Abstract
Application of chemical dispersants to oil spills in the marine environment is a common practice to disperse oil into the water column and stimulate oil biodegradation by increasing its bioavailability to indigenous bacteria capable of naturally metabolizing hydrocarbons. In the context of a spill event, the biodegradation of crude oil and gas condensate off eastern Canada is an essential component of a response strategy. In laboratory experiments, we simulated conditions similar to an oil spill with and without the addition of chemical dispersant under both winter and summer conditions and evaluated the natural attenuation potential for hydrocarbons in near-surface sea water from the vicinity of crude oil and natural gas production facilities off eastern Canada. Chemical analyses were performed to determine hydrocarbon degradation rates, and metagenome binning combined with metatranscriptomics was used to reconstruct abundant bacterial genomes and estimate their oil degradation gene abundance and activity. Our results show important and rapid structural shifts in microbial populations in all three different oil production sites examined following exposure to oil, oil with dispersant and dispersant alone. We found that the addition of dispersant to crude oil enhanced oil degradation rates and favored the abundance and expression of oil-degrading genes from a Thalassolituus sp. (that is, metagenome bin) that harbors multiple alkane hydroxylase (alkB) gene copies. We propose that this member of the Oceanospirillales group would be an important oil degrader when oil spills are treated with dispersant.
Introduction
Crude oil is introduced into the marine environment through natural geophysical processes at an estimated rate of 700 million liters per year (Kvenvolden and Cooper, 2003; Committee on Oil in the Sea: Inputs, Fates, and Effects et al., 2003). Constant exposure of native microbes to low concentrations of hydrocarbons allows the maintenance in the environment of hydrocarbonoclastic bacteria that can readily use oil-derived compounds as carbon and energy sources (Head et al., 2006; Widdel et al., 2010). In the context of the recent Deepwater Horizon (DWH) MC252 oil spill in the Gulf of Mexico, which released 750 million liters of oil into the gulf, there is now scientific consensus that hydrocarbonoclastic microorganisms played a major role in the removal of hydrocarbons from the ecosystem (American Academy of Microbiology, 2011).
Chemical dispersants act by lowering the interfacial tension between oil and water leading to oil emulsification into tiny droplets, which in turn increases bioavailability of crude oil to natural hydrocarbon degraders (Brakstad et al., 2015b). For many decades, dispersants have been used in catastrophic oil spills to help increase oil degradation rates and minimize oil delivery to shorelines (Harris and Chris, 1995; Law and Carole, 2004; Henry and Charlie, 2005; Steen et al., 2008; Bejarano et al., 2013), but because of ecological trade-offs (Smith, 1968), their deployment is controversial and their effectiveness is debated (Committee on Understanding Oil Spill Dispersants: Efficacy and Effects et al., 2005; Kleindienst et al., 2015b).
Currently, Newfoundland and Labrador is home to three active offshore oil projects: Hibernia; Terra Nova; and White Rose, while the Scotian Shelf near Sable Island has a major gas condensate-producing platform. These platforms represent the major hydrocarbon-producing systems off eastern Canada. In this study, we simulated a crude oil spill at the Hibernia and Terra Nova platforms and a gas condensate spill at the Thebaud (Sable Island) platform. Recent studies have identified bacteria from more than 79 genera that are able to degrade hydrocarbons, and several of these, including Alcanivorax, Cycloclasticus, Oleiphilus, Oleispira, Thalassolituus and some members of the genus Planomicrobium ( Mason et al., 2012; Reddy et al., 2012; Redmond and Valentine, 2012; Valentine et al., 2012; Kleindienst et al., 2016), get their carbon almost exclusively from hydrocarbons (Prince, 2010; Prince et al., 2010). Many DWH-related studies showed that microorganisms belonging to the order Oceanospirillales were involved in oil degradation (Hazen et al., 2010; Valentine et al., 2010; Kessler et al., 2011; Redmond and Valentine, 2012; Gutierrez et al., 2013) and that they possessed active oil degradation genes (Mason et al., 2012; Rivers et al., 2013), but little information was presented regarding the hydrocarbon-degrading activities of the different members of this order during an oil spill.
We applied shotgun metagenomic and metatranscriptomic analyses to evaluate the natural microbial community response following hypothetical crude oil or gas condensate spills. The biodegradation of physically and chemically dispersed gas condensate and crude oil was monitored by conducting nutrient-enhanced microcosm studies using summer and winter sea water (SW) freshly procured from the three different locations. The latest developments in bioinformatics algorithms enables the recovery of complete or draft bacterial genomes from metagenomic data sets, which is referred to as metagenome binning. In this work, we conducted a binning-centric instead of the more classical contig-based approach to analyze our sequencing data. A metagenome bin represents a draft version of a microbe’s entire genome, which allows for unprecedented population dynamics resolution in the analysis and interpretation of genomics data. This approach is gaining traction (Sangwan et al., 2016) and is increasingly being implemented for environmental sequence data analysis (Hugerth et al., 2015; Hultman et al., 2015; Evans et al., 2015). Here we found that the diversity of rare resident marine microbes of this region enabled an efficient response to experimental gas condensate and crude oil spills, and that the addition of dispersant in microcosms improved the n-alkane, but not the polycyclic aromatic hydrocarbon (PAH), degradation rate. More importantly, we provide high-resolution analyses that for the first time put in contrast the relative contribution of various members of the Oceanospirillales group. We demonstrated that the presence of dispersant favored the proliferation of a Thalassolituus metagenome bin that showed high expression levels of the multiple alkB genes it harbored.
Materials and methods
Sampling and microcosm setup
SW samples, from locations in the vicinity of oil and natural gas production platforms in offshore Newfoundland and Labrador and Nova Scotia, were collected from the sea surface (3–5 m depth), using a Sea-bird Niskin rosette frame (24 10 l bottles) (Sea-Bird Scientific, Bellevue, WA, USA) cast from the Canadian Coast Guard research vessel, Hudson, in the summer and late fall/winter of 2013. Summer (10–19 July 2013) and winter (14 November–10 December 2013) SW was sampled from Hibernia (3 m depth in summer (46.707°N and −48.785°W) and 5 m depth in winter (43.858°N and −60.157°W)), Terra Nova (3 m depth in summer (46.431°N and −48.480°W) and 5 m depth in winter (47.000°N and −48.288°W)) and Thebaud (4 m depth in summer (43.858°N and −60.157°W) and 3 m depth in winter (47.000°N and −48.471°W)) production platforms (Supplementary Information File 3—Water collection for microcosms). Microcosm studies were initiated, onboard the ship, as soon as SW was procured from the reference sites. Triplicate SW samples were recovered from three separate bottles of the Niskin rosette at each sampling site using 20 l acid-washed jerricans for microcosm preparation. For microcosms, the SW (100 ml) was transferred to 250 ml baffled flasks. Biostimulation with nutrients is an effective method for augmenting the rate of oil bioremediation (Atlas and Bartha, 1972, 1973; Bragg et al., 1994; Venosa et al., 1996; Röling et al., 2002; Coulon et al., 2007; McKew et al., 2007b). We learned from our experience that hydrocarbon degradation in the Arctic and Gulf of Mexico surface SW happens very slowly, if at all, when nutrients are limiting (Yergeau et al., 2015 and unpublished observations). For the objective of having effective operational conditions to observe the effect of dispersant in an adequate time frame, nutrients were added to all microcosms. Therefore, Bushnell-Haas (Difco, Becton Dickinson and Company, Mississauga, ON, Canada) nutrients (2 ml), weathered oils, condensate and premixed dispersant (COREXIT EC 9500 A - Nalco Energy Services, Burlington, ON, Canada)) with oil or condensate were added to each flask/bottle. The final concentrations of COREXIT EC 9500 A in microcosms were as follows: oil-with-dispersant microcosms were setup at 5.4 p.p.m. (1:186, 281 v/v); condensate-with-dispersant microcosms had 6.0 p.p.m. (1:167, 364 v/v); and dispersant-only microcosms had 7.3 p.p.m. (1:137, 461 v/v). Oil and condensate concentrations were 107.4 (1:20 v/v) and 119.5 p.p.m. (1:20 v/v), respectively.
The flasks were incubated at the ambient surface SW temperature, at the time of collection, and continually mixed on Thermo MaxQ 2000 orbital shaker tables (Thermo Scientific, Ottawa, ON, Canada) at 150 r.p.m. Microcosms were set up directly onboard ship where experiments were conducted at 6 and 11 °C for winter and summer conditions, respectively. Setup and sampling of microcosms are described in detail in Supplementary Information File 3—Microcosms experimental designs (Supplementary Table SM3). Treatments (SW control, SW+Bushnell-Haas+oil (or condensate)) and SW+Bushnell-Haas+oil (or condensate)+dispersant (COREXIT EC 9500 A), SW+dispersant were run in triplicates for each time point. Oil and dispersant concentration calculations are detailed in Supplementary Methods (Supplementary Information File 3—Supplementary Tables SM4–SM6). Two parallel sets of microcosms were prepared: one set for molecular analyses of DNA and RNA, hereby termed molecular microcosms and another set for chemistry analyses, termed chemistry microcosms. A total of 130 molecular microcosms and 234 chemistry microcosms were prepared (Supplementary Information File 3—Supplementary Table SM3).
Chemical and molecular analyses
Hydrocarbon concentrations were monitored during the course of the experiment at the specified time points (T=3, 10, 15, 28 and 42 days) from chemistry microcosms. Water samples were processed using liquid–liquid extraction (modified version of US Environmental Protection Agency Method 3510C). Further details can be found in previous studies (Cole et al., 2007; King et al., 2015b). Purified extracts of water from the microcosms were analyzed using high-resolution gas chromatography (Agilent 6890 GC) coupled to an Agilent 5973 N mass selective detector (Wilmington, DE, USA) operated in the selective ion-monitoring mode using the following GC (MDN-5 S column 30 m × 0.25 mm inner diameter, 0.25 μm film thickness, Supelco, Mississauga, ON, Canada) conditions: cool on-column injection with oven track mode (tracks 3 °C higher than the oven temperature program); 80 °C hold 2 min; ramp at 4 °C min−1 to 280 °C; hold 10 min.
Quantification criteria for PAH included retention time matching, that is, within ±0.010 min of the retention time of the standard, and comparing the relative abundance of the qualifying ion(s) (±10%), the molecular ion and one or more qualifier ions in the mass spectrum of the compound with the commercial standard. Seven levels of PAH standards were used to calibrate the system. The Auto-Quant software (Agilent, Palo Alto, CA, USA) was used to interpret and quantify analytes detected in the extracts. All processed data were compared to established limits to pass quality control. These limits have been set at ±30% of the true value compared to experimental values. If results fell outside the limits they were repeated. The same was true for reference materials used to track analytical quality.
Initial SW samples (2 l) were filtered (<10 psi) in triplicates onto Millipore 0.22 μm polyethersulfone membranes for T=0 collection of genomic material for total nucleic acid extraction. The filter was transferred into a 50 ml Falcon tube and immediately flash frozen by submersion in liquid nitrogen and stored in a −20 °C freezer onboard the ship before being transferred to a −80 °C freezer. The genomics microcosms were killed at T=5 and 42 days for the summer study and T=7 and 42 days for the winter study by filtering (⩽10 psi) the contents of each flask through a 0.22 μm filter (as described above).
Total nucleic acids were recovered using a modified version of the hexadecyl trimethyl ammonium bromide method of Ausubel (2002). The modifications were as follows: the incubation time for the Tris-EDTA/lysozyme treatment was reduced substantially from 1 h to 15 min and the temperature for this step was increased from 37 to 56 °C. The hexadecyl trimethyl ammonium bromide/NaCl incubation was followed by phenol/chloroform/isoamyl alcohol (25:24:1) then a chloroform/isoamyl alcohol extraction steps. DNA was quantified using Quant-iT PicoGreen assay (ThermoFisher Scientific, Mississauga, ON, Canada) (Invitrogen, Life Technologies, Burlington, ON, Canada) and 1 ng of gDNA was used as a template to construct the sequencing library, using the Illumina (San Diego, CA, USA) Nextera XT library preparation protocol following the manufacturer’s instructions. However, the ‘library normalization’ step was omitted and normalization was instead performed by pooling equal amounts of libraries after Quant-iT PicoGreen quantification. The quality of the pooled library was assessed (http://support.illumina.com/sequencing/sequencing_kits/nextera_xt_dna_kit/documentation.html) using an Agilent 2100 Bioanalyzer with a High Sensitivity DNA Kit. A similar methodology was used for RNA workflow with the exception that RNA was quantified using the Quant-iT RiboGreen assay and that Ribosomal RNAs were removed and library preparation was performed using ScriptSeq Complete Kit Bacteria Low-Input Library Prep protocol according to the manufacturer’s (Epicentre, Madison, WI, USA)) instructions starting with 100 ng total RNA.
Genomics and bioinformatics
Metagenomic and metatranscriptomic libraries were prepared and sequenced on an Illumina HiSeq2000 system on a 2 × 100 bp configuration. A total of 203 and 126 samples were submitted for metagenome and metatranscriptome sequencing, respectively. Sequencing data (456 Gb for metagenome and 560 Gb for metatranscriptome) was processed through our metagenomics and metatranscriptomics bioinformatics pipelines (Supplementary Information File 3—Supplementary Figure SM1). Read count summaries and insert sizes are provided for both metagenome (Supplementary Information 2 Data Sets—Supplementary Table S5) and metatranscriptome (Supplementary Information 2 Data Sets—Supplementary Table S6) sequencing libraries. Sequencing adapters were removed from each read and bases at the end of reads having a quality score <30 were cutoff (Trimmomatic v0.32; Bolger et al., 2014) to generate quality-controlled reads.
Quality control-passed reads from each sample were co-assembled using Ray software v2.3.1 (Boisvert et al., 2012) with a kmer size of 31 (see Supplementary Information 2 Data Sets—Supplementary Table S7 for assembly statistics). Gene prediction was performed by calling genes on each assembled contig using MetageneMark v1.0 (Tang et al., 2013). Genes were annotated following the JGI’s guidelines (Huntemann et al., 2016): (1) RPSBLAST (v2.2.29+) (Camacho et al., 2009) against COG database (for example, CDD v3.11); (2) RPSBLAST (v2.2.29+) against KOG database (for example, CDD v3.11). The best hit having at an e-value ⩽1e-02 was kept for each query; (3) HMMSCAN (v3.1b1) (Eddy, 2011) against PFAM-A (v27.0) database (Finn et al., 2013). Best hit having at least an e-value ⩽1e-02 was kept for each query; (4) TIGRFAM database (v15.0). Best hit having at least an e-value ⩽1e-02 was kept for each query; (5) BLASTP (v2.2.29+) against KEGG database v71.0; and (6) BLASTN (v2.2.29+) against the nucleotide database (version of 16 May 2013) of National Center for Biotechnology Information. Contig (and not gene) sequences were blasted against the NCBI’s nucleotide database as well for taxonomic assignment. For each of these database comparisons, the best hit having at least an e-value ⩽1e-02 and alignment length ⩾90 bp was kept for each query. Quality control-passed reads were mapped (BWA mem v0.7.10) (unpublished—http://bio-bwa.sourceforge.net) against contigs to assess quality of metagenome assembly and to obtain contig abundance profiles. Alignment files in bam format were sorted by read coordinates using samtools v1.1 and only properly aligned read pairs were kept for downstream steps. Each bam file (containing properly aligned paired read only) was analyzed for coverage of called genes and contigs using bedtools (v2.17.0) (Quinlan and Hall, 2010) using a custom bed file representing gene coordinates on each contig. Only paired reads both overlapping their contig or gene were considered for gene counts. Coverage profiles of each sample were merged to generate an abundance matrix (rows=contig and columns=samples) for which a corresponding counts per million (CPM) abundance matri x (edgeR v3.10.2) (Robinson et al., 2010) was generated.
Taxonomy of each contig was assigned using the NCBI taxonomy database (Sayers et al., 2009; Benson et al., 2009) (ftp://ftp.ncbi.nih.gov/pub/taxonomy/taxdump.tar.gz) (as downloaded on 5 June 2015). Each Geninfo Identifier (GI) resulting from BLASTN against nucleotide were used to retrieve full taxonomic lineages (when available) from the NCBI taxonomy database. Taxonomic lineages were integrated to the contig abundance of read counts matrix to generate an Operational Taxonomic Unit (OTU) table format file (with contigs replacing OTUs as rows). Taxonomic summaries were performed using a combination of in-house Perl and R scripts and Qiime v.1.9.0 (Caporaso et al., 2010). Bin abundance tables along with their taxonomic lineages are included in Supplementary Table S3 (Supplementary Information 2 Data Sets).
Binning was performed using Metabat (v0.26.1) (Kang et al., 2015a) and an abundance matrix was generated using the jgi_summarize_bam_contig_depths software (Kang et al., 2015b) with —minContigLength 1000 —minContigDepth 2 and —minContigIdentity 95 parameters. Bins obtained from Metabat were further processed/decontaminated by splitting each bin into three sub-bins based on the assigned taxonomic lineage at the order level as each bin typically had a significant amount of contigs associated with the same order taxon. For instance in our data set, the bin labeled ‘1’ had 947 contigs assigned to the Oceanospirillales order level, 3 contigs to the Alteromonadales and 1 contig to the Burkholderiales. Consequently, three sub-bins were generated and labeled 1-Oceanospirillales, 1-Alteromonadales and 1-Burkholderia, respectively. Sub-bins were kept only if they had a cumulative contig length of at least 100 kb and contained at least 10 contigs. To focus on the most relevant bins we kept a selection of the 20 most abundant bins out of the 601 total bins. Quality assessment of bins was done using CheckM v1.0.4 (Parks et al., 2015) and detailed in Supplementary Information 2 Data Sets (Supplementary Table S2).
For metatranscriptomic data processing, quality control-passed reads were mapped (BWA mem v0.7.10) against contigs from the metagenome assembly following alignment sorting (samtools v1.1) and gene abundance computation (bedtools v.217.0). According to our experimental design, differentially expressed genes were assessed with edgeR (v3.10.2) using its generalized linear model approach detailed by the authors (https://www.bioconductor.org/packages/3.3/bioc/vignettes/edgeR/inst/doc/edgeRUsersGuide.pdf—section 3.2.3, page 29) with metatranscriptomic raw count matrices as input. Genes having a log fold-change ratio ⩾|1.5| and false discovery rate <0.05 were considered as differentially expressed. Metadata for all samples reported in this study are available in Supplementary Table S8 (Supplementary Information 2 Data Sets).
To increase detection power in searching for alkane 1-monooxygenase genes (alkB), we made multiple alignments (MUSCLE v3.8.31) (Edgar, 2004) of each accession number previously reported (Nie et al., 2014). Markov models (named alkB, alkBmulti2 and alkBmulti3) were generated (hmmbuild followed by hmmpress, both with default parameters) from these alignments and gene sequences were compared against these training sets with hmmscan (Hmmer v3.1b1). Hits were integrated with Pfam-A database search described above and hits having an e-value ⩽1e-10 and query length ⩾100 and alignment length ⩾100 were kept. Among these matches, hits matching FA_desaturase (PF00487.19) or our newly added alkB models were considered as potential alkB genes. We applied the same procedure for genes mcp, cheR, gspE, gspF and ompS previously reported (Wang and Shao, 2014).
Availability of data
Raw sequence reads of the shotgun metagenomic and metatranscriptomic data were submitted to the sequence read archive under accession no. SRP079000 under Bio Project PRJNA329908. Metagenome bin sequences can be found under NCBI accession no. SAMN072553[31-50].
Results and discussion
Oil degradation in microcosms
The degradation rates in both summer and winter for each of the three sites (Hibernia, Terra Nova and Thebaud), in addition to the half-lives of the different hydrocarbon components (alkanes, PAHs and alkylated PAHs) are summarized in Figure 1; Supplementary Information File 1—Supplementary Figures S1 and S2; and Supplementary Information 2 Data Sets—Supplementary Table S1. Degradation rates were typically higher in the summer than in the winter for all hydrocarbon components at all three sites. On the basis of the observed degradation kinetics, alkanes were degraded faster at all three sites in comparison to PAHs and alkylated PAHs, which is in agreement with the general order of hydrocarbon compound degradation (Perry, 1984) and with what was observed during the DWH spill (Hazen et al., 2010; Valentine et al., 2010; Kessler et al., 2011; Mason et al., 2012; Redmond and Valentine, 2012; Rivers et al., 2013; Gutierrez et al., 2013). The positive effect of dispersant application was most noted for n-alkanes, especially in the winter, whereas no clear impacts were identified with PAH and alkylated PAH degradation rates.
Figure 1.
Total residual alkane concentration in Hibernia, Thebaud and Terra Nova summer and winter microcosms. Concentrations are normalized to hopane. Treatments are SW plus oil and SW plus oil with dispersant. All microcosms received Bushnell-Haas nutrients. Time of killing is shown by colored bars. Statistical significance was assessed with an analysis of variance followed by a post hoc Tukey test. ***Adjusted P-value <0.001; *Adjusted P-value <0.05.
For both seasons, dispersant had a positive effect on alkane degradation, which contains n-alkanes of various lengths (C10–C35). In the summer, conditions appeared to be more conducive to degradation even without dispersant, but the degradation rate of the alkane fraction was still positively affected by the presence of dispersant. Gas condensate (Thebaud) on the other hand, contained more readily bioavailable short n-alkanes (C15 and shorter) that were quickly degraded without the apparent need for dispersant (both seasons) and in the winter it appeared that dispersant might actually impair the degradation of condensate compounds (Supplementary Information File 1—Supplementary Figures S1 and S2). The effectiveness of chemical dispersants in low-temperature SW has shown conflicting results with studies reporting improved degradation of crude oil with dispersant, while other studies have shown a neutral to negative effect (reviewed in Cowton and Michael (2016)). More recently, dispersant was found to have a negative impact on the hydrocarbon-degrading activity of indigenous bacteria (Kleindienst et al., 2015b). We did not see a negative impact of dispersant, especially on alkane degradation at our study sites. On the contrary, our data indicated that dispersant had a positive effect on n-alkane degradation rates. However, we did not find any clear benefit for dispersant use in gas condensate where longer n-alkanes are absent. One possibility for the difference observed between our results and those of other studies may be due to the different dispersant concentrations that were used. For instance, Kleindienst and co-workers (2015b) set up their microcosms to have a final COREXIT concentration of 15 p.p.m. (1:66, 538 v/v) and 26.8 p.p.m. (1:37, 362 v/v) for their dispersant-only and oil-with-dispersant microcosms, respectively (Lewis, 2015; Kleindienst et al., 2015b). In this study, the COREXIT concentration was significantly lower with values of 7.3 p.p.m. (1:137, 461 v/v) and 5.4 p.p.m. (1: 186, 281 v/v) for dispersant and oil-with-dispersant microcosms, respectively. Except for the 42-day incubation time point in Thebaud winter microcosms, for which dispersant seemed to have a negative effect on short-chain alkane degradation, our results are generally in agreement with others (Lunel et al., 1997; Baelum et al., 2012; Lee et al., 2013; Prince and Butler, 2014) regarding the use of dispersant to stimulate oil degradation in SW. However, our data also suggest that while dispersant favors the degradation of n-alkanes, it does not affect the degradation of aromatic compounds. Our microcosms were all supplemented with inorganic nutrients (Bushnell-Haas), which was reported to select against Cycloclasticus sp. (Singh et al., 2014), an aromatic compound-degrading microbe. In consequence, presence of inorganic nutrients may also be impacting the degradation of aromatic compounds.
Effects of oil on microbial community structures
Our metagenomic analysis pipeline gave us 601 metagenome bins (Supplementary Information 2 Data Sets—Supplementary Tables S2 and S3). From a total of 943 147 contigs, 91 847 made it into these bins, representing an integration rate of 9.74%. Community taxonomic profiles were highly similar (Supplementary Information File 1—Supplementary Figure S3) between contig-centric and bin-centric approaches, which suggests that our bins captured most of the microbial diversity present in our samples as assessed by a contig-based approach. To obtain a global image of the community dynamics in our microcosms, we computed alpha diversity (observed species/bins and Shannon indexes) and beta diversity (Bray–Curtis) metrics (Supplementary Information File 1—Supplementary Figures S4 and S5). As expected, T0 conditions usually harbor more diverse communities than T5 or T42 (analysis of variance: F=18.99, P<000.1) with relatively high observed bins and Shannon index values of more than 200 and 6, respectively. Microcosms treated with oil, dispersant and oil with dispersant showed similar diversity index values. Microcosms with dispersant only presented intermediate diversity indexes: lower than the control conditions, but higher than oil or oil with dispersant. Alpha diversity indexes drop more rapidly in treatment conditions in the summer than in the winter except for the Terra Nova site, where oil with dispersant had more diverse communities in the winter. Beta diversity plots and Permanova tests performed on bin abundance (Supplementary Information File 1—Supplementary Figure S5) showed that samples cluster primarily by treatment (that is, substrate and incubation time) indicating that these two variables are the main drivers in the formation of distinct communities. Interestingly, the control conditions of all sites clustered together as well, suggesting a similar community at time zero across all sites.
We narrowed down our analysis by selecting the 20 most abundant bins across all samples for downstream analyses (Supplementary Information 2 Data Sets—Supplementary Table S2) and established their corresponding microbial community profiles (Figure 2). Colwellia bins were mostly observed at Terra Nova at day 5 in the summer and winter with oil and oil with dispersant, and were also highly enriched at day 5 in Hibernia summer microcosms with oil only. Cycloclasticus bins were highly abundant at Thebaud at 42 days in oil with dispersant in winter and to a lesser extent in the summer for the same time point. In contrast to the crude oil substrate used at the other two stations, the gas condensate from the Thebaud station contained several orders of magnitude higher PAH and alkylated PAH concentrations (Supplementary Information File 1—Supplementary Figures S1 and S2), which probably provided an ideal environment for Cycloclasticus, a well-known PAH degrader (Harayama et al., 2004; Head et al., 2006; Yakimov et al., 2007; Coulon et al., 2007; McKew et al., 2007b; Cui et al., 2008; Niepceron et al., 2010). Our results are consistent with DWH spill microbial community succession where Cycloclasticus and Colwellia were shown to be abundant (Valentine et al., 2010) and active (Redmond and Valentine, 2012; Gutierrez et al., 2013) during the spill. Colwelliaceae OTUs were also reported to increase in abundance in the later stages of microbial succession in microcosm experiments (Baelum et al., 2012). Our data suggest that Cycloclasticus is also well adapted for cold temperature PAH degradation.
Figure 2.
(a) Microbial community profiles of the most abundant 20 metagenome bins in Hibernia, Thebaud and Terra Nova summer and winter microcosms. Treatments are SW plus oil (light blue) at 5 or 7 days and 42 days; oil with dispersant (light purple) at 5 or 7 days and 42 days; and dispersant only (dark white) at 42 days. Summer microcosms were killed at 5 and 42 days and winter microcosms were killed at 7 and 42 days. Each color represent a taxon, each delimited box represent a bin and each column represent a sample. Bin identities are listed below each of their corresponding taxonomic assignment. (b) Microbial community profiles of Alcanivorax bins found in Hibernia, Thebaud and Terra Nova summer and winter microcosms. Treatments are SW plus oil (light blue) at 5 or 7 days and 42 days; oil with dispersant (light purple) at 5 or 7 days and 42 days; and dispersant only (light pink) at 42 days. Summer microcosms were killed at 5 and 42 days and winter microcosms were killed at 7 and 42 days. Each color represents an Alcanivorax metagenome bin. (c) Microbial community profiles of Marinobacter bins found in Hibernia, Thebaud and Terra Nova summer and winter microcosms. Treatments are SW plus oil (light blue) at 5 or 7 days and 42 days; oil with dispersant (light purple) at 5 or 7 days and 42 days; and dispersant only (light pink) at 42 days. Summer microcosms were killed at 5 and 42 days and winter microcosms were killed at 7 and 42 days. Each color represents a Marinobacter metagenome bin and each column represents a sample.
Glaciecola was observed in tandem with either a Pseudoalteromonas (Terra Nova; winter with dispersant) or an Alteromonas (Terra Nova and Thebaud at 5/7 days with oil and oil with dispersant) bin. Alteromonas species are metabolic generalists capable of rapidly adapting to environmental disturbances (reviewed in Dang and Lovell (2016)) and were also seen (Hazen et al., 2010; Valentine et al., 2010; Dang and Lovell, 2016) and isolated (Gutierrez et al., 2013) in the DWH water column. Glaciecola and Pseudoalteromonas are associated with biodegradation in cold environments (Yakimov et al., 2004; Deppe et al., 2005; Brakstad and Kristin, 2006), which our data supports, as Pseudoalteromonas thrived in crude oil with dispersant at Hibernia and Terra Nova in the winter. Glaciecola was also well represented in the winter at these two locations, but seemed to be more successful in the summer.
Alcanivorax is a widely distributed hydrocarbonoclastic bacterium observed in oil-impacted environments around the globe (Head et al., 2006), including the recent DWH oil spill (Joye et al., 2014). This was reflected in our metagenomic data, where Alcanivorax bins were well represented in summer microcosms (and winter microcosms to a lesser extent) containing oil and oil with dispersant at day 42 (Figure 2a). At Hibernia, Alcanivorax bins are affluent at 5 days in oil-with-dispersant, but not in oil-only summer microcosms. Alcanivorax includes five distinct bins that are differentially abundant depending on the location and treatment (Figure 2b). The most abundant bin was clearly 2-Oceanospirillales, usually followed by 36-, 82- and 1-Oceanospirillales while at Terra Nova and Thebaud, 1-Oceanospirillales stands out as the most dominant bin. A significant proportion of 2- and 36-Oceanospirillales bins were also observed at these two locations. At Thebaud, Alcanivorax bins are mainly represented by 1-Oceanospirillales in contrast to other locations that contain a more diversified Alcanivorax population, possibly because it contained gas condensate instead of crude oil. In the aftermath of the DWH oil spill, key microorganisms were identified to be intimately associated with oil degradation, one of which were members of the Oceanospirillales order. This large group comprises many subgroups at lower taxonomic levels (that is, family, genus and species) and despite many studies characterizing plume microbial populations, little information is available on the exact identity of these Oceanospirillales. Phylochip 16S rRNA gene surveys at about 1.5 km from the well head revealed that the plume microbial community was dominated by Oceanospirillales (Hazen et al., 2010). Hazen et al. (2010) also observed a Thalassolituus (also belonging to Oceanospirillales), but did not indicate its abundance; according to their results, the vast majority of their phylochip OTUs belonged to an uncultured Oceanspirillales. The 16S rRNA gene Sanger clone surveys reported the involvement of Colwellia and Cycloclasticus in propane, ethane (Valentine et al., 2010) and methane (Kessler et al., 2011) degradation. Kessler et al. (2011) also observed Oceanospirillales in low overall abundance. Mason et al. (2012) thoroughly characterized microbial communities using various nucleic acid data types. They reported 16S rRNA gene amplicon data showing high abundance of Oceanospirillales, but did not obtain enough resolution to further characterize that OTU at the genus level. They also reported a modest amount of reads associated with Colwellia and Cycloclasticus. Their shotgun metagenome data also showed that the majority of their reads were assigned to the Oceanspirillales and metatranscriptomic analyses revealed Oceanospirillales to be transcriptionally active. From their samples, they did single-cell sequencing and using partial 16S rRNA gene alignment, found that one particular single cell, carrying oil degradation gene functions, had similarity to both Oleispira antarctica and Thalassolituus oleivorans, but did not assign a definitive taxon and labeled their single-cell isolate as belonging to Oceanspirillales using 16S rRNA gene amplicons mostly classified at the class or order level. Another study observed that Oceanospirillales OTUs were very dominant in a set of plume samples (Redmond and Valentine, 2012). Rivers et al. (2013) performed metatranscriptomic and 16S rRNA gene amplicon sequencing and found Oceanspirillales OTUs in high abundance in plume samples. More specifically, they were assigned to the genera Neptuniibacter and Bermanella. From their metatranscriptome data set, transcripts of the alkB gene were found in high abundance. They reported that the majority of their transcripts mapped to six genomes, of which two were Oceanospiralles: Bermanella marisrubri and Neptuniibacter caesariensis. Alcanivorax and Marinobacter OTUs were specifically identified to be dominant in the presence of n-hexadecane and were also found in fairly high abundance in DWH plume samples (Redmond and Valentine, 2012; Gutierrez et al., 2013).
Bin abundance of Marinobacter was dependant on platform location and season, but initial communities (T0) showed remarkably similar abundances across all three sites (Figure 2c) with 16-Alteromonadales being the most abundant followed by 10- and 25-Alteromonadales. Generally, at 42 days, Marinobacter is fairly abundant in the winter in oil-only and dispersant-only microcosms, while it is in lower abundance in oil-with-dispersant microcosms. A previous study found that COREXIT mixed with either marine broth or marine broth with oil was toxic to Marinobacter hydrocarbonoclasticus(Hamdan and Fulmer, 2011). More recently, it was reported that microcosms containing relatively high dispersant concentrations prevented the dominance of Marinobacter OTUs (Kleindienst et al., 2015b; Lewis, 2015). Our results further expand on these observations and suggest that certain Marinobacter species are less competitive in a substrate containing both oil and dispersant, but can otherwise thrive when only one or the other of these two substrates is present. This is particularly apparent at Terra Nova where 10-Alteromonadales is highly abundant in oil-only microcosms at 42 days while 25- and 16-Alteromonadales are found in low abundance. In contrast, in dispersant-only microcosms for the same time point, 10- and 25-Alteromonadales are similarly abundant (Figure 2c), which suggests that certain Marinobacter species (for example, metagenome bins) may actually grow very well in the presence of dispersant.
Our results indicated a succession of microorganisms over time with more easily degradable compounds such as short-chain alkanes being the first to be depleted. This type of succession was also observed during the DWH oil spill (Atlas and Hazen, 2011; Joye et al., 2014; Kostka et al., 2014; King et al., 2015a) and more generally in marine environments (Röling et al., 2002; Head et al., 2006; Yakimov et al., 2007; Berthe-Corti* and Nachtkamp, 2010; Greer, 2010; McGenity et al., 2012). This may have been modulated by the presence of dispersant, the nature of the added substrates (gas condensate or crude oil), the sampling season and the incubation temperature. Although crude oil was added to both Hibernia and Terra Nova microcosms, they presented different successional patterns over time, which is especially noticeable in the abundance profiles of Alcanivorax bins (Figure 2b). While the community structures were very similar under control conditions (T=0) these communities were not truly identical and their initial state may have had an outcome on population composition later under the different treatments. However, Alcanivorax and Marinobacter bins had very similar abundance profiles at T0 across all sites and seasons (Figures 2b and c), which does not suggest a correlation between initial abundance profiles and later population composition. From a population dynamics perspective, bacteria appearing as dominant after oil addition were initially found to constitute <0.5% of the total bin abundance in the controls (Supplementary Information 2 Data Sets—Supplementary Table S4) and could be considered as rare microorganisms. The microbial assemblages in a given environment are thought to be governed, in part, by stochastic processes in the early phases of community succession (Dini-Andreote et al., 2015) and this could explain, to a certain degree, the community differences we observed between Hibernia and Terra Nova at 5/7 and 42 days.
Effect of dispersant on microbial community structure
The addition of dispersant to oil had an apparent effect on the structure of microbial communities at day 5 (summer), but not so much at 42 days (Figure 2; Supplementary Information File 1—Supplementary Figure S5). The addition of dispersant in winter microcosms also seemed to positively influence growth of certain microbes. In oil-only microcosms, microbes were usually in very low abundance at 7 days in the winter while they were already thriving at 5 days in the summer (cumulative CPM values in Figure 2). In contrast, in oil-with-dispersant winter microcosms, microbial communities were already in high abundance at day 7. We included nutrients and nutrients-with-dispersant controls at 42 days for all microcosm series and found microbial community profiles to be very different between these two conditions (Figure 2a). In general, nutrient-only microcosms had comparable (summer) or lower (winter) overall microbial abundance compared to nutrients-with-dispersant microcosms. Notably, at Terra Nova in the winter, the 25-Alteronomonas bin is abundant in nutrients-with-dispersant microcosms but virtually absent in nutrient-only microcosms (Figure 2c). Similarly, Marinobacter (10-, 25- and 16-Alteromonadales) are highly abundant in nutrients-with-dispersant microcosms, but in very low abundance in nutrient-only microcosms at Thebaud in the winter (Figure 2c). Conventional wisdom suggests that dispersant stimulates oil biodegradation by increasing its bioavailability in water, but it could also act as a carbon source boosting microbial growth (Mulkins-Phillips and Stewart, 1974; Lindstrom and Braddock, 2002; Chakraborty et al., 2012) and ultimately leading to faster oil degradation. Alternatively, it is possible that microbes growing on dispersant are producing metabolites that are in turn consumed by oil degraders as previously suggested (Röling et al., 2004). At Hibernia and Terra Nova in the winter, where the alkane degradation rate was clearly enhanced by dispersant, microbial profiles at 7 days are consistently dominated by a Pseudoalteromonas bin (5-Alteromonadales) in oil-with-dispersant microcosms (Figure 2). For the same winter time point in Hibernia, this Pseudoalteromonas bin is always accompanied with a Thalassolituus bin (18-Oceanospirillales) while at Terra Nova, it is associated with Colwellia, Alteromonas and Glaciecola bins, and the absence of Thalassolituus. Alcanivorax bins eventually dominated at 42 days in oil-with-dispersant winter microcosms at Hibernia while no clear picture emerged from Terra Nova because of the variation in biological replicates.
Interestingly, differences in population profiles in oil compared to oil with dispersant is mainly explained by the proliferation of a Thalassolituus bin (18-Oceanospirillales), which is consistently seen in high abundance at 5 days in oil-with-dispersant microcosms at all three sites in the summer (Figure 2). The only winter condition where the presence of a Thalassolituus bin was dominant was at Hibernia at 7 days. The near absence of Thalassolituus at other sites in the winter was possibly due to the generally slower oil degradation rate occurring in the winter (Figure 1). This bin was observed at similar abundance levels in all control microcosms regardless of the sampling season. This Thalassolituus bin would likely have been observed in the winter had our genomic microcosms been killed and sequenced at 15 or 28 days instead of 7 days. Bin 18-Oceanospirillales showed high similarity with Thalassolituus oleivorans, a microorganism that is known to have oil-degrading capabilities (Yakimov, 2004; McKew et al., 2007a) and has been observed at oil spill sites (Mason et al., 2014; Brakstad et al., 2015a). While there is strong evidence that Oceanospirillales in general are involved in oil degradation (Hazen et al., 2010; Valentine et al., 2010; Kessler et al., 2011; Mason et al., 2012; Redmond and Valentine, 2012; Rivers et al., 2013; Gutierrez et al., 2013) very little information is available regarding the precise function of Thalassolituus in oil-degrading microbial communities. A recent 16S rRNA gene amplicon survey comparing oil and oil-with-dispersant mesocosms showed shifts in microbial population structure, but did not report the presence of Oceanospirillales (Meng et al., 2016).
Alkane 1-monooxygenase gene expression
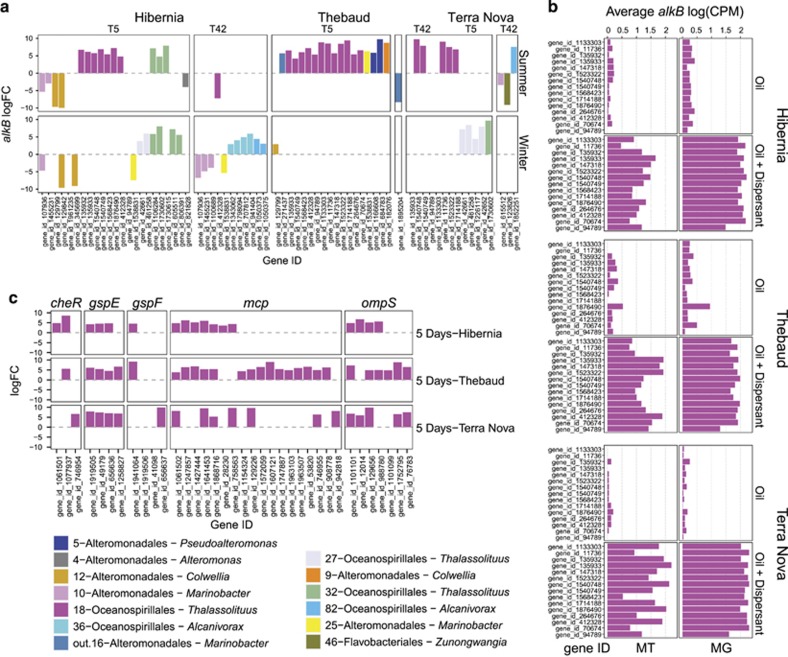
The 20 most abundant bins reported in this study were not only abundant, but transcriptionally active as well (Supplementary Information File 1—Supplementary Figure S6), which is consistent with previous findings (Mason et al., 2012; Rivers et al., 2013). Ratios of RNA:DNA for each gene of each bin were fitted with a non-parametric smoothing (Loess) and generally followed a linear or logarithmic trend: the more abundant a gene was, the more important was its expression. We focused our analyses on the expression of alkane 1-monooxygenase (alkB) a key enzyme in the degradation of n-alkanes (van Beilen et al., 2006; Rojo and Fernando, 2009) and thus an appropriate marker gene for oil degradation. We performed an extensive search of alkB genes in our assembled metagenome contigs and listed their occurrence on a per-bin basis (Supplementary Information 2 Data Sets—Supplementary Table S2). We found that out of our most 20 abundant bins, 11 harbored a total of 121 distinct alkB genes. The number of distinct alkB genes was surprisingly high: 18 for 18-Oceanospirillales, 11 for 36-Oceanospirillales and 10 for 16-Alteromonadales. A metagenome bin is an abstract unit that takes into account contig abundance covariance and tetranucleotide frequencies across samples (Kang et al., 2015b). Bins we report in this study could very well comprise two or more closely related species, explaining the high number of alkB genes we observed. However, contamination levels were relatively low (average of 5.93%) for the 20 most abundant bins we obtained in our study. Bins having 8 or more copies of alkB (16-Alteromonadales, 36-Oceanospirillale and 18-Oceanospirilalle, 3-Thiotrichales, 1-Oceanospirillale and 82-Oceanospirillales) had 17.99%, 14.96%, 11.36%, 2,7% and 1.72% contamination, respectively (Supplementary Information 2 Data Sets—Supplementary Table S2). We then obtained abundance profiles from our metatranscriptomic data, where for each microcosm location, we extracted alkB genes having a transcribed log fold change ⩾1.5 with a false discovery rate <0.05 between oil and oil-with-dispersant microcosms (Figure 3). As each treatment microcosm contained oil, it was expected to retrieve a fairly high number of upregulated alkB genes. However, because oil biodegradation was more efficient in microcosms containing oil-with-dispersant compared to oil-only microcosms, we were interested in determining what alkB gene transcripts were differentially abundant based only on the addition of dispersant. Consequently, we obtained differentially expressed potential alkB genes in oil-with-dispersant vs oil-only microcosms (Figure 3a). We found that at all locations in summer microcosms, alkB genes harbored by the 18-Oceanospirillales Thalassolituus bin were consistently more abundant in oil-with-dispersant microcosms (vs oil-only) at 5 days. At Hibernia in the summer, another Thalassolituus bin (32-Oceanospirillales) had three alkB genes with more transcripts in the oil-with-dispersant condition compared to oil-only.
Figure 3.
Fold change (log2) of significantly differentially expressed (a) alkB and (c) cheR, gspE, gspF, mcp and ompS genes in oil with dispersant vs oil alone. Values >0 means the corresponding gene is upregulated in oil with dispersant compared to the oil control. (b) Average log(CPM) values of alkB genes harbored by the 18-Oceanospirillales Thalassolituus bin at 5 days in oil and oil with dispersants in metatranscriptomics and metagenomics data types.
This Thalassolituus 18-Oceanospirillales bin clearly stands out, accounting for the majority of differentially abundant alkB genes in all oil-with-dispersant microcosms in the summer. As log fold-change values do not indicate the magnitude of gene abundance, but rather the abundance relative to another treatment, we also computed the average CPM values of each of these 18-Oceanospirillales alkB genes for the day 5 time point (Figure 3b), which confirmed that both metagenome and metatranscriptome data sets showed higher alkB read abundance in oil-with-dispersant (average values of ~1.5 log(CPM)) compared to oil-only microcosms (average values between 0 and 0.5 log(CPM)). We also looked for genes directly involved in alkane degradation (Wang and Shao, 2014). Of particular interest were genes coding for the chemotactic system components sensing n-alkanes, for which we generated hidden Markov models. We mined our metagenome data set with these models and found putative mcp, cheR and ompS genes. From our metatranscriptome data, we found that many of these genes were significantly more abundant in 18-Oceanospirillales in the presence of oil with dispersant vs oil only (Figure 3c). OmpS regulates expression of chemotactic genes, including mcp and cheR, is involved in the expression of alkB and is thought to be a signal transmitting protein and potentially the first alkane sensor outside the cell (Wang and Shao, 2014). Why Thalassolituus thrives at T5 in oil-with-dispersant microcosms is subject to speculation, but it could be that its particular OmpS and Mcp chemotactic receptors are highly responsive to emulsified oil or dispersant. From the genes reported by Wang and Shao (2014), we also found two Thalassolituus genes (gspE and gspF) showing high similarity to an ATPase involved in a type II/IV secretion system that were also positively regulated in oil-with-dispersant microcosms. Thalassolituus was found to be in competition with Alcanivorax(McKew et al., 2007a) and while the nature of this competition remains elusive, it was speculated that Thalassolituus might actively release bioactive compounds to inhibit competitors (McKew et al., 2007a; McGenity et al., 2012). Abundance of gspE and gspF transcripts will require further investigation, but it provides support that Thalassolituus actively uses its type II/IV secretion apparatus during degradation of emulsified oil.
To the best of our knowledge, this is the first time that the presence of dispersant with oil has been shown to favor Thalassolituus growth and transcription of its alkane degradation genes (alkB,cheR,ompS,mcp,gspE and gspF). Interestingly, Thalassolituus was virtually absent in microcosms that contained dispersant or oil alone: it needed both oil and dispersant to thrive. Moreover, another Thalassolituus bin (32-Oceanospirillales) had some differentially abundant alkB genes at Hibernia (summer and winter) and one at Terra Nova at 5 days in the winter. We found far fewer differentially abundant alkB genes in the winter, which is likely due to the fact that the oil degradation rate in oil-with-dispersant microcosms occurred around 15 and 28 days, later than what was observed in the summer. At this point, we can only speculate why Thalassolituus thrived in oil-with-dispersant microcosms, but our data strongly suggest an important role for this microbe. Thalassolituus was abundant near the oil degradation rate peak (5 days in summer microcosms), but did not remain as a significant part of the community by 42 days. In contrast, Alcanivorax bins were found in high abundance at both 5 and 42 days. The short window during which Thalassolituus bins were detectable might explain why this microbe has been under the radar in other studies investigating the effects of dispersants on oil degradation. In addition, the vast majority of microbial surveys in oil degradation studies were conducted with short 16S rRNA gene amplicons, which results in a lower resolution for microbial identification and has possibly contributed to its underrepresentation in the literature.
The Thalassolituus bin we identified contained 18 potential alkB genes. To gain insight into why alkB genes hosted by Thalassolituus are more efficient in an oil-with-dispersant context, we generated a multiple alignment of all 121 alkB genes contained in our 20 most abundant bins and generated a phylogenetic tree (Figure 4a), which showed that alkB genes clustered together according to their host bin. Each alkB sequence of each bin was aligned such that we obtained one representative consensus alkB sequence for each bin. We then searched for motifs in the NCBI-CDD database and found that three particular domains (NCBI-CDD:255516, 234750 and 253129) were present in the alkB representatives of the 18-Oceanospirillales Thalassolituus bin we identified, but not in alkB sequences belonging to other bins. These three motifs match the consensus sequence region located upstream of the FA_desaturase between position 482–567 (Figure 4b). Interestingly, one of these motifs (253129) belongs to the Arabidopsis phospholipase-like protein (PEARLI 4) super family. Additional work will be needed to elucidate this Thalassolituus AlkB structure, but our data suggest the possible existence of a yet uncharacterized fatty acid desaturase extended with a phospholipid domain, which could somehow show specificity toward emulsified oil, but not oil or emulsifier alone.
Figure 4.
(a) All putative AlkB amino-acid (aa) sequences were aligned into a multiple alignment (MUSCLE v3.8.31), which was used to generate a phylogenetic tree (FastTree v2.1.8). Gene IDs are colored by their associated metagenome bin. (b) A multiple sequence alignment (MUSCLE) was generated using only AlkB aa sequences belonging to the 18-Oceanospirillales Thalassolituus bin. A consensus was generated (Jalview v2.9.0b2) from this multiple sequence alignment and analyzed for its motif/domain composition. Blue: Pfam annotations; red: NCBI’s CDD annotations.
Conclusions
Dispersants have been used on a large scale for many oil spills over the last several decades, the most recent case being the DWH spill of 2010. However, there is no scientific consensus on the benefits of dispersants for stimulating hydrocarbon degradation (recently reviewed Kleindienst et al. (2015a)) with conflicting results from previous studies likely partially attributed to the absence of standardized protocols. Our data support that after 5 or 7 days of incubation in both summer and winter, the differences in oil and oil-with-dispersant microbial communities are mainly driven by the presence and activity of two Thalassolituus metagenome bins. We also found that dispersants are effective at stimulating n-alkane biodegradation in marine waters and does not act solely as a preferred substrate for microbial growth. However, regardless of the presence or absence of dispersant, under all conditions, bacteria were able to respond to oil and condensate spills and rapidly degrade hydrocarbons. Indigenous bacterial populations in the regions examined were highly diversified and included an array of known hydrocarbon degraders. Providing that there are sufficient essential nutrients, these hydrocarbon-degrading bacteria could have an important role in attenuating potential oil spills.
Acknowledgments
The NRC and COOGER thank Drs Simon Courtenay (DFO; project management), William Li (DFO; bacteria enumeration), Youyu Lu (DFO; project management), Haibo Niu (Dalhousie University; modeling work), and Yongsheng Wu (DFO; modeling work), Ms Carol Anstey (DFO; nutrients) and Mr Rod Doane (DFO; administrative support and editing) for their contributions to this study. In addition we acknowledge the excellent technical support of Brian Robinson, Gary Wolfgeschaffen, Scott Ryan, Peter Thamer, Jennifer Mason and Sylvie Sanschagrin. We acknowledge Robert Dunphy (Hibernia Management Corporation), Trudy Wells (Suncor) and Megan Tuttle (ExxonMobil) for the representative samples of crude oil and gas condensate. We thank Antoine Pagé for manuscript revision. This study was funded by the Environmental Studies Research Funds (Natural Resources Canada). We also wish to acknowledge Compute Canada for access to the McGill University High Performance Computing (HPC) infrastructure (Guillimin system).
Author contributions
JT wrote software, analyzed data and wrote manuscript. CWG and EY planned experimental design, analyzed data and edited manuscript. NF carried out the microcosms experiments. SC performed chemical analyses of microcosms. ME performed DNA extraction and prepared sequencing libraries. TLK and KL participated in the design of the study.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- American Academy of Microbiology. (2011). Microbes and oil spills. https://www.asm.org/images/stories/documents/Microbes_and_Oil_Spills.pdf. Accessed on July 10, 2017.
- Atlas RM, Bartha R. (1972). Degradation and mineralization of petroleum in sea water: limitation by nitrogen and phosphorous. Biotechnol Bioeng 14: 309–318. [DOI] [PubMed] [Google Scholar]
- Atlas RM, Bartha R. (1973). Stimulated biodegradation of oil slicks using oleophilic fertilizers. Environ Sci Technol 7: 538–541. [DOI] [PubMed] [Google Scholar]
- Atlas RM, Hazen TC. (2011). Oil biodegradation and bioremediation: a tale of the two worst spills in U.S. history. Environ Sci Technol 45: 6709–6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM. (2002). Short protocols in molecular biology: a compendium of methods from Current protocols in molecular biology. Current Protocols.
- Baelum J, Borglin S, Chakraborty R, Fortney JL, Lamendella R, Mason OU et al. (2012). Deep-sea bacteria enriched by oil and dispersant from the Deepwater Horizon spill. Environ Microbiol 14: 2405–2416. [DOI] [PubMed] [Google Scholar]
- van Beilen JB, Funhoff EG, van Loon A, Just A, Kaysser L, Bouza M et al. (2006). Cytochrome P450 alkane hydroxylases of the CYP153 family are common in alkane-degrading eubacteria lacking integral membrane alkane hydroxylases. Appl Environ Microbiol 72: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano AC, Levine E, Mearns AJ. (2013). Effectiveness and potential ecological effects of offshore surface dispersant use during the Deepwater Horizon oil spill: a retrospective analysis of monitoring data. Environ Monit Assess 185: 10281–10295. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. (2009). GenBank. Nucleic Acids Res 37: D26–D31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthe-Corti L, Nachtkamp M. (2010). Bacterial communities in hydrocarbon-contaminated marine coastal environments. In: Timmis KN, McGenity TJ, van der Meer JR, de Lorenzo V (eds), Handbook of Hydrocarbon and Lipid Microbiology. Springer-Verlag: Berlin, Heidelberg, pp 2349–2359.
- Boisvert S, Raymond F, Godzaridis E, Laviolette F, Corbeil J. (2012). Ray Meta: scalable de novo metagenome assembly and profiling. Genome Biol 13: R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg JR, Prince RC, James Harner E, Atlas RM. (1994). Effectiveness of bioremediation for the Exxon Valdez oil spill. Nature 368: 413–418. [Google Scholar]
- Brakstad OG, Kristin B. (2006). Biodegradation of petroleum hydrocarbons in seawater at low temperatures (0–5 °c) and bacterial communities associated with degradation. Biodegradation 17: 71–82. [DOI] [PubMed] [Google Scholar]
- Brakstad OG, Throne-Holst M, Netzer R, Stoeckel DM, Atlas RM. (2015. a). Microbial communities related to biodegradation of dispersed Macondo oil at low seawater temperature with Norwegian coastal seawater. Microb Biotechnol 8: 989–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakstad OG, Nordtug T, Throne-Holst M. (2015. b). Biodegradation of dispersed Macondo oil in seawater at low temperatures and different oil droplet sizes. Mar Pollut Bull 93: 144–152. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R, Romy C, Borglin SE, Dubinsky EA, Andersen GL, Hazen TC. (2012). Microbial response to the MC-252 Oil and Corexit 9500 in the Gulf of Mexico. Front Microbiol 3: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M, King T, Lee K. (2007). Analytical technique for extracting hydrocarbons from water using sample container as extraction vessel in combination with a roller apparatus. Can Tech Rep Fish Aquat Sci 2733: 1–12. [Google Scholar]
- Committee on Oil in the Sea: Inputs, Fates, and Effects, Board OS, Board M, Board TR, Division on Earth and Life Studies, National Research Council. (2003) Oil in the Sea III: Inputs, Fates, and Effects. National Academies Press: Washington, DC, USA. [PubMed] [Google Scholar]
- Committee on Understanding Oil Spill Dispersants: Efficacy and Effects, Board OS, Division on Earth and Life Studies, National Research Council. (2005) Oil Spill Dispersants: Efficacy and Effects. National Academies Press: Washington, DC, USA. [Google Scholar]
- Coulon F, McKew BA, Osborn AM, McGenity TJ, Timmis KN. (2007). Effects of temperature and biostimulation on oil-degrading microbial communities in temperate estuarine waters. Environ Microbiol 9: 177–186. [DOI] [PubMed] [Google Scholar]
- Cowton M, Michael C. (2016). Polar microbiology: the ecology, biodiversity and bioremediation potential of microorganisms in extremely cold environments. Biodiversity 17: 74–75. [Google Scholar]
- Cui Z, Lai Q, Dong C, Shao Z. (2008). Biodiversity of polycyclic aromatic hydrocarbon-degrading bacteria from deep sea sediments of the Middle Atlantic Ridge. Environ Microbiol 10: 2138–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang H, Lovell CR. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol Mol Biol Rev 80: 91–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deppe U, Uta D, Hans-Hermann R, Walter M, Garabed A. (2005). Degradation of crude oil by an arctic microbial consortium. Extremophiles 9: 461–470. [DOI] [PubMed] [Google Scholar]
- Dini-Andreote F, Stegen JC, van Elsas JD, Salles JF. (2015). Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc Natl Acad Sci USA 112: E1326–E1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. (2011). Accelerated profile HMM searches. PLoS Comput Biol 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RE. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PN, Parks DH, Chadwick GL, Robbins SJ, Orphan VJ, Golding SD et al. (2015). Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350: 434–438. [DOI] [PubMed] [Google Scholar]
- Finn RD, Alex B, Jody C, Penelope C, Eberhardt RY, Eddy SR et al. (2013). Pfam: the protein families database. Nucleic Acids Res 42: D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer CW. (2010). Bacterial diversity in hydrocarbon-polluted rivers, estuaries and sediments. In: Timmis KN, McGenity TJ, van der Meer JR, de Lorenzo V (eds), Handbook of Hydrocarbon and Lipid Microbiology. Springer-Verlag: Berlin, Heidelberg, pp 2329–2338.
- Gutierrez T, Singleton DR, Berry D, Yang T, Aitken MD, Teske A. (2013). Hydrocarbon-degrading bacteria enriched by the Deepwater Horizon oil spill identified by cultivation and DNA-SIP. ISME J 7: 2091–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan LJ, Fulmer PA. (2011). Effects of COREXIT EC9500A on bacteria from a beach oiled by the Deepwater Horizon spill. Aquat Microb Ecol 63: 101–109. [Google Scholar]
- Harayama S, Kasai Y, Hara A. (2004). Microbial communities in oil-contaminated seawater. Curr Opin Biotechnol 15: 205–214. [DOI] [PubMed] [Google Scholar]
- Harris C, Chris H. (1995). The Braer Incident: Shetland Islands, January 1993. International Oil Spill Conference Proceedings; Long Beach: CA, USA, February–March 1995, vol. 1995, No. 1, pp 813–819. [Google Scholar]
- Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N et al. (2010). Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330: 204–208. [DOI] [PubMed] [Google Scholar]
- Head IM, Jones DM, Röling WFM. (2006). Marine microorganisms make a meal of oil. Nat Rev Microbiol 4: 173–182. [DOI] [PubMed] [Google Scholar]
- Henry C, Charlie H. (2005). Review of Dispersant Use IN US Gulf of Mexico Waters Since the Oil Pollution Act of 1990. International Oil Spill Conference Proceedings; Miami Beach: FL, USA, May 2005, vol. 2005, No. 1, pp 439–442. [Google Scholar]
- Hugerth LW, Larsson J, Alneberg J, Lindh MV, Legrand C, Pinhassi J et al. (2015). Metagenome-assembled genomes uncover a global brackish microbiome. Genome Biol 16: 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman J, Waldrop MP, Mackelprang R, David MM, McFarland J, Blazewicz SJ et al. (2015). Multi-omics of permafrost, active layer and thermokarst bog soil microbiomes. Nature 521: 208–212. [DOI] [PubMed] [Google Scholar]
- Huntemann M, Ivanova NN, Mavromatis K, Tripp HJ, Paez-Espino D, Tennessen K et al. (2016). The standard operating procedure of the DOE-JGI Metagenome Annotation Pipeline (MAP v.4). Stand Genomic Sci 11: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joye SB, Teske AP, Kostka JE. (2014). Microbial dynamics following the Macondo Oil Well blowout across Gulf of Mexico environments. Bioscience 64: 766–777. [Google Scholar]
- Kang DD, Froula J, Egan R, Wang Z. (2015. a). MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3: e1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DD, Froula J, Egan R, Wang Z. (2015. b). MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3: e1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler JD, Valentine DL, Redmond MC, Du M, Chan EW, Mendes SD et al. (2011). A persistent oxygen anomaly reveals the fate of spilled methane in the deep Gulf of Mexico. Science 331: 312–315. [DOI] [PubMed] [Google Scholar]
- King GM, Kostka JE, Hazen TC, Sobecky PA. (2015. a). Microbial responses to the Deepwater Horizon oil spill: from coastal wetlands to the deep sea. Ann Rev Mar Sci 7: 377–401. [DOI] [PubMed] [Google Scholar]
- King TL, Brian R, Claire M, Patrick T, Scott R, Firas S et al. (2015. b). Fate of surface spills of Cold Lake Blend Diluted bitumen treated with dispersant and mineral fines in a wave tank. Environ Eng Sci 32: 250–261. [Google Scholar]
- Kleindienst S, Grim S, Sogin M, Bracco A, Crespo-Medina M, Joye SB. (2016). Diverse, rare microbial taxa responded to the Deepwater Horizon deep-sea hydrocarbon plume. ISME J 10: 400–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleindienst S, Paul JH, Joye SB. (2015. a). Using dispersants after oil spills: impacts on the composition and activity of microbial communities. Nat Rev Microbiol 13: 388–396. [DOI] [PubMed] [Google Scholar]
- Kleindienst S, Seidel M, Ziervogel K, Grim S, Loftis K, Harrison S et al. (2015. b). Chemical dispersants can suppress the activity of natural oil-degrading microorganisms. Proc Natl Acad Sci USA 112: 14900–14905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostka JE, Teske AP, Joye SB, Head IM. (2014). The metabolic pathways and environmental controls of hydrocarbon biodegradation in marine ecosystems. Front Microbiol 5: 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvenvolden KA, Cooper CK. (2003). Natural seepage of crude oil into the marine environment. Geo-Mar Lett 23: 140–146. [Google Scholar]
- Law RJ, Carole K. (2004). The impact of the ‘Sea Empress’ oil spill. Aquat Living Resour 17: 389–394. [Google Scholar]
- Lee K, Nedwed T, Prince RC, Palandro D. (2013). Lab tests on the biodegradation of chemically dispersed oil should consider the rapid dilution that occurs at sea. Mar Pollut Bull 73: 314–318. [DOI] [PubMed] [Google Scholar]
- Lewis A. (2015). Challenging the validity of recent claims that dispersants supress biodegradation. Int Spill Control Organ Newsl 511: 8–11. [Google Scholar]
- Lindstrom JE, Braddock JF. (2002). Biodegradation of petroleum hydrocarbons at low temperature in the presence of the dispersant Corexit 9500. Mar Pollut Bull 44: 739–747. [DOI] [PubMed] [Google Scholar]
- Lunel T, Tim L, Jan R, Nick B, Chris H, Louise D. (1997). The Net Environmental Benefit of a Successful Dispersant Operation at the Sea Empress Incident. International Oil Spill Conference Proceedings; April 1997, vol. 1997, No. 1, pp 185–194. [Google Scholar]
- Mason OU, Han J, Woyke T, Jansson JK. (2014). Single-cell genomics reveals features of a Colwellia species that was dominant during the Deepwater Horizon oil spill. Front Microbiol 5: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason OU, Hazen TC, Borglin S, Chain PSG, Dubinsky EA, Fortney JL et al. (2012). Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J 6: 1715–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGenity TJ, Folwell BD, McKew BA, Sanni GO. (2012). Marine crude-oil biodegradation: a central role for interspecies interactions. Aquat Biosyst 8: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKew BA, Coulon F, Osborn AM, Timmis KN, McGenity TJ. (2007. a). Determining the identity and roles of oil-metabolizing marine bacteria from the Thames estuary, UK. Environ Microbiol 9: 165–176. [DOI] [PubMed] [Google Scholar]
- McKew BA, Coulon F, Yakimov MM, Denaro R, Genovese M, Smith CJ et al. (2007. b). Efficacy of intervention strategies for bioremediation of crude oil in marine systems and effects on indigenous hydrocarbonoclastic bacteria. Environ Microbiol 9: 1562–1571. [DOI] [PubMed] [Google Scholar]
- Meng L, Liu H, Bao M, Sun P. (2016). Microbial community structure shifts are associated with temperature, dispersants and nutrients in crude oil-contaminated seawaters. Mar Pollut Bull 111: 203–212. [DOI] [PubMed] [Google Scholar]
- Mulkins-Phillips GJ, Stewart JE. (1974). Effect of four dispersants on biodegradation and growth of bacteria on crude oil. Appl Microbiol 28: 547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepceron M, Portet-Koltalo F, Merlin C, Motelay-Massei A, Barray S, Bodilis J. (2010). Both Cycloclasticus spp. and Pseudomonas spp. as PAH-degrading bacteria in the Seine estuary (France). FEMS Microbiol Ecol 71: 137–147. [DOI] [PubMed] [Google Scholar]
- Nie Y, Chi C-Q, Fang H, Liang J-L, Lu S-L, Lai G-L et al. (2014). Diverse alkane hydroxylase genes in microorganisms and environments. Sci Rep 4: 4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JJ. (1984) Microbial metabolism of cyclic alkanes. In: Atlas RM (ed). Petroleum Microbiology. Macmillan Publ. Co: New York, NY, USA, pp 61–98. [Google Scholar]
- Prince RC. (2010) Eukaryotic hydrocarbon degraders. In: Timmis KN, McGenity TJ, van der Meer JR, de Lorenzo V (eds), Handbook of Hydrocarbon and Lipid Microbiology. Springer-Verlag: Berlin, Heidelberg, pp 2065–2078.
- Prince RC, Butler JD. (2014). A protocol for assessing the effectiveness of oil spill dispersants in stimulating the biodegradation of oil. Environ Sci Pollut Res Int 21: 9506–9510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince RC, Gramain A, McGenity TJ. (2010). Prokaryotic hydrocarbon degraders. In: Timmis KN, McGenity TJ, van der Meer JR, de Lorenzo V (eds), Handbook of Hydrocarbon and Lipid Microbiology. Springer-Verlag: Berlin, Heidelberg, pp 1669–1692.
- Quinlan AR, Hall IM. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy CM, Arey JS, Seewald JS, Sylva SP, Lemkau KL, Nelson RK et al. (2012). Composition and fate of gas and oil released to the water column during the Deepwater Horizon oil spill. Proc Natl Acad Sci USA 109: 20229–20234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond MC, Valentine DL. (2012). Natural gas and temperature structured a microbial community response to the Deepwater Horizon oil spill. Proc Natl Acad Sci USA 109: 20292–20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers AR, Sharma S, Tringe SG, Martin J, Joye SB, Moran MA. (2013). Transcriptional response of bathypelagic marine bacterioplankton to the Deepwater Horizon oil spill. ISME J 7: 2315–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo F, Fernando R. (2009). Degradation of alkanes by bacteria. Environ Microbiol 11: 2477–2490. [DOI] [PubMed] [Google Scholar]
- Röling WFM, Milner MG, Jones DM, Fratepietro F, Swannell RPJ, Daniel F et al. (2004). Bacterial community dynamics and hydrocarbon degradation during a field-scale evaluation of bioremediation on a mudflat beach contaminated with buried oil. Appl Environ Microbiol 70: 2603–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röling WFM, Milner MG, Jones DM, Lee K, Daniel F, Swannell RJP et al. (2002). Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient-enhanced oil spill bioremediation. Appl Environ Microbiol 68: 5537–5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwan N, Xia F, Gilbert JA. (2016). Recovering complete and draft population genomes from metagenome datasets. Microbiome 4: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V et al. (2009). Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 37: D5–D15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Sherry A, Gray ND, Jones DM, Bowler BFJ, Head IM. (2014). Kinetic parameters for nutrient enhanced crude oil biodegradation in intertidal marine sediments. Front Microbiol 5: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JE. (1968). ‘Torrey Canyon’ Pollution and Marine Life: A Report by the Plymoith Laboratory of the Marine Biological Association of the United Kingdom. Cambridge University Press: Cambridge, UK.
- Steen A, Alexis S, Abigail F. (2008). Frequency of Dispersant Use Worldwide. International Oil Spill Conference Proceedings; Savannah: Georgia, USA, May 2008, vol. 2008, No. 1, pp 645–649. [Google Scholar]
- Tang S, Shiyuyun T, Mark B. (2013). Ab initio gene identification in metagenomic sequences. In: Karen EN (ed), Encyclopedia of Metagenomics. Springer New York: New York, NY, USA, pp 1–8.
- Valentine DL, Kessler JD, Redmond MC, Mendes SD, Heintz MB, Farwell C et al. (2010). Propane respiration jump-starts microbial response to a deep oil spill. Science 330: 208–211. [DOI] [PubMed] [Google Scholar]
- Valentine DL, Mezić I, Maćešić S, Črnjarić-Žic N, Ivić S, Hogan PJ et al. (2012). Dynamic autoinoculation and the microbial ecology of a deep water hydrocarbon irruption. Proc Natl Acad Sci USA 109: 20286–20291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venosa AD, Suidan MT, Wrenn BA, Strohmeier KL, Haines JR, Loye Eberhart B et al. (1996). Bioremediation of an experimental oil spill on the shoreline of Delaware Bay. Environ Sci Technol 30: 1764–1775. [Google Scholar]
- Wang W, Shao Z. (2014). The long-chain alkane metabolism network of Alcanivorax dieselolei. Nat Commun 5: 5755. [DOI] [PubMed] [Google Scholar]
- Widdel F, Knittel K, Galushko A. (2010). Anaerobic hydrocarbon-degrading microorganisms: an overview. In: Timmis KN, McGenity TJ, van der Meer JR, de Lorenzo V (eds.), Handbook of Hydrocarbon and Lipid Microbiology. Springer-Verlag: Berlin, Heidelberg, pp 1997–2021.
- Yakimov MM. (2004). Thalassolituus oleivorans gen. nov., sp. nov., a novel marine bacterium that obligately utilizes hydrocarbons. Int J Syst Evol Microbiol 54: 141–148. [DOI] [PubMed] [Google Scholar]
- Yakimov MM, Timmis KN, Golyshin PN. (2007). Obligate oil-degrading marine bacteria. Curr Opin Biotechnol 18: 257–266. [DOI] [PubMed] [Google Scholar]
- Yakimov M, Gentile G, Bruni V, Cappello S, D'Auria G, Golyshin PN et al. (2004). Crude oil-induced structural shift of coastal bacterial communities of rod bay (Terra Nova Bay, Ross Sea, Antarctica) and characterization of cultured cold-adapted hydrocarbonoclastic bacteria. FEMS Microbiol Ecol 49: 419–432. [DOI] [PubMed] [Google Scholar]
- Yergeau E, Maynard C, Sanschagrin S, Champagne J, Juck D, Lee K et al. (2015). Microbial community composition, functions, and activities in the Gulf of Mexico 1 year after the Deepwater Horizon accident. Appl Environ Microbiol 81: 5855–5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.