Abstract
Amyotrophic lateral sclerosis (ALS) is the most common adult-onset neuromuscular disorder characterised by selective loss of motor neurons leading to fatal paralysis. Current therapeutic approaches are limited in their effectiveness. Substantial advances in understanding ALS disease mechanisms has come from the identification of pathogenic mutations in dominantly inherited familial ALS (FALS). We previously reported a coding mutation in D-amino acid oxidase (DAOR199W) associated with FALS. DAO metabolises D-serine, an essential co-agonist at the N-Methyl-D-aspartic acid glutamate receptor subtype (NMDAR). Using primary motor neuron cultures or motor neuron cell lines we demonstrated that expression of DAOR199W, promoted the formation of ubiquitinated protein aggregates, activated autophagy and increased apoptosis. The aim of this study was to characterise the effects of DAOR199W in vivo, using transgenic mice overexpressing DAOR199W. Marked abnormal motor features, e.g. kyphosis, were evident in mice expressing DAOR199W, which were associated with a significant loss (19%) of lumbar spinal cord motor neurons, analysed at 14 months. When separated by gender, this effect was greater in females (26%; p< 0.0132). In addition, we crossed the DAOR199W transgenic mouse line with the SOD1G93A mouse model of ALS to determine whether the effects of SOD1G93A were potentiated in the double transgenic line (DAOR199W/SOD1G93A). Although overall survival was not affected, onset of neurological signs was significantly earlier in female double transgenic animals than their female SOD1G93A littermates (125 days vs 131 days, P = 0.0239). In summary, some significant in vivo effects of DAOR199W on motor neuron function (i.e. kyphosis and loss of motor neurons) were detected which were most marked in females and could contribute to the earlier onset of neurological signs in double transgenic females compared to SOD1G93A littermates, highlighting the importance of recognizing gender effects present in animal models of ALS.
Introduction
Amyotrophic lateral Sclerosis (ALS) is the most common adult-onset neuromuscular disorder, caused by the selective degeneration of motor neurons in the spinal cord, brain stem and motor cortex. The disease usually progresses rapidly leading to severe motor disability with most patients dying within 3–5 years of diagnosis. Understanding the pathogenic mechanisms that underlie the disease has been greatly accelerated by the identification of mutations that cause the familial form of the disease (FALS) [1]. To date at least twenty genes have been found that harbour mutations associated with FALS. The most prevalent mutations are found in C9ORF72, SOD1, TAR DNA binding protein 43 (TARDBP), and Fused in Sarcoma (FUS) and much evidence is accumulating to link these mutations with the classical neuropathological features of both familial and sporadic forms of the disease i.e. the accumulation of cytosolic ubiquitinated protein inclusions, most of which are positive for TDP-43 encoded by TARDBP [2]. Interestingly, it has emerged from these genetic studies that dysfunction of two major pathways are strongly associated with ALS pathogenesis. These effects are firstly focussed on components of RNA metabolism, through dysfunction of the regulatory properties of RNA binding proteins affecting splicing and their trafficking between the nucleus and cytosol and secondly proteostasis, affecting the endoplasmic reticulum stress response and protein degradation pathways mediated through the proteasome and autophagy [3]. A less well established area is the understanding of trigger factors that initiate this process which potentially could include glutamate excitotoxicity, impaired calcium homeostasis and oxidative stress.
Recently, our group identified a mutation in the D-amino acid oxidase gene (DAO) that was transmitted with disease [4], a substitution of arginine by tryptophan at codon 199 (DAOR199W), which has shed some light on more upstream events that may trigger disease initiation [4–6]. The selective distribution of the protein in spinal cord and brain stem nuclei suggests a previously unrecognised role for DAO in motor pathways and function [4]. The major function known for DAO is the regulation of the levels of D-serine, which has a fundamental role in excitatory pathways as a co-agonist at the N-Methyl-D-aspartic acid (NMDA) glutamate receptor (NMDAR). This important function of D-serine has now been well characterised and is known to be essential in mediating some actions of glutamate in synaptic plasticity e.g. long term potentiation [7–10]. Furthermore, we have shown that expression of DAO containing the mutation associated with FALS, DAOR199W, in primary motor neuron cultures or motor neuron cell lines promotes the formation of ubiquitinated protein aggregates, activates autophagy and increases apoptosis. Importantly, we have shown that these effects can be significantly attenuated by a selective antagonist at the D-serine binding site of the NMDA receptor, using a co-culture system where glial cells expressing DAOR199W are incubated in an insert placed above motor neuron cells lacking the mutation, which recapitulates the effects of the mutation on autophagy and apoptosis in the co-cultured motor neuron cells [6]. In addition, DAOR199W leads to impaired enzymatic DAO enzyme activity in ALS cases carrying the mutation and also when expressed in cell lines [4].
Interestingly, studies of a mouse strain (ddY/DAO−)[11] lacking DAO enzyme activity, due to a naturally occurring point mutation at position 181 leading to the substitution of glycine by arginine, revealed that while these animals grow and reproduce normally they exhibit abnormal NMDA receptor-mediated behavour [12]. Furthermore a homozygous mouse line expressing this mutation was generated (DAO-/-) that developed an abnormal limb reflex and a significant loss of motor neurons in lumbar spinal cord at 8 months [13]. The study also showed that both in this transgenic line and in the SODG93A mouse model of ALS, D-serine accumulated in the spinal cord during disease progression and a marked suppression in DAO activity was seen in the reticulospinal tract, a pathway that plays an important role in regulating motor neuron excitability [13].
The aim of this study was to determine whether the deleterious effects of DAOR199W on motor neurons detected in cell culture could be seen in vivo in transgenic mice overexpressing DAOR199W. In addition, we crossed the DAOR199W transgenic mouse line with the SOD1 mouse model of ALS to generate double transgenic mice expressing both mutations (DAOR199W/SODG93A) to determine whether their effects were synergistic. We found that significant motor neuron loss occurred in DAOR199W transgenic mice but the presence of DAOR199W in double transgenic mice (DAOR199W/SODG93A) did not potentiate the effects of SOD1 over-expression on survival.
Results
Generation of a new line of transgenic animals overexpressing DAOR199W
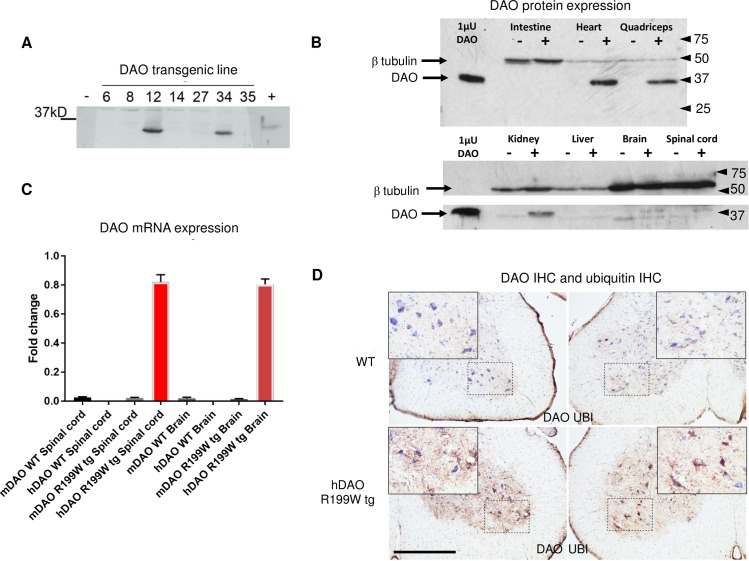
Two independent lines were generated and upon successful transmission, transgene expression was confirmed in spinal cord using western blot analysis (Fig 1A) and extended to a wider range of tissues (Fig 1B). As DAOR199W protein was under the influence of the CAGG promoter/enhancer construct (beta actin promoter with the CMV enhancer), widespread expression in many tissues was seen. The highest levels of DAO expression were found in muscle and heart, followed by kidney, spinal cord and brain, under conditions where endogenous levels of DAO were low or undetectable. Quantitative PCR using selective primer sets that detect expression of the human DAO transgene in DAOR199W transgenic mice indicated that the DAOR199W transgene was present at concentrations 40-fold greater than that of mouse DAO in the same animals (Fig 1C). In keeping with protein detection by Western blot analysis, the DAOR199W transgene was markedly over-expressed in spinal cord compared to wild-type controls and was present at even greater levels in skeletal muscle and heart. We also investigated DAO activity in DAOR199W transgenic mice compared to control littermates using the peroxidase-linked colorimetric conversion of ortho-dianisidine (protocol modified from Tedeschi et al)[14] and found a mild reduction in overall enzyme activity in the brain and spinal cord, suggesting that a degree of hetero-dimerisation with endogenous protein may occur (Figure A in S1 File). No enzyme activity was detected in the muscle of wild-type and transgenic mice despite substantial expression at the mRNA level in the transgenic mice.
Fig 1.
Transgene expression in two mouse lines (A), regional tissue expression of DAO protein (B) and mRNA (C) in DAOR199W transgenic mice and immunohistochemical localisation of DAO and ubiquitin in DAOR199W Transgenic mice (D). A. Expression of DAO in spinal cord from two DAOR199W lines. Western blot analysis of different transgenic mouse lines expressing human DAO. 200ng of spinal cord from each animal was separated by SDS-PAGE and blotted with anti-DAO antibody. Expression of DAO was present in two DAOR199W transgenic mice lines, 12 and 34 (used in the survival analysis). A porcine DAO protein standard (indicated by a + sign) was loaded in the final well. B. Regional expression of DAO protein in DAOR199W transgenic mice. The highest concentrations of DAO (37kDa) in DAOR199W transgenic mice (indicated by +) were found in heart and quadriceps muscle (upper panel), followed by kidney, spinal cord, brain (lower panel) and in both lines (shown here for line 34) compared to wild-type mice (indicated by a -). Lower levels were found in liver and small intestine. The loading control was beta tubulin (50kDa). DAO immunoreactivity was low in non-transgenic mice. C. Regional expression of DAO mRNA in DAOR199W transgenic mice. Expression of human DAO (hDAO) and mouse DAO (mDAO) in spinal cord, brain and quadriceps muscle from wild-type (WT) and DAOR199W transgenic mice (R199W tg). D. Spinal cord expression of DAO and ubiquitin in DAOR199W Transgenic mice. Lumbar spinal cord sections (L2-L4) from 8 month old non-transgenic and R199W DAO tg mice were immunostained for DAO and ubiquitin and counterstained with Nissl (blue). Ventral horns with their enlargement (boxed area) are shown for each section. Spinal cord sections from the DAOR199W transgenic showed widespread distribution of DAO staining (brown) throughout the ventral grey matter (lower left hand panel) particularly in dendrites and interneurons with lower levels present in non-transgenic littermates (upper left hand panel). Similarly, ubiquitin staining was more intense in sections from DAOR199W transgenic mice (lower right hand panel) compared to their non-transgenic littermates (upper right hand panel). Scale bar, 500um.
DAO immunoreactivity was abundant in DAOR199W transgenic mice compared to control littermates (Fig 1D) being present in large motor neurons and the surrounding grey matter of ventral spinal cord. Similarly, ubiquitin immunoreactivity was high in grey matter of the ventral spinal cord in DAOR199W mice, being most abundant in the cell bodies of large motor neurons. In contrast, levels of ubiquitin were only present at low levels in controls (Fig 1D), confirming previous studies in NSC-34 cell lines overexpressing DAOR199W compared to DAOWT where it was established that a prominent feature of the cells expressing the mutant DAOR199W allele was the presence of ubiquitinated protein aggregates [4].
Characterisation of the DAOR199W transgenic line: Effect on body weight and morphological phenotype
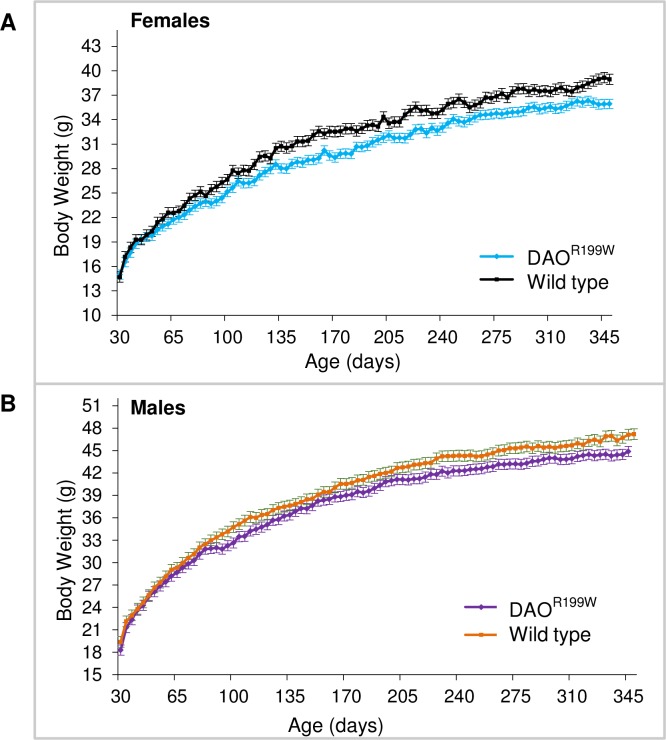
Initially, we carried out a longitudinal large cohort survival study over a period of one year, during which time body weight, motor features and gait were monitored. The assessor was blinded to the genotype of the animals. Animals expressing the transgene consistently had a lower body weight at each age compared to their wild type littermates, which was evident in both males and females (Fig 2A and 2B).
Fig 2. Longitudinal assessment of body weight in DAOR199W and wild-type littermates.
Mean group body weight plotted over time from 30 days to 350 days of age. (A) females, n = 23 DAOR199W (blue) and (n = 17) non-transgenic littermates (black). (B) males, n = 15 DAOR199W (purple) and (n = 16) non-transgenic littermates (orange). Values are means± SEMs.
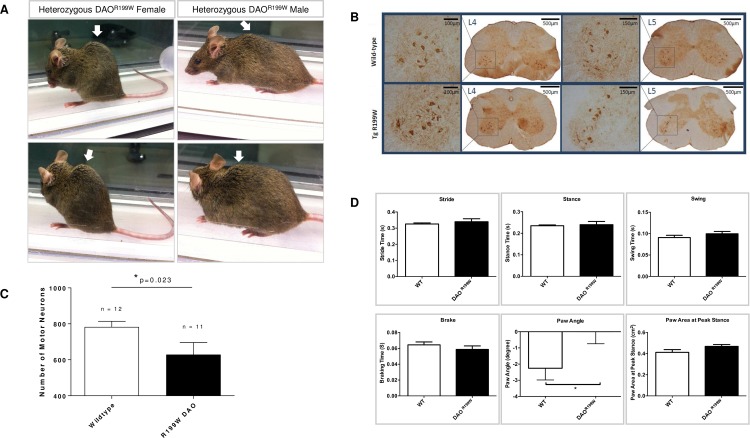
We observed some substantial and distinct features present in DAOR199W animals in terms of physical appearance that were absent from wild-type littermates (Fig 3A). All males and females bearing the mutant transgene appeared very hunched and developed progressive kyphosis, which was more conspicuous in transgenic females than the males.
Fig 3.
Phenotype of R199W DAO mouse showing (A) Kyphosis, (B) motor neuron loss in lumbar spinal cord, (C) ChAT immunohistochemisty in lumbar spinal cord (D) Assessment of various gait indices in the hindlimbs of female DAOR199W and wild-type littermates. (A) Kyphosis of the spine demonstrated in both male and female DAOR199W transgenic lines. The arrow denotes kyphosis of the spine. (B) Motor neuron counts in wildtype and R199W DAO mice aged 14–15 months. Lumbar motor neurons in regions L3-L5 were stained with anti-ChAT antibody and every third section quantified for both male and female wildtype and R199W DAO mice. The number of motor neurons in R199W DAO mice was reduced by 19.7% compared to wild-type mice (one-tailed, unpaired t-test, *p<0.05). When split by gender, motor neuron loss was significantly reduced by 25.7% in females (p = 0.0132*, one-tailed, unpaired t-test) but in males the reduction was by 16.5%, which was not significant. Histograms show means for 40 sections per animal ± SEM. (C) ChAT immunohistochemistry of L4 and L5 regions of a wild-type and transgenic animal with a DAOR199W mutation. Immunohistochemistry was performed on the lumbar spinal cord using anti-ChAT on 20μm sections of spinal cord. (A) Wild-type female L4 and L5 sections. (B) Female with DAO R199W mutation L4 and L5 sections. The number of ChAT-stained motor neurons in the female mice was 25.7% lower for the transgenic group compared to the wild-type group. (D) Assessment of various gait indices in the hind-limbs of female DAOR199W and wild-type littermates. Mean groups combined, plotted for hind-limbs of the DAOR199W (black bars) and WT (white bars) female mice. This was done through averaging the individual values generated by the DigiGait system for each hind-limb of a particular animal (n ≥ 10 per experimental group). Stride (sum of stance and swing), stance (weight bearing portion of the stride in which the paw remains in contact with the belt), swing (when the paw is not in contact with the belt) presented in the top panel, with P values of 0.5238, 0.7560 and 0.273 respectively (two-tailed, unpaired t-test, P < 0.05). Brake (time intervals between initial and maximal paw contacts with the belt), paw angle (angle of the hind-paw during peak stance in relation to the long axis of the body) and paw area at peak stance (maximal paw area in contact with the treadmill during the stance phase) presented in the bottom panel, with P values of 0.4509, 0.0435*, 0.0673 respectively (Mann-Whitney test carried out for brake time and two-tailed, unpaired t-test for the other two parameters, P < 0.05). Asterisk indicates a significant difference in term of the degree of paw angle between WT controls and DAOR199W mice. Values are standard error of the mean (±SEM).
Characterisation of the DAOR199W transgenic line: Effect on motor neuron populations
The effect of the DAOR199W mutation on motor neuron cell survival was investigated in the DAOR199W transgenic line by quantification of the motor neurons in the lumbar region of the spinal cord at the age of 14 months (13.5 to 14.5 months, Fig 3B and 3C). The quantification of the ventral horn motor neurons (L3-L5) of the transgenic animals versus their wild type littermates (n ≥ 5 per genotype, per sex) revealed a significant reduction in motor neuron numbers in mice carrying the mutant allele in comparison to their wild type littermates. When separated according to gender, this decrease (25.7%) was significant in females (P = 0.0132) though not in males (16.5% reduction).
Characterisation of the DAOR199W transgenic line: Gait analysis
The phenotypic difference between the wild type and mutant animals led us to design a gait analysis experiment. In this study, we analysed both genotypes for their gait, footfall, and mechanics covering over 40 parameters but observed no significant differences in gait and body movement between DAOR199W animals and their wild type littermates. A significant difference was however observed in terms of hind-paw angle in the DAOR199W transgenic females (P = 0.0435*, n ≥ 10) (Fig 3D). Paw angle is defined as the angle of the paw (hind or fore) during peak stance in relation to the long axis of the body. Hence, a negative value for this parameter indicates that the animal places its paw inward towards the long axis of its body every time the paw is placed on the surface of the treadmill, and a positive value is an indication of the paws being placed outwards away from the long axis of the body [15–17].
The DAOR199W females place their hind-paws in a straight line, almost parallel to the long axis of their bodies (mean value of angle, -0.008333 ± 0.7330°) in comparison to their wild type littermates (mean value of angle,-2.255 ± 0.7275°). The data obtained for the same parameter for the forepaws suggested that in order to maintain balance due the changes in the paw angle, the animal was forced to turn its forepaws inwards, giving a mean value of -1.241 ± 0.5678°, compared to -0.005000 ± 0.5485° in wild type littermates (forepaws almost parallel to the body axis) (Fig 3D). These differences may reflect an adaptation to weight loss, muscle atrophy and skeletal changes.
In summary, while no overt ALS phenotype was evident in mice expressing DAOR199W, marked structural and abnormal motor features were evident which were associated with a significant loss of lumbar motor neurons. While a mutation can cause a lethal disease in humans, it may not be possible to trigger the same phenotype in a transgenic mouse models carrying the same mutation, as seen for VAPB mutations [18], and hence we investigated whether the DAOR199W mutation would potentiate the effects of the SOD-1 mouse model of ALS.
DAOR199W and SOD1G93A survival study
Given the well-known nature of the SOD1G93A mouse model and its well-defined lifespan, we examined the effect of the DAOR199W mutation on survival of SOD1G93A mice in a longitudinal large cohort survival study to see if the mutant DAO increased the rate of disease development and progression. Thus we crossed heterozygous DAOR199W females with SOD1G93A male transgenic mice. Animals with SOD1G93A and DAOR199W/SOD1G93A genotypes were recruited in the survival study, where the operator was blind to the genotype of each mouse. The development of ALS related disabilities in both genotypes was assessed twice a week using a neurological scoring system (with a scale of 0 to 4, where 0 was normal and four represented complete paralysis of both hind-limbs) commencing from 30 days of age in conjunction with the assessment of body weight [19]. The end point was established as the time at which the mouse was unable to right itself within 20 seconds when placed on its side and at this stage the animal was culled and the final age and weight recorded [19, 20].
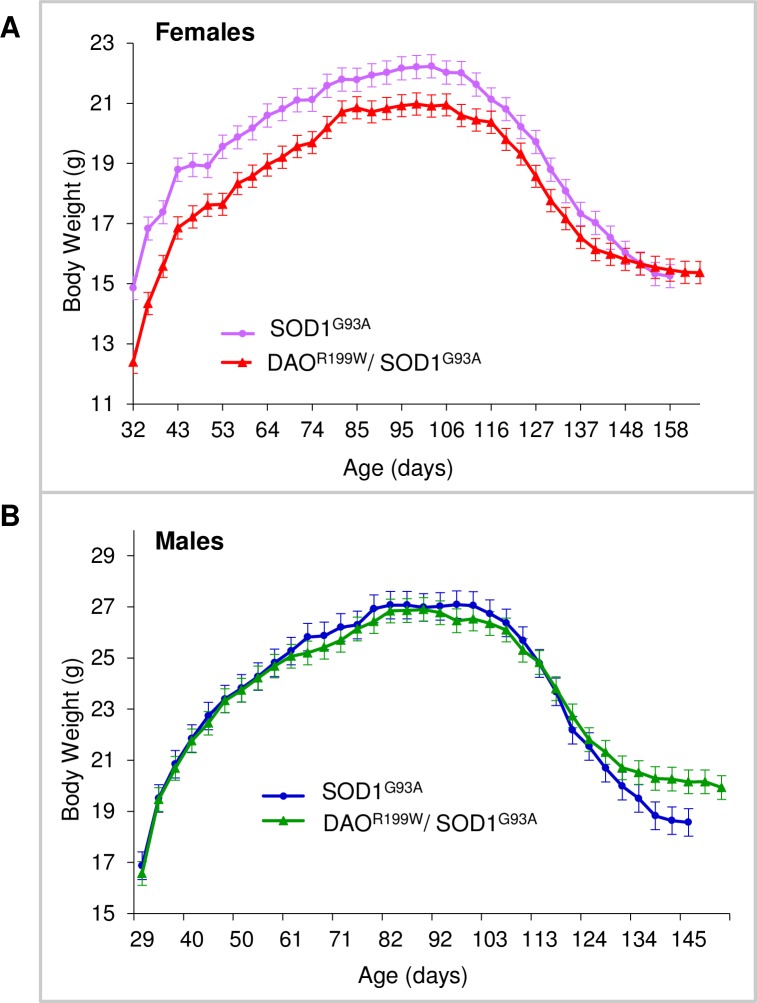
Assessment of body weight demonstrated the expected gender differences, male animals being heavier in terms of total body weight compared to their female littermates. Both genotypes, SOD1G93A and DAOR199W/SOD1G93A, increased weight steadily until all animals reached the maximum peak weight at approximately 88 days for males and 99 days for females (Fig 4A and 4B). After this point all animals experienced a marked decrease in their body weight, which continued to the humane end point. The most notable difference between genotypes was the decreased body weight seen in double transgenic female animals compared to SOD1 mice at all stages up until the peak weight was attained.
Fig 4. Longitudinal assessment of body weight in female SOD1G93A and double transgenic (SOD1G93A/DAOR199W) littermates.
Mean group body weight plotted for SOD1G93A and double transgenic (SOD1G93A/DAOR199W) mice over time from approximately 30 days of age until all the animals within a group reached the end stage and the study was terminated (n ≥ 14 per experimental group). As mice within groups died at different ages, in order to avoid decomposition of the mean at later stages, the final weights of all mice were carried forward when calculating group mean values, until the date at which the last mouse within each group died. Values are means with the standard error of the mean (SEM) indicated by error bars. (A) SOD1G93A females are presented in purple, and the double transgenic (SOD1G93A/DAOR199W) littermates are displayed in red. (B) SOD1G93A males are presented in blue, and the double transgenic (SOD1G93A/DAOR199W) littermates are displayed in green.
Assessment of neurological scores
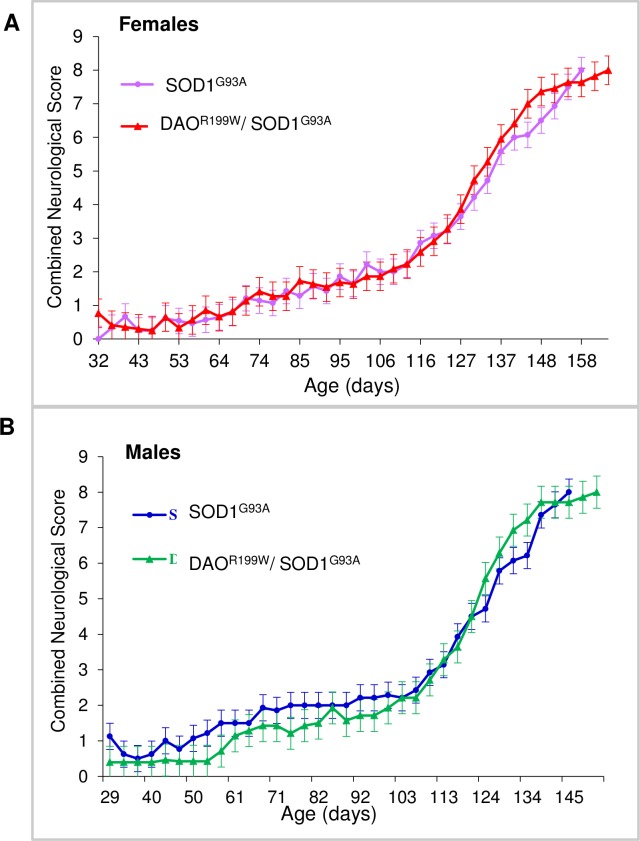
Analysis of neurological scores for both females and males demonstrates that both genotypes appeared normal early in their lifespan and did not develop any abnormalities until approximately 70 days of age, after which neurological scores consistently worsened until the animals reached the end stage. No significant differences were seen between the two genotypes (Fig 5).
Fig 5.
Longitudinal assessment of neurological scores in female SOD1G93A and double transgenic (SOD1G93A/DAOR199W) littermates (A) and for male mice (B). Mean group combined neurological score plotted for SOD1G93A and double transgenic (SOD1G93A/DAOR199W) mice over time from approximately 30 days of age until all the animals within a group died and the study was terminated (n ≥ 14 per experimental group). A combined neurological score was calculated for each mouse at each time point through summation of the neurological scores recorded for its left and right hind-limbs. Values are means with standard error of the mean (SEM) indicated by error bars. (A) SOD1G93A females are presented in purple, and the double transgenic (SOD1G93A/DAOR199W) littermates are displayed in red. (B) SOD1G93A males are presented in blue, and the double transgenic (SOD1G93A/DAOR199W) littermates are displayed in green.
Further analysis of the neurological scores was carried out as previously described [21] to yield values for onset of neurological signs, disease onset, disease progression and survival. Onset of neurological symptoms was defined as the point in time at which the animal reached a score of 2 (displayed by toe curling or dragging any part of the foot). Double transgenic females first displayed a neurological score of 2 at approximately 125 days of age, significantly earlier (p = 0.0239) than their SOD1G93A littermates (~131 days). Double transgenic males reached the score of 2 at approximately 119 days, which is slightly (but not significantly) later than their SOD1G93A littermates (~113 days). In addition, comparison of the same genotypes across the sexes indicate that while there were no differences in terms of the onset of neurological symptoms between double mutant transgenic mice, there was a significant difference in the time of onset between the SOD1G93A animals, with females exhibiting significantly later onset than male littermates (Table 1).
Table 1. Kaplan Meier time to event analyses for SOD1G93A and double transgenic (SOD1G93A/DAOR199W) in both males and females.
A. Median values obtained from Kaplan-Meier analysis for the time taken for mice to reach a score of 2 in both hindlimbs (onset of neurological scores), age at maximum weight (disease onset), disease progression and time taken for animals to reach the humane end point determined by its inability to right itself within 20 seconds of being placed on its side (survival) for both males and female littermates of either genotype groups. B. The Log-rank test assesses the null hypothesis that the Kaplan-Meier curves for all groups are identical (i.e. the presence of the DAOR199W transgene did not change the time taken to reach the event being analysed). Thus, low P-values (shown below) indicate that differences between genotype groups did not occur due to chance. Log-rank analysis of the onset of neurological symptoms in females carrying the SOD1G93A transgene versus their double transgenic littermates revealed that there were significant differences between the groups for this particular measurement. A significant difference was also detected in survival between the double transgenic males and females via the log-rank test. Significant differences are indicated in bold. Threshold of significance was set at P < 0.05.
| (A) | Median (days) | |||
| SOD1G93A | Double Transgenics (DT) | |||
| Males | Females | Males | Females | |
| Onset of Neurological Symptoms | 113 | 131 | 119 | 125 |
| Disease Onset | 89 | 99 | 88 | 99 |
| Disease Progression | 42 | 41 | 39 | 42 |
| Survival | 129 | 136 | 126 | 135 |
| (B) | Log-Rank Test (P Value) | |||
| SOD1G93A | DT | DT vs. SOD1G93A | ||
| Males vs. Females | Males vs. Females | Males | Females | |
| Onset of Neurological Symptoms | 0.0004 | 0.0518 | 0.3865 | 0.0239 |
| Disease Onset | 0.1102 | 0.1131 | 0.8183 | 0.7358 |
| Disease Progression | 0.5786 | 0.3318 | 0.2935 | 0.4750 |
| Survival | 0.0771 | 0.0241 | 0.2229 | 0.8242 |
Analysis of disease onset, defined as the point in time the animal was at its highest weight, showed an earlier age of 89 days in males compared to females (99 days of age). Onset of disease in both SOD1G93A and double transgenic mice was very similar (Table 1). Progression of disease, defined as the period of time between disease onset and the end stage, was very similar across all groups, ranging from 39 to 42 days.
Survival time, defined as the period of time between birth and the humane end point, determined by the inability of the animal to right itself within 20 seconds of being placed on its side, showed no major differences between the different genotypes and gender (Table 1), SOD1G93A and the double transgenic mice lived for a median of 129 and 126 days respectively in males and for 136 and 135 days respectively in females.
Comparison of the survival curves across the genotypes indicated that SOD1G93A females had a longer average lifespan (136 days) than their SOD1G93A male littermates (129 days) (Table 1) but these differences did not reach significance (P = 0.0771). Similar results were obtained for the double transgenic animals, with females living significantly longer (median of 135 days, P = 0.0241) than their double transgenic male littermates (median of 126 days).
Discussion
In this study we have characterized the in vivo effects of expression of a pathogenic mutation in DAO that is associated with familial ALS [4]. In earlier studies in cell culture, we showed that overexpression of DAOR199W promoted apoptosis in both primary motor neuron cultures and the motor neuron cell line, NSC-34, whereas overexpression of the wild-type protein DAOWT was without effect on cell survival. Other changes occurring in motor neurons expressing the mutant protein included the formation of ubiqutinated protein aggregates and substantial activation of autophagy [6]. Although these pathogenic effects may be mediated primarily by the accumulation of the mutant protein, other factors may be relevant, such as the impaired activity of DAO and consequential effects on D-serine metabolism. D-serine is a co-agonist at the NMDA receptor playing an essential role in mediating excitatory activity in the CNS. We showed that co-culturing motor neurons with glial cells expressing DAOR199W was sufficient to induce apoptosis in motor neurons lacking the mutant protein and that this effect was sensitive to 5,7-Dichloro-4-hydroxyquinoline-2-carboxylic acid (DCKA), an NMDAR antagonist selective for the D-serine/glycine site [6]. The increased protein aggregation and upregulation of autophagy seen in motor neuron cell lines expressing DAOR199W compared to cell lines expressing DAOWT were both also sensitive to this antagonist. These results suggest that the pathogenic effects of DAOR199W may at least be partly mediated through an excitotoxic mechanism, potentiated by the build-up of D-serine in the synapse. The high levels of DAO found in human spinal cord and brain stem motor neurons are consistent with the vital role played by DAO in preventing excitotoxic cell death in these regions.
In this study, we have generated transgenic mouse lines expressing the human DAO mutation associated with FALS in order to characterise the effects of this mutation in vivo. We monitored body weight, motor features, gait and quantified motor neurones in lumbar spinal cord in transgenic lines expressing DAOR199W in a longitudinal large cohort survival study over a period of one year to characterise effects on motor function. The most distinctive features seen were decreased body weight, marked kyphosis and loss of motor neurons in spinal cord. All of these features are characteristic of several ALS models including SOD1G93A transgenic mice [22] and conditional targeted inactivation of the Tardbp gene in spinal cord motor neurons [23]. In addition, recent studies of a naturally occurring mouse mutation in DAO [11] DAOG181R [24], bred as a homozygous line exhibited marked effects on motor phenotype, with abnormal reflexes characterised by retraction of hind limbs, as occurs in SOD1G93A mice, accompanied by a significant reduction in motor neuron number, as occurred in this study in DAOR199W mice, and axonal degeneration with muscle atrophy demonstrated at 15 months [13].
Animals expressing the DAOR199W transgene consistently had a lower body weight at each age compared to their wild type littermates (Fig 2), which was evident in both females and males (Fig 2A and 2B). This clearly shows that impaired DAO activity affects normal weight gain in heterozygous animals expressing the mutant protein in the presence of endogenous mouse DAO. The decreased body weight is consistent with studies on ddY/DAO− mice that lack DAO activity. When fed a diet in which L-phenylalanine was replaced by D-phenylalanine, these mice were unable to maintain their body weight compared to wild type animals (ddY/DAO+) [25]. This study clearly shows that impaired DAO activity affects normal weight gain both when given a D-amino acid enriched diet and also in our study on heterozygous animals expressing the mutant protein without D-amino acid enrichment but in the presence of endogenous mouse DAO. Both males and females bearing the mutant transgene also developed progressive kyphosis, which was more conspicuous in transgenic females than the males. This skeletal abnormality is also prominent in mouse models of ALS such as the SOD1 mouse carrying the human ALS mutation and may be a consequence of the weakening of the extensor thoracic paraspinal muscles as suggested by Wu et al [23] and as seen in ALS patients [26]. Further studies of gait and body movement in DAOR199W animals demonstrated a significant difference in terms of paw angle in the DAOR199W transgenic females in comparison to their wild type littermates which is likely to have been a consequence of degenerative changes resulting from motor neuron loss and muscle atrophy.
Although D-serine was not quantified in tissue homogenates in transgenic animals in this study, D-serine immunoreactivity was measured in lumbar spinal cord in transgenic and wild-type littermates using a rabbit polyclonal anti-serine antibody raised against D-serine crosslinked to bovine serum albumin with glutaraldehyde (Abcam, dilution 1:5,000), as previously used to characterise D-serine distribution in human and mouse spinal cord [6], where the full characterisation is described. Consistent with these studies, we found that D-serine immunoreactivity was predominantly concentrated in large motor neurons in transgenic mice and their non-transgenic littermate (Figure B in S1 File). In addition, serine racemase was abundant in spinal cord grey matter and strongly localised to large motor neurones (Figure C in S1 File).A similar distribution was seen in both wild-type and DAOR199W mice but this did not reflect changes occurring in extracellular and non-neuronal compartments or in the relative levels of D and L-serine. However, it is already known that D-serine is elevated in spinal cord of sporadic cases of ALS and also in the SOD1 mouse model of ALS suggesting the involvement of glutamate excitotoxicity in ALS pathogenesis [27]. Furthermore the same group also showed that D-serine was significantly elevated in spinal cord tissue homogenates from the DAOG183R homozygous mouse line at 5 months, using a sensitive and selective 2D-HPLC method, but a significant increase was not detected in the heterozygous mouse line [13].
In summary, despite the development of motor defects, within the time frame of this study, overall survival was not reduced in DAOR199W mice. This suggests that other factors must contribute to the severe phenotype seen in ALS cases expressing this mutation. We therefore looked to see if the presence of DAOR199W potentiated the development of disease in SOD1G93A transgenic mice by crossing the DAOR199W and SOD1G93A mouse lines to generate a double transgenic line (DAOR188W/SOD1G93A). A role for DAO in the pathogenicity of the SOD1G93A mouse has already been demonstrated by the observation that DAO activity is decreased by 42%, which is equivalent to the reduction seen in DAO heterozygote mice [13]. The effect of this reduced DAO activity, measured using a sensitive 2D-HPLC method, was also confirmed to result in the expected increase in levels of D-serine in spinal cord that increased with disease progression [13].
Body weight and neurological scores were monitored twice a week and survival was determined as the age at which the animal was unable to right itself within 20 seconds of being placed on its side. Overall, both the SOD1G93A and their double transgenic littermates followed the same pattern in terms of their marked reduction in body weight after 90 days. However, marked differences in body weight were seen between males and females (Fig 4). Whilst body weight was lower in female double transgenic mice compared to SOD1G93A mice, in males both SOD1G93A and double transgenic animals showed a similar body mass (Fig 4). This differential effect in females may reflect an endocrine effect, as testosterone is known to stimulate DAO activity in mouse kidney [28].
Since the DAOR199W transgenic line is heterozygous for the DAO allele on the C57B16xCBA/Ca background, as well as expressing the mutant allele they also express the wild type endogenous DAO protein. It has been demonstrated that DAO activity is significantly reduced in the lumbar region of the SOD1G93A mice, to levels approaching those of the B6DAO+/− mice [13]. It is thus likely that the effect of the endogenous DAO protein is suppressed by the presence of the SOD1G93A transgene in our double transgenic animals. As DAO is required for degradation of D-amino acids, it is therefore possible that the presence of both the SOD1G93A and DAOR199W mutant transgenes in the double transgenic females suppressed or diminished the already low levels of endogenous DAO enzyme activity, preventing animals from maintaining their overall body mass and consequently exhibiting a consistent reduction in weight when compared to their SOD1G93A littermates throughout their lifespan (Fig 2). Conversely, the marked similarity in body weight between the double transgenic males and their SOD1G93A littermates could stem from the innately higher levels of DAO enzymatic activity found in males compared to females which might buffer the influence of the double transgene, permitting a residual level of function sufficient for the animals to maintain their body weight at a level comparable to their SOD1G93A littermates. This low level of endogenous DAO enzyme activity could also account for the attenuated motor neuron loss in the lumbar region of the spinal cord that we found in males which (in contrast to the reduction observed in females) did not reach significance. This in turn may be an indication of protective properties of the wild type DAO protein against motor neuron degeneration.
Interestingly, onset of neurological symptoms in double transgenic mice also showed a differential effect in males and females (Fig 5, Table 1). Most notably, while males of both genotypes typically developed symptoms at earlier time points than females, female double transgenic animals displayed a symptomatic onset six days earlier than their female SOD1G93A littermates (125 days vs 131 days, P = 0.0239). No such potentiation was observed in male double transgenic mice. Disease progression and survival appeared largely insensitive to the presence of the DAOR199W transgene, although double transgenic females did survive significantly longer than double transgenic males (P = 0.0241).
In conclusion, our results indicate that although the different genotypes exhibit some clear differences, overall the presence of the DAOR199W transgene alongside SOD1G93A did not exert any major effects on disease onset, progression or survival in either sex. This is likely due to the aggressive and accelerated nature of the phenotype enforced by the SOD1G93A mutation, masking any effect the DAOR199W mutation may trigger. It is also important to point out that in our initial characterization of the DAOR199W, we detected enhanced ubiquitination at 8 months (Fig 1C) and motor neuron cell loss at 14 months (13.5 to 14.5 months) (Fig 3C). Such progressive changes would not have been well established within the time scale of the SOD1 pathology (which has onset at approximately 3 months with death occurring at 5 months). In future it would be useful to carry out the DAOR199W cross with a lower copy number SOD1 mouse line.
Two recent studies have further supported the relevance of DAO dysfunction to ALS pathogenesis. Firstly in a comprehensive exome sequencing study in 2,874 cases with sporadic ALS and 6,405 controls, interestingly, it was shown that DAO was the only known predisposition gene where the presence of DNA variants was significantly associated with clinical outcome, decreasing rates of survival [29]. Secondly, in a study of the effect a pathogenic mutation in a gene encoding an RNA binding protein, hnRNP A2/B2, that, in common with mutations in the FALS gene VCP causes multisystem proteinopathy, it was found to have a profound effect on the splicing of DAO [30]. The resulting abnormality was a decrease in the constitutive form of DAO, the full length transcript analysed in this study (Fig 1), accompanied by an increase in the level of a transcript lacking exon 9, that leads to truncation of the protein and severely reduces enzyme activity. In future studies, it will be important to further elucidate whether the formation of this alternative transcript contributes to the pathogenesis of ALS.
Materials and methods
All reagents purchased from Sigma unless indicated otherwise.
Ethics statement
All animal experiments were carried out under license from the Home Office (UK) in accordance with the Animals Scientific Procedures Act 1986 and were approved by Imperial College London / Royal Veterinary College ethical review committees.
Establishment of the DAOR199W transgenic mouse line
Transgenic mouse lines overexpressing DAOR199W were created using human DAOR199W, driven by a chicken β-actin promoter and cytomegalovirus enhancer, pCAGGS (Niwa et al., 1991). The linearized transgene was microinjected into pronuclei of fertilized eggs from C57BL6×CBA/Ca female mice, which were subsequently transferred to pseudopregnant recipients. Integration of the transgene into the genomic DNA was determined 3–4 weeks after birth, by collecting a small ear biopsy from the pups and screening for the transgene expression by using polymerase chain reaction (PCR). Hemizygote colonies from two independent founders expressing the transgenes were established and maintained by crossing DAOR199W males with wild-type F1 hybrid C57BL6xCBA/Ca females.
Survival studies
Two transgenic mouse lines were used, a widely used high copy number SOD1G93A and the DAOR199W mouse lines, generated in this study. Mice overexpressing human SOD1 bearing the G93A mutation causing a motor neuron phenotype [B6SJL-TgN SOD1G93A 1 Gur/J, Stock number 002726] [31] were initially purchased from the Jackson Laboratory (Bar Harbor, ME). They were maintained as hemizygotes by breeding male mice with wild type F1 hybrid C57BL6xCBA/Ca females. The SOD1G93A mice have been generated on this background for over 28 generations, and animals on this background have similar disease characteristics (as previously assessed by hindlimb electrophysiological parameters [32, 33] and longitudinal monitoring of neurological score and body weight [34, 35] and life span to the original and more widely used B6SJL hybrid genetic background [36, 37]. We did not observe any changes in terms of lifespan and disease progression in these animals with successive generations in our laboratory [35, 38].
A double transgenic (DT) mouse model was generated following breeding of heterozygous high copy number SOD1G93A males with heterozygous DAOR199W females, generating 4 different genotypes per cross. Genotypes obtained from the cross were SOD1G93A, SOD1G93A/DAOR199W, DAOR199W, and Wild-type.
Genotyping
Genotyping was carried out following weaning, using an ear biopsy (~1.5-3mm2 in size). DNA extraction was performed by overnight incubation of the samples at 55°C in 1x homogenisation ear biopsy buffer (50μl). After digestion, proteinase K (Invitrogen) was inactivated at 100°C and samples were screened for the expression of the transgenes of interest (SOD1G93A or DAOR199W) using specific PCR conditions for each genotype as detailed in S1 File.
Design for survival studies
Based on the results of Scott et al [19] who defined the optimal conditions for carrying out survival studies involving the SOD1G93A strain, we used 24 mice per experimental group per cohort (12 males and 12 females), same-gender litter matching, and excluded any animal who had experienced loss of SOD1G93A transgene copy number [19]. Disease onset and progression were established through a combination of longitudinal monitoring of weight and neurological score [19, 34, 39]. Each individual animal was assessed using a simple neurological scoring system, which provided a rapid, non-invasive and unambiguous measure of disease onset and progression in SOD1G93A mice [19, 34, 35, 38, 39]. Weight loss in high copy number SOD1G93A mice is normally a gradual process that begins at approximately 90 days of age and continues until death. Rapid losses of weight over a short period of time that does not fit the normal pattern can therefore be indicative of non ALS related problems [19]. In addition, based on the data generated from the previous survival analyses in our laboratory we determined the end point as the inability of the animal to right itself within 20 seconds of being placed on its side. From 100 days of age, the righting reflex of each mouse was examined twice daily to ensure that the end point of each mouse was accurately determined. Once mice reached the end point, they were euthanized via cervical dislocation, the date and cause of their death were recorded and an ear and tail biopsy was collected for PCR and Q-PCR respectively to reconfirm the genotype and for examination of SOD1G93A transgene copy number, as changes in the latter affect disease progression and ultimately survival. SOD1G93A transgene copy number variation was assessed by quantitative PCR (qPCR) in any animal demonstrating outlying disease progression or survival characteristics [40].
Animals participating in all the in vivo studies were housed in a minimal disease facility at the Royal Veterinary College, with a ‘12:12’ hour ‘light:dark’ cycle, food and water ad libitum. Breeding trios each consisting of one SOD1G93A transgenic male and two DAOR199W females were set up simultaneously to generate a large cohort of age matched pups with a mixture of genotypes namely SOD1G93A, DAOR199W, SOD1G93A/DAOR199W, and wild type, for enrolment in the “SOD1G93A x DAOR199W” survival study. Littermates of the same sex were housed together regardless of the genotype (not exceeding 5 animals per cage) to ensure consistency of the results in terms of age and gender, and the assessor was blinded to the genotypes of the individual mice until the termination of the study.
Survival study analysis
The development of ALS related disabilities were assessed by means of neurological scoring system for both hind legs twice weekly for each mouse from approximately 30 days of age in the case of “SOD1G93A x DAOR199W” cross. Neurological scores were assigned using a scale of 0–4, established by the ALSTDI through detailed observations of SOD1G93A mouse pathology [19, 34, 39]. Criteria used to assign each score level were:
0: Full extension of hind legs away from lateral midline when mouse is suspended by its tail and mouse can hold this for 2 seconds, suspended 2–3 times.
1: Partial or full collapse of leg extension towards lateral midline or trembling of hind legs during tail suspension.
2: Toes curl under at least twice during walking of 12 inches or any part of foot is dragging along cage bottom (a clean cage containing no bedding was used for this analysis to permit easy viewing of the toes).
3: Minimal joint movement or rigid paralysis, foot not being used for forward motion.
4: Mouse cannot right itself within 20 seconds from one or both sides when placed on its side.
Graphical representation & statistical analysis
In order to demonstrate overall changes in body weight and neurological score, mean values for both body weight and neurological score were plotted for each group over time until the point in time at which the last mouse in the group reached the humane end point. To prevent decomposition of the mean weight and neurological score values as mice reached the end point, final weight and neurological score values for individual mice that reached the end point were carried forward until the time point at which the last mouse in that particular group reached the humane end point. This is a practice commonly used when performing survival analyses with longitudinal monitoring of weight and neurological score [34, 38, 39]. In order to avoid the loss of statistical validity that can occur when performing repeated analyses on correlated serial measurements for the same subjects, relevant summary measures for weight and neurological score data were selected as advised by Matthews and colleagues and statistical analyses were performed on these summary measures [21]. Summary measures selected include: time to attain peak weight (the earliest indicator of disease onset), time from peak weight to death (an indicator of disease progression), time to attain a neurological score of 2 in both hind-limbs (defined as the onset of neurological symptoms) and time to reach a score of 4 (the uniform humane end point selected for this study). These summary measures all represent ‘time to event measures’ and were analysed via Kaplan-Meier survival analyses: the log-rank test was performed to determine the statistical significance amongst experimental groups with the null-hypothesis that presence of the DAO transgene did not change the time taken to reach the event being analysed. A low P-value thus indicates that the differences between the curves did not occur due to chance, with the threshold for significance being set at (P < 0.05) for all analyses [38]. All Kaplan-Meier survival analyses were carried out using Graphpad Prism 6 software.
In the case of assessment of overall body weight, animals were weighed and values were plotted as ‘mean group body weight’ at each particular age using Excel software. Since each genotype group generated through the crosses was considered as a new strain of mice, and taking into account the fact that the differences in body weight commenced at birth, it was concluded by our statistician advisor that differences in body weight between the different experimental groups were not statistically comparable. Thus, no statistical test was carried out for this particular variable.
Gait analysis
Upon completion of the SOD1G93A/DAOR199W survival study, animals expressing the DAOR199W mutant transgene and their wild type littermates were recruited to a gait analysis study carried out using the DigiGait Imaging System (Mouse Specifics, Inc.). An automated system equipped with transparent treadmill belt and adjustable speed, and a video camera mounted underneath the belt captured the image of the ventral side of the animals automatically pixelating and vectorizing the ventral view of the animal [15]. The collected images were automatically analysed via the DigiGait software (Mouse Specifics, Inc.) where the area of each paw was recorded in correspondence to its movement and a periodic wave curve was therefore generated describing the advance and retreat of the four limbs in comparison to the treadmill belt through consecutive strides [15]. The portions of the paw that were in contact with the treadmill belt were automatically recognized as the stance phase of the stride by the software and the portion of the paw not in contact with the treadmill belt were consequently identified as the swing phase of the stride [15]. In addition, various other gait dynamics (i.e. over 40 parameters) were determined for each individual limb by the software that were extracted from the period of time spent in various portions of the walking phase, including paw area at peak stance, paw angle, brake, swing, stride length, and stepping frequency (Text A in S1 File).
Spinal cord collection & preparation
Animals were deeply anaesthetized by intraperitoneal (IP) administration of pentobarbital (Merial, UK) overdose (approximately 120–150μl). Under terminal anaesthesia the animals were subjected to transcardial perfusion with ice-cold PBS and ice-cold 4% PFA (prepared in PBS) respectively. Upon completion of the perfusion process, the spinal column was removed and post-fixed in 4% PFA at 4°C overnight. The spinal cord was then extracted from the spinal column and cryoprotected in 30% sterilized sucrose solution (prepared in PBS) at 4°C, a process that often required a few days during which the spinal cord sank towards the bottom the container within the sucrose solution. The lumbar region of the spinal cord was then selectively dissected, and embedded in gelatin and mounted on a cork block held in place with Cryo M-bed (Bright). This process involved vertically mounting the spinal cord on the cork block with the most caudal end placed on the cork block. The ventral side of the spinal cord was marked on the cork to allow orientation of the block within the cryostat such that the ventral side met the knife first [37]. The mounted spinal cord was then snap-frozen in liquid nitrogen cooled isopentane (Fisher Scientific) (-55°C, +/-5°C) and stored at -80°C.
Lumbar spinal cord sectioning
The lumbar regions of 23 mouse spinal cords were sectioned using a cryostat (Bright, UK). Sections of 20μm were taken between the regions of L2-L6. Sections were arranged on Superfrost miscroscope slides (VWR, Belgium). Slides with sections were frozen at -80°C pending analysis.
Staining of sections
Nissl staining of nucleic acids in the tissue was performed on every 6th slide to identify the L3 to L5 regions of the animals’ spinal cords. Sections were placed in Nissl stain for 30 minutes before washing and dehydrating in graded alcohols before clearing in xylene and mounting with coverslips and DPX mountant (Sigma-Aldrich). The L3 to L5 region for each animal was determined using a Leica DM2500 microscope and descriptions of mouse spinal cord regions by Watson at al. [41]. The lumbar motor neuron pool starts at L3 and extends to L5 [42].
Immunohistochemistry for the enzyme ChAT was performed on every 2nd and 5th slide on the L3 through to L5 regions of animals. After rehydrating with phosphate buffered saline with 0.1% Tween®20 (Sigma-Aldrich) (PBS/T), endogenous peroxidase activity was quenched by incubating sections in PBS/T with 0.3% hydrogen peroxide (Sigma-Aldrich) for 5 minutes. Sections were then incubated with Avidin for 10 minutes and after washing with Biotin for 10 minutes, both from the Vector blocking kit (Vector, USA). Sections were washed again with PBS/T and blocked with 5% bovine serum albumin (Sigma-Aldrich) (BSA) in PBS/T for 30 minutes. Slides were washed with PBS/T and sections were incubated with the primary antibody, anti-ChAT (Millipore, USA), at 1:100 in PBS/T with 1% BSA overnight at 4°C.
After 3 x 5 minute washes with PBS/T sections were incubated in the secondary antibody, biotinylated anti-goat (Vector) at 1:500 in PBS/T with 1% BSA for 1 hour. Vector ABC reagent (Vector stain Elite kit) was made with 2 drops of solution A and 2 drops of solution B in 5ml PBS/T and left at room temperature for 30 minutes and then put on ice for 30 minutes before use. Sections were washed for 3 x 5 minutes with PBS/T and incubated in ABC reagent for 30 minutes. After 3 x 5 minute washes with PBS/T DAB solution was added made up using the DAB peroxidase substrate kit (Vector) of 2 drops of buffer, 4 drops DAB and 2 drops of hydrogen peroxide in 5 ml of distilled water. After 2 minutes DAB was washed off with water and dehydrated with graded alcohols before clearing in xylene for 2 x 10minutes. Slides were mounted with DPX and 60mm coverslips.
Motor neuron counts
For the ChAT stained slides the motor neurons of every section within the L3-L5 regions were counted. The motor neurons were counted that were identifiable in the structures of the lumbar ventral horn shown in Fig 3C [41]. Stained cells were included in the counts if they had a diameter of ≥20μm, specific labelling of the cytoplasm and a spherical nucleus with clear nucleolus. Cells were imaged with the Leica DM2500 microscope and the longest diameter of the cell body was measured using Q Imaging software. Counting was done blind to genotype of the animal and left and right sides of the spinal cord sections were counted separately. The 40 sections prior to the end of L5/start of L6 boundary were included in the total counts of motor neurons. Confirmatory counts were performed on every 12th section within this region of the lumbar spinal cord using Nissl stained sections. When counting was completed, the sums for the left and right side of the spinal cord sections included for each animal were separated by genotype. Unpaired, one-tailed t-tests were performed with GraphPad Prism (USA) to identify significant differences between genotype groups, where p≤0.05.
Western blot analysis
All animal tissues were collected upon euthanization of the animal via cervical dislocation, snap-frozen in liquid nitrogen and stored at -80°C. Tissues were then weighed and ground in the presence of liquid nitrogen using a pestle and mortar, and incubated on ice for 30 minutes in chilled RIPA buffer solution (150 mM sodium chloride, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0, containing 1mM PMSF, 1mM sodium orthovanadate, 5mM sodium fluoride, and a mixture of protease inhibitors (Roche). The samples were centrifuged at 4°C for 15 minutes at 15,000rpm, the supernatants were removed and used for protein quantification using the ‘BCA protein assay kit’ (Thermo Scientific) and the final concentration was determined using a ‘Nanodrop ND-1000 Spectrophotometer’ (Thermo Scientific) instrument. Samples were separated by 10% SDS/PAGE using NuPAGE LDS loading dye (Life Technologies), proteins transferred from the gel onto a PVDF membrane (Millipore) for 1 hour at a constant current of 80V, blocked in 5% non-fat milk/TBS-Tween (0.1%), and incubated with primary antibody for 1 hour at room temperature followed by secondary antibody for a further hour. All antibodies were prepared in TBS/T at the following dilutions: Anti-porcine DAO (Nordic, 1:2000); anti-beta tubulin (Abcam, 1:10000); goat anti-rabbit HRP (Bio Rad, 1:50000). Membranes were developed using enhanced chemiluminescence (‘Amersham Hyperfilm ECL’ (GE Healthcare)).
Quantitative PCR for DAO expression
Spinal cord, brain and quadriceps muscle samples were pulverised under liquid nitrogen and the powder mixed with TRIzol reagent at a ratio of 1ml TRIzol per 100mg tissue (quadriceps muscle) or 1ml per 50mg (brain and spinal cord). Samples were extracted from tissue samples using a TRIzol protocol as supplied by the manufacturers with the addition of an extra chloroform extraction step (1:1) after collection of the aqueous phase. RNA was assessed via nanodrop to ensure good 260/280 and 260/230 ratios (>1.9). Samples with 260/230 ratios lower than 1.9 were cleaned by a second round of isopropanol precipitation.
For cDNA synthesis, 800ng of RNA per reaction was reverse transcribed using the RTnanoscript2 kit (Primer Design) and final reactions were diluted 1/5 (empirically determined to be a sufficient dilution to prevent cDNA synthesis reagents having any effect on subsequent PCR efficiency). qPCR was performed in a CFX384 light cycler (Biorad) using PrecisionPLUS mastermix with SYBR green (Primer Design), with 10ul volumes of primers at a final concentration of 500nM, and with 10-15ng cDNA per well. Cycling conditions: 2mins at 95°C, then 50 cycles of: 15sec 95 oC, 20sec 60°C, 20sec 72 oC followed by a melt curve (60–95 oC in 0.5 degree steps).
The following primers were used:
hDAO qPCR Ex8F: CACCCATGACCCAGAGAGAG
hDAO qPCR Ex9R: ATTCTTCAGTGTGGGCTCCA
mDAO qPCR Ex8F: CCCATGATCCTAGCCTTGGT
mDAO qPCR Ex9R: GCTCCAGTTTACAGCAGCTC
Pak1ip1 was used as a reference gene in all tissues (Primer Design).
Determination of D-Amino Oxidase activity via spectrophotometric assay
DAO activity assays used a protocol modified from Tedeschi [14].
Non-transgenic (WT) mice and mice carrying the DAOR199W transgene were killed by cervical dislocation and tissues harvested rapidly. 3–5 animals were used per genotype. Brains, spinal cords and quadriceps muscles were flash-frozen and then pulverised under liquid nitrogen using a mortar and pestle before storage at -80°C. Tissue homogenates were prepared by combining powdered tissues with extraction buffer (75mM Sodium Pyrophosphate pH 8.5, supplemented with 0.5% NP40, complete protease inhibitor cocktail (Roche) and 1% phosphatase inhibitor cocktails 2 and 3 (Roche)) to a final concentration of approx. 50mg.ml-1 and vortexing vigorously. Homogenates were incubated on ice for 10 minutes with occasional mixing before removal of insoluble material by centrifugation (13krpm, at 4°C in a benchtop microfuge). Soluble protein fractions were collected and protein concentrations (typically 5-10mg.ml-1) determined by DC protein assay (BioRad).
DAO spectrophotometric assays were performed in duplicate in flat-bottomed 96-well plates (Corning), using an M200 Pro plate reader (TECAN) at room temperature. Soluble protein fractions (typically 100-200ug of soluble protein per well) were added to assay buffer (75mM Sodium Pyrophosphate pH 8.5, 50mM D-serine, 20uM FAD, Horseradish Peroxidase (1U.ml-1), ~1mM o-dianisidine–shaken well to oxygenate), mixed briefly, and absorbance change at 436nM measure at approximately sixty second intervals for one hour. O-dianisidine exhibits poor solubility in most aqueous solutions. For these assays, a stock solution at 33mM was prepared in a 1:1 mix of DMSO and water, then added to 75mM sodium pyrophosphate assay buffer to a final concentration of 1mM. The solution was incubated at room temperature for 10 min to allow any insoluble dye to precipitate fully, then filtered before addition of FAD, D-Serine and peroxidase. Crude tissue isolates typically exhibit a lag phase of 2–10 minutes before reaching maximal rates, with rates subsequently lowering to a basal non-specific rate after 20–30 minutes. Maximum rates were recorded and compared against a standard curve of purified porcine DAO (loaded on the same plate).
Supporting information
Text A. Methodology: Genotyping and Gait Analysis. Figure A. DAO enzyme activity in spinal cord and brain from wild-type (WT) and DAOR199W transgenic mice (R199W). Activity measured as mUnits/mg tissue/lysate in brain and spinal cord from wild-type (WT) and DAOR199W transgenic mice (R199W). Figure B. D-serine immunoreactivity in lumbar spinal cord from control and DAOR199W transgenic mice. A rabbit polyclonal Anti-D-Serine antibody (Abcam) raised against d-serine cross-linked to BSA with glutaraldehyde was used at a dilution of 1 in 5,000 as previously described in detail [6]. Figure C. Serine racemase immunoreactivity in lumbar spinal cord from control and DAOR199W transgenic mice. A mouse monoclonal Anti-serine racemase antibody (BD Biosciences) was used at a dilution of 1 in 1000 as previously described in detail [6].
(DOCX)
Acknowledgments
We are grateful to the Motor Neurone Disease Association (UK) for funding this project.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This project was funded by the Motor Neurone Disease Association (UK) following rigorous review of study design, methods for data collection and analysis provided in the grant application. They were not involved in the subsequent study or manuscript preparation.
References
- 1.Taylor JP, Brown RH, Cleveland DW. Decoding ALS: from genes to mechanism. Nature 2016; 539: 197–206 doi: 10.1038/nature20413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006; 314: 130–3 doi: 10.1126/science.1134108 [DOI] [PubMed] [Google Scholar]
- 3.Chen HJ & de Belleroche J. Endoplasmic reticulum (ER) stress in amyotrophic lateral sclerosis (ALS) In: Agostinis P, Samali A (eds) 2012. Endoplasmic reticulum stress in health and disease. Springer, Dordrecht, Heidelberg, New York, London: pp 323–337 [Google Scholar]
- 4.Mitchell J, Paul P, Chen HJ, Morris A, Payling M, Falchi M, et al. Familial amyotrophic lateral sclerosis is associated with a mutation in D-amino acid oxidase. Proc Natl Acad Sci USA. 2010; 107: 7556–7561 doi: 10.1073/pnas.0914128107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paul P & de Belleroche J. The role of D-amino acids in amyotrophic lateral sclerosis pathogenesis: a review. Amino Acids. 2012; 43: 1823–31 doi: 10.1007/s00726-012-1385-9 [DOI] [PubMed] [Google Scholar]
- 6.Paul P, Murphy T, Oseni Z, Sivalokanathan S, de Belleroche J. Pathogenic effects of amyotrophic lateral sclerosis-linked mutation in d-amino acid oxidase are mediated by d-serine. Neurobiol. Aging. 2014; 35, 876–885. doi: 10.1016/j.neurobiolaging.2013.09.005 [DOI] [PubMed] [Google Scholar]
- 7.Mothet JP, Parent AT, Wolosker H, Brady RO Jr, Linden DJ, Ferris CD, et al. D-serine is an endogenous ligand for the glycine site of the N-methyl-D-aspartate receptor. Proc Natl Acad Sci U S A. 2000; 97: 4926–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, Ge W, Chen Y, Zhang Z, Shen W, Wu C, et al. Contribution of astrocytes to hippocampal long-term potentiation through release of D-serine. Proc Natl Acad Sci U S A. 2003; 100: 15194–9. doi: 10.1073/pnas.2431073100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panatier A, Theodosis DT, Mothet JP, Touquet B, Pollegioni L, Poulain D.A. Glia-derived D-serine controls NMDA receptor activity and synaptic memory. Cell. 2006; 125: 775–84. doi: 10.1016/j.cell.2006.02.051 [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Sacchi S, Pollegioni L, Basu AC, Coyle JT, Bolshakov VY. Identity of endogenous NMDAR glycine site agonist in amygdala is determined by synaptic activity level. Nat Commun. 2013; 4: 1760 doi: 10.1038/ncomms2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konno R & Yasumura Y. Mouse mutant deficient in d-amino acid oxidase activity, Genetics. 1983; 103: 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Almond SL, Fradley RL, Armstrong EJ, Heavens RB, Rutter AR, Newman RJ, et al. Behavioral and biochemical characterization of a mutant mouse strain lacking D-amino acid oxidase activity and its implications for schizophrenia. Mol Cell Neurosci. 2006; 32: 324–34. doi: 10.1016/j.mcn.2006.05.003 [DOI] [PubMed] [Google Scholar]
- 13.Sasabe J, Miyoshi Y, Suzuki M, Mita M, Konno R, Matsuoka M, et al. D-amino acid oxidase controls motoneuron degeneration through D-serine. Proc Natl Acad Sci USA. 2012; 109: 627–32. doi: 10.1073/pnas.1114639109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tedeschi G, Pollegioni L, Negri A. Assays of D-Amino Acid Oxidases in “Unnatural Amino Acids”: Methods in Mol. Biol. 2012; Vol 794:381–395. [DOI] [PubMed] [Google Scholar]
- 15.Vincelette J, Xu Y, Zhang L N, Schaefer CJ, Vergona R, Sullivan ME, et al. Gait analysis in a murine model of collagen-induced arthritis. Arthritis Res Ther. 2007; 9 R123 doi: 10.1186/ar2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piesla MJ, Leventhal L, Strassle BW, Harrison JE, Cummons TA, Lu P, et al. Abnormal gait, due to inflammation but not nerve injury, reflects enhanced nociception in preclinical pain models. Brain Res. 2009;1295: 89–98., doi: 10.1016/j.brainres.2009.07.091 [DOI] [PubMed] [Google Scholar]
- 17.Hampton TG, Kale A, Amende I, Tang W, Mccue S, Bhagavan HN, et al. Gait disturbances in dystrophic hamsters. J Biomed Biotechnol. 2011:235354, doi: 10.1155/2011/235354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tudor EL, Galtrey CM, Perkinton MS, Lau KF, De Vos KJ, Mitchell JC, et al. Amyotrophic lateral sclerosis mutant vesicle-associated membrane protein-associated protein-B transgenic mice develop TAR-DNA-binding protein-43 pathology. Neuroscience. 2010; 167: 774–785. doi: 10.1016/j.neuroscience.2010.02.035 [DOI] [PubMed] [Google Scholar]
- 19.Scott S, Kranz JE, Cole J, Lincecum JM, Thompson K, Kelly N, et al. Design, power, and interpretation of studies in the standard murine model of ALS. Amyotroph Lateral Scler. 2008; 9: 4–15. doi: 10.1080/17482960701856300 [DOI] [PubMed] [Google Scholar]
- 20.Wang LJ, Lu YY, Muramatsu S, Ikeguchi K, Fujimoto K, Okada T, et al. Neuroprotective effects of glial cell line-derived neurotrophic factor mediated by an adeno-associated virus vector in a transgenic animal model of amyotrophic lateral sclerosis. J Neurosci, 2002; 22: 6920–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthews JN, Altman DG, Campbell MJ, Royston P. Analysis of serial measurements in medical research. Bmj.1990; 300: 230–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azzouz M., Leclerc N, Gurney M, Warter JM, Poindron P, Borg J. Progressive motor neuron impairment in an animal model of familial amyotrophic lateral sclerosis. Muscle Nerve. 1997; 20: 45–51 [DOI] [PubMed] [Google Scholar]
- 23.Wu L-S, Cheng W-C and Shen C-K. Targeted Depletion of TDP-43 Expression in the Spinal Cord Motor Neurons Leads to the Development of Amyotrophic Lateral Sclerosis-like Phenotypes in Mice. J Biol Chem. 2012; 287: 27335–344 doi: 10.1074/jbc.M112.359000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasaki M, Konno R, Nishio M, Niwa A, Yasumura Y, Enami J. A single-base-pair substitution abolishes D-amino-acid oxidase activity in the mouse. Biochim Biophys.Acta. 1992; 25: 1139(4):315–8. [DOI] [PubMed] [Google Scholar]
- 25.Konno R, Yasumura Y. Involvement of D-amino acid utilization in the mouse J. Nutrition 1984; 114:1617–21. [DOI] [PubMed] [Google Scholar]
- 26.Newman DS, Aggarwal SK, Silbergleit R. Thoracic radicular symptoms in amyotrophic lateral sclerosis. J. Neurol. Sci. 1995; 129: 38–41 [DOI] [PubMed] [Google Scholar]
- 27.Sasabe J, Chiba T, Yamada M, Okamoto K, Nishimoto I, Matsuoka M, Aiso S. D-serine is a key determinant of glutamate toxicity in amyotrophic lateral sclerosis. EMBO J. 2007; 26:4149–59. doi: 10.1038/sj.emboj.7601840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jotova I, Pavlov V, Dimitrov O, Bachrach U. Developmental aspects of polyamine-oxidizing enzyme activities in the mouse kidney. Effects of testosterone. Amino Acids. 1999; 17: 267–76. [DOI] [PubMed] [Google Scholar]
- 29.Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015; 347: 1436–41. doi: 10.1126/science.aaa3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez FJ, Pratt GA, Van Nostrand EL, Batra R, Huelga SC, Kapeli K, et al. , (2016) Protein-RNA Networks Regulated by Normal and ALS-Associated Mutant HNRNPA2B1 in the Nervous System. Neuron 2016; 92: 780–795. doi: 10.1016/j.neuron.2016.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994; 264:1772–5. [DOI] [PubMed] [Google Scholar]
- 32.Hegedus J, Putman CT, Gordon T.Time course of preferential motor unit loss in the SOD1 G93A mouse model of amyotrophic lateral sclerosis. Neurobiol Dis. 2007; 28: 154–64. doi: 10.1016/j.nbd.2007.07.003 [DOI] [PubMed] [Google Scholar]
- 33.Sharp PS, Akbar MT, Bouri S, Senda A, Joshi K, Chen H, et al. Protective effects of heat shock protein 27 in a model of ALS occur in the early stages of disease progression. Neurobiol Dis. 2008; 30: 42–55. doi: 10.1016/j.nbd.2007.12.002 [DOI] [PubMed] [Google Scholar]
- 34.Gill A, Kidd J, Vieira F, Thompson K. & Perrin S. No benefit from chronic lithium dosing in a sibling-matched, gender balanced, investigator-blinded trial using a standard mouse model of familial ALS. PLoS One 2009; 4: e6489 doi: 10.1371/journal.pone.0006489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaneb HM, Sharp PS, Rahmani-Kondori N, Wells DJ. Metformin treatment has no beneficial effect in a dose-response survival study in the SOD1(G93A) mouse model of ALS and is harmful in female mice. PLoS One. 2011; 6: e24189 doi: 10.1371/journal.pone.0024189 Epub 2011 Sep 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Azzouz M, Ralph GS, Storkebaum E, Walmsley LE, Mitrophanous KA, Kingsman S, et al. VEGF delivery with retrogradely transported lentivector prolongs survival in a mouse ALS model. Nature 2004; 429: 413–417 doi: 10.1038/nature02544 [DOI] [PubMed] [Google Scholar]
- 37.Zheng C, Nennesmo I, Fadeel B, Henter JI. Vascular endothelial growth factor prolongs survival in a transgenic mouse model of ALS. Ann Neurol. 2004; 56: 564–7. doi: 10.1002/ana.20223 [DOI] [PubMed] [Google Scholar]
- 38.Kaneb HMJ. Preclinical testing of potential therapeutics for Amyotrophic Lateral Sclerosis. 2012; [Google Scholar]
- 39.Lincecum JM, Vieira FG, Wang M Z, Thompson K, De Zutter GS, Kidd J, et al. From transcriptome analysis to therapeutic anti-CD40L treatment in the SOD1 model of amyotrophic lateral sclerosis. Nat Genet. 2010; 42: 392–9. doi: 10.1038/ng.557 [DOI] [PubMed] [Google Scholar]
- 40.Alexander GM, Erwin KL, Byers N, Deitch JS, Augelli BJ, Blankenhorn EP, et al. Effect of transgene copy number on survival in the G93A SOD1 transgenic mouse model of ALS. Molecular Brain Research. 2004; 130: 7–15 doi: 10.1016/j.molbrainres.2004.07.002 [DOI] [PubMed] [Google Scholar]
- 41.Watson C, Paxinos G, Kayaliglu G. The Spinal Cord. A Christopher and Dana Reeve Foundation Text and Atlas 2008; [Google Scholar]
- 42.McHanwell S, Biscoe TJ. The localization of motoneurons supplying the hindlimb muscles of the mouse. Philosophical Transactions of the Royal Society of London, Biological Sciences. 1981; 293 (1069): 477–508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Text A. Methodology: Genotyping and Gait Analysis. Figure A. DAO enzyme activity in spinal cord and brain from wild-type (WT) and DAOR199W transgenic mice (R199W). Activity measured as mUnits/mg tissue/lysate in brain and spinal cord from wild-type (WT) and DAOR199W transgenic mice (R199W). Figure B. D-serine immunoreactivity in lumbar spinal cord from control and DAOR199W transgenic mice. A rabbit polyclonal Anti-D-Serine antibody (Abcam) raised against d-serine cross-linked to BSA with glutaraldehyde was used at a dilution of 1 in 5,000 as previously described in detail [6]. Figure C. Serine racemase immunoreactivity in lumbar spinal cord from control and DAOR199W transgenic mice. A mouse monoclonal Anti-serine racemase antibody (BD Biosciences) was used at a dilution of 1 in 1000 as previously described in detail [6].
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.