Significance
Autophagy is a cellular process that results in the capture of cytosolic material in double-membrane vesicles, which subsequently fuse with lysosomes to degrade the captured contents. Autophagy is essential to maintain cellular homeostasis, respond to cellular stress, and prevent the accumulation of material that could damage the cell. The initiation of autophagy is carried out by the Atg1 complex. Whereas recent work has provided functional and mechanistic insight into many components of the Atg1 complex, one member of this complex—Atg20—has remained relatively uncharacterized. Here we report a detailed investigation into the structure and function of Atg20, including the identification of an amphipathic helix in Atg20 that is required for efficient autophagy and membrane tubulation.
Keywords: autophagy, vacuole, yeast
Abstract
The Atg20 and Snx4/Atg24 proteins have been identified in a screen for mutants defective in a type of selective macroautophagy/autophagy. Both proteins are connected to the Atg1 kinase complex, which is involved in autophagy initiation, and bind phosphatidylinositol-3-phosphate. Atg20 and Snx4 contain putative BAR domains, suggesting a possible role in membrane deformation, but they have been relatively uncharacterized. Here we demonstrate that, in addition to its function in selective autophagy, Atg20 plays a critical role in the efficient induction of nonselective autophagy. Atg20 is a dynamic posttranslationally modified protein that engages both structurally stable (PX and BAR) and intrinsically disordered domains for its function. In addition to its PX and BAR domains, Atg20 uses a third membrane-binding module, a membrane-inducible amphipathic helix present in a previously undescribed location in Atg20 within the putative BAR domain. Taken together, these findings yield insights into the molecular mechanism of the autophagy machinery.
Healthy cells maintain homeostasis via a vital self-cleaning mechanism, macroautophagy (hereinafter autophagy), that is conserved from yeast to mammals. Autophagy involves the sequestration of cargo by a double-membrane compartment, the phagophore, which expands and seals to form an autophagosome. Cargo selection distinguishes nonselective from selective autophagy. Nonselective autophagy engulfs random cytoplasm during starvation, whereas selective autophagy targets specific cargo (e.g., mitochondria, peroxisomes, vacuolar hydrolases) for transport to the vacuole/lysosome (1–3). Each of these processes involves what has been termed the “core” autophagy machinery, proteins that are required for both nonselective and selective autophagy. Many of the core autophagy machinery proteins have conserved homologs from yeast to mammals.
Forty-one autophagy-related (Atg) proteins have been identified in fungi, many of which have been categorized into functional groups (3, 4). The first functional group of Atg proteins assembles into the induction complex, also termed the Atg1 complex, to initiate autophagy. In budding yeast, this complex for the nonselective autophagy pathway is composed of Atg1, Atg13, Atg17, Atg29, and Atg31. In mammals, the homologous ULK1 complex is composed of ULK1 (or its homolog ULK2), ATG13, RB1CC1/FIP200, and ATG101 (5). Although the stable Atg17-Atg31-Atg29 subcomplex is required for efficient nonselective autophagy, it is not needed for selective autophagic processes. Instead, this subcomplex is replaced by Atg11, which also binds Atg1 and assembles along with Snx4 (also termed Atg24) and Atg20. The latter two proteins are sorting nexins containing a PX domain that binds membranes enriched in phosphatidylinositol-3-phosphate (PtdIns3P), thereby providing a functional connection between the Atg1 complex and the PtdIns 3-kinase complex that also plays a critical role in autophagy induction (6, 7).
Recent structural studies have partially clarified the role of some of the subunits of the Atg1/ULK1 complex; however, Snx4 and Atg20 and their role in the Atg1 complex remain unexplored. Snx4 is involved in retrieval of late-Golgi SNAREs and has a mammalian homolog, SNX4, thought to function in endocytosis and intracellular trafficking (8). Homologs of Atg20 exist in fungi from Saccharomyces cerevisiae to Schizosaccharomyces pombe; however, a human sequence-based homolog of Atg20 has not been identified, although optineurin (OPTN) has been proposed as a functional counterpart (9) and SNX30 has been proposed as a mammalian equivalent based on its dimerization pattern and phylogenetic tree (10–12). The function of Atg20 is unknown, except that it is important for an efficient Cvt pathway, for the degradation of the peroxisomal thiolase enzyme Pot1/Fox3 during pexophagy (6), and for clearance of accumulated mitochondria during mitophagy (13).
In the present study, we probed the structure and function of the least explored subunit of the Atg1 complex, Atg20. We demonstrate a facilitating role for this protein in autophagy induction, which requires a hybrid native conformation composed of structured domains mixed with intrinsically disordered regions. We also demonstrate that Atg20 forms a heterodimer with Snx4 in vitro, and characterize this complex using analytical ultracentrifugation (AUC) and small-angle X-ray scattering (SAXS). These results show how Atg20 uses distinct regions, including a unique gapped BAR domain, for optimal function.
Results
Atg20 Facilitates Autophagy Induction.
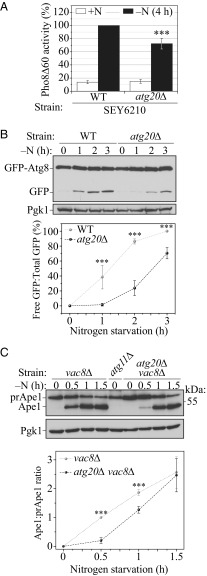
Previous studies concluded that Atg20 is not required for nonselective autophagy, because the budding yeast, S. cerevisiae, atg20∆ strain showed only a minor decrease in autophagy activity after 4 h of nitrogen starvation (6). Atg20 binds the Atg11 scaffold (2), and a recent report revealed a new facilitating function of Atg11 in the autophagy induction complex (14). This finding led us to revisit the question of whether Atg20 can also function as a facilitator in nonselective autophagy initiation. To answer this question, we first used the Pho8∆60 assay to monitor autophagy (15). This assay relies on the cytosolic Pho8∆60 zymogen, which must be targeted to the vacuole by nonselective autophagy to become active. In wild-type (WT) cells, a substantial increase in Pho8∆60 activity is observed during nitrogen starvation (Fig. 1A). In comparison, the SEY6210 atg20∆ strain showed an ∼30% decrease in Pho8∆60 activity relative to the WT strain (Fig. 1A), a result that may in fact correspond to the decrease in activity seen previously (6). To check whether this defect is background-dependent, we carried out the Pho8∆60 assay in the W303 background and did not detect a clear defect upon ATG20 deletion (Fig. 1 and SI Appendix, Fig. S1A).
Fig. 1.
Atg20 is essential for the efficient initiation of bulk autophagy. (A) WT (WLY176) and atg20Δ (HPY063) cells expressing Pho8Δ60 were shifted from nutrient-rich conditions to SD-N medium for 4 h. (B) Autophagy as measured by the GFP-Atg8 processing assay in WT (SEY6210) and atg20Δ (D3Y009) cells. Cells transformed with the plasmid (pRS426) carrying a GFP-Atg8 construct were grown in rich selective medium and then starved for 1, 2, and 3 h. The free GFP:total GFP ratio was measured and normalized to that of WT after 3 h of starvation (100%). (C) Kinetics of prApe1 maturation in nutrient-rich medium and after the shift to SD-N medium for the indicated times. The vac8∆ atg20∆ (HPY079) strain was compared with the vac8Δ (CWY230) strain. The atg11Δ (SEY6210) strain served as a negative control, and Pgk1 served as a loading control. Quantitative evaluation of western blot data was done with three to four independent experiments carried out in nutrient-rich and nitrogen starvation conditions. Error bars represent the SD from three to four independent experiments. Statistical significance was tested using the unpaired two-tailed Student’s t test: **P < 0.01; ***P < 0.005.
The Pho8Δ60 assay relies on colorimetric detection of a cleaved substrate and is typically analyzed at least 3–4 h postinduction. Accordingly, a defect manifested early in the process, such as at the stage of autophagy initiation, may be difficult to detect with this assay. Therefore, we analyzed the WT and atg20Δ cells with the GFP-Atg8 processing assay. In brief, Atg8 (or GFP-Atg8) lines both sides of the phagophore, and the population on the concave side becomes enclosed within the completed autophagosome. Following vacuolar delivery, Atg8 is readily degraded, whereas GFP is relatively stable and can be monitored as a measure of autophagy flux (16). In both the SEY6210 and W303 backgrounds, autophagy induction was significantly delayed in atg20∆ cells, with a clear difference detectable after 1–3 h of nitrogen starvation (Fig. 1B and SI Appendix, Fig. S1B).
To further test whether Atg20 is required for nonselective autophagy, we probed the transport of the precursor form of aminopeptidase I (prApe1) to the vacuole during nitrogen starvation in cells where the Cvt pathway was blocked by VAC8 deletion (17). As expected, the SEY6210 vac8Δ mutant was completely blocked for prApe1 maturation before autophagy induction, whereas processing to the mature enzyme could be readily detected within 0.5 h after shifting to starvation conditions (Fig. 1C). In comparison, the SEY6210 atg20∆ vac8∆ strain exhibited a significant delay (∼80% decrease) in prApe1 maturation after 0.5 h of nitrogen starvation. This defect gradually disappeared with the progression of autophagy. A similar result was found in the W303 background, with a clear lag in prApe1 maturation in the W303 atg20∆ vac8∆ cells relative to the W303 vac8∆ strain, although the onset of the starvation-induced transport of prApe1 to the vacuole was delayed compared with the SEY6210 background (Fig. 1C and SI Appendix, Fig. S1C). Taken together, these data reveal that Atg20 is required for the efficient induction of nonselective autophagy (Fig. 1 B and C and SI Appendix, Fig. S1 B and C).
Native Atg20 Is Predicted to Have Large Disordered Regions.
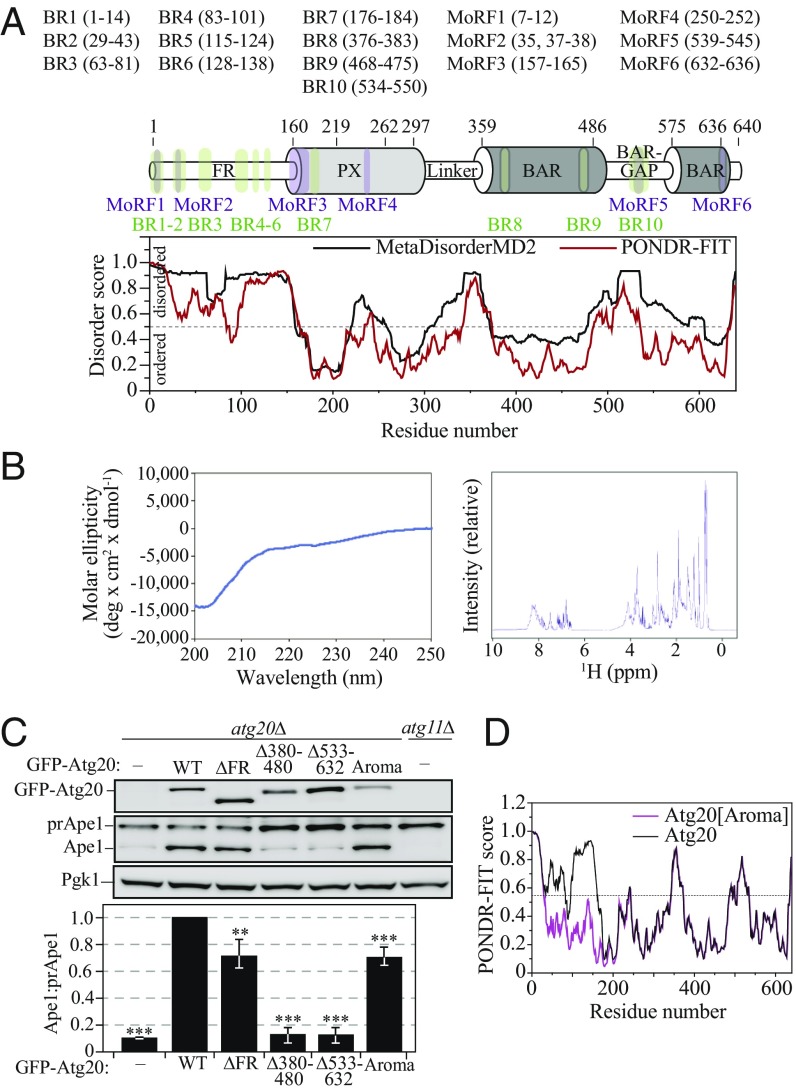
Atg20 contains a PX domain, which recruits the protein to PtdIns3P-enriched lipid membranes (6); no structure-function information is available regarding other domains in Atg20. To gain insight into the functional conformation of Atg20, we first applied a multialgorithm bioinformatics approach that analyzed the amino acid sequence of S. cerevisiae Atg20. Protein-protein BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and two metapredictors of intrinsically disordered regions (PONDR-FIT and MetaDisorderMD2) (18–20) revealed that Atg20 has a long, disordered N terminus, denoted here as the flexible region (FR; residues 1–160) (Fig. 2A). In agreement with this prediction, circular dichroism of the recombinant purified Atg20[FR] (Fig. 2B, Left) shows a negative peak at 200 nm, which is typical of disordered regions. The 1D 1H NMR spectra of Atg20[FR] (Fig. 2B, Right) also exhibits poor dispersion in the amide region, further confirming that the N terminus of Atg20 is disordered. The Atg20[FR] is followed by the PX domain (residues 160–297), which is connected to a BAR domain through a region denoted here as the linker (residues 298–358). This BAR domain (residues 359–636) has a gap in the consensus sequence, which we refer to here as the BAR-GAP (residues 487–574) (Fig. 2A). The BAR-GAP is predicted to be partially disordered, and no similar regions have been observed in any BAR domain structures deposited in the Protein Data Bank (PDB). Together, the results of the sequence-analyzing algorithms show that the native Atg20 protein is a member of the PX-BAR domain family of sorting nexins (21), and is enriched in functionally unassigned intrinsically disordered protein regions (IDPRs) at the N terminus and in the BAR-GAP.
Fig. 2.
Bioinformatics and biochemical analysis of structured and intrinsically disordered domains in Atg20. (A) Domain representation of Atg20 that incorporates the results of the protein amino acid sequence analyses by the PONDR-FIT, MetaDisorderMD2, protein-protein BLAST, MoRFPred, and ANCHOR algorithms. (B, Left) Far UV circular dichroism spectrum of the recombinant purified Atg20[FR]. (B, Right) One-dimensional 1H NMR spectrum of Atg20[FR]. (C) Empty vector, the plasmids pCuGFP-Atg20(426), pCuGFP-Atg20[ΔFR](426), pCuGFP-Atg20[Δ380–480](426), pCuGFP-Atg20[Δ533–632](426), and pCuGFP-Atg20[Aroma](426) were transformed into atg20Δ (D3Y009) cells and examined for prApe1 processing in rich selective medium. The atg11Δ (SEY6210) strain served as a negative control. Quantification of the Ape:prApe1 ratio was determined from three independent experiments. Error bars represent SDs. Statistical significance was tested using the unpaired two-tailed Student’s t test: **P < 0.05; ***P < 0.005. (D) Comparison of PONDR-FIT scores for WT (black) and Atg20[Aroma] (magenta).
IDPRs have unique physiochemical properties that render them unable to adopt a well-defined 3-dimensional structure. The biological function of IDPRs relies on various functional elements including binding modules, which undergo a disorder-to-order transition upon binding proteins, nucleic acids, or lipids (22–25). Depending on how these functional elements are identified from a protein amino acid sequence, the binding modules are termed molecular recognition features (MoRFs) or ANCHOR-based disordered binding regions (BRs). The bioinformatics tool MoRFPred (biomine-ws.ece.ualberta.ca/MoRFpred/index.html) (26) predicts MoRFs, and ANCHOR (anchor.enzim.hu/) (27, 28) predicts ANCHOR-based disordered BRs. We applied these tools to the S. cerevisiae Atg20 sequence and found 6 MoRFs and 10 predicted BRs, some of which overlap in the Atg20 protein (Fig. 2A and SI Appendix, Fig. S2). MoRF1/BR1, MoRF2/BR2, and BR3-6 are located at the flexible N terminus; MoRF3, MoRF4, and BR7 map on mobile segments of the PX domain; and BR8 and BR9 flank the first segment of the BAR domain; therefore, they exhibit relatively high sequence homology (SI Appendix, Fig. S2). MoRF5/BR10 localizes to the BAR-GAP, and MoRF6 is near the very C terminus of Atg20. Of all the MoRFs and BRs located in disordered, poorly conserved segments of Atg20, BR3 is the most highly conserved BR likely involved in protein binding, presumably via formation of a conserved secondary structure element (α-helix or β-sheet). Interestingly, BR6 contains several conserved serine or threonine residues, and could be a target for an unknown kinase. The Eukaryotic Linear Motif (ELM) database (elm.eu.org) reports the RTSLS sequence in BR6 as a canonical arginine-containing phospho motif mediating a strong interaction with YWHA/14–3-3 proteins.
Mapping Atg20 Domains Interacting with Atg11 and Snx4.
To probe how Atg20 uses its hybrid architecture, we constructed three deletion mutants—Atg20[∆FR], Atg20[∆380–480], and Atg20[∆533–632]—and one site-directed mutant, Atg20[Aroma]. Atg20[∆FR] lacks the FR and exhibits an ∼30% defect in the Cvt pathway, as determined by the prApe1 processing assay (Fig. 2C), but no defect in nonselective autophagy induced by nitrogen starvation (SI Appendix, Fig. S3 A–C). The Atg20[Δ380–480] mutant lacks the majority of the first consensus sequence for the BAR domain, whereas the Atg20[Δ533–632] mutant lacks a part of the BAR-GAP, including MoRF5/BR10 and the second BAR domain consensus sequence. These deletions produce dysfunctional proteins in the Cvt pathway (Fig. 2C), as well as in nonselective autophagy (SI Appendix, Fig. S3 A–C).
To evaluate the importance of flexibility in the FR of Atg20, we constructed Atg20[Aroma] by replacing nine Glu or Lys residues with the aromatic residues of Tyr, Phe, or Trp: K47W, K52Y, E108W, E123W, E143W, E148F, E149W, K156F, and K157W (black asterisks in SI Appendix, Fig. S2). This Atg20[Aroma] mutant has a lower propensity for disorder in its FR (Fig. 2D) and has a partial defect in the Cvt pathway (Fig. 2C) compared with the WT, but no defect in nonselective autophagy (SI Appendix, Fig. S3A). Together, the mutants demonstrate a functional role of the Atg20 FR in the Cvt pathway and an indispensable function of the Atg20 BAR in both the Cvt pathway and nonselective autophagy.
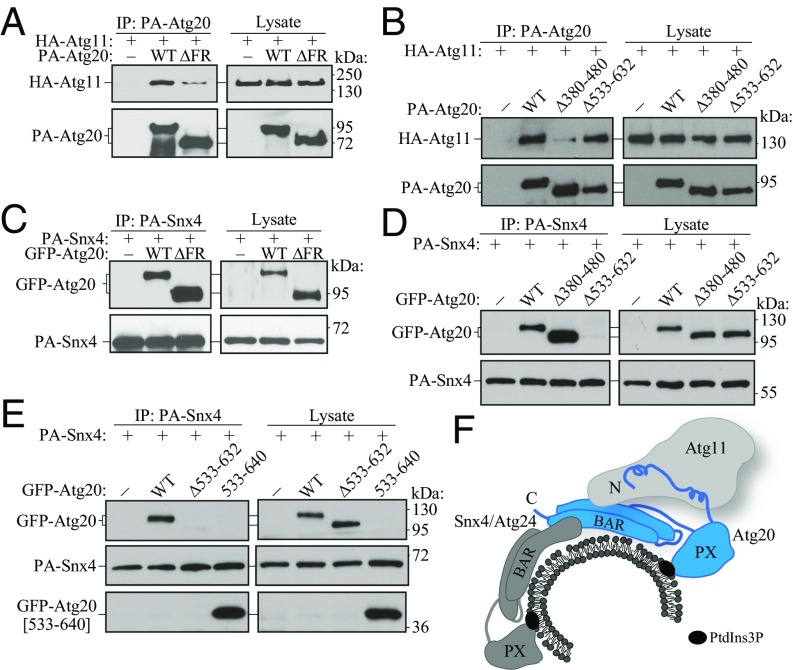
We next investigated how Atg20 binds its partner subunits in the Atg11-Atg20-Snx4 trimer. To map Atg11 and Snx4 BRs on Atg20, we carried out a coimmunoprecipitation (co-IP) analysis with three aforementioned deletion mutants. Atg20 was tagged on the N terminus with protein A (PA), and was used for affinity isolation of HA-tagged Atg11. We found that the Atg20 FR was required for efficient binding to Atg11 (Fig. 3A). The weak interaction between Atg20[∆FR] and Atg11 was completely lost when the HA tag on Atg11 was replaced with the larger GFP tag (SI Appendix, Fig. S3D). Atg20 lacking residues 380–480 also nearly failed to bind HA-tagged Atg11, revealing a second binding site for Atg11 (Fig. 3B), whereas deletion of the C-terminal amino acids 533–632 had no detectable effect. Taken together, these results suggest that Atg20 encompasses two binding sites for the Atg11 scaffold protein, the Atg20 FR domain and the 380–480 region, each of which is necessary for efficient interaction of the two proteins.
Fig. 3.
Mapping of the Atg11 and Snx4 binding sites on Atg20. (A and B) Coprecipitation of HA-Atg11 by PA-Atg20. The plasmids pCuPA(424), pCuPA-Atg20(424), pCuPA-Atg20[ΔFR](424), pCuPA-Atg20[Δ380–480](424), and pCuPA-Atg20[Δ533–632](424) were transformed into MKO (YCY123) cells and coexpressed with a plasmid encoding HA-Atg11 (pCuHA-Atg11; 416) under the CUP1 promotor. (C–E) Coprecipitation of GFP-Atg20 by PA-Snx4. The plasmids pCuGFP(426), pCuGFP-Atg20(426), pCuGFP-Atg20[ΔFR](426), pCuGFP-Atg20[Δ380–480](426), pCuGFP-Atg20[Δ533–632](426), and pCuGFP-Atg20[533-640](426) were transformed into MKO (YCY123) cells and coexpressed under the CUP1 promotor with a plasmid encoding PA-Snx4 (pCuPA-Snx4; 424). For A–E, cells were cultured in SMD, and cell lysates were prepared and incubated with IgG-Sepharose for affinity purification. The proteins were separated by SDS/PAGE and detected with the indicated antibody. (F) Schematic representation depicting the Atg11-Atg20-Snx4 trimer, based on the results presented in the figure.
Snx4 is also a member of the BAR domain family of proteins, based on the analysis of its amino acid sequence by protein-protein BLAST. Probing the same deletion mutants of Atg20 for binding with Snx4 by co-IP identified the C-terminal region 533–632 as the single binding site for Snx4 (Fig. 3 C and D). In agreement with this finding, an Atg20 peptide encompassing residues 533–640 showed a strong interaction with Snx4 (Fig. 3E). Thus, the region of Atg20 encompassed by amino acids 533–640 is both necessary and sufficient for binding Snx4.
Residues 533–632 of Atg20 contain the highly conserved motif 626NLExW (SI Appendix, Figs. S2 and S4). We analyzed the role of this motif in the interaction with Snx4 by mutating L627 and W630 to alanine (LW/AA). This mutation has no effect on the interaction of Atg20 and Snx4 (SI Appendix, Fig. S5A), but does affect the function of Atg20 in terms of prApe1 maturation (SI Appendix, Fig. S5B). Thus, the 626NLExW motif in Atg20 appears to play a structural role, perhaps in proper folding of the BAR domain, rather than serving as a binding site for Snx4. Taken together, the data presented in Fig. 3 A–E reveal a model for the Atg11-Atg20-Snx4 trimer (Fig. 3F).
Atg20 Is a Posttranslationally Modified Protein.
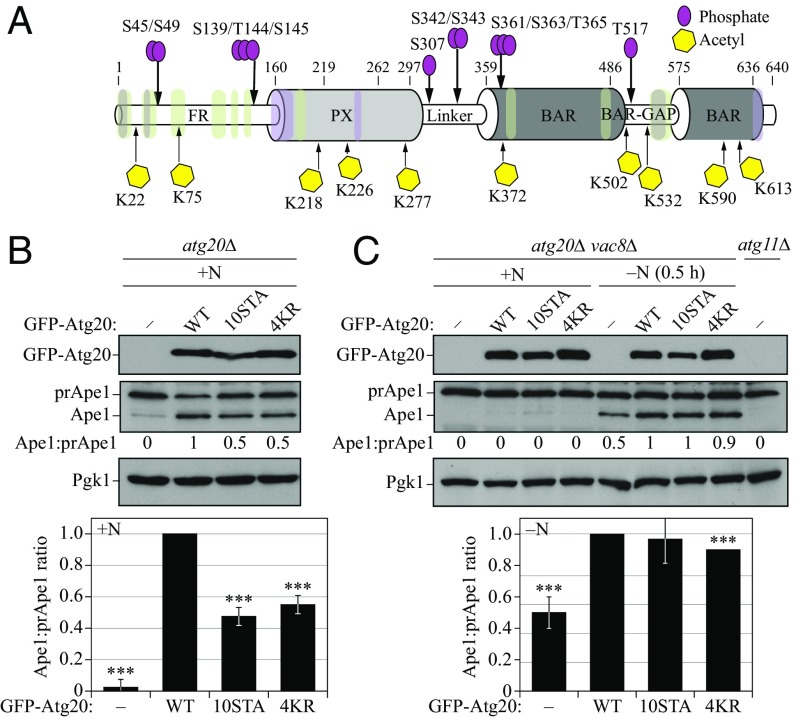
Protein function, stability, or folding efficiency can be affected by posttranslational modifications (PTMs) (29). To determine whether this mechanism fine-tunes Atg20 function, we applied LC-MS/MS analysis on the partially purified Atg20 protein and searched for PTMs (Fig. 4A). We detected 10 acetylated lysines in Atg20, specifically at residues 22, 75, 218, 226, 277, 372, 502, 532, 590, and 613. These acetylation sites, confirmed by b and/or y ions (SI Appendix, Figs. S6 and S7), are distributed evenly throughout the protein. Of the identified acetylation sites, lysine 218, located in the PX domain, is the most conserved, with 11 of the 12 yeast sequences that are aligned containing a lysine in this position (SI Appendix, Figs. S2 and S4). We detected phosphorylation of Atg20 exclusively on IDPRs, in agreement with a previous study showing correlation of phosphorylation with disordered regions (30). Atg20 is phosphorylated on Ser139, Thr144, Ser145, Ser307, Ser342, Ser343, and Thr517, all of which are previously unreported phosphorylation sites mediated by unknown kinases. In addition to these sites, our MS analysis also detected phosphorylation on Ser45 and Ser49, which are targeted by Cdc28/Cdk1 (31), and on Ser363 and Thr365, sites recognized by casein kinase 2 (CK2) (32), in agreement with previous high-throughput studies. The third site reportedly modified by CK2, Ser361, was confirmed by peptide mass only (SI Appendix, Figs. S8 and S9). The most conserved phosphorylation site is Ser307, with either a serine or a threonine present in this position in 10 of the 12 yeast sequences that were aligned (SI Appendix, Figs. S2 and S4). Ser307 is located in the linker between the PX and BAR domains.
Fig. 4.
Atg20 is a posttranslationally modified protein. (A) Schematic domain representation of Atg20 with all PTM sites that were experimentally detected by LC-MS/MS analysis. (B) Functionality of Atg20 variants in the Cvt pathway analyzed by the prApe1 maturation assay. The SEY6210 atg20Δ strain was transformed with the plasmids pCuGFP(426), pCuGFP-Atg20(426), pCuGFP-Atg20[10STA](426), and pCuGFP-Atg20[4KR](426). Cells were cultured in rich selective medium. (C) Autophagy induction examined for WT and PTM mutants of Atg20 using the prApe1 maturation assay. The SEY6210 atg20∆ vac8∆ strain was transformed with the same plasmids as indicated in B. Cells were cultured in rich selective medium and then shifted to SD-N medium for 0.5 h. The atg11Δ (SEY6210) strain served as a negative control, and Pgk1 served as a loading control. Quantification of the Ape:prApe1 ratio was determined from three independent experiments. Error bars represent SDs. Statistical significance was tested using the unpaired two-tailed Student’s t test: ***P < 0.005.
To investigate whether acetylation and phosphorylation of Atg20 are important for its function in vivo, we generated a multiple-nonphosphorylatable and multiple-nonacetylatable mutant by replacing 10 serines or threonines (Ser45, Ser49, Ser139, Thr144, Ser145, Ser342, Ser343, Ser361, Ser363, and Thr365) with alanine (Atg20[10STA]) and replacing four lysines (Lys226, Lys277, Lys372, and Lys532) with arginine (Atg20[4KR]), respectively (SI Appendix, Fig. S10). Probing these mutants using the prApe1 processing assay under growing conditions in the atg20∆ strain showed that phosphorylation and acetylation of Atg20 are necessary for an efficient Cvt pathway (Fig. 4B).
To test the importance of Atg20 PTMs in nonselective autophagy, we examined these plasmid-driven Atg20 variants using the prApe1 maturation assay in the atg20∆ vac8∆ background. We found that the Atg20[10STA] mutant exhibited no significant defect in the induction of autophagy, whereas the Atg20[4KR] mutant displayed only a 10% (albeit consistent) defect in this pathway relative to the WT (Fig. 4C). Taken together, the data in Fig. 4 suggest that phosphorylation and acetylation modulate the optimal conformation of Atg20, which is essential for an efficient Cvt pathway, but not for nonselective autophagy induction. A reason for the differing results, which were also obtained with the Atg20[∆FR] and Atg20[Aroma] mutants (Fig. 2C and SI Appendix, S3 A–C), could be that PTMs finely tune two binding sites—one of them within the Atg20 FR (Fig. 3)—for interaction with Atg11, which is critical for the Cvt pathway, but not for nonselective autophagy.
Structural Characterization of Atg20.
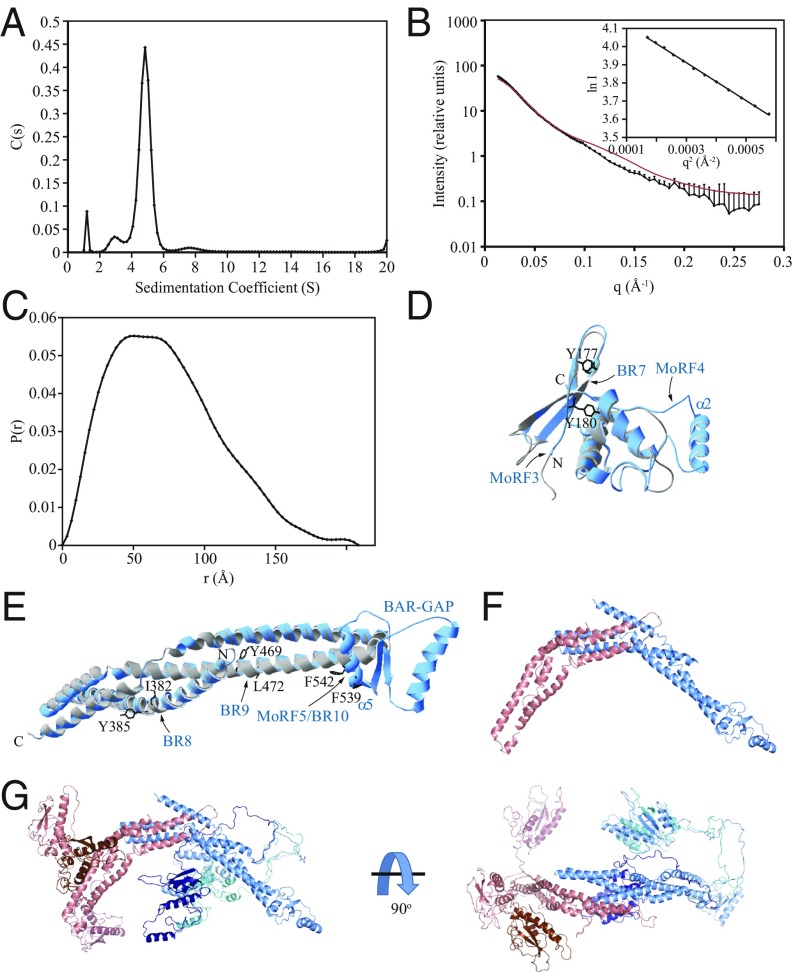
Some sorting nexins both homodimerize and heterodimerize, whereas others prefer only heterodimers. The Atg20 mammalian equivalent, SNX30, does not form homodimers, as judged by co-IP from HEK-293T cells (11). In agreement with these findings (33), we were able to produce recombinant Atg20 only in a stable heterodimeric complex with Snx4, but not as a homodimer (Fig. 5 and SI Appendix, Fig. S11). Heterodimer formation was not dependent on the FR of Atg20, as Atg20156–640-Snx421–423 was also able to form a stable heterodimer. Dynamic light scattering on Atg20156–640-Snx421–423 revealed a molecular mass of 127.6 kDa, demonstrating that the recombinant complex is indeed a heterodimer, as its expected mass is 121.5 kDa.
Fig. 5.
Structural characterization of Atg20. (A) Sedimentation velocity AUC recorded on the full-length Atg20-Snx4 heterodimer. (B) In-line SEC with SAXS data recorded on the Atg20156–640-Snx421–423 heterodimer. (Inset) The Guinier region for these data. SAXS data calculated from the Atg20-Snx4 ensemble homology model in F are shown in red. (C) Pair distance distribution function calculated from the SEC-SAXS data in B. (D and E) Homology modeling of structurally stable domains in Atg20. (D) Structural alignment of the crystal structure of the PX domain of SNX1 (gray) with the homology model of the PX domain of Atg20 (blue). (E) Structural alignment of the crystal structure of the BAR domain of SNX1 (gray) with the homology model of the BAR domain of Atg20 (blue). The amino acid residues of Atg20 mutated to Glu (Tyr177, Tyr180, Ile382, Tyr385, Tyr469, Leu472, Phe539, and Phe542) are shown (black). Phe539 and Phe542 are within MoRF5/BR10 that maps on the putative α5 helix of the BAR-GAP. (F) Atg20-Snx4 homology model of the BAR domain dimer. Atg20 is shown in blue; Snx4, in red. (G) Atg20-Snx4 ensemble model generated using BilboMD. The BAR domains of the different ensemble models are superimposed. The PX domain and linker from each model are shown in a different shade of blue (Atg20) or red (Snx4).
We next used AUC and SAXS to investigate the overall shape of the heterodimer. Both the AUC (Fig. 5A) and SAXS data (Fig. 5 B and C) demonstrated an elongated architecture for the heterodimer, as expected for a PX-BAR dimer.
We attempted to crystalize the purified Atg20-Snx4 heterodimer, but the dynamic nature of this complex rendered this attempt unsuccessful. To obtain an approximate image of the structured domains of Atg20, we used the SWISS-MODEL workspace (swissmodel.expasy.org/interactive) (34–36) and created homology models of the Atg20 PX and BAR domains based on the most similar template, human SNX1. SNX1 contains a long, disordered N terminus, as does Atg20, but the PX and BAR domains of SNX1 are more structured, with no BAR-GAP in the consensus sequence (based on the PONDR-FIT profile and protein BLAST), in contrast to Atg20. The overlap of data in Fig. 2A with the homology model of the Atg20 PX domain (I161-N297) (Fig. 5D) based on the SNX1 template (PDB ID code 2I4K) shows that the second half of MoRF3 and the short MoRF4 map onto flexible loops, and that BR7 maps onto the beginning of the β2 sheet of the PX domain. The same model also indicates that the Atg20 PX might have an additional helix (α2) compared with the SNX1 PX, which carries a flexible loop in the homologous position. The homology model of the Atg20 BAR domain (R360-E632) (Fig. 5E) based on the SNX1 template (PDB ID code 4FZS) shows α-helical rods that are typical for a BAR domain and that overlap with BAR rods of SNX1; BR8 and BR9 map onto these rods (Fig. 2A).
To generate a model for the Atg20-Snx4 heterodimer, we used SWISS-MODEL to create a homology model for Snx4 based on the most similar template, human SNX9. The BAR domains from the Snx4 and Atg20 homology models were aligned to the human SNX1 BAR domain dimer (Fig. 5F); the Atg20 PX domain model was connected to the Atg20 BAR domain model by a flexible linker. The Atg20-Snx4 homology model was used as a starting point for molecular dynamics (MD)-based fitting of the SAXS data by BilboMD (37). During the MD analysis, the linkers between the end of the PX domains and the start of the BAR domains were defined as flexible. The overall best fit to the experimental data (χ = 4.88) resulted from an ensemble of three dimeric structures (Fig. 5 B and G).
Membrane-Inducible Amphipathic Helix in Atg20.
The Atg20 BAR-GAP is a specific sequence separating the BAR domain into two segments. The BAR-GAP is predicted to be disordered by two metapredictors (18, 20) (Fig. 2A) and three unrelated, recently reported individual predictors (SI Appendix, Fig. S12). In the homology model, the MoRF5/BR10 element within the BAR-GAP maps onto a putative helix, specifically on the α5 helix (Fig. 5E). A helical wheel representation (Fig. 6A) indicates that this helix may be amphipathic. In fact, out of all predicted MoRFs and BRs in Atg20, MoRF5/BR10 is the only element with a predicted amphipathicity. Moreover, the amino acid sequences homologous to S. cerevisiae MoRF5/BR10 in other organisms that express an Atg20 homolog (Kluyveromyces lactis and Candida glabrata) are also predicted to form amphipathic helices (AHs) (SI Appendix, Fig. S13A). In general, binding-coupled AHs can mediate protein–protein interaction (38), but are common in BAR proteins (39), where they function as sensors of membrane curvature (40) or promote membrane fission (41). The most well-studied and experimentally well-proven AHs in BAR proteins are those of rat SH3GL2/endophilin A1, fruit fly Amph/amphiphysin (SI Appendix, Fig. S13B), and human SNX9 (42). In all three of these proteins, the AH is located in an IDPR, outside the BAR domain consensus sequence, and is detected by at least one of the algorithms designed to search for disordered foldable elements (MoRFs or ANCHOR BRs). The putative AH in S. cerevisiae Atg20 exhibits all these features in the comparative bioinformatics analysis (SI Appendix, Fig. S14), indicating a similar molecular mechanism of function.
Fig. 6.
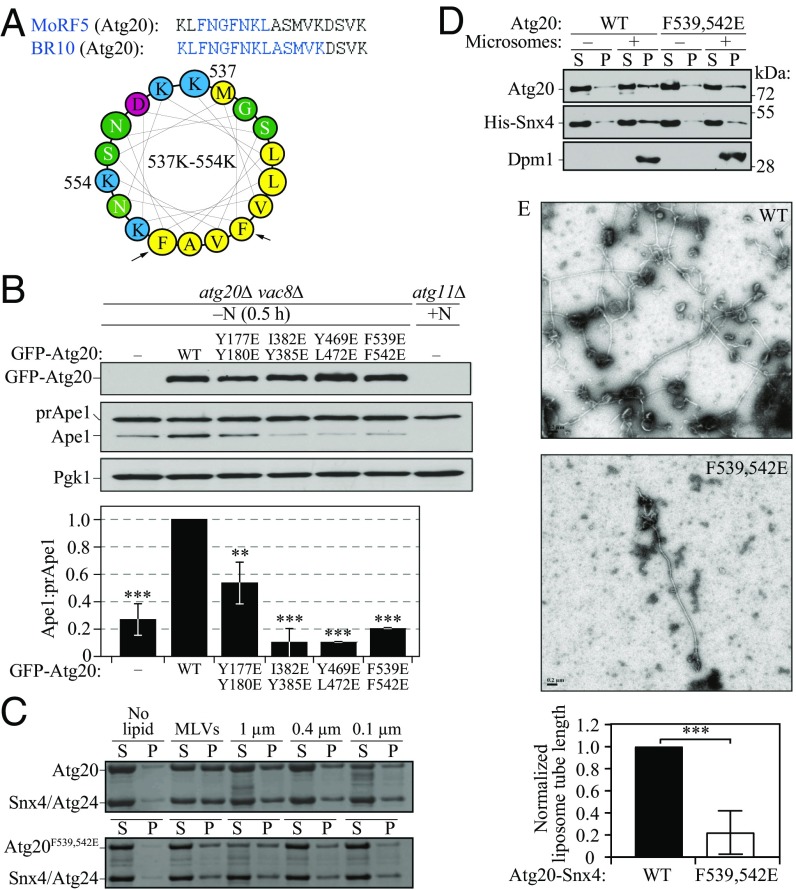
The membrane-induced AH in Atg20. (A) Helical wheel representation of the amino acid sequence in Atg20 that corresponds to MoRF5/BR10. Black arrows indicate the double mutation F539E, F542E. Yellow indicates hydrophobic amino acid residues; green, polar; red, negatively charged; blue, positively charged. (B) The vac8∆ atg20∆ (HPY079) cells transformed with the plasmids {pCuGFP(426), pCuGFP-Atg20(426), pCuGFP-Atg20[Y177E Y180/E](426), pCuGFP-Atg20[I382E Y385E](426), pCuGFP-Atg20[Y469E L472E](426), and pCuGFP-Atg20[F539E F542E](426)} were cultured in rich selective medium and then nitrogen-starved for 0.5 h. Error bars represent SD from three independent experiments. (C) Liposome sedimentation assay for the recombinant Atg20-Snx4 heterodimer, in which Atg20 was either the WT or F539E F542E mutant, with Folch liposomes of varying diameter. (D) In vitro reconstitution of yeast microsomes, isolated from SEY6210 atg20Δ snx4Δ cells, with the recombinant purified heterodimer Atg20-Snx4, in which Atg20 was either WT or mutant including the F539E F542E mutation. Supernatant (S) and pellet (P) were obtained by ultracentrifugation and analyzed by western blot analysis. (E, Upper) Representative negative stain EM images of 1.0-μm vesicles incubated with WT Atg20-Snx4 or mutant Atg20-Snx4 [F539E F542E] heterodimer. (Scale bars: 0.2 µm.) (E, Lower) Quantification of lipid tube length from grid squares, which are 484 µm2. Data from each trial were normalized to the tube length from WT. Error bars represent the SD from three independent experiments. Statistical significance was tested using the unpaired two-tailed Student’s t test: **P < 0.01; ***P < 0.005.
A typical test for the functionality of a foldable element that forms a membrane-induced AH is to replace one or two residues on its hydrophobic face—which normally becomes buried on AH insertion into the lipid bilayer—with negatively charged residue(s). We hypothesized that if disordered MoRF5/BR10 folds into a functional membrane-sensing AH in vivo, then a mutagenic disruption in its hydrophobic face should create a dysfunctional Atg20 protein. In contrast, such a mutation should have no significant effect if MoRF5/BR10 does not fold in vivo, and would retain mutagenically added negatively charged residues exposed to a hydrophilic environment.
To test our hypothesis, we mutated Phe539 and Phe542 simultaneously to glutamic acid. For comparison, we produced three double mutants as negative controls, including one in the PX domain (Y177E Y180E) and two in the BAR domain (I382E Y385E and Y469E L472E) of Atg20 (Fig. 5 D and E), reasoning that disruption of the PX or BAR domain would yield functionally defective proteins, because a structural defect induced by a mutation in one of these membrane-binding modules might prevent proper binding of the protein to the lipid membrane. The prApe1 processing assay (SI Appendix, Fig. S15A) showed that the Atg20 protein mutated in MoRF5/BR10 (F539E F542E) was significantly defective in the Cvt pathway, as were the mutants disrupting the PX and BAR domains.
To probe the importance of the putative AH in facilitating induction of autophagy by Atg20 (Fig. 2), we carried out the prApe1 processing assay in starved atg20∆ vac8∆ cells overexpressing plasmid-driven WT Atg20 or the PX, BAR, or AH mutant. Again, all these mutants were largely dysfunctional relative to the WT (Fig. 6B).
We used co-IP to test whether this dysfunction of the Atg20F539,542E mutant in the Cvt pathway (SI Appendix, Fig. S15A) and autophagy induction (Fig. 6B) are due to a weakened or lost interaction with Snx4. Our findings show that neither the Atg20F539,542E mutant nor the negative control (Atg20Y177,180E) was defective in binding to Snx4 (SI Appendix, Fig. S15B), suggesting that the Snx4 binding site does not overlap with the putative AH on Atg20, and can be narrowed down to the 546–625 region (between MoRF5 and the 626NLExW motif).
To strengthen our bioinformatics analysis and in vivo experiments suggesting the existence of the membrane-inducible AH in Atg20, we conducted liposome sedimentation assays with the recombinant Atg20-Snx4 or Atg20F539,F542E-Snx4 heterodimer against a range of Folch liposome sizes, including multilamellar vesicles and unilamellar vesicles with diameters of 1,000, 400, and 100 nm. Recombinant protein and liposomes were mixed, incubated for 30 min at room temperature, and subjected to centrifugation. The recombinant heterodimers did not pellet in the absence of lipid; therefore, the presence of protein in the pellet fractions is a result of lipid binding. The Atg20-Snx4 heterodimer bound preferentially to larger vesicles, whereas the heterodimer mutated in the Atg20 AH exhibited lower lipid binding than the WT, as manifested by a weaker band in the pellet fraction (Fig. 6C).
To further confirm a membrane-binding defect of the Atg20F539,542E mutant relative to the WT, we also carried out in vitro reconstitution of yeast microsomes, isolated from atg20Δ snx4Δ cells, with the purified full-length Atg20-Snx4 heterodimer (Fig. 6D and SI Appendix, Fig. S16). western blot analysis qualitatively showed that the Atg20-Snx4 heterodimer harboring the mutation in the Atg20 AH failed to efficiently bind to yeast membranes (i.e., with a physiological composition of lipids) compared with the WT heterodimer.
Finally, we examined the effect of the AH mutation on remodeling of membranes. In many BAR proteins, AHs are important for membrane remodeling, specifically for tubulation (11). Protein and vesicles were mixed, incubated for 1 h, and applied to EM grids. The length of membrane tubes on the grids was measured and normalized to the liposome tube length measured from the WT Atg20-Snx4 sample. Our negative-stain EM images of membrane tubulation by the Atg20-Snx4 or Atg20F539,542E-Snx4 heterodimer showed that the mutant was far less efficient in membrane remodeling than the WT (Fig. 6E).
Taken together, our data in Fig. 6 and SI Appendix, Figs. S12–S15 reveal that Atg20 encompasses a membrane-inducible AH that arises from a disordered region and is localized within the protein at an as-yet unknown position among sorting nexins, the BAR-GAP domain. Along with the PX and BAR domains, this AH in Atg20 is critical for protein function in the Cvt pathway and nonselective autophagy induction.
Discussion
Here we show that Atg20 is a dynamic protein with a hybrid architecture that combines structurally stable and dynamic domains to facilitate the efficiency of nonselective and selective autophagy (6), (Fig. 1). This hybrid conformation is modulated by phosphorylation and acetylation on numerous serines or threonines, as well as lysines. These residues form substrate motifs for mostly unknown kinases and acetyltransferases. The exception is the known substrate motif 42GVGSPKK and 46PKKSPKK of the Cdc28/Cdk1 protein kinase, which phosphorylates Ser45 and Ser49 (31), and the 362KSFTNIE motif of CK2, which phosphorylates Ser363 and Thr365 (32). The multiplicity of phosphorylation and acetylation observed in Atg20 is not unusual, and has been found in other proteins (43, 44) targeted by multisite modifications. A proposed reason for the multiplicity of modifications is a robust response to above-threshold stimulatory signals (45). This would indicate that Atg20 might function as a sensor mediating a response to signals aimed at the Atg11-Atg20-Snx4 trimer.
Although the exact purpose and interplay of phosphorylated sites in Atg20 are difficult to elucidate, several aspects can be proposed or eliminated. First, an earlier study showed that phosphorylation can induce or stabilize α-helices in cases where p-Ser or p-Thr at position i in the sequence are preceded or followed by lysines at positions i + 4 or i − 4, respectively (46). Since no p-Ser or p-Thr in Atg20 is preceded/followed by a lysine at positions i + 4 or i − 4, we can exclude the possibility that Atg20 is phosphorylated to induce/stabilize an α helix via this mechanism. Second, another study showed that p-Ser and p-Thr at N-terminal helix positions stabilize those α-helices (47). Phosphorylation on S363 and T365 in Atg20 maps on the N terminus of the BAR domain consensus sequence; thus, these residues could be phosphorylated to stabilize the N-terminal α-helical rod in the BAR domain of Atg20. Finally, phosphorylation and acetylation change the net charge in proteins; phosphate groups add a negative charge, whereas acetyl groups neutralize existing positive charges on lysines. Accordingly, these two modifications on Atg20 can modulate its overall charge and thereby a degree of compaction, which increases with decreased net charge (48). Therefore, a possible purpose of acetylation of lysines in Atg20 could be to gain compaction, especially in the Atg20 PX and BAR domains. Because BAR domains bind negatively charged membranes electrostatically using lysine and arginine residues (39), acetylated lysines in Atg20 presumably are not the lysines involved in membrane binding.
Atg20 exists only in a heterodimer with Snx4, where both these sorting nexins adopt an elongated overall conformation, as revealed by our AUC and SAXS data (Fig. 5). The PX and BAR domains in Atg20 are two membrane-binding modules that function in remodeling and sensing of PtdIns3P-enriched membranes (6, 21, 49). The BAR domain of Atg20 is unique in that it is separated into two segments by an intrinsically disordered region, the BAR-GAP (Fig. 2A). Here we present bioinformatics and experimental evidence showing that a portion of the BAR-GAP folds into a functional, membrane-inducible AH (Fig. 6 and SI Appendix, Figs. S12–S15). This helix is analogous to AHs found in other BAR proteins (11, 40, 42). Our data reveal a unique position for this type of membrane-binding module occurring among sorting nexins.
Together, the membrane-binding modules of Atg20 (PX, BAR, and AH) have dual functions, operating both in the Cvt pathway and during the induction of nonselective autophagy (Fig. 7 and SI Appendix, Fig. S15). Based on this duality, it is tempting to speculate that a membrane source recruited by Atg20 to the Cvt vesicles is also used, at least in part, for the rapid formation of the phagophore. This would explain why autophagy induction is delayed in the absence of Atg20 (Fig. 1). It is also possible that the Atg20-Snx4 BAR heterodimer stabilizes a certain curvature of the growing phagophore, which is why it preferentially binds in vitro to larger vesicles (Fig. 6C).
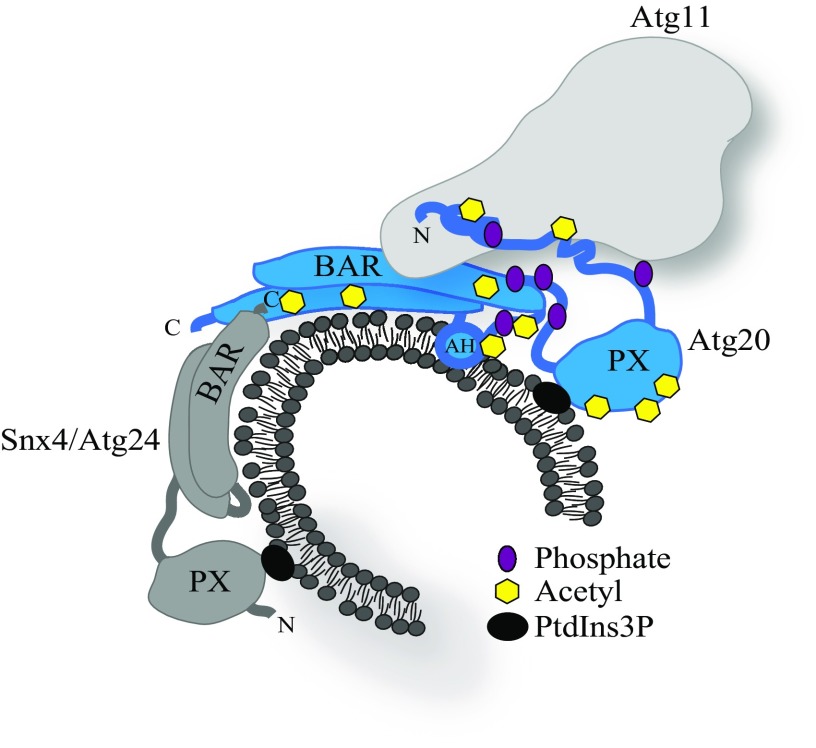
Fig. 7.
Proposed molecular mechanism of Atg20 function depicted by a schematic model of the Atg11-Atg20-Snx4 heterotrimer interacting with the PtdIns3P-enriched membrane in yeast S. cerevisiae. The PX domain of Atg20 acts as a lipid-selective module that interacts with PtdIns3P. The BAR domain and AH of Atg20 are membrane-sensing and -remodeling modules that detect and stabilize a membrane curvature. The lipid-binding modules are the main targets of acetylation, whereas phosphorylation is located predominantly in IDPRs of Atg20. Together, these two PTMs modulate the optimal Atg20 conformation that uses the disordered N terminus (FR domain) and the first segment of the BAR domain (region 380–480) to interact with Atg11. Fine-tuning of Atg20 architecture by PTMs is important for the Cvt pathway, in which Atg11 is a critical component. Snx4 forms the heterodimer with Atg20 via binding to its second segment of the BAR domain near the very C terminus. The three lipid-binding modules of Atg20 along with the PX and BAR domain of Snx4 recruit the PtdIns3P-enriched membranes. The rate of this process is critical for the efficient initiation of bulk autophagy.
The data presented in this study yield a model of the Atg11-Atg20-Snx4 heterotrimer interacting with a PtdIns3P-enriched membrane (Fig. 7). Snx4 dimerizes with Atg20 via binding to the Atg20 BAR domain near the very C terminus (in the region 546–625), in analogy to other BAR heterodimers (50), whereas Atg11 has two binding sites on Atg20, the BAR domain in the region 380–480 and the disordered N terminus. Each site is necessary for an efficient Atg11–Atg20 interaction. The overall function of Atg20 is fine-tuned by phosphorylation and acetylation, and uses membrane-binding modules (PX, BAR, and AH) to recruit PtdIns3P-enriched membranes into the forming Cvt vesicles and phagophore. Thus, a possible purpose of Atg20 in autophagy induction could be to enhance the efficiency of recruitment of PtdIns3P-enriched membranes and thereby speed buildup of the phagophore during autophagy initiation and/or stabilization of the curvature of the growing phagophore.
Our model of the Atg11-Atg20-Snx4 heterotrimer (Fig. 7) proposes a molecular mechanism of Atg20 function in budding yeast. At the same time, it raises questions in the field of mammalian autophagy of whether there is an analogous mechanism in human cells that facilitates autophagy induction via a membrane-binding protein and, if so, whether this analogous mechanism is embedded in the function of the fairly unexplored RB1CC1/FIP200 protein.
Materials and Methods
Strains, Media, and Growth Conditions.
Deletion strains created for this work were produced as described previously (51). Strains used in this study are listed in SI Appendix, Table S1. Yeast cells were grown in nutrient-rich medium [YPD; 1% (wt/vol) yeast extract, 2% (wt/vol) peptone, 2% (wt/vol) glucose] or synthetic minimal medium [SMD; 0.67% yeast nitrogen base, 2% (wt/vol) glucose, and auxotrophic amino acids and vitamins]. Autophagy was induced by shifting the cells to nitrogen starvation medium [SD-N; 0.17% nitrogen base without ammonium sulfate or amino acids and 2% (wt/vol) glucose].
Plasmids.
pCuPA-Atg20(424), pCuGFP-Atg20(426), pCu-PA-Snx4(424) (6), and pCuHA-Atg11(416) (14, 52) have been reported previously. Other plasmids were generated from pCuPA-Atg20(424) or pCuGFP-Atg20(426) by site-directed mutagenesis as described previously (53). To generate the plasmid pCuGFP-Atg20[Aroma], the DNA fragment of Atg20[Aroma] that was synthesized by GeneArt (Life Technologies) was inserted into the ClaI and BglII sites of pCuGFP-Atg20. All mutations were verified by DNA sequencing.
Precursor Ape1 Processing, GFP Processing, and Enzymatic Assays for Autophagy.
The prApe1 processing assay was carried out by culturing the cells from 0.05 OD600 units to 0.55 OD600 units in nutrient-rich medium. This allows for monitoring the Cvt pathway in mid-log phase cells that are actively transporting prApe1 to the vacuole. Cells (1 OD600 unit) were harvested, washed with 1 mL of H2O, and analyzed by western blot analysis. For the GFP processing assay, cells expressing the GFP-Atg8 construct under the CUP1 promoter on the plasmid (pRS426) were cultured from 0.25 OD600 units to 1.00 OD600 units in selective nutrient-rich medium, and then washed in 1 mL of H2O and shifted to SD-N medium for 1, 2, or 3 h. The harvested cells were washed with 1 mL of H2O and subjected to western blot analysis as described previously (54). For phosphatase assays, autophagy was induced by shifting the cells from nutrient-rich medium to SD-N medium. The harvested cells were washed in 0.85% NaCl and stored at −80 °C until the enzymatic assay.
Phosphatase Activity.
The phosphatase activity of Pho8Δ60 or mitoPho8Δ60 was assayed as described previously (15).
Immunoprecipitation.
Cells (50 OD600 units) were lysed in 4 mL of lysis buffer [1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), 0.2 M sorbitol, 1 mM MgCl2, 0.1% Tween-20, 1 mM PMSF inhibitor, Complete EDTA-Free Protease Inhibitor (Roche)] with glass beads. After centrifugation at 1,750 × g for 5 min, the resulting supernatant fraction was incubated with IgG Sepharose 6 Fast Flow (GE Healthcare) for 2 h at 4 °C. After five washes with lysis buffer, the bound proteins were eluted by incubating the Sepharose at 55 °C with SDS/PAGE buffer, followed by western blot analysis with the appropriate antibody.
GeLC-Mass Spectrometry.
PA affinity-purified Atg20 was resolved by SDS/PAGE, and the entire gel lane of the sample was divided into equal segments and excised. The gel bands were then subjected to in-gel tryptic digestion, followed by LC-MS/MS analysis with detection via collision-induced dissociation. This allowed for sequencing of the tryptic peptides while keeping the side chain modifications intact for straightforward identification and localization of the modification. The analysis was performed on an Orbitrap Velos instrument (Thermo Fisher Scientific) coupled to a Waters NanoAcquity liquid chromatography system. The proteins present in the samples were identified by database searches using the SWISS-PROT database as well as Mascot (www.matrixsciences.com) and X! Tandem (www.proteomesoftware.com) search engines. The results from the gel segments of each sample were combined and displayed using Scaffold software (www.proteomesoftware.com). PTMs of interest were specified in the database searches and displayed in the Scaffold file.
Overexpression, Purification, and Characterization of the Recombinant Atg20[FR].
S. cerevisiae Atg20 comprising residues 1–160 was subcloned into pHis2 to generate Atg20[FR] pHis2. Escherichia coli BL21 (DE3) Star cells (Invitrogen), transformed with Atg20[FR] pHis2, were grown in LB medium at 37 °C to an OD600 of 0.6, induced with 1 mM isopropyl-beta-d-thiogalactopyranoside, and grown for 18 h at 18 °C. Cells were pelleted and stored at −80 °C. Cell pellets were thawed and resuspended in 50 mM Tris, pH 8.0, 500 mM NaCl, 0.1% Triton X-100 containing 0.1 mM 4-(2-aminoethyl)benzenesulfonyl and 1 µg/mL leupeptin. Cells were lysed with three passes through a French press (Thermo Electron). Lysates were cleared by centrifugation and added to TALON resin (Clontech). The resin was washed with 50 mM Tris, pH 8.0, 500 mM NaCl, 2.5 mM imidazole, and protein was eluted using 50 mM Tris, pH 8.0, 500 mM NaCl, 200 mM imidazole.
Elutions were subjected to ion exchange chromatography using a HiTrap Q column (GE Healthcare) and eluted with a gradient from 50 mM NaCl to 1 M NaCl. Fractions containing protein were pooled and further purified using a HiLoad 16/600 Superdex 75-pg column (GE Healthcare) equilibrated in 20 mM sodium phosphate, pH 6.5, 100 mM NaCl, 0.2 mM Tris(2-carboxyethyl)phosphine (TCEP). For circular dichroism, protein was concentrated to 25 µM. Spectra were acquired with a 0.1-cm cuvette from 198 to 250 nm at 20 °C using a Jasco J-185 spectrometer. For NMR spectroscopy, protein was concentrated to 324 µM, D2O was added to a final concentration of 10%, and 1D 1H spectra were recorded on a Bruker UltraShield Plus 600-MHz magnet.
Overexpression and Purification of the Recombinant Full-Length Atg20-Snx4 or Atg20F539E, F542E-Snx4 Heterodimer.
Full-length S. cerevisiae Snx4 and Snx4 comprising residues 21–423 were each subcloned into the pET His6 tobacco etch virus (TEV) ligation-independent cloning vector (1B), a gift from Scott Gradia (29653; Addgene), to generate Snx4 1B and Snx421–423 1B. His6-TEV-Snx4 from Snx4 1B and full-length S. cerevisiae Atg20 were subcloned into the polycistronic vector pST39 to generate Atg20-Snx4 pST39 (55). Atg20F539E, F542E-Snx4 pST39 was generated from Atg20-Snx4 pST39 using QuikChange mutagenesis. His6-TEV-Snx421–423 from Snx421–423 1B and S. cerevisiae Atg20 comprising residues 156–640 were subcloned into pST39 to generate Atg20156–640-Snx421–423 pST39. E. coli BL21 (DE3) Star cells, transformed with the appropriate plasmid, were grown in LB, and protein was expressed using the same protocol as for Atg20[FR] pHis2. Cell pellets were thawed and resuspended in 50 mM Tris, pH 8.0, 500 mM NaCl, 0.1% Triton X-100 containing one Mini Complete EDTA-Free Protease Inhibitor tablet (Roche). Cells were lysed with three passes through a French press (Thermo Electron). Lysates were cleared by centrifugation and added to TALON resin (Clontech). Protein was eluted using 50 mM Tris, pH 8.0, 500 mM NaCl, 200 mM imidazole.
For Atg20-Snx4 and Atg20F539E, F542E-Snx4, elutions were further purified using a Hiload 16/60 Superdex 200 prep grade column (GE Healthcare) equilibrated in 20 mM Tris, pH 8.0, 200 mM NaCl, 0.2 mM TCEP. For Atg20156–640-Snx421–423, TEV protease was added to the elutions, the protein was cleaved overnight at 4 °C and subjected to ion exchange chromatography using a HiTrap SP column (GE Healthcare). Fractions containing Atg20156–640-Snx421–423 were further purified using a Hiload 16/60 Superdex 200 prep grade column (GE Healthcare) equilibrated in 20 mM Tris, pH 8.0, 200 mM NaCl, 0.2 mM TCEP.
Liposome Sedimentation Assay.
Folch fraction type I lipids isolated from bovine brain (Sigma-Aldrich) were dried under a nitrogen stream for 1 h and then dried overnight in a vacuum oven. Dried lipids were resuspended in 20 mM, Tris pH 8.0, 200 mM NaCl, 0.2 mM TCEP to a final concentration of 2.5 mg/mL. Lipids were subjected to freeze-thaw using dry ice and then extruded using an Avanti Mini Extruder with the appropriate size membrane. Then 25 µL of 3 µM purified Atg20-Snx4 or Atg20F539E, F542E-Snx4 was mixed with 25 µL of 2.5 mg/mL liposomes. The mixture was incubated at room temperature for 30 min, followed by centrifugation at 45,000 rpm for 50 min at 4 °C using a TLA45 rotor.
Negative Stain Electron Microscopy of Membrane Tubulation.
For the tubulation assay, 25 µL of protein (WT and F539,542E), at a concentration of 6 µM, was incubated with 1 µm extruded liposomes of Folch lipid for 60 min at room temperature. The sample was then applied to carbon-coated 400-mesh Cu/Rh grids for 1 min, followed by two washings and then staining with 2% uranyl acetate for 30 s. Images were obtained on a T12 transmission electron microscope (FEI) using low-dose conditions at 120 kV with a LaB6 filament. Images were recorded using a Gatan 2k × 2k CCD camera. Tubulation was quantified by imaging at least four grid squares (484 µm2/grid square) for each sample, WT and F539,542E. Samples were normalized to the total WT tube length produced.
AUC.
Atg20156–640-Snx421–423 (50 µM) was subjected to sedimentation velocity AUC using a Beckman Coulter ProteomeLab XL-A. Centrifugation was performed at 30,000 rpm for 22 h at 20 °C using an AN 60 Ti rotor. SEDNTERP (computer program written by D. B. Hayes, J. P. Philo, and T. M. Laue, 1994) was used to determine density, viscosity, and specific volume. SEDFIT was used to analyze raw AUC data and calculate sedimentation coefficient and frictional coefficient.
Dynamic Light Scattering.
Dynamic light scattering was performed on protein at 1 µM or liposomes at 2.5 mg/mL at 20 °C using DynaPro NanoStar (Wyatt Technology). Light scattering data were analyzed using Dynamics v7.1.8 (Wyatt Technology).
In-Line Size Exclusion Chromatography with SAXS.
In-line size exclusion chromatography (SEC) with SAXS was performed at National Synchrotron Light Source II beamline 16-ID. Atg20156–640-Snx421–423 (3.5 mg/mL) was injected onto a Superdex 200 Increase 5/150 GL column (GE Healthcare) equilibrated with 20 mM Tris, pH 8.0, 200 mM NaCl, 0.2 mM TCEP. PRIMUS was used to plot scattering data and determine the radius of gyration (Rg) using the Guinier approximation, I(q) = I(0)exp(−q2Rg2/3), where a plot of I(q) and q2 is linear for q < 1.3/Rg (56). The linearity of the Guinier plot was used to confirm that no aggregation was present in the sample. GNOM was used to determine the pair distribution function, P(r), and maximum particle dimension, Dmax (57). A starting model of Atg20156–640-Snx421–423 for MD was generated using SWISS-MODEL. BilboMD was used to generate 9,500 different conformations of Atg20156–640-Snx421–423, with the PX and BAR domains defined as rigid bodies, while regions between the PX and BAR domains (37) were defined as flexible.
Microsome Isolation and in Vitro Binding Experiment.
To isolate microsomes, atg20Δ snx4Δ SEY6210 cells (25 OD600 units) were resuspended in resuspension buffer [50 mM Tris, pH 7.5, 1 mM EDTA, 1 mM PMSF, 1 mM β−mercaptoethanol, and Complete EDTA-Free Protease Inhibitor (Roche)], mixed with glass beads, and vortexed four times for 1 min with intervals on ice between vortexing. The lysate was centrifuged at 5,900 × g for 10 min at 4 °C. Supernatant was split into two ultracentrifuge tubes and ultracentrifuged at 55,000 rpm by using a TLA 100.4 rotor (Beckman) for 15 min at 4 °C. The pellet was washed in resuspension buffer and ultracentrifuged again at 55,000 rpm for 15 min at 4 °C.
For in vitro reconstitution of yeast microsomes with the recombinant purified Atg20-Snx4 heterodimer, tubes without or with a microsomal pellet (from cells at 12.5 OD600 units) were incubated for 15 min at room temperature with the purified heterodimer (2.5 μM) in the presence of buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 2.5 mM PMSF, 200 mM MgSO4). Reconstitution samples were ultracentrifuged at 55,000 rpm for 15 min at 4 °C. Supernatant and pellet were analyzed by western blot analysis using rabbit anti-Atg20 polyclonal antibody to detect Atg20 and mouse anti-polyhistidine monoclonal antibody (H1029; Sigma-Aldrich) to detect the His-tag at the N terminus of Snx4. Dpm1 was detected by mouse anti-Dpm1 monoclonal antibody (A6429; Life Technologies).
Supplementary Material
Acknowledgments
We thank Maria Pellegrini for NMR assistance. This work was supported by National Institutes of Health Grant GM053396 (to D.J.K.) and the Protein Folding Diseases FastForward Initiative, University of Michigan. M.J.R. is supported by National Institutes of Health Grant GM113132. J.E.H. is supported by National Institute of Diabetes and Digestive and Kidney Diseases Intramural Research Program. SAXS data were collected at the life science X-ray scattering (LiX) beamline at National Synchrotron Light Source II (NSLSII). LiX operates under Department of Energy (DOE) Biological and Environmental Research Contract DE-SC0012704 and is supported by National Institutes of Health–National Institute of General Medical Sciences Grant P41GM111244. NSLSII is operated under DOE Basic Energy Sciences Contract DE-SC0012704.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708367114/-/DCSupplemental.
References
- 1.Klionsky DJ. The molecular machinery of autophagy: Unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanley RE, Ragusa MJ, Hurley JH. The beginning of the end: How scaffolds nucleate autophagosome biogenesis. Trends Cell Biol. 2014;24:73–81. doi: 10.1016/j.tcb.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Nice DC, Sato TK, Stromhaug PE, Emr SD, Klionsky DJ. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J Biol Chem. 2002;277:30198–30207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reggiori F, Klionsky DJ. Autophagic processes in yeast: Mechanism, machinery and regulation. Genetics. 2013;194:341–361. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leprince C, et al. Sorting nexin 4 and amphiphysin 2, a new partnership between endocytosis and intracellular trafficking. J Cell Sci. 2003;116:1937–1948. doi: 10.1242/jcs.00403. [DOI] [PubMed] [Google Scholar]
- 9.Steffan JS. Does Huntingtin play a role in selective macroautophagy? Cell Cycle. 2010;9:3401–3413. doi: 10.4161/cc.9.17.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Weering JRT, Verkade P, Cullen PJ. SNX-BAR proteins in phosphoinositide-mediated, tubular-based endosomal sorting. Semin Cell Dev Biol. 2010;21:371–380. doi: 10.1016/j.semcdb.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Weering JRT, et al. Molecular basis for SNX-BAR-mediated assembly of distinct endosomal sorting tubules. EMBO J. 2012;31:4466–4480. doi: 10.1038/emboj.2012.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Weering JRT, Cullen PJ. Membrane-associated cargo recycling by tubule-based endosomal sorting. Semin Cell Dev Biol. 2014;31:40–47. doi: 10.1016/j.semcdb.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 13.Mendl N, et al. Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2. J Cell Sci. 2011;124:1339–1350. doi: 10.1242/jcs.076406. [DOI] [PubMed] [Google Scholar]
- 14.Mao K, et al. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc Natl Acad Sci USA. 2013;110:E2875–E2884. doi: 10.1073/pnas.1300064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klionsky DJ. Monitoring autophagy in yeast: The Pho8Delta60 assay. Methods Mol Biol. 2007;390:363–371. doi: 10.1007/978-1-59745-466-7_24. [DOI] [PubMed] [Google Scholar]
- 16.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott SV, et al. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 18.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sickmeier M, et al. DisProt: The database of disordered proteins. Nucleic Acids Res. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozlowski LP, Bujnicki JM. MetaDisorder: A meta-server for the prediction of intrinsic disorder in proteins. BMC Bioinformatics. 2012;13:111. doi: 10.1186/1471-2105-13-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cullen PJ. Endosomal sorting and signalling: An emerging role for sorting nexins. Nat Rev Mol Cell Biol. 2008;9:574–582. doi: 10.1038/nrm2427. [DOI] [PubMed] [Google Scholar]
- 22.van der Lee R, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tompa P. Multisteric regulation by structural disorder in modular signaling proteins: An extension of the concept of allostery. Chem Rev. 2014;114:6715–6732. doi: 10.1021/cr4005082. [DOI] [PubMed] [Google Scholar]
- 24.Jakob U, Kriwacki R, Uversky VN. Conditionally and transiently disordered proteins: Awakening cryptic disorder to regulate protein function. Chem Rev. 2014;114:6779–6805. doi: 10.1021/cr400459c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uversky VN. Intrinsic disorder-based protein interactions and their modulators. Curr Pharm Des. 2013;19:4191–4213. doi: 10.2174/1381612811319230005. [DOI] [PubMed] [Google Scholar]
- 26.Disfani FM, et al. MoRFpred, a computational tool for sequence-based prediction and characterization of short disorder-to-order transitioning binding regions in proteins. Bioinformatics. 2012;28:i75–i83. doi: 10.1093/bioinformatics/bts209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mészáros B, Simon I, Dosztányi Z. Prediction of protein binding regions in disordered proteins. PLoS Comput Biol. 2009;5:e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dosztányi Z, Mészáros B, Simon I. ANCHOR: Web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009;25:2745–2746. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pejaver V, et al. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 2014;23:1077–1093. doi: 10.1002/pro.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iakoucheva LM, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ficarro SB, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 32.Albuquerque CP, et al. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma M, Burd CG, Chi RJ. Distinct complexes of yeast Snx4 family SNX-BARs mediate retrograde trafficking of Snc1 and Atg27. Traffic. 2017;18:134–144. doi: 10.1111/tra.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 35.Bordoli L, et al. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 36.Biasini M, et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small-angle X-ray scattering. Gen Physiol Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okuda M, Nishimura Y. Extended string binding mode of the phosphorylated transactivation domain of tumor suppressor p53. J Am Chem Soc. 2014;136:14143–14152. doi: 10.1021/ja506351f. [DOI] [PubMed] [Google Scholar]
- 39.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 40.Bhatia VK, et al. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. EMBO J. 2009;28:3303–3314. doi: 10.1038/emboj.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boucrot E, et al. Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell. 2012;149:124–136. doi: 10.1016/j.cell.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pylypenko O, Lundmark R, Rasmuson E, Carlsson SR, Rak A. The PX-BAR membrane-remodeling unit of sorting nexin 9. EMBO J. 2007;26:4788–4800. doi: 10.1038/sj.emboj.7601889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeh YY, et al. The identification and analysis of phosphorylation sites on the Atg1 protein kinase. Autophagy. 2011;7:716–726. doi: 10.4161/auto.7.7.15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papinski D, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 Kinase. Mol Cell. 2014;53:471–483, and erratum (2014) 53:515. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Theillet FX, et al. Physicochemical properties of cells and their effects on intrinsically disordered proteins (IDPs) Chem Rev. 2014;114:6661–6714. doi: 10.1021/cr400695p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Errington N, Doig AJ. A phosphoserine-lysine salt bridge within an alpha-helical peptide, the strongest alpha-helix side-chain interaction measured to date. Biochemistry. 2005;44:7553–7558. doi: 10.1021/bi050297j. [DOI] [PubMed] [Google Scholar]
- 47.Andrew CD, Warwicker J, Jones GR, Doig AJ. Effect of phosphorylation on alpha-helix stability as a function of position. Biochemistry. 2002;41:1897–1905. doi: 10.1021/bi0113216. [DOI] [PubMed] [Google Scholar]
- 48.Mao AH, Crick SL, Vitalis A, Chicoine CL, Pappu RV. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc Natl Acad Sci USA. 2010;107:8183–8188. doi: 10.1073/pnas.0911107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cullen PJ, Korswagen HC. Sorting nexins provide diversity for retromer-dependent trafficking events. Nat Cell Biol. 2011;14:29–37. doi: 10.1038/ncb2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Habermann B. The BAR-domain family of proteins: A case of bending and binding? EMBO Rep. 2004;5:250–255. doi: 10.1038/sj.embor.7400105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu H, Naismith JH. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guimaraes RS, Delorme-Axford E, Klionsky DJ, Reggiori F. Assays for the biochemical and ultrastructural measurement of selective and nonselective types of autophagy in the yeast Saccharomyces cerevisiae. Methods. 2015;75:141–150. doi: 10.1016/j.ymeth.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 55.Tan S. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr Purif. 2001;21:224–234. doi: 10.1006/prep.2000.1363. [DOI] [PubMed] [Google Scholar]
- 56.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: A windows PC-based system for small-angle scattering data analysis. J Appl Cryst. 2003;36:1277–1282. [Google Scholar]
- 57.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst. 1992;25:495–503. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.