Abstract
Mast cells are tissue-resident, innate immune cells present in most tissues of the body and are important effector and immunomodulatory cells. Differentiated mast cells typically are characterized by the surface expression of the receptors KIT and FcεRI, the latter especially being important for stimulation through IgE antibodies, although these cells have the ability to respond to a wide variety of environmental signals, to which they can variably react by releasing pre-stored or de novo–synthesized mediators or both. Since mast cells terminate their differentiation in their tissue of residence in response to specific microenvironmental cues, each tissue may comprise unique mast cell subtypes, and responses are tailored to the danger signals that are likely to be encountered in each anatomical location. From a transcriptional point of view, these cells therefore must be endowed with epigenetic and transcriptional programs that allow them to maintain a stable identity and at the same time allow sufficient plasticity to adapt to different environmental challenges. In this commentary, we highlight some of the recent findings that advanced our understanding of the transcriptional and epigenetic programs regulating mast cell functions.
Keywords: mast cell response, transcription factors, epigenetic control
Introduction: how many types of mast cells are there and what do they do?
Mast cells are one of the innate immune cell types involved in the first line of defense from pathogens that attempt to breach the epithelial barriers of our organism 1, 2. Indeed, these cells are most notably located in vascularized tissues, including the skin, the mucosa of the lungs, and the gastrointestinal tract, where they reside primarily at the interface with the environment, namely beneath the epithelial surface. The clearest examples of immune-related responses in which mast cells play a key role are in the context of allergy 3– 6 as well as in immunity against parasites 7. However, these cells have been involved in a plethora of processes, either protective of (tissue homeostasis and wound healing) or damaging to (chronic inflammation and cancer and autoimmune diseases 2, 8, 9) the organism, and some of the proposed functions of these cells have also become a matter of debate 1, 10. The main reasons for the difficulties in obtaining concluding evidence about mast cell functions in vivo include the complications linked to studying tissue-resident cells as well as the complexity of mast cell phenotypes and responses. Indeed, mast cells carry a wide repertoire of receptors and can be activated by an impressive number of different stimuli 11 ( Figure 1), which can lead to a battery of different responses that may occur together or independently 12. These include degranulation with release of mediators pre-stored in cytoplasmic granules 1, 2, de novo synthesis of cytokines and chemokines, release of exosomes that may act over long distances 13, and even release of DNA extracellular traps 14. These widespread possibilities of stimuli and responses, together with the fact that these are exclusively tissue-resident cells difficult to extract in sufficient number without inducing any modification to their biology, have made the task of pinpointing their main functions remarkably challenging. For example, a study aimed at defining the human mast cell transcriptome clearly showed how mast cell transcriptional responses change dramatically upon in vitro culture with interleukin-4 (IL-4) and stem cell factor 15 as compared with freshly isolated mast cells from human skin 16. Transcriptional changes reflected primarily metabolic activation, most likely linked to culture-induced cell cycle progression; however, other transcriptional changes (such as the induction of genes characteristic of other lineages) were suggestive of problems in fully maintaining cell identity in vitro. To add to this complexity, circulating human mast cell progenitors are very rare and difficult to differentiate in vitro, and so far no in vitro system has been able to recapitulate the wide variety of phenotypes or states that are likely to exist in vivo. Indeed, the complexity of signals and microenvironmental cues that lead to the migration of mast cell progenitors to specific tissues remains to be fully unraveled, although these cells are clearly influenced by changes in their cytokine milieu and by the presence of activating factors 12.
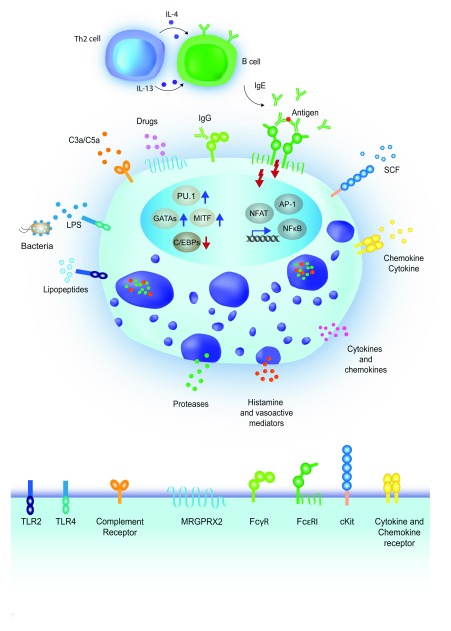
Figure 1. Mast cells and their receptors.
Mast cells are highly reactive cells expressing a plethora of receptors with specificity toward many types of stimuli. Upon activation, mast cells can release pre-formed mediators stored in cytoplasmic granules, including proteases and vasoactive mediators, while cytokines and chemokines can be either pre-stored or de novo–synthesized. The most commonly studied pathway for mast cell activation includes the engagement of the high-affinity IgE receptor (FcεRI), and is dependent on the production of antigen-specific IgE antibodies by B lymphocytes, in response to interleukin-4 (IL-4) and IL-13 produced by T helper 2 (Th2) lymphocytes 26. Crosslinking of the FcεRI-bound IgE by antigens results in mast cell degranulation and cytokine production 27. Mast cells can also express surface receptors for IgG antibodies, namely FcγR, whose engagement can lead to both activating and inhibitory signals, depending on the specific receptor involved 28. A variety of pathogen recognition receptors (PRRs), including Toll-like receptors (TLRs), are also expressed on the mast cell surface. For example, TLR2 is activated by bacterial lipopeptides, while TLR4 is activated by lipopolysaccharide (LPS) binding. Notably, TLR-mediated activation does not usually lead to mast cell degranulation, but triggers the production of de novo–synthetized mediators such as cytokines and chemokines. Mast cells also express receptors for chemokines, cytokines, and growth factors, essential not only for their maturation and differentiation but also to modulate their responses. For example, the KIT receptor binds the stem cell factor (SCF), important for mast cell proliferation and maturation 11. The MRGPRX2 receptor is activated by a range of ligands, including inflammatory peptides and drugs associated with allergic reactions 29. Finally, complement receptor-mediated activation of mast cells can be induced by different complement components, such as C3a or C5a 11. Transcription factors such as PU.1, MITF, GATA and C/EBP family members have critical roles in regulating mast cell development and in the maintenance of cell identity 30, while transcription factors such as NF-κB, NFAT and AP-1 are predominantly involved in the acute regulation of inflammatory genes 23.
Besides the challenges of studying mast cells, why should we care about these cells and how they are regulated? Studying mast cells has the obvious implication of a better understanding of mechanisms that are involved in allergy and asthma as well as in mast cell–proliferative diseases 17 ( Figure 2), and as our technological resources are improving at an unprecedented speed, we are also becoming increasingly able to gain more mechanistic details on their functions. For example, the important role of mast cells in eradicating infections by intestinal nematodes has been known for quite some time; however, the underlying mechanism was unclear. A recent study showed that mast cells respond to ATP released by intestinal epithelial cells damaged during parasite infection by secreting IL-33 18. Mast cell–derived IL-33 in turn activated group 2 innate lymphoid cells (ILC2) to produce IL-13, leading to goblet cell hyperplasia and worm expulsion. Similarly, mast cells have an established role in allergic reactions, and their contribution to such responses is dependent on the acquisition of antigen-specific IgE antibodies that are bound to the high-affinity IgE receptor (FcεRI) on the cell surface. However, it was only recently elucidated that for mast cells to acquire IgE antibodies from the blood, they must display a preferential perivascular location, which enabled direct access to the blood and sampling of the intravascular lumen through cellular projections 19. Also, mast cells turned out to have a crucial role in orchestrating the exfoliation of epithelial cells in the bacterially infected bladder, thereby inducing an important defense mechanism aimed at reducing bacterial burden 20. Indeed, IL-1β produced by bacterially exposed bladder epithelial cells potently recruited mast cells to the site of infection; uptake of the released mast cell granules by epithelial cells was followed by the release of the mast cell protease Mcpt4 and caspase-1 activation, eventually leading to cell death 20. Although the specific signals that induced mast cell degranulation in the infected bladder remain to be elucidated, this study highlighted how mast cells can influence the outcome of many different physiological responses. Mast cells were also shown to suppress humoral and cell-mediated responses in the bladder, through the production of the anti-inflammatory cytokine IL-10, most likely in an attempt to protect the organ from excessive tissue damage 21. Whether the same cell can switch from a pro- to an anti-inflammatory phenotype during the course of an infection in the bladder, or instead different subsets are involved, remains to be determined. Interestingly, mast cell–derived IL-10 was also shown to modulate contact hypersensitivity reactions, although the extent of IL-10 production (and thereby the final contribution of mast cells to the amplification or attenuation of tissue pathology) appeared to be variable depending on the severity of the model of contact hypersensitivity used 3, 5, 6.
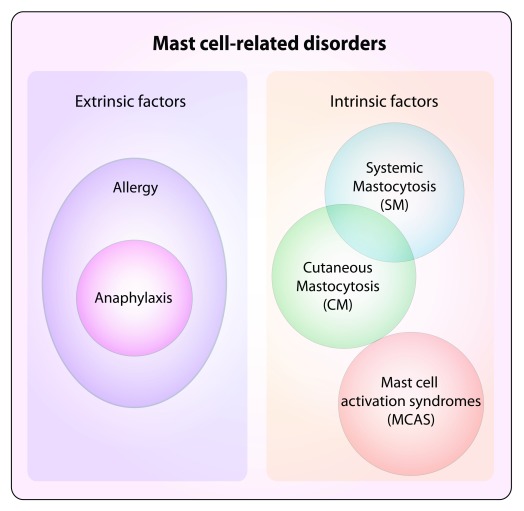
Figure 2. Mast cell–related disorders.
Diseases associated with mast cells can broadly include disorders associated with extrinsic factors, such as the ones mediated by IgE antibodies that, acting through the high-affinity IgE receptor (FcεRI) expressed on the mast cell surface, can translate into the development of allergic reactions. Allergies are detrimental immune responses against otherwise innocuous environmental antigens, which induce the production of IgE antibodies that can activate mast cells, eventually leading to, for example, allergic rhinitis, asthma, and atopic dermatitis 26. Excessive allergic reactions can translate into anaphylaxis. Other disorders can instead be cell-intrinsic, due to altered biological features of mast cells that lead to uncontrolled responses (mast cell activation syndrome, or MCAS) 31 or excessive proliferation (systemic and cutaneous mastocytosis, or SM and CM). Potentially, all mast cell disorders can display altered activation and are broadly defined as mast cell activation disorders (MCADs), although MCAS represents a subgroup displaying mast cell activation without clonal expansion 32, 33.
Transcription factors in mast cells
Many studies have assessed the transcriptional profile of mast cells, from both human and mouse, although for the most part they used cultured cells, and only in rare cases was a comparison with ex-vivo–derived tissue cells attempted 16. Since mast cells differentiate in local tissue niches, it is probable that they adaptively develop characteristic features that allow them to best function within a given context 8, 22. Indeed, the fact that tissue mast cells can be quite heterogeneous (for example, from the point of view of the content of their granules) has been recognized for quite some time 12, 22– 24. However, from a transcriptional point of view, studying the heterogeneity of tissue mast cells implies the ability to extract them directly from various tissues and perform transcriptome analyses such as RNA sequencing. In this kind of analysis, mast cell heterogeneity may be reflected in differences in their gene expression programs and this is exactly what has been observed in murine mast cells extracted from different tissues 25. Indeed, mouse mast cells derived from various anatomical locations (peritoneal cavity, ear, tongue, trachea, and esophagus) displayed a high degree of heterogeneity across the different tissues, although they clustered distinctly from other profiled lymphoid and myeloid cell types, including basophils and other granulocytes. Some signature genes that specifically characterized mast cells included a number of proteases such as Ctsg, encoding for cathepsin G; the metalloprotease gene Adamts9; and C2, encoding for the complement component C2 of the classic C3 convertase 25. Among the transcription factors, Crebl1, Smarca1, and Zfp9 appeared to be relatively specific for mast cells, although their role remains unknown, while Mitf, a transcription factor crucial for mast cell differentiation and functions 34, clearly defined mast cells from other cell types. Highlighting once again the complications associated with studies of tissue-resident cells, the authors found that incubating peritoneal mast cells in the presence of the digestion enzymes required for tissue extraction was already sufficient to alter the expression of more than 100 genes, including the gene encoding for the transcription factor Egr2. They therefore proceeded to compare enzymatically treated peritoneal mast cells with other mast cell populations enzymatically extracted from the tissues. Such analysis revealed that, overall, mast cells from the different tissues shared a core signature of 128 genes, including genes encoding for proteases or involved in metabolic pathways important for the generation of the wide repertoire of mediators that characterize mast cells. Comparative analysis of cells from the different tissues showed that mast cells from the trachea, esophagus, and tongue displayed the highest transcriptional similarity, although some specificity remained. For example, the gene encoding for the protease Mcpt1 appeared to be relatively specific for mast cells from the esophagus. Peritoneal and skin mast cells appeared to be more divergent in their transcriptional profiles, with differential expression of a number of genes, including the adhesion molecule CD34 (which was absent in skin mast cells), the transcription factor SOX7 (increased in skin mast cells), and the integrin β2, the last of which instead was preferentially detected in peritoneal mast cells. Interestingly, peritoneal mast cells were characterized by a transcriptional signature significantly associated with mitosis, and indeed these cells appeared to undergo proliferation even in the absence of inflammation 25.
Apart from MITF, other transcription factors that are known to positively or negatively impact mast cell differentiation or function (or both) belong to the GATA, STAT, and C/EBP families (reviewed in 23, 30) ( Figure 1). For instance, STAT5 expression was shown to be crucial in modulating mast cell survival in response to cytokine signals 35, and STAT5 activity in mast cells was linked to allergen-induced dermatitis 36. Interestingly, several transcription factors also showed some level of crosstalk in regulating mast cell differentiation and functions: for example, C/EBPα and MITF acted antagonistically in the specification of the basophil and mast cell lineages 37, while STAT5 acted upstream of GATA2 in the differentiation pathways leading to either mast cells or basophils 38. Other transcription factors such as HES1 39, EGR family members 40, 41, or ZEB2 42 have also been associated with at least some specific aspects of mast cell biology 23, although their exact role in vivo or their detailed mechanism of action at the genomic level requires further investigation. Of note, many of the transcription factors that are involved in mast cell activation (NFAT, NF-κB, AP-1, and so on) are also more general regulators of inflammatory genes in many immune cell types, and they will not be extensively discussed here. We refer the reader to a more comprehensive review on this topic 23.
Innate immune memory
The cell-intrinsic, short-term memory of an encounter with a pathogen or a danger signal 43 may be especially relevant for mast cells compared with very short-lived cells such as neutrophils and basophils. Mast cells are very long-lived cells, retain the ability to proliferate despite being fully differentiated 2, and can even replenish and modulate the composition of their granules after stimulation 44. The process of enhanced innate immune response against a secondary encounter with a pathogen, which was clearly defined in macrophages and other innate immune cells as “trained immunity” 45, could influence secondary mast cell responses after the first activation and could be important in the modulation of protective as well as allergic responses. Mechanistically, such innate immune memory is thought to be based mainly on epigenetic reprogramming, involving histone modifications, DNA methylation, and even the expression of selected microRNAs and other non-coding RNAs, which collectively contribute to the rewiring of the transcriptional program of the cell upon stimulation 43, 45. However, not many studies addressed the issue of cell-intrinsic, long-term changes in mast cell functional programs in response to a stimulus. For example, lipopolysaccharide (LPS) stimulation of mast cells can induce a state of unresponsiveness to a subsequent stimulation which is similar to the endotoxin tolerance described for macrophages, thereby probably representing a genuine example of mast cell short-term memory 46, and some crosstalk between IgE and LPS stimulation in mast cells was also reported 47. However, mast cells can be directly activated by many additional stimuli ( Figure 1), and whether a true trained immunity applies to mast cells for at least some of these stimuli, the relevance of such a process in vivo, and the underlying mechanisms are all aspects that require further investigation, especially in view of a potentially crucial role in modulating mast cell responses.
Epigenetic control of mast cell responses
Chromatin modifications such as covalent modifications of histone tails or DNA methylation are epigenetic mechanisms of regulation of transcription, which include all of those mechanisms that influence gene expression by modulating the accessibility of regulatory regions to transcription factors without altering the DNA sequence. Epigenetic modifications are critical mechanisms that modulate the interplay of genomic sequences with environmental signals, and indeed they have a crucial role during development, in the maintenance of cell identity, in cell differentiation, and in regulating acute responses to stimuli. Because of the complex networks in which these mechanisms act, and their ability to affect the entire genome, epigenetic studies often suffer from major obstacles that hinder our mechanistic understanding of the biological role of epigenetic modifications 48. For instance, loss-of-function studies of DNA methyltransferase (DNMT) enzymes revealed the unmistakable importance of this epigenetic modification during development 49, 50; however, the molecular mechanisms underlying the embryonic or perinatal lethality observed in the absence of DNMT enzymes remain far from being firmly established. Indeed, distinguishing causality from association or direct and indirect effects of chromatin-related processes remain major challenges 48.
What is the role of epigenetic modifications in regulating mast cell biology? In terms of histone modifications, by assessing the role of the regulatory subunit ASXL1 of a deubiquitinase complex in hematopoietic differentiation, a recent study showed that altered levels of monoubiquitination of lysine 119 on histone H2A affected mast cell differentiation 51, pointing toward a key role for histone modifications in this process. Interestingly, mast cells appear to exploit an atypical epigenetic regulatory mechanism, mediated by the endogenously produced protease tryptase that not only can be secreted by the cells to affect the extracellular milieu but also can translocate into the nucleus of the producing cell, where it mediates the cleavage of histone tails, thereby modulating gene expression 52. While some mechanistic aspects (such as the regulation of tryptase nuclear translocation) remain to be understood, this example further highlights the potential importance of histone modifications in mast cell biology. Accordingly, histone deacetylase inhibitors appeared to impact the proliferation and viability of pathogenic mast cells derived from patients with mast cell–proliferative disorders 53. But in general, very few studies have investigated the role of histone modifications or histone-modifying enzymes in regulating mast cell differentiation and function. Further studies in this direction, especially in primary mast cells obtained from tissues, would certainly provide further insights about how these cells are regulated and what goes wrong during disease.
As for DNA methylation, this process is mediated by DNMT enzymes that covalently link a methyl group to cytosines in the genomic DNA to give rise to 5-methylcytosine (5mC). Such modification can influence gene expression either by interfering with the binding of transcription factors and co-regulators to a given DNA sequence or by recruiting specific methyl-binding proteins 54. The importance of this process in transcriptional regulation is highlighted not only by the altered developmental processes observed in the absence of DNMT enzymes but also by the many examples of disease, including hematological malignancies, in which DNMT enzymes are mutated and DNA methylation is affected 55. Methylated cytosines in the genomic DNA can also be oxidized to 5-hydroxymethylcytosine (5hmC) by the action of the ten-eleven-translocation enzymes TET1-3 ( Figure 3). Such modification is usually enriched at enhancer elements and correlates with transcription 56. DNA methylation–related processes are also critical to specifically modulate mast cell differentiation and functions; indeed, we found that deletion of the mouse Tet2 gene affected primarily mast cell differentiation and proliferation 57. Interestingly, while cells lacking Tet2 were characterized by a very significant hyperproliferation compared with wild-type cells, heterozygous cells displayed an intermediate phenotype, pointing toward gene-dosage effects. These results suggest that somatic TET2 mutations on one allele may be sufficient to predispose individuals to excessive mast cell proliferation, although in the absence of additional mutations they are insufficient to cause overt disease 58, 59. Unlike Tet2 ablation, deletion of Dnmt3a in the mouse led primarily to unrestrained mast cell responses to stimuli, with significantly increased mast cell degranulation and activation both in vitro and in in vivo 60. Importantly, treatment with demethylating agents or down-modulation of the expression of another DNMT enzyme recapitulated or even exacerbated these phenotypes, at least in vitro, highlighting the key role of DNA methylation–related mechanisms in modulating mast cell activation 60, 61.
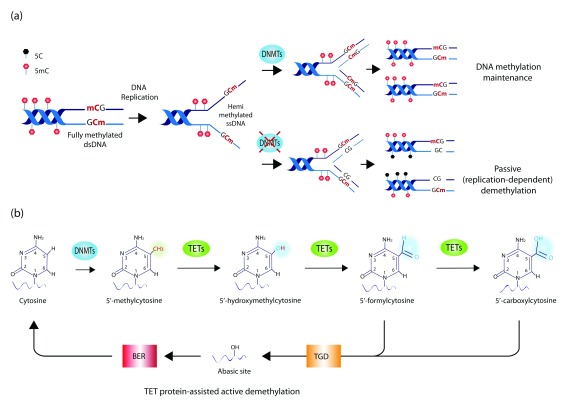
Figure 3. DNA methylation dynamics.
( a) Mechanisms of maintenance and loss of DNA methylation. During DNA replication, DNMT1 binds hemi-methylated single-stranded DNA (ssDNA) and copies DNA methylation patterns on the newly synthetized DNA strand, thereby maintaining the overall DNA methylation landscape across DNA replication and cell division. In the absence of DNMT1, such a process is impaired, resulting in the passive dilution of the methyl mark during cell division. While DNMT1 acts as the primary maintenance DNMT enzyme during cell division, DNMT3A and DNMT3B also contribute to DNA methylation as de novo DNMTs (namely they do not require a hemi-methylated DNA template but also can act on fully unmethylated DNA). ( b) DNA methylation and hydroxymethylation. DNMT enzymes methylate the 5′ carbon residue on the cytosine ring in DNA, giving rise to 5′-methylcytosine (5mC). Iterative oxidation of the methyl group mediated by ten-eleven-translocation (TET) enzymes leads to the formation of 5′-hydroxymethylcytosine, followed by 5′-formylcytosine and 5′-carboxylcytosine. The latter two modifications are recognized and excised by the thymine-DNA-glycosylase (TGD) enzyme, leaving an abasic site in the DNA, which is repaired by base excision repair (BER) mechanisms. Because this process of iterative oxidation leads to the substitution of a modified cytosine with an unmodified one, it is also termed TET protein-assisted active DNA demethylation.
DNA methylation in disease
Mast cell–related diseases manifest in a wide range of disorders displaying activation or proliferative dysregulation or both ( Figure 2) 31, 62. In particular, mastocytosis is characterized by the abnormal proliferation and accumulation of mast cells in various organs and tissues. While the exact cause of mastocytosis remains unclear, it is frequently associated with mutations in the KIT oncogene, most commonly an aspartic acid–to–valine substitution at codon 816, causing spontaneous activation of the KIT receptor 62. One of the first indications that DNA methylation–related processes might be important in mast cell biology came from the observation that decitabine, a demethylating agent used in the clinic for the treatment of myeloproliferative and myelodysplastic disorders, induced apoptosis of neoplastic mast cells, at least in vitro 63. Moreover, mutations in the gene encoding for DNMT3A were identified in 3 out of 26 patients with systemic mastocytosis 64, while TET2 mutations were identified in at least 20% of the patients 59, 64, 65, and these correlated with worse overall survival 64. Accordingly, reduced levels of cytosine modifications were observed in patients with systemic mastocytosis 66, and altered DNA methylation patterns were reported in tissues and cells of patients with asthma 67. However, patient studies are by necessity mostly correlative, and becoming able to distinguish whether such changes are cause or consequence or simply associate with a certain phenotype can be a daunting task. For example, systemic mastocytosis patients carrying TET2 mutations tended to be older and have higher monocyte counts 64; since inactivating TET2 mutations also accumulate with age in healthy individuals 68, 69, the boundaries between cause and consequence remain problematic to define. Indeed, a recent mutational analysis of more than 2,500 human subjects identified mutations in TET2 and DNMT3A as the most common age-associated mutations even in healthy individuals, and TET2 mutations had a stronger impact on the steady increase in hematopoietic clonal expansion compared with DNMT3A mutations 69. While the effects of these mutations in disease and in the normal processes of aging are being uncovered, in the future further studies will certainly lead to a better understanding of the exact role of DNA-modifying enzymes and epigenetic mechanisms in regulating mast cell functions in the context of immune responses and in disease.
Conclusions
Thanks to the development of novel mouse models and to technological advancements allowing the in vivo visualization of mast cells and their in-depth molecular analysis, we are learning more about the role of these cells in various models of disease and inflammatory responses, and we are also better able to dissect some of the mechanistic aspects of such responses. However, despite the great advancements in the field in recent years, many of the mechanisms and factors underlying mast cell differentiation and responses remain poorly defined. In the future, the combination of traditional in vitro and single-molecule studies with single-cell genomic and genome-wide approaches 70 will certainly improve our understanding of mast cell subsets and functions, potentially allowing us to analyze them within the physiological context of their microenvironment. Such developing tools will surely provide definite answers to many of the questions about mast cell biology in health and disease which we are just starting to address.
Acknowledgments
We would like to thank people in the SM lab for support and helpful discussion.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Constance Oliver, Department of Cell and Molecular Biology and Pathogenic Bioagents, Ribeirão Preto Medical School, University of São Paulo, São Paulo, Brazil
Carlo Pucillo, University of Udine, Udine, Italy
Stephen J. Galli, Department of Pathology, Stanford University School of Medicine, Stanford, CA, USA
Funding Statement
Work in the SM lab on this topic is funded by the Swiss National Science Foundation (156875).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 3 approved]
References
- 1. Wernersson S, Pejler G: Mast cell secretory granules: armed for battle. Nat Rev Immunol. 2014;14(7):478–94. 10.1038/nri3690 [DOI] [PubMed] [Google Scholar]
- 2. Abraham SN, St John AL: Mast cell-orchestrated immunity to pathogens. Nat Rev Immunol. 2010;10(6):440–52. 10.1038/nri2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dudeck A, Dudeck J, Scholten J, et al. : Mast cells are key promoters of contact allergy that mediate the adjuvant effects of haptens. Immunity. 2011;34(6):973–84. 10.1016/j.immuni.2011.03.028 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 4. Feyerabend TB, Weiser A, Tietz A, et al. : Cre-mediated cell ablation contests mast cell contribution in models of antibody- and T cell-mediated autoimmunity. Immunity. 2011;35(5):832–44. 10.1016/j.immuni.2011.09.015 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 5. Reber LL, Sibilano R, Starkl P, et al. : Imaging protective mast cells in living mice during severe contact hypersensitivity. JCI Insight. 2017;2(9): pii: 92900. 10.1172/jci.insight.92900 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 6. Grimbaldeston MA, Nakae S, Kalesnikoff J, et al. : Mast cell-derived interleukin 10 limits skin pathology in contact dermatitis and chronic irradiation with ultraviolet B. Nat Immunol. 2007;8(10):1095–104. 10.1038/ni1503 [DOI] [PubMed] [Google Scholar]
- 7. Hepworth MR, Daniłowicz-Luebert E, Rausch S, et al. : Mast cells orchestrate type 2 immunity to helminths through regulation of tissue-derived cytokines. Proc Natl Acad Sci U S A. 2012;109(17):6644–9. 10.1073/pnas.1112268109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frossi B, Mion F, Tripodo C, et al. : Rheostatic Functions of Mast Cells in the Control of Innate and Adaptive Immune Responses. Trends Immunol. 2017;38(9):648–56. 10.1016/j.it.2017.04.001 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 9. Olivera A, Rivera J: Paradigm shifts in mast cell and basophil biology and function: an emerging view of immune regulation in health and disease. Methods Mol Biol. 2014;1192:3–31. 10.1007/978-1-4939-1173-8_1 [DOI] [PubMed] [Google Scholar]
- 10. Reber LL, Marichal T, Galli SJ: New models for analyzing mast cell functions in vivo. Trends Immunol. 2012;33(12):613–25. 10.1016/j.it.2012.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marshall JS: Mast-cell responses to pathogens. Nat Rev Immunol. 2004;4(10):787–99. 10.1038/nri1460 [DOI] [PubMed] [Google Scholar]
- 12. Galli SJ, Borregaard N, Wynn TA: Phenotypic and functional plasticity of cells of innate immunity: macrophages, mast cells and neutrophils. Nat Immunol. 2011;12(11):1035–44. 10.1038/ni.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Valadi H, Ekström K, Bossios A, et al. : Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–9. 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 14. von Köckritz-Blickwede M, Goldmann O, Thulin P, et al. : Phagocytosis-independent antimicrobial activity of mast cells by means of extracellular trap formation. Blood. 2008;111(6):3070–80. 10.1182/blood-2007-07-104018 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 15. Guhl S, Artuc M, Neou A, et al. : Long-term cultured human skin mast cells are suitable for pharmacological studies of anti-allergic drugs due to high responsiveness to FcεRI cross-linking. Biosci Biotechnol Biochem. 2011;75(2):382–4. 10.1271/bbb.100745 [DOI] [PubMed] [Google Scholar]
- 16. Motakis E, Guhl S, Ishizu Y, et al. : Redefinition of the human mast cell transcriptome by deep-CAGE sequencing. Blood. 2014;123(17):e58–67. 10.1182/blood-2013-02-483792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Valent P, Akin C, Metcalfe DD: Mastocytosis: 2016 updated WHO classification and novel emerging treatment concepts. Blood. 2017;129(11):1420–7. 10.1182/blood-2016-09-731893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shimokawa C, Kanaya T, Hachisuka M, et al. : Mast Cells Are Crucial for Induction of Group 2 Innate Lymphoid Cells and Clearance of Helminth Infections. Immunity. 2017;46(5):863–874.e4. 10.1016/j.immuni.2017.04.017 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 19. Cheng LE, Hartmann K, Roers A, et al. : Perivascular mast cells dynamically probe cutaneous blood vessels to capture immunoglobulin E. Immunity. 2013;38(1):166–75. 10.1016/j.immuni.2012.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 20. Choi HW, Bowen SE, Miao Y, et al. : Loss of Bladder Epithelium Induced by Cytolytic Mast Cell Granules. Immunity. 2016;45(6):1258–69. 10.1016/j.immuni.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 21. Chan CY, St John AL, Abraham SN: Mast cell interleukin-10 drives localized tolerance in chronic bladder infection. Immunity. 2013;38(2):349–59. 10.1016/j.immuni.2012.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xing W, Austen KF, Gurish MF, et al. : Protease phenotype of constitutive connective tissue and of induced mucosal mast cells in mice is regulated by the tissue. Proc Natl Acad Sci U S A. 2011;108(34):14210–5. 10.1073/pnas.1111048108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cildir G, Pant H, Lopez AF, et al. : The transcriptional program, functional heterogeneity, and clinical targeting of mast cells. J Exp Med. 2017;214(9):2491–506. 10.1084/jem.20170910 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 24. Irani AA, Schechter NM, Craig SS, et al. : Two types of human mast cells that have distinct neutral protease compositions. Proc Natl Acad Sci U S A. 1986;83(12):4464–8. 10.1073/pnas.83.12.4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dwyer DF, Barrett NA, Austen KF, et al. : Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat Immunol. 2016;17(7):878–87. 10.1038/ni.3445 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 26. Galli SJ, Tsai M: IgE and mast cells in allergic disease. Nat Med. 2012;18(5):693–704. 10.1038/nm.2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Galli SJ, Nakae S, Tsai M: Mast cells in the development of adaptive immune responses. Nat Immunol. 2005;6(2):135–42. 10.1038/ni1158 [DOI] [PubMed] [Google Scholar]
- 28. Malbec O, Daëron M: The mast cell IgG receptors and their roles in tissue inflammation. Immunol Rev. 2007;217(1):206–21. 10.1111/j.1600-065X.2007.00510.x [DOI] [PubMed] [Google Scholar]
- 29. McNeil BD, Pundir P, Meeker S, et al. : Identification of a mast-cell-specific receptor crucial for pseudo-allergic drug reactions. Nature. 2015;519(7542):237–41. 10.1038/nature14022 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Monticelli S, Natoli G: Transcriptional determination and functional specificity of myeloid cells: making sense of diversity. Nat Rev Immunol. 2017;17(10):595–607. 10.1038/nri.2017.51 [DOI] [PubMed] [Google Scholar]
- 31. Akin C: Mast cell activation syndromes. J Allergy Clin Immunol. 2017;140(2):349–55. 10.1016/j.jaci.2017.06.007 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Valent P, Akin C, Hartmann K, et al. : Advances in the Classification and Treatment of Mastocytosis: Current Status and Outlook toward the Future. Cancer Res. 2017;77(6):1261–70. 10.1158/0008-5472.CAN-16-2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Valent P, Akin C, Arock M, et al. : Definitions, criteria and global classification of mast cell disorders with special reference to mast cell activation syndromes: a consensus proposal. Int Arch Allergy Immunol. 2012;157(3):215–25. 10.1159/000328760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shahlaee AH, Brandal S, Lee YN, et al. : Distinct and shared transcriptomes are regulated by microphthalmia-associated transcription factor isoforms in mast cells. J Immunol. 2007;178(1):378–88. 10.4049/jimmunol.178.1.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shelburne CP, McCoy ME, Piekorz R, et al. : Stat5 expression is critical for mast cell development and survival. Blood. 2003;102(4):1290–7. 10.1182/blood-2002-11-3490 [DOI] [PubMed] [Google Scholar]
- 36. Ando T, Xiao W, Gao P, et al. : Critical role for mast cell Stat5 activity in skin inflammation. Cell Rep. 2014;6(2):366–76. 10.1016/j.celrep.2013.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qi X, Hong J, Chaves L, et al. : Antagonistic regulation by the transcription factors C/EBPα and MITF specifies basophil and mast cell fates. Immunity. 2013;39(1):97–110. 10.1016/j.immuni.2013.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 38. Li Y, Qi X, Liu B, et al. : The STAT5-GATA2 pathway is critical in basophil and mast cell differentiation and maintenance. J Immunol. 2015;194(9):4328–38. 10.4049/jimmunol.1500018 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 39. Sakata-Yanagimoto M, Nakagami-Yamaguchi E, Saito T, et al. : Coordinated regulation of transcription factors through Notch2 is an important mediator of mast cell fate. Proc Natl Acad Sci U S A. 2008;105(22):7839–44. 10.1073/pnas.0801074105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li B, Power MR, Lin TJ: De novo synthesis of early growth response factor-1 is required for the full responsiveness of mast cells to produce TNF and IL-13 by IgE and antigen stimulation. Blood. 2006;107(7):2814–20. 10.1182/blood-2005-09-3610 [DOI] [PubMed] [Google Scholar]
- 41. Wu Z, Macneil AJ, Junkins R, et al. : Mast cell FcεRI-induced early growth response 2 regulates CC chemokine ligand 1-dependent CD4 + T cell migration. J Immunol. 2013;190(9):4500–7. 10.4049/jimmunol.1203158 [DOI] [PubMed] [Google Scholar]
- 42. Barbu EA, Zhang J, Berenstein EH, et al. : The transcription factor Zeb2 regulates signaling in mast cells. J Immunol. 2012;188(12):6278–86. 10.4049/jimmunol.1102660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Monticelli S, Natoli G: Short-term memory of danger signals and environmental stimuli in immune cells. Nat Immunol. 2013;14(8):777–84. 10.1038/ni.2636 [DOI] [PubMed] [Google Scholar]
- 44. Xiang Z, Block M, Löfman C, et al. : IgE-mediated mast cell degranulation and recovery monitored by time-lapse photography. J Allergy Clin Immunol. 2001;108(1):116–21. 10.1067/mai.2001.116124 [DOI] [PubMed] [Google Scholar]
- 45. Netea MG, Joosten LA, Latz E, et al. : Trained immunity: A program of innate immune memory in health and disease. Science. 2016;352(6284):aaf1098. 10.1126/science.aaf1098 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 46. Saturnino SF, Prado RO, Cunha-Melo JR, et al. : Endotoxin tolerance and cross-tolerance in mast cells involves TLR4, TLR2 and FcepsilonR1 interactions and SOCS expression: perspectives on immunomodulation in infectious and allergic diseases. BMC Infect Dis. 2010;10:240. 10.1186/1471-2334-10-240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Medina-Tamayo J, Ibarra-Sánchez A, Padilla-Trejo A, et al. : IgE-dependent sensitization increases responsiveness to LPS but does not modify development of endotoxin tolerance in mast cells. Inflamm Res. 2011;60(1):19–27. 10.1007/s00011-010-0230-4 [DOI] [PubMed] [Google Scholar]
- 48. Smale ST: Transcriptional regulation in the immune system: a status report. Trends Immunol. 2014;35(5):190–4. 10.1016/j.it.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 49. Okano M, Bell DW, Haber DA, et al. : DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–57. 10.1016/S0092-8674(00)81656-6 [DOI] [PubMed] [Google Scholar]
- 50. Li E, Bestor TH, Jaenisch R: Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–26. 10.1016/0092-8674(92)90611-F [DOI] [PubMed] [Google Scholar]
- 51. Balasubramani A, Larjo A, Bassein JA, et al. : Cancer-associated ASXL1 mutations may act as gain-of-function mutations of the ASXL1-BAP1 complex. Nat Commun. 2015;6:7307. 10.1038/ncomms8307 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 52. Melo FR, Wallerman O, Paivandy A, et al. : Tryptase-catalyzed core histone truncation: A novel epigenetic regulatory mechanism in mast cells. J Allergy Clin Immunol. 2017;140(2):474–85. 10.1016/j.jaci.2016.11.044 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 53. Lyberg K, Ali HA, Grootens J, et al. : Histone deacetylase inhibitor SAHA mediates mast cell death and epigenetic silencing of constitutively active D816V KIT in systemic mastocytosis. Oncotarget. 2017;8(6):9647–59. 10.18632/oncotarget.14181 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 54. Bird A: DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. 10.1101/gad.947102 [DOI] [PubMed] [Google Scholar]
- 55. Jaenisch R, Bird A: Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 Suppl:245–54. 10.1038/ng1089 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 56. Tsagaratou A, Lio CJ, Yue X, et al. : TET Methylcytosine Oxidases in T Cell and B Cell Development and Function. Front Immunol. 2017;8:220. 10.3389/fimmu.2017.00220 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 57. Montagner S, Leoni C, Emming S, et al. : TET2 Regulates Mast Cell Differentiation and Proliferation through Catalytic and Non-catalytic Activities. Cell Rep. 2016;15(7):1566–79. 10.1016/j.celrep.2016.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. De Vita S, Schneider RK, Garcia M, et al. : Loss of function of TET2 cooperates with constitutively active KIT in murine and human models of mastocytosis. PLoS One. 2014;9(5):e96209. 10.1371/journal.pone.0096209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Soucie E, Hanssens K, Mercher T, et al. : In aggressive forms of mastocytosis, TET2 loss cooperates with c-KITD816V to transform mast cells. Blood. 2012;120(24):4846–9. 10.1182/blood-2011-12-397588 [DOI] [PubMed] [Google Scholar]
- 60. Leoni C, Montagner S, Rinaldi A, et al. : Dnmt3a restrains mast cell inflammatory responses. Proc Natl Acad Sci U S A. 2017;114(8):E1490–E1499. 10.1073/pnas.1616420114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kelly P: DNA methylation curbs mast cell response. Science. 2017;355(6327):809. 10.1126/science.355.6327.809-a [DOI] [PubMed] [Google Scholar]
- 62. Akin C, Metcalfe DD: Systemic mastocytosis. Annu Rev Med. 2004;55:419–32. 10.1146/annurev.med.55.091902.103822 [DOI] [PubMed] [Google Scholar]
- 63. Ghanim V, Herrmann H, Heller G, et al. : 5-azacytidine and decitabine exert proapoptotic effects on neoplastic mast cells: role of FAS-demethylation and FAS re-expression, and synergism with FAS-ligand. Blood. 2012;119(18):4242–52. 10.1182/blood-2011-09-382770 [DOI] [PubMed] [Google Scholar]
- 64. Traina F, Visconte V, Jankowska AM, et al. : Single nucleotide polymorphism array lesions, TET2, DNMT3A, ASXL1 and CBL mutations are present in systemic mastocytosis. PLoS One. 2012;7(8):e43090. 10.1371/journal.pone.0043090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tefferi A, Levine RL, Lim KH, et al. : Frequent TET2 mutations in systemic mastocytosis: clinical, KITD816V and FIP1L1-PDGFRA correlates. Leukemia. 2009;23(5):900–4. 10.1038/leu.2009.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Leoni C, Montagner S, Deho' L, et al. : Reduced DNA methylation and hydroxymethylation in patients with systemic mastocytosis. Eur J Haematol. 2015;95(6):566–75. 10.1111/ejh.12537 [DOI] [PubMed] [Google Scholar]
- 67. Yang IV, Lozupone CA, Schwartz DA: The environment, epigenome, and asthma. J Allergy Clin Immunol. 2017;140(1):14–23. 10.1016/j.jaci.2017.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Busque L, Patel JP, Figueroa ME, et al. : Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44(11):1179–81. 10.1038/ng.2413 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 69. Buscarlet M, Provost S, Zada YF, et al. : DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood. 2017;130(6):753–62. 10.1182/blood-2017-04-777029 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 70. Cuvier O, Fierz B: Dynamic chromatin technologies: from individual molecules to epigenomic regulation in cells. Nat Rev Genet. 2017;18(8):457–72. 10.1038/nrg.2017.28 [DOI] [PubMed] [Google Scholar]