Kinesin-5 is required for bipolar spindle assembly; yet in the absence of kinesins-5 and -14, cells can form spindles. In fission yeast, three distinct pathways compensate for their loss. Microtubule polymerase, kinesin-6, and microtubule cross-linker execute individual roles in concert at different mitotic stages in place of the two kinesins.
Abstract
Accurate chromosome segregation relies on the bipolar mitotic spindle. In many eukaryotes, spindle formation is driven by the plus-end–directed motor kinesin-5 that generates outward force to establish spindle bipolarity. Its inhibition leads to the emergence of monopolar spindles with mitotic arrest. Intriguingly, simultaneous inactivation of the minus-end–directed motor kinesin-14 restores spindle bipolarity in many systems. Here we show that in fission yeast, three independent pathways contribute to spindle bipolarity in the absence of kinesin-5/Cut7 and kinesin-14/Pkl1. One is kinesin-6/Klp9 that engages with spindle elongation once short bipolar spindles assemble. Klp9 also ensures the medial positioning of anaphase spindles to prevent unequal chromosome segregation. Another is the Alp7/TACC-Alp14/TOG microtubule polymerase complex. Temperature-sensitive alp7cut7pkl1 mutants are arrested with either monopolar or very short spindles. Forced targeting of Alp14 to the spindle pole body is sufficient to render alp7cut7pkl1 triply deleted cells viable and promote spindle assembly, indicating that Alp14-mediated microtubule polymerization from the nuclear face of the spindle pole body could generate outward force in place of Cut7 during early mitosis. The third pathway involves the Ase1/PRC1 microtubule cross-linker that stabilizes antiparallel microtubules. Our study, therefore, unveils multifaceted interplay among kinesin-dependent and -independent pathways leading to mitotic bipolar spindle assembly.
INTRODUCTION
The microtubule plays a myriad of roles in many biological processes. During mitosis, this biopolymer forms the bipolar spindle that aligns sister chromatids in the middle of the cell before segregating them to opposite poles. Perturbation of this mitotic apparatus can lead to aneuploidy, a major risk factor for cancer development, miscarriage, birth defects, and other human diseases (Wittmann et al., 2001; Godinho and Pellman, 2014). In addition to microtubules, the mitotic spindle employs a dynamic ensemble of various microtubule-associated proteins (MAPs) and motor proteins. Not only do these proteins play a crucial role as structural components in establishing spindle bipolarity, they also regulate the dynamic properties of the spindle microtubule during distinct mitotic stages (Desai and Mitchison, 1997; Jiang and Akhmanova, 2011; Heald and Khodjakov, 2015).
One of the key MAPs regulating spindle bipolarity is the Dis1/XMAP215/TOG family, which possesses microtubule polymerase activity (Brouhard et al., 2008; Al-Bassam and Chang, 2011). Although TOG family proteins are thought to regulate microtubule plus-end dynamics, they localize to the centrosome or spindle pole body (SPB, the fungal equivalent of the centrosome), from which microtubules are nucleated (Charrasse et al., 1998; Matthews et al., 1998; Cullen et al., 1999). In fission yeast, two TOG family proteins, Alp14/Mtc1 and Dis1, have been identified and found to share an essential function in cell division (Garcia et al., 2002). Alp14/TOG forms a complex with Alp7/Mia1/TACC, where Alp7 acts as an SPB-targeting factor for this complex during mitosis (Sato et al., 2004; Sato and Toda, 2007); localization of Alp14 to the mitotic SPB is an initial, essential step for this protein to act as a microtubule polymerase for mitotic spindle assembly (Sato et al., 2004; Tang et al., 2014).
Kinesin motor proteins comprise a large protein family (kinesin-1 to kinesin-14) (Lawrence et al., 2004; Hirokawa et al., 2009) and play diverse roles in microtubule-mediated cellular activities. This family can be classified into three subgroups called N-kinesins, M-kinesins, and C-kinesins, where the motor domain is located in the N-terminal region, in the middle region or in the C-terminal region, respectively. In general, N-kinesins and C-kinesins drive microtubule plus-end– and minus-end–directed motilities respectively, while M-kinesins depolymerize microtubules.
In most eukaryotes, kinesin-5 (budding yeast Cin8 and Kip1, fission yeast Cut7, Aspergillus BimC, Drosophila Klp61F, Xenopus Eg5 and human Kif11) plays an essential role in bipolar spindle formation and successful cell division. Kinesin-5 molecules form homo-tetramers, allowing them to cross-link and subsequently slide antiparallel microtubules apart, thereby generating outward pushing force onto spindle poles (Kashina et al., 1996; Kapitein et al., 2005). In line with this notion, mutation, depletion, or chemical inhibition of kinesin-5 results in the appearance of monopolar spindles with duplicated, yet unseparated, centrosomes/SPBs (Enos and Morris, 1990; Hagan and Yanagida, 1990; Hoyt et al., 1992; Roof et al., 1992; Heck et al., 1993; Mayer et al., 1999). Interestingly, it is thought that outward force generated by kinesin-5 is antagonized by the inward force exerted by minus-end–directed motors such as kinesin-14 (budding yeast Kar3, fission yeast Pkl1 and Klp2, Aspergillus KlpA, Drosophila Ncd, Xenopus XCTK2 and human HSET/KifC1) and dynein (O’Connell et al., 1993; Gaglio et al., 1996; Pidoux et al., 1996; Saunders et al., 1997; Walczak et al., 1998; Mountain et al., 1999; Sharp et al., 1999, 2000; Mitchison et al., 2005; Tao et al., 2006; Tanenbaum et al., 2008; Ferenz et al., 2009; Gatlin et al., 2009; Bieling et al., 2010; Civelekoglu-Scholey et al., 2010; Salemi et al., 2013; Wang et al., 2015; She and Yang, 2017). Consistent with this proposition, simultaneous inactivation of kinesin-5 and kinesin-14 restores spindle formation and cell viability across the diversity of systems.
In fission yeast, Cut7Kin-5 is also essential for cell growth and its temperature-sensitive (ts) lethal mutations lead cells into mitotic arrest with characteristic V-shaped monopolar spindles at the restrictive temperature (Hagan and Yanagida, 1990, 1992). The lethality caused by cut7 ts mutations is suppressed by the deletion of Pkl1Kin-14 (Pidoux et al., 1996; Troxell et al., 2001; Rodriguez et al., 2008). We previously showed that Pkl1Kin-14 forms a ternary complex with Msd1 and Wdr8 (Toya et al., 2007; Ikebe et al., 2011) (referred to as the MWP complex) that anchors the minus ends of spindle microtubules to the mitotic SPB (Yukawa et al., 2015). This anchorage in turn generates antagonistic inward force at the SPB against Cut7Kin-5-mediated outward force (Syrovatkina and Tran, 2015; Yukawa et al., 2015). However, it is not known how cut7pkl1 double mutants can separate spindle poles and whether additional force-generating factors exist to assemble bipolar spindles. Furthermore, functional interplay between microtubule polymerases and force-generating motors during establishment and maintenance of the dynamic bipolar spindles has not been precisely addressed.
We have attempted to elucidate the kinesin-5 and -14 independent spindle assembly pathways in fission yeast. Here we show that the Alp14/TOG-Alp7/TACC microtubule polymerase complex, the Klp9Kin-6 motor, and the microtubule cross-linker Ase1/PRC1 act to promote spindle bipolarity in concert yet independently. We have found that Alp14 is absolutely essential for SPB separation and spindle bipolarity during early mitosis. In contrast, Klp9Kin-6 is required for the elongation and medial positioning of mitotic spindles during late mitosis. We discuss the implication of these findings in terms of general bipolar spindle assembly.
RESULTS
Simultaneous deletion of cut7 and pkl1 is viable but leads to SAC-dependent mitotic delay with shorter preanaphase spindles
In our previous study, we found that double mutant cells containing the ts allele cut7-21 and deletion of msd1, which encodes a subunit of the spindle-anchoring MWP complex containing Pkl1Kin-14, are viable at 36°C. However, cut7-21msd1∆ cells remain with short spindles for a prolonged period but eventually elongate them into full-length anaphase B spindles (Yukawa et al., 2015). It is known that pkl1 or msd1 deletion rescued not only cut7 ts mutants (Pidoux et al., 1996; Yukawa et al., 2015), but also complete deletion of cut7 (Olmsted et al., 2014; Syrovatkina and Tran, 2015) (Supplemental Figure S1A). We noticed that cut7∆pkl1∆ mutants produce dark pink colonies on agar plates containing Phloxine B (Figure 1A), a red dye that stains dead or sick cells (Moreno et al., 1991). To study the impact of cut7∆pkl1∆ on mitotic progression in detail, we observed the dynamic behavior of spindle microtubules and SPBs in wild-type and cut7∆pkl1∆ cells expressing mCherry-Atb2 (a microtubule marker) (Toda et al., 1984) and Cut12-GFP (an SPB marker) (Bridge et al., 1998) (Figure 1, B–F). We found that cut7∆pkl1∆ cells could assemble bipolar spindles (Figure 1, B and C) but stayed in a preanaphase stage for a much longer period of time (21 ± 5 min vs. 11 ± 3 min for wild type; Figure 1D) and that their spindle length at the onset of anaphase B was significantly shorter (1.19 ± 0.51 μm vs. 2.48 ± 0.46 μm for wild type; Figure 1E). Furthermore, spindle elongation rate during anaphase B was considerably slower in cut7∆pkl1∆ cells compared with wild-type cells (0.38 ± 0.11 μm/min vs. 0.53 ± 0.06 μm/min for wild type; Figure 1F). These results are consistent with those shown in a recent publication (Rincon et al., 2017), which appeared during the preparation of this article.
FIGURE 1:
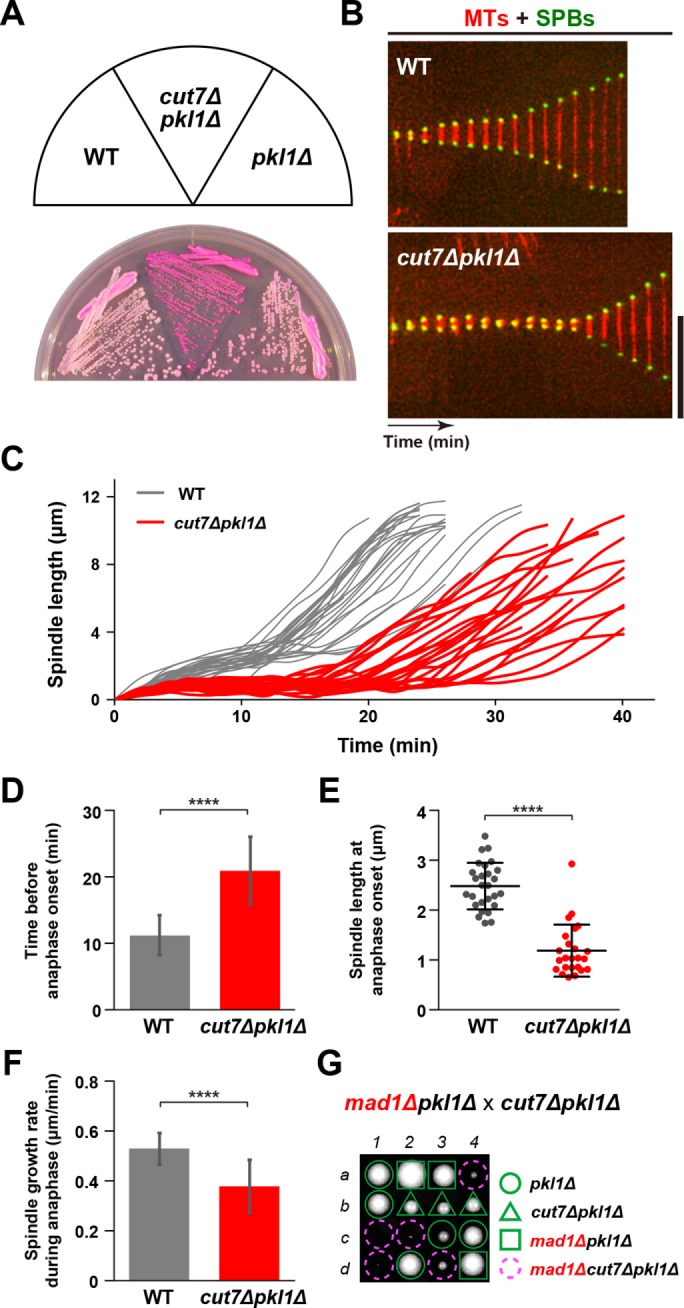
The cut7pkl1 double deletion results in SAC-dependent mitotic delay with a short preanaphase spindle. (A) Indicated strains were streaked on a rich YE5S agar plate containing Phloxine B and incubated at 33°C for 2 d. (B) Kymographic images of mitotic wild-type (top) and cut7∆pkl1∆ cells (bottom) expressing a tubulin marker (mCherry-Atb2, red) and an SPB marker (Cut12-GFP, green). Pictures were taken at 2 min intervals. Scale bar, 10 μm. (C) Profiles of mitotic progression in wild-type (gray line, n = 27) and cut7∆pkl1∆ cells (red line, n = 24). Changes of the inter-SPB distance were plotted against time. (D) The time between the initiation of SPB separation and onset of anaphase B. (E) Spindle length at anaphase onset. (F) Spindle growth rate during anaphase B. Results are given as means ± SD. Data sets were compared with a two-tailed unpaired Student’s t tests (****p < 0.0001). (G) Tetrad analysis on crosses between mad1∆pkl1∆ and cut7∆pkl1∆ strains, Individual spores (a–d) in each ascus (1–4) were dissected on YE5S plates and incubated at 27°C for 3 d. Representative tetrad patterns are shown. Circles, triangles and squares with green lines indicate pkl1∆ single mutants, cut7∆pkl1∆ double mutants and mad1∆pkl1∆ double mutants, respectively. Assuming 2:2 segregation of individual markers allows the identification of mad1∆cut7∆pkl1∆ triple mutants (indicated by dashed magenta circles).
To address the involvement of the spindle assembly checkpoint (SAC) in the mitotic delay observed in cut7∆pkl1∆ cells, we combined the deletion of genes encoding the core SAC components Mad1, Mad2, and Bub1 (Musacchio, 2015) with the cut7∆pkl1∆ mutant. Tetrad analysis showed that authentic SAC signaling is required for optimal growth or even viability of cut7∆pkl1∆ cells (Figure 1G, and Supplemental Figure S1B). These results indicate that cut7∆pkl1∆ cells manage to survive due to a SAC-mediated delay in mitotic progression, thereby providing enough time for the correction of erroneous kinetochore-microtubule attachments.
Fission yeast kinesin-6/Klp9 motor activity is essential for the viability in the absence of Cut7 and Pkl1
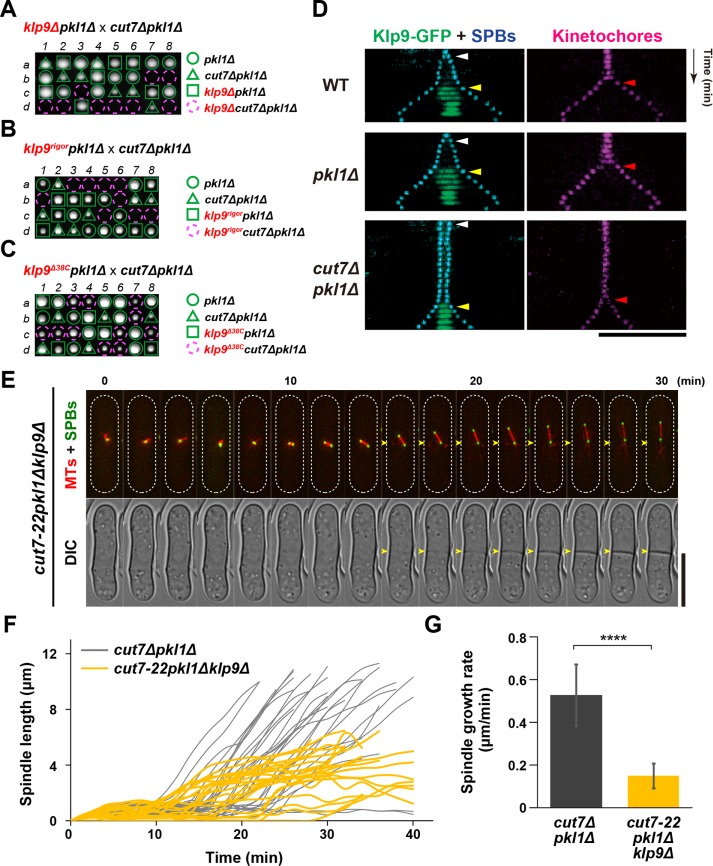
Although not completely normal, cut7∆pkl1∆ mutant cells assemble bipolar spindles and continue to proliferate. How is this possible in the absence of Cut7Kin-5 and Pkl1Kin-14, two major plus- and minus-end–directed motors? One possibility is that other kinesin motors establish and maintain spindle bipolarity in place of Cut7 and Pkl1. To investigate this possibility, we examined synthetic lethality between each deletion of other kinesin-encoding genes and cut7∆pkl1∆. Fission yeast contains seven kinesin motors (Tea2, Klp2, Klp3, Klp5, Klp6, Klp8, and Klp9) besides Cut7Kin-5 and Pkl1Kin-14; none of them is essential for cell viability on their own. Genetic crosses indicated that among them, only one kinesin, Klp9, becomes essential when combined with cut7∆pkl1∆ (Figure 2A and Supplemental Figure S2A). Klp9 belongs to the plus-end–directed kinesin-6 family and accumulates at the spindle midzone at the onset of anaphase B to promote spindle elongation during late mitosis (Fu et al., 2009).
FIGURE 2:
Motor activity of Kinesin-6/Klp9 is essential for the viability in the absence of Cut7 and Pkl1. (A, B, C) Tetrad analysis. Spores were dissected on crosses between klp9∆pkl1∆ and cut7∆pkl1∆ strains (A), between klp9rigorpkl1∆ and cut7∆pkl1∆ strains (B), or between klp9∆38Cpkl1∆ and cut7∆pkl1∆ strains (C) respectively. Manipulation was performed as in Figure 1G. Circles and triangles with green lines indicate pkl1∆ single mutants and cut7∆pkl1∆ double mutants, respectively. Squares with green lines indicate klp9∆pkl1∆ double mutants (A), klp9rigorpkl1∆ double mutants (B), or klp9∆38Cpkl1∆ double mutants (C). Assuming 2:2 segregation of individual markers allows the identification of triple mutants in each cross (indicated by dashed magenta circles). (D) Kymographic images of a wild-type, pkl1∆, or cut7∆pkl1∆ cell expressing Klp9-GFP (green) along with a kinetochore marker (Mis6-2mRFP, red) (Saitoh et al., 1997) and an SPB marker (Pcp1-CFP, blue) (Flory et al., 2002; Fong et al., 2010). Images were taken at 2-min intervals. The timing of SPB separation, anaphase onset, and Klp9-GFP accumulation at the spindle midzone is indicated with white, magenta, and yellow arrowheads, respectively. Scale bar, 10 μm. (E) Time-lapse images of a mitotic cut7-22pkl1∆klp9∆ cell. Spindle microtubules (mCherry-Atb2; red) and SPB (Cut12-GFP; green) were visualized (top). Images were taken at 2-min intervals on shifting the temperature from 27°C to 36°C for 2 h. The septation sites are indicated with yellow arrowheads in the DIC images (bottom). The cell peripheries are outlined with dotted lines (top). Scale bar, 10 μm. (F, G) Profiles of mitotic progression in cut7∆pkl1∆ (gray, n = 30) and cut7-22pkl1∆klp9∆ cells (yellow, n = 21) under the same condition as in E. Changes of the inter-SPB distance were plotted against time (F). Spindle growth rate after the inter-SPB distance reached 1.5 μm was calculated (G). Data are given as means ± SD; ****p < 0.0001 (two-tailed unpaired Student’s t test).
Recently, it was reported that Klp9 plays two key roles (Meadows et al., 2017); one involves the N-terminal motor region, thereby accelerating anaphase B spindle elongation as previously shown (Fu et al., 2009), while the other functions to control proper timing of anaphase onset and chromosome segregation (Choi and McCollum, 2012). Interestingly, the latter function is executed by the C-terminal nonmotor region independently of motor activity (Meadows et al., 2017). Given the complexity of Klp9 functions, we next addressed which activity within Klp9 makes a contribution to the viability of cut7∆pkl1∆ cells. To this end, we implemented two different types of Klp9 mutants; one is Klp9-rigor (G296A, a substitution within its motor domain), which is defective in plus-end motility and the other is Klp9-∆38C (lacking the C-terminal 38 amino acids), which abolishes the temporal role in anaphase onset (Meadows et al., 2017). Tetrad analysis clearly indicated that Klp9-rigor, but not Klp9-∆38C, is lethal in the absence of Cut7 and Pkl1 (Figure 2, B and C). This demonstrates that the plus-end motility of Klp9Kin-6 renders cut7∆pkl1∆ cells viable.
Klp9-dependent spindle elongation and positioning during anaphase B are indispensable for proper chromosome segregation in the absence of Cut7 and Pkl1
Next, we observed the timing of Klp9 accumulation on spindle microtubules in cut7∆pkl1∆ cells. As aforementioned, in wild-type cells, Klp9 is always localized to the spindle midzone only after sister chromatids separate and reach each SPB, which coincides with anaphase B onset (Fu et al., 2009; Meadows et al., 2017) (Figure 2D, top row). This was also the case in the pkl1∆ single mutant (middle row). Interestingly, the timing of Klp9 localization did not alter even in cut7∆pkl1∆ double mutant cells (bottom row). These results suggest that Klp9 localization to the spindle midzone during anaphase B is essential for cell viability in the absence of Cut7 and Pkl1; Klp9 motor does not function prior to anaphase B in cut7∆pkl1∆ as in wild-type cells.
To explore the defective phenotypes of klp9cut7pkl1 triple mutants, we used the cut7-22 ts mutant (Hagan and Yanagida, 1990, 1992). Deletion of klp9 enhanced temperature sensitivity of the cut7-22 single mutant and conferred the ts phenotype to the cut7-22pkl1∆ double mutants (Supplemental Figure S2B). To characterize the terminal phenotype of cut7-22pkl1∆klp9∆ cells, we observed spindle microtubule behavior by time-lapse live imaging at 36°C. Intriguingly, triple mutant cells appeared to assemble bipolar spindles (Figure 2, E and F), albeit shorter in length. However, spindle elongation rate during anaphase B was substantially decreased compared with the cut7∆pkl1∆ cells (0.15 ± 0.06 μm/min vs. 0.53 ± 0.14 μm/min) (Figure 2G). Notably, these anaphase B spindles did not reach cell tips (Hagan and Petersen, 2000) and instead were displaced from the cell middle (34%, n = 80, see frames after 16 min time point in Figure 2E). Moreover, the septum started to form during spindle displacement (shown by arrowheads in Figure 2E). This asymmetric positioning of shorter spindles and septation might account for the terminal chromosome “cut” phenotype in cut7∆pkl1∆ cells, in which klp9 transcription was shut off as recently described (cut7∆pkl1∆klp9Off) (Rincon et al., 2017). Taken together, these results indicate that Klp9 plays an essential role in the absence of Cut7 and Pkl1 and that its motor activity is required for spindle elongation and proper positioning during anaphase B, thereby averting the emergence of lethal chromosome segregation errors.
Temperature-sensitive klp9 mutants also lead to defective anaphase and chromosome missegregation in the absence of Cut7 and Pkl1
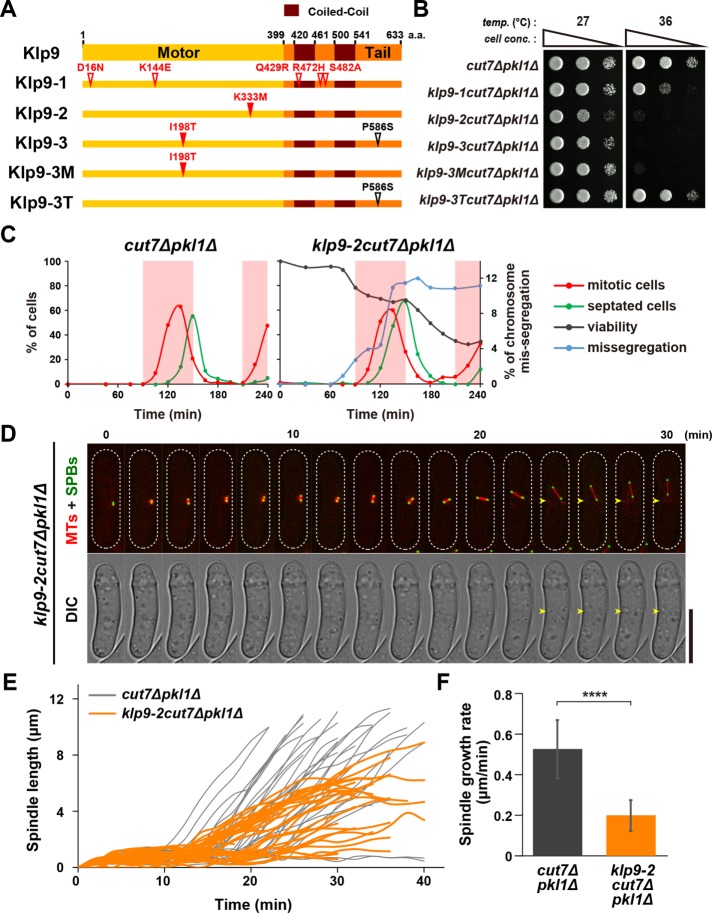
To recapitulate the terminal phenotypes of the cut7-22pkl1∆klp9∆ triple mutants and confirm the role of Klp9 in proper spindle formation, we isolated three independent klp9 mutant alleles (klp9-1, -2, -3) by PCR-based error-prone mutagenesis (see Materials and Methods). These mutants displayed ts growth in the cut7∆pkl1∆ deletion background (Figure 3, A and B); however, none of these alleles was ts in the wild-type background (Supplemental Figure S3A). While klp9-2 contained a single point mutation within the motor domain (K333M, Figure 3A), klp9-3 contained two mutations in the motor domain and the C-terminal tail region (I198T and P586S, respectively). Subsequently, we constructed klp9-3M and klp9-3T, in which each contained one of the two mutations (klp9-3M, I198T and klp9-3T, P586S, Figure 3A). The klp9-2 and klp9-3M mutants showed more severe temperature sensitivity than klp9-1 (Figure 3B). Hereafter, we sought to characterize the klp9-2 allele in more detail.
FIGURE 3:
The point mutations within the Klp9 motor domain interfere with elongation and positioning of anaphase B spindle in cut7∆pkl1∆. (A) Mutation sites in the klp9 mutants. Klp9-3 contained two separate mutations. Then two strains containing one of them were created (Klp9-3M and Klp9-3T). (B) Spot test. Indicated strains were serially (10-fold) diluted, spotted onto rich YE5S plates, and incubated at 27°C or 36°C for 2 d. cell conc., cell concentration; temp., temperature. (C) Synchronous culture analysis with centrifugal elutriation. Small G2 cells of cut7∆pkl1∆ or klp9-2cut7∆pkl1∆ grown at 27°C were collected with centrifugal elutriation and shifted to 36°C at time 0. Spindle microtubules (mCherry-Atb2) and SPB (Cut12-GFP) were visualized to analyze mitotic progression. The percentages of mitotic cells (red line) and cells with the septum (green line) were counted to monitor cell cycle progression. Relative colony-forming units at the indicated time points were assessed as cell viability (=100% at time 0; gray line). Aliquots of cell cultures at each time point were stained with Hoechst33342 to detect chromosome segregation defects (light blue line). Light red columns mark periods in mitosis. (D) Time-lapse images of mitotic klp9-2cut7∆pkl1∆ cells. Spindle microtubules (mCherry-Atb2; red) and SPB (Cut12-GFP; green) were visualized. Images were taken at 2 min intervals after incubation of cultures at 36°C for 6 h. The septation sites are indicated with yellow arrowheads. The cell peripheries are outlined with dotted lines (top). Scale bar, 10 μm. (E, F) Profiles of mitotic progression in klp9-2cut7∆pkl1∆ (orange, n = 34) cells under the same condition as in D. Changes of the inter-SPB distance are plotted against time (E). Spindle growth rate after the inter-SPB distance reached 1.5 μm was calculated (F). Data for cut7∆pkl1∆ is shown for comparison; this is the same as that presented in Figure 2, F and G. Data are given as means ± SD; ****p < 0.0001 (two-tailed unpaired Student’s t test).
To define the cell-cycle stage for which Klp9 function is essential, we performed synchronous culture analysis with centrifugal elutriation using the klp9-2cut7∆pkl1∆ mutants. Small early-G2 cells were harvested by elutriation at 27°C, shifted up to 36°C, and cell viability was monitored as cells proceeded through the cell cycle. Triple mutant cells exhibited a decline in cell viability at two time points, 90 and 165 min (black line in the right panel of Figure 3C). This viability drop paralleled the emergence of the chromosomal missegregation phenotype (blue line in the right panel of Figure 3C, representative images of mitotic cells with missegregating chromosomes are shown in Supplemental Figure S3B). Moreover, time-lapse live imaging analysis showed that klp9-2cut7∆pkl1∆ triple mutants formed shorter bipolar spindles and displayed much slower spindle elongation during anaphase B (0.20 ± 0.08 μm/min, Figure 3, D–F). It is also noteworthy that the spindle apparatus became displaced on entry into anaphase B (24 min and afterward in Figure 3D). Overall, these defective phenotypes are very similar, if not identical, to those observed in cut7-22pkl1∆klp9∆ cells described earlier (Figure 2, E–G). Taking these results together, we conclude that Klp9Kin-6 and Cut7Kin-5 coordinate to promote anaphase B spindle elongation and are also required for proper positioning of the spindle apparatus, thereby escaping from the lethal cut phenotype imposed by the cytokinetic machinery.
Several nonmotor microtubule-associated proteins are required for cell viability of cut7∆pkl1∆ mutants
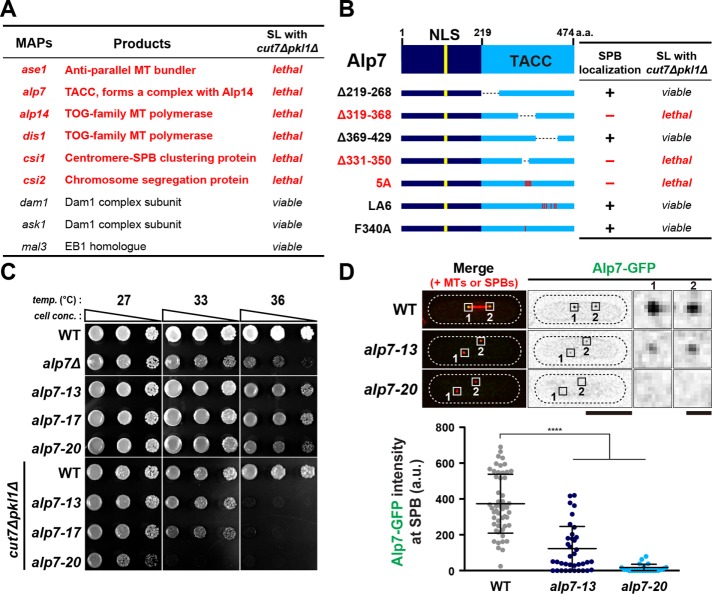
As Klp9 appears to execute its essential function in the absence of Cut7 and Pkl1 only during anaphase B, another question arises: how can preanaphase bipolar spindles, albeit shorter in length, assemble in the absence of these kinesin motors? Very recent work (Rincon et al., 2017) ascribed this role to the microtubule cross-linking/bundling factor Ase1/PRC1 (Loiodice et al., 2005; Yamashita et al., 2005; Janson et al., 2007) and the microtubule stabilizer Cls1/Peg1/CLASP (Grallert et al., 2006; Bratman and Chang, 2007). However, as these two MAPs are not required for either microtubule nucleation or polymerization per se (Janson et al., 2007; Al-Bassam et al., 2010; Al-Bassam and Chang, 2011), we presumed that some other factors/MAPs must be functional in early spindle assembly, in particular at the SPB, the microtubule-organizing center (MTOC). To identify such factors, we systematically examined synthetic lethality between each deletion of a cohort of MAPs and cut7∆pkl1∆. Random spore analysis and tetrad dissection uncovered that at least five MAPs (Alp7/Mia1, Alp14/Mtc1, Dis1, Csi1, Csi2) (Nabeshima et al., 1995; Garcia et al., 2001; Nakaseko et al., 2001; Oliferenko and Balasubramanian, 2002; Sato et al., 2004; Hou et al., 2012; Costa et al., 2014), in addition to Ase1, were required for the viability of cut7∆pkl1∆ cells (Figure 4A and Supplemental Figure S4). Notably, Alp14 and Dis1 belong to the Dis1/XMAP215/TOG family of microtubule polymerases (Al-Bassam and Chang, 2011; Al-Bassam et al., 2012; Hussmann et al., 2016; Matsuo et al., 2016).
FIGURE 4:
SPB localization of Alp7/TACC is essential for proper spindle assembly in the cut7∆pkl1∆ mutant. (A) List of MAPs tested for synthetic lethality (SL) when combined with cut7∆pkl1∆. Information of each protein function and synthetic lethality is given. (B) Truncations or point mutations of Alp7 tested for synthetic lethality with cut7∆pkl1∆. Information of SPB localization and synthetic lethality is given. See Supplemental Figure S5A for more details. (C) Spot test. Indicated strains were serially (10-fold) diluted, spotted onto rich YE5S plates and incubated at 27°C, 33°C, or 36°C for 2 d. cell conc., cell concentration; temp., temperature. (D) Intensity of Alp7-GFP localized to the mitotic SPB in the alp7 mutants. Representative images showing mitotic localization of Alp7-GFP in the indicated strains are presented at the top. Insets 1 and 2 show the magnified regions of the SPB. Cells of wild type (carrying Alp7-GFP and mCherry-Atb2) and alp7-13 or alp7-20 (carrying Alp7-GFP and Pcp1-2mRFP) were mixed in the same culture, shifted from 27°C to 36°C, and incubated for 2 h (see Supplemental Figure S5C). Quantification of signal intensities of Alp7-GFP at the SPB is shown at the bottom. All p values were obtained from the two-tailed unpaired Student’s t test. Data are presented as the means ± SD (≥20 cells). ****p < 0.0001. The cell peripheries are outlined with dotted lines. Scale bars, 10 μm (main images) and 1 μm (enlarged images of the boxed regions on the right). a.u., arbitrary unit.
Alp7/TACC is of our particular interest, as this protein forms a stable complex with Alp14/TOG (Sato et al., 2004; Al-Bassam et al., 2012; Hussmann et al., 2016) and recruits Alp14 to the mitotic SPB, thereby ensuring spindle assembly/elongation from this MTOC (Ling et al., 2009; Sato et al., 2009; Tang et al., 2014). Given these characteristics, we pursued the molecular details by which Alp7 plays an essential role in cut7∆pkl1∆ cells.
Localization of Alp7/TACC to the mitotic SPB is a prerequisite for proper spindle assembly in the cut7∆pkl1∆ mutants
Alp7/TACC plays multiple roles in cell cycle-dependent microtubule organization that are executed through interactions with several other proteins in a spatiotemporal manner (Sato et al., 2004; Tang et al., 2013, 2014; Tang and Toda, 2015; Okada et al., 2014; Zheng et al., 2014). A number of previous studies mapped functional domains within Alp7 that are required for each of its distinct roles, and hence several Alp7 mutants with defects in specific functions are available. To address which function and localization of Alp7 are important, we crossed individual alp7 mutants with a cut7∆pkl1∆ strain. This genetic analysis unequivocally showed that the regions/residues within Alp7 that are responsible for localization to the mitotic SPB are crucial for the viability of cut7∆pkl1∆ cells (Figure 4B and Supplemental Figure S5A).
To create alp7 ts mutants in the cut7∆pkl1∆ background, we implemented Alp7-5A, which contains alanine substitutions at five amino acid residues (K341, Y344, R346, K347, and Y348) (Tang et al., 2014), which is lethal in the cut7∆pkl1∆ background (Figure 4B). We introduced random mutations into these five residues by PCR-based, site-directed mutagenesis (Masuda et al., 2013) and screened for ts mutants in a cut7∆pkl1∆ strain (Materials and Methods). Three ts alleles (alp7-13, -17, and -20) were isolated, and nucleotide sequencing indicated that all three isolates indeed contained missense mutations within the five residues (alp7-13: R346I, K347R, Y348L; alp7-17: Y344T; alp7-20: Y344R) (Supplemental Figure S5B). As expected, these mutants displayed temperature sensitivity only when combined with cut7∆pkl1∆ mutations (Figure 4C). Alp7-5A is specifically defective in SPB localization during mitosis (Tang et al., 2014) and, consistent with this, Alp7-GFP intensities at the mitotic SPBs in the alp7-13 and alp7-20 mutants at the restrictive temperature showed a substantial reduction of fluorescence signals by 70 and 96%, respectively (Figure 4D and Supplemental Figure S5C). We performed immunoblotting to examine the total protein content of Alp7-GFP in these mutants and found that the levels were indistinguishable from that in wild-type cells (Supplemental Figure S5D). This indicates that Alp7-13 and Alp7-20 mutant proteins are specifically defective in localizing to the mitotic SPBs. Therefore, Alp7 localization to the mitotic SPB is essential for the viability of cut7∆pkl1∆ cells.
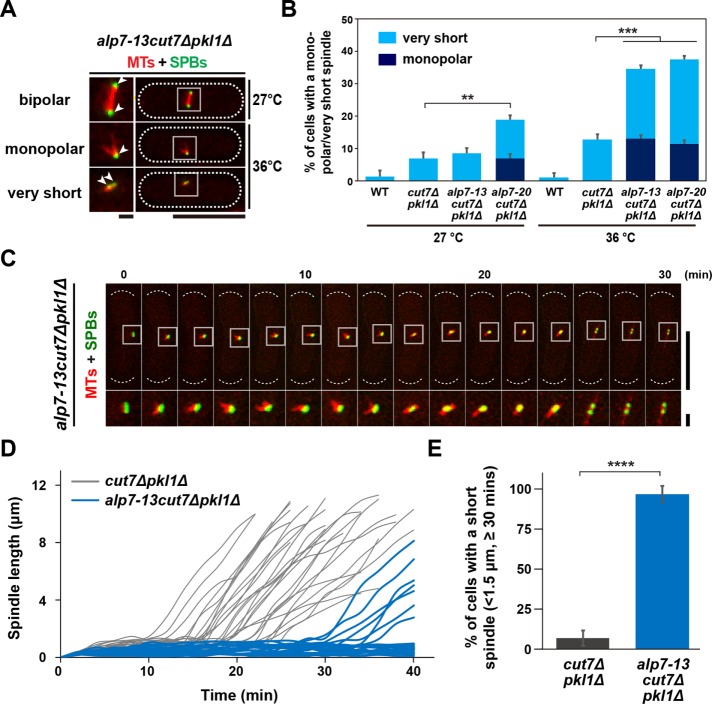
To examine the impact of these alp7 mutations on spindle assembly in the cut7∆pkl1∆ background, we observed the terminal phenotype of alp7-13cut7∆pkl1∆ or alp7-20cut7∆pkl1∆ cells expressing mCherry-Atb2 and Cut12-GFP. Notably, in both triple mutants, the frequency of cells containing monopolar or very short spindles (<0.5 μm) was dramatically increased on incubation at 36°C for 2 h compared with that of the cut7∆pkl1∆ double mutant (Figure 5, A and B). The alp7-20cut7∆pkl1∆ triple mutants showed the monopolar spindle phenotype to some extent even at 27°C (6.9% vs. <1.0% for cut7∆pkl1∆; Figure 5B). To capture mitotic progression in a single cell, we performed time-lapse live imaging. The triple mutant cells spent much longer preanaphase with monopolar or very short spindles (Figure 5C). A majority of the mutant cells (95%, n = 26) maintained a short spindle (<1.5 μm) at least for 30 min, and >70% cells (19 of 26) remained in this stage for more than 40 min (Figure 5, D and E). This result indicates that cells have the capability to assemble short bipolar spindles in the absence of Cut7Kin-5 and Pkl1Kin-14 through the SPB-localizing activity of Alp7.
FIGURE 5:
Alp7 is required for bipolar spindle assembly during early mitosis of the cut7∆pkl1∆ cells. (A) Representative spindle morphologies of the alp7-13cut7∆pkl1∆ triple mutants. Cells containing a bipolar (top), a monopolar (middle), or a very short spindle (the inter-SPB distance is <0.5 μm, bottom) are shown (Cut12-GFP, green, and mCherry-Atb2, red). Enlarged images of spindle areas (squares) are shown on the left panels. Positions of the SPB are indicated with arrowheads. The cell peripheries are outlined with dotted lines. Scale bars, 10 μm (main images) and 1 μm (enlarged images of the boxed regions). (B) The percentage of cells containing a monopolar spindle (dark blue) or a very short spindle (light blue). Each strain was grown at 27°C and shifted to 36°C for 2 h. In each experiment, more than 25 mitotic cells were observed. (C) Time-lapse images of a mitotic alp7-13cut7∆pkl1∆ cell. Spindle microtubules (mCherry-Atb2; red) and SPBs (Cut12-GFP; green) were visualized. Images were taken at 2-min intervals after 2 h incubation at 36°C. The positions of cell ends are indicated with dotted curved lines (top), and enlarged images of the boxed regions are presented at the bottom. Scale bars, 10 μm (top) and 1 μm (bottom). (D, E) Profiles of mitotic progression in alp7-13cut7∆pkl1∆ (light blue, n = 26) cells under the same condition as in C. Changes in the inter-SPB distance were plotted against time (D). The percentage of cells retaining a short spindle (the inter-SPB distance is <1.5 μm) for >30 min after SPB separation was counted (E). Data for cut7∆pkl1∆ are shown for comparison; this is the same as that presented in Figure 2, F and G. Data are given as means ± SD; **p < 0.01; ***p < 0.001; ****p < 0.0001 (two-tailed unpaired Student’s t test).
Forced tethering of Alp14/TOG to the mitotic SPB is sufficient for bipolar spindle assembly in alp7cut7pkl1 triple mutants
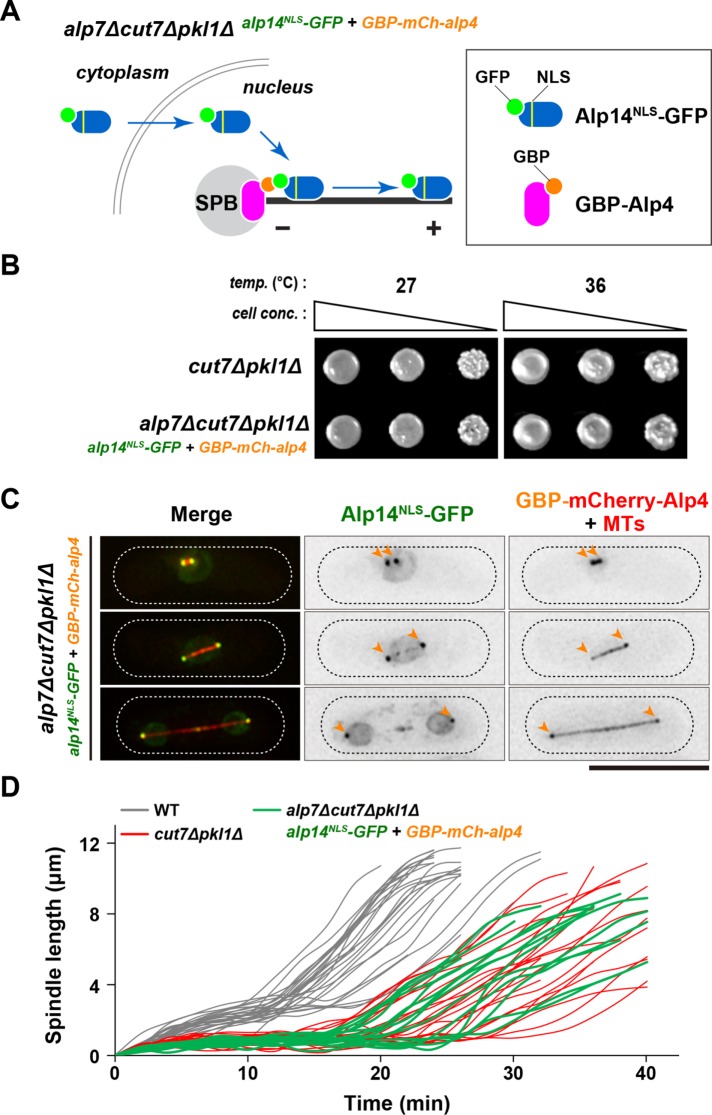
Alp7/TACC forms a complex with Alp14/TOG in the cytoplasm and facilitates the import of this complex into the nucleus through its own nuclear localization signal (NLS) (Ling et al., 2009; Sato et al., 2009; Okada et al., 2014; Okada and Sato, 2015). Alp14/TOG acts as a microtubule polymerase for mitotic spindles only when it is targeted to the SPB through Alp7 (Sato et al., 2004; Sato and Toda, 2007). Hence, we posited that if Alp14 were the sole requirement for Alp7-dependent spindle assembly, an artificial tethering of Alp14 to the mitotic SPBs might be sufficient to complement Alp7 delocalization from the SPB, thereby rescuing the lethality of alp7cut7pkl triple deletion cells. To examine this possibility, we adopted the GFP entrapment strategy based on the implementation of GFP-binding protein (GBP) (Rothbauer et al., 2008). We added a canonical NLS sequence (Kalderon et al., 1984) and GFP to the C terminus of Alp14 (Alp14NLS-GFP) so it can enter the nucleus without Alp7 (Figure 6A). When combined with GBP-tagged Alp4, which is a constitutive component of the γ-tubulin complex (Vardy and Toda, 2000), Alp14NLS-GFP is tethered to SPB in the absence of Alp7 (Figure 6A and Supplemental Figure S6A). Remarkably, cells containing SPB-tethered Alp14NLS-GFP were viable in the alp7∆cut7∆pkl1∆ background (Figure 6B), and, indeed, Alp14NLS-GFP signal was observed at the mitotic SPBs in this strain (Figure 6C). These results indicate that SPB-localizing Alp14 is capable of bypassing the requirement of Alp7 in cut7∆pkl1∆ cells. It should be noted that enforced tethering of Alp14 to the SPBs also suppressed the temperature sensitivity and microtubule poison thiabendazole (TBZ) sensitivity of an alp7 deletion strain, suggesting that targeting Alp14 to the mitotic SPBs is a major, if not the sole, function of Alp7 (Supplemental Figure S6B).
FIGURE 6:
Tethering of Alp14/TOG to mitotic SPB in the alp7∆cut7∆pkl1∆ cells enables Cut7-independent bipolar spindle formation. (A) Schematic illustration of a strategy for the forced tethering of Alp14 to the SPB in the absence of Alp7. In wild-type cells, Alp7 forms a complex with Alp14 in the cytoplasm and imports Alp14 into the nucleus and recruits it to the mitotic SPB. In the absence of Alp7, Alp14 cannot enter the nucleus nor does it localize to the SPB. However, even without Alp7, GFP- and NLS-tagged Alp14 (Alp14NLS-GFP) could enter the nucleus, where it would be tethered to the SPB through the interaction with GBP-tagged Alp4, a core component of the γ-tubulin complex (Vardy and Toda, 2000). (B) Rescue of otherwise lethal alp7∆cut7∆pkl1∆ triple mutants by forced tethering of Alp14 to the SPB. Serial dilution spot tests were performed using the indicated strains on rich YE5S media, and the cells were incubated at 27°C or 36°C for 2 d. cell conc., cell concentration; temp., temperature. (C) Visualization of Alp14NLS-GFP localization. Representative images of mitotic alp7∆cut7∆pkl1∆ cells containing Alp14NLS-GFP (green), GBP-mCherry-Alp4 (red) and mCherry-Atb2 (red) are shown. The positions of SPBs are indicated with orange arrowheads. The cell peripheries are outlined with dotted lines. Some Alp14NLS-GFP signals are detected in the nucleus and along the microtubules in addition to SPBs. Scale bar, 10 μm. (D) Profiles of mitotic progression in alp7∆cut7∆pkl1∆ cells containing SPB-tethered Alp14NLS-GFP. Changes of the inter-SPB distance are plotted against time (green lines; n = 14). Data for wild-type (thin gray lines) and cut7∆pkl1∆ (thin red lines) are the same as those shown in Figure 1C. See Supplemental Figure S7, A–C, for quantitative data.
Having obtained viable cells that contain SPB-tethered Alp14NLS-GFP in the alp7∆cut7∆pkl1∆ background, we next followed the profiles of mitotic progression and spindle assembly in this strain. It was found that these cells could assemble short bipolar spindles with delayed anaphase onset, which were very similar, if not identical, to those of cut7∆pkl1∆ cells (Figure 6D and Supplemental Figure S7, A–C). Collectively, our data indicate that in fission yeast, at least two distinct pathways (see below), one kinesin-6 mediated and the other microtubule polymerase dependent, ensure spindle assembly at different stages of mitosis in the absence of Cut7Kin-5 and Pkl1Kin-14.
DISCUSSION
In this study, we have explored the pathways that lead to bipolar spindle assembly in the absence of Cut7Kin-5 and Pkl1Kin-14 in fission yeast. Previous studies in many organisms have shown that the critical task performed by kinesin-5 in bipolar spindle assembly becomes dispensable by the depletion or inhibition of kinesin-14 (Saunders and Hoyt, 1992; O’Connell et al., 1993; Pidoux et al., 1996; Mountain et al., 1999; Sharp et al., 1999; Tanenbaum and Medema, 2010; Salemi et al., 2013). Here we have unearthed several factors that promote bipolar spindle assembly in cut7∆pkl1∆ cells. Our results show that otherwise nonessential Klp9Kin-6 and various MAPs including the Alp14/TOG microtubule polymerase are absolutely required to render these double mutant cells viable.
Roles of kinesin-6 in mitotic spindle formation and integrity
Kinesin-6 family members (fission yeast Klp9, Caenorhabditis elegans ZEN-4, Drosophila Pavarotti and Subito, and human MKLP1/CHO1/Kif23, MKLP2/Kif20A/Rab6-KIFL, and MPP1/Kif20B) are localized to the spindle midzone at anaphase (Nislow et al., 1992; Adams et al., 1998; Raich et al., 1998; Hill et al., 2000; Abaza et al., 2003; Cesario et al., 2006; Fu et al., 2009). In general, loss of this kinesin results in a disorganized midzone at anaphase and subsequent cytokinesis defects. Some of the kinesin-6 members (e.g., human MKLP1) act to directly bundle antiparallel microtubules and slide them apart, while others (e.g., human MKLP2) do so indirectly through loading the chromosome passenger complex (CPC) containing Aurora B and other regulators to the spindle midzone. As shown previously (Fu et al., 2009; Meadows et al., 2017; Rincon et al., 2017) and in this study (Figure 7), fission yeast Klp9Kin-6 plays both roles in late mitosis, that is, spindle elongation and CPC recruitment. Klp9Kin-6 may, therefore, retain features of an ancestral dual-function motor that are executed by different kinesin-6 members in human cells. It would be of great interest to test in human cells whether the depletion or inhibition of kinesin-6s leads to the lethality by simultaneous inactivation of kinesin-5 and kinesin-14 (and/or dynein).
FIGURE 7:
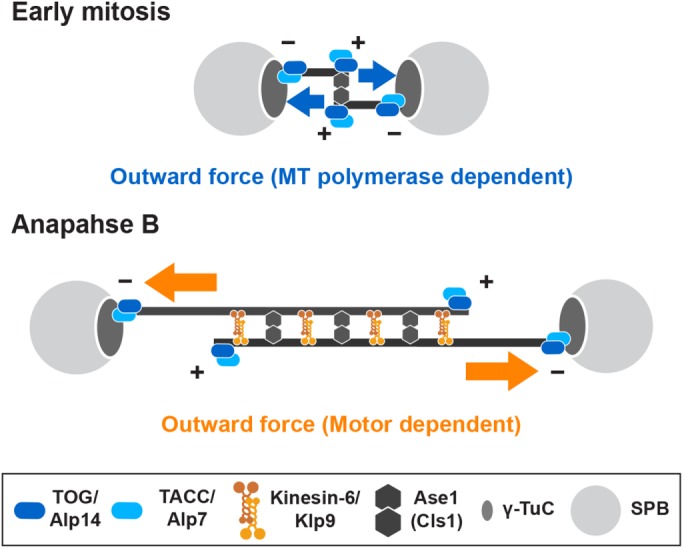
Model of bipolar spindle formation in the absence of kinesin-5/Cut7 and kinesin-14/Pkl1. In the absence of Cut7Kin-5 and Pkl1Kin-14, three distinct pathways compensate for defective spindle assembly and lethality in fission yeast. In early mitosis (top), the Alp7/TACC-Alp14/TOG microtubule polymerase complex is localized to the SPB, by which Alp14 elongates spindle microtubules. Alp14 may also promote microtubule nucleation reaction from the SPB (Reber et al., 2013; Roostalu et al., 2015; Hussmann et al., 2016). Interaction of growing microtubule plus ends with the other SPB would generate outward force in place of Cut7Kin-5. This force is sufficient to separate duplicated SPBs, thereby promoting short bipolar spindle formation in the absence of Pkl1/kinesin-14-dependent inward force. On onset of anaphase B (bottom), Klp9Kin-6 is localized to the overlapping zones within short spindles and increases spindle length by sliding them apart toward both cell ends. For simplicity, the requirement for this kinesin-6 for CPC recruitment to the spindle midzone is not depicted in this figure. Klp9Kin-6 is also important for the medial positioning of elongating spindles. Ase1/PRC1 is localized to the spindle microtubules and cross-links antiparallel microtubules to ensure the establishment of bipolarity. Cls1/CLASP may function in stabilization of bipolar spindles in cooperation with Ase1 (Bratman and Chang, 2007; Rincon et al., 2017).
Drosophila Subito was the first kinesin-6 member to be reported with a function in spindle assembly during the early stages of mitosis; Subito mutants improperly assemble metaphase microtubules, resulting in SAC activation and the emergence of lagging chromosomes (Cesario et al., 2006). As the midzone localization behavior of Klp9Kin-6 on anaphase onset is not altered in cut7∆pkl1∆ cells, we envisage that it is unlikely that Klp9Kin-6 takes part in spindle formation during early mitosis, though we cannot formally exclude this possibility at the moment. Yet, it is noteworthy that in either cut7-22pkl1∆klp9∆ or klp9-2cut7∆pkl1∆ mutant cells, spindles are frequently displaced away from the cell equator, resulting in chromosome missegregation including the cut phenotype on septation. As previously reported (Syrovatkina and Tran, 2015), it is possible that long protruding microtubules induced by the loss of Pkl1 contact one cell tip, which subsequently pushes back the nucleus toward the other end of the cell, resulting in nuclear displacement and the cut phenotype. However, no obvious microtubule protrusions were detected in the triple mutants. Klp9Kin-6 (and Cut7Kin-5) may, therefore, play a role in centering the spindle microtubule in late mitosis to prevent chromosome missegregation. An alternative explanation is that the constricting cytokinesis ring/septum pushes the nuclear envelope/spindle to one side of the cell, which is not Klp9-dependent. Future research should elucidate how bipolar spindles are positioned at the cell equator during anaphase B.
Similarities and differences of kinesin functions in mitotic spindle formation between fission yeast and humans
In human cells, it has been shown that kinesin-12/Kif15/Hklp2 functions redundantly with Kif11/Eg5Kin-5 to promote spindle bipolarity (Tanenbaum et al., 2009; Vanneste et al., 2009). Although Kif15Kin-12 is not required for bipolar spindle assembly under normal conditions, overproduction of Kif15Kin-12 can drive bipolar spindle assembly even when Eg5Kin-5 is fully inhibited, indicating that Kif15Kin-12 has the potential to take over all essential functions of Eg5Kin-5. As Klp9Kin-6 on its own is not capable of replacing the essentiality for Cut7Kin-5 under any conditions so far tested (Rincon et al., 2017), functions of fission yeast Klp9Kin-6 and human Kif15Kin-12 are likely to be different. Intriguingly, however, the simultaneous inhibition of three motors, Eg5Kin-5, Kif15Kin-12, and dynein, does prevent bipolar spindle formation (van Heesbeen et al., 2014). This situation echoes the lethality of cut7pkl1klp9 triple mutants and illuminates the conservation, diversification, and adaptability through evolution in the molecular pathways leading to kinesin-dependent bipolar spindle formation.
Strict dependence of initial spindle assembly on microtubule polymerization in the absence of kinesin-5/Cut7 and kinesin-14/Pkl1
Given the involvement of the Klp9Kin-6 motor in late mitosis and the dispensability of all other kinesin molecules, we wondered as to how cut7-22pkl1∆klp9∆ and klp9-2cut7∆pkl1∆ (and a klp9 shut-off strain recently reported [ Rincon et al., 2017]) could assemble bipolar spindles although shorter in length. Our subsequent analysis has uncovered the requirement of six MAPs in cut7∆pkl1∆ cells. Alp14 and Dis1 are microtubule polymerases, belonging to the Dis1/XMAP215/TOG family (Al-Bassam and Chang, 2011; Al-Bassam et al., 2012; Hussmann et al., 2016; Matsuo et al., 2016). We show that Alp7-mediated recruitment of Alp14 to the mitotic SPB is a critical step for this function, as forced targeting of Alp14 to this MTOC bypasses the necessity of Alp7/TACC, a loading factor for this polymerase. Dis1 primarily functions at the microtubule plus ends by physically interacting with the plus-end tracking protein Mal3/EB1 and the outer kinetochore component Ndc80 (Hsu and Toda, 2011; Matsuo et al., 2016). We previously showed that Dis1 becomes essential in the absence of either Alp7 or Alp14 (Garcia et al., 2002), implying that in cut7∆pkl1∆ cells, more microtubule polymerase activities provided by either Alp14 or Dis1 are needed than in wild-type cells for bipolar spindle assembly.
We surmise that the polymerizing microtubule plus ends push the opposite SPB, by which they provide outward force during early mitosis and that this force is sufficient to separate SPBs and promote assembly of short bipolar spindles (Figure 7). We propose that this alternative pushing force generated by Alp14-mediated microtubule polymerization could compensate for the loss of Cut7Kin-5 if antagonizing Pkl1Kin-14 is absent.
Other MAPs essential for cell viability in the absence of kinesin-5/Cut7 and kinesin-14/Pkl1
Other MAPs identified in this study include Ase1. The role of this microtubule-bundling factor was recently reported (Rincon et al., 2017), and this MAP is likely to function throughout mitosis at the antiparallel microtubule overlapping zones as a cross-linker in concert with Cls1/CLASP (Loiodice et al., 2005; Yamashita et al., 2005; Bratman and Chang, 2007; Meadows and Millar, 2008; Syrovatkina et al., 2013). We consider that during early mitosis Ase1 (and Cls1) functions in concert with Alp7 and Alp14 in SPB separation and spindle elongation and, during later stages, acts together with Klp9Kin-6 to elongate and stabilize anaphase B spindles (Figure 7).
Besides Ase1, two other proteins Csi1 and Csi2 have been identified. Previous work showed that Csi1 is important for Alp7 localization to the mitotic SPB (Zheng et al., 2014) and that this localization of Csi1 in turn requires Csi2 (Costa et al., 2014). Hence we envisage that the requirement of Csi1 and Csi2 in cut7∆pkl1∆ cells is through Alp7 localized to the SPB.
Concluding remarks
In total, our study has uncovered interplay among individual kinesins and between kinesin-dependent and -independent pathways that are required for mitotic bipolar spindle assembly. These include three mitotic kinesins, one essential (Cut7Kin-5) and two nonessential motors (Klp9Kin-6 and Pkl1Kin-14), and six nonmotor MAPs. These MAPs are further classified into two functional groups. One includes microtubule polymerases and their regulators that promote microtubule elongation from the SPB/MTOC, while the other is a microtubule-bundling factor that stabilizes antiparallel microtubules (Rincon et al., 2017). Kinesin-5 is deemed to be a promising target of cancer chemotherapeutics; however, drug-resistant cell lines often appear, which hampers the clinical usage of these inhibitors (Wacker et al., 2012; Ma et al., 2014; Dumas et al., 2016). Development of small molecule modulators against human homologues identified in this study and their combined usage with kinesin-5 inhibitors would be a complementing strategy for future tumor treatment.
MATERIALS AND METHODS
Strains, media, and genetic methods
Fission yeast strains used in this study are listed in Supplemental Table S1. Media, growth conditions, and manipulations were carried out as previously described (Moreno et al., 1991; Bähler et al., 1998; Sato et al., 2005). For most of the experiments, rich YE5S liquid media and agar plates were used. Wild-type strain (513; Supplemental Table S1), temperature-sensitive cut7-22, pkl1 deletion, klp9rigor and klp9Δ38C strains were provided by P. Nurse (The Francis Crick Institute, London, England, UK), I. Hagan (Cancer Research UK Manchester Institute, University of Manchester, Manchester, England, UK), R. McIntosh (University of Colorado, Boulder, CO), and J. B. Millar (University of Warwick, Coventry, England, UK), respectively. Spot tests were performed by spotting 5–10 μl of cells at a concentration of 2 × 107 cells/ml after 10-fold serial dilutions onto rich YE5S plates with or without a drug (TBZ). Some of the YE5S plates also contained Phloxine B, a vital dye that accumulates in dead or dying cells and stains the colonies dark pink due to a reduced ability to pump out the dye. Plates were incubated at various temperatures from 27°C to 36°C as necessary.
Preparation and manipulation of nucleic acids
Enzymes were used as recommended by the suppliers (New England Biolabs, Ipswich, MA, and Takara Bio, Shiga, Japan).
Strain construction, gene disruption, and the N-terminal and C-terminal epitope tagging
A PCR-based gene-targeting method (Bähler et al., 1998; Sato et al., 2005) was used for complete gene disruption and epitope tagging (e.g., GFP, 2mRFP, CFP, and mCherry) in the C terminus, by which all the tagged proteins were produced under the endogenous promoter. Alp14NLS-GFP was constructed by inserting in frame a canonical NLS sequence (PKKKRKV) derived from SV40 (Kalderon et al., 1984) 5′ to GFP. GBP-mCherry-Alp4 was constructed as follows: DNA fragments containing a G418-resistance gene (kanR), the alp4 promoter, GBP, and mCherry were PCR amplified and inserted into the 5′-franking region to the alp4 ORF in-frame by the fusion PCR method.
Isolation of klp9 or alp7 temperature-sensitive mutants in the cut7∆pkl1∆ background
The klp9 ts mutants were constructed by PCR-based random mutagenesis (Tang et al., 2017). The G418-resistance marker gene cassette (kanR) was inserted in the 3′ flanking region of the klp9 gene (klp9-kanR). The klp9-kanR fragment purified from this strain was amplified with PCR using TaKaRa EX taq polymerase (Takara Bio, Shiga, Japan). The alp7 ts mutants were constructed by site-directed mutagenesis using a two-step PCR amplification method (Masuda et al., 2013), by which random mutations (5′mnn3′) were introduced into the five amino acid residues corresponding to K341, Y344, R346, K347, and Y348 within Alp7. To isolate klp9 or alp7 mutants, pools of mutagenized PCR fragments were ethanol precipitated and transformed into a cut7∆pkl1∆ strain (MY990, Supplemental Table S1). Alp7-13myc was originally used for ts mutant isolation and later 13myc was replaced with GFP. G418-resistant transformants were screened for temperature sensitivity at 36°C. These mutants were crossed with a cut7∆pkl1∆ strain and random spore analysis was performed. In all segregants, the ts phenotype cosegregated with G418 resistance. Subsequently, nucleotide sequencing was performed to determine the mutated sites in the klp9 or alp7 gene of these mutants.
Fluorescence microscopy and time-lapse live cell imaging
Fluorescence microscopy images were obtained by using a DeltaVision microscope system (DeltaVision Elite; GE Healthcare, Chicago, IL) comprising a wide-field inverted epifluorescence microscope (IX71; Olympus, Tokyo, Japan) and a Plan Apochromat 60×, NA 1.42, oil immersion objective (PLAPON 60 × O; Olympus Tokyo, Japan). DeltaVision image acquisition software (softWoRx 6.5.2; GE Healthcare, Chicago, IL) equipped with a charge-coupled device camera (CoolSNAP HQ2; Photometrics, Tucson, AZ) was used. Live cells were imaged in a glass-bottomed culture dish (MatTek Corporation, Ashland, MA) coated with soybean lectin and incubated at 27°C for most of the strains or at 36°C for the ts mutants. The latter were cultured in rich YE5S media until mid-log phase at 27°C and subsequently shifted to the restrictive temperature of 36°C before microscopic observation. To keep cultures at the proper temperature, a temperature-controlled chamber (Air Therm SMT; World Precision Instruments, Sarasota, FL) was used. Time-lapse imaging was continued for further 40 min at 36°C. The sections of images acquired at each time point were compressed into a two-dimensional projection using the DeltaVision maximum intensity algorithm. Deconvolution was applied before the two-dimensional projection. Images were taken as 14–16 sections along the z-axis at 0.2 μm intervals; they were then deconvolved and merged into a single projection. Captured images were processed with Photoshop CS6 (version 13.0; Adobe, San Jose, CA).
Quantification of fluorescence signal intensities
For quantification of signal intensities of the fluorescent marker-tagged protein located at the SPB (Alp7-GFP, Figure 4D), 14 sections were taken along the z-axis at 0.2-μm intervals. Projection images of maximum intensity were obtained after deconvolution, and, on subtracting background intensities, only values of maximum fluorescence intensities covering the whole GFP signals within the z-sections were used for statistical data analysis.
Immunochemistry
Protein extracts were prepared by mechanical disruption of cells in an extraction buffer (50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 150 mM NaCl, 0.05% NP-40, 10% glycerol, 1.5 mM p-nitrophenyl phosphate, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1× protease inhibitor cocktail from Sigma-Aldrich Co., St. Louis, MO) with acid-washed glass beads in the FastPrep FP120 apparatus (2 × 25 s, power 5.5; BIO-101, Vista, CA). Protein extracts were loaded and resolved on denaturing 4–12% gradient gels (Bio-Rad Laboratories, Hercules, CA) and transferred onto polyvinylidene fluoride membranes. The membranes were blocked with 10% skim milk and then blotted with either anti-GFP (rabbit polyclonal, TP401; Torrey Pines Biolabs, Secaucus, NJ) or anti-γ-tubulin antibody (mouse monoclonal, produced GTU-88, Sigma-Aldrich Co., St. Louis, MO). After having been washed, the blots were incubated with anti-rabbit or anti-mouse horseradish peroxidase–conjugated secondary antibody (GE Healthcare, Buckinghamshire, UK) in 1% skim milk at a dilution of 1:2000. The ECL chemiluminescence kit (GE Healthcare) was used for detection.
Statistical data analysis
We used the two-tailed unpaired Student’s t test to evaluate the significance of differences in different strains. All the experiments were performed at least twice. Experiment sample numbers used for statistical testing are given in the corresponding figures and/or legends. We used this key for asterisk placeholders to indicate p values in the figures, for example, ****p < 0.0001.
Supplementary Material
Acknowledgments
We are grateful to Heinrich Leonhardt, Paul Nurse, Iain Hagan, Jonathan Millar, J. Richard McIntosh, and Gislene Pereira for providing us with strains used in this study. We thank Ken’ya Furuta for thoughtful discussion and Corinne Pinder for critical reading of the manuscript and useful suggestions. This work was supported by the Japan Society for the Promotion of Science (JSPS) (KAKENHI Scientific Research [A] 16H02503 to T.T., a Challenging Exploratory Research grant 16K14672 to T.T., Scientific Research [C] 16K07694 to M.Y.), the Naito Foundation (T.T.), and the Uehara Memorial Foundation (T.T.).
Abbreviations used:
- CPC
chromosome passenger complex
- GBP
GFP-binding protein
- MAPs
microtubule-associated proteins
- MTOC
microtubule-organizing center
- NLS
nuclear localization signal
- SAC
spindle assembly checkpoint
- SPB
spindle pole body
- ts
temperature sensitive.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-08-0497) on October 11, 2017.
REFERENCES
- Abaza A, Soleilhac JM, Westendorf J, Piel M, Crevel I, Roux A, Pirollet F. M phase phosphoprotein 1 is a human plus-end-directed kinesin-related protein required for cytokinesis. J Biol Chem. 2003;278:27844–27852. doi: 10.1074/jbc.M304522200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams RR, Tavares AA, Salzberg A, Bellen HJ, Glover DM. pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 1998;12:1483–1494. doi: 10.1101/gad.12.10.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J, Chang F. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 2011;21:604–614. doi: 10.1016/j.tcb.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J, Kim H, Brouhard G, van Oijen A, Harrison SC, Chang F. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev Cell. 2010;19:245–258. doi: 10.1016/j.devcel.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J, Kim H, Flor-Parra I, Lal N, Velji H, Chang F. Fission yeast Alp14 is a dose dependent plus end tracking microtubule polymerase. Mol Biol Cell. 2012;23:2878–2890. doi: 10.1091/mbc.E12-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Wu J, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bieling P, Kronja I, Surrey T. Microtubule motility on reconstituted meiotic chromatin. Curr Biol. 2010;20:744–749. doi: 10.1016/j.cub.2010.02.067. [DOI] [PubMed] [Google Scholar]
- Bratman SV, Chang F. Stabilization of overlapping microtubules by fission yeast CLASP. Dev Cell. 2007;13:812–827. doi: 10.1016/j.devcel.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge AJ, Morphew M, Bartlett R, Hagan IM. The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes Dev. 1998;12:927–942. doi: 10.1101/gad.12.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesario JM, Jang JK, Redding B, Shah N, Rahman T, McKim KS. Kinesin 6 family member Subito participates in mitotic spindle assembly and interacts with mitotic regulators. J Cell Sci. 2006;119:4770–4780. doi: 10.1242/jcs.03235. [DOI] [PubMed] [Google Scholar]
- Charrasse S, Schroeder M, Gauthier-Rouviere C, Ango F, Cassimeris L, Gard DL, Larroque C. The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J Cell Sci. 1998;111:1371–1383. doi: 10.1242/jcs.111.10.1371. [DOI] [PubMed] [Google Scholar]
- Choi SH, McCollum D. A role for metaphase spindle elongation forces in correction of merotelic kinetochore attachments. Curr Biol. 2012;22:225–230. doi: 10.1016/j.cub.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G, Tao L, Brust-Mascher I, Wollman R, Scholey JM. Prometaphase spindle maintenance by an antagonistic motor-dependent force balance made robust by a disassembling lamin-B envelope. J Cell Biol. 2010;188:49–68. doi: 10.1083/jcb.200908150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa J, Fu C, Khare VM, Tran PT. csi2p modulates microtubule dynamics and organizes the bipolar spindle for chromosome segregation. Mol Biol Cell. 2014;25:3900–3908. doi: 10.1091/mbc.E14-09-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen CF, Deak P, Glover DM, Ohkura H. mini spindles: a gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. J Cell Biol. 1999;146:1005–1018. doi: 10.1083/jcb.146.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Dumas ME, Sturgill EG, Ohi R. Resistance is not futile: surviving Eg5 inhibition. Cell Cycle. 2016;21:2845–2847. doi: 10.1080/15384101.2016.1204864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enos AP, Morris NR. Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. Cell. 1990;60:1019–1027. doi: 10.1016/0092-8674(90)90350-n. [DOI] [PubMed] [Google Scholar]
- Ferenz NP, Paul R, Fagerstrom C, Mogilner A, Wadsworth P. Dynein antagonizes Eg5 by crosslinking and sliding antiparallel microtubules. Curr Biol. 2009;21:1833–1838. doi: 10.1016/j.cub.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory MR, Morphew M, Joseph JD, Means AR, Davis TN. Pcp1p, an Spc110p-related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth Differ. 2002;13:47–58. [PubMed] [Google Scholar]
- Fong CS, Sato M, Toda T. Fission yeast Pcp1 links polo kinase-mediated mitotic entry to γ-tubulin-dependent spindle formation. EMBO J. 2010;29:120–130. doi: 10.1038/emboj.2009.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Ward JJ, Loiodice I, Velve-Casquillas G, Nedelec FJ, Tran PT. Phospho-regulated interaction between kinesin-6 Klp9p and microtubule bundler Ase1p promotes spindle elongation. Dev Cell. 2009;17:257–267. doi: 10.1016/j.devcel.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Bingham JB, Hasbani MJ, Gill SR, Schroer TA, Compton DA. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Koonrugsa N, Toda T. Spindle-kinetochore attachment requires the combined action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J. 2002;21:6015–6024. doi: 10.1093/emboj/cdf611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Vardy L, Koonrugsa N, Toda T. Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J. 2001;20:3389–3401. doi: 10.1093/emboj/20.13.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatlin JC, Matov A, Groen AC, Needleman DJ, Maresca TJ, Danuser G, Mitchison TJ, Salmon ED. Spindle fusion requires dynein-mediated sliding of oppositely oriented microtubules. Curr Biol. 2009;19:287–296. doi: 10.1016/j.cub.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho SA, Pellman D. Causes and consequences of centrosome abnormalities in cancer. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130467. doi: 10.1098/rstb.2013.0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A, Beuter C, Craven RA, Bagley S, Wilks D, Fleig U, Hagan IM. S. pombe CLASP needs dynein, not EB1 or CLIP170, to induce microtubule instability and slows polymerization rates at cell tips in a dynein-dependent manner. Genes Dev. 2006;20:2421–2436. doi: 10.1101/gad.381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Novel potential mitotic motor protein encoded by the fission yeast cut7+ gene. Nature. 1990;347:563–566. doi: 10.1038/347563a0. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature. 1992;356:74–76. doi: 10.1038/356074a0. [DOI] [PubMed] [Google Scholar]
- Hagan IM, Petersen J. The microtubule organizing centers of Schizosaccharomyces pombe. Curr Top Dev Biol. 2000;49:133–159. doi: 10.1016/s0070-2153(99)49007-6. [DOI] [PubMed] [Google Scholar]
- Heald R, Khodjakov A. Thirty years of search and capture: the complex simplicity of mitotic spindle assembly. J Cell Biol. 2015;211:1103–1111. doi: 10.1083/jcb.201510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck MM, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LS. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill E, Clarke M, Barr FA. The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. EMBO J. 2000;19:5711–5719. doi: 10.1093/emboj/19.21.5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- Hou H, Zhou Z, Wang Y, Wang J, Kallgren SP, Kurchuk T, Miller EA, Chang F, Jia S. Csi1 links centromeres to the nuclear envelope for centromere clustering. J Cell Biol. 2012;199:735–744. doi: 10.1083/jcb.201208001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu KS, Toda T. Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore-spindle attachment in fission yeast. Curr Biol. 2011;21:214–220. doi: 10.1016/j.cub.2010.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussmann F, Drummond DR, Peet DR, Martin DS, Cross RA. Alp7/TACC-Alp14/TOG generates long-lived, fast-growing MTs by an unconventional mechanism. Sci Rep. 2016;6:20653. doi: 10.1038/srep20653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikebe C, Konishi M, Hirata D, Matsusaka T, Toda T. Systematic localization study on novel proteins encoded by meiotically up-regulated ORFs in fission yeast. Biosci Biotechnol Biochem. 2011;75:2364–2370. doi: 10.1271/bbb.110558. [DOI] [PubMed] [Google Scholar]
- Janson ME, Loughlin R, Loiodice I, Fu C, Brunner D, Nedelec J, Tran PT. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell. 2007;128:357–368. doi: 10.1016/j.cell.2006.12.030. [DOI] [PubMed] [Google Scholar]
- Jiang K, Akhmanova A. Microtubule tip-interacting proteins: a view from both ends. Curr Opin Cell Biol. 2011;23:94–101. doi: 10.1016/j.ceb.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling YC, Vjestica A, Oliferenko S. Nucleocytoplasmic shuttling of the TACC protein Mia1p/Alp7p is required for remodeling of microtubule arrays during the cell cycle. PLoS One. 2009;4:e6255. doi: 10.1371/journal.pone.0006255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiodice I, Staub J, Gangi-Setty T, Nguyen NP, Paoletti A, Tran PT. Ase1p organizes anti-parallel microtubule arrays during interphase and mitosis in fission yeast. Mol Biol Cell. 2005;16:1756–1768. doi: 10.1091/mbc.E04-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HT, Erdal S, Huang S, Poon RY. Synergism between inhibitors of Aurora A and KIF11 overcomes KIF15-dependent drug resistance. Mol Oncol. 2014;8:1404–1418. doi: 10.1016/j.molonc.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H, Mori R, Yukawa M, Toda T. Fission yeast MOZART1/Mzt1 is an essential γ-tubulin complex component required for complex recruitment to the MTOC, but not its assembly. Mol Biol Cell. 2013;24:2894–2906. doi: 10.1091/mbc.E13-05-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Maurer SP, Yukawa M, Zakian S, Singleton MR, Surrey T, Toda T. An unconventional interaction between Dis1/TOG and Mal3/EB1 in fission yeast promotes the fidelity of chromosome segregation. J Cell Sci. 2016;129:4592–4606. doi: 10.1242/jcs.197533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews LR, Carter P, Thierry-Mieg D, Kemphues K. ZYG-9, a Caenorhabditis elegans protein required for microtubule organization and function, is a component of meiotic and mitotic spindle poles. J Cell Biol. 1998;141:1159–1168. doi: 10.1083/jcb.141.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- Meadows JC, Lancaster TC, Buttrick GJ, Sochaj AM, Messin LJ, Del Mar Mora-Santos M, Hardwick KG, Millar JB. Identification of a Sgo2-dependent but Mad2-independent pathway controlling anaphase onset in fission yeast. Cell Rep. 2017;18:1422–1433. doi: 10.1016/j.celrep.2017.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows JC, Millar JB. Latrunculin A delays anaphase onset in fission yeast by disrupting an Ase1-independent pathway controlling mitotic spindle stability. Mol Biol Cell. 2008;19:3713–3723. doi: 10.1091/mbc.E08-02-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Maddox P, Gaetz J, Groen A, Shirasu M, Desai A, Salmon ED, Kapoor TM. Roles of polymerization dynamics, opposed motors and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol Biol Cell. 2005;6:3064–3076. doi: 10.1091/mbc.E05-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Mountain V, Simerly C, Howard L, Ando A, Schatten G, Compton DA. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol. 1999;147:351–366. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr Biol. 2015;25:R1002–R1018. doi: 10.1016/j.cub.2015.08.051. [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Kurooka H, Takeuchi M, Kinoshita K, Nakaseko Y, Yanagida M. p93dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the Cdc2 target sites. Genes Dev. 1995;9:1572–1585. doi: 10.1101/gad.9.13.1572. [DOI] [PubMed] [Google Scholar]
- Nakaseko Y, Goshima G, Morishita J, Yanagida M. M phase-specific kinetochore proteins in fission yeast microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr Biol. 2001;11:537–549. doi: 10.1016/s0960-9822(01)00155-5. [DOI] [PubMed] [Google Scholar]
- Nislow C, Lombillo VA, Kuriyama R, McIntosh JR. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 1992;359:543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- O’Connell MJ, Meluh PB, Rose MD, Morris NR. Suppression of the bimC4 mitotic spindle defect by deletion of klpA, a gene encoding a KAR3-related kinesin-like protein in Aspergillus nidulans. J Cell Biol. 1993;120:153–162. doi: 10.1083/jcb.120.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada N, Sato M. Spatiotemporal regulation of nuclear transport machinery and microtubule organization. Cells. 2015;4:406–426. doi: 10.3390/cells4030406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada N, Toda T, Yamamoto M, Sato M. CDK-dependent phosphorylation of Alp7-Alp14 (TACC-TOG) promotes its nuclear accumulation and spindle microtubule assembly. Mol Biol Cell. 2014;25:1969–1982. doi: 10.1091/mbc.E13-11-0679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliferenko S, Balasubramanian MK. Astral microtubules monitor metaphase spindle alignment in fission yeast. Nat Cell Biol. 2002;4:816–820. doi: 10.1038/ncb861. [DOI] [PubMed] [Google Scholar]
- Olmsted ZT, Colliver AG, Riehlman TD, Paluh JL. Kinesin-14 and kinesin-5 antagonistically regulate microtubule nucleation by γ-TuRC in yeast and human cells. Nat Commun. 2014;5:5339. doi: 10.1038/ncomms6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, LeDizet M, Cande WZ. Fission yeast pkl1 is a kinesis-related protein involved in mitotic spindle function. Mol Biol Cell. 1996;7:1639–1655. doi: 10.1091/mbc.7.10.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raich WB, Moran AN, Rothman JH, Hardin J. Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol Biol Cell. 1998;9:2037–2049. doi: 10.1091/mbc.9.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber SB, Baumgart J, Widlund PO, Pozniakovsky A, Howard J, Hyman AA, Julicher F. XMAP215 activity sets spindle length by controlling the total mass of spindle microtubules. Nat Cell Biol. 2013;15:1116–1122. doi: 10.1038/ncb2834. [DOI] [PubMed] [Google Scholar]
- Rincon SA, Lamson A, Blackwell R, Syrovatkina V, Fraisier V, Paoletti A, Betterton MD, Tran PT. Kinesin-5-independent mitotic spindle assembly requires the antiparallel microtubule crosslinker Ase1 in fission yeast. Nat Commun. 2017;8:15286. doi: 10.1038/ncomms15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AS, Batac J, Killilea AN, Filopei J, Simeonov DR, Lin I, Paluh JL. Protein complexes at the microtubule organizing center regulate bipolar spindle assembly. Cell Cycle. 2008;7:1246–1253. doi: 10.4161/cc.7.9.5808. [DOI] [PubMed] [Google Scholar]
- Roof DM, Meluh PB, Rose MD. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J, Cade NI, Surrey T. Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat Cell Biol. 2015;17:1422–1434. doi: 10.1038/ncb3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- Saitoh S, Takahashi K, Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- Salemi JD, McGilvray PT, Maresca TJ. Development of a Drosophila cell-based error correction assay. Front Oncol. 2013;3:187. doi: 10.3389/fonc.2013.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Dhut S, Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- Sato M, Okada N, Kakui Y, Yamamoto M, Yoshida M, Toda T. Nucleocytoplasmic transport of Alp7/TACC organizes spatiotemporal microtubule formation in fission yeast. EMBO Rep. 2009;10:1161–1167. doi: 10.1038/embor.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Toda T. Alp7/TACC is a crucial target in Ran-GTPase-dependent spindle formation in fission yeast. Nature. 2007;447:334–337. doi: 10.1038/nature05773. [DOI] [PubMed] [Google Scholar]
- Sato M, Vardy L, Garcia MA, Koonrugsa N, Toda T. Interdependency of fission yeast Alp14/TOG and coiled coil protein Alp7 in microtubule localization and bipolar spindle formation. Mol Biol Cell. 2004;15:1609–1622. doi: 10.1091/mbc.E03-11-0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W, Lengyel V, Hoyt MA. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol Biol Cell. 1997;8:1025–1033. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Brown HM, Kwon M, Rogers GC, Holland G, Scholey JM. Functional coordination of three mitotic motors in Drosophila embryos. Mol Biol Cell. 2000;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Yu KR, Sisson JC, Sullivan W, Scholey JM. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat Cell Biol. 1999;1:51–54. doi: 10.1038/9025. [DOI] [PubMed] [Google Scholar]
- She ZY, Yang WX. Molecular mechanisms of kinesin-14 motors in spindle assembly and chromosome segregation. J Cell Sci. 2017;130:2097–2110. doi: 10.1242/jcs.200261. [DOI] [PubMed] [Google Scholar]
- Syrovatkina V, Fu C, Tran PT. Antagonistic spindle motors and MAPs regulate metaphase spindle length and chromosome segregation. Curr Biol. 2013;23:2423–2429. doi: 10.1016/j.cub.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrovatkina V, Tran PT. Loss of kinesin-14 results in aneuploidy via kinesin-5-dependent microtubule protrusions leading to chromosome cut. Nat Commun. 2015;6:7322. doi: 10.1038/ncomms8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, Galjart N, Medema RH. Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. EMBO J. 2008;27:3235–3245. doi: 10.1038/emboj.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, Janssen A, Geers EF, Alvarez-Fernandez M, Medema RH. Kif15 cooperates with Eg5 to promote bipolar spindle assembly. Curr Biol. 2009;19:1703–1711. doi: 10.1016/j.cub.2009.08.027. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Medema RH. Mechanisms of centrosome separation and bipolar spindle assembly. Dev Cell. 2010;19:797–806. doi: 10.1016/j.devcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Tang NH, Fong CS, Jourdain I, Masuda H, Yukawa M, Toda T. Generation and characterisation of temperature sensitive mutants of genes encoding the fission yeast spindle pole body. PeerJ Preprints. 2017;5 [Google Scholar]
- Tang NH, Okada N, Fong CS, Arai K, Sato M, Toda T. Targeting Alp7/TACC to the spindle pole body is essential for mitotic spindle assembly in fission yeast. FEBS Lett. 2014;588:2814–2821. doi: 10.1016/j.febslet.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang NH, Takada H, Hsu KS, Toda T. The internal loop of fission yeast Ndc80 binds Alp7/TACC-Alp14/TOG and ensures proper chromosome attachment. Mol Biol Cell. 2013;24:1122–1133. doi: 10.1091/mbc.E12-11-0817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang NH, Toda T. Alp7/TACC recruits kinesin-8-PP1 to the Ndc80 kinetochore protein for timely mitotic progression and chromosome movement. J Cell Sci. 2015;128:354–363. doi: 10.1242/jcs.160036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L, Mogilner A, Civelekoglu-Scholey G, Wollman R, Evans J, Stahlberg H, Schole JM. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr Biol. 2006;16:2293–2302. doi: 10.1016/j.cub.2006.09.064. [DOI] [PubMed] [Google Scholar]
- Toda T, Adachi Y, Hiraoka Y, Yanagida M. Identification of the pleiotropic cell division cycle gene NDA2 as one of two different α-tubulin genes in Schizosaccharomyces pombe. Cell. 1984;37:233–242. doi: 10.1016/0092-8674(84)90319-2. [DOI] [PubMed] [Google Scholar]
- Toya M, Sato M, Haselmann U, Asakawa K, Brunner D, Antony C, Toda T. γ-Tubulin complex-mediated anchoring of spindle microtubules to spindle-pole bodies requires Msd1 in fission yeast. Nat Cell Biol. 2007;9:646–653. doi: 10.1038/ncb1593. [DOI] [PubMed] [Google Scholar]
- Troxell CL, Sweezy MA, West RR, Reed KD, Carson BD, Pidoux AL, Cande WZ, McIntosh JR. pkl1+ and klp2+: two kinesins of the Kar3 subfamily in fission yeast perform different functions in both mitosis and meiosis. Mol Biol Cell. 2001;12:3476–3488. doi: 10.1091/mbc.12.11.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heesbeen RG, Tanenbaum ME, Medema RH. Balanced activity of three mitotic motors Is required for bipolar spindle assembly and chromosome segregation. Cell Rep. 2014;8:948–956. doi: 10.1016/j.celrep.2014.07.015. [DOI] [PubMed] [Google Scholar]
- Vanneste D, Takagi M, Imamoto N, Vernos I. The role of Hklp2 in the stabilization and maintenance of spindle bipolarity. Curr Biol. 2009;19:1712–1717. doi: 10.1016/j.cub.2009.09.019. [DOI] [PubMed] [Google Scholar]
- Vardy L, Toda T. The fission yeast γ-tubulin complex is required in G1 phase and is a component of the spindle assembly checkpoint. EMBO J. 2000;19:6098–6111. doi: 10.1093/emboj/19.22.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker SA, Houghtaling BR, Elemento O, Kapoor TM. Using transcriptome sequencing to identify mechanisms of drug action and resistance. Nat Chem Biol. 2012;8:235–237. doi: 10.1038/nchembio.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Vernos I, Mitchison TJ, Karsenti E, Heald R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr Biol. 1998;8:903–913. doi: 10.1016/s0960-9822(07)00370-3. [DOI] [PubMed] [Google Scholar]
- Wang B, Li K, Jin M, Qiu R, Liu B, Oakley BR, Xiang X. The Aspergillus nidulans bimC4 mutation provides an excellent tool for identification of kinesin-14 inhibitors. Fungal Genet Biol. 2015;82:51–55. doi: 10.1016/j.fgb.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Hyman A, Desai A. The spindle: a dynamic assembly of microtubules and motors. Nat Cell Biol. 2001;3:E28–E34. doi: 10.1038/35050669. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Sato M, Fujita A, Yamamoto M, Toda T. The roles of fission yeast ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol Biol Cell. 2005;16:1378–1395. doi: 10.1091/mbc.E04-10-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukawa M, Ikebe C, Toda T. The Msd1-Wdr8-Pkl1 complex anchors microtubule minus ends to fission yeast spindle pole bodies. J Cell Biol. 2015;290:549–562. doi: 10.1083/jcb.201412111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng F, Li T, Jin DY, Syrovatkina V, Scheffler K, Tran PT, Fu C. Csi1p recruits alp7p/TACC to the spindle pole bodies for bipolar spindle formation. Mol Biol Cell. 2014;25:2750–2760. doi: 10.1091/mbc.E14-03-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.