Abstract
In many Lactobacillales species (i.e. lactic acid bacteria), peptidoglycan is decorated by polyrhamnose polysaccharides that are critical for cell envelope integrity and cell shape and also represent key antigenic determinants. Despite the biological importance of these polysaccharides, their biosynthetic pathways have received limited attention. The important human pathogen, Streptococcus pyogenes, synthesizes a key antigenic surface polymer, the Lancefield group A carbohydrate (GAC). GAC is covalently attached to peptidoglycan and consists of a polyrhamnose polymer, with N-acetylglucosamine (GlcNAc) side chains, which is an essential virulence determinant. The molecular details of the mechanism of polyrhamnose modification with GlcNAc are currently unknown. In this report, using molecular genetics, analytical chemistry, and mass spectrometry analysis, we demonstrated that GAC biosynthesis requires two distinct undecaprenol-linked GlcNAc-lipid intermediates: GlcNAc-pyrophosphoryl-undecaprenol (GlcNAc-P-P-Und) produced by the GlcNAc-phosphate transferase GacO and GlcNAc-phosphate-undecaprenol (GlcNAc-P-Und) produced by the glycosyltransferase GacI. Further investigations revealed that the GAC polyrhamnose backbone is assembled on GlcNAc-P-P-Und. Our results also suggested that a GT-C glycosyltransferase, GacL, transfers GlcNAc from GlcNAc-P-Und to polyrhamnose. Moreover, GacJ, a small membrane-associated protein, formed a complex with GacI and significantly stimulated its catalytic activity. Of note, we observed that GacI homologs perform a similar function in Streptococcus agalactiae and Enterococcus faecalis. In conclusion, the elucidation of GAC biosynthesis in S. pyogenes reported here enhances our understanding of how other Gram-positive bacteria produce essential components of their cell wall.
Keywords: carbohydrate biosynthesis, cell wall, glycosyltransferase, polysaccharide, Streptococcus pyogenes (S. pyogenes), lipid intermediate
Introduction
The cytoplasmic membrane of Gram-positive bacteria is surrounded by a thick cell wall consisting of multiple peptidoglycan layers decorated with proteins and a variety of carbohydrate-based polymers. Rhamnose (Rha) is the main component of the carbohydrate structures in many species of the Lactobacillales order (1). These polymers play essential roles in maintaining and protecting bacterial cell envelopes and in pathogenesis of infections caused by Streptococcus pyogenes and Enterococcus faecalis (1). It is of paramount importance to understand the molecular mechanisms of Rha-containing cell wall polysaccharide biosynthesis for developing novel therapeutics against these bacterial pathogens.
S. pyogenes or group A Streptococcus (GAS)3 is associated with numerous diseases in humans ranging from minor skin and throat infections, such as impetigo and pharyngitis, to life-threatening invasive infections, such as streptococcal toxic syndrome and necrotizing fasciitis (2). The main component of GAS cell wall is the Lancefield group A carbohydrate (GAC) that includes about 40–60% of the total cell wall mass (3). Serological grouping of β-hemolytic streptococci (A, B, C, E, F, and G groups), introduced by Rebecca Lancefield in 1933, is based on the detection of carbohydrate antigens present on the cell wall (4). GAC is presumably covalently linked to N-acetylmuramic acid (MurNAc) of peptidoglycan, although the molecular structure of the linkage unit has not been established (5). All serotypes of GAS produce GAC consisting of a polyrhamnose backbone with N-acetylglucosamine (GlcNAc) side chains (6, 7). The Lancefield classification of GAS relies on serological specificity of the immunodominant GlcNAc residues in the carbohydrate antigen (7). The average molecular mass of GAC has been reported to be 8.9 ± 1.0 kDa, corresponding to an average of 18 repeating units of [→3)α-Rha(1→2)[β-GlcNAc(1→3)]α-Rha(1→] (8). Because Rha is absent in mammalian cells, GAC is an attractive candidate for a universal GAS vaccine (8, 9). Moreover, the GlcNAc side chains are an important virulence determinant in GAS (5). GAS mutants lacking GlcNAc are susceptible to innate immune clearance by neutrophils and antimicrobial agents and are significantly attenuated in animal models of GAS infection (5).
Similar to the biosynthesis of other cell-envelope polymers, peptidoglycan, lipopolysaccharide, wall teichoic acid (WTA), and capsular polysaccharide, the synthesis of Rha-containing carbohydrate structures is likely initiated on the inside of the cytoplasmic membrane and proceeds through several steps, including the attachment of the first sugar residue to a lipid carrier, undecaprenyl phosphate (Und-P), followed by elongation of the polysaccharide through stepwise addition of activated sugar residues to the lipid carrier (1). After translocation of the polysaccharide across the membrane and further modifications, it is presumably attached to peptidoglycan via a phosphate ester linkage (1). The first membrane step of O-antigen biosynthesis in enterobacteria and WTA biosynthesis in Bacillus subtilis and Staphylococcus aureus (10) is catalyzed by the UDP-GlcNAc:Und-P GlcNAc-1-phosphate transferase encoded by WecA homologs (11, 12). WecA transfers GlcNAc-phosphate from UDP-GlcNAc to Und-P, forming GlcNAc-pyrophosphoryl-undecaprenol (GlcNAc-P-P-Und). Although GAS strains do not produce WTA, all GAS genomes contain a wecA homolog, gacO (5). Significantly, GAC biosynthesis is sensitive to tunicamycin, a known inhibitor of WecA (5), and the Streptococcus mutans GacO homolog, RgpG, has been shown to complement WecA activity in E. coli (13). These observations support an essential role for GacO in the biosynthesis of GlcNAc-P-P-Und and suggest GlcNAc-P-P-Und may function as a lipid–anchor in the initiation of GAC biosynthesis.
Other genes required for GAC biosynthesis and transport are found in a separate location on the GAS chromosome and include a 12-gene locus (gacA–gacL) (Fig. 1A). The first three genes, gacA–gacC together with gacG, are conserved in many species of the Lactobacillales order (1). gacA encodes a dTDP-4-dehydrorhamnose reductase, the enzyme responsible for dTDP-rhamnose biosynthesis (14). gacB, gacC, and gacG encode putative cytoplasmic rhamnosyltransferases. In S. mutans homologous genes rgpA, rgpB, and rgpF are involved in rhamnan backbone biosynthesis (15). In GAS, gacA–gacC are essential for viability and cannot be deleted (5). However, deletion mutants have been obtained in other genes of the GAC gene cluster (5). It has been shown that gacI, gacJ, and gacK are non-essential for viability but are required for GlcNAc side-chain addition to the polyrhamnose (5). They encode a putative cytoplasmic glycosyltransferase, a small-membrane protein, and a protein with homology to the Wzx family of membrane proteins involved in the export of O-antigen and teichoic acids, respectively. In contrast, inactivation of gacD, gacE, gacF, gacG, gacH, or gacL has no effect on GAS viability, and the GAC produced by these mutants is reported to display a wild-type (WT) antigenic profile, indicating the presence of the immunodominant GlcNAc side chains (5). GacD and gacE encode the components of an ABC transport system. In S. mutans, the gacD and gacE homologs are responsible for rhamnan polysaccharide transport (15). gacF, gacH, and gacL encode a cytosolic glycosyltransferase, a putative membrane-associated glycerol phosphate transferase, and a membrane-associated glycosyltransferase, respectively.
Figure 1.
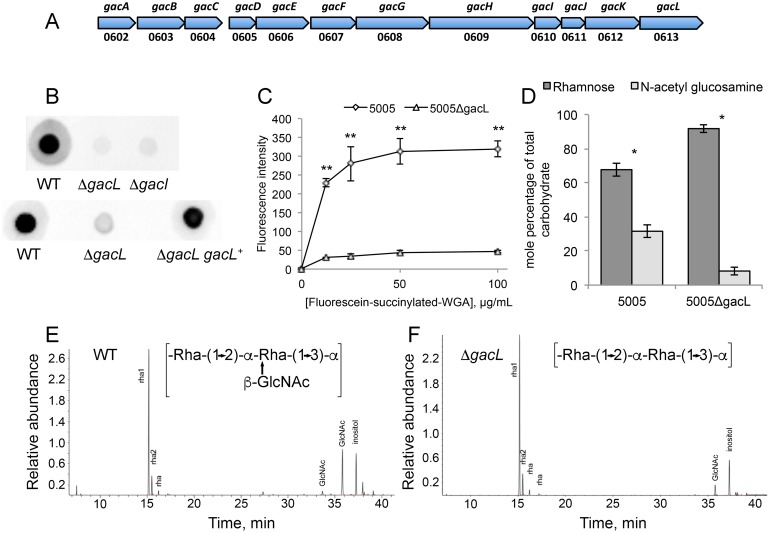
Map of S. pyogenes genes involved in GAC biosynthesis and analysis of 5005ΔgacI and 5005ΔgacL deletion mutants. A, GAC biosynthesis gene cluster. Numbers below represent MGAS5005 gene designations. B, representative immunoblot analysis of cell-wall fractions isolated from MGAS5005, 5005ΔgacI, 5005ΔgacL, and 5005ΔgacL gacL+. Data are representative of biological triplicates. C, binding of N-acetylglucosamine-specific fluorescein-succinylated WGA to whole MGAS5005 and 5005ΔgacL was measured. Data are the average of three replicates ± S.D. D, rhamnose and GlcNAc mole percentage of total carbohydrate was determined by GC-MS for cell wall material isolated from MGAS5005 and 5005ΔgacL following methanolysis as described under “Experimental procedures.” Data are the average of four replicates ± S.D. The asterisks indicate statistically different values (*, p < 0.05; **, p < 0.01) as determined by the Student's t test. E and F, GC-MS chromatograms for glycosyl composition analysis of cell wall isolated MGAS5005 and 5005ΔgacL. The deduced schematic structure of the repeating unit of GAC is shown for each strain. The chromatograms are representative of four separate analyses performed on two different cell wall preparations.
In this study we address the molecular mechanism of GlcNAc attachment to polyrhamnose. We demonstrate that GacI catalyzes formation of GlcNAc-P-Und, and GacJ stimulates the catalytic activity of GacI. Subsequently, GacL transfers GlcNAc from GlcNAc-P-Und to the polyrhamnose backbone of GAS polysaccharide. Moreover, we confirm GacO function in GlcNAc-P-P-Und formation and demonstrate the role of this GlcNAc-lipid in initiation of polyrhamnose biosynthesis.
Results
GacL is required for GlcNAc attachment to polyrhamnose
GacL, a polytopic (12–13 transmembrane segments) membrane protein, is reported to be dispensable for GlcNAc attachment to polyrhamnose (5). To investigate the function of GacL in GAC biogenesis, we disrupted gacL in the hyperinvasive S. pyogenes M1T1 serotype strain, MGAS5005 (16), creating the 5005ΔgacL mutant. In agreement with published data (5), 5005ΔgacL did not display any detectable growth phenotype in comparison with the WT strain. Surprisingly, we found that 5005ΔgacL cells failed to bind GlcNAc-specific anti-GAC antibodies, suggesting a loss of the GlcNAc antigenic epitope (Fig. 1B). The 5005ΔgacL phenotype was restored by expressing the WT copy of gacL on the mutant chromosome (Fig. 1B). To confirm that 5005ΔgacL cells are deficient in GlcNAc addition to polysaccharide, we measured the binding of fluorescently labeled succinylated wheat germ agglutinin (sWGA), a lectin that specifically binds non-reducing terminal β-GlcNAc residues, to WT and 5005ΔgacL cells. Deletion of gacL led to a significant decrease in binding of WGA to the 5005ΔgacL strain as compared with the WT (Fig. 1C), indicating that the mutant has substantially less GlcNAc-containing saccharides on the cell surface. Furthermore, direct compositional analysis of cell wall purified from WT and 5005ΔgacL cells confirmed a significant decrease of GlcNAc content in the 5005ΔgacL cell wall sample (Fig. 1, D–F). These data strongly support a role for GacL in GlcNAc side-chain attachment to polyrhamnose.
gacL deletion mutant accumulates GlcNAc-P-Und
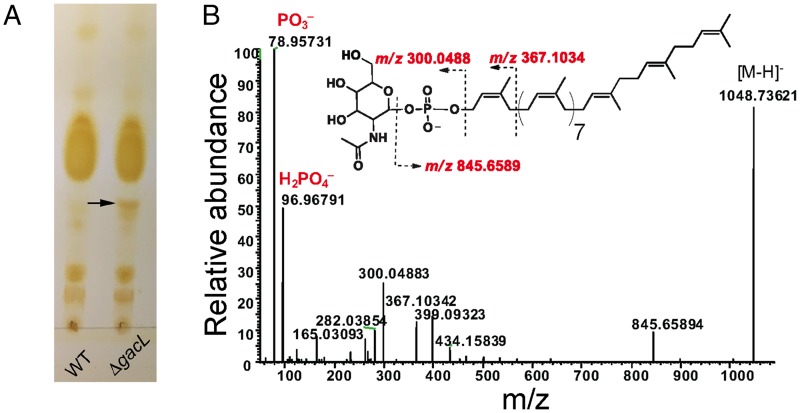
To investigate further the effect of GacL deletion in S. pyogenes, we isolated the phospholipid fractions from the membranes of WT and 5005ΔgacL cells. TLC analysis revealed accumulation of a previously unidentified phosphoglycolipid in 5005ΔgacL (Fig. 2A). The novel lipid was found to be stable to mild alkaline methanolysis, but sensitive to mild acid (0.1 n HCl, 50 °C, 50% isopropyl alcohol), and it reacted with orcinol spray (17), Dittmer-Lester phospholipid spray reagent (18), and with anisaldehyde, an isoprenol-specific reagent (19), consistent with its tentative identification as a glycophosphoprenol (data not shown). High-resolution negative-ion ESI-MS analysis of the purified lipid identified a molecular ion [M − H]−1 of m/z = 1048.73621 (Fig. 2B). In addition, prominent high-resolution fragment ions characteristic of PO3, H2PO4, N-acetylhexosamine-phosphate, and Und-P were also found (Table 1). These data are consistent with a glycolipid composed of N-acetylhexosamine-phosphate-undecaprenol. In experiments described in detail below, we found that when membrane fractions from GAS are incubated with UDP-[3H]GlcNAc, a [3H]GlcNAc-lipid that co-chromatographs on TLC with this novel lipid is rapidly formed. Importantly, the product ion spectra shown in Fig. 2B does not contain a fragment ion containing [Und-PO4-C2H2NHCOCH3]− (m/z = 929.7), arising from a cross-ring fragmentation reaction, suggesting that the hydroxyl of the anomeric carbon and the nitrogen at the 2-position of the glycosyl ring are trans to the plane of the ring (20, 21). This observation strongly suggests that the anomeric hydroxyl is present in the β configuration. Furthermore, isolated [3H]GlcNAc-P-Und phospholipid (described in detail below) is extremely sensitive to incubation with 50% phenol at 68 °C, a property that is consistent with β-GlcNAc-P-Und (supplemental Table S1) (22). Taken together, these data identify the novel glycolipid accumulating in the 5005ΔgacL strain as β-GlcNAc-P-Und and suggest that GacL is a GlcNAc transferase using β-GlcNAc-P-Und as GlcNAc donor for the addition of the GlcNAc side chains to GAC.
Figure 2.
Purification and identification of GlcNAc-phosphate-undecaprenol in 5005ΔgacL. A, TLC analysis of phospholipids isolated from MGAS5005 and 5005ΔgacL. Phospholipids extracted from bacterial strains were separated by TLC on Silica Gel G in CHCl3/CH3OH/H2O/NH4OH (65:25:4:1). Position of the novel, alkaline-resistant, and acid-labile phospholipid accumulating in 5005ΔgacL is indicated by the arrow. The results are representative of three separate experiments. B, ESI-MS/MS analysis of the novel phospholipid isolated from 5005ΔgacL. The spectrum is assigned to GlcNAc-phosphate-undecaprenol.
Table 1.
The major fragments of the precursor ion at m/z 1048
Fragment ions originating from the precursor molecular ion of 1048 were subjected to MS/MS fragmentation as described under “Experimental procedures.” The proposed molecular formulae of the product ions are supported by the observed isotope patterns for the relevant ions.
| m/z | Formula | Mass error |
|---|---|---|
| ppm | ||
| 78.95731 | PO3 | −8.19 |
| 96.96791 | H2PO4 | −6.31 |
| 300.04883 | C8H15O9NP | 3.12 |
| 845.65894 | C58H88ONP | −1.02 |
GAS synthesizes two GlcNAc-lipids in vitro
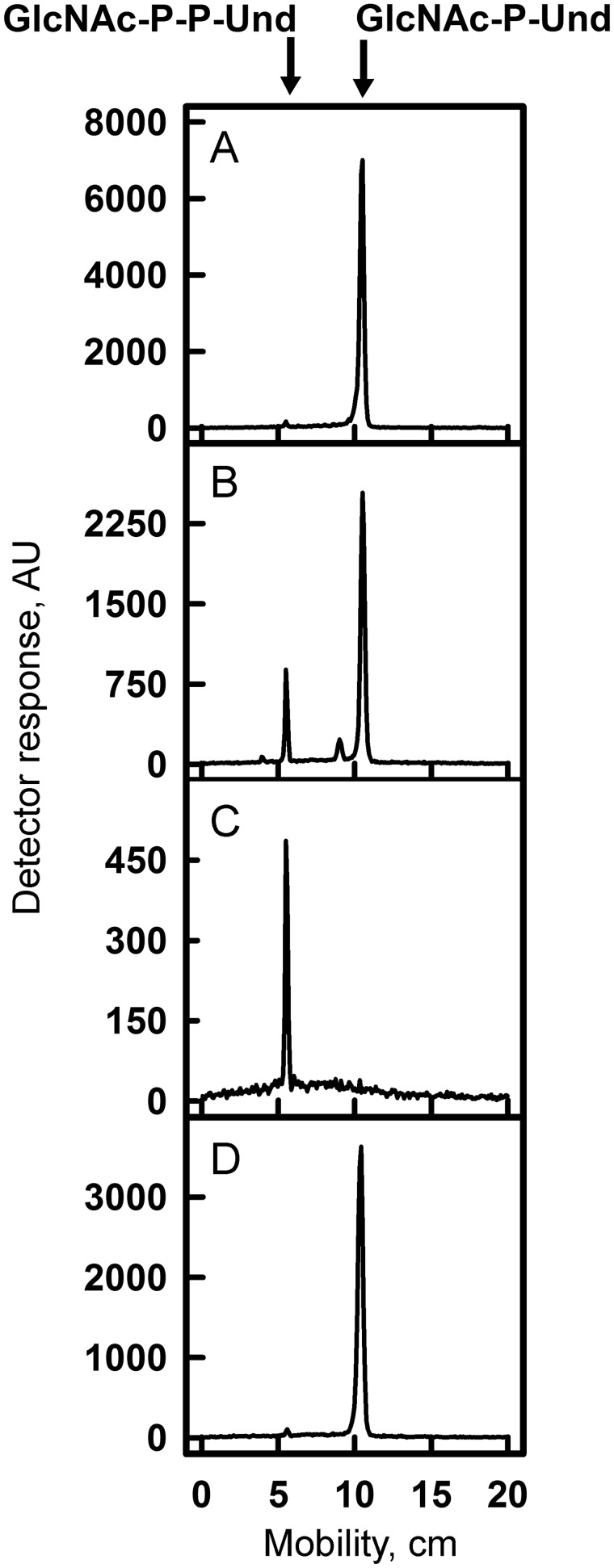
To investigate the biosynthetic origin of the novel glycolipid accumulating in 5005ΔgacL cells, membrane fractions from MGAS5005 were incubated with UDP-[3H]GlcNAc and analyzed for lipid products as described under “Experimental procedures.” Experiments showed that [3H]GlcNAc was efficiently transferred from UDP-[3H]GlcNAc into two detectable GlcNAc-lipids (Fig. 3A and Table 2). The major lipid product co-migrates on TLC with the novel lipid accumulating in the 5005ΔgacL strain and is identified as β-[3H]GlcNAc-P-Und, as described above. The minor product is assumed to be [3H]GlcNAc-P-P-Und, because it co-migrates with authentic GlcNAc-P-P-Und synthesized in Bacillus cereus membranes (Fig. 3B) (23), and its formation is potently inhibited by tunicamycin (supplemental Fig. S1).
Figure 3.
TLC of [3H]GlcNAc-lipids from in vitro incubations of GAS mutants and B. cereus membranes with UDP-[3H]GlcNAc. Membrane fractions from MGAS5005 (A), B. cereus (B), 5005ΔgacI (C), or 5005ΔgacL (D) were incubated with UDP-[3H]GlcNAc and analyzed for [3H]GlcNAc-lipid synthesis by TLC. Reaction mixtures contained 50 mm Tris-Cl, pH 7.4, 5 mm 2-mercaptoethanol, 20 mm MgCl2, 1 mm ATP, 5 μm UDP-[3H]GlcNAc (486 cpm/pmol), and bacterial membrane suspension (100–200 μg of membrane protein) in a total volume of 0.02 ml. Following a 10-min pre-incubation at 30 °C, GlcNAc-lipid synthesis was initiated by the addition of UDP-[3H]GlcNAc. After 10 min, reactions were processed for GlcNAc-lipid synthesis as described under “Experimental procedures.” The organic layers were dried and dissolved in a small volume of CHCl3/CH3OH (2:1), and a portion was removed and assayed for radioactivity by liquid scintillation spectrometry. The remainder was spotted on 10 × 20-cm plate of Silica Gel G and developed in CHCl3/CH3OH/NH4OH/H2O (65:25:1:4). [3H]GlcNAc-lipids were detected by scanning with an AR2000 Bioscan Radiochromatoscanner. AU, arbitrary units. The results are representative of three separate experiments.
Table 2.
GlcNAc-lipid synthesis in membrane fractions from various bacterial strains
Reaction mixtures contained 50 mm Tris-HCl, pH 7.4, 5 mm 2-mercaptoethanol, 20 mm MgCl2, 1 mm ATP, 5 μm UDP-[3H]GlcNAc (486 cpm/pmol), and bacterial membrane suspension (50–100 μg of membrane protein) in a total volume of 0.01 ml. Following a 10-min preincubation at 30 °C, GlcNAc-lipid synthesis was initiated by the addition of UDP-[3H]GlcNAc. After 5 min, reactions were processed for GlcNAc-lipid synthesis as described under “Experimental procedures.” The results are representative of at least three separate experiments.
| Enzyme source | GlcNAc-P-Und | GlcNAc-P-P-Und |
|---|---|---|
| pmol/mg | pmol/mg | |
| MGAS5005 | 259.4 ± 8.2 | 7.8 ± 1.8 |
| 5005ΔgacI | Not detected | 15.9 ± 2.5 |
| GBS COH1 | 62 ± 0.4 | 5.6 ± 0.8 |
| GBSΔgacI | Not detected | 12 ± 2 |
| E. coli Rosetta (DE3) | Not detected | 19.5 ± 0.3 |
| E. coli:GacI | 102.8 ± 3.3 | 5.4 ± 0.05 |
| E. coli:EpaI | 189 ± 3.9 | 7 ± 1.5 |
GacI encodes a UDP-GlcNAc:Und-P GlcNAc transferase activity
The GAS polysaccharide gene cluster contains a gene, gacI, reported to be required for GlcNAc addition to cell wall polysaccharide (5) and annotated as a GT-A type glycosyltransferase. The following studies were conducted to determine whether GAS GacI might be responsible for the synthesis of GlcNAc-P-Und. To determine whether GacI function was essential for formation of GlcNAc-P-Und, we generated a deletion of gacI in the MGAS5005. In agreement with published data (5) 5005ΔgacI lost reactivity with anti-GAC antibodies, with specificity for the immunodominant GlcNAc, indicating a loss of GlcNAc modification in polyrhamnose (Fig. 1B). When 5005ΔgacI membrane fractions were incubated with [3H]UDP-GlcNAc, no incorporation into [3H]GlcNAc-P-Und was observed, and only the minor lipid [3H]GlcNAc-P-P-Und was found (Fig. 3C, Table 2), strongly supporting an essential role for GacI in GlcNAc-P-Und synthesis. The observation that the 5005ΔgacL strain synthesizes normal levels of [3H]GlcNAc-P-Und in vitro (Fig. 3D) indicates that 5005ΔgacL does not fail to add GlcNAc side chains to polyrhamnose due to a lack of GlcNA-P-Und and supports the conclusion that GacL may be the GlcNAc-P-Und:polyrhamnan GlcNAc transferase.
To confirm the role of GacI in GlcNAc-P-Und biosynthesis, we expressed GAS GacI in Escherichia coli. When membrane fractions from E. coli carrying an empty vector were incubated with UDP-[3H]GlcNAc and analyzed by silica gel TLC, a small peak of [3H]GlcNAc-P-P-Und was found (Table 2 and supplemental Fig. S2A). In contrast, the membranes of recombinant E. coli expressing GacI accumulated two products corresponding to a small amount of GlcNAc-P-P-Und and a very large amount of GlcNAc-P-Und (Table 2 and supplemental Fig. S2B). Thus, our data strongly indicate that GacI is the GlcNAc-P-Und synthase.
GacI homologs in GBS and E. faecalis function in the biosynthesis of GlcNAc-P-Und
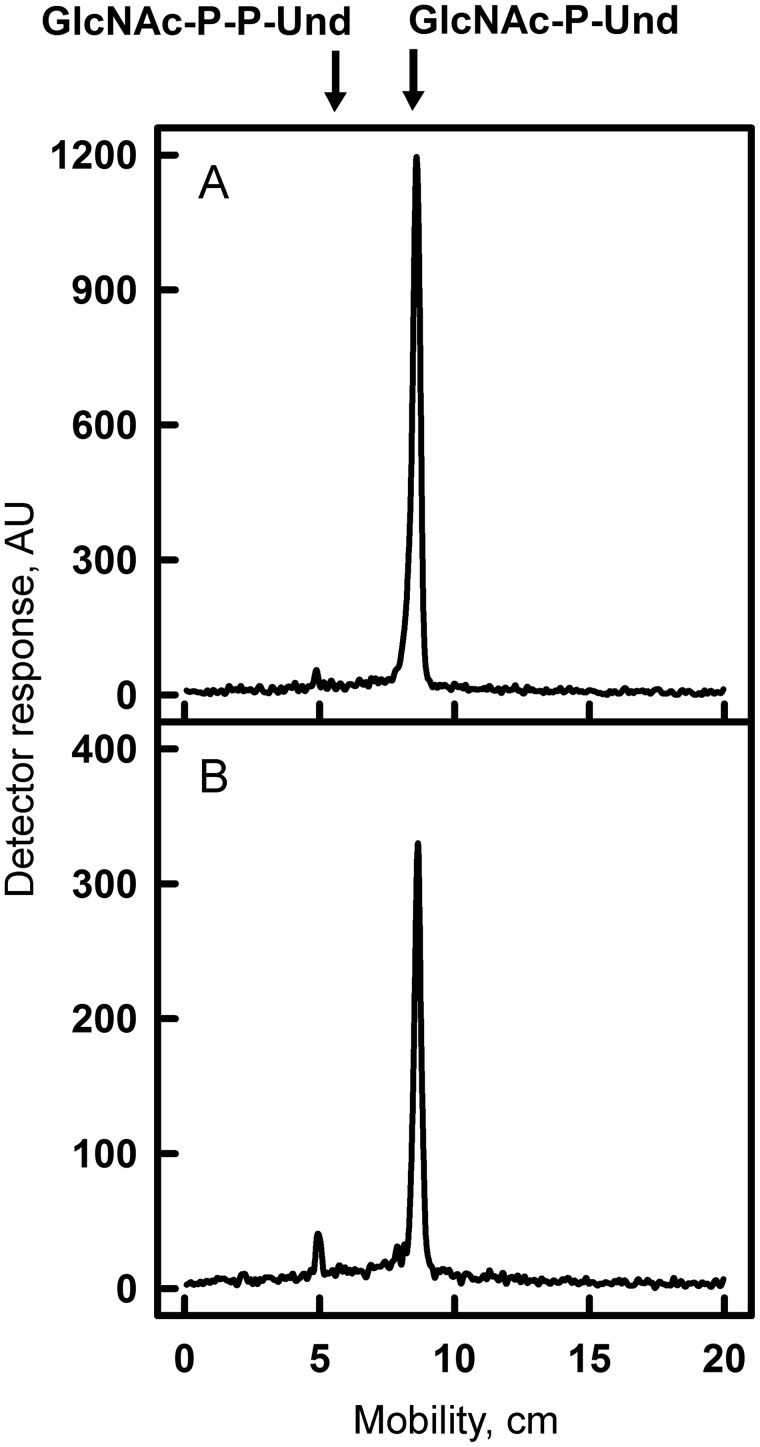
The presence of GlcNAc-P-P-Und and GlcNAc-P-Und has been previously reported in B. cereus and Bacillus megaterium (12, 23). Our in vitro studies show that the MGAS5005 strain synthesizes two [3H]GlcNAc-lipids that exactly co-migrate on TLC with the previously reported [3H]GlcNAc-lipids from B. cereus (Fig. 3, A and B). Moreover, an analysis of bacterial genomes using GacI in a BLAST search identified a GacI homolog (60% identity) in B. cereus and B. megaterium, suggesting its role in GlcNAc-P-Und biosynthesis in these bacteria (supplemental file S1). Furthermore, gacI homologs were identified in the polysaccharide biosynthesis gene clusters of the important human pathogens E. faecalis (epaI with 48% sequence identity) (24) and Streptococcus agalactiae (group B Streptococcus or GBS) (SAN_1536 gene with 46% sequence identity) (supplemental Fig. S3). To investigate whether the GBS homolog of GacI also catalyzes the formation of GlcNAc-P-Und, we engineered a knock out of the GacI homolog in GBS COH1. Although GBS COH1 membranes synthesize both [3H]GlcNAc-P-P-Und and [3H]GlcNAc-P-Und (Fig. 4B and supplemental Fig. S2D), GBS COH1ΔgacI membranes no longer synthesize [3H]GlcNAc-P-Und (Table 2 and supplemental Fig. S2E).
Figure 4.
TLC of [3H]GlcNAc-lipids from in vitro incubations of GAS and GBS membranes with UDP-[3H]GlcNAc. Membrane fractions from WT MGAS5005 (A) or GBS COH1 (B) were incubated with UDP-[3H]GlcNAc and analyzed for [3H]GlcNAc-lipid synthesis by TLC. Reaction mixtures were exactly as described in the legend to Fig. 3. After 5 min of incubation with UDP-[3H]GlcNAc, reactions were processed for GlcNAc-lipid synthesis as described under “Experimental procedures.” The organic layers were dried and dissolved in a small volume of CHCl3/CH3OH (2:1), and a portion was removed and assayed for radioactivity by liquid scintillation spectrometry. The remainder was spotted on 10 × 20-cm plate of Silica Gel G and developed in CHCl3/CH3OH/NH4OH/H2O (65:25:1:4). [3H]GlcNAc-lipids were detected by scanning with an AR2000 Bioscan Radiochromatoscanner. AU, arbitrary units. The results are representative of three separate experiments.
To confirm that the GacI homolog detected in E. faecalis, EpaI (24), also possesses GlcNAc-P-Und synthase activity, EpaI was expressed exogenously in E. coli and found to actively catalyze the formation of [3H]GlcNAc-P-Und (Table 2 and supplemental Fig. S2C). Altogether, our results are consistent with the function of GacI homologs from GAS, GBS, and E. faecalis in the transfer of GlcNAc from UDP-GlcNAc to Und-P forming GlcNAc-P-Und.
ATP stimulates GlcNAc-P-Und biosynthesis
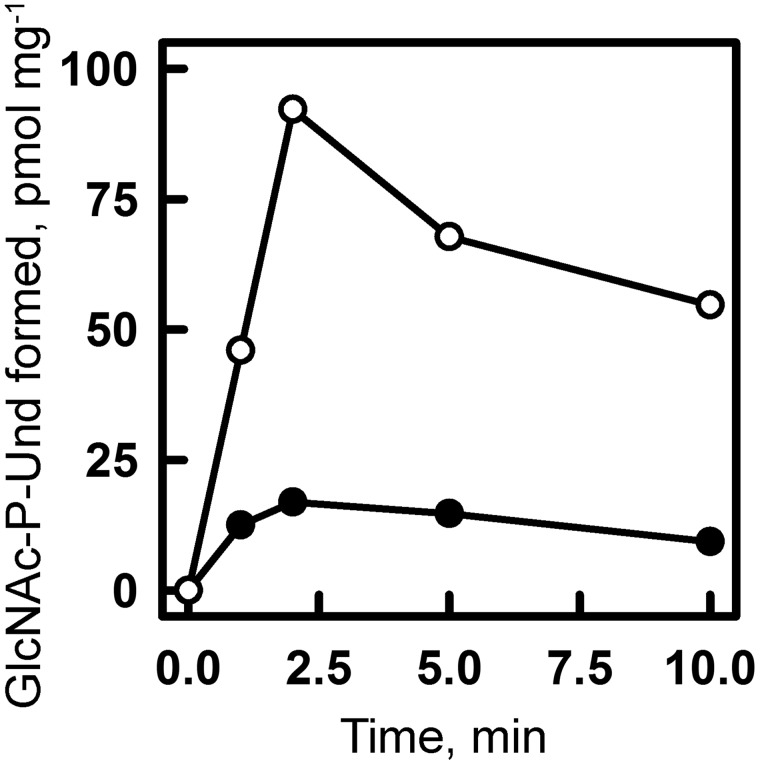
Preliminary enzymatic properties for the synthesis of [3H]GlcNAc-P-Und established that Mg2+ was the preferred divalent cation and that the formation of GlcNAc-P-Und was substantially stimulated by exogenously added Und-P (as a dispersion in CHAPS detergent). In early studies of GAC biosynthesis in GAS (25), it was shown that the addition of ATP dramatically stimulated incorporation of radioactive UDP-[3H]GlcNAc into [3H]GlcNAc membrane lipids and [3H]GlcNAc-polysaccharide. We confirmed that inclusion of 1 mm ATP significantly stimulated the incorporation of [3H]GlcNAc into [3H]GlcNAc-P-Und (Fig. 5 and supplemental Fig. S4), as well as [3H]GlcNAc-P-P-Und (supplemental Fig. S4). However, the inclusion of ATP did not stimulate GacI activity in in vitro assays of CHAPS-soluble, affinity-purified GacI solely dependent on exogenously added Und-P as acceptor. These data suggest that the effect of ATP addition is most likely due to formation of Und-P, in situ, by phosphorylation of endogenous undecaprenol by undecaprenol kinase (26).
Figure 5.
Effect of ATP on GlcNAc-P-Und synthesis in vitro in MGAS5005 membrane fractions. GlcNAc-P-Und synthesis in MGAS5005 membranes was assayed after the indicated time at 30 °C in the presence (○) or absence (●) of 1 mm ATP. Reaction mixtures were identical to those described in Fig. 3 except for the presence of ATP. Following incubation, incorporation into [3H]GlcNAc-P-Und was determined as described under “Experimental procedures.” The results are representative of three separate experiments.
GacJ forms a complex with GacI and enhances its catalytic efficiency
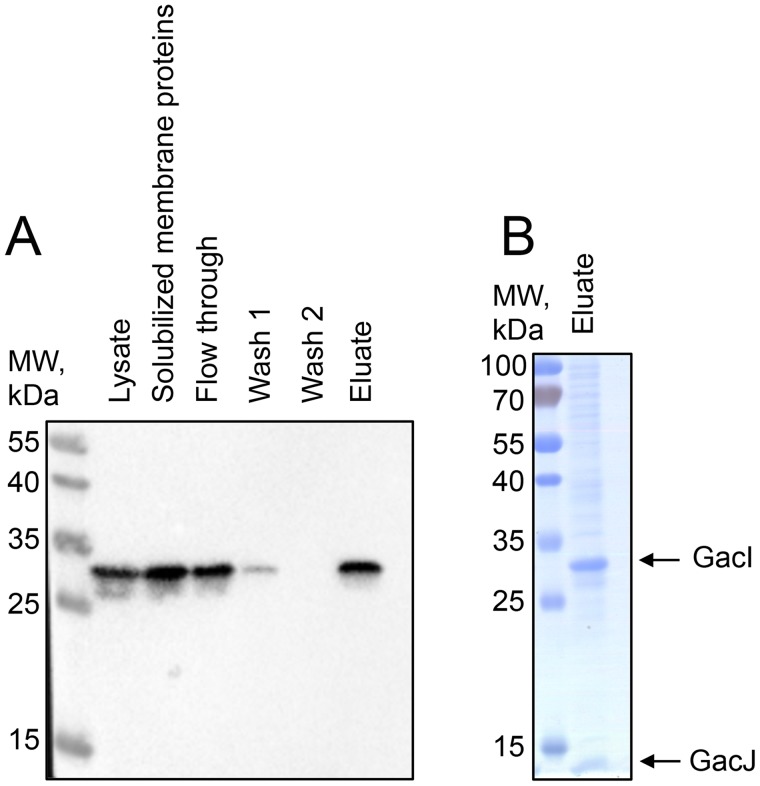
gacJ is located immediately downstream of gacI in the GAS GAC biosynthesis gene cluster and encodes a small, 113-amino acid, membrane protein (Fig. 1A). We hypothesized that GacI might form an obligate complex with GacJ. This hypothesis is based on the observation that gacJ and gacI are frequently located adjacent to each other on bacterial chromosomes and are sometimes fused to form a single polypeptide (accession numbers: GAM11018, ADH85075, ALC15489, and ADU67183). To test this hypothesis, we solubilized membranes from E. coli co-expressing GacJ and N-terminal His tagged GacI with the zwitterionic detergent CHAPS, and we isolated GacI complexes using Ni-NTA chromatography (Fig. 6A). SDS-PAGE of the affinity-purified sample revealed two bands corresponding to the anticipated molecular sizes of GacI and GacJ (Fig. 6B). Proteomics analysis of the excised protein bands confirmed the identities of the recovered proteins. This result indicates that GacI and GacJ form a stable CHAPS-soluble complex and co-purify during affinity chromatography.
Figure 6.
GacI and GacJ exist as a detergent-stable complex in the membrane. GacJ and His-tagged GacI were co-expressed in E. coli Rosetta DE3 cells and extracted from the membrane fraction in 2.5% CHAPS. The proteins were purified using Ni-NTA-agarose in the presence of 2.5% CHAPS. A, fractions collected during Ni-NTA purification were analyzed by immunoblot using anti-His antibodies. B, eluted proteins were analyzed by SDS-PAGE. The results are representative of three separate experiments.
To investigate whether GacJ performs any catalytic function in association with GacI, we tested the proteins for GlcNAc-P-Und synthase activity in vitro, individually, and in combination. When GacI was expressed singly in E. coli, enzymatically active protein was found in the membrane fraction (Table 2 and supplemental Fig. S2B), indicating that GacI does not require GacJ for activity or membrane association. GacJ was found to be catalytically inactive when expressed by itself in E. coli, consistent with the observation that 5005ΔgacI membranes show no residual GlcNAc-P-Und synthase activity. Kinetic analysis of GlcNAc-P-Und synthase activity for Und-P in membrane fractions of E. coli expressing GacI revealed an apparent Km of 18.7 μm (Table 3 and supplemental Fig. S5A). However, co-expression of GacJ dramatically lowered the apparent Km value of GacI for Und-P to 1.1 μm, suggesting a significant change in affinity for the lipid acceptor. Moreover, the Vmax of GacI activity increased from 54.2 to 18,500 pmol/min/mg in the presence of GacJ (Table 3 and supplemental Fig. S5B). A kinetic analysis of GlcNAc-P-Und synthase for Und-P in MGAS5005 strain gave an apparent Km of 6.4 μm and a Vmax of 333 pmol/min/mg (Table 3 and supplemental Fig. S5C), similar to the enzymatic parameters of GacI co-expressed with GacJ in E. coli. These enzymatic parameters are in marked contrast to those of the GlcNAc-P transferase that synthesizes GlcNAc-P-P-Und. When a similar kinetic analysis was performed in 5005ΔgacI, so that only GlcNAc-P-P-Und synthesis could be scored, an apparent Km for Und-P of 19.3 μm was obtained, and the Vmax was only 2.75 pmol/min/mg (Table 3 and supplemental Fig. S5D). Clearly, GacI displays a much greater catalytic efficiency for the synthesis of GlcNAc-P-Und than the enzyme synthesizing GlcNAc-P-P-Und, which is reflected in the dramatic difference in the rates of synthesis of the two glycolipids. In summary, our data indicate that GacJ forms a stable association with GacI and stimulates the catalytic activity of GacI.
Table 3.
Kinetic parameters of GlcNAc-lipid synthases in membrane fractions from various bacterial strains
Membrane fractions from the indicated bacterial strains were assayed for the formation of [3H]GlcNAc-lipids in the presence of increasing amounts of Und-P, added as a dispersion in 1% CHAPS. Apparent kinetic parameters for Und-P were calculated from Michaelis-Menten plots (supplemental Fig. S5) using the linear regression algorithm in Sigma Plot version 12 (Systat Software, Inc. San Jose, CA). R2 values were 0.9953 for GAS GacI expressed in E. coli; 0.983 for GAS GacI co-expressed with GacJ in E. coli, 0.9976 for GacI in MGAS5005, and 0.931 for GacO in MGAS5005. The results are representative of at least two separate experiments. ND = not detected.
| Enzyme source | GlcNAc-P-Und |
GlcNAc-P-P-Und |
||
|---|---|---|---|---|
| Km | Vmax | Km | Vmax | |
| μm | pmol/min/mg | μm | pmol/min/mg | |
| MGAS5005 | 6.4 | 333 | ND | ND |
| 5005ΔgacI | ND | ND | 19.3 | 2.75 |
| E. coli:GacI | 18.7 | 54.2 | ND | ND |
| E. coli:GacI/J | 1.1 | 18,500 | ND | ND |
GlcNAc-P-P-Und is required for initiation of polyrhamnose backbone biosynthesis
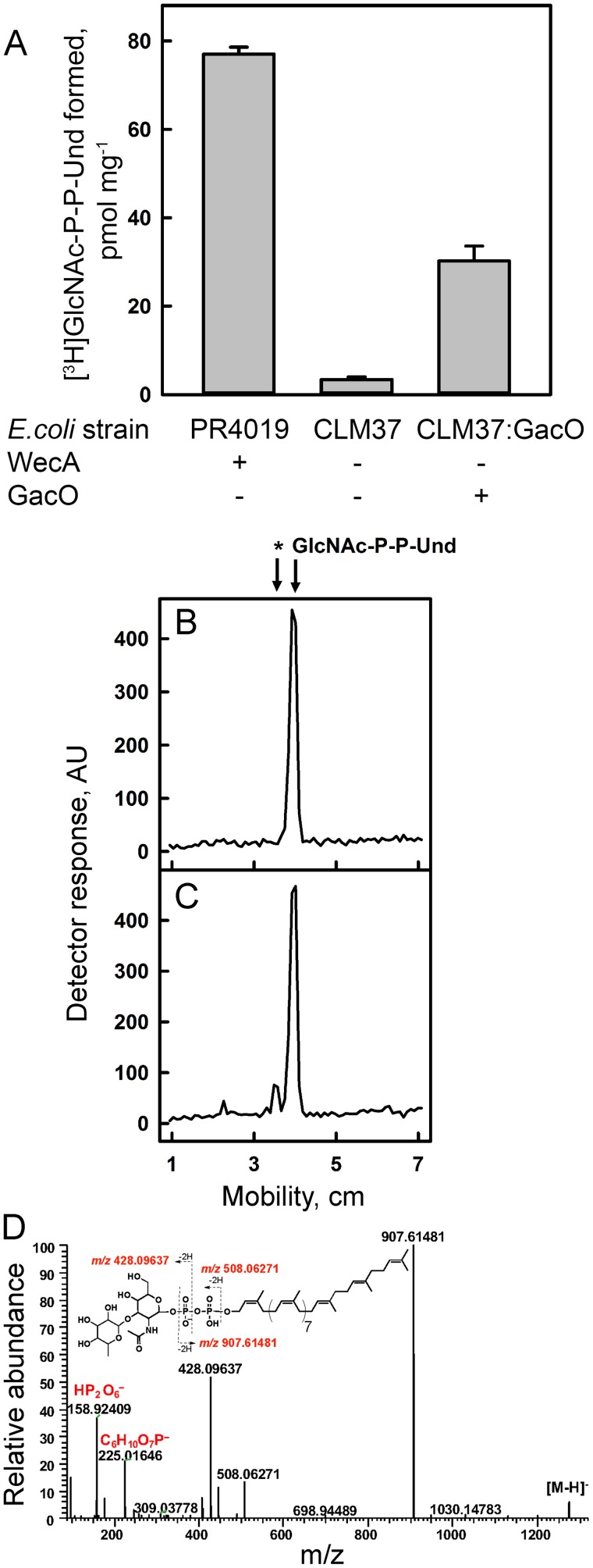
The GAS genome contains a close relative, gacO, of E. coli wecA, the tunicamycin-sensitive GlcNAc-phosphate transferase responsible for the synthesis of GlcNAc-P-P-Und in many bacteria (11, 12). Significantly, GAC synthesis in GAS is inhibited by tunicamycin (5), and a close homolog of GacO, S. mutans RpgP, is required for rhamnopolysaccharide synthesis and can complement WecA deficiency in E. coli (13). Our analysis of UDP-[3H]GlcNAc incorporation in GAS and GBS membrane lipids, revealing the presence of two GlcNAc-lipids, GlcNAc-P-Und and GlcNAc-P-P-Und, prompted us to investigate the role of GlcNAc-P-P-Und in the initiation of GAC biosynthesis. First, we investigated the role of GacO in the synthesis of GlcNAc-P-P-Und. When membrane fractions from the WecA-deficient E. coli strain (CLM37) expressing GacO, CLM37:GacO, were incubated with UDP-[3H]GlcNAc and Und-P (added as a dispersion in 1% CHAPS), [3H]GlcNAc-P-P-Und was formed at an enzymatic rate that is similar to that found in the WecA overexpressor strain, PR4019 (Fig. 7A). CLM37 carrying an empty vector synthesizes no detectable GlcNAc-P-P-Und under these conditions (Fig. 7A).
Figure 7.
Analysis of GacO function in MGAS5005. A, GacO catalyzes the synthesis of GlcNAc-P-P-Und in E. coli membranes. Reaction mixtures contained 50 mm Tris-HCl, pH 7.4, 5 mm 2-mercaptoethanol, 20 mm MgCl2, 0.5% CHAPS, 20 μm Und-P (dispersed by ultrasonication in 1% CHAPS), 5 μm UDP-GlcNAc (452 cpm/pmol), and E. coli membrane fraction from either PR4019, CLM37, or CLM37:GacO strains. Following incubation for 10 min at 30 °C, incorporation of [3H]GlcNAc into [3H]GlcNAc-P-P-Und was determined as described under “Experimental procedures.” Data are the average of three replicates ± S.D. B and C, GlcNAc-P-P-Und can function as an acceptor substrate for rhamnosylation to form rhamnosyl-GlcNAc-P-P-Und in 5005ΔgacI membranes. Incubation conditions were as described in the legend to Fig. 3. B, after 35 min of incubation with UDP-[3H]GlcNAc, the reactions were analyzed for formation of [3H]GlcNAc lipids as described in Fig. 3. C, after 5 min of incubation with UDP-[3H]GlcNAc, the reactions were incubated with 20 μm TDP-rhamnose for an additional 30 min and analyzed for formation of [3H]GlcNAc lipids. The results are representative of three separate experiments. D, ESI-MS/MS spectrum of rhamnosyl-GlcNAc-P-P-Und formed during chase with TDP-rhamnose from GlcNAc-P-P-Und synthesized in situ in membranes from S. pyogenes 5005ΔgacI.
To test whether GlcNAc-P-P-Und might function as a membrane anchor for the synthesis of the polyrhamnose chain of GAC, membrane fractions from MGAS5005 or the 5005ΔgacI mutant were pre-incubated with UDP-[3H]GlcNAc and ATP to form [3H]GlcNAc-P-P-Und in situ and chased with non-radioactive TDP-rhamnose. The formation of the resultant [3H]GlcNAc-lipids in 5005ΔgacI was analyzed by TLC on silica gel G after 30 min of incubation as shown in Fig. 7, B and C. In the absence of TDP-rhamnose, 5005ΔgacI membranes produced only [3H]GlcNAc-P-P-Und (Fig. 7B), whereas in the presence of TDP-rhamnose two radioactive products, [3H]GlcNAc-P-P-Und and an additional product with slower mobility on TLC, were observed (Fig. 7C). MS analysis of lipid extracts from the in vitro reactions revealed the presence of a molecular ion [M − H]−1 m/z = 1274.76084 with the composition, C69H114O16NP2, consistent with the expected product, rhamnosyl-GlcNAc-P-P-Und (Fig. 7D). Furthermore, the major fragment ions derived from m/z = 1274.76084 can be assigned to Rha-GlcNAc-P, Rha-GlcNAc-P-P, and P-Und, supporting the proposed structural interpretation (Fig. 7D and Table 4). Supplemental Fig. S6 shows the time-dependent accumulation of the new GlcNAc-lipid product. Significantly, parallel incubations with MGAS5005 showed that GlcNAc-P-Und was not glycosylated further. These results strongly support the possibility that GlcNAc-P-P-Und is an acceptor for rhamnosyl units in GAS and may function as the lipid anchor for the GAC polyrhamnose backbone synthesis.
Table 4.
The major fragments of the precursor ion at m/z 1274.7612
Fragment ions originating from the precursor molecular ion of 1274 were subjected to MS/MS fragmentation as described under “Experimental procedures.” The proposed molecular formulae of the product ions are supported by the observed isotope patterns for the relevant ions.
| m/z | Formula | Mass error |
|---|---|---|
| ppm | ||
| 158.92409 | HP2O6 | −1.24 |
| 225.01646 | C6H10PO7 | 2.64 |
| 428.09637 | C14H23O12NP | 2.64 |
| 508.06271 | C14H24O15NP2 | 2.25 |
| 907.61481 | C55H89O6P2 | 2.12 |
| 1274.76084 | C69H114O16NP2 | 0.08 |
GacL mutant displays increased sensitivity to peptidoglycan amidases
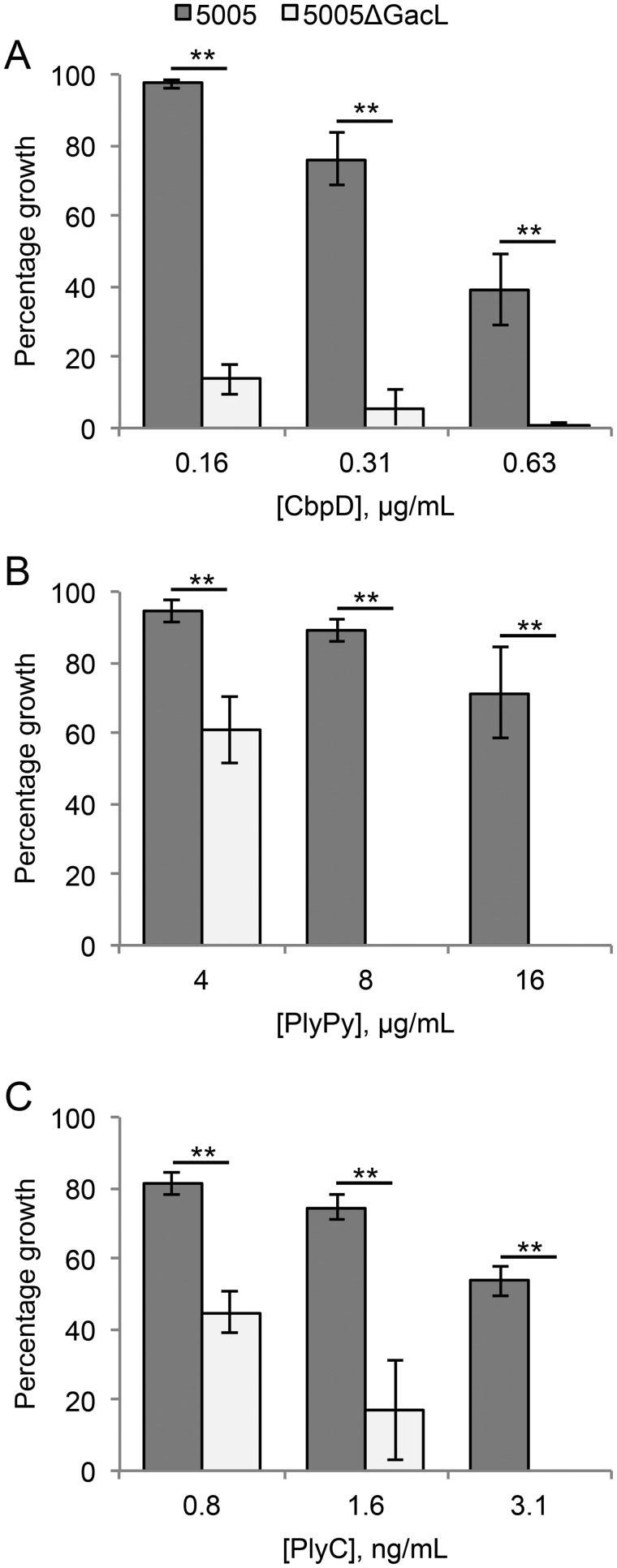
The Gram-positive cell wall protects interior structures, plasma membrane and peptidoglycan, from host-defense peptides and hydrolytic and antimicrobial enzymes. It has been shown that the GacI mutant, which is GlcNAc-deficient, is more sensitive to LL-37-induced killing (5). To test the hypothesis that the loss of GlcNAc decorations in GAC alters cell wall permeability, we investigated the sensitivity of 5005ΔgacL to peptidoglycan amidases: PlyC (27), PlyPy (28), and CbpD (29). When 5005ΔgacL cells were grown in increasing concentrations of CbpD (Fig. 8A), PlyPy (Fig. 8B), or PlyC (Fig. 8C), cellular growth was dramatically inhibited compared with MGAS5005, indicating increased sensitivity to the presence of amidases.
Figure 8.
Absence of GacL increases sensitivity to cell wall amidases. Mid-exponential phase MGAS5005 and 5005ΔgacL were grown in the indicated concentrations of CbpD (A), PlyPy (B), and PlyC (C). The change in growth is represented as a percentage of growth where no amidase was present. Data are the average of three replicates ± S.D. The asterisk indicate statistically different values (**, p < 0.01) as determined by the Student's t test.
Discussion
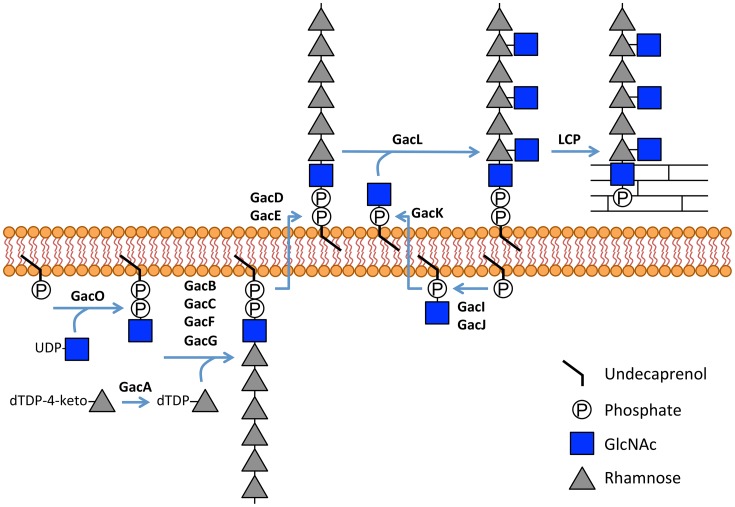
In almost all Gram-positive bacteria, cell wall-attached glycopolymers are critical for cell envelope integrity, and their depletion is lethal (30). Most streptococcal species, including two human pathogens GAS and GBS, do not synthesize WTA and instead produce Rha-containing glycopolymers as functional homologs of WTA (1). GAC is the major cell wall component of GAS and plays important roles in bacterial physiology and pathogenesis. The polyrhamnose core of GAC is modified with GlcNAc in an ∼2:1 ratio (3, 7) of rhamnose to GlcNAc. Collectively, the results of our study suggest a molecular mechanism of GAC biosynthesis in which rhamnan polymer is assembled at the cytoplasmic face of the plasma membrane, translocated to the cell surface, and modified by GlcNAc on the outer side of the membrane as illustrated in Fig. 9. We report that a lipid carrier, GlcNAc-P-P-Und, synthesized by GAS GacO, is a potential acceptor for initiation of rhamnan backbone biosynthesis. We speculate that the next step of rhamnan biosynthesis involves the action of the GacB, GacC, GacG, and GacF glycosyltransferases. The lipid-anchored polyrhamnose is then translocated across the membrane by the ABC transporter encoded by gacD and gacE. This hypothesis is supported by studies of rhamnan biosynthesis in S. mutans (13, 31, 32).
Figure 9.
Schematic diagram of GAC biosynthesis. GAC is anchored to peptidoglycan presumably via phosphodiester bond. GAC biosynthesis is initiated on the inner leaflet of the plasma membrane where GacO produces GlcNAc-P-P-Und, which serves as a membrane-anchored acceptor for polyrhamnose synthesis catalyzed by the GacB, GacC, GacF, and GacG rhamnosyltransferases. Following polymerization, polyrhamnose is transferred to the outer leaflet of the membrane presumably by the GacD/GacE ABC transporter. Also in the inner leaflet of the membrane, GacI aided by GacJ produces GlcNAc-P-Und, which then diffuses across the plasma membrane to the outer leaflet aided by GacK. Subsequently, GacL transfers GlcNAc to polyrhamnose using GlcNAc-P-Und as glycosyl donor. Finally, protein members of LytR-CpsA-Psr phosphotransferase family presumably attach GAC to peptidoglycan. Several details of this biosynthetic scheme are still speculative, and further research will be required to definitively confirm this hypothetical pathway, but the overall organization is consistent with other isoprenol-mediated capsular polysaccharide pathways.
Biosynthesis of the rhamnan backbone of GAC is likely essential for GAS viability because the GacA enzyme involved in dTDP-rhamnose biosynthesis (14) and GacB and GacC glycosyltransferases is indispensable in GAS (5). In contrast, deletion of genes required for GlcNAc attachment to the rhamnan backbone does not affect GAS viability (5). This observation supports our hypothesis that biosynthesis of polyrhamnose and its translocation to the cell surface occur separately from the pathway involved in polyrhamnose modification with GlcNAc. In bacterial and eukaryotic glycoconjugate assembly systems, structural modifications that are introduced late in the biosynthesis typically involve polyprenyl monophosphoryl donors (33). In our study, we found that GlcNAc modification of rhamnan requires GlcNAc-P-Und synthesis. Previously, GlcNAc-P-Und was isolated from various Bacilli membranes (12, 23). However the enzyme required for GlcNAc-P-Und biosynthesis was not identified, and the biological function of this lipid remained unclear. Our in vitro analysis of UDP-GlcNAc incorporation into GlcNAc-lipids by GAS membranes showed that GacI was required for the biosynthesis of GlcNAc-P-Und. Moreover, CHAPS-soluble, affinity-purified GacI protein catalyzed the transfer of GlcNAc from UDP-GlcNAc to Und-P yielding GlcNac-P-Und in an in vitro reaction system.
Interestingly, we found that, in contrast to B. cereus, GAS and GBS membranes incubated with UDP-[3H]GlcNAc synthesized primarily [3H]GlcNAc-P-Und. Because GacI and GacO utilize a common pool of Und-P, this phenomenon is presumably due to a much higher apparent affinity of GacI for Und-P compared with GacO. Significantly, we observed a measurable increase in [3H]GlcNAc incorporation into [3H]GlcNAc-P-P-Und in the GacI deletion mutant. This observation further confirmed that the relative synthetic rates of GlcNAc-P-Und and GlcNAc-P-P-Und biosynthesis are determined largely by relative affinity for Und-P. Furthermore, we found that the formation of GlcNAc-P-Und and GlcNAc-P-P-Und in GAS membranes was significantly stimulated by ATP. Because GacI activity is not stimulated directly by ATP, it is likely that the effect of ATP is due to increased formation of Und-P, in situ, via undecaprenol kinase activity. In S. mutans, membrane-associated undecaprenol kinase catalyzes the ATP-dependent phosphorylation of undecaprenol to Und-P (26). It is likely that the homolog of this enzyme encoded by M5005_Spy_0389 is responsible for Und-P biosynthesis in GAS.
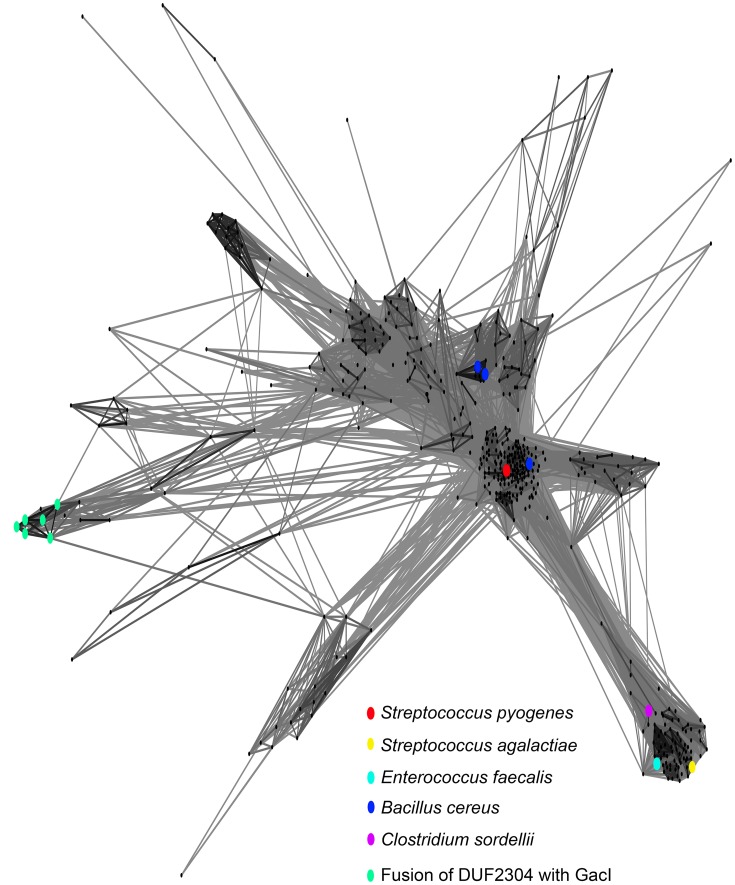
GacI is predicted by the HHpred server to have structural homology to polyisoprenyl-glycosyltransferase GtrB from Synechocystis, which is a homolog of Shigella flexneri GtrB (34). GtrB homologs in bacteria and eukaryotes belong to the GT-A superfamily of glycosyltransferases and are responsible for the transfer of glucose from UDP-glucose to the bactoprenol carrier in the cytoplasm yielding the Und-P-glucose precursor in bacteria and dolichol phosphate–glucose precursor in eukaryotes (35, 36). Our BLAST search using the GacI sequence as query found homologs of this gene in many bacterial species, with the broadest diversity observed in the phylum Firmicutes (supplemental file S1). To determine evolutionary relationships between the GacI homologs, these sequences were analyzed using CLANS (CLuster ANalysis of Sequences) (Fig. 10) (37). It is noteworthy that the B. cereus GacI has two homologs clustered in two different groups. Our bioinformatics analysis shows that these gacI homologs are located in different gene clusters encoding proteins involved in polysaccharide biosynthesis and transport. Moreover, the GAS GacI subgrouped with one B. cereus GacI. The GBS GacI and E. faecalis GacI homologs are located in the cluster well-separated from the GAS GacI subgroup. Thus, this observation points to possible horizontal gene transfer between GAS and Bacillus. Analysis of UDP-GlcNAc incorporation into GlcNAc-lipids in a GBS gacI knock-out strain confirmed the function of this gene in the biosynthesis of GlcNAc-P-Und in GBS. Additionally, we demonstrated that the E. faecalis GacI homolog (EpaI) functions in the biosynthesis of GlcNAc-P-Und. Significantly, the presence of GacI homologs in GBS and E. faecalis matches the reported occurrence of GlcNAc residues in the cell wall polysaccharides of these bacteria (38–41).
Figure 10.
Sequence relationship of GacI family of proteins. Homology between GacI homologs is graphically displayed using CLANS analysis (37). Dots correspond to individual protein sequences selected as described under “Experimental procedures” and provided as supplemental file S1. Selected homologs of S. pyogenes GacI are highlighted by colored dots.
GacJ, a small-membrane protein with three transmembrane α-helices, is required for GlcNAc side-chain attachment to polyrhamnose (5). GacJ belongs to the DUF2304 family of proteins according to the Pfam protein family database (42). In Geobacter sp., Desulfuromonas sp., Desulfurivibrio alkaliphilus, Desulfuromonas soudanensis, and Desulfurispirillum indicum, DUF2304 homologous domains are fused with gacI homologs (Fig. 10 and supplemental file S1). In Mycobacterium tuberculosis, a GacJ homolog, Rv3632, is co-transcribed with the gene encoding N-acetylgalactosaminyl-phosphate-undecaprenol synthase, PpgS, and is found to stimulate PpgS activity (43). Consistent with this finding, we showed that GacI and GacJ form a complex and co-expression of GacI with GacJ significantly enhances GacI catalytic activity. Further work is under way to characterize the mechanism of GacJ action on GacI activity.
In many bacterial systems, modification of glycoconjugates involves the export of a polyprenyl monophosphoryl-glycose molecule to the periplasm providing the glycosyl donor for the modifying reaction catalyzed by an integral membrane glycosyltransferase that possesses a GT-C fold (33). Bioinformatics searches with the HHpred program against available protein structures identified the GT-C superfamily of glycosyltransferases as the closest structural homologs of GacL. Moreover, the phenotypes of the GacL mutant, absence of the GlcNAc side-chains in GAC, and accumulation of GlcNAc-P-Und in the membrane, are consistent with GacL functioning in the transfer of GlcNAc from GlcNAc-P-Und to polyrhamnose yielding GlcNAc-modified polysaccharide.
We suggest that the Wzx flippase, GacK, may function to transport GlcNAc-P-Und to the extracellular space for utilization as GlcNAc donor in the GlcNAc modification of polyrhamnose. This hypothesis is based on the GlcNAc-deficient phenotype of the GacK mutant (5). However, further research will be required to confirm this hypothesis. The last step of GAC biosynthesis is probably similar to the last step of WTA biosynthesis; it involves attachment of GAC to certain MurNAc residues in peptidoglycan via a phosphate ester linkage (46). It is likely catalyzed by members of LytR-CpsA-Psr phosphotransferase family encoded by M5005_Spy_1099 and M5005_Spy_1474.
The identification of the mechanism of GlcNAc attachment to polyrhamnose raises the question of how this modification functions biologically in bacteria. It has been previously found that the ΔgacI mutant is hypersusceptible to human antimicrobial peptide LL-37 and the antimicrobial action of factors released by thrombin-activated platelets, suggesting a role for GlcNAc modification in protecting the plasma membrane from antimicrobial agents (5). Our study identified the importance of GlcNAc for protection of GAS peptidoglycan from amidase-induced lysis. In E. faecalis, the GacI homolog, EpaI, is involved in biosynthesis of a cell wall-attached polysaccharide (24); however, the polysaccharide structure of the mutant has not been elucidated. The ΔepaI mutant was defective in conjugative transfer of a plasmid and resistance of bacteria to detergent and bile salts (24).
In conclusion, our study provides a platform for elucidation of novel pathways of glycopolymer biosynthesis in other bacterial pathogens, including important drug-resistant bacteria such as E. faecalis and Clostridium sordellii that possess GacI and GacJ homologs (Fig. 10 and supplemental file S1). Because the enzymes involved in the biosynthesis of cell wall-attached glycopolymers are promising targets for novel antimicrobials, and the glycopolymers represent important features for diagnostics and vaccine targets, our data may provide opportunities for developing novel therapeutics against antibiotic-resistant bacterial pathogens.
Experimental procedures
Bacterial strains and growth conditions
All plasmids, strains, and primers used in this study are listed in supplemental Tables S2 and S3. The strains used in this study were GAS M1-serotype strain MGAS5005 (16), B. cereus (ATCC 14579), S. agalactiae COH1 (GBS), E. coli CLM37 (47), E. coli PR4019 (12), E. coli DH5α, and E. coli Rosetta (DE3). GAS and GBS cultures were grown in Todd-Hewitt broth (BD Biosciences) supplemented with 0.2% yeast extract (THY) or on THY agar plates at 37 °C. E. coli and B. cereus strains were grown in Luria-Bertani (LB) medium or on LB agar plates at 37 °C. When required, antibiotics were included at the following concentrations: ampicillin at 100 μg ml−1 for E. coli; streptomycin at 100 μg ml−1 for E. coli; erythromycin at 500 μg ml−1 for E. coli, and 1 μg ml−1 for GAS and GBS; chloramphenicol at 10 μg ml−1 for E. coli and 5 μg ml−1 for GAS; spectinomycin at 200 μg ml−1 for E. coli and 100 μg ml−1 for GAS and GBS.
DNA techniques
Plasmid DNA was isolated from E. coli by commercial kits (Qiagen) according to the manufacturer's instructions and used to transform E. coli, GAS, and GBS strains. Plasmids were transformed into GAS and GBS by electroporation as described previously (48). Chromosomal DNA was purified from GAS and GBS as described previously (49). To construct single-base substitutions or deletion mutations, we used the QuikChange® II XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. Constructs containing mutations were identified by sequence analysis. All constructs were confirmed by sequencing analysis (Eurofins MWG Operon).
Construction of the gacI deletion mutant in GAS
For construction of strain 5005ΔgacI, MGAS5005 chromosomal DNA was used as a template for amplification of two DNA fragments using two primers pairs: GacIm-BamHI-f/GacIdel-r and GacIdel-f/GacIm-XhoI-r (supplemental Table S3). Primer GacIdel-f is complementary to primer GacIdel-r. The two gel-purified PCR products containing complementary ends were mixed and amplified using a PCR overlap method (50) with primer pair GacIm-BamHI-f/GacIm-XhoI-r to create the deletion of gacI. The PCR product was digested with BamHI and XhoI and ligated into BamHI/SalI-digested temperature-sensitive shuttle vector pJRS233 (51). The plasmid was designated pJRS233ΔgacI (supplemental Table S2). The resulting plasmid was transformed into MGAS5005, and erythromycin-resistant colonies were selected on THY agar plates at 30 °C. Integration was performed by growth of transformants at 37 °C with erythromycin selection. Excision of the integrated plasmid was performed by serial passages in THY media at 30 °C and parallel screening for erythromycin-sensitive colonies. Mutants were verified by PCR sequencing of the loci.
Construction of the gacL deletion mutant in GAS
To create BglII and XhoI cloning sites in the polylinker region of pUC19, site-directed mutagenesis of the plasmid, using two primer pairs, pUC19-XhoI-f/pUC19-XhoI-r and pUC19-BglII-f/pUC19-BglII-f, was carried out. The plasmid was designated pUC19BX. The non-polar aadA spectinomycin resistance cassette was amplified from pLR16T (supplemental Table S2) using primers Spec-SalI-f/Spec-BamH-r (supplemental Table S3), digested with SalI/BamHI, and ligated into SalI/BamHI-digested pUC19BX to yield pUC19BXspec. MGAS5005 chromosomal DNA was used as a template for amplification of two DNA fragments using two primers pairs: gacLup-BglII-f/gacLup-SalI-r and gacLdown-BamHI-f/gacLdown-XhoI-r (supplemental Table S3). The first PCR product was digested with BglII/SalI and ligated into BglII/SalI-digested pUC19BXspec. The resultant plasmid, pUC19BXspecL1, was digested with BamHI/XhoI and used for ligation with the second PCR product that was digested with BamHI/XhoI. The resultant plasmid, pUC19BXspecL2, was digested with BglII and XhoI to obtain a DNA fragment containing aadA flanked with the gacL upstream and downstream regions. The DNA fragment was ligated into pHY304 vector (52) and digested with BamHI/XhoI to yield pHY304ΔgacL. The resulting plasmid was transformed into MGAS5005, and erythromycin-resistant colonies were selected on THY agar plates at 30 °C. The mutants were isolated as described above. 5005ΔgacL mutants were screened for sensitivity to spectinomycin and verified by PCR sequencing of the loci.
Complementation of the 5005ΔgacL mutant with gacL (5005ΔgacL gacL+)
To construct the plasmid for complementation of the 5005ΔgacL mutant, MGAS5005 chromosomal DNA was used as a template for amplification of a wild-type copy of gacL using the primer pair GacL-XhoI-f and GacL-BglII-r (supplemental Table S3). The PCR products were digested with XhoI and BglII and cloned in pBBL740 (supplemental Table S2) previously digested with the respective enzymes. The integrational plasmid pBBL740 does not have a replication origin that is functional in GAS, so the plasmid can be maintained only by integrating into the GAS chromosome through homologous recombination. The resultant plasmid pGacL was transformed into 5005ΔgacL by electroporation, and transformants were selected on agar plates containing chloramphenicol. Several chloramphenicol-resistant colonies were selected, and gacL integration into the chromosome was confirmed by sequencing a PCR fragment.
Construction of the gacI (SAN_1536) deletion mutant in GBS
GBS COH1 chromosomal DNA was used as a template for amplification of two DNA fragments using two primers pairs: gbsIup-BglII-f/gbsIup-SalI-r and gbsId-BamHI-f/gbsId-XhoI-r (supplemental Table S3). The plasmid for gacI (SAN_1536) knock-out was constructed using the same strategy described for gacL deletion (see above). The resultant plasmid, pHY304GBSΔI, was transformed into GBS COH1, and erythromycin-resistant colonies were selected on THY agar plates at 30 °C. The mutants were isolated as described above. GBS COH1 ΔgacI mutants were screened for sensitivity to spectinomycin and verified by PCR sequencing of the loci.
Construction of the plasmids for E. coli expression of GacI, GacJ, and GacO
To create a vector for expression of GacI from GAS, the gene was amplified from MGAS5005 chromosomal DNA using the primer pair GacI-NcoI-f and GacI-XhoI-r (supplemental Table S3). The PCR product was digested with NcoI and XhoI and ligated into NcoI/XhoI-digested pRSF-NT vector (supplemental Table S3). The resultant plasmid, pGacI, contained gacI fused at the N terminus with a His tag followed by a TEV protease recognition site. To create a vector for expression of GacI and GacJ, the bicistronic DNA fragment was amplified from MGAS5005 chromosomal DNA using the primer pair GacI-NcoI-f and GacJ-XhoI-r (supplemental Table S3). The vector was constructed as described above. The plasmid was designated pGacIJ. pET21_NESG plasmid for expression of the GacI homolog, EpaI, from E. faecalis was obtained from the DNASU repository (53). The construct was confirmed by sequencing analysis. The plasmids were transferred into competent E. coli Rosetta (DE3) (Novagen) using the manufacturer's protocol.
To create a vector for expression of GacO, the gene was amplified from MGAS5005 chromosomal DNA using the primer pair GacO-Xba-f and GacO-HindIII-r (supplemental Table S3). The PCR product was digested with XbaI and HindIII and ligated into XbaI/HindIII-digested pBAD33 vector. The resultant plasmid, pGacO, was transferred into competent E. coli CLM37 strain that has a deletion of the wecA gene.
Construction of the plasmids for E. coli expression of PlyPy, CbpD, and PlyC
To create a vector for expression of CbpD amidase (29), the gene was amplified from MGAS5005 chromosomal DNA using the primer pair 28-NcoI-f and 28-stop-r (supplemental Table S3). The PCR product was digested with NcoI and XhoI and ligated into NcoI/XhoI-digested pRSF-NT vector. The resultant plasmid, pCbpD, contained cbpD fused at the N terminus with a His tag followed by a TEV protease recognition site. To create a vector for expression of PlyPy amidase (28), the gene was amplified from MGAS5005 chromosomal DNA using the primer pair PlyPy-NcoI-f and PlyPy-XhoI-r. The PCR product was digested with NcoI and XhoI and ligated into NcoI/XhoI-digested pET-21d vector. The resultant plasmid, pPlyPy, contained the gene fused at the C terminus with a His tag.
To create a vector for expression of PlyC amidase (54), a DNA fragment-spanning bicistronic operon, which encodes plyCA and plyCB (27), was synthesized by Thermo Fisher Scientific. The plasmid was digested with NcoI and XhoI and ligated into NcoI/XhoI-digested pET-21d vector. The resultant plasmid pPlyC contained plyCA followed by plyCB fused at the C terminus with a His tag.
Expression and purification of GacI, EpaI, and GacI–GacJ complex
For expression of GacI, EpaI, and GacI–GacJ complex, E. coli Rosetta (DE3) cells carrying the respective plasmid were grown to an A600 of 0.4–0.6 and induced with 1 mm IPTG at 18 °C for ∼16 h. The cells were lysed in 20 mm Tris-HCl, pH 7.5, 300 mm NaCl with two passes through a EmulsiFlex-C5 microfluidizer cell disrupter (Avestin, Inc., Ottawa Ontario, Canada). The lysate was centrifuged at 7000 × g for 30 min at 4 °C. The supernatant was centrifuged at 30,000 × g for 60 min at 4 °C to isolate the membrane fraction.
To isolate GacI–GacJ complexes, the membrane proteins were solubilized in 2.5% CHAPS, 20 mm Tris-HCl, pH 7.5, 300 mm NaCl for 60 min, rotating at room temperature. Insoluble material was removed by centrifugation at 30,000 × g for 60 min at 4 °C. Solubilized GacI–GacJ were purified by Ni-NTA chromatography with washes of 2.5% CHAPS, 20 mm Tris-HCl, pH 7.5, 300 mm NaCl, 2.5% CHAPS, 20 mm Tris-HCl, pH 7.5, 300 mm NaCl, 10 mm imidazole and eluted with 2.5% CHAPS, 20 mm Tris-HCl, pH 7.5, 300 mm NaCl, 250 mm imidazole. The GacI–GacJ complex was further purified by size-exclusion chromatography on a Superdex 200 (GE Healthcare) column in 2.5% CHAPS, 20 mm HEPES, pH 7.5, 100 mm NaCl, with monitoring for protein elution at 280 nm.
Expression of GacO
For expression of GacO, E. coli CLM37 cells carrying the pertinent plasmid were grown to an A600 of 0.8 and induced with 13 mm l-arabinose at 25 °C for ∼3 h. The cells were lysed in 20 mm Tris-HCl, pH 7.5, 300 mm NaCl with two passes through a microfluidizer cell disrupter. The lysate was centrifuged at 1000 × g for 15 min at 4 °C. The supernatant was centrifuged at 40,000 × g for 60 min at 4 °C to isolate the membrane fraction.
Expression and purification of PlyPy, CbpD, and PlyC
For expression and purification of PlyPy, CbpD, and PlyC, E. coli Rosetta (DE3) cells carrying the respective plasmids were grown to an A600 of 0.4–0.6 and induced at 18 °C with 1 mm IPTG for ∼16 h. The cells were lysed in 20 mm Tris-HCl, pH 7.5, 300 mm NaCl with two passes through a microfluidizer cell disrupter. The soluble fraction was purified by Ni-NTA chromatography with washes of 20 mm Tris-HCl, pH 7.5, 300 mm NaCl, and 20 mm Tris-HCl, pH 7.5, 300 mm NaCl, 10 mm imidazole and eluted with 20 mm Tris-HCl, pH 7.5, 300 mm NaCl, 250 mm imidazole. The PlyPy and CbpD eluate was further purified by size-exclusion chromatography on a Superdex 200 column in 20 mm HEPES, pH 7.5, 100 mm NaCl for PlyPy or 20 mm MOPS, pH 6.5, 100 mm NaCl for CbpD. PlyC was further purified by anion-exchange chromatography on a Mono Q 5/50 GL (GE Healthcare) column in 10 mm sodium phosphate, pH 6.0, and a 20 column volume elution gradient of 0–500 mm NaCl.
Isolation of GAS and GBS membranes
Bacteria were grown at 37 °C to an A600 of 0.8. To obtain GAS cell membranes, cell pellet was resuspended in phosphate-buffered saline (PBS) and incubated 1 h with PlyC lysin as described previously (55). To obtain GBS membranes, the cell pellet was resuspended in acetate buffer (50 mm CH3COONa, 10 mm CaCl2, 50 mm NaCl, pH 5) and incubated with mutanolysin (200 units/ml, Sigma) for 2 h at 37 °C. After hydrolytic enzyme treatment, the bacterial suspension was sonicated using a Thermo Fisher Scientific model 505 sonic dismembrator, 15 times for 15 s. After centrifugation at 8000 × g for 10 min at 4 °C, the supernatant was collected and centrifuged at 40,000 × g for 60 min. The pellet was collected as the membrane fraction.
Mass spectrometry analysis of GacI and GacJ
LC-MS/MS for proteomic analysis was performed using an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) coupled with an Eksigent Nanoflex cHiPLC system (Eksigent) through a nanoelectrospray ionization source. The LC-MS/MS data were subjected to database searches for protein identification using Proteome Discoverer software version 1.3 (Thermo Fisher Scientific) with a local MASCOT search engine.
Dot-blot analysis
Bacterial cells (1 ml) from exponential phase cultures (A600 = 0.8) were centrifuged, washed with PBS, resuspended in 100 μl of PBS, and incubated with 1 μl of PlyC (1.5 mg/ml) for 1 h at 37 °C. After centrifugation at 16,000 × g for 2 min, 5 μl of the supernatant was spotted onto a nitrocellulose membrane. The membrane was blocked for 1 h with 7% skim milk in PBS with 0.1% Tween 20 and incubated overnight with an anti-GAC antibody diluted 1:5000 (ab9191; Abcam). Bound antibodies were detected with a peroxidase-conjugated goat anti-rabbit immunoglobulin G antibody and the Amersham Biosciences ECL (enhanced chemiluminescence) Western blotting system.
Binding of sWGA to bacteria
MGAS5005 and 5005ΔgacL cells were collected during mid-exponential phase (A600 = 0.6), washed three times with BSA/saline solution (0.5% BSA, 0.15 m NaCl), resuspended in BSA/saline solution, and incubated for 30 min at 37 °C with mixing. GlcNAc-specific fluorescein–sWGA was added to final concentrations of 0, 12.5, 25, 50, and 100 μg/ml. After 1 h of incubation at 37 °C with mixing, the cells were centrifuged, washed twice, and resuspended in BSA/saline. Bound fluorescein–sWGA was quantified in a fluorimeter SpectraMax M5 (Molecular Devices) using an excitation of 544 nm and emission of 590 nm.
Isolation of cell wall from GAS
MGAS5005 and 5005ΔgacL cell wall was prepared from exponential phase cultures (A600 = 0.8) by the SDS-boiling procedure as described for Streptococcus pneumoniae (56). Purified cell wall samples were lyophilized and used for composition analysis.
Carbohydrate composition analysis
Carbohydrate composition analysis was performed at the Complex Carbohydrate Research Center (Athens, GA) by combined gas chromatography/mass spectrometry (GC/MS) of the per-O-trimethylsilyl derivatives of the monosaccharide methyl glycosides produced from the sample by acidic methanolysis as described previously (57).
Assay of lytic activity of PlyPy, CbpD, and PlyC
Aliquots of MGAS5005 and 5005ΔgacL frozen stocks (prepared as described in Ref. 58) were inoculated into THY medium 1:20. To a 96-well plate, 200-μl aliquots of culture were dispensed in duplicate, per condition. The plates were grown at 37 °C without aeration for 2 h. An assay plate was set up containing PlyPy (8, 16, and 32 μg ml−1), CbpD (0.31, 0.63, and 1.26 μg ml−1), or PlyC (1.6, 3.1, and 6.2 ng ml−1) in a volume of 100 μl. To the assay plate, 100 μl of culture was added to each well giving final concentrations of PlyPy (4, 8, and 16 μg ml−1), CbpD (0.16, 0.31, and 0.63 μg ml−1), or PlyC (0.8, 1.6, and 3.1 ng ml−1). After mixing, the absorbance at 600 nm was measured for each well (t0), and the plate was incubated at 37 °C for 1 h. The absorbance at 600 nm was then measured again (t1). The percentage difference (t1 − t0) in growth relative to 0 mm enzyme/antimicrobial for each concentration was calculated (i.e. 0 mm PlyPy = 100%).
Extraction and characterization of bacterial glycolipids
Two liters of bacterial cells from exponential phase cultures (A600 = 0.8) were recovered by sedimentation at 10,000 × g for 30 min and washed two times with ice-cold PBS. Cells were resuspended in 50 ml of PBS, sensitized by incubation with 10 ng ml−1 PlyC for 1 h at 37 °C, and stirred vigorously with 2 volumes of CH3OH and 1 volume of CHCl3 for 30 min at room temperature. The mixture was divided into 5-ml aliquots in 12-ml glass centrifuge tubes. Insoluble material was removed by centrifugation at 200 × g, and the organic extract was transferred to a separatory funnel. The insoluble residue was further extracted with 1 ml of CHCl3/CH3OH (2:1) per tube, two times, and the extracts were combined with the previous organic phase. The organic extract was supplemented with CHCl3 and 0.9% NaCl to give a final composition of CHCl3/CH3OH/0.9% NaCl (3:2:1), mixed vigorously, and allowed to stand in the cold until phase separation was achieved. The organic phase was drained off into a second separatory funnel, and the organic layer was washed with 1/3 volume of CHCl3/CH3OH/0.9% saline (3:48:47), two times. The aqueous layers were discarded. The organic extract was dried on a vacuum rotary evaporator, dissolved in a small volume of CHCl3/CH3OH (2:1), and transferred to a 12.5 × 100-mm screw cap glass tube (with Teflon lined cap). The organic extract was dried under a stream of nitrogen gas, and the glycerolipids were destroyed by deacylation in 0.1 m KOH in toluene/CH3OH (1:3) at 0 °C, 60 min. Following deacylation, the reactions were neutralized with acetic acid, diluted with 2 volumes of CHCl3, 1 volume of CHCl3/CH3OH (2:1), and 1/5 volume of 0.9% NaCl, 10 mm EDTA. The two-phase mixture was mixed vigorously and centrifuged to separate the phases. The organic phase was dried under nitrogen, spotted on a 20 × 20-cm sheet of Silica Gel G, and developed in CHCl3/CH3OH/H2O/NH4OH (65:25:4:1). Bacterial lipids were visualized by staining with iodine vapors, and pertinent spots were scraped from the thin layer plate, eluted from the silica gel with CHCl3/CH3OH (2:1), and reserved for further analysis.
Mass spectrometry analysis of a phospholipid isolated from 5005ΔgacL
A phospholipid accumulated by 5005ΔgacL was purified as described above using preparative TLC in silica gel. The compound was analyzed by LC-MS using a Q-exactive mass spectrometer and an Ultimate 3000 ultra HPLC system (Thermo Fisher Scientific, San Jose, CA) on a Kinetex C18 reversed-phase column (2.6 × 100 mm, 2.1 μm, Phenomenex). Two solvents were used for gradient elution: solvent A, acetonitrile/water (2:3, v/v) and solvent B, isopropyl alcohol/acetonitrile (9:1, v/v). Both A and B contained 10 mm ammonium formate and 0.1% formic acid. The column temperature was maintained at 40 °C, and the flow rate was set to 0.25 ml/min. Mass spectrometric detection was performed by electrospray ionization in negative ionization mode with source voltage maintained at 4.0 kV. The capillary temperature, sheath gas flow, and auxiliary gas flow were set at 330 °C, 35 and 12 arbitrary units, respectively. Full-scan MS spectra (mass range m/z 400 to 1500) were acquired with resolution R = 70,000 and AGC target 5e5. MS/MS fragmentation was performed using high-energy C-trap dissociation with resolution R = 35,000 and AGC target 1e6. The normalized collision energy was set at 30.
Assay for incorporation of GlcNAc into polyisoprenol-linked glycolipid intermediates
Reaction mixtures for measuring the incorporation of [3H]GlcNAc into lipids contained 50 mm Tris, pH 7.4, 0.25 m sucrose, 20 mm MgCl2, 5 mm β-mercaptoethanol, 5 μm UDP-[6-3H]GlcNAc ((100–2000 cpm/pmol) American Radiolabeled Chemicals), and bacterial membrane suspension (50–250 μg bacterial membrane protein) in a total volume of 10–100 μl. In some experiments, 1 mm ATP was included, as indicated. Following incubation at 30 °C, the enzymatic reactions were terminated by the addition of 40 volumes of CHCl3/CH3OH (2:1), thoroughly mixed, and incubated for 5 min at room temperature. Insoluble material was sedimented at 200 × g, and the organic extract was transferred to a 12 × 100-mm glass tube. The residue was re-extracted with 1 ml of CHCl3/CH3OH (2:1), two times, and the organic extracts were combined. The pooled organic extracts were freed of unincorporated radioactivity by sequential partitioning with 1/5 volume of 0.9% saline, 10 mm EDTA and then with 1/3 volume of CHCl3/CH3OH/0.9% saline (3:48:47) three times, discarding the aqueous phase each time. The washed organic phases were dried under nitrogen and re-dissolved in CHCl3/CH3OH (2:1). A carefully measured aliquot was removed and analyzed for radioactivity by liquid scintillation spectrometry after drying. The remainder of the sample was analyzed by TLC on Silica Gel G, developed in CHCl3/CH3OH/H2O/NH4OH (65:25:4:1), using a BioScan AR2000 radiochromatoscanner. Incorporation of [3H]GlcNAc into individual [3H]GlcNAc-lipids was calculated using the peak integration values obtained from the BioScan.
Degradation of GlcNAc-lipids by mild acid and mild alkaline treatment
Bacterial lipids were subjected to mild acid hydrolysis (50% isopropyl alcohol, 0.1 m HCl, 50 °C, 1 h) and mild alkaline methanolysis (0.1 m KOH in toluene/CH3OH (1:3), 0 °C, 1 h). Following incubation, the reactions were neutralized with either 1 m Tris or concentrated acetic acid, diluted with CHCl3, CH3OH, and 0.9% NaCl to give a final composition of 3:2:1, respectively, and partitioned as described above. The organic phases were dried under nitrogen and either quantified for radioactivity or analyzed by thin-layer chromatography and detected by iodine staining.
Phenolysis of GlcNAc-P-Und
Purified [3H]GlcNAc-P-Und was dried under nitrogen in a 12 × 100-mm conical screw-cap tube and heated to 68 °C in 0.2 ml of 50% aqueous phenol as described by Murazumi et al. (22). Following phenolysis, 0.1 ml of water was added, and the samples were thoroughly mixed. The aqueous and phenolic layers were separated, dried, and analyzed for radioactivity by scintillation spectrometry.
Analytical methods
Membrane protein concentrations were determined using the BCA protein assay kit (Pierce) employing the method of Ruiz et al. (44). Radioactivity was quantified by liquid scintillation spectrometry on a Packard Tri-Carb Liquid Scintillation Spectrometer using Econosafe Complete Counting Mixture (Research Products International, Inc., Elk Grove, IL).
Phylogenetic analysis of GacI homologs
Sequences of GacI homologs were retrieved from the non-redundant database using BLAST (59). One thousand sequences were downloaded and were manually curated to remove sequences with shorter than 90% sequence length overlap. The redundancy was reduced using CD-HIT with 0.98 cutoff (60). Sequences were manually curated to correct misannotated translation start codons. The pairwise similarities were analyzed and visualized using CLANS with an E-value cut off 1 e−80 (37, 61, 62).
Bioinformatics analysis
The TOPCONS (http://topcons.net/)4 (63) web server was employed to predict trans-membrane regions of GacI, GacJ, and GacL. Homology detection and structure prediction were performed by the HHpred server (https://toolkit.tuebingen.mpg.de/#/tools/hhpred)4 (45).
Statistical analysis
Unless otherwise indicated, statistical analysis was carried out from at least three independent experiments. Quantitative data were analyzed using the paired Student's t test. A p value equal to or less that 0.05 is considered statistically significant.
Author contributions
J. S. R., R. J. E., H. Z., A. J. M., N. M. vS., K. V. K., and N. K. designed the experiments. J. S. R., R. J. E., and N. K. performed biochemical experiments. P. D., J. C., H. Z., and A. J. M. performed MS analysis. N. K. constructed plasmids and isolated mutants. J. S. R., R. J. E., K. V. K., and N. K. wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Charles J. Waechter for encouragement and helpful discussions. Carbohydrate composition analysis at the Complex Carbohydrate Research Center was supported by the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, United States Department of Energy Grant DE-FG02-93ER20097 to Parastoo Azadi.
This work was supported in part by National Institutes of Health Grants R21AI113253 from the NIAID (to N. K.), R01GM102129 (to J. S. R.), and 1S10OD021753 (to A. J. M.) and by the Center of Biomedical Research Excellence (COBRE) pilot grant (to K. V. K., N. K., and J. S. R.) supported by National Institutes of Health Grant P30GM110787 from the NIGMS. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1–S6, Tables S1–S3, and file S1.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- GAS
- group A Streptococcus
- GAC
- group A carbohydrate
- GlcNAc-P-P-Und
- GlcNAc-pyrophosphoryl-undecaprenol
- GlcNAc-P-Und
- GlcNAc-phosphate-undecaprenol
- Ni-NTA
- nickel-nitrilotriacetic acid
- TEV
- tobacco etch virus
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- WTA
- wall teichoic acid
- Und-P
- undecaprenyl phosphate
- sWGA
- succinylated wheat germ agglutinin
- GBS
- group B Streptococcus.
References
- 1. Mistou M. Y., Sutcliffe I. C., and van Sorge N. M. (2016) Bacterial glycobiology: rhamnose-containing cell wall polysaccharides in Gram-positive bacteria. FEMS Microbiol. Rev. 40, 464–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carapetis J. R., Steer A. C., Mulholland E. K., and Weber M. (2005) The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5, 685–694 [DOI] [PubMed] [Google Scholar]
- 3. McCarty M. (1952) The lysis of group A hemolytic streptococci by extracellular enzymes of Streptomyces albus. II. Nature of the cellular substrate attacked by the lytic enzymes. J. Exp. Med. 96, 569–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lancefield R. C. (1933) A serological differentiation of human and other groups of hemolytic streptococci. J. Exp. Med. 57, 571–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Sorge N. M., Cole J. N., Kuipers K., Henningham A., Aziz R. K., Kasirer-Friede A., Lin L., Berends E. T. M., Davies M. R., Dougan G., Zhang F., Dahesh S., Shaw L., Gin J., Cunningham M., et al. (2014) The classical lancefield antigen of group a Streptococcus is a virulence determinant with implications for vaccine design. Cell Host Microbe 15, 729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coligan J. E., Kindt T. J., and Krause R. M. (1978) Structure of the streptococcal groups A, A-variant and C carbohydrates. Immunochemistry 15, 755–760 [DOI] [PubMed] [Google Scholar]
- 7. McCarty M. (1956) Variation in the group-specific carbohydrate of group A streptococci. II. Studies on the chemical basis for serological specificity of the carbohydrates. J. Exp. Med. 104, 629–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kabanova A., Margarit I., Berti F., Romano M. R., Grandi G., Bensi G., Chiarot E., Proietti D., Swennen E., Cappelletti E., Fontani P., Casini D., Adamo R., Pinto V., Skibinski D., et al. (2010) Evaluation of a Group A Streptococcus synthetic oligosaccharide as vaccine candidate. Vaccine 29, 104–114 [DOI] [PubMed] [Google Scholar]
- 9. Sabharwal H., Michon F., Nelson D., Dong W., Fuchs K., Manjarrez R. C., Sarkar A., Uitz C., Viteri-Jackson A., Suarez R. S., Blake M., and Zabriskie J. B. (2006) Group A Streptococcus (GAS) carbohydrate as an immunogen for protection against GAS infection. J. Infect. Dis. 193, 129–135 [DOI] [PubMed] [Google Scholar]
- 10. Soldo B., Lazarevic V., and Karamata D. (2002) tagO is involved in the synthesis of all anionic cell-wall polymers in Bacillus subtilis 168. Microbiology 148, 2079–2087 [DOI] [PubMed] [Google Scholar]
- 11. Meier-Dieter U., Barr K., Starman R., Hatch L., and Rick P. D. (1992) Nucleotide sequence of the Escherichia coli rfe gene involved in the synthesis of enterobacterial common antigen. Molecular cloning of the rfe-rff gene cluster. J. Biol. Chem. 267, 746–753 [PubMed] [Google Scholar]
- 12. Rush J. S., Rick P. D., and Waechter C. J. (1997) Polyisoprenyl phosphate specificity of UDP-GlcNAc:undecaprenyl phosphate N-acetylglucosaminyl 1-P transferase from E. coli. Glycobiology 7, 315–322 [DOI] [PubMed] [Google Scholar]
- 13. Yamashita Y., Shibata Y., Nakano Y., Tsuda H., Kido N., Ohta M., and Koga T. (1999) A novel gene required for rhamnose-glucose polysaccharide synthesis in Streptococcus mutans. J. Bacteriol. 181, 6556–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van der Beek S. L., Le Breton Y., Ferenbach A. T., Chapman R. N., van Aalten D. M., Navratilova I., Boons G. J., McIver K. S., van Sorge N. M., and Dorfmueller H. C. (2015) GacA is essential for Group A Streptococcus and defines a new class of monomeric dTDP-4-dehydrorhamnose reductases (RmlD). Mol. Microbiol. 98, 946–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shibata Y., Yamashita Y., Ozaki K., Nakano Y., and Koga T. (2002) Expression and characterization of streptococcal rgp genes required for rhamnan synthesis in Escherichia coli. Infect. Immun. 70, 2891–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sumby P., Porcella S. F., Madrigal A. G., Barbian K. D., Virtaneva K., Ricklefs S. M., Sturdevant D. E., Graham M. R., Vuopio-Varkila J., Hoe N. P., and Musser J. M. (2005) Evolutionary origin and emergence of a highly successful clone of serotype M1 group a Streptococcus involved multiple horizontal gene transfer events. J. Infect. Dis. 192, 771–782 [DOI] [PubMed] [Google Scholar]
- 17. Bevenue A., and Williams K. T. (1951) Further evidence indicating the specificity of the orcinol spray reagent for ketoheptoses on paper chromatograms. Arch. Biochem. Biophys. 34, 225–227 [DOI] [PubMed] [Google Scholar]
- 18. Dittmer J. C., and Lester R. L. (1964) A simple, specific spray for the detection of phospholipids on thin-layer chromatograms. J. Lipid Res. 5, 126–127 [PubMed] [Google Scholar]
- 19. Danilov L. L., Druzhinina T. N., Kalinchuk N. A., Maltsev S. D., and Shibaev V. N. (1989) Polyprenyl phosphates: synthesis and structure-activity relationship for a biosynthetic system of Salmonella anatum O-specific polysaccharide. Chem. Physics Lipids 51, 191–203 [DOI] [PubMed] [Google Scholar]
- 20. Wolucka B. A., de Hoffmann E., Rush J. S., and Waechter C. J. (1996) Determination of the anomeric configuration of glycosyl esters of nucleoside pyrophosphates and polyisoprenyl phosphates by fast-atom bombardment tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 7, 541–549 [DOI] [PubMed] [Google Scholar]
- 21. Wolucka B. A., Rush J. S., Waechter C. J., Shibaev V. N., and de Hoffmann E. (1998) An electrospray-ionization tandem mass spectrometry method for determination of the anomeric configuration of glycosyl 1-phosphate derivatives. Anal. Biochem. 255, 244–251 [DOI] [PubMed] [Google Scholar]
- 22. Murazumi N., Yamamori S., Araki Y., and Ito E. (1979) Anomeric configuration of N-acetylglucosaminyl phosphorylundecaprenols formed in Bacillus cereus membranes. J. Biol. Chem. 254, 11791–11793 [PubMed] [Google Scholar]
- 23. Yamamori S., Murazumi N., Araki Y., and Ito E. (1978) Formation and function of N-acetyloglucosamine-linked phosphoryl- and pyrophosphorylundecaprenols in membranes from Bacillus cereus. J. Biol. Chem. 253, 6516–6522 [PubMed] [Google Scholar]
- 24. Dale J. L., Cagnazzo J., Phan C. Q., Barnes A. M., and Dunny G. M. (2015) Multiple roles for Enterococcus faecalis glycosyltransferases in biofilm-associated antibiotic resistance, cell envelope integrity, and conjugative transfer. Antimicrob. Agents Chemother. 59, 4094–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reusch V. M. Jr., and Panos C. (1977) Synthesis of “group polysaccharide” by membranes from Streptococcus pyogenes and its stabilized L-form. J. Bacteriol. 129, 1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lis M., and Kuramitsu H. K. (2003) The stress-responsive dgk gene from Streptococcus mutans encodes a putative undecaprenol kinase activity. Infect. Immun. 71, 1938–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nelson D., Schuch R., Chahales P., Zhu S., and Fischetti V. A. (2006) PlyC: a multimeric bacteriophage lysin. Proc. Natl. Acad. Sci. U.S.A. 103, 10765–10770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lood R., Raz A., Molina H., Euler C. W., and Fischetti V. A. (2014) A highly active and negatively charged Streptococcus pyogenes lysin with a rare d-alanyl-l-alanine endopeptidase activity protects mice against streptococcal bacteremia. Antimicrob. Agents Chemother. 58, 3073–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Berg K. H., Ohnstad H. S., and Håvarstein L. S. (2012) LytF, a novel competence-regulated murein hydrolase in the genus Streptococcus. J. Bacteriol. 194, 627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chu M., Mallozzi M. J., Roxas B. P., Bertolo L., Monteiro M. A., Agellon A., Viswanathan V. K., and Vedantam G. (2016) A Clostridium difficile cell wall glycopolymer locus influences bacterial shape, polysaccharide production and virulence. PLoS Pathog. 12, e1005946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsukioka Y., Yamashita Y., Nakano Y., Oho T., and Koga T. (1997) Identification of a fourth gene involved in dTDP-rhamnose synthesis in Streptococcus mutans. J. Bacteriol. 179, 4411–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamashita Y., Tsukioka Y., Tomihisa K., Nakano Y., and Koga T. (1998) Genes involved in cell wall localization and side chain formation of rhamnose-glucose polysaccharide in Streptococcus mutans. J. Bacteriol. 180, 5803–5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mann E., and Whitfield C. (2016) A widespread three-component mechanism for the periplasmic modification of bacterial glycoconjugates. Can. J. Chem. 94, 883–893 [Google Scholar]
- 34. Ardiccioni C., Clarke O. B., Tomasek D., Issa H. A., von Alpen D. C., Pond H. L., Banerjee S., Rajashankar K. R., Liu Q., Guan Z., Li C., Kloss B., Bruni R., Kloppmann E., Rost B., et al. (2016) Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis. Nat. Commun. 7, 10175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gomez-Casati D. F., Martín M., and Busi M. V. (2013) Polysaccharide-synthesizing glycosyltransferases and carbohydrate binding modules: the case of starch synthase III. Protein Pept. Lett. 20, 856–863 [DOI] [PubMed] [Google Scholar]
- 36. Lairson L. L., Henrissat B., Davies G. J., and Withers S. G. (2008) Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 [DOI] [PubMed] [Google Scholar]
- 37. Frickey T., and Lupas A. (2004) CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20, 3702–3704 [DOI] [PubMed] [Google Scholar]
- 38. Hancock L. E., and Gilmore M. S. (2002) The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proc. Natl. Acad. Sci. U.S.A. 99, 1574–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Michon F., Chalifour R., Feldman R., Wessels M., Kasper D. L., Gamian A., Pozsgay V., and Jennings H. J. (1991) The α-l-(1–2)-trirhamnopyranoside epitope on the group-specific polysaccharide of group B streptococci. Infect. Immun. 59, 1690–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pritchard D. G., Gray B. M., and Dillon H. C. Jr. (1984) Characterization of the group-specific polysaccharide of group B Streptococcus. Arch. Biochem. Biophys. 235, 385–392 [DOI] [PubMed] [Google Scholar]
- 41. Teng F., Singh K. V., Bourgogne A., Zeng J., and Murray B. E. (2009) Further characterization of the epa gene cluster and Epa polysaccharides of Enterococcus faecalis. Infect. Immun. 77, 3759–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., Potter S. C., Punta M., Qureshi M., Sangrador-Vegas A., Salazar G. A., Tate J., and Bateman A. (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Skovierová H., Larrouy-Maumus G., Pham H., Belanová M., Barilone N., Dasgupta A., Mikusová K., Gicquel B., Gilleron M., Brennan P. J., Puzo G., Nigou J., and Jackson M. (2010) Biosynthetic origin of the galactosamine substituent of arabinogalactan in Mycobacterium tuberculosis. J. Biol. Chem. 285, 41348–41355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ruiz N. (2015) Lipid flippases for bacterial peptidoglycan biosynthesis. Lipid Insights 8, 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Söding J., Biegert A., and Lupas A. N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schaefer K., Matano L. M., Qiao Y., Kahne D., and Walker S. (2017) In vitro reconstitution demonstrates the cell wall ligase activity of LCP proteins. Nat. Chem. Biol. 13, 396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Linton D., Dorrell N., Hitchen P. G., Amber S., Karlyshev A. V., Morris H. R., Dell A., Valvano M. A., Aebi M., and Wren B. W. (2005) Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol. Microbiol. 55, 1695–1703 [DOI] [PubMed] [Google Scholar]
- 48. Hoff J. S., DeWald M., Moseley S. L., Collins C. M., and Voyich J. M. (2011) SpyA, a C3-like ADP-ribosyltransferase, contributes to virulence in a mouse subcutaneous model of Streptococcus pyogenes infection. Infect. Immun. 79, 2404–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Caparon M. G., and Scott J. R. (1991) Genetic manipulation of pathogenic Streptococci. Methods Enzymol. 204, 556–586 [DOI] [PubMed] [Google Scholar]
- 50. Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., and Pease L. R. (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77, 61–68 [DOI] [PubMed] [Google Scholar]
- 51. Ruiz N., Wang B., Pentland A., and Caparon M. (1998) Streptolysin O and adherence synergistically modulate proinflammatory responses of keratinocytes to group A Streptococci. Mol. Microbiol. 27, 337–346 [DOI] [PubMed] [Google Scholar]
- 52. Chaffin D. O., Beres S. B., Yim H. H., and Rubens C. E. (2000) The serotype of type Ia and III group B streptococci is determined by the polymerase gene within the polycistronic capsule operon. J. Bacteriol. 182, 4466–4477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Seiler C. Y., Park J. G., Sharma A., Hunter P., Surapaneni P., Sedillo C., Field J., Algar R., Price A., Steel J., Throop A., Fiacco M., and LaBaer J. (2014) DNASU plasmid and PSI:Biology-Materials repositories: resources to accelerate biological research. Nucleic Acids Res. 42, D1253–D1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nelson D., Loomis L., and Fischetti V. A. (2001) Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. U.S.A. 98, 4107–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Raz A., and Fischetti V. A. (2008) Sortase A localizes to distinct foci on the Streptococcus pyogenes membrane. Proc. Natl. Acad. Sci. U.S.A. 105, 18549–18554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bui N. K., Eberhardt A., Vollmer D., Kern T., Bougault C., Tomasz A., Simorre J. P., and Vollmer W. (2012) Isolation and analysis of cell wall components from Streptococcus pneumoniae. Anal. Biochem. 421, 657–666 [DOI] [PubMed] [Google Scholar]
- 57. Santander J., Martin T., Loh A., Pohlenz C., Gatlin D. M. 3rd, and Curtiss R. 3rd (2013) Mechanisms of intrinsic resistance to antimicrobial peptides of Edwardsiella ictaluri and its influence on fish gut inflammation and virulence. Microbiology 159, 1471–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Edgar R. J., Chen J., Kant S., Rechkina E., Rush J. S., Forsberg L. S., Jaehrig B., Azadi P., Tchesnokova V., Sokurenko E. V., Zhu H., Korotkov K. V., Pancholi V., and Korotkova N. (2016) SpyB, a small heme-binding protein, affects the composition of the cell wall in Streptococcus pyogenes. Front. Cell. Infect. Microbiol. 6, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 60. Huang Y., Niu B., Gao Y., Fu L., and Li W. (2010) CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Alva V., Nam S. Z., Söding J., and Lupas A. N. (2016) The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 44, W410–W415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., and Madden T. L. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tsirigos K. D., Peters C., Shu N., Käll L., and Elofsson A. (2015) The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 43, W401–W407 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.