Gold Antifade Reagent with DAPI (Life Technologies Corporation, Carlsbad, CA, USA) for cell nucleus and Alexa Fluor 488 phalloidin (Life Technologies Corporation) for cytoskeleton. Confocal images were taken by Olympus FV1000 confocal microscope (Olympus Corporation, Tokyo, Japan).
Evaluation of Cytokines Induction by ELISA
2.5 × 105/well RAW 264.7 cells were seeded in 24-well plates and cultured overnight. RNA nanoparticles were diluted in Opti-MEM medium (Life Technologies Corporation) to final concentration of 50 nM and incubated with cells at 37°C for 8 hr. Next, cell culture supernatants were collected and stored at −80°C until assay. Concentrations of TNF-α and IL-6 in collected supernatants were examined by using Mouse ELISA MAX Deluxe sets (BioLegend, San Diego, CA, USA), following the protocol provided by the manufacturer. Experiment results were repeated for at least three independent measurements. Data were statistically analyzed by Student’s t test and were presented as mean ± SD; *p < 0.05; **p < 0.01; ***p < 0.001.
Immunoblotting Assay
4 × 105/well RAW 264.7 cells were seeded in 24-well plates and cultured overnight, followed by treatment with RNA nanoparticles (50 nM) at 37°C for 8 hr. Cells were treated with cell lysis buffer containing protease inhibitor and phosphatase inhibitor (Cell Signaling Technology, Beverly, MA, USA) for protein extraction. Total proteins were separated by 7.5% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA, USA). The membranes were blocked in 5% milk for 2 hr at room temperature and incubated overnight with antibodies (Abs) at 4°C. Immunoblots were developed using a chemiluminescent substrate and exposed to imaging films. The same membrane was re-probed with anti-actin (1:4,000) as an internal control. Antibodies used were ICAM-1 (M-19) (sc-1511) and rabbit anti-goat IgG-horseradish peroxidase (HRP) (sc-2768) from Santa Cruz (Santa Cruz, CA, USA); phospho-IκB-α (Ser32) (14D4) rabbit monoclonal antibody (mAb), IκB-α (L35A5) mouse mAb (amino-terminal antigen), β-actin (13E5) rabbit mAb (HRP conjugate), anti-rabbit IgG HRP-linked Ab, and anti-mouse IgG HRP-linked Ab from Cell Signaling Technology.
In Vivo Immunostimulation in Mice
All animal procedures were housed and performed in accordance with the Subcommittee on Research Animal Care of The Ohio State University guidelines approved by the institutional review board. Male CD-1 mice (4–5 weeks old) were purchased from Charles River Laboratories. RNA nanoparticles were injected into mice via tail vein injection at 2 mg/kg (SEQ per body weight). After 3 hr post-injection, blood samples were harvested from mice by cardiac puncture and centrifugated at 12,800 × g for 10 min. Concentrations of TNF-α and IL-6 in serum supernatant were examined by ELISA as described above.
Author Contributions
P.G. conceived, designed, and led the project (in collaboration with J.F. and M.M.). S.G. and H.L. designed and performed the experiments. M.M. contributed to part of the cytokines induction experiments and immunoblotting assay (under the supervision of J.F.). S.G., H.L., P.G., and Y.D. analyzed and interpreted the data. S.G. wrote the manuscript with input from P.G., H.L., and Y.D. and help from all authors. S.G., H.L., P.G., and Y.D. contributed to the revision of the manuscript. All authors reviewed the final version of the manuscript.
Conflicts of Interest
P.G. is a consultant of Oxford Nanopore Technologies and Nanobio Delivery Pharmaceutical Co. Ltd., as well as the cofounder of Shenzhen P&Z Bio-medical Co. Ltd. and its subsidiary US P&Z Biological Technology LLC.
Acknowledgments
This research was supported by NIH grants U01CA207946 and R01EB019036. The authors would like to thank Daniel L. Jasinski, Daniel W. Binzel, and Julia Pinsonneault for their insight during manuscript preparation. P.G.’s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation.
Footnotes
Supplemental Information includes Supplemental Materials and Methods, four figures, and one table and can be found with this article online at https://doi.org/10.1016/j.omtn.2017.10.010.
Contributor Information
Yizhou Dong, Email: dong.525@osu.edu.
Peixuan Guo, Email: guo.1091@osu.edu.
Supplemental Information
References
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science. 2012;337:1159–1161. doi: 10.1126/science.337.6099.1159. [DOI] [PubMed] [Google Scholar]
- 3.Palazzo A.F., Lee E.S. Non-coding RNA: what is functional and what is junk? Front. Genet. 2015;6:2. doi: 10.3389/fgene.2015.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo P.X., Erickson S., Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage phi 29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 5.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 6.Shu Y., Pi F., Sharma A., Rajabi M., Haque F., Shu D., Leggas M., Evers B.M., Guo P. Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv. Drug Deliv. Rev. 2014;66:74–89. doi: 10.1016/j.addr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 9.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 11.Kruger K., Grabowski P.J., Zaug A.J., Sands J., Gottschling D.E., Cech T.R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 12.Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 13.Keefe A.D., Pai S., Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breaker R.R. Complex riboswitches. Science. 2008;319:1795–1797. doi: 10.1126/science.1152621. [DOI] [PubMed] [Google Scholar]
- 15.Heil F., Hemmi H., Hochrein H., Ampenberger F., Kirschning C., Akira S., Lipford G., Wagner H., Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 16.Diebold S.S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 17.Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 18.Liu L., Botos I., Wang Y., Leonard J.N., Shiloach J., Segal D.M., Davies D.R. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlake T., Thess A., Fotin-Mleczek M., Kallen K.J. Developing mRNA-vaccine technologies. RNA Biol. 2012;9:1319–1330. doi: 10.4161/rna.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phua K.K., Nair S.K., Leong K.W. Messenger RNA (mRNA) nanoparticle tumour vaccination. Nanoscale. 2014;6:7715–7729. doi: 10.1039/c4nr01346h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hornung V., Guenthner-Biller M., Bourquin C., Ablasser A., Schlee M., Uematsu S., Noronha A., Manoharan M., Akira S., de Fougerolles A. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 22.Judge A.D., Sood V., Shaw J.R., Fang D., McClintock K., MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 23.Sledz C.A., Holko M., de Veer M.J., Silverman R.H., Williams B.R. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 24.Santulli-Marotto S., Nair S.K., Rusconi C., Sullenger B., Gilboa E. Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Res. 2003;63:7483–7489. [PubMed] [Google Scholar]
- 25.Gilboa E., McNamara J., 2nd, Pastor F. Use of oligonucleotide aptamer ligands to modulate the function of immune receptors. Clin. Cancer Res. 2013;19:1054–1062. doi: 10.1158/1078-0432.CCR-12-2067. [DOI] [PubMed] [Google Scholar]
- 26.Sharma P., Allison J.P. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 27.Sarvestani S.T., Tate M.D., Moffat J.M., Jacobi A.M., Behlke M.A., Miller A.R., Beckham S.A., McCoy C.E., Chen W., Mintern J.D. Inosine-mediated modulation of RNA sensing by Toll-like receptor 7 (TLR7) and TLR8. J. Virol. 2014;88:799–810. doi: 10.1128/JVI.01571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caskey M., Lefebvre F., Filali-Mouhim A., Cameron M.J., Goulet J.P., Haddad E.K., Breton G., Trumpfheller C., Pollak S., Shimeliovich I. Synthetic double-stranded RNA induces innate immune responses similar to a live viral vaccine in humans. J. Exp. Med. 2011;208:2357–2366. doi: 10.1084/jem.20111171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vacchelli E., Eggermont A., Sautès-Fridman C., Galon J., Zitvogel L., Kroemer G., Galluzzi L. Trial Watch: Toll-like receptor agonists for cancer therapy. OncoImmunology. 2013;2:e25238. doi: 10.4161/onci.25238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barral P.M., Sarkar D., Su Z.Z., Barber G.N., DeSalle R., Racaniello V.R., Fisher P.B. Functions of the cytoplasmic RNA sensors RIG-I and MDA-5: key regulators of innate immunity. Pharmacol. Ther. 2009;124:219–234. doi: 10.1016/j.pharmthera.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt A., Schwerd T., Hamm W., Hellmuth J.C., Cui S., Wenzel M., Hoffmann F.S., Michallet M.C., Besch R., Hopfner K.P. 5′-Triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Errett J.S., Suthar M.S., McMillan A., Diamond M.S., Gale M., Jr. The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J. Virol. 2013;87:11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uzri D., Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J. Virol. 2009;83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo P., Zhang C., Chen C., Garver K., Trottier M. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol. Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 35.Lee J.B., Hong J., Bonner D.K., Poon Z., Hammond P.T. Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat. Mater. 2012;11:316–322. doi: 10.1038/nmat3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Afonin K.A., Viard M., Koyfman A.Y., Martins A.N., Kasprzak W.K., Panigaj M., Desai R., Santhanam A., Grabow W.W., Jaeger L. Multifunctional RNA nanoparticles. Nano Lett. 2014;14:5662–5671. doi: 10.1021/nl502385k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grabow W.W., Zakrevsky P., Afonin K.A., Chworos A., Shapiro B.A., Jaeger L. Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett. 2011;11:878–887. doi: 10.1021/nl104271s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Afonin K.A., Grabow W.W., Walker F.M., Bindewald E., Dobrovolskaia M.A., Shapiro B.A., Jaeger L. Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat. Protoc. 2011;6:2022–2034. doi: 10.1038/nprot.2011.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Afonin K.A., Lindsay B., Shapiro B.A. Engineered RNA nanodesigns for applications in RNA nanotechnology. DNA RNA Nanotechnol. 2013;1:1–15. doi: 10.2478/rnan-2013-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chworos A., Severcan I., Koyfman A.Y., Weinkam P., Oroudjev E., Hansma H.G., Jaeger L. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 41.Guo P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shu D., Shu Y., Haque F., Abdelmawla S., Guo P. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat. Nanotechnol. 2011;6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H., Zhang K., Pi F., Guo S., Shlyakhtenko L., Chiu W., Shu D., Guo P. Controllable self-assembly of RNA tetrahedrons with precise shape and size for cancer targeting. Adv. Mater. 2016;28:7501–7507. doi: 10.1002/adma.201601976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H., Lee T., Dziubla T., Pi F., Guo S., Xu J., Li C., Haque F., Liang X.J., Guo P. RNA as a stable polymer to build controllable and defined nanostructures for material and biomedical applications. Nano Today. 2015;10:631–655. doi: 10.1016/j.nantod.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jasinski D., Haque F., Binzel D.W., Guo P. Advancement of the emerging field of RNA nanotechnology. ACS Nano. 2017;11:1142–1164. doi: 10.1021/acsnano.6b05737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rychahou P., Haque F., Shu Y., Zaytseva Y., Weiss H.L., Lee E.Y., Mustain W., Valentino J., Guo P., Evers B.M. Delivery of RNA nanoparticles into colorectal cancer metastases following systemic administration. ACS Nano. 2015;9:1108–1116. doi: 10.1021/acsnano.5b00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Binzel D.W., Shu Y., Li H., Sun M., Zhang Q., Shu D., Guo B., Guo P. Specific delivery of MiRNA for high efficient inhibition of prostate cancer by RNA nanotechnology. Mol. Ther. 2016;24:1267–1277. doi: 10.1038/mt.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui D., Zhang C., Liu B., Shu Y., Du T., Shu D., Wang K., Dai F., Liu Y., Li C. Regression of gastric cancer by systemic injection of RNA nanoparticles carrying both ligand and siRNA. Sci. Rep. 2015;5:10726. doi: 10.1038/srep10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee T.J., Yoo J.Y., Shu D., Li H., Zhang J., Yu J.G., Jaime-Ramirez A.C., Acunzo M., Romano G., Cui R. RNA nanoparticle-based targeted therapy for glioblastoma through inhibition of oncogenic miR-21. Mol. Ther. 2017;25:1544–1555. doi: 10.1016/j.ymthe.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shu Y., Haque F., Shu D., Li W., Zhu Z., Kotb M., Lyubchenko Y., Guo P. Fabrication of 14 different RNA nanoparticles for specific tumor targeting without accumulation in normal organs. RNA. 2013;19:767–777. doi: 10.1261/rna.037002.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haque F., Shu D., Shu Y., Shlyakhtenko L.S., Rychahou P.G., Evers B.M., Guo P. Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today. 2012;7:245–257. doi: 10.1016/j.nantod.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee T., Yagati A.K., Pi F., Sharma A., Choi J.-W., Guo P. Construction of RNA-quantum dot chimera for nanoscale resistive biomemory application. ACS Nano. 2015;9:6675–6682. doi: 10.1021/acsnano.5b03269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y., Leonard M., Shu Y., Yang Y., Shu D., Guo P., Zhang X. Overcoming tamoxifen resistance of human breast cancer by targeted gene silencing using multifunctional pRNA nanoparticles. ACS Nano. 2017;11:335–346. doi: 10.1021/acsnano.6b05910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jasinski D.L., Khisamutdinov E.F., Lyubchenko Y.L., Guo P. Physicochemically tunable polyfunctionalized RNA square architecture with fluorogenic and ribozymatic properties. ACS Nano. 2014;8:7620–7629. doi: 10.1021/nn502160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khisamutdinov E.F., Jasinski D.L., Guo P. RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS Nano. 2014;8:4771–4781. doi: 10.1021/nn5006254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shukla G.C., Haque F., Tor Y., Wilhelmsson L.M., Toulmé J.J., Isambert H., Guo P., Rossi J.J., Tenenbaum S.A., Shapiro B.A. A boost for the emerging field of RNA nanotechnology. ACS Nano. 2011;5:3405–3418. doi: 10.1021/nn200989r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grabow W.W., Jaeger L. RNA self-assembly and RNA nanotechnology. Acc. Chem. Res. 2014;47:1871–1880. doi: 10.1021/ar500076k. [DOI] [PubMed] [Google Scholar]
- 58.Shu D., Li H., Shu Y., Xiong G., Carson W.E., 3rd, Haque F., Xu R., Guo P. Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano. 2015;9:9731–9740. doi: 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khisamutdinov E.F., Li H., Jasinski D.L., Chen J., Fu J., Guo P. Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square and pentagon nanovehicles. Nucleic Acids Res. 2014;42:9996–10004. doi: 10.1093/nar/gku516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abdelmawla S., Guo S., Zhang L., Pulukuri S.M., Patankar P., Conley P., Trebley J., Guo P., Li Q.X. Pharmacological characterization of chemically synthesized monomeric phi29 pRNA nanoparticles for systemic delivery. Mol. Ther. 2011;19:1312–1322. doi: 10.1038/mt.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Halman J.R., Satterwhite E., Roark B., Chandler M., Viard M., Ivanina A., Bindewald E., Kasprzak W.K., Panigaj M., Bui M.N. Functionally-interdependent shape-switching nanoparticles with controllable properties. Nucleic Acids Res. 2017;45:2210–2220. doi: 10.1093/nar/gkx008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Afonin K.A., Viard M., Kagiampakis I., Case C.L., Dobrovolskaia M.A., Hofmann J., Vrzak A., Kireeva M., Kasprzak W.K., KewalRamani V.N., Shapiro B.A. Triggering of RNA interference with RNA-RNA, RNA-DNA, and DNA-RNA nanoparticles. ACS Nano. 2015;9:251–259. doi: 10.1021/nn504508s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobrovolskaia M.A., Shurin M., Shvedova A.A. Current understanding of interactions between nanoparticles and the immune system. Toxicol. Appl. Pharmacol. 2016;299:78–89. doi: 10.1016/j.taap.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dobrovolskaia M.A. Pre-clinical immunotoxicity studies of nanotechnology-formulated drugs: challenges, considerations and strategy. J. Control. Release. 2015;220(Pt B):571–583. doi: 10.1016/j.jconrel.2015.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fadeel B. Clear and present danger? Engineered nanoparticles and the immune system. Swiss Med. Wkly. 2012;142:w13609. doi: 10.4414/smw.2012.13609. [DOI] [PubMed] [Google Scholar]
- 66.Juliano R.L., Ming X., Carver K., Laing B. Cellular uptake and intracellular trafficking of oligonucleotides: implications for oligonucleotide pharmacology. Nucleic Acid Ther. 2014;24:101–113. doi: 10.1089/nat.2013.0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Juliano R., Alam M.R., Dixit V., Kang H. Mechanisms and strategies for effective delivery of antisense and siRNA oligonucleotides. Nucleic Acids Res. 2008;36:4158–4171. doi: 10.1093/nar/gkn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gustafson H.H., Holt-Casper D., Grainger D.W., Ghandehari H. Nanoparticle uptake: the phagocyte problem. Nano Today. 2015;10:487–510. doi: 10.1016/j.nantod.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gratton S.E., Ropp P.A., Pohlhaus P.D., Luft J.C., Madden V.J., Napier M.E., DeSimone J.M. The effect of particle design on cellular internalization pathways. Proc. Natl. Acad. Sci. USA. 2008;105:11613–11618. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Canton I., Battaglia G. Endocytosis at the nanoscale. Chem. Soc. Rev. 2012;41:2718–2739. doi: 10.1039/c2cs15309b. [DOI] [PubMed] [Google Scholar]
- 71.Champion J.A., Mitragotri S. Role of target geometry in phagocytosis. Proc. Natl. Acad. Sci. USA. 2006;103:4930–4934. doi: 10.1073/pnas.0600997103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peer D., Karp J.M., Hong S., Farokhzad O.C., Margalit R., Langer R. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 73.Shi J., Kantoff P.W., Wooster R., Farokhzad O.C. Cancer nanomedicine: progress, challenges and opportunities. Nat. Rev. Cancer. 2017;17:20–37. doi: 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ferrari M. Cancer nanotechnology: opportunities and challenges. Nat. Rev. Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 75.Jain K.K. The role of nanobiotechnology in drug discovery. Drug Discov. Today. 2005;10:1435–1442. doi: 10.1016/S1359-6446(05)03573-7. [DOI] [PubMed] [Google Scholar]
- 76.Li W., Szoka F.C., Jr. Lipid-based nanoparticles for nucleic acid delivery. Pharm. Res. 2007;24:438–449. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- 77.Gao H., Shi W., Freund L.B. Mechanics of receptor-mediated endocytosis. Proc. Natl. Acad. Sci. USA. 2005;102:9469–9474. doi: 10.1073/pnas.0503879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dobrovolskaia M.A., Aggarwal P., Hall J.B., McNeil S.E. Preclinical studies to understand nanoparticle interaction with the immune system and its potential effects on nanoparticle biodistribution. Mol. Pharm. 2008;5:487–495. doi: 10.1021/mp800032f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Merkel O.M., Urbanics R., Bedocs P., Rozsnyay Z., Rosivall L., Toth M., Kissel T., Szebeni J. In vitro and in vivo complement activation and related anaphylactic effects associated with polyethylenimine and polyethylenimine-graft-poly(ethylene glycol) block copolymers. Biomaterials. 2011;32:4936–4942. doi: 10.1016/j.biomaterials.2011.03.035. [DOI] [PubMed] [Google Scholar]
- 80.Sou K., Tsuchida E. Electrostatic interactions and complement activation on the surface of phospholipid vesicle containing acidic lipids: effect of the structure of acidic groups. Biochim. Biophys. Acta. 2008;1778:1035–1041. doi: 10.1016/j.bbamem.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 81.Moghimi S.M., Andersen A.J., Ahmadvand D., Wibroe P.P., Andresen T.L., Hunter A.C. Material properties in complement activation. Adv. Drug Deliv. Rev. 2011;63:1000–1007. doi: 10.1016/j.addr.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 82.Krieg A.M. CpG motifs: the active ingredient in bacterial extracts? Nat. Med. 2003;9:831–835. doi: 10.1038/nm0703-831. [DOI] [PubMed] [Google Scholar]
- 83.Zhang H., Endrizzi J.A., Shu Y., Haque F., Sauter C., Shlyakhtenko L.S., Lyubchenko Y., Guo P., Chi Y.I. Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA. RNA. 2013;19:1226–1237. doi: 10.1261/rna.037077.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shu Y., Shu D., Haque F., Guo P. Fabrication of pRNA nanoparticles to deliver therapeutic RNAs and bioactive compounds into tumor cells. Nat. Protoc. 2013;8:1635–1659. doi: 10.1038/nprot.2013.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Padilla R., Sousa R. Efficient synthesis of nucleic acids heavily modified with non-canonical ribose 2′-groups using a mutantT7 RNA polymerase (RNAP) Nucleic Acids Res. 1999;27:1561–1563. doi: 10.1093/nar/27.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Binzel D.W., Khisamutdinov E.F., Guo P. Entropy-driven one-step formation of Phi29 pRNA 3WJ from three RNA fragments. Biochemistry. 2014;53:2221–2231. doi: 10.1021/bi4017022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.