Significance
Toll-like receptors (TLR) are involved in pathogen recognition and defense in organisms from fruit flies to humans. Recent genomic evidence suggests that TLRs and their downstream signaling components are present in more basal phyla. We characterize a TLR in a sea anemone model and demonstrate its ability to activate NF-κB signaling when exposed to a bacterial pathogen and a known human TLR activator. Moreover, this TLR has an early developmental role in anemones. We also identify a primitive sea anemone organ that expresses components of the TLR–to–NF-κB pathway. These results demonstrate that TLRs have ancient roles in NF-κB signal transduction, pathogen detection, and development, thus providing molecular insights into how simple marine invertebrates may respond to pathogens.
Keywords: TLR, pathogen, NF-κB, development, Cnidaria
Abstract
In organisms from insects to vertebrates, Toll-like receptors (TLRs) are primary pathogen detectors that activate downstream pathways, specifically those that direct expression of innate immune effector genes. TLRs also have roles in development in many species. The sea anemone Nematostella vectensis is a useful cnidarian model to study the origins of TLR signaling because its genome encodes a single TLR and homologs of many downstream signaling components, including the NF-κB pathway. We have characterized the single N. vectensis TLR (Nv-TLR) and demonstrated that it can activate canonical NF-κB signaling in human cells. Furthermore, we show that the intracellular Toll/IL-1 receptor (TIR) domain of Nv-TLR can interact with the human TLR adapter proteins MAL and MYD88. We demonstrate that the coral pathogen Vibrio coralliilyticus causes a rapidly lethal disease in N. vectensis and that heat-inactivated V. coralliilyticus and bacterial flagellin can activate a reconstituted Nv-TLR–to–NF-κB pathway in human cells. By immunostaining of anemones, we show that Nv-TLR is expressed in a subset of cnidocytes and that many of these Nv-TLR–expressing cells also express Nv-NF-κB. Additionally, the nematosome, which is a Nematostella-specific multicellular structure, expresses Nv-TLR and many innate immune pathway homologs and can engulf V. coralliilyticus. Morpholino knockdown indicates that Nv-TLR also has an essential role during early embryonic development. Our characterization of this primitive TLR and identification of a bacterial pathogen for N. vectensis reveal ancient TLR functions and provide a model for studying the molecular basis of cnidarian disease and immunity.
Toll-like receptors (TLRs) are one of the primary mediators of the innate immune system in animals from insects to humans, and TLRs also have roles in development in several organisms (1). Innate immunity is the most evolutionarily ancient branch of the immune system and is responsible for the immediate recognition of and response to pathogens in many organisms. TLRs recognize specific microbial pathogen molecules and initiate intracellular signaling cascades to activate a variety of transcription factors, including NF-κB, which then direct the expression of key innate immune effector genes (2–4). Humans have 10 TLRs that recognize distinct pathogen ligands. The physiological responses elicited by these TLR-directed pathways are inflammation, opsonization, cell-based phagocytosis, and antimicrobial peptide production to ensure rapid pathogen clearance (3–6). In addition to the TLR pathways, mammals have two other innate immune pathways that can activate NF-κB for antiviral responses, the RIG–I/MAV and cGAS–STING pathways (7–9).
Drosophila melanogaster has nine TLRs, which activate pathways for immunity, development, and other cell functions at various life stages of the fly (10–12). The Drosophila TLR–to–NF-κB pathway is induced by fungi and Gram-positive bacteria to initiate innate immune responses, but this pathway also has an essential role in the establishment of early embryonic dorsal–ventral polarity (5, 10, 11, 13). Several Drosophila TLRs are also important for maintaining healthy tissues by inducing NF-κB–dependent apoptosis of unfit or mutant cells (12).
TLRs are transmembrane proteins with two primary domains that are required for initiating downstream signaling pathways for biological responses: (i) an ectodomain that contains a leucine-rich region (LRR) that recognizes ligands such as pathogen-associated molecular patterns (PAMPs) or endogenous ligands (e.g., Spätzle in Drosophila), and (ii) an intracellular Toll/IL-1 receptor (TIR) domain that mediates protein–protein interactions with other TIR domain-containing proteins at the plasma membrane for downstream activation of NF-κB (3–5). Although ligand engagement of the TLR often activates the NF-κB pathway, it can also activate MAP kinase and IFN pathways (2, 3).
TLRs have been subclassified by the number of cysteine clusters present in their ectodomains. Single cysteine cluster TLRs (sccTLRs), found in mammals and Drosophila Toll-9, have ectodomains with one cysteine cluster at the C-terminal end of the LRR. Multiple cysteine cluster TLRs (mccTLRs) have two or more cysteine clusters within the LRR ectodomain; mccTLRs are found in Drosophila, Caenorhabditis elegans, and some basal phyla (1, 14). Activation of mammalian sccTLRs occurs by direct engagement of the LRR by a PAMP, which causes the intracellular TIR domain to interact with adapter proteins MAL and/or MYD88, facilitating downstream activation of NF-κB (3–5, 14). In contrast, the best-characterized mccTLR, Drosophila Toll-1, does not engage PAMPs directly; instead, its LRR binds to a cleaved form of the endogenous ligand Spätzle, subsequently initiating intracellular NF-κB signaling (5, 10, 11, 14).
Genomic and transcriptomic analyses of evolutionarily primitive organisms suggest that the TLR–to–NF-κB pathway is more ancient than previously accepted (15–18). Genes encoding TLR-like proteins have been identified in the phyla Cnidaria (15, 17, 19, 20) and Porifera (21); however, many of these predicted basal TLR-like proteins lack the LRR ectodomain and consist of only a transmembrane region and a TIR domain (15, 19). To date, few studies have characterized the biological processes and downstream pathways regulated by TLR-like proteins in cnidarians or other basal phyla (15, 22).
The phylum Cnidaria is a morphologically primitive outgroup to bilaterians and comprises ∼10,000 aquatic organisms, including corals, hydras, sea anemones, and jellyfish (23). Nevertheless, many cnidarian genomes are complex and exhibit structural organization and gene-coding properties that are surprisingly similar to those in humans (23, 24). Cnidarians, especially corals, have recently attracted much research interest due to the impacts of climate change and pathogen outbreaks on marine ecosystems (25). However, little is known about how these organisms defend themselves against biological and environmental stressors at the molecular level.
One bacterial pathogen that impacts several cnidarian species is the Gram-negative bacterium Vibrio coralliilyticus. Initially described as the causative agent of white syndrome disease in corals, V. coralliilyticus has recently been shown also to cause disease in the tropical sea anemone Aiptasia pallida (26–28). Although not pathogenic at water temperatures below 24 °C, V. coralliilyticus photoinactivates coral symbionts between temperatures 24 °C and 26.5 °C (27, 29). Above 27 °C, V. coralliilyticus is highly pathogenic due to its ability to lyse coral and anemone tissues (27, 28, 30). As global ocean temperatures rise, the temperature-dependent pathogenicity of V. coralliilyticus threatens many coral species.
The starlet sea anemone Nematostella vectensis is an important model cnidarian, which is particularly intriguing because of its general resilience to pathogens and wide fluctuations in salinity, temperature, and pH (31). In fact, no microbial pathogens have been identified for N. vectensis. We previously characterized the N. vectensis NF-κB protein (Nv-NF-κB) and its proximal regulators (e.g., Nv-IκB, Nv-IKK) (32–34); however, little is known about the upstream regulation of NF-κB in cnidarians or organisms basal to arthropods (15, 22).
Herein, we characterize the single N. vectensis TLR homolog (Nv-TLR) as a mccTLR with multiple activities. We also show that V. coralliilyticus is a rapidly lethal pathogen for N. vectensis and that this bacterium and purified flagellin can activate a reconstituted Nv-TLR–to–NF-κB pathway in human cells. Nv-TLR also has a role in early development, which we hypothesize functions in an NF-κB–independent manner. Finally, we provide evidence that suggests that the nematosome, a highly motile collection of cells, plays a role in N. vectensis innate immunity. This research provides insight into the ancient roles of TLRs in pathogen detection, NF-κB signal transduction, and development, suggesting a single origin for the diverse TLRs found in insects and mammals.
Results
Cloning and Expression of the Sole N. vectensis TLR.
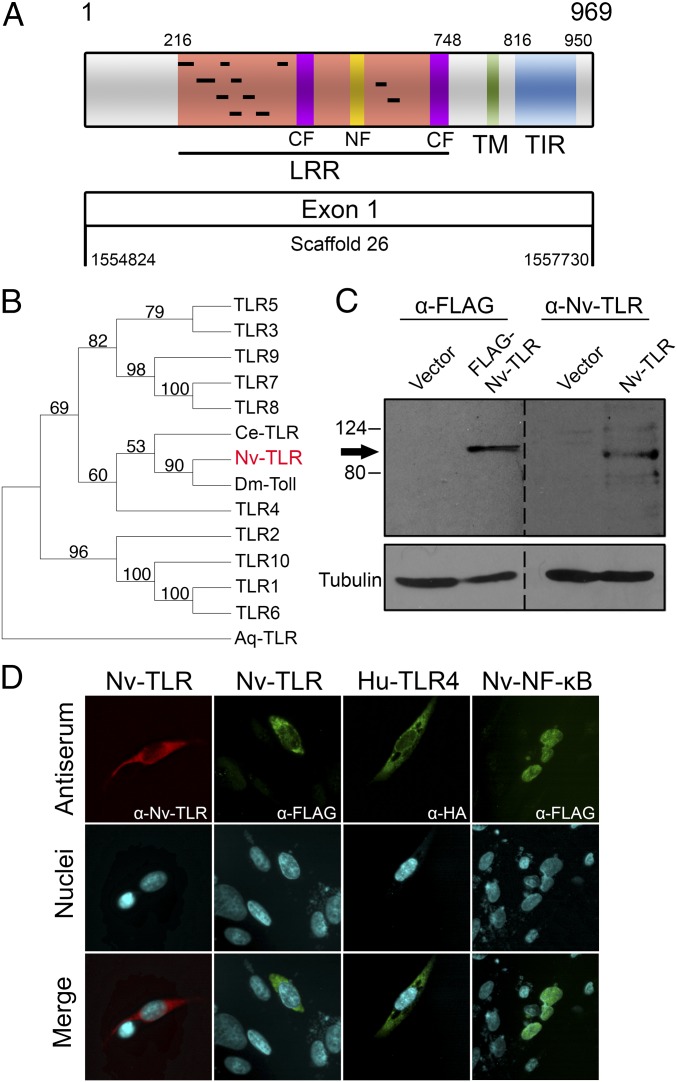
In a search of the N. vectensis genome, transcriptome, and EST databases for putative TLRs (23, 35), we identified a single TLR-like gene, which is consistent with previous bioinformatic studies mining the N. vectensis genome for TIR domain-containing proteins (17). The predicted Nv-TLR is encoded by a genomic locus that lacks introns within its coding region (Fig. 1A). We identified mRNAs corresponding to this locus in the N. vectensis transcriptome at the Joint Genome Institute (23) and in RNA-sequencing (RNA-seq) data of N. vectensis embryos (36), demonstrating that the Nv-TLR locus is expressed in anemones starting early in development. The 969-amino acid Nv-TLR is a mccTLR (14) with an ectodomain containing three cysteine clusters (two CF motifs and an NF motif), a predicted hydrophobic membrane-spanning region, and a C-terminal TIR domain (Fig. 1A). Table S1 presents the sequences of the LRR motifs, the NF and CF motifs, and the predicted transmembrane region of Nv-TLR. Phylogenetic comparison of the Nv-TLR TIR domain and the TIR domains of the 10 human TLRs, C. elegans TOL-1, and D. melanogaster Toll indicated that Nv-TLR is most similar to D. melanogaster Toll and C. elegans TOL-1 (Fig. 1B). Interestingly, the TIR domain of human TLR4 (Hu-TLR4) clusters with the TIR domains of Nv-TLR, TOL-1, and Drosophila Toll rather than with any other human TLR (Fig. 1B).
Fig. 1.
Cloning and expression of the N. vectensis TLR. (A) Nv-TLR is a 969-amino acid protein with a LRR (orange), an N-terminal LRR cysteine cluster (NF motif; yellow), two C-terminal LRR cysteine clusters (CF motifs; purple), a putative transmembrane domain (TM; green), and a TIR domain (blue). The black bars in the ectodomain indicate leucine-rich repeat motifs. See Table S1 for specific motif and domain amino acid coordinates. Shown below the protein structure are the Nv-TLR single exon structure and genomic coordinates. (B) Phylogenetic analysis of the TIR domains of all human TLRs and the TIR domains of Nv-TLR, the C. elegans TOL-1, the D. melanogaster Toll, and a TIR domain-containing protein of the sponge A. queenslandica using neighbor-joining analysis. Phylogeny was rooted with the TIR domain protein of A. queenslandica. Branches indicate bootstrap support values. (C) Western blot of 293T cells transfected with expression plasmids for vector alone, FLAG-Nv-TLR, and Nv-TLR. Membranes were probed with anti-FLAG (Left), anti–Nv-TLR (Right), or anti–β-tubulin loading control (Bottom). A dotted line indicates where the membrane was cut before Western blotting. The arrow indicates Nv-TLR. Molecular mass markers (in kDa) are indicated. (D) DF-1 chicken fibroblasts were transfected with expression plasmids for FLAG-Nv-TLR, HA-Hu-TLR4, or FLAG-Nv-NF-κB. (Top) Indirect immunofluorescence was then performed with Nv-TLR, FLAG, or HA antiserum. The antibody used for immunofluorescence in each panel is indicated. (Middle) Nuclei were stained with DAPI. (Bottom) Merged images of antibody and DAPI staining.
Initial attempts to express the native Nv-TLR cDNA in human cells were largely unsuccessful. Therefore, we subjected the Nv-TLR cDNA to human cell codon optimization (Dataset S1), and the codon-optimized Nv-TLR cDNA was subcloned into pcDNA-FLAG and non-FLAG vectors for expression in human HEK 293 cells. Western blotting of transfected cell extracts with Nv-TLR antiserum detected a band of ∼100 kDa that, as expected, migrated just below the FLAG-tagged Nv-TLR protein, which was detected by FLAG antiserum. In cells transfected with empty vector controls, no immunoreactive proteins were detected with either FLAG or Nv-TLR antiserum (Fig. 1C). Indirect immunofluorescence of transfected chicken DF-1 fibroblasts demonstrated that FLAG-Nv-TLR, detected with both FLAG and Nv-TLR antisera, showed a staining pattern outside the nucleus similar to that of HA-Hu-TLR4 (Fig. 1D). In contrast, FLAG-Nv-NF-κB showed exclusively nuclear localization in DF-1 cells, consistent with our previously published findings (32).
The Nv-TLR TIR Domain Is Required for Activation of Canonical NF-κB Signaling in Human Cells.
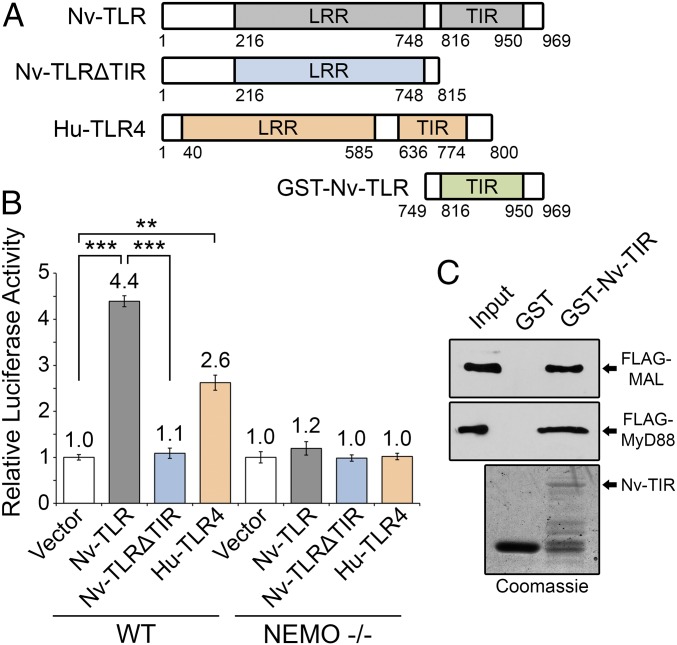
HEK 293 cells do not express human TLRs but do express all downstream NF-κB signaling pathway components, making them a useful cell line to study the ability of exogenously expressed TLRs to activate NF-κB. Hu-TLR4 has been shown to activate NF-κB when overexpressed in 293 cells, even in the absence of a stimulus (37). Under nonstimulated conditions, we found that expression of Nv-TLR in 293 cells increased the expression of an NF-κB–site luciferase reporter by 4.4-fold compared with vector control cells (Fig. 2 A and B). The level of NF-κB–site reporter gene activation by Nv-TLR was even higher than that seen with Hu-TLR4 (2.6-fold). Nv-TLRΔTIR, which lacks the TIR domain (Fig. 2A), did not induce NF-κB–site reporter gene expression above empty vector control levels (Fig. 2B).
Fig. 2.
Nv-TLR can activate NF-κB signaling in human cells through the canonical pathway. (A) Schematic indicating structures of Nv-TLR, Nv-TLRΔTIR, Hu-TLR4, and GST-Nv-TIR. (B) An NF-κB–site luciferase reporter assay was performed in wild-type 293 cells (Left) and NEMO −/− 293T cells (Right). Cells were transfected with human coreceptors CD-14 and MD-2 and with Nv-TLR, Nv-TLRΔTIR, Hu-TLR4, or the empty vector alone. Luciferase values are averages of three experiments done in triplicate and are normalized to the vector control (1.0) ± SEM. Statistical significance was determined using Student’s t test; **P < 0.01, ***P < 0.001. (C, Upper) Bacterially expressed GST or GST-Nv-TIR proteins were purified and incubated with lysates from 293T cells expressing FLAG-MAL or FLAG-MYD88. Pulldown fractions were analyzed by anti-FLAG Western blotting. The input lane contains 1% of the 293T cell lysate used in the pulldown. (Lower) An SDS-polyacrylamide gel stained with Coomassie Blue showing expression of GST and GST-Nv-TIR.
Activation of NF-κB by Hu-TLR4 proceeds through the canonical NF-κB pathway and requires the IKK scaffold protein NEMO (4). Neither Nv-TLR nor Hu-TLR4 activated the NF-κB–site luciferase reporter in 293 cells in which NEMO expression was genetically ablated (Fig. 2B).
As an early step in the activation of NF-κB, the intracellular TIR domain of Hu-TLR4 interacts with the adapter proteins MAL and MYD88 at the plasma membrane (4). To determine whether the TIR domain of Nv-TLR can also interact with these human adapter proteins, we performed a pulldown assay using bacterially expressed GST-Nv-TIR (Fig. 2A) and lysates from 293 cells expressing FLAG-MAL or FLAG-MYD88. Both FLAG-MAL and FLAG-MYD88 were detected by Western blotting of pulldown fractions using GST-Nv-TIR but not by GST alone (Fig. 2C). Expression of GST and GST-Nv-TIR was confirmed by SDS/PAGE followed by Coomassie blue staining (Fig. 2C).
The requirement of the TIR domain and NEMO for Nv-TLR–induced activation of NF-κB in 293 cells (Fig. 2B) and the ability of Nv-TIR to interact with the human MAL and MYD88 adapter proteins (Fig. 2C) indicate that Nv-TLR, like Hu-TLR4, activates NF-κB in human cells through TIR-dependent canonical NF-κB signaling.
A Reconstituted Nv-TLR–to–NF-κB Pathway Can Be Activated by V. coralliilyticus, an N. vectensis Pathogen, and by Flagellin.
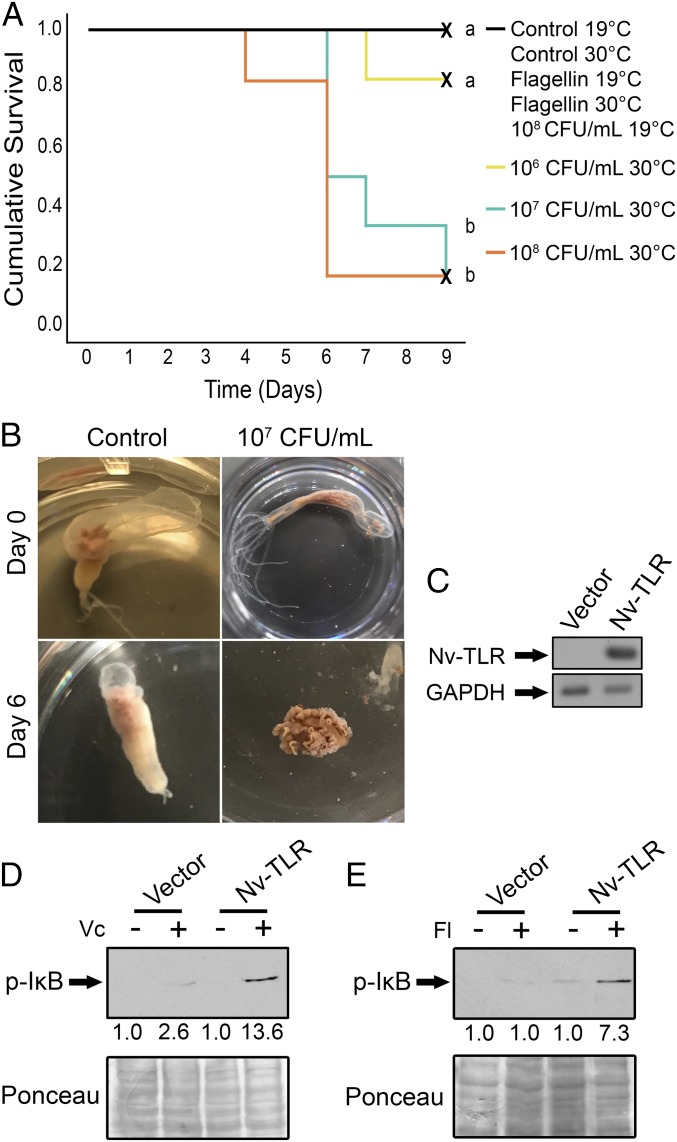
V. coralliilyticus is a coral pathogen that is of interest because of its broad host range among corals and its temperature-dependent pathogenicity (27). Recently, V. coralliilyticus was also shown to induce a lethal disease in the symbiotic sea anemone A. pallida (28). To determine whether V. coralliilyticus can induce disease in N. vectensis, anemones were exposed to increasing concentrations of V. coralliilyticus at 30 °C. V. coralliilyticus showed a dose-dependent ability to cause disease in N. vectensis (Fig. 3A). Anemones infected with 108 cfu/mL at 30 °C rapidly declined in survival between days 4 and 6, while anemones infected with 107 cfu/mL V. coralliilyticus at 30 °C showed a decline in survival starting after day 5 (Fig. 3 A and B). As in A. pallida (28), V. coralliilyticus infection at 30 °C caused N. vectensis tentacle retraction and degradation followed by extensive ectodermal tissue lysis that exposed mesentery tissues by day 6 (Fig. 3B). All control anemones (with no exposure to bacteria at 19 °C or 30 °C) and anemones infected at 19 °C with 108 cfu/mL V. coralliilyticus survived; the latter result indicates that V. coralliilyticus causes lethal disease in N. vectensis only at elevated temperatures (Fig. 3 A and B and Fig. S1). In addition, incubation of anemones at 19 °C or 30 °C with the TLR ligand flagellin had no effect on N. vectensis viability (Fig. 3A). Anemones infected with 107 cfu/mL and 108 cfu/mL V. coralliilyticus at 30 °C both demonstrated significantly decreased survival compared with anemones infected with 106 cfu/mL V. coralliilyticus at 30 °C, 108 cfu/mL V. coralliilyticus at 19 °C, and control uninfected anemones (Kaplan–Meier log-rank test, P < 0.05) (Fig. 3A).
Fig. 3.
V. coralliilyticus, an N. vectensis pathogen, and flagellin stimulate a reconstituted Nv-TLR–to–NF-κB pathway. (A) Control anemones were incubated in 1/3 ASW at 19 °C and 30 °C (black line); anemones incubated with 100 ng/mL flagellin in 1/3 ASW at 19 °C and 30 °C were a control for PAMP toxicity (black line); and anemones infected with 108 cfu/mL at 19 °C are also indicated by the black line. No death was observed for any black line condition. Anemones infected with 106 cfu/mL (yellow line), 107 cfu/mL (blue line), or 108 cfu/mL (orange line) V. coralliilyticus at 30 °C are also plotted. Nine anemones were used for each experimental condition. The different letters (a and b) next to plotted conditions represent significant differences in survivorship by Kaplan–Meier log-rank test (P < 0.05). X-marked lines represent right-censored data. (B) Disease progression in a control uninfected anemone (Left) compared with an anemone infected with 107 cfu/mL V. coralliilyticus (Right) at 30 °C after 6 d. (C) RT-PCR performed for Nv-TLR and GAPDH (an input control). (D and E) Western blotting for p-IκB in Nv-TLR 293T and vector 293T stable cell lysates (Upper) treated for 12 min with 5 × 107 cfu/mL of heat-inactivated V. coralliilyticus (Vc) (D) or for 40 min with 100 ng/mL of flagellin (Fl) (E). Total protein was stained with Ponceau as a loading control (Bottom). Fold induction of p-IκB following treatment is indicated below the Western blots and in each case is normalized to the level of p-IκB in the nontreated control (1.0).
Given that V. coralliilyticus caused lethal disease in N. vectensis, we next sought to determine whether this bacterium could stimulate Nv-TLR–dependent activation of NF-κB. To study this, we used retroviral transduction to establish 293T cells that stably express Nv-TLR. Expression of Nv-TLR in these cells was confirmed by RT-PCR (Fig. 3C) because our Nv-TLR antiserum does not readily detect the lower levels of Nv-TLR expressed by retroviral transduction. To assess Nv-TLR activity, Nv-TLR 293T stable cells were exposed to 5 × 107 cfu/mL of heat-inactivated V. coralliilyticus for 12 min, and downstream activation of the NF-κB pathway was evaluated by measuring phosphorylation of IκBα (p-IκB), which is an essential early step for activation of NF-κB (16). Nv-TLR 293T cells showed a 13.6-fold induction of p-IκB when exposed to heat-inactivated V. coralliilyticus compared with nonstimulated Nv-TLR 293T cells (Fig. 3D). In contrast, treatment of vector-293T cells with heat-inactivated V. coralliilyticus induced low (2.6-fold) levels of p-IκB.
Bacterial flagellin, but not lipopolysaccharide (LPS), has been shown to stimulate an engineered Hydra LRR protein to activate NF-κB signaling in human cells (15). We also found that treatment of Nv-TLR 293T cells with Salmonella typhimurium flagellin for 40 min (38) led to a 7.3-fold induction of p-IκB compared with nonstimulated Nv-TLR 293T cells (Fig. 3E). In contrast, treatment of vector 293T cells with flagellin did not result in increased levels of p-IκB (1.0) (Fig. 3E). Escherichia coli LPS, an activator of Hu-TLR4 (37), did not stimulate Nv-TLR to activate our NF-κB–site luciferase reporter (Fig. S2).
Taken together, these results demonstrate that the Nv-TLR can activate a downstream molecular pathway (i.e., NF-κB) in response to heat-inactivated V. coralliilyticus, a pathogenic coral bacterium that is also lethal to N. vectensis (Fig. 3 A and D). Moreover, our results identify flagellin as a PAMP that can stimulate Nv-TLR–expressing human cells to activate the NF-κB pathway (Fig. 3E).
Nv-TLR Is Coexpressed with Nv-NF-κB in Cells in Embryos and Juvenile Anemones.
To identify cells in N. vectensis that express Nv-TLR and to confirm the specificity of our Nv-TLR antiserum for immunofluorescence experiments in anemones, whole-mount immunofluorescence with Nv-TLR antiserum was performed on juvenile, four-tentacle–stage anemones. Elongated cells throughout the body column of the anemones stained positive for Nv-TLR, and there was also intense staining in the tentacles (Fig. S3A). No signal was detected after whole-mount staining of juvenile animals with preimmune serum, indicating that staining with the Nv-TLR antiserum is specific (Fig. S3A).
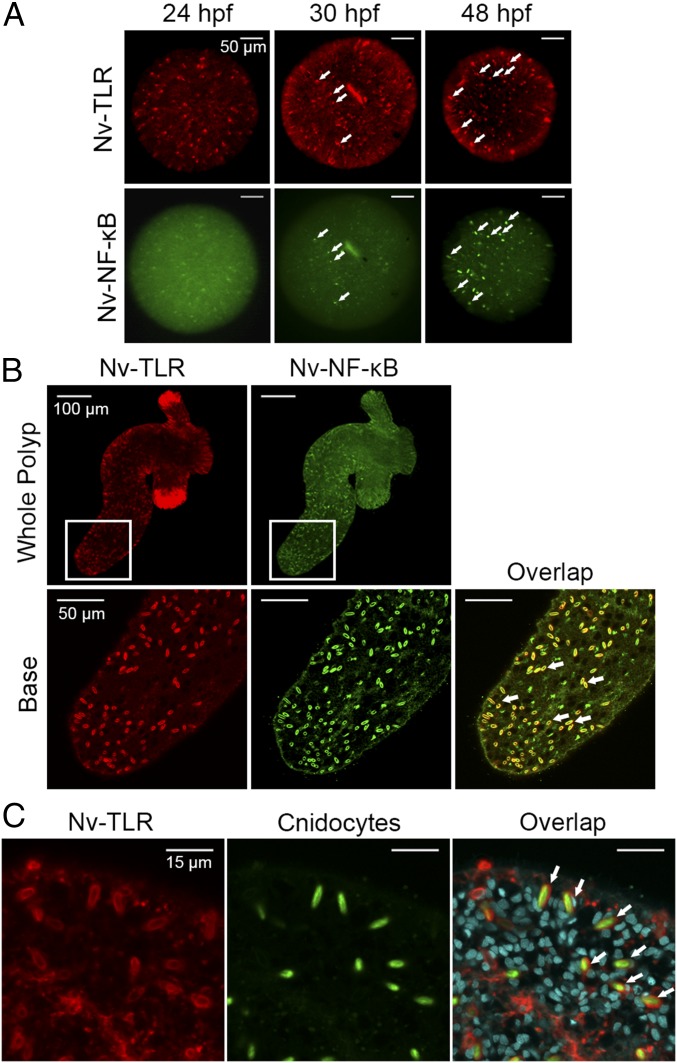
We previously showed that Nv-NF-κB can be detected by immunofluorescence in anemones as early as 30 h postfertilization and is expressed in cnidocytes starting at 40 h postfertilization (33). To determine whether Nv-TLR is also expressed early in development and in the same cells as Nv-NF-κB, whole-mount immunofluorescence of embryos at 24, 30, and 48 h postfertilization was performed with Nv-TLR and Nv-NF-κB antisera (Fig. 4A). At 24 h postfertilization, Nv-TLR was readily detected in many cells in the embryo; however, little to no Nv-NF-κB was detected (Fig. 4A). Nv-NF-κB was detected in a few cells at 30 h postfertilization, and these NF-κB positively stained cells were also positive for Nv-TLR staining (Fig. 4A, Center, white arrows); however, Nv-TLR was expressed more extensively than Nv-NF-κB at 30 h postfertilization (Fig. 4A). At 48 h postfertilization, many cells were detected that stained positive for both Nv-NF-κB and Nv-TLR, and this colocalization was observed in elongated cells (Fig. 4A, Right, white arrows).
Fig. 4.
Nv-TLR colocalizes with Nv-NF-κB in anemones. (A) Whole-mount immunofluorescence of 24-, 30-, and 48-h postfertilization embryos with Nv-TLR and Nv-NF-κB antisera. Nv-TLR is false-colored red (Upper), and Nv-NF-κB is false-colored green (Lower). White arrows indicate cells in which Nv-TLR (Upper) and Nv-NF-κB (Lower) are coexpressed in 30- and 48-h postfertilization embryos. White bars indicate scale. (B) Whole-mount immunofluorescence of a juvenile anemone with Nv-TLR and Nv-NF-κB antisera. (Upper Left) Nv-TLR (false-colored red) was detected in the body column and tentacles. (Upper Right) Nv-NF-κB (false-colored green) was detected mainly in body column cells. White squares indicate sections enlarged in the lower panels. In the base of the anemone, staining of Nv-TLR (false-colored red, Lower Left) and Nv-NF-κB (false-colored green, Lower Center) overlap; examples are indicated by white arrows (Lower Right). (C) Whole-mount immunofluorescence of a juvenile anemone was performed with Nv-TLR antiserum under calcium-free conditions. Nv-TLR is false-colored red (Left), and the internal capsules of cnidocytes are stained with DAPI (Center). Arrows in the merged image indicate Nv-TLR expression in body column cnidocytes (Right).
Our previous results have shown that Nv-NF-κB is expressed primarily in body column cnidocytes in both juvenile and adult animals (33). Based on the similar staining pattern of Nv-TLR and Nv-NF-κB at 30 and 48 h postfertilization (Fig. 4A), we determined whether Nv-TLR and Nv-NF-κB are coexpressed in cells in older anemones. Therefore, whole-mount preparations of four-tentacle–stage anemones were costained with Nv-TLR and Nv-NF-κB antisera. Indeed, there was considerable overlap of Nv-TLR and Nv-NF-κB staining in body column cells (Fig. 4B, white arrows). However, intense staining of cnidocyte-rich tentacles was observed only with Nv-TLR antiserum (Fig. 4B and Fig. S3A).
We hypothesized that Nv-TLR is expressed in cnidocytes, given that Nv-NF-κB is primarily expressed in body column cnidocytes (33) and that Nv-NF-κB and Nv-TLR are coexpressed in many cells in developing and juvenile anemones (Fig. 4 A and B). To directly demonstrate the expression of Nv-TLR in cnidocytes, we carried out indirect immunofluorescence on whole-mount preparations of juvenile anemones to determine if Nv-TLR–staining cells were also positive for the cnidocyte marker poly-γ-glutamate. Under calcium-free conditions, DAPI stains the poly-γ-glutamate found in cnidocyte capsules, and this staining can be visualized under green emission channels (39). As shown in Fig. 4C (white arrows), Nv-TLR staining surrounded the DAPI-stained cnidocyte capsule in many individual cells, indicating that Nv-TLR is expressed in cnidocytes.
Taken together, these results provide evidence that Nv-TLR and Nv-NF-κB are expressed in many of the same cells in developing embryos and juvenile anemones and that these two proteins are coexpressed in many cnidocytes. However, Nv-TLR expression is detected earlier in embryonic development than Nv-NF-κB, and Nv-TLR is highly expressed in cells in the tentacles of juvenile anemones, where little to no Nv-NF-κB can be detected (Fig. 4B and Fig. S3A).
Heterogeneity of Nv-TLR and Nv-NF-κB Expression in Tentacle and Body Column Cnidocytes.
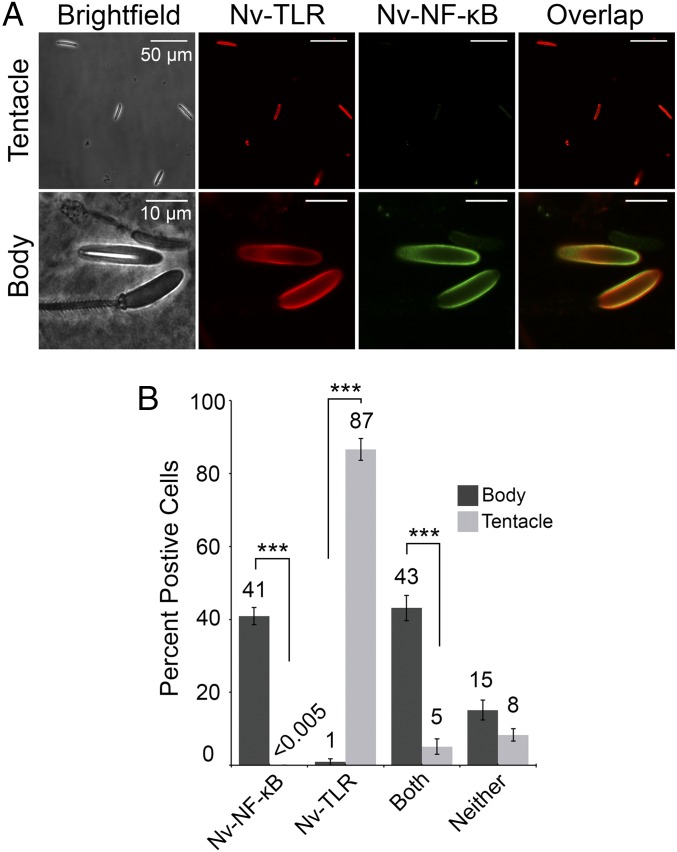
We next determined the percentage of cnidocytes in tentacles and body columns of adult anemones that express Nv-TLR and Nv-NF-κB. Cnidocytes were isolated from sectioned tentacles and body columns, fixed, and then immunostained with Nv-TLR and Nv-NF-κB antisera (Fig. 5A). We found that 43% of body column cnidocytes but only 5% of tentacle cnidocytes expressed both Nv-TLR and Nv-NF-κB (Fig. 5B). Consistent with highly concentrated Nv-TLR staining in tentacles (Fig. 4B and Fig. S3A), nearly 87% of isolated tentacle cnidocytes but only ∼1% of body column cnidocytes expressed only Nv-TLR. On the other hand, ∼41% of body column cnidocytes but less than 0.005% of tentacle cnidocytes expressed only Nv-NF-κB. Only 15% and 8% of body column and tentacle cnidocytes, respectively, expressed neither Nv-TLR nor Nv-NF-κB, and this difference was not significant (Fig. 5B). No staining was seen when isolated tentacle and body column cnidocytes were probed with the respective preimmune serum for Nv-TLR or Nv-NF-κB (Fig. S3B), indicating that the staining that we observed with isolated body column and tentacle cnidocytes is specific for Nv-TLR and Nv-NF-κB. Taken together, these results quantitatively support our whole-anemone staining, indicating that Nv-TLR is expressed in many body column cnidocytes that also express Nv-NF-κB and that high levels of Nv-TLR, but not Nv-NF-κB, are expressed in tentacles.
Fig. 5.
Nv-TLR is expressed in tentacle cnidocytes and with Nv-NF-κB in body column cnidocytes. (A, Upper) Immunofluorescence of isolated tentacle and body column cnidocytes was performed with Nv-TLR and Nv-NF-κB antisera. Four cnidocytes from tentacles (Left) were stained for Nv-TLR (false-colored red, Second from Left) and Nv-NF-κB (false-colored green, Third from Left). A merged image is shown at Right. (Lower) Three cnidocytes from body columns (Left) were stained for Nv-TLR (false-colored red, Second from Left) and Nv-NF-κB (false-colored green, Third from Left). A merged image is shown at the Right. Nv-TLR and Nv-NF-κB colocalization is observed in body column cnidocytes. (B) Expression of Nv-TLR and Nv-NF-κB in isolated tentacle and body column cnidocytes. In each case, 75 cells were counted in triplicate, and positive staining for Nv-TLR and Nv-NF-κB was recorded for each cell. Data are the means of the percent of positive cells for each marker ± SEM. Statistical significance was determined by Student’s t test; ***P < 0.001.
Nematosomes Express Components of Innate Immune Signaling Pathways and Can Engulf V. coralliilyticus.
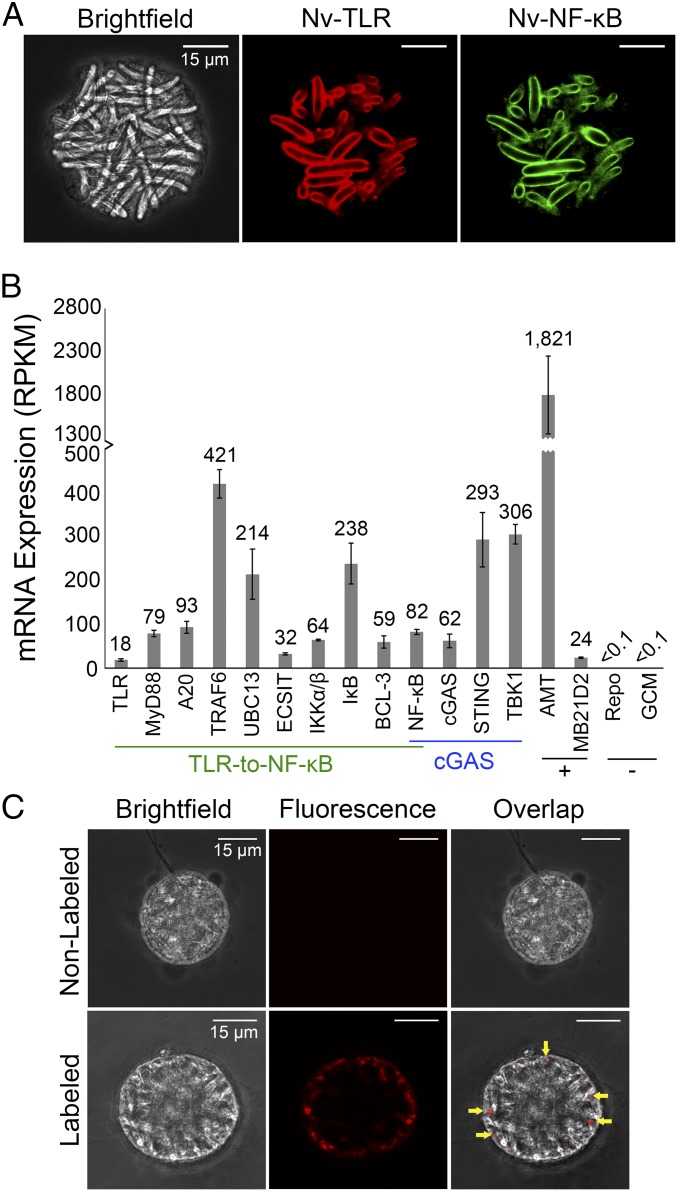
Nematosomes are highly motile multicellular structures that are unique to Nematostella species (40). Comprised of multiple cell types, including many cnidocytes, these organs circulate in the gastrovascular cavity and tentacles of sexually mature anemones (41). Previous comparative transcriptomic analyses revealed that many of the overrepresented transcripts in nematosomes are encoded by genes associated with chemical, pathogen, and wound stressors (41). To determine if Nv-TLR and Nv-NF-κB proteins are expressed in nematosomes, we isolated circulating nematosomes from live adult N. vectensis and fixed them for immunohistochemistry. Staining of these isolated nematosomes with Nv-TLR and Nv-NF-κB antisera showed that most nematosome cnidocytes express both Nv-TLR and Nv-NF-κB (Fig. 6A). Fig. S4 shows an orthogonal rendering of a nematosome stained for Nv-TLR and Nv-NF-κB.
Fig. 6.
Nematosomes express TLR–to–NF-κB and cGAS–STING innate immune pathway components and engulf V. coralliilyticus. (A) Immunofluorescence of isolated nematosomes was performed with Nv-TLR and Nv-NF-κB antisera. Nematosome cnidocytes (Left) express Nv-TLR (false-colored red, Center) and Nv-NF-κB (false-colored green, Right). (B) Expression of mRNAs for TLR–to–NF-κB (underlined in green) and cGAS–STING (underlined in blue) pathway components. Expression values were normalized as RPKM values. Data were compiled from two biological replicates of RNA-seq on isolated nematosomes (41). Nematosome-specific genes AMT and MB21D2 serve as positive controls (+), and nonnematosome neuronal markers REPO and GCM serve as negative controls (−). Data are presented as means ± SEM. (C) Confocal microscopy of optical sections of an isolated nematosome incubated with nonlabeled V. coralliilyticus (Upper Row) and a nematosome incubated with Baclight Red-labeled V. coralliilyticus (Lower Row). An overlap image (yellow arrows, Lower Right) shows labeled V. coralliilyticus inside nematosome peripheral cells. No fluorescence is detected in a nematosome exposed to nonlabeled V. coralliilyticus (Upper Row).
Given our ability to detect both Nv-TLR and Nv-NF-κB proteins in nematosome cnidocytes, we next determined whether other TLR–to–NF-κB pathway components are expressed in nematosomes using publicly available transcriptomic (23) and nematosome-specific mRNA expression data (41). To quantify the expression levels of given mRNAs, we constructed a custom Bowtie2 index of the coding sequences of homologous genes expected to be in the N. vectensis TLR–to–NF-κB pathway (TLR, MyD88, A20, TRAF6, UBC13, ECSIT, IKKα/β, IκB, BCL-3, and NF-κB) as well as those of another innate immune pathway, the cGAS–STING pathway (cGAS, STING, TBK1, and NF-κB) (Dataset S2). As expected from our immunostaining results, transcripts for Nv-TLR and Nv-NF-κB were found in the nematosome-specific expression datasets (Fig. 6B). In addition, mRNAs for homologs of all predicted TLR–to–NF-κB pathway proteins were detected in nematosomes (Fig. 6B). Interestingly, cGAS, STING, and TBK1 mRNAs were also detected in nematosomes (Fig. 6B). In humans, cGAS binds cytosolic cyclic dinucleotides, which are secondary messenger molecules in prokaryotes, and interacts with STING to ultimately activate the transcription factors IRF3 and NF-κB (7). Of note, N. vectensis does not have an IRF3 homolog (9).
To measure expression levels of TLR–to–NF-κB and cGAS–STING pathway mRNAs in nematosomes, we first compared their expression levels (in reads per kilobase of transcript per million reads mapped, RPKM) to two genes—an S-adenosylmethionine-dependent methyltransferase (AMT) homolog and a Mab-21 domain-containing 2 (MB21D2) homolog—that have been identified as nematosome-specific genes (41). AMT is more highly expressed (1,821 RPKM) in nematosomes than any other genes we measured. On the other hand, MB21D2 is expressed (24 RPKM) at a level similar to Nv-TLR (18 RPKM), and MB21D2 transcripts are less abundant than all other TLR–to–NF-κB and cGAS–STING pathway components analyzed (Fig. 6B).
As a negative control for this bioinformatic analysis and to ensure that nonnematosome tissues did not introduce contaminating mRNAs, we interrogated these datasets for mRNAs of Nv-Repo and Nv-GCM, which are neural differentiation markers in N. vectensis (42) and should not be expressed in nematosomes. Nv-Repo and Nv-GCM transcripts were not detected in the nematosome RNA-seq datasets (Fig. 6B). Taken together, these bioinformatics analyses indicate that TLR–to–NF-κB and cGAS–STING pathway mRNAs are part of the nematosome-specific transcriptome.
Recent studies have also identified peripheral phagocytes in nematosomes based on their morphology and ability to engulf heat-inactivated E. coli and dextran-labeled latex beads (41). To determine if nematosomes can engulf live V. coralliilyticus, isolated nematosomes were incubated for 14 h with cultures of this bacterium that had been labeled with the BacLight Red bacterial stain. By fluorescence microscopy, labeled V. coralliilyticus was detected in peripheral cells of isolated nematosomes (Fig. 6C, yellow arrows). No fluorescence was detected in nematosomes incubated with nonlabeled V. coralliilyticus, demonstrating that neither nematosomes nor V. coralliilyticus autofluoresce under red excitation (Fig. 6C). These data indicate that nematosomes can engulf V. coralliilyticus.
Morpholino-Based Knockdown of Nv-TLR Impairs Embryonic Development.
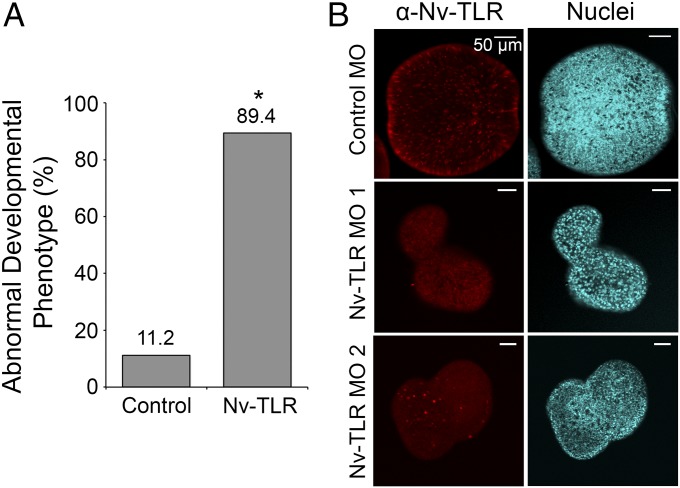
Although mostly studied in terms of innate immunity, TLR signaling is involved in other biological processes such as development (5, 10, 11, 43). To determine whether Nv-TLR also has a developmental role in N. vectensis, zygotes were microinjected with a morpholino-substituted antisense oligonucleotide specific for Nv-TLR. At 3 d postinjection, nearly 90% of Nv-TLR morpholino-injected embryos exhibited severe developmental defects (Fig. 7A), which were generally characterized by an elongated tube-like embryo with a narrowed midsection (Fig. 7B). The level of developmental abnormalities seen following injection of the Nv-TLR morpholino (∼90%) was significantly higher than the 11% seen in embryos injected with a control morpholino (the developmental defects seen at day 3 in the embryos injected with a control morpholino were likely due to microinjection manipulation) (Fig. 7A). Among the Nv-TLR morpholino-injected embryos that survived to 5 d postinjection, few Nv-TLR–expressing cells were detected by whole-mount immunostaining with Nv-TLR antiserum (Fig. 7B). Two examples of embryos with the Nv-TLR knockdown phenotype are shown in Fig. 7B. Microinjection of the control morpholino did not appear to affect embryonic development or the development of Nv-TLR–positive cells (Fig. 7B and Table S2). The average number of Nv-TLR–positive cells, as determined by analyzing optical cross-sections of morpholino-injected anemones, was lower for anemones injected with the Nv-TLR morpholino (14.5 positive cells per anemone) than for anemones injected with the control morpholino (92.3 positive cells per anemone) (Table S2). These results suggest that Nv-TLR has an essential role in early N. vectensis development.
Fig. 7.
Injection of Nv-TLR morpholino (MO) impairs embryonic development. N. vectensis zygotes were microinjected with morpholinos against a control or Nv-TLR transcript. (A) Three days postinjection, 251 live control morpholino-injected anemones and 397 live Nv-TLR morpholino-injected anemones were scored for aberrant developmental phenotypes under a dissecting microscope. Percent abnormal developmental phenotype is shown for morpholino-injected anemones. Statistical significance was determined using Fisher’s exact test, *P = 2.2 × 10−16. (B) Five days postinjection anemones were fixed and stained using Nv-TLR antiserum (Left) and DAPI for nuclei (Right). Control morpholino-injected anemones developed normally, and Nv-TLR was detected in cells throughout the embryo (Upper). Nv-TLR morpholino-injected anemones had few Nv-TLR+ cells and failed to properly develop compared with control anemones. Two Nv-TLR morpholino-injected embryos are shown: One has no cells expressing Nv-TLR (Middle), and one that has few Nv-TLR–expressing cells (Bottom).
Discussion
In this paper, we have characterized the sole TLR of the sea anemone N. vectensis. Our results indicate that this single Nv-TLR has properties that are normally distributed independently among TLRs in more complex protostomes and deuterostomes. The embryonic expression of Nv-TLR, its requirement for proper embryonic development, and its expression in specific cells in embryonic, juvenile, and adult anemones suggests that Nv-TLR has biological roles throughout the lifespan of the anemone. Moreover, Nv-TLR is a mccTLR that appears to be able to directly recognize a bacterial pathogen to initiate downstream intracellular signaling. Based on our results and previously published data (discussed below), we hypothesize that Nv-TLR has an NF-κB–independent role in embryonic development and an NF-κB–dependent role in immunity in juvenile and adult anemones.
Recent transcriptomic studies have shown that conventional TLRs and many NF-κB pathway components are expressed in several cnidarians, including the corals Acropora digitifera and Acropora millepora (17, 44); however, no studies have functionally characterized these pathways in corals, anemones, or jellyfish. Before the work presented herein, the most extensively characterized cnidarian TLR pathway is that of Hydra. Similar to N. vectensis, Hydra has genes encoding many components of a TLR–to–NF-κB pathway but lacks a conventional TLR. Instead, Hydra has separate LRR and TIR-domain proteins (15, 18). A Hydra LRR protein (HyLRR-2) fused to a human TIR domain has been shown to activate a NF-κB reporter in 293 cells in response to flagellin but not LPS (15). Those studies are consistent with our results showing that heat-inactivated V. coralliilyticus and flagellin (Fig. 3 D and E), but not LPS (Fig. S2), can activate a reconstituted Nv-TLR–to–NF-κB pathway in 293 cells. Studies in Hydra have also suggested a role for its atypical TLR pathway in pathogen defense, as silencing of a TIR domain-only protein resulted in the loss of antimicrobial peptide production (15), and MyD88-deficient Hydra showed increased sensitivity to infection by Pseudomonas aeruginosa (22). Nevertheless, it is important to note that TLR-directed activation of NF-κB has not yet been directly demonstrated in any cnidarian system. Thus, a role for TLR–to–NF-κB signaling in cnidarian immunity remains a hypothesis.
While increasing evidence suggests that cnidarians utilize ancient signaling pathways for defense, it is not known if cnidarians have specialized immune cells or organs. Previous work demonstrated that epithelial cells in Hydra and peripheral cells in the N. vectensis nematosome can engulf bacteria (15, 41). Additionally, more than 20% of nematosome-specific transcripts are involved in stress responses (chemical, pathogen, wound) (41). Based on those findings and our findings that (i) nematosomes contain TLR- and NF-κB–expressing cnidocytes, (ii) nematosome peripheral cells can engulf the pathogenic bacterium V. coralliilyticus, and (iii) mRNAs for components of the Nv-TLR–to–Nv-NF-κB and cGAS–STING pathways are expressed in nematosomes (Fig. 6), we hypothesize that the nematosome is a unique innate immune organ for Nematostella species that may combine pathogen engulfment and innate immune signal transduction in one circulating multicellular structure. We have not, however, detected Nv-TLR or Nv-NF-κB staining of the peripheral cells in the nematosome (Fig. 6A).
In animals, the overall expression of Nv-TLR and Nv-NF-κB overlaps considerably, but not completely (Figs. 4 and 5). In adult anemones, body column and nematosome cnidocytes that express Nv-TLR also often express Nv-NF-κB (Figs. 5B and 6A), but tentacle cnidocytes almost entirely express only Nv-TLR (Fig. 5B), and nearly half of body column cnidocytes express Nv-NF-κB but not Nv-TLR (Fig. 5B). These apparent differences in Nv-TLR and Nv-NF-κB staining in different populations of cnidocytes suggest that Nv-TLR functions in both Nv-NF-κB–independent (in tentacle cnidocytes) and Nv-NF-κB–dependent (in nematosome and body column cnidocytes) pathways in adult animals.
Knockdown of Nv-TLR causes early developmental defects (Fig. 7) that are not seen with Nv-NF-κB knockdown (33), and Nv-TLR expression is detected earlier than Nv-NF-κB in developing embryos (Fig. 4A). Therefore, we think it is likely that Nv-TLR has an NF-κB–independent role in early development. Although Toll-1–to–NF-κB signaling plays a key role in the development of the dorsal–ventral axis of the Drosophila embryo (11, 45), Drosophila Toll-2, Toll-6, and Toll-8 coordinate cell intercalation during convergent extension (anterior–posterior axis elongation) independent of NF-κB signaling (46). Moreover, this function of Drosophila Tolls in promoting proper embryo axis elongation is also found in basal arthropods (47). The single C. elegans mccTLR, TOL-1, also has developmental roles, in that Tol-1 mutants are either embryonic lethal or arrest as small, deformed larvae (43), and TOL-1 is required for the proper function and development of specific ciliated chemosensory neurons in adult nematodes (48). Since C. elegans lacks NF-κB (43, 48), TOL-1 must direct these developmental processes independently of NF-κB. Like Nv-TLR, the ligands that activate Drosophila Toll-2, Toll-6, and Toll-8 and C. elegans TOL-1 during development are not known (43, 46, 48), and no Spätzle-like homologs have been found in the N. vectensis genome (23). However, it has been demonstrated that these Drosophila Tolls can promote heterophilic interactions between cells, suggesting that activation occurs by engagement of an adjacent Toll (46). Whether Nv-TLR is activated during development by an endogenous ligand, by receptor–receptor interactions, or by another mechanism remains unknown.
Overall, our results show that the single TLR of N. vectensis has structural and functional characteristics that are often found in separate TLRs in organisms with multiple TLRs. The ability of Nv-TLR to activate NF-κB signaling in human cells in response to V. coralliilyticus and flagellin further suggests that an Nv-TLR–activated intracellular pathway is important for organismal pathogen detection and immune response in N. vectensis. Our findings also suggest that Nv-TLR is capable of directly recognizing pathogens and the PAMP flagellin. This challenges the conventional understanding of mccTLR activation, which had been thought to occur solely through endogenous ligands, such as Drosophila Spätzle (14). Our characterization of Nv-TLR suggests that primitive TLRs had dual functions in directing biological programs for pathogen response and embryonic development, providing an example of a single TLR that combines the independent functions of TLRs in insects and mammals.
Finally, our studies may also have relevance to the study of marine invertebrate disease. We have shown that V. coralliilyticus causes rapidly fatal disease in N. vectensis, and its effects in N. vectensis mimic the disease progression that occurs in several coral species and the anemone A. pallida, starting with tentacle degradation followed by fatal ectodermal tissue lysis (Fig. 3B and Fig. S1) (28, 49). The corals and the anemone A. pallida in which V. coralliilyticus has previously been shown to cause lethal disease are all tropical cnidarians with obligate algal symbionts (26, 28, 49). In contrast, N. vectensis does not have any known essential symbiont and can live in a variety of habitats over a wide range of environmental conditions (e.g., salinity, pH, temperature) (31). Thus, the temperature-dependent pathogenicity of V. coralliilyticus is clearly not limited to environmentally sensitive and symbiotic cnidarians. Given that N. vectensis is more amenable to genetic approaches than A. pallida or any coral species (50, 51), we believe that our findings establish a cnidarian model for the study of bacterial pathogenicity and innate immunity at a molecular level.
Materials and Methods
Plasmid Constructions, Cell Culture, and Transfection.
Genomic draft assemblies and EST databases from cnidarians.bu.edu/stellabase/index.cgi (35) and the Joint Genome Institute (23) were used to identify the cDNA encoding the 969-amino acid Nv-TLR protein. A human cell codon-optimized cDNA corresponding to the predicted amino acid sequence of Nv-TLR was synthesized (GenScript) (Dataset S1). Details about plasmids and plasmid constructions are given in Table S3. Primers used for PCR amplification, RT-PCR, and EMSAs are listed in Table S4.
HEK 293 and 293T carcinoma cells and DF-1 chicken fibroblasts were grown in DMEM (Invitrogen) supplemented with 10% FBS (Biologos), 50 U/mL penicillin, and 50 μg/mL streptomycin. HEK 293, HEK 293T, and DF-1 cells were transfected with expression plasmids using polyethylenimine (PEI) (Polysciences, Inc.) at a PEI:DNA mass ratio of 6:1 as previously described (32, 34). The medium was changed 18 h posttransfection, and whole-cell lysates were made 48 h posttransfection. If cells were used for immunofluorescence, they were passaged onto glass coverslips on the day before fixation.
Phylogenetic Analysis.
For comparative analysis of TIR domains, we used the predicted TIR domains of Nv-TLR, D. melanogaster Toll-1, C. elegans TOL-1, an Amphimedon queenslandica TLR-like protein (21), and the 10 human TLR proteins. The TIR domains of Nv-TLR and A. queenslandica TLR were identified through MEME analysis (52), and sequences were trimmed to contain only TIR domains based on motif prediction and known human TIR domains (Dataset S3). The human TLRs, D. melanogaster Toll-1, and C. elegans TOL-1 amino acid sequences, along with their annotated TIR domains, were obtained from the UniProt database (Dataset S3). Clustal Omega (53) was then used to align the trimmed and culled TIR sequence dataset, and phylogenetic comparison was performed using neighbor-joining analysis bootstrapped 1,000 times using PAUP* and rooted with A. queenslandica TLR (54).
Generation of Nv-TLR Antiserum, Preparation of Whole-Cell Lysates, and Western Blotting.
A guinea pig polyclonal antiserum was prepared against a C-terminal predicted peptide (KPLAPPQSYEGHVEMSKV) of Nv-TLR (Thermo Fisher). Whole-cell lysis of HEK 293 cells and Western blotting for FLAG proteins were performed essentially as described previously using FLAG antibody (1:1,000) (Cell Signaling Technology) (32, 34). Anti-rabbit horseradish peroxidase-linked secondary antiserum was used to bind primary immunoreactive complexes. Immunoreactive proteins were then detected with SuperSignal West Dura extended-duration substrate (Pierce).
When preparing lysates for Western blotting of 293 cells overexpressing Nv-TLR, cells were washed with PBS and then were resuspended directly in 2× SDS sample buffer [0.125 M Tris (pH 6.8), 4.6% (wt/vol) SDS, 20% (wt/vol) glycerol, 10% (vol/vol) β-mercaptoethanol] and boiled for 10–15 min. Nitrocellulose membranes were blocked overnight at 4 °C with Tris-buffered saline [100 mM Tris (pH 7.4), 9% (vol/vol) NaCl] supplemented with 8% milk, 5% normal goat serum (Gibco), 1% BSA, and 0.5% Tween 20. Membranes were then incubated for 5 h at 4 °C with Nv-TLR antiserum (1:25,000) in blocking buffer. Immunoreactive bands were visualized as described above using an anti-guinea pig horseradish peroxidase-conjugated secondary antiserum (1:50,000) in blocking buffer.
Reporter Gene Assays.
Reporter gene assays were performed in 293 cells as previously described (34). Cells were transfected with the following: 60 ng of pcDNA-FLAG (34), Hu-TLR4, Nv-TLR, or Nv-TLRΔTIR; 74 ng of the human coreceptors CD-14 and MD-2; 300 ng of a 3× NF-κB-site luciferase reporter plasmid; and 500 ng of RSV–β-gal for normalization of transfection efficiency. Before use, LPS (0111:B4; Sigma-Aldrich) was sonicated for two rounds of 1 min with 1 min rest in a water bath at level 5 in a Sonic Dismembrator 550 (Fisher Scientific Inc.). Sonicated LPS was then added to cells at a final concentration of 2.5 μg/mL in fresh medium. Six hours later, cells were lysed and analyzed according to the manufacturer’s instructions for the Luciferase Assay System (Promega).
GST Pulldown Assays.
GST and GST-Nv-TIR proteins were expressed in E. coli BL21 cells and isolated on glutathione beads as previously described (55). Five percent of the GST slurry was electrophoresed on a SDS-polyacrylamide gel and stained with Coomassie Blue to confirm the presence of each GST protein. The remaining 95% of the glutathione beads was incubated for 24 h at 4 °C with 300 μg of FLAG-MAL or FLAG-MYD88 293T cell lysates prepared as previously described (32). Beads were then washed four times with PBS to remove unbound proteins. To remove complexed proteins, the beads were boiled in 2× SDS sample buffer. As input, one percent of the 293T cell lysate used in each pulldown (3 μg) was electrophoresed on a SDS-polyacrylamide gel. After transfer to nitrocellulose, the GST pulldowns were subjected to anti-FLAG Western blotting.
N. vectensis Care and Spawning.
The care and mating of adult N. vectensis were performed as described previously (33, 56–58). Anemones were housed and spawned in 1/3 strength artificial seawater (1/3 ASW: ∼12 parts per 1,000). Spawned eggs were dejellied from their egg masses and then fertilized as previously described (56).
V. coralliilyticus Culture Preparation, Heat Inactivation, and Labeling.
V. coralliilyticus was cultured at 30 °C and prepared for anemone infection as described previously (28). For heat inactivation of V. coralliilyticus, bacteria were grown overnight at 30 °C, pelleted, washed once with PBS, resuspended in PBS, and then incubated at 56 °C for 30 min. To visualize V. coralliilyticus for nematosome engulfment experiments, 2 × 107 cfu V. coralliilyticus were washed twice with PBS and incubated in a total volume of 5 mL PBS containing 25 μL of 1 mM BacLight Red bacterial stain (Thermo Fisher) for 30 min with rotation at 30 °C. Nematosomes were then washed twice with PBS and resuspended in 1/3 ASW.
Anemone Bacterial Challenge.
For 30 °C anemone infection experiments, anemones were first acclimated at 30 °C for 24 h before infection. On the day of infection, overnight cultures of V. coralliilyticus were pelleted, washed once with 1/3 ASW, and resuspended in 1/3 ASW at the desired concentration (28). For flagellin experiments, anemones were incubated in 1/3 ASW at a final concentration of 100 ng/mL of flagellin (Salmonella typhimurium; Sigma-Aldrich). For these infection experiments, anemone mortality was assessed each day by the degree of tissue degradation and the absence of response to light and touch cues (28). To determine the statistical significance of these infection experiments, Kaplan–Meier analysis was performed to generate survival functions and log-rank significance values using the statistical software SPSS (IBM).
Generation of Stable Cell Lines, Challenge with V. coralliilyticus, and RT-PCR.
HEK 293T cells were transfected with 5 μg of the pCL10A1 retrovirus-packaging vector and 15 μg of either pMSCV-puro or pMSCV-Nv-TLR. At 18 h posttransfection, the medium for the 293T cultures was replaced. At 48 h posttransfection, transduced 293T cells were selected by adding fresh medium containing puromycin at a final concentration of 2 μg/mL.
Once stably transduced cell lines were established, 1 × 106 cells were plated in a 60-mm dish, and 24 h later fresh medium was added with 5 × 107 cfu/mL of heat-inactivated V. coralliilyticus for 12 min. For flagellin stimulation experiments, 1 × 106 cells were plated in a 60-mm dish, and 24 h later fresh medium was added with a final concentration of 100 ng/mL of flagellin (S. typhimurium; Sigma-Aldrich) for 40 min. Whole-cell lysates were made as described previously and were subjected to Western blotting (32, 34) with p-IκBα antiserum (1:1,000; Cell Signaling Technology). Fold induction of p-IκBα was determined by Fiji image processing software and was normalized to the respective nonstimulated control for each experiment.
For RT-PCR, RNA was extracted using TRIzol Reagent (Invitrogen) according to the manufacturer’s specifications. cDNA was then prepared from 500 ng RNA for each sample. RNA was combined with 1.68 µL of 15.4-µM random primers (Promega) and nuclease-free water to a final volume of 16 µL. Samples were incubated at 65 °C for 5 min and then on ice for 5 min. cDNA was synthesized by adding 6 µL 2.5 mM dNTPs, 1 µL RNasin (Promega), 6 µL 5× M-MLV buffer (Promega), and 1 µL M-MLV reverse transcriptase (Promega), and samples were incubated at 37 °C for 1 h. cDNA samples were then diluted 10-fold and used as a template for PCR. Fifty-microliter reactions were prepared with Q5 Hot Start DNA polymerase (New England Biolabs) according to the manufacturer’s protocol, subjected to 35 PCR cycles, and analyzed by agarose gel electrophoresis and staining with ethidium bromide.
Isolation of Nematosomes and Cnidocytes from Adult N. vectensis.
Nematosomes were isolated from adult N. vectensis as previously described (41). Cnidocytes were isolated from adult anemones by modification of a previously published protocol (59). For cnidocyte isolation, adult N. vectensis were relaxed for 10 min in 7% (wt/vol) MgCl2 in 1/3 ASW. Each anemone was then placed in Ca2+/Mg2+-free seawater (0.5 M NaCl, 10 mM KCl, 7 mM Na2SO4, 10 mM Hepes, pH 8.0) and tentacles or body columns were sectioned, pooled, and washed three times with fresh Ca2+/Mg2+-free seawater (with gentle centrifuging to pellet the tissue each time). After the final wash, the N. vectensis sections were placed in 250 μL of 45 °C prewarmed Ca2+/Mg2+-free seawater and incubated at 45 °C for 15 min. Samples were then vortexed for 15 s, placed in an ice bath for 30 s, and then vortexed for 15 s. The remaining intact tissue was removed with fine forceps. To separate cnidocytes by density, the tissue mixture was layered on 4 °C Percoll diluted 1:1.5 with concentrated ASW (1.12 M NaCl, 22 mM KCl, 20 mM CaCl2, 65 mM MgCl2, 2 mM NaHCO3). Samples were then centrifuged for 15 min at ∼2,000 × g at 4 °C, with the centrifuge brakes off. The aqueous layer was removed, and the pellet containing cnidocytes was washed three times with Ca2+/Mg2+-free seawater. If necessary, a second Percoll gradient was performed to purify cnidocytes. Cnidocyte isolation was visually assessed by light microscopy to ensure that mainly only cnidocytes were present in isolated fractions.
Indirect Immunofluorescence.
Indirect immunofluorescence of DF-1 cells was performed essentially as described previously (32) using primary antisera against FLAG (1:50), Nv-TLR (1:50), or HA (1:50). FLAG and HA immunoreactive complexes were detected with 488-conjugated secondary antiserum (1:80), and Nv-TLR immunoreactive complexes were detected with an anti-guinea pig Texas Red-conjugated secondary antiserum (1:80). Fixation and whole-mount indirect immunofluorescence of embryos (0–5 d postfertilization) and juvenile polyps at the four-tentacle developmental stage were performed as previously described (33, 60) using Nv-TLR (1:100) or Nv-NF-κB (1:100) antiserum. Detection of Nv-TLR and Nv-NF-κB immunoreactive complexes in N. vectensis was performed using an anti-guinea pig Texas Red-conjugated secondary antiserum (1:160) and an anti-rabbit 488-conjugated secondary antiserum (1:160), respectively. DAPI staining for poly-γ-glutamate found in intact mature cnidocytes in N. vectensis was performed as previously described (33, 60). Specimens were imaged on a FluoView FV10i confocal microscope.
For immunofluorescence of isolated N. vectensis cnidocytes, resuspended cnidocytes (in Ca2+/Mg2+-free seawater) were placed on Superfrost Plus microscope slides (Thermo Fisher) and allowed to settle for 30 min. Residual Ca2+/Mg2+-free seawater was removed, and the following procedure was performed in a humid chamber with careful attention that slides did not dry. For immunofluorescence of isolated nematosomes, nematosomes (in Ca2+/Mg2+-free seawater) were collected into a 35-mm dish, and the following procedure was performed in 35-mm dishes with rotation during each incubation. Specimens were then fixed [4% (vol/vol) paraformaldehyde, 0.2% (vol/vol) Triton X-100, PBS] for 1 h; fixative was replaced with fresh fixative after the first 30 min. Fixative was washed four times for 10 min each with PTx [0.2% (vol/vol) Triton X-100 and PBS]. Specimens were then blocked with immunofluorescence (IF) blocking buffer [5% (vol/vol) normal goat serum, 1% (wt/vol) BSA, 0.2% (vol/vol) Triton X-100 in PBS] for 2 h. Blocking buffer was removed, and specimens were incubated with Nv-TLR (1:100) or Nv-NF-κB (1:100) antiserum (32) (in IF blocking buffer) at 37 °C for 1–1.5 h. Next, unbound primary antiserum was washed four times for 10 min each with PTx. Dextran-conjugated secondary antiserum (anti-rabbit for Nv-NF-κB and anti-guinea pig for Nv-TLR) was added at 1:160 in IF blocking buffer and incubated at 37 °C for 2 h. Last, specimens were washed three times with PTx to remove unbound secondary antiserum. For immunostained cnidocytes, coverslips were mounted with VECTASHIELD mount (Vector Labs). For immunostained nematosomes, individual nematosomes were pipetted dropwise onto 35-mm optical imaging discs or Superfrost Plus microscope slides, and coverslips were mounted. Immunostained cnidocytes and nematosomes were imaged on a FluoView FV10i confocal microscope.
mRNA Expression Analysis of Nematosomes.
Paired-end reads from nematosome Fastq files from publicly available RNA-seq experiments were obtained from the European Nucleotide Archive (ERA609085 and ERA609085) (41). Control genes known to be expressed in nematosomes (AMT and MB21D2) were identified in the additional file 8 (41). Nematosome-specific gene IDs (NvecRef IDs) were obtained, and partial sequences were extrapolated from the NvecRef32742 reference transcriptome (41) and were subjected to BLAST (National Center for Biotechnology Information, NCBI) to determine specific gene identity. These control genes and known cDNA sequences of Nv-Repo, Nv-GCM, and select genes involved in either the N. vectensis NF-κB signaling or the cGAS–STING pathway (Dataset S2) were used to generate a custom Bowtie2 index for mapping. Raw reads were then mapped to both the custom Bowtie2 index and the full Nemvedraft ASM20922v1 reference transcriptome (Ensembl Metazoa) using default settings in Bowtie2. The number of reads mapped to each gene in the curated set was normalized according to both gene length and total mapped reads (in the complete transcriptome) to generate RPKM values. The relative expression of each gene is presented as the mean RPKM between the two replicates ± SEM.
Engulfment of V. coralliilyticus.
Nematosomes were isolated from adult N. vectensis as previously described (41). They were pooled into 35-mm dishes and were incubated with a final concentration of ∼5 × 106 cfu/mL of V. coralliilyticus or BacLight Red-labeled V. coralliilyticus for 14 h at room temperature in 1/3 ASW. Nematosomes were then washed four or five times with 1/3 ASW and incubated at 4 °C with rotation in a final concentration of 4% paraformaldehyde in 1/3 ASW for 30 min. Fixed nematosomes were imaged on a FluoView FV10i confocal microscope at 581 nm excitation.
Morpholino Knockdown.
Knockdown of the Nv-TLR transcript was accomplished by microinjecting fertilized N. vectensis eggs from a clonal anemone line (23) with an antisense morpholino toward a sequence at the 5′ end of the Nv-TLR mRNA. N. vectensis injections were performed as previously described (51). The sequences of the morpholinos were as follows: Control 5′-CCTCTTACCTCAGTTACAATTTATA-3′; Nv-TLR 5′-TCGAGTGCTGATATTTTTTCCGTTG-3′. Morpholinos at 700 μM were coinjected with a 10× (2 mg/mL) 488-dextran dye, and embryos that were not fluorescing at 24 h following injection were removed. Morphology was assessed 3 d postinjection, and anemones were fixed and stained for Nv-TLR at 5 d postinjection.
Supplementary Material
Acknowledgments
We thank Mark Martindale (Whitney Laboratory for Marine Science) for supplying anemones and experimental advice for knockdown experiments; Adeline Hajjar (University of Washington) for human CD-14, FLAG-MD-2, and HA-TLR4 constructs; Claudio Sette (University of Rome Tor Vergata) for the FLAG-MYD88 construct; Jan Tavernier (Ghent University) for the FLAG-MAL construct; Milad Babaei and Yuekun Liu for the NEMO-knockout 293T cells; Maria Liberti and Amy Acevedo for early contributions to this research; and Leslie Babonis, Todd Blute, Cynthia Bradham, John Finnerty, Katelyn Mansfield, Linda Nguyen, Vijendra Ramlall, Trevor Siggers, David Simmons, Naveen Wijesena, and Francis Wolenski for helpful discussions on ideas and experimental details. This research was supported by National Science Foundation (NSF) Grant IOS-1354935 (to T.D.G.). J.L.M. was supported by funds from the Arnold and Mabel Beckman Scholar Program at Boston University. L.M.W. and B.J.M. were supported by NSF Graduate Research Fellowships. M.R. was supported by the NSF-Research Experience for Undergraduates Program and by funds from the Boston University Undergraduate Research Opportunities Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711530114/-/DCSupplemental.
References
- 1.Imler JL, Zheng L. Biology of Toll receptors: Lessons from insects and mammals. J Leukoc Biol. 2004;75:18–26. doi: 10.1189/jlb.0403160. [DOI] [PubMed] [Google Scholar]
- 2.Vasselon T, Detmers PA. Toll receptors: A central element in innate immune responses. Infect Immun. 2002;70:1033–1041. doi: 10.1128/IAI.70.3.1033-1041.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Kawai T, Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med. 2007;13:460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Silverman N, Maniatis T. NF-kappaB signaling pathways in mammalian and insect innate immunity. Genes Dev. 2001;15:2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 7.Cai X, Chiu YH, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 8.Dixit E, Kagan JC. Intracellular pathogen detection by RIG-I-like receptors. Adv Immunol. 2013;117:99–125. doi: 10.1016/B978-0-12-410524-9.00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Margolis SR, Wilson SC, Vance RE. Evolutionary origins of cGAS-STING signaling. Trends Immunol. 2017;38:733–743. doi: 10.1016/j.it.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Valanne S, Wang JH, Rämet M. The Drosophila Toll signaling pathway. J Immunol. 2011;186:649–656. doi: 10.4049/jimmunol.1002302. [DOI] [PubMed] [Google Scholar]
- 11.Lindsay SA, Wasserman SA. Conventional and non-conventional Drosophila Toll signaling. Dev Comp Immunol. 2014;42:16–24. doi: 10.1016/j.dci.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer SN, et al. An ancient defense system eliminates unfit cells from developing tissues during cell competition. Science. 2014;346:1258236. doi: 10.1126/science.1258236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hetru C, Hoffmann JA. NF-kappaB in the immune response of Drosophila. Cold Spring Harb Perspect Biol. 2009;1:a000232. doi: 10.1101/cshperspect.a000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leulier F, Lemaitre B. Toll-like receptors–Taking an evolutionary approach. Nat Rev Genet. 2008;9:165–178. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- 15.Bosch TC, et al. Uncovering the evolutionary history of innate immunity: The simple metazoan Hydra uses epithelial cells for host defence. Dev Comp Immunol. 2009;33:559–569. doi: 10.1016/j.dci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Gilmore TD, Wolenski FS. NF-κB: Where did it come from and why? Immunol Rev. 2012;246:14–35. doi: 10.1111/j.1600-065X.2012.01096.x. [DOI] [PubMed] [Google Scholar]
- 17.Miller DJ, et al. The innate immune repertoire in Cnidaria–Ancestral complexity and stochastic gene loss. Genome Biol. 2007;8:R59. doi: 10.1186/gb-2007-8-4-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Augustin R, Fraune S, Bosch TC. How Hydra senses and destroys microbes. Semin Immunol. 2010;22:54–58. doi: 10.1016/j.smim.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Poole AZ, Weis VM. TIR-domain-containing protein repertoire of nine anthozoan species reveals coral-specific expansions and uncharacterized proteins. Dev Comp Immunol. 2014;46:480–488. doi: 10.1016/j.dci.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 20.Baumgarten S, et al. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc Natl Acad Sci USA. 2015;112:11893–11898. doi: 10.1073/pnas.1513318112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gauthier ME, Du Pasquier L, Degnan BM. The genome of the sponge Amphimedon queenslandica provides new perspectives into the origin of Toll-like and interleukin 1 receptor pathways. Evol Dev. 2010;12:519–533. doi: 10.1111/j.1525-142X.2010.00436.x. [DOI] [PubMed] [Google Scholar]
- 22.Franzenburg S, et al. MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proc Natl Acad Sci USA. 2012;109:19374–19379. doi: 10.1073/pnas.1213110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 24.Chapman JA, et al. The dynamic genome of Hydra. Nature. 2010;464:592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes TP, et al. Coral reefs in the Anthropocene. Nature. 2017;546:82–90. doi: 10.1038/nature22901. [DOI] [PubMed] [Google Scholar]
- 26.Sussman M, Willis BL, Victor S, Bourne DG. Coral pathogens identified for White Syndrome (WS) epizootics in the Indo-Pacific. PLoS One. 2008;3:e2393. doi: 10.1371/journal.pone.0002393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimes NE, et al. Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME J. 2012;6:835–846. doi: 10.1038/ismej.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown T, Rodriguez-Lanetty M. Defending against pathogens–Immunological priming and its molecular basis in a sea anemone, cnidarian. Sci Rep. 2015;5:17425. doi: 10.1038/srep17425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sussman M, et al. Vibrio zinc-metalloprotease causes photoinactivation of coral endosymbionts and coral tissue lesions. PLoS One. 2009;4:e4511. doi: 10.1371/journal.pone.0004511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaragoza WJ, et al. Outcomes of infections of sea anemone Aiptasia pallida with Vibrio spp. pathogenic to corals. Microb Ecol. 2014;68:388–396. doi: 10.1007/s00248-014-0397-2. [DOI] [PubMed] [Google Scholar]
- 31.Darling JA, et al. Rising starlet: The starlet sea anemone, Nematostella vectensis. BioEssays. 2005;27:211–221. doi: 10.1002/bies.20181. [DOI] [PubMed] [Google Scholar]
- 32.Wolenski FS, et al. Characterization of the core elements of the NF-κB signaling pathway of the sea anemone Nematostella vectensis. Mol Cell Biol. 2011;31:1076–1087. doi: 10.1128/MCB.00927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolenski FS, Bradham CA, Finnerty JR, Gilmore TD. NF-κB is required for cnidocyte development in the sea anemone Nematostella vectensis. Dev Biol. 2013;373:205–215. doi: 10.1016/j.ydbio.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Sullivan JC, et al. Two alleles of NF-kappaB in the sea anemone Nematostella vectensis are widely dispersed in nature and encode proteins with distinct activities. PLoS One. 2009;4:e7311. doi: 10.1371/journal.pone.0007311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan JC, et al. StellaBase: The Nematostella vectensis genomics database. Nucleic Acids Res. 2006;34:D495–D499. doi: 10.1093/nar/gkj020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tulin S, Aguiar D, Istrail S, Smith J. A quantitative reference transcriptome for Nematostella vectensis early embryonic development: A pipeline for de novo assembly in emerging model systems. Evodevo. 2013;4:16. doi: 10.1186/2041-9139-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J Biol Chem. 1999;274:10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- 38.Feuillet V, et al. Involvement of Toll-like receptor 5 in the recognition of flagellated bacteria. Proc Natl Acad Sci USA. 2006;103:12487–12492. [Google Scholar]
- 39.Szczepanek S, Cikala M, David CN. Poly-γ-glutamate synthesis during formation of nematocyst capsules in Hydra. J Cell Sci. 2002;115:745–751. doi: 10.1242/jcs.115.4.745. [DOI] [PubMed] [Google Scholar]
- 40.Hand C, Uhlinger KR. The culture, sexual and asexual reproduction, and growth of the sea anemone Nematostella vectensis. Biol Bull. 1992;182:169–176. doi: 10.2307/1542110. [DOI] [PubMed] [Google Scholar]
- 41.Babonis LS, Martindale MQ, Ryan JF. Do novel genes drive morphological novelty? An investigation of the nematosomes in the sea anemone Nematostella vectensis. BMC Evol Biol. 2016;16:114. doi: 10.1186/s12862-016-0683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marlow HQ, Srivastava M, Matus DQ, Rokhsar D, Martindale MQ. Anatomy and development of the nervous system of Nematostella vectensis, an anthozoan cnidarian. Dev Neurobiol. 2009;69:235–254. doi: 10.1002/dneu.20698. [DOI] [PubMed] [Google Scholar]
- 43.Pujol N, et al. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- 44.Rauta PR, Samanta M, Dash HR, Nayak B, Das S. Toll-like receptors (TLRs) in aquatic animals: Signaling pathways, expressions and immune responses. Immunol Lett. 2014;158:14–24. doi: 10.1016/j.imlet.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 45.Hashimoto C, Hudson KL, Anderson KV. The Toll gene of Drosophila, required for dorsal-ventral embryonic polarity, appears to encode a transmembrane protein. Cell. 1988;52:269–279. doi: 10.1016/0092-8674(88)90516-8. [DOI] [PubMed] [Google Scholar]
- 46.Paré AC, et al. A positional Toll receptor code directs convergent extension in Drosophila. Nature. 2014;515:523–527. doi: 10.1038/nature13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benton MA, et al. Toll genes have an ancestral role in axis elongation. Curr Biol. 2016;26:1609–1615. doi: 10.1016/j.cub.2016.04.055. [DOI] [PubMed] [Google Scholar]
- 48.Brandt JP, Ringstad N. Toll-like receptor signaling promotes development and function of sensory neurons required for a C. elegans pathogen-avoidance behavior. Curr Biol. 2015;25:2228–2237. doi: 10.1016/j.cub.2015.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ben-Haim Y, et al. Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int J Syst Evol Microbiol. 2003;53:309–315. doi: 10.1099/ijs.0.02402-0. [DOI] [PubMed] [Google Scholar]
- 50.Ikmi A, McKinney SA, Delventhal KM, Gibson MC. TALEN and CRISPR/Cas9-mediated genome editing in the early-branching metazoan Nematostella vectensis. Nat Commun. 2014;5:5486. doi: 10.1038/ncomms6486. [DOI] [PubMed] [Google Scholar]
- 51.Layden MJ, Röttinger E, Wolenski FS, Gilmore TD, Martindale MQ. Microinjection of mRNA or morpholinos for reverse genetic analysis in the starlet sea anemone, Nematostella vectensis. Nat Protoc. 2013;8:924–934. doi: 10.1038/nprot.2013.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bailey TL, Williams N, Misleh C, Li WW. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swofford D. Phylogenetic Analysis Using Parsimony (*and Other Methods) Sinauer Associates; Sunderland, MA: 2002. [Google Scholar]
- 55.Garbati MR, Thompson RC, Haery L, Gilmore TD. A rearranged EP300 gene in the human B-cell lymphoma cell line RC-K8 encodes a disabled transcriptional co-activator that contributes to cell growth and oncogenicity. Cancer Lett. 2011;302:76–83. doi: 10.1016/j.canlet.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Genikhovich G, Technau U. Induction of spawning in the starlet sea anemone Nematostella vectensis, in vitro fertilization of gametes, and dejellying of zygotes. Cold Spring Harbor Protoc. 2009;2009:pdb.prot5281. doi: 10.1101/pdb.prot5281. [DOI] [PubMed] [Google Scholar]
- 57.Fritzenwanker JH, Technau U. Induction of gametogenesis in the basal cnidarian Nematostella vectensis (Anthozoa) Dev Genes Evol. 2002;212:99–103. doi: 10.1007/s00427-002-0214-7. [DOI] [PubMed] [Google Scholar]
- 58.Hand C, Uhlinger KR. The unique, widely distributed, estuarine sea anemone, Nematostella vectensis Stephenson: A review, new facts, and questions. Estuaries. 1994;17:501–508. [Google Scholar]
- 59.Bouchard C, et al. Cloning and functional expression of voltage-gated ion channel subunits from cnidocytes of the Portuguese Man O’War Physalia physalis. J Exp Biol. 2006;209:2979–2989. doi: 10.1242/jeb.02314. [DOI] [PubMed] [Google Scholar]
- 60.Wolenski FS, Layden MJ, Martindale MQ, Gilmore TD, Finnerty JR. Characterizing the spatiotemporal expression of RNAs and proteins in the starlet sea anemone, Nematostella vectensis. Nat Protoc. 2013;8:900–915. doi: 10.1038/nprot.2013.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.