Significance
Salamanders have great potential to regenerate damaged organs upon injury, and thus provide an important model for understanding the mechanisms of tissue regeneration; however, genetic studies have been limited due to a lack of gene knockin strategies. In this study, we have established efficient CRISPR/Cas9 mediated gene knockin approaches in the axolotl (Ambystoma mexicanum), which has allowed us to genetically mark two critical stem cell pools for limb and spinal cord regeneration. Our genetic fate mapping establishes the role of PAX7+ satellite cells for limb muscle regeneration. This method opens up the possibility of marking and perturbing gene function inducibly in any definable cell populations in the axolotl, a key functionality required for the precise, rigorous understanding of processes such as regeneration.
Keywords: CRISPR/Cas9, knockin, neural stem cells, regeneration, satellite cells
Abstract
Salamanders exhibit extensive regenerative capacities and serve as a unique model in regeneration research. However, due to the lack of targeted gene knockin approaches, it has been difficult to label and manipulate some of the cell populations that are crucial for understanding the mechanisms underlying regeneration. Here we have established highly efficient gene knockin approaches in the axolotl (Ambystoma mexicanum) based on the CRISPR/Cas9 technology. Using a homology-independent method, we successfully inserted both the Cherry reporter gene and a larger membrane-tagged Cherry-ERT2-Cre-ERT2 (∼5-kb) cassette into axolotl Sox2 and Pax7 genomic loci. Depending on the size of the DNA fragments for integration, 5–15% of the F0 transgenic axolotl are positive for the transgene. Using these techniques, we have labeled and traced the PAX7-positive satellite cells as a major source contributing to myogenesis during axolotl limb regeneration. Our work brings a key genetic tool to molecular and cellular studies of axolotl regeneration.
The axolotl (Ambystoma mexicanum) is capable of regenerating complex biological structures throughout adulthood. One of the early events during regeneration is formation of the blastema, which contains progenitor populations that will give rise to all cell types of the newly regenerated tissue. Cell tracking has demonstrated the existence of lineage-specific progenitors giving rise to corresponding regenerated tissues (1). Concerning muscle regeneration, a previous study has shown that two salamander species, axolotl and newt (Notophthalmus viridescens), use divergent source cells during limb muscle regeneration (2). Mature muscle fiber fragmentation and dedifferentiation occur in newt, but not in axolotl. Instead, indirect evidence implicates the use of PAX7-positive satellite cells during axolotl limb regeneration. However, direct evidence of this has been lacking due to the unavailability of a specific cell label of axolotl satellite cells. Transgenesis via random integration of plasmids, although highly efficient, has remained troublesome for genetically labeling particular cell types, such as PAX7-positive satellite cells and SOX2-positive neural stem cells due to the complexity of working with the multipartite gene regulatory regions of these genes (3). Gene knockin approaches to drive gene expression from endogenous promoters would allow accurate genetic labeling and manipulation of defined cell types, and therefore represents an important goal.
Taking advantage of recently developed modern genome engineering technologies, including zinc finger nuclease, transcription activator-like effector nucleases, and clustered regularly interspaced short palindromic repeats (CRISPR), precise knockin of exogenous sequences into defined genomic loci has been successfully achieved in diverse model organisms, including mouse, rat, zebrafish, and Xenopus (4–11). The CRISPR/Cas9 genome editing system consists of two components, the endonuclease Cas9 and the guide RNA (gRNA) that can create double-strand breaks (DSB) at the targeted genomic locus (12–14). Two pathways exist to repair the DSB: nonhomologous end-joining (NHEJ) and homology-directed repair (HDR) (15). To integrate the gene of interest into the DSB locus, homologous recombination via HDR using a targeting vector harboring the 5′ and 3′ homology arms is a common approach (15, 16). More recently, Auer and colleagues established a homology-independent knockin method based on NHEJ that leads to more efficient insertion of the targeted gene at the site of gene lesion (6, 17). The targeting vector for homology-independent knockin harbors a so-called “bait” sequence that can be targeted and cut by either the same gRNA for the genomic DNA or a different gRNA. The linearized targeting vector inserts into the genomic lesion created by the Cas9 endonuclease, with concomitant indels (insertions and deletions) often being generated at the integration junctions (6, 11). Recent studies have shown that the application of purified CAS9 protein instead of Cas9 mRNA allows for the prompt formation of the gRNA-CAS9 ribonucleoprotein (RNP), which in turn leads to more efficient and rapid formation of DSBs at the targeted genomic locus (18–21). Delivery of the RNP together with the targeting construct indeed dramatically improves the knockin efficiency for generation of transgenic C. elegans, zebrafish, and primary cells in culture (7, 22–24).
In the present study, we have established highly efficient gene knockin approaches in axolotl to integrate the Cherry reporter gene and the tamoxifen-inducible ERT2-Cre-ERT2 encoding sequences into the Pax7 and Sox2 loci. Using F0 Pax7: ERT2-Cre-ERT2 transgenic axolotls, we have performed genetic fate mapping of PAX7-positive satellite cells showing that these cells robustly contribute to de novo myogenesis in axolotl limb regeneration.
Results
Knockin of a Cherry Reporter Gene into Axolotl Genomic Loci via CRISPR/Cas9-Based Homologous-Independent Integration.
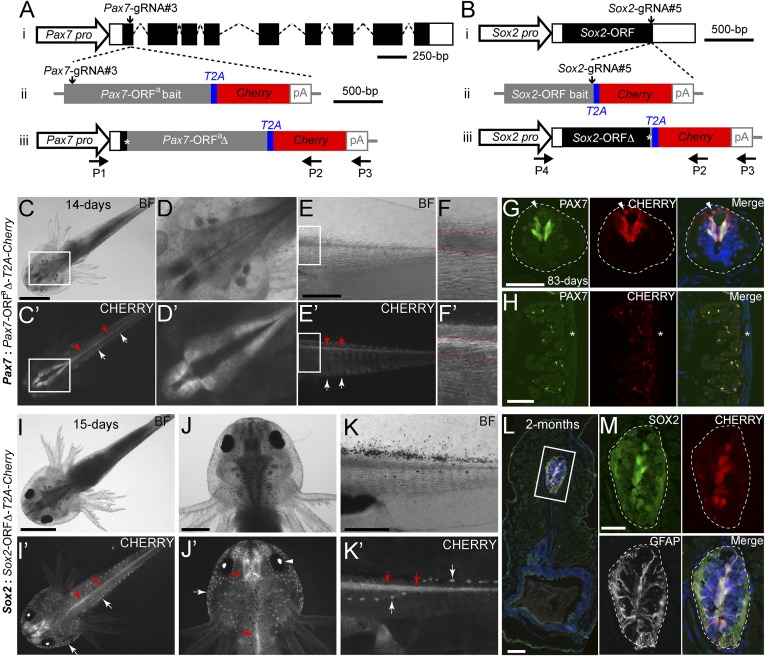
We first sought to insert the Cherry reporter gene into the axolotl Pax7 genomic locus (Fig. 1 A, i and Dataset S1). We designed and synthesized three gRNAs—Pax7-gRNA#1, #2 and #3—targeting the coding sequences in Pax7 exon1, and identified the gRNA that most efficiently induced indels (SI Appendix, Table S1). We then coinjected Pax7-gRNA#3, together with CAS9 protein and the targeting bait construct pGEMT-Pax7-ORFa-T2A-Cherry-PA, into single-cell stage axolotl eggs (SI Appendix, Fig. S1A). The targeting construct harbors sequentially the Pax7 ORF lacking the stop codon, designated Pax7-ORFa, as the bait sequence, along with the T2A viral peptide and the Cherry coding sequences (Fig. 1 A, ii). The integration of the linearized targeting construct into the Pax7 genomic locus forms a new in-frame Pax7 ORF (Pax7-ORFaΔ) in which a few nucleotide modifications often occur at the integration junction, followed by the T2A and Cherry coding sequence (Fig. 1 A, iii). In these Pax7-Cherry knockin alleles, expression of the Cherry reporter gene is directly under the control of the endogenous Pax7 regulatory sequences.
Fig. 1.
Knockin of a Cherry reporter gene into two axolotl genomic loci through CRISPR/Cas9- mediated homologous-independent integration. (A and B) The knockin strategies for the generation of the Pax7: Pax7-ORFa∆-T2A-Cherry (A) and Sox2: Sox2-ORF∆-T2A-Cherry (B) axolotl reporter lines. (i) The wild-type axolotl Pax7 (A, Dataset S1) and Sox2 (B) gene structures. Solid rectangles represent coding sequences of the exons; empty rectangles, untranslated regions; dashed lines: introns. The gRNA targeting sites are indicated by vertical arrows. (ii) The targeting constructs, pGEMT-Pax7-ORFa-T2A-Cherry-PA (A) and pGEMT-Sox2-ORF-T2A-Cherry-PA (B), contain the entire axolotl Pax7 (A; Pax7-ORFa bait) or Sox2 (B; Sox2-ORF bait) ORF of the cDNA (with the start codon, but without the stop codon) as the bait sequence, followed by T2A, the Cherry coding sequence, and the polyadenylation signal (pA). Vertical arrows indicate the gRNA targeting sites. (iii) The Pax7 (A) and Sox2 (B) alleles after knockin of the Cherry reporter gene. Asterisks indicate the junctions after the integration of the targeting constructs. The newly formed mosaic Pax7 (A) and Sox2 (B) coding sequences are designated Pax7-ORFaΔ and Sox2-ORFΔ, respectively. The horizontal arrows indicate the binding positions of the genotyping primers (P1–P4). (C–F′) Bright-field (BF; Upper) and CHERRY fluorescence (Lower) images of 14-d-old Pax7: Pax7-ORFaΔ-T2A-Cherry knockin F0 axolotls. The dorsal (C–D′) and lateral (E–F′) view images highlight the CHERRY expression in the brain (C–D′), spinal cord (red arrows), and trunk muscle (white arrows) compartments. The areas depicted by rectangles in C and C′ and E and E′ are shown at higher magnification in D and D′ and F and F′, respectively. The red dashed lines (F and F′) define the spinal cord region showing that the CHERRY expression is restricted in the dorsal domain of the spinal cord. (Scale bars: 2 mm in C, 1 mm in E.) (G and H) Immunofluorescence for PAX7 (green), CHERRY fluorescence (red), and DAPI (blue) on 10-μm tail cross-cryosections of 83-d-old Pax7: Pax7-ORFaΔ-T2A-Cherry knockin F0 axolotls shows that CHERRY expression is restricted to the PAX7-expressing domain in dorsal spinal cord (G, dashed line circles) and satellite cells (H). The arrowhead indicates inherited CHERRY in a PAX7-negative newborn neuron. The asterisk indicates tail skin. (Scale bars: 100 μm in G and H). (I–K′) Bright-field (BF, Upper) and CHERRY fluorescence (Lower) images of 15-d-old Sox2: Sox2-ORFΔ-T2A-Cherry knockin F0 axolotls. The dorsal view (I–J′) and lateral view (K and K′) images highlight the CHERRY expression in the central nervous system (red arrows), the lens (arrowhead), and the lateral line neuromasts (white arrows). (Scale bars: 2 mm in I, 1 mm in J and K.) (L and M) Immunofluorescence for SOX2 (green), GFAP (white), and CHERRY fluorescence (red) combined with DAPI (blue) on 10-μm cross-cryosections of 2-mo-old Sox2: Sox2-ORFΔ-T2A-Cherry knockin F0 axolotls shows that CHERRY expression is restricted to SOX2 positive cells in the spinal cord (dashed circles) (M). GFAP is shown to highlight the morphology of the luminal SOX2-positive radial glial cells expressing GFAP. The rectangle depicted in L is shown as separated or merged images at higher magnification in M. (Scale bars: 100 μm in L, 50 μm in M.)
Examination of live animals revealed CHERRY expression in the brain (Fig. 1 C, C′, D, and D′), spinal cord, and muscle (Fig. 1 C, C′, E, E′, F, and F′) compartments, consistent with the expression of PAX7 in other vertebrate species (25). Under higher magnification, we found that, as expected, CHERRY expression in the spinal cord was restricted specifically in the dorsal region of the cord (Fig. 1 F and F′). Based on the distribution of CHERRY expression, we could categorize the F0 Pax7: Pax7-ORFaΔ-T2A-Cherry axolotls as “low” “medium,” or “high” transgenics, based on the uniformity of CHERRY expression in the nervous system and muscles of live animals (SI Appendix, Fig. S2). Among the 157 injected eggs, 20 (12.7%) of derived F0 developing larva demonstrated CHERRY expression in the expected PAX7-expressing domains (SI Appendix, Table S2). Most of these were classified as high or medium transgenics (SI Appendix, Table S2). Implementing Cas9 mRNA instead of protein or the other gRNAs yielded a lower percentage and penetrance of reporter gene knockin (SI Appendix, Tables S1 and S2).
We then assessed the faithfulness of the Cherry transgene expression in more detail using cryosections. We examined Pax7 and Cherry mRNA localization on consecutive cross-sections by in situ hybridization and observed a very close correspondence in hybridization between the two probes (SI Appendix, Fig. S1B). At the protein level, the vast majority of CHERRY expression was found in PAX7-immunopositive cells in both the spinal cord and muscles (Fig. 1 G and H). Longitudinal sections of muscle showed the elongated morphology of satellite cells with long processes spreading over the outer surface of the muscle fibers (SI Appendix, Fig. S1D). We did identify the presence of dim CHERRY signal in some PAX7− or PAX7−/TUJ-1+ dorsal spinal cord neurons (Fig. 1G and SI Appendix, Fig. S1C), and in some muscles located particularly in the peripheral underneath the skin on tail cross-sections (Fig. 1H). Given the mRNA in situ hybridization results, these observations likely represent perdurance of protein expression in the differentiated progeny of the neural and muscle stem cells, as other experiments in our laboratory have suggested that CHERRY protein can persist for at least 2 d after extinction of mRNA. Indeed, in zebrafish, the peripheral muscle is known to be the active site of new muscle formation (26). As described below in the characterization of Sox2: Sox2-ORF∆-T2A-Cherry transgenic animals, our birth-dating studies indicate that CHERRY is found in newly differentiated progeny of stem cells. Therefore, from the combined mRNA and protein localization data, we conclude that there is faithful expression of Cherry RNA with some persistence of CHERRY protein expression in newly differentiated daughter cells (SI Appendix, Fig. S4).
To check whether the homology-independent knockin approach is generally applicable in axolotl, we followed a similar strategy to insert the Cherry gene into the 3′ end of the single-exon Sox2 genomic locus (Fig. 1 B, i). Among three designed gRNAs (Sox2-gRNA#3, #4, and #5) against the most 3′ region of the Sox2 ORF, Sox2-gRNA#5 led to the most efficient modification at the targeted sequences (SI Appendix, Table S1). The targeting construct, pGEMT-Sox2-ORF-T2A-Cherry-PA, includes the Sox2 ORF lacking the stop codon as a bait sequence, followed by the T2A and Cherry coding sequences (Fig. 1 B, ii and iii). In vertebrates, SOX2 is commonly expressed in the nervous system and is generally recognized as a neural stem cell marker (27, 28). In live Sox2: Sox2-ORF∆-T2A-Cherry F0 and F1 animals, we found CHERRY expression in the brain and spinal cord of the central nervous system, the lens, and the head/tail lateral line neuromasts (Fig. 1 I–K′ and SI Appendix, Fig. S3 A and B). In situ hybridization on tail cross-sections revealed that Cherry mRNA expression closely matched Sox2 expression in the spinal cord and the lateral line neuromasts (SI Appendix, Fig. S3C). Immunohistochemistry using an anti-SOX2 antibody on trunk cross-sections and whole-mount axolotls showed accurate CHERRY expression in the spinal cord SOX2-positive neural stem cells (Fig. 1 L and M) and head lateral line neuromasts (SI Appendix, Fig. S3D). Among 310 F0 larva deriving from the injected eggs, 45 (14.5%) harbored CHERRY expression in all or most of the SOX2-expressing regions (SI Appendix, Table S2).
Similar to the observation in Pax7: Pax7-ORFaΔ-T2A-Cherry axolotls, we also found the presence of dim CHERRY in some TUJ-1+ spinal cord neurons in Sox2: Sox2-ORF∆-T2A-Cherry animals (SI Appendix, Fig. S4C). Thus, we more closely examined whether CHERRY protein expression corresponds with Cherry and Sox2 mRNA expression. We first performed in situ hybridization using DIG-labeled Cherry or Sox2 antisense RNA probes, followed by the immunohistochemical staining with an anti-CHERRY antibody on spinal cord cross sections of Sox2: Sox2-ORF∆-T2A-Cherry animals. While the vast majority of CHERRY-positive cells showed signals for Cherry and Sox2 mRNA, we identified a few CHERRY-expressing cells that were negative for Sox2 or Cherry mRNA (SI Appendix, Fig. S4 A and B). To determine whether the CHERRY+/Cherry− cells represented neurons newly born from a stem cell, we pulse-labeled the Sox2: Sox2-ORF∆-T2A-Cherry transgenic animals with EdU to label actively dividing cells in the spinal cord, which are known to express SOX2 (29). Fixation of samples at 3 d after the EdU pulse showed that the CHERRY+/TUJ-1+ cells were labeled for EdU, indicating that the CHERRY+/TUJ-1+ cells were newly born neurons that likely inherited CHERRY expression from a SOX2-expressing stem cell (SI Appendix, Fig. S4C).
To characterize the integration sites after targeting, we carried out genomic PCR using pairs of primers such that one primer was bound in the genome either in the Pax7 or Sox2 promoter and the other was bound in the 3′ end of the targeting cassettes (Fig. 1 A, iii and 1 B, iii). Consistent with the observed expression, knockin axolotls were found to harbor the entire Cherry gene cassette (SI Appendix, Fig. S5A). Sequence analysis revealed that in most of the cases, the coding sequences were maintained in-frame at the integration junctions in the F0 Cherry transgenic axolotls (SI Appendix, Fig. S5 B and C). Moreover, the junctions were generally identical in limbs and tails in individual F0 founders, suggesting that integration occurs during early embryogenesis (three out of four; SI Appendix, Fig. S5B). We conclude that CRISPR-mediated homology-independent knockin is an efficient method for introducing a reporter gene into a defined genomic locus in axolotl, yielding faithful expression of the transgene in the targeted cells.
Knockin of a Large Gene Cassette Through CRISPR/Cas9-Mediated Homologous-Independent Targeting.
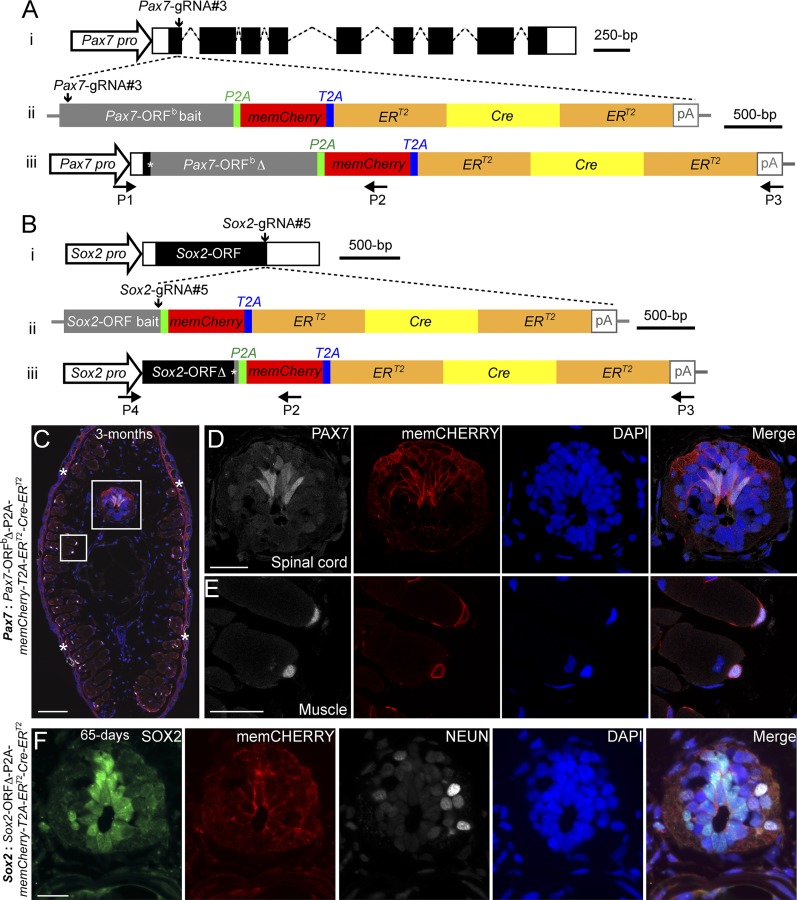
In axolotl, generation of transgenics combining a fluorescent reporter with tamoxifen- inducible ERT2-Cre-ERT2 sequences is an important means of temporal and tissue-specific control of gene expression during regeneration. Transgenesis using plasmids harboring promoters driving these sequences cannot yet access all cell types, and thus knockin of such sequences into desired genes of interest is essential (2, 3). However, targeted gene knockin becomes technically challenging with increasing size of the targeting cassette (16). Therefore, we ascertained whether we could knockin a 5-kb insert that included the bait sequence (either a Pax7 ORF or the Sox2 ORF), followed by memCherry (a GAP43 membrane-localization sequence-tagged Cherry gene)-T2A -ERT2-Cre-ERT2 (Fig. 2 A and B and Dataset S2). The same Pax7-gRNA#3 and Sox2-gRNA#5 as implemented above were used for targeting. Based on the memCHERRY expression combined with genotyping PCR, 4.3% (9 out of 211) and 5.8% (12 out of 207) of injected eggs showed integration of memCherry-ERT2-Cre-ERT2 cassette in the Pax7 and Sox2 genomic loci, respectively (SI Appendix, Table S2). Histological examination of the F0 and F1 Pax7 integrants showed memCHERRY expression in the CNS as well (Fig. 2C and SI Appendix, Fig. S6 A–B′), restricted particularly to the dorsal domain (Fig. 2 C and D and SI Appendix, Fig. S6 C, C′, and E) and in muscles (Fig. 2 C and E and SI Appendix, Fig. S6 B–C′and F). In F0 Sox2: Sox2-ORF∆-P2A-memCherry-T2A-ERT2-Cre-ERT2 axolotls, CHERRY expression was observed in the CNS, the lens, and the head/tail lateral line neuromasts (SI Appendix, Fig. S6 G–H′). Cross-sections showed colocalization of the CHERRY signal with SOX2-immunopositive spinal cord neural stem cells (Fig. 2F). Genomic PCR followed by sequence analysis showed complete integration of the entire ERT2-Cre-ERT2 cassette into the targeted loci (SI Appendix, Fig. S6I). Examples of individuals showed that three and nine nucleotide deletions resulted in one and three amino acid truncations, respectively, at integration sites (SI Appendix, Fig. S6 J and K). Our results demonstrate that large cassettes can be inserted into defined genomic loci using CRISPR/Cas9-mediated homology-independent integration to generate sophisticated transgenic axolotl strains.
Fig. 2.
Knockin of a large gene cassette into two axolotl genomic loci through a CRISPR/Cas9- mediated homologous-independent approach. (A and B) The knockin strategies for the generation of the Pax7: Pax7-ORFb∆-P2A-memCherry-T2A-ERT2-Cre-ERT2 (A) and Sox2: Sox2-ORF∆-P2A-memCherry-T2A-ERT2-Cre-ERT2 (B) axolotl transgenic lines. (i) The wild-type axolotl Pax7 (A; Dataset S1) and Sox2 (B) gene structures. Solid rectangles represent coding sequences of the exons; empty rectangles, untranslated regions; dashed lines, introns. Vertical arrows indicate the gRNA targeting sites. (ii) The targeting constructs pGEMT-Pax7-ORFb-P2A-memCherry-T2A- ERT2-Cre-ERT2-PA (A) and pGEMT-Sox2-ORF-P2A-memCherry-T2A-ERT2-Cre-ERT2 -PA (B) contain the entire axolotl Pax7 (A; Pax7-ORFb bait) and Sox2 (B; Sox2-ORF bait) ORF of the cDNA as the bait sequences, followed by the P2A, memCherry (a GAP43 membrane-localization sequence-tagged Cherry gene; Dataset S2), T2A, the ERT2-Cre-ERT2 coding sequences, and the polyadenylation signal (pA). Vertical arrows indicate the gRNA targeting sites. (iii) The Pax7 (A) and Sox2 (B) alleles after successful knockin of the P2A-memCherry-T2A-ERT2-Cre-ERT2 cassettes. Asterisks indicate the junctions after the integration of the targeting constructs. The newly formed mosaic Pax7 (A) and Sox2 (B) coding sequences were designated Pax7-ORFbΔ and Sox2-ORFΔ, respectively. The horizontal arrows indicate the binding positions of the genotyping primers (P1–P4). (C–E) Immunofluorescence for PAX7 (white) and memCHERRY (red) combined with DAPI (blue) on 10-μm tail cross-cryosections of 3-mo-old Pax7: Pax7-ORFbΔ-P2A-memCherry-T2A-ERT2-Cre-ERT2 knockin F0 axolotls shows a membrane-juxtaposed CHERRY signal surrounding PAX7-positive radial glial cells in the spinal cord (C and D) and satellite cells (C and E). The rectangles depicted in C are shown as separated or merged images at higher magnification in D and E. Asterisks in C indicate nonspecific CHERRY staining in the dermis, which is also present in the control (SI Appendix, Fig. S6D). (Scale bars: 200 μm in C, 50 μm in D and E.) (F) Immunofluorescence for SOX2 (green), CHERRY (red, memCHERRY), NEUN (white, to label neurons), and DAPI (blue) on 10-μm cross-cryosections of 65-d-old Sox2: Sox2-ORFΔ-P2A-memCherry-T2A-ERT2-Cre-ERT2 knockin F0 axolotls shows a membrane-juxtaposed CHERRY signal surrounding SOX2-positive radial glial cells in the spinal cord. (Scale bar: 50 μm.)
Satellite Cells Generate Muscle During Axolotl Limb Regeneration.
Sandoval-Guzman et al. (2) have shown that dedifferentiation of mature muscle does not occur during blastema formation in axolotl limb regeneration. Coupled with the identification of labeled muscle tissue as the source of regenerated muscle (1), the previous results indirectly suggest that satellite cells are the major source of de novo myofiber formation during regeneration in axolotl. The ability to insert the ERT2-Cre-ERT2 sequences into the Pax7 locus allowed us to directly test whether satellite cells are a major source for muscle regeneration in the axolotl. To this end, we generated F0 Pax7: Pax7-ORFbΔ-P2A-memCherry-T2A-ERT2-Cre-ERT2; CAGGS: LoxP-GFP-STOP-LoxP-Cherry double-transgenic axolotls and then validated the specificity and lack of background conversion in these animals (SI Appendix, SI Results, Figs. S7–S9).
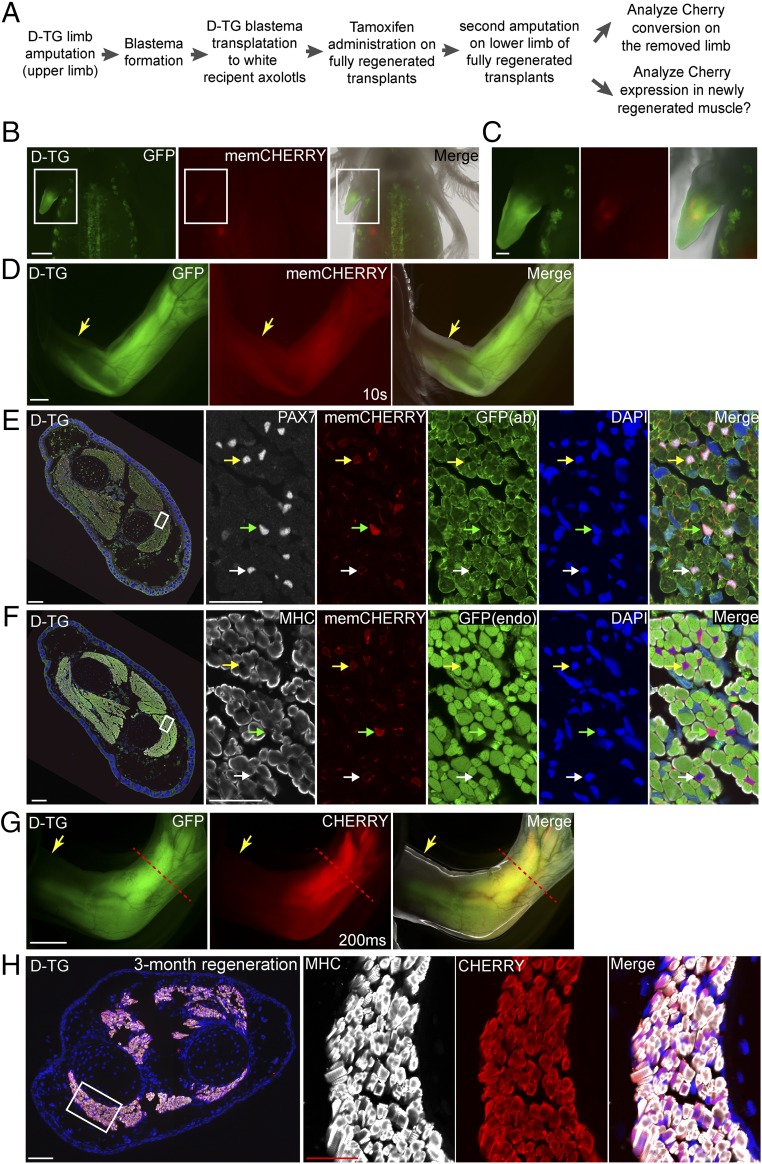
Considering the long generation time of the axolotl, the ability to perform experiments directly on genetically modified F0 animals would be a key advantage. We aimed to test the contribution of satellite cells to limb regeneration using the F0 double-transgenic animals. We obtained strong double-transgenic animals at a frequency of 1.4% (3 of 211) due to the large targeting cassette and the fact that 50% of the injected eggs expressed the LoxP reporter transgene. To perform genetic fate mapping while preserving the valuable strong double-transgenic animals for germ line transmission, we generated six double-transgenic limbs by grafting upper limb blastemas generated from a fully characterized, strong double-transgenic animal after successive rounds of limb amputation onto normal, d/d host upper limb stumps and allowing regeneration to occur to completion (Fig. 3 A–D). Another reason for performing blastema transplantation was to ensure that we were tracing PAX7-expressing cells that derive from the limb and not another distant source, since PAX7 is also expressed in several other cell lineages during development. In the transplanted chimeric axolotls, we did not observe any amputation-triggered spurious recombination of the LoxP cassette in the double-transgenic limbs before tamoxifen treatment (Fig. 3D).
Fig. 3.
Lineage tracing of satellite cells during axolotl limb regeneration. (A) Scheme of the satellite cell lineage tracing experiment. (B–D) Live images of double-transgenic limbs after blastema transplantation and before tamoxifen treatment (n = 6). The GFP, CHERRY fluorescence, and merged (with bright-field) images of the transplanted limb blastema from the double-transgenic (D-TG) (SI Appendix, Fig. S7) donor to a white recipient at 2 wk (B and C) and 3 mo (D) post-transplantation. Note that the membrane-CHERRY signal is very dim in satellite cells in live imaging. Rectangular regions in B are shown at higher magnification in C. (Scale bars: 1 mm in B and D, 500 μm in C.) (E and F) Cross-section of limbs at 2 wk after tamoxifen treatment. Immunofluorescence for PAX7 (white, E) or MHC (white, F), CHERRY (red, memCHERRY), GFP [green, from antibody (ab) staining, in E] or endogenous (endo) GFP fluorescence (green, in F) combined with DAPI (blue) on adjacent 10-μm limb cross-cryosections (showing limb muscle) (n = 3). Green and yellow arrowheads indicate strong and dim cytoplasmic CHERRY-expressing (converted) cells, respectively. White arrowhead marks unconverted, memCHERRY-expressing cells. Muscle fibers (MHC-labeled) in the converted, mature limb remain CHERRY-negative. In E and F, the areas depicted by the rectangle in the leftmost panel are shown at higher magnification at the three right panels (Scale bars: 200 μm in left and 50 μm in right.) (G) Live images of tamoxifen-converted limbs after completion of regeneration: GFP, CHERRY fluorescence, and merged (with bright-field) images (n = 3). Arrows indicate the first amputation plane from the transplantation, and the red dashed line shows the plane of amputation after tamoxifen conversion. Exposure time for CHERRY, 200 ms. Note that Cherry driven by the CAGGS promoter is more strongly expressed in muscle tissues compared with satellite cells (10 s in Fig. 3D vs. 200 ms in Fig. 3G). Note that the GFP signal observed in the digit muscles represents GFP expression from incomplete conversion of the tandem array of LoxP reporter cassettes found in this reporter strain (SI Appendix, Fig. S10). (Scale bar: 1 mm.) (H) CHERRY-expressing satellite cells contribute to muscle regeneration. Cross-section of the limb that regenerated after induction of CHERRY expression from the LoxP reporter in PAX7-positive cells. Immunofluorescence for MHC (white) and CHERRY (red) combined with DAPI (blue) of a 3-mo regenerated limb showing robust expression of cytoplasmic CHERRY in muscle cells (n = 3). The area depicted by the rectangle in the leftmost panel is shown at higher magnification in the three right panels. (Scale bars: 200 μm in left and 50 μm in right.)
Subsequently, tamoxifen was administered to induce cassette conversion in satellite cells. At 2 wk after tamoxifen treatment, we amputated the limbs at lower limb level and allowed regeneration (Fig. 3A). We first examined CHERRY expression in the removed limb piece and confirmed that cytosolic CHERRY was specifically turned on in the PAX7-positive, MHC-negative satellite cells (Fig. 3 E and F). After 3 mo of regeneration, we observed strong CHERRY expression in the regenerated lower limbs (Fig. 3G) that was clearly distinguished from the membrane CHERRY expression before tamoxifen treatment. Immunohistochemistry of the regenerated lower limbs showed that essentially all MHC-positive muscle fibers expressed cytosolic CHERRY (Fig. 3H). This result demonstrates that most, if not all, of the regenerated limb muscle fibers are derived from the initially labeled cytosolic CHERRY-positive satellite cells. These results provide direct evidence that satellite cells are the direct cell source generating muscle during axolotl limb regeneration.
Discussion
Based on CRISPR/Cas9 techniques, we have established homology-independent knockin methods in axolotl. We demonstrated the successful insertion of a Cherry reporter gene and a larger memCherry-T2A-ERT2-Cre-ERT2 (∼5 kb) cassette into the axolotl Pax7 and Sox2 genomic loci, and achieved the faithful labeling of neural stem cells and satellite cells. Furthermore, using the Pax7: memCherry-T2A-ERT2-Cre-ERT2 transgenic animals thus obtained, we traced satellite cells during regeneration and provided direct evidence of the satellite cell origin of the muscles in the regenerated limbs.
Of the injected eggs, 5–15% of the derived larvae, depending on the size of the sequences integrated into the targeted locus, were positive for the transgene based on phenotypic and genotypic analysis. Strikingly, many of the F0 transgenic embryos exhibited uniform expression of the reporter gene in the expected tissue compartments. It has been shown that CAS9 protein dramatically improves CRISPR/Cas9-mediated knockout and knockin in several model organisms and cells in culture (7, 18–21, 23, 24). Indeed, injection of CAS9 protein gave a higher rate of integrants and more uniform expression in F0 animals compared with injection of Cas9 mRNA. The first cell division in the fertilized axolotl eggs takes ∼6 h (30), and it is highly likely that transgene integration often occurs in the single cell-stage axolotl egg after injection of the CAS9 gRNA/protein complex. This idea is supported by the fact that the integration junction sequences in the tail and limb are identical in most of individual F0 knockin axolotls. The establishment of the highly penetrant knockin approach in axolotl allows us to directly study F0 transgenic axolotls for biological research.
It has been shown that the gRNA design is critical for achieving highly efficient gene knockout. Genomic DNA cleavage efficiency mediated by different gRNAs targeting the same locus varies considerably in axolotl and other species (6, 31). The mechanisms behind the variation of gRNA activity are unclear, and an effective method of predicting the best gRNA targeting to a given locus is still lacking. In zebrafish, the gRNAs that introduced a high frequency of indels were also highly efficient for knockin of exogenous sequences into the targeted locus (6). We synthesized three gRNAs at the desired targeting loci, evaluated individual gRNA activity in vivo, and selected the best gRNAs mediating at least 90% modification at the targeting sites for the knockin experiments. Consistent with the observations in zebrafish (6), our results also show that knockin efficiency is correlated with the gRNA activity. gRNAs producing a high percentage of indels also lead to highly efficient integration of exogenous sequences at the targeting loci, and vice versa.
Overall, we have established a highly penetrant knockin approach in axolotl and used the tissue-specific–inducible Cre animals to provide direct evidence of the satellite cell origin of muscles during axolotl limb regeneration. Our studies have opened the opportunities to deeply study the mechanisms of axolotl tissue regeneration with the required precision of gene expression control.
Materials and Methods
All axolotl experiments were performed in accordance with German animal laws. Immunohistochemical analysis and in situ hybridization were conducted according to established protocols. The animal experiments, immunohistochemistry, and in situ hybridization are described in detail in SI Appendix, Materials and Methods.
DNA Constructs, CAS9 Protein, and gRNAs.
Targeting constructs were obtained using Gibson assembly and classical molecular cloning methods. All gRNAs were cloned into the DR274 vector (#42250; Addgene) linearized with BbsI (32). All plasmids were verified by sequencing. CAS9 protein and gRNAs were synthesized as described previously (31, 33). gRNA activity was evaluated by collecting and pooling the tails from at least 10 axolotl embryos injected with individual gRNA RNP complexes at the single-cell stage, followed by extraction of genomic DNA as a template for PCR. The PCR products were cloned into the pGEMT vector (Promega) and then transformed into Escherichia coli, followed by sequencing of 30–40 individual clones for gRNA evaluation. The gRNAs that mediated the greatest indel production, with at least 90% of cloned PCR products harboring modifications at the targeted sites, were used for further knockin experiments. Details of cloning, gRNA-binding sequences, and gRNA evaluation are provided in SI Appendix, Materials and Methods.
Transplantation and Tamoxifen and EdU Treatment.
Before transplantation, upper limbs of donors (3- to 4-mo-old double-transgenic axolotls) were amputated and then allowed to regenerate for 6–8 d. The limb blastemas from donors were then collected and transplanted to the freshly amputated upper limb stumps of recipients of the same size, as described previously (2). To activate ERT2-Cre-ERT2, 4-hydroxytamoxifen (H7904; Sigma-Aldrich) at 40 μg/g body weight was injected i.p. into axolotls. For EdU administration, 10 μg of EdU (A10044; Thermo Fisher Scientific) per g of body weight was injected i.p. daily for 3 d into 2-mo-old Sox2: Sox2-ORFΔ-T2A-Cherry axolotls.
Imaging.
Bright-field and fluorescent axolotl larva, limb, and tail images were acquired with an Olympus stereomicroscope. Fluorescent images of sections were acquired with a Zeiss Axio Observer microscope or confocal microscope (LSM 700 or LSM 880 with Airyscan), using a 20× or 25× objective.
Supplementary Material
Acknowledgments
We thank Beate Gruhl, Anja Wagner, and Simone Kaudel for outstanding animal care. This study was supported by Human Frontier Science Program Grant RGP0016/2010; German Research Foundation (DFG) Grants 274/3-2, 274/3-3, and 274/2-3/SFB655 from Cells into Tissues; European Research Council Advanced Investigator Grant 294324; Central Funding grants from the DFG Center for Regenerative Therapies Dresden (CRTD) Grant FZ111; a CRTD Seed grant for zinc-finger nucleases; CRTD Postdoctoral Seed Grant 2015; and Research Start Grants S82111 and 8S0109 from South China Normal University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. MF996566, MF996567, and MG017611).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706855114/-/DCSupplemental.
References
- 1.Kragl M, et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- 2.Sandoval-Guzmán T, et al. Fundamental differences in dedifferentiation and stem cell recruitment during skeletal muscle regeneration in two salamander species. Cell Stem Cell. 2014;14:174–187. doi: 10.1016/j.stem.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Khattak S, et al. Germline transgenic methods for tracking cells and testing gene function during regeneration in the axolotl. Stem Cell Rep. 2013;1:90–103. doi: 10.1016/j.stemcr.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakade S, et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat Commun. 2014;5:5560. doi: 10.1038/ncomms6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Z, et al. Heritable CRISPR/Cas9-mediated targeted integration in Xenopus tropicalis. FASEB J. 2015;29:4914–4923. doi: 10.1096/fj.15-273425. [DOI] [PubMed] [Google Scholar]
- 6.Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014;24:142–153. doi: 10.1101/gr.161638.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoshijima K, Jurynec MJ, Grunwald DJ. Precise editing of the zebrafish genome made simple and efficient. Dev Cell. 2016;36:654–667. doi: 10.1016/j.devcel.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui X, et al. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol. 2011;29:64–67. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- 9.Yang H, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Y, et al. Generation of eGFP and Cre knockin rats by CRISPR/Cas9. FEBS J. 2014;281:3779–3790. doi: 10.1111/febs.12935. [DOI] [PubMed] [Google Scholar]
- 11.Kimura Y, Hisano Y, Kawahara A, Higashijima S. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep. 2014;4:6545. doi: 10.1038/srep06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li K, Wang G, Andersen T, Zhou P, Pu WT. Optimization of genome engineering approaches with the CRISPR/Cas9 system. PLoS One. 2014;9:e105779. doi: 10.1371/journal.pone.0105779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He X, et al. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 2016;44:e85. doi: 10.1093/nar/gkw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gagnon JA, et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186, and erratum (2014) 9:e106396. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang X, et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 21.Burger A, et al. Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development. 2016;143:2025–2037. doi: 10.1242/dev.134809. [DOI] [PubMed] [Google Scholar]
- 22.Sung YH, et al. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res. 2014;24:125–131. doi: 10.1101/gr.163394.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paix A, Folkmann A, Rasoloson D, Seydoux G. High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics. 2015;201:47–54. doi: 10.1534/genetics.115.179382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schumann K, et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci USA. 2015;112:10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jostes B, Walther C, Gruss P. The murine paired box gene, Pax7, is expressed specifically during the development of the nervous and muscular system. Mech Dev. 1990;33:27–37. doi: 10.1016/0925-4773(90)90132-6. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen PD, et al. Muscle stem cells undergo extensive clonal drift during tissue growth via Meox1-mediated induction of G2 cell-cycle arrest. Cell Stem Cell. 2017;21:107–119. doi: 10.1016/j.stem.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 27.Ellis P, et al. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- 28.Hernández PP, Olivari FA, Sarrazin AF, Sandoval PC, Allende ML. Regeneration in zebrafish lateral line neuromasts: Expression of the neural progenitor cell marker sox2 and proliferation-dependent and -independent mechanisms of hair cell renewal. Dev Neurobiol. 2007;67:637–654. doi: 10.1002/dneu.20386. [DOI] [PubMed] [Google Scholar]
- 29.Rodrigo Albors A, et al. Planar cell polarity-mediated induction of neural stem cell expansion during axolotl spinal cord regeneration. Elife. 2015;4:e10230. doi: 10.7554/eLife.10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schreckenberg GM, Jacobson AG. Normal stages of development of the axolotl, Ambystoma mexicanum. Dev Biol. 1975;42:391–400. doi: 10.1016/0012-1606(75)90343-7. [DOI] [PubMed] [Google Scholar]
- 31.Fei JF, et al. CRISPR-mediated genomic deletion of Sox2 in the axolotl shows a requirement in spinal cord neural stem cell amplification during tail regeneration. Stem Cell Rep. 2014;3:444–459. doi: 10.1016/j.stemcr.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fei JF, et al. Tissue- and time-directed electroporation of CAS9 protein–gRNA complexes in vivo yields efficient multigene knockout for studying gene function in regeneration. NPJ Regen Med. 2016;1:16002. doi: 10.1038/npjregenmed.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.