Abstract
The NF-κB Essential Modulator (NEMO) is the scaffolding subunit of the inhibitor of κB kinase (IKK) holocomplex and is required for the activation of the catalytic IKK subunits, IKKα and IKKβ, during the canonical inflammatory response. Though structures of shorter constructs of NEMO have been solved, efforts to elucidate the full-length structure of NEMO have proven difficult due to its apparent high conformational plasticity. To better characterize the gross dimensions of full-length NEMO, we employed in-line size exclusion chromatography – small angle X-ray scattering (SEC-SAXS). We show that NEMO adopts a more compact conformation (Dmax=320 Å) than predicted for a fully extended coiled-coil structure (>500 Å). Additionally, we map a region of NEMO (residues 112-150) in its CC1 domain that impedes the binding of linear (M1-linked) di-ubiquitin to its CC2-LZ ubiquitin binding domain. This ubiquitin binding inhibition can be overcome by a longer chain of linear, but not K63-linked polyubiquitin. Collectively, these observations suggest that NEMO may be auto-inhibited in the resting state by intramolecular interactions, and that during signaling NEMO may be allosterically activated by binding to long M1-linked polyubiquitin chains.
Graphical Abstract
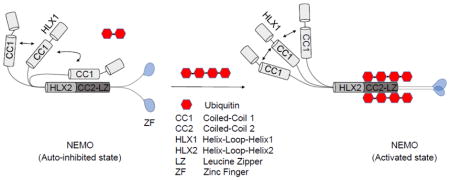
Cellular responses to various pathogenic and pro-inflammatory signaling molecules, including viral and bacterial nucleic acids, lipopolysaccharide (LPS), interleukin-1β (IL-1β), and tumor necrosis factor alpha (TNFα), depend on the activation of a family of dimeric transcription factors known as NF-κB [1]. During basal conditions, NF-κB are sequestered in the cytosol bound to inhibitor molecules called IκBs, with IκBα being the most ubiquitous. Upon binding of extracellular ligands to their cognate receptors, including members of the Toll-like receptor/interleukin-1 receptor and TNF receptor superfamilies, a central effector kinase complex, called the inhibitor of κB kinase (IKK), is activated [1, 2]. Once activated, the catalytic subunits of the IKK complex phosphorylate two N-terminal serines of IκBα leading to its K48-linked polyubiquitination and subsequent proteolytic degradation by the 26S proteasome. This frees NF-κB to translocate into the nucleus to induce transcription of pro-inflammatory and/or anti-apoptotic gene suites [1, 2].
The IKK holocomplex is composed of the catalytic subunits, IKKβ (or IKK2) and/or IKKα (or IKK1), and the helical scaffolding protein NF-κB essential modulator (NEMO) that is the central regulatory subunit [2, 3]. Genetic ablation of the 48 kDa NEMO in mice results in embryonic lethality by day 13 of development and a complete unresponsiveness to a variety of canonical NF-κB inducers in mouse embryonic fibroblast cells [4–6]. Though lacking catalytic activity of its own, NEMO articulates upstream signals to the activation of the IKK catalytic subunits. It does this mainly through its ability to bind non-degradative polyubiquitin chains (mainly M1- and K63-linked) synthesized during signaling [7–10]. NEMO has two distinct ubiquitin binding domains (UBDs). One is composed of the coiled-coil 2 (CC2) and leucine zipper (LZ) domains, together called the UBAN domain (also known as the NOA, NUB, or CoZi domain) and the other is composed of the C-terminal zinc finger motif (Fig. 1a) [11–15]. Other domains in NEMO include the HLX1, CC1 and HLX2 domains (Fig. 1a). While a part of HLX1 and CC1 interacts with IKKα or IKKβ [16], HLX2 binds the FLICE inhibitory protein (vFLIP) from Karposi sarcoma associated herpesvirus (KHSV) and Tax from the human T-lymphocyte virus (HTLV) to result in NEMO activation during viral infection [17, 18] (Fig. 1a). Mutations in various domains of NEMO have been implicated in cases of anhidrotic ectodermal dysplasia with immune deficiency (EDA-ID) and incontinentia pigmenti (IP) [19–21].
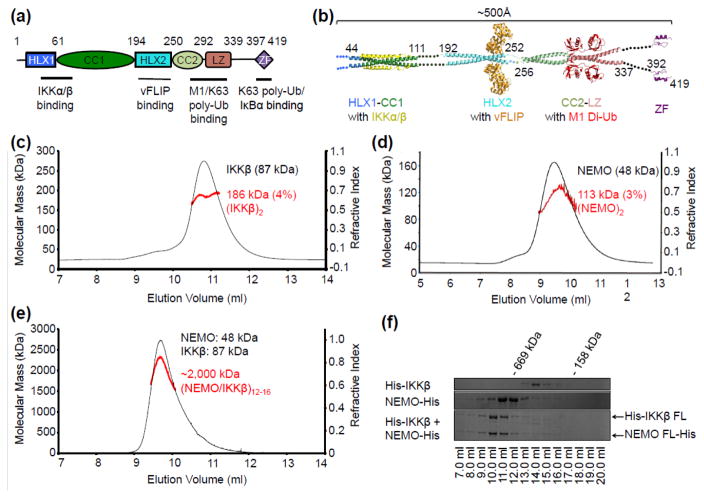
Fig. 1.
Size characterization of IKKβ, NEMO and the IKKβ/NEMO complex. (a) Domain architecture of human NEMO showing regions of protein-protein interactions. HLX1= helix-loop-helix 1, CC1= coiled-coil 1, HLX2 =helix-loop-helix 2, CC2= coiled-coil 2, LZ= leucine zipper, ZF= zinc finger. (b) A linear model of full-length NEMO with ~500 Å in length constructed from known structures of NEMO and its complexes. (c) SEC-MALS of His-IKKβ, showing a predominant dimer. (d) SEC-MALS of NEMO-His, showing a predominant dimer. (e) SEC-MALS of the IKKβ/NEMO complex, showing a megadalton higher-order oligomer. (f) Coomassie Blue-stained, 10% SDS-PAGE gel showing the Superose 6 10/300GL elution profile of IKKβ, NEMO, and the IKKβ/NEMO complex. All NEMO constructs were sub-cloned into either pET28a or pET26b vectors between NdeI and XhoI restriction sites for expression in E. coli BL21-CodonPlus® (DE3) RIPL cells (Agilent Technologies). IKKβ was sub-cloned into pFastBacHTb transfer vector for bacmid generation and expression in High Five insect cells using the Bac-to-Bac® baculoviral expression system (Thermo Fisher). All proteins were purified by nickel affinity chromatography followed by size exclusion chromatography (SEC). MALS measurements were performed with an in-line three-angle light scattering detector (mini-DAWN TRISTAR) coupled to a refractive index detector (Optilab DSP) (Wyatt Technology). Data were analyzed using ASTRA VI software.
Structural studies show that many regions of NEMO exhibit coiled-coil structures, including those responsible for binding to IKKα or IKKβ [16], vFLIP [17], and polyubiquitin [11–13], at least in the partner-bound forms (Fig. 1b). However, it is unclear if full-length NEMO is indeed an extended, long dimeric coiled-coil with a predicted length of ~500 Å (Fig. 1b). Previous studies have also reported that NEMO can exist as trimer or tetramer depending on the signaling condition, protein concentration, and the NEMO construct under study [22–24]. A recent analysis further suggests that as much as half of NEMO may be intrinsically disordered in its resting state, suggesting that partner binding may induce conformational changes in NEMO [25].
In this study, we aimed to characterize conformational or oligomeric changes introduced by the binding of NEMO interacting proteins including IKKβ, vFLIP, and polyubiquitin in the context of full-length NEMO. We show that both full-length IKKβ and full-length NEMO are primarily dimeric individually, but form a megadalton complex when together. For some NEMO constructs, a minor tetramer peak can also be detected in addition to the dimer peak in size exclusion chromatography (SEC), rationalizing previous observations of trimerization as a mixture of dimers and tetramers, and of tetramerization as dimers of dimers. We report by in-line SEC – small angle X-ray scattering (SEC-SAXS) that NEMO does not adopt a fully extended coiled-coil conformation (predicted to be >500 Å), but instead exhibits a comma-shaped form with a Dmax of ~320 Å. We also identify a region (residues 112-150) of NEMO in the CC1 domain that hinders M1-linked di-ubiquitin binding to the UBAN domain. Longer chain M1-linked polyubiquitin but not with K63-linked polyubiquitin can overcome this inhibition, suggesting polyubiquitin-induced NEMO conformational changes during signaling.
Characterization of NEMO and the IKK holocomplex
Initial biophysical characterization of the IKK holocomplex from TNFα stimulated HeLa cells showed that it purifies as a 700–900 kDa complex with IκBα phosphorylating activity [4, 26, 27]. Subsequent studies revealed that this complex composed canonically of IKKβ, IKKα, and NEMO can be reconstituted in vitro [28, 29]. Several crystal structures of nearly full-length IKKβ and a recent IKKα cryo-EM structure showed that these catalytic subunits exist primarily as dimers [30–32]. However, the oligomerization state of NEMO is controversial, and full-length NEMO has been observed to elute from SEC at a position that corresponds to a ~600 kDa globular protein [28].
To obtain a shape-independent assessment of the oligomerization state of IKKβ and full-length NEMO, we coupled multi-angle light scattering (MALS) and refractive index detection to SEC (Fig. 1c, 1d). Both IKKβ and NEMO exist primarily as dimers with a measured molecular mass of 186 kDa (4% error) and 113 kDa (3% error) respectively, in comparison with the calculated monomeric molecular weight of 87 kDa and 48 kDa. In contrast, the NEMO/IKKβ complex exhibits a measured molecular mass of ~2,000 kDa (Fig. 1e), suggesting higher-order oligomerization of the IKK holocomplex. The earlier elution volume of NEMO relative to IKKβ (Fig. 1c, 1d, 1f) despite its smaller molecular mass also suggested that NEMO is an especially elongated molecule.
Auto-inhibition in full-length NEMO shown by M1-linked di-ubiquitin binding
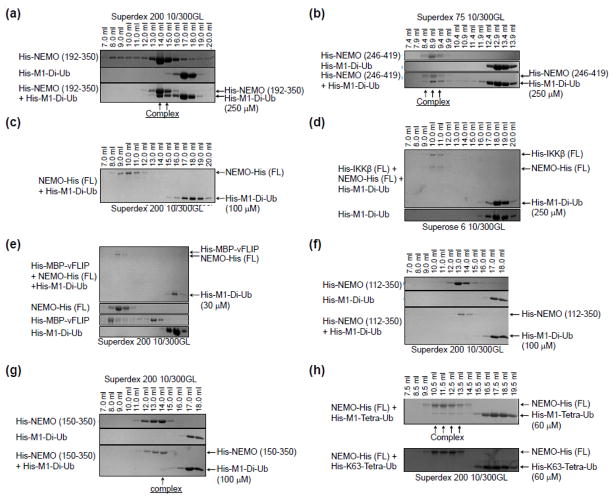
Short constructs of NEMO that encompass the UBAN domain have been shown to bind with low micromolar affinity to M1-linked polyubiquitin chains [11]. Indeed, M1-linked di-ubiquitin co-eluted with UBAN (192–350) and UBAN-ZF (246–419) by SEC (Fig. 2a, 2b). Surprisingly, this interaction was completely abrogated in the context of full-length NEMO (Fig. 2c), and this inhibition could not be rescued with the addition of other NEMO interacting proteins like IKKβ or MBP-vFLIP (Fig. 2d, 2e). We wondered if a specific region of NEMO is responsible for the interference of M1-di-ubiquitin interaction. Because the C-terminal portion of NEMO binds well to M1-di-ubiquitin, we truncated NEMO from the N-terminus. While NEMO (112–350) did not interact with M1-di-ubiquitin (Fig. 2f), NEMO (150–350) did interact with M1-di-ubiquitin (Fig. 2g), suggesting that the region between 112–150 in the CC1 domain of NEMO may be important for this inhibition.
Fig. 2.
The CC1 domain (aa 112-150) of NEMO inhibits linear di-ubiquitin binding. (a) – (h) SDS-PAGE of SEC fractions of various NEMO constructs mixed with M1-di-ubiquitin and/or IKKβ, MBP-vFLIP, or tetra-ubiquitin chains. Complex formation is indicated as well as the SEC column used in each panel. Purified NEMO constructs were incubated with purified IKKβ, MBP-vFLIP, and/or ubiquitin proteins in a roughly 1:1 or 1:2 molar ratio for 30 minutes at room temperature prior to loading onto a Superdex 75, Superdex 200 or Superose 6 column pre-equilibrated with buffer containing 20 mM Tris at pH 8.0, 150 mM NaCl, and 1 mM Dithiothreitol (DTT) run at 4 degrees celcius. The synthesis of K63-linked tetra-ubiquitin was accomplished as described in [35].
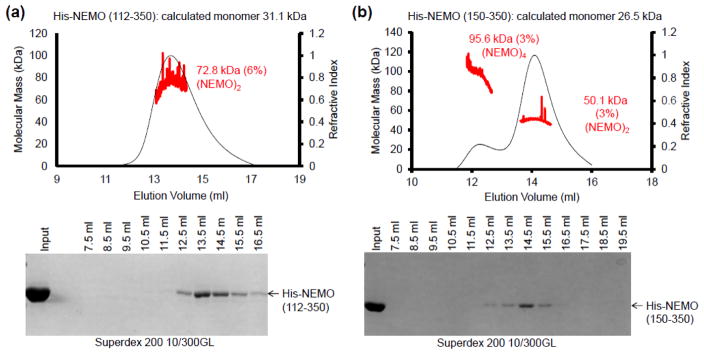
As controls, we conducted SEC-MALS to measure the molecular masses of the NEMO (112–350) (Fig. 3a) and NEMO (150–350) constructs (Fig. 3b) since it is possible that higher order oligomerization beyond the putative dimer could compete for ubiquitin binding sites. For NEMO (112–350), the measured molecular mass is 72.8 kDa (6% error), consistent with primarily a dimer of the calculated monomer molecular mass of 31.1 kDa (Fig. 3a). For NEMO (150–350), the major peak has a measured molecular mass of 50.1 kDa (3% error), which is also in agreement with a dimer of the calculated monomer molecular mass of 26.5 kDa (Fig. 3b). However, for this construct, there is also a minor peak with the measured molecular mass of 95.6 kDa (3% error) (Fig. 3b), which indicated a tetramer and suggested potential higher order oligomerization. Interestingly, it appears that only the dimeric region of the construct near 14 ml in elution volume interacted with M1-di-ubiquitin (Fig. 2g).
Fig. 3.
Size characterization of NEMO constructs. (a) SEC-MALS of NEMO (aa 112-350) along with the associated eluted fractions run on a 10% SDS-PAGE gel, showing that it behaves as a dimer with a measured molecular mass of 72,820 Daltons. (b) SEC-MALS of NEMO (aa 150-350), showing that it exists primarily as a dimer (molecular mass of 50,090 Daltons) in equilibrium with a small amount of a tetramer species (molecular mass of 95,550 Daltons). SEC-MALS was conducted as described in Fig. 1.
M1-linked tetra-ubiquitin overcomes the inhibition in full-length NEMO
Lack of binding to M1-di-ubiquitin by full-length NEMO indicates that in the resting state, NEMO exists in an auto-inhibited conformation through interactions within the dimer, either intramolecular or intermolecular. Because NEMO activation requires longer M1-linked polyubiquitin [25, 33], we tested M1-tetra-ubiquitin and K63-tetra-ubiquitin for binding to full-length NEMO. While M1-tetra-ubiquitin robustly interacted with NEMO, K63-tetra-ubiquitin did not (Fig. 2h). These data indicate that long M1-linked polyubiquitin is able to overcome NEMO auto-inhibition. Thus, our experiments revealed evidence for allosteric control of NEMO conformation including auto-inhibition in the resting state and M1-linked polyubiquitin-mediated conformational change that results in ubiquitin binding and NEMO activation.
NEMO adopts a more compact conformation than predicted for a fully extended coil-coil structure
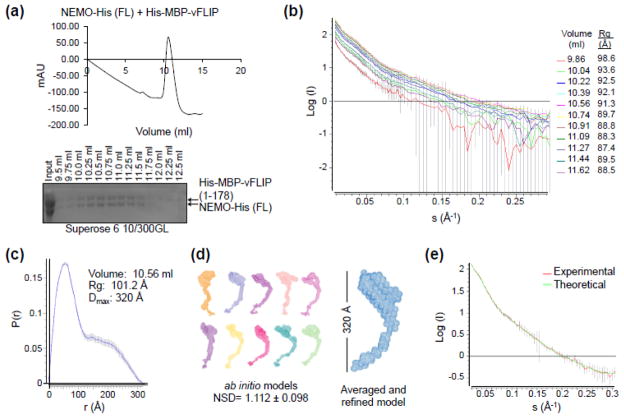
To further understand the conformational control in NEMO, we measured the gross shape and dimensions of full-length NEMO using an in-line SEC-SAXS experimental setup at the NSLS X9 beamline. The in-line SEC was aimed to eliminate large aggregates in the SAXS measurements. Small-angle X-ray scattering was measured in real-time as protein eluted off a Superose 6 10/300GL column, as shown by the chromatograph and SDS-PAGE gel of peak fractions of full-length NEMO in complex with MBP-vFLIP fusion protein (Fig. 4a). MBP-vFLIP was added to provide a large globular domain for the purpose of structure orientation since NEMO itself is not predicted to have any globular domain. Importantly, NEMO in complex with MBP-vFLIP did not appreciably change the SEC elution profile in comparison to NEMO alone suggesting that vFLIP binding does not result in any large scale conformational changes (Fig. 2e). SAXS data were collected every 35 seconds as protein eluted off the column and the raw scattering curves are shown (Fig. 4b).
Fig. 4.
SAXS analysis of the NEMO/MBP-vFLIP complex. (a) In-line Superose 6 10/300GL size exclusion chromatography of the NEMO/MBP-vFLIP complex before SAXS analysis (top). The MBP-vFLIP fusion construct was cloned into the pET28a vector for E. coli expression, and the expressed protein was purified by amylose resin followed by SEC. Peak fractions of the NEMO/MBP-vFLIP complex were run on a 10% SDS-PAGE gel (bottom). (b) Raw scattering curves of peak fractions collected every 35 seconds (flow rate = 0.3 ml/min.). The incident light wavelength was 0.92 Å and the sample to detector distance was 1.5 meters. Radial averaging and buffer subtraction was accomplished using pyXS (Brookhaven, NSLS). The radii of gyration (Rg) in angstroms were estimated from Guinier plots using PRIMUS [36] near I(0) for each scattering curve. (c) The pair-wise distribution plot, P(r), for the complex eluted at 10.56 ml. The estimated maximum intra-molecular particle distance, Dmax, is 320 Å. Both calculation of P(r) and determination of Dmax were performed using GNOM [37]. (d) Ten ab initio bead models of NEMO/MBP-vFLIP generated by DAMMIF [38] along with the averaged and refined envelope generated by DAMAVER [39]. (e) Superimposed experimental scattering curve at 10.56 ml and the calculated scattering curve from the averaged and refined model.
The particle size is relatively homogenous under the elution peak with Guinier approximations of the radii of gyration (Rg) ranging from 88.5–98.6 Å (Fig. 4b). The pairwise distribution function p(r), determined from the central peak scattering curve data, showed an Rg of 101.2 Å that is similar to those estimated from Guinier approximations (Fig. 4b, 4c), and a maximum intramolecular distance (Dmax) of 320 Å (Fig. 4c). This maximal dimension is considerably shorter than the predicted Dmax of 500 Å for fully extended NEMO (Fig. 1b). Ab initio modeling of the electron density envelope using the program DAMMIF generated ten independent models with an average normalized spatial discrepancy (NSD) value of 1.112 ± 0.098 (Fig. 4d). These models were averaged and filtered in DAMAVER to give the final refined model (Fig. 4d), which agreed well with the experimental scattering curve (Fig. 4e). The comma-shaped envelope has one end that is globular, presumably due to the presence of MBP-vFLIP, a middle section that is kinked, and the other end that is tapered. Though it is difficult to dock the various NEMO subdomain crystal structures into the envelope due to the low resolution it is obvious that NEMO does not adopt a fully extended conformation in solution.
Discussion
Our SAXS data and M1-linked di-ubiquitin binding data together provided a number of important insights into conformational regulation of NEMO. First, full-length NEMO assumes a more compact conformation with auto-inhibition in the resting state dimer. While either the N-terminal or the C-terminal region may fold back to create the comma shape of the NEMO/MBP-vFLIP complex (Fig 4d), given the apparent allosteric control in the CC1 domain (Fig. 2f–g), we propose that an N-terminal region (residues 112-150) including the CC1 domain folds back onto the vicinity of the HLX2 NEMO region where the MBP-vFLIP fusion protein binds (Fig 4d). However, our data do not exclude the possibility that the C-terminal end of NEMO may also fold back to exert auto-inhibition. Our detailed biochemical characterizations are consistent with a previous study in which NEMO was shown to be a dimeric protein that is in weak equilibrium with a tetrameric assembly [34]. They further rationalize that previously concluded trimerization of NEMO may represent mixture of dimers and tetramers.
Second, there are a number of established NEMO mutations in the proposed allosteric regulatory region of NEMO that are associated with the human disease incontinentia pigmenti [20, 21]. Two such point mutations are D113N and R123W. Since this region of NEMO has not been shown to interact with any other binding partners, a likely explanation of the disease phenotype may be related to the role of the region in NEMO allosteric regulation. Intriguingly, normal lipopolysaccharide (LPS)-induced NF-κB activation was observed in NEMO−/− cells reconstituted with these mutants [20], suggesting that this allosteric regulation may be influenced by overexpression in the reconstitution. Furthermore, the basal NF-κB activity may need to be assessed to reveal the functional phenotypes of these mutants in auto-inhibition. Finally, the higher binding affinity of M1-linked tetra-ubiquitin to NEMO can also overcome NEMO auto-inhibition. Therefore, M1-linked polyubiquitin may act as a switch to allosterically alter NEMO conformational and activate NEMO for downstream signal transduction in a physiological context.
Highlights.
Full-length NEMO is more compact than a linear coiled coil structure.
The coiled-coil 1 (CC1) domain of full-length NEMO inhibits binding to M1-linked di-ubiquitin.
Auto-inhibition of full-length NEMO can be relieved by longer M1-linked polyubiquitin.
Acknowledgments
We thank Dr. Lin Yang at the X9 beamline of National Synchrotron Light Source for assistance with data collection. This research is funded by NIH grant 5R37 AI050872 awarded to Dr. Hao Wu. Arthur V. Hauenstein is supported by an NIH T32 institutional training grant at Boston Children’s Hospital.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhang Q, Lenardo MJ, Baltimore D. 30 Years of NF-kappaB: A Blossoming of Relevance to Human Pathobiology. Cell. 2017;168:37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu F, Xia Y, Parker AS, Verma IM. IKK biology. Immunol Rev. 2012;246:239–53. doi: 10.1111/j.1600-065X.2012.01107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hinz M, Scheidereit C. The IkappaB kinase complex in NF-kappaB regulation and beyond. EMBO Rep. 2014;15:46–61. doi: 10.1002/embr.201337983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothwarf DM, Zandi E, Natoli G, Karin M. IKK-gamma is an essential regulatory subunit of the IkappaB kinase complex. Nature. 1998;395:297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 5.Rudolph D, Yeh WC, Wakeham A, Rudolph B, Nallainathan D, Potter J, et al. Severe liver degeneration and lack of NF-kappaB activation in NEMO/IKKgamma-deficient mice. Genes Dev. 2000;14:854–62. [PMC free article] [PubMed] [Google Scholar]
- 6.Yamaoka S, Courtois G, Bessia C, Whiteside ST, Weil R, Agou F, et al. Complementation cloning of NEMO, a component of the IkappaB kinase complex essential for NF-kappaB activation. Cell. 1998;93:1231–40. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 7.Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–62. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 8.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–61. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 9.Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell. 2003;115:565–76. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- 10.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–57. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 11.Lo YC, Lin SC, Rospigliosi CC, Conze DB, Wu C, Ashwell JD, et al. Structural Basis for Recognition of Diubiquitins by NEMO. Mol Cell. 2009;33:602–15. doi: 10.1016/j.molcel.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Yoshikawa A, Sato Y, Yamashita M, Mimura H, Yamagata A, Fukai S. Crystal structure of the NEMO ubiquitin-binding domain in complex with Lys 63-linked di-ubiquitin. FEBS Lett. 2009;583:3317–22. doi: 10.1016/j.febslet.2009.09.028. [DOI] [PubMed] [Google Scholar]
- 14.Grubisha O, Kaminska M, Duquerroy S, Fontan E, Cordier F, Haouz A, et al. DARPin-assisted crystallography of the CC2-LZ domain of NEMO reveals a coupling between dimerization and ubiquitin binding. J Mol Biol. 2010;395:89–104. doi: 10.1016/j.jmb.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 15.Cordier F, Grubisha O, Traincard F, Veron M, Delepierre M, Agou F. The zinc finger of NEMO is a functional ubiquitin-binding domain. J Biol Chem. 2009;284:2902–7. doi: 10.1074/jbc.M806655200. [DOI] [PubMed] [Google Scholar]
- 16.Rushe M, Silvian L, Bixler S, Chen LL, Cheung A, Bowes S, et al. Structure of a NEMO/IKK-associating domain reveals architecture of the interaction site. Structure. 2008;16:798–808. doi: 10.1016/j.str.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 17.Bagneris C, Ageichik AV, Cronin N, Wallace B, Collins M, Boshoff C, et al. Crystal structure of a vFlip-IKKgamma complex: insights into viral activation of the IKK signalosome. Mol Cell. 2008;30:620–31. doi: 10.1016/j.molcel.2008.04.029. [DOI] [PubMed] [Google Scholar]
- 18.Huang GJ, Zhang ZQ, Jin DY. Stimulation of IKK-gamma oligomerization by the human T-cell leukemia virus oncoprotein Tax. FEBS Lett. 2002;531:494–8. doi: 10.1016/s0014-5793(02)03590-1. [DOI] [PubMed] [Google Scholar]
- 19.Vinolo E, Sebban H, Chaffotte A, Israel A, Courtois G, Veron M, et al. A point mutation in NEMO associated with anhidrotic ectodermal dysplasia with immunodeficiency pathology results in destabilization of the oligomer and reduces lipopolysaccharide- and tumor necrosis factor-mediated NF-kappa B activation. J Biol Chem. 2006;281:6334–48. doi: 10.1074/jbc.M510118200. [DOI] [PubMed] [Google Scholar]
- 20.Fusco F, Bardaro T, Fimiani G, Mercadante V, Miano MG, Falco G, et al. Molecular analysis of the genetic defect in a large cohort of IP patients and identification of novel NEMO mutations interfering with NF-kappaB activation. Hum Mol Genet. 2004;13:1763–73. doi: 10.1093/hmg/ddh192. [DOI] [PubMed] [Google Scholar]
- 21.Fusco F, Pescatore A, Bal E, Ghoul A, Paciolla M, Lioi MB, et al. Alterations of the IKBKG locus and diseases: an update and a report of 13 novel mutations. Hum Mutat. 2008;29:595–604. doi: 10.1002/humu.20739. [DOI] [PubMed] [Google Scholar]
- 22.Agou F, Ye F, Goffinont S, Courtois G, Yamaoka S, Israel A, et al. NEMO trimerizes through its coiled-coil C-terminal domain. J Biol Chem. 2002;277:17464–75. doi: 10.1074/jbc.M201964200. [DOI] [PubMed] [Google Scholar]
- 23.Tegethoff S, Behlke J, Scheidereit C. Tetrameric oligomerization of IkappaB kinase gamma (IKKgamma) is obligatory for IKK complex activity and NF-kappaB activation. Mol Cell Biol. 2003;23:2029–41. doi: 10.1128/MCB.23.6.2029-2041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fontan E, Traincard F, Levy SG, Yamaoka S, Veron M, Agou F. NEMO oligomerization in the dynamic assembly of the IkappaB kinase core complex. The FEBS journal. 2007;274:2540–51. doi: 10.1111/j.1742-4658.2007.05788.x. [DOI] [PubMed] [Google Scholar]
- 25.Catici DA, Horne JE, Cooper GE, Pudney CR. Polyubiquitin Drives the Molecular Interactions of the NF-kappaB Essential Modulator (NEMO) by Allosteric Regulation. J Biol Chem. 2015;290:14130–9. doi: 10.1074/jbc.M115.640417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature. 1997;388:548–54. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 27.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell. 1997;91:243–52. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 28.Li XH, Fang X, Gaynor RB. Role of IKKgamma/nemo in assembly of the Ikappa B kinase complex. J Biol Chem. 2001;276:4494–500. doi: 10.1074/jbc.M008353200. [DOI] [PubMed] [Google Scholar]
- 29.Miller BS, Zandi E. Complete reconstitution of human IkappaB kinase (IKK) complex in yeast. Assessment of its stoichiometry and the role of IKKgamma on the complex activity in the absence of stimulation. J Biol Chem. 2001;276:36320–6. doi: 10.1074/jbc.M104051200. [DOI] [PubMed] [Google Scholar]
- 30.Xu G, Lo YC, Li Q, Napolitano G, Wu X, Jiang X, et al. Crystal structure of inhibitor of kappaB kinase beta. Nature. 2011;472:325–30. doi: 10.1038/nature09853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polley S, Huang DB, Hauenstein AV, Fusco AJ, Zhong X, Vu D, et al. A structural basis for IkappaB kinase 2 activation via oligomerization-dependent trans auto-phosphorylation. PLoS Biol. 2013;11:e1001581. doi: 10.1371/journal.pbio.1001581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polley S, Passos DO, Huang DB, Mulero MC, Mazumder A, Biswas T, et al. Structural Basis for the Activation of IKK1/alpha. Cell Rep. 2016;17:1907–14. doi: 10.1016/j.celrep.2016.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kensche T, Tokunaga F, Ikeda F, Goto E, Iwai K, Dikic I. Analysis of nuclear factor-kappaB (NF-kappaB) essential modulator (NEMO) binding to linear and lysine-linked ubiquitin chains and its role in the activation of NF-kappaB. J Biol Chem. 2012;287:23626–34. doi: 10.1074/jbc.M112.347195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivins FJ, Montgomery MG, Smith SJ, Morris-Davies AC, Taylor IA, Rittinger K. NEMO oligomerisation and its ubiquitin-binding properties. Biochem J. 2009 doi: 10.1042/BJ20090427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong KC, Helgason E, Yu C, Phu L, Arnott DP, Bosanac I, et al. Preparation of distinct ubiquitin chain reagents of high purity and yield. Structure. 2011;19:1053–63. doi: 10.1016/j.str.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 36.Konarev PV, Volkov VV, Sokolova AV, MHJK, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Cryst. 2003;36:1277–82. [Google Scholar]
- 37.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst. 1992;25:495–503. [Google Scholar]
- 38.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. Journal of Applied Crystallography. 2009;42:342–6. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. Journal of Applied Crystallography. 2003;36:860–4. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]