Abstract
Thraustochytrids are marine single-cell protists that produce large amounts of PUFAs, such as DHA. They accumulate PUFAs in lipid droplets (LDs), mainly as constituent(s) of triacylglycerol (TG). We identified a novel protein in the LD fraction of Aurantiochytrium limacinum F26-b using 2D-difference gel electrophoresis. The protein clustered with orthologs of thraustochytrids; however, the cluster was evolutionally different from known PAT family proteins or plant LD protein; thus, we named it thraustochytrid-specific LD protein 1 (TLDP1). TLDP1 surrounded LDs when expressed as a GFP-tagged form. Disruption of the tldp1 gene decreased the content of TG and number of LDs per cell; however, irregular and unusually large LDs were generated in tldp1-deficient mutants. Although the level of TG synthesis was unchanged by the disruption of tldp1, the level of TG degradation was higher in tldp1-deficient mutants than in the WT. These phenotypic abnormalities in tldp1-deficient mutants were restored by the expression of tldp1. These results indicate that TLDP1 is a thraustochytrid-specific LD protein and regulates the TG accumulation and LD morphology in A. limacinum F26-b.
Keywords: lipid droplet protein, polyunsaturated fatty acid, triacylglycerol, thraustochytrid-specific lipid droplet protein 1
Several lines of evidence indicate the benefits of n-3 PUFAs, such as DHA (22:6n-3) and EPA (20:5n-3), for human health (1–3). Thus, n-3 PUFAs are currently utilized as a medicine and in supplements; the former is provided as an FA ethyl ester and the latter as a triacylglycerol (TG), both of which are produced from marine fish oil. However, n-3 PUFAs in marine fish are considered to accumulate as a result of the food chain in the marine ecosystem (4). The primary producers of n-3 PUFAs in marine environments are bacteria, phytoplankton, and thraustochytrids. Thraustochytrids, a unicellular marine microorganism classified into Stramenopile, synthesize DHA and accumulate it as acyl chain(s) of TG in lipid droplets (LDs) and phospholipids in cellular membranes. Thraustochytrids are expected to become an alternative to fish oil because of their high productivity of n-3 PUFAs and suitable growth characteristics for industrial purposes (5–7). Thraustochytrid-derived DHA is currently provided to individuals who do not eat fish, such as vegetarians (8). Furthermore, thraustochytrids produce not only beneficial n-3 PUFAs but also useful lipids, including carotenoids and squalene (7). Basic techniques for gene manipulation, including draft genome sequences as well as the identification of metabolic enzymes responsible for DHA and DHA-containing phospholipids have been conducted for more than a decade (9–16) and thraustochytrids have emerged as a useful model for studying the metabolism and accumulation of DHA-containing lipids. We also reported the lipid profile of Aurantiochytrium limacinum F26-b (17), the strain used in this study [the former name of this strain was Schizochytrium sp. F26-b and it was renamed A. limacinum F26-b based on several taxonomic methods, including 18S rRNA gene analysis (11)].
LDs are a common intracellular lipid storage organelle in eukaryotes and prokaryotes (18–21). One of the characteristics of thraustochytrids is the possession of well-developed LDs, in which n-3 PUFAs are mainly enriched as acyl chain(s) of TG. The size of LDs may be linked to the accumulation of TG (22); however, the mechanisms by which TG accumulation and LD sizes are regulated in thraustochytrids remain unclear. LDs are coated with proteins belonging to the PAT family. PAT proteins consist of five members: perilipin (Plin)1 (formerly named perilipin), Plin2 [adipocyte differentiation-related protein (ADRP)], Plin3 [tail-interacting protein of 47 kDa (TIP47)], Plin4 (S3–12), and Plin5 (OXPAT) (23). In plants, oleosin, a major LD protein, functions to maintain the stability of LDs in plant cells (24). However, to the best of our knowledge, homologs of PAT or oleosin family proteins have yet to be identified in thraustochytrid draft genome databases.
In order to clarify the mechanisms by which TG accumulation in LDs and LD morphology are regulated in thraustochytrids, we studied an LD protein that is exclusively enriched in the LD fraction of A. limacinum F26-b. This protein, designated as thraustochytrid-specific LD protein 1 (TLDP1), localized on LDs when expressed as its green fluorescence protein (GFP)-fused form in A. limacinum F26-b. The disruption and overexpression of tldp1 in A. limacinum F26-b resulted in decreases and increases in total TG contents, respectively. The size of LDs was uniform in A. limacinum F26-b cells; however, irregular and unusually large LDs appeared in tldp1-disrupted mutants under the same conditions. These results indicate that TLDP1 regulates TG accumulation and the size/number of LDs in A. limacinum F26-b.
MATERIALS AND METHODS
Materials
Phosphatidylcholine (PC) (11:0/11:0), lysophosphatidylcholine (LPC) (13:0), phosphatidylethanolamine (PE) (12:0/12:0), diacylglycerol (DG) (12:0/12:0), and TG (12:0/12:0/12:0) were purchased from Avanti Polar Lipids. Artificial seawater (SEALIFE) was purchased from Nihonkaisui Co., Japan. An anti-tubulin antibody was purchased from MBL. TLC was purchased from Merck Millipore. BODIPY 493/503, HCS LipidTOX Red neutral lipid stain, and SYPRO Ruby protein gel stain were purchased from Thermo Fisher Scientific Inc. Immobiline DryStrips (pH 3–10, 24 cm), DryStrip cover fluid, Ampholine™ preblended (pH 3.5–9.5), Ettan DALTsixLarge electrophoresis system, and Ettan IPGphor 3 IEF system were purchased from GE Healthcare Bioscience. IC3-OSu and IC5-OSu were products of Dojindo Laboratories.
Strains and culture
A. limacinum F26-b, isolated from fallen leaves of Rhizophora mucronata collected at Ishigaki island, Okinawa, Japan, was identified by 18S rRNA gene analysis (11). It was grown in glucose-yeast extract (GY) medium [3% glucose and 1% yeast extract in 1.75% artificial sea water (SEALIFE)] with a 0.1% vitamin mixture (0.2% vitamin B1, 0.001% vitamin B2, and 0.001% vitamin B12) and 0.2% trace elements (3% EDTA di-sodium, 0.15% FeCl3·6H2O, 3.4% H3BO4, 0.43% MnCl2·4H2O, 0.13% ZnSO4·7H2O, 0.026% CoCl2·6H2O, 0.026% NiSO4·6H2O, 0.001% CuSO4·5H2O, and 0.0025% Na2MoO4·2H2O) at 25°C with rotation at 150 rpm for the period indicated. All percentages described above are expressed as weight per volume.
Measurement of cell growth (biomass)
The growth (biomass) of A. limacinum was monitored by measuring optical density at 600 nm (A600 nm). A. limacinum was generally precultured for 3 days and transferred to fresh GY liquid medium with a vitamin mixture and trace elements.
Preparation of the LD fraction
LDs were isolated from A. limacinum F26-b using a previously described method (25) with minor modifications. Briefly, cells cultivated at 25°C in 200 ml of GY medium were harvested at the middle log phase by centrifugation at 2,300 g at 4°C for 5 min and then washed with PBS twice. Cells were resuspended in 10 ml of buffer A [20 mM Tricine (pH 7.8) containing 200 mM sucrose, EDTA-free protease inhibitor tablets (cOmplete mini), and phosphatase inhibitor cocktail (PhosSTOP)] and disrupted using a probe-type sonicator four times for 30 s. The homogenates were collected, centrifuged at 3,000 g at 4°C for 10 min using a swinging bucket rotor to remove nuclei, cell debris, and unbroken cells, and the supernatant fraction was collected as the post nuclear supernatant (PNS) fraction. Three milliliters of the PNS fraction were transferred into an ultra-centrifuge tube and 2 ml of buffer B [20 mM HEPES-KOH (pH 7.4) containing 100 mM KCl and 2 mM MgCl2] was then loaded on the top of the PNS fraction. After centrifugation at 30,000 g at 4°C for 60 min using a RPS 65T swinging bucket rotor (Hitachi), the top portion of the gradient formed was collected into a 1.5 ml tube. Proteins were precipitated with chloroform-acetone (26) and resuspended in urea buffer [20 mM Tris-HCl (pH 8.5) containing 7 M urea, 2 M thiourea, and 4% CHAPS). Protein content was measured with the 2-D Quant kit (GE Healthcare).
2D-difference gel electrophoresis
The fluorescent labeling of proteins was performed according to the optimized procedure described by Uchida et al. (27). In brief, 1.1 μl of IC3-Su or IC5-Su solution (400 pmol/μl in DMSO) was added to the sample (60 μg protein/50 μl) in a light-protected microcentrifuge tube. After mixing and centrifugation, the tubes were kept on ice for 1 h in the dark. The fluorescence labeling reaction was terminated by the addition of 2 μl of lysine (10 mM). 2D-electrophoresis was performed according to the manufacturer’s protocol. The fluorescence imaging of IC3/5-labeled proteins was performed using ProXPRESS (Perkin Elmer; 540/25 nm excitation for IC3-labeled proteins and 625/30 nm excitation for IC5-labeled proteins). Proteins (60 μg each) in the LD fraction (top layer) and non-LD fraction (bottom layer) were separately labeled with IC3 (n = 3 for each analysis). Regarding internal standards, proteins in the LD-rich and non-LD fractions were mixed (60 μg in total) and labeled with IC3. In order to identify proteins, nonlabeled proteins were picked from the gel and visualized by staining with SYPRO Ruby after 2D-PAGE. Spots on 2D-PAGE were identified using Progenesis SameSpots software (version 3.3). Spots were cut off using ProXcision (Perkin Elmer) and trypsinized. The peptides obtained were analyzed using ESI-linear ion trap (LIT) MS (Finnigan LTQ; Thermo Fisher Scientific) according to a previously described protocol (28). All product ions were submitted to the computer database search analysis with the Mascot search engine (Matrix Science, Boston, MA) using the JGI genome portal database of A. limacinum ATCC MYA-1381.
Cloning of tldp1 and the preparation of recombinant TLDP1
Genomic DNA was prepared from A. limacinum F26-b using a previously described method (11). The open reading frame encoding 395 amino acids of TLDP1 was cloned from the genomic DNA of A. limacinum F26-b by PCR using the primer set of ADRP-F and ADRP-R (supplemental Table S1). The PCR product and pGEM-T Easy vector (Promega) were ligased with 2× ligation mix (TOYOBO). The 1,185 bp PCR product obtained was 100% matched with the sequence in the A. limacinum ATCC MYA-1381 genome database. The open reading frame was expressed in Escherichia coli BL21/pG-KJE8 as a His tag and Trigger factor tag-fused protein using a pCold TF vector (Takara Bio). The transformants were incubated at 37°C in Luria-Bertani (LB) medium containing 100 μg/ml ampicillin until A600nm reached ∼0.6, and the culture was kept at 15°C for 15 min. Isopropyl-D-thiogalactopyranoside was then added to the culture at a final concentration of 1 mM. After cultivation at 15°C for 24 h, cells were harvested by centrifugation (8,000 g for 15 min) and suspended in 50 mM Tris-HCl (pH 7.5) containing 150 mM NaCl and 20 mM imidazole. The suspension was kept in a sonic bath for 30 s and this procedure was repeated four times to crush the cells. Cell debris was removed by centrifugation at 5,000 g at 4°C for 15 min. After centrifugation at 17,000 g at 4°C for 15 min, the supernatant was applied to Ni Sepharose 6 Fast Flow resin (GE Healthcare) packed in Muromac mini column M (Muromachi Technos) and the column was then washed with 50 mM Tris-HCl (pH 7.5) containing 150 mM NaCl and 40 mM imidazole. Recombinant TLDP1 fused with the trigger factor was eluted with 50 mM Tris-HCl (pH 7.5) containing 150 mM NaCl and 200 mM imidazole. In order to remove the trigger factor, HRV 3C Protease (Takara Bio) was added and the digested sample was applied to a Ni Sepharose column. A single protein band showing a molecular mass of 48 kDa on SDS-PAGE was obtained.
Construction of phylogenetic tree of Plins and TLDP1
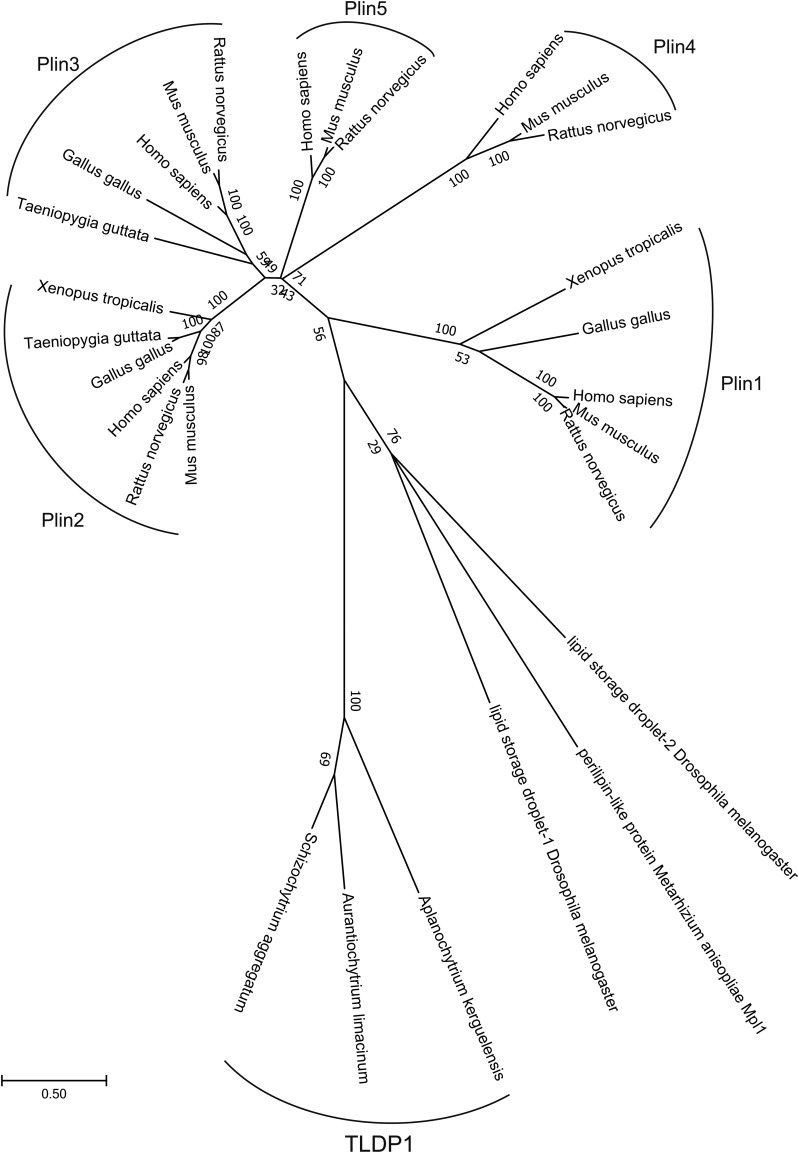
We conducted multiple alignments using MAFFT (L-INS-I) and analyzed the relationship of each protein with the ML method using MEGA7 (29, 30). Gap elimination adopted was more than 85% and amino acids were determined by the JTT+G model based on BIC. The robustness of the tree was evaluated with the bootstrap method (1,000 repeats). The accession numbers are shown in Fig. 2.
Fig. 2.
Phylogenetic tree of TLDP1 and Plin family proteins. We conducted multiple alignments using MAFFT (L-INS-I) and analyzed the relationship of each protein with the ML method using MEGA7 (29, 30). Details are presented in the Materials and Methods. Accession numbers, TLDP1 (A. limacinum, LC314401; Aplanochytrium kerguelensis, fgenesh1_kg.19_#_97_#_isotig03719; S. aggregatum, fgenesh1_pg.3_#_228), Plin1 (Homo sapiens, NP_001138783.1; Mus musculus, NP_001106942.1; Rattus norvegicus, NP_001295074.1; Gallus gallus, ADD39000.1; Xenopus tropicalis, XP_012815005.1), Plin2 (H. sapiens, NP_001113.2; M. musculus, NP_031434.3; R. norvegicus, NP_001007145.1; G. gallus, XP_015135579.1; Xenopus tropicalis, NP_988963.1; Taeniopygia guttata, XP_002192424.1), Plin3 (H. sapiens, NP_005808.3; M. musculus, NP_080112.1; R. norvegicus, XP_001061015.1; X. tropicalis, XP_004911098.1; Taeniopygia guttata, XP_012426686.1; G. gallus, XP_015152284.1), Plin4 (H. sapiens, NP_001073869.1; M. musculus, NP_065593.2; R. norvegicus, XP_008756142.1; G. gallus, NP_001304017.1), Plin5 (H. sapiens, NP_001013728.2; M. musculus, NP_001070816.1; R. norvegicus, NP_001128109.1), and others (Drosophila melanogaster Lsd-1, NP_732904.2; D. melanogaster Lsd-2, NP_572996.1; Metarhizium anisopliae Mpl1, ABI18161.1).
Preparation of an anti-TLDP1 polyclonal antibody
The preparation of an anti-rabbit polyclonal antibody was performed using purified recombinant TLDP1 (1.0 mg/ml, 750 μl) at MBL Life Science Co., Japan. The anti-serum obtained was subjected to ELISA using purified recombinant TLDP1, which confirmed that the anti-serum had a sufficient titer against TLDP1 when diluted 2,500-fold. The anti-serum reacted with the 48 kDa band (TLDP1) of the lysate from the A. limacinum F26-b (WT). However, this 48 kDa band was not detected in the lysate of the tldp1-disrupted mutant by the Western blot, indicating that the anti-serum was specific to TLDP1.
Western blotting of TLDP1
Proteins were separated on 10% SDS-PAGE, transferred to PVDF membrane, and subjected to Western blotting using anti-TLDP1 rabbit antibody (first antibody, dilution ×5,000) and anti-rabbit IgG antibody (second antibody, dilution ×10,000). The PVDF membrane, after staining with ECL Plus solution, was subjected to the Cooled CCD Camera System Ez-capture II (ATTO) and the intensity of each band on the membrane was quantified using Image J 1.49v lane analyzer.
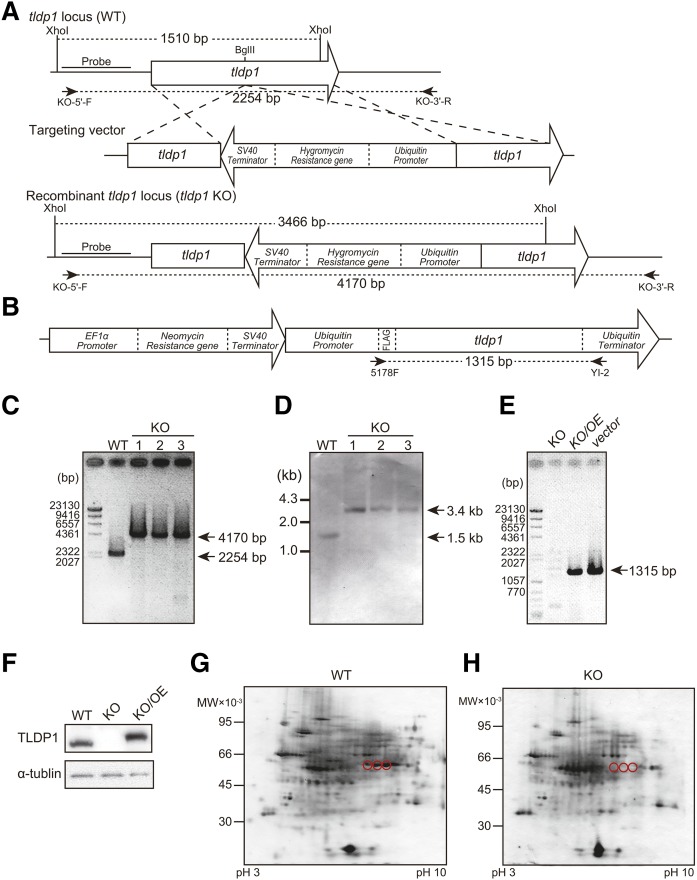
Generation of tldp1-disrupted mutants (KO)
The tldp1 of A. limacinum F26-b was disrupted by homologous recombination according to a previously described method (14). Briefly, a BglII site was added to the tldp1 KO construct by PCR-based site-directed mutagenesis using the primers ADRP-BglII-F and ADRP-BglII-R (supplemental Table S1). The hygromycin-resistant gene expression cassette (13) was inserted into the BglII site of the vector according to a previously described method (11). The KO construct was amplified by PCR using the primers ADRP-F and ADRP-R and introduced into A. limacinum F26-b by electroporation with Gene Pulser Xcell. The transformants that grew on the PDA-agar plate containing 2 mg/ml of hygromycin B were subjected to PCR screening for tldp1-disrupted mutants using the primers, KO-5′-F and KO-3′-R (Fig. 3A; supplemental Table S1). The disruption of tldp1 was also confirmed by a Southern blot analysis using genomic DNA digested with XhoI (Fig. 3A). The probes for Southern blotting were prepared using DIG DNA labeling mix (Roche Diagnostics) and the primers, KO-5′-F and KO-5′-R (supplemental Table S1).
Fig. 3.
Generation and validation of tldp1-disrupted mutants. A: Strategy for the disruption of the tldp1 gene in A. limacinum F26-b by homologous recombination using a hygromycin-resistant gene as a marker. B: Targeting vector for the overexpression of tldp1. C: PCR analysis of tldp1-desrupted mutants (KO) and the WT of A. limacinum F26-b. Sites for PCR primers are shown in A. D: Southern blotting of KO and the WT after digestion with Xhoi. Probes for Southern blotting are shown in A. E: PCR analyses of KO, overexpression of tldp1 in KO (KO/OE), and the targeting vector. Sites for PCR primers are shown in B. F: Western blotting of the WT, KO, and KO/OE. The 2D-PAGE of the LD fraction from the WT (G) and the KO (H). Three spots corresponding to TLDP1 in the WT (red circles) disappeared in KO.
Overexpression of tldp1 in KO (KO/OE)
In order to prepare the construct expressing FLAG-tag fused TLDP1 in tldp1-disrupted mutants and the WT, PCR was performed using genomic DNA as a template and the primers, YI-1 and YI-2 (supplemental Table S1). The amplified product was inserted into pEF-Neor/Ubi-EGFP, in which the egfp sequence was eliminated using an In-Fusion HD cloning kit (Takara Bio). The expression construct was introduced into KO mutants by electroporation using Gene Pulser Xcell. The transformants that grew on the PDA-agar plate containing 0.5 mg/ml of G418 were subjected to PCR screening for the tldp1 overexpressor using the primers, 5178F and YI-2 (supplemental Table S1).
Introduction of expression or KO cassettes into A. limacinum F26-b by electroporation
A. limacinum was precultured in a GY medium at 25°C for 2 days. The cells harvested were transferred into dGPY liquid medium [0.2% glucose, 0.1% polypeptone, and 0.05% yeast extraction in 1.75% artificial sea water (SEALIFE)] and cultured at 25°C for 16 h. Cells were washed with 500 μl of 1.75% SEALIFE twice and suspended in 60 μl of OPTI-MEM I (Thermo Fisher Scientific). Expression/KO cassettes (PCR products, ∼3–5 μg) were added to the cell suspension and transferred into a 1 mm gap cuvette. Electroporation was conducted with a Bio-Rad Gene Pulser Xcell electroporator (850 V, 25 μF, 200 Ω). After being pulsed twice, 800 μl of fresh medium was added. Cells were cultured at 25°C for 1 day in GY medium and then transferred to a PDA-agar plate containing suitable antibiotics.
Staining and observation of LDs
LDs, stained with BODIPY 493/503 or HCS LipidTOX Red neutral lipid stain were observed under the fluorescence microscope DMi8 with LAS X equipped with an objective lens of ×100 (numerical aperture 1.40) and a DFC3000G camera (Leica Microsystems) or confocal microscope, Nikon TE2000 with EZ-C1, equipped with an objective lens of ×60 (N.A 1.40).
Localization of TLDP1 in A. limacinum F26-b
In order to express GFP-fused TLDP1 in A. limacinum F26-b, gfp-fused tldp1 was inserted into the pEF-Neor/Ubi-EGFP vector (14). Briefly, egfp and tldp1 were separately amplified by PCR using the primers, EF1-BamHI R and Fusion R, and Bgl2-UbiP F and Fusion F, respectively (supplemental Table S1). Egfp-tldp1 was amplified by PCR using the primers EF1-BamHI R and Bgl2-UbiP F. PCR products and the pEF-Neor/Ubi-EGFP vector were digested by BamHI and BglII, and then ligased with 2× ligation mix (TOYOBO). The expression construct was amplified by PCR using the primers EFUbi556F and EFUbi5396R and introduced into A. limacinum F26-b (WT) or the tldp1-disrupted mutant (KO) by electroporation using Gene Pulser Xcell (Bio-Rad).
Preparation of cell lysates
One milliliter of cultured cells was harvested by centrifugation at 4,000 g at 4°C for 3 min and washed with 1.75% SEALIFE. Cells were suspended in 300 μl of cell lysis buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, and 1% NP-40 containing protease inhibitors cOmplete mini) and crushed at 3,000 rpm for 60 s with a beads crusher (μT-12, TAITEC) using 0.2 cm3 of glass beads that were 0.6 mm in diameter and kept on ice for 60 s in a 1.5 ml tube. This procedure was repeated three times. The supernatant was obtained by centrifugation at 800 g for 15 min at 4°C.
DG acyltransferase assay
DG acyltransferase (DGAT) activity was measured using 1,2-dipalmitoyl-sn-glycerol and 14C-palmityl-CoA as substrates according to the method described in Siloto et al. (31) with minor modifications. Briefly, the reaction mixture was composed of 15 μg of the cell lysate, 320 μM 1,2-dipalmitoyl-sn-glycerol (Sigma-Aldrich), and 0.9 μM [1-14C]palmitoyl-CoA (50 μCi, American Radiolabeled Chemicals) in 15 mM Tris-HCl (pH 7.5) containing 25 mM sucrose, 15 mM KCl, 15 mM MgCl2, and 125 μg/ml BSA (FA-free; Sigma-Aldrich). After being incubated at 30°C for 10 min, the reaction was terminated by the addition of 300 μl of chloroform/methanol (2:1, v/v). The chloroform phase was dried using a stream of nitrogen, dissolved in 10 μl of chloroform/methanol (2:1, v/v), and applied to a Silica Gel 60 TLC plate, which was developed with hexane/diethyl ether/acetic acid (50:50:1, v/v/v). The radioactivity of the corresponding TG was quantified using a Typhoon FLA 9500 Bio-imaging analyzer (GE Healthcare).
Lipase assay
Lipase activity was measured using a 4-methylumbelliferyl (4MU)-palmitate (Sigma-Aldrich), as described in (32, 33). Briefly, 100 μl of the cell lysate (10 μg protein) was mixed with 100 μl of 100 mM Tris-HCl (pH 7.5) containing 20 μM of 4MU-palmitate. The reaction mixture was kept at 37°C for the period indicated and measured by a Wallac 1420 ARVO fluorescence microplate reader set at 355 nm excitation/460 nm emission (PerkinElmer).
Protein assay
Protein content was measured using Pierce 660 nm protein assay reagent (Thermo Fisher Scientific) with BSA (Thermo Fisher Scientific) as the standard.
LC-ESI MS
Total lipids were extracted from 1 mg of dry cells by sonication for 1 min with 300 μl of chloroform/methanol (2:1, v/v) containing 10 μM PC (11:0/11:0), 10 μM LPC (13:0), 40 μM PE (12:0/12:0), 50 μM DG (12:0/12:0), and 20 μM TG (12:0/12:0/12:0) as internal standards. After the removal of cell debris by centrifugation, 75 μl of distilled water was added to the supernatant. Twenty microliters of the chloroform phase was transferred to a new tube and 480 μl of 2-propanol was then added. Phospholipids and neutral lipids were analyzed by LC-ESI MS, as described by Ikeda et al. (34) with modifications. LC-ESI MS was performed using a HPLC system (Agilent Technologies) coupled to a MS apparatus (3200 QTRAP LC/MS/MS; AB Sciex). A binary solvent gradient with a flow rate of 200 μl/min was used to separate phospholipids and neutral lipids by reverse-phase chromatography using InertSustain C18 (2.1 × 150 mm, 5 μm; GL Sciences). The gradient was started with 3% B (2-propanol with 0.1% formic acid and 0.028% ammonium) in buffer A (acetonitrile/methanol/distilled water, 19:19:2, v/v/v containing 0.1% formic acid and 0.028% ammonium) and was maintained for 3 min. The gradient reached 40% B for 21 min, then 70% B for 1 min, and was maintained for 7 min. The gradient was returned to the starting conditions for 1 min and the column was equilibrated for 7 min before the next run. For phospholipid analysis, precursor ion scan at m/z 184, corresponding to the phosphocholine head group, and neutral loss scan at m/z 141, corresponding to the phosphoethanolamine head group, were used to identify the molecular species of PC and PE, respectively (35). For neutral lipid analysis, neutral loss scans at m/z 273, 345, and 347 were used to identify DG and TG containing palmitic acid, DHA, and n-6 docosapentaenoic acid (n-6 DPA), respectively (36). The structures of phospholipids and neutral lipids were confirmed by MS/MS analysis (supplemental Figs. S1–S3). Finally, TG, DG, PC, and PE were quantified by using multiple reaction monitoring (MRM), as shown in supplemental Table S2.
GC analysis of FAs
This was done by the method described in (17).
Real-time PCR
Total RNA was extracted from A. limacinum with a Sepasol-RNA I Super G (Nacalai Tesque) and a SV Total RNA isolation system (Promega). cDNA was synthesized by a PrimeScript™ RT reagent kit with gDNA Eraser (Perfect Real Time) (Takara-Bio). Real-time PCR was performed using an Mx3000P qPCR system (Agilent Technologies) with SYBR Premix Ex Taq II (Tli RNaseH Plus) (Takara-Bio). Relative mRNA expression levels were measured using the 2−ΔΔCT method (37). Values were normalized with GAPDH mRNA (scaffold_7:1112076-1113074). Oligonucleotide primer sets are shown in supplemental Table S1.
RESULTS
Lipid profile of A. limacinum F26-b
Strain F26-b is a typical thraustochytrid that produces large amounts of PUFAs, including DHA, which are mainly found as acyl chain(s) of neutral lipids and phospholipids (17). The major FAs of strain F26-b in the neutral lipid fraction are palmitic acid (16:0, 51.5% of total FAs in the fraction), n-6 DPA (22:5n-6, 5.5%), and DHA (27.2%); while in the phospholipid fraction, those are palmitic acid (34.3%), n-6 DPA (9.6%), and DHA (46.1%), when the strain was cultured under the conditions described in the Materials and Methods. These results indicate that major FAs of the strain F26-b are palmitic acid and DHA. However, this strain (formerly named Schizochytrium sp. F26-b) produced pentadecanoic acid (C15:0) as a major saturated FA instead of palmitic acid when cultivated in a GY medium without a vitamin mixture. This odd-numbered FA was mainly incorporated into the sn-1 of PC to form 1-pentadecanoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine (17).
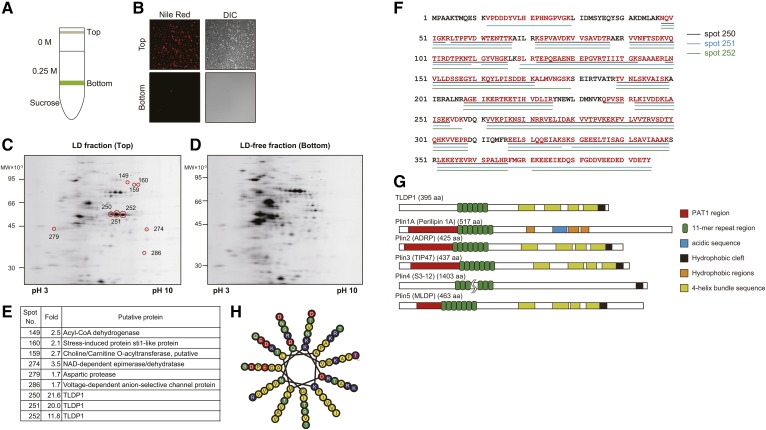
Identification of the LD protein (TLDP1) in A. limacinum F26-b
One of the characteristics of thraustochytrids is well-developed intracellular LDs, in which n-3 PUFAs containing TG accumulate. We observed a large number of LDs in A. limacinum F26-b cells when the thraustochytrid was cultured in GY medium and stained with BODIPY 493/503 or LipidTOX Red neutral lipid stain. However, we were unable to detect the highly conserved sequence of known LD proteins, such as PAT family proteins or oleosins in several thraustochytrid genome DNA databases (16), including that of A. limacinum (JGI Database, https://genome.jgi.doe.gov/Aurli1/Aurli1.home.html, US Department of Energy). Thus, we attempted to identify LD proteins in A. limacinum using 2D-difference gel electrophoresis (DIGE) followed by a LC-ESI-LIT MS analysis. We separated the LD fraction (top layer) from the LD-free fraction (bottom layer) using sucrose density gradient ultracentrifugation (Fig. 1A), as described in the Materials and Methods. It was confirmed that the top layer contained a lot of LDs, whereas the bottom layer did not, when both fractions were observed under fluorescent microscopy after staining with Nile Red (Fig. 1B). An analysis of these two fractions by 2D-DIGE revealed that nine protein spots (marked with red circles) were present at higher amounts in the LD fraction (Fig. 1C) than in the LD-free fraction (Fig. 1D). These nine protein spots were subsequently subjected to ESI-LIT MS, followed by the identification of proteins using the A. limacinum genome DNA database (Fig. 1E). Of the nine protein spots, three (#250–#252) showed the highest difference in the amounts between LD fraction and LD-free fraction (Fig. 1C vs. Fig. 1D). Three had the same molecular weights, but a different isoelectric point; however, they were considered to be derived from the same protein because the peptide sequences of the three spots matched each other at a very high rate (Fig. 1F) and covered 87% putative amino acids deduced from the genome DNA database (red letters, Fig. 1F). Furthermore, these three spots disappeared by knocking out one gene (tldp1), as described later (Fig. 3H). This LD protein was tentatively designated as TLDP1. The TLDP1 gene (tldp1) was then cloned from A. limacinum genomic DNA and sequenced (GenBank accession number LC314401). The putative TLDP1 sequence, composed of 395 amino acids, showed 16.7% and 7.3% identities at the protein level to Xenopus and human ADRPs (PLIN2), respectively, which are PAT family protein members (23, 38). TLDP1 did not possess the typical PAT1 region generally found in PAT proteins; however, TLDP1 possessed an 11-mer repeat region and 4-helix bundle sequences that are characteristic of PAT family proteins. Schematic diagrams of TLDP1 with other known PAT family proteins and the 11-mer repeat region of TLDP1 are shown in Fig. 1G and Fig. 1H, respectively.
Fig. 1.
Isolation of TLDP1 from the LD fraction of A. limacinum F26-b. A: Diagram showing the separation of the LD fraction (top layer) from the LD-free fraction (bottom layer) using sucrose density gradient ultracentrifugation. B: Validation of LD fraction. A. limacinum F26-b was cultured in GY medium containing 3% glucose at 25°C for 3 days. Cells were harvested and the cell lysate was subjected to sucrose gradient centrifugation, as shown in A. Top (LD) and bottom (LD-free) fractions were observed under fluorescent microscopy after staining with Nile Red. C, D: The 2D-PAGE of LD (C) and LD-free (D) fractions. The 2D-DIGE was performed by the method described in the Materials and Methods. The LD and LD-free fractions were separately subjected to 2D-PAGE. The proteins were stained with SYPRO Ruby solution. LD and LD-free fractions (60 μg protein/50 μl) were labeled with IC3-Su and then subjected to 2D-DIGE. Spots on 2D-PAGE were cut off using ProXcision and trypsinized. The peptides obtained were analyzed using a LIT mass spectrometer. All product ions were submitted to the analysis using the JGI genome portal database (A. limacinum ATCC MYA-1381) with the Mascot search engine. E: The table showing seven putative proteins that display the distinct differences in their protein expression levels between the LD fraction and LD-free fraction. F: The putative primary sequence of TLDP1 cloned from the genomic DNA of A. limacinum F26-b. Red characters represent peptide sequences identified by the Mascot analysis of spots 250, 251, and 252. G: Diagrams of TLDP1 and Plin family proteins. Schematic diagrams for PAT family proteins were prepared according to (51, 52). H: Schematic diagram for 11-mer repeat of TLDP1. Hydrophobic residues are in yellow, charged residues are in blue or red, and polar residues are in green. This was prepared according to (53) and visualized using HELIQUEST (54).
Phylogenetic analysis of TLDP1
The phylogenetic tree revealed that TLDP1 was evolutionarily very different from known mammalian as well as nonmammalian PAT proteins, including Plin1–5 (Fig. 2). TLDP1 also did not show any homology to the oleosins and caleosins that are major LD proteins of seed plants (39). On the other hand, TLDP1 orthologs were found in other thraustochytrids, such as Schizochytrium aggregatum and Aplanochytrium kerguelense (Fig. 2). These results indicate that TLDP1 is a thraustochytrid-specific LD protein.
Generation and validation of tldp1-disrupted mutants (KO) and their revertants (KO/OE)
In order to clarify the function of TLDP1, KOs were generated from A. limacinum F26-b (WT) by gene-targeting homologous recombination using the hygromycin resistance gene as a marker (Fig. 3A). Revertants (KO/OE) were generated from KO by the random integration of the targeting vector containing the tldp1 gene driven with the ubiquitin promoter using the neomycin resistance gene as a marker (Fig. 3B). PCR and Southern blot analyses indicated that the tldp1 gene was disrupted in KO (Fig. 3C, D) and the tldp1 gene was detected in KO/OE (Fig. 3E). The TLDP1 protein was detected in WT and KO/OE, but not in KO by Western blotting using the anti-TLDP1 antibody (Fig. 3F). The expression level of TLDP1 was 2.6-fold higher in KO/OE than in WT, suggesting that the ubiquitin promoter used for the expression of tldp1 in KO was stronger than the promoter of tldp1 in WT. The molecular mass of TLDP1 in KO/OE is somewhat larger than that in WT, possibly because the Flag-tag used in OE could influence the mobility of TDAP1 on SDS-PAGE. We confirmed that the three spots (#250–252) of the LD fraction disappeared after the disruption of the tldp1 gene, indicating that these three proteins are translated from the tldp1 gene (Fig. 3G, H).
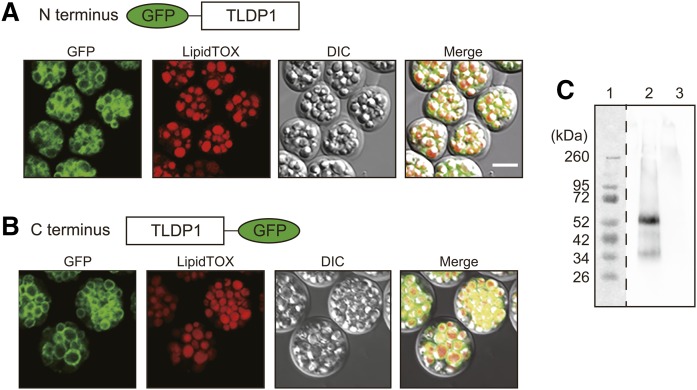
Localization of TLDP1 in A. limacinum F26-b
Although TLDP1 was isolated from the LD fraction of A. limacinum, this protein does not possess a typical PAT1 region and exhibits low identities to the LD proteins reported to date. In order to clarify whether TLDP1 is a protein that is specifically localized on LDs, tldp1 was expressed in tldp1 KO of A. limacinum as its GFP-fused form. As shown in Fig. 4A, TLDP1 that fused with GFP at the N terminus (green) surrounded the LDs visualized with LipidTOX Red neutral lipid stain (red). Even when GFP was fused to the C terminus, TLDP1 was exclusively expressed on LDs (Fig. 4B). TLDP1 was detected in the LD fraction, but not in the LD-free fraction, by Western blotting using the anti-TLDP1 antibody (Fig. 4C). These results show that TLDP1 is an LD protein that surrounds the LDs of A. limacinum and was recovered in the LD fraction when cell lysate was subjected to the sucrose-density gradient ultracentrifugation (Fig. 1A).
Fig. 4.
Localization of TLDP1 in A. limacinum F26-b. A, B: Intracellular localization of TLDP1. TLDP1 tagged with GFP at the N terminus (A) or C terminus (B) was expressed in tldp1-disrupted A. limacinum F26-b (KO) and observed under confocal fluorescent and differential interference contrast (DIC) microscopes. Cells were grown at 25°C for 4 days (A) and 5 days (B) with rotation at 150 rpm in 3 ml of GY medium and LDs were stained with a LipidTOX neutral red. Scale bar, 10 μm. C: Western blotting of TLDP1 using an anti-TLDP1 rabbit antibody. Lane 1, protein marker; lane 2, LD fraction; lane 3, LD-free fraction. Two micrograms of protein were loaded to each lane.
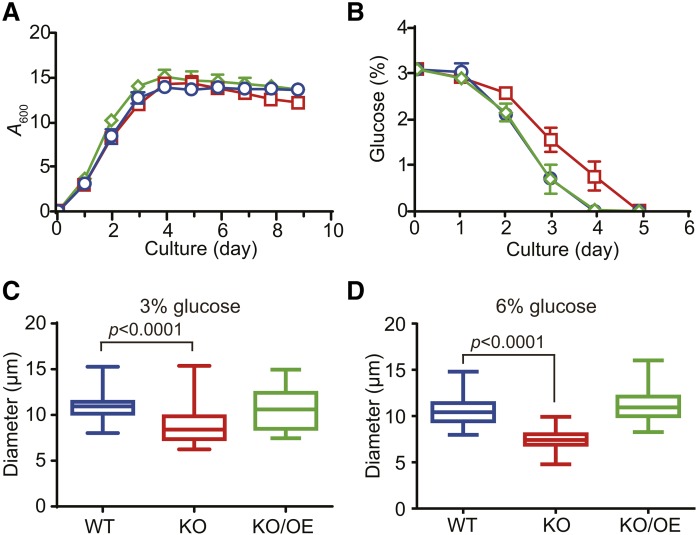
Cell growth and glucose consumption of WT, KO, and KO/OE
The cell growth (biomass) of KO was compared with that of WT and KO/OE by measuring the optical density of cell cultures at 600 nm (A600nm). Although a definite growth delay was not observed in KO until day 5 (Fig. 5A), glucose consumption was slower in KO than in WT and KO/OE (Fig. 5B). Thraustochytrids are generally considered to use glucose as an energy source and synthesize TG in the growth phase, and they use TG after the consumption of glucose. However, the delay observed in glucose consumption in KO without a significant growth defect suggests that KO uses TG efficiently as an energy source in the growth phase. This result may explain, in part, why KO does not accumulate TG in the growth phase, as described later. KO cells were slightly smaller than WT under the cultivation in a GY medium containing 3% (Fig. 5C) or 6% glucose (Fig. 5D), and this abnormality was cancelled by expression of tldp1 in KO (KO/OE). It is plausible that less accumulation of TG in KO (as described later) could make their cell size smaller.
Fig. 5.
Cell growth and glucose consumption of WT, KO, and KO/OE. Time course for cell growth (A) and glucose consumption (B) by the WT (blue open circles), KO (red open squares), and KO/OE (green open diamonds). Data represent the mean ± SE from three independent experiments (each n = 3). C, D: Box plots represent cell diameters of WT (blue), KO (red), and KO/OE (green) cultured at 25°C for 7 days with rotation at 150 rpm in the presence of 3% (C) and 6% (D) glucose. Boxes extend from 25% to 75%, whiskers represent minimum to maximum diameters, and middle lines represent the median (n = 25).
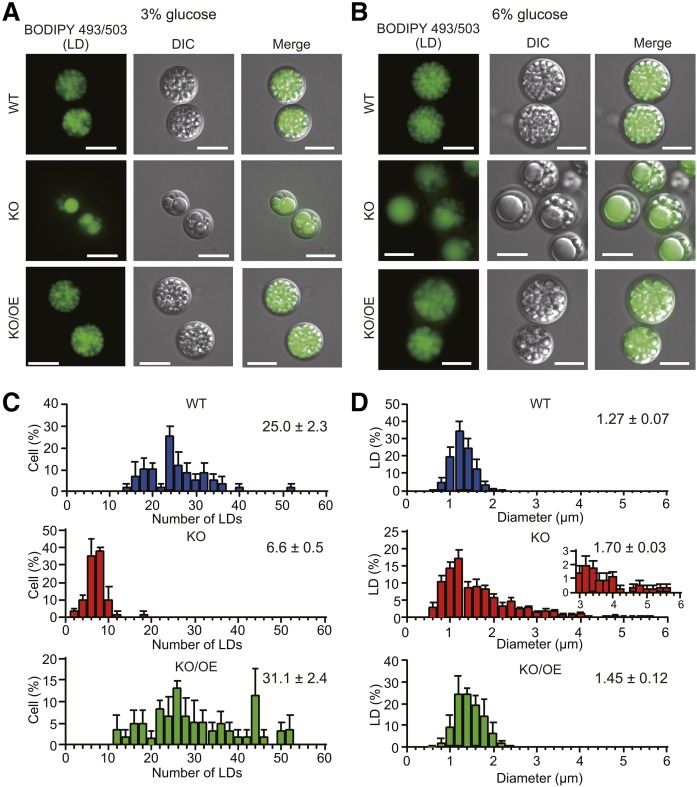
Size and number of LDs in WT, KO, and KO/OE
The most prominent difference in phenotypes between KO and WT was the morphology of LDs. The shape of individual LDs in WT was uniform when cultured in GY medium containing either 3% (Fig. 6A) or 6% (Fig. 6B) glucose, while that of KO was irregular with a decrease in the number of LDs under both conditions. In KO, we observed unusually large LDs that may have been generated by the fusion of several small LDs. These unusually large LDs disappeared with the expression of the tldp1 gene in KO (KO/OE) (Fig. 6A, B). We measured the number of LDs of cells using 20 cells and the diameter of LDs using 100 LDs (Fig. 6C, D). Based on the results obtained, we estimated the total volume of LDs per cell. The average volume of an LD was larger in KO (2.59 ± 0.13 µm3 per LD) and KO/OE (1.68 ± 0.42 µm3 per LD) than in WT (1.09 ± 0.18 µm3 per LD); however, the average number of LDs was markedly lower in KO (6.60 ± 0.50 per cell) than in WT (25.0 ± 2.3 per cell) and KO/OE (31.1 ± 2.4 per cell) and, thus, the total volume of LDs per cell was smaller in KO (17.0 ± 1.6 µm3 per cell) than in WT (26.8 ± 4.1 µm3 per cell) and KO/OE (52.5 ± 14.9 µm3 per cell).
Fig. 6.
LD phenotypes of WT, KO, and KO/OE. LD phenotypes of the WT, KO, and KO/OE cultured in 50 ml of GY medium containing 3% (A) and 6% (B) glucose at 25°C for 7 days with rotation at 150 rpm. LDs are stained with BODIPY 493/503, observed by the fluorescence microscope, DMi8, and photographed with a Leica DFC3000G camera. Scale bar, 10 μm. C: Numbers of LDs in WT (blue), KO (red), and KO/OE (green). Bars represent the distribution of the numbers of LDs in the cells. The numbers of LDs, determined for 20 each of cells after staining with BODIPY 493/503, were divided by 20. Data represent the mean ± SE from three independent experiments (each n = 20). D: Diameter of LDs in WT (blue), KO (red), and KO/OE (green). Bars represent the size distribution of LDs in cells. More than 100 LDs underwent size calculation after staining with BODIPY 493/503. Data represent the mean ± SE from three independent experiments (each n = 100). In C and D, LDs, were photographed with a Leica DMi8 and analyzed using computer software, LAS X (Leica Microsystems).
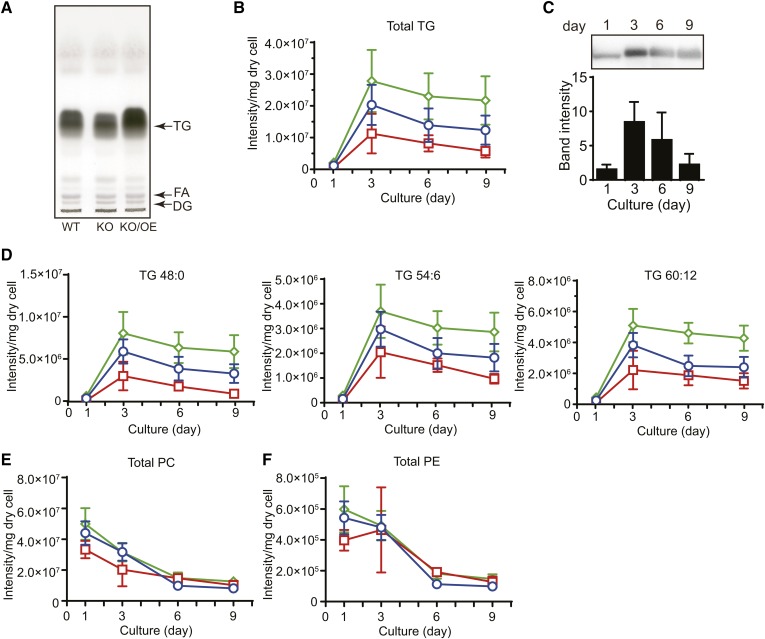
TG and phospholipid contents of WT, KO, and KO/OE
Because LDs are intracellular organelles for the storage of TG, we compared the TG content of KO with that of WT and KO/OE. As shown in TLC, the TG content of KO was lower, while that of KO/OE was higher than that of WT when cells cultured for 6 days were used in the analysis (Fig. 7A). It is important to note that TG contents are related to the total volume of LDs per cell, but not to the average diameter of LDs (Fig. 6C, D). We then investigated the accumulation of TG throughout the cultivation period of A. limacinum using LC-ESI MS. The molecular species of neutral lipids and phospholipids were identified by MS and MS/MS analyses (supplemental Figs. S1–S3) and major MRM pairs of these lipids are shown in supplemental Table S2. The total TG content (sum of major TG molecules listed) of KO was consistently lower than that of WT during the course of the culture until day 9, while that of KO/OE was higher than that of WT (Fig. 7B). The accumulation of TG peaked on day 3 and gradually decreased until day 9 for WT, KO, and KO/OE (Fig. 7B). Under the conditions used, day 3 was the end of the logarithmic growth phase (Fig. 5A) and the peak of TLDP1 expression (Fig. 7C). We subsequently examined the profiles of TG accumulation due to differences in the FA species of TG. We found that the depletion of TLDP1 decreased the content of TGs composed of three palmitic acids (TG48:0) as well as those containing DHA (TG54:6, TG60:12), while the overexpression of TLDP1 in KO increased the contents of these TGs, as shown in Fig. 7D. Simultaneously, we confirmed that the deletion and overexpression of TLDP1 did not significantly affect the total amounts of PC and PE, the two major phospholipids in A. limacinum F26-b (11, 17) (Fig. 7E, F).
Fig. 7.
Total lipid, TG, and phospholipid contents in the WT, KO, and KO/OE. A: TLC of total lipids from the WT, tldp1 KO (KO), and tldp1 KO/OE (KO/OE). Total lipids were extracted from 1 mg of dry cells (cultured for 6 days in GY medium containing 3% glucose) by chloroform/methanol (2:1, v/v) and applied to a TLC plate, which was developed with hexane/diethyl ether/acetic acid (80:20:1, v/v/v). Lipids were visualized with 3% copper sulfate in 15% phosphoric acid. B: Accumulation of total TG from the WT (blue open circles), KO (red open squares), and KO/OE (green open diamonds) during cultivation. C: Time course for the expression of TLDP1 examined by Western blotting using an anti-TLDP1 rabbit antibody. Western blotting membranes were subjected to a cooled CCD camera system, Ez-Capture II, followed by measurements of the band intensities using Image J 1.49v. Two micrograms of protein were loaded to each lane. Data represent the mean ± SD (n = 3). D: Accumulation of TG with several molecular species from the WT (blue open circles), KO (red open squares), and KO/OE (green open diamonds) during cultivation. The contents of total PC (E) and PE (F) from the WT (blue open circles), KO (red open squares), and KO/OE (green open diamonds) during cultivation. TG and phospholipid contents were measured by LC-ESI MS. The identification of TG molecular species was conducted with MS/MS (supplemental Fig. S1) and major MRM pairs of neutral lipids and phospholipids in LC-ESI MS are shown in supplemental Table S2. Cells were cultured at 25°C for the day indicated in GY medium containing 3% glucose with rotation at 150 rpm. Data of (B), (D), (E), and (F) represent the mean ± SE from three independent experiments (each n = 3).
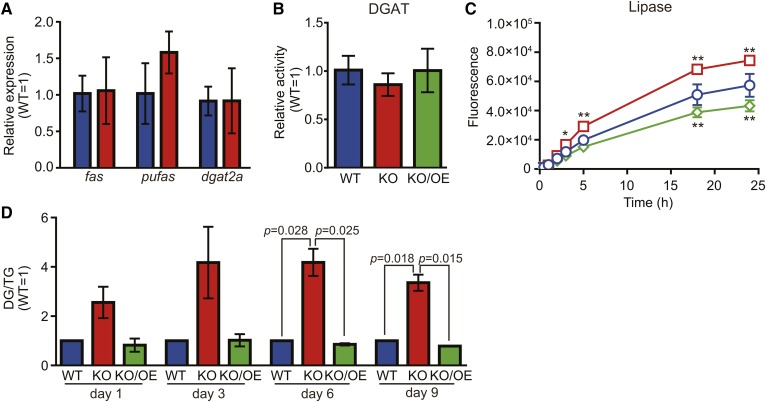
Effects of the disruption of tldp1 on the synthesis and degradation of TG
In order to elucidate the mechanisms by which TG accumulation decreased in KO, we investigated the effects of the disruption of tldp1 on the gene expression of FA synthase (fas), PUFA synthase (pufas), and TG synthase (dgat2a), which are involved in TG synthesis. We found that the disruption of tldp1 did not affect the expression of these enzymes at the mRNA level (Fig. 8A) and enzymatic activity of DGAT2 (Fig. 8B). On the other hand, the lipase activity was increased by disruption of tldp1 and suppressed by overexpression of tldp1 (Fig. 8C). We also found that the DG/TG ratio, which is used as an index for TG degradation, was higher in KO than in WT during the course of cultivation. This increase also dropped to the WT level with the expression of tldp1 in KO (KO/OE) (Fig. 8D). These results indicate that the decrease in TG accumulation in KO is due to the promotion of TG degradation, but not to the suppression of TG synthesis.
Fig. 8.
Effects of the disruption of tldp1 on the synthesis and degradation of TG. A: mRNA expression of several enzymes involved in TG metabolism. Real-time PCR was performed by the method described in the Materials and Methods. fas, fatty acid synthase (scaffold_21:887629-900072); pufas, PUFA synthase (scaffold_19:157378-169030); dgat2a, diacylglycerol acyltransferase 2A (scaffold_5:1007039-1014254). Blue and red bars represent the WT and KO, respectively. Data represent the mean ± SD (n = 3). B: DGAT activity. DGAT activity was measured at 30°C for 10 min using 15 μg of protein (cell lysate), 14C-palmitoyl-CoA, and DG (1,2-dipalmitoyl-sn-glycerol) by the method described in the Materials and Methods. Blue, red, and green bars represent the WT, KO, and KO/OE, respectively. Data represent the mean ± SD (n = 3). C: Lipase activity. Lipase activity was measured at 25°C for 30 min by a 4MU-palmitate using 10 μg of protein (cell lysate). WT, blue open circles; KO, red open squares; KO/OE, green open diamonds. Data represent the mean ± SD (n = 3). *P < 0.05; **P < 0.0001. D: DG/TG ratios. Blue, red, and green bars represent the WT, KO, and KO/OE, respectively. TG and DG were determined by LC-ESI MS as described in the Materials and Methods. Data represent the mean ± SE from three independent experiments (each n = 3).
DISCUSSION
LDs consist of a neutral lipid core and a phospholipid monolayer. The neutral lipid core is mainly composed of TG, while the phospholipid monolayer is covered by LD proteins that have structural and metabolic functions (40). In the present study, we identified seven proteins that were more strongly expressed in the LD fraction than in the LD-free fraction in A. limacinum F26-b when analyzed by 2D-DIGE (Fig. 1A–D). Among these seven proteins, TLDP1 was more strongly expressed in the LD fraction than in the LD-free fraction (approximately 20-fold stronger expression in the LD fraction than in the LD-free fraction) (Fig. 1E). Further biochemical and phylogenetic analyses revealed that: 1) TLDP1 did not possess a PAT1 region; however, it maintained its characteristics with known LD proteins, such as the 11-mer repeat region and 4-helix bundle sequences (Fig. 1G). 2) TLDP1 homologs were specifically distributed in thraustochytrids (Fig. 2). 3) TLDP1 surrounded LDs when expressed as a GFP-fused form (Fig. 4A, B). 4) The tldp1 disruption resulted in an abnormal LD morphology (Fig. 6) and a decrease in TG (Fig. 7A, B, D). These results clearly indicate that TLDP1 is an LD protein that is specifically conserved in thraustochytrids and regulates TG accumulation and LD morphology. However, further investigations are needed in order to establish whether the six other proteins are genuine LD proteins.
In spite of the universal presence of LDs from prokaryotes to plants and vertebrates, limited information is available on LDs in Stramenopiles, which is a major line of eukaryotes and has been classified into the Protista kingdom (41). Stramenopiles include photosynthetic and nonphotosynthetic taxa. Photosynthetic members include brown seaweed and diatoms, while nonphotosynthetic members comprise thraustochytrids, which were used in the present study. An LD protein (StLDP) was very recently isolated from the LD fraction of the marine diatom Phaeodactylum tricornutum belonging to Stramenopiles (42). However, StLDP did not show sequence homology to TLDP1 (identity <5%). StLDP was expressed during cultivation under nitrate deprivation and its mRNA expression increased as LD sizes became larger and, thus, this protein is considered to be associated with the LDs of Phaeodactylum tricornutum (42). In mammals, not only Plin family proteins but also the enzymes involved in TG and phospholipid metabolism, such as DGAT2 and CTP:phosphocholine cytidylyltransferase (CCT), are closely associated with LDs (43–45); however, we were not able to identify these enzymes using 2D-DIGE under the conditions used. Although 2D-DIGE is useful for identifying proteins that are more strongly expressed in the LD fraction than in the LD-free fraction, proteins that are equally expressed in both fractions must have been excluded by this method (27). Many proteins were detected at the same level in the LD and LD-free fractions (Fig. 1C, D). In order to obtain a clearer understanding of LD proteins in thraustochytrids, a total proteomic analysis of the LD fraction is needed. In addition, we crushed cells by sonication in order to obtain LDs from A. limacinum F26-b in this study, because thraustochytrids are surrounded by a rigid cell wall composed of sulfated glycans (46). Although ultrasonic treatment could damage LDs and release LD proteins from LDs, this study indicates that at least TLDP1 is recovered into the LD fraction even after preparation of LDs using sonication. However, to clear the complete protein profile of thraustochytrid LDs, the cells should be crushed by mild procedures without sonication.
We considered three protein spots (#250–252, Fig. 1C) in the LD fraction to be derived from the same protein (TLDP1) because the peptide sequences of the three protein spots matched each other very well and the three spots of TLDP1 disappeared in KO (Fig. 3H). These three proteins showed almost the same molecular masses on 2D-PAGE, whereas the isoelectric point of each protein was different (Fig. 1C), suggesting modifications with phosphate(s) on TLDP1. Although protein kinase A (PKA) consensus sequences were found on the putative primary structure of TLDP1, the phosphorylation site(s) and physiological relevance of the phosphorylation of TLDP1 remain elusive.
This study shows that TLDP1 regulates TG metabolism in LDs, possibly by protecting LDs from undergoing hydrolysis by lipase(s). This scenario may be similar to the functions of known LD proteins, such as Plin1 and its homologs. Plin1 may play a protective-barrier role in preventing the hydrolysis of TG in LDs by lipase(s), thereby inhibiting basal lipolysis (47). Disruption of the Plin1 gene in adipocytes caused an increase in basal lipolysis due to the loss of the protective barrier for lipase(s) (48). Similarly, the disruption of Mpl1, the LD protein in Metarhizium anisopliae, resulted in a lower total TG content than that in WT (49).
The mechanism by which TG degradation is suppressed by TLDP1 remains to be elucidated. A plausible scenario is that TLDP1 protects LDs from lipase attack by forming a protective barrier on LDs; however, the possibility that the suppression of lipase activity is caused by a specific interaction between TLDP1 and lipase(s) cannot be ruled out at present. In order to elucidate the specific interactions between lipase(s) and TLDP1, the lipase(s) involved in TG degradation need to be identified. However, there are at least 10 candidate lipases in the draft genome database of A. limacinum, and the lipase(s) involved in TG degradation currently remain unknown.
The KO mutant had lower LD numbers and unusually large LDs, and the expression of tldp1 in KO restored the size and number of LDs almost to the WT level (Fig. 6). Similarly, the depletion of oleosin in Arabidopsis thaliana resulted in the appearance of abnormal large LDs, which were assumed to be generated by the fusion of LDs (50). The mechanisms underlying the appearance of unusually large LDs in KO have not yet been elucidated; however, the loss of TLDP1 may result in the acceleration of LD fusion (to generate unusual large LDs and decrease LD numbers) and LD hydrolysis (to generate small LDs and decrease LD numbers).
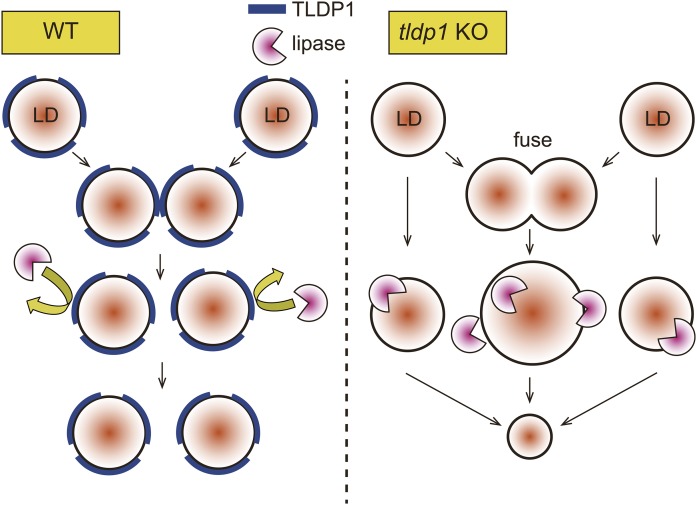
Collectively, we found that the loss of TLDP1, the first LD protein identified in thraustochytrids, decreased TG accumulation and caused a morphological abnormality in LDs in A. limacinum F26-b. These results suggest that TLDP1 inhibits TG degradation in LDs possibly by forming the protective barrier and that TLDP1 maintains LD morphology by preventing the unnecessary fusion of LDs. Two possible physiological roles of TLDP1 in A. limacinum are shown in Fig. 9.
Fig. 9.
Possible functions of TLDP1 on LDs. We observed unusually large LDs in KO, but not in WT, and the number of LDs per cell markedly decreased in KO in comparison with WT (Fig. 6C), suggesting that unusually large LDs may have been generated by the fusion of several small LDs in KO (Fig. 6D). Furthermore, the ratio of DG/TG increased in KO in comparison with WT, suggesting the increase of lipolysis in KO (Fig. 8D). These abnormal phenotypes of KO were restored by expression of tldp1 in KO. These results suggest that TLDP1 protects LDs against lipase attack and inhibits the fusion of LDs (WT, left panel), while the deletion of TLDP1 accelerates the lipolysis and fusion of LDs (KO, right panel).
The results of the present study show that thraustochytrids have a thraustochytrid-specific LD protein; however, the basic roles of LD proteins are preserved from single cell organisms to multicellular organisms, including animals and plants. To the best of our knowledge, this is the first study to describe the LD protein in thraustochytrids, which are expected to become a source of n-3 PUFAs to replace fish oils. In addition, they produced large amounts of TG containing palmitic acid (Fig. 7D), which can be used as a raw material for biofuels. Therefore, the result showing that the overexpression of tldp1 leads to the production of more TG may be useful when considering the industrial production of n-3 PUFAs and palmitic acid using thraustochytrids.
Supplementary Material
Acknowledgments
The authors thank Dr. Daiske Honda, Konan University (Japan) for his skillful and helpful comments on thraustochytrids. The authors also thank Dr. Masaki Matsumoto, Ms. Mizuho Oda, and Ms. Emiko Koba, Medical Institute of Bioregulation, Kyushu University (Japan) for the LC-MS/MS analysis and Mr. Jumpei Ito, National Institute of Genetics (Japan) for his technical advice on construction of the phylogenetic tree.
Footnotes
Abbreviations:
- ADRP
- adipocyte differentiation-related protein
- 2D-DIGE
- 2D-difference gel electrophoresis
- DG
- diacylglycerol
- DGAT
- diacylglycerol acyltransferase
- GFP
- green fluorescence protein
- GY
- glucose-yeast extract
- LD
- lipid droplet
- LIT
- linear ion trap
- LPC
- lysophosphatidylcholine
- MRM
- multiple reaction monitoring
- 4MU
- 4-methylumbelliferyl
- n-6 DPA
- n-6 docosapentaenoic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- Plin
- perilipin
- PNS
- post nuclear supernatant
- TG
- triacylglycerol
- TLDP1
- thraustochytrid-specific lipid droplet protein 1
This work was supported in part by Science and Technology Research Promotion Program Grant 26050A for Agriculture, Forestry, Fisheries, and Food Industry, Japan and Japanese Ministry of Education, Culture, Science, and Technology Basic Research B Grant 15H04488. The authors declare no competing financial interests.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Calder P. C. 2015. Marine omega-3 fatty acids and inflammatory processes: effects, mechanisms and clinical relevance. Biochim. Biophys. Acta. 1851: 469–484. [DOI] [PubMed] [Google Scholar]
- 2.Davidson M. H. 2013. Omega-3 fatty acids: new insights into the pharmacology and biology of docosahexaenoic acid, docosapentaenoic acid, and eicosapentaenoic acid. Curr. Opin. Lipidol. 24: 467–474. [DOI] [PubMed] [Google Scholar]
- 3.Lorente-Cebrián S., Costa A. G., Navas-Carretero S., Zabala M., Martínez J. A., and Moreno-Aliaga M. J.. 2013. Role of omega-3 fatty acids in obesity, metabolic syndrome, and cardiovascular diseases: a review of the evidence. J. Physiol. Biochem. 69: 633–651. [DOI] [PubMed] [Google Scholar]
- 4.Lenihan-Geels G., Bishop K. S., and Ferguson L. R.. 2013. Alternative sources of omega-3 fats: can we find a sustainable substitute for fish? Nutrients. 5: 1301–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagano N., Taoka Y., Honda D., and Hayashi M.. 2009. Optimization of culture conditions for growth and docosahexaenoic acid production by a marine thraustochytrid, Aurantiochytrium limacinum mh0186. J. Oleo Sci. 58: 623–628. [DOI] [PubMed] [Google Scholar]
- 6.Raghukumar S. 2008. Thraustochytrid marine protists: production of PUFAs and other emerging technologies. Mar. Biotechnol. (NY). 10: 631–640. [DOI] [PubMed] [Google Scholar]
- 7.Aasen I. M., Ertesvag H., Heggeset T. M., Liu B., Brautaset T., Vadstein O., and Ellingsen T. E.. 2016. Thraustochytrids as production organisms for docosahexaenoic acid (DHA), squalene, and carotenoids. Appl. Microbiol. Biotechnol. 100: 4309–4321. [DOI] [PubMed] [Google Scholar]
- 8.Geppert J., Kraft V., Demmelmair H., and Koletzko B.. 2005. Docosahexaenoic acid supplementation in vegetarians effectively increases omega-3 index: a randomized trial. Lipids. 40: 807–814. [DOI] [PubMed] [Google Scholar]
- 9.Qiu X., Hong H., and MacKenzie S. L.. 2001. Identification of a Δ4 fatty acid desaturase from Thraustochytrium sp. involved in the biosynthesis of docosahexanoic acid by heterologous expression in Saccharomyces cerevisiae and Brassica juncea. J. Biol. Chem. 276: 31561–31566. [DOI] [PubMed] [Google Scholar]
- 10.Xie Y., and Wang G.. 2015. Mechanisms of fatty acid synthesis in marine fungus-like protists. Appl. Microbiol. Biotechnol. 99: 8363–8375. [DOI] [PubMed] [Google Scholar]
- 11.Abe E., Ikeda K., Nutahara E., Hayashi M., Yamashita A., Taguchi R., Doi K., Honda D., Okino N., and Ito M.. 2014. Novel lysophospholipid acyltransferase PLAT1 of Aurantiochytrium limacinum F26-b responsible for generation of palmitate-docosahexaenoate-phosphatidylcholine and phosphatidylethanolamine. PLoS One. 9: e102377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi T., Sakaguchi K., Matsuda T., Abe E., Hama Y., Hayashi M., Honda D., Okita Y., Sugimoto S., Okino N., et al. 2011. Increase of eicosapentaenoic acid in thraustochytrids through thraustochytrid ubiquitin promoter-driven expression of a fatty acid Δ5 desaturase gene. Appl. Environ. Microbiol. 77: 3870–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuda T., Sakaguchi K., Hamaguchi R., Kobayashi T., Abe E., Hama Y., Hayashi M., Honda D., Okita Y., Sugimoto S., et al. 2012. Analysis of Δ12-fatty acid desaturase function revealed that two distinct pathways are active for the synthesis of PUFAs in T. aureum ATCC 34304. J. Lipid Res. 53: 1210–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakaguchi K., Matsuda T., Kobayashi T., Ohara J., Hamaguchi R., Abe E., Nagano N., Hayashi M., Ueda M., Honda D., et al. 2012. Versatile transformation system that is applicable to both multiple transgene expression and gene targeting for Thraustochytrids. Appl. Environ. Microbiol. 78: 3193–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohara J., Sakaguchi K., Okita Y., Okino N., and Ito M.. 2013. Two fatty acid elongases possessing C18-Δ6/C18-Δ9/C20-Δ5 or C16-Δ9 elongase activity in Thraustochytrium sp. ATCC 26185. Mar. Biotechnol. (NY). 15: 476–486. [DOI] [PubMed] [Google Scholar]
- 16.Liu B., Ertesvag H., Aasen I. M., Vadstein O., Brautaset T., and Heggeset T. M.. 2016. Draft genome sequence of the docosahexaenoic acid producing thraustochytrid Aurantiochytrium sp. T66. Genom. Data. 8: 115–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abe E., Hayashi Y., Hama Y., Hayashi M., Inagaki M., and Ito M.. 2006. A novel phosphatidylcholine which contains pentadecanoic acid at sn-1 and docosahexaenoic acid at sn-2 in Schizochytrium sp F26-b. J. Biochem. 140: 247–253. [DOI] [PubMed] [Google Scholar]
- 18.Walther T. C., and Farese R. V. Jr. 2012. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81: 687–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujimoto T., and Parton R. G.. 2011. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb. Perspect. Biol. 3: a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pol A., Gross S. P., and Parton R. G.. 2014. Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J. Cell Biol. 204: 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiam A. R., Farese R. V. Jr., and Walther T. C.. 2013. The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 14: 775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen B. C., Shamay A., and Argov-Argaman N.. 2015. Regulation of lipid droplet size in mammary epithelial cells by remodeling of membrane lipid composition-a potential mechanism. PLoS One. 10: e0121645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bickel P. E., Tansey J. T., and Welte M. A.. 2009. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta. 1791: 419–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laibach N., Post J., Twyman R. M., Gronover C. S., and Prufer D.. 2015. The characteristics and potential applications of structural lipid droplet proteins in plants. J. Biotechnol. 201: 15–27. [DOI] [PubMed] [Google Scholar]
- 25.Ding Y., Zhang S., Yang L., Na H., Zhang P., Zhang H., Wang Y., Chen Y., Yu J., Huo C., et al. 2013. Isolating lipid droplets from multiple species. Nat. Protoc. 8: 43–51. [DOI] [PubMed] [Google Scholar]
- 26.Yu J., Zhang S., Cui L., Wang W., Na H., Zhu X., Li L., Xu G., Yang F., Christian M., et al. 2015. Lipid droplet remodeling and interaction with mitochondria in mouse brown adipose tissue during cold treatment. Biochim. Biophys. Acta. 1853: 918–928. [DOI] [PubMed] [Google Scholar]
- 27.Uchida A., Sassa H., Takenaka S., Sakakibara Y., Suiko M., and Kunitake H.. 2012. Identification of self-incompatibility related proteins in the pistil of Japanese pear [Pyrus pyrifolia (Burm.f.)] by proteome analysis. Plant Omics. 5: 320–325. [Google Scholar]
- 28.Murata D., Nomura K. H., Dejima K., Mizuguchi S., Kawasaki N., Matsuishi-Nakajima Y., Ito S., Gengyo-Ando K., Kage-Nakadai E., Mitani S., et al. 2012. GPI-anchor synthesis is indispensable for the germline development of the nematode Caenorhabditis elegans. Mol. Biol. Cell. 23: 982–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar S., Stecher G., and Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones D. T., Taylor W. R., and Thornton J. M.. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- 31.Siloto R. M., Truksa M., He X., McKeon T., and Weselake R. J.. 2009. Simple methods to detect triacylglycerol biosynthesis in a yeast-based recombinant system. Lipids. 44: 963–973. [DOI] [PubMed] [Google Scholar]
- 32.Hamilton J., Jones I., Srivastava R., and Galloway P.. 2012. A new method for the measurement of lysosomal acid lipase in dried blood spots using the inhibitor Lalistat 2. Clin. Chim. Acta. 413: 1207–1210. [DOI] [PubMed] [Google Scholar]
- 33.Kelly S., and Bakhru-Kishore R.. 1979. Fluorimetric assay of acid lipase in human leukocytes. Clin. Chim. Acta. 97: 239–242. [DOI] [PubMed] [Google Scholar]
- 34.Ikeda K., Oike Y., Shimizu T., and Taguchi R.. 2009. Global analysis of triacylglycerols including oxidized molecular species by reverse-phase high resolution LC/ESI-QTOF MS/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2639–2647. [DOI] [PubMed] [Google Scholar]
- 35.Taguchi R., Houjou T., Nakanishi H., Yamazaki T., Ishida M., Imagawa M., and Shimizu T.. 2005. Focused lipidomics by tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 823: 26–36. [DOI] [PubMed] [Google Scholar]
- 36.Krank J., Murphy R. C., Barkley R. M., Duchoslav E., and McAnoy A.. 2007. Qualitative analysis and quantitative assessment of changes in neutral glycerol lipid molecular species within cells. Methods Enzymol. 432: 1–20. [DOI] [PubMed] [Google Scholar]
- 37.Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenberg A. S., Egan J. J., Wek S. A., Moos M. C. Jr., Londos C., and Kimmel A. R.. 1993. Isolation of cDNAs for perilipins A and B: sequence and expression of lipid droplet-associated proteins of adipocytes. Proc. Natl. Acad. Sci. USA. 90: 12035–12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chapman K. D., Dyer J. M., and Mullen R. T.. 2012. Biogenesis and functions of lipid droplets in plants. J. Lipid Res. 53: 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujimoto T., Ohsaki Y., Cheng J., Suzuki M., and Shinohara Y.. 2008. Lipid droplets: a classic organelle with new outfits. Histochem. Cell Biol. 130: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Derelle R., Lopez-Garcia P., Timpano H., and Moreira D.. 2016. A phylogenomic framework to study the diversity and evolution of Stramenopiles (=Heterokonts). Mol. Biol. Evol. 33: 2890–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoneda K., Yoshida M., Suzuki I., and Watanabe M. M.. 2016. Identification of a major lipid droplet protein in a marine diatom Phaeodactylum tricornutum. Plant Cell Physiol. 57: 397–406. [DOI] [PubMed] [Google Scholar]
- 43.Harris C. A., Haas J. T., Streeper R. S., Stone S. J., Kumari M., Yang K., Han X., Brownell N., Gross R. W., Zechner R., et al. 2011. DGAT enzymes are required for triacylglycerol synthesis and lipid droplets in adipocytes. J. Lipid Res. 52: 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu N., Zhang S. O., Cole R. A., McKinney S. A., Guo F., Haas J. T., Bobba S., Farese R. V. Jr., and Mak H. Y.. 2012. The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J. Cell Biol. 198: 895–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krahmer N., Guo Y., Wilfling F., Hilger M., Lingrell S., Heger K., Newman H. W., Schmidt-Supprian M., Vance D. E., Mann M., et al. 2011. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 14: 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darley W. M., Porter D., and Fuller M. S.. 1973. Cell wall composition and synthesis via Golgi-directed scale formation in the marine eucaryote, Schizochytrium aggregatum, with a note on Thraustochytrium sp. Arch. Mikrobiol. 90: 89–106. [DOI] [PubMed] [Google Scholar]
- 47.Brasaemle D. L., Rubin B., Harten I. A., Gruia-Gray J., Kimmel A. R., and Londos C.. 2000. Perilipin A increases triacylglycerol storage by decreasing the rate of triacylglycerol hydrolysis. J. Biol. Chem. 275: 38486–38493. [DOI] [PubMed] [Google Scholar]
- 48.Sztalryd C., Xu G., Dorward H., Tansey J. T., Contreras J. A., Kimmel A. R., and Londos C.. 2003. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J. Cell Biol. 161: 1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang C., and St Leger R. J.. 2007. The Metarhizium anisopliae perilipin homolog MPL1 regulates lipid metabolism, appressorial turgor pressure, and virulence. J. Biol. Chem. 282: 21110–21115. [DOI] [PubMed] [Google Scholar]
- 50.Siloto R. M., Findlay K., Lopez-Villalobos A., Yeung E. C., Nykiforuk C. L., and Moloney M. M.. 2006. The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell. 18: 1961–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brasaemle D. L. 2007. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 48: 2547–2559. [DOI] [PubMed] [Google Scholar]
- 52.Rowe E. R., Mimmack M. L., Barbosa A. D., Haider A., Isaac I., Ouberai M. M., Thiam A. R., Patel S., Saudek V., Siniossoglou S., et al. 2016. Conserved amphipathic helices mediate lipid droplet targeting of perilipins 1–3. J. Biol. Chem. 291: 6664–6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bussell R. Jr., and Eliezer D.. 2003. A structural and functional role for 11-mer repeats in alpha-synuclein and other exchangeable lipid binding proteins. J. Mol. Biol. 329: 763–778. [DOI] [PubMed] [Google Scholar]
- 54.Gautier R., Douguet D., Antonny B., and Drin G.. 2008. HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics. 24: 2101–2102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.