Diamond Blackfan Anemia (DBA) is a rare blood disorder characterized by red cell aplasia and is often accompanied by a variety of congenital abnormalities. It typically presents in infancy with macrocytic anemia and is associated with an increased risk of myelodysplastic syndrome (MDS), leukemia, and solid tumors.1 The overall actuarial survival at 40 years is 75%. Current modes of therapy include corticosteroid treatment or red cell transfusions. Treatment-related complications like iron overload (treated by chelation therapy), infections, and bone marrow (BM) transplant related complications are leading causes of death in most DBA patients rather than severe aplastic anemia and malignancy. Although the mechanisms remain unclear, approximately 20% of DBA patients enter spontaneous remission, wherein physiologically stable hemoglobin (Hb) levels are maintained in the absence of steroid therapy or transfusions.2
Alongside other inherited marrow failure disorders with overlapping clinical features caused by ribosomal gene defects like Shwachman-Diamond syndrome (SDS) and dyskeratosis congenita (DC), DBA is classified as a ribosomopathy. Most cases of DBA are due to mutations in ribosomal proteins (RP) usually resulting in haploinsufficiency. Autosomal dominant inheritance of mutations in components of the 60S subunit (RPL5, RPL11, RPL15, RPL26, RPL35A) and 40S subunit (RPS7, RPS10, RPS17, RPS19, RPS24, RPS26) cause DBA, while SDS follows autosomal recessive inheritance.3,4 Interestingly, despite the ubiquitous requirement for ribosomes, germline mutations in RP cause tissue-specific defects, the mechanisms of which are unknown. We have used whole genome sequencing (WGS) and high density single nucleotide polymorphism (SNP) arrays to solve a complex case of DBA.
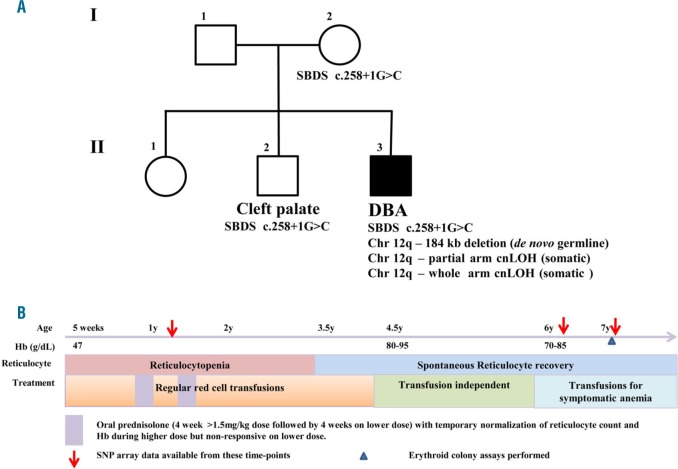
The proband ((II.3) Figure 1A), an 9-year-old boy (as of Aug 2017), is the third child of a Caucasian couple. He presented with anemia at 5 weeks (Hb 47g/L) which was treated with red cell transfusion. He required monthly transfusions for recurrent anemia and profound reticulocytopenia for the following year. BM biopsy revealed significantly reduced erythropoiesis with occasional immature erythroid cells and normal myelopoiesis and lymphopoiesis, which was consistent with a clinical diagnosis of DBA. He had mild bilateral ptosis and mild hypertelorism. No hepatosplenomegaly, renal abnormalities or bone abnormalities were seen on clinical examination. The siblings and parents had normal hematological findings. Clinical history is described in Figure 1B and hematological parameters at various time-points have been compiled in Online Supplementary Table S1. Along with anemia, he has intermittent neutropenia, monocytopenia and lymphopenia with normal lymphocyte subsets and immunoglobulin levels. He has significant growth delay, crossing height and weight centiles, and now tracking below the 1st centile. Endocrine and gastroenterological assessment is normal. He has some delays in neurocognitive development.
Figure 1.
Pedigree and disease progression of the proband with Diamond Blackfan Anemia (DBA). (A) Pedigree describing genetic lesions in the proband with DBA and his parents and siblings who display normal hematological findings. (B) Timeline depicting progress of disease and treatment strategies for the proband. Karyotype was found to be normal when tested (corresponds to red arrows).
WGS analysis of the trio (proband, parents) was carried out to identify the causal mutation in the proband, an 8- year-old boy with DBA. Of 54 candidate variants, 25 were established as benign from the literature and 28 were variants of unknown significance. A maternally inherited splice donor site variant in the Shwachman-Bodian-Diamond-Syndrome (SBDS) gene was identified; c.258+1G>C (Online Supplementary Figure S1A, Online Supplementary Table S2). This mutation is predicted to cause a frameshift (p.C84Yfs*3).5 The trio was also screened with the TruSight Cancer Panel and the maternal SBDS variant was confirmed in the proband. Sanger sequencing on samples from all family members confirmed the variant in the mother (I-2) and proband (II-3) as well as a sibling (II-2) who was born with a cleft palate (Online Supplementary Figure S1B). RT-PCR and western blot analysis revealed lower expression of SBDS in lymphoblastoid cell lines (LCL) established from peripheral blood (PB) in the family members with the variant compared to normal controls (Online Supplementary Figure S1C, S1D). However, we were unable to identify defects/imbalances pertaining to the paternal allele of SBDS using targeted next generation sequencing (NGS) on PB, BM and hair in the affected and hence continued to search for other causal mutated genes.
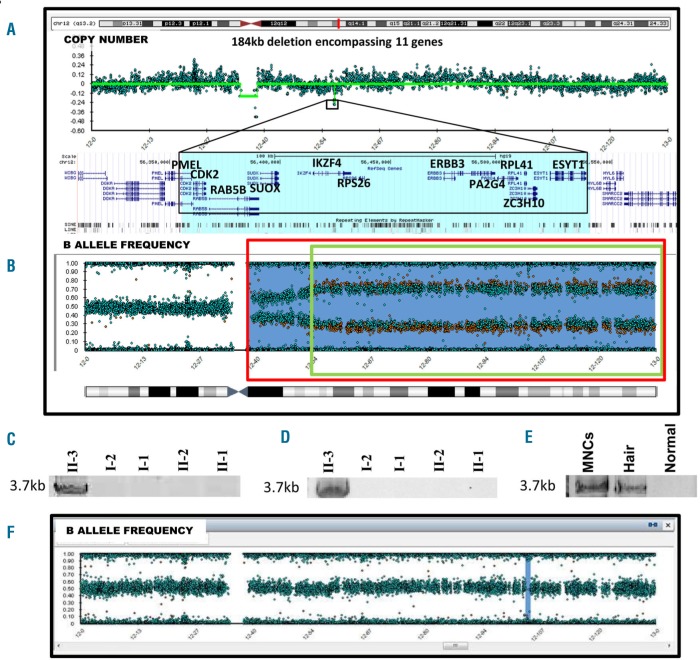
High density SNP microarrays on PB (at age 6) identified a 184 kb deletion on chromosome 12q in the proband that was not present in either of the parents (Figure 2A). This was verified by a single read from WGS spanning the breakpoints of the 184 kb deletion. This deletion was also detected in the proband’s hair sample, indicating a germline de novo mutation (Online Supplementary Figure S2B). The deleted region encompasses 11 genes (PMEL, CDK2, RAB5B, SUOX, IKZF4, RPS26, ERBB3, PA2G4, RPL41, ZC3H10 and ESYT1 – Online Supplementary Table S2). Heterozygous deletion or mutation of RPS26, a component of the small ribosomal subunit, has previously been described to cause DBA in an autosomal dominant manner.4 Intriguingly RPL41, which encodes a protein component of the large ribosomal subunit, also lies within the deleted region.
Figure 2.
Multiple defects in chr 12q in the proband. (A) SNP microarray reveals 184 kb deletion encompassing 11 genes on chr 12q and (B) copy neutral loss of heterozygosity comprising at least two regions of chr 12q - denoted by green and red boxes. (C, D) PCR across 184 kb breakpoint in peripheral blood and lymphoblastoid cell line gives product of expected size in the proband which was then confirmed to be de novo germline in a hair sample (E). (F) SNP microarray reveals copy neutral loss of heterozygosity events were absent in the proband early in the disease prior to spontaneous recovery.
The 184 kb deletion breakpoints were further defined by analysis of read depths from WGS data (Online Supplementary Figure S3). PCR using primers flanking these coordinates on gDNA yielded a 3.7 kb product (Figure 2C) in II-3 and not in other family members. Sanger sequencing using nested primers revealed that the deletion was the result of a homologous recombination event between Alu repeats (AluY: 56354900-56355009 and AluSc8: 56539460-56539754) (Online Supplementary Figure S4), which is not an uncommon mechanism contributing to germline disease.6
Moreover, SNP microarray identified at least two additional regions of copy neutral loss of heterozygosity (cnLOH) on chromosome 12 in PB (age 6), LCL (generated from PB at age 6) and BM (age 7), but not hair of the proband (Figure 2B; Online Supplementary Figure S2, Online Supplementary Figure S6). The presence of multiple somatic clones and the tendency of DBA patients to have an increased risk of myeloid malignancy raised the possibility that the patient may be pre-leukemic. To investigate this, the blood sample was interrogated for recurrent mutations associated with leukemia by targeted NGS. When analysed in comparison with hair, it was found to be free of any predicted pathogenic somatic mutations down to a 5% variant allele frequency.
Allelic imbalance with an over-representation of the paternal allele was seen on chromosome 12 across SNPs where the affected child was heterozygous, in the targeted NGS data (Online Supplementary Table S3). This was consistent with a case of two or more independent revertant mutations wherein the 184 kb deletion defect in the maternal copy of chromosome 12q was being ‘corrected’ by replacement with the intact paternal copy. The patient’s spontaneous reticulocyte recovery may be a consequence of the existence of two stably represented, non-identical clones exhibiting cnLOH spanning chromosome 12q. Absence of these cnLOH events in an earlier sample from the patient (prior to spontaneous recovery) lends further support to this hypothesis (Figure 2F). Also, the proportion of clones exhibiting cnLOH appears higher in LCL (Online Supplementary figure S2C), suggestive of a growth advantage. RPS26 expression was measured by qRT-PCR, in LCLs generated from the patient, his family members and three unrelated normal donors. Expression levels in the patient (II-3) were within the range of other family members and unrelated controls (Online Supplementary Figure S5). Although the role of RPL41 in the phenotype is unclear, a similar correction in expression levels was observed.
Revertant mosaicism has been described in several genetic diseases including skin disorders, DC and Fanconi anemia,7–9 and spontaneous remission is not uncommon in DBA.2 Moreover, mosaicism has been reported in DBA patients who attained remission.10 However, in the event of remission, the risk of developing MDS or leukemia is likely to remain in proportion to the burden of any residual disease-causing clones. The expansion of reversion events arising independently suggests a selective advantage over the defective cell population with likely contribution to correcting the phenotypic defect. We propose that revertant mosaicism is one of the mechanisms by which spontaneous remissions are attained in DBA. In such cases, where auto-correction has occurred in the hematopoietic compartment, it may be necessary to utilize alternative sources of DNA such as hair or saliva, when testing to identify the causative genetic lesion.
The DBA phenotype in the proband can primarily be attributed to haploinsufficiency of RPS26, and the effect of defects in ribosomal genes SBDS and RPL41 on disease phenotype and treatment outcome are currently unknown. We hypothesize that the patient’s developmental delay is secondary to the de novo chromosome 12q deletion. Craniofacial structures are derived from neural crest cells and, in animal models, mutations in RP can cause apoptosis of neural crest progenitors.11 Human ribosomopathies are often accompanied by craniofacial abnormalities.12 Though the proband has very mild craniofacial abnormalities, it is possible that the cleft palate in the sibling (II-2) is due to the heterozygous SBDS variant, especially considering the variable presentation of congenital abnormalities in individuals with biallelic mutations in SBDS.13 This complex molecular genetic diagnosis has been achieved by using multiple powerful technologies each with strengths and limitations. Together, however, they have provided valuable insight into disease biology of not only DBA, but likely other similar disorders.
Supplementary Material
Acknowledgements
We wish to acknowledge the patient’s family for their continuing support, cooperation and commitment to the cause of bone marrow failure research. We would like to thank Lynda Williams and Heather Thorne OAM from kConFab at Peter MacCallum Cancer Centre, Melbourne, Australia, for assistance with preparation of LCLs; and Amilia Wee for generation of initial reagents for this project. We would also like to acknowledge the Australian Diamond Blackfan Anaemia (ADBA) programme members, consisting of Sheren Al-Obaidi, Sarah CE Bray, Richard J. D’Andrea, Jianmin Ding, Amee J. George, Thomas J. Gonda, S Peter Klinken, Piyush Madhamshettiwar, Lorena Núñez Villacis, Richard B Pearson, Ben Saxon, Hamish S. Scott, Kaylene J Simpson, Adam Stephenson, Amilia Wee, Louise N. Winteringham, Mei S Wong, and Ross D. Hannan for their contribution to this work.
Footnotes
Funding: this work was supported by an Australian Diamond Blackfan Anaemia (ADBA) programme grant from the Captain Courageous Foundation and by grant funding from the National Health and Medical Research Council, Australia (APP10024215, APP1023059) and Adelaide Scholarships International from the University of Adelaide. This work has been submitted in partial fulfilment of the requirement for a PhD degree at the University of Adelaide for PV.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Vlachos A, Ball S, Dahl N, et al. Diagnosing and treating Diamond Blackfan anaemia: results of an international clinical consensus conference. Br J Haematol. 2008;142(6):859–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vlachos A, Muir E. How I treat Diamond-Blackfan anemia. Blood. 2010;116(19):3715–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gazda HT, Sheen MR, Vlachos A, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008; 83(6):769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doherty L, Sheen MR, Vlachos A, et al. Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2010;86(2):222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woloszynek JR, Rothbaum RJ, Rawls AS, et al. Mutations of the SBDS gene are present in most patients with Shwachman-Diamond syndrome. Blood. 2004;104(12):3588–3590. [DOI] [PubMed] [Google Scholar]
- 6.Deininger PL, Batzer MA. Alu repeats and human disease. Mol Genet Metab. 1999;67(3):183–193. [DOI] [PubMed] [Google Scholar]
- 7.Jonkman MF, Pasmooij AM. Realm of revertant mosaicism expanding. J Invest Dermatol. 2012;132(3 Pt 1):514–516. [DOI] [PubMed] [Google Scholar]
- 8.Hamanoue S, Yagasaki H, Tsuruta T, et al. Myeloid lineage-selective growth of revertant cells in Fanconi anaemia. Br J Haematol. 2006; 132(5):630–635. [DOI] [PubMed] [Google Scholar]
- 9.Jongmans MC, Verwiel ET, Heijdra Y, et al. Revertant somatic mosaicism by mitotic recombination in dyskeratosis congenita. Am J Hum Genet. 2012;90(3):426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrar JE, Vlachos A, Atsidaftos E, et al. Ribosomal protein gene deletions in Diamond-Blackfan anemia. Blood. 2011;118(26):6943–6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trainor PA, Merrill AE. Ribosome biogenesis in skeletal development and the pathogenesis of skeletal disorders. Biochim Biophys Acta. 2014;1842(6):769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115(16):3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myers KC, Bolyard AA, Otto B, et al. Variable clinical presentation of Shwachman-Diamond syndrome: update from the North American Shwachman-Diamond Syndrome Registry. J Pediatr. 2014; 164(4):866–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.