Abstract
Background
Plasmodium falciparum (Pf) malaria infection is suspected to cause endemic Burkitt Lymphoma (eBL), but the evidence remains unsettled. An inverse relationship between sickle cell trait (SCT) and eBL, which supports that between malaria and eBL, has been reported before, but in small studies with low power. We investigated this hypothesis in children in a population-based study in northern Uganda using Mendelian Randomization.
Methods
Malaria-related polymorphisms (SCT, IL10, IL1A, CD36, SEMA3C, and IFNAR1) were genotyped in 202 eBL cases and 624 controls enrolled during 2010–2015. We modeled associations between genotypes and eBL or malaria using logistic regression.
Findings
SCT was associated with decreased risk of eBL (adjusted odds ratio [OR] 0·37, 95% CI 0·21–0·66; p = 0·0003). Decreased risk of eBL was associated with IL10 rs1800896-CT (OR 0·73, 95% CI 0·50–1·07) and -CC genotypes (OR 0·53, 95% CI 0·29–0·95, ptrend = 0·019); IL1A rs2856838-AG (OR 0·56, 95% CI 0·39–0·81) and -AA genotype (OR 0·50, 95% CI 0·28–1·01, ptrend = 0·0016); and SEMA3C rs4461841-CT or -CC genotypes (OR 0·57, 95% CI 0·35–0·93, p = 0·0193). SCT and IL10 rs1800896, IL1A rs2856838, but not SEMA3C rs4461841, polymorphisms were associated with decreased risk of malaria in the controls.
Interpretation
Our results support a causal effect of malaria infection on eBL.
Keywords: Burkitt Lymphoma, Malaria, Plasmodium falciparum, Mendelian randomization, Sickle cell trait, Malaria resistance genes
Highlights
-
•
Mendelian randomization analysis was done to assess a causal relationship between malaria infection and endemic Burkitt lymphoma in Uganda
-
•
Carriage of the sickle cell trait was associated with decreased risk of endemic Burkitt lymphoma
-
•
Heterozygous or homozygous minor alleles of IL10 rs1800896, IL1A rs2856838, and SEMA3C rs4461841 were associated with decreased risk of endemic Burkitt lymphoma
-
•
The inverse association between sickle cell trait and endemic Burkitt lymphoma supports a causal role of malaria in endemic Burkitt lymphoma
1. Introduction
Endemic Burkitt Lymphoma (eBL) is the most common pediatric cancer in sub-Saharan Africa (Ogwang et al., 2011). Similarity in the geographical distribution of endemic eBL and Plasmodium falciparum (Pf) malaria (Haddow, 1963) and association between high titers of anti-malaria antibodies and eBL risk in case-control studies (Carpenter et al., 2008, Mutalima et al., 2008) have lent support to the hypothesis that malaria is causally-related to risk of eBL. However, concern about reverse causation, confounding(Grimes and Schulz, 2002), and limited understanding of malaria antibody responses (Teo et al., 2016), undermines the inferences that can be made using evidence from ecological and case-control studies. For example, high titer of antibodies against Pf malaria whole schizont lysate (Mutalima et al., 2008, Carpenter et al., 2008) and histidine rich protein 2 antigens are associated with high risk of eBL in children (Aka et al., 2013), whereas high titer of antibodies against Pf malaria SERA5 (SE36) (Aka et al., 2013), a blood-stage vaccine candidate, is associated with decreased risk of eBL.
The Mendelian distribution of polymorphisms related to malaria resistance, such as rs334-A/T in the hemoglobin gene (HBB) that causes the sickle cell trait (SCT) (Jallow et al., 2009) can be utilized to investigate the effect of malaria on outcomes (Smith and Ebrahim, 2003) such as stunting (Kang et al., 2013), bacteremia (Scott et al., 2011), hypertension(Etyang et al., 2016), or eBL. In 1966, Williams reported a statistically significant 2-fold lower frequency of SCT in 100 eBL cases compared to 320 hospital-based controls aged 5–15 years in Nigeria (Williams, 1966). Two studies conducted in Uganda in 1970 (Pike et al., 1970) and Ghana in 1976 (Nkrumah and Perkins, 1976) were inconclusive, but they were limited by small sample sizes with lower power, and lack of covariate data to control for confounding or population structure (Price et al., 2006). Recently, a study in Kenya reported a null association between SCT and eBL risk (Mulama et al., 2014), while a small study in Cameroon reported an inverse association with eBL risk (Hesseling et al., 2016). The Kenyan study had a large sample size (306 cases and 537 controls), but it was not population-based, was limited by selection bias of the controls sampled only from two villages, and adjustment only for sex and age (Mulama et al., 2014).
In this study, we investigated the association between malaria and eBL using a Mendelian Randomization study of SCT and other polymorphisms that potentially affect malaria risk using data from the Epidemiology of Burkitt Lymphoma in East African Children and Minors (EMBLEM) study in Northern Uganda.
2. Materials and Methods
2.1. Subjects, Design, and Participants
We investigated the causal role of malaria infection on eBL risk using Mendelian Randomization (Smith and Ebrahim, 2003). Population-based eBL cases aged 0 through 15 years and healthy controls frequency-matched on age, sex, and geographical region were enrolled in EMBLEM from November 11, 2011 to April 15, 2015 (Maziarz et al., 2017). eBL cases were clinically stable patients enrolled before starting chemotherapy at St. Mary's Hospital, Lacor, in Gulu district and Kuluva Hospital in Arua district from the north-central and northwest regions, respectively. These hospitals were the best equipped to diagnose and treat eBL in northern Uganda. eBL case identification was enhanced using eBL awareness posters, radio programs, and regular outreach to encourage referral of suspected cases to the two hospitals for histological diagnosis and treatment. We restricted case enrollment to the north-central and northwest regions because we wanted a well-defined referent population that could be efficiently targeted for case identification and sampling of population controls. The annual entomological inoculation rate in the study area varies from 397 infectious mosquito bites per year in the northwest region to 1586 infectious bites per year in the north-central region (Okello et al., 2006). The eBL incidence in these regions is also high (1–7 cases per 100,000 children) (Ogwang et al., 2008).
The controls were enrolled as previously described (Maziarz et al., 2017). Briefly, 100 villages were randomly selected from a list of all villages in the study area, stratified by geographical surrogates of malaria transmission risk, namely - rural/urban location, and village proximity to surface water (defined as a swamp, river, or lake). About three unrelated controls per case were selected from a random set of children who had lived in the study area for at least four months before enrollment, and frequency-matched on sex, age group (0–2, 3–5, 6–8, 9–11, 12–15 years), and geographical distribution of the historical eBL cases from the study area (Ogwang et al., 2008). Structured questionnaires were administered to the eBL cases at the hospital and to the controls at their home to record age, sex, the region of residence, and information about in- and out-patient malaria treatment (0–6 months, 7–12 months, or > 13 months ago). Peripheral blood samples for research were collected in 10 ml EDTA tubes from cases at the hospital, and from healthy controls at their home. Additional blood samples were collected in 4 ml EDTA tubes for clinical tests and immediately tested for malaria infection, as previously described, (Maziarz et al., 2017) and for hemoglobin level (g/dl) using QBC Star Dry Hematology Analyzer (Drucker Diagnostics, Port Matilda, PA). The research blood samples were transported in cold boxes to local EMBLEM field laboratories within two hours from sampling, and centrifuged (Eppendorf Centrifuge, Thermo Scientific, Atlanta, GA) for 15 min at 1300g to separate plasma, buffy coat, and red cell fractions. These blood fractions were stored in barcoded cryovials at − 800 °C in local temperature-monitored freezers until shipment in liquid nitrogen to the National Cancer Institute (NCI) Frederick National Cancer Laboratory, Frederick, MD, USA, for long-term storage.
2.2. Ethical Issues
The study implementation conformed to the standards indicated by the Declaration of Helsinki. Ethical approval was given by the Uganda Virus Research Institute Research and Ethics Committee, the Uganda National Council for Science and Technology, and the NCI Special Studies Institutional Review Boards. Written informed consent was obtained from the guardians of all participants, and informed assent was obtained from the participants aged ≥ 8 years old.
2.3. Procedures
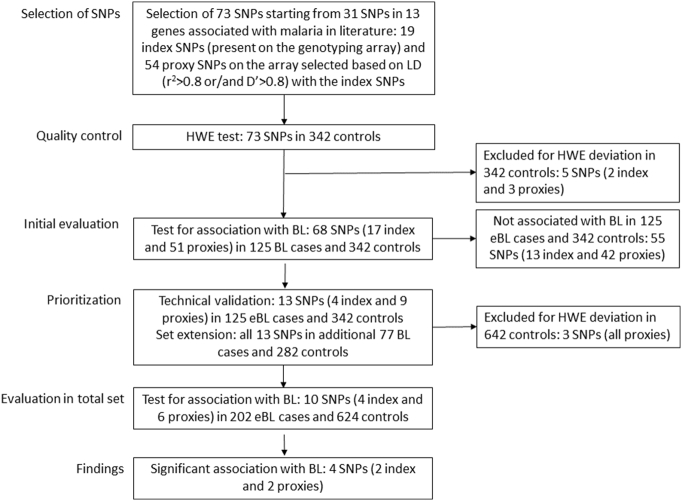
Genomic DNA was extracted from previously unthawed buffy coat samples using the Qiagen QIAsymphony automated instrument at the NCI Cancer Genomics Research (CGR) Laboratory, USA. All genotyping was performed blinded at the NCI, USA using the Infinium Omni5Exome-4 v1.3 BeadChip (Illumina, San Diego, CA, USA; 4,641,218 SNPs), TaqMan assays (Supplementary Table 3), or Sanger sequencing (for rs334 in HBB, Supplementary Fig. 1). Illumina chip genotypes available on 125 eBL cases and 342 controls were used to prioritize SNPs for evaluation in the current study (Fig. 1) and to define population structure using principal components analysis (PCA) performed on the genotypes of 700,000 genome-wide SNPs (Price et al., 2006). Significant eigenvectors were found to correlate with the region of enrollment (northwest or north-central). Thus, the region of enrollment was used to adjust for ancestry in all analyses in the total data set that also included samples without genome-wide Illumina data.
Fig. 1.
Flow chart showing strategy for selecting index single nucleotide polymorphisms (SNPs) or their proxies for evaluation in the current study.
As shown in Fig. 1, we selected 73 SNPs in 13 genes (Supplementary Table 1) to test their associations with eBL risk in 125 eBL cases and 342 controls with the Illumina genotypes. These 73 SNPs were selected by identifying a core set of 31 index SNPs (Supplementary Table 2), as instrumental variables for malaria exposure, based on published literature on the polymorphisms associated with protection against severe malaria infection or immune response to malaria. Nineteen of the 31 index SNPs were present on the Illumina chip, and 54 proxy SNPs were selected for being in strong linkage disequilibrium (LD) (r2 > 0.8 or/and D′ > 0.8) with the 19 index SNPs. Five SNPs (two index and three proxy SNPs) not in Hardy-Weinberg equilibrium (HWE) in the controls (p < 0·05) were excluded at this stage. The remaining 68 SNPs (17 index and 51 proxy) were evaluated for association with eBL in the 125 eBL cases and 342 controls (Supplementary Fig. 2 and Supplementary Table 1). Thirteen SNPs (four index and nine proxy SNPs, Supplementary Table 3) in six genes associated with eBL (p < 0·05) at this stage were prioritized for further analysis. These 13 SNPs were re-genotyped with TaqMan assays or Sanger sequencing for technical validation of the Illumina chip results, with 100% concordance, and genotyped in an additional 77 eBL cases and 282 controls to increase sample size. Three IFNAR1 SNPs (rs2253413, rs914141, and rs2856973) not in HWE in the total set of controls (p < 0·05) were excluded, leaving ten SNPs, 202 cases and 624 controls for analysis in the total set.
2.4. Statistical Analysis
HWE for each SNP was evaluated with χ2 tests in the controls. Odds ratios and 95% confidence intervals (ORs, 95% CIs) for the association of each SNP with risk of eBL, or malaria in controls, were estimated using logistic regression. We used likelihood ratio tests to compute p-values, with p < 0·05 being considered significant. Statistical tests were two-sided. SNP genotypes were coded using an additive genetic model that counts the number of minor alleles. Genotypes for uncommon alleles were coded by combining the minor homozygotes and heterozygotes in one group. Models were adjusted for possible confounders selected a priori based on their known associations with malaria infection status (Maziarz et al., 2017), including age groups (0–2, 3–5, 6–8, 9–11, 12–15 years), sex, malaria transmission risk (rural/urban and proximity to surface water based on village boundary within/more than 500 m), malaria infection status based on thick film microscopy or rapid diagnostic test (Maziarz et al., 2017), and lifetime malaria treatment as an inpatient or outpatient. As fever at the time of enrollment was rare (observed in only 2.1% of the controls), malaria test positivity in the controls was considered asymptomatic and incidental. Population structure was adjusted for by including into analysis region of enrollment (Price et al., 2006), which was correlated with eigenvectors in the PCA. Missing data was coded as a specific category, but individuals with missing data for the exposure variable of interest were excluded from the model when estimating the association. Analyses were performed using Stata SE 14 (College Park, TX). Because malaria immunity increases with age, with younger children relying more on innate immunity to control infection and older children relying more on adaptive immunity (Williams et al., 2005), analyses of eBL risk were stratified on age (< 9 years, 124 eBL cases) versus older children (9–15 years, 78 eBL cases), based on the peak age of malaria prevalence in the controls (Maziarz et al., 2017). We evaluated the associations between asymptomatic malaria infection, defined as described above, with the ten SNPs in secondary analyses restricted only to the controls. We estimated that a study of 465 subjects in 1:3 case-control ratio would have 80% power to reject a null hypothesis that SCT is randomly distributed in the cases and controls at an α = 0.05 and a prevalence of SCT of 20% in the general population in northern Uganda (Ndeezi et al., 2016). Because the index SNPs were selected for their association with malaria, the results for those SNPs were not adjusted for multiple comparisons. The results for the proxy SNPs were considered exploratory.
3. Results
Consistent with the study design, the eBL cases and controls were comparable with regard to age, sex, residence in the rural or urban villages, and region of enrollment (Table 1). Among subjects with complete data, 167 of 193 (86·5%) of the eBL cases lived in a village near surface water compared to 449 of 624 (72·0%) of the controls (p < 0·001). Similarly, 130 of 181 (71·8%) eBL cases had mild anemia compared to 144 of 604 (23·8%) controls (p < 0·001). There were 50 of 199 (25·1%) eBL cases who reported inpatient malaria treatment > 12 months before enrollment compared to 120 of 619 (19·4%) controls (p = 0·016). The corresponding numbers of participants reporting outpatient malaria treatment > 13 months before enrollment was 50 of 199 (25·1%) for eBL cases compared to 58 of 619 (9·4%) for controls (p < 0·001). Paradoxically, malaria infection was detected only in 70 of 199 (35·2%) eBL cases compared to 351 of 622 (56·3%) controls (p < 0·001).
Table 1.
Demographic and clinical characteristics of the study population.
| Characteristic | Cases, n (%) | Controls, n (%) | p value |
|---|---|---|---|
| All subjects | 202 (100) | 624 (100) | |
| Age, years (mean ± SD) | 7·70 (3·28) | 7·34 (3·40) | 0·186 |
| Age group, years | 0·568 | ||
| 0–2 | 7 (3·5) | 37 (5·9) | |
| 3–5 | 52 (25·7) | 174 (27·8) | |
| 6–8 | 65 (32·2) | 197 (31·6) | |
| 9–11 | 46 (22·8) | 134 (21·5) | |
| 12–15 | 32 (15·8) | 82 (13·1) | |
| Sexa | 0·126 | ||
| Males | 122 (61·3) | 344 (55·1) | |
| Females | 77 (38·7) | 280 (44·9) | |
| Rural/urbana | 0·56 | ||
| Rural | 118 (61·1) | 396 (63·5) | |
| Urban | 75 (38·9) | 228 (36·5) | |
| Proximity to surface watera | < 0·001 | ||
| Near | 167 (86·5) | 449 (72·0) | |
| Far | 26 (13·5) | 175 (28·0) | |
| Study region | 0·499 | ||
| Northwest | 42 (20·8) | 144 (23·1) | |
| North-central | 160 (79·2) | 480 (76·9) | |
| Anemiaa | < 0·001 | ||
| Not anemic (Hb > 11.6 g/dl) | 51 (28·2) | 460 (76·2) | |
| Anemic (Hb ≤ 11.6 g/dl) | 130 (71·8) | 144 (23·8) | |
| Inpatient malariaa | 0·016 | ||
| No | 135 (67·8) | 413 (66·7) | |
| Past 12 months | 14 (7·0) | 86 (13·9) | |
| > 13 months | 50 (25·1) | 120 (19·4) | |
| Outpatient malariaa | < 0·001 | ||
| No | 61 (30·6) | 141 (22·8) | |
| Past 12 months | 88 (44·2) | 420 (67·8) | |
| > 13 months | 50 (25·1) | 58 (9·4) | |
| Malaria infectiona, b | < 0·001 | ||
| Negative | 129 (64·8) | 271 (43·6) | |
| Positive | 70 (35·2) | 351 (56·4) |
Computation of percentages did not include categories with missing information. Three subjects with discordant gender on different forms were coded as missing for gender.
Malaria infection status was determined by thick film microscopy or rapid diagnostic test results as described in Maziarz et al., 2017. As < 2·1% of the controls had a current fever, malaria infection was considered incidental in asymptomatic subjects.
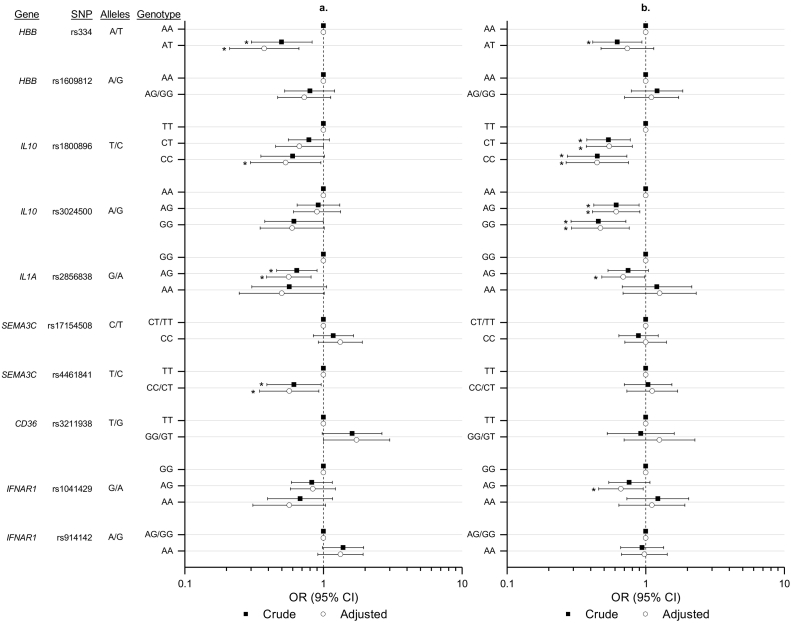
3.1. Genotype Associations With Burkitt Lymphoma
The genotype associations with eBL for 10 SNPs tested in the total sample set are shown in Fig. 2 and Supplementary Table 4. SCT (HBB rs334-AT genotype) was detected in 113 (18·2%) of 620 controls compared to 20 (10·0%) of 200 eBL cases, resulting in a significantly decreased crude OR of eBL risk (OR = 0·50, 95% CI 0·30–0·83). The significantly decreased risk of eBL associated with SCT became more pronounced after controlling for confounders (ORadj = 0·37, 95% CI 0·21–0·66). Carriage of homozygous HBB rs334-TT genotype, which corresponds to the sickle cell disease, was seen only in four controls and none of the eBL cases. These children were excluded from the analysis of SCT. In contrast, HBB rs160912 was unrelated to eBL.
Fig. 2.
Forest plots showing the associations between selected index or proxy SNPs with eBL risk (a) or with malaria infection in the controls (b). The open circles show crude odds ratios (ORs) and the black squares show ORs adjusted for covariates (see methods). The horizontal lines with vertical bars at the end depict 95% confidence intervals of the ORs. *Indicates the results that are statistically significant at p ≤ 0·05 (see text and Supplementary tables for details).
We also observed a significantly decreasing trend of eBL risk with carriage of heterozygous or homozygous minor alleles for index SNP IL10 rs1800896, and proxy SNPs IL1A rs2856838 and SEMA3C rs4461841 (Fig. 2). Compared to children with the IL10 rs1800896-TT genotype, carriage of -CT genotype (ORadj = 0·73, 95% CI 0·50–1·07) and -CC genotype (ORadj = 0·53, 95% CI 0·29–0·95) was associated with a significantly decreasing trend of the adjusted risk of eBL (ptrend = 0·0195). Compared to children with the IL1A rs2856838-GG genotype, carriage of -AG genotype (ORadj = 0·56, 95% CI 0·39–0·81) and -AA genotype (ORadj = 0·50, 95% CI 0·25–1·01) was associated with a significantly decreasing trend of the adjusted risk of eBL (ptrend = 0·0016). Compared to children with the SEMA3C rs4461841-TT genotype, carriage of the -CC and -CT genotypes were associated with a significantly decreased risk of eBL (ORadj = 0·57, 95% CI 0·35–0·93; pheterogeneity = 0·0193).
The minor allele for CD36 rs3211938-G was uncommon (4.4%) among the controls. Consistent with the literature that CD36 rs3211938-G is potentially associated with malaria susceptibility (Aitman et al., 2000) carriage of the -GG/-GT genotypes was associated with an elevated risk of eBL (ORadj = 1·73, 95% CI 1·00–3·00; pheterogeneity = 0·055) compared to carriage of the -TT genotype. Carriage of IFNAR1 rs914142-AA was associated with a non-significantly elevated risk of eBL (ORadj = 1·32, 95% CI 0·90–1·92; pheterogeneity = 0·152). Carriage of SEMA3C rs17154508 and IFNAR1 rs1041429 were not associated with significant increase or decrease in eBL risk.
The reduced risk of eBL associated with SCT, IL10 rs1800896, IL1A rs2856838, and SEMA3C rs4461841 genotypes remained statistically significant in analyses adjusting these genotypes for one another (Fig. 2 and Supplementary Table 4).
3.2. Analysis Stratified by Age
Stratified analysis of eBL risk was evaluated for in children < 9 vs. ≥ 9 years (Table 2). The risk of eBL associated with SCT was statistically significant both in younger (ORadj = 0·39, 95% CI 0·18–0·84) and older (ORadj = 0·36, 95% CI 0·15–0·89) children. The adjusted association of eBL risk with CD36 rs3211938-GG or GT genotypes increased from 1·43 (95% CI 0·68–2·98) in younger children to 2·51 (95% CI 1·05–6·04) in older children, but the ORs were not statistically different from each other (pheterogeneity = 0·197). The association of eBL risk with IFNAR1 rs914142-AA was significantly elevated in younger children (ORadj = 1·99, 95% CI 1·21–3·28), but not in older children (ORadj = 0·75, 95% CI 0·40–1·39; pheterogeneity = 0·015).
Table 2.
Association between genotypes of index malaria resistance polymorphisms or their proxies with Burkitt Lymphoma, stratified by age group.
| Gene | SNP | Genotype | < 9 years |
9–15 years |
p-Value for heterogeneity |
|---|---|---|---|---|---|
| Adjusted OR (95% CI)c | Adjusted OR (95% CI)c | ||||
| IL10 | rs1800896a | IL10b | |||
| TT | Ref. | Ref. | |||
| CT | 0·87 (0·54–1·41) | 0·55 (0·29–1·05) | |||
| CC | 0·40 (0·18–0·91) | 0·66 (0·26–1·64) | |||
| p = 0·064 | p = 0·191 | p = 0·445 | |||
| IL10 | rs3024500a | IL10 | |||
| AA | Ref. | Ref. | |||
| AG | 0·93 (0·57–1·51) | 0·80 (0·40–1·60) | |||
| GG | 0·47 (0·23–0·99) | 0·742 (0·32–1·17) | |||
| p = 0·096 | p = 0·756 | p = 0·465 | |||
| IL1A | rs2856838 | IL1A | |||
| GG | Ref. | Ref. | |||
| AG | 0·67 (0·42–1·07) | 0·48 (0·25–0·900) | |||
| AA | 0·51 (0·19–1·36) | 0·60 (0·20–1·82) | |||
| p = 0·143 | p = 0·066 | p = 0·697 | |||
| CD36 | rs3211938a | CD36 | |||
| TT | Ref. | Ref. | |||
| GG/GT | 1·43 (0·68–2·98) | 2·51 (1·05–6·041) | |||
| p = 0·354 | p = 0·040 | p = 0·197 | |||
| SEMA3C | rs17154508 | SEMA3C | |||
| CT/TT | Ref. | Ref. | |||
| CC | 0·89 (0·56–1·42) | 0·57 (0·31–1·070) | |||
| p = 0·628 | p = 0·075 | p = 0·284 | |||
| SEMA3C | rs4461841 | SEMA3C | |||
| TT | Ref. | Ref. | |||
| CC/CT | 0·61 (0·33–1·15) | 0·49 (0·21–1·16) | |||
| p = 0·115 | p = 0·092 | p = 0·567 | |||
| HBB | rs1609812 | HBB | |||
| AA | Ref. | Ref. | |||
| AG/GG | 1·57 (0·90–2·73) | 1·30 (0·59–2·85) | |||
| p = 0·116 | p = 0·514 | p = 0·587 | |||
| HBB | rs334a | HBB | |||
| AA | Ref. | Ref. | |||
| AT | 0·39 (0·18–0·84) | 0·36 (0·15–0·89) | |||
| p = 0·010 | p = 0·019 | p = 0·930 | |||
| IFNAR1 | rs1041429 | IFNAR1 | |||
| GG | Ref. | Ref.) | |||
| AG | 0·92 (0·43–1·49) | 0·76 (0·40–1·42) | |||
| AA | 0·48 (0·22–1·04) | 0·72 (0·25–2·06) | |||
| p = 0·141 | p = 0·640 | p = 0·477 | |||
| IFNAR1 | rs914142 | IFNAR1 | |||
| AG/GG | Ref. | Ref. | |||
| AA | 1·99 (1·21–3·28) | 0·75 (0·40–1·39) | |||
| p = 0·007 | p = 0·356 | p = 0·015 |
Index SNP, otherwise proxy SNP.
SNP genotypes were coded using an additive genetic model that counts the number of minor alleles. Genotypes for uncommon alleles were coded by combining the minor homozygotes and heterozygotes in one group.
Adjusted for gender, age group, current malaria infection (see text in methods), region, inpatient and outpatient malaria treatment (base model). Individuals with missing data for the exposure variable of interest were excluded from the model when estimating the association.
3.3. Genotype Associations With Asymptomatic Malaria Infection
The risks of malaria infection associated with the 10 SNPs in the controls are shown in Fig. 2 and Supplementary Table 5. SCT was associated with decreased risk of malaria infection, but in keeping with other studies (Billo et al., 2012), the effect was modest (ORadj = 0·74 (0·48–1·14). Carriage of heterozygous or homozygous minor alleles of index SNPs IL10 rs1800896 and IL10 rs3024500, and proxy SNPs IL1A rs2856838 and IFNAR1 rs1041429 was associated with decreased risk of malaria (all p < 0·05). Proxy SNP SEMA3C rs4461841, which was associated with significantly decreased risk of eBL, was not associated with malaria (Supplementary Table 5).
4. Discussion
We found that SCT was associated with a two-fold lower risk of eBL using Mendelian Randomization in a population-based study in northern Uganda, where we previously demonstrated high malaria prevalence (Maziarz et al., 2017). We observed a similar effect of SCT on eBL in younger and older children, suggesting that the effect of SCT on eBL is similar regardless of whether a child relies more on adaptive or innate malaria immune responses. Our results confirm those reported by Williams in Nigeria (Williams, 1966), by Pike and colleagues in Uganda (Pike et al., 1970), and by Hesseling and colleagues in Cameroon (Hesseling et al., 2016), albeit from smaller studies that were under powered. Our results strengthen the evidence of a causal link between malaria and eBL that was previously suggested by observational epidemiology (Bouvard et al., 2012).
We also found that the index SNP IL10 rs1800896 was significantly inversely associated with malaria infection among controls, consistent with reports that it is protective against malaria infection in the Brazilian Amazon (Da Silva Santos et al., 2012), Cameroon (Apinjoh et al., 2013), Southwest Benin (Lokossou et al., 2013), Malawi, Kenya, Ghana, and Tanzania (Kariuki et al., 2013), and that it was associated with significantly decreased risk of eBL. The inverse associations with both malaria and eBL suggest the effect of IL10 rs1800896 on eBL may be mediated by protection against malaria, and suggest that IL10 rs1800896 is an additional polymorphism to SCT that influences eBL risk by modulating malaria risk. SCT and IL10 rs1800896 may influence the risk of eBL by decreasing the frequency of malaria, the peak malaria parasite density, the duration of parasitemia, or the degree of inflammation per infection. We hypothesize that the effects of these polymorphisms lead to decreased cumulative load of malaria antigens reaching the germinal center B cells (Robbiani et al., 2015), attenuation of effects of malaria infections on Epstein-Barr infection (Rochford et al., 2005), and, plausibly, a proportionate decrease in the stimulation of germinal center B cells and decrease in the risk of developing DNA modifications necessary for initiating eBL.
Our finding of significant inverse associations between eBL risk and proxy SNPs IL1A rs2856838 and SEMA3C rs4461841 are novel, and warrant further exploration. SNP rs2856838 is located within an intron of IL1A and has no known functional significance or association with mRNA expression in human tissues (Consortium, 2013). It was selected as a proxy (r2 = 0·085; D′ = 1·00) for index SNP IL1A rs17561 (Ala114Ser), which is associated with malaria infection (Walley et al., 2004), but was not associated with eBL risk at the priortization stage (Supplementary Fig. 2), while its proxy rs2856838 was associated with eBL risk and with malaria in the controls (Fig. 2). Similarly, SEMA3C rs4461841 was selected as a proxy (r2 = 0·006 and D′ = 1·00) for index SNP CD36 rs3211938 (Tyr325STOP), which is under positive selection in endemic malaria populations, including the Yoruba (Aitman et al., 2000). CD36 mediates cytoadherence of the Pf erythrocyte membrane protein 1 (PfEMP1) virulence protein (Ockenhouse et al., 1989) which mediates the sequestration of infected erythrocytes in peripheral tissues and impairs the clearance of parasites by stopping filtration of parasites through the spleen (Smith et al., 2013). The strong D′ (1.00) but low r2 (0.006) between the SNPs suggests that they are residing on the same haplotype. Both SEMA3C rs4461841 and CD36 rs3211938 were associated with eBL risk, but the association was stronger with rs4461841 than with rs3211938.
Our questionnaire data support the hypothesis that eBL cases are more likely to be exposed to heavy malaria than controls, based on being more likely to live in a village near surface water, to report inpatient or outpatient malaria history > 13 months before enrollment, and to have mild anemia. The results also illustrate the difficulties of using observational epidemiological methods to measure complex exposures, such as malaria. For example, using blood tests for malaria and questionnaire data, we found that eBL cases were less likely to test positive for malaria and less likely to report inpatient or outpatient malaria history < 13 months before enrollment, which are paradoxical given the associations noted above suggesting greater likelihood of the eBL cases to be exposed to heavy malaria. Similar paradoxical findings of malaria parasitemia have been reported in Kenya (Asito et al., 2010) and in a prospective study in Uganda (De-The et al., 1978), but they were dismissed based on casual reports and assumptions that eBL cases were more likely to have been treated with anti-malarial drugs prior to admission than controls. We excluded this explanation using our more detailed data. For example, the results from thick film microscopy (TFM), which measures current malaria, were similar to those from rapid diagnostic tests (RDT), which remain positive for 3–4 weeks after treatment of symptomatic malaria (Grandesso et al., 2016), and the questionnaire data on inpatient and outpatient malaria history 12 months before enrollment suggested lower malaria morbidity in the eBL cases than the controls. These paradoxical results may be a clue to the nature of malaria immunity in eBL cases that increases their tolerance for acute malaria morbidity (Drakeley et al., 2005), while exposing them to heightened risk of other, later-onset, malaria complications, such as eBL. This paradox may be examined by invoking the Greek myth of Charybdis and Scylla, where children exposed to heavy malaria are evolutionarily adapted to reduce their risks of complications of acute clinical malaria infections, which are associated with higher mortality. This tolerant state to malaria may manifest as a lower frequency of malaria parasitemia and of inpatient and outpatient malaria symptoms, but with a long-term increase in the risk of eBL due to chronic antigenic stimulation (Robbiani et al., 2015), probably by low level genetically diverse malaria strains (Johnston et al., 2014).
The strengths of our study include the use of well-matched cases and controls selected from a geographically defined population-based study area with known malaria prevalence (Maziarz et al., 2017), detailed covariate data about malaria exposures, data to assess the impact of population genetic structure, and using multiple genotyping methods to assess polymorphisms. The associations with proxy SNPs may represent false-positive findings or novel findings that should be explored further. Our results encourage a more complete assessment of the effect of malaria on eBL using Mendelian Randomization analysis. For example the effects on eBL of genes whose protection against malaria has been reported in recent studies, such as blood group O (Fry et al., 2008), G6PD (Clarke et al., 2017), FREM3/GYPE (Malaria Genomic Epidemiology et al., 2015), EPB41, GYPB (Leffler et al., 2017), and ATP2B4 and MARVELD3 (Timmann et al., 2012) were not assessed in the current study because they were not covered by the Infinium Omni5Exome-4 v1.3 BeadChip used to prioritize SNPs.
In summary, we found, consistent with the literature, that SCT and IL10 rs1800896 were associated with decreased risk of asymptomatic malaria in the controls and associated with decreased risk of eBL using Mendelian Randomization analysis. We found inverse associations between IL1A SNPs rs2856838 and SEMA3C rs4461841 with eBL, which require further study. Our findings of inverse associations between SCT and IL10 rs1800896 using Mendelian Randomization support a causal effect of malaria on eBL. These results support the idea of strengthening of public health messages to stress eBL as a complication of malaria infection.
Funding Sources
The study was funded by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute (NCI) (Contracts HHSN261201100063C and HHSN261201100007I) and, in part, by the Intramural Research Program, National Institute of Allergy and Infectious Diseases (SJR), National Institutes of Health, Department of Health and Human Services. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. The content of this manuscript is the sole responsibility of the authors.
Declaration of Interests
We declare no competing interest.
Author Contributors
SMM, MDO, SJR, and PK conceived the idea, designed the study and supervised fieldwork. IDL, IO, SK, PK, HN, MDO, KB, SJR and JJG conducted and monitored field work. AB, KIU, BE, EK, AWB, MHG, MY and LPO conducted genetic testing and analyzed genetic data. RMP, AOT, KB, RJB, LA, and JFF contributed to the design, analysis, and interpretation of data. All authors contributed to the manuscript read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank the study subjects for their participation. We thank Ms. Janet Lawler-Heavner at Westat Inc. (Rockville, MD, USA) and Mr. Erisa Sunday at the African Field Epidemiology Network (Kampala, Uganda) for managing the study. We are grateful to Mr. Wilson Nyegenye at Uganda Bureau of Statistics (Kampala, Uganda) for training EMBLEM staff in field survey methods. We thank Ms. Laurie Buck, Dr. Carol Giffen, and Mr. Greg Rydzak at Information Management Services Inc. (Calverton, MD, USA) for coordinating data, preparing data analysis files. We thank Mr. David Check at Biostatistics Branch, DCEG (Bethesda, Maryland) for his help drawing graphs.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2017.09.037.
Contributor Information
Ruth M. Pfeiffer, Email: pfeiffer@mail.nih.gov.
Krizia-Ivana Udquim, Email: krizia-ivana.udquim@nih.gov.
Eric Karlins, Email: karlinser@mail.nih.gov.
Benjamin Emmanuel, Email: ben.emmanuel@umaryland.edu.
Kishor Bhatia, Email: kishor.bhatia@cgix.com.
Meredith Yeager, Email: yeagerm@mail.nih.gov.
Leona W. Ayers, Email: Leona.Ayers@osumc.edu.
Steven J. Reynolds, Email: sreynol6@jhmi.edu.
Martin D. Ogwang, Email: ogwang.martin@lacorhospital.org.
Joseph F. Fraumeni, Jr, Email: fraumenj@exchange.nih.gov.
Ludmila Prokunina-Olsson, Email: prokuninal@mail.nih.gov.
Sam M. Mbulaiteye, Email: mbulaits@mail.nih.gov.
Appendix A. Supplementary data
Supplementary material
References
- Aitman T.J., Cooper L.D., Norsworthy P.J., Wahid F.N., Gray J.K., Curtis B.R., Mckeigue P.M., Kwiatkowski D., Greenwood B.M., Snow R.W., Hill A.V., Scott J. Malaria susceptibility and CD36 mutation. Nature. 2000;405:1015–1016. doi: 10.1038/35016636. [DOI] [PubMed] [Google Scholar]
- Aka P., Vila M.C., Jariwala A., Nkrumah F., Emmanuel B., Yagi M., Palacpac N.M., Periago M.V., Neequaye J., Kiruthu C., Tougan T., Levine P.H., Biggar R.J., Pfeiffer R.M., Bhatia K., Horii T., Bethony J.M., Mbulaiteye S.M. Endemic Burkitt lymphoma is associated with strength and diversity of plasmodium falciparum malaria stage-specific antigen antibody response. Blood. 2013;122:629–635. doi: 10.1182/blood-2012-12-475665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apinjoh T.O., Anchang-Kimbi J.K., Njua-Yafi C., Mugri R.N., Ngwai A.N., Rockett K.A., Mbunwe E., Besingi R.N., Clark T.G., Kwiatkowski D.P., Achidi E.A., Malaria G.E.N.C. Association of cytokine and toll-like receptor gene polymorphisms with severe malaria in three regions of Cameroon. PLoS One. 2013;8:e81071. doi: 10.1371/journal.pone.0081071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asito A.S., Piriou E., Odada P.S., Fiore N., Middeldorp J.M., Long C., Dutta S., Lanar D.E., Jura W.G., Ouma C., Otieno J.A., Moormann A.M., Rochford R. Elevated anti-Zta IgG levels and EBV viral load are associated with site of tumor presentation in endemic Burkitt's lymphoma patients: a case control study. Infect. Agent Cancer. 2010;5(13) doi: 10.1186/1750-9378-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billo M.A., Johnson E.S., Doumbia S.O., Poudiougou B., Sagara I., Diawara S.I., Diakite M., Diallo M., Doumbo O.K., Tounkara A., Rice J., James M.A., Krogstad D.J. Sickle cell trait protects against plasmodium falciparum infection. Am. J. Epidemiol. 2012;176(Suppl. 7):S175–185. doi: 10.1093/aje/kws323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvard V., Baan R.A., Grosse Y., Lauby-Secretan B., El Ghissassi F., Benbrahim-Tallaa L., Guha N., Straif K. Carcinogenicity of malaria and of some polyomaviruses. Lancet Oncol. 2012;13:339–340. doi: 10.1016/s1470-2045(12)70125-0. [DOI] [PubMed] [Google Scholar]
- Carpenter L.M., Newton R., Casabonne D., Ziegler J., Mbulaiteye S., Mbidde E., Wabinga H., Jaffe H., Beral V. Antibodies against malaria and Epstein-Barr virus in childhood Burkitt lymphoma: a case-control study in Uganda. Int. J. Cancer. 2008;122:1319–1323. doi: 10.1002/ijc.23254. [DOI] [PubMed] [Google Scholar]
- Clarke G.M., Rockett K., Kivinen K., Hubbart C., Jeffreys A.E., Rowlands K., Jallow M., Conway D.J., Bojang K.A., Pinder M., Usen S., Sisay-Joof F., Sirugo G., Toure O., Thera M.A., Konate S., Sissoko S., Niangaly A., Poudiougou B., Mangano V.D., Bougouma E.C., Sirima S.B., Modiano D., Amenga-Etego L.N., Ghansah A., Koram K.A., Wilson M.D., Enimil A., Evans J., Amodu O.K., Olaniyan S., Apinjoh T., Mugri R., Ndi A., Ndila C.M., Uyoga S., Macharia A., Peshu N., Williams T.N., Manjurano A., Sepulveda N., Clark T.G., Riley E., Drakeley C., Reyburn H., Nyirongo V., Kachala D., Molyneux M., Dunstan S.J., Phu N.H., Quyen N.N., Thai C.Q., Hien T.T., Manning L., Laman M., Siba P., Karunajeewa H., Allen S., Allen A., Davis T.M., Michon P., Mueller I., Molloy S.F., Campino S., Kerasidou A., Cornelius V.J., Hart L., Shah S.S., Band G., Spencer C.C., Agbenyega T., Achidi E., Doumbo O.K., Farrar J., Marsh K., Taylor T., Kwiatkowski D.P., Malaria, G. E. N. C Characterisation of the opposing effects of G6PD deficiency on cerebral malaria and severe malarial anaemia. elife. 2017;6 doi: 10.7554/eLife.15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva Santos S., Clark T.G., Campino S., Suarez-Mutis M.C., Rockett K.A., Kwiatkowski D.P., Fernandes O. Investigation of host candidate malaria-associated risk/protective SNPs in a Brazilian Amazonian population. PLoS One. 2012;7:e36692. doi: 10.1371/journal.pone.0036692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-The G., Geser A., Day N.E., Tukei P.M., Williams E.H., Beri D.P., Smith P.G., Dean A.G., Bronkamm G.W., Feorino P., Henle W. Epidemiological evidence for causal relationship between Epstein-Barr virus and Burkitt's lymphoma from Ugandan prospective study. Nature. 1978;274:756–761. doi: 10.1038/274756a0. [DOI] [PubMed] [Google Scholar]
- Drakeley C.J., Corran P.H., Coleman P.G., Tongren J.E., Mcdonald S.L., Carneiro I., Malima R., Lusingu J., Manjurano A., Nkya W.M., Lemnge M.M., Cox J., Reyburn H., Riley E.M. Estimating medium- and long-term trends in malaria transmission by using serological markers of malaria exposure. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5108–5113. doi: 10.1073/pnas.0408725102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etyang A.O., Smeeth L., Cruickshank J.K., Scott J.A. The malaria-high blood pressure hypothesis. Circ. Res. 2016;119:36–40. doi: 10.1161/CIRCRESAHA.116.308763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.E., Griffiths M.J., Auburn S., Diakite M., Forton J.T., Green A., Richardson A., Wilson J., Jallow M., Sisay-Joof F., Pinder M., Peshu N., Williams T.N., Marsh K., Molyneux M.E., Taylor T.E., Rockett K.A., Kwiatkowski D.P. Common variation in the ABO glycosyltransferase is associated with susceptibility to severe Plasmodium falciparum malaria. Hum. Mol. Genet. 2008;17:567–576. doi: 10.1093/hmg/ddm331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandesso F., Nabasumba C., Nyehangane D., Page A.L., Bastard M., De Smet M., Boum Y., Etard J.F. Performance and time to become negative after treatment of three malaria rapid diagnostic tests in low and high malaria transmission settings. Malar. J. 2016;15:496. doi: 10.1186/s12936-016-1529-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes D.A., Schulz K.F. Bias and causal associations in observational research. Lancet. 2002;359:248–252. doi: 10.1016/S0140-6736(02)07451-2. [DOI] [PubMed] [Google Scholar]
- Haddow A.J. An improved map for the study of Burkitt's lymphoma syndrome in Africa. East Afr. Med. J. 1963;40:429–432. [PubMed] [Google Scholar]
- Hesseling P.B., Jam D.T., Palmer D.D., Wharin P., Tuh G.S., Bardin R., Kidd M. Burkitt's lymphoma patients in Northwest Cameroon have a lower incidence of sickle cell trait (Hb AS) than healthy controls. S. Afr. Med. J. 2016;106 doi: 10.7196/SAMJ.2016.v106i7.10693. [DOI] [PubMed] [Google Scholar]
- Jallow M., Teo Y.Y., Small K.S., Rockett K.A., Deloukas P., Clark T.G., Kivinen K., Bojang K.A., Conway D.J., Pinder M., Sirugo G., Sisay-Joof F., Usen S., Auburn S., Bumpstead S.J., Campino S., Coffey A., Dunham A., Fry A.E., Green A., Gwilliam R., Hunt S.E., Inouye M., Jeffreys A.E., Mendy A., Palotie A., Potter S., Ragoussis J., Rogers J., Rowlands K., Somaskantharajah E., Whittaker P., Widden C., Donnelly P., Howie B., Marchini J., Morris A., Sanjoaquin M., Achidi E.A., Agbenyega T., Allen A., Amodu O., Corran P., Djimde A., Dolo A., Doumbo O.K., Drakeley C., Dunstan S., Evans J., Farrar J., Fernando D., Hien T.T., Horstmann R.D., Ibrahim M., Karunaweera N., Kokwaro G., Koram K.A., Lemnge M., Makani J., Marsh K., Michon P., Modiano D., Molyneux M.E., Mueller I., Parker M., Peshu N., Plowe C.V., Puijalon O., Reeder J., Reyburn H., Riley E.M., Sakuntabhai A., Singhasivanon P., Sirima S., Tall A., Taylor T.E., Thera M., Troye-Blomberg M., Williams T.N., Wilson M., Kwiatkowski D.P., Wellcome Trust Case Control, C, Malaria Genomic Epidemiology, N Genome-wide and fine-resolution association analysis of malaria in West Africa. Nat. Genet. 2009;41:657–665. doi: 10.1038/ng.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston W.T., Mutalima N., Sun D., Emmanuel B., Bhatia K., Aka P., Wu X., Borgstein E., Liomba G.N., Kamiza S., Mkandawire N., Batumba M., Carpenter L.M., Jaffe H., Molyneux E.M., Goedert J.J., Soppet D., Newton R., Mbulaiteye S.M. Relationship between Plasmodium falciparum malaria prevalence, genetic diversity and endemic Burkitt lymphoma in Malawi. Sci Rep. 2014;4(3741) doi: 10.1038/srep03741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Kreuels B., Adjei O., Krumkamp R., May J., Small D.S. The causal effect of malaria on stunting: a Mendelian randomization and matching approach. Int. J. Epidemiol. 2013;42:1390–1398. doi: 10.1093/ije/dyt116. [DOI] [PubMed] [Google Scholar]
- Kariuki S.M., Rockett K., Clark T.G., Reyburn H., Agbenyega T., Taylor T.E., Birbeck G.L., Williams T.N., Newton C.R. The genetic risk of acute seizures in African children with falciparum malaria. Epilepsia. 2013;54:990–1001. doi: 10.1111/epi.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler E.M., Band G., Busby G.B.J., Kivinen K., Le Q.S., Clarke G.M., Bojang K.A., Conway D.J., Jallow M., Sisay-Joof F., Bougouma E.C., Mangano V.D., Modiano D., Sirima S.B., Achidi E., Apinjoh T.O., Marsh K., Ndila C.M., Peshu N., Williams T.N., Drakeley C., Manjurano A., Reyburn H., Riley E., Kachala D., Molyneux M., Nyirongo V., Taylor T., Thornton N., Tilley L., Grimsley S., Drury E., Stalker J., Cornelius V., Hubbart C., Jeffreys A.E., Rowlands K., Rockett K.A., Spencer C.C.A., Kwiatkowski D.P., Malaria Genomic Epidemiology N. Resistance to malaria through structural variation of red blood cell invasion receptors. Science. 2017:356. doi: 10.1126/science.aam6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokossou A.G., Dechavanne C., Bouraima A., Courtin D., Le Port A., Ladekpo R., Noukpo J., Bonou D., Ahouangninou C., Sabbagh A., Fayomi B., Massougbodji A., Garcia A., Migot-Nabias F. Association of IL-4 and IL-10 maternal haplotypes with immune responses to P. falciparum in mothers and newborns. BMC Infect. Dis. 2013;13(215) doi: 10.1186/1471-2334-13-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaria Genomic Epidemiology, N, Band G., Rockett K.A., Spencer C.C., Kwiatkowski D.P. A novel locus of resistance to severe malaria in a region of ancient balancing selection. Nature. 2015;526:253–257. doi: 10.1038/nature15390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maziarz M., Kinyera T., Otim I., Kagwa P., Nabalende H., Legason I.D., Ogwang M.D., Kirimunda S., Emmanuel B., Reynolds S.J., Kerchan P., Joloba M.M., Bergen A.W., Bhatia K., Talisuna A.O., Biggar R.J., Goedert J.J., Pfeiffer R.M., Mbulaiteye S.M. Age and geographic patterns of Plasmodium falciparum malaria infection in a representative sample of children living in Burkitt lymphoma-endemic areas of northern Uganda. Malar. J. 2017;16(124) doi: 10.1186/s12936-017-1778-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulama D.H., Bailey J.A., Foley J., Chelimo K., Ouma C., Jura W.G., Otieno J., Vulule J., Moormann A.M. Sickle cell trait is not associated with endemic Burkitt lymphoma: an ethnicity and malaria endemicity-matched case-control study suggests factors controlling EBV may serve as a predictive biomarker for this pediatric cancer. Int. J. Cancer. 2014;134:645–653. doi: 10.1002/ijc.28378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutalima N., Molyneux E., Jaffe H., Kamiza S., Borgstein E., Mkandawire N., Liomba G., Batumba M., Lagos D., Gratrix F., Boshoff C., Casabonne D., Carpenter L.M., Newton R. Associations between Burkitt lymphoma among children in Malawi and infection with HIV, EBV and malaria: results from a case-control study. PLoS One. 2008;3:e2505. doi: 10.1371/journal.pone.0002505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndeezi G., Kiyaga C., Hernandez A.G., Munube D., Howard T.A., Ssewanyana I., Nsungwa J., Kiguli S., Ndugwa C.M., Ware R.E., Aceng J.R. Burden of sickle cell trait and disease in the Uganda Sickle Surveillance Study (US3): a cross-sectional study. Lancet Glob. Health. 2016;4:e195–200. doi: 10.1016/S2214-109X(15)00288-0. [DOI] [PubMed] [Google Scholar]
- Nkrumah F.K., Perkins I.V. Sickle cell trait, hemoglobin C trait, and Burkitt's lymphoma. Am. J. Trop. Med. Hyg. 1976;25:633–636. doi: 10.4269/ajtmh.1976.25.633. [DOI] [PubMed] [Google Scholar]
- Ockenhouse C.F., Tandon N.N., Magowan C., Jamieson G.A., Chulay J.D. Identification of a platelet membrane glycoprotein as a falciparum malaria sequestration receptor. Science. 1989;243:1469–1471. doi: 10.1126/science.2467377. [DOI] [PubMed] [Google Scholar]
- Ogwang M.D., Bhatia K., Biggar R.J., Mbulaiteye S.M. Incidence and geographic distribution of endemic Burkitt lymphoma in northern Uganda revisited. Int. J. Cancer. 2008;123:2658–2663. doi: 10.1002/ijc.23800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogwang M.D., Zhao W., Ayers L.W., Mbulaiteye S.M. Accuracy of Burkitt lymphoma diagnosis in constrained pathology settings: importance to epidemiology. Arch. Pathol. Lab. Med. 2011;135:445–450. doi: 10.1043/2009-0443-EP.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okello P.E., Van Bortel W., Byaruhanga A.M., Correwyn A., Roelants P., Talisuna A., D'alessandro U., Coosemans M. Variation in malaria transmission intensity in seven sites throughout Uganda. Am. J. Trop. Med. Hyg. 2006;75:219–225. [PubMed] [Google Scholar]
- Pike M.C., Morrow R.H., Kisuule A., Mafigiri J. Burkitt's lymphoma and sickle cell trait. Br. J. Prev. Soc. Med. 1970;24:39–41. doi: 10.1136/jech.24.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Robbiani D.F., Deroubaix S., Feldhahn N., Oliveira T.Y., Callen E., Wang Q., Jankovic M., Silva I.T., Rommel P.C., Bosque D., Eisenreich T., Nussenzweig A., Nussenzweig M.C. Plasmodium infection promotes genomic instability and AID-Dependent B Cell Lymphoma. Cell. 2015;162:727–737. doi: 10.1016/j.cell.2015.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochford R., Cannon M.J., Moormann A.M. Endemic Burkitt's lymphoma: a polymicrobial disease? Nat. Rev. Microbiol. 2005;3:182–187. doi: 10.1038/nrmicro1089. [DOI] [PubMed] [Google Scholar]
- Scott J.A., Berkley J.A., Mwangi I., Ochola L., Uyoga S., Macharia A., Ndila C., Lowe B.S., Mwarumba S., Bauni E., Marsh K., Williams T.N. Relation between falciparum malaria and bacteraemia in Kenyan children: a population-based, case-control study and a longitudinal study. Lancet. 2011;378:1316–1323. doi: 10.1016/S0140-6736(11)60888-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G.D., Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- Smith J.D., Rowe J.A., Higgins M.K., Lavstsen T. Malaria's deadly grip: cytoadhesion of Plasmodium falciparum-infected erythrocytes. Cell. Microbiol. 2013;15:1976–1983. doi: 10.1111/cmi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo A., Feng G., Brown G.V., Beeson J.G., Rogerson S.J. Functional antibodies and protection against blood-stage malaria. Trends Parasitol. 2016;32:887–898. doi: 10.1016/j.pt.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Timmann C., Thye T., Vens M., Evans J., May J., Ehmen C., Sievertsen J., Muntau B., Ruge G., Loag W., Ansong D., Antwi S., Asafo-Adjei E., Nguah S.B., Kwakye K.O., Akoto A.O., Sylverken J., Brendel M., Schuldt K., Loley C., Franke A., Meyer C.G., Agbenyega T., Ziegler A., Horstmann R.D. Genome-wide association study indicates two novel resistance loci for severe malaria. Nature. 2012;489:443–446. doi: 10.1038/nature11334. [DOI] [PubMed] [Google Scholar]
- Walley A.J., Aucan C., Kwiatkowski D., Hill A.V. Interleukin-1 gene cluster polymorphisms and susceptibility to clinical malaria in a Gambian case-control study. Eur. J. Hum. Genet. 2004;12:132–138. doi: 10.1038/sj.ejhg.5201084. [DOI] [PubMed] [Google Scholar]
- Williams A.O. Haemoglobin genotypes, ABO blood groups, and Burkitt's tumour. J. Med. Genet. 1966;3:177–179. doi: 10.1136/jmg.3.3.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T.N., Mwangi T.W., Roberts D.J., Alexander N.D., Weatherall D.J., Wambua S., Kortok M., Snow R.W., Marsh K. An immune basis for malaria protection by the sickle cell trait. PLoS Med. 2005;2 doi: 10.1371/journal.pmed.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material