Abstract
Biological drugs like therapeutic antibodies are widely used for the treatment of various diseases like inflammatory disorders and cancer. A drawback of these novel treatments is the substantial proportion of patients experiencing adverse reactions such as loss-of-drug effect or hypersensitivity reactions. These reactions are associated with pre-existing and/or developing anti-drug antibodies. Especially IgE development is a risk factor for life-threatening systemic anaphylaxis.
Methods: In order to characterize the individual drug-specific serum IgE, an IgE cross-reactivity immune profiling (ICRIP) assay was developed. Individual IgG epitopes of anti-drug antibodies against adalimumab were identified by epitope mapping via peptide microarray.
Results: ICRIP analyses of sera from patients treated with the therapeutic antibodies adalimumab (ADL) and infliximab (IFX) reveal individual, distinct IgE binding patterns. IgG epitopes were identified mostly located in the variable region of ADL.
Conclusions: Using ICRIP and peptide microarrays for pharmacovigilance of the TNF-α blockers IFX and ADL, risk factors and biomarkers before and during therapy shall be identified. These diagnostic systems provide the basis for a safe and efficacious therapy decision for each patient in cases of adverse drug reactions mediated by different types of anti-drug antibodies.
Keywords: adalimumab, anti-drug antibodies, hypersensitivity, infliximab, precision medicine.
Introduction
Therapeutic antibodies have become a major option in the treatment of various types of cancer as well as inflammatory disorders 1. Due to their already wide-spread and growing application, pharmacovigilance of therapeutic antibodies has become very important 2-4. A well-known side effect during therapy with biologicals is the development of anti-drug antibodies, which severely interfere with therapy outcome 5, 6. In this regard, the development of drug-specific IgE associated with life-threatening anaphylaxis is a major potential risk factor. Therapeutic antibodies have various individual characteristics with immunogenic potential, e.g., amino acid sequence, production system or application procedure. Adverse effects during treatment include the development of anti-drug antibodies and associated loss-of-drug-effect due to blocking of target-binding by anti-idiotype antibodies, formation of immune complexes and enhanced clearance of biologicals as well as the induction of hypersensitivity reactions and inflammation 3, 4, 7, 8. Hypersensitivity reactions upon drug application are known adverse events during treatment with biologicals and occur in at least 5% of applications, e.g., for the TNF-α blockers adalimumab (ADL) and infliximab (IFX) 9-11. These hypersensitivity reactions are often associated with specific IgE 12-16. However, these adverse events are highly individual and do not occur in every patient. Thus, there is a high demand for precision diagnostics to identify patients at risk by identification and monitoring of pre-existing and treatment-associated anti-drug antibodies 17-19. It has been discovered that anti-drug antibodies do not only develop against chimeric antibodies (like IFX) but also during treatment with so-called “humanized” antibodies (like ADL) 20. For ADL and IFX, it has been described that these anti-drug antibodies are directed against the antigen-binding region and thus prevent binding to TNF-α, resulting in treatment failure 21-23. For IFX, the exact immunogenic peptide sequences and their localization in the TNF-α binding site have been discovered 22, 24. For ADL, the results are presented in this study. The aim of our study was the identification and characterization of pre-existing IgE and IgE development during treatment by a newly established IgE cross-reactivity immune profiling (ICRIP) assay (Fig. 1). ICRIP results for ADL and IFX are compared with IgE binding to the immunogenic/allergenic therapeutic antibody cetuximab (CTX) (registered as allergen Hom s/Mus m cetuximab by the WHO/IUIS allergen nomenclature subcommittee, www.allergen.org), which has been identified as the major α-Gal-containing allergen associated with meat allergy causing severe allergy symptoms in α-Gal-sensitized patients 12, 25. Precision diagnostics for pharmacovigilance of the TNF-α blockers IFX and ADL shall provide the basis for individual therapy optimization and a potentially necessary switch to a different therapeutic antibody due to adverse drug reactions mediated by anti-drug antibodies.
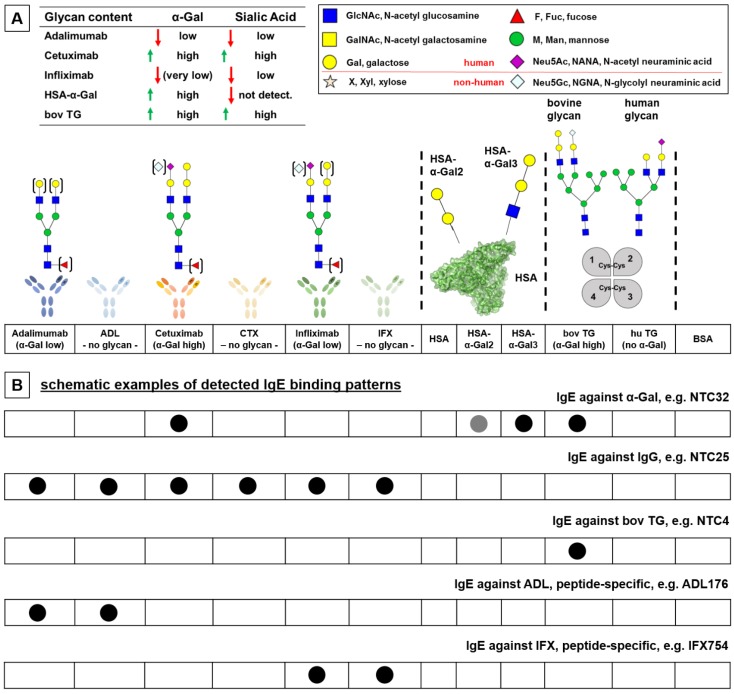
Figure 1.
Concept for IgE cross-reactivity immune profiling (ICRIP) assay: characteristics of analytes used for the detection of IgE profiles. A) Each analyte has distinct glycan and peptide characteristics for the identification of anti-drug antibodies. Functional glycan content was determined by lectin binding assay (see Fig. S2). B) Specific binding patterns point to cross-reactive, thus common, epitopes, e.g., α-Gal as indicated by IgE binding to CTX, HSA-α-Gal and bovine TG. HSA: human serum albumin; bov (hu) TG: bovine (human) thyroglobulin; BSA: bovine serum albumin.
Results
IgE cross-reactivity immune profiling to identify specific IgE epitopes on biologicals
In order to characterize serum IgE from patients treated with therapeutic antibodies, a versatile diagnostic system was designed. The array system was composed of analytes that contained peptide and glycan motifs derived from therapeutic antibodies with distinct characteristics (Fig. 1). Native and glycan-processed forms of the therapeutic antibodies ADL, CTX and IFX applied in parallel facilitated the analysis of the peptide- and glycan-dependency of serum IgE (exemplified binding patterns are depicted in Fig. 1B). All biologicals included in the assay system had a distinct glycosylation pattern that was characterized by lectin binding analysis (Fig. 1 and S2B). The CTX glycan pattern of the Fab region predominantly contained the immunogenic IgE epitope α-Gal on bisecting N-glycans, as opposed to IFX with a similar peptide sequence but no Fab N-glycosylation Sequon (Fig. S1) 26. Additionally, the non-human sialic acid N-glycolyl neuraminic acid (Neu5Gc) was present 26. The glycosylation of the Fc part is different and showed only low amounts of α-Gal, but almost 10% of high mannose N-glycan motifs 26. In the ICRIP assay system for IgE immune profiling, a conjugate of human serum albumin (HSA) and α-Gal was included to investigate the α-Gal-dependency of serum IgE. Here, α-Gal as a disaccharide as well as in its trisaccharide form facilitated the analysis of affinity aspects of α-Gal-specific IgE. Additionally, the commercial analyte for IgE against α-Gal, bovine thyroglobulin (bov TG, registered as allergen Bos d TG), in comparison with its human analogue (hu TG) was included to characterize the glycan- versus peptide-dependency of IgE against TG. By analysis of signal intensities resulting from parallel serum IgE binding to these analytes, distinct IgE patterns characteristic for every patient group and/or individual patients were obtained.
Serum IgE antibodies from infliximab- and adalimumab-treated patients show distinct binding patterns revealing glycan- and peptide-specific epitopes
Sera from IFX or ADL-treated patients with suspected loss of treatment efficacy were analyzed for anti-drug antibodies. Sera positive for anti-drug antibodies (Table 1) were further analyzed in order to reveal IgE binding patterns in the ICRIP IgE assay system established in this study (Fig. 2). Three patients treated with IFX clearly developed IgE against this therapeutic antibody (IFX438, IFX754 and IFX922; Fig. 2). This IgE against IFX in two cases was glycan-independent (thus peptide-dependent) as indicated by a comparable signal intensity for IFX and glycan-processed IFX (IFX438 and IFX754). Interestingly, IgE from IFX438 also bound strongly to bovine TG but not to HSA-α-Gal, potentially indicating an α-Gal-independent binding to bovine TG. Moreover, the IgE pattern of IFX438 contained signals for IgE against the native form of ADL. This pattern pointed to a peptide-dependent IgE cross-reaction to IgG antibodies, which, interestingly, was also detected as IgE binding pattern of serum NTC25, a non-treated control serum (Fig. 2). IgE from serum NTC25 bound to all therapeutic antibodies, including their periodate-treated forms, indicating a peptide-dependent binding. The third serum positive for anti-IFX antibodies (IFX922) showed a glycan-dependent IFX-binding as no signal for glycan-processed IFX was detected. The association of the IgE binding to bovine TG pointed to a possible common glycan IgE epitope that remains to be identified. However, no IgE binding to other α-Gal-containing analytes (CTX and HSA-α-Gal) was observed. For three IFX-treated patients, specific IgE binding to bovine TG was found (IFX452, IFX660 and IFX790). Interestingly, this IgE specificity was also found for 8 sera of the non-treated control group (NTC4, 7, 15, 16, 17, 28, 30 and 39). Thus, this pattern was most likely not associated with IFX treatment. For serum from IFX-treated patient IFX694, an interesting IgE cross-reaction with glycan-processed ADL and bovine TG was detected. It remains to be determined if this IgE binding is a peptide-specific cross-reaction. The IgE pattern of one IFX-treated patient (IFX890) showed a strong signal only for IgE binding against human TG, pointing to a possible autoimmune mechanism. In the group of ADL-treated patients, IgE from 3 out of 10 sera was directed against ADL (ADL176, ADL197 and ADL198). While IgE from patient ADL176 showed a glycan-independent (thus peptide-dependent) binding pattern, IgE antibodies from ADL197 and ADL198 did not bind to glycan-processed ADL (Fig. 2). For the two latter sera, an association with bovine TG could be observed, indicating a glycan IgE epitope. However, this glycan epitope apparently is not α-Gal as no IgE binding to CTX or HSA-α-Gal could be observed. Further IgE serum binding patterns included IgE against CTX for sera ADL86, ADL149 and ADL178 (Fig. 2). The typical pattern for IgE against α-Gal (associated with meat allergy) could be identified for serum ADL86 as IgE binding to CTX, HSA-α-Gal and bovine TG 27. No binding to glycan-processed CTX, to unconjugated HSA or human TG was detected, indicating the glycan-dependency. The IgE binding pattern of ADL149 seemed to be CTX-specific, however, no binding to the glycan-processed form of CTX was observed, which indicated a glycan- rather than a peptide-IgE-epitope. Interestingly, IgE against CTX from serum ADL178 was associated with bovine TG and also with human TG. For this pattern, it remains to be determined if the IgE cross-reaction is peptide- or glycan-dependent. The cross-reaction with human/bovine TG could be associated with peptide-specific IgE, potentially associated with autoimmunity.
Table 1.
Sera positive for anti-drug antibodies of patients treated with A) IFX or B) ADL. Patient sera were analyzed by a certified ELISA system for anti-drug antibodies (not resolving the isotype).
| A) Infliximab-treated patients | |||
| ID | anti-IFX [µg/mL] | IFX serum c [µg/mL] | ICRIP result |
| IFX438 | 54.1 | < 4.0 | ADL/IFX/IFX PI/bov TG |
| IFX452 | 63.9 | 4.9 | bov TG |
| IFX485 | 74.1 | 18.3 | neg. |
| IFX522 | 45.9 | 5.6 | neg. |
| IFX638 | 39.3 | < 4.0 | neg. |
| IFX648 | 34.2 | 11.6 | neg. |
| IFX652 | 29.7 | < 4.0 | neg. |
| IFX660 | 14.4 | 9.9 | bov TG |
| IFX694 | 45.6 | 18.9 | ADL PI/bov TG |
| IFX719 | 69.7 | 6.7 | neg. |
| IFX721 | > 80 | 11.4 | neg. |
| IFX722 | 44.8 | 4.7 | neg. |
| IFX724 | 32.6 | < 4.0 | neg. |
| IFX729 | 27.3 | < 4.0 | neg. |
| IFX752 | 18.8 | < 4.0 | neg. |
| IFX754 | 22.0 | < 4.0 | IFX/IFX PI |
| IFX767 | 67.5 | 26.1 | neg. |
| IFX790 | 34.6 | 8.3 | bov TG |
| IFX809 | 31.0 | 4.6 | neg. |
| IFX883 | 46.1 | < 4.0 | neg. |
| IFX890 | > 80 | 16.9 | hu TG |
| IFX905 | 22.6 | < 4.0 | neg. |
| IFX922 | > 80 | 21.3 | IFX/bov TG |
| IFX928 | 59.9 | 7.6 | neg. |
| IFX957 | > 80 | 12.3 | neg. |
| IFX963 | 73.9 | 28.0 | neg. |
| IFX977 | > 80 | 9.9 | neg. |
| IFX982 | 29.4 | < 4.0 | neg. |
| IFX990 | > 80 | 16.6 | neg. |
| IFX996 | > 80 | 20.5 | neg. |
| B) Adalimumab-treated patients | |||
| ID | anti-ADL [µg/mL] | ADL serum c [µg/mL] | ICRIP result |
| ADL41 | 2.4 | 13.6 | n.t. |
| ADL67 | 12.1 | > 256 | n.t. |
| ADL72 | 18.5 | > 256 | n.t. |
| ADL86 | 5.7 | > 256 | CTX/HSA-α-Gal/bov TG |
| ADL89 | 22.4 | > 256 | n.t. |
| ADL90 | 2.3 | < 4.0 | n.t. |
| ADL122 | 8.7 | 4.2 | n.t. |
| ADL123 | 14.2 | > 256 | n.t. |
| ADL134 | 34.9 | 28.3 | n.t. |
| ADL138 | < 1.25 | n.a. | neg. |
| ADL145 | < 1.25 | n.a. | neg. |
| ADL149 | 7.6 | 7.7 | CTX |
| ADL176 | 18.1 | > 256 | ADL/ADL PI |
| ADL178 | > 40 | 12.7 | CTX/bov TG/hu TG |
| ADL197 | 11.8 | < 4.0 | ADL/bov TG |
| ADL198 | 12.7 | < 4.0 | ADL/bov TG |
| ADL216 | 1.8 | < 4.0 | neg. |
| ADL242 | 3.4 | < 4.0 | n.t. |
| ADL245 | 2.9 | < 4.0 | neg. |
Figure 2.
IgE binding patterns of A) IFX-treated patients, B) ADL-treated patients and C) non-treated control sera detected by ICRIP assay. Characteristics of assay components are described in Fig. 1. A positive signal was defined as being more intense than the background signal for the negative control serum NTC1 multiplied by a factor of 2. ADL: adalimumab; ADL PI: periodate-treated adalimumab (oxidation of glycans, thus interruption of antibody-glycan-binding); CTX: cetuximab; IFX: infliximab; HSA: human serum albumin; bov (hu) TG: bovine (human) thyroglobulin. Note that bov TG is used for commercial α-Gal ImmunoCAP® test (Thermo Fisher Scientific / Phadia, o215).
Identification of immunogenic epitopes on adalimumab
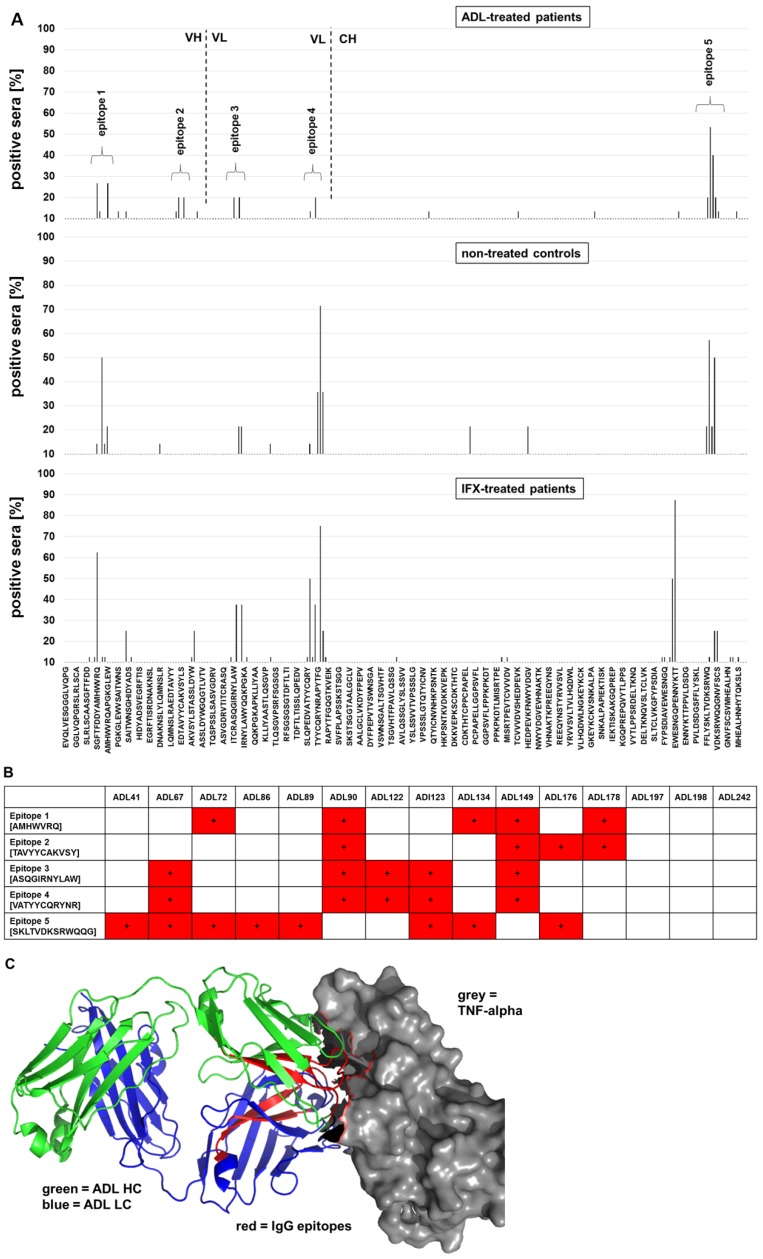
In order to reveal immunogenic epitopes on ADL, a peptide microarray was designed and probed with sera from 10 ADL-treated patients who were positive for anti-ADL antibodies (Table 1, Fig. 3 and Table S1). Four IgG epitopes were found in the variable region of ADL: epitope 1 and 2 in the heavy chain variable region (VH) and epitope 3 and 4 in the light chain variable part (VL). One epitope was detected in the constant region of ADL (CH3). Strikingly, as has been shown for anti-IFX IgG 22, epitopes of IgG against ADL are also located in the TNF-α binding regions of the variable region (Fig. 3). The individual resolution of the serum IgG epitopes on ADL indicated distinct epitope reactivity for each patient serum. Two sera (ADL90 and ADL149) showed IgG binding to every identified ADL epitope (Fig. 2). While for one serum positive for anti-ADL IgE (ADL176) IgG antibodies against epitope 2 were observed, the two remaining sera positive for anti-ADL IgE (ADL197 and ADL198) did not show IgG epitope signals above background. Interestingly, for the negative control sera from untreated individuals, distinct IgG epitopes on ADL were also detected, especially against the epitopes 1 and 4 in the variable region of ADL, but also against the prominent epitope 5 in the constant region (Fig. 3). This epitope 5, located in the (constant) Fc region of ADL, may be a risk factor for cross-reactivity of anti-drug antibodies to other therapeutic antibodies as most of them are of the IgG1 isotype, thus containing the same epitope. This cross-reactivity would not be expected for epitopes located in the variable regions of ADL and IFX. Of note, untreated control sera tested negative for anti-drug antibodies by anti-drug ELISA (data not shown). In order to determine the relevance of the oligopeptide epitopes mapped by microarray, an ELISA system with single 15-mer peptides was established with 6 peptides from the identified epitopes and 11 random peptides as controls (Table S2). In this assay system, sera from patients ADL41 and ADL134 showed especially strong signals (and thus IgG binding) for the mapped ADL peptides (Fig. S3). Sera from patient ADL72 showed low signals corresponding to lower IgG binding affinity and/or IgG serum concentration. Of note, sera from untreated individuals showed similar signal patterns (especially for peptides B1, E1 and C2), although with lower intensities corresponding to lower IgG binding affinities / concentrations (Fig. S3). A cross-reactivity analysis of serum IgG was performed with random peptides using the indicated sera containing IgG against ADL epitopes (Fig. S3). It was found that three patient sera (ADL41, 72 and 134) showed prominent cross-reactivity towards four random peptides (B4, C4, E4 and F4) while other sera as well as random oligopeptides can be considered as negative controls.
Figure 3.
Epitope mapping of serum IgG against ADL. Peptide microarrays contain the ADL sequence split into 15-mer peptides with an offset of 2 amino acids. A) Epitope mapping of anti-ADL IgG from ADL-treated patients. B) Individual anti-ADL IgG epitope binding pattern of ADL-treated patients. C) IgG epitopes on ADL are located in TNF-α binding regions. For each epitope, the minimal overlapping peptide sequences are indicated. ADL interaction with TNF-α drawn with pymol (Schrödinger).
Discussion
In this study, the TNF-α blockers IFX and ADL were investigated regarding their anti-drug antibody development with a special focus on IgE. IgE binding patterns were identified by ICRIP serum analyses. Of note, some patterns from patient sera are similar to patterns from non-treated control sera (NTC4, IgE against bovine TG, NTC25, IgE against IgG (resembling IgE rheumatoid factor), NTC32, α-Gal-associated meat allergy and a combined NTC25/NTC4 pattern) (Fig. 2).
The difference in signal strength already indicates the differences in glycan-specific IgE affinity to the different analytes. The central difficulty of anti-glycan antibody analysis is the availability of pure analytes. By the developed ICRIP assay, to a certain extent cross-reactive epitopes can be identified by assessing overlapping glycan and peptide motifs to IgE-binding. Glycan-independent (and thus peptide-dependent) IgE has been found for IFX438, IFX754 and ADL176. While sera IFX754 and ADL176 are not associated with other analytes of the ICRIP system, serum IgE against IFX from serum IFX438 is associated with native ADL and bovine TG. The latter pattern indicates cross-reactive glycans but is apparently not associated with α-Gal as no signals for CTX or HSA-α-Gal could be detected. However, there may be different glycans present on bovine TG, especially differentially branched glycans such as mannose-type N-glycan (Fig. 1). The glycan-dependent IgE of sera IFX922 and ADL197 and ADL198 are associated with IgE against bovine TG. Also, in these cases similar glycan structures on ADL and bovine TG may be the reason for this cross-reactivity. One major finding of the ICRIP serum analyses was a glycan-independent IgE immune profile that corresponds to IgE binding to IgG (resembling IgE rheumatoid factor) of potential clinical relevance in the context of treatment with biologicals 28-30. This binding pattern of IgE against IgG (IgE rheumatoid factor) was also identified in non-treated individuals (NTC25) and may be important for IgE cross-reactivity against therapeutic antibodies. It remains to be determined if a treatment with therapeutic antibodies may induce or boost these IgE against IgG, thus leading to clinical symptoms. Concerning immunogenic peptide epitopes, it was found that the epitopes of serum IgG from IFX-treated patients are located in the variable part of the Fab region 22. Thus, as may be expected from a chimeric antibody, non-human peptide sequences evoke an anti-idiotype immune response in the human host. However, it has been described that anti-drug antibodies also develop during treatment with humanized therapeutic antibodies like ADL. Interestingly, 3 out of 4 immunogenic epitopes located in the variable region of the heavy and light chain of ADL were already identified by epitope mapping of non-ADL-treated control sera (Fig. 3). It remains to be clarified if these epitopes on ADL are intrinsically immunogenic in a large part of the population because there are already significant epitopes of IgG in non-treated individuals. It remains to be determined if the identified immunogenic epitopes on ADL are responsible for IgE binding. IgG epitopes are potential IgE epitopes as it is known that IgG, especially subclass IgG1, serves as a precursor for IgE development by a B cell class switch from IgG to IgE in a Th2 environment 31. Taken together, distinct serum antibody profiles from patients treated with biologicals were identified using the newly established ICRIP assay for IgE detection. Via epitope mapping, specific, individual IgG epitopes on ADL for anti-drug antibodies were identified, which may result in loss-of-drug effect during therapy. Of note, these IgG epitopes may become IgE epitopes upon a corresponding B cell switch. As a vision for the future in the direction of precision medicine, patients treated with biologicals may be screened before and during treatment to detect antibody profiles associated with risks for certain therapies and with adverse drug effects such as IgE-mediated anaphylaxis.
Material and Methods
Patients and ethics
The study with sera from IFX- and ADL-treated patients has been approved by the local ethics committee, University of Lübeck (approval number 11-167). Sera from patients treated with the therapeutic antibodies infliximab (n=30) and adalimumab (n=19) were collected at a clinical laboratory (IPM Biotech, Hamburg) to which they were sent for the analysis of anti-drug antibodies. Sera positive for anti-drug antibodies of unknown isotype were identified by a bridging ELISA system (n=30 for anti-IFX, n=17 for anti-ADL, Table 1). Negative control sera were derived from North-German individuals who had not received any biological treatment and consumed (α-Gal-containing) red meat regularly without symptoms (n=40, Table 2).
Table 2.
Non-treated control sera analyzed for specific IgE content by singleplex allergy diagnostics. ICRIP results are shown for convenience (see Fig. 2). IgE tests were performed via a commercial ImmunoCAP system. IFX analyses for non-treated control sera were performed only for selected sera with marked IgE binding patterns. Note that bov TG was used for commercial α-Gal ImmunoCAP® test by Thermo Fisher Scientific / Phadia. Registered allergen Bos d 6 is (non-glycosylated) BSA, bovine serum albumin. n.a.: not available; n.t.: not tested.
| ID | CTX [kU/L] |
IFX [kU/L] |
bov TG [kU/L] |
Bos d 6 [kU/L] |
tryptase [µg/L] |
total IgE [kU/L] |
ICRIP result |
|---|---|---|---|---|---|---|---|
| NTC1 | < 0.1 | 0.00 | < 0.1 | < 0.1 | 4.0 | 2.8 | neg. |
| NTC2 | < 0.1 | n.t. | < 0.1 | < 0.1 | 13.2 | 6.2 | neg. |
| NTC3 | 0.14 | n.t. | < 0.1 | < 0.1 | 3.4 | 199 | neg. |
| NTC4 | n.t. | n.t. | < 0.1 | < 0.1 | 4.7 | 35.3 | bov TG |
| NTC5 | n.t. | n.t. | < 0.1 | < 0.1 | 2.9 | 5.0 | neg. |
| NTC6 | n.t. | n.t. | < 0.1 | < 0.1 | 5.4 | 9.3 | neg. |
| NTC7 | n.t. | n.t. | < 0.1 | < 0.1 | 3.1 | 27.3 | bov TG |
| NTC8 | n.t. | n.t. | < 0.1 | < 0.1 | 5.9 | 6.3 | neg. |
| NTC9 | < 0.1 | n.t. | < 0.1 | < 0.1 | 3.3 | 58.5 | neg. |
| NTC10 | n.t. | n.t. | < 0.1 | < 0.1 | 5.7 | 115 | neg. |
| NTC11 | 0.11 | n.t. | 0.18 | < 0.1 | 3.9 | 334 | neg. |
| NTC12 | n.t. | n.t. | < 0.1 | < 0.1 | 5.9 | 66.8 | neg. |
| NTC13 | n.t. | n.t. | < 0.1 | < 0.1 | 5.4 | 8.1 | neg. |
| NTC14 | n.t. | n.t. | < 0.1 | < 0.1 | 5.2 | 80.1 | neg. |
| NTC15 | < 0.1 | n.t. | < 0.1 | < 0.1 | 1.9 | < 2 | bov TG |
| NTC16 | n.t. | n.t. | < 0.1 | < 0.1 | 4.1 | 70.3 | bov TG |
| NTC17 | n.t. | n.t. | < 0.1 | < 0.1 | 4.3 | 10.5 | bov TG |
| NTC18 | n.t. | n.t. | < 0.1 | < 0.1 | 4.2 | 10.3 | neg. |
| NTC19 | n.t. | n.t. | < 0.1 | < 0.1 | 4.2 | 175 | neg. |
| NTC20 | n.t. | n.t. | < 0.1 | < 0.1 | 5.4 | 49.3 | neg. |
| NTC21 | < 0.1 | n.t. | < 0.1 | < 0.1 | 1.9 | 20.3 | neg. |
| NTC22 | n.t. | n.t. | < 0.1 | < 0.1 | 9.4 | 52.4 | neg. |
| NTC23 | n.t. | n.t. | < 0.1 | < 0.1 | 3.0 | 423 | neg. |
| NTC24 | 0.20 | n.t. | < 0.1 | < 0.1 | 6.9 | 58.8 | neg. |
| NTC25 | < 0.1 | < 0.1 | < 0.1 | < 0.1 | 3.3 | 477 | ADL/ADL PI/CTX/CTX PI/IFX/IFX PI |
| NTC26 | n.t. | n.t. | < 0.1 | < 0.1 | 3.8 | 39.2 | neg. |
| NTC27 | n.t. | n.t. | < 0.1 | < 0.1 | 8.1 | 105 | neg. |
| NTC28 | n.t. | n.t. | < 0.1 | < 0.1 | 3.1 | 26.7 | bov TG |
| NTC29 | n.t. | n.t. | < 0.1 | < 0.1 | 6.4 | 87.3 | neg. |
| NTC30 | n.t. | n.t. | < 0.1 | < 0.1 | 8.6 | 17.4 | bov TG |
| NTC31 | < 0.1 | n.t. | < 0.1 | < 0.1 | 3.9 | 41.1 | neg. |
| NTC32 | 17.3 | 0.14 | 16.1 | < 0.1 | 4.3 | 39.0 | CTX/HSA-α-Gal/bov TG |
| NTC33 | n.t. | n.t. | < 0.1 | < 0.1 | 13.2 | 46.7 | neg. |
| NTC34 | < 0.1 | n.t. | < 0.1 | < 0.1 | 6.3 | 8.4 | neg. |
| NTC35 | 0.11 | n.t. | < 0.1 | < 0.1 | 7.6 | 188 | neg. |
| NTC36 | n.t. | n.t. | < 0.1 | < 0.1 | 1.9 | 106 | neg. |
| NTC37 | < 0.1 | n.t. | < 0.1 | < 0.1 | 6.5 | 4.1 | neg. |
| NTC38 | < 0.1 | n.t. | < 0.1 | < 0.1 | 5.0 | 7.4 | neg. |
| NTC39 | < 0.1 | n.t. | < 0.1 | < 0.1 | 4.4 | 102 | bov TG |
| NTC40 | 4.46 | < 0.1 | 1.73 | < 0.1 | 3.6 | 165 | CTX |
Determination of anti-infliximab/anti-adalimumab antibodies by ELISA
The anti-IFX serum antibody titer was determined by a heterogeneous bridging ELISA test (IPM Biotech) as described before 22. Briefly, for this test a 96-well microtiter plate (Nunc maxisorp, VWR) was coated with 3 µg/mL of IFX per well. 100 µL of diluted patient sera (1:10) were acid-treated to dissolve potential immune complexes (100 mM acetic acid, 5 min, room temperature (RT)). The microtiter plate was incubated at 4 °C overnight. On the following day, wells were filled with acetic acid (100 mM) and after a 5 min incubation at RT, acid-treated fractions were transferred onto an untreated plate. After an incubation time of 60 min (RT), the wells were filled with 200 µL blocking buffer (5% BSA, 5% Casein in PBST). After another 60 min of incubation at RT, 10 µL biotinylated IFX (0.25 µg/mL) were added to each well. 100 µL streptavidin (0.5 µg/mL) conjugated with horseradish peroxidase (HRP) were added to each well. Microtiter plates were analyzed by an ELISA reader (Dynex, MRX) at wavelengths of 450 nm and 630 nm. All analyses were performed in duplicate. Anti-IFX standards (quantification range 4 - 80 µg/mL) were run on each microtiter plate. The analysis of anti-ADL antibodies was performed accordingly except for the use of a biotinylated anti-ADL reagent.
Analysis of sera by IgE cross-reactivity immune profiling (ICRIP) assay
For the analysis of the binding characteristics of serum IgE, a blot-based in vitro assay system was established. This versatile analysis system facilitates the parallel analysis of up to 12 different analytes with one serum incubation. The analysis system includes native and glycan-processed forms (see below) of the therapeutic antibodies ADL, CTX and IFX. As a pure α-Gal component, a chemical conjugate of HSA-α-Gal in the form of a disaccharide as well as a trisaccharide was included (Fig. 1). Additionally, the commercial analyte for α-Gal singleplex ImmunoCAP® diagnostics, bovine TG, as well as its human counterpart, hu TG containing the human glycan pattern, was included in the assay system. Bovine serum albumin (BSA, registered as allergen Bos d 6) represented a non-glycosylated meat allergen in the assay system. All assay components were spotted in a volume of 1 µL at a concentration of 1 µg/µL onto a nitrocellulose membrane (Amersham 0.45 µm, GE Healthcare/Thermo Fisher Scientific). If not indicated otherwise, sera were diluted 1:20 in Tris-buffered saline pH 7.4 with polysorbate (Tween20®, TBST). Serum incubation was performed overnight at RT on a horizontal shaker. IgE-binding was detected by incubation (2h, RT) with an anti-IgE antibody conjugated with horseradish peroxidase (HRP, Southern Biotech) in a dilution of 1:10,000 in TBST. For luminescence development, a kit was used that contains the substrate for the HRP enzyme (Clarity kit, Bio-Rad). Signals were analyzed by a chemiluminescence reader (Chemidoc, Bio-Rad). A positive signal for an assay component was defined as being more intense than the background signal for the negative control serum NTC1 multiplied by a factor of 2. In order to establish and verify the performance of the ICRIP assay, sera from meat allergy patients with known binding properties to different forms of α-Gal were used 27.
Processing of glycans by oxidation
Commercially available therapeutic antibodies ADL, CTX and IFX were dialyzed against water to remove drug formulation additives and freeze-dried for storage (Slide-a-Lyzer dialysis cassettes, MWCO 20 kDa, Thermo Fisher Scientific). Periodate oxidation of monosaccharides attached to proteins was performed as previously described (Thermo-Fisher protocol for sodium meta-periodate). Briefly, biologicals were incubated with 10 mM sodium periodate in phosphate-buffered saline (PBS), pH 6 for 30 min at RT. After the procedure, the periodate-treated proteins were dialyzed against water overnight at RT (Slide-a-Lyzer dialysis cassettes, 20 kDa MWCO, Thermo Fisher Scientific). Protein integrity was checked by SDS-PAGE analysis (10% acrylamide, Thermo Fisher Scientific), each lane containing 3 µg of protein (Fig. S2A). Successful oxidation of glycans was verified by lectin binding analysis with BS-I (dilution of 1:200 in TBST, pH 7.4, 0.5% Tween20®), ConA (1:10,000), SNA (1:80,000) (all from Vector Labs). Binding of biotinylated lectins was detected by incubation with alkaline phosphatase (AP)-conjugated streptavidin (Sigma-Aldrich) for 30 min at RT. After 3x washing with TBST (each time for 10 min), signals were obtained by NBT/BCIP development (Fig. S2B). As positive control, dot blot membrane was incubated with AP-anti-Fc (Sigma-Aldrich) for 2 h at RT.
Epitope mapping of anti-adalimumab serum IgG by peptide microarrays
Epitope mapping of IgG was performed with sera from ADL-treated patients as previously described for sera from IFX-treated patients 22. Briefly, the peptide sequence of ADL without the constant region of the light chain was spotted as 15-meric oligopeptides overlapping by 2 amino acids onto cellulose-coated glass slides (Celluspot, intavis). Slides were blocked with casein (5%, Hammersten grade, Sigma Aldrich) for 30 min at RT on rocking shaker. Serum was diluted 1:500 in TBST (pH 7.4, 0.5% Tween20) and incubated overnight at 4 °C on a rocking shaker. On the next day, the slides are incubated with a fluorescence-labelled anti-IgG antibody (700 nm absorption maximum, Li-Cor) and analyzed by a fluorescence imager (Li-Cor imager). Signal intensities for the secondary-antibody-only control were subtracted for each spot on the array. An epitope was defined as valid signal when fluorescence intensities are higher than the average of the signal for the included random peptides, multiplied by 3x standard deviation. This procedure results in a probability of p=0.99 32.
Analysis of mapped ADL epitopes by oligopeptide ELISA
Selected peptides (Table S2) were synthesized as described previously 22. In order to facilitate the immobilization of the oligopeptides to the ELISA plate, a biotin group was added to each peptide by using a biotinyl resin (Biotin PEG NovaTag 0.24 mmol/g, Merck, Darmstadt, Germany) for solid phase synthesis. ELISA plates pre-coated with streptavidin (ThermoFisher, Pierce, Germany) were blocked in blocking buffer (5% BSA (w/v) in TBS) overnight on a horizontal shaker at 4 °C. The wells were washed 4x with TBST. Next, the biotinylated peptides (dissolved in DMSO as stock solution) were diluted 1:1000 in TBST, applied into the corresponding wells and incubated for 1 h at RT under shaking. Sera from patients or untreated controls were applied at a dilution of 1:200 in blocking buffer and incubated overnight at 4 °C on a shaker. On the next day, the wells were washed 4x with TBST and HRP-conjugated monoclonal anti-human IgG (ThermoFisher, #054220) diluted 1:500 in blocking buffer was applied. After incubation for 90 min at RT on a shaker, wells were washed 4x with TBST. Antibody binding was visualized by addition of tetramethyl benzidine (TMB, Sigma-Aldrich/Merck, Darmstadt, Germany). After 10 min incubation at RT, the enzymatic reaction was stopped by addition of sulphuric acid (2 M), and signal intensities were measured with a microplate reader at a wavelength of 450 nm (Tecan infinite M200, Maennedorf, Switzerland). A positive signal was defined as an OD450 over 3x higher than the highest detected negative control signal (0.15, anti-IgG only against peptide C2), resulting in a threshold of OD450 = 0.45.
Supplementary Material
Characteristics for selected patients treated with adalimumab are summarized in Table S1. Selected peptides identified as IgG epitopes and examined by peptide ELISA are shown in Table S2. Sequence alignment of CTX and IFX indicates a similar peptide sequence, but a different Fab glycosylation due to the absence of a N-glycosylation Sequon in IFX (Fig. S1). Integrity analysis of periodate-treated ICRIP components are shown in figure S2A. Lectin binding analysis provide information about the accessible glycans on the ICRIP assay components (Fig. S2B). For validation of mapped IgG epitopes on ADL, a peptide ELISA assay was performed with selected peptides and selected sera (Fig. S3).
Acknowledgments
The excellent technical assistance of Lea Bendler from the Research Center Borstel with all laboratory work is gratefully acknowledged. The authors would like to thank Katja Habakuk from IPM Biotech and Angela Willanzheimer from Labor Lademannbogen for collecting sera and performing serum diagnostics. The support of the medical laboratory head of the Labor Lademannbogen, Dr. Andreas Lämmel, is very much appreciated. The project was funded by the German Ministry for Economy and Technology (BMWi) via the AiF-ZIM program, project number KF278402SB4.
Author contributions
AH performed experiments. AK provided sera from patients treated with infliximab and adalimumab. AH and UJ designed experiments. UJ obtained ethical approval. Epitope mapping procedures were developed by NR and AF. TPM developed and provided CTX and IFX singleplex discs for serum analysis. AH and UJ wrote the manuscript. NR, AF and TPM contributed to the writing process with corrections and helpful discussions. All authors read and approved the final manuscript.
Abbreviations
- ADL
adalimumab
- α-Gal2
galactosyl-α-(1,3)-galactose
- α-Gal3
galactosyl-α-1,3-galactosyl-β-1,4-N-acetyl-glucosamine
- AP
alkaline phosphatase
- BSA
bovine serum albumin
- CH
heavy chain
- CTX
cetuximab
- HRP
horseradish peroxidase
- HSA
human serum albumin
- NTC
non-treated controls
- ICRIP
IgE cross-reactivity immune profiling
- IFX
infliximab
- PBS
phosphate buffered saline
- PI
periodate
- RT
room temperature
- TG
thyroglobulin
- TNF
tumor necrosis factor
- VH
heavy chain variable region
- VL
light chain variable region
- WHO/IUIS
World Health Organisation/International Union of Immunological Societies.
References
- 1.Chan AC, Carter PJ. Therapeutic antibodies for autoimmunity and inflammation. Nat Rev Immunol. 2010 May 1;10(5):301–16. doi: 10.1038/nri2761. [DOI] [PubMed] [Google Scholar]
- 2.Pichler WJ. Adverse side-effects to biological agents. Allergy. 2006 Aug;61(8):912–20. doi: 10.1111/j.1398-9995.2006.01058.x. [DOI] [PubMed] [Google Scholar]
- 3.van Schouwenburg PA, Rispens T, Wolbink GJ. Immunogenicity of anti-TNF biologic therapies for rheumatoid arthritis. Nat Rev Rheumatol. 2013 Mar;9(3):164–72. doi: 10.1038/nrrheum.2013.4. [DOI] [PubMed] [Google Scholar]
- 4.Steenholdt C, Svenson M, Bendtzen K, Thomsen O ø, Brynskov J, Ainsworth MA. Severe infusion reactions to infliximab: aetiology, immunogenicity and risk factors in patients with inflammatory bowel disease: Severe infusion reactions to infliximab in IBD. Aliment Pharmacol Ther. 2011 Jul;34(1):51–8. doi: 10.1111/j.1365-2036.2011.04682.x. [DOI] [PubMed] [Google Scholar]
- 5.Radstake TRDJ, Svenson M, Eijsbouts AM, van den Hoogen FHJ, Enevold C, van Riel PLCM. et al. Formation of antibodies against infliximab and adalimumab strongly correlates with functional drug levels and clinical responses in rheumatoid arthritis. Ann Rheum Dis. 2009 Nov 1;68(11):1739–45. doi: 10.1136/ard.2008.092833. [DOI] [PubMed] [Google Scholar]
- 6.Yanai H, Lichtenstein L, Assa A, Mazor Y, Weiss B, Levine A. et al. Levels of Drug and Antidrug Antibodies Are Associated With Outcome of Interventions After Loss of Response to Infliximab or Adalimumab. Clin Gastroenterol Hepatol. 2015 Mar;13(3):522–530.e2. doi: 10.1016/j.cgh.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 7.Atzeni F, Talotta R, Salaffi F, Cassinotti A, Varisco V, Battellino M. et al. Immunogenicity and autoimmunity during anti-TNF therapy. Autoimmun Rev. 2013 May;12(7):703–8. doi: 10.1016/j.autrev.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 8.Vultaggio A, Castells MC. Hypersensitivity Reactions to Biologic Agents. Immunol Allergy Clin North Am. 2014 Aug;34(3):615–32. doi: 10.1016/j.iac.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 9.Corominas M, Gastaminza G, Lobera T. Hypersensitivity Reactions to Biological Drugs. J Investig Allergol Clin Immunol. 2014;24(4):212–225. [PubMed] [Google Scholar]
- 10.Murdaca G, Spanò F, Contatore M, Guastalla A, Penza E, Magnani O. et al. Immunogenicity of infliximab and adalimumab: what is its role in hypersensitivity and modulation of therapeutic efficacy and safety? Expert Opin Drug Saf. 2016 Jan 2;15(1):43–52. doi: 10.1517/14740338.2016.1112375. [DOI] [PubMed] [Google Scholar]
- 11.Steenholdt C, Svenson M, Bendtzen K, Thomsen OØ, Brynskov J, Ainsworth MA. Acute and delayed hypersensitivity reactions to infliximab and adalimumab in a patient with Crohn's disease. J Crohns Colitis. 2012 Feb;6(1):108–11. doi: 10.1016/j.crohns.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Chung CH, Mirakhur B, Chan E, Le Q-T, Berlin J, Morse M. et al. Cetuximab-Induced Anaphylaxis and IgE Specific for Galactose-α-1,3-Galactose. N Engl J Med. 2008 Mar 13;358(11):1109–17. doi: 10.1056/NEJMoa074943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fréling E, Peyrin-Biroulet L, Poreaux C, Morali A, Waton J, Schmutz J-L. et al. IgE antibodies and skin tests in immediate hypersensitivity reactions to infliximab in inflammatory bowel disease: impact on infliximab retreatment. Eur J Gastroenterol Hepatol. 2015 Oct;27(10):1200–8. doi: 10.1097/MEG.0000000000000436. [DOI] [PubMed] [Google Scholar]
- 14.Hwang SH, Yoo H-S, Yoon MK, Park H-S. Detection of IgE binding component to infliximab in a patient with infliximab-induced anaphylaxis. Ann Allergy Asthma Immunol. 2014;112(4):393–394. doi: 10.1016/j.anai.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Kato S, Kobayashi T, Kani K, Takabayashi H, Yamamoto R, Yakabi K. Elevated serum IgE prior to acute severe infusion reaction during infliximab maintenance therapy in a crohnʼs disease patient. Inflamm Bowel Dis. 2011 Dec;17(12):E156–7. doi: 10.1002/ibd.21857. [DOI] [PubMed] [Google Scholar]
- 16.Vultaggio A, Matucci A, Nencini F, Pratesi S, Parronchi P, Rossi O. et al. Anti-infliximab IgE and non-IgE antibodies and induction of infusion-related severe anaphylactic reactions. Allergy. 2010 May;65(5):657–61. doi: 10.1111/j.1398-9995.2009.02280.x. [DOI] [PubMed] [Google Scholar]
- 17.Steenholdt C, Palarasah Y, Bendtzen K, Teisner A, Brynskov J, Teisner B. et al. Pre-existing IgG antibodies cross-reacting with the Fab region of infliximab predict efficacy and safety of infliximab therapy in inflammatory bowel disease. Aliment Pharmacol Ther. 2013 Jun;37(12):1172–83. doi: 10.1111/apt.12330. [DOI] [PubMed] [Google Scholar]
- 18.Xue L, Rup B. Evaluation of Pre-existing Antibody Presence as a Risk Factor for Posttreatment Anti-drug Antibody Induction: Analysis of Human Clinical Study Data for Multiple Biotherapeutics. AAPS J. 2013 Jul;15(3):893–6. doi: 10.1208/s12248-013-9497-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steenholdt C, Ainsworth MA, Tovey M, Klausen TW, Thomsen OØ, Brynskov J. et al. Comparison of Techniques for Monitoring Infliximab and Antibodies Against Infliximab in Crohnʼs Disease. Ther Drug Monit. 2013 Aug;35(4):530–8. doi: 10.1097/FTD.0b013e31828d23c3. [DOI] [PubMed] [Google Scholar]
- 20.Harding FA, Stickler MM, Razo J, DuBridge RB. The immunogenicity of humanized and fully human antibodies. Residual Immunogenicity Resides CDR Reg MAbs. 2010;2(3):256–65. doi: 10.4161/mabs.2.3.11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frederiksen MT, Ainsworth MA, Brynskov JM, Thomsen OOM, Bendtzen KM, Steenholdt C. Antibodies Against Infliximab Are Associated with De Novo Development of Antibodies to Adalimumab and Therapeutic Failure in Infliximab-to-Adalimumab Switchers with IBD. Inflamm Bowel Dis. 2014 Oct;20(10):1714–21. doi: 10.1097/MIB.0000000000000138. [DOI] [PubMed] [Google Scholar]
- 22.Homann A, Röckendorf N, Kromminga A, Frey A, Jappe U. B cell epitopes on infliximab identified by oligopeptide microarray with unprocessed patient sera. J Transl Med. 2015 Oct 29;13(1):339. doi: 10.1186/s12967-015-0706-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu S, Liang S, Guo H, Zhang D, Li H, Wang X. et al. Comparison of the Inhibition Mechanisms of Adalimumab and Infliximab in Treating Tumor Necrosis Factor -Associated Diseases from a Molecular View. J Biol Chem. 2013 Sep 20;288(38):27059–67. doi: 10.1074/jbc.M113.491530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang S, Dai J, Hou S, Su L, Zhang D, Guo H. et al. Structural Basis for Treating Tumor Necrosis Factor (TNF )-associated Diseases with the Therapeutic Antibody Infliximab. J Biol Chem. 2013 May 10;288(19):13799–807. doi: 10.1074/jbc.M112.433961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Commins SP, Platts-Mills TAE. Delayed Anaphylaxis to Red Meat in Patients with IgE Specific for Galactose alpha-1,3-Galactose (alpha-gal) Curr Allergy Asthma Rep. 2013 Feb;13(1):72–7. doi: 10.1007/s11882-012-0315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayoub D, Jabs W, Resemann A, Evers W, Evans C, Main L. et al. Correct primary structure assessment and extensive glyco-profiling of cetuximab by a combination of intact, middle-up, middle-down and bottom-up ESI and MALDI mass spectrometry techniques. mAbs. 2013 Sep;5(5):699–710. doi: 10.4161/mabs.25423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jappe U, Minge S, Kreft B, Ludwig A, Przybilla B, Walker A, Meat allergy associated with galactosyl-α-(1,3)-galactose (α-Gal)-Closing diagnostic gaps by anti-α-Gal IgE immune profiling. Allergy; 2017. Jul 3 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gioud-Paquet M, Auvinet M, Raffin T, Girard P, Bouvier M, Lejeune E. et al. IgM rheumatoid factor (RF), IgA RF, IgE RF, and IgG RF detected by ELISA in rheumatoid arthritis. Ann Rheum Dis. 1987;46(1):65–71. doi: 10.1136/ard.46.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruber B, Ballan D, Gorevic PD. IgE rheumatoid factors: quantification in synovial fluid and ability to induce synovial mast cell histamine release. Clin Exp Immunol. 1988;71(2):289. [PMC free article] [PubMed] [Google Scholar]
- 30.Zuraw BL, O'Hair CH, Vaughan JH, Mathison DA, Curd JG, Katz DH. Immunoglobulin E-rheumatoid factor in the serum of patients with rheumatoid arthritis, asthma, and other diseases. J Clin Invest. 1981 Dec;68(6):1610–3. doi: 10.1172/JCI110418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Looney TJ, Lee J-Y, Roskin KM, Hoh RA, King J, Glanville J. et al. Human B-cell isotype switching origins of IgE. J Allergy Clin Immunol. 2016 Feb;137(2):579–586.e7. doi: 10.1016/j.jaci.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frey A, Di Canzio J, Zurakowski D. A statistically defined endpoint titer determination method for immunoassays. J Immunol Methods. 1998 Dec 1;221(1-2):35–41. doi: 10.1016/s0022-1759(98)00170-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics for selected patients treated with adalimumab are summarized in Table S1. Selected peptides identified as IgG epitopes and examined by peptide ELISA are shown in Table S2. Sequence alignment of CTX and IFX indicates a similar peptide sequence, but a different Fab glycosylation due to the absence of a N-glycosylation Sequon in IFX (Fig. S1). Integrity analysis of periodate-treated ICRIP components are shown in figure S2A. Lectin binding analysis provide information about the accessible glycans on the ICRIP assay components (Fig. S2B). For validation of mapped IgG epitopes on ADL, a peptide ELISA assay was performed with selected peptides and selected sera (Fig. S3).