Abstract
Yeast surface display, a well-established technology for protein analysis and engineering, involves expressing a protein of interest as a genetic fusion to either the N- or C-terminus of the yeast Aga2p mating protein. Historically, yeast-displayed protein variants are flanked by peptide epitope tags that enable flow cytometric measurement of construct expression using fluorescent primary or secondary antibodies. Here, we built upon this technology to develop a new yeast display strategy that comprises fusion of two different proteins to Aga2p, one to the N-terminus and one to the C-terminus. This approach allows an antibody fragment, ligand, or receptor to be directly coupled to expression of a fluorescent protein readout, eliminating the need for antibody-staining of epitope tags to quantify yeast protein expression levels. We show that this system simplifies quantification of protein-protein binding interactions measured on the yeast cell surface. Moreover, we show that this system facilitates co-expression of a bioconjugation enzyme and its corresponding peptide substrate on the same Aga2p construct, enabling enzyme expression and catalytic activity to be measured on the surface of yeast.
Keywords: Bioconjugation, Binding assay, Fluorescent protein, Sortase, Yeast surface display
1 Introduction
Recombinant proteins are expressed and tethered on the surface of cells to enable measurement of biophysical and biochemical properties in a paralleled, high-throughput manner [1, 2]. Since the first surface expression system was introduced on bacteriophage in mid-1980s [3], a variety of techniques have been developed to display proteins on the surface of bacteria [4], yeast [5], insect [6], or mammalian cells [7, 8]. In these platforms, each individual cell harbors a genetically-encoded protein variant of interest fused to a cell surface anchor protein. The protein of interest becomes accessible to the extracellular space after the fusion construct, directed via signal sequence, is transported to the cell wall or outer membrane. These methods can be used in conjunction with a library of plasmids encoding diverse protein variants to display multiple copies of a unique protein variant on the surface of each individual cell. The libraries are screened to isolate cells that express proteins with a desired phenotype, and selected protein variants are identified by sequencing the corresponding genetic material recovered from the cells. This genotype-to-phenotype linkage is central to the application of the surface display techniques in combinatorial protein engineering [9].
Over the past two decades, yeast surface display has been extensively used to analyze and engineer protein variants that possess increased binding affinity to a target of interest [5, 10, 11], higher thermal stability [12, 13], or altered catalytic properties [14–16]. Compared to the other cell surface display platforms, yeast surface display has collective advantages such as eukaryotic post-translational modifications, the ease of cell culture and genetic modification, and the compatibility with flow cytometric analysis [17]. Among various yeast surface display techniques using different anchor proteins [18, 19], a display platform pioneered by Boder and Wittrup in 1997 uses yeast cells transformed with a plasmid encoding a protein of interest genetically fused to the a-agglutinin mating protein Aga2p subunit [5]. After translation, the Aga2p subunit is covalently bound to an integrated Aga1p subunit via two disulfide bonds, processed through the yeast secretory machinery, and transported to the exterior of the yeast cell surface, where Aga1p is covalently anchored to the cell wall [20, 21] (Fig. 1A). The surface-displayed protein of interest is flanked by peptide epitope tags (i.e. c-Myc and hemagglutinin antigen (HA) tags) which enable quantification of fusion protein expression levels on each cell using fluorescent antibodies and analysis by flow cytometry. This feature allows for discrimination of protein variants with only a 2-fold difference in affinity by normalizing binding signal with expression levels on the yeast cell surface [22].
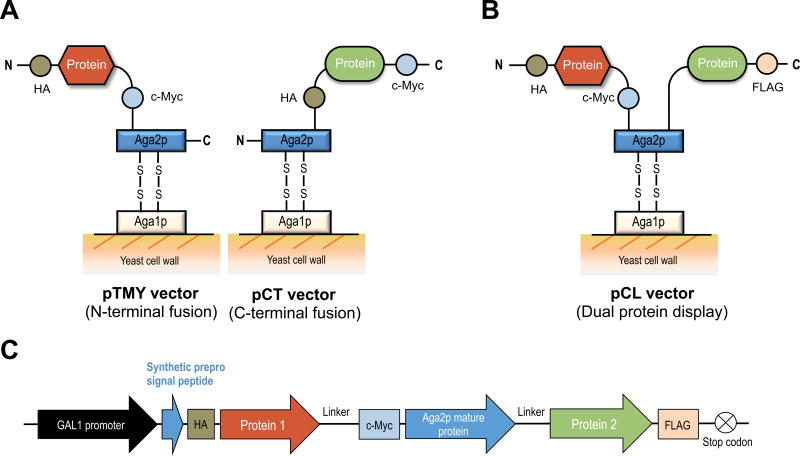
Figure 1.
Schematic of yeast surface display strategies. (A) Conventional vectors, pTMY (N-terminal Aga2p fusion) and pCT (C-terminal Aga2p fusion). (B) pCL, a vector that enables co-expression of two proteins on the N- and C-terminus of the Aga2p subunit. (C) The expression cassette of pCL in which one protein is inserted between the synthetic prepro signal peptide and Aga2p and the other protein is inserted downstream of Aga2p. Various epitope tags (HA, c-Myc and FLAG) are included to validate and compare protein expression across formats.
Conventional yeast surface display vectors are designed to display a protein of interest fused to either the N- or C-terminus of a cell wall-anchored protein [18, 19] (Fig. 1A). Alternatively, homodimeric and heterodimeric proteins have been displayed on yeast using a single vector containing two GAL1 promoters or a bidirectional GAL1–10 promoter. Using these strategies, the heavy and light chains of an anti-streptavidin Fab [23] or class II MHC α and β chains [10] have been displayed as fusions on yeast surface. As another example, homooligomeric streptavidin was functionally expressed and assembled using two yeast display vectors [24]. Such multi-protein display can also aid in enzyme engineering, where substrate channeling imparts significant kinetic advantages [25, 26].
Here, we introduce a simplified yeast display strategy that utilizes both the N- and C-termini of Aga2p to display two heterologous proteins as part of one fusion protein (Fig. 1B). We show that a number of different proteins can be anchored in this manner and retain their functional activity. In one demonstration, dual expression of a fluorescent protein along with a ligand, receptor, or antibody fragment simplifies quantification of protein expression by eliminating the need for antibody staining of epitope tags. This approach saves time and cost, allowing streamlined determination of equilibrium binding constants compared to conventional yeast surface display. In a second demonstration, we show that the dual expression of the bioconjugation enzyme Staphylococcus aureus sortase A and its corresponding peptide substrate as part of the same Aga2p construct enables measurement of catalytic activity on a non-natural substrate. This approach is simple and more generalizable compared to a previously reported method [15].
2 Materials and methods
2.1 Strains and reagents
Chemical competent TOP10 E. coli cells were used to clone, propagate, and store plasmids through culturing in LB media containing 100 µg/ml ampicillin for selection. Saccharomyces cerevisiae strain EBY100 was used for yeast surface display throughout this study [11]. Plasmids were transformed into yeast by homologous recombination using a Gene Pulser Xcell electroporation system (Bio-Rad) [11]. Transformed yeast were grown in SD-CAA media (20 g dextrose; 6.7 g Difco yeast nitrogen base; 5 g Bacto casamino acids; 5.4 g Na2HPO4; 8.56 g NaH2PO4•H2O; dissolved in deionized H2O to a volume of 1L) [11] and induced to express Aga2p fusion proteins on their surface through a galactose-inducible promoter by culturing in SG-CAA media (prepared as SD-CAA except using 20 g galactose substituted for dextrose). For the sortase bioconjugation reaction, a 10 M stock of 3-azido-1-propanamine (Azp) (Sigma, 762016) was diluted to 1 M in 1 M acetic acid immediately before use. Sulfo-dibenzocyclooctyne-biotin conjugate (Sigma, 760706) was stored as a 50 mM stock solution in dimethylformamide at −20 °C.
2.2 Construction of plasmids
The pCL vector was generally designed as shown in Figure 1C and was created by rebuilding the pTMY vector which displays the protein-of-interest, HA, and c-Myc epitope tags as a fusion to the N-terminus of Aga2p [27]. The recombinant yeast-codon-optimized enhanced GFP with mutations S65G and S72A (termed yEGFP throughout this study) was kindly provided by Prof. Eric Shusta at University of Wisconsin-Madison [28–30]. The upstream region of pTMY (N-terminal to the Aga2p mature protein) was preserved (Fig. 1C) and includes a GAL1 promoter [31], followed by a synthetic α-factor prepro signal peptide [32], a KR (Lys-Arg) KEX2 cleavage sequence [33] and an EA (Glu-Ala) peptide spacer [34]. In addition to a flexible (Gly4Ser)3 linker incorporated into pTMY upstream of the Aga2p protein, another (Gly4Ser)3 linker was introduced downstream of Aga2p to provide spatial degrees of freedom for proteins tethered at the N- and C-termini of Aga2p. To prevent homologous recombination within the plasmid DNA, nucleotide sequences were varied in the new (Gly4Ser)3 linker. The downstream region of this new linker was modified in each pCL vector and the epitope tags such as HA, c-Myc, or FLAG tags were inserted, removed, or relocated according to the design of each recombinant pCL vector as described in Supporting Information.
2.3 Binding assays
For binding assays, yeast cells were transformed with the GFP-co-expressing pCL plasmids (pCL-nGFP-Aga2p-D1.3, pCL-nGFP-Aga2p-Axl, and pCL-NK1-Aga2p-cGFP) and the corresponding pCT or pTMY plasmids (pCT-D1.3, pCT-Axl, and pTMY-NK1). After growth in SD-CAA media at 30 °C to an OD600 = 3–6, yeast cells were centrifuged and resuspended to a final OD600 of 1 in SG-CAA media followed by 24 h incubation at 20 °C for induction of protein expression. Binding affinities of each model protein on the pCL or pCT/pTMY plasmids were measured by incubating induced yeast cells with varying concentrations of target protein in PBSA (phosphate-buffered saline + 1 mg/ml BSA) for 6–17 h at room temperature. Reaction volumes and time were empirically determined to minimize ligand depletion and to ensure equilibrium was reached. After incubation, yeast cells expressing proteins using the pCL-GFP plasmids were stained with a fluorescently-labeled secondary antibody against the target protein to measure binding signals. For yeast harboring pCT or pTMY plasmids, cells were first stained with a primary antibody that binds to an epitope tag (to quantify protein expression levels) and then labeled with secondary antibodies against anti-epitope tag antibodies and the target protein. Detailed antibody-staining strategies for each model protein are described in the Supporting Information. Fluorescence values representing expression and target binding of the labeled yeast cells were measured using an Accuri C6 flow cytometer (BD Biosciences). Data were collected from 10,000 cells and analyzed using FlowJo software (Treestar Inc.). Full binding titrations were fit as a four-parameter sigmoidal curve using KaleidaGraph (Synergy Software) to calculate equilibrium dissociation constants (KD) from three technical replicates of each fit point.
2.4 Sortase bioconjugation reaction
To measure sortase bioconjugation activity and expression on yeast, the plasmids co-expressing sortase A 7M [15] and the LPETGG substrate sequence with various linkers (pCL-Srt-SS, pCL-Srt-LS, and pCL-Srt-cGFP-LS (Fig. 3B)) were transformed into EBY100 yeast cells. Single colonies were grown overnight in SD-CAA at 30 °C. Cultures were centrifuged for 2 min at 12,000 × g and resuspended to a final OD600 = 10 in fresh SD-CAA. 30 µl of this suspension was used to inoculate 270 µl of the appropriate media containing SD-CAA or SG-CAA with or without 25 mM Azp. Cells were then cultured overnight at 20 °C in 5-ml round-bottom tubes (Corning, 14-959-2). 15 µl of each of these cultures was used for analysis. Samples were washed three times in PBSA and resuspended in PBSA containing 500 µM Sulfo-dibenzocyclooctyne-biotin conjugate followed by incubation for 3 h at room temperature with agitation. These samples were washed three more times with PBSA and resuspended in a 1:500 dilution of both Avidin-PE (Thermo Fisher Scientific, A2660) and chicken anti-c-Myc (Thermo Fisher Scientific, A21281) in PBSA for 30 min at room temperature. Samples were washed three more times and used directly for analysis, or resuspended in a 1:250 dilution of AlexaFluor 488 goat anti-chicken IgY (Thermo Fisher Scientific, A11039) and incubated on ice for 20 min. Cells were then washed twice and analyzed. To detect expression and sortase activity, samples were resuspended in 200 µl PBSA and analyzed on a Guava flow cytometer (Millipore). Data were collected from 5,000 cells and analyzed using FlowJo software.
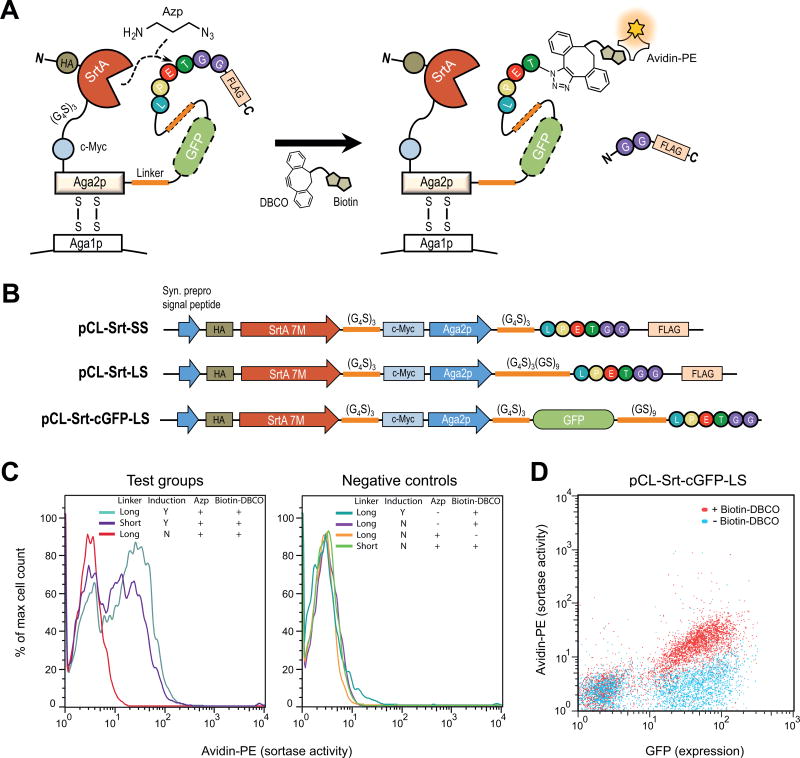
Figure 3.
Evaluation of bioconjugation enzyme activity on yeast using pCL vectors. (A, B) Schematic of protein expression and bioconjugation reactions on the yeast surface for pCL vectors co-expressing sortase A (SrtA) 7M, and its peptide substrate, LPETGG. (C) Detection of SrtA bioconjugation activity on yeast using pCL-Srt-LS (long linker) and pCL-Srt-SS (short linker) under various conditions. Samples were stained with avidin-PE to detect Azp conjugation followed by biotin Click chemistry, and also stained with AlexaFluor 488-conjugated antibodies to measure yeast expression levels. Histograms of PE signal (enzyme activity) as a percentage of the maximum cell number shows readily apparent sortase-active populations in each sample. (D) Detection of SrtA bioconjugation activity on yeast by using pCL-Srt-cGFP-LS (yEGFP plus long linker) with and without biotin-DBCO. Co-expression of yEGFP and SrtA eliminates the need for antibody-staining of epitope tags to quantify yeast surface expression levels.
3 Results
3.1 Dual protein expression vector design
Conventional yeast surface display strategies express a protein of interest as a fusion to the Aga2p subunit at either at the C-terminus (e.g. pCT [5, 11] and pYD1 (Invitrogen) vectors) or N-terminus (e.g. pTMY [27], pYD5 [34] and pCHA [35, 36] vectors) (Fig. 1A). Our dual protein yeast surface display vector (termed pCL) extends this system, displaying Aga2p flanked by two full-length proteins, one at the N-terminus and one at the C-terminus (Fig. 1B). In the pCL system, the first displayed protein is inserted between a synthetic α-factor prepro signal peptide [32] and the mature Aga2p (without its original signal peptide), fused to the N-terminus of Aga2p (Fig. 1C). The second displayed protein is inserted downstream of Aga2p and fused at the C-terminus of the mature protein. The dual protein expression cassette is led to the yeast secretory pathway by the prepro signal peptide and exported to the cell surface after being processed by the KEX2 endopeptidase that cleaves the C-terminus of Lys-Arg (KR) sequence attached at the end of the signal peptide [32, 33]. The inclusion of an EA (Glu-Ala) peptide spacer helps with efficient KEX2 cleavage of the fusion protein [34]. For the proof-of-concept studies described here, epitope tags were included both N- and C-terminal to Aga2p, as in the traditional pCT-based protein expression system, to validate expression of the fusion construct on the yeast cell surface.
3.2 A fluorescent protein fusion enables measurement of protein expression levels on the yeast cell surface
The original yeast display system uses antibody-labeling of N- or C-terminal epitope tags to quantify yeast surface expression levels by flow cytometry. In the pCL system, a fluorescent protein is fused to the N- or C-terminus of the Aga2p protein as a handle to measure expression of the entire fusion construct on yeast surface. The fluorescent protein utilized in our construct is a yeast-codon-optimized enhanced GFP (yEGFP) [28–30], although the modularity of the system theoretically allows for the use of any yeast-optimized fluorescent protein [37] to suit the characteristics (brightness, wavelength) desired by the user. A previous study showed that yEGFP is well-expressed as a C-terminal fusion to Aga2p on the widely used pCT vector [28]. To allow for a more flexible design, we confirmed that yEGFP is also functionally expressed at the N-terminus of Aga2p when it is inserted between the signal peptide and the mature protein (Supplementary Fig. 1).
Two versions of the dual protein expression vector were constructed: pCL-nGFP (yEGFP fused to the N-terminus of Aga2p) and pCL-cGFP (yEGFP fused to the C-terminus of Aga2p) (Supplementary Fig. 2). When analyzed using two-parameter flow cytometry scatter plots, both the N-terminal and the C-terminal yEGFP fusions showed distinct separation between expression-positive and expression-negative yeast cell populations (Supplementary Fig. 1). For both pCL-nGFP and pCL-cGFP, induction of fusion protein expression at 20 °C showed higher levels of GFP-positive cells compared to the 30 °C induction conditions. Therefore, all of the analyses this study were performed with yeast cells induced at 20 °C. The N-terminal yEGFP fusion exhibited lower levels of expression compared to the C-terminal yEGFP fusion, which was also confirmed by measurement of c-Myc expression levels.
3.3 Dual protein expression enables quantification of binding interactions on the yeast cell surface
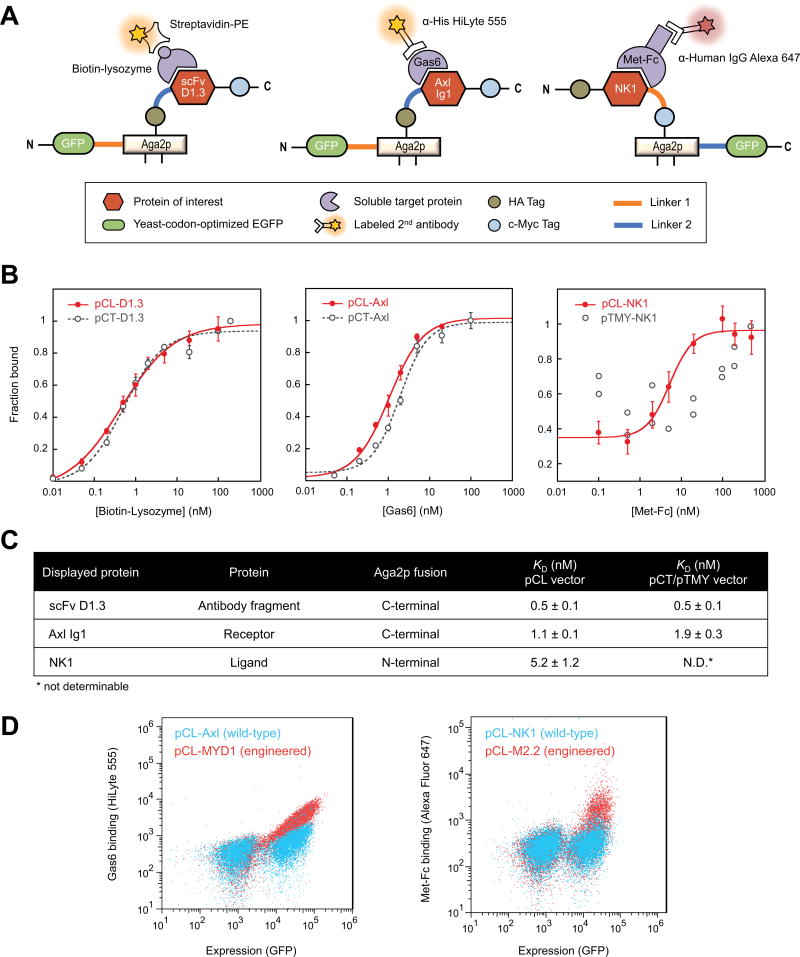
We next evaluated the reliability of the pCL-based strategy for measuring protein-protein binding interactions by testing three model proteins: 1) scFv D1.3, a murine antibody fragment that binds hen egg lysozyme [22, 38], 2) Axl Ig1, a wild-type receptor domain that binds its cognate ligand growth arrest specific 6 (Gas6) [39], and 3) NK1, a natural fragment of the N-terminal and first kringle domain of human hepatocyte growth factor ligand that binds Met receptor [27]. The scFv D1.3 and Axl Ig1 proteins were tethered through their N-terminus to the C-terminus of Aga2p, which in turn has yEGFP as an N-terminal fusion (Fig. 2A, left and middle panels, Supplementary Fig. 2A and 3). These constructs are termed pCL-nGFP-Aga2p-D1.3 (abbreviated pCL-D1.3) and pCL-nGFP-Aga2p-Axl (abbreviated pCL-Axl). The optimal construct orientation was determined by comparing expression of N-terminal and C-terminal Aga2p fusions of the protein-of-interest; for scFv D1.3, a C-terminal Aga2p fusion under induction at 20 °C was deemed the most optimal condition (Supplementary Fig. 3). To showcase the breadth of the system, NK1 was tethered through its C-terminus to the N-terminus of Aga2p, which has a C-terminal yEGFP fusion (Fig. 2A, right panel; Supplementary Fig. 2B). This construct is termed pCL-NK1-Aga2p-cGFP (abbreviated pCL-NK1). A liberated N-terminus of NK1 has been shown to be a preferable orientation for this protein [27].
Figure 2.
Quantification of protein-protein interactions on yeast using pCL vectors. (A) Schematic of protein display and antibody-staining strategies for the pCL vectors co-expressing a protein-of-interest and yEGFP on the same surface. (B) Binding curves comparing pCL and pCT/pTMY vectors for a lysozyme-binding scFv antibody fragment (left), a Gas6-binding Axl Ig1 receptor domain (middle), and the Met-binding NK1 ligand (right). Error bars correspond to the standard deviation of three independent measurements. (C) Equilibrium binding constants, KD, of yeast-displayed proteins expressed with the pCL or pCT/pTMY vectors. N.D.: Binding was not measurable for pTMY-NK1. (D) Wild-type proteins (Axl and NK1) and engineered variants (MYD1 [39] and M2.2 [27]), expressed using pCL vectors, can be differentiated at low target concentrations (0.1 nM Gas6 and 0.5 nM Met-Fc) on flow cytometry scatter plots.
Binding assays carried out with the dual protein display system are streamlined due to the constitutive fluorescence of yEGFP as a yeast surface expression read-out. After incubation with a soluble target protein, the induced yeast cells only require a single staining step with fluorophore-labeled secondary antibody against the binding target. This can be further reduced to a single-step binding assay when a soluble target is covalently labeled with a fluorescent dye. In contrast, assays involving conventional yeast display vectors require additional primary (and often secondary or sometimes tertiary), antibody staining steps for measuring protein expression levels through an N- or C-terminal epitope tag, resulting in multiple incubation and cell washing steps.
Using the simplified workflow described above, pCL-based yEGFP expression vectors were used to measure binding affinities of the three model proteins to their respective targets. The constructs pCL-D1.3, pCL-Axl and pCL-NK1 were expressed on the yeast surface and incubated with a corresponding binding partner in solution: biotinylated lysozyme (for scFv D1.3), Gas6 (for Axl Ig1), or Met-Fc (for NK1). Secondary antibodies labeled with fluorescent dyes were used to measure target-binding signals by flow cytometry (Fig. 2A). All the induced yeast cells showed a distinct GFP-positive population in scatter plots, enabling clear gating of the expressing population from which to quantify binding. Representative flow cytometry scatter plots for pCL-Axl at various Gas6 target concentrations are shown in Supplementary Figure 4.
Binding affinity measurements generated with the pCL system were compared with the results obtained from the epitope-tag-expressing conventional yeast display vectors, pCT (c-Myc expression) or pTMY (HA expression). Binding assays carried out with pCL-D1.3 (to lysozyme) and pCL-Axl (to Gas6) recapitulated equilibrium binding curves and respective affinities obtained with pCT-D1.3 and pCT-Axl (Fig. 2B, left and middle panels, and Fig. 2C). In contrast, binding of NK1 to Met-Fc was only measurable with protein expressed using the pCL-NK1 vector, and not the pTMY-NK1 construct (Fig. 2B, right panel, and Fig. 2C). NK1 protein displayed using the pTMY vector showed weak cell surface expression and negligible binding to Met-Fc (Supplementary Fig. 5), in agreement with our previous study [27].
To further validate the platform, we showed that a fusion construct displayed using the pCL vector (pCL-Axl) exhibited a linear correlation (R2 = 0.75) between the yEGFP signal and the c-Myc epitope tag expression signal on the same yeast cell (Supplementary Fig. 6A). As a final demonstration, we showed that the pCL system can qualitatively differentiate yeast-displayed wild-type proteins and previously engineered Axl or NK1 variants as measured by flow cytometry scatter plots (Fig. 2D) [27, 39]. These results demonstrate the applicability of the vectors for library construction and screening required for combinatorial protein engineering.
3.4 Additional considerations of the pCL display system
In addition to the time and cost benefit of not having to use anti-epitope antibodies to measure expression levels, the pCL-based yeast display strategy avoids a decrease in fluorescence signals which is derived from antibody or target dissociation due to multiple wash steps used in the traditional pCT system. As shown in Supplementary Figure 6B, each additional cell washing step significantly decreased c-Myc expression signals in the pCT group, demonstrating dissociation of the anti-c-Myc primary antibody from the cell yeast surface. In contrast, GFP expression signals remained constant in the pCL group, regardless of additional washing steps. Eliminating one or two washing steps for sample preparation may be especially beneficial for studying a protein with low expression or weak target binding on the yeast cell surface. Moreover, the fluorescent signal from yEGFP expression was stable at room temperature when analyzed over a 72 h period (Supplementary Fig. 6C), which spans the time window generally used for measuring kinetic dissociation rates or screening a yeast-displayed library based on kinetic off-rate parameters [40, 41].
3.5 Dual protein expression enables quantification of enzyme-substrate bioconjugation reactions on the yeast cell surface
We next investigated whether the pCL system could be applied to quantify the reaction between a bioconjugation enzyme and its substrates. In this case we developed a pCL-based enzyme-substrate system to measure sortase activity on the yeast cell surface, a simplified design from the elegant display technology previously developed by the Liu lab [15]. Sortase is a transpeptidase enzyme that has widespread use in bioconjugation reactions. The sortase enzyme from Staphylococcus aureus and its engineered derivatives 5M and 7M [15] recognize a LPXTG sequence and exchange the terminal glycine for a variety of nucleophiles, such as glycine-terminal peptides [42], hydrazides [43], and various primary amines [44, 45]. We previously showed that by using non-natural amine nucleophiles, we could label proteins in living E. coli cells [46].
We introduced the calcium-independent version of sortase, 7M, into the pCL vector and tested its ability to incorporate the non-biological amine 3-azido-1-propanamine (Azp) into LPETGG sequences on the surface of yeast. Sortase 7M was fused to N-terminus of Aga2p, followed by: 1) a short linker, 2) a long linker, or 3) yEGFP plus a long linker C-terminal to Aga2p (Fig. 3A and B). Each of these constructs contained a C-terminal LPETGG substrate sequence so that the sortase variant could carry out its enzymatic modification. The linkers were intended to test accessibility of sortase to LPETGG with different flexibilities and distances between the enzyme and the substrate.
The constructs were transformed into yeast and induced at 20 °C in the presence of Azp in the induction media. In each case, the sortase 7M variant and the linker-LPETGG were successfully expressed on yeast as shown by c-Myc staining or yEGFP fluorescence (Supplementary Fig. 7). Moreover, each enzyme-substrate construct was able to facilitate conjugation with Azp, followed by a Copper-free Click reaction between the incorporated Azp and sulfo-biotin DBCO. Biotin-conjugated 7M fusions were detected by staining with PE-labeled avidin (Fig. 3C, left). Negative controls lacking Azp or reactions carried out with yeast cultured in non-inducing media suggested that the Click reaction was specific to Azp and the LPETGG substrate sequence (Fig. 3C, right). Both the long linker variant (pCL-Srt-LS) and the short linker variant (pCL-Srt-SS) showed substantial bioconjugation, indicating that they allow sufficient proximity of the C-terminal substrate to the N-terminal enzyme (Fig. 3C, left).
Finally, a bioconjugation reaction specific to sulfo-biotin DBCO was also observed when yEGFP was included in the construct (pCL-Srt-cGFP-LS; Fig. 3B and D). 7M-expressing cells were readily distinguished by yEGFP fluorescence, and active sortase was again detected by avidin-PE using flow cytometry. Based on the close proximity of N-terminus and C-terminus of GFP, this construct does not interfere with LPETGG substrate access to the sortase active site. Similar to the binding assays described above, the GFP fusion construct has the advantage of not requiring a second antibody stain for expression, facilitating rapid analysis of cell surface chemistry.
4 Discussion
In this study, we showed that the pCL yeast display system enables co-expression of two proteins: a protein of interest and a fluorescent protein, or an enzyme and substrate pair, to facilitate protein binding assays or catalytic reactions, respectively. Considering the relatively high price of antibodies against epitope tags (such as c-Myc, FLAG, and HA), the pCL vectors containing yEGFP provide a cost-effective and simple means for measuring protein expression on the yeast cell surface. Moreover, we showed that the yEGFP signals generated from the pCL system are stable and correlate with expression levels of co-expressed proteins of interest, thus providing an alternative analysis method for yeast surface display. While direct fusion of a yEGFP to the N- or C-terminus of the displayed protein in Figure 1A is possible, the pCL system uses Aga2p as a spacer to minimize the effects of a bulky fluorescent protein on sterics and folding. Although the current vectors used in this study utilize yEGFP to quantify a protein expression, which shows optimal induction of yeast expression at 20 °C, other fluorescent proteins [37] with improved expression and stability could be used in future studies. Two pCL vectors with N-terminal or C-terminal Aga2p co-expression strategies provide flexibility for yeast surface display, and optimal expression conditions should be determined empirically for each protein-of-interest (Supplementary Fig. 3). We have also created a modified pCL vector (termed pCL2) containing codon-optimized linkers and additional restriction sites, suitable for facile and modular cloning and library construction (Supplementary Fig. 8). pCL2 comprises the C-terminal Aga2p fusion of yEGFP, which will minimize selection of false-positive fluorescent clones with frameshift mutations arising in error-prone PCR libraries.
The pCL dual protein display system also enables measurement of enzyme-mediated bioconjugation on the surface of yeast. The co-expression of sortase and the substrate sequence as a single fusion protein imparts a distinct advantage for incorporation of non-natural nucleophiles in yeast over a previously developed yeast display system for bioconjugation [15]. First, the pCL-based approach does not require solid-phase peptide synthesis of substrate sequences, and has no need for a secondary enzyme (in the previous study, phosphopantotheinyl transferase) to attach sortase substrates to the surface. Second, the pCL vector can be used with widely available EBY100 yeast, rather than the modified strain required in the previous system. Third, the platform is flexible, and a library of enzyme variants or peptide substrates could be created and screened using the pCL system. Thus, yeast display using the pCL dual protein display system provides simplicity and generalizability in the interrogation of enzymatic activity as well as protein–protein binding interactions. In addition, the pCL system has potential applications to studies where display of multiple proteins on the same yeast cell is desired, including split fluorescent protein engineering or sequential assembly of multi-enzyme cascades.
Supplementary Material
Acknowledgments
The authors thank Eric V. Shusta (U Wisconsin-Madison) for providing DNA encoding for yEGFP, K. Dane Wittrup (MIT) for the pCT plasmid encoding for scFv D1.3, and Mihalis S. Kariolis for valuable discussions on the initial construct design. This work was supported by National Institutes of Health (NIH) NCI R01 CA151706, the Howard Hughes Medical Institute International Student Research Fellowship (S.L.), a Stanford Bio-X Interdisciplinary Fellowship (S.L.), the Stanford University Medical Scientist Training Program T32GM007365 (M.F.I.), and the Stanford Medical Scholars Research Program (E.M.S.). Certain commercial equipment, instruments, or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.
Abbreviations
- yEGFP
yeast-codon-optimized GFP
- SrtA
Sortase A
- Azp
3-azido-1-propanamine
- DBCO
dibenzocyclooctyne
- PE
R-Phycoerythrin
Footnotes
Conflict of interest
The authors declare no financial or commercial conflict of interest.
References
- 1.Wittrup KD. Protein engineering by cell-surface display. Curr. Opin. Biotechnol. 2001;12:395–399. doi: 10.1016/s0958-1669(00)00233-0. [DOI] [PubMed] [Google Scholar]
- 2.Lee SY, Choi JH, Xu Z. Microbial cell-surface display. Trends Biotechnol. 2003;21:45–52. doi: 10.1016/s0167-7799(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 3.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 4.Francisco JA, Campbellt R, Iversont BL, Georgiou G. Production and fluorescence-activated cell sorting of Escherichia coli expressing a functional antibody fragment on the external surface. PNAS. 1993;90:10444–10448. doi: 10.1073/pnas.90.22.10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 6.Ernst W, Grabherr R, Wegner D, Borth N, et al. Baculovirus surface display: construction and screening of a eukaryotic epitope library. Nucleic Acids Res. 1998;26:1718–1723. doi: 10.1093/nar/26.7.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho M, Nagata S, Pastan I. Isolation of anti-CD22 Fv with high affinity by Fv display on human cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9637–9642. doi: 10.1073/pnas.0603653103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beerli RR, Bauer M, Buser RB, Gwerder M, et al. Isolation of human monoclonal antibodies by mammalian cell display. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14336–14341. doi: 10.1073/pnas.0805942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Packer MS, Liu DR. Methods for the directed evolution of proteins. Nat. Rev. Genet. 2015;16:379–394. doi: 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- 10.Boder ET, Bill JR, Nields AW, Marrack PC, et al. Yeast surface display of a noncovalent MHC class II heterodimer complexed with antigenic peptide. Biotechnol. Bioeng. 2005;92:485–491. doi: 10.1002/bit.20616. [DOI] [PubMed] [Google Scholar]
- 11.Chao G, Lau WL, Hackel BJ, Sazinsky SL, et al. Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 12.Shusta EV, Holler PD, Kieke MC, Kranz DM, et al. Directed evolution of a stable scaffold for T-cell receptor engineering. Nat. Biotechnol. 2000;18:754–759. doi: 10.1038/77325. [DOI] [PubMed] [Google Scholar]
- 13.Traxlmayr MW, Obinger C. Directed evolution of proteins for increased stability and expression using yeast display. Arch. Biochem. Biophys. 2012;526:174–180. doi: 10.1016/j.abb.2012.04.022. [DOI] [PubMed] [Google Scholar]
- 14.Lipovšek D, Antipov E, Armstrong KA, Olsen MJ, et al. Selection of horseradish peroxidase variants with enhanced enantioselectivity by yeast surface display. Chem. Biol. 2007;14:1176–1185. doi: 10.1016/j.chembiol.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Chen I, Dorr BM, Liu DR. A general strategy for the evolution of bond-forming enzymes using yeast display. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11399–11404. doi: 10.1073/pnas.1101046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuda T, Ishikawa T, Ogawa M, Shiraga S, et al. Enhancement of cellulase activity by clones selected from the combinatorial library of the cellulose-binding domain by cell surface engineering. Biotechnol. Prog. 2006;22:933–938. doi: 10.1021/bp060129y. [DOI] [PubMed] [Google Scholar]
- 17.Cherf GM, Cochran JR. Applications of yeast surface display for protein engineering. Methods Mol. Biol. 2015;1319:155–175. doi: 10.1007/978-1-4939-2748-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka T, Yamada R, Ogino C, Kondo A. Recent developments in yeast cell surface display toward extended applications in biotechnology. Appl. Microbiol. Biotechnol. 2012;95:577–591. doi: 10.1007/s00253-012-4175-0. [DOI] [PubMed] [Google Scholar]
- 19.Kuroda K, Ueda M. Arming technology in yeast—novel strategy for whole-cell biocatalyst and protein engineering. Biomolecules. 2013;3:632–650. doi: 10.3390/biom3030632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cappellaro C, Baldermann C, Rachel R, Tanner W. Mating type-specific cell-cell recognition of Saccharomyces cerevisiae: cell wall attachment and active sites of a- and alpha-agglutinin. EMBO J. 1994;13:4737–4744. doi: 10.1002/j.1460-2075.1994.tb06799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy A, Lu CF, Marykwas DL, Lipke PN, et al. The AGA1 product is involved in cell surface attachment of the Saccharomyces cerevisiae cell adhesion glycoprotein a-agglutinin. Mol. Cell. Biol. 1991;11:4196–4206. doi: 10.1128/mcb.11.8.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.VanAntwerp JJ, Wittrup KD. Fine affinity discrimination by yeast surface display and flow cytometry. Biotechnol. Prog. 2000;16:31–37. doi: 10.1021/bp990133s. [DOI] [PubMed] [Google Scholar]
- 23.van den Beucken T, Pieters H, Steukers M, van der Vaart M, et al. Affinity maturation of Fab antibody fragments by fluorescent-activated cell sorting of yeast-displayed libraries. FEBS Lett. 2003;546:288–294. doi: 10.1016/s0014-5793(03)00602-1. [DOI] [PubMed] [Google Scholar]
- 24.Furukawa H, Tanino T, Fukuda H, Kondo A. Development of novel yeast cell surface display system for homo-oligomeric protein by coexpression of native and anchored subunits. Biotechnol. Prog. 2006;22:994–997. doi: 10.1021/bp0601342. [DOI] [PubMed] [Google Scholar]
- 25.Liu F, Banta S, Chen W, Osman MH, et al. Functional assembly of a multi-enzyme methanol oxidation cascade on a surface-displayed trifunctional scaffold for enhanced NADH production. Chem. Commun. 2013;49:3766–3768. doi: 10.1039/c3cc40454d. [DOI] [PubMed] [Google Scholar]
- 26.Fujita Y, Ito J, Ueda M, Fukuda H, et al. Synergistic saccharification, and direct fermentation to ethanol, of amorphous cellulose by use of an engineered yeast strain codisplaying three types of cellulolytic enzyme. Appl. Environ. Microbiol. 2004;70:1207–1212. doi: 10.1128/AEM.70.2.1207-1212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones DS, Tsai P-C, Cochran JR. Engineering hepatocyte growth factor fragments with high stability and activity as Met receptor agonists and antagonists. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13035–13040. doi: 10.1073/pnas.1102561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang D, Shusta EV. Secretion and surface display of green fluorescent protein using the yeast Saccharomyces cerevisiae. Biotechnol. Prog. 2005;21:349–357. doi: 10.1021/bp0497482. [DOI] [PubMed] [Google Scholar]
- 29.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 30.Cormack BP, Bertram G, Egerton M, Gow NAR, et al. Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 31.Johnston M, Davis RW. Sequences that regulate the divergent GAL1–GAL10 promoter in Saccharomyces cerevisiae. Mol. Cell. Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clements JM, Catlin GH, Price MJ, Edwards RM. Secretion of human epidermal growth factor from Saccharomyces cerevisiae using synthetic leader sequences. Gene. 1991;106:267–271. doi: 10.1016/0378-1119(91)90209-t. [DOI] [PubMed] [Google Scholar]
- 33.Julius D, Brake A, Blair L, Kunisawa R, et al. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-α-factor. Cell. 1984;37:1075–1089. doi: 10.1016/0092-8674(84)90442-2. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Mathias A, Stavrou S, Neville DM. A new yeast display vector permitting free scFv amino termini can augment ligand binding affinities. Protein Eng. Des. Sel. 2005;18:337–343. doi: 10.1093/protein/gzi036. [DOI] [PubMed] [Google Scholar]
- 35.Van Deventer JA, Kelly RL, Rajan S, Wittrup KD, et al. A switchable yeast display/secretion system. Protein Eng. Des. Sel. 2015;28:317–325. doi: 10.1093/protein/gzv043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mata-Fink J, Kriegsman B, Yu HX, Zhu H, et al. Rapid conformational epitope mapping of anti-gp120 antibodies with a designed mutant panel displayed on yeast. J. Mol. Biol. 2013;425:444–456. doi: 10.1016/j.jmb.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee S, Lim WA, Thorn KS. Improved blue, green, and red fluorescent protein tagging vectors for S. cerevisiae. PLoS One. 2013;8:e67902. doi: 10.1371/journal.pone.0067902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hawkins RE, Russell SJ, Baier M, Winter G. The contribution of contact and non-contact residues of antibody in the affinity of binding to antigen: the interaction of mutant D1.3 antibodies with lysozyme. J. Mol. Biol. 1993;234:958–964. doi: 10.1006/jmbi.1993.1650. [DOI] [PubMed] [Google Scholar]
- 39.Kariolis MS, Miao YR, Jones DS, Kapur S, et al. An engineered Axl “decoy receptor” effectively silences the Gas6-Axl signaling axis. Nat. Chem. Biol. 2014;10:977–983. doi: 10.1038/nchembio.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boder ET, Wittrup KD. Optimal screening of surface-displayed polypeptide libraries. Biotechnol. Prog. 1998;14:55–62. doi: 10.1021/bp970144q. [DOI] [PubMed] [Google Scholar]
- 41.Orcutt KD, Wittrup KD. Antibody Engineering. Springer Berlin Heidelberg; Berlin, Heidelberg: 2010. Yeast display and selections; pp. 207–233. [Google Scholar]
- 42.Antos JM, Truttmann MC, Ploegh HL. Recent advances in sortase-catalyzed ligation methodology. Curr. Opin. Struct. Biol. 2016;38:111–118. doi: 10.1016/j.sbi.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li YM, Li YT, Pan M, Kong XQ, et al. Irreversible site-specific hydrazinolysis of proteins by use of sortase. Angew. Chemie - Int. Ed. 2014;53:2198–2202. doi: 10.1002/anie.201310010. [DOI] [PubMed] [Google Scholar]
- 44.Samantaray S, Marathe U, Dasgupta S, Nandicoori VK, et al. Peptide-sugar ligation catalyzed by transpeptidase sortase: A facile approach to neoglycoconjugate synthesis. J. Am. Chem. Soc. 2008;130:2132–2133. doi: 10.1021/ja077358g. [DOI] [PubMed] [Google Scholar]
- 45.Matsumoto T, Takase R, Tanaka T, Fukuda H, et al. Site-specific protein labeling with amine-containing molecules using Lactobacillus plantarum sortase. Biotechnol. J. 2012;7:642–648. doi: 10.1002/biot.201100213. [DOI] [PubMed] [Google Scholar]
- 46.Glasgow JE, Salit ML, Cochran JR. In vivo site-specific protein tagging with diverse amines using an engineered sortase variant. J. Am. Chem. Soc. 2016;138:7496–7499. doi: 10.1021/jacs.6b03836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.