Abstract
Philadelphia chromosome-positive (Ph+) leukemia is characterized by reciprocal translocation between chromosomes 9 and 22. The resultant BCR/ABL fusion protein displays constitutive tyrosine kinase activity, leading to the induction of aberrant proliferation and neoplastic transformation. The bone marrow (BM) microenvironment is tumor-promoting, and contributes to disease recurrence in Ph+ leukemia. Activity in the BM microenvironment is mediated by several cellular compartments, extracellular matrix, various soluble factors including transforming growth factor beta 1 (TGF-β1), and the hypoxic conditions in the BM niche. TGF-β1 is released during bone remodeling and plays a role in maintaining leukemic stem cells, as well as being implicated in the epithelial-mesenchymal transition (EMT) process in most solid tumors. Although EMT is largely implicated in epithelial tumors, recent findings argue for an EMT-like process in leukemia as well. The surface receptor CD44 is involved in cell adhesion, cell migration, and homing of normal and malignant hematopoietic stem cells. Elevation of CD44 expression is considered a marker for a worse prognosis in most hematological malignancies. We explored the functions of Snail and Twist1 in Ph+ leukemia. We showed that ectopic expression of Snail and, to a lesser extent, Twist1, upregulates CD44 expression that is β-catenin-dependent. Moreover, the presence of Snail or Twist1 partially blocked phorbol 12-myristate 13-acetate-induced megakaryocyte differentiation, while that of Twist significantly altered imatinib-induced erythroid differentiation. Thus EMT modulators affected proliferation, CD44 gene expression and differentiation ability of Ph+ leukemia cells.
Introduction
Philadelphia chromosome-positive (Ph+) leukemia is characterized by the t(9;22) chromosome translocation that creates the BCR/ABL oncogene. This fusion protein displays constitutive tyrosine kinase activity, leading to the induction of aberrant proliferation and neoplastic transformation. The Ph+ chromosome is found in more than 95% of chronic myeloid leukemia (CML) and in Ph+ acute lymphoblastic leukemia. Activation of BCR/ABL increases proliferation, reduces susceptibility to a variety of proapoptotic stimuli—including growth factor deprivation—and leads to neoplastic transformation 1.
ABL kinase inhibitors (AKIs) are utilized for the treatment of Ph+ leukemia. The initial response is good 2-4 but unfortunately, the clinical efficacy of this treatment decreases continuously as the disease advances. Blast crisis (BC) CML or Ph+ acute lymphoblastic leukemia patients only benefit from AKI treatment temporarily, if at all 5. Moreover, despite the remarkable success of AKIs against Ph+ leukemia, these drugs do not seem to cure the disease. This seems to be due to their failure to reliably eliminate the Ph+ leukemia stem cells (LSCs) 6. Interestingly, an increasing number of reports demonstrate that LSCs of Ph+ leukemia are dependent on BCRABL protein and not on its kinase activity, explaining the AKIs' inability to eradicate LSCs and eliminate residual disease 7-9.
The bone marrow (BM) microenvironment plays a significant role in the etiology of Ph+ leukemia. In addition, cellular adhesion of Ph+ leukemia cells to stromal cells and extracellular components within the BM niche, as well as exposure to soluble factors such as growth factors and interleukins, contribute to residual disease.
The epithelial-mesenchymal transition (EMT) encompasses a series of events leading to acquisition of motile migratory properties. It has been shown that factors regulating the development of EMT play roles in tumor progression, including TGF-β-, Wnt-, and Notch-signaling pathways, as well as Snail1, Slug, Zeb1, Twist1, and others. Although the EMT has been studied in relation to epithelium-derived tumors, increasing evidence implicates EMT activators, especially Snai/Zeb families, in hematopoietic malignancies 10. Analysis of samples from CML patients during disease progression revealed upregulation of Twist1, which correlated with AKI drug resistance, without any detectable resistance mechanism. This argues for the potential involvement of Twist1 in CML resistance and disease progression 11. Moreover, Slug contributes to apoptosis resistance, prolonged survival, and imatinib resistance of CML progenitors 12. Long-term treatment with imatinib triggers a mesenchymal-like conversion of CML cells accompanied by increased aggressiveness and associated with increased EMT-like phenotypes, adhesion and invasion 13. Moreover, Slug overexpression has been reported to be essential for the homing of CML cells to the BM 14.
CD44 is a cell-surface receptor for hyaluronic acid, involved in cell adhesion, cell matrix interaction and cell migration, and functioning as a "BM homing receptor" by directing migration of human and mouse stem cells to the BM 15, 16. Moreover, altered CD44 expression functions as a marker for worse prognosis in most hematological malignancies; expression of particular isoforms of CD44 has been associated with malignant transformation and/or the acquisition of metastatic potential. CD44 has also been implicated in LSCs, and its expression increases in several types of leukemia. Furthermore, CD44 expression increases in mouse stem/progenitor cells expressing BCR/ABL and involved in regulating LSC homing and engraftment.
In this study, we investigated the function of ectopically expressed Snail and Twist1 in Ph+ leukemia cell lines and monitored changes in the expression levels of cell-surface markers involved in cell migration and BM homing. Our data showed that ectopic expression of Snail significantly upregulates CD44 in a β-catenin-dependent manner. Moreover, ectopic expression of Twist1 and Snail compromised the ability of phorbol 12-myristate 13-acetate (PMA) to induce megakaryocyte differentiation, and ectopic expression of Twist1 had a limited effect on imatinib-induced erythroid differentiation.
Materials and Methods
Cell lines and cell culture
Breast cancer cell lines MCF7 and MDA-MB-231, and Ph+ leukemia cell lines K562, MEG-01, SupB-15, BV-173 and murine mesenchymal cells (MS-5) were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) and cells were grown in RPMI 1640 (Sigma, Rehovot, Israel) containing 2 mM L-glutamine, 10% (v/v) fetal bovine serum (FBS), and 100 IU/ml penicillin and 100 μg/ml streptomycin (PenStrep; Biological Industries, Beit Haemek, Israel). The human embryonic kidney cell line 293T (HEK293T) was obtained from ATCC and maintained in Dulbecco's Modified Eagle Medium (DMEM; Sigma) supplemented with 10% (v/v) fetal calf serum (FCS), 2 mM L-glutamine, 1 mM sodium pyruvate, and 1% (w/v) PenStrep. All cell lines were grown at 37oC in a humidified atmosphere with 5% CO2.
Trypan blue exclusion assay
Cells were plated in 6-well plates (3 x 105 cell/well) containing 3 ml of complete medium. Cells viability were monitored on day 1,2,3 and 4 after cell plating by collecting samples from the cell cultures followed by staining with 0.25% (w/v) trypan blue solution (1:1 v/v), and counting using a hemocytometer.
Construction of K562 cells overexpressing Snail and Twist1 proteins
K562 cells were infected with retrovirus carrying either Snail-ER (Snail fused to ligand binding domain of estrogen receptor) (pWZL Blast Snail ER, Addgene plasmid #18798) or Twist1-ER constructs (pWZL Blast Twist ER, Addgene plasmid #18799). Both plasmids were a gift from Bob Weinberg 17. The ER included in both constructs carried the G525R mutation, which abrogates its binding to 17β-estradiol while retaining its full sensitivity to the synthetic ligand 4'-hydroxytamoxifen. Infected cells were selected for resistance to blasticidin and were validated for the presence of Snail or Twist by RT-PCR and immunoblotting using anti-ERα antibodies.
Adhesion of K562 cells to murine mesenchymal cells (MS-5)
Luciferase reporter plasmid (CMV-Luci) was transfected into K562 cells using electroporation. MEG-01 cells were used as a positive control in this experiment. Next day, luciferase levels were determined and cell aliquots were added to empty wells or to wells containing MS-5 cells. Two hours later, wells were washed 6 times with PBS and levels of remaining luciferase levels were determined. Signals obtained from wells containing MS-5 were considered as specific adhesion signals, while signals obtained from wells that devoted of MS-5 cells were considered as non-specific signal.
Western blot analysis
Immunoblotting was performed as previously described 18. Briefly, different cells were inoculated into flasks and 48 h later, cell lysates were prepared and subjected to electrophoresis on an 8% SDS-polyacrylamide gel and western immunoblotting with the appropriate antibodies. The antibodies used in this study were rabbit polyclonal β-catenin (Cell Signaling Technologies, Danvers, MA, USA), mouse monoclonal α-tubulin (Santa Cruz Biotechnology, Santa Cruz, CA, USA), rabbit polyclonal ER (Santa Cruz Biotechnology), rabbit monoclonal Snail (Cell Signaling Technologies), rabbit monoclonal phospho c-ABL (Tyr 245) (Cell Signaling Technologies) and mouse monoclonal c-ABL (24-11) (Santa Cruz Biotechnology).
Erythroid Differentiation of K562 cells
K562 cells were stimulated with imatinib to undergo erythroid differentiation 19 as previously described 20. Briefly, erythroid differentiation was determined by monitoring levels of γ-globin expression in treated cells by real-time PCR. K562 cells were plated in 6-well plates (2 x 105 cell/ml), followed by treatment with 0.4 μM imatinib for 6 days. Cells were collected and total RNA was isolated using Tri Reagent (Sigma). Levels of γ-globin were monitored by real-time PCR and normalized to levels of β-actin. Primer sequences and annealing temperatures are provided in Supplemental Table S1.
Megakaryocyte differentiation of K562 cells
K562 cells were stimulated with PMA to undergo megakaryocyte differentiation. Megakaryocyte differentiation was determined by monitoring levels of CD41 and CD61 in treated cells 19. K562 cells were plated in 6-well plates (2 x 10 5 cell/ml), and treated with 5 ng/ml PMA for 6 days. Cells were collected, washed with PBS and subjected to FACS analysis using CD61 PC7 (#IM3716, Beckman Coulter, Nyon, Switzerland) and CD41 fluorescein isothiocyanate (FITC) (#9011-0419, eBioscience, Emeryville, CA, USA).
Silencing of β-catenin in K562 cells
The β-catenin gene was silenced in K562 cells (K562, K562/Twist1 and K562/Snail) as previously described 21. Briefly, β-catenin shRNA plasmid DNA (pLKO.1 puro shRNA β-catenin Addgene plasmid # 18803) 22, pcMV-dR8.2 dvpr and pcMV-VSV-G (Addgene plasmid #8455 and #8454, respectively) 23 were gifts from Bob Weinberg. These were co-transfected into HEK239T cells at 1.5 x 105 cell/ml using Fugene 6 (Roche Applied Science, Penzberg, Germany) according to the manufacturer's instructions. The supernatant of the infected cells was collected 48 h post-transfection, used to infect the selected K562 cells lines, and puromycin-resistant clones were selected. Levels of β-catenin transcript and protein were monitored using real-time PCR and immunoblotting, respectively.
Cellular autophosphorylation of BCR/ABL
Cellular autophosphorylation assay was used to monitor levels of phosphorylated BCRABL in treated cells following the ELISA format. Briefly, cells from the various K562 cell lines (4 x 105 cell/ml) were treated with imatinib for 1 h at the specified concentrations. Cells were collected, washed twice with cold PBS, and lysed in 1X cell lysis buffer (Cell Signaling Technologies). Using clear flat-bottom immuno nonsterile 96-well plates (Thermo Fisher Scientific), wells were coated with 50 ng/well of mouse monoclonal c-ABL (24-11) and incubated overnight at room temperature (RT). The next day, wells were washed five times with PBS + 0.05% (w/v) Tween 20 and blocking buffer (3% w/v bovine serum albumin [BSA] in PBS) was added for 2 h at RT. Next, wells were washed five times, 100 μg/well of cell lysate was diluted in 1% (w/v) NP-40, 20 mM Tris (pH 8.0), 137 mM NaCl, 10% (w/v) glycerol, 2 mM EDTA and 1 mM activated sodium orthovanadate, and plates were incubated at 4oC overnight. After washing as before, 50 ng/well of PY20 antibody (anti-pTyr conjugated to HRP; Santa Cruz Biotechnology) diluted in 0.1% BSA, 20 mM Tris, 137 mM NaCl, 0.05% Tween 20, pH 7.2-7.4 was added for 2 h at RT, followed by five washes and incubation with SIGMAFAST OPD (Sigma-Aldrich, St. Louis, MO, USA) for 30 min. Optical density was determined at 450 nm.
Flow cytometric analysis
A variety of Ph+ leukemia cells and K562 cells carrying either Snail or Twist1 were re-suspended in PBS plus 0.5% BSA to a density of 1 × 107 cell/ml. Antibodies against CD34 PE (#IM1459u, Beckman Coulter), CD117 PCy7 (#IM3698, Beckman Coulter), CD45 KRO (#A96416, Beckman Coulter), CD61 PC7 (#IM3716, Beckman Coulter), CD41 FITC (#9011-0419, eBioscience), CD44 FITC (#F7083, DAKO, Denmark), CD44 FITC (#9011-0441, eBioscience) and CD184 APC (#FAB173A, R&D Systems, Minneapolis, MN, USA ) were titrated for use or added at the volumes suggested by the manufacturer and the mixture was incubated on ice for 40 min. After washing with PBS and 0.5% BSA, the labeled cells were analyzed on a Beckman Coulter Navios cytometer.
Immunofluorescence
K562 cells (1 x 106) were washed twice with 0.05% Tween 20 in PBS, fixed with 4% paraformaldehyde for 10 min, washed with 0.05% Tween 20 in PBS and incubated in saponin buffer (Cell Signaling Technologies) for 15 min. Cells were blocked with 5% BSA in PBS for 30 min. at RT followed by incubation with a 1:50 dilution of primary rabbit anti-Snail antibody (Cell Signaling Technologies) for 30 min. at RT. Next, secondary donkey anti-rabbit-FITC antibody (Santa Cruz Biotechnology) was added at a 1:50 dilution and incubated for 30 min. at 4oC. Fluorescence levels were measured by Becton Dickinson FACS caliber using FCS express De Novo software. To localize Snail, K562 cells were incubated with Hoechst solution at a 1:5000 dilution for 3 min, followed by three washes (0.05% Tween 20 in PBS), and were observed under a confocal microscope (Zeiss, LSM700) at 20X magnification. Pictures were analyzed by Zen software.
Semi-quantitative RT-PCR
RT-PCR analysis was performed as previously described 24. Total RNA was extracted from the cells using Tri Reagent (Sigma). Single-stranded cDNA was synthesized from the total RNA. Briefly, 1 μg RNA was preincubated with 1 μl oligo(dT)17 primer, and diethylpyrocarbonate (DEPC)-treated water was added to a total volume of 15 μl at 70oC for 10 min, then the mixture was rapidly chilled on ice. To the annealed primer/template, 2 µl AMV RT 5X reaction buffer, 2 µl dNTP (25 mM), 28 units of RNasin ribonuclease inhibitor, 30 units of AMV RT, and DEPC-treated water were added to a final volume of 10 µl. The reaction was incubated at 42oC for 60 min. The resulting cDNA was amplified with a PCR kit (Bioline, Taunton, MA, USA). The primer sets used for each gene are listed in Supplemental Table S1. A total of 35 cycles of amplification were performed with an initial incubation at 94oC for 2 min and a final extension at 72oC for 15 min; each cycle consisted of denaturation at 94oC for 30 s, annealing at 55-60oC for 30 s and an extension at 72oC for 2:30 min. To ensure the use of equal amounts of cDNA from each sample in the PCR, the aliquots of the reverse transcription products were used with primers for the housekeeping gene β-actin. A total 5 µl of the PCR products was analyzed by electrophoresis on a 1.5% agarose gel.
Quantitative real-time RT-PCR
Total RNA extraction and reverse transcription were performed as described for the semi-quantitative RT-PCR. The cDNA of the mRNA transcripts was amplified by real-time quantitative PCR in a 15-μl reaction mixture containing 0.2 pmol/µl forward/reverse primers and 1X SYBR GREEN reaction mix (Kappa Biosystems, Wilmington, MA, USA). The reaction was performed in a spectrofluorometric thermal cycler (Rotor-GeneTM 6000, Corbett Research, Mortlake, Australia), with preliminary denaturation for 10 min at 95oC to activate Taq DNA polymerase, followed by 55 cycles of denaturation at 95oC for 15 s, and annealing/extension at 55-60oC for 1 min (Supplemental Table S1). Different amounts of template were used in the same reaction to ensure linear amplification of PCR products. The β-actin gene was used as an internal control amplified in the same PCR assay. The primer sets were the same as those used in the semi-quantitative RT-PCR (Supplemental Table S1).
Statistical analysis
Statistical analysis was performed using Student's t-test with significance values set at *P < 0.01 or **P < 0.001.
Results
Expression levels of EMT modulators in Ph+ leukemia cell lines
Puissant et al. 13 described an EMT-like phenotype of Ph+ leukemia cells that was associated with increased adhesion and invasion. Moreover, EMT modulator (Slug) expression has been reported to be essential for the homing of Ph+ leukemia cells to the BM 14. We therefore monitored the expression levels of EMT modulators in Ph+ leukemia cell lines by RT-PCR. As a control, we monitored their expression levels in MDA-MB-231 (mesenchymal) and MCF7 (epithelial) breast cancer cells. As expected, EMT modulators were expressed at high levels in MDA-MB-231 cells and at very low levels in MCF7 cells (Figure 1). EMT modulators were also expressed in Ph+ leukemia cells lines at varying levels: Twist1 was expressed at low levels, and Zeb1 at high levels in all Ph+ leukemia cells; Snail was expressed at comparable levels in K562 and MEG-01 cells, but its expression was low in SupB15 and BV173 cells. Interestingly, Slug was expressed at high levels in MEG-01 but was undetectable in the other Ph+ leukemia cell lines.
Figure 1.
Expression levels of EMT modulators in Ph-positive leukemia cells: Expression levels of EMT modulators in Ph+ leukemia cells in comparison to MCF7 (epithelial) and MDA-MB-231 (mesenchymal) breast cancer cells were determined by RT-PCR.
Since the EMT modulators were expressed in Ph+ leukemia cells at various levels, and expression of some of them (Snail and Zeb1) was comparable to that found in breast cancer cells, it was reasonable to speculate that the presence of EMT modulators might also be of significance in Ph+ leukemia.
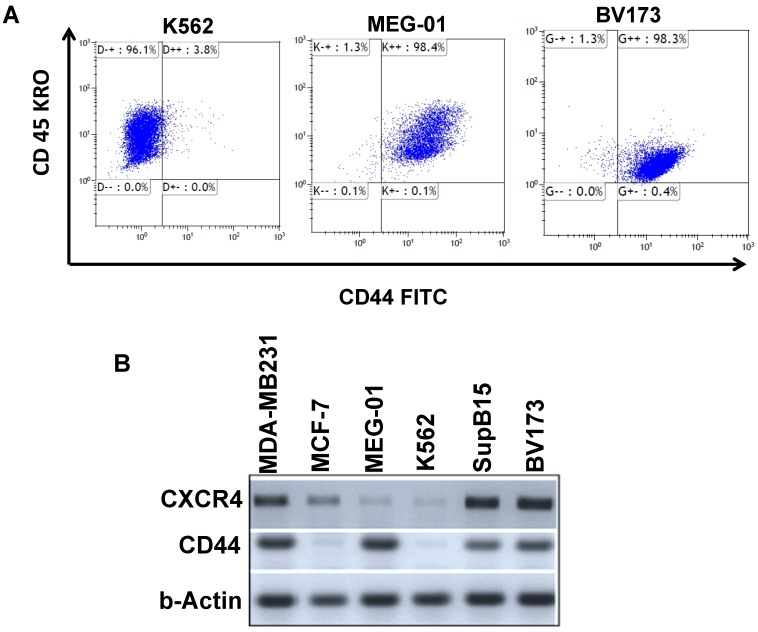
Expression of the cell-surface markers CD44 and CXCR4 in Ph+ leukemia cell lines
Ph+ LSC homing and engrafting in the BM contribute to residual disease. Homing and engrafting in Ph+ leukemia is mediated, in part, by CD44 and CXCR4 surface proteins which have been previously implicated in the etiology of Ph+ leukemia 25. We therefore monitored the expression levels of CD44 and CXCR4 (CD184) using FACS analysis and RT-PCR. Results, summarized in Table 1, demonstrated that CD44 and CXCR4 are expressed at very low levels in K562 cells (Figure 2A); in MEG-01 cells, their expression was significantly higher (Table 1, Figure 2, Supplemental Figure S1). Levels of CD44 were also high in Ph+ leukemia cell lines BV173 and SupB15 (Table 1). Interestingly, BV173 and SupB15 cells also expressed high levels of CXCR4 compared to the low levels in K562 and MEG-01 cell lines (Table 1, Supplemental Figure S2). RT-PCR analysis of CD44 and CXCR4 transcripts in the different cells (Figure 2B) was in agreement with the data presented in Table 1. Transcript levels of CD44 in the different Ph+ leukemia cells were also monitored, and CD44 levels were high in all cell lines tested except MCF7 and K562 (Figure 2B). On the other hand, transcript levels of CXCR4 were high in BV173, SupB15 and MDA-MB-231 cell lines, low in MEG-01 and K562 cell lines, and moderate in MCF 7 cell lines (Figure 2B).
Table 1.
Expression levels of CXCR4 and CD44 in Ph-positive leukemia cells: Levels of CXCR4 and CD44 in Ph-positive cells were determined using FACS analysis.
| Ph+ leukemia cell lines | % of positive cells for | |
|---|---|---|
| CD44 | CXCR4 | |
| K562 | 3.4 | 0.8 |
| MEG-01 | 99.6 | 3.3 |
| BV173 | 98.7 | 99.4 |
| SupB15 | 64.1 | 96.7 |
Figure 2.
Expression levels of CD44 and CXCR4 in Ph-positive leukemia cells: (A) FACS analysis of CD44 expression in CML cells. (B) RT-PCR analysis of CD44 and CXCR4 expression in Ph+ leukemia cell lines.
Ectopic expression of Twist1 and Snail in Ph+ leukemia cells
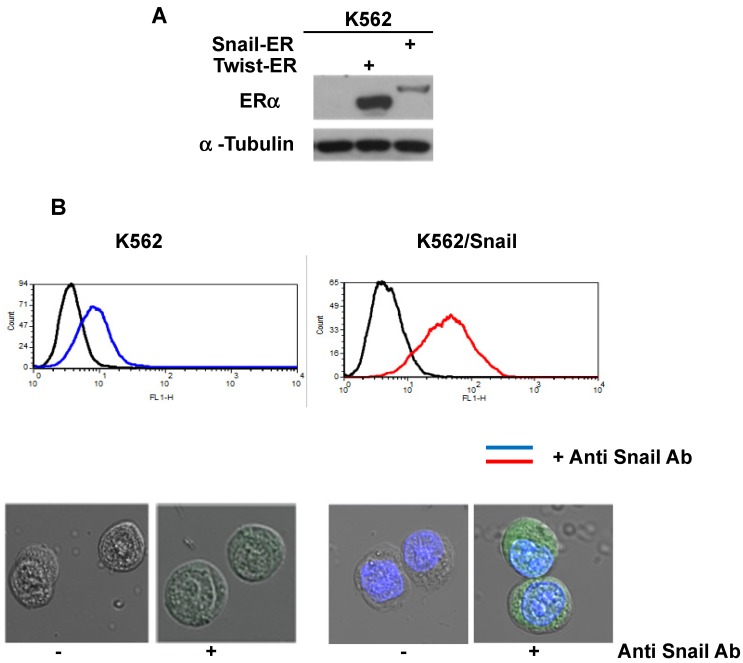
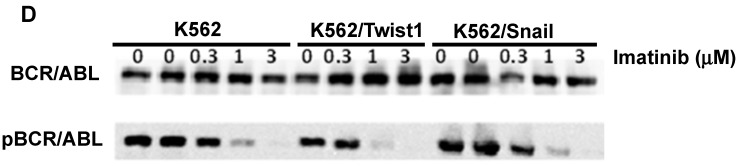
To explore the activity of EMT modulators in regulating CD44 and CXCR4 expression, we expressed them ectopically in Ph+ leukemia cell lines. For this study, we selected K562 cells which express low levels of both surface receptors (Table 1 and Figure 2) relative to other Ph+ leukemia cell lines. The expression vectors of Snail and Twist1 were introduced into K562 Ph+ leukemia cells and their expression was monitored (Figure 3). The EMT modulators used in this study lacked the endogenous 3' untranslated region of the appropriate genes to escape microRNA regulation 26. Expression was validated by RT-PCR (Supplemental Figure S1), immunoblotting (Figure 3A) and FACS (Figure 3B). Snail expression was localized to two compartments, the nucleus and the cytoplasm (Figure 3B). Next, we monitored the proliferation of K562 cells carrying Snail or Twist1 in comparison to the parental K562 cells. The presence of Snail and Twist1 conferred enhanced proliferation ability to K562 cells (Figure 3C), consistent with the finding of enhanced proliferation of BC CML cells 27. Moreover, there was no difference in the levels of BCR/ABL or phosphorylated BCR/ABL in K562 cells harboring either Snail or Twist1 (Figure 3D). Finally, the presence of Snail or Twist1 did not affect drug sensitivity to imatinib as determined by monitoring BCR/ABL auto-phosphorylation at different imatinib concentrations (Figure 3D).
Figure 3.
Ectopic expression of Snail and Twist1 in K562 cells: (A) Immunoblotting of cell lysates prepared from K562, K562/Snail and K562/Twist1 using anti-mouse ERα. (B) FACS analysis and confocal microscopy using anti-Snail antibody. (C) Proliferation of K562 cells ectopically expressing Snail or Twist1 using Trypan blue exclusion assay. (D) Immunoblotting with phosphor-ABL antibody to determine K562 cell sensitivity to imatinib.
Expression of cell-surface markers in Ph+ leukemia cells ectopically expressing Snail or Twist1
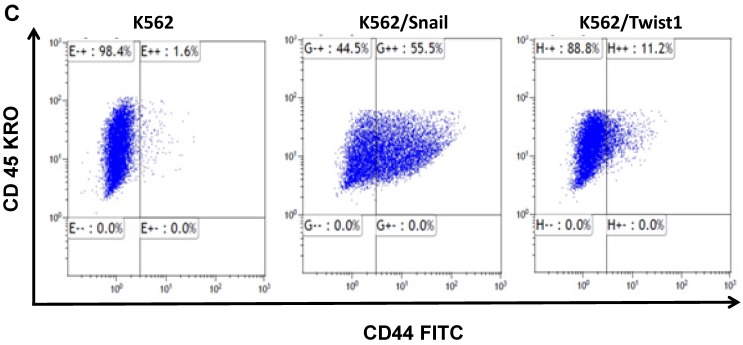
K562 cells ectopically expressing Snail or Twist1 were exposed to 20 nM 4-hydroxytamoxifen (4-OHT) for 2 weeks 17 to activate the two proteins, and levels of CD44 and CXCR4 were monitored by real-time PCR and FACS analyses. Our results, shown in Figure 4A and 4B, demonstrated low levels of CD44 expression in K562 cells relative to the significantly higher expression in SupB15 cell lines. However, the relative level of CD44 expression was significantly increased in K562 cells expressing activated Snail, by more than 10-fold (Figure 4A-4B). Interestingly, the presence of activated Twist1 in K562 cells caused only a marginal increase in CD44 levels. Moreover, FACS analysis of K562 cells harboring activated Snail or Twist1 for 4 weeks showed 55.5% and 11.2% of the cells, respectively, as positive for CD44 expression, compared to 1% in parental K562 cells (Figure 4C-4D). Interestingly, the percentage of CD44-positive K562 cells overexpressing Snail or Twist1 increased with duration of activation with 4-OHT. Supplemental Figure S3 shows increasing percentages of CD44-positive cells after 1, 2 and 4 weeks of activation.
Figure 4.
Snail and Twist1 stimulate CD44 expression in K562 cell lines: K562, K562/Twist1 and K562/Snail were treated with 4-OHT for 2 weeks and relative levels of CD44 (normalized to β-actin) were determined by real-time PCR (A). Quantitative and normalized data are shown (B). Levels of CD44 were also monitored by FACS analysis using CD45 and CD44 antibodies (C). (D) Percentage of CD44-positive cells. *, ** Significant difference at P < 0.01 and 0.001, respectively.
CXCR4 expression levels were also monitored in K562 cells and compared to those in the other Ph+ leukemia cell lines. Expression levels of CXCR4 were very low in K562 and MEG-01 cells compared to high expression levels in BV173, SubB15 and the control cells (MD-MB-231 and MCF 7) (Figure 2). Next, we tested expression levels in K562 cells harboring Snail or Twist1: the presence of Snail or Twist1 did not cause any significant change in CXCR4 expression (Table 2). Although CXCR4 levels were very low in K562 and MEG-01 cells, ectopic expression of Twist1 seemed to slightly upregulate its expression (Table 2).
Table 2.
Expression of CXCR4 in K562 ectopically expressing Twist1 and Snail: K562 cells were treated for 6-8 weeks with 4-OHT and levels of CXCR4 were determined using FACS analysis. Levels of CXCR4 were also monitored in MEG-01 cells.
| Cell | % of CXCR4-positive cells |
|---|---|
| K562 | 0.2 |
| K562/Snail | 0.4 |
| K562/Twist1 | 1.1 |
| MEG-01 | 0.4 |
Snail overexpression also stimulated the expression of other cell-surface receptors, such as CD34 (7.4% compared to 1.2% in parental K562 cells) and slightly reduced the percentage of CD117-positive cells (from 6.3 in the parental K562 cells to 3.3 in K562 cells ectopically expressing Snail). Twist1 overexpression had a different outcome: the percentages of CD34- and CD117-positive cells were 0.5 and 5.9, respectively, compared to 1.2 and 6.3, respectively, in the parental cell line. Interestingly, longer exposure to 4-OHT (about 6-8 weeks) resulted in accumulation of cells that were positive for the different surface markers (Table 3). Of special interest is that the presence of Snail was more effective than that of Twist1 in stimulating CD34 expression, whereas Twist1 was more effective at inducing CD117 expression (Table 3). Our findings for CD44 are consistent with a recent report showing enhanced homing to the BM of K562 cells ectopically expressing Slug protein 14, apparently due to upregulation of CD44. Taken together, ectopic expression of Snail, and to a lesser extent Twist1, enhanced the expression of CD44 and other surface markers that are associated with CML stemness (CD34 and CD117) and homing of human leukemia cells to the BM.
Table 3.
Expression of surface markers in K562 ectopically expressing Twist1 and Snail: K562 cells were treated for 6-8 weeks with 4-OHT and levels of different surface markers were determined using FACS analysis.
| CD Marker | K562 | K562/Snail | K562/Twist1 |
|---|---|---|---|
| CD44 | 1.6% | 55.5% | 11.2% |
| CD41 | 0.8% | 15.1% | 1.8% |
| CD34 | 0.9% | 22.1% | 16.4% |
| CD117 | 2.3% | 16.4% | 51.1% |
| CD61 | 2.8% | 28.2% | 3.6% |
Ectopic expression of Snail or Twist1 alters K562 cell line's ability to undergo erythroid and megakaryocyte differentiation
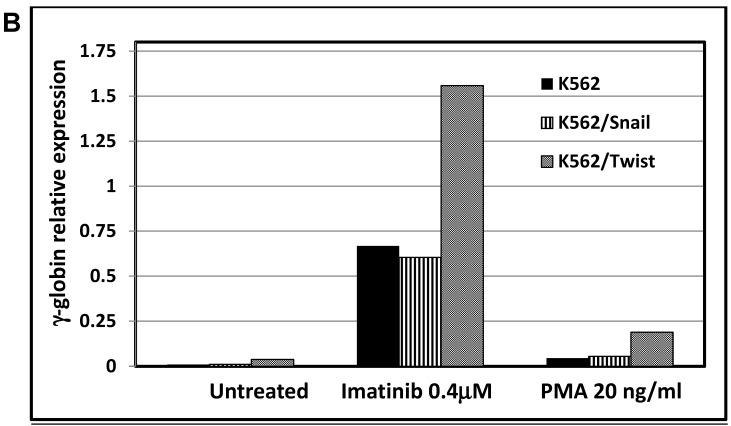
One of hallmarks of advanced stage of Ph+ leukemia and in particular BC CML is blockage of differentiation 27. We therefore determined whether ectopic expression of Snail or Twist1 would influence K562 cells' ability to undergo induced erythroid and megakaryocyte differentiations 19. We utilized hemin, butyric acid 28 and imatinib to induce erythrocyte differentiation and monitored γ-globin expression 29 as an erythroid differentiation marker by real-time PCR. Exposure to low levels of imatinib induced erythroid differentiation as reflected by an over 10-fold increase in γ-globin expression (Figure 5A and 5B). Similarly, imatinib induced erythroid differentiation in K562 cells ectopically expressing Snail to levels comparable to those of the parental K562 cells. Interestingly, K562 cells expressing Twist1 induced even higher levels of γ-globin compared to parental K562 cells (a 2- to 3-fold increase) and K562 cells ectopically expressing Snail.
Figure 5.
Effect of Snail and Twist1 on induction of K562 differentiation: (A) Real-time PCR showing relative levels of γ-globin and β-actin in K562 cells treated with imatinib for 6 days. (B) Quantitative presentation of relative γ-globin levels (normalized to β-actin) as a marker for erythrocyte differentiation. (C) Flow cytometry analysis following CD41/CD61 in K562 cells treated with 5 ng/ml PMA for 6 days compared to untreated cells. (D) Quantitative levels of CD41/CD61-positive cells after exposure to PMA (1 ng/ml) for 6 days. Experiments were repeated twice with comparable outcome.
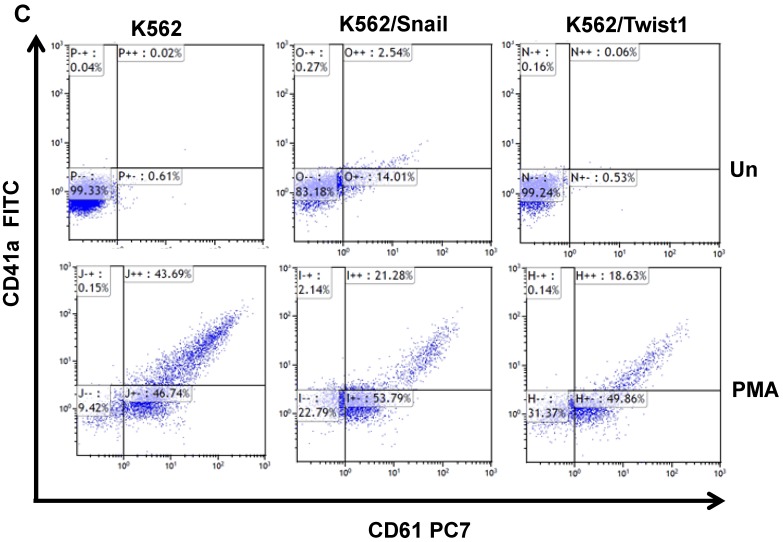
Next, we followed the ability of 1 ng/ml PMA to induce megakaryocyte differentiation by monitoring levels of the surface markers CD41/CD61 30 by FACS. Most K562 cells exposed to PMA for 6 days were positive for CD61 (46.7%) and CD41/CD61 (43.7%) (Figure 5C and 5D). Similarly, PMA induced the expression of megakaryocyte markers in K562 cells ectopically expressing Snail or Twist1, albeit at reduced efficacy. The presence of Snail or Twist1 compromised PMA's ability to induce megakaryocytic differentiation in K562 cells 19, leading to a significant reduction in the percentage of cells that were CD61- or CD41-positive to 21.28 and 18.63%, respectively, compared to parental K562 cells (43.7%). This suggested that the presence of Snail or Twist1 partially blocks megakaryocyte differentiation.
Ectopic expression of Twist1 and Snail enhance adhesion to murine mesenchymal cells (MS-5)
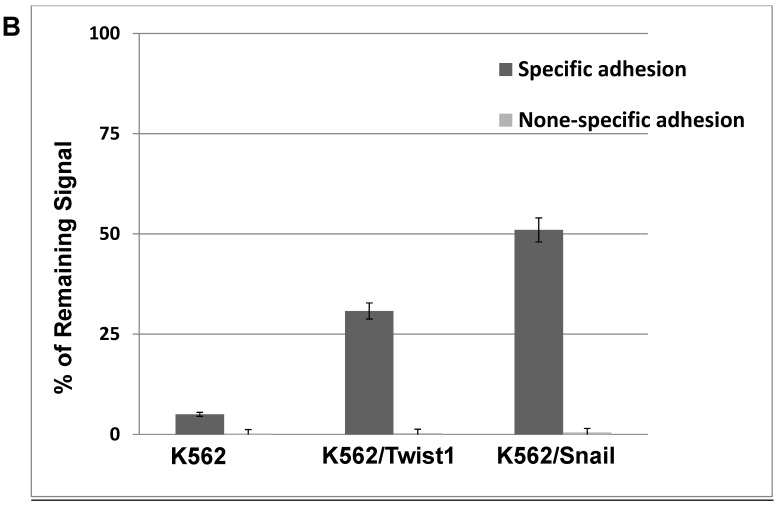
Our previous results showed significant impact of ectopic expression of Twist1 and Snail on the expression of surface molecules that in part mediate cell adhesion. Therefore, cell adhesion of K562 cells to murine mesenchymal cells (MS-5) was investigated. Results shown in Figure 6 illustrated that adhesion ability of MG-01 to MS-5 is greater compared to K562 cells (Figure 6A). However, ectopic expression of Snail or Twist1 significantly augmented adhesion ability of K562 cells to mesenchymal cells (MS-5) by causing the increase specific adhesion to MS-5 to 30.75% and 50.97% in the presence of Twist1 and Snail, respectively, compared to 5% in the parental K562 cells (Figure 6B). This argues that presence of Snail/Twist1 stimulated the adhesion ability of K562 cells that mediated by inducing the expression of CD44 and probably other adhesion molecules.
Figure 6.
Ectopic expression of Snail and Twist1 enhance adhesion to mesenchymal cells: (A) K562 and MEG-01 were transfected with Luciferase reporter plasmid and equal amounts of transfected cells were added to MS-5 pre-coated wells or empty wells. Wells were washed with PBS and levels of remaining luciferase levels were determined. (B) Comparing specific adhesion ability of transfected K562, K562/Twist1 and K562/Snail cells.
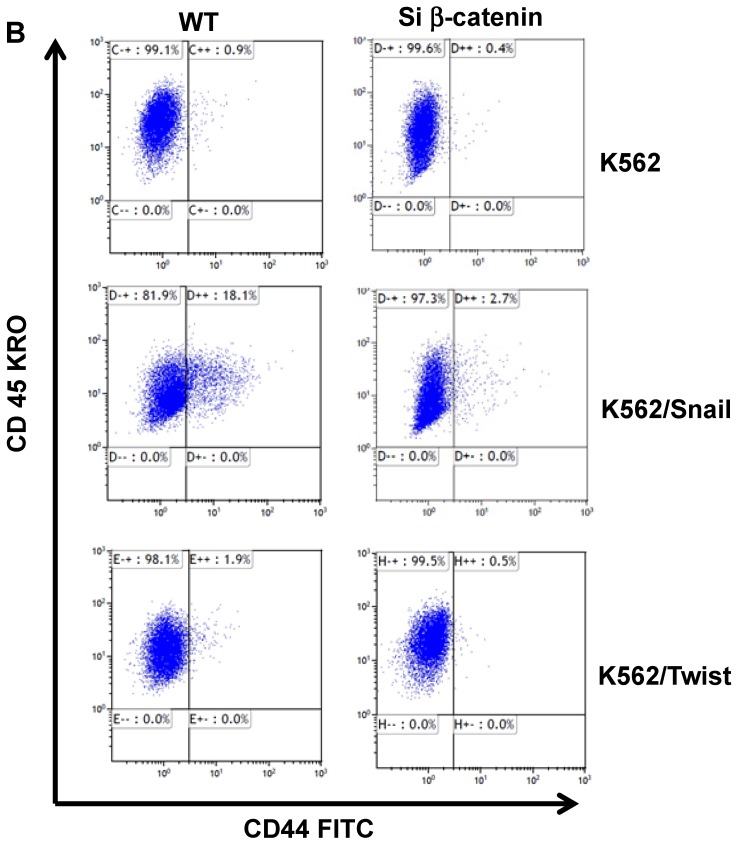
Snail-dependent upregulation of CD44 is β-catenin-dependent
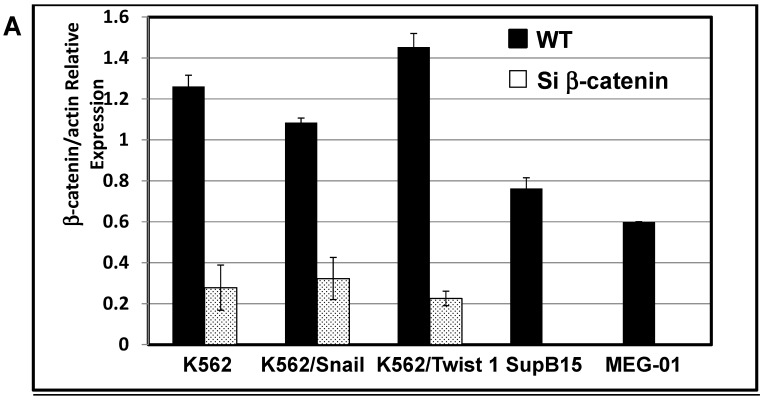
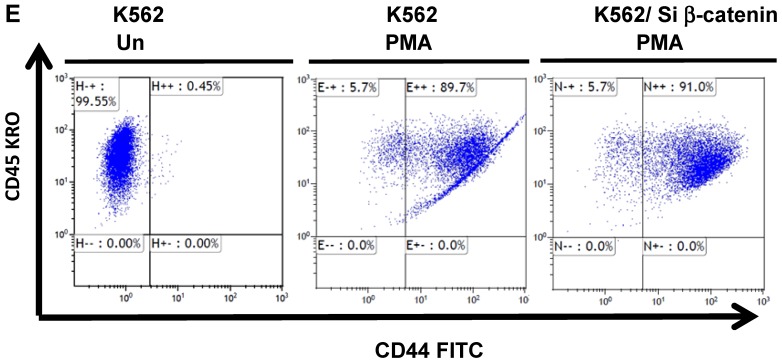
The protein β-catenin has been implicated in the regulation of CD44 expression 31. We therefore explored the cross-talk between β-catenin and Snail in the observed Snail-dependent upregulation of CD44. We silenced β-catenin in K562, K562/Snail and K562/Twist1 cells. Our results showed that in all silenced β-catenin K562 cells, there was a significant reduction in the levels of β-catenin transcript (Figure 7A). Silencing of β-catenin significantly abrogated Snail's ability to upregulate CD44, arguing that Snail-mediated upregulation of CD44 is β-catenin-dependent (Figure 7B and 7C, Supplemental Figure S4). Previous studies have shown that CD44 is also upregulated by PMA in monocytes 32. Thus, we tested whether PMA-induced CD44 is also regulated by β-catenin. Results shown in Figure 7D demonstrated that in K562 cells, exposure to PMA is sufficient to significantly upregulate levels of CD44, with about 90% of the cells being positive for CD44 compared to 0.45% in the absence of PMA. However, silencing of β-catenin in K562 cells had no effect on PMA-induced CD44 expression, in contrast to the Snail-induced CD44 expression. Moreover, we monitored levels of a variety of surface markers in response to silencing of β-catenin (Table 4).
Figure 7.
Effect of β-catenin on Snail-dependent CD44 expression. (A) Real-time PCR of β-catenin levels in wild type (WT) and β-catenin-silenced K562, K562/Snail and K562/Twist cell lines treated with 4-OHT for 2 weeks. (B) Levels of CD44 expression in K562 cells containing WT and silenced (Si) β-catenin as determined by FACS. (C) Quantitative representation of CD44 levels, as determined by FACS, in K562 cells containing WT β-catenin and Si β-catenin. (D) Quantitative representation of CD44 levels, as determined by real-time PCR, in K562 cells containing WT β-catenin and Si β-catenin. (E) FACS determination of expression levels of CD44 in PMA-induced WT and Si β-catenin K562 cells. (F) Quantitative representation of CD44 levels, as determined by FACS, of PMA-induced K562 cells in comparison to K562 Si β-catenin cells. Experiments in part E were performed twice with comparable outcomes, and the rest of the experiments in this figure were performed three times with comparable outcomes. *Significant difference at P < 0.01.
Table 4.
Effect of β-catenin in surface cell markers in K562 ectopically expressing Twist1 and Snail: K562 cells were incubated with 4-OHT for 2 weeks and expression levels of a variety of cluster of differentiations (CDs) were monitored by FACS. The experiment was performed twice with comparable outcomes.
| Cell | β-catenin | % of cells positive for | |||
|---|---|---|---|---|---|
| CD61 | CD41 | CD34 | CD44 | ||
| K562 | WT | 1.2 | 0 | 0.2 | 0.5 |
| Silenced | 5.3 | 0.1 | 1.5 | 5.3 | |
| K562/Snail | WT | 16.8 | 0.6 | 6.8 | 18.1 |
| Silenced | 0.8 | 0.1 | 0.2 | 0.5 | |
| K562/Twist1 | WT | 0.5 | 0.2 | 0.2 | 1.9 |
| Silenced | 0.8 | 0.1 | 0.1 | 0.5 | |
Results presented in Table 4 show that silencing of β-catenin reduced Snail-induced expression of CD34, CD41 and CD61. In contrast, silencing of β-catenin in K562 cells harboring Twist1 exhibited only a marginal and nonsignificant effect on the expression levels of CD34, CD41 and CD61. However, silencing of β-catenin in K562 cells caused slight (about 4-fold) upregulation of CD34, CD41, CD44 and CD61.
Discussion
Expression of EMT modulators 33 such as Snail, Slug, and Twist1 has been implicated in the induction of mesenchymal and stemness markers in epithelial cells 34 17. Moreover, there are more and more reports demonstrating the ability of EMT modulators such as Snail to increase leukemia formation and radioprotection 35. Others have implicated the Zeb2 EMT modulator in the differentiation of human stem cells and hematopoietic progenitor cells 10. To elucidate potential signaling alterations in Ph+ leukemia cells, we monitored expression levels of the EMT modulators Snail, Twist1, Slug and Zeb1 in Ph+ leukemia. Although their expression levels were correlated with the epithelial cells' transition to a mesenchymal phenotype, a few EMT modulators were also expressed in Ph+ leukemia cells at varying levels. Snail and Zeb1 were present in all Ph+ leukemia cells at comparable levels. Slug transcript was present at significant levels in MEG-01. Our hypothesis argues for functional significance of the EMT modulators in Ph+ leukemia and their possible involvement in mediating the EMT-like phenotype previously reported in leukemia cells, including Ph+ leukemia cells 13. Moreover, the EMT-like process might be involved in the development of residual disease in Ph+ leukemia.
Cell-surface receptors, and in particular CD44, have been implicated as key regulators of homing and engraftment of cancer cells to the BM stroma. Moreover, CD44 expression has been shown to increase in several types of leukemia as well as in K562 cells, and its level seems to influence proliferation and survival 36. Thus, we monitored CD44 expression levels in cell lines of different types of Ph+ leukemia by RT-PCR and FACS analysis (Table 1 and Figure 2). SupB15, BV173 and MEG-01 cell lines expressed high levels of CD44, in contrast to the K562 cell line. Our data are in agreement with results obtained by Puissant et al. 13 demonstrating that long-time exposure of CML cells to imatinib causes a mesenchymal-like conversion accompanied by increased expression of mesenchymal markers such as Snail, Osteonectin, TBX3 and β-catenin, as well as CD44.
A variety of soluble factors, and in particular TGF-β1, might take part in stimulating this EMT-like phenotype. TGF-β1 has been implicated as a regulator of EMT by upregulating levels of EMT modulators such as Snail, Twist1 and others 37. Thus, we explored the possibility that EMT modulators such as Snail and Twist1 might enhance CD44 expression in Ph+ leukemia cell lines. The K562 cell line was selected for this study because it contains minimal amounts of CD44. Our data demonstrated that ectopic expression of Snail, but not Twist1, significantly stimulates expression of CD44 as measured by real-time PCR and FACS analysis (Figure 4). In contrast, neither Snail nor Twist1 were effective at stimulating CXCR4 expression in K562 cells (Table 2).
One of hallmarks of Ph+ leukemia is blockage of differentiation 27. Thus, we explored the ability of PMA and imatinib to induce megakaryocyte and erythroid differentiation, respectively. Stimulation of γ-globin expression was used as a marker for erythroid differentiation 29. Imatinib was effective at inducing γ-globin expression in K562 as well as K562/Snail cells (Figure 5) to comparable levels. However, ectopic expression of Twist1 significantly induced γ-globin expression to 2- to 3-fold the levels obtained from K562 and K562/Snail cells. Moreover, ectopic expression of Twist1 in K562 cells compromised PMA's ability to induce megakaryocyte differentiation by about 50% compared to the parental K562 cells. Taken together, our data show that the presence of Twist1 or Snail blocks K562 cells' ability to undergo megakaryocyte differentiation, which might explain the observed phenomenon of differentiation blockage in advanced stages of CML cells, which might be the cells that undergo the EMT-like transition.
Although Ph+ leukemia cells that ectopically expressed Snail did not undergo a true EMT, they displayed an EMT-like phenotype 13, including enhanced expression of stem cell markers such as CD34 and CD117 38. An increased percentage of Ph+ leukemia cells that are positive for CD34 and also express c-kit (CD117) has been reported in non-responders to therapy and has been associated with an unfavorable outcome 39.
The Wnt/β-catenin pathway has been implicated in controlling survival, self-renewal, and differentiation of human stem cells and hematopoietic progenitor cells 40. Specifically, granulocyte-macrophage progenitors from CML patients and BC cells from patients who are resistant to therapy display activated Wnt signaling 41. To explore the cross-talk between Snail and β-catenin in our model system, we silenced β-catenin in the K562 cell lines and monitored the effect on CD44 expression. CD44 expression in K562/Snail cells was β-catenin-dependent (Figure 7). Interestingly, although MEG-01 expressed endogenous CD44 at high levels (Figure 2 and Table 1), silencing of β-catenin in this cell line (data not shown) did not cause any reduction in CD44 levels, arguing that expression of CD44 in MEG-01 is independent of β-catenin. A variety of signaling pathways have been reported to regulate CD44 expression 32. Moreover, PMA is well known to stimulate CD44 expression in a NF-κB-dependent manner 32. Thus, we investigated the ability of PMA to stimulate CD44 expression (Figure 7E-F) and found that PMA-induced CD44 expression in K562 cells is independent of β-catenin.
Taken together, Ph+ leukemia cells might acquire a mesenchymal-like phenotype in response to exposure to targeted therapy and/or growth factors such as TGF-β1 mediated by upregulation of EMT modulators. Conversion of Ph+ leukemia cells to a mesenchymal-like phenotype will accompanied by the upregulation of stemness factors such as CD34/CD117 and others, in addition to CD44, which also mediates BM homing, increased adhesion to stroma cells and engrafting and thus contributes to Ph+ leukemia residual disease.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
We thank Bob Weinberg for the gifts of plasmids pLKO.1 puro shRNA β-catenin [22], pcMV-dR8.2 dvpr and pcMV-VSV-G [23], pWZL Blast Snail ER and pWZL Blast Twist ER [17].
References
- 1.Faderl S, Talpaz M, Estrov Z, O'Brien S, Kurzrock R, Kantarjian HM. The biology of chronic myeloid leukemia. N Engl J Med. 1999;341:164–172. doi: 10.1056/NEJM199907153410306. [DOI] [PubMed] [Google Scholar]
- 2.Ottmann OG, Hoelzer D. The ABL tyrosine kinase inhibitor STI571 (Glivec) in Philadelphia positive acute lymphoblastic leukemia - promises, pitfalls and possibilities. Hematol J. 2002;3:2–6. doi: 10.1038/sj.thj.6200151. [DOI] [PubMed] [Google Scholar]
- 3.Radich JP, Dai H, Mao M, Oehler V, Schelter J, Druker B. et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2006;103:2794–2799. doi: 10.1073/pnas.0510423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talpaz M, Silver RT, Druker BJ, Goldman JM, Gambacorti-Passerini C, Guilhot F. et al. Imatinib induces durable hematologic and cytogenetic responses in patients with accelerated phase chronic myeloid leukemia: results of a phase 2 study. Blood. 2002;99:1928–1937. doi: 10.1182/blood.v99.6.1928. [DOI] [PubMed] [Google Scholar]
- 5.Gambacorti-Passerini CB, Gunby RH, Piazza R, Galietta A, Rostagno R, Scapozza L. Molecular mechanisms of resistance to imatinib in Philadelphia-chromosome-positive leukaemias. Lancet Oncol. 2003;4:75–85. doi: 10.1016/s1470-2045(03)00979-3. [DOI] [PubMed] [Google Scholar]
- 6.Graham SM, Jorgensen HG, Allan E, Pearson C, Alcorn MJ, Richmond L. et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99:319–325. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- 7.Chu S, Holtz M, Gupta M, Bhatia R. BCR/ABL kinase inhibition by imatinib mesylate enhances MAP kinase activity in chronic myelogenous leukemia CD34+ cells. Blood. 2004;103:3167–3174. doi: 10.1182/blood-2003-04-1271. [DOI] [PubMed] [Google Scholar]
- 8.Copland M, Hamilton A, Elrick LJ, Baird JW, Allan EK, Jordanides N. et al. Dasatinib (BMS-354825) targets an earlier progenitor population than imatinib in primary CML but does not eliminate the quiescent fraction. Blood. 2006;107:4532–4539. doi: 10.1182/blood-2005-07-2947. [DOI] [PubMed] [Google Scholar]
- 9.Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goossens S, Janzen V, Bartunkova S, Yokomizo T, Drogat B, Crisan M. et al. The EMT regulator Zeb2/Sip1 is essential for murine embryonic hematopoietic stem/progenitor cell differentiation and mobilization. Blood. 2011;117:5620–5630. doi: 10.1182/blood-2010-08-300236. [DOI] [PubMed] [Google Scholar]
- 11.Cosset E, Hamdan G, Jeanpierre S, Voeltzel T, Sagorny K, Hayette S. et al. Deregulation of TWIST-1 in the CD34+ compartment represents a novel prognostic factor in chronic myeloid leukemia. Blood. 2011;117:1673–1676. doi: 10.1182/blood-2009-11-254680. [DOI] [PubMed] [Google Scholar]
- 12.Mancini M, Petta S, Iacobucci I, Salvestrini V, Barbieri E, Santucci MA. Zinc-finger transcription factor slug contributes to the survival advantage of chronic myeloid leukemia cells. Cell Signal. 2010;22:1247–1253. doi: 10.1016/j.cellsig.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Puissant A, Dufies M, Fenouille N, Ben Sahra I, Jacquel A, Robert G. et al. Imatinib triggers mesenchymal-like conversion of CML cells associated with increased aggressiveness. J Mol Cell Biol. 2012;4:207–220. doi: 10.1093/jmcb/mjs010. [DOI] [PubMed] [Google Scholar]
- 14.Tanno B, Sesti F, Cesi V, Bossi G, Ferrari-Amorotti G, Bussolari R. et al. Expression of Slug is regulated by c-Myb and is required for invasion and bone marrow homing of cancer cells of different origin. J Biol Chem. 2010;285:29434–29445. doi: 10.1074/jbc.M109.089045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krause DS, Lazarides K, von Andrian UH, Van Etten RA. Requirement for CD44 in homing and engraftment of BCR-ABL-expressing leukemic stem cells. Nat Med. 2006;12:1175–1180. doi: 10.1038/nm1489. [DOI] [PubMed] [Google Scholar]
- 16.Greiner J, Ringhoffer M, Taniguchi M, Schmitt A, Kirchner D, Krahn G. et al. Receptor for hyaluronan acid-mediated motility (RHAMM) is a new immunogenic leukemia-associated antigen in acute and chronic myeloid leukemia. Exp Hematol. 2002;30:1029–1035. doi: 10.1016/s0301-472x(02)00874-3. [DOI] [PubMed] [Google Scholar]
- 17.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khateb M, Ruimi N, Khamisie H, Najajreh Y, Mian A, Metodieva A. et al. Overcoming Bcr-Abl T315I mutation by combination of GNF-2 and ATP competitors in an Abl-independent mechanism. BMC Cancer. 2012;12:563–573. doi: 10.1186/1471-2407-12-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huo XF, Yu J, Peng H, Du ZW, Liu XL, Ma YN. et al. Differential expression changes in K562 cells during the hemin-induced erythroid differentiation and the phorbol myristate acetate (PMA)-induced megakaryocytic differentiation. Mol Cell Biochem. 2006;292:155–167. doi: 10.1007/s11010-006-9229-0. [DOI] [PubMed] [Google Scholar]
- 20.Yassin M, Wasser SP, Mahajna J. Substances from the medicinal mushroom Daedalea gibbosa inhibit kinase activity of native and T315I mutated Bcr-Abl. Int J Oncol. 2008;32:1197–1204. doi: 10.3892/ijo_32_6_1197. [DOI] [PubMed] [Google Scholar]
- 21.Ratzon E, Najajreh Y, Salem R, Khamaisie H, Ruthardt M, Mahajna J. Platinum (IV)-fatty acid conjugates overcome inherently and acquired Cisplatin resistant cancer cell lines: an in-vitro study. BMC Cancer. 2016;16:140–151. doi: 10.1186/s12885-016-2182-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 23.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS. et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruimi N, Petrova RD, Agbaria R, Sussan S, Wasser SP, Reznick AZ. et al. Inhibition of TNFalpha-induced iNOS expression in HSV-tk transduced 9L glioblastoma cell lines by Marasmius oreades substances through NF-kappaB- and MAPK-dependent mechanisms. Mol Biol Rep. 2010;37:3801–3812. doi: 10.1007/s11033-010-0035-0. [DOI] [PubMed] [Google Scholar]
- 25.Hu Y, Li S. Survival regulation of leukemia stem cells. Cell Mol Life Sci. 2016;73:1039–1050. doi: 10.1007/s00018-015-2108-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abba ML, Patil N, Leupold JH, Allgayer H. MicroRNA Regulation of Epithelial to Mesenchymal Transition. J Clin Med. 2016;5:8–28. doi: 10.3390/jcm5010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103:4010–4022. doi: 10.1182/blood-2003-12-4111. [DOI] [PubMed] [Google Scholar]
- 28.Chenais B, Molle I, Trentesaux C, Jeannesson P. Time-course of butyric acid-induced differentiation in human K562 leukemic cell line: rapid increase in gamma-globin, porphobilinogen deaminase and NF-E2 mRNA levels. Leukemia. 1997;11:1575–1579. doi: 10.1038/sj.leu.2400755. [DOI] [PubMed] [Google Scholar]
- 29.Guerrasio A, Vainchenker W, Breton-Gorius J, Testa U, Rosa R, Thomopoulos P. et al. Embryonic and fetal hemoglobin synthesis in K562 cell line. Blood Cells. 1981;7:165–176. [PubMed] [Google Scholar]
- 30.Dorsey JF, Cunnick JM, Mane SM, Wu J. Regulation of the Erk2-Elk1 signaling pathway and megakaryocytic differentiation of Bcr-Abl(+) K562 leukemic cells by Gab2. Blood. 2002;99:1388–1397. doi: 10.1182/blood.v99.4.1388. [DOI] [PubMed] [Google Scholar]
- 31.Siapati EK, Papadaki M, Kozaou Z, Rouka E, Michali E, Savvidou I. et al. Proliferation and bone marrow engraftment of AML blasts is dependent on beta-catenin signalling. Br J Haematol. 2011;152:164–174. doi: 10.1111/j.1365-2141.2010.08471.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhang G, Zhang H, Liu Y, He Y, Wang W, Du Y. et al. CD44 clustering is involved in monocyte differentiation. Acta Biochim Biophys Sin (Shanghai) 2014;46:540–547. doi: 10.1093/abbs/gmu042. [DOI] [PubMed] [Google Scholar]
- 33.Knight RD, Shimeld SM. Identification of conserved C2H2 zinc-finger gene families in the Bilateria. Genome Biol. 2001;2:RESEARCH0016. doi: 10.1186/gb-2001-2-5-research0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG. et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Bio. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 35.Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang G, Zhang H, Wang J, Zhang Y, Xu H, Wang C. et al. CD44 targets Wnt/beta-catenin pathway to mediate the proliferation of K562 cells. Cancer Cell Int. 2013;13:117–130. doi: 10.1186/1475-2867-13-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peinado H, Quintanilla M, Cano A. Transforming growth factor beta-1 induces snail transcription factor in epithelial cell lines: mechanisms for epithelial mesenchymal transitions. J Biol Chem. 2003;278:21113–21123. doi: 10.1074/jbc.M211304200. [DOI] [PubMed] [Google Scholar]
- 38.Jorgensen HG, Holyoake TL. Characterization of cancer stem cells in chronic myeloid leukaemia. Biochem Soc Trans. 2007;35:1347–1351. doi: 10.1042/BST0351347. [DOI] [PubMed] [Google Scholar]
- 39.Dybko J, Haus O, Jazwiec B, Urbaniak J, Wozniak M, Kaczmar-Dybko A. et al. CD117 (c-kit) expression on CD34+ cells participates in the cytogenetic response to imatinib in patients with chronic myeloid leukemia in the first chronic phase. Acta Haematol. 2014;132:166–171. doi: 10.1159/000357373. [DOI] [PubMed] [Google Scholar]
- 40.Hu Y, Chen Y, Douglas L, Li S. beta-Catenin is essential for survival of leukemic stem cells insensitive to kinase inhibition in mice with BCR-ABL-induced chronic myeloid leukemia. Leukemia. 2009;23:109–116. doi: 10.1038/leu.2008.262. [DOI] [PubMed] [Google Scholar]
- 41.Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL. et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.