During infection with Dengue virus (DENV), host cells undergo a series of detrimental functional changes, including profound remodelling and redistribution of cell membrane structures,1,4 which gives rise to nucleus-like structures referred to as “viral factories.”2 In many cases, this phenomenon is mediated by viroporin, a family of small viral protein, which contain highly hydrophobic domains that can form amphipathic alpha helices. They are especially known for their ability to re-organize the membranes of cellular organelles.3 Indeed, viroporins may be virulence factors encoded by members of diverse families of RNA and DNA viruses of clinical interest, including hepatitis C virus (HCV), human immunodeficiency virus (HIV-1), human papilloma virus (HPV), influenza A virus (IAV), coronaviruses, picornaviruses (polioviruses), and togaviruses (see ref.4 for a comprehensive review).
While the amino acid sequences of viroporins can vary significantly, these proteins characteristically share certain common structural features and signature motifs that form basis for their identification.5 For example, after membrane insertion, oligomerization of viroporins results in the formation of a distinctive hydrophilic and aqueous channel, in which hydrophobic amino acid residues face the phospholipid bilayer and hydrophilic residues form the aqueous channel pore. Notably, these channels are typically able to transport at least one physiologically relevant ion (i.e., Na+, K+, Ca2+, Cl−, or H+).6 Moreover, basic amino acid residues and domains rich in aromatic amino acids disturb the organization of the lipid bilayer, thus contributing to membrane destabilisation.3 In addition to ion transport, viroporins have also been shown to affect vesicular trafficking,7,14 membrane remodelling,15 ion homeostasis,8,16 induction of apoptosis,9 and inflammasome activation, which might influence the innate immune response.10
Viruses of the Flaviviridae family, which includes Hepatitis C virus (HCV) and Japanese encephalitis virus (JEV), encode a variety of viroporins that exhibit distinct capabilities to permeabilise eukaryotic and prokaryotic membranes. In particular, NS2A is a virulence determinant of many flaviviruses that shares similarities with viroporins. NS2A proteins are highly hydrophobic transmembrane protein that interact with lipid bilayers and co-localize with dsRNA, as well as the viral proteins NS1, NS3, and NS5, within the replication complex,11 and play critical roles in viral assembly and release.12 Notably, the NS2A protein of Kunjin virus was shown to form pores on intracellular membranes, thereby enabling the transport of nascent RNA to the site of viral assembly.12 Furthermore, this protein plays a role in membrane remodelling, another typical characteristic of viroporins that facilitates viral assembly by transporting RNA or partially assembled nucleocapsids to subcellular compartments.13 Similarly, the NS2A protein of JEV was shown to mediate changes in bacterial permeability, in combination with other small hydrophobic non-structural proteins.14 Consistent with other viroporins, the NS2A proteins of flaviviruses exhibit 20–65% amino acid sequence similarity; however, despite this high variability, there is overall conservation of physicochemical properties. They include maintenance of the positively charged amino acids, essential for cleavage by the NS3 protease and of repeat regions enriched with hydrophobic amino acids such as leucine and valine.15
In a previous study, DENV replication and assembly were shown to be facilitated by a small hydrophobic protein that promotes cellular membrane reorganisation,16 which was later identified as NS2A.17,7 Indeed, biophysical analysis of NS2A identified a membrane-interacting region, called “dens25,” which alone is capable to modulate and affect artificial membrane structure.18 Recently, Wu et al. identified a transmembrane region of DENV NS2A, as well as specific amino acid residues within that region that are critical for the cytopathic effect during dengue infection.19 Together, these findings suggest that NS2A contains membrane interacting regions important for the cytopathic effect during dengue infection, suggesting that NS2A may behaves as viroporin during Dengue infection.18 Moreover, we recently demonstrated that the NS2B protein of DENV, which functions as a part of the viral protease NS3-2B, also behaves as a viroporin.20
The main objective of the present study was to determine whether the full-length Dengue virus serotype 2, NS2A protein possesses viroporin activity. To address this question, we performed an in silico analysis of DENV-2 NS2A to determine whether it exhibits the sequence features associated with viroporins. For these analysis, the NS2A of JEV, which possesses membrane-destabilising ability,14 was used for comparison. First, we generated hydropathic plots of the protein by the Kyte-Doolittle method21 and using the DAS Trans-membrane prediction algorithm (Fig. 1A).22 This analysis indicated presence of six putative trans-membrane helices that show significant sequence similarity with the corresponding regions in NS2A of JEV. Subsequently, a topological representation of the DENV-2 NS2A protein was generated using the Socs MEMSAT program,23 In contrast with Kyte-Doolittle method, Socs MEMSAT identified seven potential transmembrane regions which is expected due to differences in algorithm. Furthermore, this analysis indicated that the amino and carboxyl-terminal domains of DENV-2 NS2A orient toward the lumen of the endoplasmic reticulum and the cytoplasm, respectively (Fig. 1B). Notably, these findings are consistent with the topology of flavivirus NS2A proteins generated via various bioinformatics algorithms and corroborated by biochemical approaches.24
Figure 1.
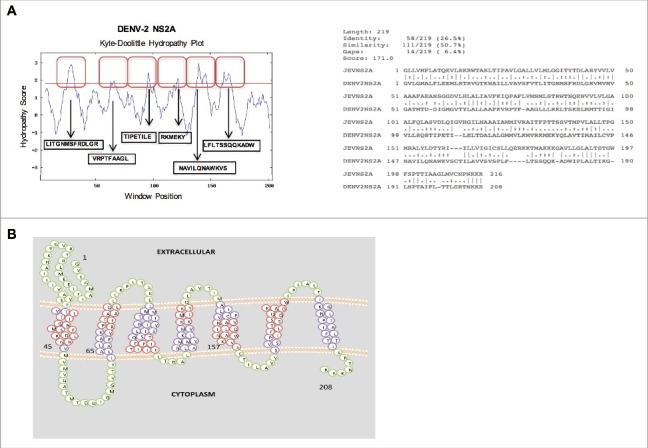
In silico analysis of the amino acid sequence (A) (Above) Hydropathy plots of the NS2A protein of Dengue virus (DENV), as generated via the Kyte-Doolittle method and DAS TM prediction algorithm using SOSUI software. Red boxes indicate the highly hydrophobic transmembrane regions of NS2A. (Below) Alignment of the amino acid sequences of NS2A from DENV and Japanese encephalitis virus (JEV), as generated using ClustalW software. Identical (*), similar (.), and low-similarity (:) amino acids were observed. (B) Proposed model of NS2A topology generated using SOCS MEMSAT software. Trans-membrane segments are shown in purple, while the green color indicates inter-transmembrane regions of NS2A. Regions highlighted in red denote highly hydrophobic, possible membrane interacting, transmembrane segments.
To examine whether DENV-2 NS2A protein, is able to modify the membrane permeability in different systems similarly to JEV NS2A,14 we compared the sequences of amino acid for these two proteins. Notably DENV and JEV NS2A exhibit 26–65% sequence identity and more than 50% sequence similarity. In addition, few amino acids within the transmembrane domains of these two proteins were identical or showed substantial similarity. This similarity suggests that the physicochemical properties of JEV and DENV-2 NS2A are comparable (Fig. 1A). In addition both proteins possess similar aromatic and basic residues that are important characteristic of viral proteins with membrane destabilising ability (Fig. 1A). Therefore, bioinformatics analysis of the DENV-2 NS2A amino acid sequence revealed a high degree of similarity with a previously reported viroporin from a member of the Flaviviridae family.14 Together, these data suggest that DENV-2 NS2A possesses membrane-altering activity.
To confirm this hypothesis, we examined whether DENV NS2A localizes to bacterial cell membranes. For these experiments, the DENV NS2A coding sequence was cloned into an inducible bacterial expression vector (pET/D-Topo) containing an N-terminal V5 epitope tag and a 6 × -histidine residue tag (His-tag) for purification and detection purposes (Fig. 2A). The resulting construct (pET- NS2A) was used to transform into Escherichia coli BL21 Star cells via transformation. The transformed cells were induced by cultivation in the presence of isopropyl β-D-1-thiogalactopyranoside (IPTG) for 0, 4, 6, and 8 hrs (Fig. 2B). Subsequently, NS2A expression was induced within E. coli for 6 h, and cell lysates were prepared and separated by SDS-PAGE. Similar to other viral proteins with viroporin activity25 DENV-2 NS2A protein was expressed in inclusion bodies. A band with the estimated molecular mass as that of NS2A (25 kDa) was then eluted from the gel (Fig. 2C) and further purified with nickel resin. Bound protein was eluted from the resin (1-mL aliquots), separated by SDS-PAGE, and examined by Coomassie Blue staining, which again revealed the presence of the 25-kDa band in the last recovered fraction. To determine whether NS2A protein associated with bacterial membranes, we fractionated bacterial lysates and then extracted membrane proteins, using TDPC detergent. Western blots were then performed using an anti-V5 antibody to detect the presence of the NS2A protein (Fig. 2D). Subsequently, the blots were washed, stripped, and re-probed using a mouse polyclonal antibody specific to E. coli OmpC (Fig. 2D), a known marker of membrane protein. Notably, DENV NS2A and OmpC (40 kDa) were present in the same fraction, indicating that NS2A targets bacterial membranes.
Figure 2.
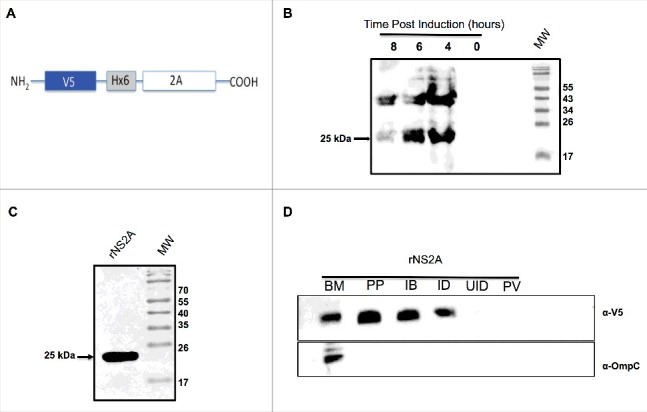
Expression and purification of NS2A protein from bacteria, and confirmation of NS2A association with bacterial membranes. (A) Schematic representation of the bacterial expression construct encoding the NS2A protein in frame with an N-terminal V5 epitope and 6 × His tag. (B) Expression and purification of NS2A protein in bacteria. Cells were induced with 1 mM IPTG at time zero, and NS2A expression was analyzed by western blot at the indicated times post-induction using an anti-V5 antibody (dilution, 1:5000). (C) Analysis of recombinant NS2A protein (rNS2A) purification following different isolation steps by SDS-PAGE and Coomassie blue staining. Preparative gel electrophoresis was used for purification. Lane 1: purified fraction of NS2A obtained from preparative gels (molecular-weight, approximately 25 kDa). (D) Localization of NS2A within Escherichia coli membranes was detected via western blot analysis using an anti-V5 antibody and a mouse polyclonal antibody specific for OmpC (a bacterial membrane protein). Lane 1: purified bacterial membranes (BM); lane 2: purified NS2A protein (PP); lane 3: inclusion bodies of bacteria transformed with NS2A (IB); lane 4: NS2A clone, induced with IPTG (ID); lane 5: NS2A clone, uninduced (UID); lane 6: bacteria transformed with the parental vector (PV).
In prokaryotic systems such as E. coli, bacterial growth can be altered directly by the expression and subsequent incorporation of the viroporins within bacterial membranes, leading to a loss of membrane integrity.14 Given that DENV and JEV are homologous, (Fig. 1A), we therefore evaluated whether DENV NS2A also possesses membrane-destabilising activity, to this end, E. coli BL21 (DE3) pLysS cells, which express bacteriophage T7 lysozyme, were transformed with a plasmid encoding NS2A (pET-NS2A) or the control plasmid (pET). In this system, disruption of the inner bacterial membrane triggers lysozyme release resulting in cell lysis. The levels of cell lysis can then be quantified by measuring the optical density of each culture. As expected, transformation of E. coli with the parental vector (pET) or, with the same vector expressing a soluble polypyrimidine-tract binding (PTB) protein (RNA-binding protein found only in the cytosol; as a negative control) had no effect on bacterial cell growth (Fig. 3A). Likewise, cells harbouring the NS2A expression vector showed no significant defect in cell growth rates when cultivated in the absence of the inducing agent IPTG. In contrast, after IPTG induction, the growth rates of cells expressing NS2A were markedly lower than those of the PTB and pET controls, indicating that NS2A protein was capable of permeabilising intracellular membranes (Fig. 3A). Further, to examine whether NS2A can also permeabilise outer cell membranes, bacterial cultures induced with IPTG were exposed to the translation inhibitor HygB and then metabolically labeled with [S35]-methionine. Notably, a complete, time-dependent abrogation of protein synthesis was observed in cells expressing NS2A, or in those harbouring the parental vector, upon HygB treatment, indicating that the bacterial membranes became permeable to HygB following NS2A induction (Fig. 3C). Together, these data provide the first evidence that DENV-2 NS2A, behaved as other viroporins,25 exhibits pore-forming activity when expressed in a bacterial system.
Figure 3.
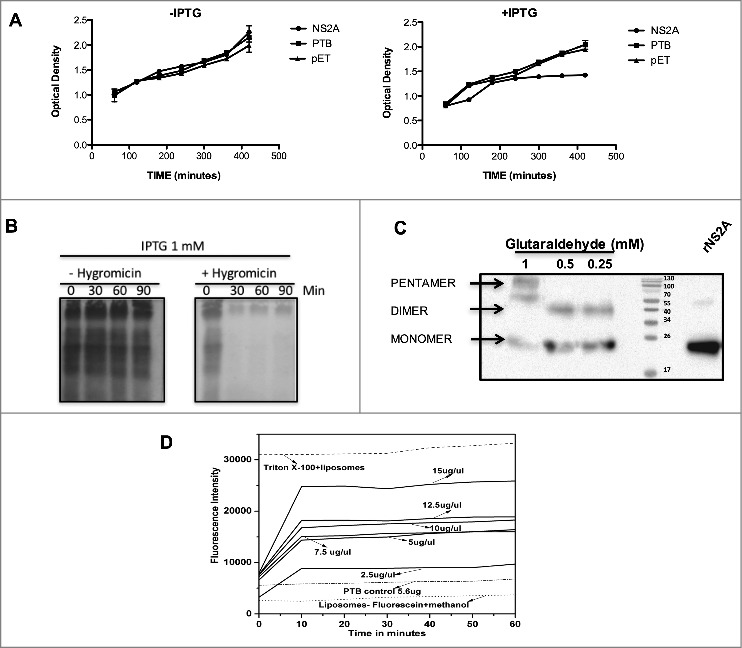
NS2A protein expression arrests the growth of Escherichia coli and affects the permeability of bacterial membranes and liposomes. (A) Bacterial membrane pET-NS2A permeability model. Growth curves of engineered E. coli BL21 pLys transformed with NS2A and PTB constructs in the absence (left) or presence (right) of IPTG. The cellular density of each bacterial culture was determined every 60 min after induction by measuring the optical density at 660 nm. (B) Cultures were induced with IPTG in the absence (−) or presence (+) of HygB, then subjected to metabolic labeling with [35S] Met-Cys and analyzed by SDS-PAGE at the indicated times. (C) Western blot analysis of NS2A oligomerisation in vitro. NS2A protein was incubated with different concentrations of glutaraldehyde (0.25, 0.5, and 1 mM) and then resolved by SDS-PAGE under reducing conditions. Oligomeric structures (dimers and pentamers) were detected using an anti-V5 antibody at a dilution of 1:5000. (D) Membrane disruption was examined by incubating liposome-encapsulated FITC (as a model for eukaryotic cell membranes) in solutions containing Triton X-100 (positive control), or NS2A (2.5–15 µg/µl). NS2A protein-mediated liposome disruption was monitored every 10 min by measuring the fluorescence emission of each mixture. Protein disruption studies were performed using 5 µL of liposome-encapsulated FITC to determine the baseline fluorescence intensity. Fluorescence was monitored using a Synergy H4 Fluorescence reader equipped with a FITC 485 excitation filter, a FITC 535 emission filter, and a 50/50 beam splitter. Fluorescence values were read every 30 s for 1 h at 37°C.
Among the conserved characteristics of viroporins are the presence of hydrophobic domains that interact with lipid membranes and the ability to oligomerise once inserted into cell membranes.26 In particular, several studies have reported viroporin oligomerisation in the presence of glutaraldehyde.20,26,27 Therefore, to further characterize whether NS2A resembles a viroporin, we evaluated the ability of this protein to form homo-oligomers when incubated in the presence of increasing concentrations of glutaraldehyde. Western blot analysis of glutaraldehyde-treated NS2A revealed the presence of two to three bands (Fig. 3C, lanes 1–3), which migrated at molecular weights consistent with monomers (25 kDa), dimers (50 kDa), and pentamers (∼125 kDa) of NS2A, suggesting that this protein oligomeriza when associated with cellular membranes. Notably, NS2A dimers were also observed in the absence of glutaraldehyde, after storage of the protein. Thus, NS2A dimer, trimer, and pentamer formation may depend on hydrophobic interactions rather than disulphide bridges, as witnessed for other viroporins.28 While NS2A oligomers likely organize into pore-like structures, which would account for the observed alterations in membrane permeability, further studies are needed to confirm this conclusion.
Lastly, we examined the ability of NS2A to induce membrane permeability in liposomes, which are artificial membrane structures that mimic eukaryotic membranes, charged with FITC.29 After purification, NS2A protein was first subjected to treatment with different solvents; the protein dissolved well in methanol and was active in liposomal mixtures. NS2A (119–714 nM) was then incubated with liposome-encapsulated FITC (25 µM). For these analysis, untreated liposomes were utilised as a control for auto-fluorescence, while FITC-labeled liposomes treated with PTB (98.25 nM) or Triton-X 100 were utilised as negative and positive controls, respectively (Fig. 3D). The maximum signal derived from positive control, was compared with the fluorescence signals obtained from liposomes incubated with the different concentrations of NS2A protein and was expressed as a percentage of the signal of the positive control. There was a protein concentration-dependent increase in fluorescence signal obtained from liposomes incubated with NS2A, with the highest level of FITC leakage (75%) being observed in those incubated with 714.29 nM NS2A (Fig. 3D). The NS2A-dependent release of FITC was also time-dependent, reaching a plateau within 10 min. Thus, these data provide further evidence that NS2A acts as a membrane permeability virulence factor (i.e., has the characteristics of a viroporin) (Fig. 3D).
In summary, the results of this study provide the first direct evidence that DENV NS2A can penetrate the hydrophobic environment of cellular membranes and exert lytic effects on the target membrane, as reported for other viroporins. This study also provides the first evidence that the DENV NS2A is capable of oligomerisation. However, future studies are necessary to assess other potential viroporin-like effects of this protein during viral infection, including apoptosis, autophagy, and modulation of innate immunity, which are features of several other viroporins.10,30
Although detailed work has been carried out to elucidate the topology and function of DENV-2 NS2A protein by studying different regions and by using various approaches, including bioinformatics (19, 23), this study, for the first time, provides the experimental data to support that the whole NS2A protein of Dengue virus exhibits membrane-destabilising ability and it could serve as a viroporin. Thus, DENV-2 NS2A protein could be critical during the DENV replication cycle.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Raul Bonilla Moreno for his technical assistance. Additionally, GS received fellowships from CONACyT. LCB, JGC, MLJ and NVS are members of the National System of Researchers, SNI.
Funding
This work was supported by CINVESTAV Mexico City.
References
- [1].Bartenschlager R, Miller S. Molecular aspects of Dengue virus replication. Future Microbiol. 2008;3:155-65. doi: 10.2217/17460913.3.2.155. PMID:18366336 [DOI] [PubMed] [Google Scholar]
- [2].Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol Cell. 2005;97:147-72. doi: 10.1042/BC20040058. PMID:15656780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gonzalez ME, Carrasco L. Viroporins. In: FEBS Letters. 2003;552(1):28-34. doi: 10.1016/S0014-5793(03)00780-4. PMID:12972148 [DOI] [PubMed] [Google Scholar]
- [4].Hyser JM, Estes MK. Pathophysiological Consequences of Calcium-Conducting Viroporins. Annu Rev Virol. 2015;2(1):473-96. doi: 10.1146/annurev-virology-100114-054846. PMID:26958925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nemésio H, Palomares-Jerez F, Villalaín J. NS4A and NS4B proteins from dengue virus: Membranotropic regions. Biochim Biophys Acta - Biomembr. 2012;1818(11):2818-30; doi: 10.1016/j.bbamem.2012.06.022. PMID:22772157. [DOI] [PubMed] [Google Scholar]
- [6].Wang K, Xie S, Sun B. Viral proteins function as ion channels. Biochim Biophys Acta. 2011;1808:510-5. doi: 10.1016/j.bbamem.2010.05.006. PMID:20478263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].De Jong AS, Visch HJ, De Mattia F, Van Dommelen MM, Swarts HG, Luyten T, Callewaert G, Melchers WJ, Willems PH, Van Kuppeveld FJ. The coxsackievirus 2B protein increases efflux of ions from the endoplasmic reticulum and Golgi, thereby inhibiting protein trafficking through the Golgi. J Biol Chem. 2006; 19;281(20):14144-50. doi: 10.1074/jbc.M511766200. PMID:16540472 [DOI] [PubMed] [Google Scholar]
- [8].Hyser JM, Collinson-Pautz MR, Utama B, Estes MK. Rotavirus disrupts calcium homeostasis by NSP4 viroporin activity. MBio. 2010;1(5):pii: e00265-10. doi: 10.1128/mBio.00265-10. PMID:21151776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Aweya JJ, Mak TM, Lim SG, Tan Y-J. The p7 protein of the hepatitis C virus induces cell death differently from the influenza A virus viroporin M2. Virus Res. 2013;172(1–2):24-34. doi: 10.1016/j.virusres.2012.12.005. PMID:23246447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Guo HC, Jin Y, Zhi XY, Yan D, Sun SQ. NLRP3 Inflammasome Activation by Viroporins of Animal Viruses. Viruses. 2015;7:3380-91. doi: 10.3390/v7072777. PMID:26114475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mackenzie JM, Khromykh aa, Jones MK, Westaway EG. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology. 1998;245:203-15. doi: 10.1006/viro.1998.9156. PMID:9636360 [DOI] [PubMed] [Google Scholar]
- [12].Leung JY, Pijlman GP, Kondratieva N, Hyde J, Mackenzie JM, Khromykh AA. Role of Nonstructural Protein NS2A in Flavivirus Assembly. J Virol. 2008;82:4731-41. doi: 10.1128/JVI.00002-08. PMID:18337583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Liu WJ, Chen HB, Khromykh AA. Molecular and Functional Analyses of Kunjin Virus Infectious cDNA Clones Demonstrate the Essential Roles for NS2A in Virus Assembly and for a Nonconservative Residue in NS3 in RNA Replication. J Virol. 2003;77:7804-13. doi: 10.1128/JVI.77.14.7804-7813.2003. PMID:12829820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chang Y-S, Liao C-L, Tsao C-H, Chen M-C, Liu C-I, Chen L-K, Lin Y-L. Membrane Permeabilization by Small Hydrophobic Nonstructural Proteins of Japanese Encephalitis Virus. J Virol. 1999;73:6257-64. PMID:10400716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Danecek P, Schein CH. Flavitrack analysis of the structure and function of West Nile non-structural proteins. Int J Bioinform Res Appl. 2010;6(2):134-46. doi: 10.1504/IJBRA.2010.032117. PMID:20223736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CKE, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. Composition and Three-Dimensional Architecture of the Dengue Virus Replication and Assembly Sites. Cell Host Microbe. 2009;5(4):365-75. doi: 10.1016/j.chom.2009.03.007. PMID:19380115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Suhy DA, Giddings TH, Kirkegaard K. Remodeling the Endoplasmic Reticulum by Poliovirus Infection and by Individual Viral Proteins: an Autophagy-Like Origin for Virus-Induced Vesicles. J Virol. 2000;74:8953-65. doi: 10.1128/JVI.74.19.8953-8965.2000. PMID:10982339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nemésio H, Villalaín J. Membrane interacting regions of dengue virus NS2A protein. J Phys Chem B. 2014;118:10142-55. doi: 10.1021/jp504911r. PMID:25119664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wu R-H, Tsai M-H, Chao D-Y, Yueh A. Scanning Mutagenesis Studies Reveal Potential Intramolecular Interaction within the C-terminal Half of Dengue Virus NS2A Involved in Viral RNA Replication and Virus Assembly/Secretion. J Virol. 2015;89(8):4281-95. doi: 10.1128/JVI.03011-14. PMID:25653435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].León-Juárez M, Martínez-Castillo M, Shrivastava G, García-Cordero J, Villegas-Sepulveda N, Mondragón-Castelán M, Mondragón-Flores R, Cedillo-Barrón L. Recombinant Dengue virus protein NS2B alters membrane permeability in different membrane models. Virol J. 2016;13:1. doi: 10.1186/s12985-015-0456-4. PMID:26728778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lesh N, Mitzenmacher M, Whitesides S, Vanetten CH, Nielsen HC, Peters JE, Beutler TC, Dill Ka, Lau KF, Bastolla U, et al.. A simple method for displaying the hydropathic character of a protein. J Chem Phys. 2012;90:3682-93. doi: 10.1016/0022-2836(82)90515-0. PMID:7108955 [DOI] [PubMed] [Google Scholar]
- [22].Cserzö M, Wallin E, Simon I. Prediction of transmembrane α-helices in prokaryotic membrane proteins: the dense alignment surface method annotations in the database were found to contain erroneous. Protein Eng. 1997;10:673-6. doi: 10.1093/protein/10.6.673. PMID:9278280 [DOI] [PubMed] [Google Scholar]
- [23].Jones DT. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics. 2007;23(5):538-44. doi: 10.1093/bioinformatics/btl677. PMID:17237066 [DOI] [PubMed] [Google Scholar]
- [24].Xie X, Gayen S, Kang C, Yuan Z, Shi P-Y. Membrane Topology and Function of Dengue Virus NS2A. Protein. 2013;87(8):4609-22. doi: 10.1128/JVI.02424-12. PMID:23408612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang Z, Yuan Y, Fan D, Yang J, Mao Z, Yan Y, Chen J. Scylla serrata reovirus p35 protein expressed in Escherichia coli cells alters membrane permeability. Virus Genes. 2015;51:69-76. PMID:26104656; doi: 10.1007/s11262-015-1218-5 [DOI] [PubMed] [Google Scholar]
- [26].Wetherill LF, Holmes KK, Verow M, Müller M, Howell G, Harris M, Fishwick C, Stonehouse N, Foster R, Blair GE, et al.. High-risk human papillomavirus E5 oncoprotein displays channel-forming activity sensitive to small-molecule inhibitors. J Virol. 2012; 86:5341-51. doi: 10.1128/JVI.06243-11. PMID:22357280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ao D, Guo HC, Sun SQ, Sun DH, Fung TS, Wei YQ, Han SC, Yao XP, Cao SZ, Liu DX, et al.. Viroporin activity of the foot-and-mouth disease virus non-structural 2B protein. PLoS One. 2015;10(5):e0125828. doi: 10.1371/journal.pone.0125828. PMID:25946195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liao Y, Yuan Q, Torres J, Tam JP, Liu DX. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology. 2006;349:264-75. doi: 10.1016/j.virol.2006.01.028. PMID:16507314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Spector AA, Yorek MA. Membrane lipid composition and cellular function. 1985; 26(9):1015-35. PMID:3906008 [PubMed] [Google Scholar]
- [30].Fung TS, Liu DX. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014;5:296. doi: 10.3389/fmicb.2014.00296. PMID:24987391 [DOI] [PMC free article] [PubMed] [Google Scholar]


