Abstract
Single-step immunoassays that can be performed directly in solution are ideally suited for point-of-care diagnostics. Our group recently developed a new platform of bioluminescent sensor proteins (LUMABS; LUMinescent AntiBody Sensor) that allow antibody detection in blood plasma. Thus far, LUMABS has been limited to the detection of antibodies recognizing natural peptide epitopes. Here, we report the development of semisynthetic LUMABS sensors that recognize nonpeptide epitopes. The non-natural amino acid para-azidophenylalanine was introduced at the position of the original antibody-recognition sites as a chemical handle to enable site-specific conjugation of synthetic epitope molecules coupled to a dibenzocylcooctyne moiety via strain-promoted click chemistry. The approach was successfully demonstrated by developing semisynthetic LUMABS sensors for antibodies targeting the small molecules dinitrophenol and creatinine (DNP-LUMABS and CR-LUMABS) with affinities of 5.8 pM and 1.3 nM, respectively. An important application of these semisynthetic LUMABS is the detection of small molecules using a competitive assay format, which is demonstrated here for the detection of creatinine. Using a preassembled complex of CR-LUMABS and an anti-creatinine antibody, the detection of high micromolar concentrations of creatinine was possible both in buffer and in 1:1 diluted blood plasma. The use of semisynthetic LUMABS sensors significantly expands the range of antibody targets and enables the application of LUMABS sensors for the ratiometric bioluminescent detection of small molecules using a competitive immunoassay format.
Keywords: BRET, NanoLuc, LUMABS, creatinine, sensor, protein engineering
The detection of antibody–antigen interactions is the basis for many bioanalytical and diagnostic assays.1 Both antibodies and antigens are important biomarkers and the specificity of their interaction has been used to develop highly specific detection strategies, either using antigens to detect antibodies or vice versa. Antibodies are often detected using heterogeneous assays such as the enzyme-linked immunosorbent assay (ELISA), which involves immobilization of antigen and multiple washing and incubation steps.2 Small molecules can be similarly detected using a competitive immunoassay. Although these traditional immunoassays are highly modular and sensitive, their multistep nature limits their integration with point-of-care diagnostic strategies. Ideally, a point-of-care test relies on a one-step assay that requires minimal expertise on the part of the end user. Most successful strategies for small molecule sensing at the point-of-care rely on the detection of analyte-specific enzymatic reactions, such as the electrochemical detection of glucose using glucose oxidase.3−5 However, this strategy is limited in scope as it relies on the availability of an enzyme to catalyze the formation of a detectable product in an analyte-specific manner.
Sensor proteins based on Bioluminescence Resonance Energy Transfer (BRET) are attractive for point of care diagnostics because they do not require external illumination and therefore allow detection of analytes directly in complex media such as blood serum. An early example is the work of Trowell and co-workers who developed BRET-based sensors based on G-protein coupled receptors that allow detection of diacetyl with fM affinity.6 Recently, two new bioluminescent sensor platforms, LUCIDs and LUMABS, have been reported that allow direct in-solution detection of small molecules and antibodies, respectively.7−9 Both sensors are designed such that binding of analyte induces a conformational change in the sensor, which modulates the energy transfer efficiency (BRET) between NanoLuc, a bright and stable luciferase emitting blue light, and an acceptor fluorophore. The LUCID sensors developed by Johnsson and co-workers are based on the intramolecular binding of a ligand(-analogue) to an analyte-specific receptor domain.7,10 The intramolecular interaction can be disrupted in a competitive manner if the small molecule of interest is present at sufficiently high concentrations, leading to a conformational change in the protein that is accompanied by a decrease in BRET efficiency between NanoLuc and a synthetic fluorophore. The LUMABS sensors developed by our group take advantage of the bivalent architecture of antibodies to modulate BRET efficiency between NanoLuc and an acceptor fluorescent domain.9 In this sensor design, NanoLuc is fused to the green fluorescent protein mNeonGreen via a semiflexible linker domain. The linker domain contains an antibody-specific epitope at both termini. In the absence of antibody, NanoLuc and mNeonGreen are kept in close proximity through helper domain interactions, which allow efficient BRET. Addition of antibody disrupts these helper interactions because the bivalent antibody–epitope interaction forces the linker into a stretched conformation. The decrease in BRET efficiency results in a color change from green to blue, which could be detected using a conventional mobile phone camera.9
An attractive feature of the modular architecture of LUMABS is that the sensor’s target specificity can be changed by simply replacing the epitopes in the semiflexible linker. Thus far, the epitopes that have been incorporated in LUMABS have been restricted to linear peptide epitopes. However, the immune system develops antibodies against a broad spectrum of molecular targets, including discontinuous conformational epitopes, post-translational modifications, sugars, ribonucleic acids, xenobiotics, and small-molecule haptens.11−16 The possibility to incorporate these more structurally diverse epitopes into LUMABS would significantly extend the system’s applicability.
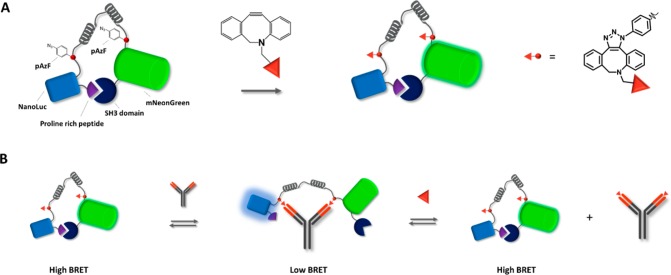
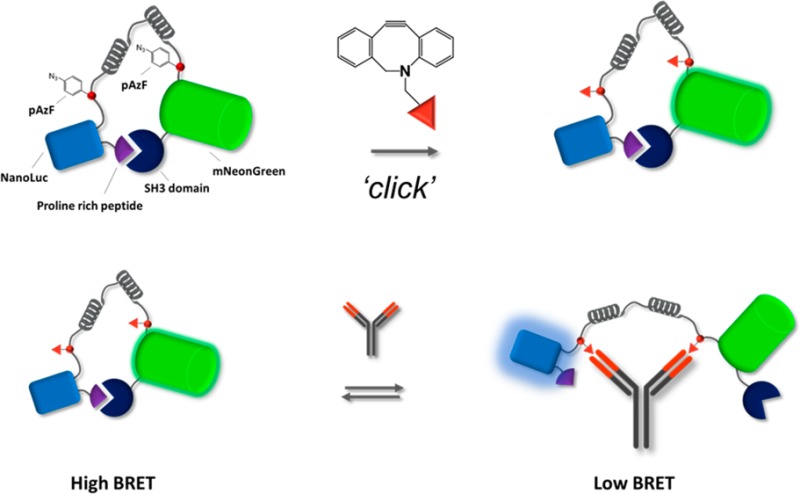
Here, we report a generic strategy to incorporate such nonpeptide epitopes in LUMABS by the introduction of para-azidophenylalanine (pAzF) as a bio-orthogonal chemical handle that allows site-specific conjugation of synthetic epitopes via strain-promoted azide–alkyne click chemistry (SPAAC). The feasibility and efficiency of this approach is demonstrated by constructing semisynthetic LUMABS variants targeting antibodies recognizing the small-molecule epitopes dinitrophenol and creatinine (Figure 1A). In addition, using creatinine as a biomedically relevant example, we demonstrate how semisynthetic LUMABS can be used for the detection of small molecules in a competitive assay format (Figure 1B).
Figure 1.
Schematic overview of pAzF-LUMABS functionalization and sensing principle. (A) Conjugation of two DBCO-functionalized antigens using strain-promoted click-chemistry. (B) Semisynthetic LUMABS can be used for the purpose of antibody detection or for antigen detection using a competitive assay format.
Experimental Section
Protein Expression and Purification
A synthetic gene encoding the flexible linker including two TAG codons was ordered from GenScript, USA and cloned into a pET28a+ vector encoding a previous LUMABS variant via restriction ligation cloning using KpnI and Spe I (New England Biolabs, USA). Correct cloning was confirmed using Sanger sequencing (StarSeq, Germany). The pET28a+ plasmid encoding TAG-LUMABS was transformed into E. coli BL21(DE3) cells (Novagen), together with a pEVOL vector encoding a tRNA/tRNA synthetase pair enabling incorporation of pAzF.17 pEVOL-pAzF was a gift from Peter Schultz (Addgene plasmid # 31186). Cells were cultured in 2YT medium (16 g peptone, 5 g NaCl, 10 g yeast extract per liter) in the presence of 30 μg/mL kanamycin and 25 μg/mL chloramphenicol and expression was induced using 0.1 mM isopropyl-β-d-thiogalactopyranoside and 0.2% arabinose in the presence of 1 mM pAzF (Bachem, catalog no. F-3075.0001). After overnight expression, cells were harvested by centrifuging for 10 min at 10000g. The resulting pellet was lysed using BugBuster (EMD Millipore) and benzonase nuclease (Sigma-Aldrich) for 30 min, and centrifuged again for 40 min at 40000g. Pure pAzf-LUMABS was obtained from the resulting supernatant using a combination of nickel-affinity and Strep-Tactin column chromatography according to the manufacturers instruction (EMD Millipore and Qiagen, respectively). Sensor purity was confirmed using SDS-PAGE and Q-ToF-MS.
Q-ToF-MS
All Q-ToF-MS analyses were performed using a High Resolution LC-MS system consisting of a Waters ACQUITY UPLC I-Class system coupled to a Xevo G2 Quadrupole Time of Flight (Q-ToF). Proteins were separated (0.3 mL/min) by the column (Polaris C18A reverse phase column 2.0 × 100 mm, Agilent) using a 15% to 75% acetonitrile gradient in water supplemented with 0.1% v/v formic acid before analysis in positive mode in the mass spectrometer. Deconvolution of the m/z spectra was done using the MaxENT1 algorithm in the Masslynx v 4.1 (SCN862) software.
Bioconjugation
DBCO–DNP was conjugated to pAzf-LUMABS (5 μM) using a 20× molar excess of DBCO–DNP in 100 μL 100 mM Tris, 150 mM NaCl, 2.5 mM desthiobiotin at pH = 8.0 (the elution buffer from the Strep-Tactin purification) for 5 h at room temperature. Excess DNP was removed by repeated dilution and concentration cycles using a 10 kDa MW cutoff Amicon filter. Product purity was confirmed using Q-ToF LC-MS. DBCO-creatinine was conjugated to pAzf-LUMABS in a similar manner, but using 7.5 μM pAzf-LUMABS. In all cases, product purity was >90% based on the LC elution profile.
Sensor Characterization
Luminescence spectra were obtained using a sensor concentration of 1 nM (DNP-LUMABS) or 5 nM (CR-LUMABS) in a black cuvette (10 mm path length) on a Varian Eclipse Spectrophotometer. Emission was recorded between 400 and 600 nm using a gate-time of 200 ms (DNP-LUMABS) or 5 ms (CR-LUMABS) and a bandwidth of 20 nm. Titrations were performed at a sensor concentration of 10 pM (antibody titrations) or 100 pM (creatinine titrations) in a total reaction volume of 100 μL in a white 96-wells plate (PerkinElmer OptiPlate, 6005299), using a Tecan Spark 10 M plate reader recording luminescence intensity between 400 and 600 nm at an interval of 15 nm. Substrate (NanoGlo assay substrate, Promega, N1110) was added at a final dilution of 5000×. Antibodies were obtained from Sigma-Aldrich (Anti-DNP, 9H8.1) and Randox (Anticreatinine, CR7.7A7). Equation 1 was used to fit titration curves for tight binding, where the Kd is similar to or lower than the sensor concentrations (DNP-LUMABS)
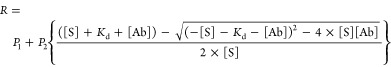 |
1 |
where P1 and P2 are a measure for the emission ratio in the absence of antibody and the total emission ratio change upon antibody saturation, respectively, S represents sensor, and Ab represents antibody.
Equation 2 was used to fit binding for cases where the sensor concentration was substantially lower than the Kd.
| 2 |
Results
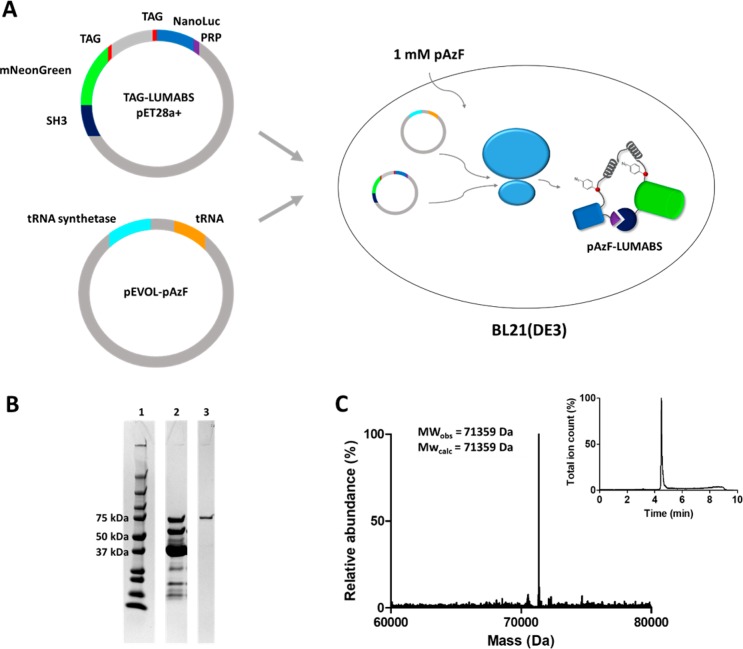
In order to incorporate pAzF at the epitope sites in LUMABS, the strategy described by Schultz and co-workers, using the amber stop codon (TAG), was followed.17 In this approach, a plasmid encoding LUMABS with amber stop codons (TAG) introduced at the epitope sites flanking the central linker was coexpressed with a plasmid (pEVOL-pAzF) encoding an orthogonal tRNA synthetase/tRNA pair that specifically introduces the pAzF at the position encoded by the TAG codon (Figure 2A). The DNA construct encoding the linker with flanking TAG codons was produced synthetically (GenScript, USA) and cloned into a previous LUMABS variant using a restriction–ligation approach, replacing the old linker with the linker containing two TAG codons, creating pTAG-LUMABS. Since some premature protein termination is typically still observed as a result of noncomplete suppression of the amber stop codon, a hexahistidine tag was incorporated on the sensor’s N-terminus, while a Strep-tag was introduced on the C-terminus. Together, these complementary purification tags should allow purification of full-length protein, eliminating truncated protein products.
Figure 2.
(A) Recombinant expression of LUMABS containing two p-azido-phenylalanine residues. A pET28a(+) plasmid encoding LUMABS with two TAG codons at the epitope is cotransformed into E. coli BL21(DE3) cells with a pEVOL plasmid encoding a tRNA/tRNA synthetase pair for the expression of tRNA encoding p-azido phenylalanine (pAzF). (B) SDS-PAGE analysis of sensor purification. 1: BioRad precision plus protein ladder. 2: Isolated protein following Ni-affinity purification. 3: pAzf-LUMABS protein obtained after subsequent Strep-Tactin-affinity purification. (C) Deconvoluted mass spectrum and LC elution profile (inset) of pAzf-LUMABS.
E. coli BL21(DE3) bacteria were co-transformed with a pEVOL-pAzf vector and the pET28a(+) vector encoding TAG-LUMABS (see Supporting Information for full DNA and amino acid sequence). After overnight expression, proteins were purified using nickel affinity and Strep-Tactin column chromatography. Sensor purity was verified using SDS-PAGE (Figure 2B). After nickel affinity purification using the N-terminal His-tag, a significant amount of truncated protein products was observed where protein production had terminated either at the first or second TAG codon. The C-terminal Strep-Tag therefore proved crucial in order to procure pure, full-length pAzF-LUMABS. Analysis using Q-ToF LC-MS revealed a single species with a molecular weight of 71359 Da, which corresponds well with the mass expected for pAzF-LUMABS with two p-azido-phenyl alanine residues (expected mass: 71358.8 Da, Figure 2C). Importantly, no evidence for the incorporation of other amino acids or the reduction of p-azido-phenyl alanine to 4-amino-phenyl alanine was observed.
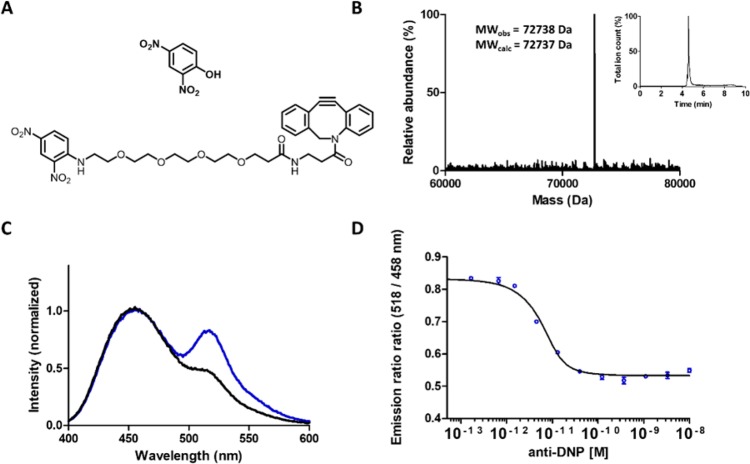
Having established the successful synthesis of LUMABS with two pAzF moieties, we next tested the feasibility of reacting these azides with DBCO-functionalized small molecule antigens. As a first target we chose anti-dinitrophenol (DNP), which is present naturally in human serum, but has also found widespread use in various bioanalytical assays.18−20 DBCO-functionalized DNP (DNP- DBCO) was synthesized in a single reaction step by reacting an amine-functionalized DBCO with a commercially available DNP-derivative consisting of a DNP linked to an NHS-ester via a short polyethylene glycol linker (Figure 3a, Scheme S1). Purification by preparative LC-MS delivered pure DNP-DBCO (>90% pure) in a 68% yield. Incubation of 20 equiv of DBCO–DNP with pAzf-LUMABS for 5 h resulted almost exclusively in the formation of DNP-LUMABS with two DNP epitopes. LC-MS analysis showed a prominent peak (>90%) with a molecular weight corresponding to the LUMABS sensor bearing two DNP groups (Figure 3b). In the absence of antibody, DNP-LUMABS showed a clearly distinguishable emission peak at 517 nm, although the BRET efficiency was somewhat decreased compared to the previously published HIV, HA, and DEN-1 sensors (Figure 3c). Nevertheless, addition of 5 nM anti-DNP antibody to DNP-LUMABS resulted in almost complete disappearance of the mNeonGreen emission peak. A titration experiment showed an antibody affinity of 1.5 ± 0.4 pM and a dynamic range of 58% (Figure 3d). This very high affinity results from the bivalent nature of the interaction between antibody and sensor and was also observed in previously developed LUMABS sensors.9
Figure 3.
(A) Molecular structure of dinitrophenol (top) and DNP-DBCO (bottom) as it was used to construct DNP-LUMABS. (B) Deconvoluted mass spectrum and LC elution profile of DNP-LUMABS. The peak represents the sensor with two DNP moieties attached (expected MW = 72737 Da). (C) Emission spectrum of 1 nM DNP-LUMABS in the absence (blue line) and presence of 5 nM anti-DNP antibody (black line). (D) Emission ratio of 10 pM DNP-LUMABS as a function of antibody concentration. Measurements were performed in a buffer consisting of 50 mM Tris-HCl, 100 mM NaCl, 10% (v/v) glycerol, and 0.05% Tween-20 at pH = 7.4. Error bars show mean ± s.e.m. (n = 2). Solid line represents fit to eq 1.
To demonstrate the use of semisynthetic LUMABS variants in a competition assay format, we next developed a LUMABS to detect anti-creatinine antibodies. The CR-LUMABS sensor incorporates the small-molecule creatinine as epitope, and should be applicable as a sensor both for anti-creatinine antibodies and, in a competitive assay format, for creatinine itself. Creatinine is produced in muscles at a relatively constant rate, and clearance of creatinine occurs through glomerular filtration. Therefore, serum creatinine levels rise if the glomerular filtration is impaired, making serum creatinine levels a valuable marker for kidney failure.21,22 In addition to the direct use of creatinine as a biomarker for kidney failure, many other urinary biomarkers are reported as a normalized ratio to urinary creatinine to correct for variations in urine production.23−25 In healthy subjects the serum concentration varies between 44 and 150 μM.26 In urine, the concentration of creatinine is around 10 mM.27 Currently, the standard method for creatinine detection is the Jaffé reaction, a colorimetric reaction between creatinine and picric acid in alkaline environments. Although the Jaffé reaction is inexpensive, it shows limited sensitivity and several interferents are known, such as metabolites and drugs that affect the readout.28 Moreover, it requires a large sample volume and is time-consuming due to sample handling and pretreatment.
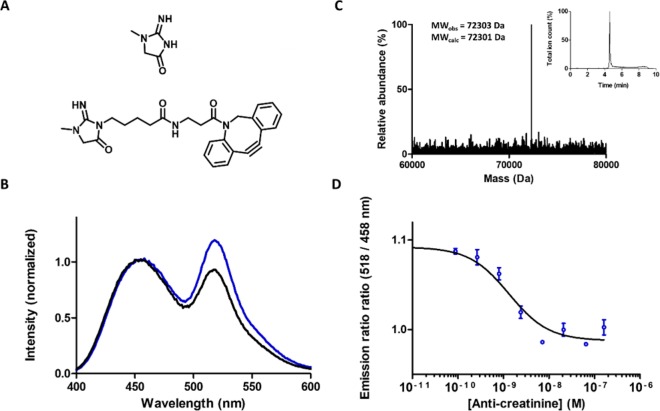
DBCO-creatinine (Figure 4a) was synthesized in three synthetic steps according to Scheme S2; details of this synthesis can be found in the Supporting Information. The regioselectivity of the first critical step in this synthesis, the reaction of creatinine with 5-bromovalerate, was assessed using 1D and 2D NMR. The correlation peaks obtained in the 1H–13C HMBC for H9 with C2 and C4 are consistent with the regioselectivities previously reported in the literature.26 The absence of cross peaks between C9–H10/H12 and C19–H1 is likely because their corresponding long-range coupling constants approach zero,29 which could be for a number of reasons.30 (see Supporting Information, Figure S9). DBCO-creatinine was conjugated to pAzf-LUMABS to create CR-LUMABS, using 20 equiv DBCO-creatinine in a 5 h reaction at room temperature. Successful coupling was confirmed using Q-ToF LC-MS, indicating that the reaction proceeded quantitatively (Figure 4b). Titration of anti-creatinine antibody to CR-LUMABS resulted in the expected decrease in BRET, revealing an affinity of 1.3 ± 0.4 nM (Figure 4d). The amount of BRET in the antibody-bound state was higher than that observed in other LUMABS sensors, resulting in a relatively modest change in emission ratio of 22%. The origin of this high amount of residual BRET is presently unclear.
Figure 4.
(A) Molecular structure of creatinine (top) and creatinine-DBCO (bottom) used to functionalize DNP-LUMABS. (B) Deconvoluted mass spectrum and LC elution profile of DNP-LUMABS. The major peak represents the sensor with two DNP moieties attached (expected MW = 72301 Da). (C) Luminescence emission spectra of 5 nM CR-LUMABS in the absence (blue line) and presence (black line) of 20 nM anticreatinine antibodies. (D) Emission ratio of 10 pM CR-LUMABS as a function of antibody concentration. Measurements were performed in a buffer composed of 50 mM Tris, 100 mM NaCl, 10% (v/v) glycerol, and 0.05% Tween-20 at pH = 7.4. Error bars show mean ± s.e.m. (n = 2). Solid line represents fit to eq 2.
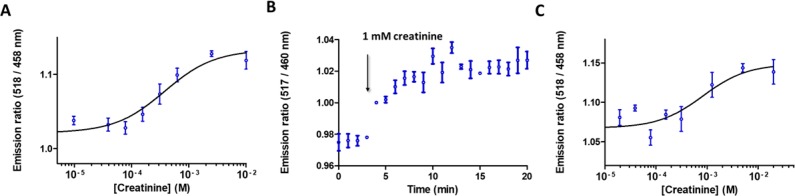
Next, we investigated whether the formation of antibody–sensor complex could be reversed by the addition of free creatinine. To ensure that most of CR-LUMABS existed in the open conformation, we first incubated 100 pM CR-LUMABS with 10 nM anti-creatinine antibody. The excess of antibody was kept as low as possible while ensuring a near-saturated sensor, as this should make the system most responsive to changes in antigen concentration. Addition of low millimolar amounts of creatinine (Kd,app = 276 ± 104 μM) was sufficient to fully disrupt the antibody–sensor complex, allowing CR-LUMABS to adopt its closed, high-BRET conformation again (Figure 5a). To establish the kinetics of the competition reaction, we monitored the BRET signal over time following addition of 1 mM creatinine to the sensor-antibody complex. Figure 5b shows that the exchange reaction is fast and equilibrium is reached within a few minutes. Finally, we investigated whether the competitive assay format could also be used to detect creatinine in more complex matrices, such as blood plasma. For this purpose, 1:1 diluted blood plasma containing the sensor–antibody complex was spiked with different concentrations of creatinine. The performance of the sensor system in blood plasma was comparable to that observed in buffer, although the LOD (0.2 mM) was in both cases limited by the relatively small change in emission ratio (Figure 5c).
Figure 5.
(A) Response of 100 pM CR-LUMABS, complexed to 10 nM anticreatinine antibody, to increasing concentrations of creatinine. (B) Emission ratio of 100 pM CR-LUMABS complexed to 10 nM anticreatinine antibody followed in time. At t = 4 min, 1 mM creatinine was added. Measurements in (A) and (B) were performed in a buffer composed of 50 mM Tris, 100 mM NaCl, 10% (v/v) glycerol, and 0.05% Tween-20 at pH = 7.4. (C) Response of 100 pM CR-LUMABS, complexed to 10 nM antibody, to increasing concentrations of creatinine in 1:1 diluted blood plasma. Error bars show mean ± s.e.m. (n = 3). Solid line represents fit to eq 2.
Discussion
In this work, the chemical diversity of epitopes that can be incorporated in LUMABS was expanded through the introduction of chemical handles at the epitope sites, enabling post-expression conjugation using bio-orthogonal click chemistry. An efficient expression and purification system was established that allowed the incorporation of the non-natural amino acid pAzF at each of the two antigen sites in sufficient yield, and excellent purity. Moreover, subsequent reaction of the p-azido-phenylalanine groups with DBCO-functionalized small molecules using SPAAC resulted in the exclusive formation of the desired LUMABS variants with two small-molecule antigens. In addition to expanding the scope of antibody targets, these newly developed semisynthetic LUMABS variants are particularly attractive for small molecule detection in a competitive assay format. A preassembled complex of CR-LUMABS sensor and an anticreatinine antibody enabled the detection of creatinine in the high micromolar regime, both in buffer and in blood plasma.
While the application of semisynthetic LUMABS for the detection of the small molecule creatinine was successfully demonstrated, the CR-LUMABS sensor displayed a relatively small ratiometric response of 22%, a result of the relatively high amount of BRET in the antibody-bound state. The reason for the high amount of residual BRET is not known. One possibility is that the large hydrophobic cyclooctyne group causes additional intramolecular interactions with either the NanoLuc of mNeonGreen domain, resulting in a more compact antibody-bound state. Alternative conjugation strategies that do not rely on the use of cyclooctyns such as Cu-catalyzed click reactions with smaller, alkyne-functionalized molecules may be explored to test this hypothesis. The modular architecture of the LUMABS platform also allows for a systematic optimization of several other parameters that could affect the amount of BRET in both the antibody-free and antibody-bound states, such as the relative order and orientation of NanoLuc, mNeonGreen and the helper domains, the length and composition of linkers between the various domains, and the type of anti-creatinine antibody that is employed. Another sensor parameter that could be adjusted is the concentration regime in which the assay is responsive to creatine. While the present system is optimal for detection of creatinine concentrations in urine, the creatinine sensitivity should ideally be a little bit higher for detection of creatinine in serum. Shifting the responsive concentration regime of the system toward lower creatinine concentrations may be achieved by incorporating weaker-binding creatinine analogues in the sensor.7
Any molecule that can be functionalized with a DBCO group or other cyclooctyns such as BCN can now be used as an epitope for LUMABS. Moreover, the expression plasmid TAG-LUMABS in principle allows the incorporation of any non-natural amino acids for which orthogonal tRNA synthetase/tRNA pairs have been developed, allowing the extension of the approach to include other conjugation strategies.31−33 Small-molecule sensing using LUMABS is therefore a generic approach with a wide scope of potential applications. An important advantage of the described sensing strategy is that it relies on the use of monoclonal antibodies, which have been generated for a very wide range of targets. The requirements for the strategy to work are the existence of an antibody that binds the analyte with sufficient affinity and the possibility to incorporate the molecule of interest (or weaker binding derivatives) in the sensor via bio-orthogonal chemistry. Although the sensor may contain weaker-binding antigen analogues, these should still bind with submicromolar affinity to the antibody to overcome the helper interactions and allow the sensor to adopt the open conformation. Because binding of the small molecule analyte has to compete with the LUMABS sensor that binds the antibody bivalently, small molecule detection using LUMABS is probably most suited for the detection of targets with concentrations in the micromolar to millimolar regime. Possible fields of application of the described small molecule sensing approach include diagnostic tests and therapeutic drug monitoring, but also field tests in a veterinary setting or food testing at production sites.
Acknowledgments
We thank Sebastian Andrei, Pim Vendrig, Camiel de Ruiter, and the TU/e SensUs team, in particular, Rafiq Lubken, Leroy Tan, and Boris Arts for their work on the synthesis of DNP-DBCO and CR-DBCO, as well as initial characterization of DNP-LUMABS and CR-LUMABS. This work was supported by ERC Starting Grant ERC-2011-StG 280255 and by ERC Proof-of-Concept Grant ERC-2013-PoC 632274 and Marie Sklodowska-Curie grant 642793.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssensors.7b00695.
TAG-LUMABS DNA sequence, supplementary methods, synthesis of DBCO-antigen conjugates, mass spectroscopy analysis, and NMR analysis (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Banala S.; Arts R.; Aper S. J. A.; Merkx M. No washing, less waiting: engineering biomolecular reporters for single-step antibody detection in solution. Org. Biomol. Chem. 2013, 11 (44), 7642–7649. 10.1039/c3ob41315b. [DOI] [PubMed] [Google Scholar]
- Lequin R. M. Enzyme Immunoassay (EIA)/Enzyme-Linked Immunosorbent Assay (ELISA). Clin. Chem. 2005, 51 (12), 2415–2418. 10.1373/clinchem.2005.051532. [DOI] [PubMed] [Google Scholar]
- Yoo E.-H.; Lee S.-Y. Glucose Biosensors: An overview of use in clinical practice. Sensors 2010, 10 (5), 4558–4576. 10.3390/s100504558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Electrochemical glucose biosensors. Chem. Rev. 2008, 108 (2), 814–825. 10.1021/cr068123a. [DOI] [PubMed] [Google Scholar]
- Wang H.-C.; Lee A.-R. Recent developments in blood glucose sensors. J. Food Drug Anal. 2015, 23 (2), 191–200. 10.1016/j.jfda.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacres H.; Wang J.; Leitch V.; Horne I.; Anderson A. R.; Trowell S. C. Greatly enhanced detection of a volatile ligand at femtomolar levels using bioluminescence resonance energy transfer (BRET). Biosens. Bioelectron. 2011, 29 (1), 119–124. 10.1016/j.bios.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Griss R.; Schena A.; Reymond L.; Patiny L.; Werner D.; Tinberg C. E.; Baker D.; Johnsson K. Bioluminescent sensor proteins for point-of-care therapeutic drug monitoring. Nat. Chem. Biol. 2014, 10 (7), 598–603. 10.1038/nchembio.1554. [DOI] [PubMed] [Google Scholar]
- Schena A.; Griss R.; Johnsson K. Modulating protein activity using tethered ligands with mutually exclusive binding sites. Nat. Commun. 2015, 6, 7830. 10.1038/ncomms8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts R.; den Hartog I.; Zijlema S. E.; Thijssen V.; van der Beelen S. H. E.; Merkx M. Detection of antibodies in blood plasma using bioluminescent sensor proteins and a smartphone. Anal. Chem. 2016, 88 (8), 4525–4532. 10.1021/acs.analchem.6b00534. [DOI] [PubMed] [Google Scholar]
- Xue L.; Yu Q.; Griss R.; Schena A.; Johnsson K. Bioluminescent antibodies for point-of-care diagnostics. Angew. Chem., Int. Ed. 2017, 56 (25), 7112–7116. 10.1002/anie.201702403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis R. Posttranslational modifications and the immunogenicity of biotherapeutics. J. Immunol. Res. 2016, 2016, e5358272. 10.1155/2016/5358272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D. R.; Cohen G. H. Interactions of protein antigens with antibodies. Proc. Natl. Acad. Sci. U. S. A. 1996, 93 (1), 7–12. 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosten T.; Domingo C.; Orson F.; Kinsey B. Vaccines against stimulants: cocaine and MA. Br. J. Clin. Pharmacol. 2014, 77 (2), 368–374. 10.1111/bcp.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M.; Olshevsky U.; Furman C.; Gabuzda D.; Posner M.; Sodroski J. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J. Virol. 1991, 65 (11), 6188–6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Venrooij W. J.; Van Beers J. J. B. C.; Pruijn G. J. M. Anti-CCP antibody, a marker for the early detection of rheumatoid arthritis. Ann. N. Y. Acad. Sci. 2008, 1143 (1), 268–285. 10.1196/annals.1443.013. [DOI] [PubMed] [Google Scholar]
- Sun Y.; Fong K.-Y.; Chung M. C. M.; Yao Z.-J. Peptide mimicking antigenic and immunogenic epitope of double-stranded DNA in systemic lupus erythematosus. Int. Immunol. 2001, 13 (2), 223–232. 10.1093/intimm/13.2.223. [DOI] [PubMed] [Google Scholar]
- Chin J. W.; Santoro S. W.; Martin A. B.; King D. S.; Wang L.; Schultz P. G. Addition of p-azido-l-phenylalanine to the genetic code of Escherichia coli. J. Am. Chem. Soc. 2002, 124 (31), 9026–9027. 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- Augustyniak E.; Adam A.; Wojdyla K.; Rogowska-Wrzesinska A.; Willetts R.; Korkmaz A.; Atalay M.; Weber D.; Grune T.; Borsa C.; et al. Validation of protein carbonyl measurement: A multi-centre study. Redox Biol. 2015, 4, 149–157. 10.1016/j.redox.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber D.; Davies M. J.; Grune T. Determination of protein carbonyls in plasma, cell extracts, tissue homogenates, isolated proteins: Focus on sample preparation and derivatization conditions. Redox Biol. 2015, 5, 367–380. 10.1016/j.redox.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsche C. E.; Parker C. G.; Tao R. N.; Kolesnikova M. D.; Douglass E. F.; Spiegel D. A. Exploring binding and effector functions of natural human antibodies using synthetic immunomodulators. ACS Chem. Biol. 2013, 8 (11), 2404–2411. 10.1021/cb4004942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levey A. S.; Bosch J. P.; Lewis J. B.; Greene T.; Rogers N.; Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Ann. Intern. Med. 1999, 130 (6), 461–470. 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- Jones C.; McQuillan G.; Kusek J.; Eberhardt M.; Herman W.; Coresh J.; Salive M.; Jones C.; Agodoa L. Serum creatinine levels in the US population: Third National Health and Nutrition Examination Survey. Am. J. Kidney Dis. 1998, 32 (6), 992–999. 10.1016/S0272-6386(98)70074-5. [DOI] [PubMed] [Google Scholar]
- Heavner D. L.; Morgan W. T.; Sears S. B.; Richardson J. D.; Byrd G. D.; Ogden M. W. Effect of creatinine and specific gravity normalization techniques on xenobiotic biomarkers in smokers’ spot and 24-h urines. J. Pharm. Biomed. Anal. 2006, 40 (4), 928–942. 10.1016/j.jpba.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Suwazono Y.; Åkesson A.; Alfvén T.; Järup L.; Vahter M. Creatinine versus specific gravity-adjusted urinary cadmium concentrations. Biomarkers 2005, 10 (2–3), 117–126. 10.1080/13547500500159001. [DOI] [PubMed] [Google Scholar]
- Jatlow P.; McKee S.; O’Malley S. S. Correction of urine cotinine concentrations for creatinine excretion: is it useful?. Clin. Chem. 2003, 49 (11), 1932–1934. 10.1373/clinchem.2003.023374. [DOI] [PubMed] [Google Scholar]
- Benkert A.; Scheller F.; Schössler W.; Hentschel C.; Micheel B.; Behrsing O.; Scharte G.; Stöcklein W.; Warsinke A. Development of a creatinine ELISA and an amperometric antibody-based creatinine sensor with a detection limit in the nanomolar range. Anal. Chem. 2000, 72 (5), 916–921. 10.1021/ac9909047. [DOI] [PubMed] [Google Scholar]
- Gaddes D.; Reeves W. B.; Tadigadapa S. Calorimetric biosensing system for quantification of urinary creatinine. ACS Sens. 2017, 2 (6), 796–802. 10.1021/acssensors.7b00161. [DOI] [PubMed] [Google Scholar]
- Bonsnes R. W.; Taussky H. H. On the colorimetric determination of creatinine by the jaffe reaction. J. Biol. Chem. 1945, 158 (3), 581–591. [Google Scholar]
- Claridge T. D. W.Applying HMBC, section 6.4.2. In High resolution NMR techniques in organic chemistry; Elsevier, 2008; Vol. 2, p 210. [Google Scholar]
- Hansen P. E. Carbon-hydrogen spin-spin coupling constants. Prog. Nucl. Magn. Reson. Spectrosc. 1981, 14, 175. 10.1016/0079-6565(81)80001-5. [DOI] [PubMed] [Google Scholar]
- Xue L.; Prifti E.; Johnsson K. A general strategy for the semisynthesis of ratiometric fluorescent sensor proteins with increased dynamic range. J. Am. Chem. Soc. 2016, 138 (16), 5258–5261. 10.1021/jacs.6b03034. [DOI] [PubMed] [Google Scholar]
- Devaraj N. K.; Weissleder R.; Hilderbrand S. A. Tetrazine-based cycloadditions: application to pretargeted live cell imaging. Bioconjugate Chem. 2008, 19 (12), 2297–2299. 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. H.; Axup J. Y.; Schultz P. G. Protein conjugation with genetically encoded unnatural amino acids. Curr. Opin. Chem. Biol. 2013, 17 (3), 412–419. 10.1016/j.cbpa.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.