Abstract
The opportunistic pathogen Pseudomonas aeruginosa forms antimicrobial resistant biofilms through sequential steps requiring several two-component regulatory systems. The sensor-regulator hybrid SagS plays a central role in biofilm development by enabling the switch from the planktonic to the biofilm mode of growth, and by facilitating the transition of biofilm cells to a highly tolerant state. However, the mechanism by which SagS accomplishes both functions is unknown. SagS harbors a periplasmic sensory HmsP, and phosphorelay HisKA and Rec domains. We used SagS domain constructs and site-directed mutagenesis to elucidate how SagS performs its dual functions. We demonstrate that HisKA-Rec and the phospho-signaling between SagS and BfiS contribute to the switch to the biofilm mode of growth, but not to the tolerant state. Instead, expression of SagS domain constructs harboring HmsP rendered ΔsagS biofilm cells as recalcitrant to antimicrobial agents as wild-type biofilms, likely by restoring BrlR production and cellular c-di-GMP levels to wild-type levels. Restoration of biofilm tolerance by HmsP was independent of biofilm biomass accumulation, RsmA, RsmYZ, HptB, and BfiSR-downstream targets. Our findings thus suggest that SagS likely makes use of a “divide-and-conquer” mechanism to regulate its dual switch function, by activating two distinct regulatory networks via its individual domains.
Keywords: phosphoproteome, tolerance, MBC, RsmZ, Gac, RsmA
INTRODUCTION
Biofilms are communities of surface-associated bacteria embedded in a self-produced, hydrated polymeric matrix (Costerton et al., 1995). An early observation of biofilms by Zobell described adherent bacterial populations forming microcolonies on glass slides immersed in sea water and noted that the number of attached cells far exceed the number of dislodged or free-floating cells (Zobell and Anderson, 1936). Biofilms have since been recognized to be the dominant mode of bacterial growth, with direct quantitation methods repeatedly showing the majority of the bacterial biomass present in natural aquatic, industrial, and medical settings being detectable in biofilms (Geesey et al., 1977; Costerton et al., 1987; Costerton et al., 1999). In addition to their widespread nature, another hallmark of biofilms is their increased resistance to antimicrobial agents, with biofilm cells having been shown to be 10–1000 fold less susceptible to various antimicrobial agents than their planktonic counterparts, a characteristic which is attributed to biofilms being extremely problematic to eradicate by conventional treatment strategies. This recalcitrance of biofilm cells to killing by antimicrobial agents has been termed "tolerance" and is believed to be attributed to a combination of different factors (Spoering and Lewis, 2001; Lewis, 2008) including restricted diffusion of antimicrobial agents, slow growth, reduced metabolic rates, increased stress tolerance, and differences in gene expression and protein production in biofilms compared to planktonic cells (Yasuda et al., 1994; Lewis, 2001; Mah and O'Toole, 2001; Stewart and Costerton, 2001; Gilbert et al., 2002; Mah et al., 2003; Keren et al., 2004; El-Azizi et al., 2005; Khan et al., 2010; Colvin et al., 2011; Nguyen et al., 2011).
Recent findings further suggest that in P. aeruginosa, drug tolerance is a function of the progression of biofilm development. Biofilm development is governed by the two-component regulatory systems (TCS) SagS, BfiSR, BfmRS, and MifRS. Activation of these four TCSs occurs in a sequential manner (SagS<BfiSR<BfmSR<MifSR), with inactivation of these systems having been shown to arrest biofilm formation at three distinct developmental stages, with ΔsagS and ΔbfiS biofilms being arrested at the irreversible attachment stage, while biofilms formed by ΔbfmR and ΔmifR arrested at the maturation-1 and -2 stages of biofilm development, respectively (Petrova and Sauer, 2009; Petrova and Sauer, 2010; Petrova and Sauer, 2011; Petrova et al., 2011, 2012). In this regulatory signaling cascade, SagS plays an important role by linking planktonic-specific Gac/Rsm-dependent signaling and the biofilm-specific TCS BfiSR to facilitate attachment-associated changes in the levels of the small regulatory RNA RsmZ and thus to enable the transition from initial attachment to the biofilm developmental progression (Petrova and Sauer, 2010; Petrova and Sauer, 2011). In addition to promoting the switch from planktonic to biofilm growth, likely via phospho-signaling to the BfiSR regulatory system, SagS was furthermore found to be required for P. aeruginosa biofilm resistance, as inactivation of sagS rendered biofilms as susceptible as planktonic cells to various antimicrobial agents, including tobramycin, norfloxacin, and hydrogen peroxide. Increased biofilm susceptibility upon sagS inactivation correlated with reduced expression of genes encoding multidrug efflux pumps and significantly reduced the expression of a gene encoding the MerR-like transcriptional regulator BrlR (Gupta et al., 2013; Gupta et al., 2014), a global regulator of biofilm resistance that modulates resistance to a broad range of antimicrobial agents in P. aeruginosa biofilms (Liao and Sauer, 2012; Liao et al., 2013). SagS contribution to biofilm resistance was consequently found to be dependent on the intracellular signaling molecule c-di-GMP and activation of BrlR. This was supported by the finding of ΔsagS mutant biofilms, unlike ΔbfiS mutant biofilms, being characterized by significantly reduced brlR transcript and BrlR protein levels, as well as cellular c-di-GMP levels reminiscent of those present in planktonic cells (Gupta et al., 2013; Gupta et al., 2014). In contrast, inactivation of BfiSR, the TCS essential for biofilm development that is activated by SagS upon surface-associated growth, did not result in increased susceptibility of mutant biofilms (Gupta et al., 2013).
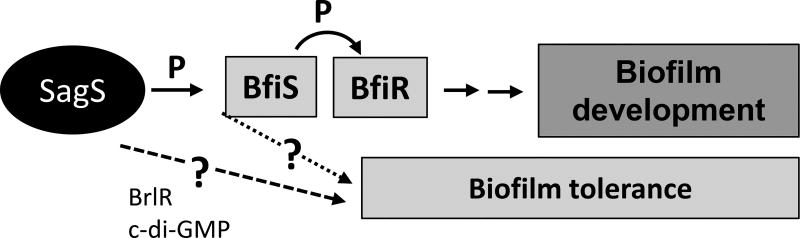
The findings support the notion of SagS having a dual function in promoting (i) the switch from the planktonic to the biofilm mode of growth, and (ii) the switch from a susceptible to a highly tolerant state (Liao and Sauer, 2012; Chambers and Sauer, 2013; Gupta et al., 2013; Liao et al., 2013; Chambers et al., 2014). However, the mechanism by which SagS carries out its dual regulatory functions is unknown (Fig. 1). While the interaction and phosphotransfer between SagS and BfiS have been implicated in the regulation of biofilm formation (Petrova and Sauer, 2009; Petrova and Sauer, 2010; Petrova and Sauer, 2011), greater details regarding the regulatory mechanism remain to be elucidated. Moreover, while previous findings suggested a role of SagS in biofilm tolerance, likely by indirectly activating BrlR and coinciding with increased c-di-GMP levels (Gupta et al., 2013; Gupta et al., 2014), the mechanism(s) by which SagS contributes to the regulation of the switch from a susceptible to a highly tolerant state is unknown. Considering the different players involved in biofilm formation and biofilm tolerance, and that both ΔsagS and ΔbfiS biofilms are arrested at the initial attachment stage, but only ΔsagS but not ΔbfiS biofilm cells are susceptible to various antimicrobial agents (Petrova and Sauer, 2011; Gupta et al., 2013), we hypothesized that SagS carries out its dual regulatory functions via two distinct pathways, with the molecular communication between SagS and BfiS via phospho signaling being independent of SagS contributing to biofilm tolerance (Fig. 1). We therefore asked whether the SagS phosphorelay contributes to biofilm development alone by determining whether the phosphotransfer domains of SagS are necessary for both of its dual regulatory functions. To address this question, we made use of truncated SagS domain constructs and site-directed mutagenesis to elucidate residues of SagS that contribute to the molecular communication between SagS and BfiS, and whether the respective residues contribute to both SagS functions.
Figure 1. Overview of the contribution of the two-component hybrid SagS to the motile-sessile and susceptible-resistance switches by P. aeruginosa cells.
Upon P. aeruginosa transition to surface-associated growth, SagS directly interacts with and phosphorylates the TCS BfiSR, thus enabling surface-associated cells to transition to the irreversible attachment stage (Petrova and Sauer, 2011). Moreover, transition to the irreversible attachment stage, regulated by SagS, marks the timing when surface-associated cells gain their heightened resistance to antimicrobial agents, with inactivation of sagS having been previously demonstrated to correlate with biofilm cells but not planktonic cells being more susceptible to antimicrobial agents (Gupta et al., 2013). SagS likely contributes to the activation of biofilm tolerance via c-gi-GMP and the transcriptional regulator BrlR. “?” indicates likely mechanism by which SagS contributes to biofilm development and/or biofilm tolerance.
RESULTS
The HisKA and Rec domains of SagS contribute to biofilm formation, but not antimicrobial tolerance
In order to elucidate how SagS carries out its dual function in promoting the switch from the planktonic to the biofilm mode of growth, and the switch from a susceptible to a highly tolerant state (Fig. 1) (Liao and Sauer, 2012; Chambers and Sauer, 2013; Gupta et al., 2013; Liao et al., 2013; Chambers et al., 2014), we first investigated the role of each of the SagS domains. Considering that SagS harbors three domains, including a centrally located histidine kinase (HisKA) domain and a C-terminal receiver (Rec) domain (Fig. 2A) likely involved in hierarchical phosphotransfer-based signaling to BfiS, we first investigated the HisKA and Rec domains to determine the role of the phosphorelay in biofilm development and biofilms gaining their heightened tolerance to antimicrobial agents. We therefore generated truncated SagS variants composed of the (i) HisKA-Rec, (ii) HisKA and (iii) Rec domain(s) (Fig. 2A). Multicopy expression of sagS in ΔsagS has previously been shown to coincide with restoration of biofilm formation and susceptibility to antimicrobial agents to wild-type levels, rather than enhanced biofilm formation or increased resistance to antimicrobial agents. Considering that multicopy expression of sagS had no adverse effect, each SagS construct, harboring a C-terminal hemagglutinin (HA) epitope, was placed into pJN105 under the control of the PBAD promoter and transformed into ΔsagS. The domain constructs were detectable at comparable levels in cell free protein extracts (Fig. S1).
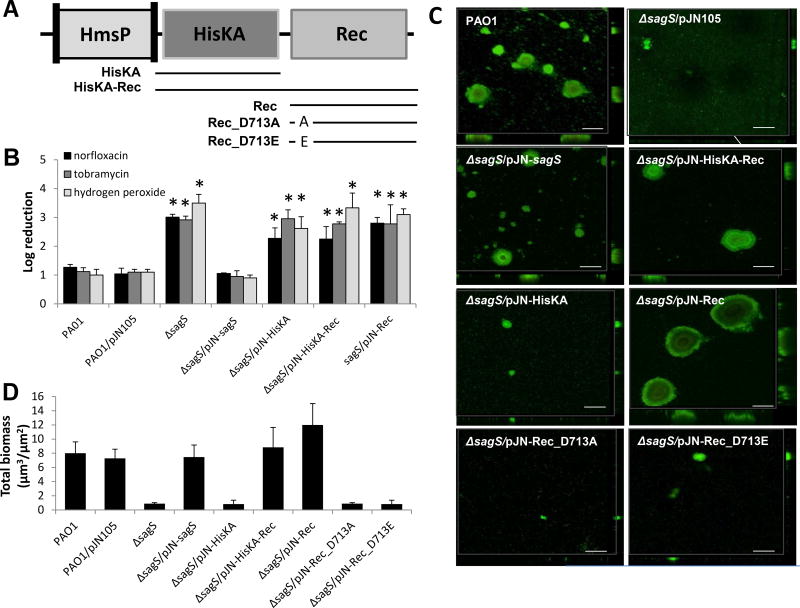
Figure 2. The HisKA and Rec domains of SagS contribute to biofilm formation, but not antimicrobial tolerance.
(A) Overview of SagS domains and SagS domain constructs. Lines underneath the domains indicate the composition of the SagS domain constructs, while the names of the resulting constructs are given next to the lines. (B) Susceptibility phenotype of ΔsagS and ΔsagS mutant strains complemented with sagS or sagS domain constructs to tobramycin, norfloxacin and hydrogen peroxide. Indicated P. aeruginosa strains were allowed to form biofilms under flowing conditions for 2 days. Biofilm cells were subsequently treated with norfloxacin (450 µg/ml), tobramycin (150 µg/ml), or hydrogen peroxide (0.6%) for 1 hr under flowing conditions. P. aerugionosa PAO1 harboring the empty plasmid pJN105 was used as a control. Biofilm susceptibility was determined by log reduction. (C) Representative confocal images showing the architecture of biofilms formed by P. aeruginosa PAO1, ΔsagS, and ΔsagS mutant strains complemented with sagS and truncated sagS constructs. Biofilms were grown for 6 days in 20-fold diluted LB medium after which time confocal images were acquired. Biofilms were stained with the LIVE/DEAD BacLight viability stain (Life Technologies). White bars = 100 µm. (D) Biofilm biomass of P. aeruginosa wild-type and mutant biofilms grown for 6 days in flow cells, as determined using confocal images and subsequent COMSTAT analysis. Error bars indicate standard deviation. *, significantly different from the values for P. aeruginosa PAO1 (p ≤ 0.01).
To elucidate the role of the specific SagS domains in promoting the switch from a susceptible to a highly tolerant state, we compared the susceptibility of biofilm cells formed by ΔsagS mutant strains overexpressing intact sagS and SagS domain constructs to that of ΔsagS biofilm cells. In agreement with previous findings (Gupta et al., 2013), ΔsagS biofilms did not develop high-level biofilm-specific drug resistance compared to the wild type, as evidenced by treatment with tobramycin (150 µg/ml), norfloxacin (450 µg/ml), or hydrogen peroxide (0.6%) for 1 hr resulting in a 3.3-fold, 3.2-fold, and 3.5-fold log reduction, respectively (Fig. 2B, S2). Multicopy expression of sagS restored the resistance phenotype of ΔsagS biofilm cells to wild-type levels, while expression of the HisKA, Rec, and HisKA-Rec domains in ΔsagS failed to do so (Fig. 2B, S2).
Given the inability of the HisKA, HisKA-Rec, and Rec domain constructs to rescue the resistance phenotype of ΔsagS mutant biofilm cells to wild-type levels, combined with previous reports of SagS contributing to BfiS activation and thus continued biofilm formation (Petrova and Sauer, 2010; Petrova and Sauer, 2011), we surmised a role of these domains in biofilm development. Therefore, we next determined whether the domain constructs are capable of restoring ΔsagS biofilm formation to wild-type levels. Considering that the HisKA and Rec domains are likely involved in hierarchical phosphotransfer-based signaling to BfiS, we anticipated the HisKA-Rec but not the HisKA or Rec domain alone to restore biofilm formation. Consistent with previous reports (Petrova and Sauer, 2011), the ΔsagS mutant formed thin biofilms lacking large cellular aggregates and harboring >5-fold-less biomass than biofilms formed by the isogenic parental strain, while overexpression of sagS in the mutant strain restored the biofilm architecture to wild-type levels (Fig. 2C,D, S3A) (Petrova and Sauer, 2009; Petrova and Sauer, 2011). As anticipated, expression of the HisKA-Rec domain, but not the HisKA domain alone, restored biofilm formation by the ΔsagS mutant strain to wild-type levels (Fig. 2C,D, S3A). However, our findings also demonstrated that expression of the Rec domain construct alone restored biofilm formation to wild-type levels (Fig. 2C,D, S3A). It is of interest to note that restoration of the biofilm architecture and thus, the biofilm biomass, to wild-type levels upon expression of HisKA-Rec coincided with c-di-GMP levels comparable to those observed in wild-type biofilms (Fig. S5). In contrast, biofilms formed by ΔsagS/pJN-HisKA were comparable to ΔsagS biofilms and harbored significantly lower c-di-GMP levels than those detected in wild-type biofilms (Fig. S5).
The HisKA-Rec and Rec domain constructs restore biofilm formation via distinct pathways
The interaction and phosphotransfer between SagS and BfiS have been implicated in the regulation of biofilm formation (Petrova and Sauer, 2009; Petrova and Sauer, 2010; Petrova and Sauer, 2011), To determine whether restoration of biofilm formation via expression of SagS domain constructs is dependent on the interaction and phosphotransfer between SagS and BfiS, we first determined whether SagS constructs are capable of interacting with BfiS. As observed using pulldown assays, the HisKA-Rec and Rec domain constructs interacted with BfiS in vivo (Fig. 3A, S4A). In contrast, little to no interaction between BfiS and the HisKA domain was noted (Fig. 3A, S4A). To determine whether the SagS domain constructs contribute to the phosphorylation status of BfiS, we made use of MOAC (Fig. 3A, S4B). Under the conditions tested and in agreement with previous findings (Petrova and Sauer, 2011), no BfiS phosphorylation was noted in ΔsagS biofilm cells, while phosphorylated BfiS was detectable in ΔsagS biofilm cells expressing full length sagS. Likewise, expression of the HisKA-Rec domain construct in ΔsagS restored BfiS phosphorylation to wild-type levels in vivo. In contrast, little to no phosphorylated BfiS was detected in ΔsagS cells expressing the Rec domain (Fig. 3A, S4B).
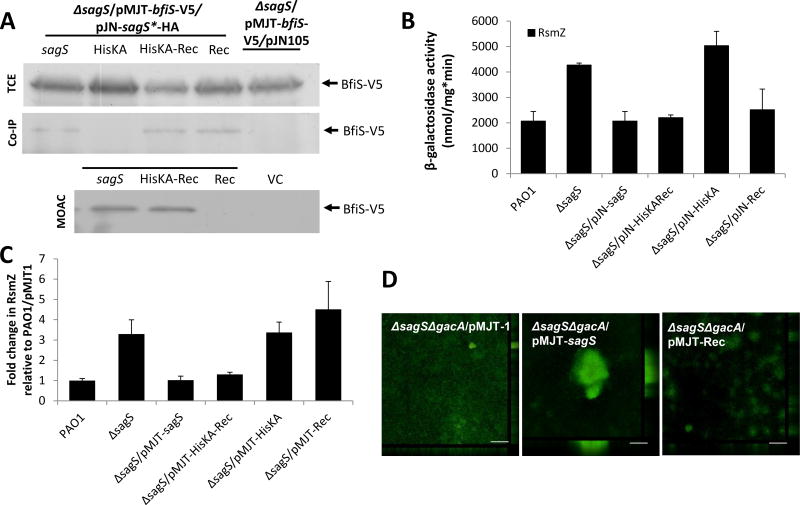
Figure 3.
The HisKA-Rec and Rec domain constructs contribute to biofilm formation via distinct pathways. (A) Detection of BfiS in total cell extracts (TCE), in pulldowns (Co-IP) using SagS as prey, and in MOAC-enriched phosphoproteomes (MOAC) of P. aeruginosa ΔsagS cells expressing bfiS and sagS domain constructs. Cell-free total cell extracts were obtained from P. aeruginosa grown as biofilms. For MOAC samples, the entire MOAC eluate concentrated using methanol/chloroform precipitation was loaded. All SagS domain constructs, including the full-length gene, harbored a C-terminal hemagglutinin (HA) tag (referred to as SagS*-HA). (B) Assessment of RsmZ promoter reporter activity as determined using the chromosomal rsmZ-lacZ reporter construct and β-galactosidase activity, and of (C) RsmZ transcript levels as determined using qRT-PCR in cells grown as biofilms for 3 days. The promoter activity is reported as nmol ONPG cleaved per total cellular protein per minute. (D) Confocal images of 6-day-old biofilms formed by the double mutant ΔsagSΔgacA harboring the empty plasmid pMJT-1 and strains expressing sagS and sagS-Rec. Biofilms were stained with the LIVE/DEAD BacLight viability stain. Representative images are shown. All experiments were carried out at least in triplicate. Error bars indicate standard deviation. *, significantly different from the values for P. aeruginosa PAO1 (p ≤ 0.01).
To determine whether phosphorylation of BfiS coincides with BfiS activity, we made use of the finding of biofilm formation correlating with the modulation of the transcript levels of the small regulatory RNAs RsmZ, wtih BfiSR-dependent arrest of biofilm formation coinciding with the posttranscriptional modulation of RsmZ levels via activation of the RNAse G CafA (Petrova and Sauer, 2010). In agreement with previous findings, inactivation of sagS coincided with increased RsmZ reporter activity and RsmZ transcript abundance, while ΔbfiS biofilm cells demonstrated increased RsmZ levels compared to wild-type biofilms but comparable RsmZ reporter activity (Fig. 3B–C). Consistent with HisKA not interacting with BfiS, expression of the construct encoding the HisKA domain alone in ΔsagS cells had little effect on RsmZ expression using rsmZ promoter reporter assays, as no difference in β-galactosidase activity was noted in ΔsagS and ΔsagS/pJN-HisKA biofilm cells (Fig. 3B). Moreover, qRT-PCR confirmed ΔsagS/pJN-HisKA biofilm cells to be characterized by elevated RsmZ transcript abundance relative to wild-type biofilms (Fig. 3C). The HisKA-Rec domain construct, however, which rescued the SagS-BfiS interaction and the biofilm formation phenotype, was capable of restoring RsmZ promoter activity and RsmZ transcript abundance to wild-type levels (Fig. 3B–C). In contrast, expression of Rec in ΔsagS biofilm cells coincided with restoration of RsmZ promoter activity (Fig. 3B), but failed to restore RsmZ transcript abundance to wild-type levels (Fig. 3C). The finding is in support of the HisKA-Rec domain contributing to BfiS and thus, CafA activation, while the Rec domain fails to do so despite the noted interaction with BfiS.
Moreover, our data suggested that HisKA-Rec and Rec restore biofilm formation by different mechanisms, with HisKA-Rec restoring biofilm formation in a manner dependent on BfiS phosphorylation (by affecting RsmZ transcript abundance and transcript processing), while the Rec domain restored biofilm formation in a manner independent of BfiS, BfiS phosphorylation, or BfiSR-dependent post-transcriptional RsmZ modulation. The difference suggested an interaction of the truncated SagS Rec domain with other regulatory networks capable of affecting RsmZ levels. The sensor hybrid SagS has been previously linked to the Gac/Rsm signaling pathway (Hsu et al., 2008). Additionally, the Rec domain of SagS has recently been demonstrated to directly bind to the cyclic di-GMP-binding adaptor protein HapZ (PA2799), with HapZ binding modulating the phosphotransfer between SagS and the downstream protein HptB (Xu et al., 2016).
To further explore the role of the Rec domain in biofilm formation, we made use of the finding that the Rec domain harbors a conserved phospho-transfer site located at the aspartate residue D713 and asked whether this residue is necessary the ability to restore biofilm-related phenotypes. A phospho-inhibitive and a phosphor-mimetic variant of the Rec domain were generated by replacing the predicted phosphorylation site D713 with alanine or glutamate, respectively (Fig. 2A). Expression of the resulting Rec variants, Rec-D713A and Rec-D713E, failed to restore the ΔsagS biofilm architecture to wild-type levels (Fig. 2C, D), suggesting that the Rec domain alone, regardless of its phosphorylation status, is unable to contribute to biofilm formation in the absence of its phosphotransfer capability. Considering the link to the Gac system, and particularly to HptB (Hsu et al., 2008; Xu et al., 2016), we furthermore tested whether the role of the Rec domain in biofilm formation is dependent on the Gac/Rsm pathway by using a ΔsagSΔgacA double mutant. The ΔsagSΔgacA biofilm architecture was comparable to that noted for ΔsagS biofilms, with overexpression of sagS restoring biofilm formation to wild-type levels (Figs. 3D, S6). In contrast, however, overexpression of the Rec-encoding construct failed to restore the ΔsagSΔgacA biofilm architecture to wild-type levels (Fig. 3D, S6) indicating a cross-talk between the Rec domain and GacA (and likely other components of the Gac/Rsm network), a cross-talk that is absent in the presence of SagS.
The SagS conserved phosphorelay residues regulate biofilm formation via BfiS in a GacA-independent manner
Considering the different contributions of the HisKA, Rec, and HisKA-Rec domains to biofilm formation (but not to biofilm tolerance), as well as the dependency of Rec domain function on its conserved phosphorylation site, we next asked whether the conserved SagS phosphorelay contributes to its dual function in the regulation of biofilm formation and tolerance of biofilm cells to antimicrobial agents. In addition to the conserved phospho-transfer site located at the aspartate residue D713 of the Rec domain, sequence alignments indicated that SagS harbors an H-box motif with a potential histidine phosphorylation site located in the HisKA domain at the histidine residue H315 (Fig. 4A–B). The potential phosphorylation sites present in intact sagS were subjected to site-directed mutagenesis, with H315 and D713 being replaced by an alanine (Fig. 4A). Additionally, the H348 residue, chosen as a positive control due to its lack of predicted conserved phosphotransfer activity and its close proximity to H315, was substituted with alanine as well. All resulting SagS variants harbored a C-terminal hemagglutinin (HA) tag to ensure production and solubility (Fig. 4C). Plasmids carrying the resulting alleles were transformed into ΔsagS.
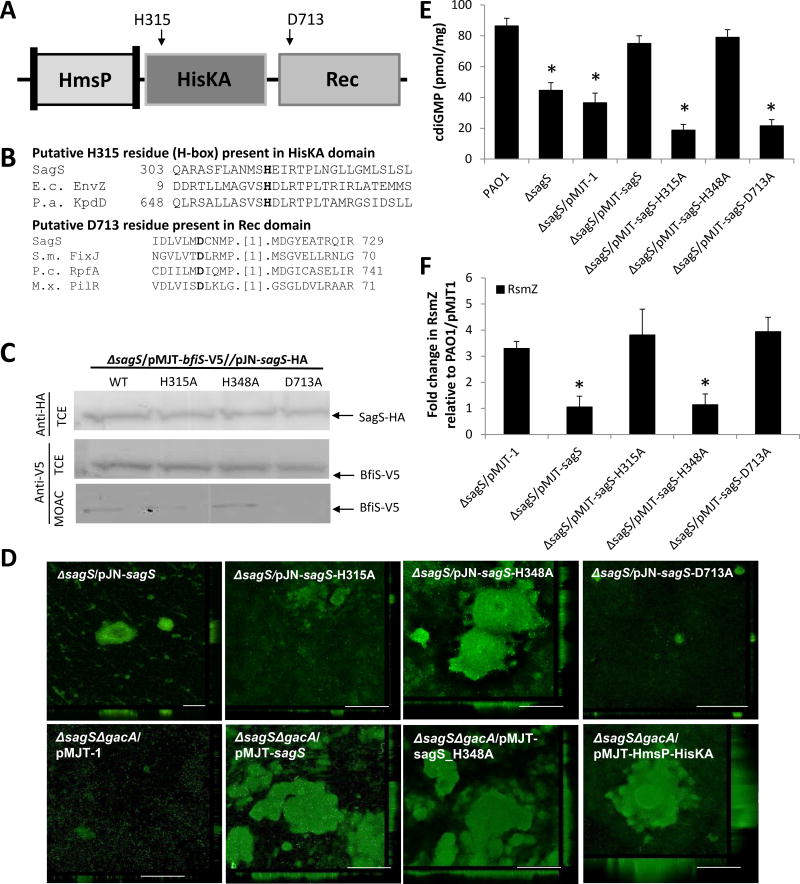
Figure 4. Conserved phosphotransfer residues of SagS contribute to biofilm development, but not antimicrobial tolerance.
(A) Overview of SagS conserved phosphorelay sites and corresponding site-directed mutagenesis targets. (B) Alignments showing the presence and location of invariant histidine and aspartate phosphorylation sites in the HisKA and Rec domains, respectively. Alignments were obtained via conserved domain BLAST; E.c. EnvZ, Escherichia coli EnvZ; P.a. KpdD, Pseudomonas aeruginosa two-component sensor KdpD; S.m. FixJ, Sinorhizobium meliloti transcriptional regulatory protein Fixj; P.c. RpfA, Pectobacterium carotovorum sensor/regulator protein RpfA; M.x. PilR, Myxococcus xanthus regulator protein PilR. (C) Detection of SagS and BfiS in total cell extracts (TCE) or MOAC-enriched phosphoproteomes (MOAC) of P. aeruginosa ΔsagS expressing wild-type sagS or sagS variants harboring alanine substitutions at amino acid residues H315, H348, and D713A by immunoblot analysis. Cell-free total cell extracts were obtained from P. aeruginosa grown biofilm cells. (D) Representative confocal images showing the architecture of biofilms formed by ΔsagS mutant strains expressing intact sagS or sagS variants harboring alanine substitutions at amino acid residues H315, H348, and D713A and the double mutant ΔsagSΔgacA harboring the empty plasmid pMJT-1 or pMJT containing sagS, sagS_H348A, or the SagS domain construct HmsP-HisKA. Biofilms were stained with the LIVE/DEAD BacLight viability stain. White bars = 100 µm. (E) c-di-GMP levels present in wild-type and ΔsagS mutant biofilm cells overexpressing sagS variants. The strains were grown in tube reactors under flowing conditions for 6 days prior to c-di-GMP extraction and quantitation by HPLC analysis. pmol/mg refers to c-di-GMP levels (pmol) per total cell protein (in mg). ΔsagS harboring the empty vector pMJT-1 was used as vector control. (F) RsmZ transcript levels as determined using qRT-PCR in cells grown as biofilms for 3 days. All experiments were carried out at least in triplicate. Error bars indicate standard deviation. *, significantly different from the values for P. aeruginosa PAO1 (p ≤ 0.01), as determined by ANOVA and SigmaStat.
Using these constructs, we first aimed to determine whether biofilm formation and tolerance of biofilm cells to antimicrobial agents rely on hierarchical phosphotransfer-based signaling from SagS to BfiS. To gauge the role of SagS phosphorylation sites in biofilm development, we first made use of the finding that SagS enhances BfiS phosphorylation under biofilm growth conditions in vivo (Petrova and Sauer, 2011) by using MOAC in conjunction with immunoblot analysis. None of the substitutions affected SagS and BfiS protein levels (Fig. 4C). Substitution of histidine H348 to alanine had no effect on the phosphorylation levels of BfiS. In contrast, however, substitution of D713 to alanine abrogated BfiS phosphorylation in vivo, while the H315A substitution significantly reduced the levels of phosphorylated BfiS (Fig. 4C, S4B), suggesting H315 and D713 to contribute to the phosphorelay transduction pathway to BfiS.
We next reasoned that if biofilm formation indeed requires BfiS phosphorylation and thus, molecular communication between SagS and BfiS, substitution of H315 and D713 to alanine will coincide with impaired biofilm formation in a manner similar to inactivation of sagS. While the ΔsagS mutant formed thin biofilms, overexpression of sagS restored the biofilm architecture to wild-type levels (Fig. 4D, S3 B–C) (Petrova and Sauer, 2009; Petrova and Sauer, 2011). Likewise, overexpressing of the positive control construct sagS-H348A in ΔsagS mutant cells resulted in biofilms comparable in biomass and height to the wild type (Fig. 4D, S3 B–C). As anticipated, however, ΔsagS mutant strains complemented with sagS-H315A and sagS-D713A failed to develop the biofilm architecture typically observed in PAO1 biofilms (Fig. 4D, S3 B–C). It is of interest to note that expression of sagS-H348A in ΔgacAΔsagS mutant cells restored biofilm formation indicating that restoration of the biofilm architecture by SagS-H348A was independent of GacA and more reliant on signaling to BfiS (Fig. 4D).
Restoration of the biofilm architecture correlated with elevated cellular c-di-GMP levels. In agreement with previous findings of ΔsagS biofilm cells harboring the secondary messenger c-di-GMP at reduced levels similar to those observed in wild-type cells grown planktonically rather than as biofilms (Gupta et al., 2014), ΔsagS biofilm cells were characterized by reduced c-di-GMP levels (Fig. 4E). Restoration of the ΔsagS mutant biofilm architecture to wild-type levels via expression of sagS or sagS-H348A coincided with restoration of cellular c-di-GMP to wild-type levels (Fig. 4E). In contrast, expression of sagS-H315A and sagS-D713A in ΔsagS mutant cells had no effect on the c-di-GMP level present in biofilm cells.
To furthermore ensure that phosphotransfer-based signaling to BfiS contributes to biofilm formation in a BfiS-dependent manner, the transcript abundance of RsmZ was determined by qRT-PCR. Overexpression of sagS-H348A in ΔsagS biofilm cells reduced RsmZ transcript levels to wild-type levels in a manner similar to overexpression of sagS. In contrast, overexpression of sagS-H315A and sagS-D713A failed to restore RsmZ abundance to wild-type levels (Fig. 4F).
Biofilm resistance to antimicrobial agents is activated independently of the SagS classical phosphorelay residues and BfiS signaling
Our findings strongly supported the notion of SagS promoting the switch from the planktonic to the biofilm mode of growth in a manner dependent on phosphotransfer-based signaling to BfiS, with restoration of the ΔsagS mutant biofilm architecture to wild-type levels relying on SagS residues H315 and D713, BfiS phosphorylation, and subsequent BfiS-dependent modulation of RsmZ transcript abundance. Considering the role of the SagS His-Asp phosphorelay in the switch from the planktonic to biofilm mode of growth, we next asked whether phosphotransfer-based signaling equally plays a role in promoting the switch from an antimicrobial susceptible to a highly tolerant state (Petrova and Sauer, 2011; Gupta et al., 2013).
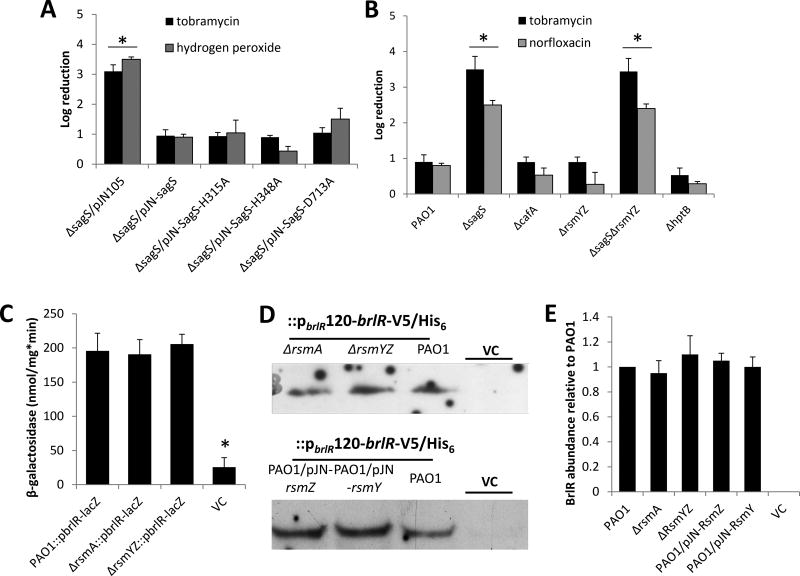
Treatment of 2-day-old biofilm cells with tobramycin or hydrogen peroxide resulted in a 3.2 and 3.3 log reduction, respectively, in viability of ΔsagS mutant biofilm cells compared to a 1-log reduction in the viability of wild-type biofilms, while overexpression of sagS restored drug tolerance to tobramycin to wild-type levels (Fig. 5A). Despite differences in biofilm architecture and biofilm biomass accumulation (Fig. 4D, S3C), expression of sagS-H315A, sagS-H348A, and sagS-D713A in ΔsagS mutant biofilms restored drug tolerance to tobramycin and hydrogen peroxide to wild-type levels (Fig. 5A). No difference in susceptibility was noted in the absence or presence of the empty vector pJN105 (Fig. 5A). The findings suggested biofilm tolerance to be independent of the conserved phosphotransfer residues and likely the phosphorelay transduction pathway to BfiS. This was further supported by the finding of biofilm tolerance being independent of the BfiSR targets CafA and RsmZ, with inactivation of cafA, rsmY and rsmZ having no effect on the susceptibility of biofilm cells to tobramycin and norfloxacin (Fig. 5B).
Figure 5. Susceptibility of biofilm cells and BrlR production is not linked to the phospho-transfer to BfiS, BfiS downstream targets or components previously linked to biofilm development.
(A–B) Susceptibility to tobramycin, hydrogen peroxide, or norfloxacin of biofilm cells of PAO1 and strains inactivated in cafA, rsmYZ, hptB, sagS and ΔsagS mutant strains expressing SagS variants. Biofilm susceptibility was determined as CFU log reduction. (C) Detection of brlR reporter activity in cells grown as biofilms for 3 days as determined using the chromosomal brlR-lacZ reporter construct and β-galactosidase activity. The promoter activity is reported as nmol ONPG cleaved per total cellular protein per minute. (D) Detection of BrlR in total cell extracts obtained from biofilm cells of P. aeruginosa PAO1 strains inactivated in RsmA or RsmYZ or overexpressing RsmY or RsmZ. All strains harbored a chromosomally located V5/His6-tagged BrlR under the control of its own promoter (PbrlR120-brlR-V5/His6), and respective protein extracts were probed for the presence of BrlR by immunoblot analysis using anti-V5 antibodies. A total of 15µg total cell extract was loaded. P. aeruginosa PAO1 harboring an empty CTX vector (VC) was used as negative control. Representative images are shown. (E) Relative BrlR abundance, based on relative intensity of protein bands detectable following probing for BrlR with anti-V5 antibodies and subsequent analysis using ImageJ (Rasband, 1997). Experiments were carried out in triplicate. Error bars denote standard deviation. *, significantly different from the values for P. aeruginosa PAO1 (p ≤ 0.01), as determined by ANOVA and SigmaStat.
Previous findings indicated that SagS contributes to biofilm resistance by indirectly activating the gene encoding the transcriptional regulator of biofilm resistance BrlR (Liao and Sauer, 2012; Gupta et al., 2013; Gupta et al., 2014). In agreement with the susceptibility assays, no difference in brlR expression was noted in wild-type, ΔrsmA and ΔrsmYZA biofilm cells using brlR-lacZ chromosomal transcriptional fusions (Fig. 5C). Likewise, no difference in BrlR production was noted in wild-type and strains inactivated in rsmA or RsmYZ or strains overexpressing RsmZ and RsmY, as indicated by immunoblot analysis using pbrlR120-brlR-V5/His6 chromosomal translational fusion (Fig. 5D–E).
The findings indicated that while phospho-signaling to BfiS, as well as the BfiS downstream targets including CafA and RsmZ, contributes to biofilm formation, the His-Asp phosphorelay of SagS, the phospho-signaling to BfiS, and ultimately RsmZ and RsmY, play no role in rendering biofilm cells resistant to antimicrobial agents (Fig. 5).
The SagS HmsP-HisKA construct promotes the transition of biofilm cells to a highly antimicrobial tolerant state in a manner independent of GacA, phosphorelay function and biofilm formation
By thus eliminating the molecular communication between SagS and BfiS, as well as two of the three SagS domains, from contributing to the switch from a susceptible to a highly tolerant state, we turned to the remaining N-terminal portion of SagS harboring the predicted HmsP domain. The HmsP domain, bounded by two transmembrane helices, is homologous to the N-terminal domains of the biofilm formation regulators BifA and HmsP in P. aeruginosa and Yersinia pestis, respectively (Fig. 6A, S7) (Kirillina et al., 2004; Bobrov et al., 2005; Kuchma et al., 2007), and represents a periplasmic sensory domain. In order to study the contribution of the HmsP domain of SagS to rendering biofilm cells resistant to antimicrobial agents, we generated a SagS variant devoid of the Rec domain. We reasoned that such a construct composed only of the HmsP and HisKA domains (referred to as HmsP-HisKA) would not be capable of phospho-signaling to BfiS, considering that the HisKA domain alone failed to restore biofilm formation, to interact with BfiS, or contribute to BfiS phosphorylation in vivo. While it is unclear how a construct devoid of the Rec domain may contribute to BfiS phosphorylation, we furthermore generated an additional variant to suppress molecular communication between SagS and BfiS (or preventing cross-talk to other TCS such as GacA) by generating a HmsP-HisKA variant harboring the H315A substitution (referred to as HmsP-HisKA_H315A, Fig. 6A).
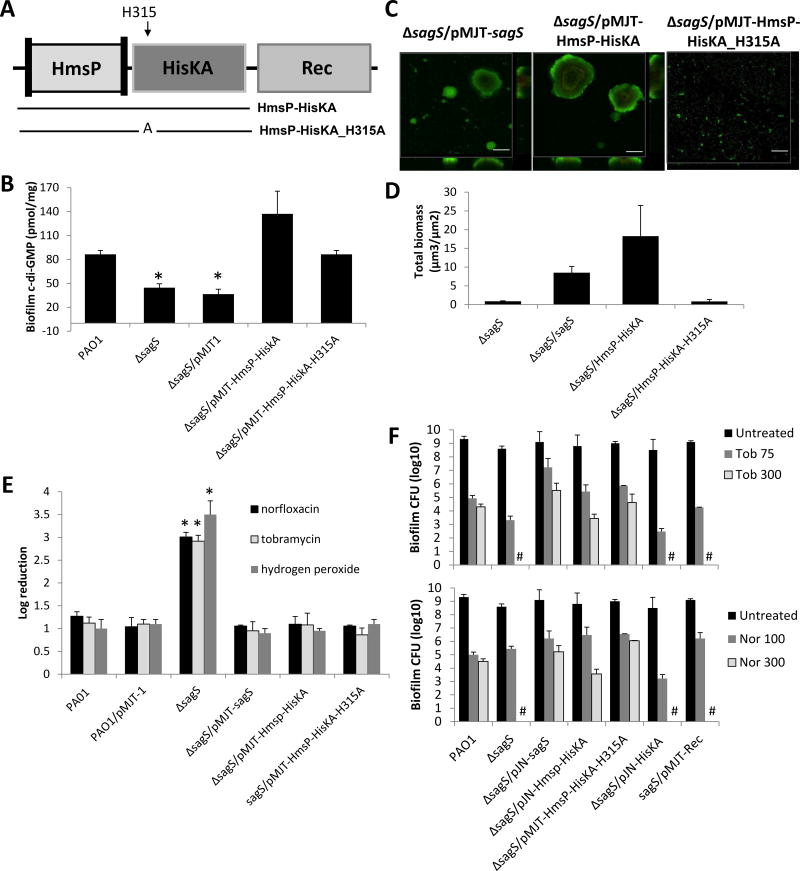
Figure 6. The HmsP domain of SagS contributes to antimicrobial tolerance, but not biofilm formation.
(A) Overview of SagS domains and SagS domain constructs. Lines underneath the domains indicate the composition of the SagS domain constructs, while the names of the resulting constructs are given next to the lines. (B) c-di-GMP levels present in wild-type and ΔsagS mutant biofilm cells expressing intact sagS or sagS domain constructs HmsP-HisKA and HmsP-HisKA_H315A variants. pmol/mg refers to c-di-GMP levels (pmol) per total cell protein (in mg). ΔsagS harboring the empty vector pMJT-1 was used as vector control. (C) Representative confocal images showing the architecture of biofilms formed by ΔsagS mutant strains expressing intact sagS or sagS domain constructs HmsP-HisKA and HmsP-HisKA_H315A. Biofilms were stained with the LIVE/DEAD BacLight viability stain. White bars = 100 µm. (D) Biofilm biomass of ΔsagS mutant strains expressing intact sagS or sagS variants, as determined using confocal images and subsequent COMSTAT analysis. (E) Susceptibility of wild-type and ΔsagS mutant biofilm cells (2 days-old) expressing intact sagS or sagS domain constructs HmsP-HisKA and HmsP-HisKA_H315A to tobramycin, norfloxacin, and hydrogen peroxide. P. aerugionosa PAO1 harboring the empty plasmid pMJT-1 was used as a control. Biofilm susceptibility was determined by log reduction. (F) Biofilm-MBC assays. P. aeruginosa PAO1, ΔsagS, and ΔsagS expressing sagS, HmsP-HisKA, HmsP-HisKA_H315A, HisKA, or Rec were grown as biofilms for 3 days and subsequently treated for 24 hr with tobramycin (75 and 300 µg/ml) and norfloxacin (100 and 300 µg/ml) under continuous flowing conditions before recovering and enumerating surviving cells. Biofilm susceptibility to tobramycin or norfloxacin was determined by viable counts (biofilm CFU, obtained from biofilm tube reactors having an inner surface area of 25 cm2). #, no viable bacteria were detected. All experiments were carried out in triplicate. Error bars denote standard deviation. *, significantly different from the values for P. aeruginosa PAO1 (p ≤ 0.01), as determined by ANOVA and SigmaStat.
Previous findings indicated that ΔsagS biofilm cells harbor the secondary messenger c-di-GMP at reduced levels, similar to those observed in wild-type cells grown planktonically rather than as biofilms, with restoration of c-di-GMP levels to wild-type biofilm-like levels not only restoring the biofilm architecture but also the recalcitrance to killing by antimicrobial agents of ΔsagS biofilm cells to wild-type levels (Gupta et al., 2014). We therefore reasoned that if the HmsP domain indeed contributes to biofilm tolerance, that expression of either SagS variant would coincide with increased tolerance and cellular c-di-GMP levels comparable to those detected in wild-type biofilms. However, we anticipated restoration of biofilm tolerance and biofilm cellular c-di-GMP levels to be independent of the restoration of the biofilm architecture to wild-type levels. As anticipated, expression of the SagS variants coincided with significantly increased c-di-GMP levels, with expression of the HmsP-HisKA_H315A variant restoring c-di-GMP to wild-type levels, while expression of the HmsP-HisKA coincided with cellular c-di-GMP levels that were significantly higher than to those present in wild-type biofilms (Fig. 6B). Despite ΔsagS biofilm cells expressing HmsP-HisKA_H315A demonstrating c-di-GMP levels comparable to wild-type biofilms, the biofilm architecture of ΔsagS/pMJT-HmsP-HisKA_H315A was comparable to that of biofilms formed by ΔsagS (Fig. 6C–D, S3A). In contrast and consistent with ΔsagS/pMJT-HmsP-HisKA biofilm cells demonstrating significantly increased c-di-GMP levels relative to wild-type biofilms, expression of HmsP-HisKA in ΔsagS mutant cells resulted in significantly enhanced biofilm biomass accumulation, average and maximum height, and microcolony formation compared to ΔsagS biofilms (Fig. 6C–D, S3A). It is of interest to note that restoration of the HmsP-HisKA variants functioned independently of GacA, as expression of HmsP-HisKA restored the biofilm architecture of ΔsagSΔgacA mutant cells to wild-type levels (Fig. 4D). Our findings suggested a role of the HmsP domain in restoring biofilm c-di-GMP levels in ΔsagS independent of restoring biofilm formation.
We next determined whether the presence of the HmsP domain promotes the switch from a susceptible to a highly tolerant state using biofilm susceptibility assays. Compared to ΔsagS biofilm cells, biofilms formed by strains ΔsagS/pMJT-HmsP-HisKA and ΔsagS/pMJT-HmsP-HisKA_H315A were as resistant to tobramycin, norfloxacin, and hydrogen peroxide as wild-type biofilms (Fig. 6E). SagS not only contributes to the resistance of biofilm cells to antimicrobial agents but also to the recalcitrance of biofilm cells to killing by bactericidal antibiotics, with inactivation of sagS correlating with complete killing of biofilm cells following 24 hr exposure to tobramycin and norfloxacin at concentrations exceeding 100 µg/ml, in contrast to survival of wild-type cells following the same treatment (Gupta et al., 2013; Gupta et al., 2014). To determine whether the HmsP-HisKA and HmsP-HisKA_H315A constructs contribute to the recalcitrance of P. aeruginosa biofilm cells to antimicrobial agents, we made use of biofilm-MBC assays. As previously reported (Gupta et al., 2013), ∆sagS biofilm cells were eradicated following 24 hr exposure to norfloxacin or tobramycin at concentrations exceeding 300 µg/ml, while expression of sagS restored the recalcitrance of ∆sagS biofilm cells to wild-type levels (Fig. 6F). Multi-copy expression of HmsP-domain-harboring constructs in ∆sagS biofilms likewise resulted in significantly reduced susceptibility to tobramycin, with expression of HmsP-HisKA and HmsP-HisKA_H315A rendering ∆sagS biofilm cells comparable in resistance to wild-type biofilms (Fig. 6F). Similar results were obtained when biofilms were treated with norfloxacin (Fig. 6F). In contrast, no difference in recalcitrance was noted upon expression of sagS-HisKA or -Rec, for which no viable cells were recovered following 24 hr exposure to norfloxacin or tobramycin at concentrations exceeding 300 µg/ml (Fig. 6F). In agreement with previous findings(Gupta et al., 2013), inactivation or overexpression of sagS had no effect on the susceptibility of planktonic cells to tobramycin (Fig. S8). Likewise, overexpression of HmsP-HisKA and HmsP-HisKA_H315A in a sagS mutant background had no effect on the susceptibility to tobramycin of planktonically grown cells (Fig. S8), further supporting the biofilm-specific nature of SagS' contribution to drug tolerance.
Multicopy expression of sagS-HmsP-HisKA in ΔsagS biofilm cells correlates with restoration of BrlR protein abundance and BrlR DNA binding to wild-type levels
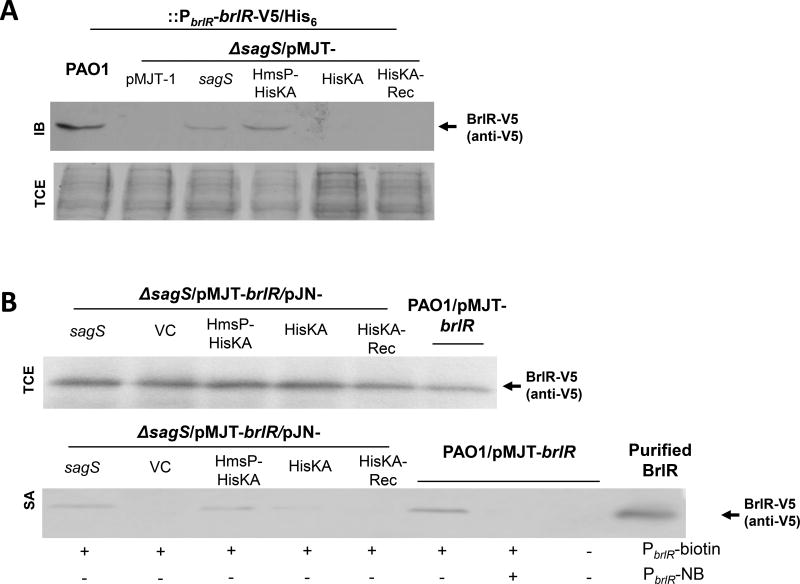
Given that the observed restoration of the biofilm resistance phenotype of ΔsagS to wild-type levels in the presence of the HmsP domain was independent of biofilm biomass accumulation and biofilm architecture, as evidenced by the expression of the HmsP-HisKA-H315A construct in ΔsagS, and considering that biofilm drug tolerance relies on the SagS-dependent production of BrlR in a c-di-GMP dependent manner (Gupta et al., 2013; Chambers et al., 2014), we next asked whether the HsmP-HisKA domain contributes to BrlR levels. To do so, we made use of a chromosomally located V5/His6-tagged BrlR variant under the control of its native promoter and immunoblot analysis (Fig. 7A). In agreement with previous findings (Gupta et al., 2013), P. aeruginosa PAO1 was found to produce BrlR, while ΔsagS mutant biofilm cells harboring the empty vector pMJT-1 lacked detectable levels of BrlR (Fig. 7A, C). Likewise, very little to no BrlR was detected in total extracts obtained from ΔsagS mutant biofilm cells overexpressing sagS-HisKA and sagS-HisKA-Rec. BrlR, however, was detectable in ΔsagS biofilm cells expressing sagS or sagS-HmsP-HisKA (Fig. 7A, C).
Figure 7. Multicopy expression of sagS-HmsP-HisKA restores BrlR levels and enables BrlR-DNA binding to wild-type levels.
(A) Detection of BrlR by immunoblot analysis. Total cell extracts (TCE) obtained from P. aeruginosa wild-type and ΔsagS mutant biofilms (3-day old) expressing a chromosomally located V5/His6-tagged BrlR under the control of its own promoter (PbrlR-brlR-V5/His6) were probed for the presence of BrlR by immunoblot analysis (IB) using anti-V5 antibodies (anti-V5). A total of 15µg total cell extract was loaded. The corresponding SDS/PAGE gel image obtained post-transfer demonstrates equal loading. (B) BrlR-DNA binding using streptavidin magnetic bead binding assays and cell extracts obtained from PAO1 and ΔsagS mutant biofilms (3-day old) overexpressing V5/His6-tagged BrlR and sagS or SagS domain constructs. Binding assays were carried out using a total of 5pmol the BrlR-V5/His6 protein obtained from the strains indicated, and 1 pmol biotinylated PbrlR. Non-biotinylated PbrlR (PbrlR-NB) was used as specific competitor DNA in 20-fold excess. BrlR binding to PbrlR was detected by immunoblot analysis using anti-V5 antibodies. +/−, indicates presence/absence of specific probe or competitor. Top image: Detection of BrlR in 15µg total cell extracts (TCE) as determined using immunoblot analysis was used as loading controls. (C) Relative BrlR abundance, based on relative intensity of protein bands detectable following probing for BrlR with anti-V5 antibodies and subsequent analysis using ImageJ (Rasband, 1997). BrlR protein production refers to BrlR abundance based on band intensity obtained following immunoblot analysis using total cell extracts. BrlR-PblR binding refers to BrlR abundance obtained following analysis of the streptavidin magnetic bead binding assays. Experiments were carried out in triplicate and representative images are shown. Error bars denote standard deviation. *, significantly different from the values for P. aeruginosa PAO1 (p ≤ 0.01), as determined by ANOVA and SigmaStat.
BrlR is a transcriptional regulator. To ensure that BrlR production correlated with BrlR function, we made use of streptavidin magnetic bead DNA binding assays using cell extracts obtained from wild-type and ΔsagS biofilm cells overexpressing sagS or sagS domain constructs and V5/His6-tagged brlR (pMJT-brlR). BrlR binding to its own promoter (PbrlR, 1 pmol) was observed using cell extracts obtained from wild-type and ΔsagS/pMJT-sagS biofilms, with addition of 20-fold excess of unlabeled competitor significantly reducing the interaction of BrlR with the brlR promoter DNA (Fig. 7B). Despite having BrlR levels comparable to wild-type biofilms due to overexpression of brlR from the pMJT-1 vector, no detectable BrlR was enriched using the brlR promoter DNA in the absence of SagS in ΔsagS/pMJT-brlR biofilms (Fig. 7B–C). Likewise, no DNA-binding was noted using cell extracts obtained from ΔsagS/pMJT-sagS-HisKA-Rec cells. Interestingly, a very low amount of BrlR binding was observed in ΔsagS/pMJT-sagS-HisKA cells (Fig. 7B–C). However, this was significantly lower than that observed for the wild-type or ΔsagS/pMJT-sagS cells, and did not translate into detectable BrlR protein levels (Fig. 7A, C). In contrast, however, higher levels of BrlR binding to PbrlR was observed using cell extracts obtained from ΔsagS/pMJT-sagS-HmsP-HisKA biofilms (Fig. 7B–C). Overall, our findings link BrlR production and activity and the restoration of tolerance and recalcitrance to killing by antimicrobial agents to the presence of the SagS HmsP-HisKA domains, but not the HisKA or Rec domains alone.
DISCUSSION
The goal of this study was to determine the mechanism by which SagS carries out its dual regulatory functions (Fig. 1). More specifically, we asked whether SagS promoting biofilm formation and biofilm tolerance is linked to its modular composition, and whether SagS regulates both functions via two distinct pathways, with the molecular communication between SagS and BfiS via phospho signaling being independent of SagS contributing to biofilm tolerance. Proteins having modular function or dual activities are not uncommon. A number of proteins linked to the regulation of surface attachment and biofilm formation are known to exhibit dual activities. C-di-GMP levels are controlled by the opposing activity of two enzymes: diguanylate cyclases containing a characteristic GGDEF domain and phosphodiesterases harboring EAL or HD-GYP domains. While most enzymes harboring both domains have been demonstrated to only possess one of the two c-di-GMP modulating activities, biochemical analysis of the P. aeruginosa MucR, the positive regulator of alginate biosynthesis, was found to possess both diguanylate cyclase and phosphodiesterase activity (Li et al., 2013). However, the opposing activities were found to be modulated in a growth mode-dependent manner, with MucR functioning as a diguanylate cyclase under planktonic growth conditions, and as a phosphodiesterase under biofilm growth conditions (Li et al., 2013). The P. aeruginosa flagella biosynthesis gene activator FleQ functions as both a repressor and an activator to control gene expression from the Pel polysaccharide biosynthesis (pel) operon promoter in response to c-di-GMP (Hickman and Harwood, 2008; Baraquet et al., 2012). FleQ represses pel gene expression by inducing DNA distortion. Binding of c-di-GMP to FleQ relieves the DNA distortion, with FleQ remaining bound to the pel promoter and being converted from a repressor to an activator (Baraquet et al., 2012).
What distinguishes SagS from other dual function proteins, however, is the finding of SagS promoting the two switch functions via two independent signaling networks, with each network being associated with a distinct portion or domain(s) of SagS (Fig. 8). This is based on the finding that SagS contributes to the transition from a planktonic to the surface-associated mode of growth via the hierarchical phosphotransfer-based signaling from SagS to BfiS, with the phosphorelay involving the conserved phospho-transfer sites located within the Rec domain at aspartate residue D713 and the histidine phosphorylation site located in the HisKA domain at histidine residue H315 (Fig. 8). Moreover, our findings indicate that the HisKA-Rec domain construct alone contributed to biofilm formation. In agreement with previous findings, HisKA-Rec-dependent biofilm development coincided with BfiSR activation and subsequent modulation of RsmZ (Goodman, 2010; Petrova and Sauer, 2010; Petrova and Sauer, 2011).
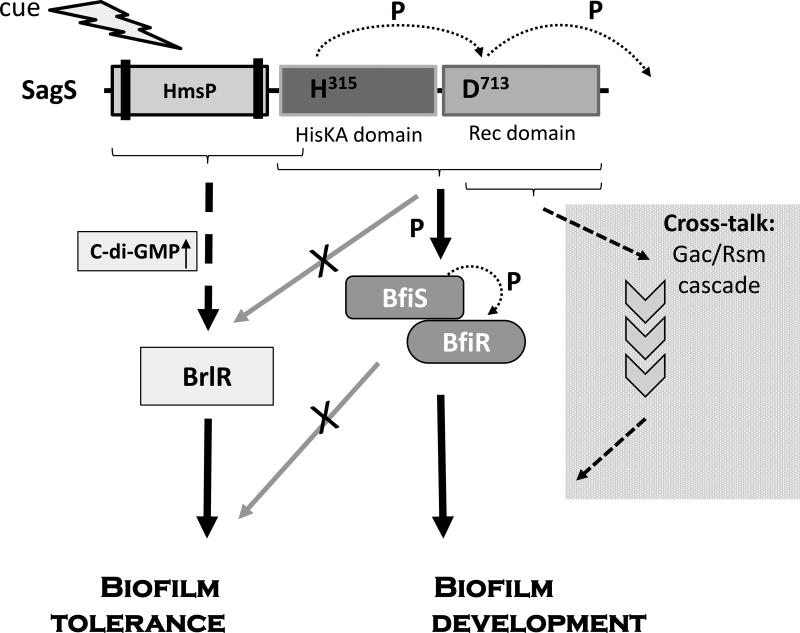
Figure 8. Proposed model of the “divide and conquer” dual regulatory function of SagS to promote biofilm development and biofilm tolerance.
The sensory protein SagS is activated via an as of yet unknown signal or cue that likely contributes to SagS autophosphorylation. Upon activation, the HmsP-HisKA and HmsP-HisKA_H315A, but not HisKA alone, contribute to biofilm tolerance. In contrast, the phosphorelay of SagS comprising H315 and D713A and its subsequent effect on the BfiS phosphorylation status and BfiS downstream targets contribute to biofilm development, but not to biofilm cells being rendered resistant to antimicrobial agents. Likewise, the HisKA and Rec domains contribute to biofilm development, but not biofilm tolerance. In addition, the Rec domain alone can contribute to biofilm development in a GacA-dependent manner suggesting cross-talk with GacA and/or components of the Gac/Rsm system. P, phosphotransfer reaction. Arrows indicate modulatory effect of domains. Lack of modulatory effect is indicated by arrows with an “X”.
While the SagS-BfiS phosphorelay contributes to biofilm development, neither the His-Asp dependent phosphotransfer-based signaling between SagS and BfiS nor the C-terminally located HisKA-Rec domains of SagS were found to play a role in biofilms being rendered tolerant to antimicrobial agents. Instead, our findings indicated that SagS contributes to biofilm cells being rendered resistant to antimicrobial agents via its N-terminally located sensory HmsP domain (Fig. 8). While our findings do not exclude a role of the HmsP domain in perceiving and relaying signals for subsequent His-Asp dependent phosphotransfer-based signaling to BfiS, biofilm tolerance was found to be independent of BfiSR downstream targets (Fig. 5). Moreover, restoration of biofilm tolerance in ΔsagS biofilm cells was independent of biofilm biomass accumulation. This is based on the finding that despite differences in the biofilm architecture and biomass accumulation, both ΔsagS/pMJT-HisKA-Rec and ΔsagS/pMJT-HisKA-Rec_H315A biofilm cells were found to be as resistant to antimicrobial agents as wild-type biofilm cells. Instead, and in agreement with previous findings, restoration of biofilm tolerance of ΔsagS biofilm cells appeared to be dependent on the presence of the transcriptional regulator BrlR (Liao and Sauer, 2012; Gupta et al., 2013; Chambers et al., 2014; Gupta et al., 2014). While the mechanism by which SagS contributes to BrlR activation has not been elucidated in full (Gupta et al., 2013; Gupta et al., 2014), restoration of biofilm tolerance and BrlR production was noted upon expression of both HmsP-HisKA and HmsP-HisKA_H315, with expression of both HmsP-variants coinciding with significantly enhanced cellular c-di-GMP levels relative to those present in ΔsagS biofilm cells. The contribution of c-di-GMP to biofilm tolerance is in agreement with previous findings demonstrating that restoration of c-di-GMP levels to wild-type biofilm-like levels not only restored BrlR production and DNA binding by BrlR to wild-type levels, but also recalcitrance to killing by antimicrobial agents of ΔsagS biofilm cells (Gupta et al., 2013; Chambers et al., 2014; Gupta et al., 2014).
Recalcitrance, or tolerance, of biofilm cells to killing by antimicrobial agents is believed to be multifactorial and dependent on factors including restricted diffusion of antimicrobial agents, slow growth, reduced metabolic rates, increased stress tolerance, and differences in gene expression and protein production in biofilms compared to planktonic cells. So how does SagS fit in? While the present study did not evaluate many of the other factors contributing to tolerance, SagS potentially contributes to diffusion limitation or differences in growth or metabolic rates. This is supported by previous findings of SagS being activated early during biofilm development, with inactivation of sagS coinciding with biofilm development being arrested at the attachment stage and the formation of thin and unstructured biofilms (Petrova and Sauer, 2011; Gupta et al., 2013). However, our findings do clearly demonstrate SagS-dependent differences in gene expression and protein production associated with BrlR, phosphotransfer events to BfiSR, subsequent BfiSR activation, and c-di-GMP modulation, with both BrlR and c-di-GMP having been linked to tolerance(Liao and Sauer, 2012; Chambers et al., 2014; Gupta et al., 2014). Our findings thus suggest that SagS contributes, at least in part, to the multifactorial nature of tolerance, likely during the initial stages of tolerance activation.
It is of interest to note that while expression of HisKA-Rec and the HmsP variant coincided restoration of c-di-GMP to wild-type levels, only HmsP variant-derived c-di-GMP levels rescued the susceptibility phenotype of ΔsagS biofilm cells to wild-type levels. Taking into account the previously described link between c-di-GMP and susceptibility to antimicrobial agents (Chambers et al., 2014; Gupta et al., 2014), and c-di-GMP and surface associated growth, and considering that expression of HmsP-HisKA_H315 restored biofilm tolerance without restoring the biofilm architecture of ΔsagS to wild-type levels, our findings hinted at SagS likely contributing to two distinct pools of c-di-GMP that separately regulate biofilm formation or biofilm tolerance. While the identity of the enzymes contributing to the distinct c-di-GMP pools is currently under investigation, it is likely that at least two diguanylate cyclases may play a role.
Taken together, our findings demonstrate that SagS carries out its dual regulatory functions via two distinct pathways, with each pathway being activated via a distinct segment within SagS. Considering that the periplasmic HmsP domain is likely involved in perceiving as of yet unknown signals or cues from the exterior of the P. aeruginosa cells, the two distinct regulatory functions of SagS are reminiscent of a “divide and conquer” regulatory switch, with HmsP relaying or dividing the cue(s) in order to (i) activate BrlR to enable a heightened resistance phenotype, while (ii) relaying signals for subsequent His-Asp dependent phosphotransfer-based signaling to BfiS to enable biofilm development. Considering the importance of the HmsP domain in biofilm tolerance, our findings may lead to strategies to impair the transition of biofilm cells from a susceptible to a resistant phenotype by targeting the HmsP domain of SagS, which may likely result in enhanced treatment options against biofilm infections.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions
All bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa strain PAO1 was used as the parental strain. All planktonic cultures were grown in Lennox broth (LB) (BD Biosciences) in flasks at 220 rpm. Biofilms were grown as described below at 22°C in 20-fold-diluted LB. Antibiotics for plasmid maintenance were used at the following concentrations: 250 µg/ml carbenicillin and 50 to 75 µg/ml gentamicin for P. aeruginosa and 100 µg/ml ampicillin and 20 µg/ml gentamicin for Escherichia coli.
Table 1.
Strains and plasmids used.
| Strains/Plasmids | Relevant genotype or description | Source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | F- φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rk−, mk+) phoA supE44 thi-1 gyrA96 relA1 tonA | Invitrogen Corp. |
| S17-1 | λpir; hsdR pro recA; RP4 2-Tc::Mu-Km::Tn7, pro, res−, mod+, StrR, TrmR | (Simon et al., 1983) |
| Pseudomonas aeruginosa | ||
| PAO1 | Wild type strain PAO1 | B.H. Holloway |
| ΔsagS | PAO1, ΔsagS (PA2824) | (Petrova and Sauer, 2011) |
| ΔsagSΔgacA | ΔsagS, ΔgacA in PAO1 | (Petrova and Sauer, 2011) |
| ΔrsmYZ | ΔrsmYΔrsmZ in PAO1 | (Kay et al., 2006) |
| ΔrsmA | ΔrsmA in PAO1 | (Kay et al., 2006) |
| ΔhptB | ΔhptB in PAO1; KmR | (Lin et al., 2006) |
| ΔcafA | ΔcafA in PAO1 | (Petrova and Sauer, 2010) |
| Plasmids | ||
| pCR2.1-TOPO | TA cloning vector; KmR; AmpR | Invitrogen Corp. |
| pRK2013 | Helper plasmid for triparental mating; mob; tra; KmR | (Figurski and Helinski, 1979) |
| pJN105 | Arabinose-inducible gene expression vector; pBRR-1 MCS; araC-PBAD; GmR | (Newman and Fuqua, 1999) |
| pMJT1 | araC-PBAD cassette of pJN105 cloned into pUCP18, AmpR (CarbR) | (Kaneko et al., 2007) |
| pminiCTX-lacZ | attB site-specific integration vector, TetR | (Becher and Schweizer, 2000) |
| pJN-sagS | C-terminal HA-tagged sagS cloned into pJN105 at NheI/SacI; GmR | (Petrova and Sauer, 2011) |
| pJN-sagS_H315A | C-terminal HA-tagged sagS with H315 mutation in pJN105 at NheI/SacI; GmR | This study |
| pJN-sagS_H348A | C-terminal HA-tagged sagS with H348A mutation in pJN105 at NheI/SacI; GmR | This study |
| pJN-sagS_D713A | C-terminal HA-tagged sagS with D713A mutation in pJN105 at NheI/SacI; GmR | This study |
| pJN-sagS-HmsP-HisKA | C-terminal HA-tagged SagS HmsP-HisKA domains cloned into pJN105 at NheI/SacI; GmR | This study |
| pJN-sagS-HisKA | C-terminal HA-tagged SagS HisKA domain cloned into pJN105 at NheI/SacI; GmR | This study |
| pJN-sagS-HisKA-Rec | C-terminal HA-tagged SagS HisKA-Rec domain cloned into pJN105 at NheI/SacI; GmR | This study |
| pJN-sagS-Rec | C-terminal HA-tagged SagS Rec domain cloned into pJN105 at NheI/SacI; GmR | This study |
| pJN-rsmY | rsmY cloned into pJN105 at EcoRI/SacI; GmR | (Petrova and Sauer, 2010) |
| pJN-rsmZ | rsmZ cloned into pJN105 at EcoRI/SacI; GmR | (Petrova and Sauer, 2010) |
| pMJT-bfiS | C-terminal His6/V5-tagged bfiS cloned into pMJT1; AmpR (CarbR) | (Petrova and Sauer, 2011) |
| pMJT-brlR | C-terminal His6/V5-tagged brlR cloned into pMJT1; AmpR (CarbR) | (Liao and Sauer, 2012) |
| pMJT-sagS | C-terminal HA-tagged sagS cloned into pMJT1 at NheI/SacI; GmR | (Petrova and Sauer, 2011) |
| pMJT-sagS-HmsP-HisKA | C-terminal HA-tagged SagS HmsP-HisKA domains cloned into pMJT1 at NheI/SacI; AmpR (CarbR) | This study |
| pMJT-sagS-HmsP-HisKA_H315A | C-terminal HA-tagged SagS HmsP-HisKA domains with H315A mutation cloned into pMJT1; AmpR (CarbR) | This study |
| pMJT-sagS-HisKA | C-terminal HA-tagged SagS HisKA domain cloned into pMJT1; AmpR (CarbR) | This study |
| pMJT-sagS-HisKA-Rec | C-terminal HA-tagged SagS HisKA-Rec domain cloned into pMJT1; AmpR (CarbR) | This study |
| pMJT-sagS-Rec | C-terminal HA-tagged SagS Rec domain cloned into pMJT1; AmpR (CarbR) | This study |
| pMJT-sagS-Rec_D713A | C-terminal HA-tagged SagS Rec domain with D713A mutation cloned into pMJT1 at NheI/SacI; GmR | This study |
| pMJT-sagS-Rec_D713E | C-terminal HA-tagged SagS Rec domain with D713E mutation cloned into pMJT1 at NheI/SacI; GmR | This study |
| pCTX-rsmY-lacZ | rsmY reporter construct in mini-CTX-lacZ; TetR | (Brencic and Lory, 2009) |
| pCTX-rsmZ-lacZ | rsmZ reporter construct in mini-CTX-lacZ; TetR | (Brencic and Lory, 2009) |
| pCTX-PbrlR-lacZ | brlR promoter reporter construct in mini-CTX-lacZ, using PbrlR-lacZF/R primers; TetR | (Liao and Sauer, 2012) |
| PbrlR-brlR-His6V5 | pMini-CTX harboring C-terminal His6/V5-tagged brlR under the control of the brlR promoter region (1–120 bp upstream of the brlR start codon) | (Gupta et al., 2014) |
Strain construction
Complementation and overexpression of full-length sagS or the different domain constructs of SagS was accomplished by amplifying the respective sequences from P. aeruginosa PAO1 genomic DNA using the oligonucleotides listed in Table 2 and placing the respective sequences under the control of an arabinose-inducible promoter in the pJN105 (Newman and Fuqua, 1999) or pMJT-1 vectors (Kaneko et al., 2007). Site-directed mutagenesis of indicated sagS sequences was accomplished by using the GeneArt Site-Directed Mutagenesis Kit (Invitrogen) according to the manufacturer’s protocol. The identity of all vector inserts was confirmed by sequencing. Plasmids were introduced into P. aeruginosa via conjugation or electroporation. Primers used for strain construction are listed in Table 2.
Table 2.
Primers used in this study.
| Oligonucleotide | Sequencea,b |
|---|---|
| Inducible Expression Constructs | |
| HmsP-HisKA-for | GCGCGCGCgctagcATGCTAGGCGGCAGAACCTCGC |
| HmsP-HisKA-HA-rev | GCGCGCGCgagctcCTAagcgtagtctgggacgtcgtatgggtaGCGCTGTTCTCCCGTGGTG |
| HisKA–for | GCGCGCGCgctagcATGCTCGGGCGCATCTCGGTAG |
| HisKA–HA-rev | GCGCGCGCgagctcCTAagcgtagtctgggacgtcgtatgggtaCAGGACCCGGGTGTTGCT |
| HisKA-Rec-for | GCGCGCGCgctagcATGCTCGGGCGCATCTCGGTAG |
| HisKA-Rec-HA-rev | GCGCGCGCgagctcCTAagcgtagtctgggacgtcgtatgggtaGTCGCTCGCGGTGAGCGG |
| Rec-for | GCGCGCgctagcATGGTCAGCCCTCCGCTGCAG |
| Rec-HA-rev | GCGCGCGCgagctcCTAagcgtagtctgggacgtcgtatgggtaGTCGCTCGCGGTGAGCGG |
| Site-directed Mutagenesis | |
| sagS_H315A_for | CTTCCTGGCCAACATGAGCGCCGAGATCCGCACGCCGCTG |
| sagS_H315A_rev | CAGCGGCGTGCGGATCTCGGCGCTCATGTTGGCCAGGAAG |
| sagS_H348_for | GCCAGCAACTGTCGATCGCCGCCGACTCCGGCAAGGTGCTGGTG |
| sagS_H348_rev | CACCAGCACCTTGCCGGAGTCGGCGGCGATCGACAGTTGCTGGC |
| sagS_D713A_for | CCATCGACCTGGTCCTGATGGCCTGCAACATGCCAGTGATGGAC |
| sagS_D713A_rev | GTCCATCACTGGCATGTTGCAGGCCATCAGGACCAGGTCGATGG |
| sagS_D713E_for | CCATCGACCTGGTCCTGATGGAGTGCAACATGCCAGTGATGGAC |
| sagS_D713E_rev | GTCCATCACTGGCATGTTGCACTCCATCAGGACCAGGTCGATGG |
| Sequencing | |
| sagScheck_rev | GCAGTACCTGGTCATGCTGG |
| sagScheck_for | CGATCTCGATCAGGCGCAG |
| qRT-PCR | |
| mreB-for | CTGTCGATCGACCTGGG |
| mreB-rev | CAGCCATCGGCTCTTCG |
| RsmZ-for | GAAGGATCGGGGAAGGGAC |
| RsmZ-rev | CGCCCACTCTTCAGTCCC |
, Restriction sites are indicated by nucleotides in lower case
, HA tag codons are underlined and indicated in lower case letters
Planktonic antibiotic susceptibility testing
To determine the role of HmsP domain constructs in antimicrobial susceptibility, P. aeruginosa PAO1 (PAO1/pMJT1) and sagS mutant strains (ΔsagS/pMJT1, ΔsagS/pMJT-sagS, ΔsagS/pMJT-HmsP-HisKA, ΔsagS/pMJT-HmsP-HisKA-H315A) were grown planktonically in LB medium at 37°C to exponential phase, then exposed to tobramycin (50 µg/ml) for 30 min, and subsequently homogenized, serially diluted and spread-plated onto LB agar. Viability was determined via CFU counts. Susceptibility is expressed as log reduction in CFU counts. Under the conditions tested, a total of 8.4×108±2.6×108 CFU/ml were detected on average for exponential phase planktonic cells.
Biofilm formation
For biofilm antibiotic susceptibility testing, biofilms were grown in a continuous flow tube reactor system (1-m-long size 13 silicone tubing; Masterflex, Cole Parmer, Inc.) with an inner surface area of 25 cm2 at a flow rate of 0.1 ml/min and in flow cell reactors (BioSurface Technologies), which also allowed for the analysis of biofilm architecture, as previously described (Sauer et al., 2002; Sauer et al., 2004; Petrova and Sauer, 2009). Biofilms were grown at 22°C in 20-fold-diluted LB medium. The same growth conditions were used to cultivate biofilms to obtain protein and RNA. For plasmid maintenance, antibiotics were used in tube reactors at the following concentrations: 2 µg/ml gentamicin and 10 µg/ml carbenicillin. Confocal laser scanning microscopy (CLSM) images of flow-cell-grown biofilms were acquired using a Leica TCS SP5 confocal microscope (Leica Microsystems, Wetzlar, Germany) and the LIVE/DEAD BacLight Bacterial Viability Kit (Life Technologies), and quantitative analysis of the images was performed using COMSTAT (Heydorn et al., 2000).
Biofilm antibiotic susceptibility assays
To assess the susceptibility of biofilm cells to antimicrobial agents, biofilms were grown for 2 days under flowing conditions (0.1 ml/min) and subsequently treated for 1 hr under flowing conditions with the following antimicrobial agents: tobramycin (150 µg/ml), norfloxacin (450 µg/ml), and hydrogen peroxide (0.3%). Following exposure of biofilms to the respective antimicrobial agents, biofilms were harvested by squeezing the tubing, followed by the extrusion of the cell paste as previously described (Sauer and Camper, 2001). To ensure complete disaggregation of cell aggregates, the resulting suspension was first homogenized using a tissue tearor, then serially diluted, and spread plated onto LB agar. Hydrogen peroxide was neutralized by the addition of 100 mM sodium thiosulfate. Viability was determined via CFU counts (Fig. S2). Susceptibility is expressed as log reduction.
The biofilm minimum bactericidal concentration (biofilm-MBC) has been defined as the concentration at which no further increase in log reduction is observed (Monzon et al., 2001; Moriarty et al., 2007; Villain-Guillot et al., 2007). To determine whether SagS and its specific domains affect the biofilm-MBC and resistance to killing, biofilms of indicated strains were grown for 3 days, after which time the medium was switched to the same medium containing increasing concentrations of tobramycin or norfloxacin, ranging from 0.5 to 400 µg/ml. Following 24 hr of exposure to the antibiotic under continuous flow at 0.1 ml/min, biofilms were harvested and the surviving bacteria enumerated by viable counts.
Immunoblot analysis and pulldowns
Abundance of HA-tagged SagS, SagS domain constructs, and V5/His6-tagged BfiS or BrlR present in P. aeruginosa cells was assessed by SDS-PAGE and immunoblot analysis. Briefly, total cell extracts from planktonic and biofilm cells were obtained by sonication as previously described (Sauer et al., 2002) followed by centrifugation for 5 min at 21,200 × g to pellet unbroken cells. The protein concentrations were determined using a modified Lowry assay and bovine serum albumin as a standard (Peterson, 1977). The samples (15 µg) were resolved on an 11% polyacrylamide gel and subsequently transferred onto PVDF membranes using a TurboTransblot apparatus (Biorad). Western blots were probed with anti-V5-HRP or anti-V5 antibodies (Invitrogen) and anti-HA antibodies (Covance). When necessary, a secondary anti-mouse IgG antibody (Cell Signaling Technologies) was used. The antibodies we used at 0.2 µg/mL. The blots were subsequently developed using ImmunStar WesternC chemiluminescent reagents (Biorad). Following transfer, SDS/PAGE gels were Coomassie-stained to ensure equal loading.
Pull-down assays were used to assess the interactions between various SagS domain constructs and BfiS in total protein cell extracts of cells co-producing V5/His6-tagged BfiS and HA-tagged bait SagS protein or SagS domain as indicated. Following immunoprecipitation of HA-tagged proteins using immobilized anti-HA antibodies at a 2 µg/mL concentration, immunoprecipitation eluates were separated by SDS/PAGE and assessed by immunoblot analysis for the presence of V5/His6-tagged prey proteins using anti-V5-HRP antibodies as described above. Pull-down assays were carried out using 200 µg protein from cellular extracts.
Phosphoprotein enrichment and detection
Analysis of phosphorylated BfiS-V5/His6 levels in protein extracts obtained from ΔsagS mutant cells expressing sagS or the various SagS domain and variant constructs was accomplished using phosphoprotein purification via metal oxide affinity chromatography (MOAC) essentially as described by Wolschin and colleagues (Wolschin et al., 2005). MOAC has been demonstrated by Krüger et al. to result in up to 20-fold enrichment of phosphoproteins and to approach 100% specificity (Krüger et al., 2007). Briefly, 750 µg of total protein cell extract were diluted with MOAC incubation buffer (30 mM MES, 0.2 M potassium glutamate, 0.2 M sodium aspartate, 0.25 % Chaps, and 8 M urea) to a final volume of 1.5 mL, and subsequently incubated for 30 min at 4°C in the presence of 80 mg of aluminum hydroxide. Unbound proteins were removed via six one-minute washes of the aluminum hydroxide slurry with 1.5mL of incubation buffer at 16,000×g and 4°C. Then, phosphoproteins were eluted from the slurry using 100 mM potassium pyrophosphate and 8M urea, desalted by methanol-chloroform precipitation, and subsequently vacuum-dried. Samples were then analyzed by SDS/PAGE, followed by the detection of BfiS-V5/His6 by immunoblotting with anti-V5 antibodies as described above. Aliquots obtained prior to MOAC were used as loading controls.
Streptavidin Magnetic bead DNA Binding Assay
BrlR binding to the promoter region of brlR was determined using the streptavidin magnetic bead DNA binding assay as previously described (Petrova et al., 2011). Briefly, a total of 1 pmol of target DNA (biotinylated PbrlR (Gupta et al., 2013; Chambers et al., 2014)) was incubated for 30 min at room temperature with 15 µg of total cell extract containing the equivalent of 5 pmol of V5/His6-tagged BrlR (as indicated by immunoblot analysis using purified V5/His6-tagged BrlR) in 25 mM Tris-Cl, pH 8, 5 mM MgCl2, 0.5 mM dithiothreitol, 1 mM EDTA, and 50 ng/uL poly(dI-dC) as nonspecific competitor DNA. For specific competition, non-biotinylated target DNA (up to 20-fold excess of target DNA) was used. Streptavidin magnetic beads (Thermo Scientific, 100 µg) were used to capture biotinylated DNA. Following four washes, the proteins co-purified with the biotinylated DNA were separated by 11% SDS/PAGE and assessed by immunoblot analysis for the presence of BrlR using anti-V5 antibodies (Invitrogen) as described above. Aliquots of total cell extracts (15 µg) were used to determine total BrlR present in each DNA binding assay (loading control).
In vivo quantification of c-di-GMP from P. aeruginosa
Cyclic di-GMP (c-di-GMP) was extracted in triplicate from wild-type and mutant strains using heat and ethanol precipitation (Morgan et al., 2006) and quantitated essentially as previously described (Petrova and Sauer, 2011; Basu Roy et al., 2013). Briefly, c-di-GMP was extracted in triplicate from wild-type and mutant strains grown planktonically to exponential phase or as biofilms for 3 days using heat and ethanol precipitation followed by centrifugation. Supernatants were combined, dried using a Speed-Vac and resuspended in 10 mM ammonium acetate. Samples (20 µl) were analyzed using an Agilent 1100 HPLC equipped with an autosampler, degasser, and detector set to 253 nm, and separated using a reverse-phase C18 Targa column (2.1×40 mm; 5 µm) at a flow rate of 0.2 ml/min with the following gradient: 0 to 9 min, 1% B; 9 to 14 min, 15% B; 14 to 19 min, 25% B; 19 to 26 min, 90% B; 26 to 40 min, 1% B (buffer A, 10 mM ammonium acetate; buffer B, methanol plus 10 mM ammonium acetate). Commercially available cyclic di-GMP was used as a reference for the identification and quantification of cyclic di-GMP in cell extracts.
Reporter assays for RsmZ and brlR transcription
Β-Galactosidase activity of strains harboring the rsmZ promoter reporter construct (Brencic and Lory, 2009; Brencic et al., 2009) and the chromosomally integrated brlR promoter reporter construct (Liao and Sauer, 2012; Chambers et al., 2014) was determined using the Miller assay (Miller, 1972) with the following modification: instead of using total cells, specific β-galactosidase activity was determined using protein extracts, obtained as previously described (Southey-Pillig et al., 2005; Petrova and Sauer, 2010). An extinction coefficient for o-nitrophenyl-β-galactoside (ONPG) cleavage at 420 nm of 4,500 nl/nmol*cm was used. Protein extracts were obtained from biofilms grown for 3 days under flowing conditions.
qRT-PCR
Transcript abundance of RsmZ was determined using qRT-PCR. To do so, isolation of mRNA and cDNA synthesis was carried out as previously described (Southey-Pillig et al., 2005; Allegrucci and Sauer, 2007; Allegrucci and Sauer, 2008; Petrova and Sauer, 2009). qRT-PCR was performed using the BioRad CFX Connect Real-Time PCR Detection System and SsoAdvanced™ SYBR® Green Supermix (BioRad) with oligonucleotides listed in Table 2. mreB was used as a control. Relative transcript quantitation was accomplished using the CFX Manager Software (BioRad), by first normalizing transcript abundance (based on the threshold cycle value (Ct)) to mreB followed by determining transcript abundance ratios. Melting curve analyses were employed to verify specific single product amplification.
Statistical analysis
Student’s t test was performed for pairwise comparisons of groups, and multivariant analyses were performed using a 1-way analysis of variance (ANOVA) followed by an a posteriori test using Sigma Stat software.
Supplementary Material
Figure S1. Detection of SagS and SagS domain constructs in cell free total cell extracts by immunoblot analysis using anti-HA antibodies. All SagS domain constructs, including the full-length gene, harbored a C-terminal hemagglutinin (HA) tag (referred to as SagS*-HA). *, SagS domain constructs vary in molecular mass.
Figure S2. Average number of viable cells, expressed as CFU, present in biofilms formed by P. aeruginosa PAO1 and mutant strains following 2 days of growth at a surface area of 25 cm2 at a flow rate of 0.1 ml/min. Experiments were repeated at least 3 times. Error bars indicate standard deviations.
Figure S3. Average and maximum thickness of P. aeruginosa wild-type and ΔsagS mutant biofilms expressing (A) SagS domain constructs or (B) SagS variants harboring site-directed mutations, as determined using confocal images and subsequent COMSTAT analysis. (C) Biofilm biomass of ΔsagS mutant strains expressing wild-type sagS or sagS variants, as determined using confocal images and subsequent COMSTAT analysis. Biofilms were grown for 6 days in flow cells. All experiments were carried out at least in triplicate. Error bars indicate standard deviation.
Figure S4. Relative BfiS abundance, based on relative intensity of protein bands detectable following probing for BfiS with anti-V5 antibodies and subsequent analysis using ImageJ (Rasband, 1997). (A) BfiS protein levels detected following Co-IP. (B) BfiS protein levels following enrichment by MOAC. Experiments were carried out in triplicate. Error bars denote standard deviation. *, significantly different from the values for P. aeruginosa PAO1 (p ≤ 0.01), as determined by ANOVA and SigmaStat.
Figure S5. c-di-GMP levels present in wild-type and ΔsagS mutant biofilm cells overexpressing sagS domain constructs. The strains were grown in tube reactors under flowing conditions for 6 days prior to c-di-GMP extraction and quantitation by HPLC analysis. pmol/mg refers to c-di-GMP levels (pmol) per total cell protein (in mg). ΔsagS harboring the empty vector pJN105 was used as vector control. All experiments were carried out at least in triplicate. Error bars indicate standard deviation.
Figure S6. Total biofilm biomass (A), and average and maximum thickness (B) of P. aeruginosa ΔsagS and ΔsagSΔgacA mutant biofilms expressing intact sagS or the sagS-Rec domain construct, as determined using confocal images and subsequent COMSTAT analysis. Biofilms were grown for 6 days in flow cells. All experiments were carried out at least in triplicate. Error bars indicate standard deviation.
Figure S7. Localization of transmembrane helices (A) present in SagS as determined using the TMHMM Prediction of transmembrane helices in proteins software (http://www.cbs.dtu.dk/services/TMHMM-2.0/). (B) Alignment of HmsP domains of P. aeruginosa SagS and BifA and Yersinia pestis HmsP (Yp_HmsP). Sec, predicted secondary structure of Yp_HmsP; H, helical regions; e, extended regions; M, membrane spanning regions, from www.blackwellpublishing.com/products/journals/.../mmi4253FigS1.doc. Identical/similar AA are shown in color and indicated by symbols (., :, *).
Figure S8. Contribution of SagS and the HmsP domain to the susceptibility of planktonic cells to tobramycin. All strains were grown planktonically to exponential phase and subsequently exposed to 50 µg/ml tobramycin for 30 min. Strain PAO1 harboring the empty plasmid pMJT1 was used as vector control. Viability was determined via CFU counts. (A) Total CFU prior to treatment. (B) Susceptibility is expressed as log reduction in CFU counts. Experiments were done at least in triplicate. Error bars denote standard deviation.
ORIGINALITY-SIGNIFICANCE STATEMENT.
The tolerance of bacterial biofilm cells to antimicrobial agents has been thought to be a function of biofilm development and biofilm biomass accumulation. Previous findings suggested that in Pseudomonas aeruginosa, the sensor-regulator hybrid SagS regulates both, (i) the transition to biofilm formation and (ii) the transition to cells gaining their enhanced tolerance to antimicrobials. However, here, we demonstrate that SagS regulates both switch functions independently, via the modular functions of its domains. Specifically, we demonstrate that while the SagS HisKA-Rec domain-mediated phosphorelay activates biofilm formation, the periplasmic sensory domain modulates the transition of cells to an antimicrobial tolerant state in a phosphotransfer-independent manner. Our findings thus, suggest that biofilm drug tolerance is not a function of biofilm development and biofilm biomass accumulation but instead regulated independently. Our findings may lead to strategies to separately impair biofilm tolerance and biofilm development for enhanced treatment options against biofilm infections.
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01 AI080710) and the Department of Defense (W81XWH-12-2-0063).
References
- Allegrucci M, Sauer K. Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms. J Bacteriol. 2007;189:2030–2038. doi: 10.1128/JB.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegrucci M, Sauer K. Formation of Streptococcus pneumoniae non-phase-variable colony variants is due to increased mutation frequency present under biofilm growth conditions. J Bacteriol. 2008;190:6330–6339. doi: 10.1128/JB.00707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraquet C, Murakami K, Parsek MR, Harwood CS. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucl Acids Res. 2012;40:7207–7218. doi: 10.1093/nar/gks384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu Roy A, Petrova OE, Sauer K. Extraction and quantification of cyclic di-GMP from Pseudomonas aeruginosa. bio-protocol. 2013 doi: 10.21769/bioprotoc.828. http://www.bio-protocol.org/wenzhang.aspx?id=828. [DOI] [PMC free article] [PubMed]
- Becher A, Schweizer HP. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques. 2000;29:948–952. doi: 10.2144/00295bm04. [DOI] [PubMed] [Google Scholar]
- Bobrov AG, Kirillina O, Perry PD. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol Lett. 2005;247:123–130. doi: 10.1016/j.femsle.2005.04.036. [DOI] [PubMed] [Google Scholar]
- Brencic A, Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol. 2009;72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol. 2009;73:434–445. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JR, Sauer K. The MerR-like regulator BrlR impairs Pseudomonas aeruginosa biofilm tolerance to colistin by repressing PhoPQ. Journal of Bacteriology. 2013;195:4678–4688. doi: 10.1128/JB.00834-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JR, Liao J, Schurr MJ, Sauer K. BrlR from Pseudomonas aeruginosa is a c-di-GMP-responsive transcription factor. Mol Microbiol. 2014;92:471–487. doi: 10.1111/mmi.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GCL, Parsek MR. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011;7:e1001264. doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Cheng KJ, Geesey GG, Ladd T, Nickel JC, Dasgupta M, Marrie JT. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- El-Azizi M, Rao S, Kanchanapoom T, Khardori N. In vitro activity of vancomycin, quinupristin/dalfopristin, and linezolid against intact and disrupted biofilms of staphylococci. Ann Clin Microbiol Antimicrob. 2005;4:2. doi: 10.1186/1476-0711-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geesey GG, Richardson WT, Yeomans HG, Irvin RT, Costerton JW. Microscopic examination of natural sessile bacterial populations from an alpine stream. Can J Microbiol. 1977;23:1733–1736. doi: 10.1139/m77-249. [DOI] [PubMed] [Google Scholar]
- Gilbert P, Maira-Litran T, McBain AJ, Rickard AH, Whyte FW. The physiology and collective recalcitrance of microbial biofilm communities. Adv Microb Physiol. 2002;46:202–256. [PubMed] [Google Scholar]
- Goodman AL. Sit and Stay a While: How BfiSR Controls Irreversible Attachment in Pseudomonas aeruginosa Biofilms. J Bacteriol. 2010;192:5273–5274. doi: 10.1128/JB.00888-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K, Marques CNH, Petrova OE, Sauer K. Antimicrobial tolerance of Pseudomonas aeruginosa biofilms is activated during an early developmental stage and requires the two-component hybrid SagS. J Bacteriol. 2013;195:4975–4987. doi: 10.1128/JB.00732-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K, Liao J, Petrova OE, Cherny KE, Sauer K. Elevated levels of the second messenger c-di-GMP contribute to antimicrobial resistance of Pseudomonas aeruginosa. Mol Microbiol. 2014;92:488–506. doi: 10.1111/mmi.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JL, Chen HC, Peng HL, Chang HY. Characterization of the histidine-containing phosphotransfer protein B-mediated multistep phosphorelay system in Pseudomonas aeruginosa PAO1. J Biol Chem. 2008;283:9933–9944. doi: 10.1074/jbc.M708836200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Thoendel M, Olakanmi O, Britigan BE, Singh PK. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J Clin Invest. 2007;117:877–888. doi: 10.1172/JCI30783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay E, Humair B, Denervaud V, Riedel K, Spahr S, Eberl L, et al. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J Bacteriol. 2006;188:6026–6033. doi: 10.1128/JB.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- Khan W, Bernier SP, Kuchma SL, Hammond JH, Hasan F, O'Toole GA. Aminoglycoside resistance of Pseudomonas aeruginosa biofilms modulated by extracellular polysaccharide. Int Microbiol. 2010;13:207–212. doi: 10.2436/20.1501.01.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirillina O, Fetherston JD, Bobrov AG, Abney J, Perry RD. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol Microbiol. 2004;54:75–88. doi: 10.1111/j.1365-2958.2004.04253.x. [DOI] [PubMed] [Google Scholar]
- Krüger R, Wolschin F, Weckwerth W, Bettmer J, Lehmann WD. Plant protein phosphorylation monitored by capillary liquid chromatography--element mass spectrometry. Biochem Biophys Res Commun. 2007;355:89–96. doi: 10.1016/j.bbrc.2007.01.108. [DOI] [PubMed] [Google Scholar]
- Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O'Toole GA. BifA, a c-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Riddle of Biofilm Resistance. Antimicrob Agents Chemother. 2001;45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol. 2008;322:107–131. doi: 10.1007/978-3-540-75418-3_6. [DOI] [PubMed] [Google Scholar]
- Li Y, Heine S, Entian M, Sauer K, Frankenberg-Dinkel N. NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by a MHYT-domain coupled phosphodiesterase. J Bacteriol. 2013;195:3531–3542. doi: 10.1128/JB.01156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Sauer K. The MerR-like transcriptional regulator BrlR contributes to Pseudomonas aeruginosa biofilm tolerance. J Bacteriol. 2012;194:4823–4836. doi: 10.1128/JB.00765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Schurr MJ, Sauer K. The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug-efflux pumps in Pseudomonas aeruginosa biofilms. J Bacteriol. 2013;195:3352–3363. doi: 10.1128/JB.00318-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CT, Huang YJ, Chu PH, Hsu JL, Huang CH, Peng HL. Identification of an HptB-mediated multi-step phosphorelay in Pseudomonas aeruginosa PAO1. Res Microbiol. 2006;157:169–175. doi: 10.1016/j.resmic.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Mah T-F, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- Mah TF, O'Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- Miller J. Experiments in molecular genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. pp. 352–355. [Google Scholar]
- Monzon M, Oteiza C, Leiva J, Amorena B. Synergy of different antibiotic combinations in biofilms of Staphylococcus epidermidis. J Antimicrob Chemother. 2001;48:793–801. doi: 10.1093/jac/48.6.793. [DOI] [PubMed] [Google Scholar]
- Morgan R, Kohn S, Hwang S-H, Hassett DJ, Sauer K. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol. 2006;188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty TF, Elborn JS, Tunney MM. Effect of pH on the antimicrobial susceptibility of planktonic and biofilm-grown clinical Pseudomonas aeruginosa isolates. Br J Biomed Sci. 2007;64:101–104. doi: 10.1080/09674845.2007.11732766. [DOI] [PubMed] [Google Scholar]
- Newman JR, Fuqua C. Broad-host-range expression vectors that carry the arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene. 1999;227:197–203. doi: 10.1016/s0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited Bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson GL. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 2009;5:e1000668. doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J Bacteriol. 2010;192:5275–5288. doi: 10.1128/JB.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation. J Bacteriol. 2011;193:6614–6628. doi: 10.1128/JB.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Schurr JR, Schurr MJ, Sauer K. The novel Pseudomonas aeruginosa two-component regulator BfmR controls bacteriophage-mediated lysis and DNA release during biofilm development through PhdA. Mol Microbiol. 2011;81:767–783. doi: 10.1111/j.1365-2958.2011.07733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Schurr JR, Schurr MJ, Sauer K. Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol Microbiol. 2012;86:819–835. doi: 10.1111/mmi.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband W. ImageJ. US National Institutes of Health; Bethesda, MD: 1997. [Google Scholar]
- Sauer K, Camper AK. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J Bacteriol. 2001;183:6579–6589. doi: 10.1128/JB.183.22.6579-6589.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Cullen MC, Rickard AH, Zeef LAH, Davies DG, Gilbert P. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol. 2004;186:7312–7326. doi: 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotech. 1983;1:784–791. [Google Scholar]
- Southey-Pillig CJ, Davies DG, Sauer K. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J Bacteriol. 2005;187:8114–8126. doi: 10.1128/JB.187.23.8114-8126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoering AL, Lewis K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol. 2001;183:6746–6751. doi: 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. The Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- Villain-Guillot P, Gualtieri M, Bastide L, Leonetti J-P. In vitro activities of different inhibitors of bacterial transcription against Staphylococcus epidermidis biofilm. Antimicrob Agents Chemother. 2007;51:3117–3121. doi: 10.1128/AAC.00343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolschin F, Wienkoop S, Weckwerth W. Enrichment of phosphorylated proteins and peptides from complex mixtures using metal oxide/hydroxide affinity chromatography (MOAC) PROTEOMICS. 2005;5:4389–4397. doi: 10.1002/pmic.200402049. [DOI] [PubMed] [Google Scholar]
- Xu L, Venkataramani P, Ding Y, Liu Y, Deng Y, Yong GL, et al. A cyclic di-GMP-binding adaptor protein interacts with histidine kinase to regulate two-component signaling. Journal of Biological Chemistry: jbc. 2016;M116:730887. doi: 10.1074/jbc.M116.730887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda H, Ajiki Y, Aoyama J, Yokota T. Interaction between human polymorphonuclear leucocytes and bacteria released from in-vitro bacterial biofilm models. J Med Microbiol. 1994;41:359–367. doi: 10.1099/00222615-41-5-359. [DOI] [PubMed] [Google Scholar]
- Zobell CE, Anderson DQ. Observations on the multiplication of bacteria in different columes of stored sea water and the influence of oxygen tenion and solid surfaces. Biol Bull. 1936;71:324–342. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Detection of SagS and SagS domain constructs in cell free total cell extracts by immunoblot analysis using anti-HA antibodies. All SagS domain constructs, including the full-length gene, harbored a C-terminal hemagglutinin (HA) tag (referred to as SagS*-HA). *, SagS domain constructs vary in molecular mass.
Figure S2. Average number of viable cells, expressed as CFU, present in biofilms formed by P. aeruginosa PAO1 and mutant strains following 2 days of growth at a surface area of 25 cm2 at a flow rate of 0.1 ml/min. Experiments were repeated at least 3 times. Error bars indicate standard deviations.
Figure S3. Average and maximum thickness of P. aeruginosa wild-type and ΔsagS mutant biofilms expressing (A) SagS domain constructs or (B) SagS variants harboring site-directed mutations, as determined using confocal images and subsequent COMSTAT analysis. (C) Biofilm biomass of ΔsagS mutant strains expressing wild-type sagS or sagS variants, as determined using confocal images and subsequent COMSTAT analysis. Biofilms were grown for 6 days in flow cells. All experiments were carried out at least in triplicate. Error bars indicate standard deviation.
Figure S4. Relative BfiS abundance, based on relative intensity of protein bands detectable following probing for BfiS with anti-V5 antibodies and subsequent analysis using ImageJ (Rasband, 1997). (A) BfiS protein levels detected following Co-IP. (B) BfiS protein levels following enrichment by MOAC. Experiments were carried out in triplicate. Error bars denote standard deviation. *, significantly different from the values for P. aeruginosa PAO1 (p ≤ 0.01), as determined by ANOVA and SigmaStat.
Figure S5. c-di-GMP levels present in wild-type and ΔsagS mutant biofilm cells overexpressing sagS domain constructs. The strains were grown in tube reactors under flowing conditions for 6 days prior to c-di-GMP extraction and quantitation by HPLC analysis. pmol/mg refers to c-di-GMP levels (pmol) per total cell protein (in mg). ΔsagS harboring the empty vector pJN105 was used as vector control. All experiments were carried out at least in triplicate. Error bars indicate standard deviation.
Figure S6. Total biofilm biomass (A), and average and maximum thickness (B) of P. aeruginosa ΔsagS and ΔsagSΔgacA mutant biofilms expressing intact sagS or the sagS-Rec domain construct, as determined using confocal images and subsequent COMSTAT analysis. Biofilms were grown for 6 days in flow cells. All experiments were carried out at least in triplicate. Error bars indicate standard deviation.
Figure S7. Localization of transmembrane helices (A) present in SagS as determined using the TMHMM Prediction of transmembrane helices in proteins software (http://www.cbs.dtu.dk/services/TMHMM-2.0/). (B) Alignment of HmsP domains of P. aeruginosa SagS and BifA and Yersinia pestis HmsP (Yp_HmsP). Sec, predicted secondary structure of Yp_HmsP; H, helical regions; e, extended regions; M, membrane spanning regions, from www.blackwellpublishing.com/products/journals/.../mmi4253FigS1.doc. Identical/similar AA are shown in color and indicated by symbols (., :, *).
Figure S8. Contribution of SagS and the HmsP domain to the susceptibility of planktonic cells to tobramycin. All strains were grown planktonically to exponential phase and subsequently exposed to 50 µg/ml tobramycin for 30 min. Strain PAO1 harboring the empty plasmid pMJT1 was used as vector control. Viability was determined via CFU counts. (A) Total CFU prior to treatment. (B) Susceptibility is expressed as log reduction in CFU counts. Experiments were done at least in triplicate. Error bars denote standard deviation.