Abstract
The apicomplexan protozoans Eimeria spp. cause coccidioses, the most common intestinal diseases in chickens. Coccidiosis is associated with significant animal welfare issues and has a high economic impact on the poultry industry. Lack of a full understanding of immunogenic molecules and their precise functions involved in the Eimeria life cycles may limit development of effective vaccines and drug therapies. In this study, immunoproteomic approaches were used to define the antigenic protein repertoire from the total proteins of unsporulated Eimeria tenella oocysts. Approximately 101 protein spots were recognized in sera from chickens infected experimentally with E. tenella. Forty-six spots of unsporulated oocysts were excised from preparative gels and identified by matrix-assisted laser desorption ionization time-of-flight MS (MALDI-TOF-MS) and MALDI-TOF/TOF-MS. For unsporulated oocysts, 13 known proteins of E. tenella and 17 homologous proteins to other apicomplexan or protozoan parasites were identified using the ‘Mascot’ server. The remaining proteins were searched against the E. tenella protein sequence database using the ‘Mascot in-house’ search engine (version 2.1) in automated mode, and 12 unknown proteins were identified. The amino acid sequences of the unknown proteins were searched using BLAST against non-redundant sequence databases (NCBI), and 9 homologous proteins in unsporulated oocyst were found homologous to proteins of other apicomplexan parasites. These findings may provide useful evidence for understanding parasite biology, pathogenesis, immunogenicity and immune evasion mechanisms of E. tenella.
Keywords: E. tenella, unsporulated oocysts, immunoproteome
Abstract
Les protozoaires Apicomplexa Eimeria spp. causent les coccidioses, les maladies intestinales les plus communes chez les poulets. La coccidiose est un problème majeur de bien-être animal et a un fort impact économique sur l'industrie de la volaille. Le manque de compréhension complète des molécules immunogènes et de leurs fonctions précises impliquées dans les cycles de vie d'Eimeria peut limiter le développement de vaccins efficaces et de pharmacothérapies. Dans cette étude, des approches immunoprotéomiques ont été utilisées pour définir le répertoire des protéines antigéniques à partir des protéines totales des oocystes non sporulés d'Eimeria tenella. Environ 101 taches protéiques ont été reconnues dans des sérums de poulets infectés expérimentalement par E. tenella. Quarante-six taches d'oocystes non sporulés ont été excisées à partir de gels et identifiées par MALDI-TOF-MS et MALDI-TOF/TOF-MS par désorption laser assistée par matrice. Pour les oocystes non sporulés, 13 protéines connues d'E. tenella et 17 protéines homologues à d'autres parasites Apicomplexa ou protozoaires ont été identifiées en utilisant le serveur “Mascot”. Les protéines restantes ont été recherchées dans la base de données de séquences protéiques d'E. tenella en utilisant le moteur de recherche “Mascot in-house” (version 2.1) en mode automatisé, et 12 protéines inconnues ont été identifiées. Les séquences d'acides aminés des protéines inconnues ont été recherchées en utilisant BLAST contre des bases de données de séquences non redondantes (NCBI), et 9 protéines homologues des oocystes non-sporulés ont été trouvées homologues à des protéines d'autres parasites Apicomplexa. Ces résultats peuvent fournir des éléments utiles pour comprendre la biologie parasitaire, la pathogenèse, l'immunogénicité et les mécanismes d'évasion immunitaire d'E. tenella.
Introduction
E. tenella is an apicomplexan protozoan parasite and one of the etiological agents of coccidiosis in poultry. Coccidiosis is the most common intestinal disease in poultry and has considerable welfare and economic implications in the poultry industry [7]. At present, coccidiosis is mainly controlled by prophylactic medication via the use of in-feed anticoccidial drugs known as coccidiostats [29]. However, the excessive use of these drugs has led to drug-resistance and involves a food safety concern. Novel approaches are urgently needed for the effective control of coccidiosis [9,31].
Understanding the pathogenesis is important for designing effective strategies to control coccidiosis. When released into the environment, the unsporulated oocysts of E. tenella undergo meiosis upon contact with oxygen and moisture [2]. Metabolism is not constant throughout the process of sporulation. Wilson and Fairbairn (1961) found that polysaccharides were utilized initially, but later lipid metabolism played a greater role [38]. Oxygen consumption and the relationship with different stages in sporulation were also studied by Wagenbach and Burns (1969) [35]. Extensive studies were performed on the sporulation processes of E. tenella oocysts by Yvoré and Coudert (1972) [10]. They separated the sporulation process into four different morphological stages related to cumulative oxygen consumption and found that the quantity of oxygen necessary at a given stage or completion of sporulation was constant and independent of temperature. Wilson & Fairbainz [38] and Wang [36] showed that respiration during sporulation initially occurred at a higher rate, but later at a lower rate when the sporulation was completed. Moreover, enzyme turnover was also studied in the unsporulated oocyst stage. Some enzymes were identified in unsporulated oocysts, but their levels decreased during the late phase of sporulation [37]. Therefore, the processing of sporulated oocysts is very important for the spread of coccidiosis in chickens. Whether the immune system of the host recognizes these key molecules in the sporulation process may affect the formation of sporulated oocysts, and could provide clues for controlling the spread of coccidiosis in chickens.
Proteomic expression studies have been proposed as a powerful method to discover new drug targets for parasitic diseases [1,33]. With advances in Eimeria genomics, the protein repertoire of the microneme organelles and refractile body of E. tenella has been identified via proteomic approaches [7,11]. Some protein repertoires of microneme secretory organelles were identified in micronemes. Immunoproteomics was also used to identify immunogenic molecules and pathogenicity factors in the sporozoite stage of E. tenella. Approximately 50 spots of the 130 analyzed spots were defined as antigens on the basis of the anti-E. tenella chicken sera used [11]. Liu et al. studied the whole proteins of the second-generation merozoite of E. tenella by two-dimensional gel electrophoresis (2-DE) and western blotting using sera isolated from chickens infected experimentally with E. tenella, and approximately 640 spots were detected on the proteome map, while 85 spots were recognized [24].
Mass spectrometry (MS)-based proteomic profiling and protein identification is a powerful tool for the discovery of new disease biomarkers. Among the MS platforms, matrix-assisted laser desorption/ionization time-of-flight/time-of-flight (MALDI-TOF/TOF) MS offers high sample throughput and flexibility in qualitative and quantitative analysis of amino and organic acids. In order to detect the immunogenic proteins of unsporulated oocysts, an immunoproteomic approach coupled with MALDI-TOF/TOF-MS was used in this study to identify the antigenic proteins from the total proteins of unsporulated oocysts. Defining the immunogenic proteins will make it possible to design potential vaccine targets to effectively control coccidiosis, and these results could also enrich the knowledge on the immunogenic proteins of unsporulated oocysts.
Materials and methods
Ethics statement
The Ethics Committee of Jiangxi Agricultural University approved the animal protocol for this study (protocol number P-2013-03). All the procedures involving animals in this study were carried out in accordance with The Care and Use Guidelines of Experimental Animals established by the Ministry of Agriculture of China.
Parasite collection
E. tenella (Nanchang strain) were maintained by passage through coccidia-free two-week-old chickens (Sanhuang Breeding Farm, Nanchang, Jiangxi, China). Unsporulated oocysts were obtained by passing the cecal contents through a 100-mesh sieve at 7 days post-infection, and the oocysts were then sterilized using 20% sodium hypochlorite and washed several times in tri-distilled water. All procedures were performed on ice. After centrifugation at 5,000 rpm for 5 min at 4°C, oocysts were stored in liquid nitrogen, as described elsewhere [32].
Protein sample preparation
Unsporulated oocysts were broken down in lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, 40 mM dithiothreitol (DTT), 0.2% Bio-Lyte 3-10 ampholytes and 1 mM PMSF) by the ultrasonic approach in an ice bath (200 W, work time 5s, interval time 10s, 100 cycles). The content was centrifuged at 15,000 rpm for 2 min at 4°C, and the soluble proteins were cleaned with the 2D-cleanup kit and quantified using Plus One TM2-D Quant Kit (Amersham Pharmacia Biotech, Little Chalfont, UK).
Two-dimensional electrophoresis (2-DE)
Isoelectric focusing (IEF). The pellet of proteins was dissolved in IPG rehydration/sample buffer (8 M urea, 2% CHAPS, 50 mM dithiothreitol (DTT), 0.2% Bio-Lyte 3-10 ampholytes, 0.001% bromophenol blue; BioRad, Hercules, CA, USA) by incubating for at least 1h at room temperature and centrifuged at 15,000 rpm for 15 min at room temperature to remove undissolved materials. 320 µL of 600 µg protein were adsorbed onto a 17 cm Immobiline DryStrip (IPG, pH range 3-10; BioRad) and IEF was performed in the BioRad PROTEAN IEF cell. Twelve hours of positive rehydration preceded zone electrophoresis at 20°C under the following conditions: S1 250 V, 1h; S2 1000 V 1h; S3 4000 V 2.5 h; and S4 8000 V 50000 V.h. Strips could be used immediately for the second dimension electrophoresis, or stored at −80 °C for a short period.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Prior to SDS-PAGE, each IPG strip was incubated in 6 mL of equilibration buffer I (375 mM Tris-HCl (pH 8.8), 6 M urea, 2% SDS, 2% DTT; BioRad) for 15 min and in 6 mL of equilibration buffer II (375 mM Tris-HCl (pH 8.8), 6 M urea, 2.5% iodoracetamide, 2% SDS; BioRad) for an additional 15 min. IPG strips and SDS-PAGE molecular weight standards (11–170 kDa; Fermentas Inc., Waltham, MA, USA) were loaded into homogeneous 12.5% polyacrylamide gels and sealed with 1% agarose solution. A Tris-glycine buffer system was used, as described by Laemmli (1970). Electrophoresis was performed in two steps at 16 °C: 3 w/gel for 45 min and 15 w/gel until the tracking dye reached the bottom of the gels. One gel was stained with Coomassie Brilliant Blue (CBB) G-250 and another was electrophoretically transferred onto a polyvinylidene fluoride (PVDF) membrane (Amersham Pharmacia Biotech) for western-blotting.
Analysis of the stained gel. The stained gels were scanned, and images were then analyzed using ImageMaster™ 2D Platinum Software (Version 5.0, Swiss Institute of Bioinformatics, Geneva, Switzerland) for spot detection, quantification, as well as comparison and analyses.
Immunoblot analysis
Preparation of immunized sera. Chickens reared in coccidian-free conditions until 2 weeks of age were orally infected with 1 × 104 sporulated oocysts of E. tenella per chicken. At 3 days post-first infection, the chickens were challenged four times at 3-day intervals with 5,000 sporulated oocysts per chicken. Ten negative control birds were maintained under the same conditions in an adjacent room and were inoculated by gavage with distilled water only. At 5 weeks after the last infection, blood samples were obtained from chickens by cardiac puncture. The blood was allowed to clot for 1h at 37 °C, and then overnight at 4 °C. After centrifugation at 4,000 rpm for 10 min, serum was aliquoted in 1 mL vials and stored at −20 °C until use [24].
Western blotting. After SDS-PAGE, proteins were transferred from the 2-DE gel onto a polyvinylidene difluoride (PVDF) membrane with blotting buffer (39 mM glycine, 48 mM Tris-base, 20% methanol, and 0.037% SDS) using a semidry blotting apparatus (TE77, Amersham Pharmacia Biotech). Electrotransfer was carried out for 2h with a current/area of 0.70 mA/cm2. The membrane was blocked with 5% w/v skim milk in phosphate-buffered saline (PBS) (pH 7.4) containing 0.05% Tween-20 (PBST) for 2h at room temperature, incubated with chicken serum (1:100 dilution) for 1h at room temperature and subsequently washed with PBST. It was then incubated with a horseradish peroxidase-conjugated anti-chicken IgG (a dilution of 1:2000, Bio-Rad) for 1h at room temperature. After washing with PBST, the membrane was incubated with 50 mM Tris-HCl buffer (pH 7.4) and developed with 3,3'-diaminobenzidine (DAB, Sigma, St Louis, MO, USA) substrate until optimum color development was observed. The anti-E. tenella serum was used to detect the immunogenic spots and the normal chicken serum was used to test another blotting membrane as a negative control.
Spot excision and in-gel trypsin digestion. After Western blotting, the matched protein spots on the gel were identified by comparison with the molecular size ladder using a gel documentation system (Gene Genius, Syngene, Frederick, MD, USA) and related software (GeneSnap 6.08 and GeneTools, Syngene). The matched protein spots were manually excised from the gel, destained with 30% acetonitrile (ACN) /100 mM NH4HCO3 until the gels were completely destained. The gels were dried in a vacuum centrifuge. The in-gel proteins were reduced with dithiothreitol (10 mM DTT/ 100 mM NH4HCO3) for 30 min at 56 °C, then alkylated with iodoacetamide (200 mM IAA/100 mM NH4HCO3) in the dark at room temperature for 30 minutes. Gel pieces were briefly rinsed with 100 mM NH4HCO3 and ACN, respectively. Gel pieces were digested overnight in 12.5 ng/µL trypsin in 25 mM NH4HCO3. The peptides were extracted three times with 60% ACN/0.1% trifluoroacetic acid (TFA). The extracts were pooled and dried completely using a vacuum centrifuge.
MS analysis of protein spots and database searches
MS data for protein identification were obtained by using a MALDI-TOF/TOF instrument (5800 proteomics analyzer; Applied Biosystems, Foster City, CA, USA). Instrument parameters were set using the 4000 Series Explorer software (Applied Biosystems). The MS spectra were recorded in reflector mode in a mass range from 800 to 4000 with a focus mass of 2000. CalMix5 standard for MS was used to calibrate the instrument (ABI 4700 Calibration Mixture). For one main MS spectrum, 25 subspectra with 125 shots per subspectrum were accumulated using a random search pattern. For MS calibration, autolysis peaks of trypsin ([M + H] + 842.5100 and 2,211.1046) were used as internal calibrates, and up to 10 of the most intense ion signals were selected as precursors for MS/MS acquisition, excluding the trypsin autolysis peaks and the matrix ion signals. In MS/MS positive ion mode, 50 subspectra with 50 shots per subspectrum for one main MS spectrum were accumulated using a random search pattern. Parameters were set up as follows: Collision energy as 2 kV, collision gas as air, and default calibration as Glu1-Fibrino-peptide B ([M + H] + 1,570.6696) spotted onto Cal 7 positions of the MALDI target. Combined peptide mass fingerprinting PMF and MS/MS queries were performed by using the MASCOT search engine 2.2 (Matrix Science, Ltd.) embedded into GPS-Explorer Software 3.6 (Applied Biosystems) on the Eimeria database of Uniprot and NCBI, with the following parameter settings: 100 ppm mass accuracy, trypsin cleavage one missed cleavage allowed, carbamido methylation set as fixed modification, oxidation of methionine allowed as variable modification, MS/MS fragment tolerance was set to 0.4 Da. A GPS Explorer protein confidence index ≥ 95% was used for further manual validation.
Results
Two-dimensional gel electrophoresis of unsporulated oocyst proteins
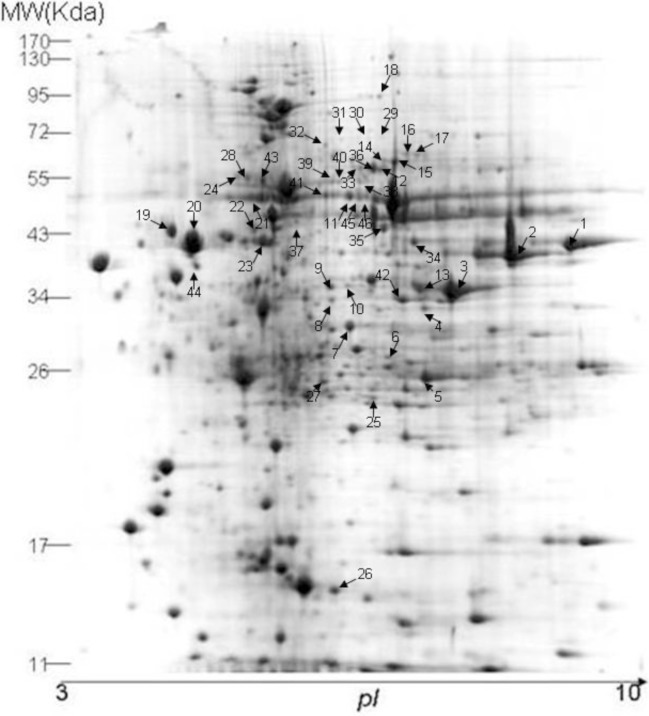
About 600 µg/gel soluble proteins of unsporulated oocysts were separated by 2-DE over an isoelectric point (pI) range of 3-10 (IPG linear gradient). CBB G-250 stained gels revealed about 656 spots of unsporulated oocysts according to the protocol described by Candiano et al. [8]. The molecular weights of most proteins were between 11 and 130 kDa (Fig. 1).
Figure 1.
Identification of proteins of unsporulated E. tenella oocysts by 2-D gel electrophoresis. Isoelectro-focusing was performed with 600 μg protein for 50 kVh using a pI 3-10 strip. SDS-PAGE was performed on a 12.5% gel and then stained with CBB. Numbers are used to mark the spots analyzed.
Analysis and identification of immunogenic proteins of unsporulated oocysts via the NCBI database
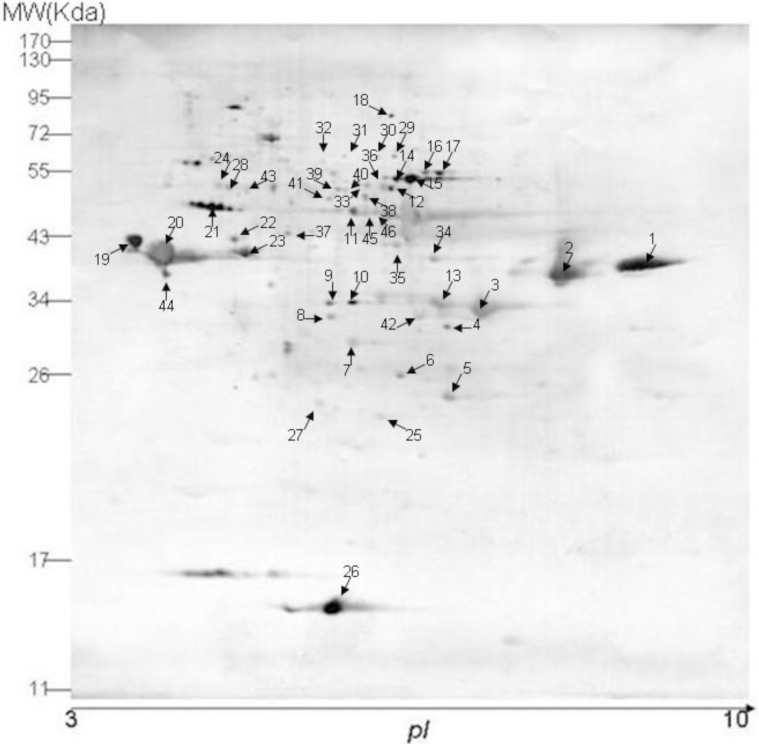
The western blot profiling of 2-DE gels is shown in Figure 2. Approximately 101 spots were observed on the PVDF membrane (Fig. 2). Among them, 46 matched closely to the protein spots and were characterized by MS. No spots were detected with normal chicken serum. Data were compared to those in the NCBI database using the ‘Mascot’ server. The probability score for the match, molecular weight (MW), pI, number of peptide matches and the sequence coverage were used for confidence of spot identification. The results are summarized in Table 1.
Figure 2.
Immunoblot profiling of unsporulated E. tenella oocysts using chicken anti-E. tenella serum.
Table 1.
Identification of unsporulated Eimeria tenella oocyst proteins in non-redundant sequence databases (NCBI) using data from MALDI-TOF-MS and MALDI-TOF/TOF-MS.
| Spot | Accession number | Score | Peptide matched | Sequence coverage (%) | Mr/pI | Protein |
|---|---|---|---|---|---|---|
| 3 | gi|25989637 | 105 | 17/79 | 38 | 36232/7.00 | lactate dehydrogenase [E. acervulina] |
| 4 | gi|145535758 | 72 | 26/93 | 22 | 155774/5.61 | hypothetical protein GSPATT00019908001 (P. t) |
| 6 | gi|156097314 | 65 | 19/83 | 19 | 159966/6.14 | 6-phosphofructokinase, putative (P. v) |
| 7 | gi|118490906 | 154 | 12/37 | 50 | 27839/9.31 | hypothetical protein (E. tenella) |
| 8 | gi|118490770 | 317 | 42/96 | 40 | 84827/8.27 | hypothetical protein (E. tenella) |
| 10 | gi|124804156 | 64 | 16/86 | 19 | 84877/9.22 | hypothetical protein PF11_0247 (P .f) |
| 11 | gi|50400241 | 230 | 32/100 | 69 | 48458/5.62 | Enolase (E. tenella) |
| 13 | gi|25989639 | 136 | 22/100 | 54 | 35399/6.54 | lactate dehydrogenase (E. tenella) |
| 16 | gi|118397285 | 79 | 21/96 | 27 | 126785/7.55 | hypothetical protein TTHERM_00947400 (T. t) |
| 20 | gi|401829 | 427 | 46/76 | 56 | 70814/5.16 | heat-shock protein [E. acervulina] |
| 21 | gi|145517049 | 71 | 16/91 | 35 | 46979/9.32 | hypothetical protein GSPATT00012541001 (P. t) |
| 22 | gi|401829 | 86 | 19/98 | 27 | 70814/5.16 | heat- shock protein [E. acervulina] |
| 23 | gi|401829 | 605 | 66/98 | 67 | 70814/5.16 | heat-shock protein [E. acervulina] |
| 24 | gi|50400241 | 260 | 30/89 | 64 | 48458/5.62 | Enolase (E. tenella) |
| 26 | gi|124054715 | 215 | 17/50 | 66 | 13343/5.92 | actin depolymerizing factor (E. tenella) |
| 28 | gi|1037176 | 269 | 39/78 | 29 | 77516/5.22 | immunoglobulin heavy chain binding protein (E. tenella) |
| 31 | gi|118379641 | 66 | 17/88 | 20 | 106631/6.01 | hypothetical protein TTHERM_00348310 (T. t) |
| 32 | gi|118377447 | 74 | 16/92 | 22 | 87148/6.14 | hypothetical protein TTHERM_01207560 (T. t) |
| 33 | gi|4033429 | 112 | 22/92 | 43 | 58238/5.95 | Pyruvate kinase (PK) (E. tenella) |
| 34 | gi|145494414 | 69 | 10/96 | 42 | 22764/9.18 | hypothetical protein GSPATT00006303001 (P. t) |
| 35 | gi|118359856 | 77 | 14/94 | 23 | 70549/8.46 | polyadenylate-binding protein 2 (T. t) |
| 36 | gi|4033429 | 146 | 24/72 | 46 | 58238/5.95 | Pyruvate kinase (PK) (E. tenella) |
| 38 | gi|145519139 | 77 | 25/93 | 18 | 192289/5.98 | hypothetical protein GSPATT00013486001 (P. t) |
| 39 | gi|124805406 | 70 | 11/97 | 34 | 37233/9.54 | mitochondrial phosphate carrier protein (P. f) |
| 40 | gi|145498869 | 76 | 15/96 | 38 | 47850/10.02 | hypothetical protein GSPATT00036807001 (P. t) |
| 41 | gi|50400241 | 132 | 23/100 | 48 | 48458/5.62 | Enolase (E. tenella) |
| 43 | gi|21542248 | 214 | 33/94 | 62 | 50377/4.78 | Tubulin beta chain (Beta-tubulin) (E. tenella) |
| 44 | gi|156098346 | 67 | 10/52 | 14 | 123864/9.42 | hypothetical protein PVX_091055 (P. v) |
| 45 | gi|50400241 | 204 | 26/63 | 59 | 48458/5.62 | Enolase (E. tenella) |
| 46 | gi|50400241 | 197 | 26/67 | 59 | 48458/5.62 | Enolase (E. tenella) |
Note: P. t: Paramecium tetraurelia strain d4-2; T. t: Tetrahymena thermophila SB210; P. f: Plasmodium falciparum 3D7; P. v: Plasmodium vivax SaI-1.* common protein in two stages
Thirteen spots were identified as known proteins of E. tenella. A group of proteins shown as antigenic proteins on immunoblots was identified as enolase and pyruvate kinase (PK), which may be involved in glycolysis. Enolases were detected in Spots 11, 24, 41, 45 and 46. PKs with an MW of 58 kDa were also detected in Spots 33 and 36. Other proteins, such as lactate dehydrogenase (Spot 13), known to be involved in glycolysis, actin depolymerizing factor (Spot 26), immunoglobulin heavy chain binding protein (Spot 28), and tubulin beta chain (Spot 43) were detected as immunogenic proteins of E. tenella. In addition, two hypothetical proteins of E. tenella were identified on Spots 7 and 8 which showed immunogenic with chicken anti-E. tenella serum. However, their functions were unknown.
Seventeen spots were identified as homologous proteins to other protozoa. Proteins on Spots 4, 21, 34, 38 and 40 were similar to hypothetical proteins of Paramecium tetraurelia (P. tetraurelia) strain d4-2. Proteins on Spots 16, 31 and 32 were found to be homologous to hypothetical proteins of Tetrahymena thermophila (T. thermophila) SB210. The protein on Spot 35 was identified as similar to polyadenylate-binding protein 2 of T. thermophila SB210. The proteins on Spots 10 and 39 were identified as hypothetical protein and mitochondrial phosphate carrier protein of Plasmodium falciparum (P. falciparum, PfmpC) 3D7, respectively. A 6-phosphofructokinase and a hypothetical protein of Plasmodium vivax (P. vivax) SaI-1 were also identified for the proteins on Spots 6 and 44, respectively. The proteins on Spots 20, 22 and 23 matched the heat-shock protein (HSP70), and the protein on Spot 3 matched the lactate dehydrogenase of Eimeria acervulina (E. acervulina).
Identification of proteins of unsporulated E. tenella using the E. tenella protein sequence database
The results obtained by searching against the E. tenella protein database using the ‘Mascot in-house’ search engine (version 2.1) indicated that the proteins on 12 spots were unknown proteins of E. tenella. After comparison of amino acid sequences with those in the NCBI databases, 9 spots were found to be significantly homologous to other apicomplexan parasites, except Spots 14, 19 and 30 (Table 2).
Table 2.
Identification of unsporulated Eimeria tenella oocyst proteins by E. tenella protein database from E. tenella genome assembly using ‘Mascot in-house’ and then amino acid sequence BLAST against non-redundant sequence databases (NCBI).
| Spot | Accession number |
Score | Peptide matched | Sequence coverage (%) | Mr/pI | Related contig | E. tenella genome assembly | Contig BLAST against NCBI database |
|---|---|---|---|---|---|---|---|---|
| 2 | XXX12652.tmp3 | 91 | 14/64 | 43 | 36760/7.59 | Contig12652 | complement (60072..61332) | |
| gb|AAK20420.1 | glyceraldehyde-3-phosphate dehydrogenase (T. g) | |||||||
| ref|XP-001348772.1 | glyceraldehyde-3-phosphate dehydrogenase (P. f 3D7) | |||||||
| 5 | XXX2043.tmp6 | 200 | 25/78 | 42 | 32616/8.14 | Contig2043 | complement (13545..17007) | |
| emb|CAH03718.2 | putative secretory protein (T. a) | |||||||
| 9 | XXX2256.tmp1 | 90 | 14/69 | 27 | 44109/6.53 | Contig2256 | complement (2339..7012) | |
| gb|EAN34194.1 | pyruvate dehydrogenase E1 component beta subunit, mitochondrial, putative (T. p) | |||||||
| 12 | XXX4578.tmp1 | 265 | 37/87 | 52 | 77965/8.48 | Contig4578 | complement (414..7001) | |
| gb|AAC47725.3 | myosin-B (T. g) | |||||||
| ref|XP-766628.1 | myosin B (T. p strain Muguga) | |||||||
| 14 | XXX6171.tmp6 | 204 | 27/80 | 30 | 54407/4.97 | Contig6171 | complement (38747..40753) | –––––––––– |
| 15 | XXX12652.tmp25 | 239 | 35/96 | 40 | 55757/6.46 | Contig12652 | complement (7093..9803) | |
| gb|ABB00911.1 | eukaryotic translation initiation factor 5 (T. g) | |||||||
| 17 | XXX2989.tmp10 | 84 | 16/96 | 38 | 59063/6.62 | Contig2989 | complement (41668..45596) | |
| gb|EDO05336.1 | fumarate hydratase, putative (B. b) | |||||||
| 18 | XXX607.tmp12 | 228 | 49/93 | 26 | 174714/8.27 | Contig607 | complement (49066..58084) | |
| gb|EDL46529.1 | SNF2 family N-terminal domain containing protein (P. v) | |||||||
| 19 | XXX1454.tmp24 | 71 | 10/86 | 60 | 33007/ 4.46 | Contig1454 | complement (127070..127972) | ––––––––––- |
| 27 | XXX12511.tmp24 | 76 | 14/98 | 55 | 27814/6.31 | Contig12511 | complement (12140..13529) | |
| gb|EAL35019.1 | proteasome subunit (C. h TU502) | |||||||
| 29 | XXX3535.tmp1 | 327 | 39/65 | 44 | 93367/8.44 | Contig3535 | complement (3785..8777) | |
| gb|AAC47724.1 | myosin-A (T. g) | |||||||
| 30 | XXX6171.tmp6 | 204 | 27/80 | 30 | 54407/4.97 | Contig6171 | complement (38747..40753) | ––––––––––- |
Note: T. g: Toxoplasma gondii; P. f 3D7: Plasmodium falciparum 3D7; T. a: Theileria annulata; T. p: Theileria parva; T. g: Toxoplasma gondii; T. p strain Muguga: Theileria parva strain Muguga; B. b: Babesia bovis; P. v: Plasmodium vivax; C. h TU502: Cryptosporidium hominis TU502.
The amino acid sequence of Spot 2 shared similarity of 76% and 74% to the glyceraldehyde-3-phosphate dehydrogenase of Toxoplasma gondii (T. gondii) and P. falciparum 3D7, respectively. For Spots 5 and 9, the amino acid sequences showed similarities of 52% and 68% to the putative secretory protein and pyruvate dehydrogenase E1 component beta subunit of mitochondria from Theileria annulata (T. annulata) and Theileria parva (T. parva), respectively. Similarly, the amino acid sequence of Spot 12 shared similarity of 43% and 35% to myosin B from T. parva strain Muguga and T. gondii, respectively. The protein on Spot 15 showed high homology to eukaryotic translation initiation factor 5 (62% identity) of T. gondii. The protein on Spot 17 matched 65% identity to fumarate hydratase of Babesia bovis (B. bovis). The protein on Spot 18 was close to SNF family N-terminal domain containing protein (52% identity) of P. vivax.
The proteins on Spots 27 and 29 showed high homology to proteasome subunit (64% identity) and myosin A (59% identity) of Cryptosporidium hominis (C. hominis) TU502 (Fig. 3) and T. gondii, respectively. Spots 14, 19 and 30 were neither identified nor related to known parasite proteins. Detailed results are listed in Table 2.
Figure 3.
Spot 27's amino acid sequence homologous to Cryptosporidium hominis proteasome searched against the E. tenella genome using the ‘Mascot in-house’ server. A: E. tenella amino acid sequences obtained by MALDI-TOF-MS were searched using BLAST against the NCBI database. Deduced amino acid sequences on Spot 27 were aligned with the deduced sequence from the E. tenella genome Contig 12511: complements (12140…13529) are in bold red script. B: Alignment of the predicted amino acid sequence from Spot 27 (A) with the C. hominis proteasome subunit. The alignments of sequences are in bold red script.
Discussion
At least seven Eimeria species parasitizing chickens have been reported [34]. Among them, E. tenella is the most virulent species in chickens [27]. E. tenella has a complicated life cycle with extracellular invasive stages that infect host cells in the ceca and intracellular multiplication stages [15]. After three asexual multiplication stages in the chicken cecum, the parasites undergo sexual differentiation to produce unsporulated oocysts and shed into feces of infected chickens [19].
To our knowledge, this study is the first report on unsporulated oocyst soluble proteins of E. tenella by an immunoproteomics method. In this study, the map showed the presence of many abundant spots between pI 3-10 in Fig. 1 and about 656 spots were separated on a 2D gel.
Database search results of the immunogenic proteins indicated that several different spots were matched to the same protein. This was probably attributed to the isoforms produced by post-translational proteolytic processing and modification, the cleavage of alkaline regions or phosphorylation of multiple residues. Similar results have been described in other reports [20,39].
Immunogenic proteins identified included: lactate dehydrogenase, enolase, HSP70, actin depolymerizing factor (ADF), immunoglobulin heavy chain binding protein (BiP), PK, tubulin beta chain (Beta-tubulin), hypothetical proteins and other homologous proteins from other Apicomplexa parasites or protozoa. Moreover, enolase, lactate dehydrogenase, and Beta-tubulin were detected in the other stages including sporozoite [11] and the second merozoite stage [24], and showed immunogenicity.
Enolase and PK were identified, which seems relevant to an adaptation of the metabolism in the anaerobic development of the intracellular stage as glycolytic enzymes [14,22,30]. De Venevelles et al. found the two enzymes as antigens in the sporozoite stage of E. tenella [11]. All these results would further support the theory that enolase may be an important immunogenic protein of E. tenella.
Beta-tubulin, as a subcellular structural protein, was also identified from unsporulated E. tenella oocysts. In other experiments, this protein was shown to induce immunity protective against natural infection [21].
The protein on Spot 29 was identified as homologous to myosin A of T. gondii. The head domain of myosin A of T. gondii demonstrated excellent conservation of the ATP and actin-binding domains, as well as other motifs consistently conserved among all myosins [3,5,18]. Myosins are actin-dependent molecular motors that play important roles in muscle contraction, cell motility and organelle transport [6]. ADF was identified as a specific immunogenic protein as Spot 26 in the unsporulated oocyst stage. ADF plays an important role in remodeling the actin cytoskeleton and contributes greatly to the invasion of host cells by the apicomplexan parasite [25,40].
Several spots (20, 22 and 23) were identified as heat-shock proteins by their homologies with the HSP70 of E. acervulina. Parasite invasion often induced an increase in the synthesis of parasite HSP70, either in response to an increase in the host's temperature or due to stress from the invasive process [26,28].
The proteins on Spot 28 matched the BiP of E. tenella in unsporulated oocysts. As a protein in the endoplasmic reticulum (ER), BiP plays a role in the assembly and the folding of proteins [12,17]. Two hypothetical proteins of E. tenella (Spots 7 and 8) were found to be highly immunogenic, but we were not able to find their function in the databases of NCBI and UniProt [23].
The protein on Spot 39 of unsporulated oocysts matched PfmpC. The PfmpC as a mitochondrial carrier protein belonged to one of nine members of the mitochondrial carrier family present in P. falciparum, including an ATP/ADP exchanger and a di/tri-carboxylate exchanger, probably involved in transport of TCA cycle intermediates across the mitochondrial membrane [4,16].
The protein on Spot 2 was found to be highly similar to glyceraldehyde-3-phosphate dehydrogenase of T. gondii (76% identity) and P. falciparum (71% identity). It is a tetrameric NAD-binding enzyme involved in glycolysis and glyconeogenesis [13]. Similarly, the Spot 5 protein was highly homologous to putative secretory protein of T. annulata (52% identity). The Spot 15 protein was similar to eukaryotic translation initiation factor 5 (62% identity) of T. gondii, but function was still unclear. The Spot 17 protein was similar to fumarate hydratase (65% identity) of B. bovis. The fumarate hydratase (fumarase) is a component of the citric acid cycle, and also engages in the reductive pathway from oxaloacetate to succinate during anaerobic growth [6].
A proteasome subunit of C. hominis TU502 showed high similarity to the Spot 27 protein. The high homology (64% identity) indicated the close relation between the two species. Proteasome, as a complex of proteins, could exert the function of the central enzyme of nonlysomal protein degradation in both the cytosol and nucleus [41].
Comparing the substances with those identified in Liu et al. (2009) [24], lactate dehydrogenase, enolase and heat-shock proteins were found in both studies. Some surface secretory proteins, such as surface antigen and microneme proteins, and regulatory proteins, such as 14-3-3 protein, were recognized by the sera isolated from chickens infected experimentally with E. tenella in the second-generation merozoite stage of E. tenella. In addition, some glycolytic enzymes, such as 6-phosphofructokinase and PK, and microfilamentous proteins, such as actin depolymerizing factor, were identified in the unsporulated oocyst stage of E. tenella.
In conclusion, this is the first report on the immunoproteomic profiling of unsporulated E. tenella oocysts. Forty-six spots were identified as immunogenic proteins. These new findings enrich knowledge of the immunogenic proteins of the unsporulated oocyst stages of E. tenella. The newly identified antigens are of great interest in terms of understanding pathogen-host interactions and in the search for novel vaccine targets. Further investigations into the characterization and functions of the immunogenic proteins would be beneficial to clarify the mechanisms of immune responses, immune evasion, immunopathology, and in the development of vaccines against Eimeria.
Acknowledgements
This work was funded by a grant from the National Natural Science Foundation of the People's Republic of China (No. 31560691).
Cite this article as: Zhang Z, Wang S, Li C, Liu L. 2017. Immunoproteomic analysis of the protein repertoire of unsporulated Eimeria tenella oocysts. Parasite 24, 48
References
- 1. Barrett J, Jefferies JR, Brophy PM. 2000. Parasite proteomics. Parasitology Today, 16(9), 400-403. [DOI] [PubMed] [Google Scholar]
- 2. Belli SI, Smith NC, Ferguson DJ. 2006. The coccidian oocyst: a tough nut to crack! Trends in Parasitology, 22(9), 416-423. [DOI] [PubMed] [Google Scholar]
- 3. Bement WM, Hasson T, Wirth JA, Cheney RE, Mooseker MS. 1994. Identification and overlapping expression of multiple unconventional myosin genes in vertebrate cell types. Proceedings of the National Academy of Sciences of the United States of America, 91(24), 11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhaduri-McIntosh S, Vaidya AB. 1996. Molecular characterization of a Plasmodium falciparum gene encoding the mitochondrial phosphate carrier. Molecular and Biochemical Parasitology, 78(1-2), 297-301. [DOI] [PubMed] [Google Scholar]
- 5. Bhuiyan MS, McLendon P, James J, Osinska H, Gulick J, Bhandary B, Lorenz JN, Robbins J. 2016. In vivo definition of cardiac myosin-binding protein C's critical interactions with myosin. European Journal of Physiology, 468(10), 1685-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brayton KA, Lau AO, Herndon DR, Hannick L, Kappmeyer LS, Berens SJ, Bidwell SL, Brown WC, Crabtree J, Fadrosh D, Feldblum T, Forberger HA, Haas BJ, Howell JM, Khouri H, Koo H, Mann DJ, Norimine J, Paulsen IT, Radune D, Ren Q, Smith RK, Jr., Suarez CE, White O, Wortman JR, Knowles DP, Jr., McElwain TF, Nene VM. 2007. Genome sequence of Babesia bovis and comparative analysis of apicomplexan hemoprotozoa. PLoS Pathogens, 3(10), 1401-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bromley E, Leeds N, Clark J, McGregor E, Ward M, Dunn MJ, Tomley F. 2003. Defining the protein repertoire of microneme secretory organelles in the apicomplexan parasite Eimeria tenella. Proteomics, 3(8), 1553-1561. [DOI] [PubMed] [Google Scholar]
- 8. Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Righetti PG. 2004. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis, 25(9), 1327-1333. [DOI] [PubMed] [Google Scholar]
- 9. Chapman HD, Cherry TE, Danforth HD, Richards G, Shirley MW, Williams RB. 2002. Sustainable coccidiosis control in poultry production: the role of live vaccines. International Journal for Parasitology, 32(5), 617-629. [DOI] [PubMed] [Google Scholar]
- 10. Coudert P, Licois D, Streun A. 1979. Characterization of Eimeria species. Parasitology research, 59(3), 227-234. [DOI] [PubMed] [Google Scholar]
- 11. De VP, Chich JF, Faigle W, Loew D, Labbé M, Girardmisguich F, Péry P. 2004. Towards a reference map of Eimeria tenella sporozoite proteins by two-dimensional electrophoresis and mass spectrometry. International Journal for Parasitology, 34(12), 1321-1331. [DOI] [PubMed] [Google Scholar]
- 12. Dunn PP, Bumstead JM, Tomley FM. 1996. Primary structure of a BiP homologue in Eimeria spp. Parasitology Research, 82(6), 566-568. [DOI] [PubMed] [Google Scholar]
- 13. Fast NM, Kissinger JC, Roos DS, Keeling PJ. 2001. Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Molecular Biology and Evolution, 18(3), 418-26. [DOI] [PubMed] [Google Scholar]
- 14. Ferguson DJ, Parmley SF, Tomavo S. 2002. Evidence for nuclear localisation of two stage-specific isoenzymes of enolase in Toxoplasma gondii correlates with active parasite replication. International Journal for Parasitology, 32(11), 1399-1410. [DOI] [PubMed] [Google Scholar]
- 15. Fernando MA, Rose ME, Millard BJ. 1987. Eimeria spp. of domestic fowl: the migration of sporozoites intra- and extra-enterically. Journal of Parasitology, 73(3), 561-567. [PubMed] [Google Scholar]
- 16. Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature, 419(6906), 498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haas IG, Wabl M. 1983. Immunoglobulin heavy chain binding protein. Nature, 306(5941), 387-389. [DOI] [PubMed] [Google Scholar]
- 18. Heintzelman MB, Schwartzman JD. 1997. A novel class of unconventional myosins from Toxoplasma gondii. Journal of Molecular Biology, 271(1), 139-146. [DOI] [PubMed] [Google Scholar]
- 19. Jeurissen SH, Janse EM, Vermeulen AN, Vervelde L. 1996. Eimeria tenella infections in chickens: aspects of host-parasite: interaction. Veterinary Immunology and Immunopathology, 54(1-4), 231-238. [DOI] [PubMed] [Google Scholar]
- 20. Jianzhen H, Haitian M, Liming Y, Sixiang Z. 2007. Developmental changes of protein profiles in the embryonic Sanhuang chicken liver. Journal of Veterinary Medicine A, Physiology, Pathology, Clinical Medicine, 54(9), 464-469. [DOI] [PubMed] [Google Scholar]
- 21. Kundu K, Banerjee PS, Garg R, Kumar S, Mandal M, Maurya PS, Tomley F, Blake D. 2015. Cloning and sequencing of beta-tubulin and internal transcribed spacer-2 (ITS-2) of Eimeria tenella isolate from India. Journal of Parasitic Diseases, 39(3), 539-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Labbe M, Peroval M, Bourdieu C, Girard-Misguich F, Pery P. 2006. Eimeria tenella enolase and pyruvate kinase: a likely role in glycolysis and in others functions. International Journal for Parasitology, 36(14), 1443-1452. [DOI] [PubMed] [Google Scholar]
- 23. Ling KH, Rajandream MA, Rivailler P, Ivens A, Yap SJ, Madeira AM, Mungall K, Billington K, Yee WY, Bankier AT, Carroll F, Durham AM, Peters N, Loo SS, Isa MN, Novaes J, Quail M, Rosli R, Nor Shamsudin M, Sobreira TJ, Tivey AR, Wai SF, White S, Wu X, Kerhornou A, Blake D, Mohamed R, Shirley M, Gruber A, Berriman M, Tomley F, Dear PH, Wan KL. 2007. Sequencing and analysis of chromosome 1 of Eimeria tenella reveals a unique segmental organization. Genome Research, 17(3), 311-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu L, Xu L, Yan F, Yan R, Song X, Li X. 2009. Immunoproteomic analysis of the second-generation merozoite proteins of Eimeria tenella. Veterinary Parasitology, 164(2-4), 173-182. [DOI] [PubMed] [Google Scholar]
- 25. Liu Z, Yin L, Li Y, Yuan F, Zhang X, Ma J, Liu H, Wang Y, Zheng K, Cao J. 2016. Intranasal immunization with recombinant Toxoplasma gondii actin depolymerizing factor confers protective efficacy against toxoplasmosis in mice. BMC Immunology, 17(1), 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maresca B, Carratu L. 1992. The biology of the heat shock response in parasites. Parasitology Today, 8(8), 260-266. [DOI] [PubMed] [Google Scholar]
- 27. Marugan-Hernandez V, Cockle C, Macdonald S, Pegg E, Crouch C, Blake DP, Tomley FM. 2016. Viral proteins expressed in the protozoan parasite Eimeria tenella are detected by the chicken immune system. Parasites & Vectors, 9, 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Newport G, Culpepper J, Agabian N. 1988. Parasite heat-shock proteins. Parasitology Today, 4(11), 306-312. [DOI] [PubMed] [Google Scholar]
- 29. Peek HW, Landman WJ. 2011. Coccidiosis in poultry: anticoccidial products, vaccines and other prevention strategies. Veterinary quarterly, 31(3), 143-161. [DOI] [PubMed] [Google Scholar]
- 30. Qian X, Xu W, Xu J, Shi Q, Li J, Weng Y, Jiang Z, Feng L, Wang X, Zhou J, Jin H. 2017. Enolase 1 stimulates glycolysis to promote chemoresistance in gastric cancer. Oncotarget, 8(29), 47691-47708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shirley MW, Smith AL, Tomley FM. 2005. The biology of avian Eimeria with an emphasis on their control by vaccination. Advances in Parasitology, 60(60), 285-330. [DOI] [PubMed] [Google Scholar]
- 32. Song X, Zhang Z, Liu C, Xu L, Yan R, Li X. 2016. Evaluation of the persistence, integration, histopathology and environmental release of DNA vaccine encoding Eimeria tenella TA4 and chicken IL-2. Veterinary Parasitology, 229, 22-30. [DOI] [PubMed] [Google Scholar]
- 33. Suarez-Cortes P, Sharma V, Bertuccini L, Costa G, Bannerman NL, Sannella AR, Williamson K, Klemba M, Levashina EA, Lasonder E, Alano P. 2016. Comparative proteomics and functional analysis reveal a role of Plasmodium falciparum osmiophilic bodies in malaria parasite transmission. Molecular & Cellular Proteomics, 15(10), 3243-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vrba V, Pakandl M. 2015. Host specificity of turkey and chicken Eimeria: controlled cross-transmission studies and a phylogenetic view. Veterinary Parasitology, 208(3-4), 118-124. [DOI] [PubMed] [Google Scholar]
- 35. Wagenbach GE, Burns WC. 1969. Structure and respiration of sporulating Eimeria stiedae and E. tenella oocysts. Journal of Protozoology, 16(2), 257-263. [DOI] [PubMed] [Google Scholar]
- 36. Wang CC. 1976. Inhibition of the respiration of Eimeria tenella by quinolone coccidiostats. Biochemical Pharmacology, 25(3), 343-349. [DOI] [PubMed] [Google Scholar]
- 37. Wang CC, Stotish RL. 1978. Multiple leucine aminopeptidases in the oocysts of Eimeria tenella and their changes during sporulation. Comparative Biochemistry & Physiology B Comparative Biochemistry, 61(2), 307-313. [Google Scholar]
- 38. Wilson PAG, Fairbairnz D. 1961. Biochemistry of sporulation in oocysts of Eimeria acervulina. Journal of Eukaryotic Microbiology, 8(4), 410-416. [Google Scholar]
- 39. Wu Z, Zhang W, Lu C. 2008. Immunoproteomic assay of surface proteins of Streptococcus suis serotype 9. Pathogens and Disease, 53(1), 52-59. [DOI] [PubMed] [Google Scholar]
- 40. Xu JH, Qin ZH, Liao YS, Xie MQ, Li AX, Cai JP. 2008. Characterization and expression of an actin-depolymerizing factor from Eimeria tenella. Parasitology Research, 103(2), 263-270. [DOI] [PubMed] [Google Scholar]
- 41. Xu P, Widmer G, Wang Y, Ozaki LS, Alves JM, Serrano MG, Puiu D, Manque P, Akiyoshi D, Mackey AJ, Pearson WR, Dear PH, Bankier AT, Peterson DL, Abrahamsen MS, Kapur V, Tzipori S, Buck GA. 2004. The genome of Cryptosporidium hominis. Nature, 431(7012), 1107-1112. [DOI] [PubMed] [Google Scholar]