SUMMARY
A major challenge in understanding the cellular diversity of the brain has been linking activity during behavior with standard cellular typology. For example, it has not been possible to determine whether principal neurons in prefrontal cortex active during distinct experiences represent separable cell types, and it is not known whether these differentially active cells exert distinct causal influences on behavior. Here, we develop quantitative hydrogel-based technologies to connect activity in cells reporting on behavioral experience with measures for both brain-wide wiring and molecular phenotype. We find that positive and negative-valence experiences in prefrontal cortex are represented by cell populations that differ in their causal impact on behavior, long-range wiring, and gene expression profiles, with the major discriminant being expression of the adaptation-linked gene NPAS4. These findings illuminate cellular logic of prefrontal cortex information processing and natural adaptive behavior and may point the way to cell-type-specific understanding and treatment of disease-associated states.
INTRODUCTION
The many regions and layers of the mammalian prefrontal cortex are known to contain cells with a rich diversity of activity patterns. Indeed, otherwise-indistinguishable populations of principal cells exhibiting profoundly distinct changes in activity in response to stimulus have been characterized by electrophysiological recording and cellular-resolution fluorescence Ca2+ imaging (Hayden et al., 2008; Insel and Barnes, 2015; Ito et al., 2003; Matsumoto et al., 2007; Pinto and Dan, 2015; Warden et al., 2012). At the same time, datastreams of anatomical and molecular information on prefrontal cell typology have emerged from a variety of methods, also pointing toward a rich diversity of principal neurons despite the traditional view that these cells were more homogenous than the highly diverse and readily separable interneurons (Hof et al., 1996; Lee et al., 2014; Soloway et al., 2002). These findings have highlighted the morphological, wiring, and electrophysiological diversity of principal neurons even within individual layers and subregions.
However, the mapping and correspondences (if any) among these different domains of diversity (activity during behavior, long-range wiring, and molecular phenotype) have remained largely unclear within prefrontal cortex. Addressing this issue would have fundamental implications for elucidating the cellular logic of prefrontal cortex function; moreover, differences in wiring, role in behavior, and molecular signatures among differentially responsive cells could provide long-elusive insight into the mechanisms of action of current neuromodulation therapies and even lay the foundation for developing new kinds of cell-targeted disease treatment. But the unique and non-stereotyped nature of each mammalian nervous system has prevented experimental resolution of this fundamental question, since cellular-resolution activity in a brain during behavior would have to be directly linked to quantitative local and global wiring, as well as to molecular analysis of the very same cells in the same mammalian brain, which has not yet been possible. Here, we address this question by developing approaches to quantify long-range anatomy, molecular signatures, and the causal impact of prefrontal cortical cells, defined by activity during distinct experiences.
RESULTS
Behavioral Cohort-Scale Whole-Brain Activity Mapping
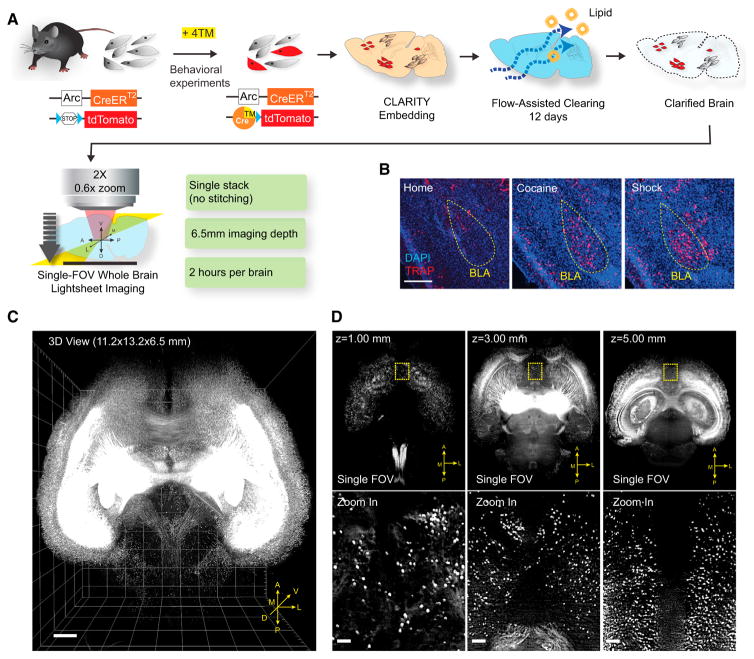
We began with development and integration of enhanced Arc-TRAP and CLARITY technologies. The ArcTRAP transgenic mouse line was previously shown to translate temporally delimited neuronal activity into fluorescence via CreER/tamoxifen-mediated recombination (Figure 1A) (Guenthner et al., 2013). While the ArcTRAP line had been reported to have high background recombination, we developed an aqueous formulation of the fast-acting tamoxifen (4-hydroxytamoxifen, 4TM) enabling dosage reduction by an order of magnitude; this approach to ArcTRAP was found to eliminate the background recombination (Figures 1B and S1B). ArcTRAP mice can then be exposed to a rewarding or aversive experience (e.g., cocaine dosing at 15 mg/kg or a series of 0.5 mA/2 s foot shocks, respectively) with resulting robust and faithful labeling of neurons (Figure S1A); for example, histology confirmed that both experience types (with comparable but opposite conditioning value; Figures S1C and S1D) recruited neurons in basolateral amygdala (BLA; which is known to be involved in both reward and aversion processing) (Hsiang et al., 2014) (Figure 1B).
Figure 1. Behavioral Cohort-Scale Brain-wide Activity Mapping.
(A) Schematic of ArcTRAP labeling and the enhanced cohort-scale CLARITY pipeline for rapid whole-brain clearing and imaging. CreER expression is driven by the activity-dependent Arc promoter to mediate 4TM-dependent recombination that permanently labels the active neurons with tdTomato.
(B) Representative confocal images from 40-μm sections showing TRAP labeling in BLA (yellow circle). Scale bar, 400 μm.
(C) 3D rendering of a CLARITY-processed whole-mouse brain (ArcTRAP) imaged by light-sheet microscopy. Scale bar, 500 μm.
(D) Top: single field of view (FOV) images at the indicated imaging depths. Bottom: zoomed-in images from the yellow-boxed regions in the top row showing cellular resolution. Scale bar, 100 μm.
See also Figure S1.
This approach brings untapped potential for obtaining deeper perspective on the cells activated, including insight into brain-wide patterns and molecular properties of cells recruited. Traditional thin-section histology would be prohibitively labor-intensive for whole-brain analysis in large behavioral-scale cohorts and also subject to sampling loss and bias, but the recent emergence of tissue transparency techniques, such as CLARITY, has, in principle, enabled lossless high-resolution brain-wide imaging compatible with molecular analysis (Chung et al., 2013; Tomer et al., 2014), if not yet in large cohorts of subjects. We therefore developed an enhanced CLARITY process suitable for this challenge of large-cohort analysis linked to behavioral experience; key capabilities achieved included much faster speed and lower cost, as well as automated analysis algorithms adapted to both TRAP labeling and widely available commercial light-sheet microscope hardware (Figures 1A, 1C, 1D, and S1E–S1H; see the Experimental Procedures).
Employing this integrated TRAP/CLARITY approach, multiple cohorts (with temporally delimited exposure to appetitive cocaine dosing, aversive foot shock, or neither experience; n = 5 mice per group) were processed using two-color imaging on the light-sheet microscope. Image volumes taken in the green (autofluorescence) channel were iteratively co-registered, using an intensity-based non-linear registration algorithm, to generate an average reference brain. The registration based on the green channel from each brain was then used to warp the red channel (consisting of the TRAP signal) for each brain into this reference space (Figure S1I).
We first manually delineated potentially relevant regions in the reference brain, including mPFC, NAc (nucleus accumbens), BLA, BNST (bed nucleus of the stria terminalis), LH (lateral hypothalamus), PVN (paraventricular nucleus of hypothalamus), and CeA (central nucleus of the amygdala). At this level of inspection, the singly defined regional levels of neuronal activation were found to be largely similar between the two very different kinds of experience (Figures 2A–2D and S2H–S2J; Table S1), highlighting the need for a more refined analysis. To search for patterns in cell labeling that could differentiate the two conditions despite this apparent similarity, we manually registered the Allen Brain Atlas (ABA) (Oh et al., 2014) to the CLARITY reference. Locations of experience-recruited cells were also mapped into the reference, so that the number of recruited cells in every brain region could be quantified in an unbiased fashion (Menegas et al., 2015); changes in TRAP cell numbers (relative to home cage-animal mean values) in ~200 brain regions were then automatically quantified (Figures S2A–S2G).
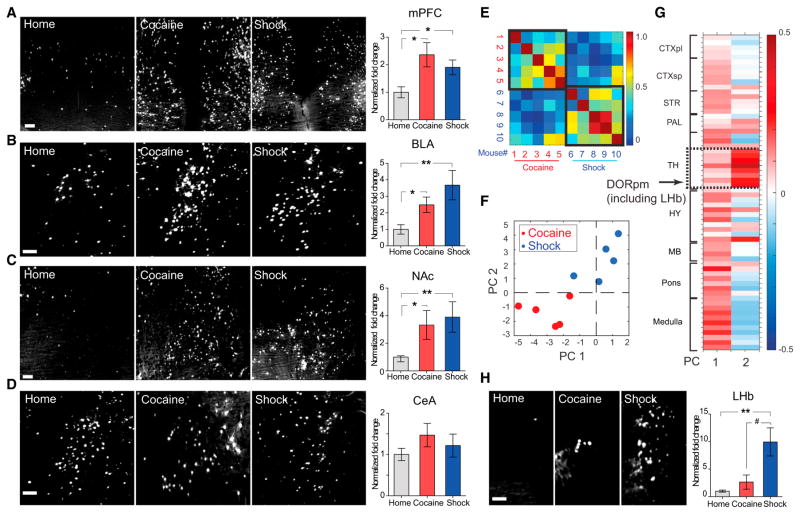
Figure 2. Cocaine and Shock Recruit Overlapping Brain Regions.
(A–D) TRAP cells in manually annotated regions. Left: representative images taken at the center of the indicated regions (max-projection of 100 μm volume). Scale bars, 100 μm. Right: fold change in TRAP cell numbers (normalized to home cage).
(E) Pearson correlation among the ten mice, based on the r-value computed from fold-activation changes relative to home cage across all non-zero-containing brain regions. Note the higher brain-wide correlation values within behavioral groups (black bounding boxes) compared to across-groups.
(F) Locations of individual mice projected into the 2D space of the two principal components (PCs) comprising the majority of the variance (in arbitrary PC units), where the position of each mouse corresponds to the extent to which a particular principal component accounts for that mouse’s variance across all brain regions.
(G) Principal component coefficients (in arbitrary PC weight units) across brain areas—the contribution of each brain area to each principal component—were summarized as clusters of proximal regions. Note the distinct region-selective contribution to PC 2 (dashed box; detailed in text). CTXpl/sp, cortical plate/subplate; SRT, striatum; PAL, pallidum; TH, thalamus; HY, hypothalamus; MB, midbrain; DORpm, polymodal association cortex-related dorsal thalamus.
(H) Representative image and quantification of TRAP cells in LHb. Scale bar, 100 μm. For all panels, n = 5 per group, *p < 0.05, **p < 0.01, unpaired t test comparing behavioral group to home cage; # p < 0.05, unpaired t test comparing cocaine versus shock group. All p values were adjusted for multiple comparisons using the false discovery rate method. Error bars, mean ±SEM.
Individual single-region activation patterns consistent across subjects within a group largely agreed with the manual analysis. For example, robust increases in TRAPped cells were observed in both settings within mPFC, NAc, BLA, pallidal regions (containing BNST), and hypothalamus (Figure S2K). However, the change in active cell count across all regions revealed common multi-region correlation patterns in mice from the same experimental group, suggesting experience-specific brain-wide patterns (Figure 2E). To investigate these differences, we applied principal component analysis (PCA) to the matrix of activated cell-count changes in cocaine- or shock-labeled mice. The first two principal components (PCs) comprised >75% of the variance for the dataset (Figure S2L) and thus were examined more closely. Plotting the position of each mouse in the space defined by the first two PCs revealed a clear separation between the behavioral conditions (Figure 2F).
To better understand the source of this separation, we examined the PC loadings across all of the included brain areas (Figure 2G). The first PC was found to reflect more general differences in activated cell recruitment across all regions (e.g., higher mean change across all regions in shock versus control mice; p < 0.05, t test). The second PC, by contrast, revealed a specific correlated group of DORpm (polymodal association cortex-related dorsal thalamus) regions, containing the lateral habenula (LHb), a key nucleus for aversive behaviors (Li et al., 2011, 2013). These regions were more strongly recruited by shock than by cocaine, revealing a selective responsiveness to the aversive experience. Consistently, manual quantification also found that shock more than cocaine, activated the LHb (p < 0.01 for shock versus home-cage experience; p < 0.05 for shock versus cocaine experience; Figure 2H). Thus, this behavioral cohort-scale TRAP/CLARITY labeling revealed brain-wide patterns of neuronal activation, as well as quantitative region-specific activation differences at the cellular level between different experimental conditions.
Resolving mPFC Populations and Projections Activated by Appetitive or Aversive Experience
Similarity in activation pattern by appetitive and aversive experiences has been reported in individually selected brain regions (verified broadly, though not in all regions, by the brain-wide analysis here) (Johnson et al., 2010; Xiu et al., 2014). A falsifiable hypothesis would be that the same neuron-type distribution was recruited by the two stimuli. In mPFC, the existing literature does not support or falsify this hypothesis, though mPFC is associated with specific reward and aversion processes (including cocaine-conditioned place preference, (Tzschentke and Schmidt, 1998, 1999), fear and anxiety behaviors (Burgos-Robles et al., 2009; Lammel et al., 2011; Shah et al., 2004; Shah and Treit, 2003), and more general functions potentially relevant to the single-population hypothesis (including attention, salience- and novelty-detection, and working memory) (Dalley et al., 2004; Miller, 2000). The region-specific brain-wide analysis here may open the door to testing a distinct hypothesis—that appetitive and aversive experience recruit distinct neuron-type populations. Axonal projection anatomy is one of the most important features that might resolve principal cell population types involved in such distinct processes (Jennings et al., 2013; Kim et al., 2013; Lammel et al., 2012; Namburi et al., 2015), but this feature has been difficult to explore in a brain-wide fashion, while remaining linked (at the cellular level) to function during behavior.
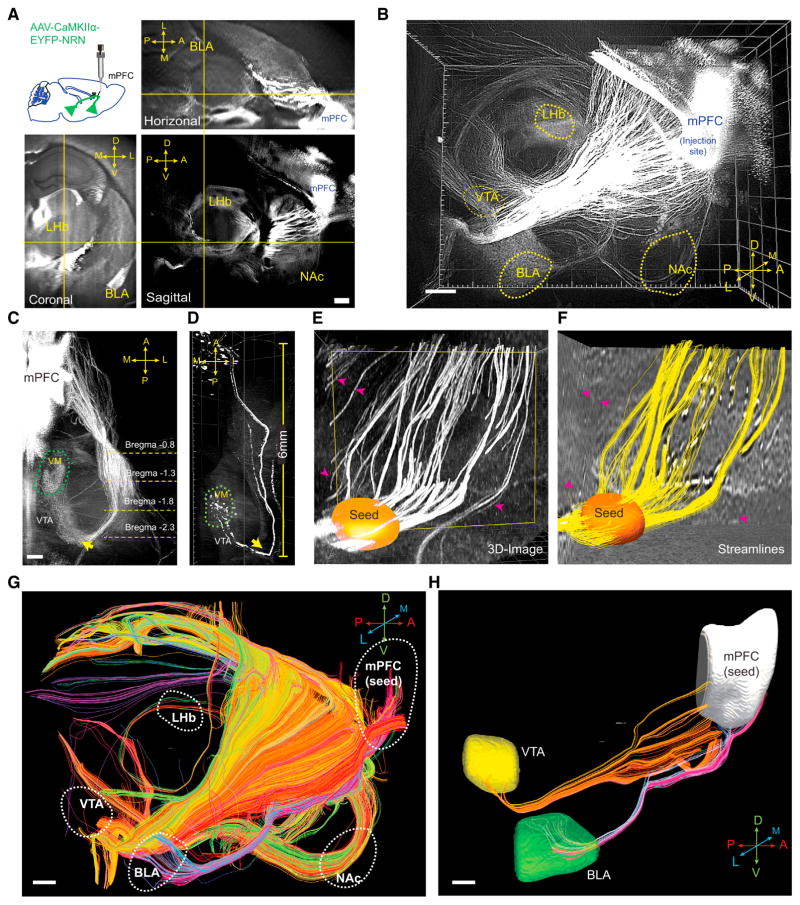
A very strongly expressed activity-dependent cell-filling label (unlike traditional nuclear c-fos immunostaining or transiently or transgenically expressed fluorophores) in principle might allow for acquisition of this crucial wiring information as well from the same experimental subjects, provided that axon tracts of labeled neurons could be robustly imaged and quantified in this context. We therefore developed a CLARITY-optimized axonal-filling fluorescent protein, engineered, in part, by inserting the 3′ UTR of neuritin (NRN) RNA (Akten et al., 2011) at the C terminus of EYFP (Figures S3A and S3B). We found that this construct could be packaged into high-titer adeno-associated virus (AAV) capsids that indeed enabled focal injection-defined projection labeling in CLARITY; for example, efferent mPFC projections could be readily followed throughout the entire adult mouse brain after a single stereotaxic injection (Figures 3A and 3B). Visualizing axonal tracks in 3D revealed key topographical features that were difficult, if not impossible, to detect in thin 2D sections (Figures 3A and S3C); for example, a prominent axon bundle traveling from mPFC to ventral medial thalamus was observed to carry out a sharp U turn near the VTA (Figures 3C and 3D), a potentially important feature that has not been described in existing atlases (Figure S3D).
Figure 3. CLARITY Enables Brain-wide Origin/Target-Defined Projection Mapping.
(A) 2D orthogonal views of a mouse brain. Insert shows schematic for location of viral injection. D, dorsal; V, ventral; A, anterior; P, posterior; L, lateral; M, medial.
(B) 3D rendering of CLARITY hemisphere, visualizing outgoing mPFC projections (imaged by 2× objective at 0.8× zoom with a single FOV, step size: 4 μm, 1,000 steps).
(C) 3D visualization of the axonal bundle projecting from mPFC to VM (ventral medial thalamus), showing tracts turning near the VTA (indicated by arrows).
(D) Visualizing the same projection in (C) with sparse labeling by injecting a smaller volume of the same virus (25 nL).
(E) Raw image from a CLARITY volume. Orange: user-defined “seed region” so that only the fibers passing this region were tracked.
(F) Streamlines reconstructed from (E), using structural tensor-based tractography. Note that fibers in the CLARITY image that did not pass the user-defined seed region were excluded in the reconstruction (indicated by the magenta arrows).
(G) Reconstructed brain-wide streamlines from CLARITY image in (B). The streamlines are color-coded for orientation. A–P, red; D–V, green; L–M, blue.
(H) Representative computational isolation of mPFC fibers that project to VTA (yellow) or BLA (green).
Scale bars, 500 μm.
See also Figure S3.
Quantifying these tracts was also an unresolved challenge, and we therefore developed a method to compute 3D structure tensors from CLARITY images for tractography (Figures S3E–S3H). Faithful reconstruction of calculated streamlines was achieved (using tools adapted from diffusion tractography; Supplemental Experimental Procedures); these streamlines mapped onto fibers from CLARITY images (Figures 3E and 3F), and importantly, the streamline count in each bundle tightly correlated with the ground-truth physical diameter of the axonal bundles (Figure S3I). Using this method, we reconstructed whole-brain projections based on CLARITY images (Figure 3G); connectivity between a seed region (defined by stereotaxic injection site) and any specified target could be readily visualized and assessed by counting streamlines (Figures 3H and S3J–S3M).
To integrate this capability, with the needed additional capability of projection-labeling in cells defined by their use during experience, we sought to develop a viral CreER/4TM strategy to translate time-locked activity to sustained transgene expression. We therefore engineered a c-Fos promoter combining minimal promoter and regulatory elements in intron-1 (Figure S4A) (Barth et al., 2004; Schilling et al., 1991; Smeyne et al., 1992) that can be packaged into AAV particles and is specific enough to capture neuronal activity (Figures S4B–S4D). We also inserted a destabilized ER-Cre-ER-PEST cassette under this promoter (Kawashima et al., 2013; Li et al., 1998; Matsuda and Cepko, 2007); when injected into a reporter mouse, this viral CreER/4TM system reliably enabled activity- and tamoxifen-dependent cell-body and projection labeling (Figures S4E and S4F).
A final essential feature for quantitative activity-dependent projection mapping was normalization on an individual level to the absolute tract labeling strength independent of activity; this normalization is crucial in a virus-based approach to control for variation in injection efficacy. We enabled this feature (Figure 4A) by building in simultaneous two-color activity-independent (structural, EYFP) labeling and activity-dependent (tdTomato) labeling of projections from the same injection site, in which these two viruses infected cells with a stable relative ratio across animals (Figures S4G and S4H). Dual-color quantification of projections to multiple downstream regions is then achieved by counting the number of streamlines ending in these regions, and the activity-dependence is corrected for anatomical and injection variability from the red/green streamline ratio at the individual animal level. This quantification of projection use across the brain from behaviorally defined neuronal populations is (for brevity) termed here CLARITY-based activity projection tracking upon recombination, or CAPTURE (Figure 4A).
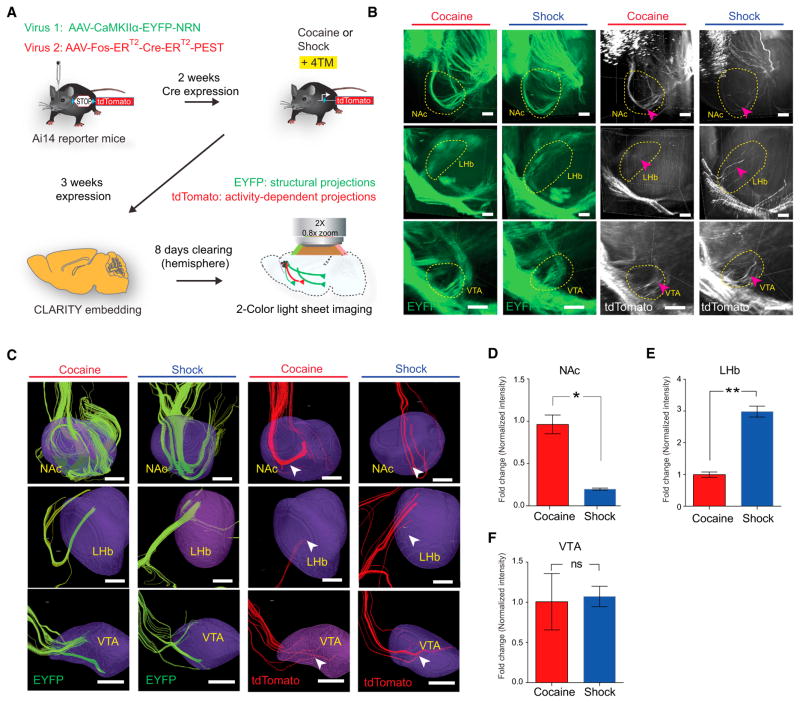
Figure 4. Distinct Projection Targets of Cocaine and Shock-Activated mPFC Populations.
(A) Summary of CAPTURE workflow (described in text).
(B) Representative CLARITY images of the structural projections (green, EYFP) and activity-dependent projections (white, tdTomato) from cocaine- and shock-labeled mice in NAc (top), LHb (middle), and VTA (bottom). Arrowheads indicate axon bundles terminating in the circled region. Scale bar, 200 μm.
(C) Reconstructed streamlines from(B), showing streamlines terminating in the 3D brain regions (purple). Green streamlines, reconstructed from EYFP fibers; red streamlines, reconstructed from tdTomato fibers. Scale bars, 200 μm.
(D–F) Quantification of projection intensity from cocaine- and shock-activated mPFC populations in three regions. Behavior-specific projection intensity was quantified using the ratio between red and green fibers (i.e., the number of red streamlines, divided by the number of green streamlines) terminating in indicated regions and plotted as fold change (relative to the cocaine group, n = 6 per group; ns, p > 0.05, *p < 0.05, **p < 0.01, unpaired t test). Error bars, mean ±SEM.
See also Figure S4.
Distinct Projection Patterns among Behavioral Experience-Defined mPFC Populations
We applied CAPTURE to quantify projections from cocaine- and shock-recruited mPFC populations. Ai14 reporter mice were co-injected with CaMKIIα-EYFP-NRN and cFos-ER-Cre-ER-PEST AAVs and subjected to 4TM-mediated cocaine and shock labeling. With CAPTURE, projections from all CaMKIIα(principally excitatory glutamatergic) neurons are labeled with EYFP and projections from behaviorally recruited populations are labeled with tdTomato. Importantly, EYFP fibers in the NAc, BLA, and VTA were found to be indistinguishable between the cocaine- and shock-labeled animals, indicating minimal variation in viral injection, transduction, and expression between the two groups (Figure 4B).
In the very same animals, significantly more projections from behaviorally activated mPFC neurons were observed targeting the NAc in cocaine-exposed animals compared to shock-exposed animals. Conversely, we observed significantly more behaviorally activated mPFC fibers to the LHb in shock-exposed animals (Figures 4C–4F). No significant difference in red/green (activity/structure) ratio was observed between the two groups in the VTA, showing no detectable systematic difference in efficiency or targeting of viral labeling. The cocaine-activated mPFC population thus preferentially projects to the NAc, whereas the shock-activated population projects more strongly to LHb, revealing that the neurons recruited in mPFC by distinct-valence experience are not simply different in terms of the patterns of input that they happen to receive, but represent anatomically distinct cell populations in terms of projection pattern across the brain.
Molecular Signature of Experience-Defined mPFC Neurons: Pre-existing NPAS4 Population
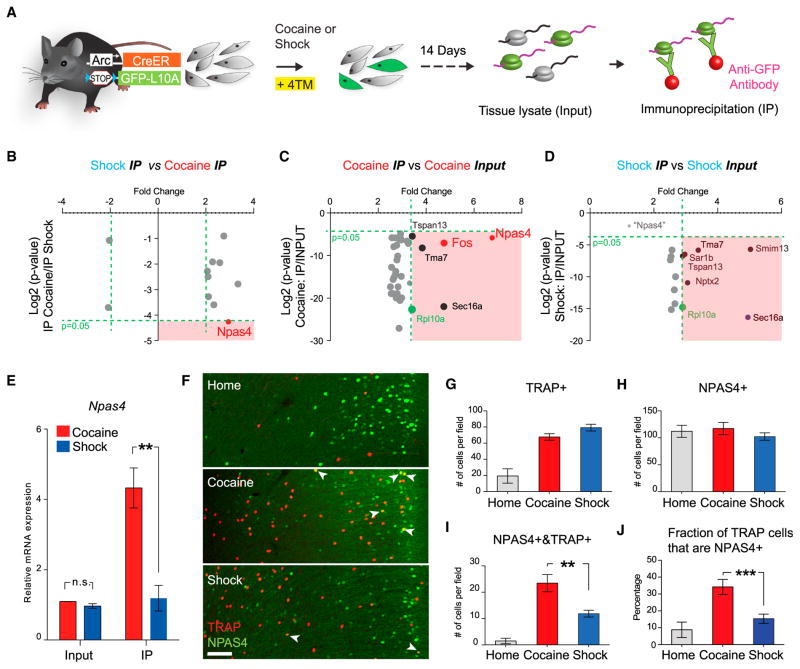
Next, we sought to identify molecular distinctions among cocaine- and shock-responsive populations in mPFC. We began by crossing ArcCreER mice to a Cre-dependent ribosome-tagged mouse (Long et al., 2014) to allow for labeling the ribosomes from neuronal populations activated by well-defined stimuli (Figure 5A). With this approach, the tagged ribosome can be immunoprecipitated (IP) from tissue lysate using an anti-GFP antibody, isolating translating mRNA for analysis (Ekstrand et al., 2014; Heiman et al., 2008; Sanz et al., 2009). These mice were exposed to either cocaine or shocks with 4TM. After labeling, mice were returned to the home cage for 2 weeks before initiation of ribosome purification, a design focused on identifying intrinsic signatures of these populations rather than on immediate mRNA expression changes associated with the exposure to stimuli.
Figure 5. Cocaine Preferentially Activates the NPAS4+ Population in mPFC.
(A) Schematic of activity-dependent ribosome profiling. Green, activated neurons; gray, non-activated neurons.
(B–D) Scatter plot of the most-enriched genes comparing cocaine- versus shock-activated cells (cocaine IP versus shock IP) (B), cocaine IP versus cocaine input (C) and shock IP versus shock input (D). The bottom right quadrant (pink) of each scatter plot denotes genes with p < 0.05 and fold change >2 for the indicated comparisons. As a positive control, the enrichment of RpL10a is highlighted in green. Black dots denote genes that were non-specifically enriched by IP (as shown enrichment in both cocaine and shock IP fraction). In (D), Npas4 (light-gray dot, fold change = 1.35, p value = 0.385) did not approach the statistically significant (pink) quadrant.
(E) Quantitative PCR analysis of Npas4 mRNA expression in the input and IP fractions.
(F) Representative images showing the overlap between TRAP+ and NPAS4+ cells in the mPFC. Scale bar, 100 μm. Arrowheads indicate double-positive cells.
(G–I) Quantification of numbers of TRAP+ (G), NPAS4+ (H), and TRAP+&NPAS4+ (I) cells in the mPFC.
(J) Percentage of NPAS4+ cells in TRAP+ cells under three conditions. n = 4 per group, *p < 0.05, **p < 0.01, unpaired t test. Error bars, mean ±SEM.
Equal enrichment of GFP transcripts under these conditions was first confirmed by qPCR (Figure S5A). Four groups of mRNA (“IP fraction” for ribosome-associated transcripts from cocaine and shock and “input” for whole-tissue lysate from cocaine and shock; see the Experimental Procedures) were then analyzed by microarray. For both stimuli, IP fraction groups were enriched 3-fold in the ribosomal-component mRNA (Rpl10a) compared with input groups (Figures 5C and 5D), further demonstrating similar levels of mRNA enrichment between treatments. Gene expression profiles from cells recruited under the two conditions were highly similar upon initial examination (<10 genes showed >2-fold difference; Figure S5C); however, a transcript of the activity-dependent transcription factor Npas4 (Bloodgood et al., 2013; Lin et al., 2008; Shamloo et al., 2006) was uniquely enriched in cocaine-activated cells compared to shock-activated cells (Figure 5B). This enrichment was confirmed by comparing the IP fraction to the input from the cocaine-labeled cells (Figure 5C) and finally validated by qPCR (Figure 5E); in contrast, this enrichment was not observed by comparing the IP and input fractions from the shock-labeled population (Figure 5D).
Because Npas4 itself is an immediate early gene that can be transiently induced by neuronal activity, we first tested if its enrichment in the cocaine-recruited population could relate to a lasting general cocaine-elevated Npas4 transcription. Both cocaine and shock caused a rapid increase of Npas4 expression in mPFC within 30 min, as expected; however, this induction under both conditions, (also as expected) completely returned to baseline after 10 days before the ribosome was profiled (Figure S5B). Indeed, no difference was observed when we examined Npas4 expression in whole-tissue lysates from cocaine- and shock-labeled mPFC (Figure 5E), indicating that exposure to cocaine did not lead to an overall long-term increase of Npas4 expression in mPFC.
At the single-cell level, we discovered that the number of mPFC cells with NPAS4 immunoreactivity did not differ between behavioral groups or between the home-cage and behavior-challenged animals (Figures 5F–5H). However, TRAP/NPAS4 double-positive cell counts were significantly higher in cocaine-exposed mice (Figure 5I), consistent with activity recruitment of an NPAS4 population. Similarly, the percentage of NPAS4+ cells among TRAP+ cells was also significantly higher in cocaine-labeled mPFC than in shock-labeled mPFC (Figure 5J). Importantly, we found that another positive-valence experience (highly palatable food consumption), but not another negative-valence experience (restraint stress), also selectively recruited this mPFC NPAS4+ population (Figures S5D and S5E), further illustrating that a pre-existing NPAS4+ population is preferentially activated by salient stimuli associated with positive valence rather than negative valence. Finally, the molecular overlap between the positive valence-associated population and the NPAS4+ population led us to further examine the spatial distribution of valence-specific populations, since the NPAS4+ cells appeared to be enriched in superficial layers of the mPFC (Figures 5F and S5E); indeed, the positive-valence experiences were associated with a significantly larger fraction of TRAPped layer 2/3 neurons in the mPFC than in the negative-valence experiences (Figure S5F).
Cocaine- and Shock-Activated Populations Control Appetitive and Aversive Behaviors
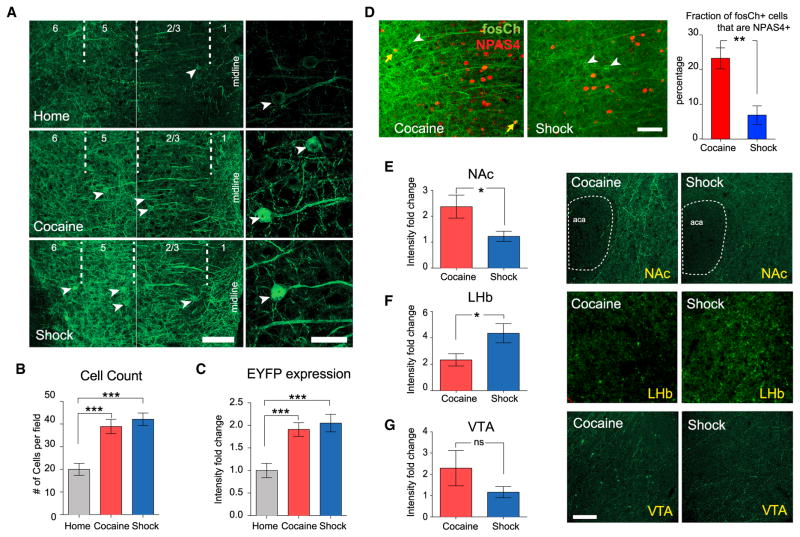
Next, we tested if electrical activity in these behavioral activity-defined populations had distinct positive or negative conditioning valence for the same animals that had experienced the stimulus, assessed by causal impact on behavior during the place preference task. To maintain robust, low-background, and direct region-specific control in mPFC, we designed a viral approach employing channelrhodopsin tagged with EYFP (ChR2-EYFP) under the control of the AAV-cFos backbone described earlier (termed fosCh; Figure 4A). Animals injected with this virus (in mPFC) were exposed to daily cocaine dosing or shock over 5 consecutive days, after which time robust increases in the number of fosCh-labeled cells and in EYFP expression were observed (Figure 6A–6C).
Figure 6. Use of fosCh for Targeting Cocaine- and Shock-Activated mPFC Populations.
(A) Representative images showing fosCh expression in mPFC following the indicated behaviors. Left, two 40×images were stitched for visualizing lamina across the cortical depth. Arrowheads indicate fosCh+ neurons. Scale bars, 100 μm. Right, high-magnification images of individual fosCh neurons. Scale bars, 25 μm.
(B) Fold change in fosCh cell numbers.
(C) Fold change in mean EYFP fluorescence intensity. n = 11–14 per group, ***p < 0.001, unpaired t test.
(D) Representative images and quantification of fosCh and NPAS4+ cells. White arrowheads indicate fosCh-positive neurons. Yellow arrows indicate NPAS4/fosCh double-positive cells. n = 5 per group, **p < 0.01, unpaired t test.
(E–G) Left: comparing density of fosCh projections for cocaine and shock groups. Right: representative images showing the density of fosCh projections in indicated regions. aca, anterior part of anterior commissure. Scale bars, 100 μm. n = 11–14 per group, *p < 0.05, unpaired t test. Error bars, mean ±SEM.
See also Figure S6.
We first quantified Npas4 expression in fosCh cells, hypothesizing that cocaine-labeled fosCh cells would exhibit higher Npas4 expression compared with shock-labeled fosCh cells. This was indeed the case (Figure 6D); importantly, expression of general excitatory or inhibitory neuronal markers did not differ between those two populations (Figures S6A and S6B). Consistent with CAPTURE findings, cocaine-labeled fosCh cells were found to project strongly to NAc, while the LHb contained significantly denser fibers arising from the shock-labeled cells (Figures 6E–6G). Crucially, this method of targeting was sufficient to enable optical control over the resulting sparsely distributed neuronal subsets; fosCh-labeled cells displayed robust light-evoked firing assessed by in vivo electrophysiological recording (Figures 7A and 7B). Together, these data demonstrated resolution with the fosCh strategy of the same pattern that had been characterized molecularly and anatomically and enabled the final test of whether these neuronal subsets were capable of differentially controlling behavior.
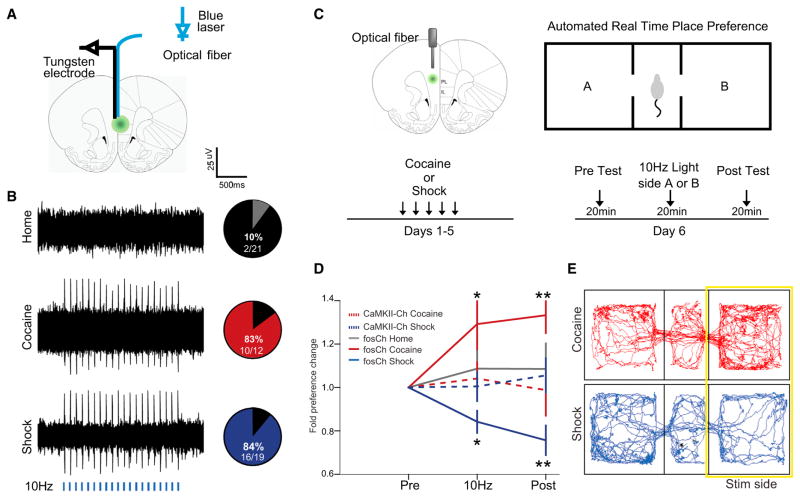
Figure 7. Differential Behavioral Influence of Cocaine- and Shock-Activated mPFC Populations.
(A) Schematic to illustrate the placement of the recording electrode and optical fiber for in vivo recording. The optrode was lowered in 100 μm steps along the dorsal-ventral axis of mPFC.
(B) Left: representative extracellular recordings showing neural response to a 10 Hz light train (5 ms pulses for 2 s, every 5 s, 5 mW 473 nm light, indicated by blue bars). Right: pie charts indicate percentage of recording sites showing light-evoked action potential firing for the home cage (gray), cocaine (red), and shock (blue) groups.
(C) Schematic shows the location of the optical fiber positioned above the injection site in green. After 5 days of training, mice were tested by a real time place preference test, which consisted of three consecutive 20-min trials.
(D) Behavioral results plotted as fold change in preference for the light stimulated side (normalized by baseline preference) across each of the trials. n = 10–14 per group, *p < 0.05, **p < 0.01, ANOVA, followed by Tukey’s multiple comparison test. Error bars, mean ±SEM.
(E) Movement tracking data from representative cocaine- and shock-labeled animals during the light-stimulation trial.
See also Figure S7.
To address this question, we employed the real-time place preference paradigm in which 10 Hz light-pulse trains were automatically triggered on entry into one side of a behavioral chamber. Place preference was monitored over three consecutive trials: before, during, and after light delivery for reactivation of fosCh-defined neuronal ensembles (Figure 7C). To control for the possibility that behavior could be biased by randomly labeled neurons, an additional group of animals was included in which comparable expression of ChR2 was driven by the CaMKIIα promoter without link to prior activity (Figures S7A and S7B). Optogenetic stimulation of these non-activity-specific neuronal populations did not influence place preference, nor were homecage-recruited fosCh-population animals observed to exhibit preference or aversion for the chamber in which the cells were optically activated. Remarkably, however, reactivation of the shock- or cocaine-defined fosCh populations induced significant (and opposite-direction) shifts in place preference, with cocaine-exposed mice demonstrating preference, and shock-exposed mice demonstrating aversion, for the photostimulation-paired side (Figures 7D and 7E; mean preference change at post-test for cocaine: 1.3x ±0.1, Wilcoxon p = 0.0006; for shock: 0.8x ±0.1, Wilcoxon p = 0.002). These data reveal that the activity-defined mPFC neural populations differ not only anatomically and molecularly but also in functional impact in modulating behavior.
DISCUSSION
It has been remarked (building on Dobzhansky’s comment that “nothing in biology makes sense except in the light of evolution” (Dobzhansky, 1973) that “nothing in neurobiology makes sense except in the light of behavior” (Shepherd, 1988). This perspective may be directly relevant to efforts in multiple subfields of neuroscience to describe the cellular diversity in nervous systems. While these efforts are pioneering and informative, it may be limiting to collect cellular typology datastreams relating to anatomy, genetics, and membrane biophysics, without knowledge of the importance of activity patterns in those same cells during behavior. However, forging such direct linkages has been difficult, especially in vertebrates. Indeed no system may be more challenging in this regard than the mammalian mPFC, which does not have well-defined direct sensory input or motor output pathways, and wherein principal cells with similar spike properties and anatomical positioning exhibit divergent or even opposing responses during behavior. Here, we have addressed these challenges, focusing on the specific problem of whether mPFC cells that respond differently to positive or negative valence experiences are, in fact, separable cell types as definable by molecular or anatomical measures, as well as by causal impact on behavior.
We identified not only brain-wide projection patterns separating mPFC cells responding to positive- and negative-valence experience but also a unique and unexpected enrichment of NPAS4 expression characteristic of mPFC cells responding to appetitive experience (Figure 4; Movie S1) that was not observed in cells responding to salient aversive experiences with comparable conditioning value. In illustrating that a pre-existing NPAS4+ population is preferentially activated by salient stimuli associated with positive valence rather than with negative valence, these mPFC results may be contrasted with elegant memory-engram models in which cells expressing a key activity marker may be recruited into diverse stimulus representations, as with CREB-expressing cells in amygdala fear conditioning (Han et al., 2007). NPAS4 has been linked to cellular and micro-circuit processes relating to the balance of excitation and inhibition, including a form of adaptive plasticity in which inhibitory synapses are formed on cells in response to high levels of excitation (Bloodgood et al., 2013; Lin et al., 2008; Maya-Vetencourt et al., 2012; Spiegel et al., 2014). This previously known role of NPAS4, together with the observation here that the positive-valence, but not negative-valence, experience recruited the mPFC NPAS4 population may bear relevance to previous observations among behaviorists that appetitive-learning experiences may be more vulnerable to adaptation or extinction, whether with natural stimuli or in pathological situations seen in drug dependence (Bouton, 2004).
The CAPTURE approach (the integration of enhanced CLARITY for behavioral cohort-scale work with cell and projection quantification linked to activity during behavior) is provided here in the form of tools, software, and datasets that may be readily applied across systems. The intact-brain approach is not only scalable in a practical manner to include large cohorts of experimental subjects and enabling of quantitative projection and population analysis but also helps address key confounds relating to selection bias and loss of continuity in traditional sectioning approaches widely used across neuroscience and biology. Moreover, improved 3D understanding of the path of behaviorally relevant axon bundles through the intact brain may be critical for the expanding field of neuromodulation, which is increasingly dependent on accurate targeting of white matter tracts (Henderson, 2012).
These tools are also readily adaptable for broader applications. For example, the NRN construct allowing rapid axonal projection labeling may be useful along with recombinase- or tetracycline-controlled (Liu et al., 2012) promoters for driving axon-filling fluorophores in approaches to multiplexed cell-type-specific CLARITY projection mapping or for driving opsins (Gore et al., 2015) to enhance functional assays of long-range projections (for example, to test synaptic release at different points along an axonal projection in slice assays or to test behavioral significance of cells defined by projection target). The fos-CreER virus (Figure 4) might be used in conjunction with future generations of high-potency opsin expression systems to enable optogenetic control based on experience temporally defined with 4TM, in strategies that could be eventually developed to reduce the number of repeated exposures currently required for sustained expression using the fosCh construct (Figure 7C). Lastly, recombinase-dependent constructs expressing Ca2+ indicators (such as G/RCaMPs) may be used to resolve temporal dynamics and individual-neuron activity magnitudes within experience-defined cells; these features are not accessible by conventional binary IEG-based methods but may be important, for example, in positive versus negative reinforcement in the PFC (Pi et al., 2013; Pinto and Dan, 2015).
In future work, it will be important to build on these results with additional investigation of the mPFC populations. For example, exploring long-term changes in gene expression and structural properties in these assemblies after experience (continuing to leverage the activity-tagging and intact-brain methods) may help to identify cell-population-resolved processes important in plasticity of adaptive or maladaptive circuit dynamics. And iden-tification of these separable mPFC populations, along with analysis of their interactions and modulations during adaptive and pathological conditions (in the setting of rapid parallel advances in high-content molecular phenotyping, targeted drug screening and design, and genome editing) may point the way toward cell-population-targeted interventions in diseases of addiction, anxiety, and fear.
EXPERIMENTAL PROCEDURES
Animals
Male and female C57BL/6J mice were group-housed on a reverse 12-hr light/dark cycle. Mice were 6–8 weeks old at viral infusion. Ai14 and wild-type C57BL/6 mice were purchased from JAX. Rosa26loxp-stop-loxp-eGFP-L10 mice were from Dr. Evan Rosen at Harvard Medical School. Male mice were used in behavioral assays. Both male and female mice were used for anatomy assays. All experimental protocols were approved by the Stanford University Institutional Animal Care and Use Committee and were in accordance with the guidelines from the NIH.
CAPTURE Labeling
Ai14 mice were injected with a 1-μl mixture of AAV8-CaMKIIα-EYFP-NRN and AAV8-cFos-ER-Cre-ER-PEST in the mPFC. 2 weeks later, the mice were given 15 mg/kg cocaine (IP) or 20 random foot shocks (2 s/0.5 mA, two shocks/minute on average) for 2 consecutive days. The control group remained in their home cage for the whole period. 10 mg/kg 4-TM was given to all mice 3 hr after the last behavior section to enable CreER-mediated recombination. The mice were returned to their home cages for an additional 4 weeks to allow the full expression of fluorophores.
CLARITY and Whole-Brain Imaging
Three key features of the new CLARITY platform were (1) accelerated clarification through parallelized flow-assisted clearing crucial for large cohorts and independent of specialized equipment, such as electrophoresis or perfusion chambers, (2) >90% cost reduction using a new refractive index-matching process, and (3) optical properties such that the whole mouse brain can be imaged using a commercial light-sheet microscope (Lavision) under a single field of view and as a single stack in less than 2 hr with single-cell resolution (this speed and simplicity is also useful for large behavioral cohorts). The COLM system (described earlier in Tomer et al., 2014; Lerner et al., 2015) is optimized for high-resolution imaging and is most useful for studies requiring sub-cellular information, while the approach outlined here is optimized for speed and practicality in cellular resolution work, most useful for robust application to large cohorts in CLARITY (see the Supplemental Experimental Procedures). All custom software, including the scripts used for atlas building, registration, and analysis, as well as code for CLARITY-based tractography, are freely available in a unified package at http://capture-clarity.org. The Web site also provides detailed protocols, sample images, and other CLARITY-related resources.
Supplementary Material
Acknowledgments
We thank M. Greenberg (Harvard) for the NPAS4 antibody, E. Rosen (Harvard) for ribosome tag mice, N. Renier and M. Tessier-Lavigne (Rockefeller) for valuable discussions during lab visits, Christina Kim for discussions, and S. Pak, M. Lo, and C. Perry for technical assistance. W.E.A. is supported by a Fannie & John Hertz Foundation Fellowship. K.D. is supported by NIMH, NIDA, the Wiegers Family Fund, HHMI (HCIA support to L.L. and K.D.), and the U.S. Army Research Laboratory and Defense Advanced Research Projects Agency (Cooperative Agreement Number W911NF-14-2-0013); nothing in this material represents the official views or policies of these funders. The authors have disclosed these findings to the Stanford Office of Technology Licensing. K.D. is a founder and scientific advisor of ClearLight Diagnostics, a startup exploring improvement of cancer diagnostics using CLARITY-related methods; all software, clones, sequences, protocols, and computational resources are freely distributed and supported (http://capture-clarity.org, http://clarityresourcecenter.org, and http://www.stanford.edu/group/dlab/optogenetics).
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, one table, one movie, and one data file and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2016.05.010.
AUTHOR CONTRIBUTIONS
L.Y. and K.D. designed the project. L.Y. performed TRAP, CLARITY, and light-sheet imaging, surgery, viral tracing, molecular profiling, and optogenetics experiments, and collected and analyzed all associated data with contributions from A.-C.W., C.R., J.J., and A.A. W.E.A. developed custom code for TRAP registration, and W.E.A., L.Y., and B.H. performed computational analysis on the TRAP brains. K.R.T., C.R., and I.B.W. developed and validated the fosCh construct in the K.D. lab with contributions from A.L.B. and performed optogenetic experiments. L.Y. developed the CAPTURE pipeline, and Q.T., L.Y., C.H.H., and J.M. developed the CLARITY-based tractography. L.L. provided unpublished TRAP mice. L.Y. and K.D. wrote the paper with input from W.E.A. and K.R.T. K.D. supervised all aspects of the work.
References
- Akten B, Kye MJ, Hao T, Wertz MH, Singh S, Nie D, Huang J, Merianda TT, Twiss JL, Beattie CE, et al. Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc Natl Acad Sci USA. 2011;108:10337–10342. doi: 10.1073/pnas.1104928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth AL, Gerkin RC, Dean KL. Alteration of neuronal firing properties after in vivo experience in a FosGFP transgenic mouse. J Neurosci. 2004;24:6466–6475. doi: 10.1523/JNEUROSCI.4737-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodgood BL, Sharma N, Browne HA, Trepman AZ, Greenberg ME. The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature. 2013;503:121–125. doi: 10.1038/nature12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouton ME. Context and behavioral processes in extinction. Learn Mem. 2004;11:485–494. doi: 10.1101/lm.78804. [DOI] [PubMed] [Google Scholar]
- Burgos-Robles A, Vidal-Gonzalez I, Quirk GJ. Sustained conditioned responses in prelimbic prefrontal neurons are correlated with fear expression and extinction failure. J Neurosci. 2009;29:8474–8482. doi: 10.1523/JNEUROSCI.0378-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalley JW, Cardinal RN, Robbins TW. Prefrontal executive and cognitive functions in rodents: neural and neurochemical substrates. Neurosci Biobehav Rev. 2004;28:771–784. doi: 10.1016/j.neubiorev.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Nothing in biology makes sense except in the light of evolution. Am Biol Teach. 1973;35:125–129. [Google Scholar]
- Ekstrand MI, Nectow AR, Knight ZA, Latcha KN, Pomeranz LE, Friedman JM. Molecular profiling of neurons based on connectivity. Cell. 2014;157:1230–1242. doi: 10.1016/j.cell.2014.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore F, Schwartz EC, Brangers BC, Aladi S, Stujenske JM, Likhtik E, Russo MJ, Gordon JA, Salzman CD, Axel R. Neural representations of unconditioned stimuli in basolateral amygdala mediate innate and learned responses. Cell. 2015;162:134–145. doi: 10.1016/j.cell.2015.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, Luo L. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron. 2013;78:773–784. doi: 10.1016/j.neuron.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JH, Kushner SA, Yiu AP, Cole CJ, Matynia A, Brown RA, Neve RL, Guzowski JF, Silva AJ, Josselyn SA. Neuronal competition and selection during memory formation. Science. 2007;316:457–460. doi: 10.1126/science.1139438. [DOI] [PubMed] [Google Scholar]
- Hayden BY, Nair AC, McCoy AN, Platt ML. Posterior cingulate cortex mediates outcome-contingent allocation of behavior. Neuron. 2008;60:19–25. doi: 10.1016/j.neuron.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suárez-Fariñas M, Schwarz C, Stephan DA, Surmeier DJ, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson JM. “Connectomic surgery”: diffusion tensor imaging (DTI) tractography as a targeting modality for surgical modulation of neural networks. Front Integr Nuerosci. 2012;6:15. doi: 10.3389/fnint.2012.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof PR, Ungerleider LG, Webster MJ, Gattass R, Adams MM, Sailstad CA, Morrison JH. Neurofilament protein is differentially distributed in subpopulations of corticocortical projection neurons in the macaque monkey visual pathways. J Comp Neurol. 1996;376:112–127. doi: 10.1002/(SICI)1096-9861(19961202)376:1<112::AID-CNE7>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Hsiang HL, Epp JR, van den Oever MC, Yan C, Rashid AJ, Insel N, Ye L, Niibori Y, Deisseroth K, Frankland PW, Josselyn SA. Manipulating a “cocaine engram” in mice. J Neurosci. 2014;34:14115–14127. doi: 10.1523/JNEUROSCI.3327-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel N, Barnes CA. Differential activation of fast-spiking and regular-firing neuron populations during movement and reward in the dorsal medial frontal cortex. Cereb Cortex. 2015;25:2631–2647. doi: 10.1093/cercor/bhu062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Stuphorn V, Brown JW, Schall JD. Performance monitoring by the anterior cingulate cortex during saccade countermanding. Science. 2003;302:120–122. doi: 10.1126/science.1087847. [DOI] [PubMed] [Google Scholar]
- Jennings JH, Sparta DR, Stamatakis AM, Ung RL, Pleil KE, Kash TL, Stuber GD. Distinct extended amygdala circuits for divergent motivational states. Nature. 2013;496:224–228. doi: 10.1038/nature12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZV, Revis AA, Burdick MA, Rhodes JS. A similar pattern of neuronal Fos activation in 10 brain regions following exposure to reward- or aversion-associated contextual cues in mice. Physiol Behav. 2010;99:412–418. doi: 10.1016/j.physbeh.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T, Kitamura K, Suzuki K, Nonaka M, Kamijo S, Takemoto-Kimura S, Kano M, Okuno H, Ohki K, Bito H. Functional labeling of neurons and their projections using the synthetic activity-dependent promoter E-SARE. Nat Methods. 2013;10:889–895. doi: 10.1038/nmeth.2559. [DOI] [PubMed] [Google Scholar]
- Kim SY, Adhikari A, Lee SY, Marshel JH, Kim CK, Mallory CS, Lo M, Pak S, Mattis J, Lim BK, et al. Diverging neural pathways assemble a behavioural state from separable features in anxiety. Nature. 2013;496:219–223. doi: 10.1038/nature12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel S, Ion DI, Roeper J, Malenka RC. Projection-specific modulation of dopamine neuron synapses by aversive and rewarding stimuli. Neuron. 2011;70:855–862. doi: 10.1016/j.neuron.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel S, Lim BK, Ran C, Huang KW, Betley MJ, Tye KM, Deisseroth K, Malenka RC. Input-specific control of reward and aversion in the ventral tegmental area. Nature. 2012;491:212–217. doi: 10.1038/nature11527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AT, Gee SM, Vogt D, Patel T, Rubenstein JL, Sohal VS. Pyramidal neurons in prefrontal cortex receive subtype-specific forms of excitation and inhibition. Neuron. 2014;81:61–68. doi: 10.1016/j.neuron.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner TN, Shilyansky C, Davidson TJ, Evans KE, Beier KT, Zalocusky KA, Crow AK, Malenka RC, Luo L, Tomer R, Deisseroth K. Intact-brain analyses reveal distinct information carried by SNc dopamine subcircuits. Cell. 2015;162:635–647. doi: 10.1016/j.cell.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR. Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- Li B, Piriz J, Mirrione M, Chung C, Proulx CD, Schulz D, Henn F, Malinow R. Synaptic potentiation onto habenula neurons in the learned helplessness model of depression. Nature. 2011;470:535–539. doi: 10.1038/nature09742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Zhou T, Liao L, Yang Z, Wong C, Henn F, Malinow R, Yates JR, 3rd, Hu H. βCaMKII in lateral habenula mediates core symptoms of depression. Science. 2013;341:1016–1020. doi: 10.1126/science.1240729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, Tonegawa S. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JZ, Svensson KJ, Tsai L, Zeng X, Roh HC, Kong X, Rao RR, Lou J, Lokurkar I, Baur W, et al. A smooth muscle-like origin for beige adipocytes. Cell Metab. 2014;19:810–820. doi: 10.1016/j.cmet.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci USA. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Matsumoto K, Abe H, Tanaka K. Medial pre-frontal cell activity signaling prediction errors of action values. Nat Neurosci. 2007;10:647–656. doi: 10.1038/nn1890. [DOI] [PubMed] [Google Scholar]
- Maya-Vetencourt JF, Tiraboschi E, Greco D, Restani L, Cerri C, Auvinen P, Maffei L, Castrén E. Experience-dependent expression of NPAS4 regulates plasticity in adult visual cortex. J Physiol. 2012;590:4777–4787. doi: 10.1113/jphysiol.2012.234237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegas W, Bergan JF, Ogawa SK, Isogai Y, Umadevi Venkataraju K, Osten P, Uchida N, Watabe-Uchida M. Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass. eLife. 2015;4:e10032. doi: 10.7554/eLife.10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EK. The prefrontal cortex and cognitive control. Nat Rev Neurosci. 2000;1:59–65. doi: 10.1038/35036228. [DOI] [PubMed] [Google Scholar]
- Namburi P, Beyeler A, Yorozu S, Calhoon GG, Halbert SA, Wichmann R, Holden SS, Mertens KL, Anahtar M, Felix-Ortiz AC, et al. A circuit mechanism for differentiating positive and negative associations. Nature. 2015;520:675–678. doi: 10.1038/nature14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi HJ, Hangya B, Kvitsiani D, Sanders JI, Huang ZJ, Kepecs A. Cortical interneurons that specialize in disinhibitory control. Nature. 2013;503:521–524. doi: 10.1038/nature12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto L, Dan Y. Cell-type-specific activity in prefrontal cortex during goal-directed behavior. Neuron. 2015;87:437–450. doi: 10.1016/j.neuron.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci USA. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling K, Luk D, Morgan JI, Curran T. Regulation of a fos-lacZ fusion gene: a paradigm for quantitative analysis of stimulus-transcription coupling. Proc Natl Acad Sci USA. 1991;88:5665–5669. doi: 10.1073/pnas.88.13.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AA, Treit D. Excitotoxic lesions of the medial prefrontal cortex attenuate fear responses in the elevated-plus maze, social interaction and shock probe burying tests. Brain Res. 2003;969:183–194. doi: 10.1016/s0006-8993(03)02299-6. [DOI] [PubMed] [Google Scholar]
- Shah AA, Sjovold T, Treit D. Inactivation of the medial prefrontal cortex with the GABAA receptor agonist muscimol increases open-arm activity in the elevated plus-maze and attenuates shock-probe burying in rats. Brain Res. 2004;1028:112–115. doi: 10.1016/j.brainres.2004.08.061. [DOI] [PubMed] [Google Scholar]
- Shamloo M, Soriano L, von Schack D, Rickhag M, Chin DJ, Gonzalez-Zulueta M, Gido G, Urfer R, Wieloch T, Nikolich K. Npas4, a novel helix-loop-helix PAS domain protein, is regulated in response to cerebral ischemia. Eur J Neurosci. 2006;24:2705–2720. doi: 10.1111/j.1460-9568.2006.05172.x. [DOI] [PubMed] [Google Scholar]
- Shepherd GM. Neurobiology. 2. Oxford University Press; 1988. [Google Scholar]
- Smeyne RJ, Schilling K, Robertson L, Luk D, Oberdick J, Curran T, Morgan JI. fos-lacZ transgenic mice: mapping sites of gene induction in the central nervous system. Neuron. 1992;8:13–23. doi: 10.1016/0896-6273(92)90105-m. [DOI] [PubMed] [Google Scholar]
- Soloway AS, Pucak ML, Melchitzky DS, Lewis DA. Dendritic morphology of callosal and ipsilateral projection neurons in monkey prefrontal cortex. Neuroscience. 2002;109:461–471. doi: 10.1016/s0306-4522(01)00507-3. [DOI] [PubMed] [Google Scholar]
- Spiegel I, Mardinly AR, Gabel HW, Bazinet JE, Couch CH, Tzeng CP, Harmin DA, Greenberg ME. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157:1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc. 2014;9:1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzschentke TM, Schmidt WJ. Discrete quinolinic acid lesions of the rat prelimbic medial prefrontal cortex affect cocaine- and MK-801-, but not morphine- and amphetamine-induced reward and psychomotor activation as measured with the place preference conditioning paradigm. Behav Brain Res. 1998;97:115–127. doi: 10.1016/s0166-4328(98)00034-5. [DOI] [PubMed] [Google Scholar]
- Tzschentke TM, Schmidt WJ. Functional heterogeneity of the rat medial prefrontal cortex: effects of discrete subarea-specific lesions on drug-induced conditioned place preference and behavioural sensitization. Eur J Neurosci. 1999;11:4099–4109. doi: 10.1046/j.1460-9568.1999.00834.x. [DOI] [PubMed] [Google Scholar]
- Warden MR, Selimbeyoglu A, Mirzabekov JJ, Lo M, Thompson KR, Kim SY, Adhikari A, Tye KM, Frank LM, Deisseroth K. A pre-frontal cortex-brainstem neuronal projection that controls response to behavioural challenge. Nature. 2012;492:428–432. doi: 10.1038/nature11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiu J, Zhang Q, Zhou T, Zhou TT, Chen Y, Hu H. Visualizing an emotional valence map in the limbic forebrain by TAI-FISH. Nat Neurosci. 2014;17:1552–1559. doi: 10.1038/nn.3813. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.