Abstract
Bovine γδ T cells are amongst the first cells to accumulate at the site of Mycobacterium bovis infection; however, their role in the developing lesion remains unclear. We utilized transcriptomics analysis, in situ hybridization, and a macrophage/γδ T cell co-culture system to elucidate the role of γδ T cells in local immunity to M. bovis infection. Transcriptomics analysis revealed that γδ T cells upregulated expression of several novel, immune-associated genes in response to stimulation with M. bovis antigen. BCG-infected macrophage/γδ T cell co-cultures confirmed the results of our RNAseq analysis, and revealed that γδ T cells from M. bovis-infected animals had a significant impact on bacterial viability. Analysis of γδ T cells within late-stage M. bovis granulomas revealed significant expression IFN-γ and CCL2, but not IL-10, IL-22, or IL-17. Our results suggest γδ T cells influence local immunity to M. bovis through cytokine secretion and direct effects on bacterial burden.
Keywords: Mycobacterium bovis, γδ T cell, granuloma, bovine
1. Introduction
Tuberculosis (TB) is among the most important infectious diseases worldwide. In 2015 1.8 million people died from this disease [1]. Mycobacterium bovis (M. bovis) is a member of the Mycobacterium tuberculosis complex (Mtbc), and is the causative agent of TB in cattle (BTB). M. bovis is an aerobic pathogen capable of causing zoonosis in most mammals, including humans. This disease has a significant detrimental impact on the livestock industry; costing billions of dollars in losses each year due to disease testing and control efforts [2]. Eradication attempts have been successful in some countries; however, the broad host range and low infective dose of BTB makes worldwide eradication difficult.
Cattle are a natural host for M. bovis, and BTB parallels human TB in several aspects of disease pathogenesis and the development of innate and adaptive immune responses [3, 4]. Historically, the study of bovine and human TB has been closely intertwined, and our understanding of disease in animals has been instrumental in our understanding of that in humans. For example, the vaccine strain that is widely administered to infants and people at high risk for TB is actually M. bovis Bacille Calmette Guerin (BCG). This vaccine was tested in cattle before being administered to humans. Similarly, IFN-γ release assays were first implemented in the bovine TB eradication program, and are now widely used in human diagnostics. Thus, the study of virulent M. bovis infection in cattle represents an excellent model for understanding Mycobacterium tuberculosis (M. tb) infection in humans, and for testing novel vaccine strategies and therapeutics [2].
Granulomas are characteristic of TB infections, and are the body’s attempt to protect the host by containing the invading mycobacteria. They are an organized structure of immune cells that form around the invading bacterium and are comprised of macrophages, neutrophils and lymphocytes. The structures undergo a process of ordered maturation during the course of disease, and can be staged (I-IV) based upon cellular composition and amount of fibrosis and necrosis [5–8]. Importantly, simple formation of a granuloma is not sufficient alone to control or eliminate the disease. The ability of the host to establish well-organized granulomas, with an appropriate balance of pro- and anti-inflammatory immune responses is crucial to controlling the infection [9, 10]. Despite the importance of the granuloma structure in dictating the outcome of infection, we understand very little about the dynamics of the immune response at the site of infection. Specifically, the cells and cytokine production necessary for formation and maintenance of an effective granuloma.
γδ T cells are a unique subset of CD3+ T cells that possess functions characteristic of both innate and adaptive immunity, and are therefore thought to bridge the two arms of the immune system. γδ T cells constitute a significant proportion of the immune cells found in the mucosal and epithelial surfaces of the respiratory tract. These cells are generally recognized to be critical in the first line of defense against invading pathogens and in shaping the downstream adaptive immune response [11]. However, the frequency of γδ T cells circulating in mice, humans, and non-human primates is low, representing 1–5% of the circulating peripheral lymphocyte population [12], making it difficult to experimentally dissect the role of the γδ T cells in the immune response. In contrast, γδ T cells circulate at significantly increased frequencies in ruminant species, where they constitute 30–60% of the peripheral blood lymphocytes in young animals [13, 14]. The increased incidence of these cells in blood makes the bovine an excellent model for studying γδ T cells and for understanding their role in innate and adaptive immunity.
γδ T cells in mice and cattle accumulate in the lungs and lung-associated lymph nodes after either M. bovis infection or BCG vaccination administered via respiratory routes [15, 16]. These cells are also among the first cells to arrive at the site of infection [17]. γδ T cells have been shown to accumulate within all stages of lesions in cattle infected with M. bovis, and are often found localizing to the lymphoid mantle surrounding the periphery of the lesions [18]. Mice deficient in γδ T cells develop large and poorly organized granulomas during M. tb infection [19]. Similarly, mice and rodents depleted of γδ T cells show alterations in granuloma architecture with increases in neutrophil infiltration and necrosis [20]. These findings suggest that γδ T cells may be an important source of cytokines and chemokines which aid in the recruitment of other immune cells to the site of infection. In vitro, γδ T cells have been shown to produce significant amounts of IFN-γ, similar to that of CD4+ T cells, in response to mycobacterial antigens [21]. However, less is known about their capacity to secrete IFN-γ in vivo, particularly at the site of infection. The ability of these cells to secrete chemotactic molecules or other immune factors in response to M. bovis infection is not well defined. Therefore, in this study, we used RNASeq analysis to further define the M. bovis-specific γδ T cell response. This approach allowed us to identify previously unrecognized immunologic factors that may contribute to the γδ T cell’s capacity to establish and maintain granuloma structures in vivo. To correlate the in vitro responses measured by our RNASeq analysis with those that occur in vivo at the site of infection, we also used in situ hybridization. This allowed us to assess the expression of multiple cytokines by γδ T cells accumulating in the chronic, granulomatous lesions of cattle infected with virulent M. bovis. We also developed a novel, in vitro macrophage/γδ T cell co-culture system. This system allowed us to model the interactions that may occur in the lungs between tissue-resident γδ T cells and M. bovis-infected macrophages in the early stages of BTB infection. Our hypothesis is that γδ T cells influence immune cell recruitment and granuloma formation, and shape the adaptive M. bovis-specific immune response by producing inflammatory and regulatory cytokines and chemokines at the site of M. bovis infection. Determining the role that γδ T cells play in the localized immune response to M. bovis infection is expected to further our understanding of basic γδ T cell biology. Our findings may also aid in developing effective ways in which to manipulate protective responses to TB in both humans and animals.
2. Materials and methods
2.1 Animals
Tissues samples were collected from animals used in a previous study [22]. Briefly, 23 Holstein steers approximately 6 months of age were obtained from a tuberculosis-free herd in Sioux Center, Iowa and housed in a biosafety level-3 (BSL-3) facility at the National Animal Disease Center (NADC), Ames, Iowa, USA, according to Institutional Biosafety and Animal Care and Use Committee guidelines. Treatment groups consisted of non-infected steers (n = 7) and animals receiving 104 colony forming units (cfu) of M. bovis 95–1315 (n = 8) or 104 cfu M. bovis 10-7428 (n = 8) by aerosol as described by Palmer et al. [23].
Animals used for co-culture experiments were 10 Holstein steer calves that were housed at the NADC in Ames IA. Calves were experimentally infected with 104 cfu of virulent M. bovis 10-7428 as above and peripheral blood was collected at ~12 weeks after challenge. Prior to sample collection, the calves were confirmed BTB positive by skin test and whole blood IFN-γ release assay. Blood samples were also obtained from 19 Holstein steer calves maintained in an M. bovis-free herd housed at the Kansas State University Dairy Facility in Manhattan, KS. Animals from both herds were age, breed, and sex matched in order to minimize possible herd-to-herd variation.
All animal procedures were conducted in strict accordance with federal and institutional guidelines and were approved by the NADC Institutional Animal Care and Use Committee or the Kansas State University Institutional Animal Care and Use Committee.
2.2 Preparation of PBMCs
Peripheral blood was collected from the jugular vein into 2X acid citrate dextrose. Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coat fractions and overlaid onto Histopaque 1077 (Sigma Aldrich, St. Louis, MO). Contaminating red blood cells were removed using a hypotonic lysis. Cells were washed and re-suspended in complete RPMI (cRPMI) composed of RPMI 1640 (Life Technologies, Carlsbad, CA) supplemented with 2mM L-glutamine, 1% antibiotic-antimycotic solution, 1% nonessential amino acids, 2% essential amino acids, 1% sodium pyruvate, 50µM 2-mercaptoethanol (ME), and 10% fetal bovine serum (FBS).
2.3 γδ RNA Sequencing
PBMCs from 5 M. bovis-infected animals were collected, and stimulated samples were cultured with purified protein derivative of bovine tuberculin (PPD-b) (Prionics AG, Schlieren, Switzerland) at 200 U/mL in CRPMI. Unstimulated control samples were cultured with cRPMI only for 18 hours. γδ T cells were then sorted (as described in section 2.5) to >90% purity by magnetic activated cell sorting (MACS) according to manufacturer’s instructions (Miltenyi Biotec, Auburn, CA). γδ T cell RNA was extracted using Trizol Reagent (Invitrogen, Life Technologies) according to manufacturer’s instructions. Sample quality and quantity was confirmed by Tape Station analysis and Qubit quantification, and then Truseq RNA Libraries prepared per manufacturer’s instructions. The libraries were sequenced on an Illumina HiSeq 2500 System.
Sequencing reads were aligned to the most recently annotated version of the bovine genome (Bos_taurus_UMD_3.1.1). Single end reads were obtained and FastQC was ran to identify overrepresented reads. Differential expression analysis was performed using RNA Sequencing by Expectation Maximization (RSEM) and empirical Bayes sequencing (EBSeq) [24, 25].
A list of commonly differentially expressed genes resulting from the RNA sequencing analysis was submitted to Ingenuity Pathway Analysis (IPA; Ingenuity Systems, USA) in order to identify the most significant canonical pathways [26].
2.4 RNA Scope
Visualization of γδ T cells, cytokine, and chemokine mRNA transcripts was done according to manufacturer’s instructions for RNAScope 2.0 (Advanced Cell Diagnostics, Hayward, CA, USA). Samples were sectioned from formalin-fixed, paraffin embedded tissues from animals 3 months post-infection. The tissue sample slides were baked for 1 hour at 60°C in a HybEZ ™ hybridization oven (Advanced Cell Diagnostics). Tissues were then de-paraffinized in xylene followed by rehydration in ethanol and dried at room temperature (RT) for 5 minutes. Slides were treated with an endogenous peroxidase block for 10 minutes at RT. Slides were rinsed in double distilled water (ddH2O), and immersed in an antigen retrieval citrate buffer for 22 minutes at boiling (210°). Slides were washed in ddH20 followed by an ethanol rinse and allowed to air dry. A hydrophobic barrier was created around the tissue sections and the slides were then allowed to dry at RT overnight.
The following day, slides were incubated with a protease for 30 minutes at 40°C in the HybEZ oven. Slides were then washed in ddH2O, and target or control probes applied, and incubated at 40°C for 2 hours. Bos taurus-specific probe combinations (Advanced Cell Diagnostics) were used; γδ T cell TCR (Cat. No. 407481-C2), IFN-γ (Cat. No. 315581), IL-10 (Cat. No. 420941), IL-17A (Cat. No. 406601), IL-22 (Cat. No. 420931) and CCL2 (Cat. No. 14314A). The positive control probe consisted of a proprietary 2-plex probe for Bos taurus cyclophilin B (Cat. No. 319451-C2), while the negative control probe targeted dapB of Bacillus subtilis strain SMY (Cat. No. 320751). The slides were then rinsed in wash buffer (Advanced Cell Diagnostics) for 2 minutes. Proprietary signal amplification reagents 1 through 6 were serially applied for 30 minutes, 15 minutes, 30 minutes, 15 minutes, 30 minutes and 15 minutes, respectively, with a 2 minute rinse in wash buffer between each amplification reagent. Incubations with amplifier reagents 1 through 4 were done in the HybEZ oven at 40°C, while incubations with amplifier reagents 5 and 6 were done at RT. Slides were then incubated with a proprietary Red A and B mixture for 30 minutes at RT, followed by a rinse in wash buffer. Slides were incubated in a Green A and B mixture and RT for 10 minutes, and washed in ddH20. Slides were immersed in Gill’s hematoxylin for 30 seconds at RT, washed in ddH20 and then briefly immersed into ammonia, followed by a wash in ddH20. Finally, slides were dried for 15 minutes at 60°C, and cover-slipped using mounting media (EcoMount, Biocare Medical, Concord, CA, USA).
In order to quantify the amount of cytokine or chemokine being expressed by γδ T cells, 10 representative images at 100X magnification from 5 M. bovis-infected calves were taken around the periphery of each late-stage granuloma. Granulomas were determined to be late-stage, meaning stage III or stage IV, based on descriptions by previous groups [5–8]. Stage III and IV granulomas are described to have full fibrous encapsulation, and a necrotic center surrounded by a zone of epithelioid macrophages, multinucleated giant cells, and lymphocytes [5–8]. The images were then used to quantify the number of the cells that were expressing the cytokine or chemokine of interest, and what percentage of those cells were γδ T cells.
2.5 γδ T cell and Monocyte Sorting and co-cultures
Cells for co-cultures were collected from either M. bovis-infected calves or from calves with no prior exposure to M. bovis as described in 2.1. Monocytes and γδ T cells were enriched from PBMCs using Magnetic Activated Cell Sorting (MACS) according to the manufacturer’s instructions. Briefly, PBMCs were re-suspended at 107 cells/mL in MACS buffer (0.5% BSA, 2mM EDTA in PBS) and labeled with 10µg/mL mouse anti-bovine CD14 (Clone CAM36A) or 10µg/mL mouse anti-bovine γδ T cell receptor (Clone GB21A), both from Washington State Monoclonal Antibody Center (Pullman, WA, USA), for 20 minutes at 4°C. Monocytes and γδ T cells were washed then labeled with anti-mouse IgG1 or IgG2a+b Microbeads (Miltenyi Biotech) respectively. CD14+ and γδ T cell populations were purified over magnetic columns by positive selection.
Isolated monocytes were allowed to differentiate into macrophages by plating at 5×105 monocytes per well in 24 well plates and cultured in cRPMI media with GM-CSF (Kingfischer Biotech, St. Paul, MN, USA) at 4ng/mL for 7 days at 37°C in 5% CO2, and the culture media was changed every 3 days [27]. After 7 days of culture, monocyte-derived macrophages (MDM) were treated with media alone or infected with BCG Danish strain 1331 at a multiplicity of infection (MOI) of either 1 or 10 for 4 hours in cRPMI without antibiotics and antimycotics at 37°C in 5% CO2. After infection, MDM were washed twice with cRPMI to remove any extracellular BCG. Finally, BCG-infected macrophages were cultured either alone or with 2.5×106 autologous γδ T cells for 24 and 72 hours. Control wells included MDM with no BCG infection, cultured alone or together with autologous γδ T cells.
2.6 BCG culture
BCG Danish strain 1331 was cultured in Middlebrook 7H9 broth supplemented with 10% OADC (oleic acid, albumin, dextrose, catalase) enrichment (BD Biosciences), and 0.05% Tween 80 (Sigma Aldrich) (7H9-OADC-T). BCG was maintained in 7H9-OADC-T at 37°C in 5% CO2 until used in co-cultures. Optical density (OD) was measured with a SmartSpec™ 3000 spectrophotometer (Bio Rad, Hercules, CA, USA).
2.7 BCG viability
BCG viability assay was performed as previously described Baquero and Plattner [28]. Briefly, after 72 hours, MDM cell cultures were collected in 7H9-OADC-T media and frozen at −80°C until ready for analysis. After thawing, cells were vortexed vigorously for 10 seconds and centrifuged at 400 × g for 2 minutes. Pellets were re-suspended in 7H9-OADC-T and incubated in 24-well plates for 24 h at 37°C in 5% CO2. Contents of each well were centrifuged at 4200 × g for 10 minutes and pellets were re-suspended in 50 μL of sterile saline solution in 2 mL conical tubes. 1 μL of fluorescein diacetate (FDA) (Sigma Aldrich) at a concentration of 2 mg/mL was added to each tube. After 30 min of incubation at 37°C, samples were analyzed by flow cytometry using a BD LSRFortessa X-20 (BD Biosciences, Franklin Lakes, New Jersey, USA). Standardization of the procedure and determination of gates was generated from known proportions of live and heat-killed BCG (Figure S2).
2.8 Cytokine Profile Secretion
Cell culture supernatants were collected after 72 hours of incubation in co-cultures, and stored at −80°C until thawed for ELISA analysis. Commercial bovine VetSet™ ELISA kits (Kingfisher Biotech, Saint Paul, MN, USA) were used to quantify bovine IFN-γ (detection range 0.125–8 ng/mL), IL-17 (detection range 0.188–12 ng/mL), and CXCL10 (detection range 0.188–12 ng/mL) according to manufacturer’s instructions. Optical density was measured using an Epoch microplate spectrophotometer (BioTek, Winooski, VT, USA).
2.9 Real-time PCR
Total RNA was extracted using the RNeasy Mini RNA Isolation Kit (Qiagen, Germantown, MD, USA), according to manufacturer’s instructions. Contaminating genomic DNA was removed using RNase-Free DNase digestion set (Qiagen), according to manufacturer’s instructions. The RNA concentration in each sample was measured by using a NanoDrop 8000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Total eluted RNA was reverse transcribed into cDNA using Random Primers and Superscript III Reverse Transcriptase per the manufacturer’s instructions (Invitrogen, Life Technologies). Quantitative real-time PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems). Forward and Reverse primers from previous published work [29–32] were used or the Integrated DNA Technologies PrimerQuest Tool was used to create custom primers that have not been previously published (listed in Table 1). Reactions were performed on Mx2005P qPCR System (Agilent Technologies). The following amplification conditions were used: 2 minutes at 50°, 10 minutes at 95°, 40 cycles of 15 seconds at 95°, and 1 minute at 60°, followed by a dissociation step, 15 seconds at 95°, 1 minute at 60°, 15 seconds at 95°, and 15 seconds at 60°. Relative gene expression was determined using the 2−ΔΔCt method with RPS9 as the reference housekeeping gene [33, 34].
Table 1.
Primers used for qPCR
| Gene | Accession Number | Strand | Sequence (5′-3′) | Reference |
|---|---|---|---|---|
| RPS-9 | NM_001101152.1 | Forward | CGCCTCGACCCAGAGCTGAAG | [29] |
| Reverse | CCTCCAGACCTCACGTTTGTTCC | |||
| IFN-γ | NM_174086.1 | Forward | AGAATCTCTTTCGAGGCCGGAG | [29] |
| Reverse | TATTGCAGGCAGGAGGACCATTAC | |||
| IL-17 | NM_001008412 | Forward | CACAGCATGTGAGGGTCAAC | [30] |
| Reverse | GGTGGAGCGCTTGTGATAAT | |||
| IL-10 | NM_174088.1 | Forward | TTACCTGGAGGAGGTGATG | [29] |
| Reverse | GTTCACGTGCTCCTTGATG | |||
| TNF-α | NM_173966.3 | Forward | CGGGGTAATCGGCCCCCAGA | [29] |
| Reverse | GGCAGCCTTGGCCCCTGAAG | |||
| IL-22 | NM_001098379 | Forward | GAGGTGCTGTTCCCCCAAT | [31] |
| Reverse | GAAGGGCACCACCTTTTC | |||
| IL-1β | X54796 | Forward | ATGGGTGTTCTGCATGAG | [29] |
| Reverse | AAGGCCACAGGAATCTTG | |||
| CCL1 | XM_002695632.3 | Forward | CCTCTGCCAGTGAAAGGAAA | |
| Reverse | GGAAGAAGGACTGGTGTGAAG | |||
| CCL2 | NM_174006.2 | Forward | CGCCTGCTGCTATACATTCA | |
| Reverse | GCTCAAGGCTTTGGAGTTTG | |||
| CCL4 | NM_001075147.2 | Forward | ATGGCTGCCTTCTGTTCTC | |
| Reverse | GGAATCTTCCGCAGAGTGTAA | |||
| CCL8 | NM_174007.1 | Forward | GCTCAGCCAGATTCAGTTTCT | |
| Reverse | GGTGATTCTCGTGTAGCTGTC | |||
| CCL24 | NM_174007.1 | Forward | TCCAACTCACAGGTTGCATAA | |
| Reverse | GCCTCACTACAAGACCAGAAG | |||
| CXCL10 | NM_001046551.2 | Forward | ACACCGAGGCACTACGTTCT | |
| Reverse | TAAGCCCAGAGCTGGAAAGA | |||
| GNLY | NM_001075143.1 | Forward | CTGCTGCTCCAAGGAGAAGA | [32] |
| Reverse | GCAGTGGAGGGAGTTTGGT | |||
| GZMB | NM_174296.2 | Forward | TATGCCCTCTACAGACAATCTA | |
| Reverse | CTTGGATCTCCAGCACATATC | |||
| NOS2 | NM_174296.2 | Forward | GGGAGATTGGAGGGAGATTA | |
| Reverse | TTGGTAGCAGGTCAAGTAAAG |
2.10 Statistical Analysis
ΔCt values were used in the statistical analysis of relative gene expression. ΔΔCt values were transformed (2−ΔΔCt) and are shown as expression relative to uninfected control samples, as appropriate. Data were analyzed using a paired one-way analysis of variance (ANOVA) and Tukey’s multiple comparisons test using Prism v7.0 (GraphPad Software, La Jolla, CA, USA). The mean and standard error of the mean (SEM) were calculated in experiments containing multiple data points. A P value of ≤0.05 was considered statistically significant.
3. Results
3.1 RNA Sequencing of M. bovis-specific γδ T cells
We have previously published that γδ T cells accumulate in the pulmonary lesions of chronic, M. bovis infected cattle. γδ T cells are hypothesized to play a role in skewing the adaptive immune response towards Th1 responses in the early stages of disease [35]; however, the role of γδ T cells in local immunity during the later stages of disease are not defined. Therefore, the first objective of our study was to identify novel functions for M. bovis-specific γδ T cells that are participating in the immune response during the later stages of disease. PBMC were prepared from the peripheral blood of 5 virulent M. bovis-infected calves. PBMC were re-stimulated in vitro for 18 hours with M. bovis PPD-b to the ensure cells were activated and responding specifically to M. bovis antigen. In control wells, PBMC were cultured with cRPMI and remained unstimulated. γδ T cells were then purified from both the stimulated and unstimulated cultures and mRNA was isolated for whole-transcriptome RNA sequencing. The γδ T cell response to M. bovis antigen was determined by comparing transcripts expressed in unstimulated control cultures with M. bovis PPD-b stimulated cultures from the same animal. Our transcriptomics analysis revealed a range of 299-2,240 genes that were > 2-fold differentially expressed (DE) between the unstimulated and PPD-b-stimulated γδ T cells for all five animals. Of all of the genes that were DE, 132 of those genes were common between all five animals. Sixty of the common DE genes were up-regulated, 60 genes were down-regulated, and 12 genes had mixed regulation between animals. Importantly, both IFN-γ and IL-17A were amongst the genes identified as being significantly upregulated in response to PPD-b. We and others have previously demonstrated that γδ T cells produce both cytokines in response to mycobacterial antigens [21, 36–38], therefore corroborating these previous results and confirming that the results of our RNASeq analysis were in agreement with previously published observations.
We selected the 132 genes that were commonly DE between γδ T cells from all five calves, and subjected them to a pathways analysis (Figure 1) where the most significant canonical pathways were revealed. Genes encoding for chemokines, regulatory cytokines, and cytotoxic factors, such as CXCL10, TNF, SOCS1 and Granzyme B represented a significant proportion of the genes involved in the immune system related pathways (Table 2). The next most significantly represented pathway was G protein signaling which included genes such as: adrenomedullin (ADM), endothelin β receptor (EDNRB), and platelet-activating factor receptor (PTAFR). Other genes relating to the metabolic pathway such as, apolipoprotein E (APOE), inhibin beta A chain (INHBA), and syntaxin-binding protein 1 (STXBP1) also comprised a significant portion of the pathways identified (Table 3). A complete list of the 132 genes that were DE amongst all five animals is presented in Table S1. Several immune-related genes had not been previously described in γδ T cells responding to M. bovis infection. These genes were selected analyzed by qRT-PCR as a means to validate our RNA sequencing results (Table 4).
Figure 1. Pathways related to the immune response are most significantly modulated in M. bovis-specific γδ T cells.
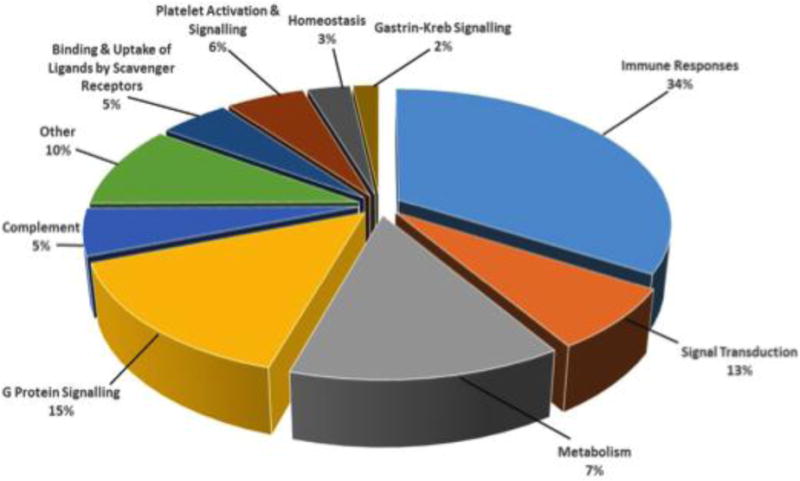
PBMCs were collected from calves (n=5) infected with virulent M. bovis and stimulated over-night with PPD-b or culture media alone. γδ T cells were isolated by magnetic separation and RNA was extracted and subjected to transcriptomics analysis. Differential gene expression values were calculated for each gene as the fold change of stimulated γδ T cells over mock treated γδ T cells for each animal. Genes commonly expressed between all five calves were subjected to a pathways analysis to reveal the most significant canonical pathways being represented by RNA sequencing results.
Table 2.
Immune system related genes that were differentially expressed by γδ T cells from all five M. bovis-infected calves in response to stimulation with PPD-b
| Gene | Fold Change | Function |
|---|---|---|
| IFN-γ | 99646.37 (± 69587.71) | Inflammatory cytokine |
| IL-17A | 88228.55 (± 52531.11) | Pro-inflammatory cytokine |
| CXCL10 | 80726.06 (± 31605.93) | Chemoattractant |
| GZMB | 27526.51 (± 20565.8) | Cytotoxicity |
| IL-15RA | 6464.67 (± 4269.81) | Proliferation |
| SOCS1 | 67.1 (± 19.08) | Negative feedback cytokine signaling |
| IL-1β | 26.11 (± 6.21) | Inflammatory cytokine |
| NOS2 | 25.99 (± 11.85) | Antimicrobial |
| TNF | 13.1 (± 3.16) | Inflammatory cytokine |
| CCL2 | 1.06 (± 0.1) | Immune cell recruitment |
Values are ± SEM
Table 3.
Genes relating to G protein signaling, Metabolism and Signal Transduction that were differentially expressed by γδ T cells from all five M. bovis-infected calves in response to stimulation with PPD-b
| Pathway | Gene | Name | Fold Change |
|---|---|---|---|
| G Protein Signaling | CXCL10 | Interferon gamma induced protein 10 | 80726.06 ± 31605.93 |
| LOC504773 | Regakine-1 | 6.87E-02 ± 3.74E-02 | |
| PTAFR | Platelet-activating factor receptor | 0.0145 ± 0.009 | |
| EDNRB | Endothelin B receptor | 0.0014 ± 0.001 | |
| ADM | ADM Adrenomedullin-11-26 Proadrenomedullin N-20 terminal peptide | 7285.56 ± 4096.94 | |
| Metabolism | APOE | Apolipoprotein E | 3.01E-02 ± 1.95E-02 |
| STXBP1 | Syntaxin-binding protein 1 | 5310.15 ± 2220.27 | |
| HMOX1 | Heme oxygenase 1 | 2.25E-02 ± 1.91E-02 | |
| CD36 | Platelet glycoprotein 4 | 5.18E-05 ± 1.14E-05 | |
| HBA | Hemoglobin subunit alpha | 19829.58 ± 7702.57 | |
| INHBA | Inhibin beta A chain | 7737.023 ± 8867.88 | |
| TNF | Tumor necrosis factor | 13.09 ± 3.16 | |
| Signal Transduction | |||
| CISH | Cytokine-inducible SH2-containing protein | 10123.22± 8427.81 | |
| ISG15 | Ubiquitin-like protein ISG15 | 34.81 ± 12.26 | |
| TYROBP | TYRO protein tyrosine kinase binding protein | 0.045 ± 0.016 |
Values are ± SEM
Table 4.
Confirming RNA sequencing with qPCR.
| Gene | RNA Sequencing | RT-qPCR |
|---|---|---|
| IFN-γ | 99646.37 (± 69587.71) | 2145.75 (± 1994.26) |
| IL-17 | 88228.55 (± 52531.11) | 1546.28 (± 1343.04) |
| TNF-α | 13.1 (± 3.16) | 523.49 (± 493.45) |
| CCL2 | 1.06 (± 0.1) | 100.79 (±68.69) |
| CXCL10 | 80726.06 (± 31605.93) | 247.42 (± 196.03) |
| NOS2 | 25.99 (± 11.85) | 87.38 (± 57.96) |
| SOCS1 | 67.11 (± 19.08) | 24.16 (± 9.03) |
| Granzyme B | 27526.51 (± 20565.78) | 29.8 (± 20.77) |
γδ T cells from calves that were subjected to RNA sequencing were also analyzed by qPCR to confirm sequencing results. Values indicate average fold change ± SEM in gene expression between unstimulated and PPD-b stimulated γδ T cells isolated from M. bovis-infected calves (n=5).
3.2 Evaluation of γδ T cell cytokine and chemokine production in situ in response to M. bovis infection
Our transcriptomics analysis gave us a better understanding of the M. bovis-specific response of peripheral γδ T cells; however, systemic immune responses are often not an accurate measure of the response at the site of BTB infection. We next wanted to compare the peripheral and localized responses, as well as gain a better understanding of cytokine and chemokine production by γδ T cells at the site of M. bovis infection. Tissue samples from the lungs and mediastinal lymph nodes were collected from 5 Holstein calves, at 3 months post infection with virulent M. bovis. The tissue samples were evaluated by RNAScope, an in situ hybridization assay that allows for the detection of target RNA within intact cells. This assay was utilized as it is much more sensitive and specific than other in situ assays. Proprietary probes were used to stain for mRNA transcripts of the γδ TCR, IFN-γ, IL-10, IL-17, IL-22 and CCL2 (Figure 2). All granulomas were determined to be late-stage based on exhibition of necrotic centers with numerous lymphocytes, including γδ T cells, surrounding the periphery [5–8]. Ten representative images at 100X magnification were taken around the periphery of each late-stage granuloma and were used to quantify instances of co-expression between γδ T cells and the cytokine/chemokine of interest (Table 5).
Figure 2. γδ T cells express various cytokines and chemokines within late-stage granulomas.
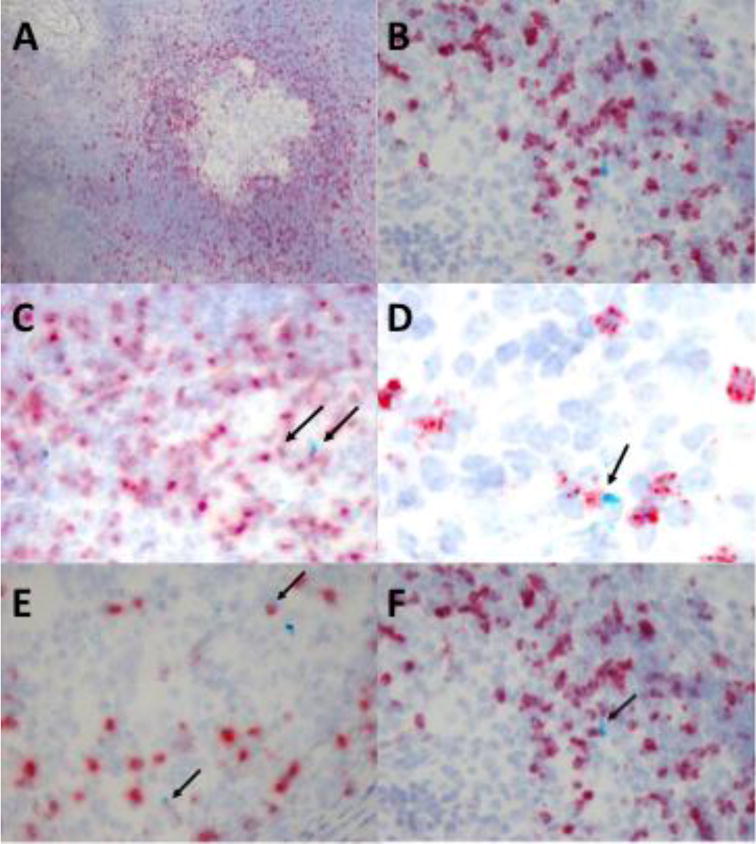
Tissue samples from granulomatous lesions in the lungs and mediastinal lymph nodes were harvested from calves (n=5) 3 months post-infection with virulent M. bovis. Tissue sections were preserved onto slides by formalin fixation and paraffin embedding. RNAScope was used for in situ analysis of mRNA transcripts of various cytokines/chemokines (turquoise) and the γδ TCR (red). Characteristic TB granuloma at 10X magnification (A) and at 40X magnification (B) within the lymph node of a chronically infected animal. Arrows indicate instances of γδ T cell and IFN-γ (C) IL-10 (D) CCL2 (E) IL-17 (F) co-expression.
Table 5.
Quantitative measure of mRNA labeling using RNAScope for various cytokines and chemokines expressed by γδ T cells within bovine pulmonary tuberculoid granulomas.
| Lung (n=5) | Lymph node (n=5) | |
|---|---|---|
| IFN-γ | 6.94% (± 4%) | 22.82% (± 9%) |
| IL-10 | 4.68% (± 3%) | 16.45% (± 1%) |
| IL-17 | 12.00% (± 8%) | 12.87% (± 8%) |
| CCL2 | 38.22% (± 9%) | 21.37% (± 6%) |
Tissue samples from granulomatous lesions in the lungs and mediastinal lymph nodes were harvested from calves (n=5) 3 months post-infection with virulent M. bovis. Tissue sections were preserved onto slides by formalin fixation and paraffin embedding. RNAScope was used for in situ analysis of mRNA transcripts of various cytokines/chemokines and the γδ TCR. Ten representative images at 100X magnification from around the periphery of the granuloma were used for quantification. Values represent the percentage of γδ T cells expressing the cytokine or chemokine of interest out of the total number of cells expressing the cytokine or chemokine of interest ± SEM (n=4 for CCL2 in the lung).
Fewer than 10% of the cells in the lungs expressing transcripts for IFN-γ at this time point were γδ T cells (Figure 2C and Table 5). In contrast, γδ T cells comprised greater than 20% of the cells expressing IFN-γ within the lymph nodes of these same animals. CCL2 expression is known to be upregulated in TB granulomas, and has been associated with increased disease severity in both humans and animals, although the source of CCL2 within the granuloma has not been defined [39–42]. We observed that γδ T cells comprise a significant proportion of the cells expressing transcripts for CCL2 within both the lungs and the lymph nodes of infected animals (Figure 2E, Table 5). Expression of IL-10, IL-17, and IL-22 is also upregulated during TB, and γδ T cells have been implicated as a significant source of these cytokines at the site of infection [43–45]. However, in contrast to published reports, we found that fewer than 20% of cells producing IL-10 or IL-17 were γδ T cells (Figure 2 and Table 5), and we observed no γδ T cells expressing IL-22 in the tissues at this stage of infection (data not shown). Our in situ results confirm that γδ T cells are a significant population surrounding the periphery of late-stage lesions, and that these cells are an important source of immune factors that play a role in shaping the local response to M. bovis.
3.3 Cytokine expression during initial interactions between γδ T cells and BCG-infected macrophages
Measuring the early immune response to TB at the site of pulmonary infection is challenging due to the difficulty locating and isolating granulomas prior to the development of grossly apparent lesions. These lesions are often undetectable until 2–4 weeks after infection [6]. Further, in situ analysis of tissues collected at the time of necropsy in a chronically infected animal allows for assessment of only a single time point after infection. Measuring a single time point does not allow for functional analysis of the viable immune cells that accumulate at the site of infection. M. bovis is primarily transmitted to cattle via the aerosol route [46]. Therefore, the first cells likely to encounter the infection are macrophages residing in the lungs and upper respiratory tract, followed by other lung-resident sentinel cells. γδ T cells represent a significant proportion of the sentinel cells lining the respiratory mucosa [47]. Our next objective was to develop an in vitro model that allowed us to assess the initial interactions that may occur between M. bovis-infected macrophages and sentinel γδ T cells during the early stages of infection. Due to technical and logistical difficulties, we were unable to utilize alveolar macrophages for our in vitro co-culture studies. As an alternative, we developed a γδ T cell and MDM co-culture system using γδ T cells from M. bovis-infected or M. bovis-naïve animals, cultured with autologous MDM. Peripheral blood monocytes were isolated from naïve or virulent M. bovis-infected calves, and cultured for 7 days with GM-CSF to generate MDM [27].
On day 7, MDM were infected at 1 or 10 MOI with M. bovis BCG Danish Strain 1331. Control cells remained uninfected. 4 hours later, autologous γδ T cells were added at a ratio of 1:5 with MDM. The co-cultures were collected for qPCR analysis after 18 hours of incubation. Cell culture supernatants were collected after 72 hours of co-culturing and preserved for measurements of cytokine secretion by ELISA. We measured expression of IL-17 and IFN-γ, cytokines known to be upregulated by γδ T cells responding to M. bovis infection [38, 41, 44], and utilized the results from our M. bovis-specific γδ T cell RNA sequencing data to select the biological factors to assess in our co-culture experiments. Co-cultured γδ T cells and MDM from virulent M. bovis-infected animals expressed higher levels of IFN-γ and IL-1β than did co-cultured γδ T cells and MDM from M. bovis-naïve animals, regardless of MOI (Figure 3). Inter-animal variability was a complication in our co-culture experiments; however, general trends were observed. Co-cultured cells from naïve animals tended to express increased levels of IL-10, IL-22, and TNF-α compared to co-cultured cells from infected animals. In the naïve animals, a lower MOI correlated with increased expression of IL-22 and TNF-α, while a MOI of 10 was correlated with increased IL-10 expression (Figure 3). No significant differences in IL-17 expression were detected between γδ T cells/MDM co-cultures from naïve compared to M. bovis-infected animals.
Figure 3. Cytokine expression in γδ T cell/MDM co-cultures from naïve and M. bovis-infected calves stimulated with BCG.
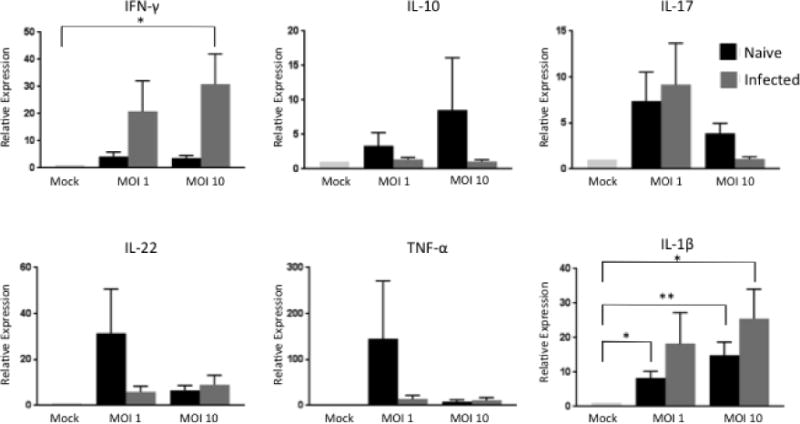
MDM and autologous γδ T cells isolated from M. bovis-infected or naïve animals were cultured together for 24 hours after a 4 hour infection with BCG at an MOI of 1 or 10. RNA was extracted and reverse transcribed into cDNA and qPCR was performed for various inflammatory, anti-inflammatory, and regulatory cytokines. Results were normalized to the housekeeping gene RPS-9, and expressed relative to uninfected γδ/MDM co-culture (mock) samples. Data represent means ± SEM (n=19 for naïve group and n=10 for infected group) (* P≤ 0.05; ** P≤0.01; ANOVA).
3.4 Chemokine expression during initial interactions between γδ T cells and BCG-infected MDM
γδ T cells are thought to contribute to the establishment and maintenance of well-organized granulomas [19, 48]. The results of our RNA sequencing analysis suggested that γδ T cells express transcripts for a number of chemokines that may play a role in recruiting other immune cells to the site of infection. Therefore, we used qPCR to measure expression of CCL1, CCL2, CCL4, CCL8, CCL24 and CXCL10 in our BCG-infected γδ T cell/MDM co-cultures. As seen in Figure 4, co-cultures from virulent M bovis-infected animals tended to express increased levels of CCL2, CCL8, and CXCL10 compared to co-cultures from naïve animals. CCL2 and CCL8 expression were greatest when MDM were infected at an MOI of 1, while CXCL10 expression was greatest at the higher MOI (Figure 4). Co-cultures from naïve animals tended to express increased levels of CCL1 and CCL24 compared to M. bovis-infected animals. CCL4 expression was significantly increased in co-cultures from both naïve and infected animals at an MOI of 10; however, co-cultured cells from M. bovis-infected animals expressed more CCL4 than the naïve animals at the lower MOI. Our results show γδ T cells are capable of expressing numerous chemokines in response to M. bovis infection, and it is likely that these chemokines play an important role in immune cell recruitment to the site of infection.
Figure 4. Chemokine expression in γδ T cell/MDM co-cultures from naïve and M. bovis-infected calves stimulated with BCG.
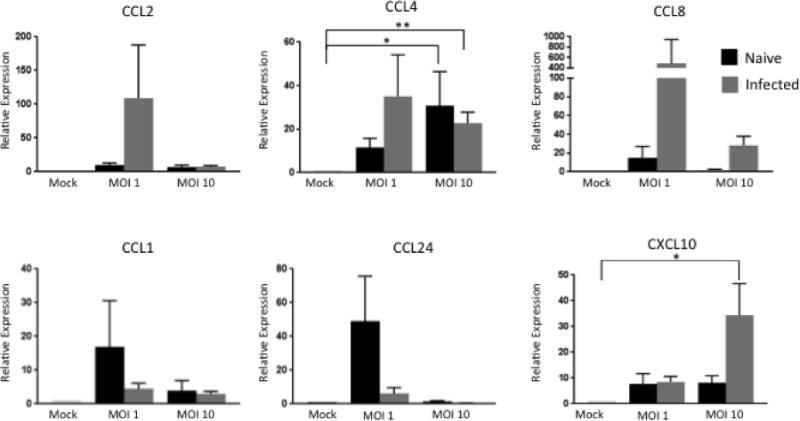
MDM and autologous γδ T cells isolated from M. bovis-infected or naïve animals were cultured together for 24 hours after a 4 hour infection with BCG at an MOI of 1 or 10. RNA was extracted and reverse transcribed into cDNA and qPCR was performed on various chemokines. Results were normalized to the housekeeping gene RPS-9, and expressed relative to uninfected γδ/MDM co-culture (mock) samples. Data represent means ± SEM (n=19 for naïve group and n=10 for infected group) (* P≤ 0.05; ** P≤0.01; ANOVA).
3.5 Cytokine and chemokine production during initial interactions between γδ T cells and BCG-infected MDM
To confirm the results of our qPCR analysis, commercial ELISA kits were used to measure the concentration of selected cytokines and chemokines in cell culture supernatants isolated from γδ T cell/MDM co-cultures after 72 hours. Consistent with our qPCR analysis, co-culturing γδ T cells from M. bovis-infected animals in direct contact with BCG-infected MDM resulted in increased production of IFN-γ, IL-17, and CXCL10 compared to BCG-infected MDM cultured alone (Figure 5). A higher MOI induced increased production of IFN-γ and CXCL10 in γδ T cell/MDM co-cultures from virulent M. bovis-infected animals, while the MOI did not seem to have an effect on IL-17 production. No significant IFN-γ, IL-17 or CXCL10 production was detected in cell culture supernatants from naïve animal co-cultures. Our ELISA results corroborate the results of our qPCR analysis and confirm that γδ T cells are an important source of IFN-γ, IL-17, and CXCL10 during M. bovis infection.
Figure 5. γδ T cells from M. bovis-infected calves are the main source of IL-17, IFN-γ and CXCL10 when in contact with BCG-infected MDM.
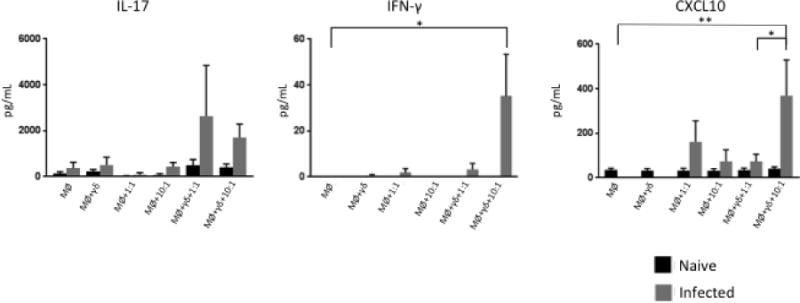
Commercial ELISA kits were used to measure IL-17A, IFN-γ, and CXCL10 from the supernatants of uninfected and BCG-infected co-cultures of MDM in direct contact with autologous γδ T cells. Data represent mean ± SEM (n=19 for naïve group and n=10 for infected group) (* P≤ 0.05; ** P≤0.01; ANOVA).
3.6 Expression of cytotoxic factors during initial interactions between γδ T cells and BCG-infected MDM
γδ T cells have been found to have cytotoxic capabilities during early TB infection. One group found that γδ T cells in cattle are able to directly kill macrophages that are infected with M. bovis [49]. In support of this previous report, our transcriptomics analysis also revealed that γδ T cells upregulate expression of both Granzyme B and nitric oxide synthase (NOS2) in response to stimulation with PPD-b. Therefore, we evaluated expression of the cytotoxic factors granzyme B, NOS2 and granulysin in our γδ T cell/MDM co-cultures. As seen in Figure 6, co-cultures from M. bovis-infected animals tended to express higher levels of granulysin and granzyme B compared to the co-cultured cells from naïve animals, and this expression was most pronounced at an MOI of 10 compared to the lower MOI. NOS2 was more highly expressed by infected animals at the lower MOI, with no difference between groups at the higher MOI.
Figure 6. Cytotoxic factors expressed in γδ T cell/MDM co-cultures from naïve and M. bovis-infected calves stimulated with BCG.
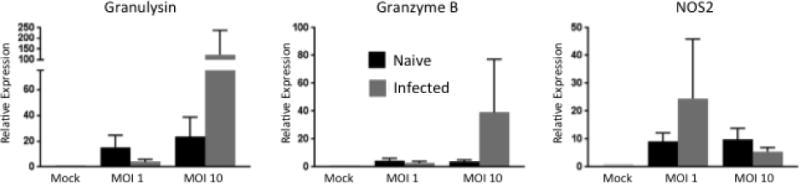
MDM and autologous γδ T cells isolated from M. bovis-infected or naïve animals were cultured together for 24 hours after a 4 hour infection with BCG at an MOI of 1 or 10. RNA was extracted and reverse transcribed into cDNA and qPCR was performed on various cytolytic factors. Results were normalized to the housekeeping gene RPS-9, and expressed relative to uninfected γδ/MDM co-culture (mock) samples. Data represent means ± SEM (n=19 for naïve group and n=10 for infected group).
To determine the functional cytotoxic capacity of γδ T cells interacting with MDM, we analyzed BCG viability within our co-culture systems. Cell cultures were collected, stained with FDA, and analyzed by flow cytometry to quantify amounts of live BCG present using a protocol adapted from Baquero and Plattner [28]. As seen in Figure 7, BCG viability was significantly reduced in γδ T cell/MDM co-cultures established from virulent M. bovis-infected calves, compared to co-cultures established from M. bovis-naïve animals. This reduction was most apparent in cultures infected with the higher MOI. Together, our results suggest that in addition to a role in recruitment of immune cells to developing granulomas, that γδ T cells may contribute to the control of M. bovis infection through their ability to directly impact mycobacterial viability.
Figure 7. BCG viability is reduced when cultured with cells from M. bovis-infected animals.
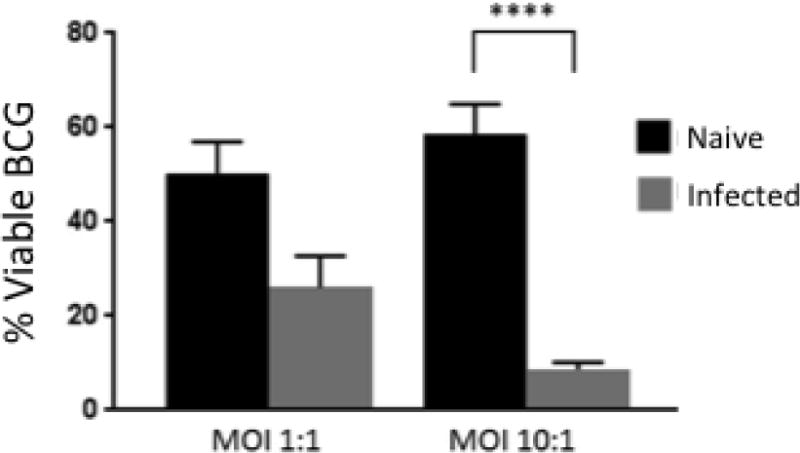
Cells were collected 72 hours after infection with BCG and frozen until ready for analysis. After thawing, cells were re-suspended in 7H9-OADC-T and incubated in for 24 h. Cells were centrifuged and re-suspended in saline solution, and 1 μL of FDA at a concentration of 2 mg/mL was added to each tube. After 30 min of incubation at 37°C, samples were analyzed by flow cytometry. Data represent means ± SEM (n=19 for naïve group and n=8 for infected group) (**** P≤0.0001; ANOVA).
4. Discussion
γδ T cells are thought to play an important role during M. bovis infection in cattle primarily through their production of IFN-γ, which is critical for promoting the development of a strong Th1 immune response [21]. In addition to production of IFN-γ, γδ T cells have the capacity for a variety of other innate and adaptive immune functions, such as regulatory cytokine production and chemokine production [50–53]. As a significant population in the respiratory mucosa, it is highly likely that γδ T cells participate in local immunity to M. bovis infection. However, their specific role at the site of infection, particularly with regard to these so-called alternative functions, has not been well characterized. To the best of our knowledge, we are the first to perform transcriptomics analysis on γδ T cells responding to M. bovis infection. Through this approach, we identified thousands of genes that were DE between unstimulated and PPD-b stimulated peripheral γδ T cells. Many of these genes have not been previously described, thus providing significant insight into the diverse functions of these cells during Mycobacterium infection. Of particular interest, we observed upregulation of a number of cytokine genes, some of which were expected (IFN-γ, IL-17A, IL-10) and some that have not been previously described. TNF-α was identified in our transcriptomics analysis as a gene that was upregulated by γδ T cells in response to M. bovis antigen. In our in vitro co-culture assay, TNF-α appeared to be more predominantly expressed by γδ T cells isolated from naïve animals compared to γδ T cells from M. bovis-infected animals. The initial stages of granuloma formation are dependent upon the production of TNF-α, as its signaling is crucial in maintaining chemokine concentrations to mediate early immune cell recruitment [54–56]. Human γδ T cells have been shown to produce TNF-α in response to M. tb infection [57]; however the expression of TNF-α by bovine γδ T cells responding to M. bovis infection has not been previously described. Its significant upregulation in our in vitro co-culture system, particularly in co-cultures from naïve animals, supports an important role for this cytokine during the early stages of infection.
IL-1β was also identified as being significantly up-regulated by M. bovis-specific γδ T cells in our transcriptomics analysis, and in our γδ T cell/macrophage co-cultures. IL-1β has been found to play an important role in the anti-mycobacterial response by aiding in macrophage destruction of mycobacteria [58, 59]; however, its expression by γδ T cells has not been previously characterized during M. bovis infection. One caveat to our transcriptomics analysis is the approach used to prepare our γδ T cells. The cells were stimulated in a mixed PBMC culture and then purified by magnetic beads. Positive selection of αβ T cells is know to impact gene expression and T cell activation [60]. The impact of positive selection is less clear for γδ T cells. Our experiments were controlled by comparing gene expression to unstimulated γδ T cells that were cultured and sorted in the same manner. However, it is possible that crosslinking of the γδ TCR during positive magnetic cell sorting had an impact on the activation status of the cell and IL-1β expression may be an artifact of the isolation process, rather than a component of the physiologic γδ T cell response. Another possible explanation for our observation is the purity of our γδ T cell preparations. Our magnetic sorting protocol yielded γδ T cell purities of >90%. However, lllumina sequencing is highly sensitive and it is possible that this low level of contamination may have included a myeloid population that was expressing IL-1β. In our co-culture experiments, we analyzed gene expression by MDM cultured together with γδ T cells; therefore, we cannot rule out the possibility that the increased IL-1β expression observed in our co-cultures was also due to contributions from the co-cultured MDM.
It is hypothesized that the chemokine response by infected cells during the initial infection are crucial for the control of the invading mycobacteria. The chemokine response during chronic infection likely aids in granuloma formation and maintenance, in attempt to physically wall-off the pathogens [44]. Our results suggest a role for γδ T cells in promoting the formation and maintenance of a well-organized granuloma. Our sequencing and co-culture system allowed us to identify several novel chemokines that were upregulated by γδ T cells responding to M. bovis antigen, including CCL4, CCL8 and CXCL10. CCL4 production in the context of TB has been previously described [61], however not in the bovine model and not by γδ T cells. We observed increased CCL4 expression in the co-cultures from both M. bovis-infected and naïve animals, suggesting a potential role for this chemokine throughout BTB disease progression. Consistent with our results in BTB, γδ T cells have been previously shown to express CCL8 in response to Anaplasma marginale infection [52]. Our findings suggest γδ T cell production of CCL8 may be an important mediator in cell recruitment during chronic BTB infection. CXCL10 was identified as differentially upregulated in our transcriptomics analysis, and was up-regulated in co-cultures from M. bovis-infected animals compared to M. bovis-naïve animals. CXCL10 is known to be increased in the stage I and stage IV granulomas in cattle infected with M. bovis, however the source of CXCL10 has not been identified [44, 45]. Our results suggest that γδ T cells may be one important source of CXCL10 at the site of infection.
Interestingly, through our in situ analysis, we identified CCL2 as a chemokine that was highly expressed by γδ T cells at the site of M. bovis infection. However, we did not observe significant CCL2 expression in our in vitro co-cultures, nor was this chemokine identified as significantly DE in our transcriptomics analysis. CCL2 expression is significantly upregulated in M. tb granulomas, and has been associated with increased disease severity in both humans and animals, although the source of CCL2 within the granulomas was not been defined in these previous reports [39–41]. In the report by Alvarez et al., depletion of γδ T cells from M. bovis- infected mice resulted in a significant reduction in CCL2 expression. However, the authors concluded that γδ T cells themselves were not a significant source of the chemokine, but indirectly contributed to its production. The reasons for this disparity are unclear; however, our results rely on the more physiologic model of M. bovis infection in cattle, while the results by Alvarez et al. employed a murine model of BTB infection. The increased expression of CCL2 by γδ T cells at the site of infection may support effective granuloma formation, leukocyte recruitment, or bacterial containment within the granuloma.
γδ T cells from non-human primates are a significant source of IL-22 during M. tb infection, both in the peripheral blood and at the site of infection [62]. In agreement, Steinbach et al. recently described bovine γδ T cells from the periphery as being major producers of IL-22 during infection with M. bovis [43]. Consistent with these previous reports, we measured significant expression of IL-22 in our γδ T cell/macrophage co-cultures; in contrast, however, we observed no expression of IL-22 by γδ T cell within late-stage M. bovis granulomas. This is in agreement with Aranday-Cortes et al., who observed a decrease in IL-22 levels by qPCR in late-stage granulomas from M. bovis-infected cattle [43, 44]. Similarly, Palmer et al. observed very low levels of expression of IL-22 by RNAScope in any stage of lesion collected at 150 days post M. bovis infection [63]. In human patients chronically infected with M. tb, cellular immune responses become depressed [64]; therefore, it may not be surprising that decreased levels of IL-22 were found at the later stages of infection.
The disparities we observed in both CCL2 and IL-22 expression between our in vitro re-stimulation assay and our in situ analysis highlight possible differences between the systemic and mucosal immune responses during TB [65].and underline the importance of analyzing the local immune response rather than relying solely on in vitro assays with peripheral blood populations. Our in vitro co-culture experiments were designed to model the interactions between M. bovis infected macrophages and γδ T cells that may occur during the early stages of BTB infection. Unfortunately, due to technical challenges, we were unable to utilize alveolar macrophages or lung-resident γδ T cell populations for our in vitro co-culture experiments. While we believe that many aspects of our co-culture system are an accurate reflection of the interactions that may occur during BTB infection, the use of peripheral blood populations is an important caveat that may impact the interpretation of our results. Our future experiments will address the interactions between tissue-resident γδ T cells and M. bovis-infected alveolar macrophages, and determine how these responses differ compared to circulating immune cells.
In mice, CD27neg γδ T cells are an important source of innate IL-17 production during mycobacterium infection [reviewed in 66]. In humans, there is no clear evidence of innate IL-17 production by γδ T cells. Instead, human Vγ2Vδ2 T cells need to be polarized in the periphery to express IL-17, much like CD4 T cells [67]. Interestingly, we did not observe innate IL-17 production by bovine γδ T cells in response to BCG infection our co-culture system, suggesting that cattle may be more similar to human γδ T cells with regard to innate IL-17 production. Interestingly, however, we have previously observed innate IL-17 production by bovine γδ T cells in response to viral infection and in response to TLR stimulation [53, 68]. M. bovis employs a number of immune evasion strategies, including altering inflammatory and regulatory cytokine production [69]. Thus, active suppression by the live BCG used in our co-cultures may be one possible explanation for the lack of innate IL-17 production by γδ T cells in our co-culture assays.
After discovering multiple cytotoxicity genes upregulated in our γδ T cell RNA sequencing results, we evaluated the expression of cytotoxic genes in our co-culture systems. In agreement with our RNASeq analysis, we measured increased expression of NOS2, granzyme B, and granulysin in our co-cultures from both M. bovis-infected and naïve animals. Although the expression of these genes in our co-cultures could be attributed to either MDM or γδ T cells, the results of our transcriptomics analysis were highly suggestive that γδ T cells were a significant contributing source for all three transcripts. Granulysin expression in our co-culture system was expected, as a bovine homologue of granulysin has been previously identified, and production of granulysin by γδ T cells has been previously described in the context of human TB [32, 70]. Until recently, NOS2 expression has been primarily attributed to myeloid cells, and the production of iNOS has been well documented in human and murine macrophages in response to TB and BCG infection [71–73]. However, recent studies have also established that lymphoid cells have the capacity to express NOS2 [74, 75]. A report in mice has also shown that NOS2 expression has a significant impact on the viability and proliferative capacity of γδ T cells in vivo [72]. To our knowledge, ours is the first report describing NOS2 expression by γδ T cells in cattle, and the first to demonstrate this expression in the context of BTB. Importantly, in addition to expressing molecules associated with cytotoxicity, we determined that γδ T cells had the functional capacity to eliminate M. bovis-infected cells in our in vitro co-culture system. This result is in agreement with a previous report by Martino et al., showing that BCG-infected human monocyte-derived dendritic cells induce the development of a functionally cytotoxic central memory Vγ9Vδ2 T cell population that is highly efficient at killing infected monocytes in vitro [76]. Similarly, a report by Skinner et al., demonstrated that in vitro, bovine γδ T cells in cattle can directly kill macrophages that are infected with M. bovis [49]. In vivo, γδ T cells from non-human primates have been shown to express both perforin and granulysin [77]. The capacity of bovine γδ T cells for cytotoxicity at the site of M. bovis infection is unknown and will be the subject of future research in our laboratory.
Measuring the function of immune cells within developing TB granulomas remains challenging. This is primarily due to technical difficulties in locating lesions during the early stages of infection, and in isolating viable immune cells from lesions during the later stages of infection. However, utilizing RNAScope allowed us to look at mRNA expression of cytokines and chemokines by γδ T cells directly at the site of late BTB infection. Consistent with our own previous work [42], and reports from others [8, 44], we observed γδ T cells accumulating in the lymphoid mantle surrounding the periphery of late-stage lesions. In our RNAscope analysis, γδ T cells comprised a significant portion of the immune cells expressing IFN-γ within lymph node granulomas, although this proportion was much reduced within lung granulomas. This difference in tissue cytokine expression is in agreement with recent results by Palmer et al., who observed significant differences in inflammatory cytokine expression between lung and lymph node lesions [63]. Although the biological significance of these findings is unclear, it suggests that anatomic location has a significant impact on the host immune response to M. bovis. We were surprised to observe relatively few cells, γδ T cells or otherwise, expressing transcripts for any of the inflammatory cytokines that we measured. However, our findings are in agreement with another recent report by Palmer et al., which used RNAscope to quantify overall expression of inflammatory cytokines, including IFN-γ, within virulent M. bovis granulomas at various stages of development [45]. In this study, Palmer et al. observed no significant expression of IL-10 and only low levels of IL-17, while the most highly expressed transcripts included IFN-γ and the chemokines CXCL9 and CXCL10. It is important to recall that in situ analyses are limited to only a single time point, often belying the complex and dynamic interactions that are occurring during an active M. bovis infection.
In summary, our findings show that γδ T cells are extremely dynamic and are capable of producing a number of different cytokines and chemokines, suggesting an important role for these cells throughout disease progression at the site of M. bovis infection. Although peripheral γδ T cells were initially used for RNAseq, the results allowed us to identify novel immune factors that could potentially play a role at the site of M. bovis infection. Utilizing RNAScope allowed us to look directly at the site of infection. However, it is important to note that the tissue samples collected for this study depict only a brief snap-shot of γδ T cell function within granulomas during chronic infection. Future studies should be aimed further describing γδ T cell functions at the site of infection over the full course of M. bovis infection. Moreover, our unique co-culture approach to mock initial interactions of γδ T cells with infected macrophages at the site of infection gave us insight into γδ T cell expression of immune factors during the early stages of granuloma development. Future studies utilizing alveolar macrophages in the co-culture system would allow for a more physiologic model of the initial interactions between γδ T cells and infected macrophages at the site of infection. Further studies utilizing trans-well systems or blocking antibodies would also allow for a better understanding of γδ T cell interactions with innate immune cells. Taken together, our findings strongly support the hypothesis that γδ T cells play a dynamic role in immune cell recruitment and granuloma maintenance during the immune response to BTB infection
Highlights.
γδ T cells accumulate at the site of Mycobacterium bovis infection
γδ T cells express CCL2 and IFN-γ within late-stage TB granulomas
In vitro, γδ T cells express CCL4, CCL8 and CXCL10 in response to infected MDM
In vitro, γδ T cells express cytotoxic factors and reduce BCG burden in infected MDM
Acknowledgments
Research reported in this publication was made possible in part by Jenny Hackett and the services of the KU Genome Sequencing Core. The authors would like to also thank Dr. Susan Brown from the KSU Bioinformatics Core for the assistance and expertise. This project was supported by the KU Center for Molecular Analysis of Disease Pathways COBRE P20 GM103638, which is funded by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health and Kansas State University Start-up Funds to J.L. McGill.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interests
The authors declare no conflict of interests
References
- 1.World Health Organization. Global Tuberculosis Report. World Health Organization; Geneva: 2016. [Google Scholar]
- 2.Waters WR, Palmer MV, Buddle BM, Vordermeier HM. Bovine tuberculosis vaccine research: Historical perspectives and recent advances. Vaccine. 2012;30:2611–2622. doi: 10.1016/j.vaccine.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 3.VanRhijn, Godfroid J, Michel A, Rutten V. Bovine tuberculosis as a model for human tuberculosis: advantages over small animal models. Microbes and Infection. 2008;10:711–715. doi: 10.1016/j.micinf.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Waters WR, Palmer MV, Thacker TC, Davis WC, Sreevatsan S, Coussens P, Meade KG, Hope JC, Estes DM. Tuberculosis Immunity: Opportunities from Studies with Cattle. Clinical and Developmental Immunology. 2011;2011:768542. doi: 10.1155/2011/768542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thoen CO, Steele JH, Kaneene JB. Zoonotic Tuberculosis: Mycobacterium bovis and Other Pathogenic Mycobacteria. John Wiley & Sons; 2014. [Google Scholar]
- 6.Palmer MV, Waters WR, Thacker TC. Lesion Development and Immunohistochemical Changes in Granulomas from Cattle Experimentally Infected with Mycobacterium bovis. Veterinary Pathology. 2007;44:863–874. doi: 10.1354/vp.44-6-863. [DOI] [PubMed] [Google Scholar]
- 7.Rhoades ER, Frank AA, Orme IM. Progression of chronic pulmonary tuberculosis in mice aerogenically infected with virulent Mycobacterium tuberculosis. The International Journal of Tuberculosis and Lung Disease. 1997;78:57–66. doi: 10.1016/s0962-8479(97)90016-2. [DOI] [PubMed] [Google Scholar]
- 8.Wangoo A, Johnson L, Gough J, Ackbar R, Inglut S, Hicks D, Spencer Y, Hewinson G, Vordermeier M. Advanced Granulomatous Lesions in Mycobacterium bovis-infected Cattle are Associated with Increased Expression of Type I Procollagen, γδ (WC1+) T Cells and CD 68+ Cells. Journal of Comparative Pathology. 2005;133:223–234. doi: 10.1016/j.jcpa.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Flynn J, Chan J, Lin P. Macrophages and control of granulomatous inflammation in tuberculosis. Mucosal Immunology. 2011;4:271–278. doi: 10.1038/mi.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gideon HP, Phuah J, Myers AJ, Bryson BD, Rodgers MA, Coleman MT, Maiello P, Rutledge T, Marino S, Fortune SM, Kirschner DE, Lin PL, Flynn JL. Variability in Tuberculosis Granuloma T Cell Responses Exists, but a Balance of Pro- and Anti-inflammatory Cytokines Is Associated with Sterilization. PLoS Pathogens. 2015;11:e1004603. doi: 10.1371/journal.ppat.1004603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayday AC. γδ cells: a right time and a right place for a conserved third way of protection. Annual Review of Immunology. 2000;18:975–1026. doi: 10.1146/annurev.immunol.18.1.975. [DOI] [PubMed] [Google Scholar]
- 12.Kabelitz D. γδ T-cells: cross-talk between innate and adaptive immunity. Cellular and Molecular Life Sciences. 2011;68:2331. doi: 10.1007/s00018-011-0696-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hein WR, Mackay CR. Prominence of γδ T cells in the ruminant immune system. Immunology Today. 1991;12:30–34. doi: 10.1016/0167-5699(91)90109-7. [DOI] [PubMed] [Google Scholar]
- 14.Jutila MA, Holderness J, Graff JC, Hedges JF. Antigen-independent priming: a transitional response of bovine γδ T-cells to infection. Animal Health Research Reviews. 2008;9:47–57. doi: 10.1017/S1466252307001363. [DOI] [PubMed] [Google Scholar]
- 15.Price S, Davies M, Villarreal-Ramos B, Hope J. Differential distribution of WC1+ γδ TCR+ T lymphocyte subsets within lymphoid tissues of the head and respiratory tract and effects of intranasal M. bovis BCG vaccination. Veterinary Immunology and Immunopathology. 2010;136:133–137. doi: 10.1016/j.vetimm.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Dieli F, Ivanyi J, Marsh P, Williams A, Naylor I, Sireci G, Caccamo N, Di Sano C, Salerno A. Characterization of Lung γδ T Cells Following Intranasal Infection with Mycobacterium bovis Bacillus Calmette-Guérin. J Immunol. 2003;170:463. doi: 10.4049/jimmunol.170.1.463. [DOI] [PubMed] [Google Scholar]
- 17.Doherty ML, Bassett HF, Quinn PJ, Davis WC, Kelly AP, Monaghan ML. A sequential study of the bovine tuberculin reaction. Immunology. 1996;87:9–14. [PMC free article] [PubMed] [Google Scholar]
- 18.Cassidy JP, Bryson DG, Pollock JM, Evans RT, Forster F, Neill SD. Early lesion formation in cattle experimentally infected with Mycobacterium bovis. Journal of Comparative Pathology. 1998;119:27–44. doi: 10.1016/S0021-9975(98)80069-8. [DOI] [PubMed] [Google Scholar]
- 19.D’Souza CD, Cooper AM, Frank AA, Mazzaccaro RJ, Bloom BR, Orme IM. An anti-inflammatory role for gamma delta T lymphocytes in acquired immunity to Mycobacterium tuberculosis. J Immunol. 1997;158:1217. [PubMed] [Google Scholar]
- 20.Smith RA, Kreeger JM, Alvarez AJ, Goin JC, Davis WC, Whipple DL, Estes DM. Role of CD8+ and WC-1+ gamma/delta T cells in resistance to Mycobacterium bovis infection in the SCID-bo mouse. Journal of Leukocyte Biology. 1999;65:28–34. doi: 10.1002/jlb.65.1.28. [DOI] [PubMed] [Google Scholar]
- 21.Lee J, Choi K, Olin MR, Cho SN, Molitor TW. γδ T Cells in Immunity Induced by Mycobacterium bovis Bacillus Calmette-Guérin Vaccination. Infection and Immunity. 2004;72:1504–1511. doi: 10.1128/IAI.72.3.1504-1511.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waters WR, Thacker TC, Nelson JT, DiCarlo DM, Maggioli MF, Greenwald R, Esfandiari J, Lyashchenko KP, Palmer MV. Virulence of Two Strains of Mycobacterium bovis in Cattle Following Aerosol Infection. Journal of Comparative Pathology. 2014;151:410–419. doi: 10.1016/j.jcpa.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Palmer MV, Ray Waters W, Whipple DL. Aerosol delivery of virulent Mycobacterium bovis to cattle. Tuberculosis. 2002;82:275–282. doi: 10.1054/tube.2002.0341. [DOI] [PubMed] [Google Scholar]
- 24.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BMG, Haag JD, Gould MN, Stewart RM, Kendziorski C. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, Winsor GL, Hancock REW, Brinkman FSL, Lynn DJ. InnateDB: systems biology of innate immunity and beyond—recent updates and continuing curation. Nucleic Acids Research. 2013;41:D1228–D1233. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Werling D, Hope JC, Howard CJ, Jungi TW. Differential production of cytokines, reactive oxygen and nitrogen by bovine macrophages and dendritic cells stimulated with Toll-like receptor agonists. Immunology. 2004;111:41–52. doi: 10.1111/j.1365-2567.2004.01781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baquero MM, Plattner BL. Bovine WC1+ γδ T lymphocytes modify monocyte-derived macrophage responses during early Mycobacterium avium subspecies paratuberculosis infection. Veterinary Immunology and Immunopathology. 2016;170:65–72. doi: 10.1016/j.vetimm.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Sacco RE, Nonnecke BJ, Palmer MV, Waters WR, Lippolis JD, Reinhardt TA. Differential Expression of Cytokines in Response to Respiratory Syncytial Virus Infection of Calves with High or Low Circulating 25-Hydroxyvitamin D3. PLOS ONE. 2012;7:e33074. doi: 10.1371/journal.pone.0033074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thacker TC, Palmer MV, Waters WR. T-Cell mRNA Expression in Response to Mycobacterium bovis BCG Vaccination and Mycobacterium bovis Infection of White-Tailed Deer. Clinical and Vaccine Immunology. 2009;16:1139–1145. doi: 10.1128/CVI.00424-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rainard P, Cunha P, Bougarn S, Fromageau A, Rossignol C, Gilbert FB, Berthon P. T Helper 17-Associated Cytokines Are Produced during Antigen-Specific Inflammation in the Mammary Gland. PLOS ONE. 2013;8:e63471. doi: 10.1371/journal.pone.0063471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Endsley JJ, Furrer JL, Endsley MA, McIntosh MA, Maue AC, Waters WR, Lee DR, Estes DM. Characterization of Bovine Homologues of Granulysin and NK-lysin. J Immunol. 2004;173:2607. doi: 10.4049/jimmunol.173.4.2607. [DOI] [PubMed] [Google Scholar]
- 33.Janovick-Guretzky NA, Dann HM, Carlson DB, Murphy MR, Loor JJ, Drackley JK. Housekeeping Gene Expression in Bovine Liver is Affected by Physiological State, Feed Intake, and Dietary Treatment1. Journal of Dairy Science. 2007;90:2246–2252. doi: 10.3168/jds.2006-640. [DOI] [PubMed] [Google Scholar]
- 34.Nelson CD, Reinhardt TA, Thacker TC, Beitz DC, Lippolis JD. Modulation of the bovine innate immune response by production of 1α,25-dihydroxyvitamin D3 in bovine monocytes. Journal of Dairy Science. 2010;93:1041–1049. doi: 10.3168/jds.2009-2663. [DOI] [PubMed] [Google Scholar]
- 35.Kennedy HE, Welsh MD, Bryson DG, Cassidy JP, Forster FI, Howard CJ, Collins RA, Pollock JM. Modulation of Immune Responses to Mycobacterium bovis in Cattle Depleted of WC1(+) γδ T Cells. Infection and Immunity. 2002;70:1488–1500. doi: 10.1128/IAI.70.3.1488-1500.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smyth AJ, Welsh MD, Girvin RM, Pollock JM. In Vitro Responsiveness of γδ T Cells from Mycobacterium bovis-Infected Cattle to Mycobacterial Antigens: Predominant Involvement of WC1(+) Cells. Infection and Immunity. 2001;69:89–96. doi: 10.1128/IAI.69.1.89-96.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rhodes SG, Hewinson RG, Vordermeier HM. Antigen Recognition and Immunomodulation by γδ T Cells in Bovine Tuberculosis. J Immunol. 2001;166:5604. doi: 10.4049/jimmunol.166.9.5604. [DOI] [PubMed] [Google Scholar]
- 38.Lockhart E, Green AM, Flynn JL. IL-17 Production Is Dominated by γδ T Cells rather than CD4 T Cells during Mycobacterium tuberculosis Infection. J Immunol. 2006;177:4662. doi: 10.4049/jimmunol.177.7.4662. [DOI] [PubMed] [Google Scholar]
- 39.Hasan Z, Cliff JM, Dockrell HM, Jamil B, Irfan M, Ashraf M, Hussain R. CCL2 Responses to Mycobacterium tuberculosis Are Associated with Disease Severity in Tuberculosis. PLOS ONE. 2010;4:e8459. doi: 10.1371/journal.pone.0008459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mastroianni CM, Lancella L, Mengoni F, Lichtner M, Santopadre P, D’agostino C, Ticca F, Vullo V. Chemokine profiles in the cerebrospinal fluid (CSF) during the course of pyogenic and tuberculous meningitis. Clinical and Experimental Immunology. 1998;114:210–214. doi: 10.1046/j.1365-2249.1998.00698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alvarez AJ, Endsley JJ, Werling D, Mark Estes D. WC1+γδ T Cells Indirectly Regulate Chemokine Production During Mycobacterium bovis Infection in SCID-bo Mice. Transboundary and Emerging Diseases. 2009;56:275–284. doi: 10.1111/j.1865-1682.2009.01081.x. [DOI] [PubMed] [Google Scholar]
- 42.McGill JL, Sacco RE, Baldwin CL, Telfer JC, Palmer MV, Ray Waters W. The role of gamma delta T cells in immunity to Mycobacterium bovis infection in cattle. Veterinary Immunology and Immunopathology. 2014;159:133–143. doi: 10.1016/j.vetimm.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 43.Steinbach S, Vordermeier HM, Jones GJ. CD4+ and γδ T Cells are the main Producers of IL-22 and IL-17A in Lymphocytes from Mycobacterium bovis-infected Cattle. Scientific Reports. 2016;6:29990. doi: 10.1038/srep29990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aranday-Cortes E, Bull NC, Villarreal-Ramos B, Gough J, Hicks D, Ortiz-Peláez Á, Vordermeier HM, Salguero FJ. Upregulation of IL-17A, CXCL9 and CXCL10 in Early-Stage Granulomas Induced by Mycobacterium bovis in Cattle. Transbound Emerg Dis. 2013;60:525–537. doi: 10.1111/j.1865-1682.2012.01370.x. [DOI] [PubMed] [Google Scholar]
- 45.Palmer MV, Thacker TC, Waters WR. Analysis of Cytokine Gene Expression using a Novel Chromogenic In-situ Hybridization Method in Pulmonary Granulomas of Cattle Infected Experimentally by Aerosolized Mycobacterium bovis. Journal of Comparative Pathology. 2015;153:150–159. doi: 10.1016/j.jcpa.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 46.Neill SD, Pollock JM, Bryson DB, Hanna J. Pathogenesis of Mycobacterium bovis infection in cattle. Veterinary Microbiology. 1994;40:41–52. doi: 10.1016/0378-1135(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 47.Vantourout P, Hayday A. Six-of-the-best: unique contributions of γδ T cells to immunology. Nature Reviews Immunology. 2013;13:88–100. doi: 10.1038/nri3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plattner BL, Doyle RT, Hostetter JM. Gamma–delta T cell subsets are differentially associated with granuloma development and organization in a bovine model of mycobacterial disease. International Journal of Experimental Pathology. 2009;90:587–597. doi: 10.1111/j.1365-2613.2009.00679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skinner MA, Parlane N, McCarthy A, Buddle BM. Cytotoxic T-cell responses to Mycobacterium bovis during experimental infection of cattle with bovine tuberculosis. Immunology. 2003;110:234–241. doi: 10.1046/j.1365-2567.2003.01731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guzman E, Hope J, Taylor G, Smith AL, Cubillos-Zapata C, Charleston B. Bovine γδ T Cells Are a Major Regulatory T Cell Subset. The Journal of Immunology Author Choice. 2014;193:208–222. doi: 10.4049/jimmunol.1303398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoek A, Rutten VP, Kool J, Arkesteijn GJ, Bouwstra RJ, Van Rhijn I, Koets AP. Subpopulations of bovine WC1(+) γδ T cells rather than CD4(+)CD25(high)Foxp3(+) T cells act as immune regulatory cells ex vivo. Veterinary Research. 2009;40:06. doi: 10.1051/vetres:2008044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lahmers KK, Hedges JF, Jutila MA, Deng M, Abrahamsen MS, Brown WC. Comparative gene expression by WC1+ γδ and CD4+ αβ T lymphocytes, which respond to Anaplasma marginale, demonstrates higher expression of chemokines and other myeloid cell-associated genes by WC1+ γδ T cells. Journal of Leukocyte Biology. 2006;80:939–952. doi: 10.1189/jlb.0506353. [DOI] [PubMed] [Google Scholar]
- 53.McGill JL, Nonnecke BJ, Lippolis JD, Reinhardt TA, Sacco RE. Differential chemokine and cytokine production by neonatal bovine γδ T-cell subsets in response to viral toll-like receptor agonists and in vivo respiratory syncytial virus infection. Immunology. 2013;139:227–244. doi: 10.1111/imm.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scott Algood HM, Lin PL, Flynn JL. Tumor Necrosis Factor and Chemokine Interactions in the Formation and Maintenance of Granulomas in Tuberculosis. Clinical Infectious Diseases. 2005;41:S189–S193. doi: 10.1086/429994. [DOI] [PubMed] [Google Scholar]
- 55.Kindler V, Sappino AP, Grau GE, Piguet PF, Vassalli P. The inducing role of tumor necrosis factor in the development of bactericidal granulomas during BCG infection. Cell. 1989;56:731–740. doi: 10.1016/0092-8674(89)90676-4. [DOI] [PubMed] [Google Scholar]
- 56.Roach DR, Bean AGD, Demangel C, France MP, Briscoe H, Britton WJ. TNF Regulates Chemokine Induction Essential for Cell Recruitment, Granuloma Formation, and Clearance of Mycobacterial Infection. J Immunol. 2002;168:4620. doi: 10.4049/jimmunol.168.9.4620. [DOI] [PubMed] [Google Scholar]
- 57.Tsukaguchi K, de Lange B, Boom WH. Differential Regulation of IFN-γ, TNF-α, and IL-10 Production by CD4+ αβTCR+ T Cells and Vδ2+ γδ T Cells in Response to Monocytes Infected with Mycobacterium tuberculosis-H37Ra. Cellular Immunology. 1999;194:12–20. doi: 10.1006/cimm.1999.1497. [DOI] [PubMed] [Google Scholar]
- 58.Zhou Y, Shah SZA, Yang L, Zhang Z, Zhou X, Zhao D. Virulent Mycobacterium bovis Beijing Strain Activates the NLRP7 Inflammasome in THP-1 Macrophages. PLoS ONE. 2016;11:e0152853. doi: 10.1371/journal.pone.0152853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Master SS, Rampini SK, Davis AS, Keller C, Ehlers S, Springer B, Timmins GS, Sander P, Deretic V. Mycobacterium tuberculosis Prevents Inflammasome Activation. Cell Host & Microbe. 2008;3:224–232. doi: 10.1016/j.chom.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stanciu LA, Shute J, Holgate ST, Djukanović R. Production of IL-8 and IL-4 by positively and negatively selected CD4+ and CD8+ human T cells following a four-step cell separation method including magnetic cell sorting (MACS) Journal of Immunological Methods. 1996;189:107–115. doi: 10.1016/0022-1759(95)00240-5. [DOI] [PubMed] [Google Scholar]
- 61.Sutherland JS, Mendy J, Gindeh A, Walzl G, Togun T, Owolabi O, Donkor S, Ota MO, Kon Fat ET, Ottenhoff THM, Geluk A, Corstjens PLAM. Use of lateral flow assays to determine IP-10 and CCL4 levels in pleural effusions and whole blood for TB diagnosis. Tuberculosis. 2016;96:31–36. doi: 10.1016/j.tube.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 62.Yao S, Huang D, Chen CY, Halliday L, Zeng G, Wang RC, Chen ZW. Differentiation, Distribution and γδ T Cell-Driven Regulation of IL-22-Producing T Cells in Tuberculosis. PLOS Pathogens. 2010;6:e1000789. doi: 10.1371/journal.ppat.1000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Palmer MV, Thacker TC, Waters WR. Differential Cytokine Gene Expression in Granulomas from Lungs and Lymph Nodes of Cattle Experimentally Infected with Aerosolized Mycobacterium bovis. PLOS ONE. 2016;11:e0167471. doi: 10.1371/journal.pone.0167471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang M, Lin Y, Iyer DV, Gong J, Abrams JS, Barnes PF. T-cell cytokine responses in human infection with Mycobacterium tuberculosis. Infection and Immunity. 1995;63:3231–3234. doi: 10.1128/iai.63.8.3231-3234.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brighenti S, Andersson J. Local Immune Responses in Human Tuberculosis: Learning From the Site of Infection. The Journal of Infectious Diseases. 2012;205:S316–S324. doi: 10.1093/infdis/jis043. [DOI] [PubMed] [Google Scholar]
- 66.Chien Y, Zeng X, Prinz I. The natural and the inducible: IL-17 producing gamma delta T cells. Trends in Immunology. 2013;34:151–154. doi: 10.1016/j.it.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ness-Schwickerath KJ, Jin C, Morita CT. Cytokine Requirements for the Differentiation and Expansion of IL-17A- and IL-22-producing Human Vγ2Vδ2 T Cells. Journal of Immunology (Baltimore, Md. : 1950) 2010;184:7268–7280. doi: 10.4049/jimmunol.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McGill JL, Rusk RA, Guerra-Maupome M, Briggs RE, Sacco RE. Bovine Gamma Delta T Cells Contribute to Exacerbated IL-17 Production in Response to Co-Infection with Bovine RSV and Mannheimia haemolytica. PLOS ONE. 2016;11:e0151083. doi: 10.1371/journal.pone.0151083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schluger NW, Rom WN. The Host Immune Response to Tuberculosis. Am J Respir Crit Care Med. 1998;157:679–691. doi: 10.1164/ajrccm.157.3.9708002. [DOI] [PubMed] [Google Scholar]
- 70.Dieli F, Poccia F, Lipp M, Sireci G, Caccamo N, Di Sano C, Salerno A. Differentiation of Effector/Memory Vδ2 T Cells and Migratory Routes in Lymph Nodes or Inflammatory Sites. The Journal of Experimental Medicine. 2003;198:391–397. doi: 10.1084/jem.20030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sciorati C, Rovere P, Ferrarini M, Paolucci C, Heltai S, Vaiani R, Clementi E, Manfredi AA. Generation of Nitric Oxide by the Inducible Nitric Oxide Synthase Protects γδ T Cells from Mycobacterium tuberculosis-Induced Apoptosis. J Immunol. 1999;163:1570. [PubMed] [Google Scholar]
- 72.Douguet L, Cherfils-Vicini J, Bod L, Lengagne R, Gilson E, Prévost-Blondel A. Nitric Oxide Synthase 2 Improves Proliferation and Glycolysis of Peripheral γδ T Cells. PLOS ONE. 2016;11:e0165639. doi: 10.1371/journal.pone.0165639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saito S, Nakano M. Nitric oxide production by peritoneal macrophages of Mycobacterium bovis BCG-infected or non-infected mice: regulatory role of T lymphocytes and cytokines. Journal of Leukocyte Biology. 1996;59:908–915. doi: 10.1002/jlb.59.6.908. [DOI] [PubMed] [Google Scholar]
- 74.Yang J, Zhang R, Lu G, Shen Y, Peng L, Zhu C, Cui M, Wang W, Arnaboldi P, Tang M, Gupta M, Qi CF, Jayaraman P, Zhu H, Jiang B, Chen S, He JC, Ting AT, Zhou MM, Kuchroo VK, Morse HC, Ozato K, Sikora AG, Xiong H. T cell–derived inducible nitric oxide synthase switches off T(H)17 cell differentiation. The Journal of Experimental Medicine. 2013;210:1447–1462. doi: 10.1084/jem.20122494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Obermajer N, Wong JL, Edwards RP, Chen K, Scott M, Khader S, Kolls JK, Odunsi K, Billiar TR, Kalinski P. Induction and stability of human Th17 cells require endogenous NOS2 and cGMP-dependent NO signaling. J Exp Med. 2013;210:1433. doi: 10.1084/jem.20121277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martino A, Casetti R, Sacchi A, Poccia F. Central Memory Vγ9Vδ2 T Lymphocytes Primed and Expanded by Bacillus Calmette-Guérin-Infected Dendritic Cells Kill Mycobacterial-Infected Monocytes. J Immunol. 2007;179:3057. doi: 10.4049/jimmunol.179.5.3057. [DOI] [PubMed] [Google Scholar]
- 77.Chen CY, Yao S, Huang D, Wei H, Sicard H, Zeng G, Jomaa H, Larsen MH, Jacobs WR, Jr, Wang R, Letvin N, Shen Y, Qiu L, Shen L, Chen ZW. Phosphoantigen/IL2 Expansion and Differentiation of Vγ2Vδ2 T Cells Increase Resistance to Tuberculosis in Nonhuman Primates. PLOS Pathogens. 2013;9:e1003501. doi: 10.1371/journal.ppat.1003501. [DOI] [PMC free article] [PubMed] [Google Scholar]


