Abstract
Plants have developed multiple strategies to sense the external environment and to adapt growth accordingly. Delay of germination 1 (DOG1) is a major quantitative trait locus (QTL) for seed dormancy strength in Arabidopsis thaliana that is reported to be expressed exclusively in seeds. DOG1 is extensively regulated, with an antisense transcript (asDOG1) suppressing its expression in seeds. Here, we show that asDOG1 shows high levels in mature plants where it suppresses DOG1 expression under standard growth conditions. Suppression is released by shutting down antisense transcription, which is induced by the plant hormone abscisic acid (ABA) and drought. Loss of asDOG1 results in constitutive high‐level DOG1 expression, conferring increased drought tolerance, while inactivation of DOG1 causes enhanced drought sensitivity. The unexpected role of DOG1 in environmental adaptation of mature plants is separate from its function in seed dormancy regulation. The requirement of asDOG1 to respond to ABA and drought demonstrates that antisense transcription is important for sensing and responding to environmental changes in plants.
Keywords: abscisic acid signalling, DOG1, drought stress, non‐coding antisense RNA regulation
Subject Categories: Plant Biology, RNA Biology, Signal Transduction
Introduction
Delay of germination 1 (DOG1) was initially characterized as a QTL for seed dormancy variability between selected Arabidopsis thaliana accessions 1. In agreement, DOG1 has been reported to be exclusively expressed in seeds and in Arabidopsis dog1 loss‐of‐function mutants displaying only seed dormancy‐related phenotypes 1, 2. Subsequent studies have demonstrated that the seed dormancy function of DOG1 is conserved in many different plant species 3, 4, 5, 6. Recent work has also shown that multiple independently evolved DOG1 alleles are responsible for the adaptation of Arabidopsis to local conditions, explaining the surprisingly high proportion of naturally occurring variability in dormancy 7.
DOG1 expression is highly controlled in seeds, with regulators targeting DOG1 alternative splice site selection 8, alternative polyA site selection 9 and DOG1 expression 10, 11, 12, 13. In addition, we recently described the regulatory activity of a long, presumably non‐coding antisense transcript that suppresses DOG1 expression in “cis” and thereby dormancy strength in seeds 14.
Despite its predominant seed‐specific expression, DOG1 has been repeatedly identified in genomewide association studies (GWAS) as a candidate for controlling flowering time in Arabidopsis 15, 16, 17. In agreement with this notion, RNAi‐based silencing of DOG1 in wheat and lettuce revealed not only defects in seed dormancy, but also in flowering phenotype 3, 5. However, flowering defects have not been observed in Arabidopsis dog1 knockout plants 1, 5.
All tested DOG1 mRNA isoforms show primarily seed‐specific expression 1, 9, 18, whereas the DOG1 antisense transcript (asDOG1) is most strongly expressed in seedlings 14. The asDOG1 transcript originates from the 3′ end of DOG1 close to the major polyA site 14, 18, and it appears to be a member of a group of long, presumably non‐protein‐coding, RNAs (lncRNA). For recent reviews, see 19, 20.
Antisense transcription from within terminators is a widespread and conserved phenomenon 21. Yeast terminators that serve as promoters for antisense transcription share features that are typical for canonical sense promoters, including high H3K4me3 levels and the presence of TATA‐like elements 22, 23, but the functions of these transcripts are not always clear. Antisense transcription in yeast has been shown to permit the sensing of inorganic phosphate, lithium and many other stimuli 24, 25, 26, 27, 28.
Antisense transcription is well known in plants 29, 30. We recently demonstrated that some of these transcripts are initiated from within terminators which, as in yeast, show similarity to canonical promoters, including the presence of TATA boxes 31. Importantly, many of the ncRNAs found in plants, including antisense transcripts, are extensively regulated by external and internal inputs. One example is COOLAIR, the antisense partner of FLC, which is upregulated by low temperature 32. Thus, data from both yeast and Arabidopsis indicate that antisense transcripts can serve to regulate the expression of linked protein‐coding genes in response to environmental conditions. For example, FLC antisense, redundantly with other players, was shown to be important for control of FLC regulation in response to cold 32, 33.
Here, we show that asDOG1 expression is strongly suppressed by both the plant hormone abscisic acid (ABA) and drought, resulting in the release of antisense‐dependent silencing of DOG1. This discloses a role of Arabidopsis seed dormancy QTL DOG1 in drought response as dog1 mutants are drought‐sensitive, and asDOG1‐deficient plants (with constitutively high DOG1 expression) are drought‐resistant. Finally, we demonstrate that the ability of the antisense promoter to respond to ABA is absolutely required for the regulation of DOG1 expression by this hormone. In summary, by dissecting the regulation of asDOG1 by external stimuli, we have uncovered a novel and unexpected function of the major Arabidopsis seed dormancy QTL in drought response. Moreover, this study provides further evidence that antisense‐mediated regulation of gene expression is important in plant responses to environmental cues.
Results
DOG1 antisense is highly expressed in Arabidopsis leaves
We have previously shown that DOG1 antisense (asDOG1) negatively regulates DOG1 sense expression in seeds. Notably, asDOG1 is relatively weakly expressed in seeds and shows much higher expression in seedlings 14.
To thoroughly analyse asDOG1 expression in different organs of Arabidopsis, we examined luciferase (LUC) activity in plants containing the IRES‐LUC cassette driven by the asDOG1 promoter—p AS DOG1::LUC 14. We found that the asDOG1 promoter is highly active in the apical meristem, flowers and young leaves, but shows progressively diminishing activity in older leaves (Fig EV1). A complementary analysis using plants expressing a transgene containing the LUC reporter gene fused with the full‐length DOG1 locus—p DOG1 LUC::DOG1—revealed that DOG1 mRNA is very weakly expressed in tissues other than seeds, with expression detected in the meristem, flowers and young leaves and diminishing activity in older leaves (Fig EV1). Analysis of plants expressing p DOG1 shDOG1::LUC, with deletion of the 3′ DOG1 region (Δ antisense), revealed a slightly different expression pattern, with LUC activity detected mainly in older leaves, only a very weak signal in young leaves and no signal in flowers (Fig EV1B and D).
Figure EV1. Tissue‐specific expression of DOG1 sense and antisense.
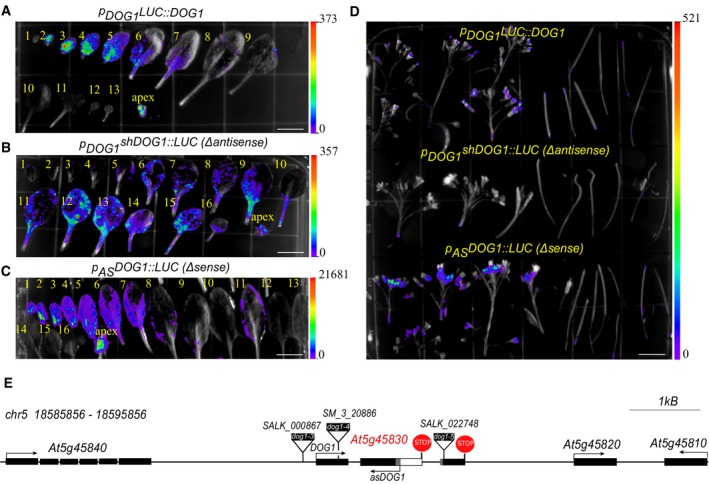
- DOG1 sense mRNA expression driven by the genomic construct. Numbers represent consecutive leaf number, from young to old. Scale bar: 2 cm.
- DOG1 sense mRNA expression driven by DOG1 genomic construct with antisense promoter removed. Numbers represent consecutive leaf number, from young to old. Scale bar, 2 cm.
- DOG1 antisense expression analysis. Numbers represent consecutive leaf number, from young to old. Scale bar, 2 cm.
- LUC expression analysis in flowers and siliques of plants expressing p DOG1 LUC::DOG1, p DOG1 shDOG1::LUC and p AS DOG1::LUC. Scale bar: 2 cm.
- Schematic diagram of the DOG1 gene at genomic scale: black boxes, exon sequences; grey boxes, alternative exonic regions; white box region included in alternatively polyadenylated DOG1 short transcript; arrows show the sense and antisense transcripts TSS and expression direction of DOG1; red circles indicate TTS for short and long mRNA of DOG1, correspondingly. The positions of T‐DNA insertions are indicated by black rectangles: dog1‐3 (SALK_000867), dog1‐4 (SM_3_20886) and dog1‐5 (SALK_022749).
Thus, in agreement with our previous report, asDOG1 shows strong expression in the meristem and leaves 14. Furthermore, its expression is highly tissue‐specific and similar to that of DOG1 sense mRNA. The tissue specificity of antisense transcripts, or more broadly lncRNA, has been often reported in plants 29, 30 and in other organisms 34, 35, 36. The expression of many of these non‐protein‐coding transcripts is not only tissue‐specific, it is also extensively regulated by the external environment 37, 38. This prompted us to examine the ability of the asDOG1 promoter to respond to external stimuli. Importantly, DOG1 antisense has been detected in Arabidopsis leaves in high‐throughput direct RNA sequencing analysis 39, 40, as shown previously 14.
DOG1 antisense is strongly downregulated by the hormone ABA
We challenged p AS DOG1::LUC plants with the plant hormones ABA and gibberellin (GA), which both perform essential functions in seed dormancy and vegetative growth 41, 42, 43, 44. Treatment with ABA resulted in nearly complete silencing of the antisense promoter activity in leaves, compared to mock‐ or GA‐treated plants (Figs 1A and B, and EV2A and B, Appendix Fig S1). ABA plays an important role in the response of plants to stress, including water deprivation where it acts as part of a signalling cascade leading to drought resistance 45, 46, 47. Subsequently, we showed that the DOG1 antisense promoter was also strongly downregulated in response to 5 days of water withdrawal (Figs 1C and D, and EV3A–C), a result validated by strand‐specific RT–qPCR analysis in Col‐0 (WT) plants (Fig 1E).
Figure 1. DOG1 antisense expression is selectively regulated by ABA and drought in Arabidopsis leaves. Application of the plant hormone ABA but not GA affects luciferase activity driven by the antisense promoter.
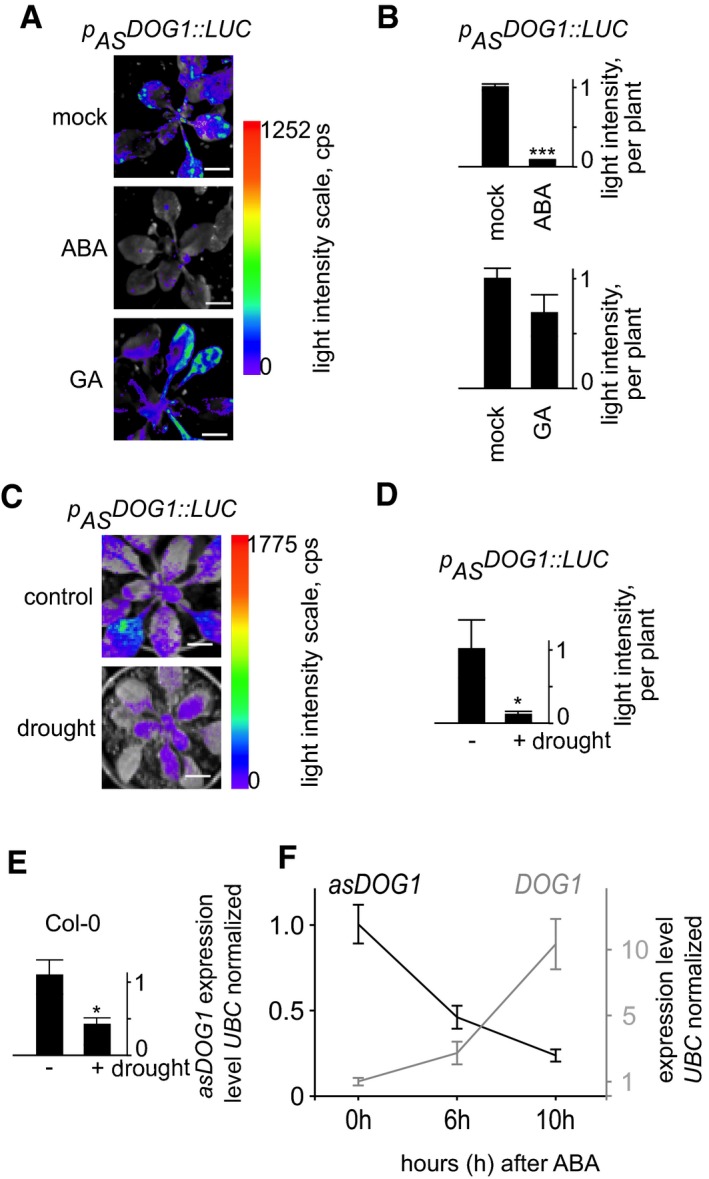
- Representative picture of plants carrying p AS DOG1::LUC sprayed with mock solution, ABA or GA and imaged 24 h later. The full pictures are shown in Fig EV2 and Appendix Fig S1. Scale bar, 1 cm.
- Quantification of emitted light intensity per plant after treatment with ABA and GA, compared with mock treatment.
- Antisense expression is reduced in response to drought conditions. The full pictures are shown in Fig EV3. Scale bar, 1 cm.
- Quantification of emitted light intensity per plant for control and drought‐treated plants.
- Strand‐specific RT–qPCR asDOG1 expression analysis of 3‐day Col‐0 (WT) drought‐treated vs. control plants.
- The asDOG1 expression level (black line) is reduced while DOG1 sense expression (grey line) is increased after application of ABA. Leaves of mature Col‐0 (WT) 40‐day‐old plants were sprayed with ABA and collected 0, 6 and 10 h later for RNA extraction and RT–qPCR. Signals were normalized against the level of the UBC transcript.
Figure EV2. ABA response of 20‐day‐old transgenic lines expressing pASDOG 1::LUC .
-
A, BLUC pictures taken after 24 h of treatment and LUC quantification per seedlings of 20‐day‐old transgenic lines expressing p AS DOG1::LUC. Scale bar, 2 cm. ***t‐test‐derived P‐value < 0.001. Error bars represent standard deviation, n = 9 for each of three independent lines.
-
CLUC pictures of 40‐day‐old transgenic Arabidopsis lines expressing p DOG1 shDOG1::LUC. Scale bar, 2 cm.
-
Dp DOG1 LUC::DOG1 constructs after mock and ABA treatments. Scale bar, 2 cm.
Figure EV3. Analysis of drought responses in pASDOG 1::LUC transgenic plants and dog1 mutants.
-
A, BDownregulation of antisense transcription by drought of two independent p AS DOG1::LUC lines. Scale bar, 1 cm.
-
CGraph represents quantification of light intensity per plant. Error bars represent standard deviation. *t‐test‐derived P‐value < 0.05, n = 12 for each of two lines.
-
DDrought susceptibility of dog1‐3 and dog1‐4 mutants in comparison with Col‐0 (WT) plants after 5 days of treatment, independent repeat. Scale bar, 2 cm.
-
EGrowth phenotype of Col‐0 (WT) and dog1‐3 mutant plants used for RT–qPCR analysis of DOG1 sense/antisense and marker genes at 3 days of drought. Scale bar, 2 cm.
We next analysed how asDOG1 expression responds to ABA in Col‐0 (WT) plants by using RT–qPCR to monitor mRNA levels. In agreement with our initial observations (Fig 1A and B), we detected a strong (approx. 80%) reduction in asDOG1 transcript abundance 10 h after ABA treatment. Simultaneously, we saw a ten‐fold increase in the level of the short functional form of DOG1 mRNA (Fig 1F).
Thus, asDOG1 displays a tissue‐specific expression pattern and is regulated by external signals. This response is selective, since the antisense expression was strongly downregulated by ABA but was not affected by GA (Fig 1B and Appendix Fig S1). The concomitant downregulation of antisense and upregulation of sense expression in response to ABA suggest that these processes are interconnected, but do not allow the causative factor to be singled out (Fig 1F).
Mutation of the DOG1 gene causes enhanced sensitivity to drought
The strong upregulation of DOG1 expression following ABA treatment suggests that this gene may perform some unrecognized novel functions during stress. This notion received support from the finding that DOG1 knockout plants (dog1‐3 and dog1‐4, described in 1, 9, 14 and shown in Fig EV1E), displayed a drought‐sensitive phenotype in comparison with Col‐0 (WT) plants. In the 15 plants of each line subjected to water deprivation, 14 of the WT recovered, while, respectively, none and one of the dog1‐3 and dog1‐4 mutants recovered (Figs 2A and B, and EV3D). To confirm this observation by other means, watering was withheld for a short time (2.5–3 days), and at this time point, there was no major visible phenotypical difference between WT and dog1 mutant (Fig EV3E). This resulted in DOG1 sense mRNA upregulation (Fig 2C). Subsequently, the expression of stress response marker genes KIN1, RD29A, RD29B, RD22 and RAB18 was analysed by RT–qPCR. In agreement with a previous report 48, we observed strong upregulation of all tested marker genes upon drought treatment in Col‐0 (WT) plants. In contrast, upregulation of four out of the five tested genes (KIN1, RD29B, RD29A, RAB18) was diminished in the dog1‐3 mutant (Fig 2D). For example, we observed > 147‐fold induction of KIN1 in WT plants, but only ~ 34‐fold upregulation in the dog1‐3 mutant in response to drought (Fig 2D). Furthermore, ABA treatment triggered similar responses in the expression of the selected marker genes (Fig EV4).
Figure 2. dog1 mutant plants are susceptible to drought stress.
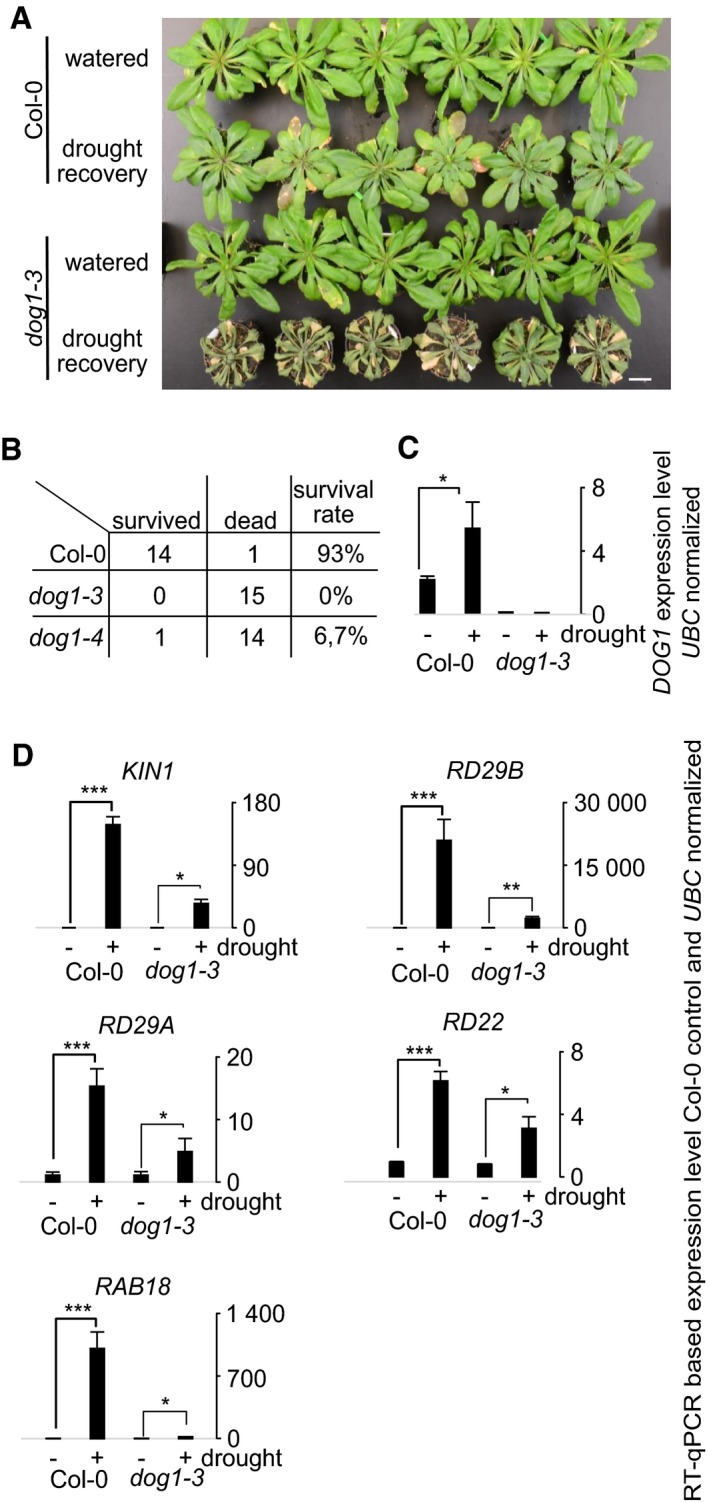
- Col‐0 (WT) and dog1‐3 mutant plants were either watered normally or subjected to water withdrawal for 5 days and then watered again and allowed to recover for 2 days. A picture of a representative experiment is shown. Scale bar, 2 cm.
- Quantification of recovered and dead Col‐0 (WT), dog1‐3 and dog1‐4 plants; plants were scored as dead when unable to grow and produced offspring after an extended recovery period.
- Analysis of DOG1 sense expression in drought‐treated vs. control Col‐0 (WT) and dog1‐3 mutant plants, n = 4.
- dog1‐3 mutant plants are defective in the regulation of the majority of the tested drought marker genes. Col‐0 (WT) and dog1‐3 mutant plants were subjected to drought for 3 days, and the expression of five marker genes was determined using RT–qPCR, n = 4.
Figure EV4. RT–qPCR analysis of selected marker gene expression in Col‐0 (WT) and dog1‐3 mutant after 10 h of 100 μM ABA spray‐inoculation.
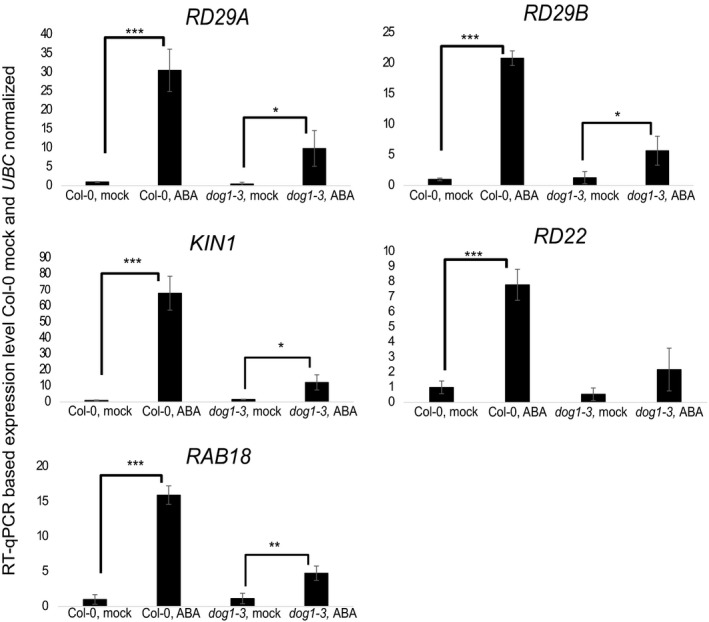
Error bars represent standard deviation. *, ** and *** represent t‐test P‐values of < 0.05, < 0.01 and < 0.001, respectively, n = 4.
Thus, by examining the response of asDOG1 expression to plant hormones, we discovered a novel and unexpected function of the major seed dormancy QTL DOG1 in drought response. DOG1 mRNA expression was induced by ABA and drought (Figs 1F and 2C) and DOG1 knockout plants showed weaker induction of stress marker genes following water deprivation as well as enhanced susceptibility to drought (Fig 2 and Appendix Fig S2).
ABA‐dependent DOG1 regulation requires antisense transcription
In response to ABA and drought, we observed upregulation of DOG1 and downregulation of asDOG1 (Figs 1, 2, EV2 and EV3, Appendix Fig S2). To further characterize the DOG1 response to ABA, we studied plants expressing a full‐length DOG1 transgene fused with the LUC reporter gene and driven under control of DOG1 sense promoter (p DOG1 LUC::DOG1). These plants were subjected to ABA treatment at different stages of development and LUC activity measured (Fig 3A and B). This analysis showed that DOG1 transcription was significantly increased at 10 days, but the strongest induction was observed in 40‐day‐old plants, in our growing conditions corresponding to the stage just before bolting. Given the ability of the antisense promoter to respond to ABA when separated from sense transcription (Fig 1A and B), we tested whether the inverse was the case for the DOG1 sense promoter. We found that removal of DOG1 antisense transcription rendered the truncated construct p DOG1 shDOG1::LUC insensitive to ABA at all tested stages of development (Fig 3C and D). This Δ antisense version of DOG1 was expressed at levels similar to or higher than those observed after ABA treatment of p DOG1 LUC::DOG1 plants (Figs 3C and D, and EV2C and D). Importantly, RT–qPCR analysis for sense and strand‐specific analysis for antisense confirmed that DOG1 is induced and asDOG1 is reduced at all tested developmental stages, with the strongest effect observed at older stages (Fig 3E and F).
Figure 3. DOG1 upregulation by ABA is strongest 40 days after germination and requires asDOG1 .
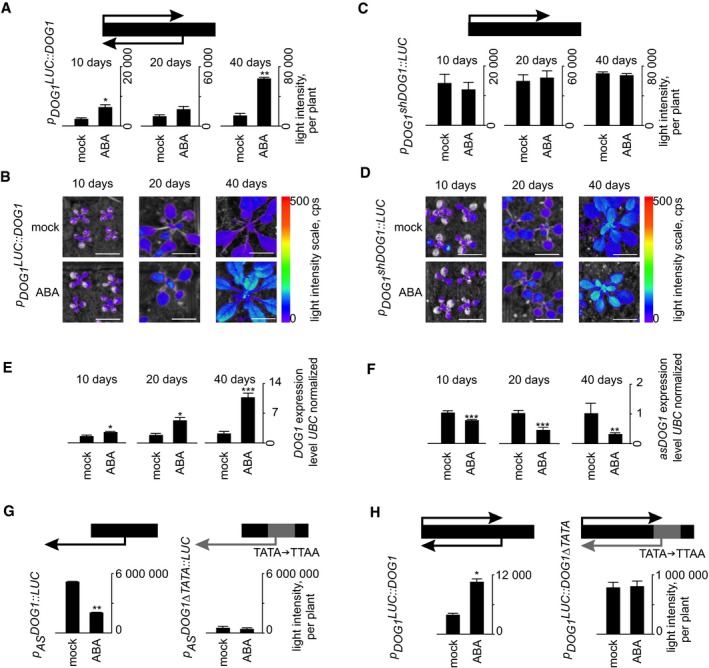
- Plants expressing LUC fused with full‐length DOG1 (p DOG1 LUC::DOG1) were sprayed with mock solution or ABA 10, 20 and 40 days after germination and analysed 24 h after treatment. The graphs show mean emitted light intensity per plant, n = minimum 9 per each of two lines.
- Representative picture of mock‐ and ABA‐treated p DOG1 LUC::DOG1 plants. The full picture is shown in Fig EV2C and D. Scale bar, 1 cm.
- A truncated construct that lacks the DOG1 antisense promoter region (p DOG1 shDOG1::LUC) is not induced by ABA and is highly expressed throughout development. The graphs show mean emitted light intensity per plant, n = minimum 9 per each of two lines.
- Representative picture of mock‐ and ABA‐treated p DOG1 shDOG1::LUC plants. Scale bar: 1 cm.
- RT–qPCR analysis of DOG1 sense mRNA level, n = 4.
- Strand‐specific RT–qPCR analysis of antisense levels after 10 h of ABA treatment in 10‐, 20‐ and 40‐day‐old Col‐0 plants, n = 4.
- The p AS DOG1::LUC reporter is silenced by the application of ABA, while mutation of TATA elements in the asDOG1 promoter (p AS DOG1∆TATA::LUC) leads to attenuation of expression in the presence and absence of ABA, n = 8 per each of three lines.
- Mutation of TATA elements in the asDOG1 promoter leads to high‐level DOG1 sense expression and non‐responsiveness to ABA, n = minimum 8 per each of three lines.
This indicated that the deleted 3′ region contains elements required for the ability of DOG1 to respond to ABA and drought. The construct p DOG1 shDOG1::LUC extends to the end of DOG1 exon 2 and lacks not only the antisense promoter but also the alternative splice sites used to generate the long three‐exon version of DOG1 mRNA, as well as the proximal and distal polyA sites 14. RT–qPCR analysis showed that neither alternative splicing nor use of the alternative polyA sites was affected in the response to ABA (Appendix Fig S2C and D). This indicates asDOG1 as a candidate for mediating upregulation of DOG1 mRNA in response to ABA.
To confirm the role of asDOG1 in DOG1 regulation in response to drought, we took advantage of the occurrence of canonical promoter‐like features in antisense promoters of Arabidopsis previously demonstrated by us 31. In agreement with the reported overrepresentation of TATA box elements in terminators producing antisense transcripts 31, a number of such elements were detected in the DOG1 antisense promoter. To test their significance in the regulation of asDOG1 expression, we mutated 32 predicted TATA elements (defined using the PLACE 49 and PlantCARE 50 web tools) in the antisense promoter driving the IRES‐LUC reporter gene (p AS DOG1::LUC). These sequences were converted to TTAA or TTTA to create the construct p AS DOG1∆TATA::LUC (Appendix Fig S3). Transgenic plants with this construct produced very weak LUC activity in comparison with those transformed with the non‐mutated control p AS DOG1::LUC construct, indicating the importance of the mutated TATA boxes in DOG1 antisense promoter function (Fig 3G). Moreover, the application of ABA strongly suppressed expression from the control construct, but had no effect on LUC activity produced by the p AS DOG1∆TATA::LUC‐transformed plants (Fig EV5).
Figure EV5. Bioluminescence analysis of transgenic lines expressing pASDOG 1::LUC and pASDOG 1::LUCΔTATA lines after 24 h.
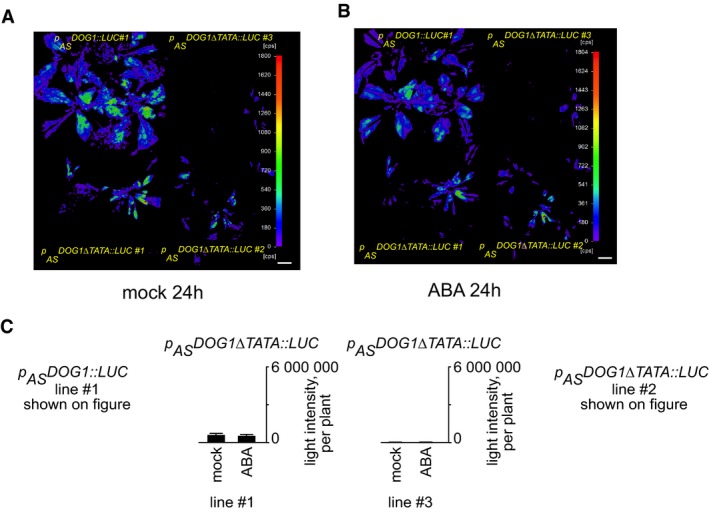
-
A, BPlants were mock treated and imagined after 24 h, subsequently sprayed with 100 mM ABA and imaged 24 h later. Scale bar, 2 cm.
-
CQuantification of LUC pictures as relative light intensity per plant. Error bars represent standard deviation, n = 8 for each of three lines.
The mutation of TATA boxes in the DOG1 antisense promoter appeared to effectively suppress its activity. Next, we introduced the TATA box mutations into the context of the genomic DOG1 locus (p DOG1 LUC::DOG1∆TATA), which allowed us to directly test the effect of antisense on DOG1 expression and its responsiveness to ABA. The p DOG1 LUC::DOG1∆TATA construct was expressed at a much higher level than the control construct p DOG1 LUC::DOG1, and it had lost the ability to respond to ABA (Fig 3H).
In summary, removal of the DOG1 3′ region containing the antisense promoter or mutation of TATA box elements required for DOG1 antisense transcription rendered the DOG1 gene unresponsive to ABA. These data are consistent with a model in which under standard conditions DOG1 expression is continuously silenced by antisense. In response to ABA and drought, antisense promoter activity is suppressed resulting in release of silencing and upregulation of DOG1 sense expression.
Drought resistance phenotype of plants expressing antisense‐deficient DOG1
Our data showed that DOG1 antisense transcription is crucial for the ABA/drought response that results in the release of antisense‐mediated silencing of DOG1 sense expression. Notably, dog1 mutants showed enhanced sensitivity to drought (Figs 2 and EV3D), which indicated that constitutive high‐level expression of DOG1 caused by the removal of antisense expression might lead to drought resistance. To test this hypothesis, we examined transgenic lines expressing the short DOG1::LUC fusion construct under the control of the native promoter (p DOG1 shDOG1::LUC). This construct was shown to be functional in seeds, as judged by its ability to partially complement the dog1‐3 seed dormancy phenotype 9. All three independent transgenic lines tested showed increased drought resistance when compared to Col‐0 (WT) and transgenic plants containing p DOG1 LUC::DOG1 or p AS DOG1::LUC (Fig 4A and Appendix Fig S4). RT–qPCR analysis of selected stress marker genes showed a significant increase in the expression of four of the five tested genes (RD29A, RD29B, RD22 and KIN1) in two p DOG1 shDOG1::LUC lines compared to Col‐0 (WT) in the absence of ABA treatment (Fig 4B). However, following treatment with ABA, there was no clear difference in the expression of these four stress marker genes between the p DOG1 shDOG1::LUC lines and Col‐0 (WT). This suggests that enhanced drought tolerance of plants with constitutive high DOG1 expression results from partial activation of stress response in Arabidopsis even in the absence of drought.
Figure 4. High‐level DOG1 expression due to antisense removal causes enhanced drought resistance in Arabidopsis .
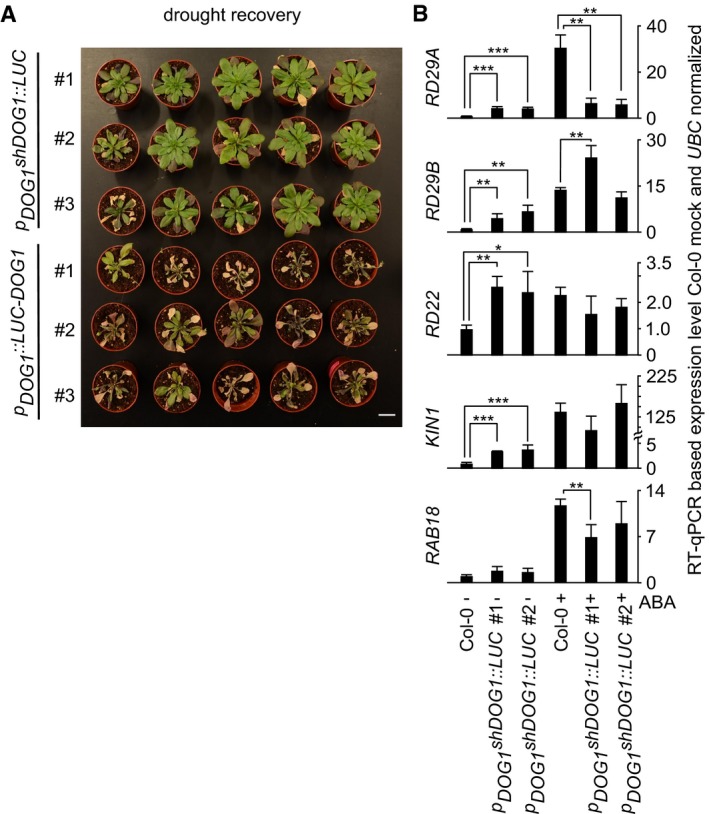
- Plants expressing the truncated asDOG1 promoter‐deficient version p DOG1 shDOG1::LUC are more resistant to drought than plants expressing the full‐length genomic version p DOG1 ::LUC‐DOG1. Three independent lines are shown for each construct, following 10 days of water deprivation. Scale bar, 2 cm.
- RT–qPCR quantification of selected drought marker genes in mock‐ and ABA‐treated Col‐0 (WT) and two independent p DOG1 shDOG1::LUC transgenic lines. Error bars represent standard deviation, and *, ** and *** represent t‐test P‐values of < 0.05, < 0.01 and < 0.001, respectively, n = 4.
Discussion
DOG1 is a novel player in the drought response of Arabidopsis
We have shown that Arabidopsis dog1 mutants are sensitive to drought, while DOG1‐overexpressing plants are more resistant to this stress than the wild type (Figs 2 and 4). Therefore, apart from its well‐characterized function in seed dormancy 1, 51, 52, DOG1 is also an important player in the Arabidopsis drought response.
Interestingly, seed dormancy and drought share many similarities. Most striking is the water deficit caused by the developmental programme of seed desiccation and by external conditions affecting plants 53. The similarities also extend to the molecular players involved, including the plant hormone ABA and its transduction pathway, which are required for both strong seed dormancy and drought resistance 2, 54, 55, 56, 57. Our findings now add DOG1 to this list. DOG1 antisense acts as a suppressor of DOG1 expression in seeds and also in leaves, where it suppresses seed dormancy and drought response, respectively.
DOG1 was characterized as an important player in Arabidopsis seed dormancy regulation over a decade ago 1. However, despite numerous subsequent studies employing metabolomic and proteomic approaches 2, 58, 59, how the DOG1 protein mechanistically controls seed dormancy is still one of the most challenging questions in the field 19. DOG1 protein is a plant‐specific protein that has neither extensive homology to known proteins outside the plant kingdom nor contains any domain of known function, but its dimerization has been shown to be required for its ability to enforce seed dormancy 18.
Our analysis of stress response marker genes showed that dog1 mutant and DOG1‐overexpressing plants have contrasting patterns of expression for the majority of the tested genes (Figs 2D and 4B). The selected marker genes are all downstream effectors of the ABA signal transduction pathway 48, suggesting that drought resistance is mediated by DOG1 either directly or indirectly via the ABA pathway. In support of this notion, a recent study demonstrated misregulation of ABI5, an ABA pathway component, in dog1 mutant seeds 2. Genetic interaction between DOG1 and ABI3 or nced9, an ABA pathway component and ABA biosynthesis mutant, respectively, has also been reported 2, 5. Together these data suggest that DOG1 protein may control both seed dormancy and drought resistance through similar mechanisms involving the ABA pathway. We also showed that DOG1 expression is upregulated in leaves by exogenous ABA or drought treatment (Figs 1F, 2 and 3E). This implies that DOG1 expression is under the control of the ABA signalling pathway. It has been reported that ABA has a positive effect on expression of the Arabidopsis DOG1 Cvi allele 59 and DOG1 homolog in Lepidium sativum and Sisymbrium officinale 4, 60 during seed imbibition by an unknown mechanism. Recent efforts have also shown that DOG1 protein directly interacts with a number of PP2C phosphatases and genetically requires PP2C phosphatases to impose dormancy on developing seeds 61.
On the one hand, our data indicate that DOG1 acts upstream of the ABA stress response pathway, while on the other, it suggests that it is regulated by ABA. In the absence of mechanistic data concerning the function of DOG1 protein, it is currently not possible to precisely place this protein within the drought response and dormancy establishment pathways.
DOG1 was initially characterized as an important player in seed dormancy regulation. In agreement with this function, DOG1 mRNA is highly expressed in seeds but is nearly undetectable in seedlings 1, 14. Analysis of available high‐throughput data show that DOG1 expression is upregulated under osmotic stress conditions 62, 63. Despite this, DOG1 has been repeatedly identified as a GWAS for flowering time 15, although no flowering time phenotype has been observed in Arabidopsis dog1 mutants grown under standard conditions. However, RNAi‐based silencing of the DOG1 homolog in wheat and lettuce results in early flowering 3, 5. We have shown that mutants in DOG1 are more susceptible to drought (Figs 2A and EV3D). Therefore, it is possible that under mild conditions of water deprivation the enhanced drought sensitivity of dog1 mutants alters the flowering time. This remains to be tested directly, but drought has been shown to cause early flowering as part of a stress‐escape response 64. Interestingly, this effect is at least partially dependent on the ABA pathway 65, 66. In addition, recently a number of drought‐associated QTLs have been identified between Kas‐1 and Tsu‐1, one of which maps close to DOG1 loci 67.
Antisense requirement for the response of DOG1 to changes in the external environment
In plants grown under standard conditions, DOG1 mRNA is exclusively found in seeds, where its expression is extensively regulated 1. This includes the recently described inhibition of DOG1 expression by a long non‐coding antisense transcript asDOG1, also known as 1GOD 14. Our analysis using RT–qPCR showed that asDOG1 is highly abundant in seedlings and mature leaves, where its expression is very efficiently suppressed by drought and ABA (Figs 1, 3F, EV2 and EV4). Moreover, using multiple approaches, we demonstrated that asDOG1 is required for the response of the DOG1 gene to drought/ABA signals. In the absence of this antisense transcript, DOG1 is constitutively highly expressed in leaves (Fig 3C and D). Our data define the inhibition of asDOG1 promoter activity by ABA as an early step in the DOG1 response to drought, which is an example of ncRNA‐dependent sensing of external conditions. This observation corroborates the reported importance of ncRNA transcripts for environmental response, including COOLAIR, an antisense transcript generated from the FLC locus 32, 68, 69, 70.
We recently described an antisense‐based mechanism for the regulation of gene expression involving 3′ end‐bound SWI/SNF complexes 31. Interestingly, the majority of the 3′ bound SWI/SNF targets that we identified are genes that are extensively regulated by the external and internal environment 31, 71. This suggests that the requirement for antisense of DOG1, in its response to environmental changes, is a common but underappreciated mechanism for regulating gene expression in plants. In agreement with this notion, multiple reports have shown that plant ncRNAs including antisense transcripts are extensively regulated by external cues 30, 32, 37. Antisense transcription has been implicated in the regulation of gene expression in response to environmental cues in other eukaryotes including yeast and mammals 38, 72, 73. However, in contrast to FLC and examples from yeast, DOG1 antisense not only facilitates the environmental response, but seems to be absolutely required for it (Fig 3). This may reflect differences in the physiological nature of the processes controlled by antisense, and the lack of parallel pathways for the upregulation of DOG1 in response to drought.
Based on our data, we propose a model integrating DOG1 regulation and function in the establishment of drought resistance in Arabidopsis (Fig 5). In this model, the antisense transcript asDOG1 limits DOG1 expression in vegetative tissue. Increased ABA levels inhibit antisense expression, and the silencing of sense expression is released. The proximally polyadenylated short DOG1 (shDOG1) mRNA is transcribed and translated to produce functional DOG1 protein. Based on our marker gene analysis and available genetic data from seeds, we speculate that DOG1 protein modulates the ABA pathway to implement the final level of drought resistance.
Figure 5. Model describing the drought response of DOG1, which depends on the function of the antisense transcript asDOG1 .
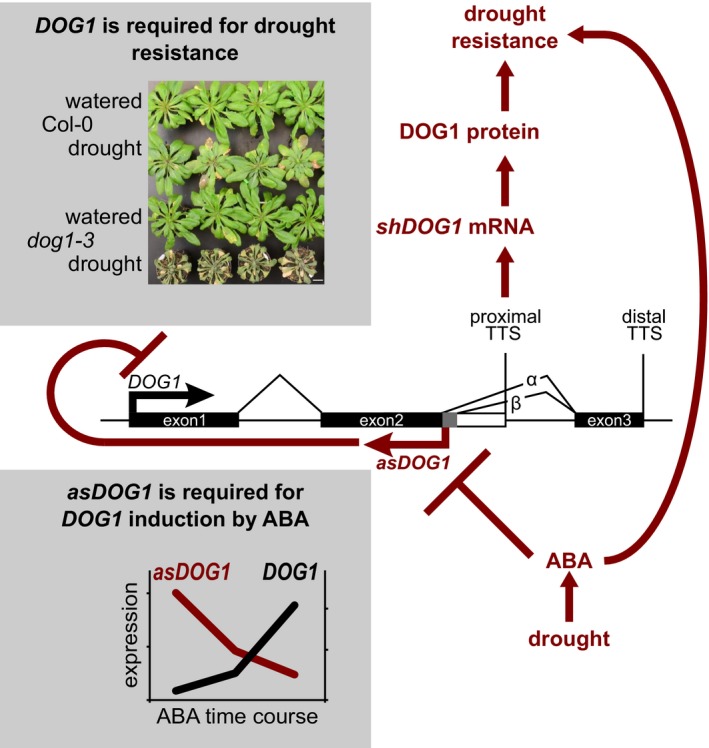
For a detailed description see the Discussion. Drought causes ABA induction that leads to drought resistance. Part of this response is due to silencing of asDOG1, which results in the induction of DOG1 transcription as shown in the bottom panel (adapted from Fig 1F). The induction of DOG1 expression is required for and contributes to the final ABA‐induced drought resistance, as shown in the top panel (adapted from Fig 2A).
We have previously shown that in seeds, asDOG1/1GOD suppresses DOG1 expression in “cis” but the molecular mechanism of that suppression is currently not clear. Here we show that in response to ABA and drought asDOG1 levels are reduced, releasing DOG1 expression. The fact that the asDOG1 deficient lines p DOG1 shDOG1::LUC and p DOG1 DOG1∆TATA::LUC are constitutively highly expressed in the presence and absence of ABA (Fig 3) suggests that the asDOG1 originating from the endogenous copy is unable to silence in “trans” DOG1 sense expression in leaves as shown before by us in seeds 14. The molecular mechanism of asDOG1‐mediated DOG1 suppression is currently not clear. Importantly, the DOG1 locus is devoid of DNA methylation, small RNA or high H3K9me2, suggesting that the molecular mechanism may not involve RNA interference but may be based on cis‐acting mechanisms linked more directly to antisense transcription 74.
Our data demonstrated that DOG1 antisense suppresses DOG1 expression not only in seeds but also in mature Arabidopsis plants. By studying asDOG1 responsiveness to stimuli, we have discovered a novel unexpected function of DOG1 in drought response. Finally, our data showed an absolute requirement for asDOG1 in the DOG1 response to drought and ABA.
Materials and Methods
Plant materials, growth conditions
Arabidopsis seeds were sterilized as described 14 then plated to ½ MS plates and grown in long‐day (LD) conditions at 22°C/18°C. Arabidopsis thaliana plants were grown in soil in a greenhouse with an LD photoperiod (16 h light/8 h dark) at 22°C/18°C. For all experiments, Col‐0 was used as the WT background. The DOG1 T‐DNA insertion mutants dog1‐3 (SALK_000867) and dog1‐4 (SM_3_20886) were previously described and characterized 1, 9, 14. To analyse asDOG1 function in seeds we used the dog1‐5 mutant which has a T‐DNA insertion in the DOG1 exon 3 region, resulting in low antisense expression 14. However, in seedlings of this mutant, the level of asDOG1 was only slightly affected compared to Col‐0 (WT) control seedlings (Appendix Fig S5), precluding its use.
Cloning of genetic constructs and generation of Arabidopsis transgenic plants
Several bioinformatics tools were used to predict full‐length sense and antisense DOG1 promoters by analysing the DNA sequence for the presence of potential cis‐regulatory elements and transcription initiation sites specific for DOG1 and surrounding genes. According to bioinformatics analysis of genomic regions, we amplified DOG1 sense (p DOG1 shDOG1::LUC, −1,155; +1,900 from ATG) and antisense (p AS DOG1::LUC, +1,143; −996 from TGA) promoters from Arabidopsis Col‐0 (WT) plants and cloned into a pGWB635‐LUC expression vector. The full‐length LUC::DOG1 genomic construct (p DOG1 LUC::DOG1) used in our study was previously described and characterized 14, 31. All constructs were transformed into Agrobacterium tumefaciens GV3101 strain by electroporation and subsequently used for generation of stable transgenic lines, as described 75. T3 homozygous lines were used for analysis.
RNA extraction, cDNA synthesis and adapter‐mediated RT–qPCR assay
Total RNA was extracted from seedlings or leaves using TRIsure (Bio‐Rad). Samples were treated with TURBO DNase (Ambion) according to the standard manufacturer protocol and efficiency of DNA removal was analysed using PCR with PP2A primers 14. The quality and amount of RNA samples were tested on 1.2% agarose gel and a NanoDrop 2000 spectrophotometer; 2–2.5 μg RNA was used for cDNA synthesis (sense and antisense, correspondingly). cDNA for sense transcript was synthesized using oligo dT primers. cDNA for antisense analysis was synthesized using a gene‐specific primer with an adapter, followed by qPCR with a tag‐specific primer (AS_SS_RT) and DOG1 primers (AS_F, AS_R) as described in 14. RT–qPCR was performed using a LightCycler 480 real‐time system (Roche) with SYBR Green mix (Roche). RT–qPCR results were normalized against the expression level of the Arabidopsis UBC21 (PEX4) gene as described previously 76. P‐values presented on graphs indicate *P < 0.05, **P < 0.01 and ***P < 0.001, calculated using a two‐tailed t‐test in Microsoft Office Excel. Error bars represent standard deviation.
Drought stress
Arabidopsis Col‐0 (WT), dog1‐3 and dog1‐4 mutants and the T3 generation of Arabidopsis transgenic lines expressing p DOG1 shDOG1::LUC, p DOG1 LUC::DOG1 and p AS DOG1::LUC subjected to drought test were grown in soil. All seeds were imbibed at 4°C for 3 days and then planted into 7‐cm pots with perlite‐supplemented soil. Seven to ten days later, individual seedlings were transferred into 5‐cm G‐type TEKU pots (Poeppelmann, USA) which facilitate drying, and were grown for 4–5 weeks under normal water conditions. Watering was withheld just before the bolting stage, for 4.5–5 days in the case of Col‐0 (WT), dog1‐3 and dog1‐4 mutants and up to 9–10 days in case of transgenic lines expressing DOG1 sense, antisense and full‐length genomic constructs. Plants were re‐watered and assessed for survival on the second day after re‐watering based on the protocol adapted from 77. Drought experiments were repeated at least three times.
Hormonal treatments and luciferase measurement
Hormonal treatment with ABA and GA4 hormones was performed on soil‐grown Arabidopsis seedlings or adult transgenic plants. Arabidopsis plants (10‐, 20‐ and 40‐day‐old) were spray‐inoculated with 100 μM ABA 78, 79 as well as 10 μM 80 and 50 μM GA4 (Appendix Fig S1) hormones using an EcoSprayer (www.sirchie.com, France). After spraying, plants were covered for an hour, transferred to growing chambers and subjected to either RNA extraction and RT–qPCR analysis or LUC imaging at defined time points. Luciferase treatment was performed as previously described 14; briefly, plants were kept in darkness for 10 min, sprayed with 0.5 mM luciferin, kept in darkness for 15–20 min, and then, emitted light was measured by a NightSHADE LB985 camera, with an exposure time of 10 min. Further, LUC data were processed and calculated using IndiGo software (ver. 2.0.5.0, Berthold Technology, Germany). Light intensity units were calculated as the sum of emitted light (cps—count per second) detected by camera per plant. Error bars represent standard deviation.
Data quantification and statistics
Data were quantified as described in the relevant sections. For luminescence quantification, the light signal intensity was normalized for plant area, and error bars represent the standard deviation of the signal between individual plants. All statistical tests were done using a two‐tailed t‐test, implemented in Microsoft Office Excel.
Promoter mutagenesis
Bioinformatics analysis of DOG1 antisense promoter revealed significant enrichment of classical TATA boxes in this region, mainly defined in the second (last) intron. We mutagenized all predicted TATA boxes in the antisense direction by introducing a point mutation in each TATA box element (TATA → TTAA or TTTA in case the former would create a new TATA box with the surrounding sequence). To mutate the identified 32 TATA boxes located in and around the second intron of the DOG1 gene, two independent DNA fragments of the DOG1 gene carrying corresponding mutations were synthesized by GeneArt Gene Synthesis Company (Invitrogen). Two fragments were amplified using the primers listed in Appendix Table S1, fused using fusion PCR and cloned into a pJET 1.2 vector and sequenced. Subsequently, a DNA fragment with mutations in the TATA boxes was introduced into the p DOG1 LUC::DOG1 construct via BamHI‐NcoI restriction sites and p AS DOG1::LUC via EcoRI‐SalI sites. Generated transgenic T2 lines were analysed for expression and for response to hormones as described above.
Author contributions
RY and SS conceived the study and designed the experiments; RY and KK cloned constructs and generated the LUC‐tagged DOG1 transgenic plants; RY and KK performed LUC analysis of DOG1 transgenic lines; RY, HF and AC performed drought experiments and RT–qPCR experiments; AK and GD were involved in analysis of drought/ABA signalling; RY and SS wrote the article.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
Grant funding numbers: This work was funded by Polish National Science Centre grant number 2011/01/D/NZ8/03690 to SS, Polish National Science Centre grant 2011/03/B/NZ3/00297 to GD and Polish National Science Centre grant 2014/13/D/NZ3/03101 to AK.
EMBO Reports (2017) 18: 2186–2196
References
- 1. Bentsink L, Jowett J, Hanhart CJ, Koornneef M (2006) Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis . Proc Natl Acad Sci USA 103: 17042–17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dekkers BJW, He H, Hanson J, Willems LAJ, Jamar DCL, Cueff G, Rajjou L, Hilhorst HWM, Bentsink L (2016) The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J 85: 451–465 [DOI] [PubMed] [Google Scholar]
- 3. Ashikawa I, Mori M, Nakamura S, Abe F (2014) A transgenic approach to controlling wheat seed dormancy level by using Triticeae DOG1‐like genes. Transgenic Res 23: 621–629 [DOI] [PubMed] [Google Scholar]
- 4. Graeber K, Linkies A, Müller K, Wunchova A, Rott A, Leubner‐Metzger G (2010) Cross‐species approaches to seed dormancy and germination: conservation and biodiversity of ABA‐regulated mechanisms and the Brassicaceae DOG1 genes. Plant Mol Biol 73: 67–87 [DOI] [PubMed] [Google Scholar]
- 5. Huo H, Wei S, Bradford KJ (2016) DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc Natl Acad Sci USA 113: E2199–E2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rikiishi K, Maekawa M (2014) Seed maturation regulators are related to the control of seed dormancy in wheat (Triticum aestivum L.). PLoS One 9: e107618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kerdaffrec E, Filiault DL, Korte A, Sasaki E, Nizhynska V, Seren Ü, Nordborg M (2016) Multiple alleles at a single locus control seed dormancy in Swedish Arabidopsis . eLife 5: e22502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dolata J, Guo Y, Koowerzo A, Smolinski D, Brzyzek G, Jarmoowski A, Swiezewski S (2015) NTR1 is required for transcription elongation checkpoints at alternative exons in Arabidopsis . EMBO J 34: 544–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cyrek M, Fedak H, Ciesielski A, Guo Y, Sliwa A, Brzezniak L, Krzyczmonik K, Pietras Z, Kaczanowski S, Liu F et al (2016) Seed dormancy in Arabidopsis is controlled by alternative polyadenylation of DOG1. Plant Physiol 170: 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Y, Koornneef M, Soppe WJJ (2007) The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19: 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grasser M, Kane CM, Merkle T, Melzer M, Emmersen J, Grasser KD (2009) Transcript elongation factor TFIIS is involved in Arabidopsis seed dormancy. J Mol Biol 386: 598–611 [DOI] [PubMed] [Google Scholar]
- 12. Zheng J, Chen F, Wang Z, Cao H, Li X, Deng X, Soppe WJJ, Li Y, Liu Y (2012) A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol 193: 605–616 [DOI] [PubMed] [Google Scholar]
- 13. Miatton E (2012) Characterization of PDF1 and its interaction with DELAY OF GERMINATION1 (DOG1) in the control of seed dormancy in Arabidopsis thaliana . PhD Thesis, University of Köln
- 14. Fedak H, Palusinska M, Krzyczmonik K, Brzezniak L, Yatusevich R, Pietras Z, Kaczanowski S, Swiezewski S (2016) Control of seed dormancy in Arabidopsis by a cis‐acting noncoding antisense transcript. Proc Natl Acad Sci USA 113: E7846–E7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT et al (2010) Genome‐wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Heerwaarden J, van Zanten M, Kruijer W (2015) Genome‐wide association analysis of adaptation using environmentally predicted traits. PLoS Genet 11: e1005594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alonso‐Blanco C, Andrade J, Becker C, Bemm F, Bergelson J, Borgwardt KM, Cao J, Chae E, Dezwaan TM, Ding W et al (2016) 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana . Cell 166: 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nakabayashi K, Bartsch M, Ding J, Soppe WJJ (2015) Seed dormancy in Arabidopsis requires self‐binding ability of DOG1 protein and the presence of multiple isoforms generated by alternative splicing. PLoS Genet 11: e1005737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nonogaki H (2014) Seed dormancy and germination‐emerging mechanisms and new hypotheses. Front Plant Sci 5: 233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nonogaki H (2017) Seed biology updates – highlights and new discoveries in seed dormancy and germination research. Front Plant Sci 8: 524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin S, Zhang L, Luo W, Zhang X (2015) Characteristics of antisense transcript promoters and the regulation of their activity. Int J Mol Sci 17: 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murray SC, Mellor J (2016) Using both strands: the fundamental nature of antisense transcription. Bioarchitecture 6: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Murray SC, Serra Barros A, Brown DA, Dudek P, Ayling J, Mellor J (2011) A pre‐initiation complex at the 3′‐end of genes drives antisense transcription independent of divergent sense transcription. Nucleic Acids Res 40: 2432–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Uhler JP, Hertel C, Svejstrup JQ (2007) A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci USA 104: 8011–8016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M (2008) A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell 32: 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Camblong J, Beyrouthy N, Guffanti E, Schlaepfer G, Steinmetz LM, Stutz F (2009) Trans‐acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae . Genes Dev 23: 1534–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gelfand B, Mead J, Bruning A, Apostolopoulos N, Tadigotla V, Nagaraj V, Sengupta AM, Vershon AK (2011) Regulated antisense transcription controls expression of cell‐type‐specific genes in yeast. Mol Cell Biol 31: 1701–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Dijk EL, Chen CL, d'Aubenton‐Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix‐Né P, Loeillet S et al (2011) XUTs are a class of Xrn1‐sensitive antisense regulatory non‐coding RNA in yeast. Nature 475: 114–117 [DOI] [PubMed] [Google Scholar]
- 29. Chekanova JA (2015) Long non‐coding RNAs and their functions in plants. Curr Opin Plant Biol 27: 207–216 [DOI] [PubMed] [Google Scholar]
- 30. Yamada M (2017) Functions of long intergenic non‐coding (linc) RNAs in plants. J Plant Res 130: 67–73 [DOI] [PubMed] [Google Scholar]
- 31. Archacki R, Yatusevich R, Buszewicz D, Krzyczmonik K, Patryn J, Iwanicka‐Nowicka R, Biecek P, Wilczynski B, Koblowska M, Jerzmanowski A et al (2016) Arabidopsis SWI/SNF chromatin remodeling complex binds both promoters and terminators to regulate gene expression. Nucleic Acids Res 45: 3116–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold‐induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802 [DOI] [PubMed] [Google Scholar]
- 33. Csorba T, Questa JI, Sun Q, Dean C (2014) Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci USA 111: 16160–16165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM et al (2006) Experimental validation of the regulated expression of large numbers of non‐coding RNAs from the mouse genome. Genome Res 16: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP et al (2009) Chromatin signature reveals over a thousand highly conserved large non‐coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon‐Vega B, Regev A, Rinn JL (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ben Amor B, Wirth S, Merchan F, Laporte P, d'Aubenton‐Carafa Y, Hirsch J, Maizel A, Mallory A, Lucas A, Deragon JM et al (2009) Novel long non‐protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res 19: 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Contreras‐Cubas C, Palomar M, Arteaga‐Vázquez M, Reyes JL, Covarrubias AA (2012) Non‐coding RNAs in the plant response to abiotic stress. Planta 236: 943–958 [DOI] [PubMed] [Google Scholar]
- 39. Duc C, Sherstnev A, Cole C, Barton GJ, Simpson GG (2013) Transcription termination and chimeric RNA formation controlled by Arabidopsis thaliana FPA. PLoS Genet 9: e1003867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sherstnev A, Duc C, Cole C, Zacharaki V, Hornyik C, Ozsolak F, Milos PM, Barton GJ, Simpson GG (2012) Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat Struct Mol Biol 19: 845–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Finch‐Savage WE, Leubner‐Metzger G (2006) Seed dormancy and the control of germination. New Phytol 171: 501–523 [DOI] [PubMed] [Google Scholar]
- 42. Hilhorst HWM, Karssen CM (1992) Seed dormancy and germination: the role of abscisic acid and gibberellins and the importance of hormone mutants. Plant Growth Regul 11: 225–238 [Google Scholar]
- 43. Millar AA, Jacobsen JV, Ross JJ, Helliwell CA, Poole AT, Scofield G, Reid JB, Gubler F (2006) Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′‐hydroxylase. Plant J 45: 942–954 [DOI] [PubMed] [Google Scholar]
- 44. Miransari M, Smith DL (2014) Plant hormones and seed germination. Environ Exp Bot 99: 110–121 [Google Scholar]
- 45. Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI (2010) Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev 24: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fujita Y, Fujita M, Shinozaki K, Yamaguchi‐Shinozaki K (2011) ABA‐mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res 124: 509–525 [DOI] [PubMed] [Google Scholar]
- 47. Fernando VCD, Schroeder DF (2016) Role of ABA in Arabidopsis salt, drought, and desiccation tolerance In Abiotic and biotic stress in plants – recent advances and future perspectives, Shanker AK, Shanker C. (eds), pp 507–524. Rijeka: InTech; [Google Scholar]
- 48. Saez A, Robert N, Maktabi MH, Schroeder JI, Serrano R, Rodriguez PL (2006) Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol 141: 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Higo K (1998) PLACE: a database of plant cis‐acting regulatory DNA elements. Nucleic Acids Res 26: 358–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis‐acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Graeber K, Nakabayashi K, Miatton E, Leubner‐Metzger G, Soppe WJJ (2012) Molecular mechanisms of seed dormancy. Plant Cell Environ 35: 1769–1786 [DOI] [PubMed] [Google Scholar]
- 52. Kronholm I, Picó FX, Alonso‐Blanco C, Goudet J, de Meaux J (2012) Genetic basis of adaptation in Arabidopsis thaliana: local adaptation at the seed dormancy QTL DOG1. Evolution 66: 2287–2302 [DOI] [PubMed] [Google Scholar]
- 53. Angelovici R, Galili G, Fernie AR, Fait A (2010) Seed desiccation: a bridge between maturation and germination. Trends Plant Sci 15: 211–218 [DOI] [PubMed] [Google Scholar]
- 54. Kermode AR (2005) Role of abscisic acid in seed dormancy. J Plant Growth Regul 24: 319–344 [Google Scholar]
- 55. Lim CW, Baek W, Jung J, Kim J‐H, Lee SC (2015) Function of ABA in stomatal defense against biotic and drought stresses. Int J Mol Sci 16: 15251–15270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sah SK, Reddy KR, Li J (2016) Abscisic acid and abiotic stress tolerance in crop plants. Front Plant Sci 7: 571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vishwakarma K, Upadhyay N, Kumar N, Yadav G, Singh J, Mishra RK, Kumar V, Verma R, Upadhyay RG, Pandey M et al (2017) Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front Plant Sci 8: 161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Holdsworth MJ, Bentsink L, Soppe WJJ (2008) Molecular networks regulating Arabidopsis seed maturation, after‐ripening, dormancy and germination. New Phytol 179: 33–54 [DOI] [PubMed] [Google Scholar]
- 59. Teng S, Rognoni S, Bentsink L, Smeekens S (2008) The Arabidopsis GSQ5/DOG1 Cvi allele is induced by the ABA‐mediated sugar signalling pathway, and enhances sugar sensitivity by stimulating ABI4 expression. Plant J 55: 372–381 [DOI] [PubMed] [Google Scholar]
- 60. Carrillo‐Barral N, Matilla AJ, García‐Ramas C, Rodríguez‐Gacio Mdel C (2015) ABA‐stimulated SoDOG1 expression is after‐ripening inhibited during early imbibition of germinating Sisymbrium officinale seeds. Physiol Plant 155: 457–471 [DOI] [PubMed] [Google Scholar]
- 61. Née G, Kramer K, Nakabayashi K, Yuan B, Xiang Y, Miatton E, Finkemeier I, Soppe WJJ (2017) DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat Commun 8: 72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg‐Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV‐B light, drought and cold stress responses: AtGenExpress global abiotic stress data set. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- 63. Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An ‘electronic fluorescent pictograph’ browser for exploring and analyzing large‐scale biological data sets. PLoS One 2: e718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Riboni M, Robustelli Test A, Galbiati M, Tonelli C, Conti L (2014) Environmental stress and flowering time: the photoperiodic connection. Plant Signal Behav 9: e29036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Riboni M, Galbiati M, Tonelli C, Conti L (2013) GIGANTEA enables drought escape response via abscisic acid‐dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1. Plant Physiol 162: 1706–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Riboni M, Robustelli Test A, Galbiati M, Tonelli C, Conti L (2016) ABA‐dependent control of GIGANTEA signalling enables drought escape via up‐regulation of FLOWERING LOCUS T in Arabidopsis thaliana . J Exp Bot 67: 6309–6322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lovell JT, Mullen JL, Lowry DB, Awole K, Richards JH, Sen S, Verslues PE, Juenger TE, McKay JK (2015) Exploiting differential gene expression and epistasis to discover candidate genes for drought‐associated QTLs in Arabidopsis thaliana . Plant Cell 27: 969–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang Z‐W, Wu Z, Raitskin O, Sun Q, Dean C (2014) Antisense‐mediated FLC transcriptional repression requires the P‐TEFb transcription elongation factor. Proc Natl Acad Sci USA 111: 7468–7473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Angel A, Song J, Yang H, Questa JI, Dean C, Howard M (2015) Vernalizing cold is registered digitally at FLC. Proc Natl Acad Sci USA 112: 4146–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rosa S, Duncan S, Dean C (2016) Mutually exclusive sense–antisense transcription at FLC facilitates environmentally induced gene repression. Nat Commun 7: 13031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Li C, Gu L, Gao L, Chen C, Wei C‐Q, Qiu Q, Chien C‐W, Wang S, Jiang L, Ai L‐F et al (2016) Concerted genomic targeting of H3K27 demethylase REF6 and chromatin‐remodeling ATPase BRM in Arabidopsis . Nat Genet 48: 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang X, Song X, Glass CK, Rosenfeld MG (2011) The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harb Perspect Biol 3: a003756–a003756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Beck ZT, Xing Z, Tran EJ (2016) LncRNAs: bridging environmental sensing and gene expression. RNA Biol 13: 1189–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Krzyczmonik K, Wroblewska‐Swiniarska A, Swiezewski S (2017) Developmental transitions in Arabidopsis are regulated by antisense RNAs resulting from bidirectionally transcribed genes. RNA Biol 14: 838–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- 76. Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W‐R (2005) Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis . Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhao Y, Chan Z, Gao J, Xing L, Cao M, Yu C, Hu Y, You J, Shi H, Zhu Y et al (2016) ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci USA 113: 1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang Y, Li L, Ye T, Lu Y, Chen X, Wu Y (2013) The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis . J Exp Bot 64: 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Xiong L, Lee H, Ishitani M, Zhu J‐K (2002) Regulation of osmotic stress‐responsive gene expression by the LOS6/ABA1 locus in Arabidopsis . J Biol Chem 277: 8588–8596 [DOI] [PubMed] [Google Scholar]
- 80. Andrés F, Porri A, Torti S, Mateos J, Romera‐Branchat M, García‐Martínez JL, Fornara F, Gregis V, Kater MM, Coupland G (2014) SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc Natl Acad Sci USA 111: E2760–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File


