ABSTRACT
Acinetobacter baumannii is an important human pathogen and considered as a major threat due to its extreme drug resistance. In this study, the genome of a hyper-virulent MDR strain PKAB07 of A. baumannii isolated from an Indian patient was sequenced and analyzed to understand its mechanisms of virulence, resistance and evolution. Comparative genome analysis of PKAB07 revealed virulence and resistance related genes scattered throughout the genome, instead of being organized as an island, indicating the highly mosaic nature of the genome. Many intermittent horizontal gene transfer events, insertion sequence (IS) element insertions identified were augmenting resistance machinery and elevating the SNP densities in A. baumannii eventually aiding in their swift evolution. ISAba1, the most widely distributed insertion sequence in A. baumannii was found in multiple sites in PKAB07. Out of many ISAba1 insertions, we identified novel insertions in 9 different genes wherein insertional inactivation of adeN (tetR type regulator) was significant. To assess the significance of this disruption in A. baumannii, adeN mutant and complement strains were constructed in A. baumannii ATCC 17978 strain and studied. Biofilm levels were abrogated in the adeN knockout when compared with the wild type and complemented strain of adeN knockout. Virulence of the adeN knockout mutant strain was observed to be high, which was validated by in vitro experiments and Galleria mellonella infection model. The overexpression of adeJ, a major component of AdeIJK efflux pump observed in adeN knockout strain could be the possible reason for the elevated virulence in adeN mutant and PKB07 strain. Knocking out of adeN in ATCC strain led to increased resistance and virulence at par with the PKAB07. Disruption of tetR type regulator adeN by ISAba1 consequently has led to elevated virulence in this pathogen.
KEYWORDS: Acinetobacter baumannii, biofilm, efflux pumps, insertion sequence element, international clone II, SNPs
Introduction
Acinetobacter baumannii is one of the most significant nosocomial pathogens and considered as a major threat to the global medical community.1 The reasons for its success as a human pathogen are its ability to develop resistance against almost all the classes of antibiotics and its capacity to survive under extremely adverse conditions.2 Several typing methods including MLST were used to study the global epidemiology of A. baumannii that determines the dynamics of disease transmission at local hospital levels, within the country level or across different nations. Next generation sequencing methods are more efficient recent epidemiological tools to analyze whole genomes of A. baumannii clones and different lineages of international clones. Moreover, a complete knowledge of clonal population structure of A. baumannii and dominant international clonal lineages is prerequisite to understand the importance of high-risk clones in the dynamics of international dissemination of antibiotic resistant strains.3 Further, whole genome analysis of multiple resistant clones may also decipher the role of mobile genetic elements in emergence of resistance in A. baumannii.
Mobile genetic elements, such as Insertion Sequence (IS) elements are known to be present copiously in A. baumannii, among which ISAba1 of class IS6 is the predominant one that has been shown to modulate the expression of resistance associated genes such as ampC, blaOXA-51 like and blaOXA-23 like –genes.4-6 The presence of additional promoters with ISAba1 intensifies the expression of OXA-23 carbapenemase.7 Such findings indicate that resistance mechanisms in A. baumannii are constantly co-evolving not only with a single IS-element, but also with multiple IS-elements to cope up with the antibiotic pressure.7 Although, there have been reports on other IS elements present in A. baumannii such as ISAba2, ISAba3, ISAba4, ISAba125 and ISAba825, the copy number of these elements are sparse when compared with ISAba1.
Resistance of A. baumannii to various antimicrobial agents is predominantly mediated by mechanisms such as enzymatic inactivation, target site modification, active efflux and decreased influx of drugs.8 Previously, it was shown that overexpression of resistance nodulation cell division (RND)-type efflux systems contributing to drug resistance in A. baumannii for multiple antibiotics.8-10 Among RND-type efflux systems, AdeIJK pump seems to be mainly contributing to intrinsic resistance of A. baumannii toward many major antibiotics.11 However, the exact mechanisms of the efflux systems involved are not completely understood.
While there is ample literature available regarding antibiotic resistance in A. baumannii and its underlying mechanisms, not much is known about the role of IS-elements in assisting the evolution, pathogenesis and successive progression of this nosocomial pathogen. Previously, it was shown that ISAba1 disruped genes related to two component systems (adeS) and membrane porins (carO) escalate the genome plasticity of A. baumannii.7,12 According to another work, ISAba1 mediated disruption of the global repressor proteins such as H-NS in clinical strains had resulted in elevated virulence.13 Recently a few attempts have been made to understand the virulence potential of A. baumannii using whole genome analysis. However, numerous previous molecular experiments contemplate that there may not be any individual factor that can contribute to the organism's virulence.14-16
The virulence determining genes of A. baumannii are believed to be multiple in nature and not specific to any international clones (IC)/ non-international clones.16 Although there have been numerous virulence genes identified in A. baumannii, their mere presence alone do not confer virulence as they have been found intact in less-virulent strains too.17 Analysis of genome sequence helps to predict and identify the virulence genes and their putative functions based on their homologies with well-studied genes from closely related bacteria. There have been a limited number of research works on genome perspectives of A. baumannii through performing whole genome analysis of clinical strains originated from India. The present study analyzed the genome of A. baumannii PKAB07, an MDR strain that was a carbapenem-resistant isolated from a wound infection in a hospitalized patient. Besides, in vitro studies revealed that PKAB07 is a hyper-invasive strain that was exhibiting high induction of early apoptosis in maximum number of human epithelial cells. Whole genome analysis of PKAB07 helped in understanding the nature of this genome and also assisted us in identifying various resistance and virulence related traits present in it.
Materials and methods
Illumina NGS - Assembly and annotation
Genomic paired-end reads of 2 × 100 nucleotides were generated on an Illumina Genome analyzer II platform. A total of 9.4 million reads of insert size between 180 to 220 bp were generated. The adaptor sequences were trimmed from the reads using FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). The trimmed sequence reads were assembled denovo using appropriate assembly software and annotated as described in our previous draft release.18
Additional tools in genome annotation
Accessory softwares used in addition to regular NGS analysis tools were: PHAST server for identifying phage related genes, manual search in BASys annotation server to look for resistance and stress related genes.19,20 MLST analysis of assembled genome using CGE server21; CRISPR finder for finding regular interspaced palindromic repeats17; and CGview for making circular genome map and locating IS elements.22
Identification of IS-disrupted genes
The position of IS-elements throughout the genome was predicted by IS-finder and a map showing the loci of ISAba1 in the genome was generated with CG view.23,24 The genes preceding and following ISAba1 were identified using ACT Artemis tool.25 The disruption of identified genes and their copy number were verified by performing BLAST analysis of the intact full-length gene from other reference strains with the assembled genome files.
Comparative genomics
A total of 43 genomes of A. baumannii including PKAB07 were downloaded from NCBI for whole genome multiple sequence alignment by ProgressiveMauve.26 From the ‘backbone’ file generated by this software, the fragments that are shared by all the 43 genomes were segregated from the rest in MS-Excel to generate the core and the accessory genome. This data was used to construct the Circos map to represent the affinities among the genomes with respect to the accessory genome.27 Regions <5kb were ignored. The genomic content distance matrix generated by it was used to prepare the heat map by R-program and dendrogram by FigTree (http://tree.bio.ed.ac.uk/software/figtree/). Clustering of orthologous genes was performed using the MBGD server.28 Methods adopted to identify genes of two component systems (TCS) are described in supplementary file SM1.
Comparative analysis of efflux pumps genes
Whole genomes along with the plasmids belonging to the 43 genomes were uploaded to the MBGD server in GenBank format, which clustered the protein products of all the genes. Two genes were clustered together if their protein products align with a cut-off value of > 75% coverage and share >60% of identity. From the list of orthologous genes generated, genes belonging to RND-type efflux pumps and efflux pumps from other families, such as ATP-binding cassette transporters, heavy-metal transporters that confer resistance to copper, zinc, major facilitator super family (MFS) and multidrug and toxic compound extrusion (MATE) were selected (total 22 clusters) and used to create a map depicting the presence or absence of each gene belonging to efflux pump system among the genomes analyzed.
Generation of adeN knockout
PCR primers for the forward flanking (FF) and reverse flanking (RF) regions of A. baumannii adeN gene and kanamycin cassette were designed using GeneTool software. The primer sequences are given in Table 1. Genomic DNA isolated from A. baumannii ATCC 17978 strain was used as a template for amplification of FF and RF regions of adeN. Kanamycin resistance cassette was amplified using PCR TOPO II vector (Invitrogen, USA) as a template. Purification of the amplified products was performed using GeneJET PCR clean up kit (Thermo scientific, USA) following the manufacturer's instructions.
Table 1.
List of primers used for nested overlap extension PCR and real-time qPCR analysis.
| Sl. No | Primer Name | Sequence | Amplicon size (bp) |
|---|---|---|---|
| 1 | adeN | F-5′ CCGGGAATTCATGCATGATCCAGTCCTTGAG 3′ | 654 |
| R- 5′ ATCG GAATTC TTAGACTTTATGATGGCCCTTT 3′ | |||
| 2 | adeN FF | F- 5′ ATGCATGATCCAGTCCTTGAG 3′ | 130 |
| R – 5′CCCAGCTGGCAATTCCGGCAGCA | |||
| TCATATCCTTTTTCCA | |||
| 3 | adeN RF | F -5′CTTGACGAGTTCTTCTGAATCCTGA | 154 |
| TCATTCCTCTCTTAT 3′ | |||
| R - 5′ TTAGACTTTATGATGGCCCTTT | |||
| 4 | Kanamycin Cassette | F-5′CCGGAATTGCCAGCTGGG 3′ | 934 |
| F -5′ TTCAGAAGAACTCGTCAAG 3′ | |||
|
Reel-time PCR Primers | |||
| 5 | Outer membrane receptor cat. (OMRC) | F- 5′GCCGACTGGACTAGCT 3′ | 123 |
| R- 5′GCAGCGGCCTATACAGC 3′ | |||
| 6 | Putative Membrane Protein (PTP) | F- 5′TGCGGTGTTAACGTTCTGGATT 3′ | 140 |
| R-5′ACCCAGCCAATAGCAGTCCAG 3′ | |||
| 7 | Putative transcript regulator (PTR) | F-5′TCGTCAATCATGCTGGTGGT 3′ | 224 |
| R-5′CCAAACGTAAAAAGGCAATGTG 3′ | |||
| 8 | Putative dehydrogenase (PDH) | F-5′AAGCGTACCGTGGTTTTGTTCT 3′ | 127 |
| R-5′TCCCTCAATTTTTTTACGCAGTC 3′ | |||
| 9 | adeJ | F 5′CATCGGCTGAAACAGTTGAA 3′ | |
| R 5′GCCTGACCATTACCAGCACT 3′ | |||
| 10 | Dehydrogenase with different specificities (DDS) | F- 5′CGCCGCATAGGTCGATAAAT 3′ | 189 |
| R-5′GGTCGCCGTATTGGTAAAGGT 3 | |||
| 11 | Outer membrane receptor protein Fe transporter (OMRIR) | F-5′CCGCCTGATGAATTTCCA 3′ | 177 |
| R-5′TGGTTTTGGTGCACGTTCTAC 3′ | |||
| 12 | Putative acetyl transferase (PAT) | F-5′GCGATGAGCGAGAAGAAGATA 3′ | 175 |
| R- 5′GCCACATGCCCAACAATAGTA 3′ | |||
| 13 | rpoB | F- 5′TCCGCACGTAAAGTAGGAAC | 154 |
| R- 5′ATGCCGCCTGAAAAAGTAAC | |||
Overlap extension PCR
At their 5′ ends the FF adeNR and RF adeNF primers (Table 1) were added with an extension of around 20 nucleotides homologous to the KNCF and KNCR primers, respectively. The amplicons obtained in the aforementioned PCR reactions were added at equimolar concentrations in a PCR mixture and subjected to nested overlap extension PCR with adeNF, adeNR primers using pfu DNA polymerase (Thermo scientific, USA). The reaction conditions adopted for overlap extension PCR were 94°C for 30 s, 40°C for 1 min, 72°C for 2 min (10 cycles) followed by 94°C for 30 s, 55°C for 1 min, 72°C for 3 min (20 cycles) with a final extension at 68°C for 10 min. The PCR amplicons generated (ΔadeN) with the kanamycin resistance cassette flanked by 130 bp upstream and 154bp downstream fragments of adeN were purified using GeneJET PCR purification kit (Thermo Scientific, USA) following manufacturer's instructions. The purified ΔadeN amplicons were used for further electroporation experiments.
Preparation of electrocompetent cells and electroporation
Two micrograms of the ΔadeN amplicons were added to the electrocompetent cells prepared from an overnight grown culture of A. baumannii ATCC 17978, mixed thoroughly by gentle tapping and kept on ice for 30 min. The electrocompetent cells were subjected to electroporation at 1.8 KV for 3.2 ms. Nine hundred microlitres of fresh LB broth was immediately added to the cuvette and this preparation was transferred to a sterile 1.5 ml microfuge tube and kept in an orbital shaker at 37° C for 1 h. The cells were then pelleted at 4000 rpm for 5 min at 4°C and plated on LB agar plates supplemented with 100µg/ml kanamycin. Confirmation of transformed colonies was performed by PCR using forward and reverse primers of adeN and kanamycin cassette. The colonies with amplicon sizes of ∼1.2 kb i.e. larger than the gene size (> 654 bp) were categorized to be transformants with disrupted adeN. This disruption was also confirmed by sequencing of adeN amplicons.
adeN complementation using pWH1266
Intact full length adeN was amplified from the A. baumannii ATCC 17978 genomic DNA using adeN full length primers (Table 1). The amplicons were purified using GeneJET PCR clean up kit. Purified amplicon and shuttle vector pWH1266 were digested with fast digest EcoRI (Thermo scientific, USA) at 37°C and ligation reactions were performed using T4 DNA ligase (Thermo scientific, USA) at 22°C. The ligated product was then transformed into E.coli DH5α cells. The isolated plasmid pWH1266:adeN was electroporated into A. baumannii ATCC 17978:ΔadeN electrocompetent cells by electroporation. The complemented A. baumannii ATCC 17978: ΔadeNΩadeN strains were selected on LB agar with tetracycline 10µg/ml and confirmed by PCR.
Growth curve analysis of A. baumannii strains
Growth rate of all the 4 strains namely PKAB07, ATCC 17978, ΔadeN and ΔadeNΩadeN was monitored for 24h. Briefly, overnight cultures grown in LB were diluted in fresh LB medium to an OD600 of 0.2. Two hundred microlitres of fresh media were distributed into 96 well plate and to this 2 µl of OD set culture were inoculated. The growth experiments were performed at 37° C using the Spectra-Max M Series multi-mode microplate reader with continuous shaking. The OD600 values were measured at every 30 min for 24 h. The averages were calculated from values obtained for the bacteria grown in 6 parallel wells. The results of this experiment has been illustrated in Fig. S1 (and supplementary file SM1)
MIC determination and biofilm analysis
Antimicrobial susceptibility testing for determination of MIC for multiple antimicrobials was performed using E-strips and by microbroth dilution method as per CLSI guidelines.29 MIC values were determined for tigecycline, ceftazidime and meropenem using the MIC E-strips (Hi-Media laboratories, Mumbai, India) and for chloramphenicol, ciprofloxacin, tobramycin and amikacin MIC determination was by micro-broth dilution method in 96 well microtiter plate. Hundred micro-liter of Muller-Hinton broth was serially diluted from highest concentration of antibiotics (1024µg/ml) to lowest concentration (0.5µg/ml) in 96 well microtiter plate. 2µl of 4–6 h grown culture were inoculated in each well. Wells without inoculation were maintained as negative control along with positive control. Microtiter plates were incubated for 16–18 hours at 37°C and MIC value was determined based on the turbidity in each well.
The ability of the A. baumannii strains ATCC 17978, ATCC 17978ΔadeN, ATCC 17978 ΔadeNΩadeN and PKAB07 to form biofilm was tested quantitatively using the micro-titer plate assay.30 Briefly, strains were grown overnight at 37°C in brain heart infusion (BHI) broth. Two hundred micro liters of sterile BHI broth was added in the wells of a 96-well plate and 2µl of each of the overnight grown cultures were inoculated into the wells in triplicates followed by 24 h incubation. The biofilm formed by individual strains were quantified according to the standard protocol.30
Quantitative RT-PCR
Total RNA extraction
The total RNA of bacteria grown at 37°C was isolated using the TRI reagent (Sigma- Aldrich). The quality of the isolated RNA was determined using Nano-Drop (Thermo Scientific). For each strain with similar growth condition 3 biological replicates were included. Overnight cultures of all strains were diluted to OD600 equivalent to 0.1 and grown at 37°C to an OD600 equivalent to 0.6 in LB. The extracted total RNA was diluted to the final concentration of 25 ng/µl. Equal amount of RNA were used for quantitative RT-PCR analysis. The reactions were performed using the 7500 Real-Time PCR System (Applied Biosystems), with the following cycle profile: 1 cycle at 50°C for 2 min, followed by 40 cycles at 95°C for 5 s, 60°C for 34 s and 72°C for 30s. All the experiments were performed in triplicates. Relative quantification was used to compare the amount of a target nucleic acid present in the samples. The ratio between the reference and the test sample was calculated as follows: Ratio (Reference/Target) = 2Cq(Ref) - Cq(Target). Each result is presented as the mean value of three independent results with their standard deviation. All the primers used for qPCR are listed in Table 1.
Acridine orange and ethidium bromide (AO/EtBr) staining and Annexin V analysis
A confluent layer of A549 cells in a 12-well plate was co-cultured with overnight grown cultures of A. baumannii ATCC 17978, ATCC 17978ΔadeN, ATCC 17978 ΔadeNΩadeN complemented strain and PKAB07 at an MOI of 100. The plates were then incubated for 24 h following which the medium was removed and cells were washed thrice with PBS. AO/EB staining method followed was described in SM1. The percentages of cell death induced by these four strains were quantified using flow cytometer (Guava Easycyte 8HT Millipore). A confluent monolayer of A549 cells was infected with A. baumannii strains for 6 h following which the cells were harvested and stained with Annexin V – FITC. (Annexin V - FITC Apoptosis detection kit, Sigma Aldrich, USA). The late apoptotic and early-apoptotic population of A549 cells were analyzed using bench top flow cytometer (Guava Easycyte 8HT Millipore). The staining procedures were performed by following manufacturer's instructions.
Galleria mellonella survival analysis
The degree of virulence of the wild type ATCC 17978 strain, PKAB07, ΔadeN and adeN complemented strain were evaluated in an in vivo infection model, Galleria melonella, the greater wax moth. G. mellonella larvae maintained at Prof. Rakesh Seth's laboratory at Department of Zoology, University of Delhi, were obtained and cultivated in laboratory for survival analysis. The larvae weighed in the range of 250 to 300 mg at the late instar stage, were used for infection assays. Phosphate buffered saline (PBS) and wild type reference strain were used as negative and non-virulent controls respectively. Overnight grown cultures of the strains were pelleted, washed twice with PBS and adjusted to 1 OD at 600 nm by re-suspending in PBS. The cultures were plated on LB agar and the CFU was evaluated after overnight incubation. 10 µl of the 1000 fold diluted cultures of each strain were injected into the left proleg of the larvae and each set of injections were performed on 16 larvae of uniform size. The larvae were maintained at 37°C after injection and observed for 6 d following injection. Since the lethal dose standardized for ATCC 17978 killed the larvae in our setup, sub-lethal dose of the bacterial inoculum determined by serial dilution followed by CFU calculation were injected to evaluate the elevated virulence potential due to adeN disruption. The results were documented and the survival rates were plotted by using Kaplan-Meier method.31 The differences in the virulence potential based on the survival rates were assessed and the statistical significance was analyzed by unpaired 2-tailed student's t-test. All the analyses were performed on two or more independent experiments using Graphpad Prism software version 6.0.
Results and discussion
Molecular epidemiology of A. baumannii PKAB07
Multi-locus sequence typing (MLST) of strain PKAB07 revealed that it belongs to the sequence type 195 (ST 195), clonal complex (CC) 92B according to MLST scheme of Bartual and designated as International Clone II world over. MLST assigned to the representative sequenced genomes of A. baumannii and their clonal complex affiliation according to Bartual and Pasteur schemes is given in Table 2. Most of the previous studies have chosen IC-II strains for genome analysis, indicating the global dissemination of a successful clone (IC-II) that has the ability to cause severe nosocomial infections.
Table 2.
A. baumannii genomes used in comparative whole genome analysis and MLST analysis.
| Isolate | STOXF STPAS | CCOXF CCPAS | IC | GenBank ID |
|---|---|---|---|---|
| 3207 | 1321/422 | NA/79 | NA | CP015364.1 |
| 6200 | 1161/464 | NA/85 | NA | CP010397.1 |
| 1656–2 | 423/2 | 92/2 | II | CP001921.1 |
| 3027STDY5784958 | 136/2 | 92/2 | II | LT594095.1 |
| A1 | 231/1 | 109/1 | I | CP010781.1 |
| AB0057 | 207/1 | 109/1 | I | CP001182.1 |
| AB030 | 758/79 | 636/79 | NA | CP009257.1 |
| AB031 | 1000/638 | NA/NA | NA | CP009256.1 |
| Ab04-mff | 447/10 | 447/10 | VIII | CP012006.1 |
| AB307–0294 | 231/1 | 109/1 | I | CP001172.1 |
| AB5075-UW | 945/1 | 109/1 | I | CP008706.1 |
| AbH12O-A2 | 924/79 | 636/79 | NA | CP009534.1 |
| AC29 | 195/2 | 92/2 | II | CP007535.2 |
| AC30 | 195/2 | 92/2 | II | CP007577.1 |
| ACICU | 437/2 | 92/2 | II | CP000863.1 |
| ATCC17978 | 112/437 | NA/92 | II | CP018664.1 |
| AYE | 231/1 | 109/1 | I | CU459141.1 |
| BJAB07104 | 368/2 | 92/2 | II | CP003846.1 |
| BJAB0715 | 642/23 | 447/10 | VIII | CP003847.1 |
| BJAB0868 | 218/2 | 92/2 | II | CP003849.1 |
| D1279779 | 942/267 | NA/NA | NA | CP003967.2 |
| D36 | 498/81 | 109/1 | I | CP012952.1 |
| DU202 | 423/2 | 92/2 | II | CP017152.1 |
| IOMTU433 | 919/622 | 862/NA | NA | AP014649.1 |
| KBN10P02143 | 191/2 | 92/2 | II | CP013924.1 |
| LAC-4 | 447/10 | 447/10 | VIII | CP007712.1 |
| MDR-TJ | 369/2 | 92/2 | II | CP003500.1 |
| MDR-ZJ06 | 643/2 | 92/2 | II | CP001937.1 |
| NCGM237 | 455/2 | 92/2 | II | AP013357.1 |
| ORAB01 | 208/2 | 92/2 | II | CP015483.1 |
| PKAB07 | 195/NA | 92/NA | II | CP006963.1 |
| R2090 | 942/267 | NA/NA | NA | LN868200.1 |
| SDF | NA/17 | NA/NA | NA | CU468230.2 |
| TCDC-AB0715 | 218/2 | 92/2 | II | CP002522.2 |
| TYTH-1 | 455/2 | 92/2 | II | CP003856.1 |
| XH386 | 208/2 | 92/2 | II | CP010779.1 |
| XH856 | 381/2 | 92/2 | II | CP014541.1 |
| XH857 | 806/215 | 254/215 | NA | CP014540.1 |
| XH858 | 642/23 | 447/10 | VIII | CP014528.1 |
| XH859 | 368/2 | 92/2 | II | CP014539.1 |
| XH860 | 457/2 | 92/2 | II | CP014538.1 |
| YU-R612 | 191/2 | 92/2 | II | CP014215.1 |
| ZW85–1 | 378/639 | 109/NA | I | CP006768.1 |
Note. NA= not available.
A. baumannii ACICU genome was used as a reference for the PKAB07 genome assembly. ACICU is an IC-II clone which had caused outbreak in Rome in 2005.32 It has a genome size of 3.9 Mb, as compared with 4.23 Mb of PKAB07.33 Since no ISAba1 elements were found in ACICU, as compared with 26 copies in PKAB07, this may be one of the causes for its shorter genome. Notably, a large 86Kb island AbaR1 found in ACICU, which is not present in PKAB07 and this could be one of the reasons for the fact that PKAB07 is more virulent although its degree of resistance is comparatively lower than former. Nevertheless, ACICU has 2 large plasmids pACICU1 (28.2kb) and pACICU2 (64.3kb), as compared with a small plasmid pPKAB07 (8.8kb) in PKAB07 that is devoid of any antibiotic resistance or virulence gene.
Genome sequencing and assembly
The trimmed and filtered sequence reads were assembled denovo using velvet, ABySS and A5 pipeline. All the contigs from different assemblies were pooled and a total of 284 contigs were reordered against available (during 2015) 17 complete genomes of A. baumannii using ProgressiveMAUVE.26 The highest level of similarity was observed against A. baumannii ACICU genome, which was thereafter used as the reference sequence by abacas.pl (http://abacas.sourceforge.net/index.html) to generate genome assembly of 4,304,370 bp (4.3 Mb).
Annotation of PKAB07 genome
Resistance and stress tolerance related genes
In Gram-negative bacterial pathogens, multidrug resistance is likely to be mediated by the production of antibiotic hydrolysing enzymes, over-production of RND type efflux pumps, through modification of outer membrane porins or by modifying the target and making them inaccessible to the antibiotic binding. Annotation performed on PKAB07 genome using RAST server predicted around 3877 protein coding genes that included 59 genes related to resistance to antibiotics and toxic compounds.34 ResFinder identified resistance genes encoding aminoglycoside modifying enzymes (strA and strB),16S rRNA methyl transferase gene (armA), β-lactamases (blaOXA-66, ampC), macrolide resistance (mphE and msrE) and tetracycline resistance (tetB).35 Some of the known and putative antibiotic resistance genes in the A. baumannii genome specifying the resistance mediated through different and varying mechanisms against the β-lactams, aminoglycosides and macrolides are given in Table 3.
Table 3.
Important genome segment with genomic coordinates 2875478 – 2905039 present only in 21 genomes including A. baumannii PKAB07 but absent in many other genomes.
| Protein ID | Predicted functions | Function characterized |
|---|---|---|
| AHJ94053 | TetR family Transcriptional Regulator | — |
| AHJ94054 | TetR family Transcriptional Regulator | — |
| AHJ94055 | Alcohol Dehydrogenase | — |
| AHJ94056 | Muconate-Lactonizing Protein | Degradation of Aromatic Compounds in Β-Ketoadipate pathway |
| AHJ94057 | Major Facilitator Transporter | — |
| AHJ94058 | Prevent-Host-Death Protein | Antitoxin of toxin-Antitoxin stability System |
| AHJ94059 | Transporter | Permeases of the Drug/Metabolite transporter (Dmt) superfamily |
| AHJ94060 | Membrane Transport Protein | — |
| AHJ94061 | Membrane Protein Marc | Multiple Antibiotic Transporter (Intracellular Trafficking And Secretion) |
| AHJ94062 | Signal Peptide Protein | — |
| AHJ94063 | Ethanolamine Utilization Protein Eutq | — |
| AHJ94064 | Oxidoreductase | — |
| AHJ94065 | Rubredoxin/Rubredoxin Reductase | — |
| AHJ94066 | Aldehyde Dehydrogenase | — |
| AHJ94067 | 2Fe-2S Ferredoxin | — |
| AHJ94068 | Choline Transporter | — |
| AHJ94069 | Choline Transporter | — |
| AHJ94070 | LysR Family Transcriptional Regulator | — |
| AHJ94071 | GABA Permease | — |
| AHJ94072 | Methylmalonate-Semialdehyde Dehydrogenase | — |
| AHJ94073 | Hypothetical Protein | — |
| AHJ94074 | Amino Acid Oxidase | — |
| AHJ94075 | Polyketide Synthase Regulator | — |
| AHJ94076 | TetR Family Transcriptional Regulator | — |
Additional manual search performed using BASys20 revealed the presence of various classes of efflux pump coding genes such as RND type, ATP binding cassette type (ABC), small multidrug resistance (SMR), major facilitator super family (MFS) and multidrug and toxic compound extrusion (MATE). In addition, genes responsible for metabolising/modifying antibiotics of various classes such as β-lactams, aminoglycosides, glycopeptides, polypeptides, macrolides and sulfa drugs were found to be dispersed throughout the assembled genome. A total of 18 predicted genes responsible for providing tolerance to metals such as arsenic, copper, cobalt, zinc, cadmium, tellurium, mercury and chromium were present across the genome. Further, numerous other genes responsible for stress tolerance, resistance to inorganic compounds and dyes as well as multi-drug resistance mediating putative transcription regulators were also detected that could contribute significantly to the persistence of A. baumannii. RAST annotation has revealed a total of 102 RNA coding genes. Twenty-one rRNA genes were present (7 each of 5SrRNAs, 16S rRNAs, and 23S rRNAs) in PKAB07 that was demonstrated through RNAmmer analysis. Lastly, CRISPR related genes were not found in this genome.
Comparative genomics
A. baumannii PKAB07 may not apparently harbour any unique islands, as revealed by our comparative genome analysis. Nevertheless the mosaic genomic structure of PKAB07 is evident by the scattered core genome interlaced by 202 long and short accessory genome segments (Fig. 1). One of the most important gene segments 29561 bp (genomic coordinates 2875478 – 2905039, Table 3), is present in 21 other genomes including PKAB07. This segment has 23 ORFs, which include many coding DNA sequences for various genes such as different transcriptional regulators (mainly TetR family of regulators), genes that facilitate transporting antibiotics, small molecules and toxins. It is interesting to note that, this region is always associated with CC-92 of IC-II. This could be a significant factor in making this clone successful globally.
Figure 1.

Circos plot showing SNP-distribution in the genome of A. baumannii PKAB07 and its correlation with accessory genome and the GC-skew (From inside out) Track-1 (green) - regions that are absent in at least one of the genomes (the accessory genome). Track-2 (red) – The chromosome of A. baumannii PKAB07. Track-3 (purple/orange) – Histogram showing GC-skew. Track-4 (red) – Scatter plot of SNP count/kb. Note the scattering of the plot outward at genomic regions where either a steep deviation from the GC-skew occurs or a syntenic link appears, or both (particularly, regions around 0.1 –0.3 Mb, 0.7–1 Mb, 1.2 Mb, 2–2.3 Mb; the outer numbers showing genomic coordinates in mb)
Two component systems (TCS)
PKAB07 has at least 21 TCSs, one of the highest numbers for A. baumannii revealed through analyzing P2CS database.36 According to the database the histidine kinases (HKs) were classified as classic, hybrid and unorthodox or CheA family, and those lack HATPase or receiver domains were classified as phospho-transfer proteins. The response regulators (RRs) fell into OmpR, NarL, NtrC, LytR and CheY families. We found 18 HKs and 17 RRs and 2 hybrids in A. baumannii PKAB07, with the HisKA, HATPase and the receiver domains residing in the same protein molecule. Together, these constituted 14 cognate pairs of HK-RRs; whereas no detectable partner was found for the rest of the 7 TCS
Genome plasticity and evolution
A total of 43 fully sequenced genomes representing isolates from diverse geographical regions such as Asia (India, China and Malaysia), European (France, Italy and Korea), the Americas (USA) and Australia, which made the study demographically unbiased were analyzed. PKAB07 is one of the largest among the genomes analyzed and some of main feature of the genome is given (supplementary file SM1). Strain PKAB07, stands out from the rest as shown in the unrooted tree (Fig. 2). The resistance, virulence and stress response modules were found to be scattered throughout the core and accessory genomes which is very significant and is not fully understood. But this does indicate that the organism had acquired genomic regions by recombination or horizontal transfers37 albeit intermittently, resulting in a scattered accessory genome devoid of any island. Moreover, the presence of numerous insertion elements had further augmented the genome rearrangements. It is interesting to note that due to the tremendous selective pressure A. baumannii is subjected to while thriving in hostile environments, the organism successfully managed to disperse the resistance, virulence and stress response modules throughout the core and accessory genome. This may give A. baumannii immense flexibility to re-model a sub-set of its entire repertoire of genes rather than just organizing these modules into a ‘patch’ of the island and just gaining or losing it as a whole. This phenomenon might explain how it helps A. baumannii in driving forward to a positive selection and evolution. Finally, the evolutionary potential of this bacterium is evident due to the presence of a large number of transposases (n = 60) that includes both full length and remnant transposases, though the significance of remnants is unclear.
Figure 3.
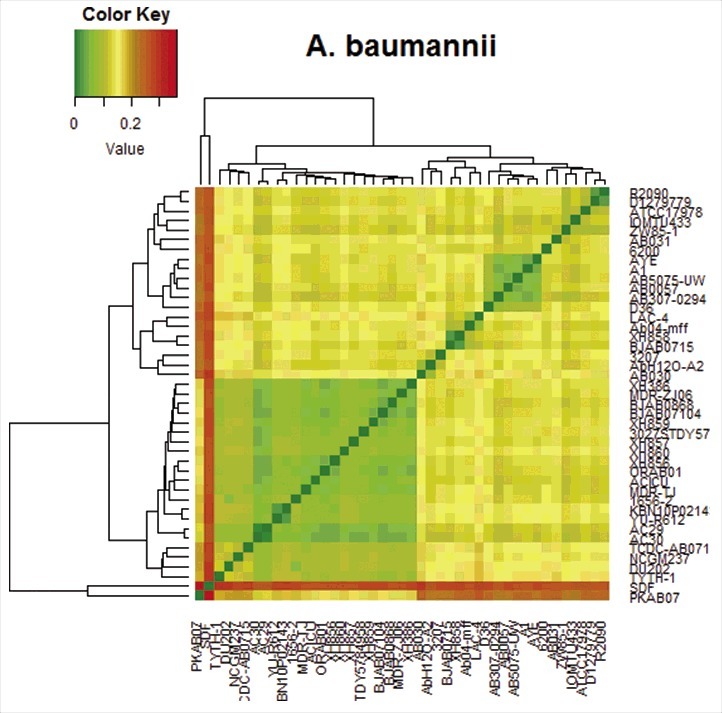
Heat map based on the distance matrix generated by ProgressiveMauve.26 The pair-wise distance matrix was used to generate the heatmap using R program (http://www.r-project.org/). The legend shows the gradient color-code to the values in the distance matrix.
Figure 2.
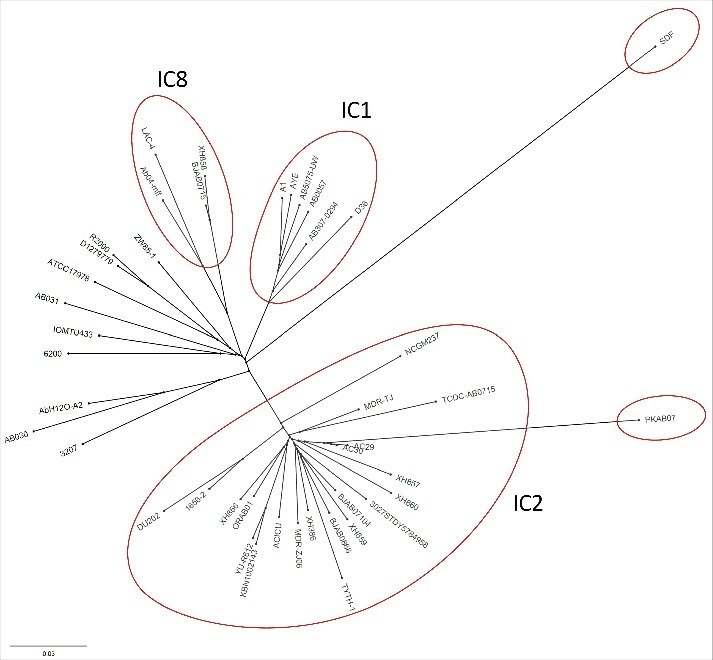
Unrooted phylogenetic tree representing the relatedness among the genomes under study. The tree file was generated by ProgressiveMauve and was visualized in FigTree (Version 1.4.1) (http://tree.bio.ed.ac.uk/software/figtree/). The PKAB07, despite closest to the clonal AC29, stands out in the tree, where as the rest of the strains clustered into 3 clades, a tree similar to that of Tan et al., (2013).
Single nucleotide polymorphisms (SNPs)
The distribution of SNPs was found to be elevated around the accessory genome as depicted in circus plot (Fig. 1). Recent studies reveal that IC-II lineage have accumulated an elevated number of SNPs, which are located preferentially around regions that are subjected to frequent homologous recombination events38-40 These SNPs are probably the products of homologous recombination events and represent recombination hot-spots. This rapid accumulation of SNPs is indicative of rapidness at which A. baumannii clonal lineage has been evolving.40 GC-content has always been the gold standard for identifying HGT in prokaryotes41 but this study did not find any shift in the GC content associated with elevated SNPs (Fig. 1). Since the elevated SNPs distribution around accessory genome did not always represent a deviation in the GC-skew while mining for the regions of HGT one should also take SNPs into consideration.
Gene expression modulation due to ISAba1 insertion
ISAba1 of A. baumannii was previously shown to modulate the expression of resistance related genes by providing hybrid promoters or by disrupting the global transcriptional repressor protein encoding genes such as H-NS through insertional inactivation leading to increased resistance and virulence.4,7,12,13 IS-Finder identified ISAba1 to be present in 26 copies in PKAB07, the highest number among the sequenced A. baumannii genomes and their exact positions were also located using ACT Artemis (Fig. 4). Interestingly, we identified 7 novel ISAba1 mediated gene disruptions in 7 coding DNA sequences (CDS) and 3 ISAba1 insertions in the upstream of genes namely TonB-dependent receptor plug domain, ampC and putative acetyl transferase resulting in overexpression of these genes. Except ampC, all the upstream insertions and gene disruptions observed were in novel regions which codes for transcriptional regulators, repressors, outer membrane proteins and receptors, which was not reported hitherto. The insertions were confirmed by PCR wherein genes encoding transcription regulator (AdeN), cAMP binding protein, TonB dependent siderophore receptor (OMRC), putative outer membrane protein (PTP or OMPX), a hypothetical protein (HYP), dehydrogenase with different specificities (DDS) and putative dehydrogenase (PDH),which are disrupted, generated the amplicons of larger size that included the size of IS element along with gene size.
Figure 4.

Location of ISAba1 loci in the genome of A. baumannii PKAB07 illustrated using CGview. Manual input through numerals for displaying the positions of ISAba1 throughout the genome was fed in CG view tool and the image was generated.
Over expression due to ISAba1 insertion
Insertion of ISAba1 in upstream of the genes such as ampC, putative acetyl transferase (PAT) and TonB-dependent receptor plug was observed in PKAB07. Over-expression of ampC has been previously reported attributing to ISAba1 insertion upstream of this gene.42 PAT of PKAB07 is an acetyltransferase belonging to GCN5-related N-acetyltransferase (GNAT) super family that was first identified as aminoglycoside acetyltransferases in bacteria responsible for the development of resistance to the aminoglycoside antibiotics.43 Though very few GNATs have been characterized till date there are large number of known substrates for them that include antibiotics, small molecules and homoserine lactones that help in bacterial survival and pathogenesis.43 ISAba1 insertion in PKAB07 is in the upstream of PAT gene probably might have created a hybrid promoter that was found to be responsible for overexpression of this gene leading to aminoglycoside resistance in PKAB07. qRT-PCR experiments revealed 10-fold increase in expression of PAT compared with wild type in this study, which is significant (Fig. 5). In the same manner acylation of other antibiotics and homoserine lactones can also be anticipated since these factors are required for its survival and invasion of host.
Figure 5.
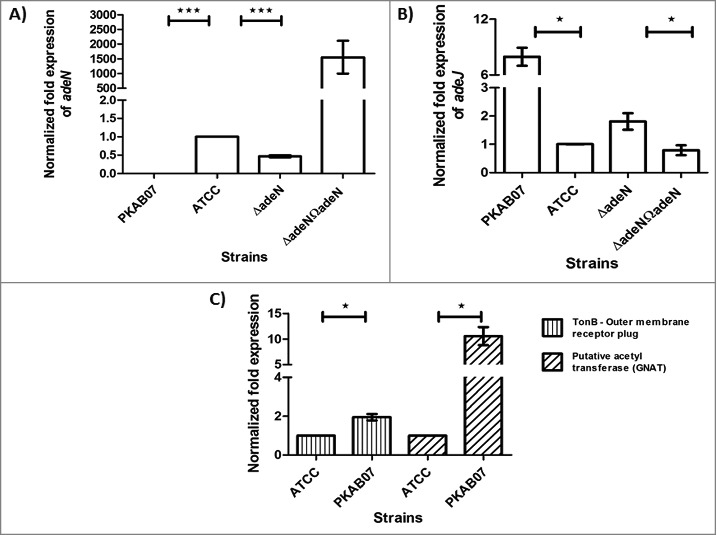
Quantitative Real-time PCR experiments for determining the expression levels of genes that are affected by ISAba1. Expression levels of adeN, adeJ, PAT and TonB receptor plug domain genes in A. baumannii strains ATCC 17978, ATCC ΔadeN, adeN complement and PKAB07. mRNA levels were determined by quantitative real-time PCR and normalized to the expression of rpoB in each sample. All data are representatives of at least 3 independent experiments. Statistical significance was derived using 2-tailed paired Student's t-test *, P < 0.05; **, P < 0.01; ***, P< 0.00 1. Error bars represent the standard error (SE) of the mean.
TonB dependent siderophore receptor mutations results in avirulent phenotypes in Gram-negative bacteria according to earlier studies.44 TonB dependent siderophore receptor gene in PKAB07 is disrupted by ISAba1, showed 41% similarity and 23% identity with fpvA(a TonB receptor homolog) found in Pseudomonas syringae.45 Mutants of fpvA showed reduced ability to produce EPS, increased tolerance to antimicrobial agents, accelerated swarming ability and increased biosurfactant production, indicating that swarming motility and biosurfactant production might be negatively controlled by siderophore synthesized.45,46 In addition, TonB-dependent receptor plug domain has also been identified in PKAB07, which has sequence similarity with the plug domains from other structurally characterized TonB-dependent transporters.47 The receptor plug domains of TonB-dependent transporters can fold independently after translocation across the inner membrane, with subsequent insertion of the folded plug domain into the L-barrel. However, extracellular loops of the L-barrel are required for ligand binding.47 This domain structure suggests that such a plug might be stable even when expressed in the absence of the transmembrane L-barrel. We found ISAba1 insertion upstream of TonB-dependent receptor plug domain gene in PKAB07, which is likely leading to overexpression of this plug that locks the L-barrel possibly blocking the transport of siderophores as reported elsewhere.47 Indeed, this study experimentally proved that overexpression of plug domain was happening in PKAB07 by performing by qPCR experiment and quantifying the transcripts of plug domain (Fig. 5).
ISAba1 mediated control of multidrug efflux systems
When we analyzed multiple genomes for studying efflux pump genes and majority of them appeared to be conserved. However, a few efflux pump gene losses were found. The most apparent was the loss of 5 efflux pump genes by the strain SDF (Fig. 6), which was isolated from body lice.48 SDF is a commensal strain that was found to be highly susceptible to antibiotics. In this regard, we believe that it is more appropriate for the strain to lose many of these efflux pump genes as it is not required in a niche it thrives. This study more importantly observed ISAba1 insertion in a negative regulator gene of adeIJK efflux pump.
Figure 6.
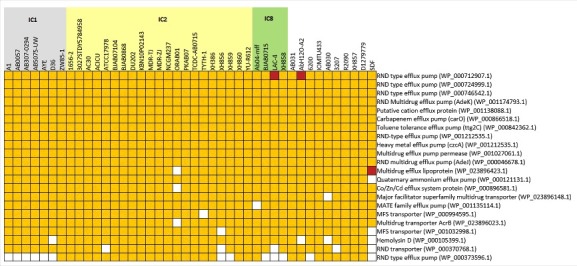
RND-type efflux pump genes among the sequenced genomes. 22 clusters of orthologous genes belonging to RND-type efflux pumps and efflux pumps that cause antibiotic resistance were mapped against 43 genomes that were analyzed, showing the presence (yellow) or absence (white) of the orthologous genes. Brown colored squares indicate the presence of 2 paralogs in the same genome. Strains that belong to IC1, IC2 and IC8 are shaded gray, yellow and green, respectively.
Among the 3 major diversely regulated multidrug efflux systems reported so far in A. baumannii, AdeIJK efflux system is controlled by a TetR regulator namely AdeN.49,50 The TetR family is one of the commonest transcriptional regulator families in bacteria that are often involved in the regulation of efflux systems by acting as repressors.51,52 It has been established that interaction of a ligand with the LBD domain of the transcription regulator leads to a conformational change that interferes with the DNA-binding ability of the protein, thus relieving transcription repression.52 We identified ISAba1 mediated disruption of adeN in PKAB07 that had eased out adeN repression of AdeIJK and thereby allowing the uncontrolled expression of genes of the adeIJK operon.53 The results of qRT-PCR for adeJ, one of the major component of AdeIJK efflux pump in this study showed a significant overexpression in PKAB07 (8-fold) and experimental adeN knockout (2-fold) when compared with control (Fig. 5). Abrogation of adeN repressor activity due to ISAba1 disruption could have led to multiple drug resistance in PKAB07 (Table 4). Moreover, adeN has been shown to be responsible for the repression of the adeIJK operon only in susceptible strains of A. baumannii.53
Table 4.
Determination of MIC for 7 antibiotics in 4 strains included in the study.
| Antimicrobial agents* |
|||||||
|---|---|---|---|---|---|---|---|
| Strain tested | CIP | CHL | CAZ | TGC§ | MRP | TOB | AMK |
| PKAB07 | 32 | 16 | <0.16 | 1.5 | >256 | >256 | >256 |
| ΔadeN | <0.5 | 16 | >256 | 0.19 | >256 | <0.5 | <0.5 |
| ΔadeNΩadeN | <0.5 | <0.5 | >256 | 0.125 | >256 | <0.5 | <0.5 |
| ATCC 17978 | <0.5 | <0.5 | >256 | 0.125 | >256 | <0.5 | <0.5 |
CIP-Ciprofloxacin, CHL- Chloramphenicol, CAZ-Ceftazidime, TGC- Tigecycline, MRP- Meropenem, TOB-Tobramycin, AMK- Amikacin.
E- strips were used for determination of MIC
AdeIJK pump appears to be contributing to intrinsic resistance toward many antibiotics including β-lactams and tigecycline.11 Since, overexpression of the adeIJK operon has been shown to be toxic in E. coli and Acinetobacter it is suggested that expression of this gene cluster is tightly regulated.11 Additionally, adeN repression of adeIJK only observed in susceptible strains of A. baumannii.53 Insertional inactivation of adeN might have led to diminished susceptibility to antibiotics that are substrates for the AdeIJK. Natural overexpression of adeIJK efflux pump is unlikely since there exists a threshold for toxicity. However, disruption of adeN, which seemed to be lethal leading to overexpression of adeIJK initially, is appeared to be slowly evolving into an intrinsic low level of expression tolerable to A. baumannii. Such observations confirm tight control of AdeN over AdeIJK efflux pump and for the first time we have observed naturally occurring insertional inactivation of adeN by ISAba1.
Role of adeN in regulating virulence of A. baumannii
This study attempted to investigate any other important functions that can be attributed to adeN that might help in pathogenesis of A. baumannii. Since, transcription regulator adeN appeared to have significant additional physiologic roles, other than the controlling efflux mechanisms such as virulence and persistence, adeN gene knockouts were investigated initially for their abilities to form biofilm and induce apoptosis in co-culturing conditions.
Construction of adeN knockout in A. baumannii ATCC 17978
The adeN knockout strain was constructed through homologous recombination followed by gene replacement as described elsewhere.54 ΔadeN construct was prepared by placing kanamycin cassette in the middle with flanking upstream and downstream regions of adeN. Electroporation of competent cells with the ΔadeN construct formed around 80 colonies on the LB agar plate containing kanamycin (100µg/ml). Subsequently, the colonies were screened by PCR for the presence of disrupted adeN with a amplicon size of ∼1.2 kb that is larger than the adeN gene size (654bp). All the colonies were found to possess this amplicon i.e. ΔadeN construct, thereby confirming to be mutant colonies with defective adeN.
adeN disruption affects biofilm formation
When the ΔadeN and wild type strain ATCC 17978 were tested for the extent of biofilm production through microtiter plate assay, adeN mutation nearly abolished the biofilm forming ability of A. baumannii. When compared with the wild type strain, more than threefold reduction in the amount of biofilm formed by mutant strains was witnessed. A similar scenario was noted in PKAB07, which had natural disruption of adeN because of ISAba1 insertion. To further evaluate the importance of adeN in biofilm formation, a replicative plasmid of Acinetobacter pWH1266 carrying full-length intact adeN (pWH1266::adeN) was electroporated into ΔadeN mutant strain for complementation. Biofilm level in the complemented strain was significantly restored though it was slightly lesser than the wild type strain (Fig. 7). Our results reveal that adeN knockout fails to form biofilm, which was restored in the complemented strain. Such results provide concrete evidence that adeN could be a positive regulator in controlling the process of biofilm formation in A. baumannii. Moreover, complemented strain showed very high level expression of adeN, which is expected. Similar results of overexpression was also found by Tomaras et al., 200855 and Mussi et al., 201056 when they complemented bfmR and blsA alleles of A. baumannii in pWH1266 respectively. Indeed, they suggest such overexpression is most probably due to high copy number of the allele (in our case it is adeN) in pWH1266 derived complemented mutant generated from parental ATCC 17978 strain.
Figure 7.

Biofilm levels in adeN mutant and wild type strains - Microtitre plate biofilm assay was performed for A. baumannii strains ATCC 17978, ATCC ΔadeN, adeN complement and PKAB07 and the results were analyzed. Statistical significance was derived using 2-tailed Student's t-test (**, p < 0.005; ***, p < 0.001). Experiments were performed in triplicates and error bars represent the standard deviations.
AO/EB staining and Annexin V assay revealed ΔadeN to be hyper virulent
Based on the results observed for the PKAB07 strain which was hypervirulent as well harboured mutated adeN, this study looked into the role of adeN in conferring virulence to A. baumannii. When the wild type and adeN mutant strains were co-cultured with the A549 cells for 24 h, there was a drastic increase in the virulence potential of the adeN mutant strain, which reduced the viability of A549 cells significantly (Fig. 8A). Conversely, the wild type and complemented strains did not induce any notable cell death (Fig. 8A). Early apoptotic and the late apoptotic population of A549 cells after co-culturing with A. baumannii were analyzed using flow cytometer. Flow cytometry data was analyzed by combining the plots from both the upper right and lower right quadrants, which shows the population of both apoptotic and early apoptotic cells. About 5,000 events were captured for each sample and the percentage of annexin V positive cells were calculated. ΔadeN strain induced more cell death (60%) when compared with the wild type ATCC (38%) and ATCC 17978 ΔadeNΩadeNstrain (14%) (Fig. 8B). The results of ΔadeN strain and PKAB07 (58%) that had adeN disrupted by ISAba1 were almost similar (Fig. 9).
Figure 10.
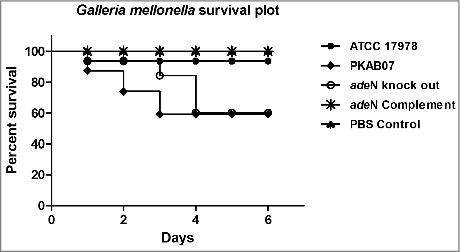
Survival Plot of Galleria mellonella larvae infected with the adeN knockout, wild type and complemented strains. PKAB07 and ΔadeN strains were found to be hyper- virulent and killed relatively higher number of larvae when compared with controls, ATCC 17978 and PBS. The virulence was compromised when G. mellonella infected with the complemented strain of ΔadeN with pWH1266::adeN plasmid.
Figure 8.

(A) Co-culturing followed by AO/EB stainingA549 cells co-cultured with A. baumannii strains were added with AO/EB viability check stain and observed under fluorescent microscope B-2A filter (Nikon Eclipse TS100, Japan). PKAB07 and ΔadeN strains were found to be hyper invasive and induced more cell death as evidenced by the cellular uptake of ethidium bromide by A549 cells (B & C) when compared with the uninfected control (A). The virulence was compromised when the ΔadeN strain was complemented with pWH1266::adeN plasmid (D). Scale bar represents 50µm at 40X magnification.
Figure 9.

Annexin (V)expression analysis through flow-cytometer. Expression of Annexin V was evaluated using Annexin V-FITC Apoptosis Detection Kit (Sigma Aldrich, USA). A549 cells infected with A. baumannii strains ATCC 17978, ATCC ΔadeN, adeN complement and PKAB07 were analyzed after incubation for 24 h at 37°C, and statistical significance was derived using 2-tailed Student's t test (*x, P < 0.05; ***, P < 0.001). Experiments were performed in triplicates and error bars represent the standard deviations.
Galleria mellonella killing assays
The degree of virulence in different strains of A. baumannii was determined by G. mellonella killing assays. The sub-optimal dose of ATCC 17978 which did not affect the survival of the larvae up to 6 d of infection was found to be 6.4 × 104, 5.2 × 104, 5.7 × 104 and 5.9 × 104 for PKAB07, ATCC, ΔadeN and complemented strain respectively. The dose of the A. baumannii inoculum was fixed based on the earlier observations.31 Among the injected strains, PKAB07 and ΔadeN were found to be more virulent and causing increased mortality by killing more number of larvae than the ATCC 17978 and ATCC 17978 ΔadeNΩadeN strain (Fig. 9).Previously, it was known that adeN represses the expression of AdeIJK efflux pump and it was also speculated that AdeIJK efflux pump proteins could be important to manifest virulence as their absence abrogated the virulence in A. baumannii.57 Lack or down regulation of AdeIJK genes may cause defective effluxing, wherein defense related toxins of G. mellonella that have entered the bacteria are likely to be retained within leading to subsequent killing of bacteria thereby helping the insect survival.57 Besides, our findings substantiate that the overexpression of adeIJK has direct influence on elevation of virulence, in addition to increased efficiency of effluxing. This result confirms the fact that AdeN has a definitive role in controlling the expression of virulence related genes in A. baumannii.
Elevated resistance due to adeN disruption by ISAba1
AdeIJK efflux system has a wide range of substrates, which is very similar to those of the AcrAB-TolC and MexAB-OprM systems.58,59 AdeIJK efflux pump system contributes to resistance toward β-lactams, chloramphenicol, tetracycline, erythromycin, lincosamides, fluoroquinolones, fusidic acid, novobiocin, rifampin, trimethoprim, acridine, safranin, pyronine and sodium dodecyl sulfate.11 Besides, it works in an additive fashion to bring tigecycline resistance along with AdeABC efflux system. It is also suggested that AdeIJK may have more important physiologic roles other than efflux pumping.11 One recent investigation identified AdeN repressing the expression of the adeIJK operon.53 Rosenfeld et al. (2012) experimentally deleted the adeN gene partially in A. baumannii by insertion-inactivation to check the regulation of AdeIJK expression.53 Experimental deletion of adeN resulted in elevated resistance to multiple antibiotics that are likely substrates of the AdeIJK efflux pump. The present study, through whole-genome sequencing approach, for the first time identified naturally occurring ISAba1 insertion in adeN gene in PKAB07 strain of A. baumannii leading to insertion-inactivation of adeN, which may have resulted in diminished susceptibility of this strain to various classes of antibiotics. The antibiotic susceptibility results of PKAB07, ΔadeN and complement of knockout proved this point (Table 4).
Based on the in vitro assay results observed for the PKAB07 strain, which was hypervirulent that harboured a disrupted adeN, we intended to study the role of adeN in conferring virulence to A. baumannii. Interestingly, when the wild type and adeN mutant strains were co-cultured with the A549 cells for 24 h, there was a drastic increase in the virulence potential of the adeN mutant strain which reduced the viability of A549 cells. Conversely, the wild type strain did not induce any notable cell death (Fig. 8 and 9). This experimental result confirms that adeN could be a negative regulator of virulence, and adeN disruption in A. baumannii causes this bacteria to behave abnormally with elevated virulence properties.
The regulatory networks controlling the expression of A. baumannii virulence factors such as metal acquisition, motility, adhesion, invasion and biofilm formation remains largely unknown. A recent study compared the genome sequences of hyper motile strains and wild type strains, and revealed that IS mediated disruption of H-NS like genes resulted in enhanced virulence/motility.13 It was also shown that this H-NS encoding gene is a common homolog of histone-like nucleotide structuring protein of E. coli, which is also known to be a major global transcriptional repressor. It is interesting to note that similar ISAba1mediated disruption of TetR regulator adeN through insertional inactivation in PKAB07 resulted in various alterations including increased virulence potential, hyper invasiveness and global transcriptional changes.13
Many earlier studies show positive correlation between biofilm formation and virulence in A. baumannii wherein the components of biofilm have been associated with host inflammatory response. In our study there exists an interesting scenario wherein the strain was found to be a moderate biofilm producer but possess high virulence. One previous study has observed increased expression of AdeIJK efflux pump when adeN was experimentally disrupted53 and such an overexpression might also lead to reduced fitness.10 Another complementing study showed AdeIJK efflux pump defective strains were compromised in their virulence potential when compared with their wild type counterparts and these findings clearly indicate that normal functioning of AdeIJK efflux pump is pivotal requirement for causing host cell damage.57 It has also been shown that in MDR clinical isolates, a functional AdeIJK is likely to be responsible for the intrinsic low-level resistance for different antibiotics at the same time having high capacity to form biofilms with increased adhering ability to respiratory epithelial cells, leading to increased virulence.11,60 Contradictorily, one recent study revealed that proteomic analysis of bacterial membranes in overexpressing mutants of adeIJK had resulted in under expression of proteins belonging to chaperone-usher (CU) pilus assembly systems (CsuA/B and CsuC & FimA fimbrial protein). Both pilins CsuA/B and FimA have been detected in the matrix of different A. baumannii pellicles,61 and the disruption of both CU systems results in a severe decrease in biofilm formation on abiotic surfaces.62,63 Interestingly, such a negative correlation between overexpression of efflux systems leading to antibiotic resistance and reduction in biofilm formation has also been recently observed in a transcriptomic analysis of imipenem-selected A. baumannii.64 Taken together, there is fair chance of occurrence of phenomenon wherein biofilm formation and antibiotic resistance could be controlled antagonistically by certain negative regulators such as adeN as it is seen in our case. Therefore, adeN disruption in PKAB07 and adeN knock out leading to reduction in the amount of biofilm signifies that biofilm may not enhance the virulence in all circumstances.
The present study, for the first time describes the alternative physiologic roles of adeN where in it possibly appears to be a smart negative regulator of genes associated with virulence such as biofilm formation, host cell killing, which are not its primary role. Moreover, naturally occurring adeN disruptions by mobile genetic elements may help A. baumannii to evolve in the stressful micro-environment of the host. In summary, the phenotypic and genotypic analysis performed using strains such as ATCC 17978, PKAB07, adeN mutants, complemented mutant strain revealed a significant role for adeN in the regulation of A. baumannii virulence associated genes.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are very grateful to Dr. Chung-Yu Lan, Institute of Molecular and Cellular Biology, National Tsing Hua University, Hsin-Chu City, Taiwan for providing us pWH1266 plasmid construct that helped us in complementation experiments. Authors also would like to thank Dr. Arunkumar Dhayalan, Assistant Professor, Department of Biotechnology, Pondicherry University for helping us in analysis of real-time PCR data. Faculty members of Microbiology Departments of Pondicherry Institute of Medical Sciences, Puducherry and Adichunchanagiri Institute of Medical Sciences, Bellur are gratefully acknowledged for providing clinical isolates of Acinetobacter baumannii.
Funding
KP thank the University Grant Commission (UGC) (F No. 36–190/2008(SR)) and Indian Council of Medical Research (ICMR) (No: 5/3/3/14/2007-ECD-I), Government of India for project funding.
References
- [1].Dent LL, Marshall DR, Pratap S, Hulette RB. Multidrug resistant Acinetobacter baumannii: a descriptive study in a city hospital. BMC Infect Dis 2010; 10:196; PMID:20609238; https://doi.org/ 10.1186/1471-2334-10-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Katsaragakis S, Markogiannakis H, Toutouzas KG, Drimousis P, Larentzakis A, Theodoraki EM, Theodorou D. Acinetobacter baumannii infections in a surgical intensive care unit: predictors of multi-drug resistance. World J Surg 2008; 32:1194-202; PMID:18408967; https://doi.org/ 10.1007/s00268-008-9571-3 [DOI] [PubMed] [Google Scholar]
- [3].Zarrilli R, Pournaras S, Giannouli M, Tsakris A. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents 2013; 41:11-9; PMID:23127486; https://doi.org/ 10.1016/j.ijantimicag.2012.09.008 [DOI] [PubMed] [Google Scholar]
- [4].Adams MD, Chan ER, Molyneaux ND, Bonomo RA. Genome wide analysis of divergence of antibiotic resistance determinants in closely related isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 2010; 54:3569-77; PMID:20530228; https://doi.org/ 10.1128/AAC.00057-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Corvec S, Caroff N, Espaze E, Giraudeau C, Drugeon H, Reynaud A. AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J Antimicrob Chemother 2003; 52:629-35; PMID:12951337; https://doi.org/ 10.1093/jac/dkg407 [DOI] [PubMed] [Google Scholar]
- [6].Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, Pitt TL. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett 2006; 258:72-7; PMID:16630258; https://doi.org/ 10.1111/j.1574-6968.2006.00195.x [DOI] [PubMed] [Google Scholar]
- [7].Lee Y, Kim CK, Lee H, Jeong SH, Yong D, Lee K. A novel insertion sequence, ISAba10, inserted into ISAba1 adjacent to the blaOXA-23 gene and disrupting the outer membrane protein gene carO in Acinetobacter baumannii. Antimicrob Agents Chemother 2011; 55:361-63; PMID:20937784; https://doi.org/ 10.1128/AAC.01672-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Durante-Mangoni E, Zarrilli R. Global spread of drug-resistant Acinetobacter baumannii: molecular epidemiology and management of antimicrobial resistance. Future Microbiol 2011; 6:407-22; PMID:21526942; https://doi.org/ 10.2217/fmb.11.23 [DOI] [PubMed] [Google Scholar]
- [9].Yoon EJ, Chabane YN, Goussard S, Snesrud E, Courvalin P, Dé E, Grillot-Courvalin C. Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. MBio 2015; 6:e00309-15; https://doi.org/ 10.1128/mBio.00309-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yoon EJ, Balloy V, Fiette L, Chignard M, Courvalin P, Grillot-Courvalin C. Contribution of the Ade resistance-nodulation-cell division-type efflux pumps to fitness and pathogenesis of Acinetobacter baumannii. MBio 2016; 7:e00697-16; https://doi.org/ 10.1128/mBio.00697-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Damier-Piolle L, Magnet S, Brémont S, Lambert T, Courvalin P. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob Agents Chemother 2008; 52:557-62; PMID:18086852; https://doi.org/ 10.1128/AAC.00732-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sun JR, Perng CL, Chan MC, Morita Y, Lin JC, Su CM, Wang WY, Chang TY, Chiueh TS. A truncated AdeS kinase protein generated by ISAba1 insertion correlates with tigecycline resistance in Acinetobacter baumannii. PLoS One 2012; 7:e49534; PMID:23166700; https://doi.org/ 10.1371/journal.pone.0049534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Eijkelkamp BA, Stroeher UH, Hassan KA, Elbourne LD, Paulsen IT, Brown MH. H-NS plays a role in expression of Acinetobacter baumannii virulence features. Infect Immun 2013; 81:2574-83; PMID:23649094; https://doi.org/ 10.1128/IAI.00065-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Antunes LC, Imperi F, Carattoli A, Visca P. Deciphering the Multifactorial Nature of Acinetobacter baumannii pathogenicity. Plos One 2011; 6:e22674; PMID:21829642; https://doi.org/ 10.1371/journal.pone.0022674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Penwell WF, Arivett BA, Actis LA. The Acinetobacter baumannii entA gene located outside the acinetobactin cluster is critical for siderophore production, iron acquisition and virulence. PLoS One 2012; 7:e36493; PMID:22570720; https://doi.org/ 10.1371/journal.pone.0036493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tayabali AF, Nguyen KC, Shwed PS, Crosthwait J, Coleman G, Seligy VL. Comparison of the virulence potential of Acinetobacter strains from clinical and environmental sources. PLoS One 2012; 7:e37024; PMID:22655033; https://doi.org/ 10.1371/journal.pone.0037024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Antunes LC, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis 2014; 71:292-301; PMID:24376225; https://doi.org/ 10.1111/2049-632X.12125 [DOI] [PubMed] [Google Scholar]
- [18].Saranathan R, Tomar A, Sudhakar P, Arunkumar KP, Prashanth K. Draft genome sequence of a multidrug-resistant Acinetobacter baumannii PKAB07 clinical strain from India belonging to sequence type 195. Genome Announc 2014; 20:2; https://doi.org/ 10.1128/genomeA.00184-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res 2011; 39:W347-52; https://doi.org/ 10.1093/nar/gkr485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Van Domselaar GH, Stothard P, Shrivastava S, Cruz JA, Guo A, Dong X, Lu P, Szafron D, Greiner R, Wishart DS. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res 2005; 33:W455-W459; PMID:15980511; https://doi.org/ 10.1093/nar/gki593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, et al.. Multilocus sequence typing of total genome sequenced bacteria. J Clin Micobiol 2012; 50:1355-61; https://doi.org/ 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Grissa I, Vergnaud G, Pourcel C. CRISPR Finder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 2007; 35:W52-7; PMID:17537822; https://doi.org/ 10.1093/nar/gkm360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. IS-finder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 2006; 34:D32-6; PMID:16381877; https://doi.org/ 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics 2005; 21:537-539; PMID:15479716; https://doi.org/ 10.1093/bioinformatics/bti054 [DOI] [PubMed] [Google Scholar]
- [25].Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. ACT: the Artemis Comparison Tool. Bioinformatics 2005; 21:3422-23; PMID:15976072; https://doi.org/ 10.1093/bioinformatics/bti553 [DOI] [PubMed] [Google Scholar]
- [26].Darling AE, Mau B, Perna NT. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 2010; 5:e11147; PMID:20593022; https://doi.org/ 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res 2009; 19:1639-45; https://doi.org/ 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Uchiyama I. MBGD: microbial genome database for comparative analysis. Nucleic acids res 2003; 31:58-621; PMID:12519947; https://doi.org/ 10.1093/nar/gkg109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing: twenty-third informational supplement M100-S23 CLSI 2013; CLSI, Wayne, PA, USA [Google Scholar]
- [30].Rao RS, Karthika RU, Singh SP, Shashikala P, Kanungo R, Jayachandran S, Prashanth K. Correlation between biofilm production and multiple drug resistance in imipenem resistant clinical isolates of Acinetobacter baumannii. Indian J Med Microbiol 2008; 26:333-7; PMID:18974485; https://doi.org/ 10.4103/0255-0857.38876 [DOI] [PubMed] [Google Scholar]
- [31].Peleg AY, Jara S, Monga D, Eliopoulos GM, Jr Moellering RC, Mylonakis E. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob Agents Chemother 2009; 53:2605-9; PMID:19332683; https://doi.org/ 10.1128/AAC.01533-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Longo B, Pantosti A, Luzzi I, Agapito T, Di Sora F, Gallo S, Placanica P, Monaco M, Dionisi AM, Volpe I, Montella F, Cassone A, and Rezza G. Molecular findings and antibiotic-resistance in an outbreak of Acinetobacter baumannii in an intensive care unit. Ann Ist Super Sanita 2007; 43:83-8; PMID:17536158 [PubMed] [Google Scholar]
- [33].Iacono M, Villa L, Fortini D, Bordoni R, Imperi F, Bonnal RJ, Sicheritz-Ponten T, De Bellis G, Visca P, Cassone A and Carattoli A. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob agent chemother 2008; 52:2616-25; https://doi.org/ 10.1128/AAC.01643-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F et al.. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008; 9:75; PMID:18261238; https://doi.org/ 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67:2640-44; PMID:22782487; https://doi.org/ 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Barakat M, Ortet P, Jourlin-Castelli C, Ansaldi M, Méjean V, Whitworth DE. P2CS: a two-component system resource for prokaryotic signal transduction research. BMC Genomics 2009; 15:315; https://doi.org/ 10.1186/1471-2164-10-315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Traglia GM, Chua K, Centrón D, Tolmasky ME, Ramírez MS. Whole-Genome Sequence Analysis of the Naturally Competent Acinetobacter baumannii Clinical Isolate A118. Genome Biol Evol 2014; 6:2235-39; PMID:25164683; https://doi.org/ 10.1093/gbe/evu176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Snitkin ES, Zelazny AM, Montero CI, Stock F, Mijares L; NISC Comparative Sequence Program, Murray PR, Segre JA et al.. Genome-wide recombination drives diversification of epidemic strains of Acinetobacter baumannii. PNAS 2011; 108:13758-63; PMID:21825119; https://doi.org/ 10.1073/pnas.1104404108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wen H, Wang K, Liu Y, Tay M, Lauro FM, Huang H, Wu H, Liang H, Ding Y, Givskov M et al.. Population dynamics of an Acinetobacter baumannii clonal complex during colonization of patients. J Clin Microbiol 2014; 52:3200-08; PMID:24951812; https://doi.org/ 10.1128/JCM.00921-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Tan SY, Chua SL, Liu Y, Høiby N, Andersen LP, Givskov M, Song Z, Yang L. Comparative genomic analysis of rapid evolution of an extreme-drug-resistant Acinetobacter baumannii clone. Genome Biol Evol 2013; 5:807-18; PMID:23538992; https://doi.org/ 10.1093/gbe/evt047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].McLean MJ, Wolfe KH, Devine KM. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J Mol Evol 1998; 47:691-96; PMID:9847411; https://doi.org/ 10.1007/PL00006428 [DOI] [PubMed] [Google Scholar]
- [42].Hamidian M, Hall RM. ISAba1 targets a specific position upstream of the intrinsic ampC gene of Acinetobacter baumannii leading to cephalosporin resistance. J Antimicrob Chemother 2013; 68:2682-3; PMID:23788477; https://doi.org/ 10.1093/jac/dkt233 [DOI] [PubMed] [Google Scholar]
- [43].Vetting MW, S de Carvalho LP, Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys 2005; 433:212-26; https://doi.org/ 10.1016/j.abb.2004.09.003 [DOI] [PubMed] [Google Scholar]
- [44].Torres AG, Redford P, Welch RA, Payne SM. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect Immun 2001; 69:6179-85; PMID:11553558; https://doi.org/ 10.1128/IAI.69.10.6179-6185.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Reeves SA, Torres AG, Payne SM. TonB is required for intracellular growth and virulence of Shigella dysenteriae. Infect Immun 2000; 68:6329-36; PMID:11035742; https://doi.org/ 10.1128/IAI.68.11.6329-6336.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Taguchi F, Suzuki T, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y. The siderophore pyoverdine of Pseudomonas syringae pv. tabaci 6605 is an intrinsic virulence factor in host tobacco infection. J Bacteriol 2010; 192:117-26; PMID:19854904; https://doi.org/ 10.1128/JB.00689-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Oke M, Sarra R, Ghirlando R, Farnaud S, Gorringe AR, Evans RW, Buchanan SK. The plug domain of a neisserial TonB-dependent transporter retains structural integrity in the absence of its transmembrane beta-barrel. FEBS Lett 2004; 564:294-300; PMID:15111112; https://doi.org/ 10.1016/S0014-5793(04)00196-6 [DOI] [PubMed] [Google Scholar]
- [48].Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, Bataille E, Dossat C, Gas S, Kreimeyer A, Lenoble P, et al.. Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS One 2008; 3:e1805; https://doi.org/ 10.1371/journal.pone.0001805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Coyne S, Rosenfeld N, Lambert T, Courvalin P, Périchon B. Over expression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 2010; 54:4389-93; PMID:20696879; https://doi.org/ 10.1128/AAC.00155-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Marchand I, Damier-Piolle L, Courvalin P, Lambert T. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob Agents Chemother 2004; 48:3298-304; PMID:15328088; https://doi.org/ 10.1128/AAC.48.9.3298-3304.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ramos JL, Martínez-Bueno M, Molina-Henares AJ, Terán W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 2005; 69:326-56; https://doi.org/ 10.1128/MMBR.69.2.326-356.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yu Z, Reichheld SE, Savchenko A, Parkinson J, Davidson AR. A comprehensive analysis of structural and sequence conservation in the TetR family transcriptional regulators. J Mol Biol 2010; 400:847-64; https://doi.org/ 10.1016/j.jmb.2010.05.062 [DOI] [PubMed] [Google Scholar]
- [53].Rosenfeld N, Bouchier C, Courvalin P, Périchon B. Expression of the resistance-nodulation-cell division pump AdeIJK in Acinetobacter baumannii is regulated by AdeN, a TetR-type regulator. Antimicrob Agents Chemother 2012; 56:2504-10; PMID:22371895; https://doi.org/ 10.1128/AAC.06422-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Aranda J, Poza M, Pardo BG, Rumbo S, Rumbo C, Parreira JR, Rodríguez-Velo P, Bou G. A rapid and simple method for constructing stable mutants of Acinetobacter baumannii. BMC Microbiol 2010; 10:279; PMID:21062436; https://doi.org/ 10.1186/1471-2180-10-279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiol 2008; 154:3398-409; https://doi.org/ 10.1099/mic.0.2008/019471-0 [DOI] [PubMed] [Google Scholar]
- [56].Mussi MA, Gaddy JA, Cabruja M, Arivett BA, Viale AM, Rasia R, Actis LA. The opportunistic human pathogen Acinetobacter baumannii senses and responds to light. J Bacteriol 2010; 192:6336-45; PMID:20889755; https://doi.org/ 10.1128/JB.00917-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Gebhardt MJ, Gallagher LA, Jacobson RK, Usacheva EA, Peterson LR, Zurawski DV, Shuman HA. Joint transcriptional control of virulence and resistance to antibiotic and environmental stress in Acinetobacter baumannii. MBio 2015; 6:e01660-15; PMID:26556274; https://doi.org/ 10.1128/mBio.01660-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Nikaido H, Basina M, Nguyen V, Rosenberg EY. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those beta-lactam antibiotics containing lipophilic side chains. J Bacteriol 1998; 180:4686-92; PMID:9721312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Li XZ, Barré N, Poole K. Influence of the MexA-MexB-oprM multidrug efflux system on expression of the MexC-MexD-oprJ and MexE-MexF-oprN multidrug efflux systems in Pseudomonas aeruginosa. J Antimicrob Chemother 2000; 46:885-93; PMID:11102405; https://doi.org/ 10.1093/jac/46.6.885 [DOI] [PubMed] [Google Scholar]
- [60].Lee HW, Koh YM, Kim J, Lee JC, Lee YC, Seol SY, Cho DT, Kim J. Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin Microbiol Infect 2008; 14:49-54; PMID:18005176; https://doi.org/ 10.1111/j.1469-0691.2007.01842.x [DOI] [PubMed] [Google Scholar]
- [61].Nait Chabane Y, Marti S, Rihouey C, Alexandre S, Hardouin J, Lesouhaitier O, Vila J, Kaplan JB, Jouenne T, Dé E. Characterisation of pellicles formed by Acinetobacter baumannii at the air-liquid interface. PLoS One 2014; 9:e111660; PMID:25360550; https://doi.org/ 10.1371/journal.pone.0111660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiol 2003; 149:3473-3484; https://doi.org/ 10.1099/mic.0.26541-0 [DOI] [PubMed] [Google Scholar]
- [63].Rumbo-Feal S, Gómez MJ, Gayoso C, Álvarez-Fraga L, Cabral MP, Aransay AM, Rodríguez-Ezpeleta N, Fullaondo A, Valle J, Tomás M, Bou G, Poza M. Whole transcriptome analysis of Acinetobacter baumannii assessed by RNA-sequencing reveals different mRNA expression profiles in biofilm compared to planktonic cells. PLoS One 2013; 8:e72968; PMID:24023660; https://doi.org/ 10.1371/journal.pone.0072968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Chang KC, Kuo HY, Tang CY, Chang CW, Lu CW, Liu CC, Lin HR, Chen KH, Liou ML. Transcriptome profiling in imipenem-selected Acinetobacter baumannii. BMC Genomics 2014; 15:815; PMID:25260865; https://doi.org/ 10.1186/1471-2164-15-815 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


