Significance
Epigenetic modifications, such as histone methylation, are crucial for gene expression, chromatin organization, and cellular identity. These modifications can be faithfully transmitted to daughter cells during the cell cycle. How epigenetic marks are inherited through DNA replication remains poorly understood. Histone hypoacetylation and histone H3 lysine 9 (H3K9) methylation are two conserved epigenetic marks of heterochromatin, a transcriptionally repressive form of chromatin. Here we demonstrate that the two conserved small histone-fold subunits of the DNA polymerase epsilon complex, Dpb3 and Dpb4, form a heterodimer and play an important role in coordinating the inheritance of histone hypoacetylation and H3K9 methylation during replication. This study provides mechanistic insights into how epigenetic marks in heterochromatin are transmitted through the cell cycle.
Keywords: epigenetic inheritance, heterochromatin, replication, DNA polymerase epsilon, Schizosaccharomyces pombe
Abstract
During DNA replication, chromatin is disrupted ahead of the replication fork, and epigenetic information must be restored behind the fork. How epigenetic marks are inherited through DNA replication remains poorly understood. Histone H3 lysine 9 (H3K9) methylation and histone hypoacetylation are conserved hallmarks of heterochromatin. We previously showed that the inheritance of H3K9 methylation during DNA replication depends on the catalytic subunit of DNA polymerase epsilon, Cdc20. Here we show that the histone-fold subunit of Pol epsilon, Dpb4, interacts an uncharacterized small histone-fold protein, SPCC16C4.22, to form a heterodimer in fission yeast. We demonstrate that SPCC16C4.22 is nonessential for viability and corresponds to the true ortholog of Dpb3. We further show that the Dpb3–Dpb4 dimer associates with histone deacetylases, chromatin remodelers, and histones and plays a crucial role in the inheritance of histone hypoacetylation in heterochromatin. We solve the 1.9-Å crystal structure of Dpb3–Dpb4 and reveal that they form the H2A–H2B-like dimer. Disruption of Dpb3–Dpb4 dimerization results in loss of heterochromatin silencing. Our findings reveal a link between histone deacetylation and H3K9 methylation and suggest a mechanism for how two processes are coordinated during replication. We propose that the Dpb3–Dpb4 heterodimer together with Cdc20 serves as a platform for the recruitment of chromatin modifiers and remodelers that mediate heterochromatin assembly during DNA replication, and ensure the faithful inheritance of epigenetic marks in heterochromatin.
Histone modifications are major carriers of epigenetic information and contribute to gene expression and genome organization (1). When DNA replicates, chromatin is disrupted ahead of the replication fork, and chromatin structure and epigenetic information are restored behind the fork (2, 3). A central unresolved question in the field of epigenetics is how the histone marks are maintained through replication.
Chromatin is divided into two distinct domains: euchromatin and heterochromatin. Whereas euchromatin is loosely packed and transcriptionally active, heterochromatin contains highly condensed and silenced DNA. During replication, the tightly packed structure of heterochromatin provides an additional challenge for replicating DNA and transmitting epigenetic information (3–5). Histone hypoacetylation and the histone H3 lysine 9 (H3K9) methylation are two conserved epigenetic hallmarks of heterochromatin. Both modifications play crucial roles in heterochromatin assembly and are stably transmitted through the cell cycles (5). How histone hypoacetylation and H3K9 methylation are faithfully inherited and how these two processes are coordinately regulated during replication remain poorly understood.
Fission yeast (Schizosaccharomyces pombe) has emerged as an excellent model for studying heterochromatin. Heterochromatin in fission yeast comprises peri-centromeres, telomere, and mating-type regions (6, 7). As in multicellular organisms, H3K9 methylation and hypoacetylation are enriched in heterochromatin in fission yeast. H3K9 methylation in fission yeast is mediated by the CLCR complex, which contains the H3K9 methyltransferase Clr4/Suv39 and the silencing proteins Rik1, Dos1/Raf1, Dos2/Raf2, and Cul4 (8–12). RNA interference (RNAi) plays an important role in H3K9 methylation and heterochromatin silencing (13). During replication, Cdc20/Pol2, the DNA polymerase (Pol) epsilon catalytic subunit, interacts with the CLRC complex. Cdc20 also associates with a transcription regulator to promote the transcription of heterochromatin (14, 15). Heterochromatin transcripts are processed into small interference RNAs (siRNAs) by RNAi machinery (16). siRNAs, together with Cdc20, target the CLRC complex to the next-generation histones to mediate H3K9 methylation (14). H3K9 methylation is bound by a human HP1 homolog, Swi6, that assembles the chromatin into a repressive structure. Hypoacetylation of histones in S. pombe heterochromatin is mediated by histone deacetylases, including Sir2, Clr3, and Clr6 (17–19). However, little is known about how the histone hypoacetylation is coupled with DNA replication.
DNA Pol epsilon is largely responsible for leading-strand synthesis during replication and consists of the four conserved subunits: Cdc20/Pol2, Dpb2, Dpb3, and Dpb4 (20, 21). Dpb3 and Dpb4 are two small histone-fold proteins that are not essential for viability in budding yeast (22, 23). In this organism, Dpb3 and Dpb4 have been shown to be involved in heterochromatin silencing, although the underlying mechanism remains unclear (24). In fission yeast, homologs of Dpb3 and Dpb4 have been identified; however, the Dpb3 homolog was found to be essential for viability, whereas Dpb4 was not (25).
Here, using mass spectrometry, we identified that in fission yeast, Dpb4 interacts with an uncharacterized small histone-fold protein, SPCC16C4.22. We demonstrated that SPCC16C4.22 is a true ortholog of Dpb3 and exists in a complex with Dpb4. We further showed that the Dpb3–Dpb4 complex is important for the recruitment of histone deacetylases to heterochromatin, suggesting a mechanism for how H3K9 methylation and histone deacetylation are coordinately inherited during replication. On solving the 1.9-Å crystal structure of the Dpb3–Dpb4 complex, we observed that Dpb3 and Dpb4 form an H2A–H2B-like heterodimer and gained further insight into the role of this dimer in chromatin regulation. Together, our findings indicate that the Pol epsilon complex provides a platform for the recruitment of chromatin modifiers and remodelers during DNA replication, which in turn ensures the accurate perpetuation of heterochromatin marks.
Results
Dpb4 Is Important for Heterochromatin Silencing.
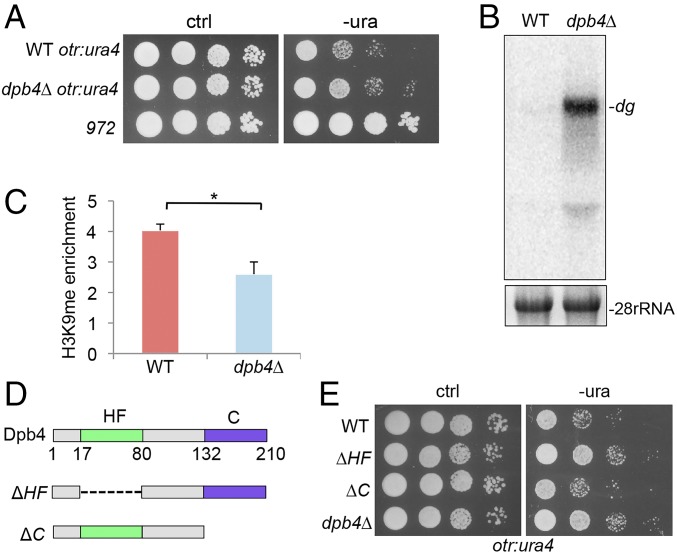
Dpb4 is the only DNA polymerase epsilon subunit in fission yeast identified as not essential for viability (25). To determine its role on heterochromatin formation, we created a dpb4 deletion mutant. The integrity of heterochromatin in dpb4∆ cells was analyzed using the ura4+ reporter gene inserted at an otr region within pericentromeric heterochromatin. ura4+ is required for the production of uracil. Our growth assays showed that dpb4∆ cells carrying the reporter grew consistently faster than wild type, indicating that the heterochromatin silencing is disrupted in the mutant (Fig. 1A). The loss of silencing was further confirmed by using media containing 5-fluoroorotic acid, a drug that kills cells expressing ura4+ (SI Appendix, Fig. S1), and northern blotting (Fig. 1B). Deletion of dpb4+ results in partial loss of silencing compared with dos1∆ mutant, reminiscent of that observed in sir2∆ (SI Appendix, Fig. S2). Consistent with the silencing defect, we found that histone H3K9 methylation is partially reduced in the dpb4∆ mutant, using chromatin immunoprecipitation (ChIP; Fig. 1C).
Fig. 1.
Dpb4 is required for heterochromatin formation. (A) Serial dilutions of indicated cells harboring ura4+ at the otr region were plated in minimal medium without uracil (-ura). Strain 972, wild-type strain harboring ura4+ at the endogenous locus. (B) Northern blotting analysis of transcript levels from the pericentromeric dg repeats. 28S rRNA was used as a loading control. (C) Analysis of H3K9 methylation in the otr region by ChIP. ChIP assays were performed using an antibody against H3K9 methylation and primers specific for ura4+. act1+ was used as a control. Three independent experiments were performed. Error bars indicate SD. *P < 0.05. (D) A schematic diagram of Dpb4 deletion mutants. C, coiled–coil domain; HF, histone-fold domain. (E) Growth assays for dpb4-ΔHF and dpb4-ΔC mutants carrying ura4+ in the pericentromeric otr regions. Serial dilutions of cells were plated on -ura medium.
Dpb4 contains a prominent histone-fold domain at its N terminus, whereas its C terminus has a coiled–coil domain. The histone-fold domain, which is shared by all core histones and histone variants (26, 27), often serves as a dimerization motif (26). To investigate the role of the domain, we constructed the histone-fold domain-deleted versions of dpb4+ (Fig. 1D), dpb4-∆HF, and replaced the endogenous dpb4+. We found that the silencing in otr region in dpb4-∆HF mutant is reduced to a similar level to the dpb4-null mutant (Fig. 1E). The loss of silencing in otr region in the mutant was confirmed by northern blotting (SI Appendix, Fig. S3). These data indicate that the histone-fold domain is important for the function of Dpb4 in heterochromatin silencing. We also created dpb4-∆C, a coiled–coil domain-deleted version of dpb4+ (Fig. 1D). However, we found that deletion of the coiled–coil domain has little effect on heterochromatin silencing (Fig. 1E and SI Appendix, Fig. S3).
Dpb4 Associates with Key Silencing Factors.
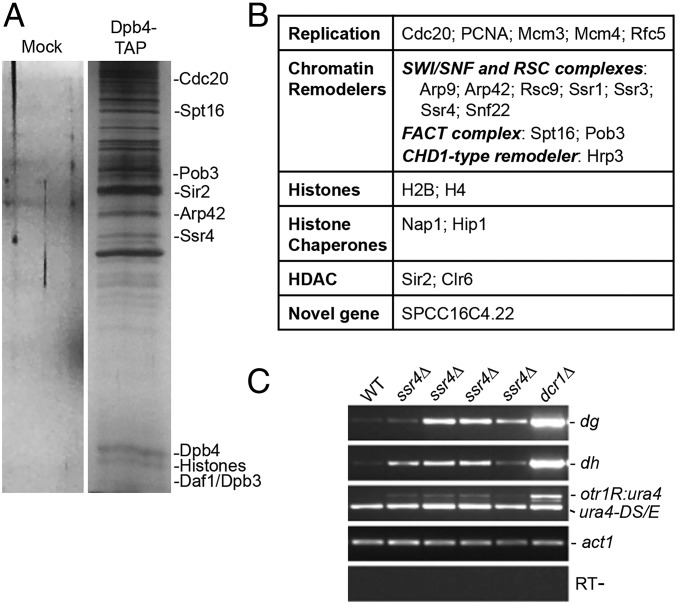
To better understand the role of Dpb4 in heterochromatin, we performed a TAP (tandem affinity purification) purification assay using cell extracts from wild-type cells carrying TAP-tagged Dpb4, and followed by mass spectrometry (spec) analysis (Fig. 2A and Dataset S1). Cells carrying Dpb4-TAP show no detectable silencing defects in our growth assays, indicating that the fusion protein is functional. Our mass spec analysis identified several replication factors in the Dpb4-TAP purified fraction, including Cdc20, PCNA, Rfc5, and replication licensing factors, including Mcm3 and Mcm4 (Fig. 2B), consistent with Dpb4 being a subunit of the Pol epsilon. We also found that Dpb4 associates with chromatin remodeling complexes, including FACT complex subunits (Spt16, Pob3) and the CHD1-type remodeler, Hrp3 (Fig. 2B). FACT complex and Hrp3 have been shown to be required for heterochromatin assembly in fission yeast (28, 29).
Fig. 2.
Mass spectrometry analysis of TAP-tagged purified Dpb4 revealed that Dpb4 interacts with silencing factors and histones. (A) Silver staining of TAP-tagged purified Dpb4 and a control purification from an untagged strain. (B) The proteins identified by mass spectrometry analysis of purified Dpb4 were grouped on the basis of their functions. (C) Heterochromatin transcripts are accumulated in ssr4Δ mutant. Transcripts derived from dg and dh peri-centromeric transcripts (Top two panels) and from ura4+ reporter inserted at otr region (Middle) were analyzed by RT-PCR from indicated strains. act1, actin; RT-, no reverse transcriptase control.
In addition, our mass spec results revealed that Dpb4 associates with the subunits of the SWI/SNF chromatin remodeling complex, including Snf22, Arp9, Arp42, Ssr1, Ssr3, and Ssr4 (Fig. 2B). Ssr4 in the complex was annotated as an essential gene on Pombase. However, we were able to isolate a total deletion mutant of ssr4+ using homologous recombination, showing that the gene is not essential. The mutant cells grow slower than wild-type and have minor morphological defects (SI Appendix, Fig. S4A). Using RT-PCR, we found that heterochromatin transcripts from pericentromeric regions accumulate in ssr4∆ cells, indicating that ssr4+ is required for heterochromatic silencing (Fig. 2C). Transcripts from the ura4+ reporter inserted in the pericentromeric otr region in the mutant are also increased (Fig. 2C). We also detected minor reduction of siRNAs from the otr region in ssr4∆ cells (SI Appendix, Fig. S4B).
Aside from replication factors and chromatin remodelers, histone proteins, including H4 and H2B, were identified in the Dpb4-TAP purified fraction. We also found the histone chaperones, Nap1, and the HIRA protein, Hip1. Importantly, we identified histone deacetylases Sir2 and Clr6 in the complex (Fig. 2B). All these proteins were not found in the control purifications. Together, our findings demonstrate that Dpb4 associates with factors involved in heterochromatin assembly.
SPCC16C4.22 Is the Ortholog of Dpb3.
We failed to capture protein SPAC17G8.03c in our pull-down assays. This protein was previously identified as the homolog of budding yeast Dpb3 in fission yeast (25). We found instead a protein, SPCC16C4.22, that copurified with Dpb4. We named this protein Daf1 (Dpb4 associated factor 1). Daf1 is a small protein, containing 87 amino acids with a prominent histone-fold domain at its N terminus from amino acids 10–72 (SI Appendix, Fig. S5). We found that Daf1shows high homology with human Dpb3/Pole4 (40% identity, 63% positives) and budding yeast Dpb3 (37% identity, 51% positives). In contrast, protein SPAC17G8.03c, previously identified as Dpb3 in fission yeast, shares 34% and 26% homology with human and yeast Dpb3, respectively.
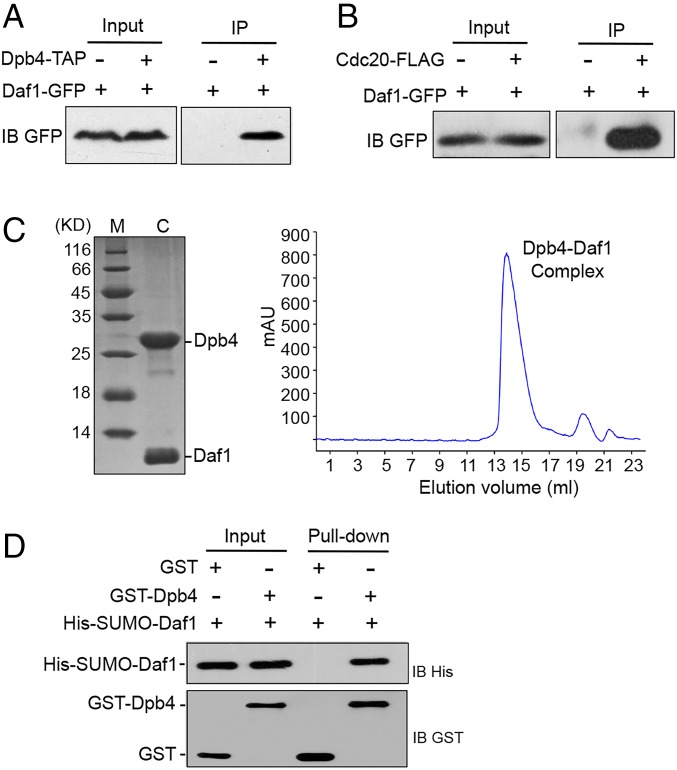
Our coimmunoprecipitation (Co-IP) analysis indicated that Daf1-GFP coimmunoprecipitated with Dpb4-TAP, confirming that the two proteins interact in vivo (Fig. 3A). However, previously identified GFP-tagged Dpb3 was unable to coimmunoprecipitate with Dpb4-TAP (SI Appendix, Fig. S6). We also confirmed that Daf1-GFP interacts with FLAG-tagged Cdc20 (Fig. 3B). To verify direct interaction between Dpb4 and Daf1, the proteins were coexpressed in Escherichia coli. We found that they were purified as a stable complex, as evidenced by the single elution peak in the gel-filtration profile (Fig. 3C). This was further confirmed by our in vitro pull-down assay (Fig. 3D). In contrast, we found that the previously identified Dpb3 was unable to pull down Dpb4 in vitro (SI Appendix, Fig. S7).
Fig. 3.
Daf1/Dpb3 and Dpb4 form a heterodimer. (A) Cell lysates from cells expressing Dpb4–TAP and Daf1–GFP and from control cells expressing Daf1–GFP only were subjected to immunoprecipitation with an antibody specific for TAP. Precipitated proteins were analyzed by immunoblotting (IB), using the indicated antibody. (B) Cell lysates from cells expressing Cdc20–FLAG and Daf1–GFP were immunoprecipitated with a FLAG antibody and analyzed by immunoblotting using a GFP antibody. Cells expressing Daf1–GFP only were used as a control. (C) Dpb4 and Daf1 form a stable complex in vitro, as evidenced by the elution profile of gel-filtration chromatography (Right). The purified Dpb4-Daf1 complex was analyzed on SDS–PAGE (Left). C, Dpb4–Daf1 complex; M, molecular weight. (D) GST pull-down assays demonstrated physical binding between Daf1 and Dpb4. Both input and pull-down samples were subjected to immunoblotting with anti-His and anti-GST antibodies.
To assess the function of Daf1 in growth and chromatin regulation, we deleted endogenous daf1+ and were able to isolate a viable mutant, daf1∆, indicating that daf1+ is not essential for viability, such as DPB3 in Saccharomyces cerevisiae. The daf1∆ mutant displays no significant growth defect or temperature-sensitive phenotype. Also, similar to dpb3 and dpb4 mutants in S. cerevisiae (23, 30), our fluorescence-activated cell sorting analysis indicated that daf1∆ and dpb4∆ in fission yeast exhibited only minor S-phase progression defects (SI Appendix, Fig. S8). Together, our results demonstrated that Daf1 is the true ortholog of Dpb3. We thus refer to the gene as dpb3+ from now on.
Structure of the Dpb3–Dpb4 Heterodimer.
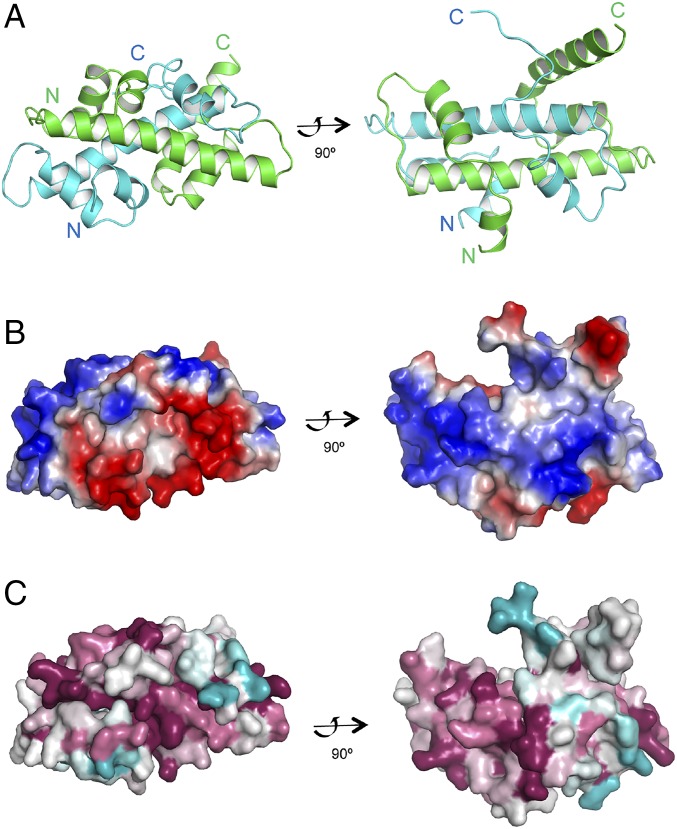
To further characterize the interaction of Dpb3/Daf1 and Dpb4, we purified and crystallize the Dpb3–Dpb4 complex. The structure was determined to 1.9 Å by single-wavelength anomalous diffraction. The final model was refined to Rwork/Rfree values of 19.2%/24.4%, containing residues 1–87 of Dpb3, residues 9–106 of Dpb4, and 220 water molecules, with very good deviations from ideal geometry (SI Appendix, Table S1). The flexible C-terminal coiled–coil tail (∼100 aa) of Dpb4 is missing in the structure because of unexpected proteolysis during crystallization. As expected, each individual polypeptide of Dpb3 and Dpb4 shares a common histone-like fold (helix–turn–helix–turn–helix), consisting of a core of three helices, with a long central helix flanked on either side by a loop segment and a shorter helix (Fig. 4A and SI Appendix, Fig. S9A). Dpb3 and Dpb4 interact with each other in a head-to-tail fashion, burying ∼4,869 Å2 of total surface area within dimer interface. Dpb3 and Dpb4 in budding yeast have also been suggested to form a heterodimer (30).
Fig. 4.
Crystal structure of the Dpb3–Dpb4 complex. (A) Ribbon drawing of the Dpb3–Dpb4 complex. Dpb3 is colored in cyan, and Dpb4 is colored in green. Right viewed as 90° rotation from Left. (B) Surface electrostatic properties of the Dpb3–Dpb4 complex. The contour level is at ±5 kT/e; red for negative potential and blue for positive potential. Right viewed as 90° rotation from Left. (C) Corresponding molecular surface of the Dpb3–Dpb4 complex in A is colored by sequence conservation as computed by ConSurf program based on sequences alignment from 500 nonredundant Dpb3 or Dpb4 homologous proteins.
The overall structure and electrostatic character on the surface of the Dpb3–Dpb4 complex are highly similar to that of other known histone-fold pairs, such as NF-YC/NF-YB (31), and H2A/H2B (32) (Fig. 4 B and C and SI Appendix, Fig. S9B). Similar to other histone-fold pairs, the protein surface of the Dpb3–Dpb4 complex is positively charged, suggesting a role in DNA binding and chromatin remodeling, consistent with previous findings on Dpb4 in budding yeast (24, 30, 33) (Fig. 4B). Dpb3–Dpb4 has been previously characterized as the H2A–H2B family protein. This is confirmed by the presence of a long fourth helix, αC, in Dpb4 extended from the core histone motif helix α1–loop L1–helix α2-loop L2–helix α3, characteristic for the H2B family. The αC in Dpb4 is even longer and followed by a flexible coil–coil domain (Fig. 4A). The histone-fold domain is essential for Dpb4’s function in heterochromatin silencing (Fig. 1E), and the extended αC of the histone-fold domain in Dpb4 appears to be ideally suitable for the interaction with the chromatin modifiers and other factors identified earlier. Also, similar to H2A-related proteins, Dpb3 contains a similar C-terminal helix followed by the helix alpha3, but it is much shorter. The Dpb3–Dpb4 complex has also features of the histone H3/H4 dimer. The topology of the overall fold of Dpb3–Dpb4 is similar to that of H3/4. Also similar to H3, Dpb3 has a helix at the N terminus, although it is shorter and adopts a different orientation. However, unlike H4, Dpb4 lacks the long N-terminal helix before the histone motif.
Dpb3–Dpb4 Is Important for Coordination of Histone Hypoacetylation and H3K9 Methylation.
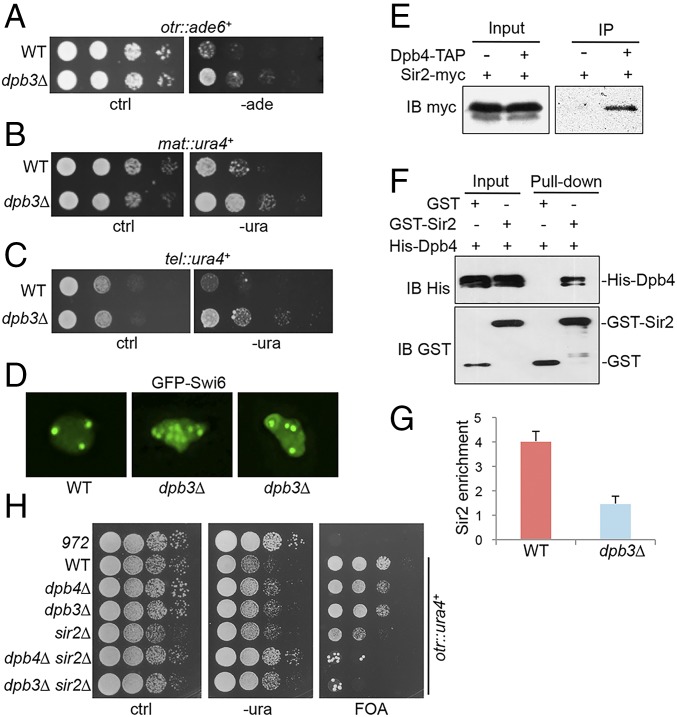
We found that similar to dpb4∆ mutants, dpb3∆ displays partial loss of silencing in otr region (Fig. 5A and SI Appendix, Fig. S2). We also found that silencing in mating-type and telomere is also significantly lost in the mutant (Fig. 5 B and C). Consistent with the silencing defects, H3K9 methylation is decreased more than 50% in dpb3∆ (SI Appendix, Fig. S10A), whereas histone acetylation at the pericentromeric region is significantly increased (SI Appendix, Fig. S10B). GFP-Swi6 also appears to be more diffusely distributed throughout the nucleus in these cells (Fig. 5D), suggesting that deletion of dpb3+ results in partial dissociation of Swi6 from heterochromatin. In S. pombe interphase cells, heterochromatin is organized into three to four clusters underneath the nuclear envelope; as a result, in cells expressing GFP-Swi6, three to four fluorescent foci can be visualized (8). Interestingly, dpb3∆ cells frequently displays an excessive number of GFP-Swi6 spots (Fig. 5D), suggesting that heterochromatin may be formed at ectopic regions in the mutant. Lagging chromosomes is a common feature of heterochromatin mutants (8). We found that ∼12% of dpb3∆ mutant cells displayed severely disorganized nuclei, including uneven, lagging, or fragmented chromosomes. In contrast, these phenotypes are rarely seen in wild-type cells (<1%) (SI Appendix, Fig. S11).
Fig. 5.
Dpb3–Dpb4 coordinates histone hypoacetylation and H3K9 methylation. (A) Serial dilutions of dpb3Δ cells harboring ade6+ at the otr region were plated in -ade medium. (B) Growth assays for dpb3Δ mutant carrying ura4+ in the mating-type region. Serial dilutions of cells were plated on -ura medium. (C) Growth assays for dpb3Δ mutant containing ura4+ next to telomere in -ura medium. (D) GFP–Swi6 distribution in dpb3Δ mutant. (E) Cell lysates from cells expressing Sir2–myc and Dpb4–TAP were immunoprecipitated with an antibody specific for TAP. Precipitated proteins were analyzed by Western blotting using a myc antibody. Cells expressing Sir2–myc only were used as a control. (F) Direct interaction between Dpb4 and Sir2 revealed by GST pull-down assays. Both input and pull-down samples were subjected to IB with indicated antibodies. (G) Association of Sir2 with the pericentromeric otr region is reduced in dpb3Δ mutant. ChIP assays were conducted in cells expressing Sir2–myc using an anti-myc antibody. act1+ was used as a control. Error bars indicate SD. (H) Serial dilutions of indicated cells harboring ura4+ at the otr region were plated in -ura medium or medium containing 5-fluoroorotic acid.
To validate the interaction between Dpb4 and Sir2, we performed Co-IP and showed that Sir2 copurified with Dpb4 (Fig. 5E). A direct interaction between GST-Sir2 and His-Dpb4 was detected by our in vitro GST pull-down assays (Fig. 5F). In addition, ChIP assays revealed that the level of Sir2-myc at otr region is substantially reduced in dpb3∆ mutant relative to wild-type (Fig. 5G), suggesting that Dpb3 is required for the recruitment of Sir2 to heterochromatin. Given that Cdc20 mediates H3K9 methylation by recruiting CLRC, our data suggest that Cdc20 and Dpb3–Dpb4 dimer may function together to coordinately regulate the inheritance of H3K9 methylation and histone hypoacetylation during replication. Consistent with this, we found that the double mutants of sir2∆ with dpb3∆ and dpb4∆ exhibited synthetic defects in heterochromatin silencing (Fig. 5H).
Dimerization of Dpb3 and Dpb4 Is Required for Heterochromatin Silencing.
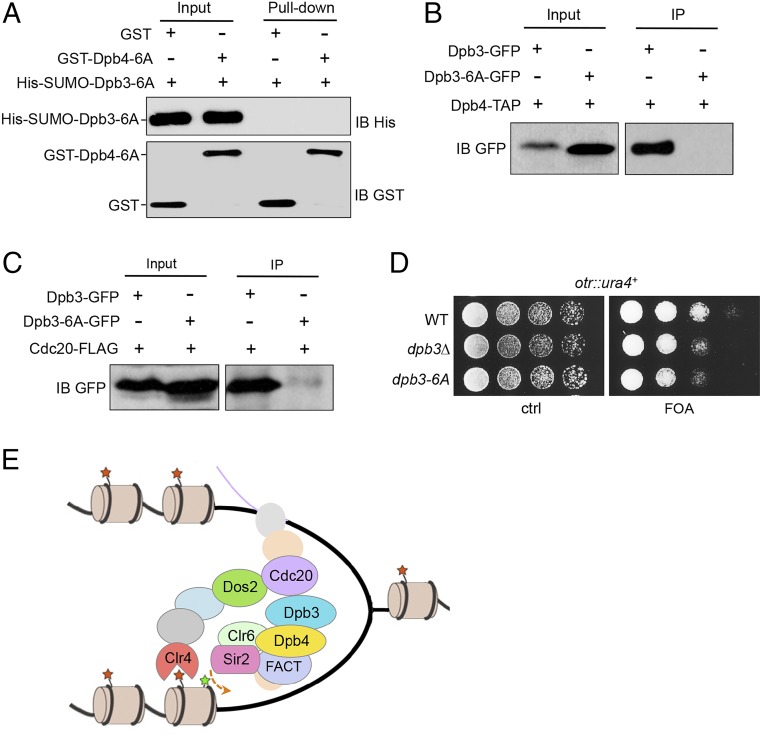
To disrupt the dimerization of Dpb3 and Dpb4, a series of residues at the dimer interface were selected for mutagenesis, including Leu10, Ile18, Leu35, Phe43, Leu47, and Phe80 in Dpb3 and Leu17, Leu30, Phe51, Phe54, Phe88, and Leu92 in Dpb4 (SI Appendix, Fig. S12). We found that the Dpb3-6A mutant (L10A/I18A/L35A/F43A/L47A/F80A) was unable to dimerize with the Dpb4-6A mutant (L17A/L30A/F51A/F54A/F88A/L92A), as shown by the in vitro pull-down assays (Fig. 6A). We then generated a strain expressing Dpb3-6A–GFP and Dpb4–TAP under their endogenous promoters at the native loci. Co-IP experiments showed that Dpb3-6A–GFP was unable to coimmunoprecipitate Dpb4–TAP (Fig. 6B), indicating that the mutations in Dpb3 are sufficient to disrupt the Dpb3–Dpb4 interaction in vivo. Furthermore, Co-IP experiments showed that Dpb3-6A–GFP failed to interact with Cdc20-FLAG (Fig. 6C). Importantly, the dimerization mutant showed the same loss of heterochromatin silencing as dpb3∆ (Fig. 6D), indicating that the dimerization of Dpb3 and Dpb4 is important for heterochromatin assembly.
Fig. 6.
Dimerization of Dpb3 and Dpb4 is essential for heterochromatin silencing. (A) GST pull-down assays demonstrated that physical binding between Dpb3-6A and Dpb4-6A was abolished. Input and pull-down samples were subjected to immunoblotting with anti-His and anti-GST antibodies. (B) Cell lysates from cells expressing Dpb3-6A–GFP and Dpb4–TAP were immunoprecipitated with an antibody specific for TAP and analyzed by immunoblotting using a GFP antibody. Cells expressing Dpb3–GFP and Dpb4–TAP were used as a control. (C) Cell lysates from cells expressing Dpb3-6A–GFP and Cdc20–FLAG were subjected to immunoprecipitation with an antibody specific for FLAG. Precipitated proteins were analyzed by immunoblotting using a GFP antibody. Cells expressing Dpb3–GFP and Cdc20–FLAG were used as a control. (D) Heterochromatin silencing is lost in the dpb3-6A mutant. Serial dilutions of indicated cells carrying ura4+ in the otr region were plated on 5-fluoroorotic acid medium. (E) Model. During DNA replication, the Dpb3–Dpb4 dimer at replication forks recruits histone deacetylases to histones for histone deacetylation (green star), whereas Cdc20 mediates the recruitment of the CLRC complex for H3K9 methylation (red star). Chromatin remodelers, including FACT, CHD1, and SWI/SNF, and histone chaperones facilitate the reassembly of heterochromatin.
Discussion
Here we demonstrated that in fission yeast, Dpb4, the histone-fold subunit of Pol epsilon, interacts with a histone-fold protein, SPCC16C4.22. We further showed that SPCC16C4.22 is the true ortholog of Dpb3/Pole4. Same as DPB3 in budding yeast, SPCC16C4.22 is not essential for viability. The protein forms a stable heterodimeric complex with Dpb4, and physically associates with the Pol epsilon catalytic subunit, Cdc20. Another histone-fold protein SPAC17G8.03c has been previously identified as Dpb3 in fission yeast. However, unlike SPCC16C4.22, SPAC17G8.03c is an essential gene (25). We also were unable to detect the interaction between SPAC17G8.03c and Dpb4 in vitro and in vivo. In fact, we found that SPAC17G8.03c more closely resembles budding yeast BUR6 (40% identity, 65% similarity) and human DR1-associated protein 1 (DRAP1; 38% identity, 61% similarity). BUR6/DRAP1 is a subunit of transcription regulator complex binding to TATA-binding protein to prevent the assembly of the preinitiation complex (34).
Histone hypoacetylation and H3K9 methylation are conserved epigenetic marks essential for heterochromatin structure and function (5). To date, the molecular mechanism underlying the perpetuation of the state of histone hypoacetylation during replication remains elusive. Furthermore, little is known about how the inheritance of these two heterochromatin hallmarks is coordinated through replication. We showed that the Dpb3–Dpb4 complex is important for the recruitment of the histone deacetylase, Sir2, to heterochromatin regions. Cells defective in Dpb3 failed to recruit Sir2 to heterochromatin and display strong silencing defects. Our study thus suggests a mechanism for the inheritance of histone hypoacetylation during replication. In a previous study, we showed that Cdc20, the catalytic subunit of the Pol epsilon complex, regulates the transcription of siRNA precursors and recruitment of the CLRC complex to ensure the inheritance of H3K9 methylation (15). Together, these results uncover a previously unrecognized link between the two conserved heterochromatin marks (Fig. 6E) and provide insight into the mechanism for how the H3K9 methylation and histone deacetylation processes are coupled during DNA replication.
We showed that the Dpb3–Dpb4 complex also interacts with FACT and the CHD1-type remodelers, both of which are required for heterochromatin assembly in fission yeast (28, 29). In budding yeast, the FACT complex has been shown to cooperate with the replisome to reestablish the chromatin state during replication (35). We also found that Dpb4–Dpb3 complex interacts with the SWI/SNF remodeling complex, and that Ssr4 in the complex is essential for heterochromatin silencing. It has been shown recently that the mammalian SWI/SNF-like protein SMARCAD1 in human cells promotes the re-establishment of repressive chromatin during replication by interacting with histone deacetylase 1/2 and G9a/GLP (36). In addition, Dpb4 interacts with the histone chaperones, including Hip1 and Nap1. Hip1 is a subunit of the HIRA complex and has been reported to be important for heterochromatin silencing in fission yeast (37). Nap1 is a well-characterized histone chaperone involved in nucleosome assembly (38).
We reported the first crystal structure of the Dpb3–Dpb4 dimer at 1.9 Å. Our data revealed that Dpb3 and Dpb4 form the H2A–H2B-like dimer. Disruption of Dpb3–Dpb4 dimerization leads to loss of heterochromatin silencing. Our mass spectrometry data showed that Dpb3–Dpb4 associates with histones. The mouse counterpart of Dpb4 has also been found to directly interact with histones (39). It is likely that Dpb3–Dpb4 may bind the histone H3–H4 complex after the disassembly of nucleosomes during DNA replication, and subsequently recruits silencing factors to these histones to mediate histone modifications. Our structural study sheds light on how the complex is involved in chromatin regulation.
Our previously published results (14), combined with the findings presented here, suggest that, in addition to its well-characterized role in DNA replication, the Pol epsilon complex also serves as a platform for recruiting chromatin remodeling factors and modifiers to ensure the faithful inheritance of epigenetic state in heterochromatin. We propose the following model that integrates our current and previous observations (Fig. 6E): During DNA replication, the Dpb3–Dpb4 dimer at replication forks recruits histone deacetylases to facilitate histone deacetylation, whereas Cdc20 is responsible for the recruitment of the CLRC complex for H3K9 methylation. FACT, CHD1, SWI/SNF remodelers and histone chaperones associated with Pol epsilon further promote the reassembly of heterochromatin.
Materials and Methods
Details of the materials and methods used in this study, including media and genetic analysis, mass spectrometry, Co-IP, crystallization, and structure determination; in vitro pull-down assays; mutational studies on dimer interface; fluorescence-activated cell sorting; ChIP; northern blot; and RT-PCR, are provided in SI Appendix, SI Materials and Methods. Fission yeast strains used in this study are listed in SI Appendix, Table S2. Primers used for ChIP and RT-PCR are listed in SI Appendix, Table S3.
Supplementary Material
Acknowledgments
We thank R. Allshire for providing strains; M. Gonzalez and J. Yang for critical reading of the manuscript; the Cold Spring Harbor Laboratory mass spectrometry facility for the mass spectrometry analysis; and J. Qi for data collection at the Shanghai Synchrotron Radiation Facility (BL9U). F.L. is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts. This project was supported in part by NIH Grant R01 GM106037 (to F.L.), National Science Foundation Grant MCB-1330557 (to F.L.), the National Key R&D Program of China 2016YFA0500503 (to Y.-h.C.), the CAS Strategic Priority Research Program XDB08020301 (to Y.-h.C.), the National Natural Science Foundation of China (NSFC) Grant 31470728 (to Y.-h.C.), and the NSFC Grant 31728010 (to F.L. and Y.-h.C).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5Y26 and 5Y27).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712961114/-/DCSupplemental.
References
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 3.Budhavarapu VN, Chavez M, Tyler JK. How is epigenetic information maintained through DNA replication? Epigenetics Chromatin. 2013;6:32. doi: 10.1186/1756-8935-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallace JA, Orr-Weaver TL. Replication of heterochromatin: Insights into mechanisms of epigenetic inheritance. Chromosoma. 2005;114:389–402. doi: 10.1007/s00412-005-0024-6. [DOI] [PubMed] [Google Scholar]
- 5.Dillon N. Heterochromatin structure and function. Biol Cell. 2004;96:631–637. doi: 10.1016/j.biolcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Li F. Are all repeats created equal? Understanding DNA repeats at an individual level. Curr Genet. 2016;63:57–63. doi: 10.1007/s00294-016-0619-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li F, et al. Two novel proteins, dos1 and dos2, interact with rik1 to regulate heterochromatic RNA interference and histone modification. Curr Biol. 2005;15:1448–1457. doi: 10.1016/j.cub.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 9.Jia S, Kobayashi R, Grewal SI. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol. 2005;7:1007–1013. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- 10.Horn PJ, Bastie JN, Peterson CL. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 2005;19:1705–1714. doi: 10.1101/gad.1328005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong EJ, Villén J, Gerace EL, Gygi SP, Moazed D. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2005;2:106–111. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- 12.Thon G, et al. The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics. 2005;171:1583–1595. doi: 10.1534/genetics.105.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 14.Li F, Martienssen R, Cande WZ. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature. 2011;475:244–248. doi: 10.1038/nature10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaratiegui M, et al. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature. 2011;479:135–138. doi: 10.1038/nature10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motamedi MR, et al. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 17.Shankaranarayana GD, Motamedi MR, Moazed D, Grewal SI. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol. 2003;13:1240–1246. doi: 10.1016/s0960-9822(03)00489-5. [DOI] [PubMed] [Google Scholar]
- 18.Grewal SI, Bonaduce MJ, Klar AJ. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics. 1998;150:563–576. doi: 10.1093/genetics/150.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freeman-Cook LL, et al. Conserved locus-specific silencing functions of Schizosaccharomyces pombe sir2+ Genetics. 2005;169:1243–1260. doi: 10.1534/genetics.104.032714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 21.Asturias FJ, et al. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat Struct Mol Biol. 2006;13:35–43. doi: 10.1038/nsmb1040. [DOI] [PubMed] [Google Scholar]
- 22.Araki H, et al. Cloning DPB3, the gene encoding the third subunit of DNA polymerase II of Saccharomyces cerevisiae. Nucleic Acids Res. 1991;19:4867–4872. doi: 10.1093/nar/19.18.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohya T, Maki S, Kawasaki Y, Sugino A. Structure and function of the fourth subunit (Dpb4p) of DNA polymerase epsilon in Saccharomyces cerevisiae. Nucleic Acids Res. 2000;28:3846–3852. doi: 10.1093/nar/28.20.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iida T, Araki H. Noncompetitive counteractions of DNA polymerase epsilon and ISW2/yCHRAC for epigenetic inheritance of telomere position effect in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:217–227. doi: 10.1128/MCB.24.1.217-227.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spiga MG, D’Urso G. Identification and cloning of two putative subunits of DNA polymerase epsilon in fission yeast. Nucleic Acids Res. 2004;32:4945–4953. doi: 10.1093/nar/gkh811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arents G, Moudrianakis EN. The histone fold: A ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc Natl Acad Sci USA. 1995;92:11170–11174. doi: 10.1073/pnas.92.24.11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Pursell ZF, Linn S. Identification and cloning of two histone fold motif-containing subunits of HeLa DNA polymerase epsilon. J Biol Chem. 2000;275:31554. [PubMed] [Google Scholar]
- 28.Lejeune E, et al. The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi. Curr Biol. 2007;17:1219–1224. doi: 10.1016/j.cub.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jae Yoo E, et al. Hrp3, a chromodomain helicase/ATPase DNA binding protein, is required for heterochromatin silencing in fission yeast. Biochem Biophys Res Commun. 2002;295:970–974. doi: 10.1016/s0006-291x(02)00797-0. [DOI] [PubMed] [Google Scholar]
- 30.Tsubota T, Maki S, Kubota H, Sugino A, Maki H. Double-stranded DNA binding properties of Saccharomyces cerevisiae DNA polymerase epsilon and of the Dpb3p-Dpb4p subassembly. Genes Cells. 2003;8:873–888. doi: 10.1046/j.1365-2443.2003.00683.x. [DOI] [PubMed] [Google Scholar]
- 31.Romier C, Cocchiarella F, Mantovani R, Moras D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J Biol Chem. 2003;278:1336–1345. doi: 10.1074/jbc.M209635200. [DOI] [PubMed] [Google Scholar]
- 32.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 33.Dang W, Kagalwala MN, Bartholomew B. The Dpb4 subunit of ISW2 is anchored to extranucleosomal DNA. J Biol Chem. 2007;282:19418–19425. doi: 10.1074/jbc.M700640200. [DOI] [PubMed] [Google Scholar]
- 34.Kim S, Na JG, Hampsey M, Reinberg D. The Dr1/DRAP1 heterodimer is a global repressor of transcription in vivo. Proc Natl Acad Sci USA. 1997;94:820–825. doi: 10.1073/pnas.94.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foltman M, et al. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep. 2013;3:892–904. doi: 10.1016/j.celrep.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 36.Rowbotham SP, et al. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol Cell. 2011;42:285–296. doi: 10.1016/j.molcel.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 37.Blackwell C, et al. The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres. Mol Cell Biol. 2004;24:4309–4320. doi: 10.1128/MCB.24.10.4309-4320.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park YJ, Luger K. The structure of nucleosome assembly protein 1. Proc Natl Acad Sci USA. 2006;103:1248–1253. doi: 10.1073/pnas.0508002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolognese F, Imbriano C, Caretti G, Mantovani R. Cloning and characterization of the histone-fold proteins YBL1 and YCL1. Nucleic Acids Res. 2000;28:3830–3838. doi: 10.1093/nar/28.19.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.