ABSTRACT
Sae2 promotes the repair of DNA double-strand breaks in Saccharomyces cerevisiae. The role of Sae2 is linked to the Mre11/Rad50/Xrs2 (MRX) complex, which is important for the processing of DNA ends into single-stranded substrates for homologous recombination. Sae2 has intrinsic endonuclease activity, but the role of this activity has not been assessed independently from its functions in promoting Mre11 nuclease activity. Here we identify and characterize separation-of-function mutants that lack intrinsic nuclease activity or the ability to promote Mre11 endonucleolytic activity. We find that the ability of Sae2 to promote MRX nuclease functions is important for DNA damage survival, particularly in the absence of Dna2 nuclease activity. In contrast, Sae2 nuclease activity is essential for DNA repair when the Mre11 nuclease is compromised. Resection of DNA breaks is impaired when either Sae2 activity is blocked, suggesting roles for both Mre11 and Sae2 nuclease activities in promoting the processing of DNA ends in vivo. Finally, both activities of Sae2 are important for sporulation, indicating that the processing of meiotic breaks requires both Mre11 and Sae2 nuclease activities.
KEYWORDS: DNA damage response, DNA repair, double-strand breaks, recombination
INTRODUCTION
The Sae2 endonuclease of Saccharomyces cerevisiae plays important roles in DNA double-strand break (DSB) repair. Sae2 promotes the resection of 5′ strands at DNA ends that initiate homologous recombination as well as the removal of protein-DNA conjugates from genomic DNA and the processing of DNA secondary structures that arise during replication (1–10). Yeast cells lacking Sae2 are deficient in these functions to different degrees; however, the mechanistic basis of these functions is not well understood. Three primary biochemical activities have been attributed to Sae2 through in vitro studies using recombinant proteins. First, Sae2 promotes Exo1- and Dna2-mediated end processing in a manner that is cooperative with Mre11/Rad50/Xrs2 (MRX) (5, 6); second, Sae2 promotes Mre11 nuclease activity on protein-blocked DNA ends (10, 11); and third, Sae2 exhibits intrinsic endonuclease activity on branched structures and single-stranded DNA (ssDNA)/double-stranded DNA (dsDNA) junctions (12), similar to the functional ortholog of Sae2 in human cells, CtIP (13, 14). How these biochemical activities map to the biological roles of Sae2 is not fully understood. While Sae2 has functional orthologs in higher eukaryotes (15), the primary sequence of Sae2 is poorly conserved between species, and the distinct activities attributed to the protein have not been successfully separated by mutagenesis.
The roles of Sae2 are also unclear relative to other nucleases that are known to function in DNA repair. Yeast strains expressing nuclease-deficient forms of Mre11 have been shown to be similar to strains with deletions of SAE2, most notably in meiosis and the processing of cruciform structures in vivo (3, 4, 9, 16, 17), leading to the suggestion that Sae2 likely promotes the nuclease activity of Mre11, which was subsequently demonstrated in vitro (10, 11). However, other studies indicated that the DNA damage sensitivity of SAE2 deletion strains to the Top1 poison camptothecin is significantly higher than that of Mre11 nuclease-deficient strains (2), suggesting that Sae2 performs additional functions apart from stimulating Mre11 nuclease activity.
Like Sae2 and CtIP, Dna2 nuclease activity is also specific for single-stranded DNA and 5′-flap structures (18, 19). The Dna2 enzyme functions in Okazaki fragment processing during replication and is essential in budding yeast (20). Dna2 is a helicase as well as a nuclease and functions in the long-range resection of double-strand breaks, a role which was shown to be downstream of MRX/Sae2 and redundant with the 5′-to-3′ exonuclease Exo1 (21, 22). The MRX complex stimulates the recruitment of both Exo1 and Dna2 to DNA ends in vivo (6) and promotes the activity of both enzymes in vitro (5, 23, 24). Finally, the nuclease activity of Dna2 was shown to be functionally redundant with Mre11 nuclease activity in cellular responses to radiation damage, indicating the possibility of shared substrates (25).
To clarify the roles of Sae2 in DNA repair, we sought to identify separation-of-function mutations that eliminate either the ability of Sae2 to promote Mre11 activity or the intrinsic nuclease activity of Sae2. Since Sae2 does not have easily identifiable domains, we used a structure-based strategy of targeted mutagenesis that utilizes predictions of helical propensity and disorder to identify sites that are likely to nucleate the transient secondary structure in the Sae2 polypeptide. We find that one of these mutants is specifically deficient in promoting Mre11 nuclease activity, while another mutant is deficient in the intrinsic nuclease activity of Sae2. In vivo, these mutants show very different phenotypes in combination with an Mre11 nuclease-deficient allele or Dna2 hypomorphic strains, suggesting that the roles of Mre11 nuclease activity and Sae2 nuclease activity are functionally distinct. The intrinsic nuclease activity of Sae2 is not required for DNA damage survival in cells when the stimulation of Mre11 activity by Sae2 is functional; however, both activities are essential during meiosis.
RESULTS
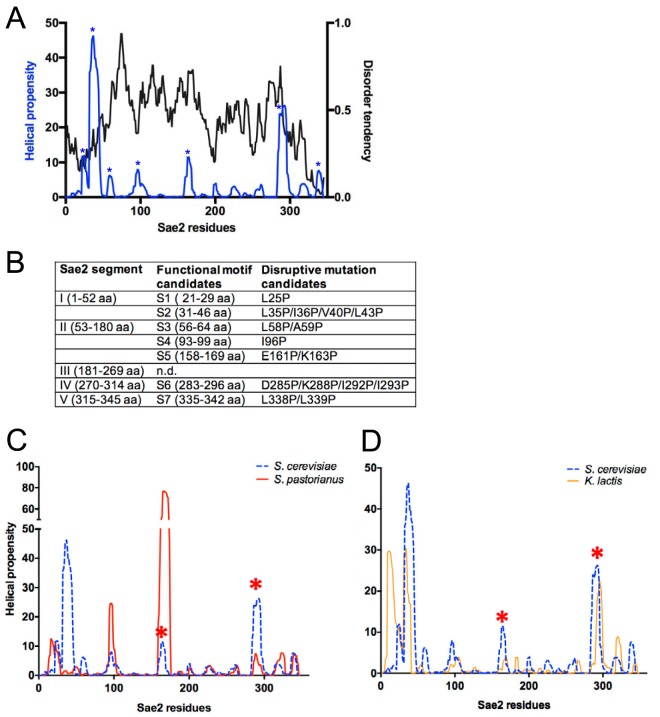
The primary amino acid sequence of Sae2 is minimally conserved, and there are no predicted motifs or domains apart from the C terminus, which shares a very short “FPSTQ” motif with orthologs in other organisms and the proposed DNA-binding “RHR” motif (26, 27). The N terminus of this protein contains a self-interaction domain that is important for the function of Sae2 and also for its stability (28). Equivalent regions in the Schizosaccharomyces pombe Ctp1 and human CtIP proteins were shown previously to be tetrameric (29, 30). Besides this N-terminal region, the majority of Sae2 is predicted to be disordered based on the output of the IUPred algorithm, which estimates local interresidue interactions (31) (Fig. 1A). This analysis is based on existing structural data for a large set of alpha-helical structures and predicts disorder based on amino acid composition and sequence complexity. It is possible that Sae2 adopts a functional structure through interactions between the monomers in the complex, by binding to another protein, or by binding to DNA.
FIG 1.
Structure prediction-guided mutagenesis of Sae2. (A) Disorder tendency of Sae2 residues determined with IUPred (31). Values with a disorder score of >0.5 are predicted to be disordered. Helical propensity was predicted with AGADIR (64); segments of increased helical propensity are marked with stars. Based on order/disorder and the location of putative helical motifs, Sae2 may be divided into five structurally distinct segments (segments I to V). (B) Segments I, II, IV, and V contain motifs of increased helical propensity (S1 to S7) that may contribute to Sae2 function. Proline mutations were designed to disrupt putative α-helices S1 to S7. aa, amino acids. (C and D) Prediction of the helical propensities of the Saccharomyces pastorianus and Kluyveromyces lactis Sae2 proteins compared to that of Sae2 of S. cerevisiae. Asterisks indicate positions of E161/K163 and D285/K288 peaks.
Many disordered proteins undergo transient incursions into secondary structures that are thought to be nucleated at specific amino acids that are most favorable for alpha-helical interactions (32, 33). To investigate this, we analyzed helical propensity along the Sae2 polypeptide using the AGADIR prediction algorithm based on the helix-coil transition theory (34). This analysis suggested several sites in Sae2 that are likely sites of helical transitions, a strategy that was used previously to predict critical residues in the Sgs1 and Rmi1 components of the Sgs1/Top3/Rmi1 complex that also contain unstructured domains (35, 36). We identified several structural motifs of this type in Sae2 (Fig. 1A, asterisks, and B) and predicted mutations that would disturb helices initiated at these sites. We focused on the motifs in regions II through V because the L25P mutation was identified previously in a genetic screen and is known to result in a loss of function in vivo (28) but no change in nuclease activity in vitro (data not shown). Mutations in residues of peak helical propensity in regions II through V to proline residues were made, since prolines are the most disruptive amino acids in stable alpha-helices.
While the primary amino acid sequence of Sae2 is not well conserved among eukaryotes, the overall pattern of helical propensity appears to be generally conserved. Examples of this are shown with comparisons between Sae2 nucleases of S. cerevisiae, Saccharomyces pastorianus, and Kluyveromyces lactis in Fig. 1C and D.
Phenotypes of Sae2 mutants alone and in combination with DNA2 nuclease or MRE11 nuclease deficiency reveal functions of intrinsic nuclease and stimulatory activities of Sae2.
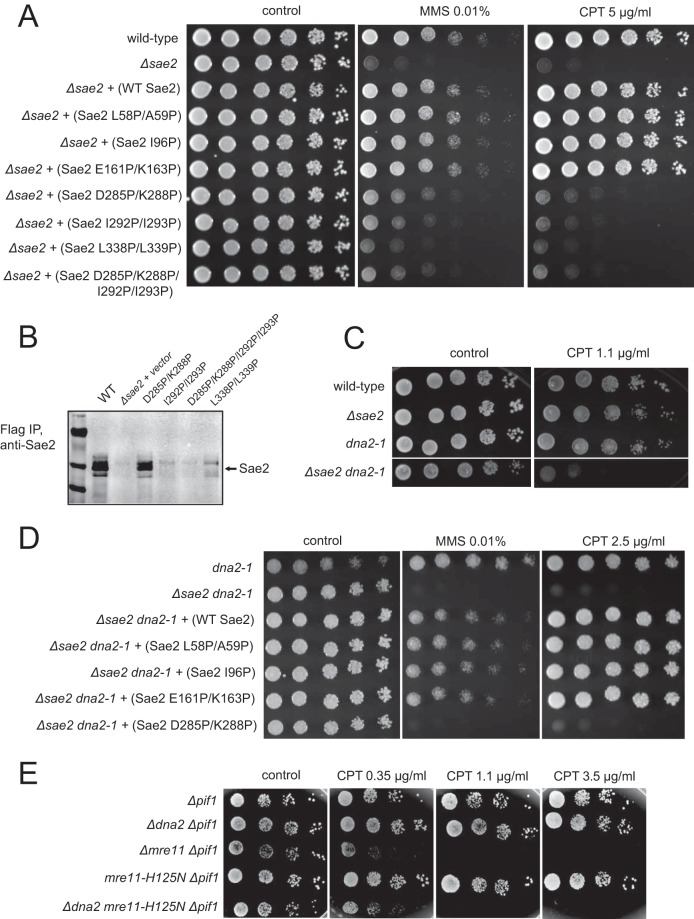
Each combination of target residues was mutated in the Sae2 open reading frame and expressed from a low-copy-number CEN plasmid under the control of the native Sae2 promoter in sae2Δ yeast cells (Fig. 2A). Serial dilutions of cells expressing the indicated SAE2 alleles were grown on medium containing camptothecin (CPT) or methyl methanesulfonate (MMS), which showed that most of the alleles partially or completely rescued the DNA damage sensitivity of sae2Δ cells; however, the D285P/K288P, I292P/I293P, and L338P/L339P combinations failed to fully complement the deletion strain for MMS or CPT sensitivity. Analysis of the expression of the mutants in yeast by immunoprecipitation and Western blotting showed that the D285P/K288P allele was expressed similarly to wild-type Sae2, while the other mutants were not stably expressed (Fig. 2B); thus, we focused our attention on the D285P/K288P mutant.
FIG 2.
Mutation of helix-nucleating segments increases the sensitivity of Sae2-deficient yeast cells to DNA damage. (A) FLAG-tagged Sae2 was expressed from a CEN plasmid under the control of the native Sae2 promoter in sae2Δ yeast cells. Fivefold serial dilutions of cells expressing the indicated Sae2 alleles were plated onto nonselective medium (control) or medium containing methyl methanesulfonate (MMS) (0.01%) or camptothecin (CPT) (5.0 μg/ml) and grown for 48 h. WT, wild type. (B) FLAG-tagged Sae2 C-terminal mutants were expressed from a 2μ plasmid in sae2Δ yeast cells and isolated by immunoprecipitation (IP) with anti-FLAG antibody. Sae2 levels in the immunoprecipitates were determined by Western blotting with anti-FLAG antibody. (C) sae2Δ yeast cells were compared to a strain with the nuclease-deficient Dna2 allele dna2-1 or a Δsae2 dna2-1 double mutant using serial dilutions of the cells as described above for panel A. (D) The dna2-1 or sae2Δ dna2-1 yeast strain complemented with FLAG-tagged Sae2 alleles expressed from a CEN plasmid under the control of the native Sae2 promoter, as indicated, was tested for MMS and CPT sensitivity as described above for panel A. Plates with DNA-damaging agents were incubated for 90 h, while the control plate was incubated for 70 h. (E) Δpif1, Δdna2 Δpif1, Δmre11 Δpif1, mre11-H125N Δpif1, and Δdna2 mre11-H125N Δpif1 strains were analyzed for CPT sensitivity as described above for panel A.
The multifunctional helicase/nuclease Dna2 functions primarily in Okazaki fragment processing during replication but also participates in long-range 5′-strand resection at DNA double-strand breaks (21, 22, 37) and functionally overlaps the Mre11 nuclease (25). Here we show that a null allele of SAE2 combined with a nuclease-deficient allele of DNA2 (sae2Δ dna2-1) is hypersensitive to DNA-damaging agents (Fig. 2C), suggesting that the Dna2 nuclease has functions that are at least partially redundant with those of Sae2. To examine the Sae2 mutants in the absence of Dna2 nuclease activity, we expressed the mutant alleles in the sae2Δ dna2-1 strain and found that region II mutants of Sae2 fully rescued CPT and MMS sensitivity, whereas the D285P/K288P mutant did not (Fig. 2D). We conclude that this mutant likely lacks a functional activity that is partially redundant with Dna2 nuclease activity. It is likely that this activity is the stimulation of Mre11 nuclease function based on the synthetic CPT sensitivity observed with the MRE11 deletion or nuclease-deficient alleles combined with dna2 mutations (Fig. 2E). Previous work demonstrated redundancy between Dna2 and Mre11 nuclease activities (25), and Sae2 has been shown to stimulate Mre11 nuclease activity in vitro (10).
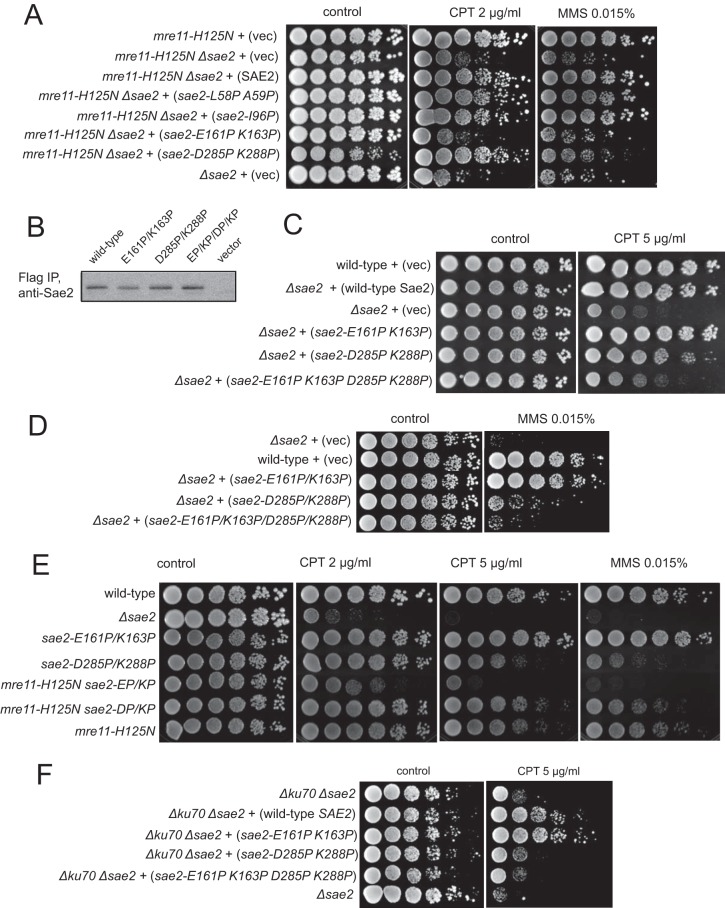
We also analyzed the DNA damage sensitivity conferred by the sae2 mutant alleles in a sae2Δ mre11-H125N background because Sae2 and Mre11 nuclease activities have been suggested to have overlapping roles in vivo (38, 39). Yeast cells expressing the mre11-H125N allele are deficient in Mre11 nuclease activity and fail to sporulate but are proficient in promoting the 5′-strand resection of restriction enzyme-generated ends as well as nonhomologous end joining (NHEJ) and telomere maintenance (16). A sae2Δ mre11-H125N double mutant strain has CPT sensitivity similar to that of the sae2Δ single mutant strain (data not shown), but here we find that the new sae2 mutants show phenotypes in this mre11-H125N background that are remarkably different from that of either the sae2Δ or the sae2Δ dna2-1 strain (Fig. 3A). The strain expressing the E161P/K163P mutant, which rescues strains similarly to the wild-type allele in the sae2Δ or sae2Δ dna2-1 background, is extremely sensitive to CPT and MMS. In contrast, the strain expressing the D285P/K288P allele shows much better survival. To determine if the E161P/K163P mutant form of Sae2 is expressed normally, we examined the levels of the mutants expressed from low-copy-number plasmids by immunoprecipitation and Western blotting and found that the protein levels are slightly lower than the levels of wild-type Sae2 and the D285P/K288P mutant (Fig. 3B), but this does not appear to affect its function, since DNA damage survival promoted by this mutant is similar to or exceeds that of the wild-type strain (Fig. 2A).
FIG 3.
Deficiency in Mre11 nuclease generates synthetic sensitivity to DNA-damaging agents in combination with central domain SAE2 mutations. (A) Yeast strains deficient in Mre11 nuclease activity, mre11-H125N (16) and the mre11-H125N sae2Δ double mutant, were complemented with FLAG-tagged Sae2 alleles expressed from a CEN plasmid under the control of the native SAE2 promoter as indicated and tested for MMS and CPT sensitivity as described in the legend to Fig. 2A. (B) FLAG-tagged Sae2 mutants were expressed from low-copy-number CEN plasmids in sae2Δ yeast cells and isolated by using immunoprecipitation with anti-FLAG antibody. Sae2 protein levels in the immunoprecipitates were determined by Western blotting with anti-Sae2 antibody. (C) FLAG-tagged Sae2 was expressed from a low-copy-number plasmid under the control of the native SAE2 promoter in sae2Δ yeast cells. Fivefold serial dilutions of cells expressing the indicated Sae2 alleles were plated onto nonselective medium (control) or medium containing camptothecin (CPT) (5 μg/ml). (D) Yeast strains with the indicated genotypes (mutant alleles integrated into the endogenous SAE2 locus, in the W303 background) were tested for MMS sensitivity as described above for panel A. (E) Yeast strains of the indicated genotypes (mutant alleles integrated into the endogenous SAE2 locus, in the W303 background) were tested for CPT and MMS sensitivity as described above for panel A. (F) ku70Δ sae2Δ yeast strains were complemented with FLAG-tagged SAE2 alleles expressed from a CEN plasmid under the control of the native SAE2 promoter as indicated and tested for CPT sensitivity as described above for panel A.
The D285P/K288P and E161P/K163P alleles of Sae2 appear to confer very different phenotypes in sae2Δ mre11-H125N and sae2Δ dna2-1 backgrounds (Fig. 2 and 3), suggesting that they alter different functions of Sae2. With this in mind, the mutations were combined in a single allele and compared in a sae2Δ strain, which indeed showed that the E161P/K163P mutations confer marked sensitivity to CPT and MMS when tested in the context of the D285P/K288P allele (Fig. 3C and D). Together, the two groups of mutations reduce the survival of the sae2Δ strain to the level of the vector-complemented control, consistent with a complete loss of function in the sae2-E161P/K163P/D285P/K288P mutant.
The phenotypes of the sae2-E161P/K163P and the sae2-D285P/K288P Sae2 mutant alleles with respect to CPT sensitivity were also recapitulated in strains containing knock-in mutations at the genomic loci, confirming that the expression level does not affect this phenotype (Fig. 3D and E). The survival of a yeast strain expressing the sae2-D285P/K288P allele from the genomic locus is very similar to that of a strain expressing the mre11-H125N allele from the genomic locus in the same strain background, while the sae2-E161P/K163P expression strain shows wild-type levels of survival (Fig. 3E). Similar to the observations with overexpression strains, combining the sae2-E161P/K163P allele with mre11-H125N results in dramatic sensitivity to DNA-damaging agents compared to either of the alleles separately.
The Ku heterodimer blocks DNA end resection in vivo and in vitro, and the deletion of Ku in yeast has been shown to partially rescue mre11Δ strains for DNA damage sensitivity (6, 7, 40–45). To determine if the absence of Ku affects the phenotypes of the Sae2 mutants, we analyzed CPT sensitivity in a ku70Δ sae2Δ background (Fig. 3F). Although the sensitivity of this strain is overall lower than that of a sae2Δ strain, the SAE2 mutant alleles appear to have the same phenotypes; thus, the differences between the mutants are not attributable to the presence of Ku on DNA ends.
The D285P/K288P Sae2 mutant lacks the ability to stimulate Mre11.
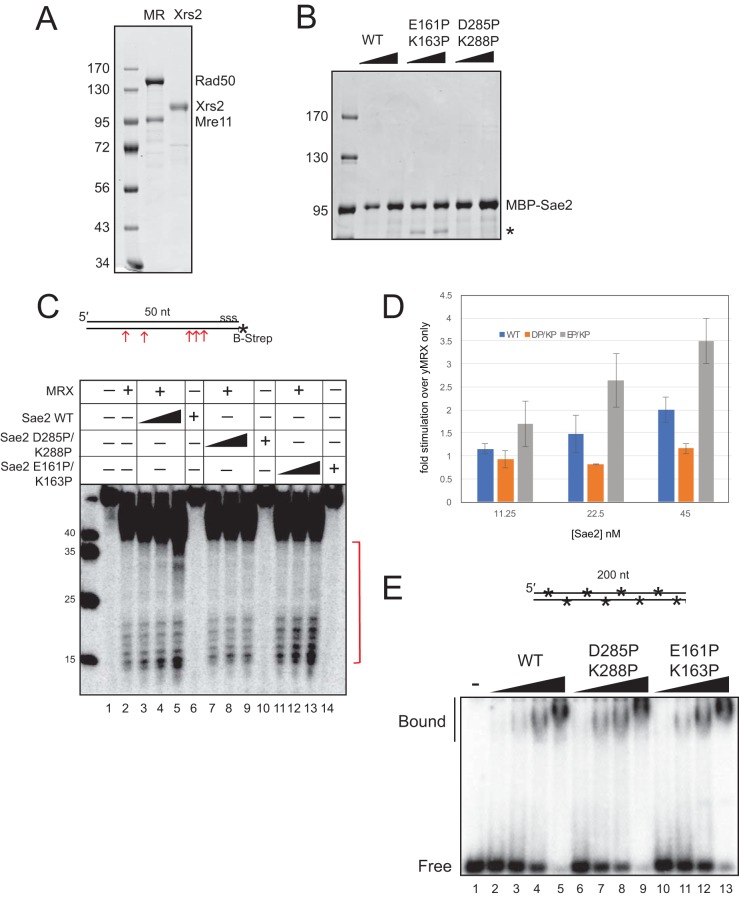
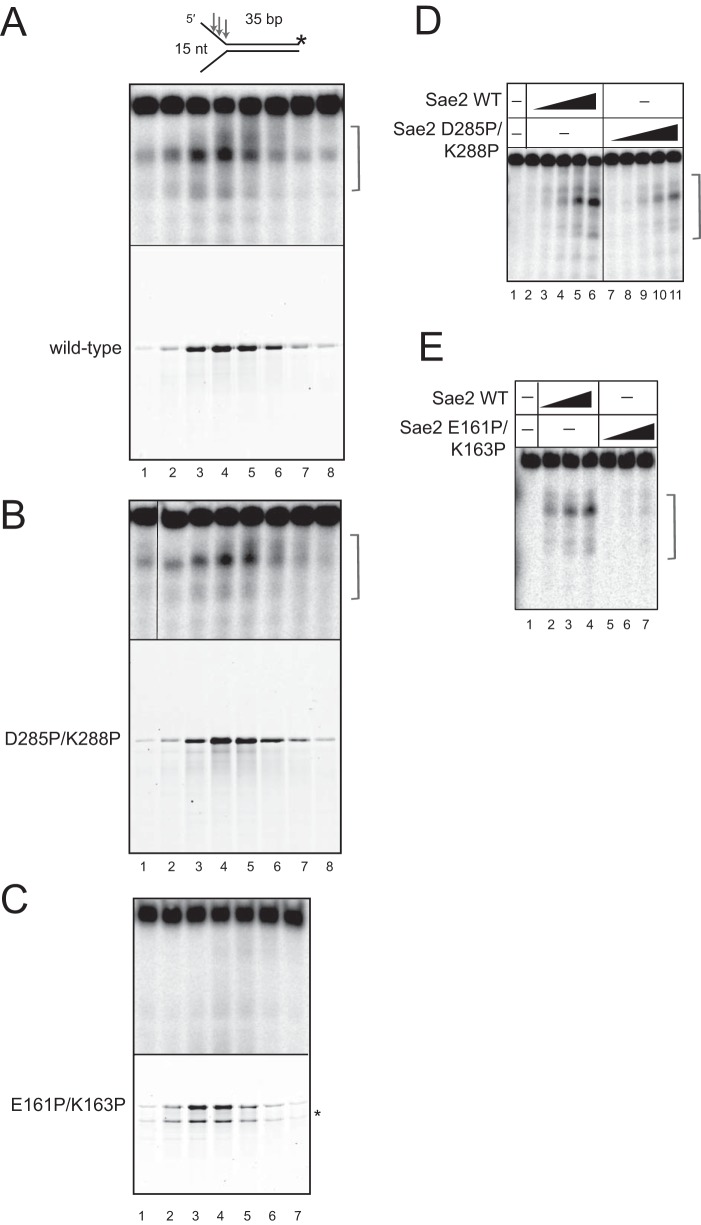
To test for specific deficiencies associated with the D285P/K288P and E161P/K163P mutants, maltose binding protein (MBP)-tagged Sae2 proteins and MRX were expressed and purified from insect cells (Fig. 4A and B). Both mutants were purified similarly to the wild-type protein and were predominantly dimeric, with minor oligomeric and monomeric peaks, also similar to the wild-type protein (data not shown). Cannavo and Cejka previously showed that wild-type Sae2 made in insect cells stimulates Mre11 nuclease activity on model DNA substrates containing a streptavidin-biotin adduct that mimics a covalent protein attachment (10). Here we use a similar substrate containing a biotin-streptavidin adduct on the 5′ end of the bottom strand, which is also labeled with 32P, as indicated (Fig. 4C). The recombinant MRX complex cleaves this substrate on the bottom strand with the protein conjugate but is stimulated severalfold by the addition of Sae2 (Fig. 4C, lanes 3 to 5). The addition of the D285P/K288P mutant, however, did not yield any stimulation (Fig. 4C, lanes 7 to 9), indicating a deficiency in this function. Quantitation of these results is shown in Fig. 4D. Since this could be due to a lack of DNA binding by this mutant, we examined the ability of the Sae2D285P/K288P protein to bind to DNA in a gel mobility shift assay, which showed that the mutant binds to a linear DNA fragment similarly to the wild type (Fig. 4E).
FIG 4.
Sae2 C-terminal mutations disrupt the stimulation of MRX endonuclease activity. (A) Recombinant His6-tagged Mre11/Flag-tagged Rad50 (MR) and Flag-tagged Xrs2 expressed in Sf21 insect cells in an SDS-PAGE gel stained with Coomassie blue. (B) Recombinant His-MBP-Sae2 proteins expressed in Sf21 insect cells in an SDS-PAGE gel stained with Coomassie blue, as indicated. * indicates the degradation product of the E161P/K163P (EP/KP) mutant; this is a C-terminal degradation product of the full-length protein since it is recognized by an anti-His antibody (data not shown). (C) The recombinant wild-type MRX complex (25 nM) was incubated with a 50-bp dsDNA substrate containing a 5′ biotin-streptavidin block (B-Strep) and a 5′ 32P label, as indicated. “sss” on the top strand denotes 5 phosphorothioate bonds at the 3′ end to prevent exonucleolytic degradation. Reaction mixtures contained wild-type or mutant Sae2 (11.25, 22.5, and 45 nM), 5 mM MgCl2, 1 mM MnCl2, and 1 mM ATP and were incubated at 30°C for 30 min. Reaction products were separated on a gel containing 16% acrylamide, 20% formamide, and 6 M urea and analyzed by using a phosphorimager. The arrows and bracket indicate sites of cleavage. (D) Data from two replicates of the experiment in panel C were quantified for products (bands within the bracket in panel C), and the fold increase in stimulation by Sae2 over yeast Mre11/Rad50/Xrs2 (yMRX) alone was calculated. Error bars indicate standard deviations. (E) Gel mobility shift assays were performed with wild-type or mutant recombinant Sae2 proteins, as indicated, with 4.3, 8.7, 17.5, and 35 nM protein and an internally 32P-labeled 197-bp DNA substrate. Protein-DNA complexes are indicated (“bound”) in comparison to the free DNA (“free”). nt, nucleotides.
In contrast to the Sae2D285P/K288P mutant, the recombinant Sae2E161P/K163P mutant protein stimulated MRX nuclease activity on the blocked end substrate (Fig. 4C, lanes 11 to 13, and D) and even exhibited higher-than-wild-type activity on this substrate. DNA binding by this mutant was also similar to that of the wild type (Fig. 4E).
The Sae2E161P/K163P mutant lacks endonuclease activity.
We have previously shown that recombinant MBP-Sae2 expressed in Escherichia coli is active as a 5′-flap endonuclease (12, 13). Here we tested the wild-type MBP-Sae2 protein expressed in insect cells and purified through a 4-column purification protocol and found that the recombinant protein exhibits the same endonuclease activity on a branched DNA substrate, as we previously observed, and the pattern of activity matches well with the levels of protein across the heparin column fractions stained with Coomassie (Fig. 5A). We also removed the MBP tag with tobacco etch virus (TEV) protease, confirmed that the endonuclease activity of Sae2 is not dependent on the presence of the MBP tag, and again demonstrated that a mock preparation made from insect cells without the Sae2 virus shows no activity when treated similarly by using the same purification protocol (data not shown).
FIG 5.
Sae2 mutations in the central domain inactivate Sae2 endonuclease activity. (A) Recombinant Sae2 proteins were purified by using 4 chromatographic steps, the last one being heparin sulfate resin. Wild-type Sae2-containing fractions from this purification are shown (bottom). Nuclease assays were performed with the corresponding fractions, using a 32P-labeled DNA substrate, as shown. Reaction products were separated on a gel containing 15% acrylamide and 7 M urea and analyzed by using a phosphorimager; products are indicated with the arrows and bracket. (B) Heparin fractions from the purification of the D285P/K288P mutant of Sae2 were analyzed as described above for panel A. (C) Heparin fractions from the purification of the E161P/K163P Sae2 mutant were analyzed as described above for panel A. * indicates the degradation product of Sae2-E161P/K163P identified by mass spectrometry analysis of this band. (D) Wild-type and D285P/K288P mutant Sae2 proteins (1.25, 2.5, 5, 10, and 20 nM) were tested by using nuclease assays as described above for panel A. Reaction products were separated by denaturing polyacrylamide gel electrophoresis and analyzed by using a phosphorimager. (E) Wild-type and E161P/K163P mutant Sae2 proteins (5, 10, and 20 nM) were tested by using nuclease assays as described above for panel A.
The D285P/K288P and E161P/K163P mutant forms of recombinant Sae2 were tested in the nuclease assay on the branched substrate, which showed that the D285P/K288P mutant exhibits activity comparable to that of the wild type in this assay, while the E161P/K163P mutant appears to be completely nuclease deficient (Fig. 5B to E). The deficiency with the E161P/K163P mutant is not attributable to a DNA-binding deficiency, as the mutant shows DNA binding comparable to that of the wild-type protein (Fig. 4D).
The sae2-D285P/K288P and sae2-E161P/K163P mutants are deficient in resection in vivo.
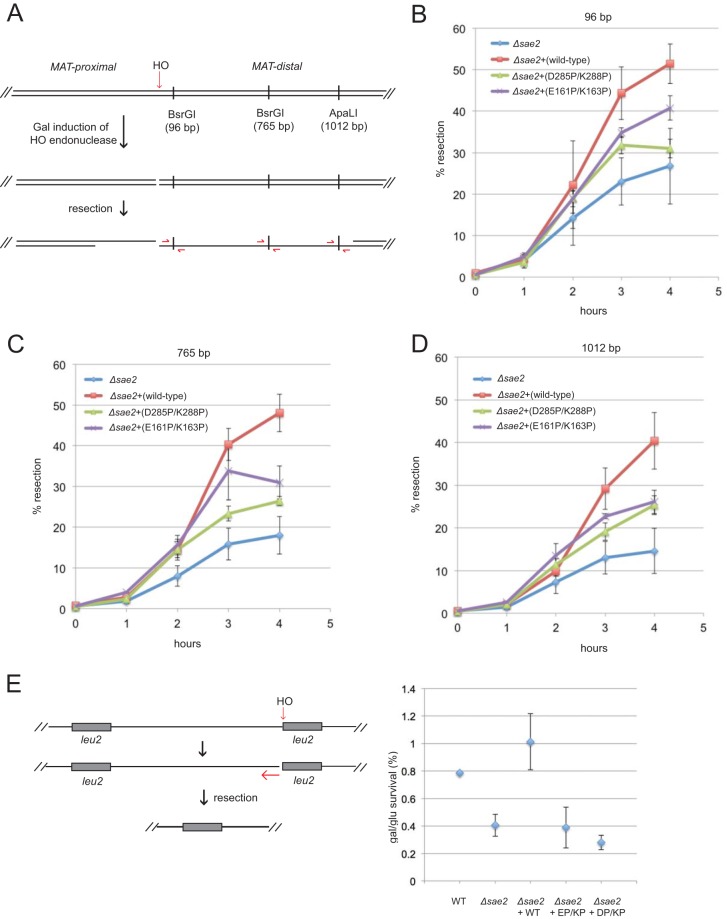
Yeast strains deficient in Sae2 exhibit a reduced efficiency of DNA end resection such that the levels of 3′ ssDNA intermediates generated from DNA breaks are lower than those in wild-type strains and the rate of ssDNA production is reduced (8). To test the resection ability of the Sae2 mutants, we used a yeast strain containing deletions of HML and HMR mating-type donor loci and examined resection from a galactose-induced HO endonuclease break at the MAT locus (46) (Fig. 6A). The sae2Δ strain complemented with wild-type SAE2 showed up to 50% resection of genomic DNA by 4 h after the addition of galactose, by analyzing ssDNA using quantitative PCR at sites located 96 bp, 765 bp, and 1,012 bp from the HO breakpoint (Fig. 6B to D) (47). The uncomplemented sae2Δ strain exhibited much lower resection levels, ranging from 25% at sites close to the HO cut site to as low as 15% when measured 1,012 bp from the HO site. The strains complemented with the D285P/K288P and E161P/K163P mutants showed levels of resection intermediates between the deletion strain and the wild-type expression strain, indicating that both mutants are impaired in the stimulation of DSB resection in vivo but that they both retain partial function. However, the sae2-D285P/K288P strain consistently showed lower resection rates than those of the strain expressing the E161P/K163P allele, consistent with the decreased DNA damage survival observed with the D285P/K288P allele.
FIG 6.
Sae2 C-terminal and central domain mutations reduce resection efficiency in vivo. (A) Diagram of the MAT locus showing the site of HO endonuclease cleavage and the locations of the BsrGI and ApaLI restriction sites used to determine ssDNA levels, as shown. (B) Quantification of ssDNA at the BsrGI site located 96 bp from the HO-generated DSB in sae2Δ yeast cells expressing the vector only, wild-type Sae2, or the D285P/K288P or E161P/K163P Sae2 mutant on 2μ plasmids at various times after the addition of galactose. Error bars indicate standard errors of the means from 3 biological replicates. (C) Quantification of ssDNA at the BsrGI site located 765 bp from the HO-generated DSB in sae2Δ yeast cells as described above for panel B. (D) Quantification of ssDNA at the ApaLI site located 1,012 bp from the HO-generated DSB in sae2Δ yeast cells as described above for panel B. (E) Diagram of the HO endonuclease site adjacent one of two partial alleles of LEU2 (his4::leu2 and leu2::cs) on chromosome III. Resection from the cut site through ∼25 kb of the intervening sequence generates a LEU2 repair product by single-strand annealing (48). Wild-type or sae2Δ strains expressing the mutant alleles as indicated were plated in dilutions onto galactose, which induces HO endonuclease, or on glucose. The percentage of survivors was determined. Error bars indicate standard errors of the means from 3 biological replicates.
To examine long-range resection, we also utilized a yeast strain containing a galactose-inducible HO endonuclease site on chromosome III (Fig. 6E). The survival of this strain requires the resection of ∼25 kb of genomic DNA to expose homologous regions of the LEU2 gene that promote DNA repair by single-strand annealing (48). Yeast strains deficient in Sae2 were previously shown to have very low viability in comparison to wild-type strains after induction with HO (8). Here we tested strains carrying the D285P/K288P and E161P/K163P mutant Sae2 alleles for survival on galactose compared to glucose solid media and found that both mutant alleles confer defects in long-range resection.
MRX stimulation by Sae2 is important for DNA end processing.
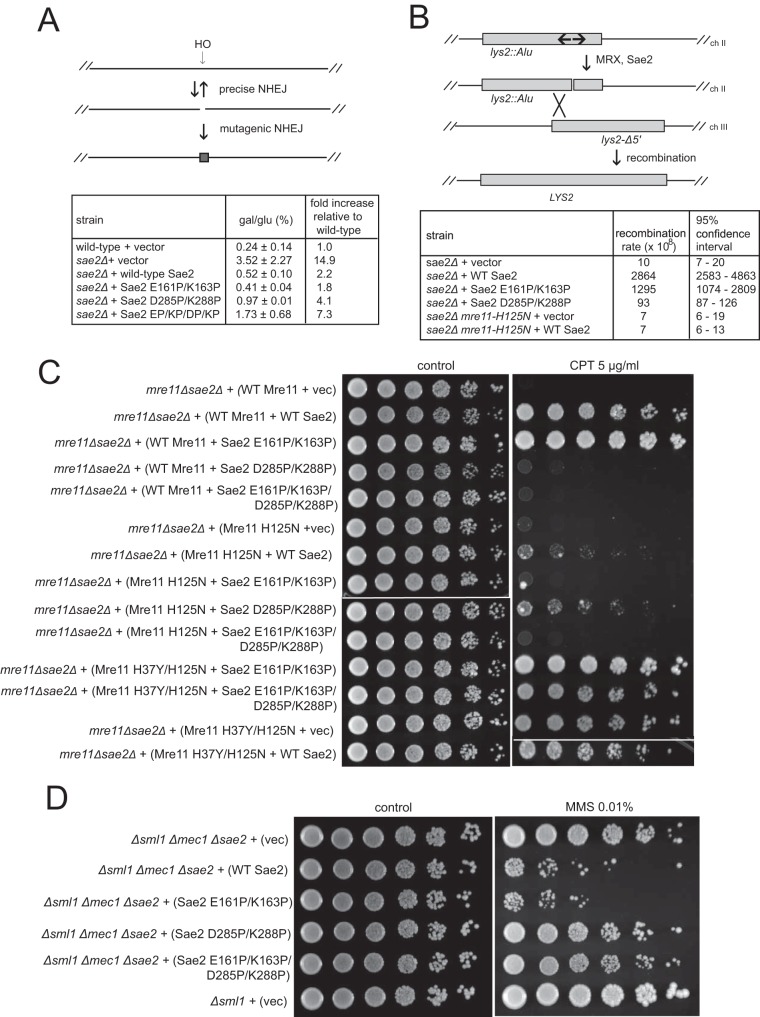
Homologous recombination and nonhomologous end joining are considered to be competing pathways for the processing and repair of DNA double-strand breaks (49). Previous work indicated that Sae2 plays an antagonistic role in Ku-dependent end joining while promoting microhomology-mediated end joining (50), consistent with its positive role in end resection. Here we used strains expressing galactose-induced HO endonuclease in the absence of HML and HMR donor cassettes (Fig. 6A to D) but measured viability on galactose versus glucose media. Survival under these circumstances was previously shown to be dependent on mutagenic NHEJ (51). We found that the deletion of SAE2 resulted in an ∼15-fold increase in viability, which was suppressed by the expression of wild-type SAE2 or the E161P/K163P mutant but only partially suppressed by the expression of the D285P/K288P mutant or the D285P/K288P/E161P/K163P combination (Fig. 7A).
FIG 7.
Sae2 C-terminal mutations reduce Sae2 function in end joining and cruciform processing. (A, top) Diagram showing the HO endonuclease cleavage site at the MAT locus and mutagenic end joining that removes the recognition site for cleavage. (Bottom) Wild-type and sae2Δ yeast strains expressing the vector or various SAE2 alleles on 2μ plasmids, as indicated, were plated onto glucose (glu)- or galactose (gal)-containing plates, and the percentage of survivors (gal/glu) was calculated. The error shown is the standard deviation from 3 biological replicates. (B, top) Diagram showing an inverted repeat on chromosome II before and after a spontaneous DSB promoted by MRX and Sae2, as previously shown (3). Recombination between the DSB and a homologous region on chromosome III can generate a LYS2 prototroph. (Bottom) Spontaneous rates of this recombination event were measured in wild-type and sae2Δ yeast strains expressing the vector or various Sae2 alleles on CEN plasmids using fluctuation analysis. The measured rates as well as the 95% confidence intervals are shown. (C) Yeast strains deficient in MRE11 and SAE2 were complemented with FLAG-tagged sae2 alleles expressed from a 2μ plasmid under the control of the native SAE2 promoter and with yeast Mre11 expressed from the native promoter on CEN plasmids, as indicated, and were tested for CPT sensitivity as described in the legend to Fig. 2A. (D) Flag-tagged Sae2 was expressed from a CEN plasmid under the control of the native Sae2 promoter in Δsml1 Δmec1 Δsae2 yeast cells as indicated. Fivefold serial dilutions of cells expressing the indicated Sae2 alleles were plated onto nonselective medium (control) or medium containing MMS (0.01%).
Sae2 and the MRX complex also regulate the processing of DNA secondary structures such as cruciforms and hairpins in budding yeast (3), similar to the SbcC/D orthologs of Rad50 and Mre11 in E. coli (52). Lobachev et al. established an in vivo assay for palindrome-induced DNA processing in S. cerevisiae by creating a strain with an inverted repeat located in a nonfunctional LYS2 gene, which is resolved by DSB formation and gene conversion by utilizing a homologous sequence on another chromosome (3). We tested the activity of our Sae2 mutants in this strain by determining the frequency of spontaneously occurring LYS+ prototrophs and found that sae2 strains exhibit a rate of gene conversion that is at least 130-fold lower than that of a strain complemented with wild-type Sae2 (Fig. 7B). Similar to the data from the NHEJ assay described above, the sae2-D285P/K288P allele only partially complements the deletion strain, while the sae2-E161P/K163P allele confers nearly wild-type levels of recombination. Mre11 nuclease-deficient strains were previously found to be deficient in this activity (3), as we confirm in our study, and a double mutant expressing the mre11-H125N nuclease-deficient allele in an mre11Δ sae2Δ background shows recombination rates similar to that of the mre11Δ sae2Δ strain complemented with mre11-H125N and wild-type SAE2 (Fig. 7B). Thus, in this context, the nuclease activity of Mre11 appears to play a central role in processing the DNA lesion.
Reduced DNA binding by Mre11 alleviates SAE2 mutant phenotypes.
Yeast strains deficient in Sae2 were previously shown to have a slower release of the MRX complex from sites of DNA double-strand breaks (7, 53, 54). The sensitivity of sae2 strains to MMS and CPT can be suppressed by mutations in Mre11 that cause a partial loss of the DNA-binding ability (1, 55), suggesting that Sae2 promotes the release of MRX from DNA and that this is an important function of Sae2. To examine whether mutations in Mre11 alter the effects of the sae2 mutations characterized in this study, we used a sae2Δ mre11Δ strain complemented with MRE11 or SAE2 alleles on plasmids under the control of the native yeast promoters (Fig. 7C). We found that with wild-type Mre11 expressed, the D285P/K288P Sae2 mutant causes increased sensitivity compared to that in a sae2Δ background, whereas the expression of the sae2-E161P/K163P allele appears to increase CPT resistance. In contrast, when the Mre11 nuclease-deficient H125N mutant is expressed from the plasmid in the sae2Δ mre11Δ strain, the expression of the sae2-E161P/K163P mutant is very toxic with CPT exposure (Fig. 7C), consistent with the results in the sae2Δ background.
We also tested the ability of the H37Y mutation, one of the Mre11 suppressor mutations reported previously (1, 55), to suppress the phenotypes of the sae2-D285P/K288P and sae2-E161P/K163P alleles in order to determine if the lower DNA-binding capacity of Mre11 can suppress the effects of losing either Mre11 nuclease-promoting activity or intrinsic Sae2 nuclease activity. This is a possibility since the suppression of sae2Δ DNA damage sensitivity was shown previously to be independent of Mre11 nuclease activity (1, 55). We found that the sensitivity of strains expressing sae2-E161P/K163P in addition to mre11-H125N can be completely suppressed by the addition of the mre11-H37Y mutation (Fig. 7C), and a similar suppression was observed with the combined D285P/K288P/E161P/K163P mutant. Thus, the defects in Sae2 caused by these separation-of-function mutations, at least with respect to DNA damage sensitivity, can also be attributed to toxic MRX occupancy at DSB sites or ssDNA intermediates. Consistent with these results, we also observed that the deletion of Mre11 is epistatic to all the Sae2 mutants analyzed in this study (data not shown).
The deletion of SAE2 in a mec1Δ strain was previously shown to rescue the signaling defect caused by the loss of Mec1 as well as the survival of exposure to MMS (56), a phenomenon attributed to the increased lifetime of intact DNA ends that occurs in a sae2Δ strain that can further activate Tel1 (ATM). Here we confirmed the increased MMS survival of sae2Δ strains in a mec1Δ background and also found that the effect of the expression of the sae2-D285P/K288P mutant allele is equivalent to that of a sae2Δ strain in promoting the survival of mec1Δ cells (Fig. 7D). In contrast, the sae2-E161P/K163P mutant allele behaves like wild-type sae2. In this case, the combined D285P/K288P/E161P/K163P mutant is equivalent to D285P/K288P mutant; thus, only Mre11 nuclease activity and not intrinsic Sae2 activity is important for this function.
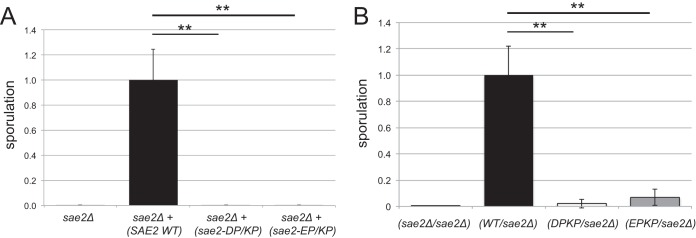
The SAE2 gene was initially identified as an essential component of the meiotic recombination pathway, as the null mutant is deficient in sporulation and accumulates covalent Spo11 adducts similarly to a rad50S strain (4, 9). Here we constructed diploid strains deficient in both alleles of the SAE2 gene and expressing either wild-type or sae2-D285P/K288P or sae2-E161P/K163P mutant alleles from a low-copy-number plasmid. The strains were induced to sporulate, and viable spores were quantitated by using random-spore analysis (Fig. 8A). These results show that both the D285P/K288P and E161P/K163P mutants are deficient in sporulation; thus, both the stimulation of Mre11 activity and intrinsic Sae2 activity are essential during the processing of covalent Spo11 conjugates. We also analyzed sporulation using diploids constructed from sae2Δ strains and strains with sae2 mutant alleles integrated at the genomic locus. Random-spore analysis of independently derived diploids also showed a striking deficiency in viability associated with both mutant alleles, although the level of sporulation was higher than that observed for the sae2Δ strain (Fig. 8B).
FIG 8.
Both Mre11 stimulation and intrinsic nuclease activity are required for sporulation. (A) Quantitation of sporulation by diploid strains of the indicated genotypes with low-copy-number plasmids expressing wild-type Sae2 or the D285P/K288P (DP/KP) or E161P/K163P (EP/KP) Sae2 mutant, showing the fraction of sporulation as measured by random-spore analysis relative to the wild-type SAE2-expressing strain. Three independent strain isolates were analyzed, and error bars indicate standard deviations. (B) Sporulation was measured as described above for panel A with strains containing integrated mutant alleles, as indicated. Three independent strain isolates were analyzed. **, P < 0.005.
DISCUSSION
The Sae2 protein is an important component of the DNA repair machinery and plays roles in initiating resection, resolving protein-DNA adducts, and modulating DNA damage signaling (28, 49). It is a structure-specific endonuclease that cleaves ssDNA/dsDNA junctions and 5′ flaps and promotes the MRX-dependent cleavage of hairpin-containing structures (12). Sae2 has also been shown to stimulate Mre11 nuclease activity on protein-blocked DNA ends (10), which we observed and demonstrated with human CtIP and Mre11/Rad50/Nbs1 (11). In this study, we identify mutants that separate these two biochemical activities of Sae2, providing tools to study the roles of these activities in different biological functions of Sae2.
Disruption of central helices in Sae2 blocks its intrinsic nuclease activity.
The in vitro and in vivo experiments described here indicate that the intrinsic nuclease activity of Sae2 is genetically separable from the activity of Sae2 in promoting Mre11 activity. This is important since the intrinsic nuclease activity of Sae2 has been questioned (10), yet CtIP, the human ortholog of Sae2, has been shown to possess nearly identical flap endonuclease activity (13, 14). The purified recombinant Sae2E161P/K163P protein lacks intrinsic nuclease activity but is competent in stimulating Mre11 nuclease activity in vitro. The two mutations are predicted to disrupt a helix in the central region of Sae2, which we identified as being the region critical for Sae2 nuclease activity in previous studies (data not shown) and is also analogous to the location of the nuclease domain in CtIP (13, 14). Our observation that the nuclease-deficient sae2-E161P/K163P mutant shows severe sensitivity to DNA-damaging agents when combined with an Mre11 nuclease-deficient mutation suggests that, with respect to survival of DNA damage, intrinsic Sae2 nuclease activity and Mre11 nuclease activity work in parallel in vegetative cells.
Recent studies of Ctp1, the ortholog of Sae2 and CtIP in fission yeast, yielded mutations in this protein that were shown to eliminate the “clipping” of Rec12 (Spo11) from DNA ends during meiosis but to support the resection of DNA ends similarly to wild-type strains (57). Further analysis of these mutants showed that they were rescued by a deletion of Ku, consistent with a resection defect (58). However, it is not clear whether the Ctp1 mutants identified previously by Ma et al. are defective in the stimulation of Mre11 or in an intrinsic catalytic activity.
Sae2 C-terminal helix disruption blocks noncatalytic stimulation of Mre11.
The D285P/K288P mutant is competent for Sae2 endonuclease activity but completely deficient in stimulating Mre11 nuclease activity. This C-terminal region of Sae2 shows some degree of sequence conservation in Sae2 orthologs in other organisms (27), suggesting an essential and conserved function. Helical structure prediction analysis predicts a strong and persistent helix including these residues that is hydrophilic in nature. This exposed helix can potentially interact with different proteins to mediate various functions.
In vivo, the D285P/K288P mutant shows sensitivity to CPT and MMS in a sae2Δ background, but a striking loss of viability is observed with these agents in a sae2Δ dna2-1 strain in which the activity of the Dna2 nuclease is reduced to 5% of the wild-type activity (37) (Fig. 2). The synthetic lethality with sae2Δ and dna2-1 alleles upon exposure to DNA damage and the lack of complementation by the D285P/K288P mutant suggest that Mre11 nuclease activity and Dna2 nuclease activity share overlapping roles in the processing of topoisomerase adducts. Consistent with this hypothesis, an mre11-H125N dna2Δ pif1Δ strain is also hypersensitive to CPT (25). In contrast, the E161P/K163P mutant fully rescues a sae2Δ dna2-1 strain, suggesting that the intrinsic nuclease activity of Sae2 does not function in parallel with that of Dna2 and that Sae2 nuclease activity is not required for Mre11 nuclease functions in vivo.
Consistent with the hypothesis that there are two distinct activities of Sae2 that contribute to its effect on the DNA damage response, mutation of both clusters of sites in the combined E161P/K163P/D285P/K288P mutant reduces DNA damage survival to the level of a SAE2 deletion. While sae2Δ deletion strains expressing the sae2-E161P/K163P allele do not show a damage phenotype, the mutations have a strong effect when tested in the context of the sae2-D285P/K288P allele, consistent with a dominant role of Mre11 nuclease stimulation and a requirement for Sae2 nuclease activity in cells deficient in Mre11 activity.
Resection of DNA ends into single-stranded DNA intermediates is a critical step regulating DNA repair by homologous recombination. We found that both the sae2-E161P/K163P and the sae2-D285P/K288P mutants failed to fully suppress the resection deficiency of a sae2Δ strain in comparison to a wild-type allele. This is surprising considering the complete suppression of sae2Δ strains by the sae2-E161P/K163P allele in DNA damage survival. One possibility is that there is a threshold effect with resection such that the higher level of resection promoted by the sae2-E161P/K163P mutant than that promoted by the sae2-D285P/K288P mutant is high enough to completely restore function. Alternatively, it is possible that processes other than resection play a more important role in DNA damage survival. For instance, the ability to induce the removal of MRX from DNA ends may be a stronger determinant of viability, as long as a minimal level of resection takes place.
It is also striking that the mre11-H125N sae2Δ strain expressing the sae2-E161P/K163P allele shows enhanced CPT sensitivity compared to that of the vector-complemented strain, indicating a dominant negative effect (Fig. 3). We attribute this to the requirement for Sae2 for the removal of Mre11 (1, 7, 53–55), and the inability of Sae2 to cleave DNA, in the context of Mre11 nuclease deficiency, may generate long-lived MRX complexes on DNA that are cytotoxic. The suppression of this toxicity with the mre11-H37Y mutation (Fig. 7C) confirms that increased MRX occupancy on DNA contributes to the phenotype of the mre11-H125N sae2-E161P/K163P strain.
The intrinsic nuclease activity of Sae2 is not required for some Sae2 functions, namely, the processing of cruciform structures (3) and the processing of DNA strands that has been suggested to block Tel1 activity in the absence of Mec1 (56). The exclusive role for Mre11 nuclease activity in the processing of hairpin structures is also suggested by an analysis of the Rad50 and Mre11 proteins SbcC and SbcD in hairpin processing in E. coli, where there is no apparent Sae2 ortholog (52). In this case, the loss of SbcC/D strongly promotes the stability of inverted repeats, similar to the situation for MRX-deficient strains of yeast. The observation that Mre11 nuclease-deficient strains show no additional deficit in cruciform processing with a SAE2 deletion indicates that the intrinsic activity of Sae2 is not a contributor in this context. Another situation in which Sae2 nuclease activity is dispensable is that for strains lacking DNA2 or Dna2 nuclease activity. The D285P/K288P Sae2 mutant that is deficient for Mre11 stimulation completely fails to complement sae2Δ dna2-1 cells, whereas the sae2-E161P/K163P mutant, which is proficient in Mre11 stimulation, fully rescues the sensitivity of the strain to DNA damage (Fig. 2). Taken together, these data suggest that Sae2-promoted Mre11 nuclease activity and Dna2 nuclease function are redundant with each other, and the requirement for Sae2 in the absence of Dna2 stems solely from its stimulatory effect on Mre11 nuclease activity.
In contrast to our observations with DNA-damaging agents in vegetatively growing cells, we found that both the noncatalytic and catalytic functions of Sae2 are essential for sporulation, based on the inability of the sae2-D285P/K288P and sae2-E161P/K163P mutants to complement a sae2Δ strain during meiosis. This generally correlates with the absolute requirement for Sae2 during the processing of Spo11 adducts compared to its less essential phenotype in vegetatively growing cells. However, why the intrinsic activity of Sae2 is needed in addition to that of Mre11 during meiosis is unknown and likely awaits the reconstitution of meiotic double-strand break formation and processing in vitro to be fully understood.
MATERIALS AND METHODS
S. cerevisiae strains.
See Table S1 in the supplemental material for S. cerevisiae strains. Genomic mutations at the SAE2 locus were made via a 2-step PCR-based method (59). Mutant alleles were fully sequenced at both steps. Yeast strains deficient in MEC1 were first grown with wild-type MEC1 on a URA3-based plasmid, which was removed on 5-fluoroorotic acid (5-FOA)-containing plates before growth of the strains.
Yeast Sae2 expression constructs.
The S. cerevisiae wild-type SAE2 gene was cloned into the low-copy-number pRS313 vector (60) under the control of the native SAE2 promoter with the coding sequence for a 2× FLAG tag at the N terminus (cloning details are available upon request) to create pTP1496. Mutant alleles of SAE2 were made from pTP1496 by QuikChange mutagenesis (Agilent Technologies). A high-copy-number vector containing the wild-type SAE2 gene with a 2× FLAG tag in pRS425 (61), “FLAG-SAE2/2μ” (28), was a gift from John Petrini. Mutant versions of this plasmid or of pTP1496 are listed in Table S1 in the supplemental material.
Recombinant protein expression.
Baculovirus expression constructs for wild-type and mutant Sae2 proteins were made by cloning His-MBP-Sae2 from an E. coli expression vector into pFastBac1 (Thermo Fisher) to create pTP3211 and the corresponding bacmid pTP3213. The D285P/K288P and E161P/K163P mutants were made by using QuikChange mutagenesis (Agilent Technologies) according to the manufacturer's instructions to create pTP3759 and pTP3845, respectively, and the corresponding bacmids pTP3769 and pTP3846.
His-MBP-Sae2 was purified from Sf21 insect cells by affinity chromatography on amylose (NEB) and nickel-nitrilotriacetic acid (Ni-NTA) (Qiagen) resin, followed by ion-exchange chromatography on a HiTrap SP column (GE) and a HiTrap heparin column (GE). All steps were done at 0°C to 4°C, and all buffers contained 0.5% Tween 20 unless otherwise specified. The His-MBP-Sae2 complex was expressed in Sf21 insect cells, and the cells were lysed in buffer A (25 mM Tris [pH 8.0], 100 mM NaCl, 10% glycerol, 1 mM dithiothreitol [DTT]) supplemented with 10 mM EDTA, using homogenization followed by sonication. The lysate was cleared by ultracentrifugation, applied to amylose resin, washed, and eluted in buffer A containing 10 mM maltose and 14 mM β-mercaptoethanol instead of DTT. The eluate was then applied to Ni-NTA resin, washed with 0.5 M LiCl, and eluted in Ni-B buffer (50 mM KH2PO4, 10% glycerol, 250 mM imidazole, 20 mM β-mercaptoethanol, 50 mM KCl). The eluate was diluted 3-fold with zero-salt buffer A and loaded onto a HiTrap sulfopropyl (SP) column using a fast protein liquid chromatography (FPLC) system. The column was washed and eluted with 60% buffer B (25 mM Tris [pH 8.0], 1 M NaCl, 10% glycerol, 1 mM DTT). The peak SP fractions were combined, diluted 15-fold with zero-salt buffer A, and injected onto a HiTrap heparin column by using an FPLC system. The column was washed extensively with 10% buffer B. MBP-Sae2 was eluted in 0.5-ml fractions using a gradient of 10 to 70% buffer B in 10 ml. Fractions 23 and 24 had the highest protein concentrations and were flash frozen and stored in 10-μl aliquots at −80°C. A mock prep was conducted with Sf21 insect cells expressing no recombinant protein. For some Sae2 nuclease assays, the peak elution fraction after SP HiTrap column treatment was loaded onto a Superdex-200 gel filtration column (GE) to separate monomeric, dimeric, and multimeric His-MBP-Sae2.
The yeast Mre11/Rad50/Xrs2 complex containing FLAG-tagged Rad50 and His6-tagged Mre11 was purified by using Ni-NTA, SP, and anti-FLAG resin. Insect cells expressing complexes were lysed in Ni-A buffer (50 mM KH2PO4, 10% glycerol, 2.5 mM imidazole, 20 mM β-mercaptoethanol, 0.5 M KCl) using homogenization followed by sonication. Lysates were cleared by ultracentrifugation, applied to 3-ml Ni-NTA resin (Qiagen), washed, and eluted with Ni-B buffer (50 mM KH2PO4, 10% glycerol, 250 mM imidazole, 20 mM β-mercaptoethanol, 50 mM KCl). The eluate was then loaded onto a Hi-Trap SP column (GE), washed with buffer A (25 mM Tris [pH 8.0], 100 mM NaCl, 10% glycerol, 1 mM DTT), and eluted with 60% buffer B (25 mM Tris [pH 8.0], 1 M NaCl, 10% glycerol, 1 mM DTT). The eluate was diluted with buffer A to 150 mM NaCl, applied to 1 ml of anti-FLAG M2 antibody resin (Sigma), washed with 5 volumes of 0.5 M LiCl and buffer A, and eluted with 0.1 mg/ml FLAG peptide (Sigma) in buffer A. Aliquots were flash frozen and stored at −80°C FLAG-Xrs2 was purified as described by Deshpande et al. (11).
Protein expression analysis in yeast.
Yeast cells at an optical density (OD) of 100 and containing appropriate high-copy-number Sae2 expression plasmids were collected and used to test the protein expression level. The cells were resuspended in lysis buffer (25 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride [PMSF]) and lysed by using a bead beater (1 min) with 0.3-ml glass beads. The beads and insoluble material were removed by centrifugation. Sae2 was immunoprecipitated by using anti-FLAG M2 magnetic beads (Invitrogen), washed 3 times, and eluted from the beads by using 1× SDS loading buffer. The eluted material was separated on a 10% SDS-PAGE protein gel and analyzed by Western blotting with anti-Sae2 antibody (custom polyclonal antibody made in mouse; Precision Antibody).
In vitro endonuclease assay with Sae2.
The DNA substrate was made by using oligonucleotides TP2622 (5′-CTGCAGGGTTTTTGTTCCAGTCTGTAGCACTGTGTAAGACAGGCCAGATG-3′) and TP1152 (5′-CATCTGGCCTGTCTTACACAGTGCTACAGACTGGAGTCCTCATCAGACTG-3′). TP2622 was radiolabeled at the 3′ end by using terminal deoxynucleotidyl transferase (TdT) and α-32P-cordycepin and then annealed to the complement TP1152 (1.2-fold molar excess). Both oligonucleotides were purified on denaturing acrylamide gels prior to making the assay substrate.
The DNA substrate (0.1 nM) was incubated with MBP-Sae2 in nuclease buffer (25 mM morpholinepropanesulfonic acid [MOPS] [pH 7.0], 60 mM NaCl, 1 mM ATP, 1 mM dithiothreitol [DTT], 5 mM MgCl2, 0.1 mg/ml bovine serum albumin [BSA], 6% glycerol) in Lo-Bind Eppendorf tubes at 30°C for 2 h. The MBP-Sae2 fractions eluted from the heparin column were diluted with zero-salt buffer A before being added to the nuclease assay reaction mixture to achieve equivalent NaCl concentrations in a 10-μl reaction mixture volume. Reactions were stopped by adding 2 μl of stop solution (0.5% SDS, 20 mM EDTA [pH 8.0], 5 μM oligonucleotide TP2622) to the mixture, and the mixture was lyophilized, resuspended in formamide loading buffer, and resolved on a 15% acrylamide–urea gel at a constant wattage (40 W) for 2.5 h. Gels were analyzed by phosphorimager analysis (GE). For TEV cleavage of the MBP tag, the desired aliquots of MBP-Sae2 were incubated with 10 U of TEV protease (Promega) overnight at 16°C. The cleaved aliquot was used for nuclease assays directly. A TEV-only aliquot was used as a negative control in this assay.
In vitro endonuclease assay with MRX and Sae2.
The DNA substrate was made by using oligonucleotides TP5124 (5′-TGGGTCAACGTGGGCAAAGATGTCCTAGCAATGTAATCGTCTATGACGTT-3′), containing 5′ dT-biotin (where “dT” stands for deoxythymidine), and TP4559, containing 5 consecutive phosphorothioate bonds at the 3′ end. TP5124 was radiolabeled at the 5′ end by using T4 polynucleotide kinase (NEB) and [γ-32P]ATP and was then annealed to the complement TP4559 (1.2-fold molar excess). Both oligonucleotides were purified on denaturing acrylamide gels prior to making the assay substrate.
The radiolabeled TP5124-TP4559 substrate was incubated with a 20-fold molar excess of streptavidin at room temperature (RT) for 10 min prior to addition to the reaction mixture. Yeast Mre11/Rad50 (yMR) and Xrs2 were mixed and incubated on ice for 10 min prior being added to the assay mixture. In a 10-μl reaction mixture volume, 25 nM yMR and 25 nM Xrs2 were incubated with 1 nM DNA in a solution containing 25 mM MOPS (pH 7.0), 20 mM Tris (pH 8.0), 80 mM NaCl, 8% glycerol, 1 mM DTT, 1 mM ATP, 20 nM streptavidin, 5 mM MgCl2, 1 mM MnCl2, and 0.2 mg/ml BSA in Protein Lo-Bind Eppendorf tubes (Millipore) at 30°C for 30 min. Reactions were stopped with 0.2% SDS and 10 mM EDTA, and the mixture was lyophilized; dissolved in formamide; boiled at 100°C for 4 min; loaded onto denaturing polyacrylamide gels containing 16% acrylamide, 20% formamide, and 6 M urea; and separated at 40 W for 1.5 h, followed by phosphorimager analysis.
Gel mobility shift assay for Sae2 DNA binding.
The DNA substrate was prepared by PCR amplification of a 197-bp fragment from the pFastbac1 plasmid using oligonucleotides TP347 (5′-TATTCCGGATTATTCATACCGTCCC-3′) and TP1621 (5′-CCTCTACAAATGTGGTATGGCTG-3′) as primers. Amplification was done in the presence of [α-32P]dATP to internally radiolabel the PCR product. The PCR product was separated on a 1% agarose gel and purified with Ultrafree-DA columns (Millipore). MBP-Sae2 was incubated with the DNA substrate in the presence of a solution containing 25 mM MOPS (pH 7.0), 20 mM Tris (pH 8.0), 8% glycerol, 2 mM DTT, 70 mM NaCl, and 0.2 mg/ml BSA. The reaction mixtures were incubated at 30°C for 10 min. The reaction products were resolved on a 4% native acrylamide gel in 0.5× Tris-borate-EDTA (TBE) at a constant voltage of 80 V for 75 min. The gel was dried and exposed overnight to a phosphorimager screen for analysis.
In vivo analysis of resection.
Yeast strains based on JKM179 (46) were grown in synthetic defined medium lacking leucine (SD-leucine medium) in the presence of glucose overnight and then washed twice with water before being resuspended in SD-leucine medium containing 2% raffinose. After growth overnight, cells were diluted and grown to log phase in yeast extract-peptone-dextrose-adenine (YPDA) medium until the OD at 600 nm (OD600) was ∼0.5. Galactose was added to 2%, and cells at an OD600 of 5 were removed at each time point, harvested, washed with water, and frozen at −20°C. The MasterPure yeast DNA purification kit (Epicentre) was used to isolate genomic DNA according to the manufacturer's instructions. A total of 350 ng genomic DNA per sample was added with the appropriate restriction enzyme buffer in a volume of 35 μl, which was divided equally into a mock sample and a sample to which 2 U of the restriction enzyme (BsrGI or ApaLI; NEB) was added. Reaction mixtures were incubated for 3 h at 37°C before serial dilution and quantitative PCR (qPCR). Two microliters of each enzyme digestion mixture (or dilution) was used per 10 μl TaqMan qPCR mixture with TaqMan Universal master mix (Applied Biosystems). The following primers and probes were used: primers TP3896 (5′-TACGTGGTGACGGATATTGGG-3′) and TP3897 (5′-GGGAACAAGAGCAAGACGATG-3′) and probe TP3898 (5′-6-carboxyfluorescein [FAM]-CAACCTCCGCCACGACCACACTC-6-carboxytetramethylrhodamine [TAMRA]-3′) for the 96-bp BsrGI site, primers TP3681 (5′-TCATATCATCGACGTAATGACCACTTA-3′) and TP3682 (5′-GTTTGGATACCATAAGTGACGATATTAAGT-3′) and probe TP3680 (5′-FAM-CCTCCGTCCAATCTGTGCACAATGAAGTT-TAMRA-3′) for the 765-bp ApaLI site, and primers TP3919 (5′-ACCTTCTTCATTACTATTCATCTTCGC-3′) TP3920 (5′-CTTAGCTTGTACCAGAGGAAGCAA-3′) and probe TP3921 (5′-FAM-CACAAGTCTTCTCTCCCTTGGTGTTTCCA-TAMRA-3′) for the 1,012-bp BsrGI site. Threshold cycle (CT) values obtained from the analysis were used to calculate the percentage of resected DNA, as previously described (47).
LYS2 palindrome resolution assays.
Yeast strains based on ALE94 containing the inverted Alu repeat (3) were first tested on plates to confirm lysine auxotrophy. Fourteen colonies were grown overnight in SD-uracil-histidine, and dilutions were plated onto SD-lysine and on SD-uracil-histidine plates to determine the percentage of LYS+ cells. These data were used to calculate conversion rates using fluctuation analysis, as described previously (62).
NHEJ assays.
Yeast strains based on JKM179 (46) were grown in appropriate selective media, and dilutions were plated onto glucose- or galactose-containing plates. The percentage of survivors on galactose was calculated relative to the number of survivors on glucose plates from 3 independent biological replicates.
Sporulation.
Diploid yeast strains heterozygous at the CAN1 locus were sporulated, and the efficiency of viable spore formation was determined on SD-arginine plates containing canavanine as described previously (63).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for yeast strains and plasmids from James Haber, Lorraine Symington, Kirill Lobachev, John Petrini, Sang Eun Lee, Katsunori Sugimoto, and Xiaolan Zhao.
Work in the Paull laboratory was supported in part by the Cancer Prevention and Research Institute of Texas and grants RP110465 and R01GM081425 from the National Institutes of Health to K.H.S.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00156-17.
REFERENCES
- 1.Chen H, Donnianni RA, Handa N, Deng SK, Oh J, Timashev LA, Kowalczykowski SC, Symington LS. 2015. Sae2 promotes DNA damage resistance by removing the Mre11-Rad50-Xrs2 complex from DNA and attenuating Rad53 signaling. Proc Natl Acad Sci U S A 112:E1880–E1887. doi: 10.1073/pnas.1503331112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng C, Brown JA, You D, Brown JM. 2005. Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics 170:591–600. doi: 10.1534/genetics.104.028795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lobachev KS, Gordenin DA, Resnick MA. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108:183–193. doi: 10.1016/S0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 4.McKee AH, Kleckner N. 1997. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics 146:797–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, Paull TT. 2010. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat Struct Mol Biol 17:1478–1485. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G, Lee SE. 2010. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J 29:3370–3380. doi: 10.1038/emboj.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langerak P, Mejia-Ramirez E, Limbo O, Russell P. 2011. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet 7:e1002271. doi: 10.1371/journal.pgen.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clerici M, Mantiero D, Lucchini G, Longhese MP. 2005. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem 280:38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- 9.Prinz S, Amon A, Klein F. 1997. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics 146:781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannavo E, Cejka P. 2014. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature 514:122–125. doi: 10.1038/nature13771. [DOI] [PubMed] [Google Scholar]
- 11.Deshpande RA, Lee JH, Arora S, Paull TT. 2016. Nbs1 converts the human Mre11/Rad50 nuclease complex into an endo/exonuclease machine specific for protein-DNA adducts. Mol Cell 64:593–606. doi: 10.1016/j.molcel.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 12.Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. 2007. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell 28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makharashvili N, Tubbs AT, Yang SH, Wang H, Barton O, Zhou Y, Deshpande RA, Lee JH, Lobrich M, Sleckman BP, Wu X, Paull TT. 2014. Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection. Mol Cell 54:1022–1033. doi: 10.1016/j.molcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Li Y, Truong LN, Shi LZ, Hwang PY, He J, Do J, Cho MJ, Li H, Negrete A, Shiloach J, Berns MW, Shen B, Chen L, Wu X. 2014. CtIP maintains stability at common fragile sites and inverted repeats by an end resection-independent endonuclease activity. Mol Cell 54:1012–1021. doi: 10.1016/j.molcel.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makharashvili N, Paull TT. 2015. CtIP: a DNA damage response protein at the intersection of DNA metabolism. DNA Repair 32:75–81. doi: 10.1016/j.dnarep.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 16.Moreau S, Ferguson JR, Symington LS. 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol 19:556–566. doi: 10.1128/MCB.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rattray AJ, McGill CB, Shafer BK, Strathern JN. 2001. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics 158:109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart JA, Campbell JL, Bambara RA. 2010. Dna2 is a structure-specific nuclease, with affinity for 5′-flap intermediates. Nucleic Acids Res 38:920–930. doi: 10.1093/nar/gkp1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fortini BK, Pokharel S, Polaczek P, Balakrishnan L, Bambara RA, Campbell JL. 2011. Characterization of the endonuclease and ATP-dependent flap endo/exonuclease of Dna2. J Biol Chem 286:23763–23770. doi: 10.1074/jbc.M111.243071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budd ME, Campbell JL. 1995. A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc Natl Acad Sci U S A 92:7642–7646. doi: 10.1073/pnas.92.17.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mimitou EP, Symington LS. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, Ira G, Sung P. 2010. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. 2010. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Budd ME, Campbell JL. 2009. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS One 4:e4267. doi: 10.1371/journal.pone.0004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P. 2007. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell 28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. 2007. Human CtIP promotes DNA end resection. Nature 450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HS, Vijayakumar S, Reger M, Harrison JC, Haber JE, Weil C, Petrini JH. 2008. Functional interactions between Sae2 and the Mre11 complex. Genetics 178:711–723. doi: 10.1534/genetics.107.081331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies OR, Forment JV, Sun M, Belotserkovskaya R, Coates J, Galanty Y, Demir M, Morton CR, Rzechorzek NJ, Jackson SP, Pellegrini L. 2015. CtIP tetramer assembly is required for DNA-end resection and repair. Nat Struct Mol Biol 22:150–157. doi: 10.1038/nsmb.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andres SN, Appel CD, Westmoreland JW, Williams JS, Nguyen Y, Robertson PD, Resnick MA, Williams RS. 2015. Tetrameric Ctp1 coordinates DNA binding and DNA bridging in DNA double-strand-break repair. Nat Struct Mol Biol 22:158–166. doi: 10.1038/nsmb.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dosztanyi Z, Csizmok V, Tompa P, Simon I. 2005. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 32.Namba K. 2001. Roles of partly unfolded conformations in macromolecular self-assembly. Genes Cells 6:1–12. doi: 10.1046/j.1365-2443.2001.00384.x. [DOI] [PubMed] [Google Scholar]
- 33.Dyson HJ. 2011. Expanding the proteome: disordered and alternatively folded proteins. Q Rev Biophys 44:467–518. doi: 10.1017/S0033583511000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munoz V, Serrano L. 1994. Elucidating the folding problem of helical peptides using empirical parameters. Nat Struct Biol 1:399–409. doi: 10.1038/nsb0694-399. [DOI] [PubMed] [Google Scholar]
- 35.Kennedy JA, Daughdrill GW, Schmidt KH. 2013. A transient alpha-helical molecular recognition element in the disordered N-terminus of the Sgs1 helicase is critical for chromosome stability and binding of Top3/Rmi1. Nucleic Acids Res 41:10215–10227. doi: 10.1093/nar/gkt817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kennedy JA, Syed S, Schmidt KH. 2015. Structural motifs critical for in vivo function and stability of the RecQ-mediated genome instability protein Rmi1. PLoS One 10:e0145466. doi: 10.1371/journal.pone.0145466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Budd ME, Choe W, Campbell JL. 2000. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J Biol Chem 275:16518–16529. doi: 10.1074/jbc.M909511199. [DOI] [PubMed] [Google Scholar]
- 38.Mimitou EP, Symington LS. 2009. DNA end resection: many nucleases make light work. DNA Repair 8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paull TT. 2010. Making the best of the loose ends: Mre11/Rad50 complexes and Sae2 promote DNA double-strand break resection. DNA Repair 9:1283–1291. doi: 10.1016/j.dnarep.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foster SS, Balestrini A, Petrini JH. 2011. Functional interplay of the Mre11 nuclease and Ku in the response to replication-associated DNA damage. Mol Cell Biol 31:4379–4389. doi: 10.1128/MCB.05854-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, Iwasaki H, Mizuno K, Ohta K, Uritani M, Ushimaru T, Yoshinaga K, Ueno M. 2003. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol 23:5186–5197. doi: 10.1128/MCB.23.15.5186-5197.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mimitou EP, Symington LS. 2010. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J 29:3358–3369. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wasko BM, Holland CL, Resnick MA, Lewis LK. 2009. Inhibition of DNA double-strand break repair by the Ku heterodimer in mrx mutants of Saccharomyces cerevisiae. DNA Repair 8:162–169. doi: 10.1016/j.dnarep.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang SH, Zhou R, Campbell J, Chen J, Ha T, Paull TT. 2013. The SOSS1 single-stranded DNA binding complex promotes DNA end resection in concert with Exo1. EMBO J 32:126–139. doi: 10.1038/emboj.2012.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Y, Paull TT. 2013. DNA-dependent protein kinase regulates DNA end resection in concert with Mre11-Rad50-Nbs1 (MRN) and ataxia telangiectasia-mutated (ATM). J Biol Chem 288:37112–37125. doi: 10.1074/jbc.M113.514398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399–409. doi: 10.1016/S0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 47.Zierhut C, Diffley JF. 2008. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J 27:1875–1885. doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaze MB, Pellicioli A, Lee SE, Ira G, Liberi G, Arbel-Eden A, Foiani M, Haber JE. 2002. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell 10:373–385. doi: 10.1016/S1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- 49.Symington LS, Gautier J. 2011. Double-strand break end resection and repair pathway choice. Annu Rev Genet 45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 50.Lee K, Lee SE. 2007. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics 176:2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moore JK, Haber JE. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol 16:2164–2173. doi: 10.1128/MCB.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Connelly JC, Leach DR. 1996. The sbcC and sbcD genes of Escherichia coli encode a nuclease involved in palindrome inviability and genetic recombination. Genes Cells 1:285–291. doi: 10.1046/j.1365-2443.1996.23024.x. [DOI] [PubMed] [Google Scholar]
- 53.Lisby M, Barlow JH, Burgess RC, Rothstein R. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 54.Clerici M, Mantiero D, Lucchini G, Longhese MP. 2006. The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep 7:212–218. doi: 10.1038/sj.embor.7400593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Puddu F, Oelschlaegel T, Guerini I, Geisler NJ, Niu H, Herzog M, Salguero I, Ochoa-Montano B, Vire E, Sung P, Adams DJ, Keane TM, Jackson SP. 2015. Synthetic viability genomic screening defines Sae2 function in DNA repair. EMBO J 34:1509–1522. doi: 10.15252/embj.201590973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Usui T, Ogawa H, Petrini JH. 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol Cell 7:1255–1266. doi: 10.1016/S1097-2765(01)00270-2. [DOI] [PubMed] [Google Scholar]
- 57.Ma L, Milman N, Nambiar M, Smith GR. 2015. Two separable functions of Ctp1 in the early steps of meiotic DNA double-strand break repair. Nucleic Acids Res 43:7349–7359. doi: 10.1093/nar/gkv644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jensen KL, Russell P. 2016. Ctp1-dependent clipping and resection of DNA double-strand breaks by Mre11 endonuclease complex are not genetically separable. Nucleic Acids Res 44:8241–8249. doi: 10.1093/nar/gkw557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reid RJ, Lisby M, Rothstein R. 2002. Cloning-free genome alterations in Saccharomyces cerevisiae using adaptamer-mediated PCR. Methods Enzymol 350:258–277. doi: 10.1016/S0076-6879(02)50968-X. [DOI] [PubMed] [Google Scholar]
- 60.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119–122. doi: 10.1016/0378-1119(92)90454-W. [DOI] [PubMed] [Google Scholar]
- 62.Spell RM, Jinks-Robertson S. 2004. Determination of mitotic recombination rates by fluctuation analysis in Saccharomyces cerevisiae. Methods Mol Biol 262:3–12. [DOI] [PubMed] [Google Scholar]
- 63.Adams A, Gottschling DE, Kaiser C. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 64.Munoz V, Serrano L. 1997. Development of the multiple sequence approximation within the AGADIR model of alpha-helix formation: comparison with Zimm-Bragg and Lifson-Roig formalisms. Biopolymers 41:495–509. doi:. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.