Abstract
Membrane curvature is a fundamental feature of cells and their organelles. Much of what we know about how cells sense curved surfaces comes from studies examining nanometer-sized molecules on nanometer-scale curvatures. We are only just beginning to understand how cells recognize curved topologies at the micron scale. In this review, we provide the reader with an overview of our current understanding of how cells sense and respond to micron-scale membrane curvature.
Keywords: Septin, membrane curvature, BAR domain, SpoVM
Cellular Membranes and Curvature
Shape is a common feature used to describe cells. Consider the transformation of a platelet from a smooth disc to a protrusion-rich, activated state; the distinct morphologies of cells such as neurons and podocytes; and the variety of cell shapes found in the microbial world. The ability to make and change shapes is central to specialized cell functions and adaptations of cells to changing environments. This link between form and function is exemplified in a dendritic spine or the dramatic switches between budding and filamentation of pathogenic fungi in hosts. Shape depends on cells spatially controlling networks of proteins and regulating the physical properties of membranes. While it is somewhat understood how cells generate different shapes, comparatively little is known about how cells recognize their own shape and use information about their geometry.
One way cell shape can be described is in terms of membrane curvature. Cell membranes are thin, lipid-protein mosaics that function primarily to compartmentalize the cell and provide a selective barrier to the extracellular environment [1]. Additional functions of membranes include responding to physical forces, serving as signaling platforms, regulating the transport of nutrients and ions into the cell, and the storage of lipids and proteins [2–5]. Membrane composition between cell and organelle membranes can vary extensively, with differences arising from the combinations of particular lipid species, fatty acyl chains and saturation. In vitro studies have shown that these differences can alter membrane physicochemical characteristics such as thickness, curvature, fluidity, surface charge and what proteins reside in or on specific membranes [6–9]. Ultimately, cell and organelle shape can be influenced by properties of the membrane.
Cellular membrane curvature can span from nanometer to micrometer scales (Figure 1A). Membrane curvature sensing is best understood for nanometer-sized molecules binding nanometer curvatures such as ArfGAP1 [10–12], α-synuclein [13], and BAR proteins [14, 15]. When compared to typical globular proteins, curvatures at the micron scale would have a thousand-fold larger area (Figure 1A). How do nanometer sized proteins perceive membrane curvatures that are several orders of magnitude their scale? In this review, we discuss identified micron-scale curvature sensors and how they may relate to nanometer curvature sensors. We then go on to highlight recent insights into how cells perceive and use micron-scale membrane curvature using nanometer-sized proteins.
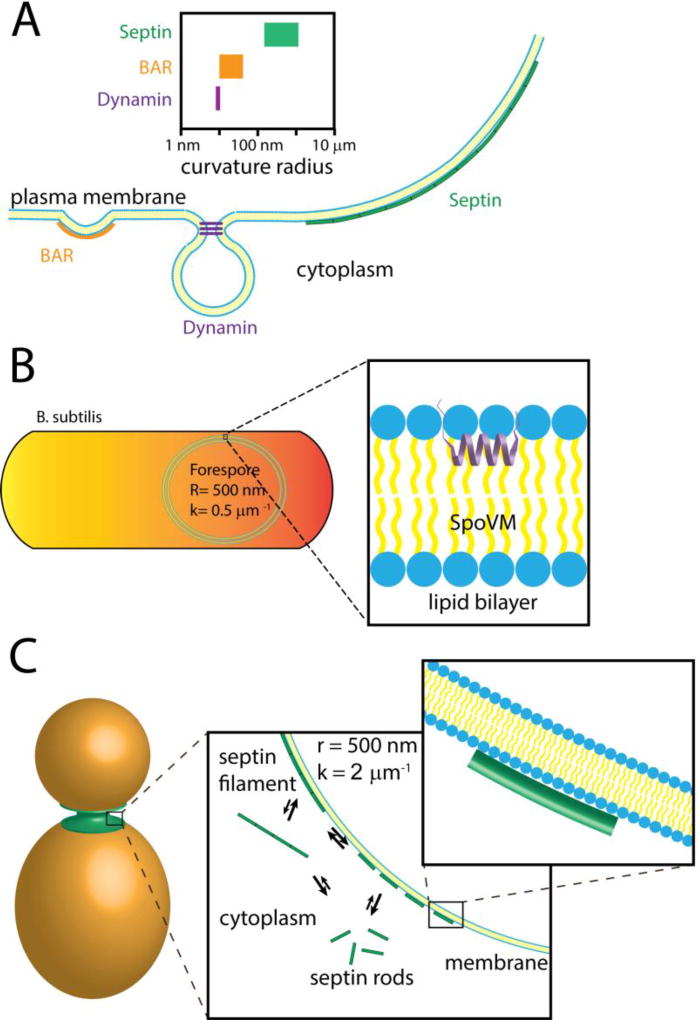
Figure 1. The disparity between nanometer molecules and micron-scale curvature at the membrane.
(A) Cells possess a variety of proteins that can bind and recognize curved biological membranes. Some of the best understood membrane curvature “sensors” are BAR domain proteins (left) and dynamin (center), which recognize steep, nanometer-scale curvatures. Comparatively, the septin cytoskeleton (right) recognizes much shallower curvatures at a micron-scale. The range of curvatures recognized by these proteins are depicted above.
(B) During sporulation in the bacterium Bacillus subtilis SpoVM preferentially binds to the forespore membrane, the only site of positive curvature in the cell, which has a curvature of about 0.5 µm−1 (or a radius of 500 nm). The amphipathic helix of SpoVM is thought to insert into the lipid bilayer of the forespore membrane (inset). Across the length of SpoVM (~ 4 nm), the perceived membrane curvature is nearly flat.
(C) After bud emergence in the budding yeast Saccharomyces cerevisiaie, septins are localized to the mother-bud neck, which is the only area of continuous positive curvature at the plasma membrane. Septins are thought to be in an equilibrium between soluble and assembled states at the plasma membrane (first inset). Septins may cooperatively associate with lipid supported bilayers on beads in vitro suggesting that septin polymerization into filaments stabilizes their association to the membrane. Across the length of septin rod (17–32 nm), the perceived membrane curvature is nearly flat (second inset).
Sensing Micron-Scale Curvatures
To date, only SpoVM and the septins have been identified to be able to sense positive micron-scale membrane curvature. SpoVM was identified in the bacterium Bacillus subtilis as essential for spore formation [16]. Septins were identified in the budding yeast as essential gene products during cytokinesis, and have since been found to be conserved throughout eukaryotes but missing from land plants [17–20]. For both SpoVM and septins, the ability to sense curvature at this scale is central to their biological function. In the case of SpoVM, curvature sensing is likely essential for spore maturation [21]. For septins, their best-established curvature role is in the morphogenesis checkpoint where they act to sense the presence of a bud in yeast to coordinate cell shape with cell cycle progression [22, 23]. Before elaborating on these cell biological functions, we will discuss experiments that established these proteins as micron-scaled curvature sensors, and the current thinking on how these relatively small proteins recognize such shallow curvatures.
Bacterial Micron-Scale Curvature Sensors: SpoVM
In response to environmental stresses, B. subtilis sporulate to generate a dormant endospore [24]. The endospore is metabolically inactive and can withstand harsh environmental conditions [25, 26]. Sporulation is dependent on the formation of an intracellular double membrane structure called the forespore. At 1-micron in diameter, the forespore is the only site of shallow positive membrane curvature in the cell [27] (Figure 1B). SpoVM, a 26 amino acid polypeptide, localizes to the forespore where it recruits SpoIVA [21, 28]. SpoIVA can then recruit a network of forespore proteins necessary for forespore maturation. spoVM mutants form very thin, immature spores that are not resistant to environmental stresses [16]. This indicates SpoVM is essential for proper B. subtilis sporulation. However, the possibility remained that some other protein is the true curvature sensor, and not SpoVM.
To determine whether SpoVM can distinguish between different curvatures, recombinant SpoVM was incubated with lipid bilayers supported on spherical silica beads of defined curvatures [21, 29]. Saturation binding curves for SpoVM adsorption onto 2 µm and 8 µm diameter surfaces revealed that at low concentrations, SpoVM preferentially binds the 2 µm beads. Increasing the SpoVM concentration showed binding to both bead sizes; however, SpoVM adsorption onto 2 µm beads occurred more rapidly. Saturation binding curves for both bead sizes further revealed that although the number of binding sites for SpoVM were similar on both curvatures, SpoVM had a threefold-higher affinity for the 2 µm-sized beads compared to 8 µm-sized beads. ese current data suggest that SpoVM has intrinsic, micron-scale curvature sensing abilities. Interestingly, replacing the proline as the 9th residue with an alanine (SpoVMP9A) abolishes its curvature preference to 2 µm beads, leading to equal binding to both bead sizes, with a two-fold increase in affinity when compared to wild-type SpoVM. This raises the question; how does a single amino acid residue contribute to curvature sensitivity?
Analysis of the primary sequence followed by solution state NMR of SpoVM revealed that an amphipathic helix extends from amino acid residues 11 to 23, flanked by flexible N and C termini [21, 29]. Amphipathic helices harbor hydrophobic and polar regions on opposite faces of the helix and are a recurring theme of membrane curvature sensors. Interestingly, in contrast to SpoVM, SpoVMP9A was helical along the entire length of the protein and had an increase in positive charge along its polar face. Probing the interaction of SpoVM with the membrane identified that the helical residues of SpoVM were less accessible to a water-soluble probe than the N or C termini. Molecular dynamics (MD) simulations show that both SpoVM and SpoVMP9A may submerge within the bilayer, well below the phospholipid head groups, oriented parallel to the membrane. Although the wild-type SpoVM N terminus was found to be highly flexible, the SpoVMP9A N terminus was more rigid. It was proposed that the flexible N terminus is important for recruiting additional SpoVM molecules and other forespore proteins to positively curved membranes in a cooperative manner, consistent with the obtained Hill coefficients from saturation binding curves [29]. Cooperativity may emerge from oligomerization and thus it will be important to determine if SpoVM can oligomerize (perhaps mediated by its flexible N-terminus), and if oligomerization is critical for membrane curvature sensing. It will also be important to determine the specificity of SpoVM membrane curvature recognition between nanometer and micrometer-scale curvatures.
Interestingly, similar themes emerge for a sensor of negative membrane curvature from prokaryotes, DivIVA. This protein preferentially assembles at sites of micron-scale, negative membrane curvature and binds membranes using an amphipathic helix. Several prokaryotic proteins, including DivIVA, localize to sites of membrane curvature (Table 1), however, their intrinsic curvature sensing capacity in vitro remains to be determined.
Table 1.
Prokaryotic proteins localizing to sites of membrane curvature.
| Protein | Curvature localized to in vivo |
Membrane binding motif |
Deform liposomes in vitro? |
Formation of higher order structures |
Primary Function | References |
|---|---|---|---|---|---|---|
| FtsZ | Negative | − | + when complexed with FtsA* or when fused to a membrane targeting sequence | + | Cytokinesis | [109–111] |
| FtsA | Negative | Amphipathic helix | + when complexed with FtsZ | + | Tether FtsZ to the membrane, Z-ring assembly | [111–113] |
| MreB | Negative and Positive | Amphipathic Helix | + | + | Cell shape maintenance/ cell wall synthesis | [114–117] |
| CrvA | Low positive to negative | Unknown. Data suggests CrvA localizes in the periplasm | Unknown. Shown to be required for the curved shape of V. cholerae | + | Cell wall insertion, generates cell curvature | [118] |
| CreS | Positive | − | Unknown. Shown to be required for the curved shape in C. crescentus | + | Cell shape maintenance, surface colonization | [119–122] |
Septins Recognize Micron-Scale Curvature in Eukaryotic Cells
Septins are filament forming, GTP-binding proteins conserved from yeast to humans. Recombinant and immunoprecipitated septins from budding yeast form octameric rod complexes, as seen using negative stain transmission electron microscopy (TEM) [30]. Structural analysis of septins from worms, and mammalian cells show that rod complex formation is an intrinsic feature of septins [31, 32]. The septin rod contains two copies of each septin polypeptide, and depending on the organism, can vary in length from ~ 17 nm in C. elegans [31] to ~32 nm in S. cerevisiae [30]. Septin rods can associate and diffuse at the membrane, where they can bind end-on to one another to form micron-scaled filaments as demonstrated by total internal reflection fluorescence (TIRF) microscopy [33]. Septin filaments can be arranged to a variety of higher-order structures, some of which are at sites of micron-scale membrane curvature, including at the bud-neck in S. cerevisiae, and at the bases of dendritic spines and hyphal branches in neurons and filamentous fungi, respectively [22, 34–36]. At these sites, septins coordinate cell cycle progression [22], influence the organization of actin [37] and microtubules [38], and may function as a lateral diffusion barrier in the membrane [39–41]. The fact that septin structures are often localized to sites of micron-scale membrane curvature suggested that the septins might be able to “sense” these shallow curvatures.
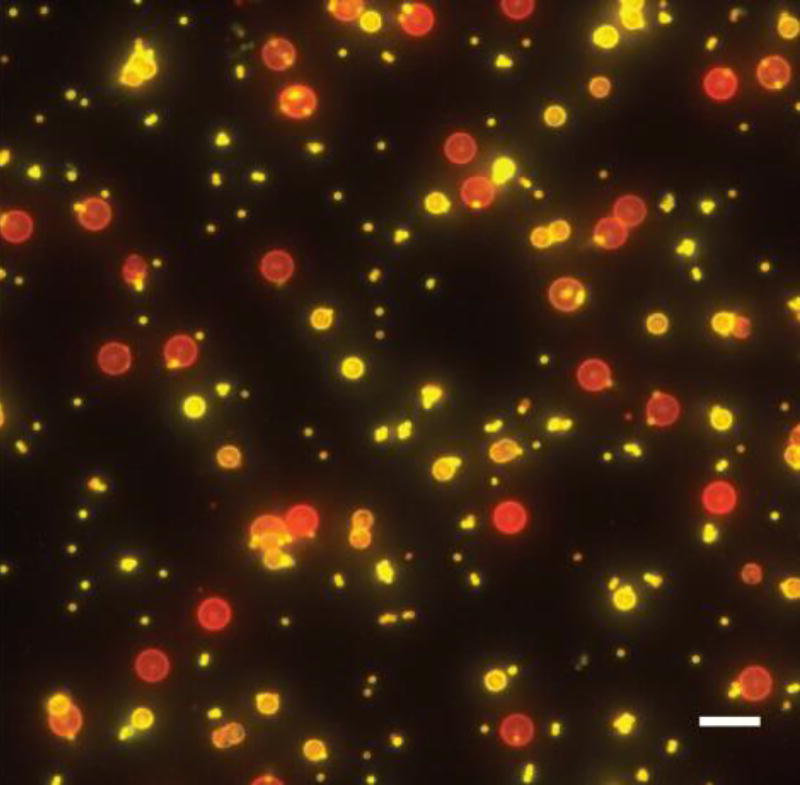
To test if septins could sense curvature, recombinant septins were incubated with supported lipid bilayers on beads of different diameters, similar to experiments done with SpoVM detailed above. Purified septins from S. cerevisiae and mammals preferentially bound to membranes with micron-scale curvatures, comparable to what is seen at the bud neck and at the cytokinetic furrow [36]. Specifically, septins were enriched on 1 µm and 3 µm beads, and less so on beads above and below these sizes (Figure 2) [36]. Furthermore, time-lapse microscopy showed that septins accumulated faster onto 1 µm beads than 5 µm beads, suggesting that differences in maximum adsorption alone cannot explain preferential septin binding to different curvatures. Interestingly, septin binding to 5 µm beads had a bimodal distribution such that some beads were coated with septins while other beads were devoid of septins, suggestive of a cooperative mechanism for septin adsorption. These data suggest that membrane affinity and binding cooperativity - rather than the number of available membrane binding sites - are what determine septin binding preference to curved membranes, akin to SpoVM.
Figure 2. Septins have a preference for micron-scale membrane curvatures.
In vitro reconstitution of septins with lipid bilayer-coated beads (red) of various diameters shows septins enrichment on 1 µm and 3 µm beads over 5 µm beads (yellow). Scale bar, 10 µm
How do septins distinguish different curvatures? Is septin filament polymerization required to sense micron-scale curvature? Or can a single septin rod sense shallow membrane curvature? It is tempting to speculate that membrane curvature is recognized at the level of the septin filament which can extend hundreds of nanometers, and not necessarily at the level of the rod. To test this hypothesis, Bridges et al. assessed whether individual, non-polymerizable mutant septin rods preferentially associated to supported lipid bilayers on beads of varying sizes [36]. Non-polymerizable rods did not stably associate to beads of any size, indicating septin association with membranes is likely driven by the collective and cooperative affinity of many subunits of a filament. To bypass this limitation, histidine-tagged, non-polymerizable septin rods were recruited to lipid coated beads containing Ni2+-NTA headgroups. Remarkably, these rods preferentially adsorbed onto 1 µm diameter beads, albeit much less well than the filament-forming septins. These data suggest that at some level a single septin rod can recognize micron scale curvature, possibly through the flexibility of the rod itself [30, 32]. Future work should address how the length scale of septin filaments affects curvature sensing, what surface of the septin binds to curved surfaces, and whether lateral interactions (bundling) of septin filaments and different membrane curvatures affect septins’ cooperative affinity to the membrane.
Sensing micron-scale curvature in the cell: An open question
How might SpoVM or septins “detect” micron-scale membrane curvature at the individual protein level? To address this question, we will discuss mechanisms employed by nanometer-scale curvature sensors and how those mechanisms may relate to micron-scale curvature sensors. Nanometer-scale curvature sensors utilize amphipathic helices [11], tune protein-membrane affinity through electrostatic interactions [42], and formation of polymeric scaffolds [15] to sense curvature (a subset of nanometer sensors summarized in Table 2). Interestingly, some evidence suggests that both SpoVM and septins might also use amphipathic helices and electrostatic interactions.
Table 2.
Nanometer sensors that sense curvature.
| Protein | Curvature localized to in vitro |
Scale | BAR domain |
Insertional amphipathic helix |
Deform liposomes in vitro? |
Formation of higher ordered structures |
Membrane binding sensitive to electrostatics |
Membrane binding sensitive to lipid packing defects |
Undergo PTM |
Primary Function |
References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Endophilin | Positive | nm | + N-BAR | + | + | + | + | + | + | Endocytosis | [15, 44, 123–126] |
| Amphiphysin | Positive | nm | + N-BAR | + | + | + | + | Unknown | Unknown | Endocytosis | [14, 104, 127] |
| IRSp53 | Negative | nm | + I-BAR | − | + | Unknown | + | Unknown | Unknown | Filopodia formation | [42, 128–131] |
| ESCRT-III | Negative | nm | − | + | + | + | + | Unknown | Unknown | Multivesicular body formation | [132–134] |
| ArfGAP1 | Positive | nm | − | + | Unknown | Unknown | − | + | + | Vesicular trafficking | [10, 12, 135, 136] |
| SpoVM | Positive | µm | − | + | Unknown | Unknown | Unknown | Unknown. Suggested by [29] | − | Forespore formation in B. subtilis | [21, 29] |
| DivIVa | Negative | µm | − | + | − | + | Unknown | Unknown | − | Cell division site selection | [137] |
| Septin | Positive | µm | − | Unknown | + | + | + | Unknown | + | Cytokinesis | [30, 33, 36, 91, 103] |
How are amphipathic helices utilized to “sense” curvature? The expanding leaflet of a membrane in response to high curvature at the nanometer scale can drive phospholipid head groups apart. This reveals open sites for amphipathic helix insertion into the membrane [43]. On the micron-scale, defects in lipid packing are likely to manifest differently, and may be at the level of the fatty acyl chain rather than with the phospholipid head group [29]. This could explain why SpoVM penetrates the bilayer and likely localizes well below the phospholipid head group. Altering the charge of an amphipathic helix affects both nanometer and micrometer curvature sensing. Increasing positive charge on the polar face of the SpoVMP9A helix resulted in increased affinity for lipid-coated beads and impaired curvature sensing. Similarly, adding only two lysine residues to the polar face of the otherwise uncharged amphipathic helix of ArfGAP1, a nanometer-scale curvature sensor, increased membrane binding and completely abolished curvature sensitivity [11]. We speculate amphipathic helices may be a conserved feature for both nanometer- and micrometer-scale membrane curvature recognition.
Another common feature among nanometer-scale curvature sensors is their cooperative association with curved membranes. Cooperativity could arise through oligomerization of proteins. For example, the H0 amphipathic helix of endophilin is important in forming lateral contacts with neighboring endophilin molecules to form scaffolds/lattices on lipid tubules in vitro [44]. Alternatively, cooperativity could arise through protein-mediated changes in the membrane, thereby favoring the recruitment of additional proteins. For example, coarse-grained simulations of an N-BAR domain on flat membranes show additional protein recruitment upon the initial adherence and curvature induction via individual proteins (see Box 1) [45]. On the micron scale, both SpoVM and septin adsorption onto micron-scale curvatures seem to be reliant on effective affinity differences for various curved surfaces and cooperativity in binding. Adjusting cooperativity and affinity in Monte Carlo simulations of SpoVM adsorption onto beads highlighted how these two parameters synergistically combine to affect curvature sensitivity. Small differences in affinity combined with modest cooperativity alterations yield clear curvature preferences for SpoVM [29]. For septins, filament polymerization is essential for its stable association to membranes; its bimodal adsorption onto 5 µm beads suggest a cooperative mechanism is particularly important on curved surfaces [36].
Box 1 anometer and micrometer scale curvature induction.
Curvature sensing is often linked to curvature induction, however, the degree to which the two are separable can be challenging to study. For the BAR-domain protein amphiphysin, it has been seen that at low protein densities the proteins are generally in a curvature sensing regime and at high protein densities crowding and potentially lateral associations that build a rigid scaffold then induce membrane curvature [104]. Similar density-sensitive behavior has also been seen for dynamin that also can assemble and act in a curvature-sensitive manner [105]. Thus, the role of a protein as a sensor vs an inducer may be highly dependent on the concentration of protein.
Septins can bind and tubulate giant unilamellar vesicles (GUV) to micron-scale tubules [106]. This work suggests that curvature sensing and induction could be tightly connected for septins and potentially sensing the local geometry can be used to recruit more protein and drive membrane bending. However, artificially recruiting high concentrations of his-GFP to Ni2+-NTA containing GUVs also results in tabulation [107], suggesting that protein crowding alone can drive membrane bending. Thus, whether tubulation is a specific function or by-product of crowding is not clear.
The fact that septins induce micron-scale tubules as opposed to highly variable or nanometer-scale tubules would support that induction of curvature may be physiologically relevant but it would be useful to assess septin curvature sensing and induction of a range of protein densities as has been done with amphiphysin and dynamin. Septin recruitment to cell blebs in mammalian cells [108] may be a context where septins both sense and induce curvature changes. In the case of experiments with SpoVM and septins using beads, induction is not possible because the bilayers are supported and similarly in fungal cells, the cell wall limits the role of septins in inducing curvature [36]. In cells without such rigid extracellular domains, further work is required to disentangle the relationship between micron scale-curvature sensing and induction.
Although additional work needs to be done on the molecular basis of cooperativity, it seems central feature of micron-scale curvature recognition. Could the local curvature regulate the recruitment of proteins on the membrane through some cooperative mechanism? It is tempting to speculate that the local curvature could promote the formation of different higher-ordered septin structures. However, this is a hypothesis that remains to be tested. The knowledge of nanometer curvature sensors can serve as a powerful guide for understanding micrometer-scale sensing but further work will be required to determine how the mechanisms established for nanometer perception are also exploited for micrometer sensing. Many questions remain regarding how these mechanisms first described at the nanometer scale transfer to the micron scale, if at all. The depth and breadth of these questions reveal what a ripe problem this is for further study.
How cells use micron scale curvature
Micron scale curvature throughout the cell cycle
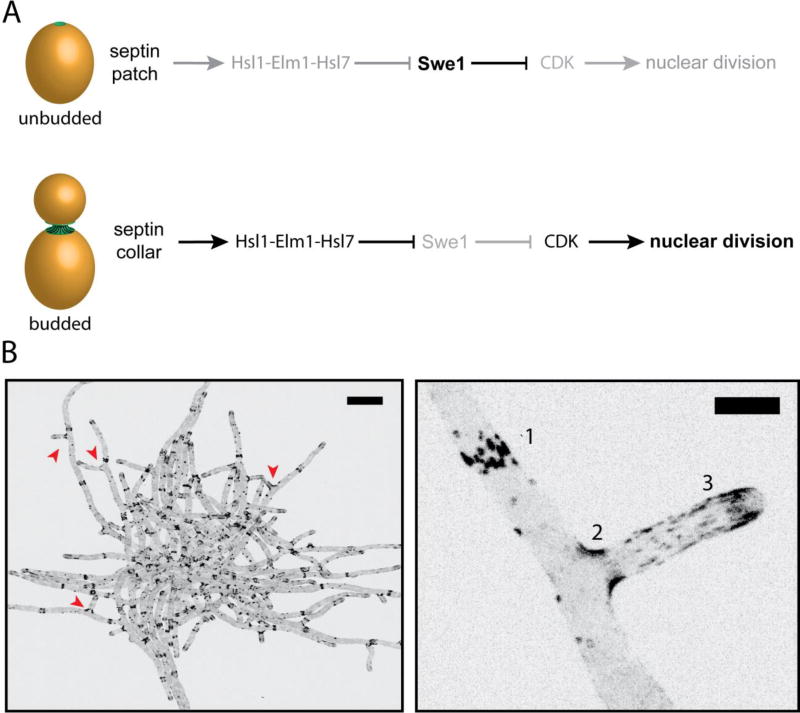
Reconstitution of recombinant SpoVM and septins in vitro demonstrate these proteins by themselves are sufficient to preferentially associate with micron-scale membrane curvature. How do cells use these micron-scale curvature sensors? One of the best examples of a cell using information about its shape to inform a decision is the morphogenesis checkpoint in budding yeast. The morphogenesis checkpoint effectively monitors whether the cell has bud, delaying nuclear division until after bud emergence [46]. However, this raises the question: how does the cell “know” whether it has a bud? Stresses that prevent bud formation lead to an increase and stabilization in the levels of the inhibitory kinase, Swe1 (wee1 homologue), which phosphorylates the mitotic CDK, delaying the cell cycle [47–49]. After bud emergence, the septin collar at the mother-bud neck recruits a series of effectors (Hsl1, Elm1, and Hsl7) that lead to the hyperphosphorylation and degradation of Swe1 [22, 23, 50–54]. The point at which Hsl1, Elm1, and Hsl7 are localized to the septin collar coincides with when septin filaments are arranged in a parallel array aligned with the positive membrane curvature of the bud neck [23, 55]. In cells that have not properly formed a bud, septins are unable to stably recruit Hsl1, Elm1, and Hsl7, thus allowing Swe1 levels to rise (Figure 3A). Thus, the ability of septins to recognize the positive membrane curvature of the mother-bud neck is critical to the morphogenesis checkpoint.
Figure 3. Septin structures and localization in budding yeast and the filamentous fungus Ashbya gossypii.
A. Morphogenesis checkpoint in S. cerevisiae: Hsl1-Elm1-Hsl7 are not recruited to the septin patch in unbudded cells, thereby allowing the CDK-inhibitor Swe1 levels to rise. As soon as the bud emerges, septin localization transitions to an hourglass shape coincident with the generation of micron-scale curvature. Swe1-regulators are recruited after this transition to initiate the degradation of Swe1 and promote progression through the cell cycle.
B. An inverted, maximum z-stack projection of a A. gossypii cell expressing Cdc11-GFP. Scale bar, 20 µm. (Bottom right) Zoomed-in image of A. gossypii hyphae expressing Cdc11-GFP from above. Septins are organized into various higher-ordered structures in Ashbya: [1] Thick bars structures, [2] basal collars at branch points, and [3] thin, flexible filaments. Scale bar, 5 µm.
Septins also localize to the developing forespore membranes during sporulation in both S. cerevisiae [56] and S. pombe [57]. During sporulation, membranous structures engulf and partition nuclear material into four separate haploid nuclei spores [58, 59]. These spores display curvatures from approximately 1 µm−1 – 0.5 µm−1 (compared to 2 µm−1 at the mother-bud neck, Figure 1C). During spore formation expression of spore specific septins, SPR3 and SPR28 increase [60]. An analogous process occurs within S. pombe [57, 61]. These spore specific septins are essential for sporulation, as mutants for either of these genes have misshapen forespore membranes, their forespore membranes fail to encapsulate nuclei, and cell walls do not mature properly [57, 62]. Mechanistically, how septins contribute to sporulation, remains unclear, but is presumably related to micron-scale membrane shape changes. It is interesting to note that spore-specific septins and mitotic-specific septins appear recognize different membrane curvatures (the forespore membrane vs. the bud neck), possibly owing a different composition of the respective septin oligomer. This would suggest that different septin complexes may have different intrinsic curvature preferences, a possibility that is also interesting to consider in mammalian cells where a more diverse suite of septin proteins and isoforms can coexist in the same cell. The problems of how septin curvature sensing is tuned developmentally and via expression of distinct heteromeric complexes is wide open for exploration.
Closely related to budding yeast, the filamentous fungus, Ashbya gossypii, has been a powerful, emerging model to examine the varied forms and functions of the septin cytoskeleton. During hyphal growth septins polymerize at the plasma membrane where they can form three distinct higher-ordered structures: 1) interregion rings, which are straight septin bundles organized as bars circumferentially around hyphae, 2.) basal collars that localize to newly emergent hyphae, 3.) thin, flexible filaments that run parallel to the hyphal axis [35] (Figure 3B). Septin accumulation to the base of branches suggested their preferential localization to saddle points or sites of positive curvature. When quantified, it became clear that septins are recruited in proportion to the degree of local positive curvature. In contrast, both the thick bars and thin flexible septin filaments align along the hyphal tube, as to seemingly avoid the negative curvature that exists orthogonal to the hyphal long axis. Interestingly, Hsl7, was found to localize exclusively to the septin bar structures and not curved structures [36]. This suggests that the local geometry of the membrane may dictate the local organization of septin filaments, which in turn could control recruitment of downstream signaling proteins.
Septins are also found at the cleavage furrow of most dividing animal cells [63]. Loss of septins leads to varying degrees of cytokinesis defects, depending on the organism and distinct septin complexes that participate in early cytokinesis and abscission [64]. In many cells, septin enrichment is coincident with ingression, where the membrane curvature is increasing. This raises the possibility that as a cleavage furrow ingresses, septin recruitment is enhanced in a curvature-dependent manner, potentially to stabilize or regulate the actomyosin contractile ring. In this scenario membrane curvature might supply a source of positive feedback in septin recruitment during cytokinesis. Interestingly, there are also F-BAR proteins found to associate with the cytokinetic furrow in a variety of systems [65, 66]. During cytokinesis, some F-BAR proteins are thought to target vesicles from the late secretory pathway, whereas others are thought to participate in membrane curvature induction. It is worth investigating whether F-BAR proteins and septins coordinate the different length scales of curvature changes at the plasma membrane during cytokinesis.
Micron scale curvature in autophagy and lipid droplets
Autophagy is a cellular process where cytoplasmic components are degraded under stress or aging. During autophagy aggregated protein complexes, ribosomes [67], organelles including mitochondria [68], and invading microorganisms including Listeria monocytogenes [69] and Shigella flexneri [70] can be targeted for degradation suggesting broad size distribution among autophagosomes. Indeed, these structures have been observed to range in curvature from 2.2 µm−1 to 4 µm−1 in yeast and 1.6 µm−1 to 4 µm−1 in mammalian cells [71–73]. Recent studies showed that septins assemble into cage-like structures around intracellular bacteria in a manner promoted by mitochondria [70, 74]. The topology and positive membrane curvature of rod-shaped bacteria and mitochondria are ideal for septins, raising the possibility that a septin-mediated immune response is induced by this ideal curvature.
Another subcellular structure with a wide variety of curvatures are lipid droplets. Lipid droplets (LD), amalgams of neutral lipids surrounded by a phospholipid monolayer, have been shown to contribute to lipid and membrane homeostasis in several organisms and cell types including S. cerevisiae [75], adipocytes [76], and hepatocytes [77]. Lipid droplets display curvatures ranging from 0.4 µm−1 to 50 µm−1 in non-adipocytes [5]. Knockdown of Sept9 in Huh7 cells resulted in a decrease in LD number and size [78]. Additionally, Sept9 was found to colocalize with neutral lipid biosynthetic enzyme, diacylglycerol acyltransferase-1 (DGAT-1), further supporting that septins may be involved in regulating lipid and membrane homeostasis. Further work needs to be done to determine whether septins are in fact directly binding to and sensing the curvature of the surface of lipid droplets, or whether septins are recruited by other factors to the surface of droplets.
Micron scale curvature and lateral compartmentalization and function of membrane-interacting proteins
Biological membranes can be compartmentalized into specific domains with distinct protein compositions. One way to compartmentalize membranes is through membrane diffusion barriers [79–81]. Diffusion barriers have been postulated at the base of cilia [41], dendritic spines [82], and at the tip of mating projections during chemotropism [83], at the mother-bud neck in S. cerevisiae [39, 84], and the annulus in spermatozoa [85]. Interestingly, septins localize to each of these structures, and have been proposed to contribute to the membrane diffusion barrier [86].
In the budding yeast, there is even evidence that septins act as a diffusion barrier for membranes that are adjacent to the plasma membrane. A septin-ER tether limits the diffusion of ER transmembrane proteins Pho88 and Ist2 between the mother and bud [40, 84]. One component of the barrier appears to be through a formation of a sphingolipid-dependent compartment at the mother-bud neck. This is thought to create a specialized membrane domain to restrict the mobility of transmembrane proteins [87].
Interestingly, septins were shown to induce local domains of supported lipid bilayers consisting of dimyristol- and dioylphosphatidylcholne (DMPC and DOPC, respectively) and phosphatidylinositol (PI) [88]. It has been postulated that septins can act as a membrane diffusion barrier either by acting as physical hindrance for other membrane associated proteins, through altering the lipid abundance, by affecting lipid localization, or by some combination of the three. Future work is needed to understand the mechanisms by which septins influence mobility in membranes and the degree to which this function is due to the local topology of the membrane.
Regulation of micron-scale curvature sensors
Given the possibility of electrostatics driving curvature recognition, a potential way for cells to tune curvature preferences is through post-translational modifications that modulate the affinity of protein complexes for the membrane. Genetic and biochemistry experiments have revealed numerous potential septin regulatory mechanisms, including direct protein-protein interactions, post-translational modifications (PTMs) and lipid-protein interactions [32, 35, 89–95]. Septin filaments can be assembled into a variety of different organizations and patterns as higher-order structures [35, 55, 89, 96–98]. It is tempting to speculate that septins are tightly regulated to specify different higher-order organizations of septin filaments and possibly even fine tune septin membrane curvature recognition. Although very little is known regarding how septins or other micron-scale curvature sensors are regulated, next we will discuss what is known about septin regulation as it pertains to their ability to sense micron-scale membrane curvature.
Post-translational modifications as a means to regulate protein localization to micron scale membrane curvature
Proteins often undergo dynamic transitions between an insoluble membrane bound state and soluble cytoplasmic state. Post-translational modifications (PTMs) of proteins is an attractive mechanism to spatiotemporally control the localization and functions of proteins, including those that act on micron-scale membrane curvatures. Throughout the cell cycle of S. cerevisiae, the septin cytoskeleton is subject to changes in organization [55, 90, 99]. Not surprisingly, all five mitotic septins can be phosphorylated at multiple sites [91, 92]. Septin-associated kinases Gin4, Elm1, and Cla4 have been shown to be important for regulating higher-order septin structures. Cla4 is required for septin collar formation in S. cerevisiae [93], whereas loss of either Gin4 or Elm1 in A. gossypii results in a loss of thick septin bar structures [35]. Nonetheless, septin localization to sites of positive curvature still persist. This suggests that straight septin bars require phosphorylation whereas assembly on curved membranes does not. However, a direct a link between septin phosphorylation and curvature sensing has yet to be established. Recently, Shen et. al. showed that phosphorylation within the GTP-binding domain of Sept12 abolished septin ring formation at the annulus in sperm [85], further highlighting the importance of septin regulation by phosphorylation. Interestingly, an acetylation/SUMO-ylation switch was found to regulate Cdc11 localization from the bud scar to the new incipient bud site in S. cerevisiae [94]. Despite this, it is still unclear whether septin phosphorylation, acetylation, and SUMO-ylation contribute to curvature sensing. We speculate that cells may tune the specific curvature preference of septins using PTMs that change the affinity of septins for different membrane shapes.
The role of the membrane composition in regulating micron scale membrane curvature
Lipid asymmetry across the bilayer is an inherent feature of cellular membranes; phosphatidylcholine and sphingolipids are predominately localized to the outer leaflet of lipid bilayers, whereas phosphatidylserine, phosphatidylethanolamine, phosphoinositols and their phosphoinositide derivatives localize to the inner leaflet [95]. This provides a means to localize membrane binding proteins to particular compartments on the basis of electrostatics, lipid packing, and local geometry. Phosphoinositides (PI) in particular, seem to have an emerging role in regulating cellular processes where micron-scale curvature is present including forespore membrane extension in S. pombe [57], autophagy [100], and micropinocytosis [101] in mammalian cells. Septins may be locally recruited to membranes based on the local lipid composition.
Mammalian septin filaments comprised of, Sept2, Sept6, and Sept7 were found to localize to mature macropinocytic vesicles in a PI(3,5)P2-dependent manner and septins are found in what are thought to be PI(4,5)P2-rich sites in yeast at mating projections and the neck [101, 102]. PI(4,5)P2-containing lipid monolayers were shown to drive septin filament assembly even in high salt conditions where septin polymerization is perturbed in solution [103] These data suggest that phosphoinositides play a key role in regulating protein adsorption onto a wide range of membrane curvatures and may play a role in modulating curvature preferences.
Concluding Remarks
How do SpoVM and septins recognize micron-scale positive membrane curvature? Interestingly, both SpoVM and the septins cooperatively associate membranes with shallow positive curvature; hinting that polymerization may be important. Septins are known to polymerize into filaments hundreds of nanometers to micrometers in length. For septins it is tempting to speculate that membrane curvature is recognized at the level of the filament, and not necessarily at the level of the rod. Nonetheless, even non-polymerizable septin mutants preferentially associate with shallow curved membranes, albeit much weaker than septins that can polymerize. How then might SpoVM and septins sense shallow curvature at the protein level? Many nanometer-scale curvature sensors utilize an amphipathic helix that inserts into the lipid bilayer. SpoVM, and possibly septins, also have amphipathic helixes. Perhaps both insertion of an amphipathic helix to the membrane is conserved mechanism for sensing curvature, even at shallow curvatures.
The fact that individual septin rods can intrinsically detect shallow membrane curvature raises an interesting question. If septin filament polymerization is unnecessary for curvature sensing, why polymerize at all? To answer this question, first consider the kinetics of an individual septin rod on and off the membrane (Figure 1C). Free septin rods that are not incorporated into filaments are favored to dissociate from the membrane (high koff relative to kon). Stable septin association to the membrane depends whether it is incorporated into a polymerized septin filament. This is likely critical for septins to reliably and correctly “sense” persistent micron-scale curvature. Otherwise, if individual septin rods could tightly associate with shallow curved membranes without polymerizing, septins would presumably localize to dispersed regions across the cortex, anywhere shallow curvature might incidentally arise (such as a relatively flat, yet slightly undulating membrane). Thus, septin filament polymerization at the membrane ensures that septins curvature recognition is limited only to persistent, shallow membrane curvatures that define cellular shapes at the micron scale.
We are just beginning to understand how cells perceive cellular membrane curvature, and by extension how cells “sense” their shape (see Outstanding Questions). It will be interesting to determine whether there are analogous proteins to SpoVM that can recognize shallow positive membrane curvature in other prokaryotes. Despite their variety of shapes and forms, both animal and fungal cells utilize the same cytoskeletal proteins, the septins, to sense micron-scale cellular membrane curvature. Oligomeric septin rods can assemble into micron-scaled filaments at the membrane, and these filaments can then assemble into various larger structures depending where they polymerize in cell. Interestingly, the organization of these larger assemblies appear to be dependent on the local membrane curvature. In budding yeast, the ability of septins to “sense” positive curvature at the mother-bud neck is critical to ensure that both the mother and future daughter cell inherit a nucleus before cytokinesis. In many different cell types, septins localize to the cytokinentic furrow and are required for cytokinesis. Further work will hopefully elucidate roles of septins in different cell types, and determine whether the ability of septins to recognize membrane curvature is essential for these cell types.
Trends:
Cells can recognize their own shape and local geometries on the micron scale.
SpoVM and the septins have been identified as micron-scale curvature sensors.
Modes of protein association with the membrane and oligomerization enable curvature sensing on the micron scale.
Cells likely tune the curvature preference of sensors using post-translational modifications and varying local lipid composition.
Outstanding questions:
What alternative mechanisms enable micron-scale curvature sensing using nanometer proteins?
What is the relationship between curvature sensing and curvature induction?
How do cells tune the curvature preference of sensors such as septins so as to sense a wide range of geometries within the same cell or in different cell types?
How do membrane composition and organization contribute to micron-scale curvature sensing?
What other families of proteins are capable of sensing micron scale-curvature?
Acknowledgments
We would like to thank members of the Gladfelter Lab for discussion and constructive comments on this draft. This work is supported by the National Science Foundation (MCB-0719126, to ASG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Singer SJ, GLN The Fluid Mosaic Model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 2.Walker RG, et al. A Drosophila mechanosensory transduction channel. Science. 2000;287(5461):2229–34. doi: 10.1126/science.287.5461.2229. [DOI] [PubMed] [Google Scholar]
- 3.Brown MF. Curvature forces in membrane lipid-protein interactions. Biochemistry. 2012;51(49):9782–95. doi: 10.1021/bi301332v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bretscher A. Polarized growth and organelle segregation in yeast: the tracks, motors, and receptors. J Cell Biol. 2003;160(6):811–6. doi: 10.1083/jcb.200301035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujimoto T, Parton RG. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb Perspect Biol. 2011;3(3) doi: 10.1101/cshperspect.a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis BA, Engelman DM. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J Mol Biol. 1983;166(2):211–7. doi: 10.1016/s0022-2836(83)80007-2. [DOI] [PubMed] [Google Scholar]
- 7.Budvytyte R, et al. Structure and properties of tethered bilayer lipid membranes with unsaturated anchor molecules. Langmuir. 2013;29(27):8645–56. doi: 10.1021/la401132c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szule JA, et al. The effects of acyl chain length and saturation of diacylglycerols and phosphatidylcholines on membrane monolayer curvature. Biophys J. 2002;83(2):977–84. doi: 10.1016/s0006-3495(02)75223-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, et al. Membrane cholesterol modulates the fluid shear stress response of polymorphonuclear leukocytes via its effects on membrane fluidity. Am J Physiol Cell Physiol. 2011;301(2):C451–60. doi: 10.1152/ajpcell.00458.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bigay J, et al. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 2005;24(13):2244–53. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drin G, et al. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14(2):138–46. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 12.Vanni S, et al. A sub-nanometre view of how membrane curvature and composition modulate lipid packing and protein recruitment. Nat Commun. 2014;5:4916. doi: 10.1038/ncomms5916. [DOI] [PubMed] [Google Scholar]
- 13.Pranke IM, et al. alpha-Synuclein and ALPS motifs are membrane curvature sensors whose contrasting chemistry mediates selective vesicle binding. J Cell Biol. 2011;194(1):89–103. doi: 10.1083/jcb.201011118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peter BJ, et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303(5657):495–9. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 15.Simunovic M, et al. How curvature-generating proteins build scaffolds on membrane nanotubes. Proc Natl Acad Sci U S A. 2016;113(40):11226–11231. doi: 10.1073/pnas.1606943113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levin PA, et al. An unusually small gene required for sporulation by Bacillus subtilis. Mol Microbiol. 1993;9(4):761–71. doi: 10.1111/j.1365-2958.1993.tb01736.x. [DOI] [PubMed] [Google Scholar]
- 17.Mostowy S, Cossart P. Septins: the fourth component of the cytoskeleton. Nat Rev Mol Cell Biol. 2012;13(3):183–94. doi: 10.1038/nrm3284. [DOI] [PubMed] [Google Scholar]
- 18.Hartwell LH, et al. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants. Genetics. 1973;74(2):267–86. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan F, et al. Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol Biol. 2007;7:103. doi: 10.1186/1471-2148-7-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onishi M, Pringle JR. The nonopisthokont septins: How many there are, how little we know about them, and how we might learn more. Methods Cell Biol. 2016;136:1–19. doi: 10.1016/bs.mcb.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Ramamurthi KS, et al. Geometric cue for protein localization in a bacterium. Science. 2009;323(5919):1354–7. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longtine MS, et al. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20(11):4049–61. doi: 10.1128/mcb.20.11.4049-4061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang H, et al. Sensing a bud in the yeast morphogenesis checkpoint: a role for Elm1. Mol Biol Cell. 2016;27(11):1764–75. doi: 10.1091/mbc.E16-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan IS, Ramamurthi KS. Spore formation in Bacillus subtilis. Environ Microbiol Rep. 2014;6(3):212–25. doi: 10.1111/1758-2229.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henriques AO, Moran CP., Jr Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol. 2007;61:555–88. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- 26.McKenney PT, et al. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol. 2013;11(1):33–44. doi: 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev. 2012;36(1):131–48. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramamurthi KS, et al. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol Microbiol. 2006;62(6):1547–57. doi: 10.1111/j.1365-2958.2006.05468.x. [DOI] [PubMed] [Google Scholar]
- 29.Gill RL, Jr, et al. Structural basis for the geometry-driven localization of a small protein. Proc Natl Acad Sci U S A. 2015;112(15):E1908–15. doi: 10.1073/pnas.1423868112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertin A, et al. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc Natl Acad Sci U S A. 2008;105(24):8274–9. doi: 10.1073/pnas.0803330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.John CM, et al. The Caenorhabditis elegans septin complex is nonpolar. EMBO J. 2007;26(14):3296–307. doi: 10.1038/sj.emboj.7601775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sirajuddin M, et al. Structural insight into filament formation by mammalian septins. Nature. 2007;449(7160):311–5. doi: 10.1038/nature06052. [DOI] [PubMed] [Google Scholar]
- 33.Bridges AA, et al. Septin assemblies form by diffusion-driven annealing on membranes. Proc Natl Acad Sci U S A. 2014;111(6):2146–51. doi: 10.1073/pnas.1314138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho SJ, et al. Septin 6 regulates the cytoarchitecture of neurons through localization at dendritic branch points and bases of protrusions. Mol Cells. 2011;32(1):89–98. doi: 10.1007/s10059-011-1048-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeMay BS, et al. Regulation of distinct septin rings in a single cell by Elm1p and Gin4p kinases. Mol Biol Cell. 2009;20(8):2311–26. doi: 10.1091/mbc.E08-12-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bridges AA, et al. Micron-scale plasma membrane curvature is recognized by the septin cytoskeleton. J Cell Biol. 2016;213(1):23–32. doi: 10.1083/jcb.201512029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mavrakis M, et al. Septins promote F-actin ring formation by crosslinking actin filaments into curved bundles. Nat Cell Biol. 2014;16(4):322–34. doi: 10.1038/ncb2921. [DOI] [PubMed] [Google Scholar]
- 38.Nolke T, et al. Septins guide microtubule protrusions induced by actin-depolymerizing toxins like Clostridium difficile transferase (CDT) Proc Natl Acad Sci U S A. 2016;113(28):7870–5. doi: 10.1073/pnas.1522717113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barral Y, et al. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol Cell. 2000;5(5):841–51. doi: 10.1016/s1097-2765(00)80324-x. [DOI] [PubMed] [Google Scholar]
- 40.Luedeke C, et al. Septin-dependent compartmentalization of the endoplasmic reticulum during yeast polarized growth. J Cell Biol. 2005;169(6):897–908. doi: 10.1083/jcb.200412143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu Q, et al. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science. 2010;329(5990):436–9. doi: 10.1126/science.1191054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takemura K, et al. Salt Bridge Formation between the I-BAR Domain and Lipids Increases Lipid Density and Membrane Curvature. Sci Rep. 2017;7(1):6808. doi: 10.1038/s41598-017-06334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hatzakis NS, et al. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat Chem Biol. 2009;5(11):835–41. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- 44.Mim C, et al. Structural basis of membrane bending by the N-BAR protein endophilin. Cell. 2012;149(1):137–45. doi: 10.1016/j.cell.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simunovic M, et al. Linear aggregation of proteins on the membrane as a prelude to membrane remodeling. Proc Natl Acad Sci U S A. 2013;110(51):20396–401. doi: 10.1073/pnas.1309819110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lew DJ. The morphogenesis checkpoint: how yeast cells watch their figures. Curr Opin Cell Biol. 2003;15(6):648–53. doi: 10.1016/j.ceb.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 47.Sia RA, et al. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol Biol Cell. 1996;7(11):1657–66. doi: 10.1091/mbc.7.11.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McMillan JN, et al. Determinants of Swe1p degradation in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13(10):3560–75. doi: 10.1091/mbc.E02-05-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sia RA, et al. Control of Swe1p degradation by the morphogenesis checkpoint. EMBO J. 1998;17(22):6678–88. doi: 10.1093/emboj/17.22.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakchaisri K, et al. Coupling morphogenesis to mitotic entry. Proc Natl Acad Sci U S A. 2004;101(12):4124–9. doi: 10.1073/pnas.0400641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Asano S, et al. Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast. EMBO J. 2005;24(12):2194–204. doi: 10.1038/sj.emboj.7600683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raspelli E, et al. Budding yeast Dma1 and Dma2 participate in regulation of Swe1 levels and localization. Mol Biol Cell. 2011;22(13):2185–97. doi: 10.1091/mbc.E11-02-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cid VJ, et al. Dynamic localization of the Swe1 regulator Hsl7 during the Saccharomyces cerevisiae cell cycle. Mol Biol Cell. 2001;12(6):1645–69. doi: 10.1091/mbc.12.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shulewitz MJ, et al. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19(10):7123–37. doi: 10.1128/mcb.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ong K, et al. Architecture and dynamic remodelling of the septin cytoskeleton during the cell cycle. Nat Commun. 2014;5:5698. doi: 10.1038/ncomms6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fares H, et al. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J Cell Biol. 1996;132(3):399–411. doi: 10.1083/jcb.132.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Onishi M, et al. Role of septins in the orientation of forespore membrane extension during sporulation in fission yeast. Mol Cell Biol. 2010;30(8):2057–74. doi: 10.1128/MCB.01529-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neiman AM. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics. 2011;189(3):737–65. doi: 10.1534/genetics.111.127126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kopecka M. Sporulation in protoplasts of the yeast, Saccharomyces cerevisiae. J Gen Microbiol. 1974;83(0):171–8. doi: 10.1099/00221287-83-1-171. [DOI] [PubMed] [Google Scholar]
- 60.Kaback DB, Feldberg LR. Saccharomyces cerevisiae exhibits a sporulationspecific temporal pattern of transcript accumulation. Mol Cell Biol. 1985;5(4):751–61. doi: 10.1128/mcb.5.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mata J, et al. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet. 2002;32(1):143–7. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- 62.Heasley LR, McMurray MA. Roles of septins in prospore membrane morphogenesis and spore wall assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2016;27(3):442–50. doi: 10.1091/mbc.E15-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spiliotis ET, et al. A mitotic septin scaffold required for Mammalian chromosome congression and segregation. Science. 2005;307(5716):1781–5. doi: 10.1126/science.1106823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Estey MP, et al. Distinct roles of septins in cytokinesis: SEPT9 mediates midbody abscission. J Cell Biol. 2010;191(4):741–9. doi: 10.1083/jcb.201006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arasada R, Pollard TD. A role for F-BAR protein Rga7p during cytokinesis in S. pombe. J Cell Sci. 2015;128(13):2259–68. doi: 10.1242/jcs.162974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takeda T, et al. Drosophila F-BAR protein Syndapin contributes to coupling the plasma membrane and contractile ring in cytokinesis. Open Biol. 2013;3(8):130081. doi: 10.1098/rsob.130081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kraft C, et al. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10(5):602–10. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 68.Kanki T, et al. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17(1):98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoshikawa Y, et al. Listeria monocytogenes ActA is a key player in evading autophagic recognition. Autophagy. 2009;5(8):1220–1. doi: 10.4161/auto.5.8.10177. [DOI] [PubMed] [Google Scholar]
- 70.Mostowy S, et al. Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe. 2010;8(5):433–44. doi: 10.1016/j.chom.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 71.Baba M, et al. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139(7):1687–95. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fengsrud M, et al. Ultrastructural characterization of the delimiting membranes of isolated autophagosomes and amphisomes by freeze-fracture electron microscopy. Eur J Cell Biol. 2000;79(12):871–82. doi: 10.1078/0171-9335-00125. [DOI] [PubMed] [Google Scholar]
- 73.Nguyen N, et al. Sensing Membrane Curvature in Macroautophagy. J Mol Biol. 2017;429(4):457–472. doi: 10.1016/j.jmb.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krokowski S, et al. Mitochondria promote septin assembly into cages that entrap Shigella for autophagy. Autophagy. 2016:0. doi: 10.1080/15548627.2016.1228496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang PL, et al. Lipid droplets maintain lipid homeostasis during anaphase for efficient cell separation in budding yeast. Mol Biol Cell. 2016;27(15):2368–80. doi: 10.1091/mbc.E16-02-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heid H, et al. On the formation of lipid droplets in human adipocytes: the organization of the perilipin-vimentin cortex. PLoS One. 2014;9(2):e90386. doi: 10.1371/journal.pone.0090386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ajat M, et al. Hepatic stellate cells retain the capacity to synthesize retinyl esters and to store neutral lipids in small lipid droplets in the absence of LRAT. Biochim Biophys Acta. 2017;1862(2):176–187. doi: 10.1016/j.bbalip.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 78.Akil A, et al. Septin 9 induces lipid droplets growth by a phosphatidylinositol-5-phosphate and microtubule-dependent mechanism hijacked by HCV. Nat Commun. 2016;7:12203. doi: 10.1038/ncomms12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boettcher B, et al. Nuclear envelope morphology constrains diffusion and promotes asymmetric protein segregation in closed mitosis. J Cell Biol. 2012;197(7):921–37. doi: 10.1083/jcb.201112117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trimble WS, Grinstein S. Barriers to the free diffusion of proteins and lipids in the plasma membrane. J Cell Biol. 2015;208(3):259–71. doi: 10.1083/jcb.201410071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gehlen LR, et al. Nuclear geometry and rapid mitosis ensure asymmetric episome segregation in yeast. Curr Biol. 2011;21(1):25–33. doi: 10.1016/j.cub.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 82.Ewers H, et al. A Septin-Dependent Diffusion Barrier at Dendritic Spine Necks. PLoS One. 2014;9(12):e113916. doi: 10.1371/journal.pone.0113916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kelley JB, et al. RGS proteins and septins cooperate to promote chemotropism by regulating polar cap mobility. Curr Biol. 2015;25(3):275–85. doi: 10.1016/j.cub.2014.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chao JT, et al. Polarization of the endoplasmic reticulum by ER-septin tethering. Cell. 2014;158(3):620–32. doi: 10.1016/j.cell.2014.06.033. [DOI] [PubMed] [Google Scholar]
- 85.Shen YR, et al. SEPT12 phosphorylation results in loss of the septin ring/sperm annulus, defective sperm motility and poor male fertility. PLoS Genet. 2017;13(3):e1006631. doi: 10.1371/journal.pgen.1006631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Caudron F, Barral Y. Septins and the lateral compartmentalization of eukaryotic membranes. Dev Cell. 2009;16(4):493–506. doi: 10.1016/j.devcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 87.Clay L, et al. A sphingolipid-dependent diffusion barrier confines ER stress to the yeast mother cell. Elife. 2014;3:e01883. doi: 10.7554/eLife.01883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yamada S, et al. Septin Interferes with the Temperature-Dependent Domain Formation and Disappearance of Lipid Bilayer Membranes. Langmuir. 2016;32(48):12823–12832. doi: 10.1021/acs.langmuir.6b03452. [DOI] [PubMed] [Google Scholar]
- 89.Garcia G, 3rd, et al. Subunit-dependent modulation of septin assembly: budding yeast septin Shs1 promotes ring and gauze formation. J Cell Biol. 2011;195(6):993–1004. doi: 10.1083/jcb.201107123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.DeMay BS, et al. Septin filaments exhibit a dynamic, paired organization that is conserved from yeast to mammals. J Cell Biol. 2011;193(6):1065–81. doi: 10.1083/jcb.201012143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meseroll RA, et al. Septin phosphorylation and coiled-coil domains function in cell and septin ring morphology in the filamentous fungus Ashbya gossypii. Eukaryot Cell. 2013;12(2):182–93. doi: 10.1128/EC.00251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Egelhofer TA, et al. The septins function in G1 pathways that influence the pattern of cell growth in budding yeast. PLoS One. 2008;3(4):e2022. doi: 10.1371/journal.pone.0002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Versele M, Thorner J. Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J Cell Biol. 2004;164(5):701–15. doi: 10.1083/jcb.200312070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim SW, et al. Genetic incorporation of Nepsilon-acetyllysine reveals a novel acetylation-sumoylation switch in yeast. Biochim Biophys Acta. 2017 doi: 10.1016/j.bbagen.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 95.Suetsugu S, et al. Dynamic shaping of cellular membranes by phospholipids and membrane-deforming proteins. Physiol Rev. 2014;94(4):1219–48. doi: 10.1152/physrev.00040.2013. [DOI] [PubMed] [Google Scholar]
- 96.Kinoshita M, et al. Self- and actin-templated assembly of Mammalian septins. Dev Cell. 2002;3(6):791–802. doi: 10.1016/s1534-5807(02)00366-0. [DOI] [PubMed] [Google Scholar]
- 97.Hernandez-Rodriguez Y, et al. The septin AspB in Aspergillus nidulans forms bars and filaments and plays roles in growth emergence and conidiation. Eukaryot Cell. 2012;11(3):311–23. doi: 10.1128/EC.05164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hernandez-Rodriguez Y, Momany M. Posttranslational modifications and assembly of septin heteropolymers and higher-order structures. Curr Opin Microbiol. 2012;15(6):660–8. doi: 10.1016/j.mib.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 99.DeMay BS, et al. Rapid and quantitative imaging of excitation polarized fluorescence reveals ordered septin dynamics in live yeast. Biophys J. 2011;101(4):985–94. doi: 10.1016/j.bpj.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tan X, et al. PtdIns(4,5)P2 signaling regulates ATG14 and autophagy. Proc Natl Acad Sci U S A. 2016;113(39):10896–901. doi: 10.1073/pnas.1523145113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dolat L, Spiliotis ET. Septins promote macropinosome maturation and traffic to the lysosome by facilitating membrane fusion. J Cell Biol. 2016;214(5):517–27. doi: 10.1083/jcb.201603030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Garrenton LS, et al. Pheromone-induced anisotropy in yeast plasma membrane phosphatidylinositol-4,5-bisphosphate distribution is required for MAPK signaling. Proc Natl Acad Sci U S A. 2010;107(26):11805–10. doi: 10.1073/pnas.1005817107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bertin A, et al. Phosphatidylinositol-4,5-bisphosphate promotes budding yeast septin filament assembly and organization. J Mol Biol. 2010;404(4):711–31. doi: 10.1016/j.jmb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sorre B, et al. Nature of curvature coupling of amphiphysin with membranes depends on its bound density. Proc Natl Acad Sci U S A. 2012;109(1):173–8. doi: 10.1073/pnas.1103594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Roux A, et al. Membrane curvature controls dynamin polymerization. Proc Natl Acad Sci U S A. 2010;107(9):4141–6. doi: 10.1073/pnas.0913734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tanaka-Takiguchi Y, et al. Septin-mediated uniform bracing of phospholipid membranes. Curr Biol. 2009;19(2):140–5. doi: 10.1016/j.cub.2008.12.030. [DOI] [PubMed] [Google Scholar]
- 107.Stachowiak JC, et al. Membrane bending by protein-protein crowding. Nature Cell Biology. 2012;14(9):944-+. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- 108.Gilden JK, et al. The septin cytoskeleton facilitates membrane retraction during motility and blebbing. J Cell Biol. 2012;196(1):103–14. doi: 10.1083/jcb.201105127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Osawa M, et al. Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 2009;28(22):3476–84. doi: 10.1038/emboj.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Osawa M, Erickson HP. Liposome division by a simple bacterial division machinery. Proc Natl Acad Sci U S A. 2013;110(27):11000–4. doi: 10.1073/pnas.1222254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol. 2005;55(6):1722–34. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 112.Krupka M, et al. Escherichia coli FtsA forms lipid-bound minirings that antagonize lateral interactions between FtsZ protofilaments. Nat Commun. 2017;8:15957. doi: 10.1038/ncomms15957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Loose M, Mitchison TJ. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat Cell Biol. 2014;16(1):38–46. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ursell TS, et al. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc Natl Acad Sci U S A. 2014;111(11):E1025–34. doi: 10.1073/pnas.1317174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Renner LD, et al. Studying biomolecule localization by engineering bacterial cell wall curvature. PLoS One. 2013;8(12):e84143. doi: 10.1371/journal.pone.0084143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Garner EC, et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333(6039):222–5. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.van den Ent F, et al. Bacterial actin MreB forms antiparallel double filaments. Elife. 2014;3:e02634. doi: 10.7554/eLife.02634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bartlett TM, et al. A Periplasmic Polymer Curves Vibrio cholerae and Promotes Pathogenesis. Cell. 2017;168(1–2):172–185. e15. doi: 10.1016/j.cell.2016.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Charbon G, et al. Bacterial intermediate filaments: in vivo assembly, organization, and dynamics of crescentin. Genes Dev. 2009;23(9):1131–44. doi: 10.1101/gad.1795509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ausmees N, et al. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 2003;115(6):705–13. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- 121.Cabeen MT, et al. Mutations in the Lipopolysaccharide biosynthesis pathway interfere with crescentin-mediated cell curvature in Caulobacter crescentus. J Bacteriol. 2010;192(13):3368–78. doi: 10.1128/JB.01371-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Persat A, et al. The curved shape of Caulobacter crescentus enhances surface colonization in flow. Nat Commun. 2014;5:3824. doi: 10.1038/ncomms4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gallop JL, et al. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25(12):2898–910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Capraro BR, et al. Kinetics of endophilin N-BAR domain dimerization and membrane interactions. J Biol Chem. 2013;288(18):12533–43. doi: 10.1074/jbc.M112.435511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shi Z, Baumgart T. Membrane tension and peripheral protein density mediate membrane shape transitions. Nat Commun. 2015;6:5974. doi: 10.1038/ncomms6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ambroso MR, et al. Endophilin A1 induces different membrane shapes using a conformational switch that is regulated by phosphorylation. Proc Natl Acad Sci U S A. 2014;111(19):6982–7. doi: 10.1073/pnas.1402233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Isas JM, et al. Tubulation by amphiphysin requires concentration-dependent switching from wedging to scaffolding. Structure. 2015;23(5):873–81. doi: 10.1016/j.str.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Prevost C, et al. IRSp53 senses negative membrane curvature and phase separates along membrane tubules. Nat Commun. 2015;6:8529. doi: 10.1038/ncomms9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Saarikangas J, et al. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol. 2009;19(2):95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 130.Mattila PK, et al. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol. 2007;176(7):953–64. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lim KB, et al. The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J Biol Chem. 2008;283(29):20454–72. doi: 10.1074/jbc.M710185200. [DOI] [PubMed] [Google Scholar]
- 132.Lee IH, et al. Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proc Natl Acad Sci U S A. 2015;112(52):15892–7. doi: 10.1073/pnas.1518765113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Buchkovich NJ, et al. Essential N-terminal insertion motif anchors the ESCRT-III filament during MVB vesicle formation. Dev Cell. 2013;27(2):201–14. doi: 10.1016/j.devcel.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 134.Chiaruttini N, et al. Relaxation of Loaded ESCRT-III Spiral Springs Drives Membrane Deformation. Cell. 2015;163(4):866–79. doi: 10.1016/j.cell.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Drin G, Antonny B. Amphipathic helices and membrane curvature. FEBS Lett. 2010;584(9):1840–7. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 136.Dephoure N, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105(31):10762–7. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lenarcic R, et al. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 2009;28(15):2272–82. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]