Abstract
Lung cancer is a malignant lung tumor with various histological variants that arise from different cell types, such as bronchial epithelium, bronchioles, alveoli, or bronchial mucous glands. The clinical course and treatment efficacy of lung cancer depends on the histological variant of the tumor. Therefore, accurate identification of the histological type of cancer and respective protein biomarkers is crucial for adequate therapy. Due to the great diversity in the molecular-biological features of lung cancer histological types, detection is impossible without knowledge of the nature and origin of malignant cells, which release certain protein biomarkers into the bloodstream. To date, different panels of biomarkers are used for screening. Unfortunately, a uniform serum biomarker composition capable of distinguishing lung cancer types is yet to be discovered. As such, histological analyses of tumor biopsies and immunohistochemistry are the most frequently used methods for establishing correct diagnoses. Here, we discuss the recent advances in conventional and prospective aptamer based strategies for biomarker discovery. Aptamers like artificial antibodies can serve as molecular recognition elements for isolation detection and search of novel tumor-associated markers. Here we will describe how these small synthetic single stranded oligonucleotides can be used for lung cancer biomarker discovery and utilized for accurate diagnosis and targeted therapy. Furthermore, we describe the most frequently used in-clinic and novel lung cancer biomarkers, which suggest to have the ability of differentiating between histological types of lung cancer and defining metastasis rate.
Keywords: lung cancer, biomarker, histological type, aptamers, diagnostics, targeted therapy
1. Introduction
Lung cancer is the most common cancer and the leading cause of cancer-related deaths worldwide, with approximately 1.8 million new cases and 1.6 million deaths in 2012 [1,2,3,4]. The five-year survival rate of patients with lung cancer is approximately 13–15% [5]. Diagnosis of lung cancer at late stages is a determining factor of high mortality from the disease. It is primarily explained by metastases from lung into the central nervous system as observed in 54% cases [6,7]. Therefore, early lung cancer detection is necessary for reducing the high mortality rate. Understanding the biological mechanisms of tumor development and biomarker expression typical for lung cancer and specific for all histological types is crucial for accurate diagnosis, treatment, and drug development.
2. Histology of Lung Cancer Types
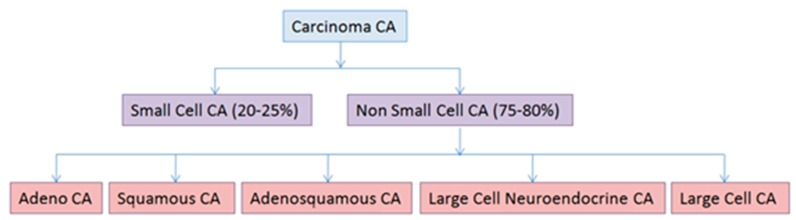
Lung cancer is a malignant lung tumor that may stem from bronchial epithelium, bronchioles, alveoli, and bronchial mucous glands. It is characterized by post-treatment relapses, metastasis, and a variety of histological types. In 2015, the new World Health Organization (WHO) classification of lung tumors was published [8]. The two main lung cancer types are small-cell lung carcinoma (SCLC) and non-small-cell lung carcinoma (NSCLC) (Figure 1). Approximately 80% of lung cancer cases are NSCLC, which have diverse molecular-biological features and clinical course forms of the disease [8,9]: adenocarcinoma, adenosquamous carcinoma, squamous cell carcinoma, large cell carcinoma, and large cell neuroendocrine carcinoma.
Figure 1.
The new World Health Organization (WHO) classification of lung cancer histological types. The various types of lung cancer have different origins and histological features (Figure 2). Small-cell lung carcinoma (SCLC) is characterized by small size cells, absence of differentiation, fast tumor growth, metastasis at early stages, and release of specific biomarkers and hormones. At present, there are two points of view on SCLC histogenesis. According to the first hypothesis, SCLC arises from cells of the diffuse endocrine system, i.e., the amine precursor uptake decarboxylation (APUD)-system (Figure 2); the second suggests this type of lung cancer originates from the endodermbronchial lining layer [10]. CA: carcinoma.
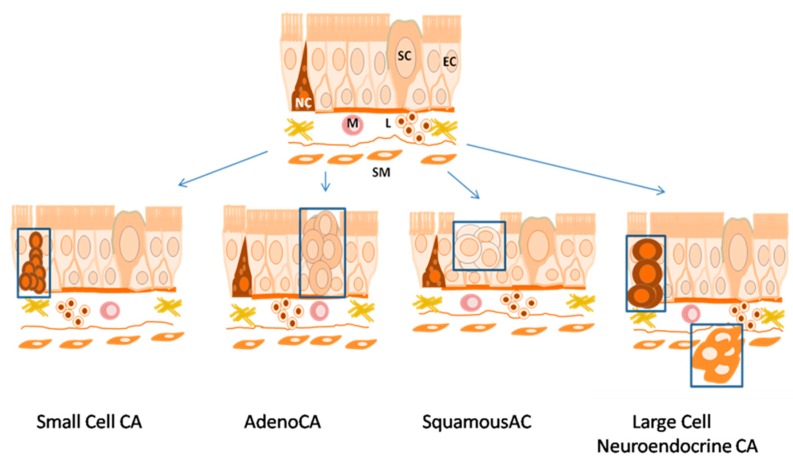
Among the other subtypes of NSCLC, adenocarcinoma arises from glandular cells of bronchial mucosa and now represents the dominant histological subtype among the other lung cancer types (Figure 2). Squamous lung cancer arises from the modified bronchial epithelial cells (Figure 2) and is characterized by one of the following specific differentiation features: keratinization, keratin pearl formation, or the presence of intercellular bridges. Аdenosquamous carcinoma is a type of cancer that contains two types of cells: squamous cells (thin, flat cells that line certain organs) and gland-like cells [9]. Large cell neuroendocrine carcinoma is a malignant epithelial tumor, which is comprised of large polygonal cells that do not show any obvious evidence of histological differentiation. The cases include large cell neuroendocrine carcinoma, basaloid carcinoma, lymphoepithelioma-like carcinoma, and clear cell carcinoma. The tumor arises from neuroendocrine cells of the respiratory tract lining layer or smooth muscle cells of its wall (Figure 2). Large-cell carcinoma is a heterogeneous group of undifferentiated malignant neoplasms that lack the cytologic and architectural features of small cell carcinoma and glandular or squamous differentiation. Large-cell carcinoma is categorized as a subtype of NSCLC that originates from epithelial cells of the lung [11].
Figure 2.
Histogenesis of histological types of lung cancer. SM—Smooth Muscle; M—Macrophage; L—Lymphocyte; NC—Neuroendocrine Cell; EC—Epithelial Cell; SC—Secretory Cell.
It is well known that a unique combination of exogenous and endogenous factors influences the occurrence and development of lung cancer in each individual. Therefore, lung cancer, like other oncological diseases, is heterogeneous. Thus, in addition to various histological types, this disease also has many molecular and pathological subtypes characterized by heterogeneous cellular genetic and epigenetic changes and a different combination of protein biomarkers. However, at present, data on protein signatures of molecular subtypes of histological types of lung cancer is extremely limited, but a large number of genetic studies reflecting the probability of certain mutations in genes are presented. In particular, mutations of EGFR (epidermal growth factor receptor) in lung adenocarcinoma have been well studied. It was found that in patients with lung adenocarcinoma, the probability of EGFR mutations increases linearly from age 3.7% (18–30 years) to 18.5% (81–100 years), and in female non-smokers, the probability of mutations is higher than in men [12,13]. In male non-smokers, the probability of EGFR mutation is much higher than in smokers [12].
Identification of the correct histological type of lung cancer and their molecular subtypes is necessary due to different treatment strategies. Tumor cells of each histological type release certain protein biomarkers into the bloodstream and therefore play a key role in cancerogenesis. The use of blood plasma to determine the origin and nature of the malignant cells for diagnosis requires knowledge about expression of protein biomarkers, their specificity, sensitivity, and their release by different types of lung cancer cells [14,15,16].
3. Methods Currently Employed to Diagnose Lung Cancer
Currently, lung cancer is detected mostly in the late stages due to such symptoms as coughing, coughing up blood, shortness of breath, and chest pains. Unfortunately, the early stages of this disease are often detected only by accident. Chest radiography and computer tomography are the most commonly used methods for lung cancer diagnosis. However, as they can only identify visible and irreversible changes in lung, there is a need for additional methods for early diagnosis. In order to overcome this challenge, it is essential to discover novel, highly sensitive, and specific biomarkers [15].
4. Circulating Biomarkers of Carcinogenesis
Diagnostic significance of protein biomarkers is defined by their sensitivity and specificity. Biomarker sensitivity is determined by the percentage of true positive results of analysis in a group of oncological patients, while biomarker specificity is determined by a percentage of true negative results of analysis in a group of healthy people and patients with benign diseases. Unfortunately, to date, 100% sensitive and specific biomarkers have not been found. Moreover, some cancer-specific biomarkers were also found in plasma of healthy people.
For non-invasive detection of lung cancer biomarkers, biological materials such as tumor tissues, blood, exhaled breath condensate, sputum, and urine are usually utilized. Exhaled breath condensate is liquid received from the respiratory tract that consists of cytokines, proteins, and DNA [17]. It has been established that the content of condensate from lung cancer patients differs from condensate of healthy people, nevertheless, specific protein biomarkers have not been detected. Similarly, in sputum, one of the most attractive non-invasive source of lung cancer biomarkers, specific markers are also yet to be detected [18].
Thus, blood remains the most perspective source for biomarker discovery because cellular debris penetrate into the bloodstream from the tumor. As a result, blood may be used as a minimally invasive liquid biopsy. Blood is a complex matrix containing tumor-associated circulating lipids, proteins, RNAs, miRNAs, DNAs, as well as cancer, immune, stromal, and endothelial cells [19].
Tumor-associated biomarkers are biological molecules that can be detected and serve as indicators of pathogenic processes or pharmacological/pharmacodynamic response to treatment [20]. Different oncomarkers can be used to distinguish normal and pathogenic processes [4]. An ideal biomarker originates from neoplastic cells, is indiscernible in healthy and benign tissues, and can be identified by simple methods in the available biological material (biological fluids). It should be sensitive, specific and cost-saving.
Tumor biomarkers are divided into several types: genetic (mutations, changes in number of copies, matrix RNA expression), epigenetic (changes in DNA methylation profile), proteomic (changes in level and profile of protein expression), metabolic (changes in level and spectrum of low molecular weight metabolites), DNAs and RNAs circulating in blood plasma, exosomal microRNAs (miRNAs), synthesis profile and level of miRNAs, protein biomarkers, circulating tumor cells (CTCs), and immune, stromal, and endothelial cells [17,18,20,21,22,23,24,25,26]. Overall, proteins are the most suitable biomarkers for lung cancer diagnosis because of their involvement in cellular processes. A panel of biomarkers (in particular, CYFRA 21-1 (cytokeratins), EPCAM (epithelial cell adhesion molecule), ProGRP (pro-gastrin-releasing peptide), CEACAM (carcinoembryonic antigen), and others are used for screening various malignancies including lung cancer. However, in practice, this system often fails in providing sufficient sensitivity and information of value for optimal screening. For example, for the diagnosis of lung cancer, CEACAM sensitivity and specificity is 69% and 68% respectively, while that for CYFRA 21-1 was 43% and 89%, respectively [27]. Diagnostic sensitivity of plasma ProGRP in distinguishing SCLC was estimated to be approximately 84%, and specificity 95% [28]. For comparison, CT has a sensitivity is about 94% with low specificity and high false-positive rate in the detection of lung cancers [29].
Various approaches are used for biomarker discovery; in particular, mass spectrometry analysis is commonly used for protein profiling of tumors and another popular method, which will be discussed, is affinity-based enrichment using aptamers and other molecules.
5. Proteomic-Based Lung Cancer Biomarker Search
Mass spectrometry methods allow identification and analysis of thousands of proteins in biological systems [30]. Mass spectrometry can provide valuable information such as the differences between protein profiles of normal and tumor lung tissue. As such, Kang and coauthors established that the main lung cancer biomarker is the β-chain of human HP (haptoglobin) [31], others also classified SAA (serum amyloid A) [4], APOA1 (apolipoprotein A-1) [32], ANXA (annexin), VIM (vimentin), NM (non-muscle myosin), CALM (calmodulin), CFL (cofilin),TMS (thymosin), and EGFR (epidermal growth factor receptor) as lung cancer biomarkers [30] (Table 1).
Table 1.
Protein biomarkers of lung cancer defined using proteomic studies.
| Lung Cancer Type | Protein Biomarkers of Lung Cancer | Reference |
|---|---|---|
| NSCLC, SCLC | AGER, C10orf116, ADD2, PRX, LAMB3, SYNM, SPTA1, ANK1, HBE1, HBG1, CA1, TNXB, MMRN2, HBA1, CAV1, HBB, COL6A6, C1orf198, CLIC2, SDPR, EHD2, APOA2, NDUFB7, PRKCDBP, LAMA3, LBN | [33,34,35] |
| ACT, 3 IGFBP3, L-PGDS | [35] | |
| SAA | [36] | |
| SAA, HAP, HGF | [36,37] | |
| TTR | [38,39] | |
| SAA, AAG1/2, CLU, SSA, AAG1, SAA, TTR | [6,35,37,40,41] | |
| APOA4, FIBA, LBN, SAA, CP, HP, TTR, KRT2A, GLT1B, CK1, AKT, MBL2, AAG1-2, FGA | [42] | |
| GSN, HP, FCN3, CNDP1 | [43] | |
| Lung adenocarcinoma | CALCA, CPS1, CHGB, IVL, AGR2, NASP, PFKP, THBS2, TXNDC17, PCSK1, CRABP2, ACBD3, DSG2, LRBA, STRAP, VGF, NOP2, LCN2, CKMT1B, AKR1B10, PCNA, CPD, PSME3, VIL1 | [44,45,46,47] |
| Squamous lung cancer | SERPINB5, RPL5, PKP1, RPL10, AKR1B10, AKR1C1, PCNA, RPS2, AKR1C3, THBS2, ACBD3, VSNL1, AHCY, IMMP10, PAK2, IVL, IARS, PSMD2, GBP5, MCM6, NDRG1, NOP58, S100A2, NRG1-2, CNDP1 | [45,47] |
| UCRP, CER, UPA, MT1-MMP, SFN, TF, ALB, S100A9, STMN, ENO, PLAU, IGFBP7, MMP14, THBS1, TTR | [48] |
NSCLC: non-small-cell lung carcinoma; SCLC: small-cell lung carcinoma.
Mass spectrometry helped discriminate the differences between protein profiles of adenocarcinoma and squamous lung cancer. Nevertheless, despite numerous known tumor-associated biomarkers, sensitivity of proteomic studies is insufficient—only 79% for the first and second stage of lung cancer [49]. Low sensitivity is most likely related to the loss of a number of low copy proteins which are present in tissues and blood in trace amount.
6. Protein Biomarkers Used in Lung Cancer Diagnosis
Early diagnosis of lung cancer could be based on detection of protein markers and autoantibodies specific for each type of cancer [50]. In particular, screening studies conducted in the United States where 1613 patients used EarlyCDT®-Lung test revealed lung cancer at stage I. The blood test detected early lung cancer in asymptomatic patients with higher specificity than imaging tests [50]. A similar level of specificity and sensitivity for lung cancer diagnosis using autoantibodies was achieved in the studies conducted by Caroline J. Chapman [51]. Thus, high sensitivity and specificity levels make the panel of autoantibodies an important addition to the standard methods for early diagnosis of lung cancer.
Despite the great advances made in lung cancer biomarker discovery, no data on biomarkers with high enough specificity and sensitivity has been found. This is related to a number of reasons: (1) inefficiency of techniques applied for biomarker search, (2) genetic heterogeneity of tumors, (3) poor reproducibility of laboratory tests, (4) poor research design, (5) low concentration of analyzed biomarkers, and (6) insufficient number of tissue banks for screening [25].
Nevertheless, lung cancer therapy has advanced mostly due to target treatment approaches. However, it was found that effective therapy for each histological type of lung cancer requires prior knowledge about specific molecular targets. This establishes the importance of both the diagnosis of lung cancer as well as further identification of the histological type. Several biomarkers are currently used for clinical lung cancer detection. However, a variety of proteins that may act as novel tumor-associated markers are yet to be proven (Table 2).
Table 2.
Conventional protein biomarkers of lung cancer.
| № | Protein Biomarkers of Lung Cancer | Reference |
|---|---|---|
| 1 | CEACAM (Carcinoembryonic Antigen) | [5,34,38,40,43,55,56,57,58,59,60,61,62] |
| 2 | CYFRA21-1 (Cytokeratin-19 fragments) | [5,8,26,34,38,43,55,56,58,61,62,63,64,65,66,67] |
| 3 | CA125 (Cancer Antigen 125) | [68] |
| 4 | PKLK (Plasma kallikrein) | [51] |
| 5 | ProGRP (Pro-gastrin-releasingpeptide) | [17,56,69,70] |
| 6 | NSE (Neuron-specific enolase) | [56,61,70,71,72] |
| 7 | ТРА 6, 7, 8 | [23,43] |
| 8 | NRG2, 100 | [43] |
| 9 | CNDP | [43] |
| 10 | APOВ100 | [43] |
| 11 | SCC (Squamous cell carcinoma antigen) | [73,74] |
| 12 | VEGF (Vascularendothelial growth factor) | [56] |
| 13 | EGFR (Epidermal Growth Factor) | [50] |
| 14 | PIK3CA, HER2, BRAF, ROS, RET, NRAS, MET, MEK1 | [6,7] |
| 15 | HER2 | [75] |
| 17 | C4.4A | [20] |
| 18 | PSF3 | [76,77] |
| 19 | FAM83B | [54] |
| 20 | ECD, CTNNB , VIM, S100A4 | [47] |
| 21 | S100A7 | [32] |
| 22 | COX2 | [78] |
| 23 | MUC1 | [74,79] |
Some clinically used biomarkers such as CEACAM, CYFRA 21-1, and ProGRP have low concentrations in serum, and therefore, each biomarker alone cannot be used for early lung cancer diagnosis [52,53]. As such, they are used as a mixture. In particular, the combination of CEACAM and CYFRA 21-1 could be used for detection of adenocarcinoma [54]. Statistically significant differences between lung cancer patients and healthy people were found using a panel of CEACAM, CA125, CYFRA 21-1, and NY-ESO (cancer-testis antigen) [45]. Other authors also used NSE (neuron specific enolase), CEACAM, and CYFRA 21-1 for differentiation of histological subtypes of lung cancer [29].
The use of serum markers, namely LDH (lactate dehydrogenase), CRP (C-reactive protein), CEACAM, NSE, and CYFRA 21-1, has enhanced accuracy of lung cancer diagnosis to 94.8% [42]. William L. Bigbee and coauthors suggest a panel of biomarkers—PRL (prolactin), TTR (transthyretin), THBS1 (thrombospondin 1), SELE (selectin E), MIF (macrophage migration inhibitory factor), PLAT (plasminogen activator, tissue type), EGFR, ERBB2 (erb-b2 receptor tyrosine kinase 2), CYFRA 21-1, and serum APBA (2-dehydropantoate 2-reductase)for early lung cancer diagnosis with 77.1% sensitivity and 76.2% specificity [80]. The combination of CEACAM, RBP (retinol-binding proteins), SERPIN (serpin peptidase inhibitors), and SART (U4/U6.U5 tri-snRNP-associated protein 1)was also used to diagnose lung cancer and classify patients in the independent validation set (sensitivity—77.8%; specificity—75.4%) [81].
An overview of recent publications on early lung cancer diagnosis showed that combinations of a variety of tumor-associated biomarkers could be more useful than using each of them separately [82]. Nevertheless, no composition for detection of lung cancer at early or premalignant stages has been found [83].
7. Aptamer-Based Affinity Enrichment Methods for Lung Cancer Biomarker Discovery
In recent years, a new specific technique of biomarker discovery using aptamers, i.e., AptaBID, has been developed [64]. Aptamers are small single-stranded DNA or RNA (30–100 nt) oligonucleotides that form three-dimensional structures capable of binding certain targets. Specific binding of the aptamers is conditioned by the dimensional structure, spatial charges distribution, phosphates and the mismatch of bases, capable of electrostatic and van der Waals interactions and forming hydrogen bonds [84]. Being highly selective, aptamers can quickly distinguish small differences in thousands of proteins and therefore can be used in a wide variety of applications including molecular imaging, drug delivery, therapy, diagnostics and biomarker discovery.
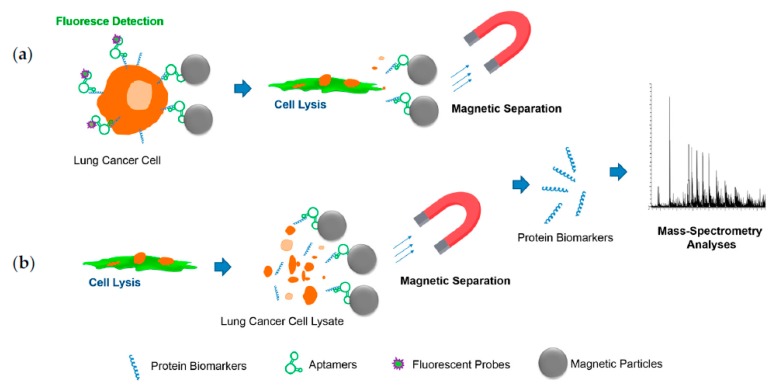
Despite the promising qualities aptamers possess for diagnostic and therapeutic purposes, their widespread application is still limited due to problems and pitfalls that currently plague the technology [85,86]. Conventional SELEX (Systematic Evolution of Ligands by EXponential Enrichment) selection and its various modifications are time and labor consuming, and no universal and automatic aptamer generation procedure has been established [85]. The main challenge is generation of efficient aptamers for in vivo applications with high affinity to viable cells and tissues, long-lasting stability in bloodstream, high selectivity, and low cross-reactivity. These “bottlenecks” could be overcome by restricting selection to in vivo like conditions and increasing the chemical diversity of oligonucleotides by addition of modified bases and chemical modifications, which introduce new functionalities to aptamers. Nucleotide chemical modification can also help prevent aptamer degradation and excretion from the bloodstream by renal filtration, increase circulation time, improve aptamer binding, and potentially expand their use for therapy and diagnostics [85,86,87]. Current selection processes became more efficient due to novel bio-separation technologies and high-throughput screening technologies. Often, aptamers generated against purified or recombinant proteins do not show good binding in vivo and sometimes have cross-reactivity [86,87]. Aptamers for diagnostic and therapeutic applications are more effective when selected against complex targets (cells, viruses, bacteria, etc.), but identification of their exact binding partner is often complicated and expensive. Specific biomarkers are isolated by aptamer-mediated affinity purification using magnetic separation from whole cells (Figure 3a) or cell lysates (Figure 3b). Purified proteins are identified using mass-spectrometry analyses (Figure 3).
Figure 3.
Schematic representation of aptamer based biomarker discovery. Affinity purification of aptamer protein targets: (a) from whole cells; (b) form cell lysates.
Modifications of AptaBID approach were applied for aptamer facilitated detection of several lung cancer protein biomarkers such as CTSD (cathepsin D) [61], VIM (vimentin), DEF (defensin) [62,88], ANXA2 (annexin A2), ANXA5 (annexin A5), H2B (H2B histone family member M), and CLU(clusterin) [62], LMN (lamin), and TUB (tubulin), ACT (actin) [88].
CTSD is implicated in tumorigenesis and is hyper-expressed in lung cancer tissues and plasma [89]. ANX (annexin) are considered as targets of breast cancer, pancreatic cancer and laryngeal carcinoma therapy alone and/or synergistically [90]. Ubiquitylated H2B in cancer cells plays an important role in human malignancy [91]. VIM, a major constituent of the intermediate filament family of proteins is involved in cancer initiation and progression, tumorigenesis, metastasis formation, and epithelial-to-mesenchymal transition [92,93,94]. Its expression is increased in moderately and well-differentiated adenocarcinoma and in giant cell carcinoma [92,95]. LMN of the nuclear lamina modulate cell proliferation, differentiation, as well as epithelial-mesenchymal transition and migration [96,97]. It serves as a marker of good or poor patient survival depending on tumor subtype [98,99]. TUB hyperexpression and their post-translational modifications are correlated with poor prognosis and chemotherapy resistance of various cancers [100]. DEF through EGFR activation and downstream signaling pathways, influence cell migration and proliferation [101] and are associated with cancer invasiveness [102].
Some proteins expressed by circulating tumor cells (CTC) have lung origin, and thus, aptamers for these markers could be used to isolate specific CTC from blood [62]. Some of these cancer-related targets were found in crude blood plasma of patients with NSCLC and SCLC and could be detected with an electrochemical aptamer–based sensor [103]. Lung cancer cells express the same biomarkers, and therefore corresponding aptamers could be used for histological structure characterization of lung adenocarcinoma. The selected DNA aptamers showed binding to various tumor structures, such as elastic fibers, tumor cells, blood vessels, and elastin, which play important roles in tumor formation and growth [88].
Another powerful aptamer-based proteomic technology allowing large-scale comparison of proteome profiles in small volumes of biological samples with low limits of detection, a broad dynamic range, and high reproducibility has been suggested by Larry Gold and SomaLogic [104]. This approach enables the discovery of novel biomarkers using Slow Off-rate Modified Aptamers (SOMAmers) for affinity enrichment [104]. SOMAmers engage proteins and increase the range of epitopes available for binding because of their more hydrophobic surfaces compared with conventional aptamers [82].
A highly multiplexed proteomic technology SOMAscan technology demonstrated the possibility of sensitive proteomic assay of protein expression signatures in NSCLC using healthy adjacent and distant tissues from surgical resections [82]. Thirty-six proteins with the largest mean fold-change in protein expression between tumor and non-tumor tissue samples have been suggested as novel protein biomarkers of NSCLC and were classified into biological processes associated with important hallmarks of cancerogenesis: angiogenesis, growth and metabolism, inflammation and apoptosis, and invasion and metastasis [82].
8. Lung Cancer Diagnosis Using Aptamers
Сancer-related proteins can be detected using various sensors, most of them rely on antibody-antigen interaction in a sandwich-like system that requires two different types of antibodies for target identification. The main challenge in the development of reliable diagnostic sensors based on antibodies is due to the fluctuation of their affinity depending on manufacture and batch. Synthetic aptamers overcome these limitations as their properties at the same conditions depend only on nucleotide sequence [105,106]. Another advantage of using aptamers over the conventional antibodies is the possibility to modify them chemically with various labels, active groups, and nanoparticles, which is important for biosensors design [105]. Some aptamers change their conformation after binding to their target molecules, which makes the development of switchable aptasensors and fluorescence quenching sensors possible, which could not be achieved with antibodies [107]. Thus, aptamers are widely used for development of various diagnostic tools (optical, colorimetric, fluorescence, electrochemical, microfluidic, PET (positron-emission tomography), CT (computed tomography), NMR (nuclear magnetic resonance), MRI (magnetic resonance imaging), or ultrasound imaging etc.) [105,106,107].
More than 20 different aptamers selected by a number of research groups all over the world demonstrated their high sensitivity for lung cancer diagnosis in different sensor systems as well as a unique potential for a targeted therapy (Table 3, Figure 4).
Table 3.
Aptamers for lung cancer diagnostics and therapy.
| Aptamers | Target Cells | Protein Target | Application | Reference |
|---|---|---|---|---|
| Small cell lung cancer | ||||
| HCA12 HCC03 HCH07 HCH01 |
Cell lines: NCI–H69 NCI–H146 NCI–H128 |
Not determined | Formalin-fixed, Paraffin-embedded Tissue Array; Extraction and Detection with Aptamer Conjugated Magnetic/Fluorescent Nanoparticles using fluorescence microscopy and flow cytometry | [108] |
| 16-1 | SBC3 cell line | Not determined | Fluorescence microscopy and flow cytometry | [109] |
| Lung Adenocarcinoma | ||||
| EJ7 ADE2 | H23 cell line H23, A549 cell line | Not determined | Flow cytometry | [110] |
| S13, S50 | EGFR-transfected A549 cell line | EGFR | Antiproliferative activity | [111] |
| R50 | A549 cells transfected with EGFR-GFP | NCL | Apoptosis induction | [112] |
| LC-17 | Post-operative tissue | TUB | Aptahistochemical analyses of tissues Isolation of circulating tumor cells |
[62,88] |
| LC-18, | Post-operative tissue | VIM, LMN | Aptahistochemical analyses of tissues Isolation of circulating tumor cells Electrochemical detection of protein biomarkers in human blood plasma |
[62,88,103] |
| LC-224 | Post-operative tissue | ACT methylated at position 73 | Aptahistochemical analyses of tissues | [88] |
| LC-110 | Post-operative tissue | CLU H2B | Isolation of circulating tumor cells | [62] |
| LC-183 | Post-operative tissue | CTSD | Isolation of circulating tumor cells Inhibition of growth of primary cancer cell cultures |
[62,113] |
| MA3 | Сell lines: A549, MCF-7 | MUC1 | Targeted delivery of doxorubicin | [114] |
| MUC-1 aptamer | A549 cell line | MUC1 | Targeted delivery of plasmid DNA | [115] |
| GL21.T | A549 (Axl+) cell line | AXL | Aptamer used as carriers for cell-targeted delivery of a miRNA with tumor suppressor function, let-7g; miR-212 | [116,117], |
| Other | ||||
| aptNCL | CL1-5 cell line | NC L | Targeted delivery of siRNA chimeras | [118] |
| AS1411 | Multiple cancer cell types | NC L | PET imaging of lung cancer with Cu-64 labeled aptamer | [119] |
| S1, S6, S11e, S15 | NSLC | Not determined | [120] | |
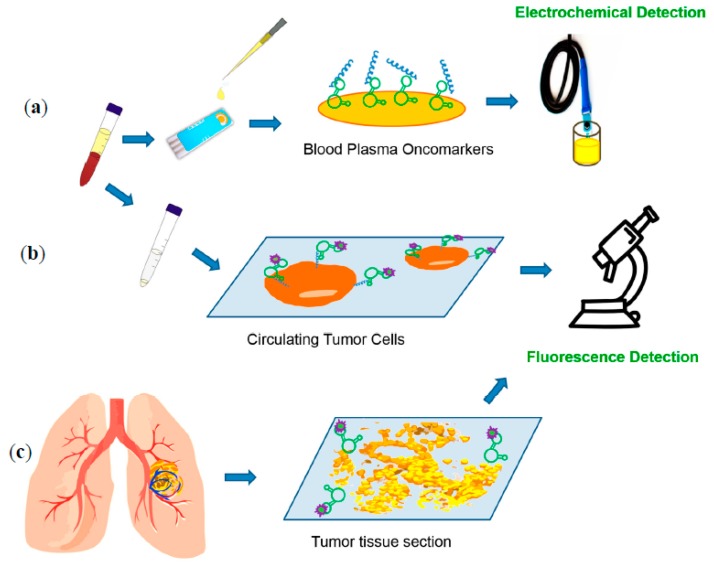
Figure 4.
Schematic representation of aptamer-based lung cancer diagnostic tools. (a) analyses of blood plasma oncomarkers using electrochemical detection; (b) circulating tumor cells capture and fluorescence detection ; (c) aptamer based immunohistochemistry-like characterization of lung cancer histological structure.
Aptamers selected to postoperative adenocarcinoma tissues have been utilized for detection of circulating tumor cells in blood [62], characterization of histological structure of lung adenocarcinoma [88], and electrochemical sensing of blood plasma biomarkers [103].
Aptamers to lung cancer were developed not only for diagnostics, but also for cancer treatment. Aptamers alone demonstrate antitumor activity in cell cultures; S13, S50 inhibit proliferation [111], LC-183 suppress cancer cell growth [113], and R50 cause apoptosis [112]. Aptamers are effective targeting ligands; anti mucin-1 aptamer is suitable for carrying doxorubicin [114] and plasmid DNA [115] to cancer cells in most adenocarcinomas. Aptamer GL21.T are used as carriers for selective delivery of a miRNA to A 549 cells, processing by the RNA interference machinery, and silencing let-7 g target genes, thus suppressing let-7 g function. This conjugate reduced tumor growth in vivo in a xenograft lung adenocarcinoma model [116]. Aptamers to NCL (nucleolin) have been used for targeted delivery of siRNA chimeras for lung cancer therapy [118] and PET imaging in vivo in a xenograft lung adenocarcinoma model [119].
Several aptamers against SCLC cells have high affinity and specificity in different assay formats for cell lines and tissues from the patient samples. Conjugates of these aptamers with magnetic and fluorescent nanoparticles effectively extracted SCLC cells from mixed cell media for isolation, enrichment, and sensitive detection [108]. Other aptamers to SCLC SBC3 cell line with good selectivity are suitable for fluorescence microscopy and flow cytometry analyses. Unfortunately, their exact protein targets have not yet been determined.
Aptahistochemistry for Identification of Lung Cancer Biomarkers
An important clinical method often used for diagnosis of tumor biomarkers is immunohistochemistry. A commonly used marker to identify adenocarcinoma is TTF-1 (transcription termination factor 1) [121], but in 70–90% cases of small cell lung cancer, expression of this marker is present. Squamous cell carcinoma biomarkers such as TP63 (tumor protein p63), CK5/6 (cytokeratin 5/6), 34βE12 (high molecular weight cytokeratins) could also be identified in adenocarcinoma [121]. Immunohistochemistry of small cell lung cancer is required only in problematic cases; usually, hematoxilin and eosin staining is sufficient for diagnosis. AE1/AE3 (pancytokeratin) is used to demonstrate that the tumor is a carcinoma rather than a lymphoid lesion [9].
Immunostaining is based on histological identification of tumor biomarkers and abnormal blood vessels by specific agents such as antibodies, but this method has some limitations such as: relatively high cost, difficulties in quantifying results, probes in tissue immunohistochemistry-like staining, as described in the research by Galina S. Zamay et al. [88]. They have shown that DNA-aptamers previously selected to postoperative lung cancer tissue specifically bind to different structures of tumor tissue including elastic fibers, tumor cells, blood vessels and elastin, having an important role in the formation of tumors. Protein binding partners of the aptamers were identified using affinity purification followed by mass spectrometry analyses, and validated with correspondent antibodies. According to this data, LMN (lamin), VIM (vimentin), TUB (tubulin), and ACT (actin) detected with the help of aptamers LC-18, LC-17, and LC-24 are involved in cancer progression and could act as lung adenocarcinoma biomarkers [88].
9. Biomarkers of Different Histological Lung Cancer Types
In addition to the numerous biomarkers that are currently used for clinical lung cancer detection, other proteins of new tumor-associated markers and their respective roles are also investigated (Table 2). For example, CEA (carcino embryionic antigen) is a 180-kDa glycoprotein as well as a carcinoembryonic antigen of fetal embryonic development, in addition to this, it is also a biomarker as its concentration level is increased in blood of patients with all lung cancer types (Figure 5) [48,79]. CEA is involved in cell adhesion and modulation processes [57]. As a result, tumors with high expression of CEA have high metastatic potential that may be caused by cell-cell adhesion between tumor and vessels because CEA is involved in homo- and heterotypic interactions with other cells [72]. High levels of CEA in serum is also correlated with brain metastases [122,123]. Serum levels of CEA may be useful for assessment of prognostic information about the risk of recurrence and death from lung cancer [76,124]. Importantly, the level of CEA does not correlate with the stage of the disease [72].
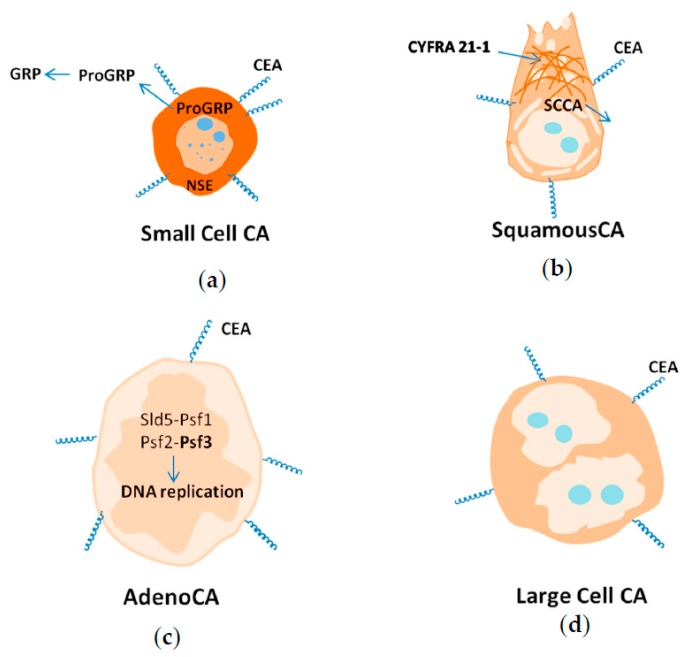
Figure 5.
Biomarkers of Small Cell Lung Cancer (a), Squamous Lung Cancer (b), Lung Adenocarcinoma (c), Large Cell Lung Cancer (d). GRP: gastrin-releasing peptide ; CEA: carcinoembryonic antigen ; NSE: neuron specific enolase; SCCA: squamous cell carcinoma antigen ; CYFRA 21-1: cytokeratins ; Sid5 : Systemic RNA interference defective protein 5 ; Psf1-Psf3: GINS complex subunits 1-3.
9.1. Small Cell Lung Cancer
SCLC arises from neuroendocrine cells of the APUD-system (amine precursor uptake and decarboxylation system) [67] and has two of the main biological features of these cells—production of L-DOPA-decarboxylase (L-3,4-dihydroxyphenylalanine- decarboxylase) and NSE (Figure 5a). L-DOPA decarboxylase is the gene encoding for the enzyme that catalyzes the biosynthesis of dopamine in humans [125]. NSE is a glycolytic neuron specific isoenzyme of enolase with two almost identical 39-kDa polypeptides produced in the central and peripheral neurons and malignant tumors of neuroectodermal origin; NSE is specific only for SCLC [42]. Adrenocorticotropic hormone, serotonin, antidiuretic hormone, calcitonin, growth hormone, melanocyte-stimulating hormone, and estrogen are also produced in SCLC.
The other well-known biomarker of SCLC is ProGRP (pro-gastrin-releasing peptide). High levels of ProGRP were found in the blood of patients with SCLC and medullary thyroid cancer (>200 pgmL−1). Blood plasma of healthy people and patients with benign diseases have ProGRP concentrations of 35 pgmL−1 and 45–103 pgmL−1 respectively. ProGRP has organ specificity and does not correlate with the stage of lung cancer. ProGRP is more specific than NSE; unfortunately, the use of this biomarker for further studies is complicated due to its instability and difficulty of identification. Sensitivity and specificity of ProGRP were 80% and 90%, respectively, while NSE showed fewer rates of sensitivity and specificity—64% and 43%. However, 27% of patients with SCLC had increased levels of NSE and normal levels of ProGRP [22]. According to this data, the simultaneous detection of ProGRP and NSE should improve the sensitivity and specificity of SCLC diagnosis (Figure 5a).
9.2. Squamous Lung Cancer
Squamous lung cancer arises from modified bronchial epithelial cells. One of the most distinctive features of squamous lung cancer is high levels of fragmented cytokeratin CK-19 subunit—CYFRA 21-1 (Figure 5b). CK-19 is a protein component of intermediate fibers of epithelial cells [126]. The level of CYFRA 21-1 is increased during the malignization process of normal epithelial cells. CYFRA 21-1 is highly expressed in serum of patients with a metastatic form of squamous lung cancer. In contrast to this, high concentrations of CYFRA 21-1 are not typical for SCLC [48,60,127].
The other specific protein for squamous lung cancer is SCCA (squamous cell carcinoma antigen), a 48-kDa protein which is found in increased levels in squamous lung cancer [58,127,128]. SCCA is an inhibitor of serine proteases such as human CELA (chymotrypsin), CAPN1 (calpain 1), and CTSL (cathepsin L) [129]. It also inhibits apoptosis of tumor cells and stimulates invasion and metastasis [130].
9.3. Adenocarcinoma
Adenocarcinoma arises from glandular cells of bronchial mucosa and expresses several protein markers (Figure 5c).
Diagnosis of adenocarcinoma is often based on identification of molecular markers of mutations, in particular EGFR, ERCC (DNA excision repair protein), RRM 1 (ribonucleoside-diphosphate reductase), KRAS (KRAS proto-oncogene), TS (thymidylate synthetase), and EML4-Alk (anaplastic lymphoma kinase receptor tyrosine kinase) [78]. Recently, protein PSF3 (DNA replication complex GINS) has become popular as a biomarker of adenocarcinoma [75,77,131]. PSF3 is a member of the heterotetrameric complex GINS (“go-ichi-ni-san” complex, from the first letters of the Japanese numbers 5-1-2-3) comprising SLD5 (Systemic RNA interference defective protein 5), PSF1 (GINS complex subunit 1), PSF2 (GINS complex subunit 2), and PSF3 (GINS complex subunit 3). This complex associates with proteins, which in turn regulate both the initiation and the progression of DNA replication [132]. To date, an overexpression of PSF3 in adenocarcinoma has been clearly established, which leads us to conclude that its level should be higher in blood plasma. However, data on the level of PSF3 in blood has yet to be reported. In addition to these biomarkers, several novel lung adenocarcinoma-associated proteins have been found using aptamers, such as LMN (lamin) and VIM (vimentin), DEF (neutrophil defensin) and TUB (tubulin), ACT (cytoplasmic actin), CTSD (cathepsin D), CLU (clusterin), NCL (nucleolin), and MUC1 (mucin-1). According to recent studies, identification of such proteins would improve the diagnosis of adenocarcinoma.
9.4. Large Cell Carcinoma
Large cell carcinoma is a malignant epithelial tumor that comprises large polygonal cells showing no obvious evidence of histological differentiation. Large cell carcinoma is characterized by small, scattered groups of large non-differentiated, polimorphic, and often dual- or multi-core cells [11]. Data on specific biomarkers of this histological type of lung cancer have not been found (Figure 5d).
9.5. Adenosquamous Carcinoma
Adenosquamous carcinoma is characterized by the features of squamous cell carcinoma and adenocarcinoma simultaneously. Consequently, it has a protein biomarker of both histotypes—MUC (mucin) [58].
9.6. Large Cell Neuroendocrine Carcinoma
Large cell neuroendocrine carcinoma (LCNEC) is extremely rare. There are difficulties related to its diagnosis and treatment. LCNEC showed overexpression of TOP SST (topoisomerasis somatostatin precursor), and ERCC1 (excision repair 1, endonuclease non-catalytic subunit) [133].
9.7. Protein Biomarkers to Main Histological Types of Lung Cancer
Despite the length at which clinically used protein biomarkers have been studied, the data shows that their levels in patients’ blood with different histological types of lung cancer varies. Table 4 presents comparative levels of well-known lung cancer biomarkers in blood plasma of patients with NSCLC, SCLC, and healthy people.
Table 4.
Comparative levels of lung cancer biomarkers in blood plasma of patients with non-small-cell lung carcinoma (NSCLC) and small-cell lung carcinoma (SCLC) and healthy people.
| Tumor-Associated Protein | NSCLC | SCLC | Normal |
|---|---|---|---|
| LDH | 525.079 ± 24.817 ng mL−1 [134] | 209.880 ± 161.322 ng mL−1 [134] | <245 ng mL−1 [134] |
| CRP | 25.079 ± 24.817 ng mL−1 [134] | 14.935 ± 21.078 ng mL−1 [134] | <8 ng mL−1 [134] |
| CEA | 51.493 ± 77.529 ng mL−1 [134] 78.5 ng mL−1 [23] ≥ 100 ng mL−1 [65] |
25.074 ± 40.957 [134] | <5.0 ng mL−1 5.0 ng mL−1 [23,61] <20.9 ng mL−1 6.5 ng mL−1 [66] |
| NSE | 13.638 ± 5.571 ng mL−1 [134] >6.4 ng mL−1 [19] 5–35 ng mL−1 17.95 ng mL−1 [61] 0–170 ng mL−1 [23] |
62.972 ± 63.012 [134] 50.8 ng mL−1 [61] 15–173 ng mL−1 [23] |
15.7–17.1 ng mL−1 15.2 ng mL−1 13 ng mL−1 [65] |
| CYFRA21-1 | 12.447 ± 15.814 ng mL−1 [134] 81.7 ng mL−1 [23] |
6.418 ± 9.567 ng mL−1 [134] | <3.3 ng mL−1 [134] 3.3 ng mL−1 [35] 3.3 ng mL−1 [61,65] 0.5 ng mL−1 [65] 2.0 ng mL−1 [23] |
| SCCA | 0.22–3.79 ng mL−1 [61] 0.5–1.7 >2 ng mL−1 [135] |
0.15 ng mL−1 [61] | 1.5 ng mL−1 [23] |
| TPS | 0–3842 ng mL−1 [136] | 12.5–773 ng mL−1 [23] | 34.9 ng mL−1 UL−1 [23] |
| ProGRP | <35 pg mL−1 [22] | >200 pg mL−1 [22] | <35 pg mL−1 [22] |
Protein biomarkers of two main histological types of NSCLC, adenocarcinoma and squamous lung cancer and their respective levels in blood plasma are compared to a healthy control group and summarized in Table 5.
Table 5.
Comparative levels of well-known lung cancer biomarkers in blood plasma of patients with adenocarcinoma and squamous lung cancer and healthy people.
| Tumor-Associated Protein | Adenocarcinoma | Squamous Carcinoma | Normal |
|---|---|---|---|
| CEA | 30.76 ng mL−1 [61] 0.6–588 ng mL−1 [23] 3.5–11.1 ng mL−1 [66] |
4.49 ng mL−1 [134] 0.8–587 ng mL−1 [23] |
<5.0 ng mL−1 5.0 ng mL−1 [23,61] <20.9 ng mL−1 6.5 ng mL−1 [66] |
| NSE | 17.95 ng mL−1 [134] | 16.83 ng mL−1 [134] | 15.7–17.1 ng mL−1 15.2 ng mL−1 13 ng mL−1 [23] |
| CYFRA21-1 | 4.00 ng mL−1 [61] 5.79 ± 6.75 ng mL−1 [63] 1.3–4.4 ng mL−1 [66] |
10.34 ng mL−1 [61] | <3.3 [134] 3.3 ng mL−1 [23,61] 0.5 ng mL−1 [23] 2.0 ng mL−1 [66] |
| SCCA | 0.22 ng mL−1 [61] 0.5–1.7 >2 ng mL−1 [66] |
3.79 ng mL−1 [61] | 1.5 ng mL−1 [66] |
| TPS | 10–3842 ng mL−1 [23] | 0–3000 ng mL−1 [23] | 34.9 ng mL−1 [23] |
Thus, the analysis of clinical biomarkers has shown that the use of six of the most specific protein biomarkers will help improve early diagnosis of lung cancer and allow differentiating between lung cancer histological types. These are summarized in Table 6.
Table 6.
A panel of biomarkers specific for SCLC, adenocarcinoma, squamous lung cancer, and large cell lung cancer.
| Biomarker | CEA ngmL−1 | NSE ngmL−1 | ProGRP pgmL−1 | PSF3 | CYFRA21-1 ngmL−1 | SCCA ngmL−1 |
|---|---|---|---|---|---|---|
| Small Cell CA | 25.07 ± 41.1 | 50.8–173 | >200 | normal | 6.42 ± 9.57 | 0.15 |
| AdenoCA | 0.6–588 | 17.95 | ~35 | overexpression | 1.3–5.79 | 0.22–2.0 |
| SquamousCA | 0.8–587 | 16.83 | ~35 | normal | 10.34 | 3.79 |
| Large Cell CA | 51.5–100 | 4.6–17.95 | ~35 | normal | 1.3–5.79 | 0.22–2.0 |
| Healthy | 5–20.9 | 13–17.1 | ~35 | normal | 0.5–1.3 | 1.5 |
CEA is a biomarker specific for all lung cancer types;
NSE is a biomarker of NSCLC, and a marker of metastasis;
CYFRA21-1 is a general biomarker for screening for lung cancer, and a biomarker of squamous lung cancer in metastatic form;
SCCA is a biomarker of squamous lung cancer;
PSF3 is a biomarker of adenocarcinoma;
ProGRP is a biomarker of SCLC;
SCCA and mucin are biomarkers of adenosquamous carcinoma;
SST is a biomarker of large cell neuroendocrine carcinoma.
10. Conclusions
The great phenotypic diversity of each histological lung cancer type and the absence of highly specific and sensitive biomarkers make lung cancer diagnosis rather difficult. Panels of various biomarkers have been recently applied and are becoming more popular as this technique improves early lung cancer detection. In this review, we suggest the use of a panel consisting of eight tumor-associated biomarkers—CEA, CYFRA21-1, ProGRP, CEA, PSF3, MUC, SCCA, and SST—allow us to differentiate between each histological type of lung cancer and to define the metastasis rate. In addition to conventional biomarker discovery methods, aptamer-based detection of several lung cancer biomarkers, such as LMN and VIM, DEF, and TUB, could be helpful for accurate diagnostics.
Acknowledgments
The study was funded by the Russian Foundation for Basic Research, Government of Krasnoyarsk Territory, Krasnoyarsk Region Science and Technology Support Fund to the research project № 16-42-240662р_а.
Conflicts of Interest
Authors declare no conflicts of interest.
References
- 1.Arya S., Bhansali S. Lung cancer and its early detection using biomarker-based biosensors. Chem. Rev. 2011;111:6783–6809. doi: 10.1021/cr100420s. [DOI] [PubMed] [Google Scholar]
- 2.Brambilla E., Travis W.D. Lung cancer. In: Stewart B.W., Wild C.P., editors. World Cancer Report. World Health Organization; Geneva, Switzerland: 2014. [Google Scholar]
- 3.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D., Forman D., Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 4.Sung H., Cho J. Biomarkers for the lung cancer diagnosis and their advances in proteomics. BMB Rep. 2008;41:615–625. doi: 10.5483/BMBRep.2008.41.9.615. [DOI] [PubMed] [Google Scholar]
- 5.Fiorentino F., Macaluso M., Miranda F., Montanari M., Russo A., Bagella L., Giordano A. CTCF and BORIS regulate Rb2/p130 gene transcription: A novel mechanism and a new paradigm for understanding the biology of lung cancer. Mol. Cancer Res. 2011;9:225–233. doi: 10.1158/1541-7786.MCR-10-0493. [DOI] [PubMed] [Google Scholar]
- 6.Cho L., Dowell J., Garwood D., Spangler A., Choy H. Prophylactic cranial irradiation with combined modality therapy for patients with locally advanced non-small cell lung cancer. Semin. Oncol. 2005;32:293–298. doi: 10.1053/j.seminoncol.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 7.Gaspar L., Scott C., Rotman M., Asbell S., Phillips T., Wasserman T., McKenna W., Byhardt R. Recursive partitioning analysis (RPA) of prognostic factors in three radiation therapy oncology group (RTOG) brain metastases trials. Int. J. Radiat. Oncol. Biol. Phys. 1997;37:745–751. doi: 10.1016/S0360-3016(96)00619-0. [DOI] [PubMed] [Google Scholar]
- 8.Schnabel P., Junker K. Pulmonary neuroendocrine tumors in the new WHO 2015 classification. Start of breaking new grounds? Pathologe. 2015;36:283–292. doi: 10.1007/s00292-015-0030-2. [DOI] [PubMed] [Google Scholar]
- 9.Travis W. Update on small cell carcinoma and its differentiation from squamous cell carcinoma and other non-small cell carcinomas. Mod. Pathol. 2012;25:S18–S30. doi: 10.1038/modpathol.2011.150. [DOI] [PubMed] [Google Scholar]
- 10.Weynants P., Humblet Y., Canon J., Symann M. Biology of small-cell lung-cancer—An overview. Eur. Respir. J. 1990;3:699–714. [PubMed] [Google Scholar]
- 11.Muller K. Histological classification and histogenesis of lung-cancer. Eur. J. Respir. Dis. 1984;65:4–19. [PubMed] [Google Scholar]
- 12.Dogan S., Shen R., Ang D., Johnson M., D’Angelo S., Paik P., Brzostowski E., Riely G., Kris M., Zakowski M., et al. Molecular epidemiology of EGFR and KRAS mutations in 3026 lung adenocarcinomas: Higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin. Cancer Res. 2012;18:6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imyanitov E., Demidova I., Gordiev M., Filipenko M., Kekeyeva T., Moliaka Y., Gervas P., Kozhemyako V., Vodolazhskiy D., Sergeyeva L., et al. Distribution of EGFR mutations in 10,607 Russian patients with lung cancer. Mol. Diagn. Ther. 2016;20:401–406. doi: 10.1007/s40291-016-0213-4. [DOI] [PubMed] [Google Scholar]
- 14.Capelozzi V. Role of immunohistochemistry in the diagnosis of lung cancer. Jornal Brasileiro De Pneumologia. 2009;35:375–382. doi: 10.1590/S1806-37132009000400012. [DOI] [PubMed] [Google Scholar]
- 15.Marshall H., Bowman R., Yang I., Fong K., Berg C. Screening for lung cancer with low-dose computed tomography: A review of current status. J. Thorac. Dis. 2013;5:S524–S539. doi: 10.3978/j.issn.2072-1439.2013.09.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vazquez M., Koizurni J., Henschke C., Yankelevitz D. Reliability of cytologic diagnosis of early lung cancer. Cancer Cytopathol. 2007;111:252–258. doi: 10.1002/cncr.22767. [DOI] [PubMed] [Google Scholar]
- 17.Rabinowits G., Gercel-Taylor C., Day J., Taylor D., Kloecker G. Exosomal microrna: A diagnostic marker for lung cancer. Clin. Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 18.Mitas M., Hoover L., Silvestri G., Reed C., Green M., Turrisi A., Sherman C., Mikhitarian K., Cole D., Block M., et al. Lunx is a superior molecular marker for detection of non-small lung cell cancer in peripheral blood. J. Mol. Diagn. 2003;5:237–242. doi: 10.1016/S1525-1578(10)60480-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan H., Lewis C., Thomas P. Exhaled breath analysis: Novel approach for early detection of lung cancer. Lung Cancer. 2009;63:164–168. doi: 10.1016/j.lungcan.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 20.Jantus-Lewintre E., Usó M., Sanmartín E., Camps C. Update on biomarkers for the detection of lung cancer. Lung Cancer Targ. Ther. 2012;3:21–29. doi: 10.2147/LCTT.S23424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andre F., Schartz N., Movassagh M., Flament C., Pautier P., Morice P., Pomel C., Lhomme C., Escudier B., Le Chevalier T., et al. Malignant effusions and immunogenic tumour-derived exosomes. Lancet. 2002;360:295–305. doi: 10.1016/S0140-6736(02)09552-1. [DOI] [PubMed] [Google Scholar]
- 22.Montani F., Marzi M., Dezi F., Dama E., Carletti R., Bonizzi G., Bertolotti R., Bellomi M., Rampinelli C., Maisonneuve P., et al. Mir-test: A blood test for lung cancer early detection. Cancer Res. 2015;75 doi: 10.1158/1538-7445.AM2015-1573. [DOI] [PubMed] [Google Scholar]
- 23.Nagrath S., Sequist L., Maheswaran S., Bell D., Irimia D., Ulkus L., Smith M., Kwak E., Digumarthy S., Muzikansky A., et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sozzi G., Boeri M., Rossi M., Verri C., Suatoni P., Bravi F., Roz L., Conte D., Grassi M., Sverzellati N., et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: A correlative MILD trial study. J. Clin. Oncol. 2014;32:768–773. doi: 10.1200/JCO.2013.50.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sozzi G., Conte D., Leon M., Cirincione R., Roz L., Ratcliffe C., Roz E., Cirenei N., Bellomi M., Pelosi G., et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J. Clin. Oncol. 2003;21:3902–3908. doi: 10.1200/JCO.2003.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Valenti R., Huber V., Filipazzi P., Pilla L., Sovena G., Villa A., Corbelli A., Fais S., Parmiani G., Rivoltini L. Human tumor-released microvesicles promote the differentiation of myeloid cells with transforming growth factor-β-mediated suppressive activity on T lymphocytes. Cancer Res. 2006;66:9290–9298. doi: 10.1158/0008-5472.CAN-06-1819. [DOI] [PubMed] [Google Scholar]
- 27.Paci M., Rapicetta C., Maramotti S. New biomarkers for lung cancer. Expert Opin. Med. Diagn. 2010;4:201–224. doi: 10.1517/17530051003725113. [DOI] [PubMed] [Google Scholar]
- 28.Kato H. Expression and function of squamous cell carcinoma antigen. Anticancer Res. 1996;16:2149–2153. [PubMed] [Google Scholar]
- 29.Doseeva V., Colpitts T., Gao G., Woodcock J., Knezevic V. Performance of a multiplexed dual analyte immunoassay for the early detection of non-small cell lung cancer. J. Transl. Med. 2015;13:55. doi: 10.1186/s12967-015-0419-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Indovina P., Marcelli E., Maranta P., Tarro G. Lung cancer proteomics: Recent advances in biomarker discovery. Int. J. Proteom. 2011 doi: 10.1155/2011/726869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang S., Sung H., Ahn J., Park J., Lee S., Park C., Cho J. The haptoglobin β chain as a supportive biomarker for human lung cancers. Mol. Biosyst. 2011;7:1167–1175. doi: 10.1039/c0mb00242a. [DOI] [PubMed] [Google Scholar]
- 32.Maciel C., Junqueira M., Kawamura M., Paschoal M., Duarte R., Carvalho M., Domone G. Differential proteomic serum pattern of low molecular weight proteins expressed by adenocarcinoma lung cancer patients. Mol. Cell. Proteom. 2004;3:S215. [PubMed] [Google Scholar]
- 33.Kokkonen N., Ulibarri I., Kauppila A., Luosujarvi H., Rivinoja A., Pospiech H., Kellokumpu I., Kellokumpu S. Hypoxia upregulates carcinoembryonic antigen expression in cancer cells. Int. J. Cancer. 2007;121:2443–2450. doi: 10.1002/ijc.22965. [DOI] [PubMed] [Google Scholar]
- 34.Ueda K., Saichi N., Takami S., Kang D., Toyama A., Daigo Y., Ishikawa N., Kohno N., Tamura K., Shuin T., et al. A comprehensive peptidome profiling technology for the identification of early detection biomarkers for lung adenocarcinoma. PLoS ONE. 2011;6:e18567. doi: 10.1371/journal.pone.0018567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng X., Hood B., Sun M., Conrads T., Day R., Weissfeld J., Siegfried J., Bigbee W. Lung cancer serum biomarker discovery using glycoprotein capture and liquid chromatography mass spectrometry. J. Proteome Res. 2010;9:6440–6449. doi: 10.1021/pr100696n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bharti A., Ma P.C., Mauliketal G. Haptoglobin α-subunit and hepatocyte growth factor can potentially serve as serum tumor biomarkers in small cell lung cancer. Anticancer Res. 2004;24:1031–1038. [PubMed] [Google Scholar]
- 37.Dai S., Wang X., Liu L., Liu J., Wu S., Huang L., Xiao X., He D. Discovery and identification of serum amyloid a protein elevated in lung cancer serum. Sci. China Ser. C Life Sci. 2007;50:305–311. doi: 10.1007/s11427-007-0053-x. [DOI] [PubMed] [Google Scholar]
- 38.Liu L., Liu J., Dai S., Wang X., Wu S., Wang J., Huang L., Xiao X., He D. Reduced transthyretin expression in sera of lung cancer. Cancer Sci. 2007;98:1617–1624. doi: 10.1111/j.1349-7006.2007.00576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L., Liu J., Wang Y., Dai S., Wang X., Wu S., Wang J., Huang L., Xiao X., He D. A combined biomarker pattern improves the discrimination of lung cancer. Biomarkers. 2011;16:20–30. doi: 10.3109/1354750X.2010.521257. [DOI] [PubMed] [Google Scholar]
- 40.Howard B., Wang M., Campa M., Corro C., Fitzgerald M., Patz E. Identification and validation of a potential lung cancer serum biomarker detected by matrix-assisted laser desorption/ionization-time of flight spectra analysis. Proteomics. 2003;3:1720–1724. doi: 10.1002/pmic.200300514. [DOI] [PubMed] [Google Scholar]
- 41.Liu G., Wu Q., Liu G., Song X., Zhang J. Psoriasin (S100A7) is a novel biomarker for lung squamous cell carcinoma in humans (retracted article. See vol. 16, 40, 2016) Cancer Cell Int. 2015;15:18. doi: 10.1186/s12935-014-0154-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Yu Z., Chen X., Cui L., Si H., Lu H., Liu S. Prediction of lung cancer based on serum biomarkers by gene expression programming methods. Asian Pac. J. Cancer Prev. 2014;15:9367–9373. doi: 10.7314/APJCP.2014.15.21.9367. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H., Liu J., Yue D., Gao L., Wang D., Zhang H., Wang C. Clinical significance of E-cadherin, β-catenin, vimentin and S100A4 expression in completely resected squamous cell lung carcinoma. J. Clin. Pathol. 2013;66:937–945. doi: 10.1136/jclinpath-2013-201467. [DOI] [PubMed] [Google Scholar]
- 44.Diamandis E., Goodglick L., Planque C., Thomquist M. Pentraxin-3 is a novel biomarker of lung carcinoma. Clin. Cancer Res. 2011;17:2395–2399. doi: 10.1158/1078-0432.CCR-10-3024. [DOI] [PubMed] [Google Scholar]
- 45.Goetsch C.M. Genetic tumor profiling and genetically targeted cancer therapy. Semin. Oncol. Nurs. 2011;27:34–44. doi: 10.1016/j.soncn.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 46.Kim H., Oh I., Shin M., Park J., Choi H., Ban H., Kim K., Kim Y., Shin J., Ryang D., et al. Plasma proGRP concentration is sensitive and specific for discriminating small cell lung cancer from nonmalignant conditions or non-small cell lung cancer. J. Korean Med. Sci. 2011;26:625–630. doi: 10.3346/jkms.2011.26.5.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Landi M., Zhao Y., Rotunno M., Koshiol J., Liu H., Bergen A., Rubagotti M., Goldstein A., Linnoila I., Marincola F., et al. Microrna expression differentiates histology and predicts survival of lung cancer. Clin. Cancer Res. 2010;16:430–441. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foa P., Fornier M., Miceli R., Seregni E., Santambrogio L., Nosotti M., Cataldo I., Sala M., Caldiera S., Bombardieri E. Tumour markers CEA, NSE, SCC, TPA and CYFRA 21.1 in resectable non-small cell lung cancer. Anticancer Res. 1999;19:3613–3618. [PubMed] [Google Scholar]
- 49.Tessitore A., Gaggiano A., Cicciarelli G., Verzella D., Capece D., Fischietti M., Zazzeroni F., Alesse E. Serum biomarkers identication mass spectrometry in high-mortality tumors. Int. J. Proteom. 2013;2013:1–15. doi: 10.1155/2013/125858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jett J., Peek L., Fredericks L., Jewell W., Pingleton W., Robertson J. Audit of the autoantibody test, EarlyCDT®-Lung, in 1600 patients: An evaluation of its performance in routine clinical practice. Lung Cancer. 2014;83:51–55. doi: 10.1016/j.lungcan.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 51.Chapman C., Thorpe A., Murray A., Parsy-Kowalska C., Allen J., Stafford K., Chauhan A., Kite T., Maddison P., Robertson J. Immunobiomarkers in small cell lung cancer: Potential early cancer signals. Clin. Cancer Res. 2011;17:1474–1480. doi: 10.1158/1078-0432.CCR-10-1363. [DOI] [PubMed] [Google Scholar]
- 52.Mizuguchi S., Nishiyama N., Iwata T., Nishida T., Izumi N., Tsukioka T., Inoue K., Uenishi T., Wakasa K., Suehiro S. Serum Sialyl Lewis(x) and cytokeratin 19 fragment as predictive factors for recurrence in patients with stage I non-small cell lung cancer. Lung Cancer. 2007;58:369–375. doi: 10.1016/j.lungcan.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 53.Pujol J., Grenier J., Daures J., Daver A., Pujol H., Michel F. Serum fragment of cytokeratin subunit-19 measured by CYFRA-21-1 immunoradiometric assay as a marker of lung-cancer. Cancer Res. 1993;53:61–66. [PubMed] [Google Scholar]
- 54.Okada M., Nishio W., Skaamoto T., Uchino K., Yuki T., Nakagawa A., Tsubota N. Effect of hystologic type and smoking status on interpretation of serum carcinoembryonic antigen value in non-small cell lung carcinoma. Ann. Thorac. Surg. 2004;78:1004–1009. doi: 10.1016/j.athoracsur.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 55.Arrieta O., Saavedra-Perez D., Kuri R., Aviles-Salas A., Martinez L., Mendoza-Posada D., Castillo P., Astorga A., Guzman E., de la Garza J. Brain metastasis development and poor survival associated with carcinoembryonic antigen (CEA) level in advanced non-small cell lung cancer: A prospective analysis. BMC Cancer. 2009;9:119. doi: 10.1186/1471-2407-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ginsberg M., Grewal R., Heelan R. Lung cancer. Radiol. Clin. N. Am. 2007;45:21–43. doi: 10.1016/j.rcl.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 57.Kuespert K., Pils S., Hauck C. Ceacams: Their role in physiology and pathophysiology. Curr. Opin. Cell Biol. 2006;18:565–571. doi: 10.1016/j.ceb.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lakshmanan I., Ponnusamy M., Macha M., Haridas D., Majhi P., Kaur S., Jain M., Batra S., Ganti A. Mucins in lung cancer diagnostic, prognostic, and therapeutic implications. J. Thorac. Oncol. 2015;10:19–27. doi: 10.1097/JTO.0000000000000404. [DOI] [PubMed] [Google Scholar]
- 59.Okabe N., Ezaki J., Yamaura T., Muto S., Osugi J., Tamura H., Imai J., Ito E., Yanagisawa Y., Honma R., et al. FAM83B is a novel biomarker for diagnosis and prognosis of lung squamous cell carcinoma. Int. J. Oncol. 2015;46:999–1006. doi: 10.3892/ijo.2015.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okamura K., Takayama K., Izumi M., Harada T., Furuyama K., Nakanishi Y. Diagnostic value of CEA and CYFRA 21-1 tumor markers in primary lung cancer. Lung Cancer. 2013;80:45–49. doi: 10.1016/j.lungcan.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 61.Wojcik E., Kulpa J., Sas-Korczynska B., Korzeniowski S., Jakubowicz J. ProGRP and NSE in therapy monitoring in patients with small cell lung cancer. Anticancer Res. 2008;28:3027–3033. [PubMed] [Google Scholar]
- 62.Zamay G., Kolovskaya O., Zamay T., Glazyrin Y., Krat A., Zubkova O., Spivak E., Wehbe M., Gargaun A., Muharemagic D., et al. Aptamers selected to postoperative lung adenocarcinoma detect circulating tumor cells in human blood. Mol. Ther. 2015;23:1486–1496. doi: 10.1038/mt.2015.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bastawisy A., Elazzouny M., Mohammed G., Awadallah A., Behiry E. Serum cytokeratin 19 fragment in advanced lung cancer: Could we eventually have a serum tumor marker? Ecancer. 2014;394:1–9. doi: 10.3332/ecancer.2014.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berezovski M., Lechmann M., Musheev M., Mak T., Krylov S. Aptamer-facilitated biomarker discovery (AptaBiD) J. Am. Chem. Soc. 2008;130:9137–9143. doi: 10.1021/ja801951p. [DOI] [PubMed] [Google Scholar]
- 65.Li X., Hayward C., Fong P., Dominguez M., Hunsucker S., Lee L., McLean M., Law S., Butler H., Schirm M., et al. A blood-based proteomic classifier for the molecular characterization of pulmonary nodules. Sci. Transl. Med. 2013;5:207ra142. doi: 10.1126/scitranslmed.3007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Molina R., Filella X., Auge J. Progrp: A new biomarker for small cell lung cancer. Clin. Biochem. 2004;37:505–511. doi: 10.1016/j.clinbiochem.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 67.Stovold R., Blackhall F., Meredith S., Hou J., Dive C., White A. Biomarkers for small cell lung cancer: Neuroendocrine, epithelial and circulating tumour cells. Lung Cancer. 2012;76:263–268. doi: 10.1016/j.lungcan.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 68.Gandara D., Li T., Lara P., Mack P., Kelly K., Miyamoto S., Goodwin N., Beckett L., Redman M. Algorithm for codevelopment of new drug-predictive biomarker combinations: Accounting for inter- and intrapatient tumor heterogeneity. Clin. Lung Cancer. 2012;13:321–325. doi: 10.1016/j.cllc.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miyake Y., Kodama T., Yamaguchi K. Pro-gastrin-releasing peptide(31–98) is a specific tumor-marker in patients with small-cell lung-carcinoma. Cancer Res. 1994;54:2136–2140. [PubMed] [Google Scholar]
- 70.Shimizu K., Yukawa T., Okita R., Saisho S., Maeda A., Nojima Y., Nakata M. Cyclooxygenase-2 expression is a prognostic biomarker for non-small cell lung cancer patients treated with adjuvant platinum-based chemotherapy. World J. Surg. Oncol. 2015;13:21. doi: 10.1186/s12957-014-0426-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chee J., Naran A., Misso N., Thompson P., Bhoola K. Expression of tissue and plasma kallikreins and kinin B1 and B2 receptors in lung cancer. Biol. Chem. 2008;389:1225–1233. doi: 10.1515/BC.2008.139. [DOI] [PubMed] [Google Scholar]
- 72.Pachter J., de Vries H., Fabry Z. The blood-brain barrier and its role in immune privilege in the central nervous system. J. Neuropathol. Exp. Neurol. 2003;62:593–604. doi: 10.1093/jnen/62.6.593. [DOI] [PubMed] [Google Scholar]
- 73.Fortes D., Allen M., Lowe V., Shen K., Wigle D., Cassivi S., Nichols F., Deschamps C. The sensitivity of 18F-fluorodeoxyglucose positron emission tomography in the evaluation of metastatic pulmonary nodules. Eur. J. Cardio Thorac. Surg. 2008;34:1223–1227. doi: 10.1016/j.ejcts.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 74.Kikuchi T., Hassanein M., Amann J., Liu Q., Slebos R., Rahman S., Kaufman J., Zhang X., Hoeksema M., Harris B., et al. In-depth proteomic analysis of nonsmall cell lung cancer to discover molecular targets and candidate biomarkers. Mol. Cell. Proteom. 2012;11:916–932. doi: 10.1074/mcp.M111.015370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tane S., Sakai Y., Hokka D., Okuma H., Ogawa H., Tanaka Y., Uchino K., Nishio W., Yoshimura M., Maniwa Y. Significant role of Psf3 expression in non-small-cell lung cancer. Cancer Sci. 2015;106:1625–1634. doi: 10.1111/cas.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hotta K., Segawa Y., Takigawa N., Kishino D., Saeki H., Nakata M., Mandai K., Eguchi K. Evaluation of the relationship between serum carcinoembryonic antigen level and treatment outcome in surgically resected clinical-stage I patients with non-small-cell lung cancer. Anticancer Res. 2000;20:2177–2180. [PubMed] [Google Scholar]
- 77.Tauchi S., Sakai Y., Fujimoto S., Ogawa H., Tane S., Hokka D., Tanaka Y., Nishio W., Yoshimura M., Yanagita E., et al. Psf3 is a prognostic biomarker in lung adenocarcinoma: A larger trial using tissue microarrays of 864 consecutive resections. Eur. J. Cardio Thorac. Surg. 2016;50:758–764. doi: 10.1093/ejcts/ezw077. [DOI] [PubMed] [Google Scholar]
- 78.Sholl L. Biomarkers in lung adenocarcinoma a decade of progress. Arch. Pathol. Lab. Med. 2015;139:469–480. doi: 10.5858/arpa.2014-0128-RA. [DOI] [PubMed] [Google Scholar]
- 79.Lee D., Kim S., Kang J., Hong S., Jeon E., Kim Y., Yoo I., Park J., Jang H., Lee H., et al. Serum carcinoembryonic antigen levels and the risk of whole-body metastatic potential in advanced non-small cell lung cancer. J. Cancer. 2014;5:663–669. doi: 10.7150/jca.9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bigbee W.L., Gopalakrishnan V., Weissfeld J.L., Wilson D.O., Dacic S., Lokshin A.E., Siegfried J.M. A multiplexed serum biomarker immunoassay panel discriminates clinical lung cancer patients from high-risk individuals found to be cancer-free by CT screening. J. Thorac. Oncol. 2012;7:698–708. doi: 10.1097/JTO.0b013e31824ab6b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Patz E., Campa M., Gottlin E., Kusmartseva I., Guan X., Herndon J. Panel of serum biomarkers for the diagnosis of lung cancer. J. Clin. Oncol. 2007;25:5578–5583. doi: 10.1200/JCO.2007.13.5392. [DOI] [PubMed] [Google Scholar]
- 82.Mehan M., Ayers D., Thirstrup D., Xiong W., Ostroff R., Brody E., Walker J., Gold L., Jarvis T., Janjic N., et al. Protein signature of lung cancer tissues. PLoS ONE. 2012;7:e35157. doi: 10.1371/journal.pone.0035157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jantus-Lewintre E., Uso M., Sanmartin E., Gallach S., Sirera R., Hernando A., Martinez N., Figueroa S., Casimiro E., Camps C. Ratios between VEGF ligands and receptors in tumor and stroma have impact on the outcome in resectable NSCLC. J. Clin. Oncol. 2013;31:e22147. [Google Scholar]
- 84.Patel D.J., Suri A.K. Structure, recognition and discrimination in RNA aptamer complexes with cofactors, amino acids, drugs and aminoglycoside antibiotics. J. Biotechnol. 2000;74:39–60. doi: 10.1016/S1389-0352(99)00003-3. [DOI] [PubMed] [Google Scholar]
- 85.Jiang F., Liu B., Lu J., Li F., Li D., Liang C., Dang L., Liu J., He B., Badshah S., et al. Progress and challenges in developing aptamer-functionalized targeted drug delivery systems. Int. J. Mol. Sci. 2015;16:23784–23822. doi: 10.3390/ijms161023784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lakhin A., Tarantul V., Gening L. Aptamers: Problems, solutions and prospects. Acta Nat. 2013;5:34–43. [PMC free article] [PubMed] [Google Scholar]
- 87.Rozenblum G., Lopez V., Vitullo A., Radrizzani M. Aptamers: Current challenges and future prospects. Expert Opin. Drug Discov. 2016;11:127–135. doi: 10.1517/17460441.2016.1126244. [DOI] [PubMed] [Google Scholar]
- 88.Zamay G.S., Ivanchenko T., Zamay T.N., Grigorieva V.L., Glazyrin Y.E., Kolovskaya O.S., Garanzha I., Barinov A., Krat A.V., Mironov G., et al. DNA-aptamers for characterization of histological structure of lung adenocarcinoma. Mol. Ther. Nucleic Acid. 2016;6:150–162. doi: 10.1016/j.omtn.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lou X., Xiao T., Zhao K., Wang H., Zheng H., Lin D., Lu Y., Gao Y., Cheng S., Liu S., et al. Cathepsin D is secreted from M-B cells: Its potential role as a biomarker of lung cancer. J. Proteome Res. 2007;6:1083–1092. doi: 10.1021/pr060422t. [DOI] [PubMed] [Google Scholar]
- 90.Deng S., Wang J., Hou L., Li J., Chen G., Jing B., Zhang X., Yang Z. Annexin A1, A2, A4 and A5 play important roles in breast cancer, pancreatic cancer and laryngeal carcinoma, alone and/or synergistically. Oncol. Lett. 2013;5:107–112. doi: 10.3892/ol.2012.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cole A., Clifton-Bligh R., Marsh D. Histone h2b monoubiquitination: Roles to play in human malignancy. Endocrine-Related Cancer. 2015;22:T19–T33. doi: 10.1530/ERC-14-0185. [DOI] [PubMed] [Google Scholar]
- 92.Havel L., Kline E., Salgueiro A., Marcus A. Vimentin regulates lung cancer cell adhesion through a VAV2-RAC1 pathway to control focal adhesion kinase activity. Oncogene. 2015;34:1979–1990. doi: 10.1038/onc.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kidd M., Shumaker D., Ridge K. The role of vimentin intermediate filaments in the progression of lung cancer. Am. J. Respir. Cell Mol. Biol. 2014;50:1–6. doi: 10.1165/rcmb.2013-0314TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Richardson F., Young G., Sennello R., Wolf J., Argast G., Mercado P., Davies A., Epstein D., Wacker B. The evaluation of E-Cadherin and vimentin as biomarkers of clinical outcomes among patients with non-small cell lung cancer treated with erlotinib as second- or third-line therapy. Anticancer Res. 2012;32:537–552. [PubMed] [Google Scholar]
- 95.Upton M., Hirohashi S., Tome Y., Miyazawa N., Suemasu K., Shimosato Y. Expression of vimentin in surgically resected adenocarcinomas and large cell carcinomas of lung. Am. J. Surg. Pathol. 1986;10:560–567. doi: 10.1097/00000478-198608000-00006. [DOI] [PubMed] [Google Scholar]
- 96.Chow K., Factor R., Ullman K. The nuclear envelope environment and its cancer connections. Nat. Rev. Cancer. 2012;12:196–209. doi: 10.1038/nrc3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Friedl P., Wolf K., Lammerding J. Nuclear mechanics during cell migration. Curr. Opin. Cell Biol. 2011;23:55–64. doi: 10.1016/j.ceb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Machiels B., Ramaekers F., Kuijpers H., Groenewoud J., Oosterhuis J., Looijenga L. Nuclear lamin expression in normal testis and testicular germ cell tumours of adolescents and adults. J. Pathol. 1997;182:197–204. doi: 10.1002/(SICI)1096-9896(199706)182:2<197::AID-PATH823>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 99.Foster C.R., Przyborski S.A., Wilson R.G., Hutchison C.J. Lamins as cancer biomarkers. Biochem. Soc. Trans. 2010;38:297–300. doi: 10.1042/BST0380297. [DOI] [PubMed] [Google Scholar]
- 100.Parker A.L., Kavallaris M., McCarroll J.A. Microtubules and their role in cellular stress in cancer. Front. Oncol. 2014;4:1–19. doi: 10.3389/fonc.2014.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aarbiou J., Verhoosel R., van Wetering S., de Boer W., van Krieken J., Litvinov S., Rabe K., Hiemstra P. Neutrophil defensins enhance lung epithelial wound closure and MUCIN gene expression in vitro. Am. J. Respir. Cell Mol. Biol. 2004;30:193–201. doi: 10.1165/rcmb.2002-0267OC. [DOI] [PubMed] [Google Scholar]
- 102.Holterman D., Diaz J., Blackmore P., Davis J., Schellhammer P., Corica A., Semmes O., Vlahou A. Overexpression of α-defensin is associated with bladder cancer invasiveness. Urol. Oncol. Semin. Orig. Investig. 2006;24:97–108. doi: 10.1016/j.urolonc.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 103.Zamay G.S., Zamay T.N., Kolovskii V.A., Shabanov A.V., Glazyrin Y.E., Veprintsev D.V., Krat A.V., Zamay S.S., Kolovskaya O.S., Gargaun A., et al. Electrochemical aptasensor for lung cancer-related protein detection in crude blood plasma samples. Sci. Rep. 2016;6:34350. doi: 10.1038/srep34350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gold L., Ayers D., Bertino J., Bock C., Bock A., Brody E.N., Carter J., Dalby A.B., Eaton B.E., Fitzwater T., et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Prakash J., Rajamanickam K. Aptamers and their significant role in cancer therapy and diagnosis. Biomedicines. 2015;3:248–269. doi: 10.3390/biomedicines3030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang Y., Chen Y., Han D., Ocsoy I., Tan W. Aptamers selected by cell-selex for application in cancer studies. Bioanalysis. 2010;2:907–918. doi: 10.4155/bio.10.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang R., Zhang Y., Cai J., Cai W., Gao T. Aptamer-based fluorescent biosensors. Curr. Med. Chem. 2011;18:4175–4184. doi: 10.2174/092986711797189637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen H., Medley C., Sefah K., Shangguan D., Tang Z., Meng L., Smith J., Tan W. Molecular recognition of small-cell lung cancer cells using aptamers. Chemmedchem. 2008;3:991–1001. doi: 10.1002/cmdc.200800030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kunii T., Ogura S., Mie M., Kobatake E. Selection of DNA aptamers recognizing small cell lung cancer using living cell-selex. Analyst. 2011;136:1310–1312. doi: 10.1039/c0an00962h. [DOI] [PubMed] [Google Scholar]
- 110.Jimenez E., Sefah K., Lopez-Colon D., Van Simaeys D., Chen H., Tockman M., Tan W. Generation of lung adenocarcinoma DNA aptamers for cancer studies. PLoS ONE. 2012;7:e46222. doi: 10.1371/journal.pone.0046222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hu J., Zhao Z., Liu Q., Ye M., Hu B., Wang J., Tan W. Study of the function of G-Rich aptamers selected for lung adenocarcinoma. Chem. Asian J. 2015;10:1519–1525. doi: 10.1002/asia.201500187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu L., Zhang Z., Zhao Z., Liu Q., Tan W., Fang X. Cellular internalization and cytotoxicity of aptamers selected from lung cancer cell. Am. J. Biomed. Sci. 2013;5:47–58. doi: 10.5099/aj130100047. [DOI] [Google Scholar]
- 113.Zamay A., Zamay G., Glazyrin Y., Zamay T., Krat A., Modestov A., Zubkova O., Spivak E., Sukhovolskaia M., Kuznetsova S., et al. DNA aptamer to human lung adenocarcinoma shows antitumor effect. Oligos Pept. Chim. Oggi Chem. Today. 2014;32:24–28. [Google Scholar]
- 114.Hu Y., Duan J., Zhan Q., Wang F., Lu X., Yang X. Novel MUC1 aptamer selectively delivers cytotoxic agent to cancer cells in vitro. PLoS ONE. 2012;7:e31970. doi: 10.1371/journal.pone.0031970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kurosaki T., Higuchi N., Kawakami S., Higuchi Y., Nakamura T., Kitahara T., Hashida M., Sasaki H. Self-assemble gene delivery system for molecular targeting using nucleic acid aptamer. Gene. 2012;491:205–209. doi: 10.1016/j.gene.2011.09.021. [DOI] [PubMed] [Google Scholar]
- 116.Esposito C., Cerchia L., Catuogno S., De Vita G., Dassie J., Santamaria G., Swiderski P., Condorelli G., Giangrande P., de Franciscis V. Multifunctional aptamer-miRNA conjugates for targeted cancer therapy. Mol. Ther. 2014;22:1151–1163. doi: 10.1038/mt.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Iaboni M., Russo V., Fontanella R., Roscigno G., Fiore D., Donnarumma E., Esposito C., Quintavalle C., Giangrande P., de Franciscis V., et al. Aptamer-miRNA-212 conjugate sensitizes NSCLC cells to trail. Mol. Ther. Nucleic Acids. 2016;5:e289. doi: 10.1038/mtna.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lai W., Wang W., Chang Y., Chang C., Yang P., Peck K. Synergistic inhibition of lung cancer cell invasion, tumor growth and angiogenesis using aptamer-siRNA chimeras. Biomaterials. 2014;35:2905–2914. doi: 10.1016/j.biomaterials.2013.12.054. [DOI] [PubMed] [Google Scholar]
- 119.Li J., Zheng H., Bates P., Malik T., Li X., Trent J., Ng C. Aptamer imaging with CU-64 labeled AS1411: Preliminary assessment in lung cancer. Nucl. Med. Biol. 2014;41:179–185. doi: 10.1016/j.nucmedbio.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 120.Zhao Z., Xu L., Shi X., Tan W., Fang X., Shangguan D. Recognition of subtype non-small cell lung cancer by DNA aptamers selected from living cells. Analyst. 2009;134:1808–1814. doi: 10.1039/b904476k. [DOI] [PubMed] [Google Scholar]
- 121.Rekhtman N., Ang D., Sima C., Travis W., Moreira A. Immunohistochemical algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on large series of whole-tissue sections with validation in small specimens. Mod. Pathol. 2011;24:1348–1359. doi: 10.1038/modpathol.2011.92. [DOI] [PubMed] [Google Scholar]
- 122.Reiber H. Dynamics of brain-derived proteins in cerebrospinal fluid. Clin. Chim. Acta. 2001;310:173–186. doi: 10.1016/S0009-8981(01)00573-3. [DOI] [PubMed] [Google Scholar]
- 123.Reiber H., Jacobi C., Felgenhauer K. Sensitive quantitation of carcinoembrionic antigen in cerebrospinal fluid and its barrier-dependent differentiation. Clin. Chim. Acta. 1986;156:259–269. doi: 10.1016/0009-8981(86)90069-0. [DOI] [PubMed] [Google Scholar]
- 124.Grunnet M., Sorensen J. Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung Cancer. 2012;76:138–143. doi: 10.1016/j.lungcan.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 125.Papadopoulos E., Petraki C., Gregorakis A., Chra E., Fragoulis E., Scorilas A. l-Dopa decarboxylase mRNA levels provide high diagnostic accuracy and discrimination between clear cell and non-clear cell subtypes in renal cell carcinoma. Clin. Biochem. 2015;48:590–595. doi: 10.1016/j.clinbiochem.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 126.Stieber P., Dienemann H., Hasholzner U., Muller C., Poley S., Hofmann K., Fatehmoghadam A. Comparison of cytokeratin fragment 19 (CYFRA 21-1), tissue polypeptide antigen (TPA) and tissue polypeptide specific antigen (TPS) as tumor-markers in lung-cancer. Eur. J. Clin. Chem. Clin. Biochem. 1993;31:689–694. doi: 10.1515/cclm.1993.31.10.689. [DOI] [PubMed] [Google Scholar]
- 127.Wang J., Yi Y., Li B., Wang Z., Sun H., Zhang P., Huang W. CYFRA 21-1 can predict the sensitivity to chemoradiotherapy of non-small-cell lung carcinoma. Biomarkers. 2010;15:594–601. doi: 10.3109/1354750X.2010.504308. [DOI] [PubMed] [Google Scholar]
- 128.Ferrigno D., Buccheri G., Biggi A. Serum tumor-markers in lung-cancer: History, biology and clinical-applications. Eur. Respir. J. 1994;7:186–197. doi: 10.1183/09031936.94.07010186. [DOI] [PubMed] [Google Scholar]
- 129.Kato H., Tamai K., Magaya T. Clinical value of SCC-antigen, a subfraction of tumor antigen TA-4, in the manage-ment of cervical cancer. Gan No Rinsho. 1985;31:594–599. [PubMed] [Google Scholar]
- 130.Suminami Y., Nawata S., Kato H. Biological role of SCC antigen. Tumor Biol. 1998;19:488–493. doi: 10.1159/000030042. [DOI] [PubMed] [Google Scholar]
- 131.Hokka D., Maniwa Y., Tane S., Nishio W., Yoshimura M., Okita Y., Ohbayashi C., Sakai Y., Chen X., Hayashi Y. Psf3 is a prognostic biomarker in lung adenocarcinoma. Lung Cancer. 2013;79:77–82. doi: 10.1016/j.lungcan.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 132.Bermudez V., Farina A., Raghavan V., Tappin I., Hurwitz J. Studies on human DNA polymerase epsilon and GINS complex and their role in DNA replication. J. Biol. Chem. 2011;286:28963–28977. doi: 10.1074/jbc.M111.256289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Makino T., Mikami T., Hata Y., Otsuka H., Koezuka S., Isobe K., Tochigi N., Shibuya K., Homma S., Iyoda A. Comprehensive biomarkers for personalized treatment in pulmonary large cell neuroendocrine carcinoma: A comparative analysis with adenocarcinoma. Ann. Thorac. Surg. 2016;102:1694–1701. doi: 10.1016/j.athoracsur.2016.04.100. [DOI] [PubMed] [Google Scholar]
- 134.Yuan C., Yang K., Tang H., Chen D. Diagnostic values of serum tumor markers CYFRA 21-1, SCCAg, ferritin, CEA, CA19-9, and AFP in oral/oropharyngeal squamous cell carcinoma. Clin. Oncol. 2016;9:3381–3386. doi: 10.2147/OTT.S105672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pastor A., Menendez R., Cremades M., Pastor V., Llopis R., Aznar J. Diagnostic value of SCC, CEA and CYFRA 21.1 in lung cancer: A bayesian analysis. Eur. Respir. J. 1997;10:603–609. [PubMed] [Google Scholar]
- 136.Nakamura H., Kawasaki N., Taguchi M., Kabasawa K. Survival impact of epidermal growth factor receptor overexpression in patients with non-small cell lung cancer: A meta-analysis. Thorax. 2006;61:140–145. doi: 10.1136/thx.2005.042275. [DOI] [PMC free article] [PubMed] [Google Scholar]