ABSTRACT
Neisseria meningitidis, typically a resident of the oro- or nasopharynx and the causative agent of meningococcal meningitis and meningococcemia, is capable of invading and colonizing the urogenital tract. This can result in urethritis, akin to the syndrome caused by its sister species, N. gonorrhoeae, the etiologic agent of gonorrhea. Recently, meningococcal strains associated with outbreaks of urethritis were reported to share genetic characteristics with the gonococcus, raising the question of the extent to which these strains contain features that promote adaptation to the genitourinary niche, making them gonococcus-like and distinguishing them from other N. meningitidis strains. Here, we analyzed the genomes of 39 diverse N. meningitidis isolates associated with urethritis, collected independently over a decade and across three continents. In particular, we characterized the diversity of the nitrite reductase gene (aniA), the factor H-binding protein gene (fHbp), and the capsule biosynthetic locus, all of which are loci previously suggested to be associated with urogenital colonization. We observed notable diversity, including frameshift variants, in aniA and fHbp and the presence of intact, disrupted, and absent capsule biosynthetic genes, indicating that urogenital colonization and urethritis caused by N. meningitidis are possible across a range of meningococcal genotypes. Previously identified allelic patterns in urethritis-associated N. meningitidis strains may reflect genetic diversity in the underlying meningococcal population rather than novel adaptation to the urogenital tract.
KEYWORDS: Neisseria gonorrhoeae, Neisseria meningitidis, bacterial genomics, urethritis
INTRODUCTION
The genus Neisseria comprises many commensal organisms as well as two primarily pathogenic species. Pathogenic Neisseria species include Neisseria gonorrhoeae (also known as the gonococcus), the etiologic agent of gonorrhea, and N. meningitidis (also known as the meningococcus), the etiologic agent of meningococcal meningitis and meningococcemia. However, the distinction between commensal and pathogen is imprecise: Neisseria strains classically defined as commensals can cause disease, and N. meningitidis strains defined as pathogenic routinely persist asymptomatically in carriers (1). Accordingly, the meningococcus inhabits the nasopharynx commensally in about 10% of the population; this carriage state is likely the source for symptomatic cases of meningococcal disease (1). Furthermore, although N. meningitidis and N. gonorrhoeae were conventionally thought to occupy distinct human ecological niches, case reports in the literature across several decades have indicated that N. meningitidis is capable of invading and colonizing the urethra and in doing so results in urethritis akin to that caused by gonococcal infection (2–4). Oral sex has been strongly associated with many such cases, suggesting that pathogenesis depends on orogenital contact (2, 5, 6).
N. meningitidis strains isolated from cases of urethritis serve as natural experiments well-suited for advancing our understanding of how Neisseria strains diverge and specialize for ecological niches within their human hosts. Investigating the dynamics and mechanisms by which these atypical isolates have potentially adapted could also improve epidemiological characterization of the transmission networks of pathogenic Neisseria. Whole-genome sequencing offers one approach for both understanding the epidemiology of N. meningitidis-associated urethritis and interrogating the genetic basis of possible adaptation of these meningococcal lineages. Recent studies employing genomics have suggested that particular alleles of nitrite reductase (AniA), the factor H-binding protein (fHbp), and the capsule are associated with N. meningitidis strains isolated from genitourinary infections (2, 3, 7, 8). However, because these studies focused primarily on genetically related isolates, their power to distinguish genuine adaptive features from shared features due to population structure was limited. We thus assembled and sequenced a diverse collection of urethritis-associated meningococcal strains to assess whether any previously identified or novel genetic signals could explain the unusual pathogenesis of these lineages.
RESULTS AND DISCUSSION
Isolate metadata and population structure.
Our sample set included 39 isolates of urethritis-associated N. meningitidis and 1 isolate of urethritis-associated N. lactamica, all collected from male patients over a decade across eight countries (Table 1). The average age of the patients for whom metadata were available (n = 24) was 30.8 years, with a range of 21 to 52 years. All except one of the patients had symptoms of urethritis (Table 1). The core genome of all 40 isolates comprised 1,237 genes, of which 1,177 were included in the core genome alignment, whereas the core genome of the 39 meningococcal isolates comprised 1,384 genes. The maximum likelihood phylogeny constructed using a concatenated core genome alignment and rooted using the N. lactamica isolate (NlUS07-1) indicated a diverse sampling of urethritis-associated neisserial strains (Fig. 1), with computational sequence typing revealing seven clonal complexes and 19 sequence types (STs) in this data set (Table 1).
TABLE 1.
Patient metadata and strain fine typing
| Isolatea | Source, yr (reference) | Country of origin | Yr of isolation | Patient age (yr) | Symptoms | Additional patient informationb | Fine typec |
|---|---|---|---|---|---|---|---|
| NmIR13-1 (12028_13) | PubMLST | Ireland | 2013 | 50 | Urethritis | B:P1.22,14:F5-5:ST-10981 | |
| NmIT14-1 (PE5) | PubMLST | Italy | 2014 | Urethritis | C:P1.5-1,10-8:F3-6:ST-11 | ||
| NmIT14-2 (PE6) | PubMLST | Italy | 2014 | Urethritis | C:P1.5-1,10-8:F3-6:ST-11 | ||
| NmIT14-3 (PE7) | PubMLST | Italy | 2014 | Urethritis | C:P1.5-1,10-8:F3-6:ST-11 | ||
| NmJP04-1 | NIID | Japan | 2004 | 21 | Urethritis | C. trachomatis positive | Y:P1.5-2,10-1:F4-1:ST-23 |
| NmJP05-1 | NIID | Japan | 2005 | Urethritis | B:P1.21-2,2-33:F1-7:ST-687 | ||
| NmJP05-2 | NIID | Japan | 2005 | Urethritis | Y:P1.5-2,10-1:F4-1:ST-23 | ||
| NmJP05-3 | NIID | Japan | 2005 | 30 | Urethritis | −:P1.18-1,3:F4-1:ST-35 | |
| NmJP06-1 | NIID | Japan | 2006 | 29 | Urethritis | Y:P1.5-2,10-1:F4-1:ST-23 | |
| NmJP12-1 | NIID | Japan | 2012 | Urethritis | W:P1.5,2:F1-1:ST-10651 | ||
| NmJP12-2 | NIID | Japan | 2012 | 28 | Urethritis | C. trachomatis negative | Y:P1.5-2,10-40:F4-1:ST-23 |
| NmJP12-3 | NIID | Japan | 2012 | 33 | Balanoposthitis | C. trachomatis negative | Y:P1.18,25-34:F5-5:ST-198 |
| NmJP14-1 | NIID | Japan | 2014 | 34 | Urethritis | Y:P1.5-2,10-15:F4-1:ST-23 | |
| NmJP14-2 | NIID | Japan | 2014 | Urethritis | C:P1.5-1,10-8:F3-6:ST-11 | ||
| NmJP14-3 | NIID | Japan | 2014 | 29 | Urethritis | C. trachomatis negative | Y:P1.5-2,10-1:F4-1:ST-11120 |
| NmJP14-4 | NIID | Japan | 2014 | Urethritis | C. trachomatis negative | −:P1.5-2,10-1:F4-1:ST-23 | |
| NmJPb05-1 | NIID | Japan | Before 2005 | Urethritis | −:P1.5-1,2-5:F5-5:ST-823 | ||
| NmJPb05-2 | NIID | Japan | Before 2005 | 21 | Urethritis | Y:P1.5-1,2-2:F5-8:ST-23 | |
| NmJPb05-3 | NIID | Japan | Before 2005 | 25 | Urethritis | −:P1.5-1,1:F1-7:ST-2045 | |
| NmJPb05-4 | NIID | Japan | Before 2005 | 52 | Urethritis | Y:P1.5-2,10-1:F4-1:ST-23 | |
| NmJPb05-5 | NIID | Japan | Before 2005 | 32 | Urethritis | Y:P1.5-2,10-1:F4-1:ST-23 | |
| NmJPb05-6 | NIID | Japan | Before 2005 | 25 | Urethritis | Y:P1.5-2,10-1:F4-1:ST-23 | |
| NmJPb09-1 | NIID | Japan | Before 2009 | Urethritis | Y:P1.5-2,10-1:F4-1:ST-23 | ||
| NmSL13-1 | WHO | Slovenia | 2013 | 46 | Urethritis | MSM | B:P1.19,15:F3-6:ST-3091 |
| NmSL13-2 | WHO | Slovenia | 2013 | 20 | Urethritis | MSM, C. trachomatis negative | −:P1.18-1,*:F5-7:ST-5953 |
| NmSL13-3 | WHO | Slovenia | 2013 | 30 | Urethritis | B:P1.7-2,4:F1-5:ST-41 | |
| NmSL14-1 | WHO | Slovenia | 2014 | 28 | Carriage | MSM/MSW, C. trachomatis negative | B:P1.22,14:F5-5:ST-213 |
| NmSL15-1 | WHO | Slovenia | 2015 | 32 | Urethritis | MSW, C. trachomatis negative | −:P1.18,25-1:F5-5:ST-198 |
| NmUS02-1 | CDC | USA | 2002 | 23 | Urethritis | MSW | Y:P1.5-2,10-1:F4-1:ST-23 |
| NmUS03-1 | CDC | USA | 2003 | 40 | Urethritis | MSW | C:P1.5,2:F3-6:ST-11 |
| NmUS03-2 | CDC | USA | 2003 | 30 | Urethritis | MSW | −:P1.19,15:F1-18:ST– |
| NmUS04-1 | CDC | USA | 2004 | 24 | Urethritis | MSW | C:P1.7-2,13-2:F1-7:ST-278 |
| NlUS07-1 | CDC | USA | 2007 | 33 | Urethritis | MSW | |
| NmFR12-1 (LNP26948) | Taha et al., 2016 (7) | France | 2012 | 25 | Urethritis | C:P1.5-1,10-1:F3-6:ST-10482 | |
| NmUKb13-1 (NM9853) | Harrison et al., 2017 (3) | UK | 2011–2013 | Urethritis | B:P1.7-2,4:F1-5:ST-41 | ||
| NmUKb13-2 (NM8525) | Harrison et al., 2017 (3) | UK | 2011–2013 | Urethritis | B:P1.19-1,15-11:F5-1:ST-269 | ||
| NmUKb13-3 (NM10492) | Harrison et al., 2017 (3) | UK | 2011–2013 | Urethritis | Z:P1.18,25-15:F5-7:ST-3882 | ||
| NmUKb13-4 (NM10763) | Harrison et al., 2017 (3) | UK | 2011–2013 | Urethritis | Z:P1.22-4,14-13:F5-7:ST-10866 | ||
| NmUS16-1 (NM-1) | Toh et al., 2017 (2) | USA | 2016 | Urethritis | C:P1.5-1,10-8:F3-6:ST-11 | ||
| NmUS16-2 (NM-2) | Toh et al., 2017 (3) | USA | 2016 | Urethritis | C:P1.5-1,10-8:F3-6:ST-11 |
Isolate names in parentheses are the original names of the isolates from the source database or publication.
Patient sexual orientation and results of testing for Chlamydia trachomatis are indicated when available. MSM, men who have sex with men; MSW, men who have sex with women.
−, computational serogrouping yielded no results; *, novel porA sequence.
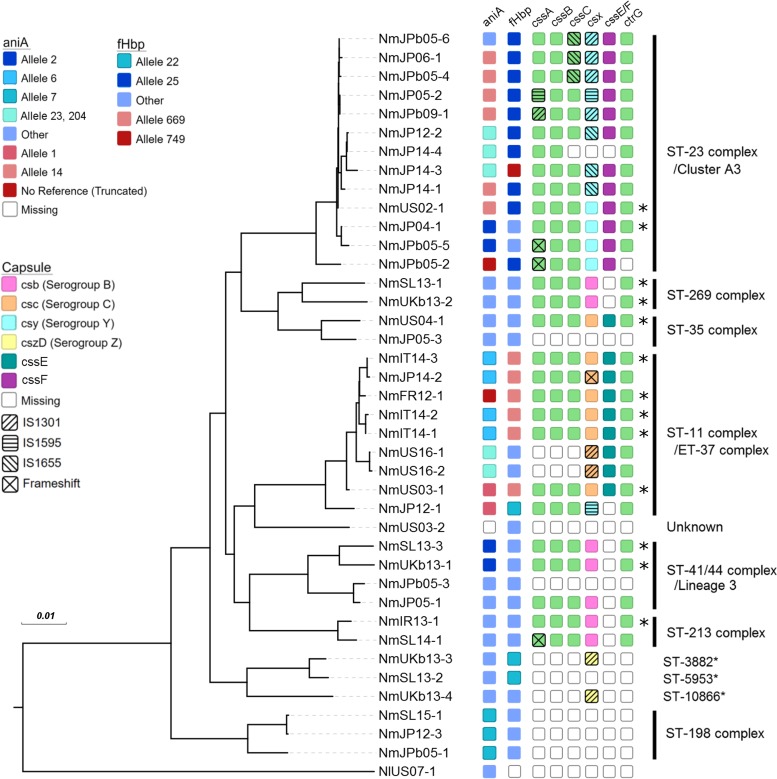
FIG 1.
Core genome phylogeny and genomic characterization of aniA, fHbp, and capsular region A of urethritis-associated N. meningitidis and N. lactamica isolates. For aniA and fHbp, blue shading indicates a full gene product and red shading indicates a truncated gene product. Alleles associated with truncated genes and alleles associated with full genes found in three or more isolates are specifically denoted in the key on the left; all other alleles were grouped into the “other” category. aniA alleles 23 and 204 are putatively gonococcal in origin. For capsule region A, csx variants which determine the serogroup (where csx can be csb, csc, csy, or cszD), the presence of either cssE or cssF, and capsule gene disruption via insertion sequence or frameshift are specified by the colors and symbols in the key. *, strains predicted to encode capsules; unshaded boxes, missing genes. Clonal complexes are indicated along the right, with asterisks indicating no associated clonal complex.
Of the meningococcal strains, 13 isolates (33.3%) belonged to the hyperinvasive lineage ST-11 (electrophoretic type 37 [ET-37]) or ST-41/44 complex (lineage 3), and 17 (43.6%) belonged to the emerging invasive lineage ST-23 (cluster A3), ST-213, or ST-269. The hyperinvasive ST-11 (ET-37) lineage included previously reported urethritis-associated N. meningitidis strains, such as NmUS16-1 and NmUS16-2 collected by Toh et al. in 2017 (2) and the men who have sex with men (MSM)-associated strain NmFR12-1 collected by Taha et al. in 2016 (7). Three isolates from Italy obtained through PubMLST (NmIT14-1, NmIT14-2, NmIT14-3) were also part of this clonal complex. The emerging invasive lineage ST-23 was represented by a cluster of 12 isolates from Japan and 1 isolate from the United States (NmUS02-1). One isolate (NmUS03-2) harbored an unknown sequence type. In this isolate, every gene queried as part of multilocus sequence typing (MLST) was associated with an allele in the PubMLST database (abcZ, n = 7; adk, n = 16; aroE, n = 55; fumC, n = 10; gdh, n = 3; pdhC, n = 56; pgm, n = 16); however, the combination of alleles together was novel. The remaining isolates belonged to rare clonal complexes. The three N. meningitidis urethritis isolates (NmJP12-3, NmSL15-1, NmJPb05-1) phylogenetically the closest to the gonococcus formed a distinct clade and were all grouped under ST-198 (see Fig. S1 in the supplemental material). Prior literature has indicated that, out of cnl meningococcal strains, ST-198 isolates in particular have been able to cause invasive meningococcal disease, despite being unencapsulated (9). The presence of three such unusual isolates in our data set raises the possibility that urethritis is another manifestation of this clade's pathogenicity.
Nitrite reductase gene (aniA) and factor H-binding protein gene (fHbp).
The nitrite reductase gene (aniA) of N. meningitidis is often frameshifted in clinical isolates but is intact in N. gonorrhoeae, suggesting that AniA may play an important role for homeostasis in the microaerophilic environment of the urogenital tract (7). In keeping with this hypothesis, four of the five isolates in a collection of urethritis-associated N. meningitidis isolates collected in France and Germany from 2006 to 2012 harbored an intact aniA gene and exhibited active nitrite reductase activity in vitro (7). However, because these strains were closely related, the association of intact aniA with urogenital infection could instead be due to the population structure. We thus investigated the allelic diversity of aniA and characterized the prevalence of frameshifted variants to understand whether intact aniA is associated with urogenital colonization. We found that the aniA gene was present in all isolates except NmUS03-2; mapping the sequencing reads of this isolate to the MC58 reference genome (GenBank accession number AE002098.2) showed a drop in coverage to zero for aniA and the loci immediately adjacent. Excluding NmFR12-1 (original name, LNP26948) from Taha et al. (7), we found that 24.3% (9/37) of the isolates harbored a truncated aniA, indicating that intact aniA is not necessary for urogenital colonization (Fig. 1). Full-length aniA exhibited a range of alleles, whereas truncated aniA largely belonged to allele 14 (6/9 isolates); this was also evident in the nucleotide phylogeny, where most of the truncated aniA sequences clustered together (Fig. S2). Intriguingly, 12.8% (5/39) of the isolates harbored genes with suspected gonococcal origins (as determined by BLAST analysis of the nucleotide sequences against the sequences in the nr/nt database). These five isolates were split into two clades, each harboring a distinct allele: NmJP14-3, NmJP14-4, and NmJP12-2 contained allele 23, and NmUS16-1 and NmUS16-2 from Toh et al. (2) contained allele 204 (Fig. 1). Tzeng et al. recently reported that NmUS16-1, NmUS16-2, and additional urethritis-associated isolates in the same lineage contained gonococcal nitric oxide reductase genes (norB), in addition to gonococcal aniA (8). In NmJP12-2, NmJP14-3, and NmJP14-4, we similarly found that norB is gonococcal in origin, implying that the entire norB-aniA cassette was acquired via horizontal recombination.
Frameshifted fHbp has been associated with urethritis isolates and results in significant effects on virulence in a mouse model (7). In our data set, all isolates contained the fHbp gene, and considerable diversity was present, with all three Novartis variants being represented. Only a minority of samples (6/38, excluding NmFR12-1) harbored truncated fHbp (alleles 669 and 749 in Fig. 1), indicating that this particular genotype is not strongly associated with urogenital colonization. Furthermore, 5/6 isolates with truncated fHbp (NmIT14-1, NmIT14-2, NmIT14-3, NmUS03-1, and NmJP14-2) contained the same allele (allele 669) and belonged to the same clonal complex (the ST-11 complex) as NmFR12-1, suggesting that the association of urethritis with fHbp truncation is likely due to relatedness between strains (Fig. 1).
Additional genetic features present in N. gonorrhoeae but not generally found in N. meningitidis may also be responsible for the potential urogenital adaptation undergone by our urethritis isolates. We therefore examined the allelic distribution in our sample set of the class 1 outer membrane porin PorA and the membrane-bound c-type cytochrome CcoP, the genes for both of which were previously characterized as divergent between N. meningitidis and N. gonorrhoeae (10, 11). Feavers and Maiden found that in the gonococcus, the expression of the PorA protein was inactivated via frameshift mutations and deletions of parts of the TATAAT box and the poly(G) residue portions of the promoter (11), and Aspholm et al. found that in the meningococcus, a single nucleotide polymorphism resulting in CcoP truncation was present (10). In both cases, N. meningitidis strains associated with urethritis in our data set harbored genes with wild-type meningococcal characteristics (i.e., an intact porA gene and promoter and truncated ccoP), suggesting that a gonococcus-like porA pseudogene and ccoP are not strictly necessary for neisserial colonization of the urogenital tract.
Capsule.
The meningococcal capsule is important in the pathogenesis of meningococcal disease, in that it facilitates bacterial survival by promoting evasion of the immune system; however, capsule expression also appears to detrimentally impact adhesion and entry into human cells (12). Because capsule disruptions have previously been found in urethritis-associated N. meningitidis strains (2), we characterized the biosynthetic locus in our isolates, with a particular focus on genes in region A, which comprises capsule biosynthesis enzymes. For serogroups B, C, W, and Y, region A genes include cssA, cssB, and cssC, which function in cytidine-5′-monophosphate-N-acetylneuraminic acid synthesis, as well as a csx (where csx can be csb, csc, csw, or csy) gene which encodes a serogroup-specific polymerase (13). We also characterized genes in region B, comprising capsule translocation genes ctrE and ctrF, and genes in region C, comprising capsule export genes ctrABCD. We considered present, intact genes in regions A through C to be necessary for production of the capsule.
Isolates NmSL15-1, NmJPb05-1, NmJP12-3, and NmJPb05-3 contained cnl allele 2, where cnl is defined to be an approximately 113-bp intergenic region that has replaced regions A and C of the capsule (Fig. 1) (13). Three of these isolates (excluding NmJPb05-3) belonged to the ST-198 clade; among the rare cases of unencapsulated N. meningitidis isolates associated with invasive disease, isolates from the ST-198 clade are frequently found (9). Isolates in the ST-198 clade also lacked region B (ctrE, ctrF) genes, whereas NmJPb05-3 possessed them. Three other isolates (NmSL13-2 in ST-5953, NmUS03-2 with an unknown sequence type, and NmJP05-3 in the ST-35 complex) also lacked capsule region A genes but did not possess any of the characteristic cnl alleles present in the PubMLST database. While NmSL13-2 and NmUS03-2 belonged to unusual sequence types and possessed intact region B and C genes, NmJP05-3 lacked region C genes and was closely related to an encapsulated meningococcus in the ST-35 complex (NmUS04-1), suggesting that loss of the capsule occurred due to recombination.
Out of the isolates in which capsule genes were present, 17 harbored some type of disruption. Four of these 17 strains contained a frameshift mutation in a capsule region A gene: NmJPb05-2, NmJPb05-5, and NmSL14-1 contained various insertions in nonhomopolymeric regions of the cssA gene, resulting in a truncated peptide product, and NmJP14-2 contained a 1-nucleotide deletion resulting in truncation of the csc gene. The remaining 13 isolates contained insertion sequence-mediated disruptions. In NmUS16-1, NmUS16-2, NmUKb13-3, and NmUKb13-4, IS1301-mediated disruption of the csx (where csx is csb, csc, csy, or cszD) gene was also associated with complete disruption of the upstream cssABC operon. In the ST-23 complex, a cluster of eight related isolates from Japan harbored one or two insertion sequence disruptions in the middle of the cssA, cssC, or csy gene. Within this clade, the distribution of insertion sequence families and the pattern of genes disrupted appeared to mirror the core genome maximum likelihood phylogeny (Fig. 1).
The remaining 14 isolates harbored an intact capsule region A; 6 of these isolates (NmSL13-1, NmSL13-3, NmJP05-1, NmIR13-1, NmUKb13-1, and NmUKb13-2) belonged to serogroup B, 6 (NmUS04-1, NmUS03-1, NmFR12-1, NmIT14-1, NmIT14-2, and NmIT14-3) belonged to serogroup C, and 2 (NmJP04-1 and NmUS03-1) belonged to serogroup Y. While the assemblies of three of these isolates contained frameshift mutations in either the csb (NmSL13-1 and NmJP05-1) or csy (NmUS01-2) gene, these mutations were found in homopolymeric tracts of C or A residues that play a role in reversible phase-variable expression, hindering exact prediction of the capsular phenotype (14). With the exception of NmJP05-1, which contained a frameshift mutation in ctrA, all 13 of these isolates also harbored intact region B and C genes; thus, in our sample set, up to one-third of the urethritis-associated N. meningitidis strains are predicted to be encapsulated. Isolate NmFR12-1 from Taha et al. (originally denoted LNP26948) was previously confirmed to produce capsule serogroup C (7), and isolate NmSL13-3 from this study was confirmed to produce capsule serogroup B via slide agglutination. Although we cannot indicate for certain capsule production for the other strains solely from a genomic assessment, in keeping with our observation, we found other reports of encapsulated urethritis-associated N. meningitidis strains throughout the literature (15–17).
Horizontal gene transfer.
Horizontal gene transfer offers a critical source of genetic diversity for neisserial adaptation to environmental pressures, especially with respect to antibiotics. Reduced susceptibility and resistance to the third-generation cephalosporins arises primarily through interspecies horizontal recombination of the penA locus (18); the most common of these mosaic variants is known as the penA XXXIV allele. Deghmane et al. identified this allele in invasive and urethritis-associated meningococci with decreased susceptibility to cefotaxime and ceftriaxone, two of the drugs used to treat meningococcal infection (19). In our sample set, we identified two isolates (NmIT14-3 and NmJP14-2) that contain the penA XXXIV allele. These isolates were within the ST-11 complex and clustered closely together on the phylogenetic tree; furthermore, they shared the same fine type (C:P1.5-1,10-8:F3-6:ST-11) as the invasive meningococcal isolates identified by Deghmane et al. (19). The absence of the penA XXXIV allele in other urethritis-associated isolates suggests that the acquisition of this allele is not related exclusively to urogenital infection and can instead be explained by the clonal spread and diversification of a C:P1.5-1,10-8:F3-6:ST-11 ancestral strain that acquired the penA XXXIV allele from N. gonorrhoeae.
Recombination of other genomic fragments from the gonococcus, which has adapted for sexual transmission, may confer advantages for meningococcal colonization of the genitourinary niche. To undertake a systematic approach for investigating horizontal gene transfer events, we analyzed the core genome shared between our 39 isolates and 13 of the World Health Organization (WHO) gonococcal reference strains (20). Genes that contained signals of gonococcal-to-meningococcal recombination, as detected by the fastGEAR method, in multiple urethritis-associated meningococcal isolates were further investigated. Overall, we found that of the 1,237 identified meningococcal core genes, the majority (1,198, or 96.8%) contained no signal of gonococcal-to-meningococcal recombination. Of the remaining 39 genes, 33 contained only one or two recombination events. dtpT and infB each contained 10 instances of detected recombination; however, the detected recombination loci for dtpT were only weakly supported (Bayes factors, 1.2 to 2.0). Recombinations in infB were more strongly supported (Bayes factors, 10.6 to 61.9) but were generally found in meningococcal lineages phylogenetically the closest to gonococcal lineages, suggesting that identity by descent could also be a possible explanation. Because signals of recombination spanning over the entire gene (e.g., as found in aniA and norB as described above) may be overlooked by fastGEAR, we also examined genes for which at least five meningococcus-derived genes were clustered a priori into gonococcal lineages. Out of these results, we identified only aniA and norB to be hits after filtering out genes with low levels of diversity (as defined by genes with 90% of sites or greater identical). Thus, recombination in the core genome does not appear to be a necessary component of adaptation to the urogenital environment.
Conclusion.
An increase in N. meningitidis-associated cases of urethritis raises the question of an emerging urethrotropic meningococcal clade (4, 6). Recent genomic analyses of N. meningitidis strains isolated from cases of urethritis have suggested that particular alleles of aniA and fHbp and disruptions in the capsule biosynthetic enzymes may be associated with the atypical pathogenesis of these strains (2, 7). To further investigate these associations, we assembled and sequenced a broad convenience sample collection of urethritis-associated N. meningitidis strains collected independently over a decade and across three continents. We found that most isolates belonged to hyperinvasive or emerging invasive clonal complexes, with the remainder being associated with either no clonal complex or unusual ones, such as the cnl invasive ST-198 complex. We found that the nitrite reductase gene aniA is generally intact, but the presence of frameshifts resulting in truncated proteins in nearly a quarter of our isolates implies that N. meningitidis can survive in the microaerophilic environment of the urethra without nitrite reduction. We observed two instances of putative gonococcal-to-meningococcal recombination of the norB-aniA gene cassette, which may promote anaerobic growth in the urogenital tract (8). The previously reported association of an fHbp disruption with urethritis-associated N. meningitidis appears to be a result of the population structure, as the majority of our non-ST-11 complex strains harbored intact fHbp. Finally, we observed substantial diversity in the capsule, including intact open reading frames, insertion sequence disruptions, frameshift mutations, and cnl, showing that urogenital colonization is possible across a range of capsular phenotypes.
On the basis of our findings, a phylogenetically diverse array of N. meningitidis strains can cause urethritis, affirming that the textbook niche specifications of Neisseria species are too narrow. We note several questions, unanswered in this study, that may be promising avenues for future investigation. First, these results do not address whether certain lineages of N. meningitidis may be better adapted to growth and transmission once they are within the urogenital niche. Second, because the sample used in this study was a convenience sample, population-level prevalence and mutational diversity could not be inferred. Epidemiological studies that evaluate the incidence of meningococcal urethritis, meningococcal nasopharyngeal carriage, and sexual behaviors will help distinguish whether increased rates of urethritis may be due to spillover from individuals with higher carriage rates or a higher frequency of orogenital contact or lineage-specific adaptation to the urogenital niche. Subsequent functional analyses will then be required to characterize the role of candidate genes and alleles (8). Third, the association of N. meningitidis with cases of urethritis does not necessarily imply a causal link; for instance, coinfection with other sexually transmitted organisms that cause nongonococcal urethritis (e.g., Chlamydia trachomatis, Mycoplasma genitalium) can occur. In some but not all urethritis cases described here, testing was done to rule out coinfection with C. trachomatis (Table 1). Future studies confirming the causation of urethritis by N. meningitidis should likewise aim to rule out other causes of nongonococcal urethritis. With the increased incidence of N. meningitidis-associated cases of urethritis, as reported by Bazan et al. (4, 6) and Toh et al. (3), leading to concern about the emergence of meningococcal lineages adapted to urogenital infection and transmission, such studies will be critical for informing the appropriate clinical and public health responses.
MATERIALS AND METHODS
Sample collection.
Nongonococcal Neisseria isolates associated with men with urethritis (39 N. meningitidis isolates and 1 N. lactamica isolate) were obtained from the U.S. Centers for Disease Control and Prevention's (CDC's) Gonococcal Isolate Surveillance Project (GISP) (n = 5), the World Health Organization (WHO) Collaborating Centre (CC) for Gonorrhoea and Other Sexually Transmitted Infections (n = 5), the Japanese National Institute of Infectious Diseases (NIID) (n = 19), and from prior studies (n = 11). GISP isolates were strains presumed on collection to be gonococcus but after sequencing were identified by Kraken (21) to be nongonococcal. WHO CC and Japanese isolates were identified through routine culture-based diagnosis by the use of samples from men with urethritis. Isolates were sequenced as described below. Previously published sequences included urethritis-associated N. meningitidis genome sequences described in prior publications (n = 7) and sequences found in the online PubMLST database (n = 4) (Table 1). PubMLST isolates were located by querying the database for isolates by the use of Neisseria meningitidis for the species and urethral for the source. Isolates were named (or renamed for previously published isolates; Table 1) according to the country of origin and year of isolation.
DNA sequencing and analysis.
DNA was prepared from the isolates, and Nextera libraries were constructed using standard protocols, with sequencing being performed on an Illumina platform. FASTQ format reads were quality trimmed using the Sickle tool (https://github.com/najoshi/sickle), underwent quality control in FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and de novo assembled using the SPAdes (version 3.6.2) genome assembler (22). A genus-level phylogeny was constructed using the PhyloPhlAn method (23), incorporating reference strains selected from a Neisseria-wide taxonomy (24). A species-level maximum likelihood phylogeny was constructed with the RAxML (version 8.2.8) tool (25) using a core genome alignment generated by Roary analysis (version 3.6.0) with a minimum BLASTP identity of 90% (26). Meningococcal fine typing was conducted using the meningotype (v0.7-beta) program, and computational serogrouping results were corrected when additional information was available (see “Candidate gene analysis” below) (27). Phylogenetic trees were visualized and annotated in FigTree software (http://tree.bio.ed.ac.uk/software/figtree/).
Candidate gene analysis.
An aniA reference nucleotide sequence from MC58 (GenBank accession number AE002098.2) was used in a BLASTn query with default parameters against a BLAST database built using the contigs from the N. meningitidis urethritis isolate genomes. Top hits were extracted from the contigs. Samples with no BLAST hits were denoted as missing. Each nucleotide sequence was queried against the PubMLST database to identify its allelic number. Sequences which did not match a PubMLST allele were translated to assess whether the peptide was truncated. Nucleotide sequences were aligned using the MAFFT (version 7.017) multiple-sequence alignment program (28) and phylogenetic trees constructed in Geneious (version 9.1.7) software (29) using the neighbor-joining method.
fHbp allele 1 (Pfizer subfamily B, Novartis variant family 1), as defined in the PubMLST database (locus fHbp), was used in a BLASTn query with default parameters against the N. meningitidis urethritis isolate contig database. Top BLASTn results were extracted from the contigs and queried against the PubMLST database to identify its allelic number.
Capsule region A genes were first analyzed using the PubMLST genome query tool (30). Isolates with all genes for a particular capsule locus, as defined elsewhere (13), were denoted as complete. Isolates that lacked capsule genes but matched a particular capsule null locus (cnl) allele, as defined by PubMLST, were indicated accordingly. Many isolates possessed a subset of the capsule genes or lacked capsule genes but did not match a cnl allele in the PubMLST database. These isolates were further characterized using a combination of the BLASTn program to detect capsule genes with novel alleles not in the PubMLST database and the ISmapper program using reference insertion sequences from the ISfinder database (31, 32). Non-cnl isolates which lacked capsule genes were analyzed by mapping sequencing reads to the capsule region A reference sequences in the work of Harrison et al. (13) in the Geneious (version 9.1.7) program. Genes in capsule regions B and C were analyzed by use of BLASTn and the Geneious (version 9.1.7) program.
Recombination detection.
The pangenome and core genome of the urethritis isolates and gonococcal isolates selected from the WHO reference panel (20) were constructed using Roary analysis (26) with 95% identity and the MAFFT (version 7.017) program (28) core genome alignment parameters. Genes in the core genome were selected for further analysis using fastGEAR (33). fastGEAR under default settings was run on each core gene alignment individually, and each gene was scored on the basis of the number of meningococcal isolates which harbored at least one recent recombination of 200 bp or more in size and a log(Bayes factor) greater than 0.5 from the gonococcal lineage. Genes with the highest number of putative directional recombinations were further analyzed via the MAFFT (version 7.017) (28) alignments and nucleotide neighbor-joining trees in Geneious (version 9.1.7) (29). Genes present in meningococcal strains that were initially clustered into gonococcal lineages via fastGEAR were also examined, as these could also represent instances of horizontal recombination.
Accession number(s).
Newly determined sequences have been deposited in GenBank, and the accession numbers can be found via BioProject record number PRJNA384053 (see also Table S1 in the supplemental material).
Supplementary Material
ACKNOWLEDGMENTS
This publication made use of the PubMLST website (http://pubmlst.org/), developed by Keith Jolley (30) and sited at the University of Oxford. The development of that website was funded by the Wellcome Trust.
We thank David Nelson (Indiana University) and Cecilia Fazio (Istituto Superiore di Sanità) for helpful discussions. We also thank the members of the Y. H. Grad lab for helpful advice and assistance.
This work was funded by the Smith Family Foundation (to Y.H.G.), the Harvard Global Health Institute (to K.C.M.), and the Research Committee of Örebro County and the Örebro University Hospital Foundation, Örebro, Sweden (to M.U.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01018-17.
REFERENCES
- 1.Yazdankhah SP, Caugant DA. 2004. Neisseria meningitidis: an overview of the carriage state. J Med Microbiol 53:821–832. doi: 10.1099/jmm.0.45529-0. [DOI] [PubMed] [Google Scholar]
- 2.Toh E, Gangaiah D, Batteiger BE, Williams JA, Arno JN, Tai A, Batteiger TA, Nelson DE. 2017. Neisseria meningitidis ST11 complex isolates associated with nongonococcal urethritis, Indiana, USA, 2015-2016. Emerg Infect Dis 23:336–339. doi: 10.3201/eid2302.161434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison OB, Cole K, Peters J, Cresswell F, Dean G, Eyre DW, Paul J, Maiden MC. 2017. Genomic analysis of urogenital and rectal Neisseria meningitidis isolates reveals encapsulated hyperinvasive meningococci and coincident multidrug-resistant gonococci. Sex Transm Infect 93:445–451. doi: 10.1136/sextrans-2016-052781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazan JA, Peterson AS, Kirkcaldy RD, Briere EC, Maierhofer C, Turner AN, Licon DB, Parker N, Dennison A, Ervin M, Johnson L, Weberman B, Hackert P, Wang X, Kretz CB, Abrams AJ, Trees DL, Del Rio C, Stephens DS, Tzeng Y-L, DiOrio M, Roberts MW. 2016. Notes from the field: increase in Neisseria meningitidis-associated urethritis among men at two sentinel clinics—Columbus, Ohio, and Oakland County, Michigan, 2015. MMWR Morbid Mortal Wkly Rep 65:550–552. doi: 10.15585/mmwr.mm6521a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urra E, Alkorta M, Sota M, Alcala B, Martinez I, Barron J, Cisterna R. 2005. Orogenital transmission of Neisseria meningitidis serogroup C confirmed by genotyping techniques. Eur J Clin Microbiol Infect Dis 24:51–53. doi: 10.1007/s10096-004-1257-7. [DOI] [PubMed] [Google Scholar]
- 6.Bazan JA, Turner AN, Kirkcaldy RD, Retchless AC, Kretz CB, Briere E, Tzeng YL, Stephens DS, Maierhofer C, Del Rio C, Abrams AJ, Trees DL, Ervin M, Licon DB, Fields KS, Roberts MW, Dennison A, Wang X. 8 May 2017. Large cluster of Neisseria meningitidis urethritis in Columbus, Ohio, 2015. Clin Infect Dis. doi: 10.1093/cid/cix215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taha M-K, Claus H, Lappann M, Veyrier FJ, Otto A, Becher D, Deghmane A-E, Frosch M, Hellenbrand W, Hong E, Parent du Châtelet I, Prior K, Harmsen D, Vogel U. 2016. Evolutionary events associated with an outbreak of meningococcal disease in men who have sex with men. PLoS One 11:e0154047. doi: 10.1371/journal.pone.0154047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzeng YL, Bazan JA, Turner AN, Wang X, Retchless AC, Read TD, Toh E, Nelson DE, Del Rio C, Stephens DS. 2017. Emergence of a new Neisseria meningitidis clonal complex 11 lineage 11.2 clade as an effective urogenital pathogen. Proc Natl Acad Sci U S A 114:4237–4242. doi: 10.1073/pnas.1620971114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schork S, Schluter A, Blom J, Schneiker-Bekel S, Puhler A, Goesmann A, Frosch M, Schoen C. 2012. Genome sequence of a Neisseria meningitidis capsule null locus strain from the clonal complex of sequence type 198. J Bacteriol 194:5144–5145. doi: 10.1128/JB.01099-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aspholm M, Aas FE, Harrison OB, Quinn D, Vik A, Viburiene R, Tonjum T, Moir J, Maiden MC, Koomey M. 2010. Structural alterations in a component of cytochrome c oxidase and molecular evolution of pathogenic Neisseria in humans. PLoS Pathog 6:e1001055. doi: 10.1371/journal.ppat.1001055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feavers IM, Maiden MC. 1998. A gonococcal porA pseudogene: implications for understanding the evolution and pathogenicity of Neisseria gonorrhoeae. Mol Microbiol 30:647–656. doi: 10.1046/j.1365-2958.1998.01101.x. [DOI] [PubMed] [Google Scholar]
- 12.Spinosa MR, Progida C, Tala A, Cogli L, Alifano P, Bucci C. 2007. The Neisseria meningitidis capsule is important for intracellular survival in human cells. Infect Immun 75:3594–3603. doi: 10.1128/IAI.01945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison OB, Claus H, Jiang Y, Bennett JS, Bratcher HB, Jolley KA, Corton C, Care R, Poolman JT, Zollinger WD, Frasch CE, Stephens DS, Feavers I, Frosch M, Parkhill J, Vogel U, Quail MA, Bentley SD, Maiden MC. 2013. Description and nomenclature of Neisseria meningitidis capsule locus. Emerg Infect Dis 19:566–573. doi: 10.3201/eid1904.111799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammerschmidt S, Müller A, Sillmann H, Mühlenhoff M, Borrow R, Fox A, van Putten J, Zollinger WD, Gerardy-Schahn R, Frosch M. 1996. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol Microbiol 20:1211–1220. doi: 10.1111/j.1365-2958.1996.tb02641.x. [DOI] [PubMed] [Google Scholar]
- 15.Beck A, Fluker JL, Platt DJ. 1974. Neisseria meningitidis in urogenital infection. Br J Vener Dis 50:367–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagman M, Forslin L, Moi H, Danielsson D. 1991. Neisseria meningitidis in specimens from urogenital sites. Is increased awareness necessary? Sex Transm Dis 18:228–232. doi: 10.1097/00007435-199110000-00006. [DOI] [PubMed] [Google Scholar]
- 17.Judson FN, Ehret JM, Eickhoff TC. 1978. Anogenital infection with Neisseria meningitidis in homosexual men. J Infect Dis 137:458–463. doi: 10.1093/infdis/137.4.458. [DOI] [PubMed] [Google Scholar]
- 18.Grad YH, Harris SR, Kirkcaldy RD, Green AG, Marks DS, Bentley SD, Trees D, Lipsitch M. 2016. Genomic epidemiology of gonococcal resistance to extended-spectrum cephalosporins, macrolides, and fluoroquinolones in the United States, 2000-2013. J Infect Dis 214:1579–1587. doi: 10.1093/infdis/jiw420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deghmane AE, Hong E, Taha MK. 2017. Emergence of meningococci with reduced susceptibility to third-generation cephalosporins. J Antimicrob Chemother 72:95–98. doi: 10.1093/jac/dkw400. [DOI] [PubMed] [Google Scholar]
- 20.Unemo M, Golparian D, Sanchez-Buso L, Grad Y, Jacobsson S, Ohnishi M, Lahra MM, Limnios A, Sikora AE, Wi T, Harris SR. 2016. The novel 2016 WHO Neisseria gonorrhoeae reference strains for global quality assurance of laboratory investigations: phenotypic, genetic and reference genome characterization. J Antimicrob Chemother 71:3096–3108. doi: 10.1093/jac/dkw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segata N, Bornigen D, Morgan XC, Huttenhower C. 2013. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun 4:2304. doi: 10.1038/ncomms3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett JS, Jolley KA, Earle SG, Corton C, Bentley SD, Parkhill J, Maiden MC. 2012. A genomic approach to bacterial taxonomy: an examination and proposed reclassification of species within the genus Neisseria. Microbiology 158:1570–1580. doi: 10.1099/mic.0.056077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwong J, Gonçalves da Silva A, Stinear T, Howden B, Seemann T. 2017. meningotype: in silico typing for Neisseria meningitidis. https://github.com/MDU-PHL/meningotype. [DOI] [PMC free article] [PubMed]
- 28.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawkey J, Hamidian M, Wick RR, Edwards DJ, Billman-Jacobe H, Hall RM, Holt KE. 2015. ISMapper: identifying transposase insertion sites in bacterial genomes from short read sequence data. BMC Genomics 16:667. doi: 10.1186/s12864-015-1860-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mostowy R, Croucher NJ, Andam CP, Corander J, Hanage WP, Marttinen P. 2017. Efficient inference of recent and ancestral recombination within bacterial populations. Mol Biol Evol 34:1167–1182. doi: 10.1093/molbev/msx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.