The role of proteins with sorbin and Src homology 3 domains in living organisms is poorly understood, despite the high degree of conservation of these proteins in metazoans. The single member of this protein family in Caenorhabditis elegans plays roles in muscle contraction at the worm equivalent of Z-disks and in positioning organelles.
Abstract
We have identified and characterized sorb-1, the only sorbin and SH3 domain–containing protein family member in Caenorhabditis elegans. SORB-1 is strongly localized to integrin adhesion complexes in larvae and adults, including adhesion plaques and dense bodies (Z-disks) of striated muscles and attachment plaques of smooth muscles. SORB-1 is recruited to the actin-binding, membrane-distal regions of dense bodies via its C-terminal SH3 domains in an ATN-1(α-actinin)– and ALP-1(ALP/Enigma)–dependent manner, where it contributes to the organization of sarcomeres. SORB-1 is also found in other tissues known to be under mechanical stress, including stress fibers in migratory distal tip cells and the proximal gonad sheath, where it becomes enriched in response to tissue distention. We provide evidence for a novel role for sorbin family proteins: SORB-1 is required for normal positioning of the mitochondrial network in muscle cells. Finally, we demonstrate that SORB-1 interacts directly with two other dense body components, DEB-1(vinculin) and ZYX-1(zyxin). This work establishes SORB-1 as a bona fide sorbin family protein—one of the late additions to the dense body complex and a conserved regulator of body wall muscle sarcomere organization and organelle positioning.
INTRODUCTION
Focal adhesions are asymmetric adhesive structures that transmit force between the extracellular matrix and the cytoskeleton, allowing generation of cellular traction and mechanotransduction (Ciobanasu et al., 2013). Because these functions impinge upon diverse cellular processes, including proliferation, apoptosis, adhesion, extracellular matrix remodeling, and migration, focal adhesions are crucial mechanical and signaling complexes during normal development, wound repair, and oncogenesis. Focal adhesions typically bind the extracellular matrix via α/β-integrin heterodimers, which themselves are bound intracellularly by a host of kinases, actin-binding proteins, and intermolecular scaffolds (Zaidel-Bar et al., 2007; Case and Waterman, 2015). Among the latter are sorbin family proteins, which are characterized by an N-terminal sorbin homology (SoHo) domain and three C-terminal Src homology 3 (SH3) domains.
The mammalian sorbin family consists of CAP/Ponsin, ArgBP2, and vinexin, which are further diversified by alternative splicing variants, including isoforms lacking the third SH3 domain (Wang et al., 1997) or the SoHo domain (Kioka et al., 1999; Murase et al., 2012). Sorbin itself, which is produced by an alternate transcript from the ArgBP2 locus (Hand and Eiden, 2005), lacks all SH3 domains (Vagne-Descroix et al., 1991) and plays nonadhesive roles in the duodenal lumen, instead promoting absorption of water and sodium ions (Pansu et al., 1981; Charpin-Elhamri et al., 2000). With the exception of sorbin, these proteins are widely expressed and particularly abundant in striated muscles, and they localize to focal adhesions, stress fibers, adherens junctions, and the nucleus (Wang et al., 1997; Kioka et al., 1999; Mandai et al., 1999; Lebre et al., 2001), suggesting that they may play a role in cross-talk between adhesive structures.
Sorbin family proteins have been proposed to strengthen focal adhesions through multiple pathways: by promoting local F-actin accumulation and stress fiber formation (Kioka et al., 1999), by inhibiting focal adhesion disassembly (Zhang et al., 2006), and by impeding integrin receptor endocytosis (Cestra et al., 2005; Tosoni and Cestra, 2009). By stabilizing focal adhesions, sorbin family proteins may inhibit cell spreading and migration (Zhang et al., 2006; Taieb et al., 2008; Fernow et al., 2009; Kioka et al., 2010; Roignot et al., 2010), although there is also evidence that sorbin proteins may inhibit these processes by negatively regulating lamellipodial actin dynamics (Cestra et al., 2005).
Given the considerable overlap in expression and function of vertebrate sorbin family proteins, simpler model systems with less redundancy could be useful for studying their functions in vivo. The Drosophila sorbin homologue, DCAP/rexin, has been reported to play roles in glucose metabolism (Yamazaki and Yanagawa, 2003), sensation of vibration and sound, and maintenance of muscle morphology (Bharadwaj et al., 2013). Here we identify and characterize the sole Caenorhabditis elegans sorbin homologue, SORB-1, which localizes to focal adhesion–like structures in myocytes. In nematode body wall muscle cells, SORB-1 is recruited to the actin-binding region of dense bodies, which serve as a type of focal adhesion and, as a homologous structure to Z-disks, transmit the force of muscle contraction (Francis and Waterston, 1985; Moerman and Williams, 2006; Gieseler et al., 2017). Additionally, we have found that SORB-1 is recruited to stress fibers in two cell types of the somatic gonad, and it performs a previously unidentified function for sorbin family proteins in the positioning of mitochondria in body wall muscle cells.
RESULTS
sorb-1 encodes the sole sorbin family gene in C. elegans
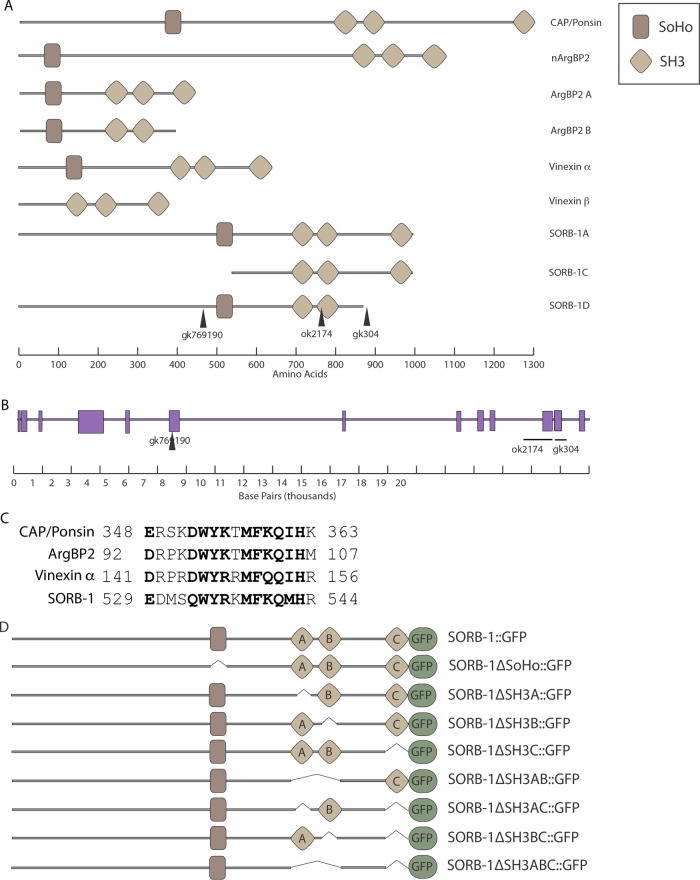
Sequencing of C. elegans cDNA clones yk357a12 and yk22a10 revealed two splicing variants of a single open reading frame (ORF); the first of these, sorb-1a, encodes a product with an N-terminal SoHo domain and three C-terminal SH3 domains. Sequence alignment of the predicted SORB-1A protein against the human proteome showed roughly equivalent amino acid identity to CAP/Ponsin, ArgBP2, and vinexin (32, 33, and 28%, respectively), although vinexin was ranked as the most significant match by E value (8 × 10–33 for vinexin vs. 9 × 10–24 and 2 × 10–30 for CAP/Ponsin and ArgBP2, respectively). In keeping with the Human Genome Organization Nomenclature Committee classification of sorbin and SH3 domain–containing family genes, we have named this homologue sorb-1. cDNA yk22a10 provides evidence for an alternatively spliced variant, sorb-1d, that lacks the third SH3 domain and is therefore structurally similar to ArgBP2 B (http://www.wormbase.org/get?name=CB102708;class=clone; Figure 1).
FIGURE 1:
SORB-1 is homologous to human SoHo proteins. (A) Sorbin family proteins contain an N-terminal SoHo domain and three C-terminal SH3 domains, with splicing variants that lack either the SoHo domain (as in vinexin β) or the most C-terminal SH3 domain (as in ArgBP2 B). SORB-1A and SORB-1D structures were predicted from cDNAs yk357a12 and yk22a10, respectively. Lesion gk769190 is an amber nonsense mutation. gk304 and ok2174 are complex deletions/insertions, which cause frame shifts that are predicted to truncate SORB-1 where indicated. (B) The sorb-1 genomic region; exons are indicated as purple rectangles, and the genetic lesions used in this study are indicated. (C) Conservation within the core region of the SoHo domain establishes C. elegans as a sorbin family member. Conserved residues are in bold. Numbers indicate N- and C-terminal residue positions. (D) Single and multiple domain deletions of SORB-1::GFP were used to assay SORB-1 localization in this study.
SORB-1 localizes to several types of integrin adhesion sites of both striated and smooth muscles
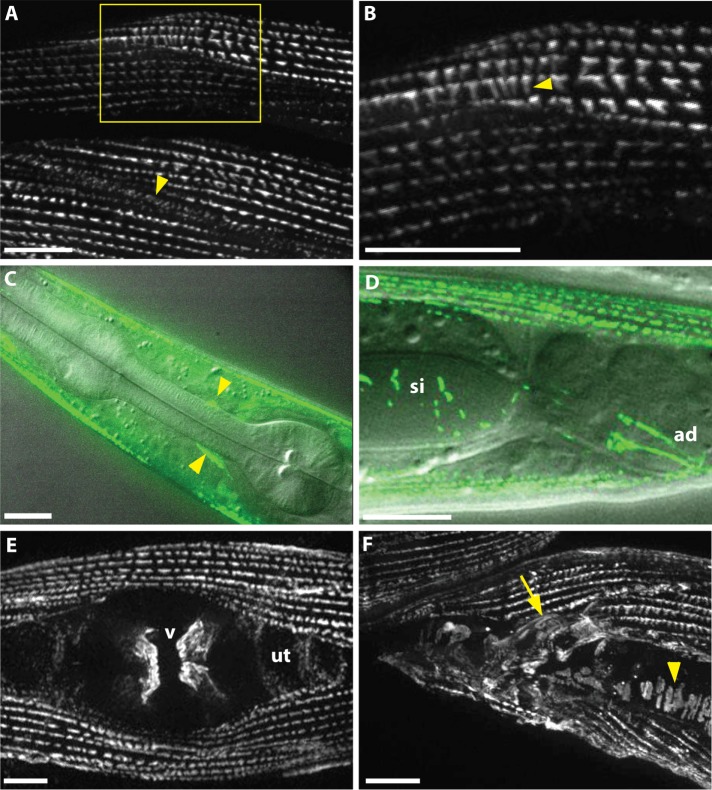
A translational sorb-1::gfp fusion driven by 2 kb of endogenous promoter was used to determine expression and subcellular localization of SORB-1 protein in C. elegans. sorb-1::gfp was strongly expressed in all body wall muscles, where the protein tagged with green fluorescent protein (GFP) localized to adhesion plaques between myocytes and to dense bodies (Figure 2, A and B). To confirm the dense body localization and determine its depth, we costained sorb-1::gfp animals with antibodies to known dense body components. As shown in Figure 3, SORB-1::GFP colocalizes with both DEB-1(vinculin) and ATN-1(α-actinin). However, vinculin is known to be located at the membrane-proximal base of dense bodies (Barstead and Waterston, 1989), whereas α-actinin is known to be located in the deeper, membrane-distal portion of the finger-like dense body structure. As indicated from a confocal Z-series, SORB-1 colocalizes with both DEB-1 and ATN-1, although it seems to be more associated with DEB-1 (Supplemental Figure S1): as confocal sections progress from the outer muscle cell membrane to deeper into the cell, the SORB-1::GFP signal attenuates at the same position as DEB-1, whereas the ATN-1 signal persists deeper into the cell.
FIGURE 2:
SORB-1::GFP localizes to muscle dense bodies and other muscle attachment sites. (A) SORB-1 localizes to dense bodies and adhesion plaques between myocytes (arrowhead). Image was created from a Z-projection of 10 focal planes. (B) Zoom of boxed region in A. Fluorescent signal localizes along the depth of the dense body structures (arrowhead). (C) Composite of fluorescence and DIC imaging, identifying SORB-1::GFP in muscle arm attachments at the nerve ring (arrowheads). (D) Composite of fluorescence and DIC imaging, lateral view. SORB-1::GFP signal localizes to intestinal attachment sites of the stomatointestinal (si) and anal depressor (ad) muscles. (E) Hermaphrodite, ventral view. Signal localizes to attachments of the vulval muscles (v) and is diffusely punctate in the underlying uterus (ut). (F) Male, lateral view. SORB-1 localizes to attachments of the diagonal (arrowhead) and spicule-associated muscles (arrow) of the male tail. Scale bars = 10 μm.
FIGURE 3:
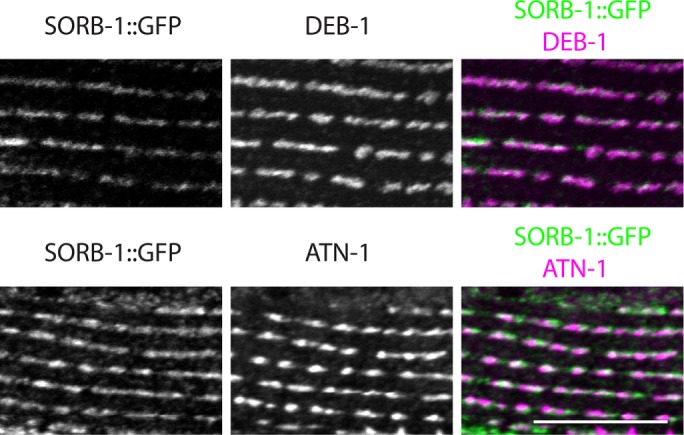
SORB-1::GFP colocalizes with both DEB-1 and ATN-1. sorb-1::gfp animals were fixed and costained with antibodies to GFP and antibodies to DEB-1 (top row) or antibodies to ATN-1 (bottom row). A portion of a single body wall muscle cell is shown. Scale bar = 10 μm.
SORB-1::GFP was also observed in muscle arm attachment sites at the nerve ring (Figure 2C), where protrusions from head muscles form contacts with the basement membrane (White et al., 1986). SORB-1::GFP was also observed in all nonstriated muscles of C. elegans with the exception of the pharynx, localizing exclusively to integrin adhesion sites. Fluorescent signal localized strongly to the origins and insertions of the vulval and anal depressor muscles (Figure 2, D and E) as well as to the spicule-associated and diagonal muscles of the male tail (Figure 2F). In myocytes more closely resembling vertebrate smooth muscle, such as the uterus, stomatointestinal muscle, and proximal gonad sheath, GFP signal was present in small puncta present throughout the tissue (Figure 2, D and E, and Supplemental Figure S4B). In both smooth and striated muscles in C. elegans, SORB-1 localizes strongly at sites subject to mechanical stress, suggesting a role in strengthening the adhesive structures at these sites and/or cell signaling in response to tensile force.
SORB-1 localization in body wall muscles requires its SH3 domains and the membrane-distal dense body components ATN-1 and ALP-1, but not FRG-1
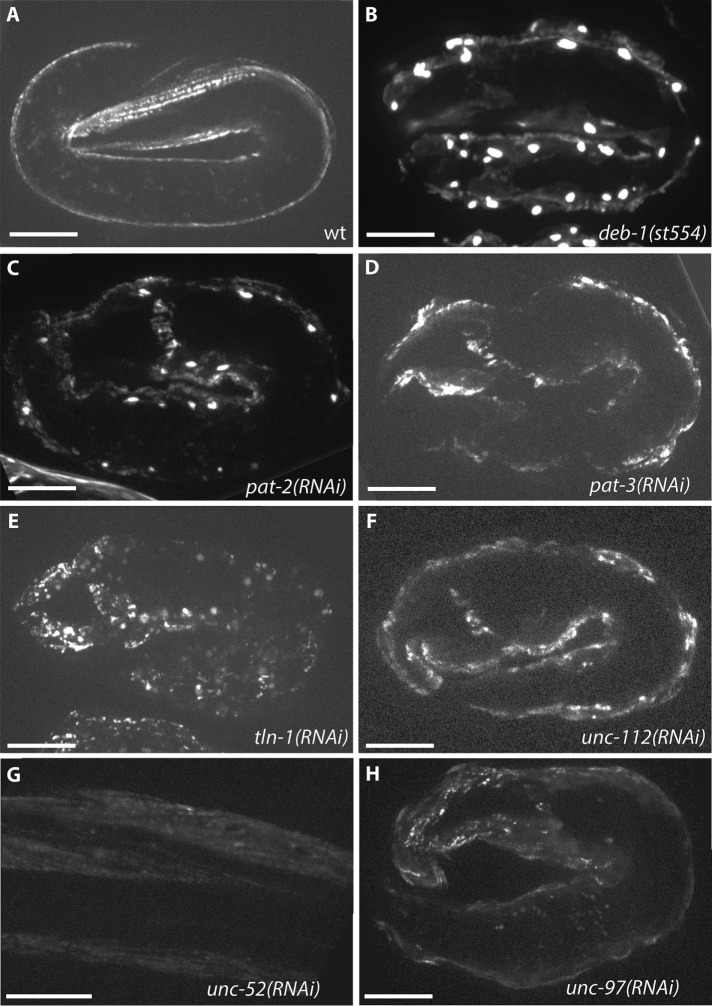
Because the localization of sorbin family proteins in cultured mammalian cells depends on integrins and other proteins in integrin adhesion complexes, we next examined the genetic requirements for dense body recruitment of SORB-1 in C. elegans. Dense bodies are the functional and anatomical equivalents of Z-disks in vertebrate striated muscle, and because of their anchorage to the outer sarcolemma and what is known about their protein composition, they are one form of focal adhesion (the other being the M-line; Moerman and Williams, 2006; Lecroisey et al., 2007; Qadota and Benian, 2010; Gieseler et al., 2017). Proper localization of SORB-1 begins at approximately the twofold stage of embryonic elongation (Figure 4A) and depends on core dense body proteins and early organizers of the dense body, including PAT-2(α-integrin), PAT-3(β-integrin), UNC-112(kindlin), DEB-1(vinculin), UNC-52(perlecan), UNC-97(PINCH), and TLN-1(talin). Single RNA interference (RNAi) knockdown or genomic mutation of each of these components did not abrogate sorb-1::gfp expression, but fluorescent signal was mislocalized to broad patches at the membrane of Pat (Paralyzed, Arrested at Twofold) embryos (Figure 4). SORB-1::GFP appears to accumulate in the nucleus of deb-1(st554) and pat-2(RNAi) embryos (Figure 4, B and C, and Supplemental Figure S2), whereas no punctate accumulation was observed with knockdown of the core dense body component pat-3 (Figure 4D).
FIGURE 4:
Early SORB-1 localization requires dense body components. (A) SORB-1::GFP accumulates at dense bodies beginning at the twofold stage of embryogenesis and persists throughout adulthood. wt, wild type. (B–H) Mislocalization of SORB-1::GFP in a twofold embryo resulting from loss of indicated dense body components. Nuclear accumulation of SORB-1 was observed in deb-1(st554) embryos (B) and was observed less strongly in pat-2(RNAi) and tln-1(RNAi) embryos (C and E). Nuclear localization of SORB-1::GFP was not observed in pat-3(RNAi), unc-112(RNAi), or unc-97(RNAi) embryos (D, F, and H) or in unc-52(RNAi) L1 larvae (G). Scale bars = 10 μm.
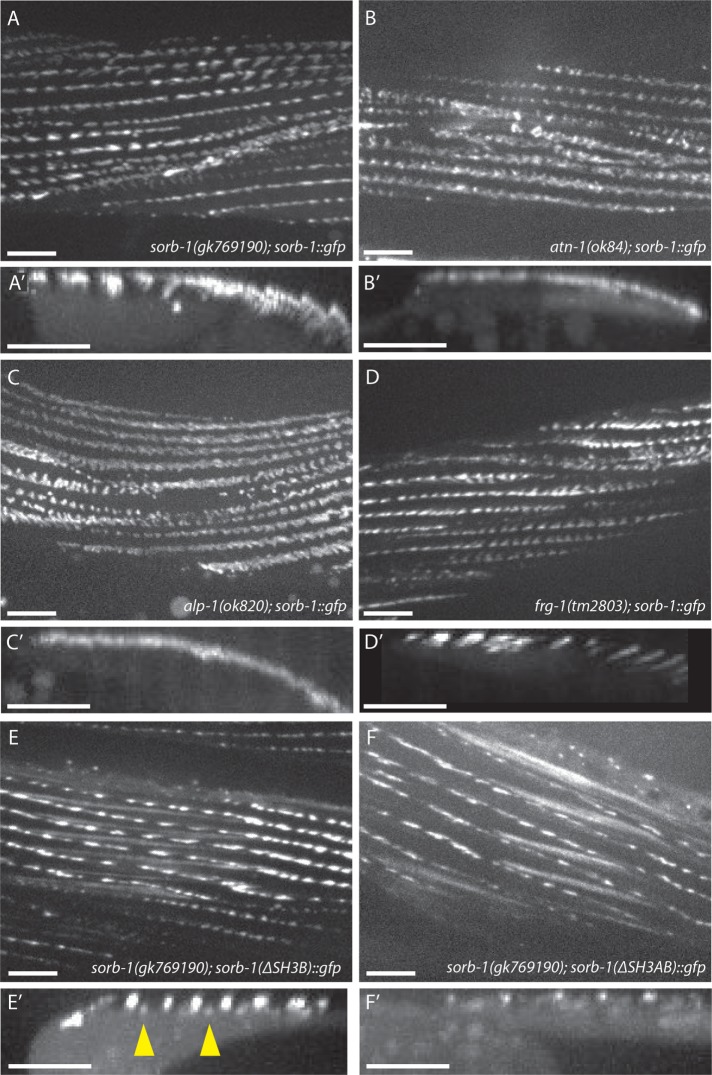
To date, few proteins have been identified as distal components of the dense body. One such protein, ATN-1, is the sole nematode orthologue of the actin-binding protein α-actinin (Moulder et al., 2010), which is a direct binding partner for the SoHo region of ArgBP2 and the first two SH3 domains of vinexin in vertebrates (Kioka et al., 1999; Ronty et al., 2005). atn-1(ok84)-null homozygotes survive to adulthood and show loss of SORB-1::GFP localization in the membrane-distal region of dense bodies; although membrane-proximal signal is not abrogated, it is less spatially restricted around the core of the dense body in ok84 animals (compare A and A′ to B and B′ in Figure 5). This is consistent with electron microscopy observations of atn-1(ok84) mutant muscle, which showed short but broad dense bodies and near-normal localization of the membrane-proximal components β-integrin and vinculin (Moulder et al., 2010).
FIGURE 5:
ATN-1, ALP-1, and the N-terminal SH3 domains of SORB-1 are required for correct dense body localization of SORB-1. (A, A’) SORB-1::GFP localization at dense bodies in the predicted SORB-1 protein-null background, sorb-1(gk769190) (A), and a projection of 100 Z-stack reslices showing localization to dense bodies (A’, arrowhead). (B, B’) Recruitment of SORB-1::GFP to the membrane-distal region of dense bodies is abolished in atn-1(ok84) mutants: fluorescent signal is restricted to the cell surface in projections of Z-stack reslices (B′). (C, C’) alp-1(ok820); sorb-1::gfp adults display no membrane-distal dense body GFP, and signal is less spatially restricted around the membrane-proximal dense body. (D, D’) SORB-1::GFP localization is unperturbed in a frg-1(tm2803) mutant background. (E, E’) SORB-1(∆SH3B)::GFP is mislocalized to M-line structures (arrowheads) in sorb-1(gk769190) worms. (F, F’) SORB-1::GFP(∆SH3AB)::GFP is not recruited to the membrane-distal dense body, similar to wild-type SORB-1::GFP in atn-1(ok84) and alp-1(ok820) backgrounds. Scale bars = 10 μm.
The membrane-distal dense body component ALP-1 (McKeown et al., 2006) also displays expanded membrane-proximal signal in atn-1(ok84) mutants (Han and Beckerle, 2009). Therefore, we investigated whether SORB-1 localization was dependent on the presence of ALP-1. Similarly to atn-1 mutants, alp-1(ok820) mutants exhibit loss of SORB-1::GFP signal from the deep portion of the dense body and less spatial restriction at the membrane-proximal region (Figure 5, C and C’). This redistribution of SORB-1::GFP is not likely due to perturbations of ATN-1 or DEB-1 themselves, as these proteins are localized normally in alp-1 mutants (Han and Beckerle, 2009). The remaining known component of the membrane-distal portion of dense bodies, FRG-1(FRG1) (Liu et al., 2010), does not appear to be required for SORB-1 recruitment, as SORB-1::GFP localization is unperturbed in frg-1(tm2803) mutants (Figure 5, D and D′).
To ascertain which domains of SORB-1 are responsible for its localization, domain dropout constructs were expressed in both a wild-type background and sorb-1(gk769190), which contains a nonsense mutation before the SoHo domain (Figure 1A). The second SH3 domain appears to help exclude SORB-1 from the other focal adhesion–like structure in nematode striated muscle, the M-line, as SORB-1(∆SH3B)::GFP remained present at proximal and distal dense bodies but was partially redistributed to the deep region of M-lines in sorb-1(gk769190) animals (Figure 5, E and E′). Deletion of the first two SH3 domains was sufficient to prevent localization to the deep region of dense bodies (Figure 5, F and F′). Removing the SoHo or SH3C domains did not appreciably perturb SORB-1::GFP localization in any of the tissues studied (unpublished data). Together, these data place SORB-1 downstream of ATN-1 and ALP-1, but not FRG-1, in distal dense body assembly and show that proper SORB-1 localization relies primarily on its two most N-terminal SH3 domains.
sorb-1 mutants exhibit defects in sarcomere organization but normal motility
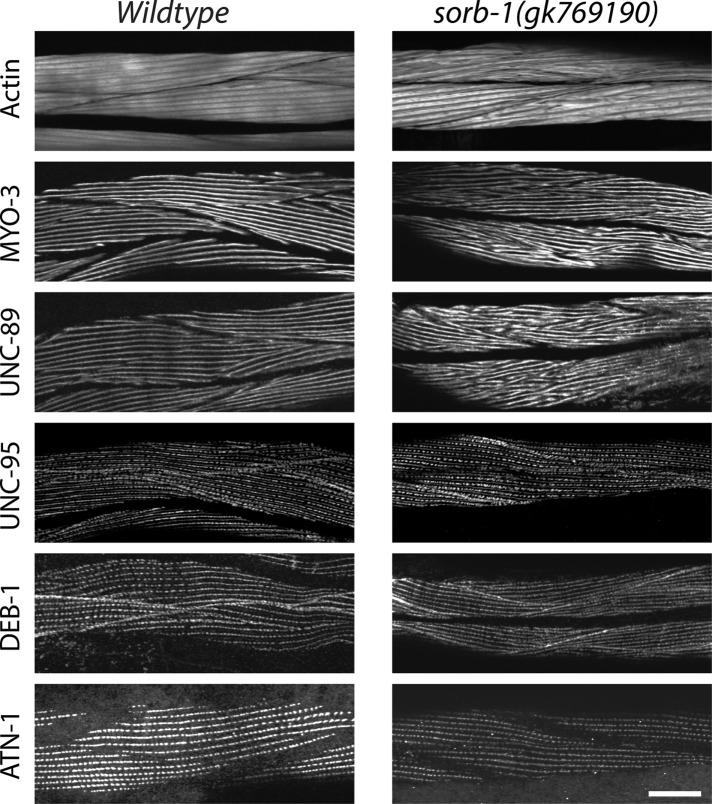
Because SORB-1 localizes to the actin-binding region of dense bodies in body wall muscles, we investigated whether sorb-1 loss of function could produce defects in sarcomeric organization and/or function. sorb-1(gk769190) is a nonsense mutation, W461amber, obtained from the Million Mutation Project collection (Thompson et al., 2013). We compared the sarcomere organization of wild-type and sorb-1(gk769190) adult body wall muscle, examining the organization of thin filaments (detected with fluorescent phalloidin), thick filaments (MYO-3 [myosin heavy chain A]), M-lines (UNC-89 [obscutin]), the membrane-proximal portions of M-lines and dense bodies (UNC-95), and dense bodies (DEB-1 [vinculin] and ATN-1 [α-actinin]). As shown in Figure 6, remarkably, although SORB-1::GFP is localized to dense bodies, we detected no obvious abnormalities in the pattern or shape of dense bodies when reacted with antibodies to ATN-1 or DEB-1. Also, UNC-95 is normally located near the base of dense bodies and M-lines in sorb-1(gk769190). Nevertheless, actin thin filaments are disorganized in most cells as assessed via phalloidin staining. In addition, both thick filaments (detected by antibodies to MYO-3 [myosin heavy chain]) and their organizing centers, the M-lines (detected by antibodies to UNC-89 [obscurin]), are disorganized in sorb-1(gk769190) worms. Instead of the parallel uniform lines of staining for MYO-3 and UNC-89 seen in wild type, sorb-1(gk769190) shows V-shaped patterns and some degree of aggregation for both proteins. The muscle structural defects in sorb-1(gk769190) animals did not manifest as a gross reduction in the quantity or quality of sinusoidal waves produced during movement compared with young adult or 2-d-old adult wild-type controls (Supplemental Table S1). Similar defects in sarcomere structure were found for two additional sorb-1 mutant alleles, sorb-1(ok2174) and sorb-1(gk304), each of which is an intragenic deletion resulting in early protein truncation (Supplemental Figure S3). Of particular note, however, the effects on thin filaments, thick filaments, and M-lines are less severe in sorb-1(gk304) worms. sorb-1(gk304) is the most C-terminal of the mutants, which could account for this weaker phenotype. SORB-1(ΔSH3C)::GFP localizes normally (Figure 5), and sorb-1(gk304) deletes SH3C. Thus, sorb-1(gk304) might produce a protein that localizes properly but has a mild functional defect.
FIGURE 6:
sorb-1 loss of function results in disorganization of thick filaments, M-lines, and, to some extent, thin filaments. Wild-type or sorb-1(gk769190) young adults were immunostained with antibodies to MYO-3 (thick filaments), UNC-89 (M-lines), UNC-95 (base of dense bodies and M-lines), DEB-1 (base of dense bodies), and ATN-1 (major and deeper portion of dense bodies) and reacted with fluorescent phalloidin (thin filaments). The sorb-1 mutant shows disorganization of thin filaments in most body wall muscle cells. The sorb-1 mutant shows disorganization of thick filaments and M-lines. Despite SORB-1 localizing to dense bodies, other dense body components (UNC-95, DEB-1, and ATN-1) show no obvious disorganization. Scale bar = 20 μm.
In addition to muscle attachment sites, GFP-tagged SORB-1 localized to distal tip cells (DTCs) of the gonad, beginning in the late L4 larval stage and persisting throughout adulthood, including within cytoneme projections; however, knockdown of sorb-1 did not perturb DTC migration or cytoneme morphology (Supplemental Figure S4A), nor did sorb-1 overexpression or knockdown significantly affect brood size compared with wild type (Supplemental Table S2). Depletion of ina-1, an α-integrin required for proper DTC migration and gonad morphology (Baum and Garriga, 1997), occasionally led to a redistribution of SORB-1::GFP in the proximal gonad (Supplemental Figure S4B).
Mitochondria become disorganized in the absence of SORB-1
Mitochondria within C. elegans body wall muscles normally align in tubular tracks parallel to actin filaments. This alignment requires membrane-proximal and distal dense body components (Etheridge et al., 2012), as well as components of the LINC (Linker of Nucleoskeleton and Cytoskeleton) complex of organelle-tethering proteins, such as ANC-1, UNC-83, and UNC-84 (Hedgecock and Thomson, 1982; Starr and Han, 2002). We therefore examined muscles in sorb-1 mutants for mitochondrial mispositioning defects. Live mitochondrial imaging of sorb-1(gk769190) and atn-1(ok84) homozygotes revealed reproducible defects in mitochondrial organization (Figure 7) consisting of nonparallel mitochondrial tracks, sizable discontinuities within tracks, and accumulation of mitochondrial signal into rounded masses. The observed defects were commensurate with organization and alignment phenotypes previously described for RNAi-mediated knockdown of ATN-1(α-actinin) and DEB-1(vinculin) (Etheridge et al., 2012). While mitochondrial aggregation or nonparallel branching were observed in 31.8% of wild-type myocytes scored (n = 22), sorb-1(gk769190) and atn-1(ok84) displayed a 63.4% (n = 71) and 55.1% (n = 69) incidence of these defects, respectively (each significantly different from wild type, p < 0.01, Fisher's exact test). These results suggest a novel role for sorbin family proteins in organizing mitochondria, although the mechanistic link between SORB-1 and mitochondrial positioning remains to be elucidated.
FIGURE 7:
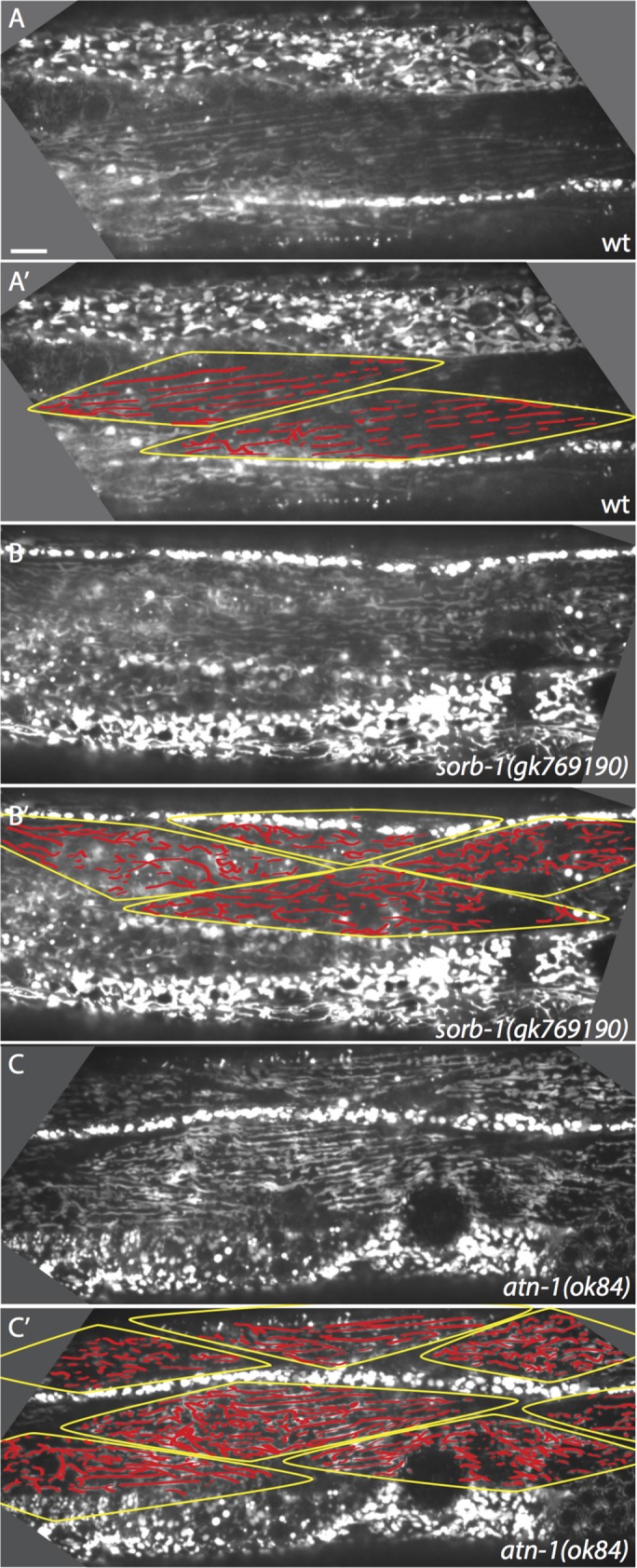
Mitochondrial organization is disrupted in the absence of SORB-1. (A) Alignment of mitochondria parallel to thick and thin filaments in a wild-type (wt) young adult. (A’) Myocytes in A are outlined, and mitochondrial tracks are highlighted. (B, B’) Mitochondrial organization is modestly disrupted in sorb-1(gk769190) worms. There is a greater proportion of branched tracks and small puncta that do not lie in a clear linear track. (C, C’) Mitochondria are also modestly disorganized in atn-1(ok84) worms. Longitudinal tracks are frequently interrupted, and signal accumulates in small puncta. Scale bar = 10 μm.
SORB-1 interacts with DEB-1(vinculin) and with ZYX-1(zyxin)
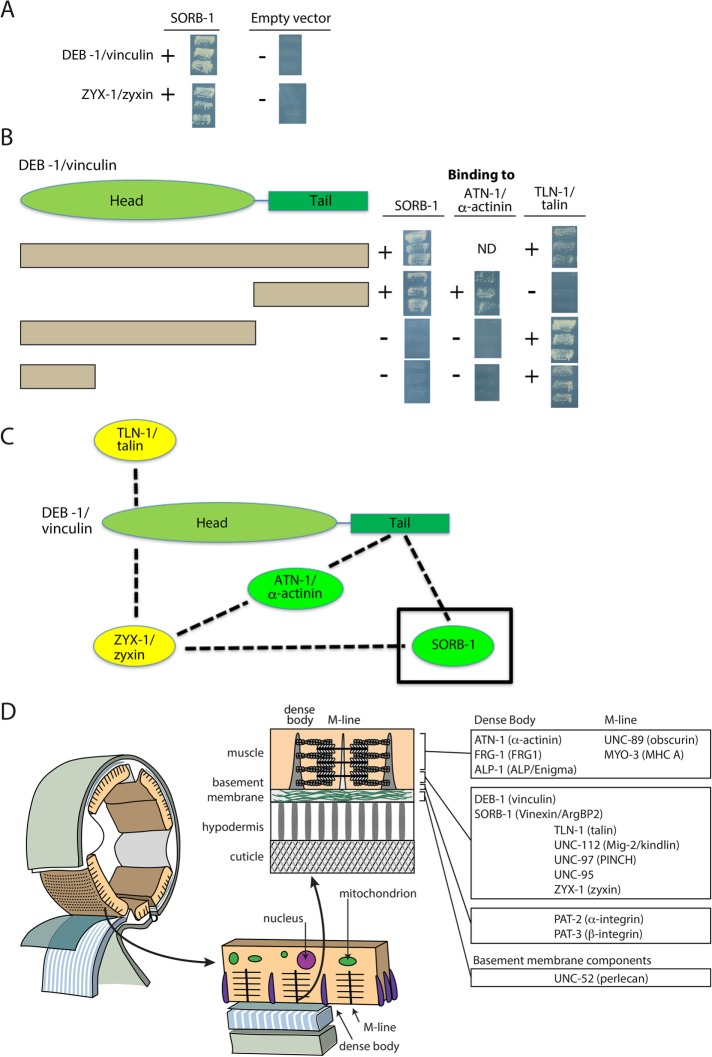
To help understand how SORB-1 is localized to dense bodies and performs its functions, we used SORB-1 to screen a collection of 16 known dense body proteins (Supplemental Table S3) with a yeast two-hybrid assay. As shown in Supplemental Table S3 and Figure 8A, SORB-1 was found to interact only with DEB-1 and ZYX-1. ZYX-1 is the unique zyxin-like protein in C. elegans that is orthologous to the vertebrate zyxin subfamily composed of zyxin, migfilin, TRIP6, and LPP (Lecroisey et al., 2013).
FIGURE 8:
Yeast two-hybrid assays show SORB-1 interacts with DEB-1 and ZYX-1. (A) Screening a collection of dense body proteins revealed interaction of SORB-1 with DEB-1 and ZYX-1. The images show the growth of yeast from three independent colonies on plates lacking adenine. (B) The indicated full-length and deletion derivatives of DEB-1 were used to map interaction regions for SORB-1, ATN-1, and TLN-1. To the right of each row are images of the growth of yeast colonies, as described for A. The DEB-1 C-terminal tail region interacts with SORB-1 and ATN-1, whereas the most N-terminal part of the DEB-1 head region interacts with TLN-1. ND, no data. (C) Summary of interactions among SORB-1, ZYX-1, ATN-1, DEB-1, and TLN-1. Green denotes dense body–specific proteins; yellow denotes proteins found at both dense bodies and M-lines. (D) Summary of localization of conserved dense body and M-line components examined in this and other studies. Schematic adapted from Hardin (2011).
DEB-1 is composed of an N-terminal head and a C-terminal tail. As indicated in Figure 8B, the C-terminal tail of DEB-1, but not its N-terminal head, binds to SORB-1, consistent with vertebrate vinculin and vinexin β (Takahashi et al., 2005). The DEB-1 C-terminal tail also binds to ATN-1 but not TLN-1. However, a portion of the N-terminal head of DEB-1 binds to TLN-1. These interactions are summarized in Figure 8C. In contrast, the localization of SORB-1 does not depend on the presence of ZYX-1; SORB-1::GFP localizes properly in a zyx-1-null mutant, zyx-1(gk190) (Supplemental Figure S5). Similarly, the localization of ZYX-1 does not depend on the presence of SORB-1; in the loss-of-function sorb-1(gk769190) mutant, ZYX-1 is properly localized (Supplemental Figure S5). The localization of SORB-1 when ZYX-1 is absent is probably explained by the interaction of SORB-1 with DEB-1 (Figure 8C). That the SORB-1/DEB-1 interaction is crucial is shown by the mislocalization of SORB-1::GFP in the absence of DEB-1 (Figure 4).
DISCUSSION
SORB-1 is a newly identified component of muscle dense bodies in C. elegans
Sorbin family proteins are versatile components of integrin adhesion sites, often referred to as focal adhesions in cultured cells, which regulate adhesion to the extracellular matrix and therefore key oncogenic processes such as cell-matrix adhesion, migration, and the ability to survive and proliferate independently of substrate anchorage (Suwa et al., 2002; Yuan et al., 2005; Mitsushima et al., 2006; Mizutani et al., 2007). This study establishes sorb-1 as a sorbin and SH3 domain–containing family member in C. elegans. SORB-1 protein is most homologous to vertebrate vinexin in both amino acid sequence and cellular function, localizing to integrin adhesion sites in the musculature and somatic gonad as well as along stress fibers in the somatic gonad. Mutational analysis indicates that proper localization of GFP-tagged SORB-1 requires core dense body proteins PAT-2(α-integrin), PAT-3(β-integrin), UNC-112(kindlin), and TLN-1(talin), which are the sine qua non for dense body formation (Gieseler et al., 2017), as well as the more membrane-distal dense body components DEB-1(vinculin), ATN-1(α-actinin), and ALP-1(ALP/Enigma) (Figures 4 and 5). Although a previous screen identified a physical interaction between DEB-1 and SORB-1 (Xin et al., 2009), our directed yeast two-hybrid assays elucidate this interaction: the C-terminal tail domain of DEB-1 interacts with both ATN-1 and SORB-1, and the N-terminal head domain of DEB-1 interacts with TLN-1 (Figure 8). In contrast to the known interaction between ArgBP2 and α-actinin (Ronty et al., 2005; Anekal et al., 2015), we did not find that SORB-1 interacts directly with ATN-1, although negative results from yeast two-hybrid assays do not necessarily rule out interactions. If our yeast two-hybrid results reflect in vivo interactions, then the apparent mislocalization of SORB-1::GFP in atn-1(ok84) may simply result from the dense bodies of atn-1(ok84) having an abnormally short and broad structure (Moulder et al., 2010).
SORB-1 contributes to sarcomeric organization
We have shown that in C. elegans, SORB-1 localizes to dense bodies and adhesion plaques, two types of integrin-mediated cell-matrix adhesion sites in nematode body wall muscle. However, despite this localization to dense bodies, three independently isolated loss-of-function alleles of sorb-1 (Figure 6 and Supplemental Figure S3) show no effect on the localization of basal (UNC-95 and DEB-1) or deeper (ATN-1) dense body components. One of these components, DEB-1, interacts with SORB-1 (Figure 8). Nevertheless, the disorganization of thin filaments found in sorb-1 mutants (sorb-1(gk769190) and sorb-1(ok2174); Figure 6 and Supplemental Figure S3) is consistent with a function at dense bodies, since dense bodies anchor actin thin filaments in nematode muscle. However, the other effect of sorb-1 mutants on the sarcomere appears to be at thick filaments (MYO-3) and its associated organizing center in the middle of the A-band, the M-line (UNC-89). That both MYO-3 and UNC-89 are affected is expected for several reasons. First, unc-89 mutants show disorganized thick filaments by immunostaining with anti–MYO-3 (Qadota et al., 2008, 2016). Second, both UNC-89 and MYO-3 reside in the middle of sarcomeric A-bands (Miller et al., 1983; Benian et al., 1996; Small et al., 2004). However, the effect on thick filaments and M-lines is somewhat unexpected, as we have shown here that SORB-1 does not reside at thick filaments or M-lines. There are two possibilities: first, given the close and extensive physical interactions between actin thin filaments and myosin thick filaments, it is possible that the primary effect of loss of sorb-1 function is on actin thin filaments, which then secondarily causes disruption of thick filaments and their attachment structures but leaves the membrane-associated portion of M-lines (e.g., UNC-95) intact. This explanation is consistent with the abnormal organization of thin filaments observed in mutants of genes encoding M-line–specific proteins, for example, UNC-98 (Mercer et al., 2003), UNC-96 (Mercer et al., 2006), and UNC-89 (our unpublished observations). A second, more exotic explanation for the effect of sorb-1 mutations on thick filaments and the M-line is that SORB-1 has a role in signaling between the dense body and the nucleus, which affects the expression of thick filament/M-line proteins (discussed below).
Normal organelle positioning requires SORB-1
We have identified a novel role for SoHo domain proteins in organelle positioning. In C. elegans muscle, loss of sorb-1 function leads to mitochondrial mispositioning. While this work was in review, results of a large-scale screen were reported with data similar to ours regarding the effects of loss of sorb-1 function on mitochondrial positioning (Etheridge et al., 2015). The alignment of mitochondria in apical tracks parallel to contractile fibers probably allows for optimal respiration in highly ATP-demanding tissues such as muscle (Hardy et al., 2009; Kinsey et al., 2011). These mitochondrial tracks depend on prior organization of the sarcomere: knockdown of early dense body organizers in C. elegans produces severely disorganized contractile filaments and severely fragmented mitochondria, while knockdown of late dense body components, which are largely nonessential, results in modestly disorganized and fragmented mitochondrial networks (Etheridge et al., 2012). When mitochondria become disorganized in the absence of distal dense body components such as SORB-1, body wall muscles may not fuel ATP-driven actomyosin contraction at maximum efficiency, resulting in diminished muscle flexion.
The mechanism bridging distal dense body components to mitochondria remains unclear, but one candidate is the SUN (Sad-1/UNC-84) and KASH (Klarsicht/ANC-1/Syne/homology) system of LINC proteins, which tether organelles to the cytoskeleton and contribute to body wall muscle mitochondrial organization. The giant KASH protein ANC-1 localizes to C. elegans dense bodies (Starr and Han, 2002), and loss of ANC-1 or one of its binding partners, the SUN protein UNC-84, results in severe mitochondrial disorganization similar to loss of essential dense body proteins (Hedgecock and Thomson, 1982; Malone et al., 1999; Starr and Han, 2002). ANC-1 possesses two N-terminal calponin domains (Starr and Han, 2002) that may link mitochondria to the actin cytoskeleton, although direct actin-binding activity has not been demonstrated. It is possible that dense body components such as the SORB-1/DEB-1/ZYX-1/ATN-1/TLN-1 complex may subtly organize ANC-1–bound actin at the membrane-distal portion of the dense body.
SORB-1 may participate in mechanotransduction
Unlike other distal dense body components, which are constitutively found in the nucleus (Hobert et al., 1999; Mercer et al., 2003; Broday et al., 2004; Liu et al., 2010; Lecroisey et al., 2013), SORB-1 appears to be excluded from the nucleus even when overexpressed, displaced from the dense body by SH3 domain deletion, or displaced by loss of distal dense body components. This suggests that increased cytoplasmic availability of SORB-1 is not sufficient for nuclear translocation, despite the presence of a putative nuclear localization signal (as determined by cNLS Mapper [ Kosugi et al., 2009]). Rather, this appears to be a specific response to the reduction of core dense body proteins PAT-2(α-integrin), UNC-112(kindlin), DEB-1(vinculin), and TLN-1(talin) (Figure 4). The one exception is PAT-3(β-integrin). The conspicuous lack of nuclear SORB-1::GFP signal in pat-3(RNAi) embryos suggests that nuclear transport of SORB-1 is a PAT-3–dependent process. Interestingly, one of the two dense body proteins that we have shown to interact directly with SORB-1, ZYX-1(zyxin), is found constitutively in the nucleus and mainly in the middle of dense bodies (Lecroisey et al., 2013). Perhaps in response to a reduction in core dense body proteins, SORB-1 carries additional ZYX-1 into the nucleus.
Although vertebrate ArgBP2 has been observed in cultured cell nuclei (Wang et al., 1997), no nuclear roles for sorbin family proteins are currently known. We hypothesize that SORB-1 may participate in a feedback mechanism between dense bodies and the nucleus, which could serve to detect whether adequate tension is produced by muscle contractions and regulate expression of relevant genes accordingly. Periodic muscle contractions are necessary for complete C. elegans embryonic elongation (Williams and Waterston, 1994), a morphogenetic event in which circumferential F-actin filament bundles in the hypodermis compress the embryo to fourfold its original length (Chisholm and Hardin, 2005). Beyond ∼1.5-fold, failure to produce muscle contractions or to transmit contractile force to the hypodermis (e.g., due to muscle detachment from the extracellular matrix) prevents further elongation (Williams and Waterston, 1994; Zhang et al., 2011). Failure to produce adequate muscle contractions during this time could feasibly initiate a feedback signal to up-regulate genes responsible for the underperforming component.
If SORB-1 participates in nuclear feedback from dense bodies during elongation, it may do so in conjunction with the Abl orthologue ABL-1. Vertebrate ArgBP2 orthologues have been observed in nuclei of cultured cells and have been demonstrated to associate with, and be phosphorylated by, both of the closely related nonreceptor tyrosine kinases Abl and Arg (Wang et al., 1997). Additionally, Abl is capable of shuttling between the nuclear and integrin-associated pools in response to changes in integrin-mediated adhesion (Lewis et al., 1996; Taagepera et al., 1998). Nuclear Abl is capable of altering gene transcription via phosphorylation of RNA polymerase II (Baskaran et al., 1993) and histone acetyltransferases (Kaidi and Jackson, 2013). Taken together, these results suggest ABL-1 as a likely nuclear effector of SORB-1. Future experiments targeting this and other possibilities should further elucidate SORB-1’s functions in muscle cells.
MATERIALS AND METHODS
Strains
Unless otherwise noted, strains were maintained at 20°C on nematode growth medium (NGM) plates seeded with OP50 (Brenner, 1974). All fluorescent sorb-1 constructs were injected into the germline of Bristol N2 or sorb-1(gk169190) at 20 ng/μl, together with coinjection marker pRF4(rol-6(su1006)) at 80 ng/μl and noncoding F35D3 DNA at 20 ng/μl. All strains containing the temperature-sensitive allele hlh-1(cc561ts) were maintained at 15°C.
The following strains were used in this study: jcEx153(sorb-1::gfp; rol-6(su1006)), jcEx171(Psorb-1::gfp; rol-6(su1006)), jcEx172(sorb-1a(∆SH3BC)::gfp; rol-6(su1006)), jcEx173(sorb-1a(∆SH3ABC)::gfp; rol-6(su1006)), jcEx175(sorb-1a(∆SoHo)::gfp; rol-6(su1006)), hlh-1(cc561ts) II, ina-1(gm39) III, ina-1(gm144) III, deb-1(st554)/unc-5(e53) unc-24(e138) IV; jcEx243(sorb-1::gfp), alp-1(ok820) IV; jcEx256((sorb-1a::gfp; rol-6(su1006)), sorb-1(gk769190) IV, sorb-1(gk769190) IV; jcEx153, sorb-1(gk769190) IV; jcEx161, sorb-1(gk769190) IV; jcEx175, sorb-1(gk769190) IV; jcEx172, sorb-1(gk769190) IV; jcEx173, atn-1(ok84) V; jcEx239((sorb-1a::gfp; rol-6(su1006)) myo-3(st386) V; and qIs19(Plag-2::gfp; rol-6(su1006)) V.
Creation of DNA constructs
To generate the sorb-1::gfp construct pTDL06, cDNA yk357a12 (provided by Y. Kohara, National Institute of Genetics, Japan) was first PCR amplified using primers ponF07 (GCCGCTCTGAACTAGTGGAT) and ponR07 (ATCTAGATAATGACGTTTAACATATTTC) and then cloned into the GFP expression vector pPD95.75 by PstI/XbaI digestion (New England Biolabs [NEB]). Approximately 2.2 kb of promoter sequence and 2.1 kb of genomic sorb-1 sequence were then inserted by PCR amplification of the sorb-1 5′ region from N2 genomic DNA using primers ponF15 (AGCATGCCCGTTTTTCAGCCAGTTGTT) and ponR16 (CCGTCGAGCTTGTAGAGTAGGGAG). This amplicon and pTDL06 were SphI/DraIII digested (NEB), and the products were ligated to generate pTDL07 (Psorb-1::sorb-1::gfp).
Domain deletion constructs were created using dropout PCR mutagenesis with the following primer pairs. To delete the SORB-1 ORF from pTDL07, creating the transcriptional reporter pTDL25, ponF21 (ATCTAGAGGATCCCCGGGAT) and ponR23b (TCCTGGAATATGGGAGTTTGAAGA) were used. To delete the SH3C domain from pTDL07, creating pTDL24, ponF27 (TATCTAGAGGATCCCCGGGAT) and ponR21 (ACATTTGCAAATTAGAATCCGAT) were used. To delete the SH3A domain, creating pTDL26 (sorb-1ΔSH3A::gfp from pTDL07 template) and pTDL28 (sorb-1ΔSH3AC::gfp from pTDL24 template), ponF24 (ATTAATACTGGAAATCAAGGGGATAGTCAA) and ponR24 (TGTTGCTGTCATAACTGGCT) were used. To delete the SH3B domain, creating pTDL27 (sorb-1ΔSH3B::gfp from pTDL07 template) and pTDL32 (sorb-1ΔSH3BC::gfp from pTDL24 template), ponF25 (GAACAAGTGGAGCAACACAT) and ponR25 (TCTCATCTTTTGACTATCCCCTTGA) were used. To delete the SH3A and SH3B domains, creating pTDL29 (sorb-1ΔSH3AB::gfp from pTDL07 template) and pTDL31 (sorb-1ΔSH3ABC::gfp from pTDL24 template), ponF25 (GAACAAGTGGAGCAACACAT) and ponR24 (TGTTGCTGTCATAACTGGCT) were used. To delete the SoHo domain from pTDL07, creating pTDL30, ponF26 (GGACTACACCGACTGCATCAAATTT) and ponR26 (TTGATGTGCTCATTGGCTGCT) were used.
The sorb-1 feeding RNAi construct pTDL04 was created by ligating the 1.1-kb BglII/StyI (NEB) digestion product of the yk357a12 into the feeding RNAi vector L4440.
To construct yeast two-hybrid plasmids (bait and prey) containing sorb-1 full-length cDNA, the following three cloning steps were performed. 1) sorb-1 N-terminus (1–583 base pairs) was amplified from a cDNA library (RB2; a gift from Robert Barstead, Oklahoma Medical Research Foundation) using PCR with primers sorb-1-N1 (GCGCTCGAGCCCGGGATGATGCATCATCCTCATCCATTC) and sorb-1-N2 (TGATGCACTGTCATCAATGTCGAATTC). 2) After DNA sequence confirmation, the EcoRI fragment from cDNA yk357a12 was ligated to the sorb-1 N-terminal coding sequence, resulting in a full-length cDNA for sorb-1 in a cloning vector. 3) The SmaI/SalI fragment of sorb-1 full-length cDNA was cloned into pGBDU-C1 and pGAD-C1 SmaI and SalI sites, resulting in pGBDU-sorb-1 (bait) and pGAD-sorb-1 (prey).
Genotyping primers used
To genotype sorb-1(gk769190), the following primers were used to create a 359–base pair amplicon from worm lysate, which could be DdeI digested into 279– and 80–base pair bands if the gk769190 allele was present: ponF31 (CATAAGAACGAGCGGAAGTTGG) and ponR02 (CCTTCATTCGAACCGTCTTCT).
To determine the 5′ extent of the sorb-1 ORF, the following primers were used with genomic cDNA library template (sorb-1 5′ R01 was the reverse primer for all reactions):
sorb-1 5′ R01 (GACAAAGATACACACGGTGTCTC),
sorb-1 5′ F01 (ATGATGCATCATCCTCATCCATT),
sorb-1 5′ F02 (ATGCCTACAACTTTGACACCT),
sorb-1 5′ F03 (ATGTGGCGGCGTTGAGAAAT),
sorb-1 5′ F04 (ATGTGATACATGGGCAGTTGGAT),
sorb-1 5′ F05 (TGAAGACACCAACTGGAA ATAGTT),
sorb-1 5′ F06 (ATGTGGATCCACATCTTAACAAGCA),
sorb-1 5′ F07 (ATGAGTCGAAACCACCAG TG),
sorb-1 5′ F08 (ATGACAGTGCATCAATTATT TCCTCG), and
sorb-1 5′ F09 (CTTCAAACTCCCATATTCCA GGA).
RNA interference
Double-stranded RNAs for injection RNAi were generated from Kohara cDNAs in vitro (Ambion), which were then injected into the posterior pseudocoelomic cavity of L4 worms at 2 μg/μl. After a minimum of 24 h, injected worms were singled, and phenotypes were assayed in F1 progeny. Efficacy of sorb-1 RNAi was assayed by loss of GFP signal in sorb-1::gfp animals.
Feeding RNAi experiments in strains carrying hlh-1(cc561ts) were performed at both 15 and 20°C, using isopropyl β-d-1-thiogalactopyranoside (IPTG; Sigma) to induce double-stranded RNA production.
Live worm imaging
Differential interference contrast (DIC) imaging of live animals was performed on 5% agar pads prepared on glass microscope slides. Worms were first picked to a small drop of M9 buffer on the pad. The majority of the buffer was then removed by pipette and replaced with 4 μl of 30 μM levamisole. Slides were then sealed with a coverslip and imaged immediately using a Nikon Optiphot-2 microscope and a QImaging QICAM camera controlled by ImageJ software. Slides prepared in this manner were imaged for a maximum of 20 min in order to minimize morphological artifacts. GFP images were collected using a Yokagawa CSU-10 spinning-disk scanhead mounted on a Nikon Eclipse E600 confocal microscope with a Prior motorized stage and a Hamamatsu ORCA-ER Camera controlled by Perkin Elmer-Cetus UltraView or Micro-Manager software.
Scoring of mitochondrial morphology
Mitochondrial staining was adapted from previously described methods (Artal-Sanz and Tavernarakis, 2009). Briefly, a 1-mM DMSO stock solution of Mitotracker Red CMXRos (Molecular Probes) was diluted to 10 nM in water. Dilute solution (250 μl per 60-mm NGM plate) was spread over the bacterial lawn and allowed to dry overnight. L2 larvae were singled to these plates and allowed to feed for 48 h. For consistency, only body wall muscles oriented en face to the objective were scored. Myocytes were scored wild type if mitochondria predominantly aligned in longitudinal tracks with few small puncta or aggregations of signal; modestly disorganized if they displayed numerous small puncta, extensive branching, or aggregation; or severely disorganized if signal was largely confined to puncta and aggregates with no discernable longitudinal track. Imaging of fluorescently labeled mitochondria was performed using spinning-disk microscopy as described above.
Immunostaining for muscle components
For immunostaining with antibodies to various muscle components, worms were fixed and immunostained according to the method described in Nonet et al. (1993) and described in further detail in Wilson et al. (2012). The following primary antibodies were used at 1:200 dilution: anti–UNC-89 (mouse monoclonal MH42; Benian et al., 1996), anti–MYO-3 (mouse monoclonal 5-6; Miller et al., 1983), anti–ATN-1 (mouse monoclonal MH35; Francis and Waterston, 1991), anti–DEB-1 (mouse monoclonal MH24; Francis and Waterston, 1991), anti-GFP (rabbit polyclonal; Life Technologies/Thermo Fisher Scientific; A11122). anti–UNC-95 (rabbit polyclonal Benian-13; Qadota et al., 2007) was used at 1:100 dilution. MH35 and MH42 were kindly provided by Pamela Hoppe (Western Michigan University). Secondary antibodies, also used at 1:200 dilution, included anti-rabbit Alexa 488 (Invitrogen), anti-rat Alexa 594 (Invitrogen), and anti-mouse Alexa 594 (Invitrogen). Images were captured at room temperature with a Zeiss confocal system (LSM510) equipped with an Axiovert 100M microscope and an Apochromat ×63/1.4 numerical aperture oil immersion objective in ×2.5 zoom mode. The color balances of the images were adjusted using Adobe Photoshop (Adobe, San Jose, CA). To confirm nuclear localization of SORB-1::GFP in deb-1 embryos, deb-1(st554); sorb-1::GFP embryos were fixed as described previously (Costa et al., 1998) and stained with 0.2 μg/ml anti-GFP (rabbit polyclonal; Life Technologies). Before the final wash with phosphate-buffered saline, 1 mM of 4',6-diamidino-2-phenylindole (Molecular Probes) was added. Images were acquired with an Olympus Fluoview FV1000 microscope and software.
Locomotion assays
Mobility was assayed by singling worms to a fresh lawn of Escherichia coli and allowing them to crawl for 60 s. The number of waves in each track was measured and compared versus wild-type or control RNAi animals. Wave quality was binned as wild type if they were smoothly sinusoidal or defective if they contained either low-amplitude or sawtooth waves, indicating hypoflexion and tail dragging, respectively.
Somatic gonad and brood size assays
DTC morphology was assayed in sorb-1(RNAi) experiments using integrated Plag-2::gfp. Each cytoneme was scored for length, number of branches, and frequency of breaks, defined as discontinuities in fluorescent signal. Gonad morphology was assayed by DIC microscopy of live worms. Brood sizes were scored by singling L4 larvae and moving them to new plates every 24 h. After 3 d, total brood size for each worm was calculated.
Yeast two-hybrid assays
Yeast two-hybrid assays using SORB-1 bait and prey plasmids were performed as described previously (Mackinnon et al., 2002; Miller et al., 2006). A “dense body bookshelf” collection of 15 proteins was previously described (Warner et al., 2013), except for the addition of PKN-1 (Supplemental Table S3).
Supplementary Material
Acknowledgments
We thank Y. Kohara for providing sorb-1 cDNA clones, the Caenorhabditis Genetics Center for providing strains, and Gary Williams and the staff of WormBase (www.wormbase.org) for discussions related to the structure of the sorb-1 locus. We also thank Brandon Fields and Nathan Szewczyk for consulting on experiments. This work was supported by National Institutes of Health Grants no. GM-058038 (to J.H.) and AR-064307 (to G.M.B.).
Abbreviations used:
- DIC
differential interference contrast
- DTC
distal tip cell
- GFP
green fluorescent protein
- KASH
Klarsicht/ANC-1/Syne/homology
- LINC
Linker of Nucleoskeleton and Cytoskeleton
- NGM
nematode growth media
- ORF
open reading frame
- RNAi
RNA interference
- SH3
Src homology 3
- SoHo
sorbin homology
- SUN
Sad-1/UNC-84
- wt
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-06-0455) on October 4, 2017.
REFERENCES
- Anekal PV, Yong J, Manser E. Arg kinase-binding protein 2 (ArgBP2) interaction with alpha-actinin and actin stress fibers inhibits cell migration. J Biol Chem. 2015;290:2112–2125. doi: 10.1074/jbc.M114.610725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artal-Sanz M, Tavernarakis N. Prohibitin couples diapause signalling to mitochondrial metabolism during ageing in C. elegans. Nature. 2009;461:793–797. doi: 10.1038/nature08466. [DOI] [PubMed] [Google Scholar]
- Barstead RJ, Waterston RH. The basal component of the nematode dense-body is vinculin. J Biol Chem. 1989;264:10177–10185. [PubMed] [Google Scholar]
- Baskaran R, Dahmus ME, Wang JY. Tyrosine phosphorylation of mammalian RNA polymerase II carboxyl-terminal domain. Proc Natl Acad Sci USA. 1993;90:11167–11171. doi: 10.1073/pnas.90.23.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum PD, Garriga G. Neuronal migrations and axon fasciculation are disrupted in ina-1 integrin mutants. Neuron. 1997;19:51–62. doi: 10.1016/s0896-6273(00)80347-5. [DOI] [PubMed] [Google Scholar]
- Benian GM, Tinley TL, Tang X, Borodovsky M. The Caenorhabditis elegans gene unc-89, required fpr muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J Cell Biol. 1996;132:835–848. doi: 10.1083/jcb.132.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj R, Roy M, Ohyama T, Sivan-Loukianova E, Delannoy M, Lloyd TE, Zlatic M, Eberl DF, Kolodkin AL. Cbl-associated protein regulates assembly and function of two tension-sensing structures in Drosophila. Development. 2013;140:627–638. doi: 10.1242/dev.085100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broday L, Kolotuev I, Didier C, Bhoumik A, Podbilewicz B, Ronai Z. The LIM domain protein UNC-95 is required for the assembly of muscle attachment structures and is regulated by the RING finger protein RNF-5 in C. elegans. J Cell Biol. 2004;165:857–867. doi: 10.1083/jcb.200401133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Waterman CM. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat Cell Biol. 2015;17:955–963. doi: 10.1038/ncb3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestra G, Toomre D, Chang S, De Camilli P. The Abl/Arg substrate ArgBP2/nArgBP2 coordinates the function of multiple regulatory mechanisms converging on the actin cytoskeleton. Proc Natl Acad Sci USA. 2005;102:1731–1736. doi: 10.1073/pnas.0409376102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpin-Elhamri G, Elbaba M, Descroix-Vagne M, Pansu D, Perret JP. Inhibitory effect of sorbin on pepsin secretion in conscious cats and rabbits. Peptides. 2000;21:65–72. doi: 10.1016/s0196-9781(99)00175-8. [DOI] [PubMed] [Google Scholar]
- Chisholm AD, Hardin J. Epidermal morphogenesis. WormBook. 2005:1–22. doi: 10.1895/wormbook.1.35.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciobanasu C, Faivre B, Le Clainche C. Integrating actin dynamics, mechanotransduction and integrin activation: the multiple functions of actin binding proteins in focal adhesions. Eur J Cell Biol. 2013;92:339–348. doi: 10.1016/j.ejcb.2013.10.009. [DOI] [PubMed] [Google Scholar]
- Costa M, Raich W, Agbunag C, Leung B, Hardin J, Priess JR. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge T, Oczypok EA, Lehmann S, Fields BD, Shephard F, Jacobson LA, Szewczyk NJ. Calpains mediate integrin attachment complex maintenance of adult muscle in Caenorhabditis elegans. PLoS Genet. 2012;8:e1002471. doi: 10.1371/journal.pgen.1002471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge T, Rahman M, Gaffney CJ, Shaw D, Shephard F, Magudia J, Solomon DE, Milne T, Blawzdziewicz J, Constantin-Teodosiu D, et al. The integrin-adhesome is required to maintain muscle structure, mitochondrial ATP production, and movement forces in Caenorhabditis elegans. FASEB J. 2015;29:1235–1246. doi: 10.1096/fj.14-259119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernow I, Tomasovic A, Siehoff-Icking A, Tikkanen R. Cbl-associated protein is tyrosine phosphorylated by c-Abl and c-Src kinases. BMC Cell Biol. 2009;10:80. doi: 10.1186/1471-2121-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis GR, Waterston RH. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J Cell Biol. 1985;101:1532–1549. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, Waterston RH. Muscle cell attachment in Caenorhabditis elegans. J Cell Biol. 1991;114:465–479. doi: 10.1083/jcb.114.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieseler K, Qadota H, Benian GM. Development, structure, and maintenance of C. elegans body wall muscle. WormBook. 2017:1–59. doi: 10.1895/wormbook.1.81.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han HF, Beckerle MC. The ALP-Enigma protein ALP-1 functions in actin filament organization to promote muscle structural integrity in Caenorhabditis elegans. Mol Biol Cell. 2009;20:2361–2370. doi: 10.1091/mbc.E08-06-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand D, Eiden LE. Human sorbin is generated via splicing of an alternative transcript from the ArgBP2 gene locus. Peptides. 2005;26:1278–1282. doi: 10.1016/j.peptides.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Hardin J. Mechanotransduction: getting morphogenesis down pat. Curr Biol. 2011;21:R309–R311. doi: 10.1016/j.cub.2011.03.052. [DOI] [PubMed] [Google Scholar]
- Hardy KM, Dillaman RM, Locke BR, Kinsey ST. A skeletal muscle model of extreme hypertrophic growth reveals the influence of diffusion on cellular design. Am J Physiol Regul Integr Comp Physiol. 2009;296:R1855–R1867. doi: 10.1152/ajpregu.00076.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock EM, Thomson JN. A gene required for nuclear and mitochondrial attachment in the nematode Caenorhabditis elegans. Cell. 1982;30:321–330. doi: 10.1016/0092-8674(82)90038-1. [DOI] [PubMed] [Google Scholar]
- Hobert O, Moerman DG, Clark KA, Beckerle MC, Ruvkun G. A conserved LIM protein that affects muscular adherens junction integrity and mechanosensory function in Caenorhabditis elegans. J Cell Biol. 1999;144:45–57. doi: 10.1083/jcb.144.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaidi A, Jackson SP. KAT5 tyrosine phosphorylation couples chromatin sensing to ATM signalling. Nature. 2013;498:70–74. doi: 10.1038/nature12201. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kinsey ST, Locke BR, Dillaman RM. Molecules in motion: influences of diffusion on metabolic structure and function in skeletal muscle. J Exp Biol. 2011;214:263–274. doi: 10.1242/jeb.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioka N, Ito T, Yamashita H, Uekawa N, Umemoto T, Motoyoshi S, Imai H, Takahashi K, Watanabe H, Yamada M, Ueda K. Crucial role of vinexin for keratinocyte migration in vitro and epidermal wound healing in vivo. Exp Cell Res. 2010;316:1728–1738. doi: 10.1016/j.yexcr.2010.03.019. [DOI] [PubMed] [Google Scholar]
- Kioka N, Sakata S, Kawauchi T, Amachi T, Akiyama SK, Okazaki K, Yaen C, Yamada KM, Aota S. Vinexin: a novel vinculin-binding protein with multiple SH3 domains enhances actin cytoskeletal organization. J Cell Biol. 1999;144:59–69. doi: 10.1083/jcb.144.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebre AS, Jamot L, Takahashi J, Spassky N, Leprince C, Ravise N, Zander C, Fujigasaki H, Kussel-Andermann P, Duyckaerts C, et al. Ataxin-7 interacts with a Cbl-associated protein that it recruits into neuronal intranuclear inclusions. Hum Mol Genet. 2001;10:1201–1213. doi: 10.1093/hmg/10.11.1201. [DOI] [PubMed] [Google Scholar]
- Lecroisey C, Brouilly N, Qadota H, Mariol MC, Rochette NC, Martin E, Benian GM, Segalat L, Mounier N, Gieseler K. ZYX-1, the unique zyxin protein of Caenorhabditis elegans, is involved in dystrophin-dependent muscle degeneration. Mol Biol Cell. 2013;24:1232–1249. doi: 10.1091/mbc.E12-09-0679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecroisey C, Segalat L, Gieseler K. The C. elegans dense body: anchoring and signaling structure of the muscle. J Muscle Res Cell Motil. 2007;28:79–87. doi: 10.1007/s10974-007-9104-y. [DOI] [PubMed] [Google Scholar]
- Lewis JM, Baskaran R, Taagepera S, Schwartz MA, Wang JY. Integrin regulation of c-Abl tyrosine kinase activity and cytoplasmic-nuclear transport. Proc Natl Acad Sci USA. 1996;93:15174–15179. doi: 10.1073/pnas.93.26.15174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Jones TI, Tang VW, Brieher WM, Jones PL. Facioscapulohumeral muscular dystrophy region gene-1. FRG-1) is an actin-bundling protein associated with muscle-attachment sites. J Cell Sci. 2010;123:1116–1123. doi: 10.1242/jcs.058958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinnon AC, Qadota H, Norman KR, Moerman DG, Williams BD. C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr Biol. 2002;12:787–797. doi: 10.1016/s0960-9822(02)00810-2. [DOI] [PubMed] [Google Scholar]
- Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- Mandai K, Nakanishi H, Satoh A, Takahashi K, Satoh K, Nishioka H, Mizoguchi A, Takai Y. Ponsin/SH3P12: an l-afadin- and vinculin-binding protein localized at cell-cell and cell-matrix adherens junctions. J Cell Biol. 1999;144:1001–1017. doi: 10.1083/jcb.144.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown CR, Han HF, Beckerle MC. Molecular characterization of the Caenorhabditis elegans ALP/Enigma gene alp-1. Dev Dyn. 2006;235:530–538. doi: 10.1002/dvdy.20633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer KB, Flaherty DB, Miller RK, Qadota H, Tinley TL, Moerman DG, Benian GM. Caenorhabditis elegans UNC-98, a C2H2 Zn finger protein, is a novel partner of UNC-97/PINCH in muscle adhesion complexes. Mol Biol Cell. 2003;14:2492–2507. doi: 10.1091/mbc.E02-10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer KB, Miller RK, Tinley TL, Sheth S, Qadota H, Benian GM. Caenorhabditis elegans UNC-96 is a new component of M-lines that interacts with UNC-98 and paramyosin and is required in adult muscle for assembly and/or maintenance of thick filaments. Mol Biol Cell. 2006;17:3832–3847. doi: 10.1091/mbc.E06-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, 3rd, Ortiz I, Berliner GC, Epstein HF. Differential localization of two myosins within nematode thick filaments. Cell. 1983;34:477–490. doi: 10.1016/0092-8674(83)90381-1. [DOI] [PubMed] [Google Scholar]
- Miller RK, Qadota H, Landsverk ML, Mercer KB, Epstein HF, Benian GM. UNC-98 links an integrin-associated complex to thick filaments in Caenorhabditis elegans muscle. J Cell Biol. 2006;175:853–859. doi: 10.1083/jcb.200608043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsushima M, Ueda K, Kioka N. Vinexin beta regulates the phosphorylation of epidermal growth factor receptor on the cell surface. Genes Cells. 2006;11:971–982. doi: 10.1111/j.1365-2443.2006.00995.x. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Ito H, Iwamoto I, Morishita R, Deguchi T, Nozawa Y, Asano T, Nagata KI. Essential roles of ERK-mediated phosphorylation of vinexin in cell spreading, migration and anchorage-independent growth. Oncogene. 2007;26:7122–7131. doi: 10.1038/sj.onc.1210512. [DOI] [PubMed] [Google Scholar]
- Moerman DG, Williams BD. Sarcomere assembly in C. elegans muscle. WormBook. 2006:1–16. doi: 10.1895/wormbook.1.81.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder GL, Cremona GH, Duerr J, Stirman JN, Fields SD, Martin W, Qadota H, Benian GM, Lu H, Barstead RJ. α-actinin is required for the proper assembly of Z-disk/focal-adhesion-like structures and for efficient locomotion in Caenorhabditis elegans. J Mol Biol. 2010;403:516–528. doi: 10.1016/j.jmb.2010.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K, Ito H, Kanoh H, Sudo K, Iwamoto I, Morishita R, Soubeyran P, Seishima M, Nagata K. Cell biological characterization of a multidomain adaptor protein, ArgBP2, in epithelial NMuMG cells, and identification of a novel short isoform. Med Mol Morphol. 2012;45:22–28. doi: 10.1007/s00795-010-0537-9. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Grundahl K, Meyer BJ, Rand JB. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- Pansu D, Vagne M, Bosshard A, Mutt V. Sorbin, a peptide contained in porcine upper small intestine which induces the absorption of water and sodium in the rat duodenum. Scand J Gastroenterol. 1981;16:193–199. doi: 10.3109/00365528109181955. [DOI] [PubMed] [Google Scholar]
- Qadota H, Benian GM. Molecular structure of sarcomere-to-membrane attachment at M-Lines in C. elegans muscle. J Biomed Biotechnol. 2010;2010:864749. doi: 10.1155/2010/864749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Blangy A, Xiong G, Benian GM. The DH-PH region of the giant protein UNC-89 activates RHO-1 GTPase in Caenorhabditis elegans body wall muscle. J Mol Biol. 2008;383:747–752. doi: 10.1016/j.jmb.2008.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Mayans O, Matsunaga Y, McMurry JL, Wilson KJ, Kwon GE, Stanford R, Deehan K, Tinley TL, Ngwa VM, Benian GM. The SH3 domain of UNC-89 (obscurin) interacts with paramyosin, a coiled-coil protein, in Caenorhabditis elegans muscle. Mol Biol Cell. 2016;27:1606–1620. doi: 10.1091/mbc.E15-09-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Mercer KB, Miller RK, Kaibuchi K, Benian GM. Two LIM domain proteins and UNC-96 link UNC-97/pinch to myosin thick filaments in Caenorhabditis elegans muscle. Mol Biol Cell. 2007;18:4317–4326. doi: 10.1091/mbc.E07-03-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignot J, Taieb D, Suliman M, Dusetti NJ, Iovanna JL, Soubeyran P. CIP4 is a new ArgBP2 interacting protein that modulates the ArgBP2 mediated control of WAVE1 phosphorylation and cancer cell migration. Cancer Lett. 2010;288:116–123. doi: 10.1016/j.canlet.2009.06.030. [DOI] [PubMed] [Google Scholar]
- Ronty M, Taivainen A, Moza M, Kruh GD, Ehler E, Carpen O. Involvement of palladin and alpha-actinin in targeting of the Abl/Arg kinase adaptor ArgBP2 to the actin cytoskeleton. Exp Cell Res. 2005;310:88–98. doi: 10.1016/j.yexcr.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Small TM, Gernert KM, Flaherty DB, Mercer KB, Borodovsky M, Benian GM. Three new isoforms of Caenorhabditis elegans UNC-89 containing MLCK-like protein kinase domains. J Mol Biol. 2004;342:91–108. doi: 10.1016/j.jmb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- Suwa A, Mitsushima M, Ito T, Akamatsu M, Ueda K, Amachi T, Kioka N. Vinexin beta regulates the anchorage dependence of ERK2 activation stimulated by epidermal growth factor. J Biol Chem. 2002;277:13053–13058. doi: 10.1074/jbc.M108644200. [DOI] [PubMed] [Google Scholar]
- Taagepera S, McDonald D, Loeb JE, Whitaker LL, McElroy AK, Wang JY, Hope TJ. Nuclear-cytoplasmic shuttling of C-ABL tyrosine kinase. Proc Natl Acad Sci USA. 1998;95:7457–7462. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taieb D, Roignot J, Andre F, Garcia S, Masson B, Pierres A, Iovanna JL, Soubeyran P. ArgBP2-dependent signaling regulates pancreatic cell migration, adhesion, and tumorigenicity. Cancer Res. 2008;68:4588–4596. doi: 10.1158/0008-5472.CAN-08-0958. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Mitsushima M, Okada N, Ito T, Aizawa S, Akahane R, Umemoto T, Ueda K, Kioka N. Role of interaction with vinculin in recruitment of vinexins to focal adhesions. Biochem Biophys Res Commun. 2005;336:239–246. doi: 10.1016/j.bbrc.2005.08.064. [DOI] [PubMed] [Google Scholar]
- Thompson O, Edgley M, Strasbourger P, Flibotte S, Ewing B, Adair R, Au V, Chaudhry I, Fernando L, Hutter H, et al. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 2013;23:1749–1762. doi: 10.1101/gr.157651.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosoni D, Cestra G. CAP (Cbl associated protein) regulates receptor-mediated endocytosis. FEBS Lett. 2009;583:293–300. doi: 10.1016/j.febslet.2008.12.047. [DOI] [PubMed] [Google Scholar]
- Vagne-Descroix M, Pansu D, Jornvall H, Carlquist M, Guignard H, Jourdan G, Desvigne A, Collinet M, Caillet C, Mutt V. Isolation and characterisation of porcine sorbin. Eur J Biochem. 1991;201:53–59. doi: 10.1111/j.1432-1033.1991.tb16254.x. [DOI] [PubMed] [Google Scholar]
- Wang B, Golemis EA, Kruh GD. ArgBP2, a multiple Src homology 3 domain-containing, Arg/Abl-interacting protein, is phosphorylated in v-Abl-transformed cells and localized in stress fibers and cardiocyte Z-disks. J Biol Chem. 1997;272:17542–17550. doi: 10.1074/jbc.272.28.17542. [DOI] [PubMed] [Google Scholar]
- Warner A, Xiong G, Qadota H, Rogalski T, Vogl AW, Moerman DG, Benian GM. CPNA-1, a copine domain protein, is located at integrin adhesion sites and is required for myofilament stability in Caenorhabditis elegans. Mol Biol Cell. 2013;24:601–616. doi: 10.1091/mbc.E12-06-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Williams BD, Waterston RH. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KJ, Qadota H, Mains PE, Benian GM. UNC-89 (obscurin) binds to MEL-26, a BTB-domain protein, and affects the function of MEI-1 (katanin) in striated muscle of Caenorhabditis elegans. Mol Biol Cell. 2012;23:2623–2634. doi: 10.1091/mbc.E12-01-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X, Rual JF, Hirozane-Kishikawa T, Hill DE, Vidal M, Boone C, Thierry-Mieg N. Shifted Transversal Design smart-pooling for high coverage interactome mapping. Genome Res. 2009;19:1262–1269. doi: 10.1101/gr.090019.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Yanagawa S. Axin and the Axin/Arrow-binding protein DCAP mediate glucose-glycogen metabolism. Biochem Biophys Res Commun. 2003;304:229–235. doi: 10.1016/s0006-291x(03)00582-5. [DOI] [PubMed] [Google Scholar]
- Yuan ZQ, Kim D, Kaneko S, Sussman M, Bokoch GM, Kruh GD, Nicosia SV, Testa JR, Cheng JQ. ArgBP2gamma interacts with Akt and p21-activated kinase-1 and promotes cell survival. J Biol Chem. 2005;280:21483–21490. doi: 10.1074/jbc.M500097200. [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat Cell Biol. 2007;9:858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Landmann F, Zahreddine H, Rodriguez D, Koch M, Labouesse M. A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature. 2011;471:99–103. doi: 10.1038/nature09765. [DOI] [PubMed] [Google Scholar]
- Zhang M, Liu J, Cheng A, Deyoung SM, Chen X, Dold LH, Saltiel AR. CAP interacts with cytoskeletal proteins and regulates adhesion-mediated ERK activation and motility. EMBO J. 2006;25:5284–5293. doi: 10.1038/sj.emboj.7601406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.