Proper positioning of the chromosomal passenger complex (CPC) at the cell division plane is required for cytokinesis. We show here that CPC targeting to the equatorial cortex depends on both the kinesin MKlp2 and a direct interaction with actin. These recruitment mechanisms converge to promote successful cleavage furrow ingression.
Abstract
During cytokinesis, the chromosomal passenger complex (CPC) promotes midzone organization, specifies the cleavage plane, and regulates furrow contractility. The localizations of the CPC are coupled to its cytokinetic functions. At the metaphase-to-anaphase transition, the CPC dissociates from centromeres and localizes to midzone microtubules and the equatorial cortex. CPC relocalization to the cell middle is thought to depend on MKlp2-driven, plus end–directed transport. In support of this idea, MKlp2 depletion impairs cytokinesis; however, cytokinesis failure stems from furrow regression rather than failed initiation of furrowing. This suggests that an alternative mechanism(s) may concentrate the CPC at the division plane. We show here that direct actin binding, via the inner centromere protein (INCENP), enhances CPC enrichment at the equatorial cortex, thus acting in tandem with MKlp2. INCENP overexpression rescues furrowing in MKlp2-depleted cells in an INCENP-actin binding–dependent manner. Using live-cell imaging, we also find that MKlp2-dependent targeting of the CPC is biphasic. MKlp2 targets the CPC to the anti-parallel microtubule overlap of the midzone, after which the MKlp2-CPC complex moves in a nondirected manner. Collectively, our work suggests that both actin binding and MKlp2-dependent midzone targeting cooperate to precisely position the CPC during mitotic exit, and that these pathways converge to ensure successful cleavage furrow ingression.
INTRODUCTION
Cytokinesis (C phase), the final step in cell division, individualizes two cells from one. The completion of cytokinesis requires the chromosomal passenger complex (CPC), a heterotetramer composed of the Aurora B kinase (ABK), the scaffolding protein INCENP (inner centromeric protein), and the small regulatory subunits Survivin and Borealin (Carmena et al., 2012). At the metaphase-to-anaphase transition, the CPC relocalizes from the centromeres of mitotic chromosomes to the spindle midzone, an anti-parallel array of microtubules (MTs) that forms in between poleward-moving chromosomes during anaphase (Cooke et al., 1987). The midzone localization of the CPC is distinct in that it occupies a narrow, medial zone of MT plus-end overlap, similar to Kif4A, centralspindlin, and other factors that are critical for central spindle assembly. The CPC also enriches at the equatorial cortex, a region of the plasma membrane where the cleavage furrow will assemble (Earnshaw and Cooke, 1991; Murata-Hori and Wang, 2002). Finally, furrow constriction causes the CPC to concentrate at the midbody, an intracellular bridge that connects dividing cells. The CPC has key functions at each of these locations; these functions range from stabilizing the central spindle to controlling the timing of abscission (Green et al., 2012; Mierzwa and Gerlich, 2014; Kitagawa and Lee, 2015). Mechanisms that properly localize the CPC are therefore crucial to its cell-division functions.
In somatic cells, the kinesin-6 MKlp2 (Kif20A) is important for the CPC to localize on the spindle midzone (Gruneberg et al., 2004). The reverse is also true, as the CPC is required to target MKlp2 to the spindle midzone (Hummer and Mayer, 2009). Interestingly, the ability of INCENP to bind MTs directly (Wheatley et al., 2001; Rosasco-Nitcher et al., 2008; Noujaim et al., 2014) appears to be essential for this midzone localization of MKlp2 (van der Horst et al., 2015), suggesting that the CPC may play a role in mediating the MKlp2–midzone interaction. Given that other kinesin-6 motors (MKlp1, MPP1) exhibit MT plus end–directed motility in vitro (Nislow et al., 1992; Abaza et al., 2003; Mishima et al., 2004; Hizlan et al., 2006), the predominant model in the field is that MKlp2 delivers the CPC to MT plus ends of the central spindle through a stepping-based mechanism like other kinesin motors. Work in Xenopus oocyte extracts, however, suggests that Kif4A is the transport motor for the CPC, and that Kif20A simply allows the CPC to engage MTs (Nguyen et al., 2014). Whether this model applies to somatic cells is not known, since oocytes use an embryonic form of Kif20A (Kif20AE) to position the CPC during cytokinesis.
How the CPC targets the equatorial cortex (Earnshaw and Cooke, 1991) is not well understood. Selective disassembly of astral MTs with a low dose of nocodazole eliminates cortical Aurora B (Murata-Hori and Wang, 2002), suggesting that the CPC decorates astral MTs that run laterally along the cell cortex. However, several lines of evidence support an alternative model wherein MKlp2 and the CPC target the equatorial cortex through direct interactions with actomyosin. First, MKlp2 interacts with nonmuscle myosin II (Kitagawa et al., 2013). Second, INCENP binds actin directly in vitro (Chen et al., 2007; Landino and Ohi, 2016). Third, an INCENP mutant that cannot bind MTs still localizes to the equatorial cortex (van der Horst et al., 2015). Finally, the CPC strongly decorates F-actin rather than MTs at the aster-aster interzone in Xenopus oocytes (Nguyen et al., 2014) and in cells undergoing monopolar cytokinesis (Hu et al., 2008). Because the equatorial cortex represents an intersection between actin and MTs, cells may use both cytoskeletons to fine-tune CPC localization at the cell cortex.
Although it is well known that a crucial function of the CPC on the central spindle is to stabilize it (Green et al., 2012), the role of the CPC at the equatorial cortex is less clear. Work in Xenopus oocytes has demonstrated that it is the CPC rather than centralspindlin that promotes the formation of cleavage apparatus (Nguyen et al., 2014). In Drosophila, the loss of MKlp2 (Subito) does not impair cytokinetic progression (Cesario et al., 2006), suggesting that the midzone and the cortically localized population of the CPC are nonequivalent. Similarly, in human cells, MKlp2-depleted cells begin to furrow but then regress, suggesting that the cortical CPC population is specifically required for sustained ingression of the cleavage furrow (Kitagawa et al., 2013). Additionally, ingression fails in cells expressing a mutant of MKlp2 that is defective in its ability to enrich on the equatorial cortex but not the midzone (Kitagawa et al., 2013). An important implication of these studies is that the CPC may use an MKlp2-independent mechanism to target the equatorial cortex. This is in agreement with our previous finding that INCENP binds actin directly (Landino and Ohi, 2016), leading us to test the possibility that actin binding underlies cortical recruitment of the CPC. In addition to reporting the results of these findings here, we also provide evidence that actin-based recruitment of the CPC to the equatorial cortex can sufficiently promote cleavage furrow ingression independent of MKlp2-dependent targeting.
RESULTS
CPC localization to the division plane requires F-actin and MTs
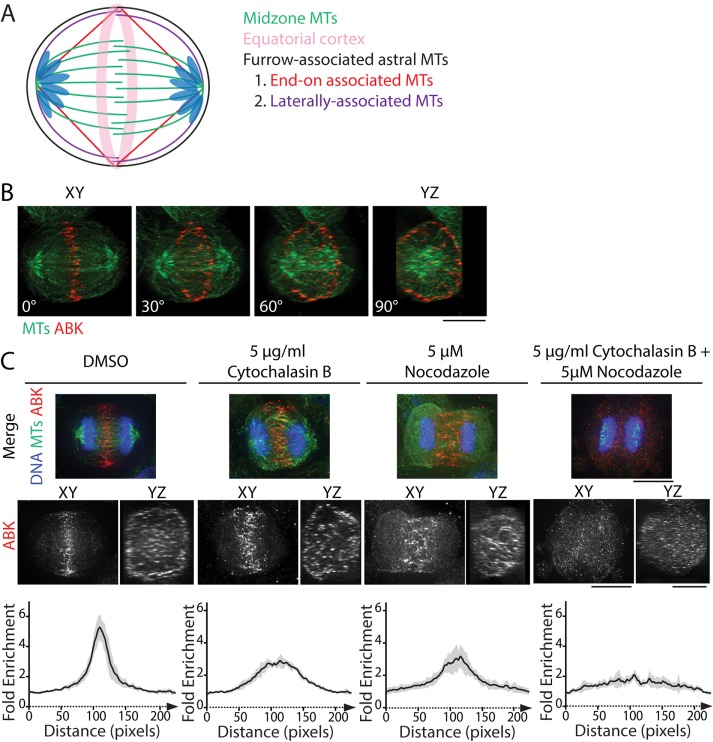
At the metaphase-to-anaphase transition, the CPC dissociates from centromeres and relocates to the division plane, the site where cleavage furrow ingression will occur (Carmena et al., 2012). The division plane includes the MT plus-end overlap of the spindle midzone, furrow-associated astral MTs, and the actomyosin-rich equatorial cortex (Figure 1A). Localization of the CPC at the division plane is complex. The CPC is concentrated at the overlap region of the spindle midzone, but it can also be seen at the equatorial cortex (Earnshaw and Cooke, 1991; Kitagawa et al., 2013). Using immunofluorescence of fixed cells, we observed ABK localization at the midzone and the cell periphery in whole-cell volume projections of C-phase HeLa cells, as expected (Figure 1B and Supplemental Figure S1).
FIGURE 1:
CPC localization to the division plane requires both MTs and actin. (A) Schematic of MT populations within the division plane of a bipolar C-phase cell. Midzone MTs (green) create a barrier between segregated chromosomes, whereas furrow-associated astral MTs extend toward the equatorial cortex (pink). These MTs can associate end-on (red) or laterally (purple) with the cortex. (B) Volume projection of an anaphase HeLa cell rotated 0° (XY dimension), 30°, 60°, and 90° (YZ dimension) and fixed and stained for tubulin (green) and ABK (red). Scale bar, 10 μm. (C) Maximum z-projections of the XY dimensions (top) and volume projections of ABK in the XY and YZ dimensions (middle) of cells treated with DMSO, 5 μg/ml cytochalasin B, 5 μM nocodazole, or both drugs simultaneously. Cells were stained with antibodies to tubulin (green) and ABK (red). DNA was counterstained with Hoechst 33342 (blue). Scale bars, 10 μm. Bottom, fold enrichment of ABK immunofluorescence parallel to the spindle axis for each condition. Immunofluorescence was measured using line scans 100 pixels wide (∼6 µm) and 230 pixels long (∼14 µm). Data represent the mean ± SD; n = 7 (DMSO), 6 (cytochalasin B), 7 (nocodazole), and 7 (cytochalasin + nocodazole) cells.
Because INCENP binds actin (Chen et al., 2007; Landino and Ohi, 2016) and MKlp2 binds myosin II (Kitagawa et al., 2013), we hypothesized that cortical localization of the CPC is mediated by both actin and MTs. To test this, we treated cells briefly (10 min) with 5 μM nocodazole to destabilize MTs, 5 μg/ml cytochalasin B to disassemble F-actin, or both drugs simultaneously and analyzed the localization of the CPC component ABK. In cells treated with dimethyl sulfoxide (DMSO), ABK localized to a narrowly focused band at the division plane and prominently to the equatorial cortex, similar to untreated cells (Figure 1, B and C). Pharmacological perturbation of either MTs or actin displaced ABK in two ways. First, ABK localized broadly along the spindle axis as opposed to a narrow band at the division plane. Second, the cortical localization of ABK was less pronounced (Figure 1C). Notably, ABK localization to the cell middle was only lost when both actin filaments and MTs were simultaneously disrupted (Figure 1C). This result suggests that the integrity of both filament systems is important for maintaining CPC localization at the division site during mitotic exit.
ABK localizes to the division plane in cells depleted of MKlp2
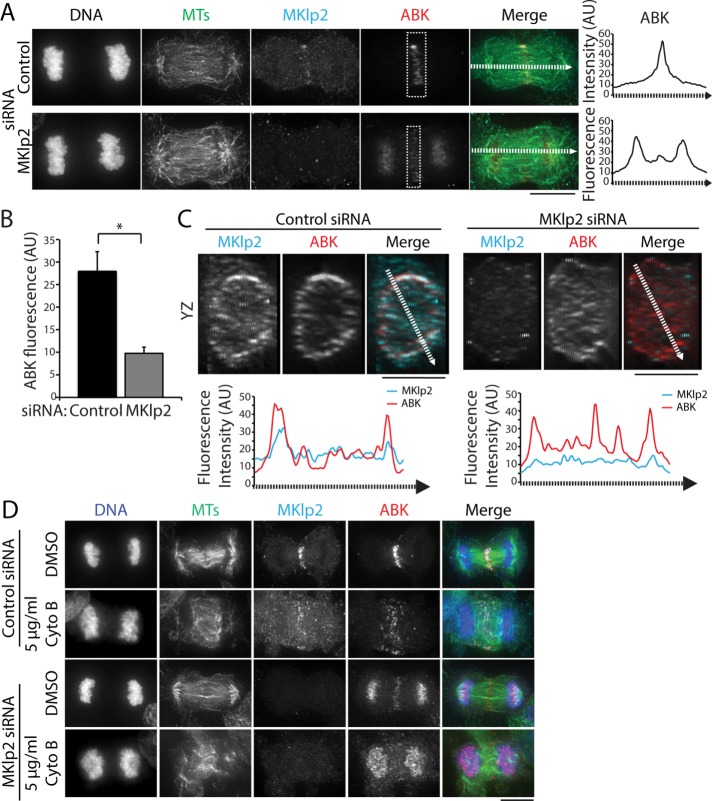
CPC targeting to the division plane is thought to occur through MKlp2-dependent MT plus end–directed transport (Gruneberg et al., 2004; Kitagawa et al., 2013, 2014). In agreement with previous work (Gruneberg et al., 2004; Kitagawa et al., 2013), we found that MKlp2 depletion using RNA interference (RNAi) led to an enrichment of ABK on chromosomes in C-phase cells (Figure 2A). However, we also observed that a low level of ABK was able to localize to the division plane despite marked reduction of MKlp2 immunofluorescence on the midzone (Figure 2A). At the population level, knockdown efficiency was heterogeneous, and we therefore restricted our analysis to cells with no detectable midzone-localized MKlp2. To quantify ABK levels at the division plane, we measured the integrated fluorescence intensity in cells treated with a control or MKlp2-targeting small interfering RNA (siRNA; boxes in Figure 2A). MKlp2 RNAi treatment decreased the mean ABK fluorescence approximately threefold at the division plane compared with cells transfected with control siRNA (Figure 2, A and B). This suggests that MKlp2 depletion decreases the amount of CPC that targets the division plane and that another mechanism(s) targets the CPC to the cell middle. We also observed that ABK was present at the equatorial cortex in MKlp2-depleted cells, as viewed in volume projections (YZ dimensions) of the division plane (Figure 2C), suggesting that ABK can localize to the equatorial cortex after MKlp2 depletion.
FIGURE 2:
The CPC localizes to the division site in the cells depleted of MKlp2. (A) Left, maximum z-projections of HeLa cells transfected with control (top) or MKlp2 (bottom) siRNA. Cells were stained with antibodies to tubulin (green), MKlp2 (teal), and ABK (red). DNA was counterstained with Hoechst 33342. Dashed boxes indicate sample regions used for quantitation of ABK fluorescence intensity. Dashed lines represent 10-pixel-wide (∼0.5 µm) line scans. Scale bar, 10 μm. Right, line scans of ABK fluorescence intensity along the spindle axis in cells treated with control or MKlp2 siRNA. AU, arbitrary units. (B) Quantification of ABK fluorescence intensity at the division plane in cells treated as described in A. Data represent the mean ± SE, n > 50 cells from three independent experiments, *p < 0.05. (C) Top, volume projections (YZ dimensions) of the division plane of cells shown in A. Dashed lines represent 10-pixel-wide (∼0.5 µm) line scans. Scale bars, 10 μm. Bottom, line scans of MKlp2 and ABK fluorescence intensity across the YZ projection of the division plane. (D) Maximum z-projections of HeLa cells transfected with control or MKlp2 siRNA and treated with DMSO or 5 μg/ml cytochalasin B (Cyto B). Cells were stained with antibodies to tubulin (green), MKlp2 (teal), and ABK (red). DNA (blue) was counterstained with Hoechst 33342. Scale bar, 10 μm.
Because actin perturbations, such as cytochalasin B treatment (Figure 1B) or mutation of the INCENP-actin binding site (Landino and Ohi, 2016), disrupt the C-phase localization of the CPC, we hypothesized that the CPC might accumulate at the division plane in MKlp2-depleted cells in a manner that requires F-actin. To test this possibility, we treated MKlp2-depleted cells briefly (10 min) with 5 μg/ml cytochalasin B and observed that endogenous ABK was only detectable on chromosomes and no longer enriched in the division plane (Figure 2D). This observation suggests that C-phase recruitment of the CPC to the division plane after MKlp2 depletion depends on F-actin.
Cortical enrichment of the CPC requires MKlp2 and INCENP-actin binding
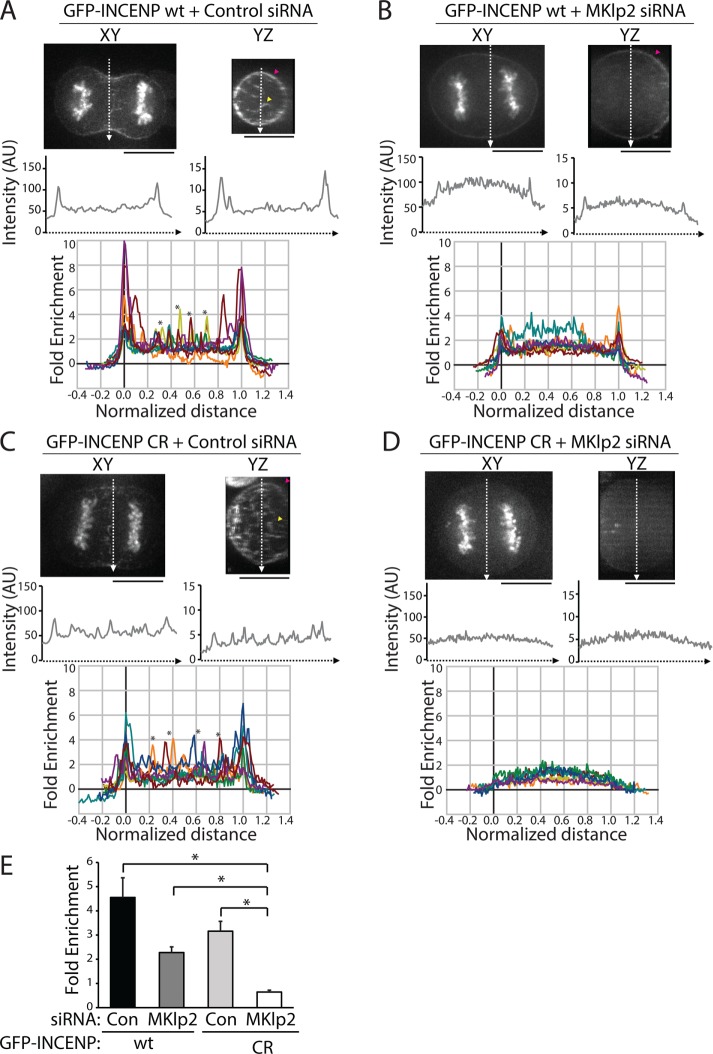
To quantitatively investigate the relative contribution of MKlp2-dependent versus actin-based cortical targeting of the CPC during C phase, we analyzed the cortical localization of transiently expressed INCENP tagged with green fluorescent protein (GFP) in live cells. This approach ensured that the cortical population was not affected by fixation methods used for immunofluorescence. Similar to previous experiments, volume projections of the division plane were used to visualize cortical enrichment. In a single optical section in the XY plane, line scans along the division plane revealed peaks of GFP-INCENP fluorescence at the cell edges (Figure 3A). Similarly, line scans across YZ volume projections of the division plane showed peaks of GFP-INCENP at the cortex. We analyzed the cortical enrichment of GFP-INCENP from multiple cells using line scans across YZ volume projections and by plotting the fold enrichment relative to cytoplasmic fluorescence (Figure 3A; see Materials and Methods) in order to control for variable expression levels. Notably, overexpression of GFP-INCENP resulted in an increased level of CPC on the chromosomes as cells entered anaphase, and we therefore excluded this region of the cell from our analysis. In cells expressing GFP-INCENP, the peak cortical intensity was 4.5 ± 0.8-fold higher than the cytoplasmic intensity (mean ± SD, n = 10 cells; Figure 3E). These cells also showed peaks of fluorescence in the cell center that likely correspond to midzone-localized CPC (marked by asterisks in Figure 3A).
FIGURE 3:
Cortical enrichment of GFP-INCENP at the division plane requires MKlp2 and INCENP-actin binding. (A) Top, single z-plane micrographs in the XY and YZ dimensions of the division plane in a HeLa cell expressing GFP-INCENP treated with control siRNA. Pink arrowhead denotes cortical GFP-INCENP fluorescence; yellow arrowhead marks GFP-INCENP fluorescence on a midzone bundle. Scale bars, 10 μm. Middle, line scans across XY or YZ projections of the division plane in the cell shown above. Bottom, fold enrichment of GFP-INCENP fluorescence intensity, relative to cytoplasmic fluorescence, from line scans across the YZ projection of cells treated with control siRNA. Asterisks mark peaks of GFP-INCENP localized to midzone bundles. n = 10 cells. (B) Top, single z-plane micrographs in the XY and YZ dimensions of the division plane in a cell expressing GFP-INCENP treated with MKlp2 siRNA. Pink arrowhead denotes cortical GFP-INCENP fluorescence. Scale bars, 10 μm. Middle, line scans across XY or YZ projections of the division plane in the cell shown above. Bottom, fold enrichment of GFP-INCENP fluorescence intensity, relative to cytoplasmic fluorescence, from line scans across the YZ projection of cells treated with MKlp2 siRNA. (C) Top, single z-plane micrographs in the XY and YZ dimensions of the division plane in a HeLa cell expressing GFP-INCENP CR treated with control siRNA. Pink arrowhead denotes cortical GFP-INCENP CR fluorescence; yellow arrowhead marks GFP-INCENP CR fluorescence on a midzone bundle. Scale bars, 10 μm. Middle, line scans across XY or YZ projections of the division plane in the cell shown above. Bottom, fold enrichment of GFP-INCENP CR fluorescence intensity, relative to cytoplasmic fluorescence, from line scans across the YZ projection of cells treated with control siRNA. Asterisks mark peaks of GFP-INCENP CR localized to midzone bundles. n = 10 cells. (D) Top, single z-plane micrographs in the XY and YZ dimensions of the division plane in a HeLa cell expressing GFP-INCENP CR treated with MKlp2 siRNA. Scale bars, 10 μm. Middle, line scans across XY or YZ projections of the division plane in the cell shown above. Bottom, fold enrichment of GFP-INCENP CR fluorescence intensity, relative to cytoplasmic fluorescence, from line scans across the YZ projection of cells treated with MKlp2 siRNA. n = 9 cells. (E) Quantification of cortical peak intensities of GFP-INCENP or GFP-INCENP CR from line scans across the YZ projection as described in A–D. Three adjacent peak intensity values were averaged for each individual cell. n ≥ 9 cells,*p < 0.05.
We next tested whether MKlp2 was required for cortical enrichment of the CPC by repeating this analysis in MKlp2-depleted cells. Again, due to cell-to-cell variability in knockdown efficiency, we were careful to restrict our analysis to only cells that lacked midzone-localized GFP-INCENP. Peaks of cortical GFP-INCENP were reduced but not eliminated by MKlp2 depletion (Figure 3B). The cortical enrichment of GFP-INCENP in YZ projections of the division plane was 2.3 ± 0.2-fold higher than cytoplasmic GFP-INCENP (Figure 3E). The partial reduction of cortical GFP-INCENP in MKlp2-depleted cells suggests that there are multiple pathways that recruit the CPC to the cortex.
We hypothesized that cortical accumulation of GFP-INCENP after MKlp2 depletion may depend on INCENP-actin binding, and to test this idea we utilized our previously described charge reversal INCENP mutant (GFP-INCENP CR), which disrupts actin binding in vitro (Landino and Ohi, 2016). We quantified the cortical enrichment of GFP-INCENP CR in cells treated with a control or MKlp2-targeting siRNA. Similar to wild-type GFP-INCENP (GFP-INCENP wt), GFP-INCENP CR is enriched at the cell cortex, with peaks of fluorescence in the cell center that likely correspond to midzone localization (marked by asterisks in Figure 3C). Quantification of cortical enrichment revealed that GFP-INCENP CR fluorescence was 3.2 ± 0.4-fold higher than cytoplasmic fluorescence (Figure 3E). The cortical enrichment of GFP-INCENP CR is lower than that of GFP-INCENP wt, suggesting that actin binding enhances CPC localization at the equatorial cortex in the presence of MKlp2. To test whether MKlp2 and INCENP-actin binding act in tandem to target the CPC to the equatorial cortex, we next tested whether GFP-INCENP CR was enriched at the equatorial cortex in cells depleted of MKlp2. We found that MKlp2-depleted cells expressing GFP-INCENP CR showed no cortical enrichment of the CPC in XY sections or YZ projections of the division plane (Figure 3D). Quantification of GFP-INCENP CR cortical fluorescence after MKlp2 depletion revealed that the cortical intensity was lower than the cytoplasmic signal (0.6 ± 0.1-fold enrichment, Figure 3E). Collectively, these observations indicate that maximal localization of the CPC to the equatorial cortex in unperturbed cells requires both direct actin binding and MKlp2-dependent targeting. Moreover, they suggest that the INCENP–actin interaction is sufficient to recruit the CPC to the equatorial cortex in cells depleted of MKlp2.
The CPC and MKlp2 decorate the midzone immediately after anaphase onset
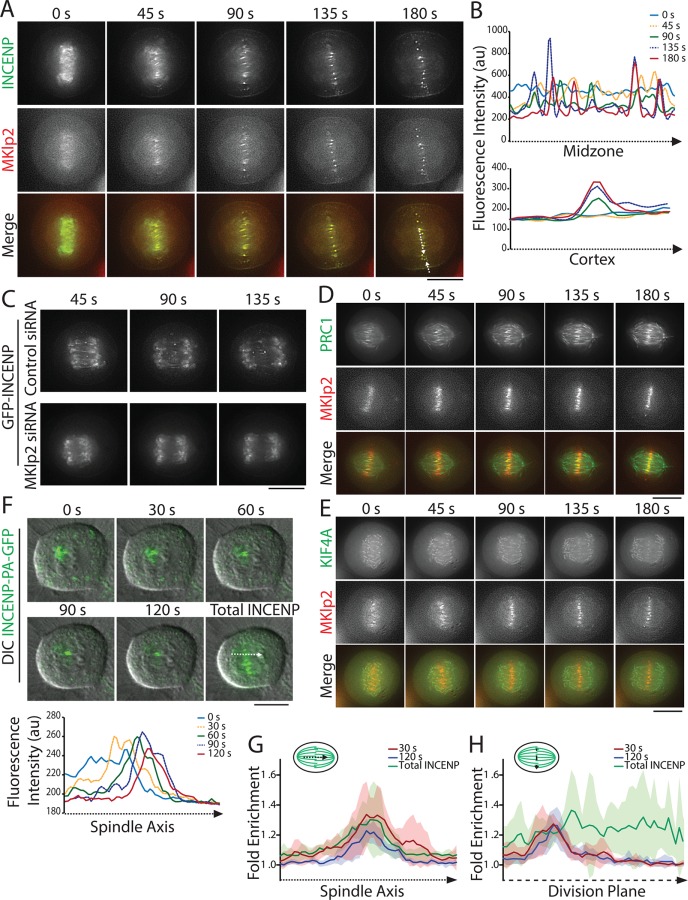
Previous analysis of CPC localization during C phase has relied primarily on protein localizations in fixed cells (Gruneberg et al., 2004; Hummer and Mayer, 2009; Kitagawa et al., 2013). To investigate CPC and MKlp2 dynamics with improved spatial and temporal resolution, we used live-cell imaging to track the movements of transiently expressed GFP-INCENP and MKlp2-mCherry at the metaphase-to-anaphase transition. As cells entered anaphase, we observed that GFP-INCENP and MKlp2-mCherry were colocalized in short, discrete stripes in between sister chromatids that had just disjoined (∼45 s after anaphase onset; Figure 4A and Supplemental Movie S1). These stripes were parallel to the spindle axis, suggesting that they correspond to the anti-parallel MT overlap region of the early midzone (Figure 4B). Shortly thereafter (∼135 s after anaphase onset), both GFP-INCENP and MKlp2-mCherry coalesced into radially symmetric spots of varying fluorescence intensity that moved erratically within the division plane (Figure 4A and Supplemental Movie S1). Over the same time period, we observed that GFP-INCENP became gradually enriched at the equatorial cortex (Figure 4B), indicating that cortical accumulation of the CPC occurs shortly after anaphase onset. As expected from previous work (Gruneberg et al., 2004; Hummer and Mayer, 2009), we did not observe the appearance of stripes or midzone-localized spots of GFP-INCENP in cells depleted of MKlp2 (Figure 4C and Supplemental Movie S2).
FIGURE 4:
The CPC localizes to the early midzone at anaphase onset. (A) Single z-plane micrographs taken from a time-lapse movie of a HeLa cell transiently expressing GFP-INCENP (green) and MKlp2-mCherry (red). Time is indicated in seconds relative to anaphase onset (0 s). Dashed lines indicate regions used to generate line scans. Scale bar, 10 μm. (B) Line scans of GFP-INCENP fluorescence intensity on the midzone and the cortex as indicated in A. (C) Single z-plane micrographs taken from a time-lapse movie of HeLa cells transiently expressing GFP-INCENP and treated with control or MKlp2-targeting siRNA. Time is indicated in seconds relative to anaphase onset (0 s). Scale bar, 10 μm. (D) Single z-plane micrographs taken from a time-lapse movie of a HeLa cell transiently expressing GFP-PRC1 (green) and MKlp2-mCherry (red). Time is indicated in seconds relative to anaphase onset (0 s). Scale bar, 10 μm. (E) Single z-plane micrographs taken from a time-lapse movie of a HeLa cell transiently expressing GFP-KIF4A (green) and MKlp2-mCherry (red). Time is indicated in seconds relative to anaphase onset (0 s). Scale bar, 10 μm. (F) Top, maximum intensity projections of three 1-µm optical sections taken from a time-lapse movie of a HeLa cell stably expressing INCENP-PA-GFP (green) after 48 h in 125 ng/ml doxycycline. Chromosomes were visualized using DIC. Time is indicated in seconds relative to photo activation (0 s). The final frame shows total photoactivation of INCENP. Dashed line represents region used to generate line scans. Bottom, line scans represent INCENP-PA-GFP fluorescence over time along the spindle midzone after anaphase onset. (G) Fold enrichment of INCENP-PA-GFP fluorescence parallel to the spindle axis at 30 s (red) or 120 s (blue) after photoactivation and total cellular INCENP (green). Schematic indicates the orientation of the line scan. Data represent the mean ± SD, n = 6 cells. (H) Fold enrichment of INCENP-PA-GFP fluorescence parallel to the division plane at 30 s (red) or 120 s (blue) after photoactivation and total cellular INCENP (green). Schematic indicates the orientation of the line scan. Data represent the mean ± SD, n = 6 cells.
Localization of the CPC to the region of anti-parallel MT overlap on the midzone is thought to occur via MKlp2-dependent transport. We were therefore somewhat surprised to observe a sudden relocation of both proteins to the midzone immediately after sister chromatid separation. We speculated that the complex might instead recognize a unique feature of the central portion of the midzone, such as the anti-parallel geometry of midzone MTs. To examine this possibility, we coimaged MKlp2-mCherry with GFP-PRC1 (Mollinari et al., 2002) or GFP-KIF4A (Stumpff et al., 2012), proteins that specifically localize to the midzone plus-end overlap. We found that stripes of MKlp2-mCherry colocalized with regions of the spindle most enriched for GFP-PRC1 (Figure 4D and Supplemental Movie S3), suggesting that MKlp2 preferentially targets to anti-parallel MTs after anaphase onset. Similarly, MKlp2-mCherry colocalized with GFP-KIF4A in early anaphase, although we observed that stripes of MKlp2-mCherry targeted to the early midzone before GFP-KIF4A was detectable (Figure 4E and Supplemental Movie S4). Collectively, these findings lead us to propose that the MKlp2-CPC complex colocalizes with the anti-parallel MT overlap immediately after anaphase onset.
The transfer of the CPC from centromeres to the midzone could occur in two ways: first, the CPC could directly load onto the MTs immediately adjacent to its respective centromere. Alternatively, the CPC could diffuse away from the centromere, form a complex with MKlp2, and rebind the midzone. To differentiate between these possibilities, we fused INCENP to photoactivatable GFP (INCENP-PA-GFP) and locally activated it on chromosomes immediately before anaphase onset. This allowed us to track a subset of centromeric CPC at the metaphase-to-anaphase transition. Photoactivated INCENP moved as a discrete mass from the centromere to the midzone MT overlap at anaphase onset and did not distribute across the midzone (Figure 4F and Supplemental Movie S5). Additionally, we observed that the position and intensity of photoactivated INCENP remained largely unchanged after loading onto the midzone, suggesting that the interaction between MKlp2-CPC and the anti-parallel overlap is stable (Figure 4, G and H). This result argues against a diffusion-based model of CPC midzone localization and instead suggests that MKlp2-CPC targets the nearest anti-parallel MT overlap after the complex dissociates from the centromere upon anaphase onset.
The CPC and MKlp2 move together during C phase
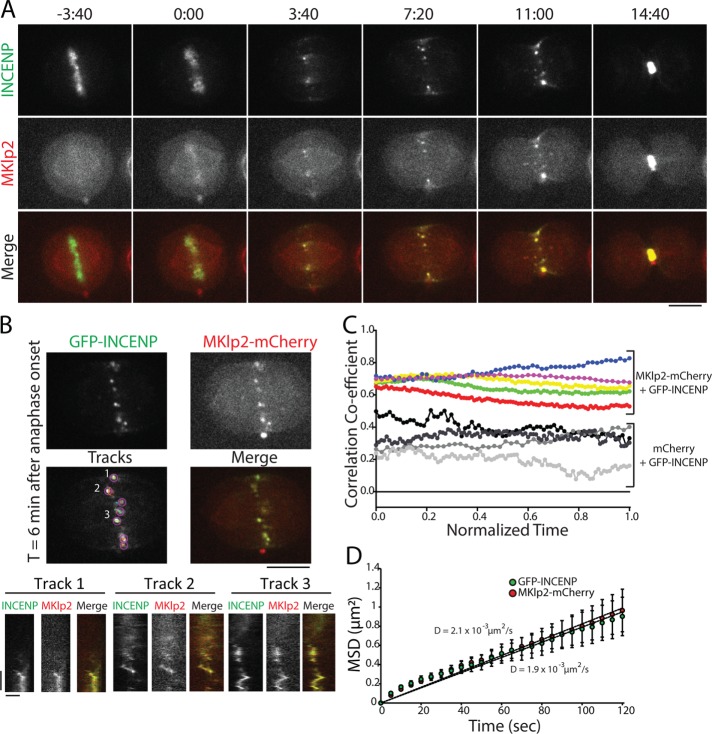
Our analysis of GFP-INCENP cortical enrichment demonstrates that the CPC can target the equatorial cortex in cells depleted of MKlp2 (Figure 3B). Cortical targeting depends on INCENP-actin binding, but maximal recruitment requires both actin and MKlp2 (Figure 3E). To better understand the relationship between MKlp2 and the CPC during cortical targeting, we followed the movement of GFP-INCENP and MKlp2-mCherry throughout C phase using near-simultaneous live-cell imaging within the midzone and in the plane of the cell cortex. Spots of GFP-INCENP and MKlp2-mCherry were always colocalized on the midzone during C phase (Figure 5A and Supplemental Movie S6) and moved in unison when tracked over time by kymograph (Figure 5B). It is important to point out that we did not measure copy numbers of INCENP and MKlp2, but the spots that we tracked almost certainly do not represent single molecules. Therefore, our conclusions do not consider the possibility that single molecules behave fundamentally different from spots. To quantify the association of GFP-INCENP and MKlp2-mCherry across multiple cells, we tracked the Pearson’s r on the midzone during C phase (Figure 5C). GFP-INCENP and MKlp2-mCherry were positively correlated on the midzone at anaphase onset (0.68 ± 0.02, mean ± SD, t = 0) and immediately before cleavage furrow ingression (0.72 ± 0.11, t = 1, n = 5 cells), in agreement with fixed-cell observations (Gruneberg et al., 2004). Cells coexpressing GFP-INCENP and a control mCherry fluorophore did not show a strong positive correlation on the midzone (0.26 ± 0.02, t = 0; 0.30 ± 0.14, t = 1; n = 3 cells), indicating that the colocalization between the CPC and MKlp2 is specific. This finding indicates that midzone-localized CPC is predominantly associated with MKlp2 for the duration of C phase.
FIGURE 5:
GFP-INCENP and MKlp2-mCherry are colocalized on the midzone throughout mitotic exit. (A) Single z-plane micrographs of the midzone taken from a time-lapse movie of a HeLa cell transiently expressing GFP-INCENP and MKlp2-mCherry plated on 100 µg/ml fibronectin. Time is indicated in minutes:seconds relative to anaphase onset (0:00). Scale bar, 10 μm. (B) Top, single z-plane micrograph taken from the time-lapse movie shown in A representing a time point 6 min after anaphase onset. The “tracks” panel highlights examples of spots and tracks used to quantify MSD. Numbers indicate tracks that correspond to kymographs below. Scale bar, 10 μm. Bottom, kymographs of GFP-INCENP and MKlp2-mCherry fluorescence generated from the spots indicated above. Scale bars, 100 s (y-axis) and 1 μm (x-axis). (C) Pearson’s r of GFP-INCENP and MKlp2-mCherry on the midzone (colored circles). Colocalization was quantified from immediately after anaphase onset (t = 0) to immediately before cleavage furrow ingression (t = 1). Control cells expressing GFP-INCENP and mCherry are represented by grayscale circles. n = 3 cells (control) and 5 cells (experimental) from three independent experiments. (D) MSD of GFP-INCENP (green circles) and MKlp2-mCherry (red circles) spots in the cortical plane and in the midzone during anaphase. Data represent mean ± SE. Linear regression represents best fit used to calculate the diffusion coefficient. Diffusion coefficient = 1.9 × 10−3 µm2/s (GFP-INCENP) or 2.1 × 10−3 µm2/s (MKlp2-mCherry). n = 626 tracks (GFP-INCENP) or 424 tracks (MKlp2-mCherry) from six cells from three independent experiments.
We next investigated the dynamic properties of GFP-INCENP and MKlp2-mCherry spots on the midzone during C phase. We used automated tracking to determine the XY position of fluorescent spots over time (Tinevez et al., 2016) and then quantified the mean-squared displacement (MSD) of these tracks (Tarantino et al., 2014). Spots of GFP-INCENP and MKlp2-mCherry were tracked independently in order to account for all fluorescent spots and not just those that were strongly colocalized. The MSD of GFP-INCENP and MKlp2-mCherry were both best fit using linear regression, indicating that midzone movement is largely nondirected (Figure 5D). MSD analysis of spot movement on the midzone yielded nearly identical diffusion coefficients: GFP-INCENP diffused at a rate of 1.9 × 10–3 µm2/s (n = 626 tracks from six cells) and MKlp2-mCherry diffused at a rate of 2.1 × 10–3 µm2/s (n = 424 tracks from six cells; Figure 5D), demonstrating that the two proteins have the same dynamic properties during C phase and confirming persistent colocalization during mitotic exit. Our analysis did reveal a small number of events that were consistent with directed movement; however, these events comprised ∼2–6% of the total time tracked in each cell. Additionally, this analysis represents time points after which the CPC has coalesced into discrete spots and does not include stripes of GFP-INCENP and MKlp2-mCherry that appear immediately after anaphase onset (Figure 4A). That movement of MKlp2-CPC is nondirected once the complex coalesces into spots suggests that motor-directed movement does not dominate CPC dynamics during C phase.
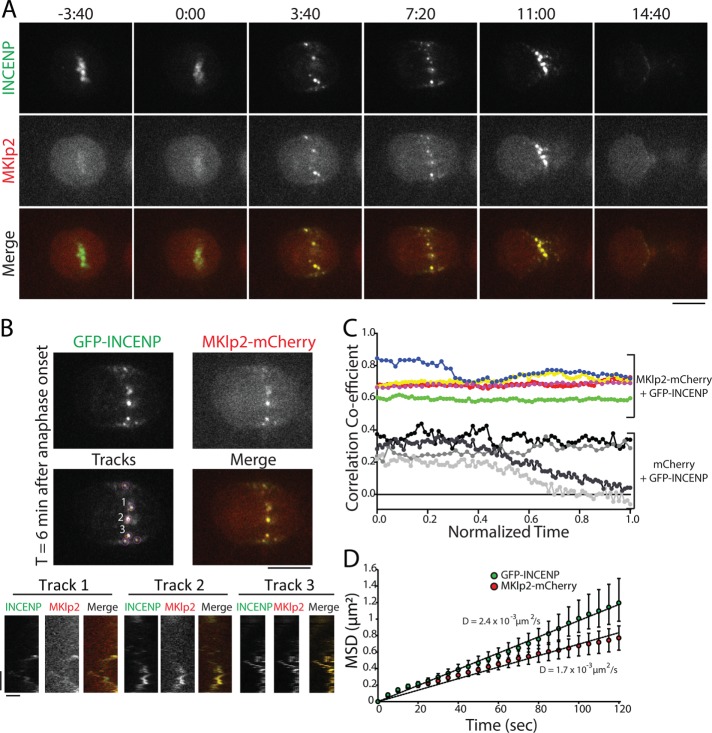
The midzone and the cortex have distinct cytoskeletal environments, and we therefore performed the same analysis of MKlp2-CPC spot movement in the plane of the actin-rich cortex. We created a large surface on the ventral side of the cell for imaging by plating cells on 100 μg/ml fibronectin (Taneja et al., 2016) and imaged the focal plane nearest the coverslip. Similar to our analysis of the midzone, spots of GFP-INCENP were colocalized with MKlp2-mCherry in the plane of the cell cortex throughout C phase (Figure 6A and Supplemental Movie S7), and these spots moved together when tracked by kymograph (Figure 6B). Spots of GFP-INCENP and MKlp2-mCherry were positively correlated at anaphase onset (0.68 ± 0.09, t = 0) and immediately before cleavage furrow ingression (0.68 ± 0.05, t = 1, n = 5 cells), whereas control cells expressing GFP-INCENP and mCherry were weakly positively correlated (0.25 ± 0.03, normalized time = 0; 0.07 ± 0.15, normalized time = 1; n = 3 cells; Figure 6C). The colocalization of GFP-INCENP and MKlp2-mCherry on the midzone and in the cortical plane suggests that MKlp2 does not deposit the CPC on the cell cortex and then unbind; instead, it suggests that the CPC and MKlp2 are always associated throughout C phase, regardless of location.
FIGURE 6:
GFP-INCENP and MKlp2-mCherry are colocalized in the plane of the cell cortex. (A) Single z-plane micrographs in the plane of the cell cortex taken from a time-lapse movie of a HeLa cell transiently expressing GFP-INCENP and MKlp2-mCherry plated on 100 µg/ml fibronectin. Time is indicated in minutes:seconds relative to anaphase onset (0:00). Scale bar, 10 μm. (B) Top, single z-plane micrographs in the plane of cell cortex taken from a time-lapse movie of a HeLa cell transiently expressing GFP-INCENP and MKlp2-mCherry plated on 100 μg/ml fibronectin. Micrographs represent a time point 6 min after anaphase onset. The “tracks” panel highlights examples of spots and tracks used to quantify MSD. Numbers indicate tracks that correspond to kymographs below. Scale bar, 10 μm. Bottom, kymographs of GFP-INCENP and MKlp2-mCherry generated from the spots indicated above. Scale bars, 100 s (y-axis) and 1 μm (x-axis). (C) Pearson’s r of GFP-INCENP and MKlp2-mCherry in the plane of the cell cortex (colored circles). Colocalization was quantified from immediately after anaphase onset (t = 0) to immediately before cleavage furrow ingression (t = 1). Control cells expressing GFP-INCENP and mCherry are represented by grayscale circles. n = 3 (control) and 5 cells (experimental) from three independent experiments. (D) MSD of GFP-INCENP (green circles) and MKlp2-mCherry (red circles) spots in the cortical plane during C phase. Data represent mean ± SE. Linear regression represents best fit used to calculate the diffusion coefficient. n = 625 tracks (GFP-INCENP) or 537 tracks (MKlp2-mCherry) from six cells from three independent experiments.
To characterize the behavior of spot movement, we again used automated tracking to follow the XY position of GFP-INCENP and MKlp2-mCherry in the plane of the cortex and calculated the MSD of these tracks. Like midzone-localized spots, the MSD of GFP-INCENP spots in the cortical plane was best fit using a linear regression, as was the MSD of MKlp2-mCherry, indicating that movement in the plane of the cortex, like midzone spot movement, is nondirected. The diffusion coefficient of GFP-INCENP was 2.5 × 10–3 µm2/s (n = 625 tracks from six cells, Figure 6D), and the diffusion coefficient of MKlp2-mCherry was 1.7 × 10–3 µm2/s (n = 537 tracks from six cells). The small difference between GFP-INCENP and MKlp2-mCherry diffusion coefficients is likely due to the relatively dim fluorescence of mCherry, which affects the accuracy of automated tracking. Even so, the MSD of GFP-INCENP and MKlp2-mCherry were not significantly different at the terminal time point (120 s, p > 0.05, two-sample Student’s t test; Figure 6D). This analysis reveals that the dynamic movement of MKlp2-CPC in the cortical plane mirrors the behavior of the complex within the midzone, and neither population moves in a manner that resembles motor-directed stepping.
INCENP overexpression rescues cleavage furrow ingression failure in MKlp2-depleted cells
Our findings indicate that the CPC can target the equatorial cortex after MKlp2 depletion, but the function of this population during C phase is not clear. Because the CPC is known to regulate cytokinesis, we tested whether CPC at the equatorial cortex could promote cleavage furrow ingression. To investigate this possibility, we took advantage of the observation that cytokinesis fails in cells that are depleted of MKlp2 (Kitagawa et al., 2013), working under the hypothesis that this defect is due to reduced levels of cortical CPC (Figure 3B). We reasoned that INCENP overexpression may cause cortical CPC to accumulate, via actin binding, to levels that are required for the completion of furrowing. To test this, we expressed GFP-INCENP or the actin binding–deficient GFP-INCENP CR using either transient overexpression or stable cell lines generated using the recombination-mediated cassette exchange (RMCE) system (Khandelia et al., 2011) that uniformly express GFP, GFP-INCENP, or GFP-INCENP CR in a doxycycline-inducible manner.
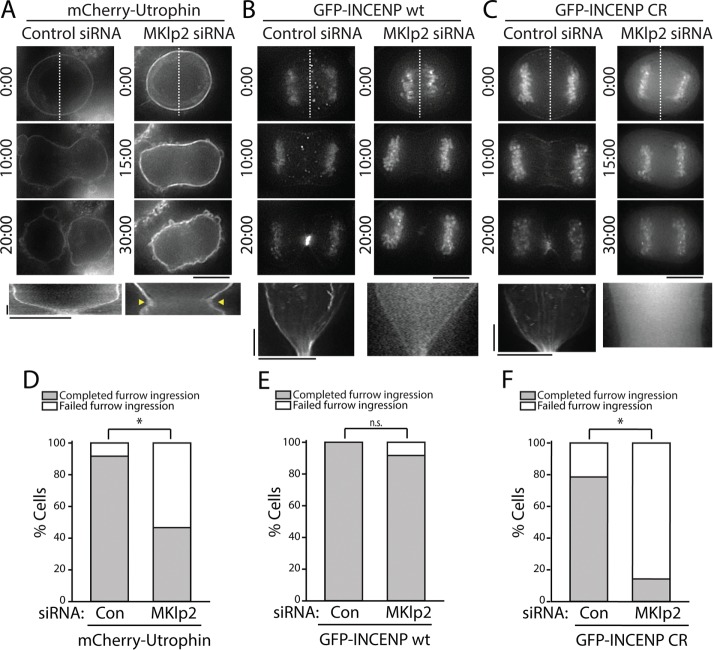
In agreement with previous findings (Kitagawa et al., 2013), we observed that approximately half of MKlp2-depleted cells successfully completed cleavage furrow ingression (seven out of 15 cells; Figure 7C), as judged by imaging of mCherry-Utrophin (Figure 7D). Those that failed cytokinesis did so as a consequence of furrow regression rather than a failure to initiate ingression (arrowheads in Figure 7A and Supplemental Movie S8), which is consistent with the previously described phenotype exhibited in MKlp2-depleted cells (Kitagawa et al., 2013). A majority of control cells completed furrow ingression (11/12 cells; Figure 7, A and D), resulting in the formation of a midbody. Similar results were obtained in cells stably expressing GFP (Supplemental Figure S2).
FIGURE 7:
Overexpression of GFP-INCENP rescues cleavage furrow ingression in MKlp2-depleted cells. (A) Top, single z-plane micrographs taken from time-lapse movies of cells transiently expressing mCherry-Utrophin after treatment with a control or MKlp2-targeting siRNA. Time is indicated in minutes:seconds. Dashed lines were used to generate kymographs. Scale bar, 10 μm. Bottom, kymographs of mCherry-Utrophin fluorescence across the division plane. Yellow arrows represent cleavage furrow regression. Scale bars, 2.5 min (y-axis) and 10 μm (x-axis). (B) Top, single z-plane micrographs taken from time-lapse movies of cells transiently expressing GFP-INCENP after treatment with a control or MKlp2-targeting siRNA. Time is indicated in minutes:seconds. Dashed lines were used to generate kymographs. Scale bar, 10 μm. Bottom, kymographs of GFP-INCENP fluorescence across the division plane. Scale bars, 2.5 min (y-axis) and 10 μm (x-axis). (C) Top, single z-plane micrographs taken from time-lapse movies of cells transiently expressing GFP-INCENP CR after treatment with a control or MKlp2-targeting siRNA. Time is indicated in minutes:seconds. Dashed lines were used to generate kymographs. Scale bar, 10 μm. Bottom, kymographs of GFP-INCENP CR fluorescence across the division plane. Scale bars, 2.5 min (y-axis) and 10 μm (x-axis). (D) Quantification of the percentage of cells from A that successfully completed furrow ingression. n = 12 (control) and 15 (MKlp2 siRNA) cells, *p < 0.05. (E) Quantification of total percentage of cells from B that successfully completed furrow ingression. n = 14 cells for each condition, n = 8 (control) and 12 (MKlp2 siRNA) cells. (F) Quantification of total percentage of cells from C that successfully completed furrow ingression. n = 14 cells for each condition, *p < 0.05.
In contrast to cells expressing mCherry-Utrophin, we found that a transient overexpression of GFP-INCENP rescued cleavage furrow ingression in MKlp2-depleted cells (furrow ingression complete in 11/12 cells; Figure 7, B and E, and Supplemental Movie S9) without impacting C-phase progression in control-depleted cells (eight out of eight cells). Stable expression of GFP-INCENP also rescued furrowing after MKlp2 depletion (Supplemental Figure S2). Importantly, only cells that did not show any GFP-INCENP localized on the midzone were included in this analysis, suggesting that successful furrow ingression was mediated through CPC localized to the equatorial cortex.
We next tested whether this rescue phenotype required actin-dependent recruitment of the CPC using our charge reversal mutant. Like GFP-INCENP wt, transient overexpression of GFP-INCENP CR did not interfere with cleavage furrow ingression because cells transfected with control siRNA completed furrowing (11/14 cells; Figure 7, C and F, and Supplemental Movie S10). Depletion of MKlp2 coupled with overexpression of GFP-INCENP CR caused the majority of cells to fail in cytokinesis (furrowing complete in two out of 14 cells; Figure 7, C and F). Importantly, furrow ingression failed to initiate in these cells. Stable expression of GFP-INCENP CR after MKlp2 depletion also resulted in cytokinetic failure, indicating that this result is specific to the loss of CPC-actin binding (furrowing complete in three out of 13 cells; Supplemental Figure S2). A portion of these cells were able to initiate furrowing (four out of 13 cells), but this was soon followed by furrow regression. Other cells were unable to initiate furrowing (six out of 13 cells), in agreement with our observations from transient transfection. These findings demonstrate that overexpression of INCENP can rescue cleavage furrow ingression after MKlp2 depletion in a manner that requires the INCENP–actin interaction. Taken together, our data suggest that the key function of cortically localized CPC is to promote the initiation and successful completion of furrowing.
DISCUSSION
In this work, we investigated the role of actin, MTs, and MKlp2 in recruiting the CPC to the equatorial cortex of dividing cells. Although midzone targeting of the CPC has been the subject of many studies, comparably little has been done to address cortical targeting mechanism(s). Because the CPC requires the kinesin-6 MKlp2 to concentrate at the MT plus-end overlap of the midzone (Gruneberg et al., 2004; Hummer and Mayer, 2009; Kitagawa et al., 2013), the field-accepted view is that the CPC accumulates at MT plus ends via motor-dependent directed transport. With respect to the furrow, this model is appealing given that MT plus ends can be observed near the furrow cortex (Canman et al., 2003). During monopolar cytokinesis, however, the CPC does not concentrate at MT plus ends of monopolar midzones, and it instead localizes to an actin-rich zone that underlies the cortex (Hu et al., 2008). One possible explanation for these data is that the CPC-MKlp2 complex accumulates at regions of anti-parallel MT overlap rather than at MT plus ends.
Consistent with this notion, PRC1, a MT-associated protein that preferentially binds anti-parallel MT overlaps (Bieling et al., 2010), has been hypothesized to localize several kinesin motors to the midzone, including MKlp2 (Gruneberg et al., 2006). Indeed, our live-cell imaging indicates that MKlp2 and PRC1 are colocalized in the center of the midzone during mitotic exit (Figure 4D). Photoactivation of INCENP suggests that MKlp2 targets the CPC to this location immediately upon leaving the centromere (Figure 4F). If the MKlp2-CPC complex diffused significantly after removal from centromeres, one would expect the midzone to be more broadly decorated by MKlp2-CPC. Importantly, GFP-INCENP fluorescence did not diminish significantly over ∼2 min, suggesting a stable interaction with the midzone. Finally, we observed that the movement of MKlp2-CPC spots exhibited diffusive behavior rather than directed motion throughout C phase, supporting the idea that MKlp2 targets the CPC to the cell middle through the recognition of anti-parallel MTs, as opposed to plus end–directed transport. Consistent with this possibility, recent investigations of MKlp2’s mechanochemistry suggest that the protein may not function as a classical transport motor and instead may work to organize MT bundles (Atherton et al., 2017). We cannot of course exclude a minor contribution from MKlp2 motility to midzone positioning of the complex; such motility could focus the CPC at MT plus ends at the overlap region. A final note is that a conformational change in PRC1 induced by MT cross-linking (Subramanian et al., 2010) is likely required to facilitate a PRC1–MKlp2 interaction, as PRC1 is present at the plus ends of monopolar midzone MTs, which are parallel in orientation (Hu et al., 2008).
In principle, cortical targeting of the CPC could stem from PRC1–MKlp2–CPC interactions that occur on anti-parallel MT overlaps of filaments juxtaposed with the cell cortex (Figure 1A). This could explain the effect of nocodazole in ablating cortical Aurora B-GFP in experiments by Murata-Hori and Wang (2002). However, it is also possible that the equatorial cortex itself reinforces the cortical association of MKlp2-CPC via interactions with actomyosin. Indeed, it is known that MKlp2 binds nonmuscle myosin II (Kitagawa et al., 2013) and that INCENP binds F-actin (Chen et al., 2007; Landino and Ohi, 2016). The interaction between MKlp2 and myosin II may explain our observation that GFP-INCENP CR targets to the cortex despite disrupting CPC-actin binding (Figure 3C). Furthermore, the CPC colocalizes with actin during monopolar cytokinesis (Hu et al., 2008) and also in a cell-free system that reconstitutes the cytokinetic furrow (Nguyen et al., 2014). Here, using an INCENP mutant that cannot bind F-actin (Landino and Ohi, 2016), we demonstrated that the ability of INCENP to associate with actin promotes the accumulation of the CPC at the equatorial cortex, even in cells depleted of MKlp2. Pharmacological disruption of actin and MTs mirrors these findings: loss of either MTs or actin alone is not sufficient to disrupt CPC localization at the cell middle, but disassembly of both cytoskeletal systems is. We quantified the relative contribution of each filament system using live-cell analysis to avoid fixation artifacts that could impact cortical CPC accumulation. In agreement with our immunofluorescence experiments, cortical localization of the CPC in live cells expressing the INCENP-actin binding mutant is nullified upon depletion of MKlp2, leading us to speculate that the CPC can only reach the division plane through either direct actin binding or MKlp2-dependent targeting.
Because cortical actin is only moderately enriched at the site of the presumptive furrow in mammalian cells (Murthy and Wadsworth, 2005; Rankin and Wordeman, 2010), how a general interaction of INCENP with F-actin can concentrate the CPC at the cell middle independent of MKlp2 is not clear. For example, we do not observe localization of GFP-INCENP at the polar cortices. It is reasonable to speculate that the mechanism of actin-based recruitment to the cell equator depends on more than just the INCENP–actin interaction. One possibility is that cortical flow delivers the CPC toward the division site (Bray and White, 1988), although our live-cell imaging of GFP-INCENP at the cell cortex did not detect such biased movements. We cannot rule out the possibility that the cortical movements of CPC switch to directed flow after MKlp2 depletion; unfortunately, cells depleted of MKlp2 do not have enough cortically localized GFP-INCENP to allow accurate tracking and characterization of spot movement. A second possibility is that the CPC reaches the cortex through additional interactions with factors at the division site such as furrow-specific lipids (Fung et al., 2017).
From a functional perspective, we found that the actin-dependent CPC recruitment pathway is a viable mechanism for promoting successful cell division. Furrows normally “bounce back” after MKlp2 depletion, resulting in failed cytokinesis and binucleate cells (Kitagawa et al., 2013). This phenotype has been attributed to decreased levels of CPC at the division plane. In agreement with this idea, we found that overexpression of GFP-INCENP, but not an INCENP mutant incapable of interacting with actin, rescues furrowing in MKlp2-depleted cells (Figure 7). Importantly, these cells lack midzone-associated GFP-INCENP and only show enrichment at the cell cortex, indicating that cortically localized CPC is sufficient to promote furrow closure in the absence of midzone-localized CPC. Cells expressing an INCENP-actin binding mutant, depleted of MKlp2, exhibit a more penetrant cytokinesis failure phenotype than cells depleted of MKlp2 alone, confirming that cortical CPC is capable of promoting furrow ingression. Owing to the cell-to-cell variability we observe in MKlp2 knockdown efficiency, we cannot rule out the possibility that low levels of MKlp2 are involved in targeting the CPC to the cortex, even when GFP-INCENP is not detectable on the midzone. However, previous work has shown that an INCENP-MT binding mutant, which abolishes MKlp2 midzone localization, localizes to the cell cortex and is sufficient to promote cytokinesis (van der Horst et al., 2015), a finding consistent with our results. Also consistent with our results is the observation that directly impairing MKlp2’s ability to target the midzone does not completely abolish CPC targeting to the cell middle (Cullati et al., 2017). Collectively, our work strongly suggests that cortically localized CPC promotes cleavage furrow ingression and that MKlp2 is not directly required for cytokinesis. This is in agreement with previous studies showing that while Aurora B is required for cytokinesis in Drosophila, Subito is not (Cesario et al., 2006). Cortical CPC targets that initiate cytokinesis and ensure furrow closure will be important to identify and provide fertile grounds for future studies.
MATERIALS AND METHODS
Cell culture and transfections
Cell lines were cultured in media supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin at 37°C with 5% CO2. HeLa “Kyoto” cells were cultured in DMEM. Stable HeLa cells expressing GFP-INCENP under the control of a doxycycline-inducible promoter were cultured in DMEM containing 1 µg/ml puromycin during selection and then cultured in DMEM. Transfections were performed using Lipofectamine 2000 (Invitrogen) for plasmid DNA or HiPerFect (Qiagen) for siRNAs according to manufacturer instructions. For both, cells were cultured in Opti-MEM supplemented with 10% FBS after transfection. siRNA sequences used in this study were as follows: Stealth Lo GC negative control (Life Technologies) and MKlp2, 5′-AACCACCTATGTAATCTCATG (Qiagen; Kitagawa et al., 2013).
To generate HeLa cells that stably express GFP-INCENP in a doxycycline-inducible manner, we used the high-efficiency, low-background RMCE system (Khandelia et al., 2011). Acceptor HeLa cells (Sturgill et al., 2016) were cotransfected with pEM784 and pEM791 containing either GFP-INCENP or GFP-INCENP CR as previously described (Landino and Ohi, 2016). After transfection (24 h), cells were cultured in the presence of 1 µg/ml puromycin and were then transferred to media containing 2 µg/ml puromycin 48 h post-transfection to select for transgenic cells. Cells were pooled and cultured in DMEM containing 10% FBS, penicillin, and streptomycin. GFP-INCENP expression was induced using 2 µg/ml doxycycline for 24 h.
For pharmacological perturbations, drug stocks in DMSO were diluted in DMEM immediately before use. Cells were treated with the following small molecules at 37°C for the lengths of time indicated: 5 μg/ml cytochalasin B (Sigma) for 30 min and 5 μM nocodazole (Sigma) for 10 min. For simultaneous treatment, cells were treated with 5 μg/ml cytochalasin B and 5 μM nocodazole for 10 min.
Immunofluorescence and fixed-cell imaging
For most experiments, HeLa cells were fixed with methanol at –20°C for 3 or 10 min. The following primary antibodies were used in this study: mouse anti-tubulin (DM1α; Vanderbilt Antibody and Protein Resource), 1:500; rat anti-tubulin (YL1/2; Accurate Chemical and Scientific Corporation), 1:500; anti-AIM (BD Transduction Labs), 1:500; anti-INCENP (Abcam), 1:500; and anti-MKlp2 (Abnova), 1:50. Secondary antibodies conjugated to Alexa 488, Alexa 594, or Alexa 647 (Invitrogen) were used at 1:1000. Primary and secondary antibody incubations were for 1 h each at room temperature. To visualize actin, cells were fixed with 1% glutaraldehyde in cytoskeleton buffer (10 mM 2-(N-morpholino)ethanesulfonic acid, pH 6.1; 138 mM KCl; 3 mM MgCl2; 2 mM ethylene glycol tetraacetic acid) plus 11% sucrose at room temperature for 10 min. Actin was stained with phalloidin–tetramethyl rhodamine isothiocyanate (Sigma) at 500 ng/μl. DNA was counterstained with 5 μg/ml Hoechst 33342. Stained cells were mounted in Prolong Gold (Invitrogen).
Cells were visualized using either a 60 × 1.4 numerical aperture (NA) or 100 × 1.4 NA objective (Olympus) on a DeltaVision Elite imaging system (GE Healthcare) equipped with a Cool SnapHQ2 charge-coupled device (CCD) camera (Roper). Optical sections were collected at 200-nm intervals and processed using ratio deconvolution in SoftWorx (GE Healthcare). Images were prepared for publication using ImageJ and NIS-Elements AR.
Averaged line scans of ABK immunofluorescence were generated using a 6-µm-wide and 14-µm-long line along the spindle axis in maximum intensity projections. Peak intensity values were manually aligned, and fluorescence intensity was plotted as fold enrichment over cellular background fluorescence.
To measure ABK fluorescence after MKlp2 depletion, a boxed region at the division plane was used to determine the integrated fluorescence of a sum intensity projection. An equivalently sized region outside the division plane was used to measure background fluorescence, and this was subtracted from the division plane fluorescence. MKlp2– cells were selected for analysis based on the loss of MKlp2 immunofluorescence on the midzone.
Live-cell imaging
Widefield live-cell imaging was performed at 37°C with 5% CO2 and a 60 × 1.4 NA objective on a DeltaVision Elite imaging system equipped with a WeatherStation Environmental Chamber. For live imaging of HeLa cells transiently expressing GFP-INCENP and MKlp2-mCherry (Figure 4), cells were plated on glass-bottom dishes (MatTek Corporation) 48 h before imaging and transfected 24 h before imaging with plasmid DNA and/or siRNA as described above. Immediately before imaging, cells were incubated in movie medium (L-15 medium without phenol red supplemented with 10% FBS, penicillin/streptomycin, and 7 mM potassium-4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.7). One optical section was selected, and cells were imaged at 3-s intervals. For live imaging of HeLa cells expressing MKlp2-mCherry and GFP-PRC1, or MKlp2-mCherry and GFP-KIF4A, one optical section was selected, and cells were imaged at 5-s intervals. For live-cell imaging of transiently expressed GFP-INCENP constructs and mCherry-Utrophin (Supplemental Figure S2), HeLa cells were plated on glass-bottom dishes (MatTek Corporation) 48 h before imaging and transfected with plasmid DNA and/or siRNA 24 h before imaging. Culture media were exchanged for movie medium immediately before imaging. To image probes during C phase, one optical section was selected, and cells expressing GFP-INCENP were imaged at 5-s intervals. Cells expressing mCherry-Utrophin were imaged at 30-s intervals to minimize photobleaching. To image MKlp2-depleted HeLa cells stably expressing GFP-INCENP CR (Figure 7), cells were plated on glass-bottom dishes 48 h before imaging. Before imaging (24 h), siRNA was transfected as described above, and 2 μg/ml doxycycline was added. Cells were imaged in movie medium, GFP-INCENP was imaged at 10-s intervals, and differential interference contrast (DIC) imaging was used to image the cell cortex.
Live-cell confocal imaging was performed at 37°C with 5% CO2 and a 60 × 1.4 NA in movie medium. Cells were imaged using a Nikon spinning-disk confocal microscope equipped with a stage top incubator (Tokai Hit) and an Andor DU-897 electron-multiplier CCD camera. Fluorophores were excited using 488- and 561-nm diode laser lines. To generate volume projections of the division plane in cells transiently expressing GFP-INCENP or GFP-INCENP CR (Figure 3), 200-nm optical sections were collected using triggered acquisition to reduce temporal delay (Figure 3). Cells were transfected with plasmid DNA and siRNA 24 h before imaging. To image transiently expressed GFP-INCENP and MKlp2-mCherry in the midzone and cortical planes (Figures 5 and 6), cells were plated on glass-bottom dishes coated with 100 µg/ml fibronectin. Without this treatment, HeLa cells are rounded during mitosis, impeding cortical imaging. Cells were transfected with plasmid DNA 24 h before imaging. Two optical sections, corresponding to the cortex and the midzone, were imaged at 5-s intervals using triggered acquisition to reduce temporal delay. The focal plane nearest the coverslip where GFP-INCENP and MKlp2-mCherry puncta were in focus was selected as the cortical plane. For live imaging of C phase in GFP-INCENP stable HeLa cells (Figure 7), cells were plated on glass-bottom dishes 48 h before imaging, and siRNA transfection and doxycycline addition occurred 24 h before imaging. A single optical section was imaged at 15-s intervals.
Photoactivation experiments were performed using a stable HeLa cell line that inducibly expressed INCENP-PA-GFP and a microRNA to deplete endogenous INCENP, using the RMCE system (using plasmid pERB140), as described above. Cells were induced with doxycycline (125 ng/ml) 48 h before imaging and seeded on 22 × 22 mm glass coverslips (no. 1.5; Fisher Scientific) coated with poly-d-lysine (Sigma-Aldrich) 24 h before imaging. Coverslips were mounted in magnetic chambers (Chamlide CM-S22-1, LCI) and maintained in L-15 medium without phenol red (Invitrogen) supplemented with 10% FBS and penicillin/streptomycin. Temperature was maintained at ∼35°C using an environmental control chamber (Incubator BL; PeCon GmbH). Images were acquired using a spinning-disk confocal microscope (DM4000; Leica) with a 100 × 1.4 NA objective, an XY Piezo-Z stage (Applied Scientific Instrumentation), a spinning disk (Yokogawa), an electron-multiplier CCD camera (ImageEM; Hamamatsu Photonics), and a 488-nm laser for imaging (LMM5; Spectral Applied Research), controlled by MetaMorph software (Molecular Devices). This system was equipped with an iLas targeted laser illuminator (Roper Scientific) with a 405-nm laser (CrystaLaser LC model no. DL405-050-O, 27 mW output after fiber coupling), controlled using the iLas2 software module within MetaMorph. Metaphase cells were identified in DIC and monitored until anaphase onset. Shortly after anaphase onset, a region of chromatin near the outer edge of the metaphase plate, away from the cell center, was photoactivated by rasterizing 10 times with the laser at 15% power. Three 1-µm optical sections were imaged every 30 s for 2 min, and then the entire cell was photoactivated (10× raster, 15% power) to reveal the total cellular INCENP.
Analysis of live-cell imaging
To quantify cortical enrichment of GFP-INCENP at the cell cortex, volume projections of the YZ dimensions of the division plane were generated using ImageJ. Line scans across the YZ projection of the division plane were normalized for length, aligning peak intensity values or cell edges where applicable. Background fluorescence was measured outside the cell, and this value was subtracted from all intensity measurements along the line scan. Fluorescence intensity values were then plotted as fold enrichment over cytoplasmic background, to control for variable protein expression. To quantify the relative increase of fluorescence intensity at the cortex across multiple cells, three adjacent values from each peak of fluorescence intensity were averaged.
Averaged line scans of INCENP-PA-GFP were generated using a 6-µm-long line along the spindle axis or along the division plane in maximum intensity projections of three 1-µm optical sections. Line scans were generated from micrographs at 30 or 120 s after photoactivation, or immediately after photoactivation of total cellular INCENP. Peak intensity values were manually aligned, and fluorescence intensity was plotted as fold enrichment over nonactivated cellular background fluorescence.
GFP-INCENP and MKlp2-mCherry spot tracking was performed using the TrackMate plug-in in ImageJ (Tinevez et al., 2016). GFP-INCENP and MKlp2-mCherry were each tracked individually for both the cortex and the midzone. Parameters used were as follows: a Laplacian of Gaussian detector, an estimated blob diameter of 1.0 µm, a threshold of 10.0, a simple linear assignment problem tracker (for nonbranching tracks), a linking max distance of 1.0 µm, a gap closing max distance of 1.0 µm, and a gap closing frame of 1. Only tracks with a lifetime of ≥15 s (or three frames) were considered. MSD plots were obtained using the MSDanalyzer MATLAB plug-in (Tarantino et al., 2014), where only the first 120 s were used for MSD analysis. Diffusion coefficients were calculated by dividing the slope of the MSD plot (obtained by linear regression) by 4 to reflect two-dimensional space. To quantify the Pearson’s r of GFP-INCENP and MKlp2-mCherry on the midzone and the cortex, images were cropped in time only to analyze the correlation from anaphase onset to the initiation of furrowing. Only the region corresponding to the division plane was included so as not to analyze areas beyond the chromosomes plate, or outside the cell. The r was measured using the Genome Damage and Stability Centre stack correlation analyzer with no additional thresholding or corrections. To normalize for the duration of C phase, the time cells began furrowing was normalized to 1 and anaphase onset was normalized to 0.
Immunoblotting
For preparation of whole-cell lysates, cells were washed three times with phosphate-buffered saline (PBS) and resuspended in 2× Laemmli buffer. After heating to 95°C for 5 min, proteins were resolved by SDS–PAGE and transferred to nitrocellulose (Whatman). Immunoblots were blocked with 5% wt/vol milk in Triton X-100 in PBS and then probed with primary antibodies (anti-INCENP, anti-AIM, or DM1α to detect tubulin) at 1:500 for 1 h. Blots were then probed with species-appropriate fluorescently tagged secondary antibodies for 45 min. Fluorescence was measured using an Odyssey fluorescence detection system (LI-COR Biosciences).
Statistical analysis
Statistically relevant differences in experimental data were determined using the T.TEST function in Excel (Microsoft). In all cases, p values report the two-tailed distribution of a two-sample Student’s t test assuming unequal variance.
Supplementary Material
Acknowledgments
We thank Jason Stumpff (University of Vermont) for the plasmid encoding GFP-KIF4A and Bob Margolis (Sanford Burnham Prebys) for the plasmid encoding GFP-PRC1, as well as members of the Ohi lab for insightful discussions. This work was supported by National Institutes of Health Grant no. R01GM086610 to R.O. and R01GM083988 to M.L. R.O. is a scholar of the Leukemia and Lymphoma Society.
Abbreviations used:
- ABK
Aurora B kinase
- AU
arbitrary units
- CCD
charge-coupled device
- CPC
chromosomal passenger complex
- CR
charge reversal
- DIC
differential interference contrast
- DMSO
dimethyl sulfoxide
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- INCENP
inner centromeric protein
- MSD
mean-squared displacement
- MT
microtubule
- NA
numerical aperture
- PA
phosphoactivatable
- PBS
phosphate-buffered saline
- RMCE
recombination-mediated cassette exchange
- RNAi
RNA interference
- siRNA
small interfering RNA
- wt
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-06-0399) on September 27, 2017.
REFERENCES
- Abaza A, Soleilhac JM, Westendorf J, Piel M, Crevel I, Roux A, Pirollet F. M phase phosphoprotein 1 is a human plus-end-directed kinesin-related protein required for cytokinesis. J Biol Chem. 2003;278:27844–27852. doi: 10.1074/jbc.M304522200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton J, Yu IM, Cook A, Muretta JM, Joseph A, Major J, Sourigues Y, Clause J, Topf M, Rosenfeld SS, et al. The divergent mitotic kinesin MKLP2 exhibits atypical structure and mechanochemistry. Elife. 2017;6, e27793 doi: 10.7554/eLife.27793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Telley IA, Surrey T. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 2010;142:420–432. doi: 10.1016/j.cell.2010.06.033. [DOI] [PubMed] [Google Scholar]
- Bray D, White JG. Cortical flow in animal cells. Science. 1988;239:883–888. doi: 10.1126/science.3277283. [DOI] [PubMed] [Google Scholar]
- Canman JC, Cameron LA, Maddox PS, Straight A, Tirnauer JS, Mitchison TJ, Fang G, Kapoor TM, Salmon ED. Determining the position of the cell division plane. Nature. 2003;424:1074–1078. doi: 10.1038/nature01860. [DOI] [PubMed] [Google Scholar]
- Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012;13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesario JM, Jang JK, Redding B, Shah N, Rahman T, McKim KS. Kinesin 6 family member Subito participates in mitotic spindle assembly and interacts with mitotic regulators. J Cell Sci. 2006;119:4770–4780. doi: 10.1242/jcs.03235. [DOI] [PubMed] [Google Scholar]
- Chen Q, Lakshmikanth GS, Spudich JA, De Lozanne A. The localization of inner centromeric protein (INCENP) at the cleavage furrow is dependent on Kif12 and involves interactions of the N terminus of INCENP with the actin cytoskeleton. Mol Biol Cell. 2007;18:3366–3374. doi: 10.1091/mbc.E06-10-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke CA, Heck MM, Earnshaw WC. The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J Cell Biol. 1987;105:2053–2067. doi: 10.1083/jcb.105.5.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullati SN, Kabeche L, Kettenbach AN, Gerber SA. A bifurcated signaling cascade of NIMA-related kinases controls distinct kinesins in anaphase. J Cell Biol. 2017;216:2339–2354. doi: 10.1083/jcb.201512055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Cooke CA. Analysis of the distribution of the INCENPs throughout mitosis reveals the existence of a pathway of structural changes in the chromosomes during metaphase and early events in cleavage furrow formation. J Cell Sci. 1991;98(Pt 4):443–461. doi: 10.1242/jcs.98.4.443. [DOI] [PubMed] [Google Scholar]
- Fung SY, Kitagawa M, Liao PJ, Wong J, Lee SH. Opposing activities of Aurora B Kinase and B56-PP2A phosphatase on MKlp2 determine abscission timing. Curr Biol. 2017;27:78–86. doi: 10.1016/j.cub.2016.10.042. [DOI] [PubMed] [Google Scholar]
- Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Annu Rev Cell Dev Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- Gruneberg U, Neef R, Honda R, Nigg EA, Barr FA. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J Cell Biol. 2004;166:167–172. doi: 10.1083/jcb.200403084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U, Neef R, Li X, Chan EH, Chalamalasetty RB, Nigg EA, Barr FA. KIF14 and citron kinase act together to promote efficient cytokinesis. J Cell Biol. 2006;172:363–372. doi: 10.1083/jcb.200511061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizlan D, Mishima M, Tittmann P, Gross H, Glotzer M, Hoenger A. Structural analysis of the ZEN-4/CeMKLP1 motor domain and its interaction with microtubules. J Struct Biol. 2006;153:73–84. doi: 10.1016/j.jsb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Hu CK, Coughlin M, Field CM, Mitchison TJ. Cell polarization during monopolar cytokinesis. J Cell Biol. 2008;181:195–202. doi: 10.1083/jcb.200711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummer S, Mayer TU. Cdk1 negatively regulates midzone localization of the mitotic kinesin Mklp2 and the chromosomal passenger complex. Curr Biol. 2009;19:607–612. doi: 10.1016/j.cub.2009.02.046. [DOI] [PubMed] [Google Scholar]
- Khandelia P, Yap K, Makeyev EV. Streamlined platform for short hairpin RNA interference and transgenesis in cultured mammalian cells. Proc Natl Acad Sci USA. 2011;108:12799–12804. doi: 10.1073/pnas.1103532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Fung SY, Hameed UF, Goto H, Inagaki M, Lee SH. Cdk1 coordinates timely activation of MKlp2 kinesin with relocation of the chromosome passenger complex for cytokinesis. Cell Rep. 2014;7:166–179. doi: 10.1016/j.celrep.2014.02.034. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Fung SY, Onishi N, Saya H, Lee SH. Targeting Aurora B to the equatorial cortex by MKlp2 is required for cytokinesis. PLoS One. 2013;8:e64826. doi: 10.1371/journal.pone.0064826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Lee SH. The chromosomal passenger complex (CPC) as a key orchestrator of orderly mitotic exit and cytokinesis. Front Cell Dev Biol. 2015;3:14. doi: 10.3389/fcell.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landino J, Ohi R. The timing of midzone stabilization during cytokinesis depends on myosin II activity and an interaction between INCENP and actin. Curr Biol. 2016;26:698–706. doi: 10.1016/j.cub.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzwa B, Gerlich DW. Cytokinetic abscission: molecular mechanisms and temporal control. Dev Cell. 2014;31:525–538. doi: 10.1016/j.devcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- Mishima M, Pavicic V, Gruneberg U, Nigg EA, Glotzer M. Cell cycle regulation of central spindle assembly. Nature. 2004;430:908–913. doi: 10.1038/nature02767. [DOI] [PubMed] [Google Scholar]
- Mollinari C, Kleman JP, Jiang W, Schoehn G, Hunter T, Margolis RL. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J Cell Biol. 2002;157:1175–1186. doi: 10.1083/jcb.200111052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata-Hori M, Wang YL. The kinase activity of aurora B is required for kinetochore-microtubule interactions during mitosis. Curr Biol. 2002;12:894–899. doi: 10.1016/s0960-9822(02)00848-5. [DOI] [PubMed] [Google Scholar]
- Murthy K, Wadsworth P. Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Curr Biol. 2005;15:724–731. doi: 10.1016/j.cub.2005.02.055. [DOI] [PubMed] [Google Scholar]
- Nguyen PA, Groen AC, Loose M, Ishihara K, Wuhr M, Field CM, Mitchison TJ. Spatial organization of cytokinesis signaling reconstituted in a cell-free system. Science. 2014;346:244–247. doi: 10.1126/science.1256773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow C, Lombillo VA, Kuriyama R, McIntosh JR. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 1992;359:543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- Noujaim M, Bechstedt S, Wieczorek M, Brouhard GJ. Microtubules accelerate the kinase activity of Aurora-B by a reduction in dimensionality. PLoS One. 2014;9:e86786. doi: 10.1371/journal.pone.0086786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin KE, Wordeman L. Long astral microtubules uncouple mitotic spindles from the cytokinetic furrow. J Cell Biol. 2010;190:35–43. doi: 10.1083/jcb.201004017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosasco-Nitcher SE, Lan W, Khorasanizadeh S, Stukenberg PT. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science. 2008;319:469–472. doi: 10.1126/science.1148980. [DOI] [PubMed] [Google Scholar]
- Stumpff J, Wagenbach M, Franck A, Asbury CL, Wordeman L. Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Dev Cell. 2012;22:1017–1029. doi: 10.1016/j.devcel.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill EG, Norris SR, Guo Y, Ohi R. Kinesin-5 inhibitor resistance is driven by kinesin-12. J Cell Biol. 2016;213:213–227. doi: 10.1083/jcb.201507036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Wilson-Kubalek EM, Arthur CP, Bick MJ, Campbell EA, Darst SA, Milligan RA, Kapoor TM. Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell. 2010;142:433–443. doi: 10.1016/j.cell.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja N, Fenix AM, Rathbun L, Millis BA, Tyska MJ, Hehnly H, Burnette DT. Focal adhesions control cleavage furrow shape and spindle tilt during mitosis. Sci Rep. 2016;6:29846. doi: 10.1038/srep29846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarantino N, Tinevez JY, Crowell EF, Boisson B, Henriques R, Mhlanga M, Agou F, Israel A, Laplantine E. TNF and IL-1 exhibit distinct ubiquitin requirements for inducing NEMO-IKK supramolecular structures. J Cell Biol. 2014;204:231–245. doi: 10.1083/jcb.201307172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2016;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- van der Horst A, Vromans MJ, Bouwman K, van der Waal MS, Hadders MA, Lens SM. Inter-domain cooperation in INCENP promotes Aurora B relocation from centromeres to microtubules. Cell Rep. 2015;12:380–387. doi: 10.1016/j.celrep.2015.06.038. [DOI] [PubMed] [Google Scholar]
- Wheatley SP, Carvalho A, Vagnarelli P, Earnshaw WC. INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr Biol. 2001;11:886–890. doi: 10.1016/s0960-9822(01)00238-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.