ABSTRACT
Intraflagellar transport (IFT) particles are composed of polyprotein complexes IFT-A and IFT-B as well as cargo adaptors such as the BBSome. Two IFT-B subunits, IFT25 and IFT27 were found to form a heterodimer, which is essential in exporting the BBSome out of the cilium but not involved in flagellar assembly and cytokinesis in vertebrates. Controversial results were, however, recorded to show that defects in IFT, flagellar assembly and even cytokinesis were caused by IFT27 knockdown in Chlamydomonas reinhardtii. Using C. reinhardtii as a model organism, we report that depletion of IFT25 has no effect on flagellar assembly and does not affect the entry of the BBSome into the flagellum, but IFT25 depletion did impair BBSome movement out of the flagellum, clarifying the evolutionally conserved role of IFT25 in regulating the exit of the BBSome from the flagellum cross species. Interestingly, depletion of IFT25 causes dramatic reduction of IFT27 as expected, which does not cause defects in flagellar assembly and cytokinesis in C. reinhardtii. Our data thus support that Chlamydomonas IFT27, like its vertebrate homologues, is not involved in flagellar assembly and cytokinesis.
KEY WORDS: Chlamydomonas reinhardtii, Intraflagellar transport, IFT protein, Flagellar assembly, BBSome
Summary: Chlamydomonas IFT25 is not involved in flagellar assembly, but is required for export of the BBSome from the flagellum.
INTRODUCTION
Cilia and flagella project from the surface of most eukaryotic cells in interphase and share the same organelle structure that consists of a microtubule extension called axoneme surrounded by a specialized plasma membrane (Dutcher, 2003; Rosenbaum and Witman, 2002). Although these organelles differ in performing signaling and motility-based functions they are assembled by an evolutionally conserved process called intraflagellar transport (IFT), where the linear IFT trains of large protein complexes undergo robust bidirectional motility along the axoneme (Kozminski et al., 1993; Pigino et al., 2009). During IFT, IFT trains are carried to the flagellar tip by the heterotrimeric kinesin-2 motor and back to the cell body by the cytoplasmic dynein-1b motor (Cole et al., 1998; Pazour et al., 1999, 1998; Porter et al., 1999; Rosenbaum and Witman, 2002). IFT trains are composed of IFT particles and adaptor complexes like the BBSome. IFT particles consist of two different complexes, called IFT-A and IFT-B (Cole et al., 1998; Follit et al., 2009; Ou et al., 2007). The function of IFT-A and -B has been dissected to be responsible for retrograde and anterograde IFT, respectively (Iomini et al., 2001; Piperno et al., 1998; Tsao and Gorovsky, 2008). As a result, mutations in IFT-B subunits and the kinesin motor typically block the anterograde IFT, resulting in loss of flagella (Brazelton et al., 2001; Brown et al., 2015; Deane et al., 2001; Hou et al., 2007; Kobayashi et al., 2007; Miller et al., 2005; Mueller et al., 2005; Pazour et al., 2000); whereas defects in IFT-A subunits and the dynein motors typically hamper the retrograde IFT, leading to the production of stumpy flagella that accumulates materials in flagella (Bizet et al., 2015; Engel et al., 2012; Hou et al., 2004; Iomini et al., 2009; Lin et al., 2013; Niwa, 2016; Pazour et al., 1999, 1998; Piperno et al., 1998; Porter et al., 1999). The BBSome is characteristic of an IFT cargo adaptor and null mutants lacking individual BBS proteins hardly disrupt IFT and flagellar assembly but cause accumulations of abnormal membrane-associated proteins in the flagellum of C. reinhardtii (Lechtreck et al., 2009a) or defects in promoting ciliary targeting of membrane proteins in mammals (Berbari et al., 2008; Jin et al., 2010). Although how IFT-A, IFT-B and the BBSome interact to assemble functional IFT trains remains largely unknown, recent studies have shown that the IFT-B subunit IFT74 is required for the coupling between IFT-A and IFT-B, at least in C. reinhardtii (Brown et al., 2015), and a second IFT-B subunit, the small GTPase IFT27, plays a role in linking the BBSome to IFT-B as found in the mouse model (Eguether et al., 2014; Liew et al., 2014).
Among the 16 IFT-B particle proteins identified thus far, two IFT-B subunits, IFT25 (Follit et al., 2009; Keady et al., 2012; Lechtreck et al., 2009b; Wang et al., 2009) and the small Rab-like GTPase IFT27 (Qin et al., 2007), are unique in that the two proteins are conserved in vertebrates C. reinhardtii and Trypanosoma brucei, but lack orthologues in some ciliated organisms such as Caenorhabditis elegans and Drosophila (Aldahmesh et al., 2014; Eguether et al., 2014; Follit et al., 2009; Huet et al., 2014; Iomini et al., 2009; Keady et al., 2012; Lechtreck et al., 2009b; Liew et al., 2014; Qin et al., 2007; Wang et al., 2009). Both proteins differ from other conventional IFT-B subunits in that depletion of either of two proteins in mouse or mammalian cells blocked the export of the BBSome out of the cilium but did not cause defects in flagellar assembly (Eguether et al., 2014; Keady et al., 2012; Liew et al., 2014). This is easy to understand because mammalian IFT25 acts as a binding partner of IFT27 and is essential to maintain the stability of IFT27 (Eguether et al., 2014; Keady et al., 2012; Liew et al., 2014). As a result, knockout of IFT25 resulted in almost complete loss of IFT27 and eventually caused the same phenotype as that caused by IFT27 knockout (Eguether et al., 2014; Keady et al., 2012; Liew et al., 2014). Interestingly, controversial results were recorded in the literature that knockdown of IFT27 caused the dissociation of IFT particles, loss of flagella and even defects in cytokinesis in C. reinhardtii (Qin et al., 2007), or led to failure to import IFT-A and IFT dynein into flagella in T. brucei (Huet et al., 2014). Although the underlying molecular mechanisms seem different, both cases gained a common outcome that loss of IFT27 causes defects in IFT and flagellar assembly. Taken together, these results suggest that IFT25 and IFT27 probably play a role in IFT and flagellar assembly in a species-dependent manner (Eguether et al., 2014; Huet et al., 2014; Keady et al., 2012; Liew et al., 2014; Qin et al., 2007). Chlamydomonas IFT25 was also proven to be essential to maintain the stability of IFT27 (Bhogaraju et al., 2011) which is supposed to cause the same defects in IFT, flagellar assembly and cytokinesis as that caused by IFT27 knockdown (Qin et al., 2007), and depletion of Chlamydomonas IFT25 thus is supposed to cause depletion of IFT27. However, it was noted that the specificity of the IFT27 knockdown phenotype was not proven in C. reinhardtii in the previous study, as a strict functional rescue assay was not performed and an off-target effect thus cannot be excluded (Qin et al., 2007). Therefore, for the first time, we aim to clarify the role of IFT25 in IFT and flagellar assembly in C. reinhardtii to investigate if the role of IFT25 in IFT and flagellar assembly was evolutionally conserved or species specific.
RESULTS
IFT25 has a similar cellular distribution pattern as IFT proteins and undergoes IFT in flagella
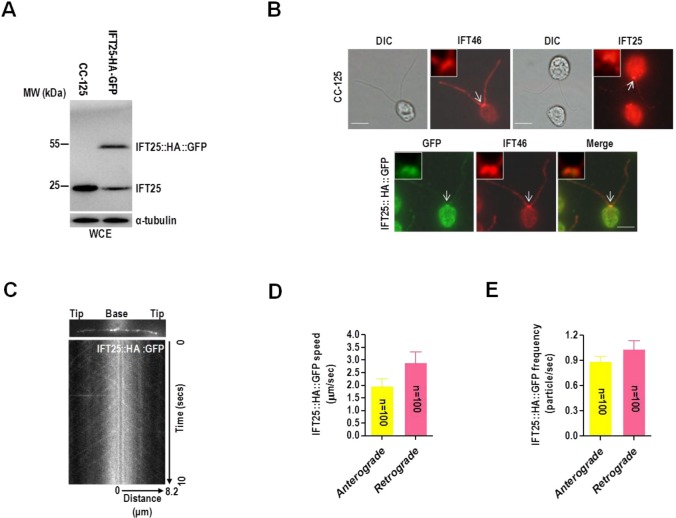
Our previous study showed that IFT25 has spotted distribution along the flagellum, thus showing as a typical pattern of IFT protein (Wang et al., 2009). However, its localization pattern in the basal body is unique in that IFT25 sits right above the basal body but not inside of it, as shown by other IFT-B subunits (Wang et al., 2009). This was probably caused by the poor quality of the anti-IFT25 antibody used. In this study, a full-length C. reinhardtii IFT25 was expressed, purified, and used to produce antisera in two rabbits (Fig. S1A,B). Western blotting assay showed that the anti-IFT25 antibody detected one single band of approximately 20 kDa in wild-type cells (Fig. 1A). One additional fusion protein band with a predicted size of approximately 50 kDa was also detected by the same antibody in a transgenic strain expressing IFT25::HA::GFP (Fig. 1A). The transgenic stain remained totally IFT25 (the native IFT25 plus the IFT25::HA::GFP fusion protein), the same as wild-type cells, indicating that the level of IFT25 in wild-type cells is probably tightly controlled.
Fig. 1.
IFT25 localizes in the basal body and flagella and undergoes IFT in flagella. (A) Western blots of wild-type CC-125 cells and a GFP-expressing CC-125 transgenic strain with antibodies to IFT25 and α-tubulin. WCE, whole cell extracts. (B) Immunofluorescence images of wild-type CC-125 and IFT25::HA::GFP strains. As detected in wild-type CC-125 cells, IFT25 has a similar basal body and flagella localization pattern as IFT46 (upper panel). When checked in the IFT25-HA-GFP transgenic cells, IFT25-HA-GFP and IFT46 co-localized at the basal bodies and flagella (lower panel) (green, α-GFP; red, α-IFT25 and α-IFT46). DIC, phase contrast photos. White arrows indicate the basal body to be enlarged in the top-left corner of each picture. Scale bars: 5 µm. (C) Single frame and kymograph from TIRF microscopy video imaging of IFT25::HA::GFP transgenic strain (Movie 1, ∼10 fps). (D) The speeds of anterograde and retrograde IFT25::HA::GFP in the IFT25::HA::GFP transgenic strain. Anterograde IFT25::HA::GFP speed is 1.97±0.28 μm/s (n=100, P=0.037) and retrograde IFT25::HA::GFP speed is 2.90±0.43 μm/s (n=100, P=0.052). Speeds are mean values±95% confidence interval and P-values were calculated from a student's t-test. (E) The frequencies of anterograde and retrograde IFT25::HA::GFP in the IFT25::HA::GFP transgenic strain. The anterograde frequency of IFT25::HA::GFP is 0.89±0.06 particle/sec (n=100, P=0.042) and its retrograde frequency is 1.04±0.10 particle/sec (n=100, P=0.028). Frequencies are mean values±95% confidence interval and P-values were calculated from a student's t-test. In D and E, data are presented as mean±s.d.
In wild-type cells, the anti-IFT25 antibody detected signals along the flagellum (Fig. 1B). At the basal body region, IFT25 had a distribution pattern similar to IFT46 (Fig. S2A,B for anti-IFT46 antiserum generation) as a band perpendicular to each basal body (Fig. 1B). To confirm this observation, the transgenic strain expressing IFT25::HA::GFP was applied to determine the cellular localization pattern of the native IFT25 by using a monoclonal anti-GFP antibody. Immunofluorescence staining showed that IFT25::HA::GFP exhibited a similar localization as IFT46, with the presence of the proteins in the basal body and inside flagella (Fig. 1B). As a result, both the native and fusion IFT25 proteins thus showed typical IFT protein distribution pattern in the basal body and flagella. In addition, total internal reflection fluorescence microscopy (TIRFM) analysis performed on living IFT25::HA::GFP transgenic cells showed that IFT25::HA::GFP underwent typical bidirectional IFT (Movie 1), similar to IFT trafficking observed for C. reinhardtii IFT27::GFP (Qin et al., 2007) and KAP (Mueller et al., 2005). Once kymographs (Fig. 1C) were made to quantify IFT train speed the mean anterograde speed was determined to be 1.97±0.28 μm/s (n=100) and mean retrograde speed was measured at 2.90±0.43 μm/s (n=100) (Fig. 1D). In addition, the mean frequencies of anterograde and retrograde IFT25::HA::GFP were measured to be 0.89±0.06 particle/sec (n=100) and 1.04±0.10 particle/sec (n=100), respectively, also similar to the frequencies observed for IFT56-GFP (Ishikawa et al., 2014) (Fig. 1E). Overall, these results show that IFT25 distribution and movement in flagella are similar to the other IFT proteins studied to date in C. reinhardtii, and the N-terminal GFP-tagged IFT25 was functional in IFT and flagellar assembly and can be used to replace the native IFT25.
IFT25 and IFT27 form a heterodimer to exist both in IFT-B and outside of it in vivo
Our previous study had shown that IFT25 and IFT27 co-sediment with IFT-B proteins in sucrose density gradients of both the whole cell and flagellar extracts of C. reinhardtii (Wang et al., 2009). Different from other IFT-B complex proteins, the majority of IFT25 and IFT27 are also observed to appear in a discrete IFT25- and IFT27-containing small peak in sucrose density gradients of both whole cell and flagellar extracts of C. reinhardtii, and the two proteins physically interact with each other (Wang et al., 2009). To determine if there were any other components than IFT25 and IFT27 in the small complex in vivo, we purified IFT25- and IFT27-associated protein complexes from whole cell extracts of two transgenic strains expressing either IFT27::HA::6×His or IFT25::HA::6×His. To detect the appearance of IFT27 in the complex by western blotting assay, the N-terminal 18 amino acids of C. reinhardtii IFT27 were synthesized and used to produce antisera in two rabbits (Fig. S3). Gel filtration chromatography of the eluate from the Ni-NTA resin showed cofractionation of IFT27::HA::6×His and IFT25 or IFT25::HA::6×His and IFT27 with a molecular weight of 60 kDa, indicating that the small complex only contains IFT25 and IFT27 as its protein components (Fig. 2A,B). Based on this observation, we conclude, along with our previous result (Wang et al., 2009), that IFT25 and IFT27 form a heterodimeric complex we called IFT25/27 that exists both within the IFT-B complex and outside of it in vivo.
Fig. 2.
IFT25 and IFT27 form an IFT-A- and IFT-B-independent heterodimer in vivo. Ni-NTA resin elute from whole cell lysates of the IFT25::HA::6×His (A) and IFT27::HA::6×His (B) transgenic strains were fractionated by size exclusion chromatography (S200 sizing column). As detected by western blotting, IFT25::HA::6×His and IFT27 cofractionated at ∼60 kDa, independent of IFT-A protein IFT139 and IFT-B protein IFT46 (A) and IFT27::HA::6×His and IFT25 also cofractionated at ∼60 kDa, independent of IFT139 and IFT46 (B). For both panels, relative total protein concentration of the fractions is shown as absorbance at 280 nm. The elution volume of one standard marker (60 kDa) is indicated on the western blots. WCE, whole cell extracts.
IFT25 does not affect the stability of IFT-A and -B complexes
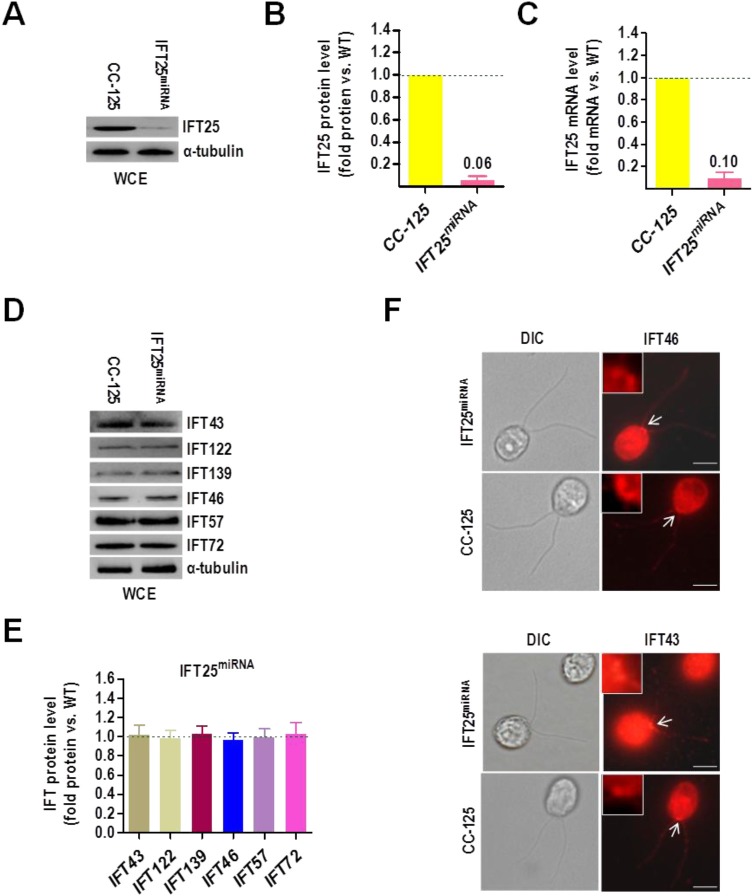
Each IFT-B complex protein contributes to the stability of IFT-B complex. Loss of a single IFT-B complex protein typically causes partial or complete dissociation of IFT-B complex, depending on which protein is depleted (Brazelton et al., 2001; Fan et al., 2010; Hou et al., 2007; Pazour et al., 2000; Qin et al., 2007). To date, IFT27 is the only IFT-B subunit that, once partially depleted by interfering RNA technology, was shown to cause dissociation of both IFT-A and -B complexes in C. reinhardtii (Qin et al., 2007). To test whether IFT25 influences the stability of IFT-A and -B complexes, we created a vector-based IFT25 miRNA strain called IFT25miRNA (Fig. 3A) and measured the protein level of the IFT-B components IFT46, IFT57 (Fig. S4A,B for anti-IFT57 antiserum generation) and IFT72 (Fig. S5A,B for anti-IFT72 antiserum generation) and the IFT-A components IFT43, IFT122 (Fig. S6 for anti-IFT122 antiserum generation) and IFT139 (Fig. S7A,B for anti-IFT139 antiserum generation) in these cells. Western blotting assay showed that IFT25miRNA cells experienced reductions of IFT25 to ∼6% of wild-type level (Fig. 3B). The IFT25 mRNA of IFT25miRNA cells was reduced to ∼10% of wild type (Fig. 3C), indicating that depletion of IFT25 protein was caused by miRNA interference. However, IFT25miRNA cells did not see a significant depletion of IFT-A and -B proteins (Fig. 3D,E), revealing that IFT25 has no influence on the stability of IFT-A and -B complexes, thus consistent with the observation that knockout of IFT25 in mouse largely does not affect the stability of IFT particle proteins (Eguether et al., 2014; Keady et al., 2012). In addition, the IFT-A protein IFT43 and the IFT-B protein IFT46 of the IFT25miRNA cells, similar to that of the wild-type CC-125 cells, showed typical basal body and flagellar distribution patterns, suggesting that IFT25 depletion does not affect the localization of the IFT-A and -B complexes (Fig. 3F).
Fig. 3.
IFT25 does not affect the stability of IFT-A and -B complexes. (A) Western blots comparing the cellular levels of IFT25 in wild-type CC-125 and IFT25miRNA cells. Alpha-tubulin was used as a loading control. WCE, whole cell extracts. (B) IFT25 protein levels were normalized to the α-tubulin housekeeping protein and presented as percentage-change relative to wild-type CC-125 IFT25. The data shown was calculated from three western blots (n=3 repeats). Error bars indicate s.d. As calculated, IFT25miRNA cells contained approximately 6% as much IFT25 as CC-125 cells. (C) Real-time PCR comparing the IFT25 mRNA in wild-type CC-125 and IFT25miRNA cells. Transcript levels were normalized to the GBLP housekeeping gene and presented as percentage-change relative to wild-type CC-125 mRNA. The data shown was calculated from three independent assays (n=3 repeats). Error bars indicate s.d. As calculated, IFT25miRNA cells contained approximately 10% as much IFT25 mRNA as CC-125 cells. (D) Western blots of wild-type CC-125 and IFT25miRNA whole cell extracts with antibodies against the IFT-A proteins IFT43, IFT122 and IFT139 and the IFT-B proteins IFT46, IFT57 and IFT72 as indicated. Alpha-tubulin was used as a loading control. WCE, whole cell extracts. (E) Protein levels of the IFT-A proteins IFT43, IFT122 and IFT139 and the IFT-B proteins IFT46, IFT57 and IFT72 were normalized to the α-tubulin housekeeping protein and presented as percentage-change relative to wild-type CC-125 counterparts. As calculated, both IFT-A and IFT-B components remained unchanged in IFT25miRNA as compared to CC-125 cells. The data shown was calculated from three western blots (n=3 repeats). Error bars indicate s.d. (F) Immunofluorescence images of wild-type CC-125 and IFT25miRNA cells. As detected in both wild-type CC-125 and IFT25miRNA cells, the IFT-A protein IFT43 and the IFT-B protein IFT46 both show a typical basal body and flagella localization pattern. DIC, phase contrast photos. White arrows indicate the basal body to be enlarged in the top-left corner of each picture. Scale bars: 5 µm.
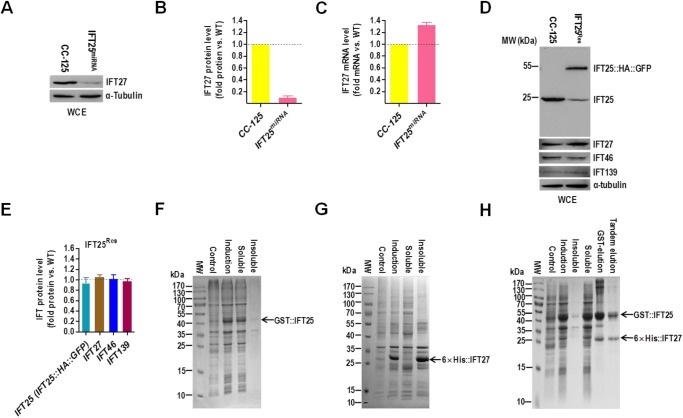
IFT25 is essential to maintain IFT27 stability in vivo
To investigate the influence of IFT25 on IFT27 stability, we measured the protein and transcript levels of IFT27 in the vector-based IFT25 miRNA strain (IFT25miRNA). IFT25 depletion in IFT25miRNA cells caused a dramatic reduction of IFT27 protein to a level of as low as ∼7% of wild-type level (Fig. 4A,B), and a significant increase in IFT27 mRNA to a level of as high as ∼133% of wild-type level (Fig. 4C), indicating that cells respond to the loss of IFT27 protein by increasing IFT27 transcript level. In addition, expression of the IFT25::HA::GFP transgene in IFT25miRNA cells did not cause change on IFT46 and IFT139 protein levels (strain IFT25Res) but IFT27 protein abundance was restored upon rescue of IFT25 (IFT25::HA::GFP) expression in the IFT25Res strain (Fig. 4D,E). After continuously passaging IFT25miRNA for six months, the recovered IFT25Rec strain maintained wild-type levels of IFT25, IFT46 and IFT139 proteins as well as IFT27 (Fig. S8A,B). Thus, the stability of IFT27 protein strictly relies on its partner protein IFT25 in vivo. By performing an in vitro experiment, we found that an N-terminal GST-tagged IFT25 (GST::IFT25) was completely soluble when expressed in bacteria alone (Fig. 4F). However, N-terminal 6×His-tagged IFT27 (6×His::IFT27) was almost completely insoluble when expressed alone in bacteria (Fig. 4G). In contrast, when 6×His::IFT27 and GST::IFT25 were co-expressed in bacteria, the majority of both proteins were present in the soluble fraction after tandem purification (Fig. 4H). By having these in vitro and in vivo results, we therefore conclude that IFT25 is an obligate binding partner of IFT27 and protects IFT27 from aggregation through a direct interaction (also see Bhogaraju et al., 2011).
Fig. 4.
IFT25 is essential to maintain IFT27 stability in vivo. (A) Western blots comparing the cellular levels of IFT27 in wild-type CC-125 and IFT25miRNA cells. (B) IFT27 protein levels were normalized to the housekeeping protein α-tubulin and presented as percentage-change relative to wild-type CC-125 IFT27. The data shown was calculated from three independent assays (n=3 repeats). Error bars indicate s.d. As calculated, IFT25miRNA cells contained approximately 7% as much IFT27 as CC-125 cells. (C) Real-time PCR comparing the IFT27 mRNA in wild-type CC-125 and IFT25miRNA cells. Transcript levels were normalized to the GBLP housekeeping gene and presented as percentage-change relative to wild-type CC-125 mRNA. As calculated, IFT25miRNA cells contained approximately 133% as much IFT27 mRNA as CC-125 cells. The data shown was calculated from three independent assays (n=3 repeats). Error bars indicate s.d. (D) Western blots of wild-type CC-125 and IFT25Res whole cell extracts with antibodies to IFT25, IFT27, IFT46 and IFT139. (E) IFT proteins including IFT25, IFT27, IFT46 and IFT139 in IFT25Res cells were normalized to the housekeeping protein α-tubulin and presented as percentage-change relative to their wild-type CC-125 counterparts. When IFT25 protein (IFT25 plus IFT25::HA::GFP) level was rescued in the IFT25Res strain IFT27 level was also rescued in these cells. IFT46 and IFT139 remained unchanged in IFT25Res cells as compared to CC-125. The data shown was calculated from three western blots (n=3 repeats). Error bars indicate s.d. (F-H) Coomassie Blue-stained gels, showing that bacterial-expressed recombinant GST::IFT25 is mostly soluble (F), bacterial-expressed recombinant 6×His::IFT27 is mostly insoluble (G) and co-expression of 6×His::IFT27 and GST::IFT25 is primarily present in the soluble fraction (H). For all panels, the lanes indicated as ‘Control’ and ‘Induction'contained the bacterial lysates before and after IPTG induction, respectively. The lanes labeled with ‘Insoluble’ and ‘Soluble’ are the insoluble and soluble fractions recovered after centrifugation of cell lysates. The soluble fractions were further purified by glutathione-agarose resin (F), Ni-NTA resin (G) or ‘tandem’ purification with both resins (H). For panels A and D, alpha-tubulin was used as a western blot loading control. WCE, whole cell extract.
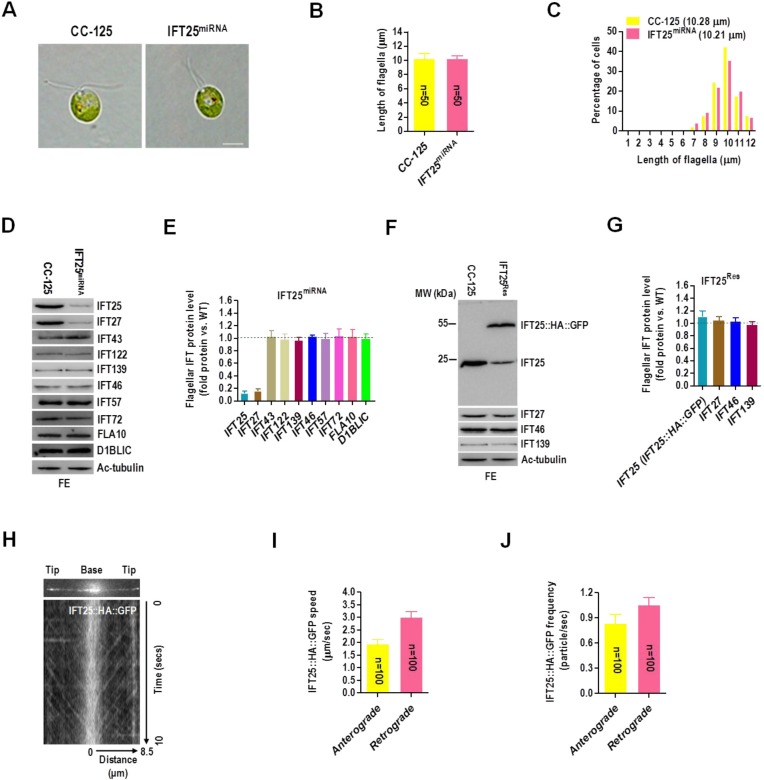
IFT25 is not required for flagellar assembly
Given that depletion of IFT25 has no effect on IFT-A and -B stability, we wondered whether depletion of IFT25 could cause defects in IFT and flagellar assembly. To test this hypothesis, we examined the IFT25miRNA knockdown strain and found that the majority of IFT25miRNA cells were individual cells with wild-type swimming behavior (Fig. 5A). IFT25miRNA cells had flagella with normal length (mean length=10.21 µm) as compared to the wild-type cells (mean length=10.28 µm) (Fig. 5B) and the flagella length distribution of IFT25miRNA cells was also similar to that of CC-125 cells (Fig. 5C). We next checked the levels of IFT25 and IFT27 as well as the IFT-A proteins IFT43, IFT122 and IFT139, the IFT-B proteins IFT46, IFT57 and IFT72, the kinesin subunit FLA10 (Fig. S9 for anti-FLA10 antiserum generation) and the dynein subunit D1BLIC (Fig. S10 for anti-D1BLIC antiserum generation) in flagella isolated from IFT25miRNA cells. When gel loading was normalized for equal Ac-tubulin, the strain showed the same levels of IFT-A, IFT-B and motor proteins as wild-type flagella (Fig. 5D,E). As expected, the amount of IFT25 and IFT27 both dramatically decreased in IFT25miRNA flagella to a level of as low as ∼13% and 17% of wild-type level (Fig. 5D,E). Since the C-terminal HA- and GFP-tagged IFT25 (IFT25::HA::GFP) was functional to participate in IFT (Fig. 1), IFT25::HA::GFP was then used to rescue the authentic IFT25 in IFT25Res strain. When IFT25 (IFT25::HA::GFP) protein level was rescued in the IFT25miRNA strain, the levels of IFT25 (IFT25::HA::GFP) and IFT27 in IFT25Res flagella were determined to be the same as that of the wild-type flagella (Fig. 5F,G). As expected, the recovered strain IFT25Rec maintained wild-type level of IFT25, IFT46 and IFT139 proteins as well as IFT27 in flagella (Fig. S11A,B). TIRFM analysis performed on living IFT25Res cells showed that IFT25::HA::GFP underwent bidirectional IFT movements similar to that in the IFT25::HA::GFP transgenic cells (see Movie 2). After the kymograph was made to quantify IFT train speed (Fig. 5H) the mean anterograde speed was measured at 1.93±0.19 μm/s (n=100) and mean retrograde speed was calculated as 3.01±0.23 μm/s (n=100) (Fig. 5I). The mean frequencies of anterograde and retrograde IFT25::HA::GFP were measured to be 0.83±0.11 particle/sec and 1.05±0.09 particle/sec, respectively (Fig. 5J), also similar to the frequencies observed for IFT25::HA::GFP under a CC-125 background (Fig. 1E). These observations showed that depletion of IFT25 does not affect IFT protein content (except for IFT27) in flagella and flagella length, suggesting that IFT25 is not required for flagellar assembly in C. reinhardtii.
Fig. 5.
C. reinhardtii IFT25 is not required for flagella assembly. (A) DIC images of wild-type CC-125 and IFT25miRNA cells. Scale bars: 5 µm. (B) CC-125 and IFT25miRNA cells had a mean flagella length of 10.28 µm and 10.21 µm, respectively. The data shown was calculated from 50 flagella (n=50 repeats). Error bars indicate s.d. (C) Histograms showing that IFT25miRNA cells had a similar flagella length distribution pattern as CC-125 strain. (D) Western blots of wild-type CC-125 and IFT25miRNA flagellar extracts with antibodies listed on the right. (E) The flagella of IFT25miRNA cells contained approximately 13% and 17% as much IFT25 and IFT27 as flagella of CC-125 cells. (F) Western blots of wild-type CC-125 cells and IFT25Res flagella extracts with antibodies listed on the right. (G) When IFT25 protein (IFT25 plus IFT25::HA::GFP) level was rescued in the IFT25Res strain IFT27 level was also rescued in these cells. IFT46 and IFT139 remained unchanged in IFT25Res flagella as compared to CC-125. (H) Single frame and kymograph from TIRF microscopy video imaging of IFT25Res transgenic strain (Movie 2, ∼10 fps). (I) The speeds of anterograde and retrograde IFT25::HA::GFP in the IFT25::HA::GFP transgenic strain. Anterograde IFT25::HA::GFP speed is 1.93±0.19 μm/s (n=100, P=0.033) and retrograde IFT25::HA::GFP speed is 3.01±0.23 μm/s (n=100, P=0.025). Speeds are mean values±95% confidence interval and P-values were calculated from a student's t-test. (J) The frequencies of anterograde and retrograde IFT25::HA::GFP in the IFT25Res transgenic strain. The anterograde frequency of IFT25::HA::GFP is 0.83±0.11 particle/sec (n=100, P=0.057) and its retrograde frequency is and 1.05±0.09 particle/sec (n=100, P=0.089). Frequencies are mean values±95% confidence interval and P-values were calculated from a student's t-test. For panels D and F, acetylated α-tubulin (Ac-tubulin) was used as a western blot loading control. FE, flagellar extract. For panels E and G, IFT proteins including IFT25, IFT27, the IFT-B component IFT46 and the IFT-A component IFT139 in IFT25Res flagella were normalized to the Ac-tubulin housekeeping protein and presented as percentage-change relative to their wild-type CC-125 counterparts. IFT46 and IFT139 remained unchanged in flagella of IFT25res cells as compared to that of CC-125. The data shown was calculated from three western blots (n=3 repeats). Error bars indicate s.d.
IFT25 is required for the export of the BBSome from flagella
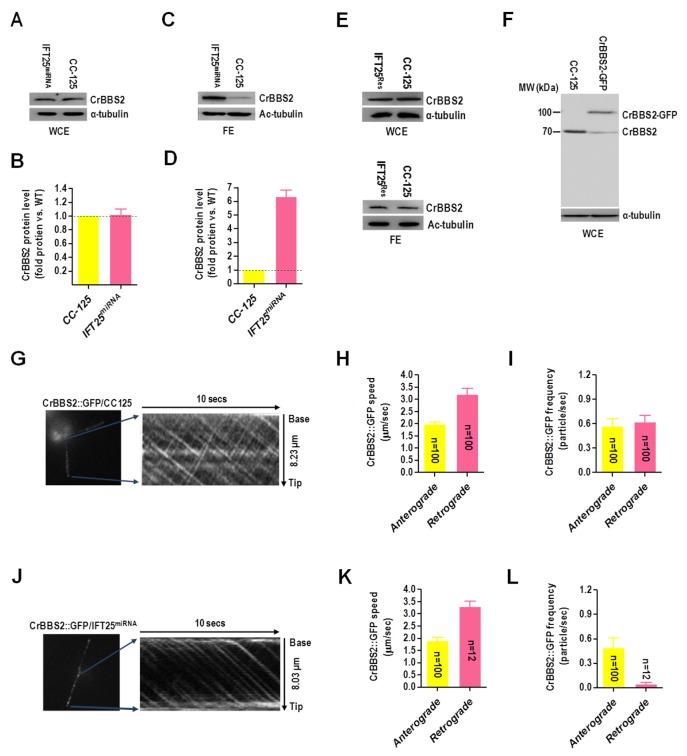
By generating a polyclonal antibody against the BBSome subunit, CrBBS2 (Fig. S12A,B), we identified that IFT25miRNA cells contained CrBBS2 at a similar level as wild-type cells (Fig. 6A,B), indicating that IFT25 had no effect on the BBSome stability. However, the amount of CrBBS2 in the flagellum of IFT25miRNA cells was dramatically increased as compared to that in wild-type cells (Fig. 6C,D). Given that the IFT25-expression recused IFT25Res cells contained CrBBS2 both in whole cell and flagellar extracts at a similar level as wild-type cells (Fig. 6E), these results all together confirmed that the BBSome was accumulated in the flagellum due to IFT25 depletion. To measure its movement in flagella, a transgenic strain expressing CrBBS2::GFP was created (Fig. 6F). TIRFM analysis showed that CrBBS2::GFP fusion protein underwent typical bidirectional IFT (Fig. 6G; Movie 3). The anterograde and retrograde velocities of CrBBS2-GFP were determined to be 1.98±0.10 μm/s (n=100) and 3.21±0.23 μm/s (n=100), respectively, which all seem similar to that of other IFT proteins (Fig. 6H). This observation was in agreement with previous studies that the trafficking of the BBSome in flagella is an IFT-dependent process (Eguether et al., 2014; Lechtreck et al., 2009a; Liew et al., 2014; Ou et al., 2005; Williams et al., 2014). Frequencies of the anterograde and retrograde CrBBS2::GFP were measured to be 0.57±0.09 particle/sec (n=100) and 0.62±0.08 particle/sec (n=100), respectively (Fig. 6I). This was also in agreement with the previous observation showing that not all IFT particles were loaded with the BBSome complex in the flagellum of C. reinhardtii (Lechtreck et al., 2009a). To clarify if depletion of IFT25 can cause uncoupling of the BBSome from the IFT machinery a transgenic strain expressing CrBBS2::GFP fusion protein was created on an IFT25miRNA cell background and the movement of the CrBBS2::GFP in the flagellum was measured by TIRFM. The kymograph showed that CrBBS2::GFP fusion protein underwent typical bidirectional IFT (Fig. 6J; Movie 4). Interestingly, although CrBBS2::GFP moved in an anterograde direction at a velocity of 1.91±0.12 μm/s (n=100), only a few retrograde-trafficking CrBBS2-GFP with normal speed (3.30±0.23 μm/s, n=12) were detected (Fig. 6K), showing that the retrograde movement of the BBSome was obviously impaired without IFT25. In addition, anterograde frequency of CrBBS2::GFP is 0.49±0.12 particle/sec (n=100), however its retrograde frequency is only 0.04±0.02 particle/sec (n=12), indicating that the retrograde CrBBS2-GFP was nearly undetectable (Fig. 6L). Taken together, these observations clearly showed that depletion of C. reinhardtii IFT25 leads to the hyper-accumulation of the BBSome in flagella by affecting its export from flagella.
Fig. 6.
IFT25 is required for the export of the BBSome from flagella. (A) Western blots of wild-type CC-125 and IFT25miRNA whole cell extract with α-CrBBS2. Alpha-tubulin was used as a western blot loading control. WCE, whole cell extract. (B) CrBBS2 protein levels were normalized to the α-tubulin housekeeping protein and presented as percentage-change relative to wild-type CC-125 CrBBS2. The data shown was calculated from three independent assays (n=3 repeats). Error bars indicate s.d. (C) Western blots of wild-type CC-125 and IFT25miRNA flagella extracts with α-CrBBS2. Acetylated α-tubulin (Ac-tubulin) was used as a western blot loading control. FE, flagella extract. (D) CrBBS2 protein levels were normalized to the Ac-tubulin housekeeping protein and presented as percentage change relative to wild-type CC-125 CrBBS2. The data shown was calculated from three independent assays (n=3 repeats). Error bars indicate s.d. (E) Western blots of wild-type CC-125 and IFT25Res whole cell and flagellar extracts with α-CrBBS2. Alpha-tubulin was used as a western blot loading control. WCE, whole cell extract; FE, flagellar extract. (F) Western blots of wild-type CC-125 cells and a CrBBS2-GFP-expressing CC-125 transgenic strain with antibodies to CrBBS2 and α-tubulin. (G) Single frame and kymograph from TIRF microscopy video imaging of CrBBS2::GFP transgenic strain (Movie 3, ∼10 fps). (H) The speeds of anterograde and retrograde CrBBS2::GFP in the CrBBS2::GFP transgenic strain. Anterograde CrBBS2::GFP speed is 1.98±0.10 μm/s (n=100, P=0.062) and retrograde CrBBS2::GFP speed is 3.21±0.23 μm/s (n=100, P=0.078). (I) The frequencies of anterograde and retrograde CrBBS2::GFP in the CrBBS2::GFP transgenic strain. The anterograde frequency of CrBBS2::GFP is 0.57±0.09 particle/sec (n=100, P=0.091) and its retrograde frequency is and 0.62±0.08 particle/sec (n=100, P=0.054). (J) Single frame and kymograph from TIRF microscopy video imaging of CrBBS2::GFP transgenic strain on an IFT25miRNA cell background (Movie 4, ∼10 fps). (K) The mean speeds of anterograde and retrograde CrBBS2::GFP in the CrBBS2::GFP transgenic strain on an IFT25miRNA cell background. Anterograde CrBBS2::GFP speed is 1.91±0.12 μm/s (n=100, P=0.013) and retrograde CrBBS2::GFP speed is retrograde CrBBS2::GFP speed is 3.30±0.23 μm/s (n=12, P=0.096) on an IFT25miRNA cell background. (L) Mean frequencies of anterograde and retrograde CrBBS2::GFP in the IFT25miRNA transgenic strain. The anterograde frequency of CrBBS2::GFP is 0.49±0.12 particle/sec (n=100, P=0.027) and its retrograde frequency is only 0.04±0.02 particle/sec (n=12, P=0.087). For panels H, I, K and L, speeds and frequencies are mean values±95% confidence interval and P-values were calculated from a student's t-test.
DISCUSSION
IFT particles in C. reinhardtii consist of two complexes, IFT-A (at least 6 subunits) and -B (at least 16 subunits) that separate in sucrose gradients (Cole et al., 1998; Fan et al., 2010; Follit et al., 2009; Ishikawa et al., 2014; Lechtreck et al., 2009b; Qin et al., 2007; Wang et al., 2009). Most recently, biochemical and recombinant reconstruction studies further divide IFT-B into two biochemically stable sub-complexes, the core IFT-B1 (IFT88/81/74/70/56/52/46/27/25/22) and the peripheral IFT-B2 (IFT172/80/57/54/38/20) (Katoh et al., 2016; Lucker et al., 2005; Taschner et al., 2016). Both sub-complexes act as potential IFT cargoes to carry tubulin/MT blocks and flagella signaling molecules necessary for the buildup of functional flagella (Taschner et al., 2016). Functional studies performed with a combination of several model organisms have shown that depletion of even a single core IFT-B1 subunit (Brazelton et al., 2001; Brown et al., 2015; Deane et al., 2001; Fan et al., 2010; Hou et al., 2007; Kobayashi et al., 2007; Pazour et al., 2000) can cause the complete disassembly of IFT-B, eventually leading to the occurrence of so-called ‘bald’ mutants. However, this is not always true because the core IFT-B1 subunit IFT56, once depleted, does not cause the instability of IFT-B and severe defects in flagella assembly (only slightly shorter flagella observed for IFT56 mutant) but instead leads to motility-associated defects of flagella in C. reinhardtii (Ishikawa et al., 2014) and hedgehog signaling defects in mouse (Swiderski et al., 2014). The core IFT-B1 subunits IFT25 and IFT27 are also not involved in maintaining the stability of IFT-B and flagellar assembly but required for the exchange of hedgehog signaling molecules and the BBSome between the cell body and flagella as tested in mouse model (Eguether et al., 2014; Keady et al., 2012). Taken together, the data in the literature showed that, even for some core IFT-B1 subunits, they are not structurally required for the maintenance of IFT-B stability and flagellar assembly but rather, more likely, act as IFT cargo proteins to carry flagella signaling molecules.
The IFT-B subunits IFT25 and IFT27 are unique among the IFT-B subunits since two proteins are not conserved in all ciliated organisms, suggesting that they are probably not required for flagellar assembly. Our previous Chlamydomonas study noticed that two proteins exist both in IFT-B complex and outside of it, demonstrating that IFT25 and IFT27 behavior is biochemically different from other IFT-B subunits (Wang et al., 2009). Two subsequent mouse studies showed that depletion of rodent IFT25 or IFT27 alone does not cause disruption of IFT particles and flagellar assembly but results in defects in signal-dependent movement of hedgehog components (Eguether et al., 2014; Keady et al., 2012). In this study, we identified that Chlamydomonas IFT25 and IFT27 form a heterodimeric complex in vivo, which physically exist both in IFT-B complex and outside of it. Both this study and other (Bhogaraju et al., 2011) in vitro studies confirmed that Chlamydomonas IFT25 is an obligate binding partner of IFT27 and its main function appears to protect IFT27 from aggregation. This notion was strengthened when knockdown of Chlamydomonas IFT25 was observed to eventually cause dramatic depletion of IFT27 in vivo. Different from the conventional IFT-B subunits, once depleted, typically causing the occurrence of ‘bald’ mutant, IFT25 depletion does not cause apparent disruption of IFT particles and the disassembly of flagella, indicating that IFT25 is independently structured within IFT-B. IFT25 was then uncoupled from other conventional IFT-B subunits in their function in flagella assembly. Interestingly, partial depletion of Chlamydomonas IFT27 had been reported to cause the instability of both IFT-A and -B and eventually lead to a complete absence of flagella and even defects in cytokinesis, suggesting that IFT27 is a critical factor required for not only IFT and flagella assembly but also for cell cycle control in C. reinhardtii (Qin et al., 2007). This could be true as IFT27 was probably not deprived to a level low enough to cause the reported phenotype in this study. However, ift27 knockout in mouse and cultured cells also did not find its role in controlling IFT, flagella assembly and cell cycle (Eguether et al., 2014; Liew et al., 2014). Other than this, the specificity of the IFT27 knockdown phenotype has not been proven in C. reinhardtii in the previous study as a strict functional rescue assay was not performed and an off-target effect thus cannot be excluded (Qin et al., 2007).
Chlamydomonas and mouse BBSome is not required for IFT and flagellar assembly and its protein subunits are of relatively low abundance in flagella (Eguether et al., 2014; Keady et al., 2012; Lechtreck et al., 2009a). Disruption of the BBSome proteins causes dramatic accumulation of many signaling membrane proteins, indicating its role in coupling specific cargoes to the IFT machinery (Eguether et al., 2014; Keady et al., 2012; Lechtreck et al., 2009a). Mouse model has shown that depletion of IFT25/27 causes accumulation of the BBSome and signaling membrane proteins in the cilium, demonstrating that it is the IFT25/27 that acts as an adaptor between the BBSome and IFT machinery (Eguether et al., 2014; Keady et al., 2012; Liew et al., 2014). In this study, knockdown of Chlamydomonas IFT25 led to depletion of IFT27 and abnormal accumulation of the BBSome proteins in flagella. The BBSome accumulation in flagella is caused by disrupting the movement of the BBSome out of the flagella but not by interrupting the entry of the BBSome into the flagella, suggesting that IFT25/27 is an evolutionally conserved adaptor between the BBSome and IFT machinery across species. Together with the data shown in this study and the data in the literature (Aldahmesh et al., 2014; Eguether et al., 2014; Keady et al., 2012; Lechtreck et al., 2009a; Liew et al., 2014; Wang et al., 2009), we conclude that IFT25 behaves unlike a conventional IFT-B subunit and is dispensable for flagella assembly but required to export the BBSome from flagella in C. reinhardtii. Next we will further investigate if direct depletion of IFT27 can cause phenotype(s) the same as depletion of IFT25 in C. reinhardtii.
MATERIALS AND METHODS
Antibodies
Rabbit-raised polyclonal antibodies are shown in the supplementary Figs S1-7,9,10 and 12. Antibodies against HA (rat 3F10, Roche), GFP (mAbs 7.1 and 13.1, Roche), α-tubulin (mAb B512, Sigma-Aldrich) and Ac-α-tubulin (mAb 6-11B-1, Sigma-Aldrich) were commercially available. Rabbit-raised polyclonal antibody against Chlamydomonas IFT43 was reported previously (Zhu et al., 2017). For immunoblotting analysis, a dilution was used for IFT25 (1:500), IFT27 (1:200), IFT43 (1:1000), IFT46 (1:1000), IFT57 (1:1000), IFT72 (1:1000), IFT122 (1:1000), IFT139 (1:500), FLA10 (1:1000), D1BLIC (1:1000) and CrBBS2 (1:2000).
Strains and culture conditions
C. reinhardtii wild-type strain CC-125 was used throughout this study and was obtained from the Chlamydomonas Genetic Center at the University of Minnesota, Twin Cities, MN, USA (www.chlamy.org). The IFT25 miRNA strain was generated as described below. For experiments, the strains were cultured in liquid or solid TAP medium at 23°C under continuous white light (∼80 μE/m2/s) with constant aeration when desired.
Plasmids
Plasmids for the miRNA experiment were designed following a previously described method (Hu et al., 2014). Briefly, the miRNA targeting the C. reinhardtii IFT25 gene was designed using WMD3 software (http://wmd3.weigelworld.org). The output oligonucleotide was combined with the miRNA cre-MIR1157 (accession number MI0006219), resulting in a 167 bp IFT25 miRNA precursor sequence (atcaggaaaccaaggcgcgctagcttcctgggcgcagtgttccagctgcagtacGGGGGTAATGCCTAAGGATTActcgctgatcggcaccatgggggtggtggtgatcagcgctaTAATCCTTAGGCATTACCCCCtactgcagccggaacactgccaggagaatt). The sequences were synthesized by Genewiz (China), and ligated to the pHK263 plasmid (Hu et al., 2014). The resulting vectors were then named as pMi-IFT25. Expression vectors were created on a pBluescript KS(+) backbone and contained either HA::6×His or HA::GFP coding sequences followed immediately downstream by a sequence encoding the Rubisco 3′-UTR and the aphVIII cassette (paromomycin resistant gene). To express IFT25::HA::6×His and IFT25::HA::GFP, a 1.9 kb genomic DNA fragment composed of an 870 bp promoter sequence and the coding region of ift25 was amplified from genomic DNA by using primer pair (5′-GGTGGATCCGGAAGATATGGCAAGCGTGC-3′ and 5′-GCTGAATTCGGCGTAGTCGGGCACGTCGTAGGGGTAGAACTCGTCCTCGAAGCCG-3′) and inserted into the corresponding expression vector. To generate IFT27::HA::6×His vector, a 2 kb IFT27 fragment consisting of the 1.2 kb promoter sequence and its coding region was amplified from genomic DNA by using primer pair 5′-GTACTAGTATGTACGACACTGCGTTAC-3′ and 5′-AAGAATTCGTAGTTGCGGCAAGCGACC-3′ and inserted into the corresponding expression vector. The rescue construct pBKS-gIFT25::HA::GFP-Ble was a derivative of pBKS-gIFT25::HA::GFP-Paro, created by replacing the aphVIII gene with a zeocin resistant gene (ble) amplified from pSP124S by suing primer pair 5′-AAGGGCCCGCCAGAAGGAGCGCAGCC-3′ and 5′-TTGGTACCGCTTCAAATACGCCCAGC-3′ (Stevens et al., 1996). To express CrBBS2::GFP, CrBBS2 cDNA were synthesized by Genewiz (China). The HSP70A-RBCS2 promoter was amplified from pBKS-gIFT25::HA::GFP-Paro by using primer pair 5′-GCTCTAGAGCCAGAAGGAGCGCAGCCAA-3′ and 5′-GCCATATGTTTAAGATGTTGAGTGACTTCT-3′. Then, the pBKS-HSP70A-RBCS2-CrBBS2::GFP-Paro and the pBKS-HSP70A-RBCS2-CrBBS2::GFP-Ble were constructed by replacing the gIFT25::HA::GFP DNA fragment in pBKS-gIFT25::HA::GFP-Paro and pBKS-gIFT25::HA::GFP-Ble with the CrBBS2::GFP fragment by a three-way ligation. The new constructs were verified by direct nucleotide sequencing. Details about constructs are available upon request.
Chlamydomonas transformation
Transformation of the C. reinhardtii strain was performed using the electroporation method as described previously (Shimogawara et al., 1998). An exponential electric pulse of 2000 V/cm was applied to the cells using a Gene Pulser Xcell electroporation apparatus (Bio Rad). The capacitance was set at 50 μF and no shunt resistor was used. The transformants were selected on TAP plates with 20 µg/ml paromomycin (Sigma-Aldrich), 15 µg/ml bleomycin (Invitrogen), or both antibiotics. The plot showing the transformants and transformation efficiency was created with Prism 5 (GraphPad Software).
SDS-PAGE and western blotting
Preparation of whole cell and flagellar extracts was performed as detailed previously (Wang et al., 2009), except that sonication method was instead used to open up the cell body and flagella. SDS-PAGE electrophoresis and western blotting was performed as previously described (Fan et al., 2010). If not otherwise specified, 10 µg of total protein from each sample was loaded for SDS-PAGE electrophoresis. To quantify the expression levels of proteins, the western blots were quantified with the histogram function in Mini Chemiluminescent Imaging apparatus software (Sage Creation) and defined as (MIband-MIbackground)*Pixelsband. MI represents the mean intensity. MIbackground is the mean intensity of the area around a target band. The intensity of the bands was normalized to the intensity of a loading control protein.
DNA and RNA analysis
Genomic DNA was prepared using a Genomic DNA Prep kit following the kit's protocol (Solarbio, Beijing, China). Total cellular RNA was extracted from 1×108 C. reinhardtii cells in the logarithmic phase of growth with Trizol reagent (Invitrogen) according to manufacturer's protocol. 5 µg of RNA from each sample was reverse transcribed at 42°C for 4 h followed by 95°C for 10 min using SuperScript® III Reverse Transcriptase (Invitrogen) and oligo(dT)20 primers. Relative transcript amounts of specific IFT proteins were measured by SYBR-green quantitative PCR using primer pairs that span either introns or exons of the target genes with an A&B 7500 Fast Real-Time PCR System (Applied Biosystems). The PCR reactions were performed at 94°C for 2 min followed by 40 cycles of 94°C for 10 s, 60°C for 30 s, and 68°C for 2 min. Samples were normalized using guanine nucleotide-binding protein subunit beta-like protein (GBLP) as a housekeeping gene internal control. For each sample, three sets of mRNA were independently isolated and quantified three times each. Data was analyzed with GraphPad Prism 5 (GraphPad Software) and presented as mean±s.d. Primer pairs used were: 5′-AGTCTTGCATTCGGGTGTCT-3′ and 5′-CGAGCTCGACTTCAAAGACC-3′ for ift25; 5′-TCTCGGTGGAGCTCTTTCTG-3′ and 5′-GCTCACATCGAACACGAGAA-3′ for ift27; and 5′-GTCATCCACTGCCTGTGCTTCT-3′ and 5'-GGCCTTCTTGCTGGTGATGTT-3′ for GBLP.
In vivo pull-down and gel filtration chromatography
In vivo pull-down of IFT25- and IFT27-associated protein complexes was performed using transgenic strains expressing C-terminal HA and 6×His double-tagged IFT25 and IFT27. The cells were lysed by sonication and the protein complex was purified by batch method using the Ni-NTA purification system (GE Healthcare) according to the company's protocol. 5 µg of each protein sample was analyzed by SDS-PAGE electrophoresis and western blotting as described above. To isolate IFT25/27, the eluate containing the Ni-NTA resin captured proteins was fractionated on a Superdex 200 10/300 GL column (GE Healthcare). Fractions (0.5 ml) were collected from the top of the gradient and subjected to SDS-PAGE electrophoresis and western blotting as described above. The molecular weight of the purified protein complexes were estimated by comparing to standard protein makers.
Immunofluorescence microscopy
Immunofluorescence staining assay was performed as described in our precious study (Wang et al., 2009). Healthy sample cells grown under continuous illumination were seeded to 0.1% polyethyleneimine-coated coverslips for 8 min under strong light. The cells were then permeablized and fixed with ice-cold methanol twice each for 10 min. Thereafter, cells were rehydrated with phosphate buffered saline (PBS) and incubated overnight in blocking buffer (5% BSA, 1% cold fish gelatin, and 10% goat serum in PBS) at 4°C. The next day, primary antibodies were diluted in the blocking buffer and incubated with the coverslips for 4 h. After washing the cells ten times with PBS, Alexa-Fluor 488 conjugated goat anti-mouse IgG secondary antibody and Alexa-Fluor 542 conjugated goat anti-rabbit IgG secondary antibody (Molecular Probes, Invitrogen, USA) was diluted in the blocking buffer at a dilution of 1:200 and incubated with the coverslips for 2 h. After washing the cells an additional ten times with PBS, they were viewed on an Olympus BX53F fluorescent microscope at 400× amplification. Images were captured with an Olympus DP72 CCD camera (Olympus) with an exposure time between 0.002 and 2 s. The images were processed with ImageJ (version 1.42 g; NIH). The primary antibodies against IFT25 (1:5 dilution), IFT43 (1:50 dilution), IFT46 (1:50 dilution), GFP (1:5 dilution) have been described in the antibodies section above.
Screening of the IFT25miRNA cells and flagellar length analysis of the transgenic cells
The screening of IFT25miRNA cells was initiated by checking the cellular level of the target protein through western blots of whole cell extracts with IFT25 antibody. IFT25miRNA strains showing a reduced level of the target protein were selected for further phenotypic analysis. Measurement of flagellar length was performed as previously described (Fan et al., 2010). The data was processed with GraphPad Prism 5 (GraphPad Software). For each strain, over 50 cells were measured and flagellar lengths were presented as mean value (µm).
Purification of bacterial-expressed proteins
A dual expression vector was created on a pGEX2T backbone (GE Healthcare) and three plasmids were generated for expression of IFT25 or IFT27 alone and for co-expression of IFT25 and IFT27. IFT25 and IFT27 were expressed with N-terminal GST (GST::IFT25) or 6×His (6×His::IFT27) tags. Bacterial expression of the recombinant proteins has been described previously (Fan et al., 2010). Clarified cell extract was applied to 0.4 ml glutathione-agarose beads (GST::IFT25), Ni-NTA resin (6×His::IFT27) or both for tandem purification of GST::IFT25 and 6×His::IFT27. Aliquots (8 μl) from each step of the purification were resolved on 10% SDS-PAGE gels and visualized with Coomassie Blue staining.
Prokaryotic expression and purification of recombinant IFT protein antigens
Full-length C. reinhardtii IFT25, IFT46, IFT72 cDNAs and partial CrBBS2 (1-299 bp), IFT139 (1-459 bp) and IFT57 (955-1200 bp) cDNA fragments were cloned into pET-28a, pGEX-2T and pMAL-C2X vectors and resulted expression vectors were then expressed in Escherichia coli BL21(DE3) strain. Bacteria transformed with the desired plasmids were grown at 37°C in LB medium to an OD600 of 0.6, added 0.2 mM IPTG and reduced the growth temperature to 22°C to induce the protein expression for 6 h. For MBP tag purification: cells were collected, washed twice with PBS buffer (50 mM Tris-HCl, 500 mM NaCl, pH 7.4) and resuspended in 20 ml lysis buffer (20 mM Tris-HCl, 300 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol, pH 7.4), after being completely sonicated, centrifuged at 12,000 rpm/min (19802 xg) for 10 min, and the insoluble materials removed. The soluble proteins were then applied to 1 ml of Dextrin Sepharose™ High Performance column equilibrated with lysis buffer for 2 h at 4°C overnight and subsequently washed extensively by lysis buffer. At last, the bounded proteins were eluted from the resin using elution buffer (20 mM Tris, 300 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethano, 10 mM Maltose, pH 7.4). Samples were quantified with Brandford method and then subjected to SDS-PAGE electrophoresis and Coomassie Blue-staining. For 6×His tag purification: cells were collected, washed twice with PBS buffer (50 mM Tris-HCl, 500 mM NaCl, pH 7.4) and resuspended in 20 ml lysis buffer (50 mM Tris-HCl, 500 mM NaCl, 5 mM Imidazole, pH 7.4), being completely sonicated, centrifuged at 12,000 rpm/min (19802 xg) for 10 min, and removed the soluble buffer. The pellets were washed with lysis buffer (50 mM Tris-HCl, 500 mM NaCl, 5 mM Imidazole, pH 7.4) containing 2 M urea three times, and centrifuged at 12,000 rpm/min (19802 xg) for 10 min, and the insoluble materials collected. Pellets were dissolved with lysis buffer (50 mM Tris-HCl, 500 mM NaCl, 5 mM Imidazole, pH 7.4) containing 8 M urea, and were then applied to 1 ml of Ni Sepharose High Performance column equilibrated with lysis buffer for 2 h at room temperature or 4°C overnight and subsequently washed extensively by lysis buffer (pH 7.4). At last, the bounded proteins were eluted from the resin using elution buffer (50 mM Tris-HCl, 500 mM NaCl, 500 mM Imidazole, pH 7.4) containing 8 M urea. Samples were quantified with Brandford method and then subjected to SDS-PAGE electrophoresis and Coomassie Blue staining. For GST tag purification: cells were collected, washed twice with PBS buffer (50 mM Tris-HCl, 500 mM NaCl, pH 7.4) and resuspended in 20 ml lysis buffer (50 mM Tris-HCl, 500 mM NaCl, pH 7.4), after being sonicated completely, centrifuged at 12,000 rpm/min (19802 xg) for 10 min, and the insoluble materials removed. The soluble proteins were then applied to 1 ml of Glutathione Sepharose™ 4B High Performance column equilibrated with lysis buffer for 2 h at 4°C overnight and subsequently washed extensively by lysis buffer. At last, the bounded proteins were eluted from the resin using elution buffer (50 mM Tris-HCl, 300 mM NaCl, 2 mM MgCl2, 10 mM Glutathione, pH 7.4). Samples were quantified with Brandford method and then subjected to SDS-PAGE electrophoresis and Coomassie Blue staining.
Preparation of polyclonal antibody
Two micrograms of purified-protein antigen, the N-terminal 17 amino acids (MVKKEVKPIDITATLRC) of IFT27, N-terminal 19 amino acids (MPPAGGGSESVKVVVRCRP) of FLA10, N-terminal 17 amino acids (MAAPAMLPGQAVKTPGS) of D1BLIC and N-terminal 19 amino acids (MRTVVAWQETPPEKDGVRN) of IFT122 (synthesized by ChinaPeptides, Shanghai, China) were mixed with Freund's Adjuvant (complete) (Sigma F5881, USA) for the first injection, and the subsequent two injections used Freund's Adjuvant (incomplete) (Sigma F5506, USA). The rabbits were raised in the animal service center of Tianjin University of Science and Technology and injected every 10 days. After collecting the antiserum, affinity purified with Protein A Sepharose™ CL-4B to specific bound IgG as per the manufacturer's instructions. Briefly, 1 ml antiserum was mixed with 14 ml binding buffer (12 mM Na2HPO4, 8 mM NaH2PO4, pH 7.0), and binding with Protein A resin equilibrated with binding buffer for 2 h at room temperature or 4°C overnight, washed the resin with binding buffer 3 times and eluted with elution buffer (0.1 M Glycine, pH 2.7). Experiments with rabbits are approved by Tianjin University of Science and Technology and performed according to the relevant regulatory guide for laboratory animals.
IFT video imaging and speed measurements
The flagellar motility of GFP-tagged proteins was imaged at ∼10 frames per second (fps) using total internal reflection fluorescence (TIRF) microscopy on an inverted microscope (Eclipse Ti, Nikon) equipped with a through-the-objective TIRF system, a 100×/1.49 NA TIRF oil immersion objective (Nikon), and a cooled EM charge coupled device (CCD) camera (QuantEM:512SC, Photometrics) as detailed previously (Engel et al., 2009a,b). To quantify IFT speeds, kymographs were generated with Metamorph (version 7.7, MDS Analytical Technologies) and measured with NIS-Elements AR (version 3.2, Nikon) and ImageJ (version 1.42 g) as previously described (Engel et al., 2009a,b).
Statistical analysis
All the data were presented as mean±s.d. Statistical analysis performed with GraphPad Prism 5 (GraphPad Software). For comparisons on speeds and frequencies of the GFP-labeled IFT proteins, one-sample unpaired t-test was used.
Supplementary Material
Acknowledgements
We thank Hai-Yue Ren, Wei Tian, Zhen Wang and Peng-Fei Han for help with making the polyclonal IFT antibodies, and Dr Junmin Pan at Tsinghua University, Beijing, China for kindly providing the α-IFT43 polyclonal antibody.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: Z.-C.F.; Methodology: K.H., M.W.; Investigation: B.D., S.W., J.W., Y.-X.L., Z.P., D.-M.M.; Writing - original draft: B.D.; Writing - review & editing: Z.-C.F.; Supervision: Z.-C.F.; Funding acquisition: Z.-C.F.
Funding
This study was supported by the grant from International Center for Genetic Engineering and Biotechnology (ICGEB) (grant number CRP/CHN15-01 to Z.-C.F.).
Supplementary information
Supplementary information available online at http://bio.biologists.org/lookup/doi/10.1242/bio.026278.supplemental
References
- Aldahmesh M. A., Li Y., Alhashem A., Anazi S., Alkuraya H., Hashem M., Awaji A. A., Sogaty S., Alkharashi A., Alzahrani S. et al. (2014). IFT27, encoding a small GTPase component of IFT particles, is mutated in a consanguineous family with Bardet-Biedl syndrome. Hum. Mol. Genet. 23, 3307-3315. 10.1093/hmg/ddu044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari N. F., Lewis J. S., Bishop G. A., Askwith C. C. and Mykytyn K. (2008). Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc. Natl. Acad. Sci. USA 105, 4242-4246. 10.1073/pnas.0711027105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S., Taschner M., Morawetz M., Basquin C. and Lorentzen E. (2011). Crystal structure of the intraflagellar transport complex 25/27. EMBO J. 30, 1907-1918. 10.1038/emboj.2011.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizet A. A., Becker-Heck A., Ryan R., Weber K., Filhol E., Krug P., Halbritter J., Delous M., Lasbennes M.-C., Linghu B. et al. (2015). Mutations in TRAF3IP1/IFT54 reveal a new role for IFT proteins in microtubule stabilization. Nat. Commun. 6, 8666 10.1038/ncomms9666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazelton W. J., Amundsen C. D., Silflow C. D. and Lefebvre P. A. (2001). The bld1 mutation identifies the Chlamydomonas osm-6 homolog as a gene required for flagellar assembly. Curr. Biol. 11, 1591-1594. 10.1016/S0960-9822(01)00485-7 [DOI] [PubMed] [Google Scholar]
- Brown J. M., Cochran D. A., Craige B., Kubo T. and Witman G. B. (2015). Assembly of IFT Trains at the ciliary base depends on IFT74. Curr. Biol. 25, 1583-1593. 10.1016/j.cub.2015.04.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D. G., Diener D. R., Himelblau A. L., Beech P. L., Fuster J. C. and Rosenbaum J. L. (1998). Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141, 993-1008. 10.1083/jcb.141.4.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane J. A., Cole D. G., Seeley E. S., Diener D. R. and Rosenbaum J. L. (2001). Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 11, 1586-1590. 10.1016/S0960-9822(01)00484-5 [DOI] [PubMed] [Google Scholar]
- Dutcher S. K. (2003). Elucidation of basal body and centriole functions in Chlamydomonas reinhardtii. Traffic 4, 443-451. 10.1034/j.1600-0854.2003.00104.x [DOI] [PubMed] [Google Scholar]
- Eguether T., San Agustin J. T., Keady B. T., Jonassen J. A., Liang Y., Francis R., Tobita K., Johnson C. A., Abdelhamed Z. A., Lo C. W. et al. (2014). IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev. Cell 31, 279-290. 10.1016/j.devcel.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel B. D., Ishikawa H., Wemmer K. A., Geimer S., Wakabayashi K., Hirono M., Craige B., Pazour G. J., Witman G. B., Kamiya R. et al. (2012). The role of retrograde intraflagellar transport in flagellar assembly, maintenance, and function. J. Cell Biol. 199, 151-167. 10.1083/jcb.201206068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel B. D., Lechtreck K.-F., Sakai T., Ikebe M., Witman G. B. and Marshall W. F. (2009a). Total internal reflection fluorescence (TIRF) microscopy of Chlamydomonas flagella. Methods Cell Biol. 93, 157-177. 10.1016/S0091-679X(08)93009-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel B. D., Ludington W. B. and Marshall W. F. (2009b). Intraflagellar transport particle size scales inversely with flagellar length: revisiting the balance-point length control model. J. Cell Biol. 187, 81-89. 10.1083/jcb.200812084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z.-C., Behal R. H., Geimer S., Wang Z., Williamson S. M., Zhang H., Cole D. G. and Qin H. (2010). Chlamydomonas IFT70/CrDYF-1 is a core component of IFT particle complex B and is required for Flagellar assembly. Mol. Biol. Cell 21, 2696-2706. 10.1091/mbc.E10-03-0191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J. A., Xu F., Keady B. T. and Pazour G. J. (2009). Characterization of mouse IFT complex B. Cell Motil. Cytoskeleton 66, 457-468. 10.1002/cm.20346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Pazour G. J. and Witman G. B. (2004). A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol. Biol. Cell 15, 4382-4394. 10.1091/mbc.E04-05-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Qin H., Follit J. A., Pazour G. J., Rosenbaum J. L. and Witman G. B. (2007). Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J. Cell Biol. 176, 653-665. 10.1083/jcb.200608041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Deng X., Shao N., Wang G. and Huang K. (2014). Rapid construction and screening of artificial microRNA systems in Chlamydomonas reinhardtii. Plant J. 79, 1052-1064. 10.1111/tpj.12606 [DOI] [PubMed] [Google Scholar]
- Huet D., Blisnick T., Perrot S. and Bastin P. (2014). The GTPase IFT27 is involved in both anterograde and retrograde intraflagellar transport. eLife 3, e02419 10.7554/eLife.02419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C., Babaev-Khaimov V., Sassaroli M. and Piperno G. (2001). Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J. Cell Biol. 153, 13-24. 10.1083/jcb.153.1.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C., Li L., Esparza J. M. and Dutcher S. K. (2009). Retrograde intraflagellar transport mutants identify complex A proteins with multiple genetic interactions in Chlamydomonas reinhardtii. Genetics 183, 885-896. 10.1534/genetics.109.101915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Ide T., Yagi T., Jiang X., Hirono M., Sasaki H., Yanagisawa H., Wemmer K. A., Stainier D. Y., Qin H. et al. (2014). TTC26/DYF13 is an intraflagellar transport protein required for transport of motility-related proteins into flagella. eLife 3, e01566 10.7554/eLife.01566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., White S. R., Shida T., Schulz S., Aguiar M., Gygi S. P., Bazan J. F. and Nachury M. V. (2010). The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141, 1208-1219. 10.1016/j.cell.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y., Terada M., Nishijima Y., Takei R., Nozaki S., Hamada H. and Nakayama K. (2016). Overall architecture of the intraflagellar transport (IFT)-B complex containing Cluap1/IFT38 as an essential component of the IFT-B peripheral subcomplex. J. Biol. Chem. 291, 10962-10975. 10.1074/jbc.M116.713883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keady B. T., Samtani R., Tobita K., Tsuchya M., San Agustin J. T., Follit J. A., Jonassen J. A., Subramanian R., Lo C. W. and Pazour G. J. (2012). IFT25 Links the Signal-Dependent Movement of Hedgehog Components to Intraflagellar Transport. Dev. Cell 22, 940-951. 10.1016/j.devcel.2012.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Gengyo-Ando K., Ishihara T., Katsura I. and Mitani S. (2007). IFT-81 and IFT-74 are required for intraflagellar transport in C. elegans. Genes Cells 12, 593-602. 10.1111/j.1365-2443.2007.01076.x [DOI] [PubMed] [Google Scholar]
- Kozminski K. G., Johnson K. A., Forscher P. and Rosenbaum J. L. (1993). A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA 90, 5519-5523. 10.1073/pnas.90.12.5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K.-F., Johnson E. C., Sakai T., Cochran D., Ballif B. A., Rush J., Pazour G. J., Ikebe M. and Witman G. B. (2009a). The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell Biol. 187, 1117-1132. 10.1083/jcb.200909183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K.-F., Luro S., Awata J. and Witman G. B. (2009b). HA-tagging of putative flagellar proteins in Chlamydomonas reinhardtii identifies a novel protein of intraflagellar transport complex B. Cell Motil. Cytoskeleton 66, 469-482. 10.1002/cm.20369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew G. M., Ye F., Nager A. R., Murphy J. P., Lee J. S., Aguiar M., Breslow D. K., Gygi S. P. and Nachury M. V. (2014). The intraflagellar transport protein IFT27 promotes BBSome exit from cilia through the GTPase ARL6/BBS3. Dev. Cell 31, 265-278. 10.1016/j.devcel.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Nauman N. P., Albee A. J., Hsu S. and Dutcher S. K. (2013). New mutations in flagellar motors identified by whole genome sequencing in Chlamydomonas. Cilia 2, 14 10.1186/2046-2530-2-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucker B. F., Behal R. H., Qin H., Siron L. C., Taggart W. D., Rosenbaum J. L. and Cole D. G. (2005). Characterization of the intraflagellar transport complex B core: direct interaction of the IFT81 and IFT74/72 subunits. J. Biol. Chem. 280, 27688-27696. 10.1074/jbc.M505062200 [DOI] [PubMed] [Google Scholar]
- Miller M. S., Esparza J. M., Lippa A. M., Lux F. G. III, Cole D. G. and Dutcher S. K. (2005). Mutant kinesin-2 motor subunits increase chromosome loss. Mol. Biol. Cell 16, 3810-3820. 10.1091/mbc.E05-05-0404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller J., Perrone C. A., Bower R., Cole D. G. and Porter M. E. (2005). The FLA3 KAP subunit is required for localization of kinesin-2 to the site of flagellar assembly and processive anterograde intraflagellar transport. Mol. Biol. Cell 16, 1341-1354. 10.1091/mbc.E04-10-0931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa S. (2016). The nephronophthisis-related gene ift-139 is required for ciliogenesis in Caenorhabditis elegans. Sci. Rep. 6, 31544 10.1038/srep31544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G., Blacque O. E., Snow J. J., Leroux M. R. and Scholey J. M. (2005). Functional coordination of intraflagellar transport motors. Nature 436, 583-587. 10.1038/nature03818 [DOI] [PubMed] [Google Scholar]
- Ou G., Koga M., Blacque O. E., Murayama T., Ohshima Y., Schafer J. C., Li C., Yoder B. K., Leroux M. R. and Scholey J. M. (2007). Sensory ciliogenesis in Caenorhabditis elegans: assignment of IFT components into distinct modules based on transport and phenotypic profiles. Mol. Biol. Cell 18, 1554-1569. 10.1091/mbc.E06-09-0805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Wilkerson C. G. and Witman G. B. (1998). A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J. Cell Biol. 141, 979-992. 10.1083/jcb.141.4.979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Dickert B. L. and Witman G. B. (1999). The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 144, 473-481. 10.1083/jcb.144.3.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Dickert B. L., Vucica Y., Seeley E. S., Rosenbaum J. L., Witman G. B. and Cole D. G. (2000). Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151, 709-718. 10.1083/jcb.151.3.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G., Geimer S., Lanzavecchia S., Paccagnini E., Cantele F., Diener D. R., Rosenbaum J. L. and Lupetti P. (2009). Electron-tomographic analysis of intraflagellar transport particle trains in situ. J. Cell Biol. 187, 135-148. 10.1083/jcb.200905103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Siuda E., Henderson S., Segil M., Vaananen H. and Sassaroli M. (1998). Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J. Cell Biol. 143, 1591-1601. 10.1083/jcb.143.6.1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter M. E., Bower R., Knott J. A., Byrd P. and Dentler W. (1999). Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell 10, 693-712. 10.1091/mbc.10.3.693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Wang Z., Diener D. and Rosenbaum J. (2007). Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr. Biol. 17, 193-202. 10.1016/j.cub.2006.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J. L. and Witman G. B. (2002). Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3, 813-825. 10.1038/nrm952 [DOI] [PubMed] [Google Scholar]
- Shimogawara K., Fujiwara S., Grossman A. and Usuda H. (1998). High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148, 1821-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D. R., Rochaix J. D. and Purton S. (1996). The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol. Gen. Genet. 251, 23-30. [DOI] [PubMed] [Google Scholar]
- Swiderski R. E., Nakano Y., Mullins R. F., Seo S. and Bánfi B. (2014). A mutation in the mouse ttc26 gene leads to impaired hedgehog signaling. PLoS Genet. 10, e1004689 10.1371/journal.pgen.1004689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M., Weber K., Mourão A., Vetter M., Awasthi M., Stiegler M., Bhogaraju S. and Lorentzen E. (2016). Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin-binding IFT-B2 complex. EMBO J. 35, 773-790. 10.15252/embj.201593164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao C.-C. and Gorovsky M. A. (2008). Tetrahymena IFT122A is not essential for cilia assembly but plays a role in returning IFT proteins from the ciliary tip to the cell body. J. Cell Sci. 121, 428-436. 10.1242/jcs.015826 [DOI] [PubMed] [Google Scholar]
- Wang Z., Fan Z.-C., Williamson S. M. and Qin H. (2009). Intraflagellar transport (IFT) protein IFT25 is a phosphoprotein component of IFT complex B and physically interacts with IFT27 in Chlamydomonas. PLoS ONE 4, e5384 10.1371/journal.pone.0005384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. L., McIntyre J. C., Norris S. R., Jenkins P. M., Zhang L., Pei Q., Verhey K. and Martens J. R. (2014). Direct evidence for BBSome-associated intraflagellar transport reveals distinct properties of native mammalian cilia. Nat. Commun. 5, 5813 10.1038/ncomms6813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Zhu X., Wang L., Liang Y., Feng Q. and Pan J. (2017). Functional exploration of the IFT-A complex in intraflagellar transport and ciliogenesis. PLoS Genet. 13, e1006627 10.1371/journal.pgen.1006627 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.