Summary
Plant defense to microbial pathogens is often accompanied by significant growth inhibition. How plants merge immune system function with normal growth and development is still poorly understood. Here, we investigated the role of target of rapamycin (TOR), an evolutionary conserved serine/threonine kinase, in the plant defense response.
We used rice as a model system and applied a combination of chemical, genetic, genomic and cell-based analyses.
We demonstrate that ectopic expression of TOR and Raptor (regulatory-associated protein of mTOR), a protein previously demonstrated to interact with TOR in Arabidopsis, positively regulates growth and development in rice. Transcriptome analysis of rice cells treated with the TOR-specific inhibitor rapamycin revealed that TOR not only dictates transcriptional reprogramming of extensive gene sets involved in central and secondary metabolism, cell cycle and transcription, but also suppresses many defense-related genes. TOR overexpression lines displayed increased susceptibility to both bacterial and fungal pathogens, whereas plants with reduced TOR signaling displayed enhanced resistance. Finally, we found that TOR antagonizes the action of the classic defense hormones salicylic acid and jasmonic acid.
Together these results indicate that TOR acts as a molecular switch for activation of cell proliferation and plant growth at the expense of cellular immunity.
Keywords: growth-defense trade-offs, jasmonic acid, plant defense, rice (Oryza sativa), salicylic acid, target of rapamycin (TOR)
Introduction
Plants live in complex environments where they are continuously subjected to attack by microbial pathogens and herbivorous insects. To survive these conditions, plants have evolved a multilayered immune system that is composed of two interconnected branches, termed pathogen associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) (Jones & Dangl, 2006). PTI is triggered by perception of invariant pathogen- or microbe-associated molecular patterns (PAMPs/MAMPs). In most cases, PTI is sufficient to impede pathogen colonization and suppress disease development. Successful pathogens, however, can disrupt PTI by injecting small effector proteins into the apoplast or the cytosol of host cells. Plants, in turn, have adapted to recognize these attacker-specific effectors by means of transmembrane or intracellular resistance proteins, triggering ETI (Jones & Dangl, 2006; Schwessinger & Ronald, 2012).
Since both PTI and ETI are associated with the mobilization of a diverse arsenal of energy-consuming biochemical and physical defenses, plant immune activation needs to be tightly regulated. Indeed, ecological and physiological studies have shown that activation of stress-adaptive responses often compromises growth and yield (Herms & Mattson, 1994; Huot et al., 2014). This so-called growth-defense trade-off is based on the premise that plants possess a limited pool of resources that demand prioritization towards either growth or immunity, depending on the prevailing conditions. As plants must both grow and defend themselves to survive and reproduce, maintaining and optimizing the balance between growth and immunity holds extensive agricultural importance. However, apart from the involvement of several hormone pathways and leucine-rich repeat receptor kinases (Belkhadirl et al., 2014; Huot et al., 2014; Lozano-Duran & Zipfel, 2015; Reitz et al., 2015; Campos et al., 2016), little is known about the molecular mechanisms and upstream regulatory components orchestrating growth-defense trade-offs.
One protein that may serve to balance plant growth and defense is the target of rapamycin (TOR) kinase. Originally identified in yeast through genetic mutant screens for resistance to the anti-proliferative drug rapamycin (Heitman et al., 1991), TOR is conserved in all eukaryotes and functions as a central command element that drives cellular and organismal growth and metabolism in response to endogenous and environmental cues (Dobrenel et al., 2016). The TOR signaling pathway is the focus of intensive research efforts in animals due to its role in health, disease, cancer and aging (Laplante & Sabatini, 2012; Fruman & Rommel, 2014). TOR proteins are atypical Ser/Thr kinases that belong to the family of phosphoinositide 3-kinase-related kinases. In yeast and mammals, TOR functions in two structurally and functionally distinct multiprotein complexes, named TORC1 and TORC2 (Laplante & Sabatini, 2012). Although TORC1 and TORC2 are present in a diverse range of organisms, plants only possess components of the TORC1 complex (Xiong & Sheen, 2015). Recent work in the model plant Arabidopsis thaliana has revealed that TORC1, which consists of TOR, Raptor (regulatory-associated protein of mTOR) and LST8 (lethal with SEC13 protein 8), promotes plant growth and cell proliferation by modulating a myriad of biological processes including transcription, translation, ribosome biogenesis, autophagy, and primary and secondary metabolism (Dobrenel et al., 2011, 2016; Henriques et al., 2014; Xiong & Sheen, 2014, 2015; Rexin et al., 2015; Xiong et al., 2016; Zhang et al., 2016). Ectopic expression of Arabidopsis TOR improves water use efficiency and yield potential in transgenic rice (Bakshi et al., 2017). However, whether TORC1-steered growth affects plant immunity and, hence, whether TOR orchestrates growth-defense trade-offs in plants remains to be tested.
Using rice as a model system and applying a combination of chemical, genetic, genomic and cell-based analyses, we demonstrate that TOR signaling not only coordinates plant growth and development but also alters susceptibility to fungal and bacterial pathogens. Moreover, our findings offer novel insights into the biological functions and regulatory mechanisms of TOR and suggest that TOR suppresses PTI at least in part by antagonizing the action of the defense hormones salicylic acid (SA) and jasmonic acid (JA). We propose that TOR acts as a molecular switch for activation of cell proliferation and plant growth at the expense of cellular immunity.
Materials and Methods
Plant materials and growth conditions
Unless noted otherwise, rice (Oryza sativa L.) plants (cultivar Kitaake) were maintained in the glasshouse (28 ± 2°C, 12 h photoperiod). Plants were grown in commercial potting soil (Structural; Snebbout, Kaprijke, Belgium) and fertilized weekly until flowering with a solution containing 0.2% iron sulfate and 0.1% ammonium sulfate. For in vitro experiments, rice seeds were surface sterilized by agitation in 2% sodium hypochlorite for 20 min, rinsed three times with sterile demineralized water and cultivated under growth chamber conditions (12 h 28°C : 12 h 24°C, day : night photoperiod, and 70% relative humidity) on Gamborg B5/1% plant agar medium (Duchefa Biochemie, Haarlem, the Netherlands).
Gene cloning and plasmid constructs
The coding sequences of OsTOR (LOC_Os05g14550) and OsRaptor1 (LOC_Os12g01922) were amplified using cDNA isolated from 2-wk-old Kitaake plants as PCR template. Total RNA was isolated using Trizol (Invitrogen) and treated with Turbo DNA-free DNAse (Ambion) following the manufacturer’s protocol. 2 μg of total RNA was used for cDNA synthesis using Superscript II Reverse Transcriptase (Thermo) and gene-specific RT primers (Supporting Information Table S1). Given the large size of the full length TOR cDNA (7.4 kB), we amplified seven DNA fragments with c. 25 bp overlap using Phusion polymerase (Thermo). Primer combinations used were TOR_F1-F7 (Table S1). For cloning OsRaptor1, four separate fragments were amplified using the primer combinations Raptor1_F1-F4 (Table S1). PCR products of the expected size were gel purified and 2 μl of each purified PCR product combined for a chimeric PCR reaction without primers using the following conditions: denaturation 95°C for 1 min, annealing 42°C for 30 s, extension 72°C for 30 s kB−1, 12 cycles. The chimeric PCR reaction was diluted 1 : 1000 and used as template in a PCR reaction using the flanking primer combinations TOR_FL and Raptor1_FL (Table S1). PCR products of the correct size were gel purified and cloned into pCR8/GW/TOPO vector (Invitrogen). We verified the DNA sequence of the inserts by standard Sanger sequencing and recombined the full length OsTOR and OsRaptor1 coding sequences into the Gateway-compatible pNC1300::Ubi destination vector (Chern et al., 2005) using LR clonase reactions (Invitrogen).
To construct vectors for RNAi-mediated gene silencing, we used primers TOR_RNAi and Raptor1_RNAi (Table S1) to amplify a 440 and 401-bp fragment of OsTOR and OsRaptor1 respectively. The PCR products were cloned into pCR8/GW/TOPO and confirmed by sequencing. The inserts were then recombined into the Gateway-compatible pANDA vector (Miki & Shimamoto, 2004) using LR Clonase (Invitrogen).
Rice transformation
Rice transformation was performed as described previously (Chern et al., 2005). Agrobacterium tumefaciens strain EHA105 was used to infect rice callus from cultivar Kitaake for transformation. Transgenic plants were selected on growth media containing hygromycin (40 mg l−1) and genotyped by PCR using primers Hyg-1 and Hyg-2 targeting the hygromycin selection marker.
Pharmacological experiments
All chemicals were stored and dissolved in dimethyl sulfoxide (DMSO) according to the manufacturer’s instructions. Aliquots of stock solution were stored at −20°C at a concentration of 10–30 mM. Plates containing Torin2 or Rapamycin were generally prepared a couple of days before use. For hormone treatments, the two youngest fully developed leaves from 4-wk-old seedlings were detached, cut into 2 cm pieces and incubated overnight on sterilized distilled water to remove any residual wounding stress. Hormone treatment was carried out in six-well plates by floating detached leaf pieces on aqueous solutions containing the respective hormones. If not stated otherwise, hormones and TOR inhibitors were administered throughout the entire duration of the experiments.
Rice pathogen culture and inoculation assays
Xanthomonas oryzae pv. oryzae (Xoo)
Bacterial inoculations were performed with Xoo strain PXO99 (Philippine race 6) (Song et al., 1995) using the standard leaf-clipping method. Before inoculation the pathogen was grown on Sucrose Peptone Agar (SPA) and incubated at 28°C. For inoculation experiments, single colonies were transferred to liquid SP medium and grown for 24–48 h at 28°C, until the OD600 had reached 0.5. Six-week-old plants were inoculated by clipping the fifth and sixth stage leaves of each tiller with scissors dipped in the Xoo suspension. Twelve days after inoculation, disease severity was assessed by measuring the length of the water-soaked lesions. For bacterial growth analysis, inoculated leaves from three plants were pooled, ground up thoroughly using mortar and pestle and resuspended in 5 to 10 ml water. The leaf suspensions were diluted accordingly and plated on SPA. Plates were incubated at 28°C in the dark and colonies were counted within 2–3 d.
Smear inoculation was carried out as described previously (Nozue et al., 2011). Rice leaves were smeared three times with Xoo-soaked gauze. For mock treatments, water was used instead of Xoo suspension.
Cochliobolus miyabeanus
Cochliobolus miyabeanus strain Cm988 (De Vleesschauwer et al., 2010) was grown for sporulation at 28°C on PDA. Seven-day-old mycelium was flattened onto the medium using a sterile spoon and exposed to blue light for 3 d under the same conditions mentioned above. Upon sporulation, conidia were harvested exactly as stated in De Vleesschauwer et al. (2016). Plants were inoculated at the five-leaf-stage by spraying a final concentration of 2×104 spores per ml in 0.5% gelatin (type B bovine skin) until run-off. Inoculated plants were kept in a dew chamber (≥92% relative humidity; 28 ± 2°C) for 24 h and thereafter transferred to growth chamber conditions (12 h 28°C : 12 h 24°C, day : night photoperiod, and 70% relative humidity) for disease development. Disease development was assessed 4 d post inoculation using digital image analysis (APS assess software; Lakhdar Lamari, Winnipeg, Canada) for quantification of symptomatic leaf areas.
Rhizoctonia solani
Rhizoctonia solani isolate 1–16 belonging to anastomosis group AG-1 IA (De Vleesschauwer et al., 2016), was maintained on potato dextrose agar (PDA). Detached leaves of 6-wk-old plants were inoculated by carefully placing 0.6-cm-diameter agar plugs taken from 5-d-old PDA cultures on the adaxial side of the leaves. Five days after incubation under growth chamber conditions (12 h 28°C : 12 h 24°C, day : night photoperiod, and 70% relative humidity), the area of the water-soaked disease lesions was quantified using APS assess software.
Rice suspension cell cultures
Suspension cultures of rice cells (cultivar Kitaake) were grown in modified AA medium on a rotary shaker (120 rpm) at 28°C in the dark. The cells were subcultured in fresh medium every week (10 ml old culture to 40 ml new AA), and all experiments were performed 3 or 4 d after transfer. The alkalinization response of the cells was measured using a semi-micro pH electrode.
RNA extraction and quantitative RT-PCR
Total leaf RNA was extracted using TRIZOL reagent according to the manufacturer’s protocol (Invitrogen) and subsequently treated with Turbo DNase (Ambion) to remove genomic DNA contamination. First-strand cDNA was synthesized from 2 mg of total RNA using Multiscribe reverse transcriptase (Applied Biosystems) and random primers following the manufacturer’s instructions. Quantitative PCR amplifications were conducted exactly as described in De Vleesschauwer et al. (2016). Nucleotide sequences of all primers used are listed in Table S1.
Microarray analysis and data processing
Rice suspension cells (cultivar Kitaake) were grown and treated with 100 μM rapamycin as described before. Samples from mock-infected and brown spot-inoculated plants were taken 24 h post treatment. Three independent biological repetitions were included in the analysis. For microarray analysis, we used a previously described two-dye method allowing direct comparison between two samples on the same microarray (Satoh et al., 2010). In brief, cyanine 3- or cyanine 5-labelled cRNA samples were synthesized from 850 ng of total RNA using a low-input RNA labelling kit (Agilent Technologies, Santa Clara, CA, USA) and hybridized to commercially available 60-mer 44K Agilent rice arrays according to the manufacturer’s protocols (Agilent Technologies). A full list of the expressed sequence tags (ESTs) represented on the array can be found at http://ricexpro.dna.affrc.go.jp/rice-44k-microarray.html. Following washing, slide image files were generated using a DNA microarray scanner (Agilent Technologies) and signal intensities were extracted and normalized within each array using Feature Extraction version 9.5 (Agilent Technologies). Signal intensities among all arrays were normalized according to the quantile method for standardization (global scaling) using EXPANDER 6. All data are available in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) database under the reference GSE93872. Significance analysis was performed using a fixed linear model (LIMMA) implemented in Multiple Experiment Viewer (MeV). Differentially expressed genes were defined as genes with a false discovery rate of <0.05 and at least three-fold difference in signal ratio. Gene ontology analysis was performed with MapMan version 3.5.1 (Usadel et al., 2009).
SA and JA measurements
Collected leaf samples were homogenized using liquid nitrogen; and 100 mg of plant material was extracted for 24 h at −80°C using 5 ml of the modified Bieleski solvent. After filtration (30 kDa Amicon® Ultra centrifugal filter unit, 30 min, 2700 g, 4°C) and evaporation (Turbovap®, 10°C, until dryness), the extract was reconstituted with 0.5 ml methanol/water (20 : 80 v/v) with 0.1% formic acid. Chromatographic separation was performed on a U-HPLC system (Thermo Fisher Scientific) equipped with a Nucleodur C18 column (50 × 2 mm; 1.8 μm dp) and using a mobile phase gradient (300 μl min−1; solvent A: 0–1 min at 20%, 1–2.5 min from 20 to 45%; 2.5–9 min from 45 to 100%; 9–10 min at 100%; 10–14 min at 20%) consisting of acidified (A) methanol (0.01% formic acid) and (B) water (0.1% formic acid). Mass spectrometric analysis was carried out in selected-ion monitoring (SIM; isolation window 1.0 m/z) mode with a Q Exactive™ Orbitrap mass spectrometer (Thermo Fisher Scientific), operating in negative electrospray ionization mode at a resolution of 70000 full width at half maximum.
Statistical analyses
All statistical analyses were performed using the software package SPSS Statistics v22. Normality of the data was verified by means of the Kolmogorov–Smirnov test (α = 0.05). Homoscedasticity of the data was verified by means of the modified Levene test (α = 0.05). When conditions of normality and homoscedasticity were fulfilled, data were compared by means of One-way ANOVA. In case these conditions were not met, nonparametric tests (Kruskal–Wallis and Mann–Whitney) were used.
Results
OsTOR drives transcriptional reprogramming of extensive gene sets involved in central metabolism and defense signaling
In a first attempt to shed light on the TOR signaling network in crop plants, we investigated the global transcriptome changes in rice suspension cells treated with the TOR-specific inhibitor rapamycin. In yeasts and animals, rapamycin has been used extensively to dissect the TOR signaling. Although plant TOR is considered to be poorly sensitive to rapamycin due to mutations in the FKBP12 protein that prevent the assembly of the inhibitory complex TOR-rapamycin-FKBP12 (Ren et al., 2011, 2012), recent studies showed that tomato and liquid-medium A. thaliana seedlings are sensitive to micromolar doses of rapamycin (Xiong & Sheen, 2012; Xiong et al., 2013, 2016). Consistent with these findings, we identified 1948 up- and 1911 downregulated rice genes in response to 100 μM rapamycin (Tables S2, S3). Gene ontology analysis confirmed that OsTOR directs myriad metabolic and regulatory processes ranging from primary and secondary metabolism to stress signaling, cell cycle control and organization, metabolite and nutrient transport, protein and cell wall modifications, and hormone metabolism (Fig. 1; Tables S4–S6). Much like in other organisms, OsTOR signaling appears to activate predominantly anabolic and biosynthetic processes while repressing catabolism-associated reactions (Tables S5, S6).
Fig. 1.
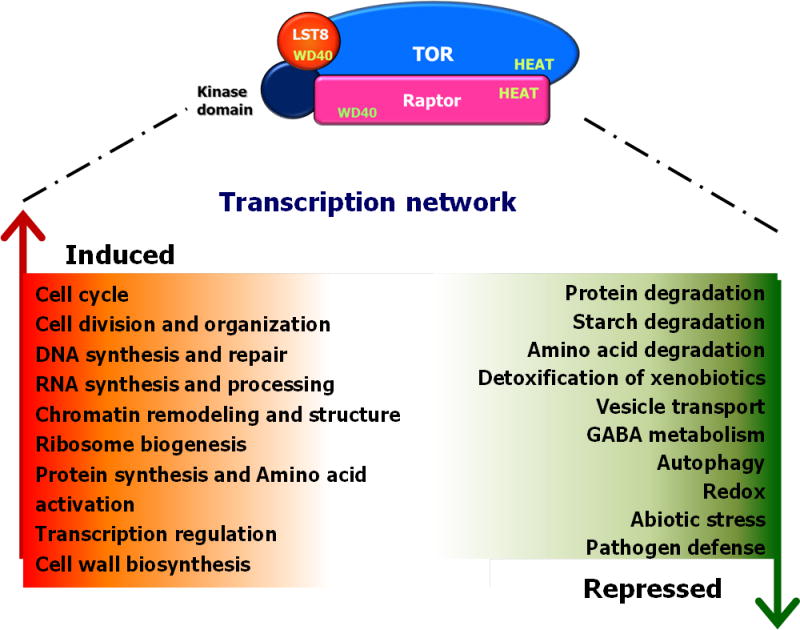
Overview of biological processes regulated by OsTOR signaling as determined by microarray analysis. Kitaake rice suspension cells were treated with 100 μM rapamycin for 24 h and subjected to genome-wide transcriptome analysis using 44k rice GeneChips. The results showed that OsTOR positively regulates anabolic processes while suppressing catabolic metabolism. In addition, OsTOR drives transcriptional reprogramming of extensive gene sets implicated in stress tolerance and plant defense responses.
From this analysis, we identified >120 genes encoding histones, chromatin modulators, ribosomal proteins and protein synthesis machineries that were significantly suppressed by rapamycin treatment, consistent with the previously established role of TOR in orchestrating transcriptional and translational processes in other eukaryotic species (Table S4). Similarly, genes implicated in cell cycle control, cell division and cell organization, and DNA and RNA repair and processing were significantly repressed. Rapamycin treatment also downregulated genes involved in key anabolic processes, including amino acid, lipid and nucleotide synthesis, but activated many genes mediating the degradation of sugars, proteins, amino acids, lipids and xenobiotics. Notably, several starch-degrading enzyme isoforms (e.g. alpha and beta amylases) were among the most strongly upregulated targets whereas starch synthases were mostly repressed, implying an important role of TOR signaling in regulating starch metabolism. Consistent with previous studies in A. thaliana (Ren et al., 2012; Caldana et al., 2013; Xiong et al., 2013, 2016), rapamycin-sensitive TOR signaling also affected a number of genes involved in glycolysis, TCA cycle and the electron transport chain, and activated broad gene sets coding for the synthesis and modification of plant cell walls and cell wall proteins such as arabinogalactans, extensins and expansins. Coupled with the extensive TOR regulation of primary, secondary and cell wall metabolism was the differential expression of a large set of genes coding for nutrient and metabolite transporters (sucrose, inositole, sulfate and ammonium transporters, and glucose-6-phosphate translocators), lipid transfer proteins, protein secretion and targeting, and vesicle trafficking.
Also a large number of stress-associated genes are activated upon rapamycin treatment, including genes encoding heat shock proteins, glutathione-S-transferases and redox-associated enzymes such as glutaredoxins, thioredoxins, catalases, ascorbate peroxidases and glutathione reductases (Table S4). These changes were observed alongside induction of several pathogenesis-related transcripts and activation of numerous defense-associated WRKY and MYB transcription factors. Most tellingly in this regard, disruption of TOR signaling by rapamycin led to a >30-fold induction of the SA master regulatory gene OsWRKY45. Finally, rapamycin treatment triggered significant upregulation of several genes implicated in the biosynthesis of defense-associated secondary metabolites such as lignin, carotenoids, flavonoids, tocopherol and phenylpropanoids. Given the widespread implication of these metabolites in plant defense responses, these findings suggest that TOR not only regulates growth and development but also may impinge on the plant’s innate immune system.
Identification of TOR and Raptor genes from rice
To further interrogate the potential function of TOR in orchestrating growth-defense trade-offs, we first identified the TOR and Raptor genes in the rice genome. While two TOR genes have been found in yeast, plants and mammals typically contain a single TOR gene (Laplante & Sabatini, 2012). Consistent with a recent study (Maegawa et al., 2015), we found a single rice TOR homolog present on chromosome 5. The rice TOR gene (OsTOR) spans 25.933 kB genomic region with 58 exons and 57 introns (Fig. S1a), which is similar to the number of introns found in TOR of A. thaliana (55) and Homo sapiens (56). The gene codes for a predicted 2465 amino acid-long protein containing all domains characteristic of eukaryotic TOR proteins, including the N-terminal HEAT, focal adhesion target (FAT) and FKBP12/rapamycin binding (FRB) motifs, as well as the kinase and C-terminal FAT (FATC) domain. Alignment of OsTOR with TOR protein sequences from other eukaryotes showed a high degree of sequence identity, especially within the kinase and FATC domains (Fig. S1b,c). Phylogenetic analysis revealed that plant TOR proteins resolve to two clades, clearly separating monocot and dicot species (Fig. S1d).
We found two loci encoding Raptor on chromosomes 12 and 11, designated OsRaptor1 and OsRaptor2. These genes are 8.527 and 5.969 kB in length (from translation initiation site to stop codon) and contain 22 and 21 introns, respectively (Fig. S2a). The predicted OsRaptor proteins contain the Raptor N-terminal Conserved (RNC) domain as well as the HEAT and WD40 motifs and show 99% identity. OsRaptor2 lacks the N-terminal 142 amino acids of OsRaptor1, suggesting it may be a truncated paralog. Alignment of these sequences with mammalian Raptor, the budding yeast Raptor homolog KOG1 and Raptor proteins from A. thaliana and other plant species showed that there is a striking degree of conservation among all Raptor homologs (Fig. S2b,c). OsRaptor1 and the Saccharomyces cereviseae Raptor homolog KOG1, the most divergent member included in the analysis, show 37% identity throughout their length.
TOR positively regulates growth and development in rice
We cloned the full length OsTOR and OsRaptor1 genes by PCR and generated transgenic rice lines expressing the cloned genes under the control of a constitutive maize ubiquitin promoter. Seven independent T0 lines for TOR OX and three for Raptor OX were obtained, out of which 49 T1 lines were randomly selected for further analysis (Table S7).
Consistent with previous reports in A. thaliana (Deprost et al., 2007), overexpression of full-length OsTOR positively influenced plant growth compared with wild-type Kitaake plants, causing phenotypes including longer shoots, increased root growth, enhanced tillering, earlier flowering and increased seed yield (Fig. 2; Table S7). In addition, TOR-overexpressing lines also showed an increase in epidermal and mesophyll cell size relative to nontransformed Kitaake (Figs 2i, S3). These phenotypes correlated well with the presence of the transgene as verified by PCR genotyping (Table S7). Moreover, qRT-PCR analysis demonstrated that many of the lines showing improved growth also exhibited increased expression of OsTOR relative to wild-type and azygous controls (Table S7).
Fig. 2.
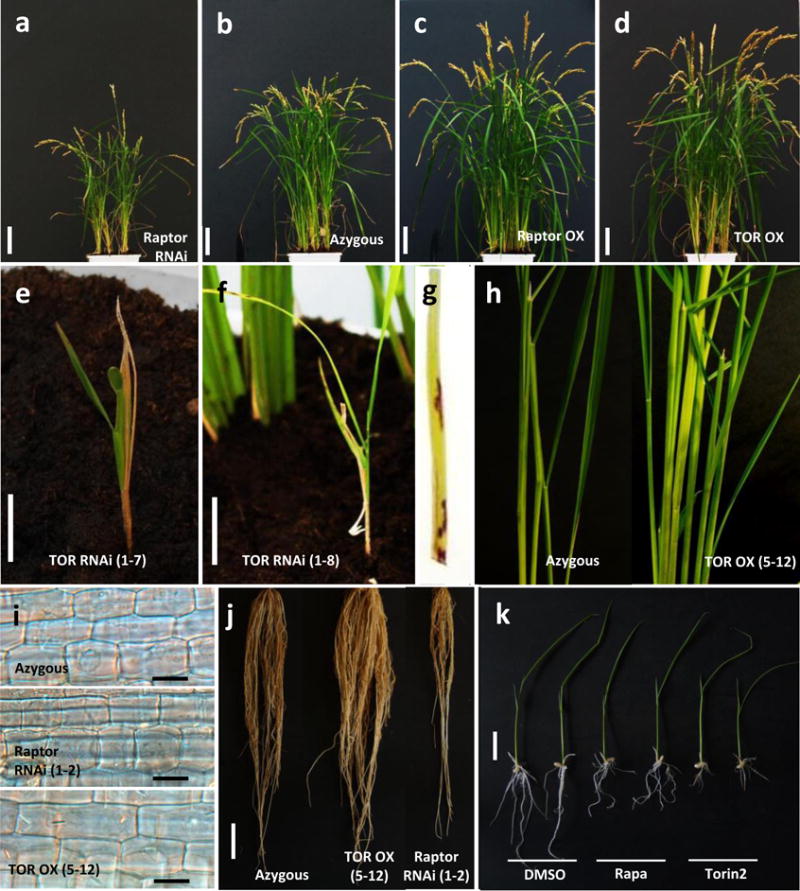
Target of rapamycin (TOR) positively regulates growth and development in rice plants. (a–g) Phenotypes of transgenic rice lines with increased or decreased TOR signaling output. From left to right: (a) Raptor RNAi (progeny of line Raptor RNAi 1–2), (b) azygous control, (c) Raptor-overexpressing (progeny of line Raptor OX 5–15), (d) TOR-overexpressing (progeny of line TOR OX 5–12), and (e, f) TOR RNAi plants. Pictures were taken 12 wk after germination. Silencing of TOR results in dwarfed growth, sterility and formation of necrotic lesion mimics (as shown in g), whereas TOR overexpression enhances tiller number, seed yield, epidermal and mesophyllic cell elongation, and root growth (h–j). (k) Chemical disruption of TOR signaling with the TOR-specific inhibitors rapamycin (100 μM) and Torin2 (30 μM) slows vegetative growth of in vitro grown Kitaake plants. Pictures were taken 10 d after imbibition. Bars: (a–f) 5 cm. (i) 50 μM; (j, k) 2 cm.
Overexpressing OsRaptor1 reproduced most of the phenotypes observed for the OsTOR OX transgenics, whereas reduced root and shoot growth was observed in rice lines silenced for OsRaptor1 (Fig. 2). Despite several attempts, only three T0 OsRaptor1 RNAi lines were obtained. Analysis of T1 progeny revealed a clear correlation between genotype and phenotype with lines carrying the transgene and displaying strongly reduced expression of OsRaptor1 growing smaller and slower compared to wild-type and azygous controls (Fig. 2; Table S7). Consistent with earlier work showing that reduced TOR signaling extends lifespan in a number of model organisms (Kapahi et al., 2004; Robida-Stubbs et al., 2012), silencing OsRaptor1 also affected senescence in detached leaf experiments. In contrast to the phenotypes observed for the OsTOR OX lines, OsRaptor1 RNAi plants showed downregulated expression of the senescence-associated marker gene OsSAG39 and, accordingly, delayed and reduced signs of visible yellowing as compared to wild-type plants and azygous controls (Fig. S3e,g).
To further extend these findings, we attempted to disrupt TOR signaling by generating OsTOR RNAi plants. However, only two T0 lines could be obtained and analysis of T1 progeny identified only two plants with substantially reduced (>40%) OsTOR expression relative to wild-type (Table S7). Interestingly, these plants showed strong phenotypes, including severe dwarfism, defects in leaf number, color and shape, reduced tillering, delayed flowering and the rapid development of necrotic lesion mimics (Fig. 2e,g). Moreover, none of the plants expressing the OsTOR RNAi construct and showing reduced OsTOR expression produced viable seeds. These data indicate a strong counterselection for the complete constitutive silencing of OsTOR and, hence, are consistent with previous reports identifying TOR as an essential gene in plants (Menand et al., 2002; Deprost et al., 2007; Ren et al., 2011, 2012; Caldana et al., 2013).
To circumvent abovementioned lethality and gain additional insight into the role of plant TOR signaling, we next pursued a pharmacological approach whereby wild-type rice plants (cultivar Kitaake) were grown in vitro on medium containing TOR-specific inhibitors. Treating rice seedlings with increasing concentrations of rapamycin reduced root elongation in a dose-dependent manner (Figs 2k, S3b,c). Consistent with the reported role of AtTOR in auxin signal transduction (Schepetilnikov et al., 2013; Deng et al., 2016), rapamycin treatment also led to a partial loss of root gravitropism (Fig. S3c). Reduced root and shoot elongation was also observed in response to the TOR ATP-competitive inhibitor, Torin2 (Fig. S3d). Together with the strong phenotypes of the OsTOR and OsRaptor1 transgenics, these data establish TOR as a positive regulator of rice growth and development.
TOR promotes disease susceptibility independent of the pathogen’s lifestyle
To test whether TOR also molds the outcome of rice–pathogen interactions, we challenged our transgenic lines with the hemibiotrophic bacterium Xoo, the causal agent of bacterial leaf blight disease. Six-week-old T1 progeny were examined for co-segregation of genotype with phenotype by PCR analysis and measurement of the length of the Xoo-induced leaf blight lesions (Fig. 3). Interestingly, both OsTOR and OsRaptor1 OX plants displayed significantly enhanced susceptibility to the virulent Xoo strain PXO99 at 12 dpi, with average lesions of 8–12 cm as compared to 5–6 cm on nontransformed Kitaake and azygous controls (Fig. 3a,b,d). By contrast, OsRaptor1 RNAi plants exhibited an c. 50% reduction in lesion length relative to wild-type and azygous controls (Fig. 3c). In OsTOR OX lines, growth parameters such as tiller number, plant height, biomass and seed weight were inversely correlated with Xoo resistance (Fig. S4). Bacterial growth analyses correlated well with lesion length developments. At 16 dpi, PXO99 titers reached c. 2 × 1010 colony forming units (CFU) per leaf in OsTOR and OsRaptor1 OX lines, a greater than 10-fold increase compared to non-transformed Kitaake and azygous controls. In OsRaptor1 RNAi plants, however, PXO99 grew 10-fold less than in the controls with populations leveling off to <1 × 108 CFU per leaf (Fig. 3e).
Fig. 3.
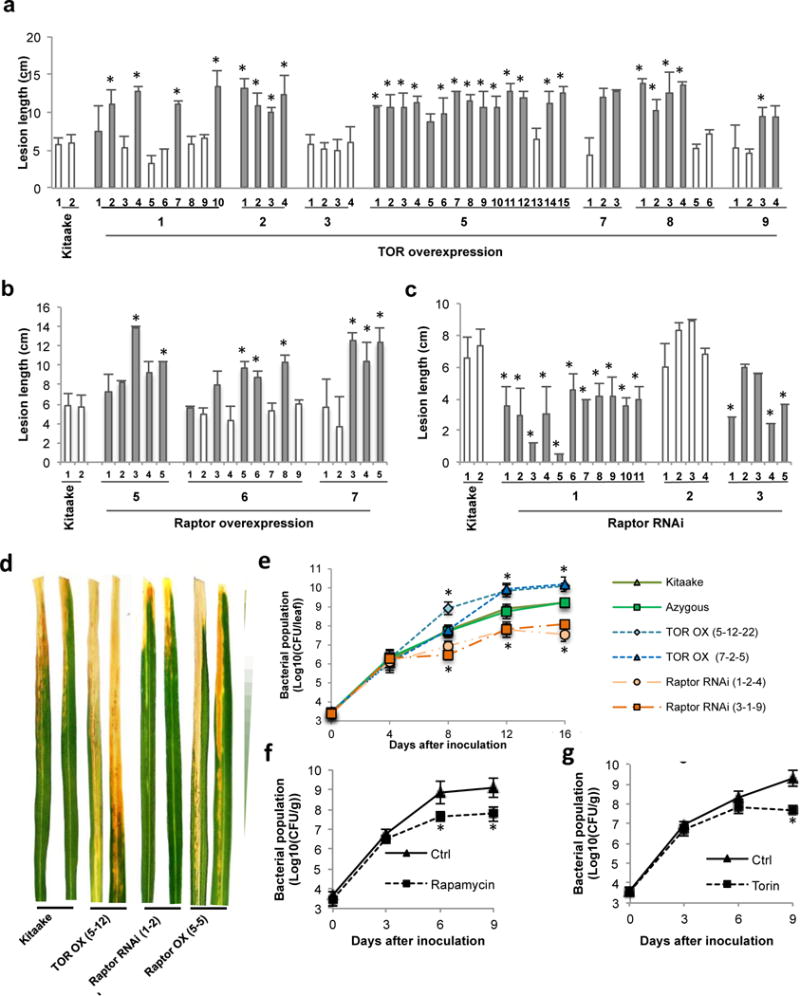
OsTOR induces susceptibility against the bacterial leaf blight pathogen Xanthomonas oryzae pv. oryzae (Xoo). (a–c) Segregating T1 progeny were genotyped for the presence of the transgene using hygromycin-specific primers. Closed bars represent plants carrying the transgene, open bars indicate azygous controls. All progeny plants were inoculated with virulent Xoo strain PXO99 using the leaf clipping method and lesion lengths were measured 12 d after inoculation. Data are means ± SD of at least three leaves. Asterisks indicate statistically significant differences compared to nontransformed Kitaake controls (Mann–Whitney, α = 0.05). These results showed that target of rapamycin (TOR) signaling output is inversely correlated with resistance to Xoo. Photos showing representative disease symptoms were taken 12 d after inoculation and are shown in (d). (e) Xoo PXO99 populations in Kitaake, a null transgene control and two independent T2 TOR OX and Raptor RNAi lines at 0–16 d after inoculation. Data shown are means ± SD of three replicates of at least two pooled leaves. Asterisks indicate statistically significant differences compared to Kitaake samples for each time point separately (Duncan, α = 0.05). (f, g) Blocking TOR signaling with rapamycin (100 μM) or Torin2 (30 μM) restricts Xoo multiplication in wild-type plants. The two youngest fully developed leaves of 6-wk-old Kitaake plants were detached, smear-inoculated with PXO99, and floated on aqueous solutions containing DMSO or TOR inhibitors. Bacterial densities were determined at 3-d intervals. Data are means ± SD of three replicates of four pooled leaves. Asterisks indicate statistically significant differences compared to control Kitaake samples (Duncan, α = 0.05). One repetition of the experiment yielded similar results. CFU, colony forming units.
Because it cannot be ruled out that the enhanced disease resistance seen in the OsRaptor1 RNAi plants is due to pleiotropic effects associated with the semi-dwarf phenotype of these lines, we also tested the effect of transient TOR inhibition on Xoo resistance. To this end, detached leaves of wild-type Kitaake plants were smear-inoculated with PXO99 as described previously (Nozue et al., 2011), and floated on aqueous solutions of rapamcyin or Torin2. Xoo multiplication was monitored at 3-d intervals. Consistent with the results from our OsRaptor1 RNAi lines, both rapamycin and Torin2 treatment significantly restricted Xoo multiplication (Fig. 3f,g). Together these findings strongly suggest that TOR acts as a negative regulator of resistance to Xoo.
To analyze whether TOR also conditions susceptibility to pathogens other than Xoo, we inoculated PCR-positive T3 progeny derived from two TOR OX lines, one Raptor OX line and two Raptor RNAi lines with the necrotrophic fungi R. solani and C. miyabeanus. Overexpression of TOR and Raptor1 rendered plants hyper-susceptible to both pathogens (Fig. 4a,b). Interestingly, bioassays with previously described Arabidopsis TOR mutant plants (Deprost et al., 2007) revealed a similar picture with TOR-overexpressing and raptor1-1 mutants showing increased and decreased susceptibility, respectively, against the hemibiotrophic bacterium Pseudomonas syringae pv. tomato, the necrotrophic leaf fungus Botrytis cinerea and the soil-borne vascular pathogen Verticillium longisporum (Fig. S3; Methods S1, S2). Moreover, treating young lettuce seedlings with rapamycin strongly reduced infection by the biotrophic oomycete pathogen Bremia lactucae (Fig. S5; Methods S3).
Fig. 4.
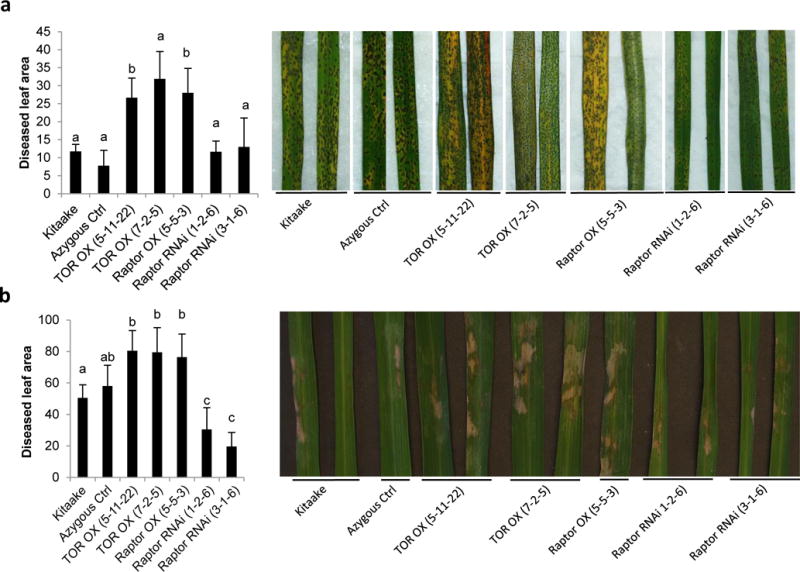
OsTOR conditions susceptibility towards the necrotrophic fungal pathogens Cochliobolus miyabeanus and Rhizoctonia solani. (a) T3 lines overexpressing target of rapamycin (TOR) or Raptor are less resistant to infection with C. miyabeanus strain Cm988 than wild-type Kitaake and null segregating control plants. Overexpressing TOR or Raptor also favors infection by the sheath blight pathogen R. solani (b). Data shown are means ± SD. Bars with different letters are significantly different (Mann Whitney, n ≥ 6, α = 0.05). Disease development was assessed using digital image analysis for quantification of symptomatic leaf areas. Leaves showing representative disease symptoms were photographed (a) 4 and (b) 5 d post inoculation, respectively. Experiments were repeated twice with similar results.
TOR antagonizes PAMP-triggered immunity
The ability of TOR to enhance susceptibility against pathogens exhibiting diverse lifestyles prompted us to test whether TOR affects PTI. To this end, we investigated the effect of rapamycin and Torin2 treatment on two PTI-associated immune responses occurring at different time points after PAMP exposure; medium alkalinization (a very early response indicative of membrane depolarization) and defense gene activation (a later response). Chitin and xylanase were selected as fungal PAMPs while Pseudomonas aeruginosa-derived lipopolysaccharides (LPS) were used as bacterial PAMP.
As shown in Fig. S6, neither rapamycin nor Torin2 significantly influenced the alkalinization response of rice suspension cells treated with any of these PAMPs. However, both TOR inhibitors strongly boosted chitin-inducible activation of the rice PTI marker genes MLO, β-glucanase, PAL, PAL1 and PAL2, with combined treatments resulting in a four- to 10-fold higher expression compared to cells treated with inhibitors or chitin only (Figs 5a, S7). Similar effects were observed in response to both xylanase and LPS (Figs 5b, S7). Given the inability of rapamycin and Torin2 to affect MAMP-triggered medium alkalinization, these findings suggest that TOR does not interfere with the perception of MAMPs per se but rather alleviates PTI by interfering downstream in the pathway. To test this hypothesis, we next investigated the effect of TOR on the expression of PTI-driven pathogen resistance. Five-week-old T3 TOR OX and Raptor RNAi lines were treated with chitin for 8 h and subsequently inoculated with C. miyabeanus. Pretreatment of detached leaves of wild-type Kitaake plants with 100 μg/ml chitin reduced the diseased leaf area by almost 30%, demonstrating the effectiveness of chitin-inducible PTI under our experimental conditions (Fig. 5c,d). Similar reductions in disease severity were obtained in a Raptor RNAi background (Fig. 5d). However, overexpressing TOR almost completely attenuated the chitin-inducible resistance observed in wild-type seedlings (Fig. 5d), further suggesting a negative effect of TOR on downstream PTI signaling.
Fig. 5.
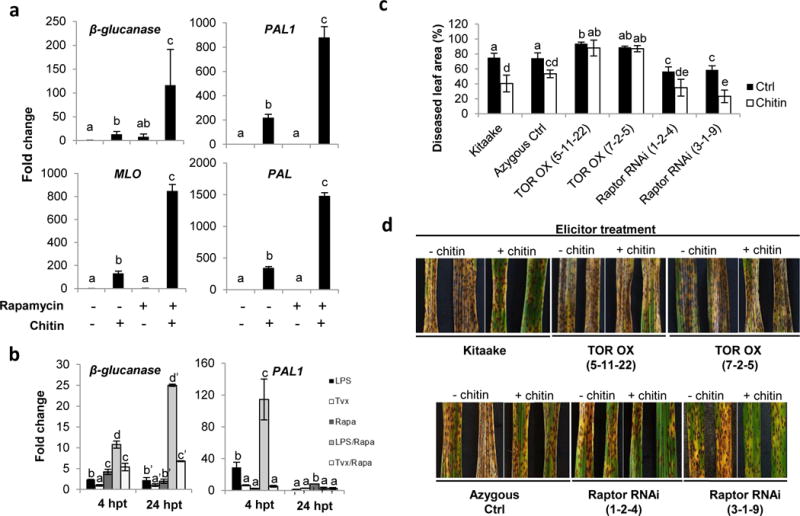
OsTOR signaling suppresses pathogen associated molecular pattern-triggered immunity (PTI). (a, b) Effect of rapamycin treatment (100 μM) on the expression of PTI marker genes in rice suspension cells treated with insoluble crab shell chitin (100 μg ml−1), Pseudomonas aeruginosa-derived lipopolysaccharides (LPS, 100 μg ml−1) or fungal xylanase (Tvx, 30 μg ml−1). Chitin-treated samples were harvested 4 h post treatment (hpt), LPS and Tvx-treatments were sampled at 4 and 24 hpt. Expression levels were measured by quantitative reverse transcription (qRT)-PCR and normalized to actin reference gene expression. Data shown are normalized to dimethyl sulfoxide (DMSO) (solvent control)-treated samples. Bars depict average expression level ± SD of two technical and two biological replicates. Different letters indicate statistically significant differences (T-test, α = 0.05). (c, d) Target of rapamycin (TOR) overexpression compromises chitin-induced resistance to the brown spot pathogen Cochliobolus miyabeanus. Detached leaves of wild-type Kitaake, an azygous control, and two independent TOR-overexpressing and Raptor RNAi lines were pretreated with insoluble chitin (200 μg ml−1) for 6 h before being challenged with C. miyabeanus strain Cm988. Disease development was assessed 4 d post inoculation using digital image analysis for quantification of symptomatic leaf areas. Data are from a representative experiment that was repeated twice with similar results. Bars with different letters are significantly different (Mann–Whitney, n ≥ 7, α = 0.05).
TOR negatively interacts with the defense hormones SA and JA
Recent advances in plant immunity research have underscored the central importance of plant hormone signaling in triggering downstream immune responses (Pieterse et al., 2009; Robert-Seilaniantz et al., 2011). In view of these findings and given the ability of TOR inhibition to amplify late but not early PTI responses, we hypothesized that TOR may subdue immunity at least in part by interfering with the plant’s hormone signaling network. Consistent with this hypothesis, we found several hormone pathways including those related to JA, ethylene, abscisic acid, auxins and brassinosteroids to be statistically over-represented among the differentially expressed genes in our microarray study (Tables S4–S6). Moreover, gene ontology terms associated with the same hormone pathways were significantly enriched within the top 100 OsTOR interacting proteins as predicted by the genome-scale network server RiceNet (Tables S8–S11). RiceNet is a probabilistic functional gene network for rice constructed using a modified Bayesian integration of many different data types from several different organisms, with each data type weighted according to how well it links genes that are known to function together in rice (Lee et al., 2011, 2015). RiceNet analysis also uncovered lipoxygenase, a domain characteristic of JA biosynthetic enzymes, as the most significantly enriched motif among the putative OsTOR interactors (Table S12). JA, along with SA, are central immunity hormones and their importance in the plant defense network is well established (Pieterse et al., 2009; De Vleesschauwer et al., 2014). Therefore, to further elucidate how OsTOR affects rice immunity, we focused on exploring its interaction with the rice SA and JA pathways.
Consistent with TOR antagonizing JA signaling, rapamycin treatment potentiated the growth-repressive effect of exogenously administered MeJA and boosted expression of the JA-marker genes JiPR10 and JaMYB in rice suspension cells inoculated with the supernatant of virulent Xoo cultures (Fig. 6a,b). Note that cells were infected with culture supernatant to prevent the rapid cell death seen in preliminary experiments using living Xoo bacteria. Treating inoculated rice cells with rapamycin also resulted in a strong upregulation of the SA marker genes NPR1 and WRKY45, suggesting that endogenous TOR antagonizes both SA and JA signaling in rice (Fig. 6a). Supporting this hypothesis, we observed increased expression of JA- and SA-responsive genes in leaves of 3-wk-old TOR and Raptor RNAi plants, whereas overexpressing TOR resulted in constitutive downregulation of JA and SA marker genes (Fig. 6c). Moreover, TOR overexpressing plants exhibited a significant decrease in endogenous JA content relative to wild-type Kitaake and azygous controls, whereas OsRaptor RNAi accumulated approximately three times the amount of JA and SA found in the non-transgenic controls (Fig. 6d). Finally, silencing OsRaptor1 primed SA- and JA-dependent gene expression in response to exogenous benzothiadiazole (BTH) and MeJA treatments (Fig. 6e,f). Together these findings support the hypothesis that TOR enhances disease susceptibility, at least in part, through antagonistic crosstalk with SA and JA.
Fig. 6.
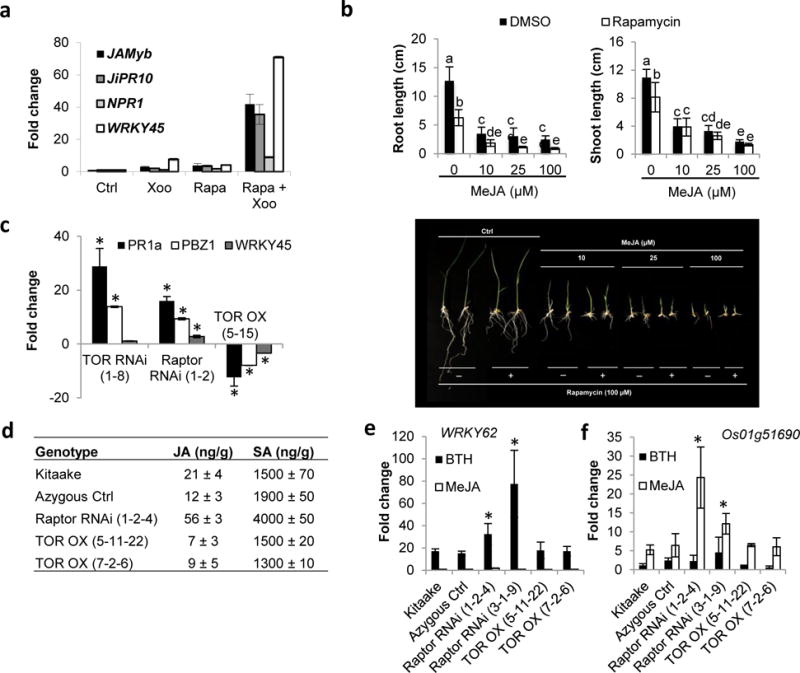
OsTOR signaling attenuates the action of the classic immune hormones salicylic acid (SA) and jasmonic acid (JA). (a) Pharmacological inhibition of target of rapamycin (TOR) signaling primes expression of SA and JA marker genes in rice cell cultures treated with supernatant of Xanthomonas oryzae pv. oryzae (Xoo) PXO99 cultures. Two milliliters suspension cells were treated with 100 μM rapamycin and/or 50 μl Xoo supernatant. Samples were harvested 6 h after treatment and analyzed by quantitative reverse transcription (qRT)-PCR. Data shown are expressed relative to dimethyl sulfoxide (DMSO) (solvent control)-treated samples. Bars depict average expression level ± SD of two technical and two biological replicates. Asterisks indicate statistically significant differences relative to DMSO-treated samples (T-test, α = 0.05). (b) Rapamycin treatment potentiates methyljasmonate (MeJA)-induced growth restriction. Pregerminated Kitaake seedlings were grown on medium containing increasing doses of MeJA in combination or not with 100 μM rapamycin. Root and shoot lengths were recorded 7 d after germination. Bars depict means ± SD. Data are from a representative experiment that was repeated twice with similar results. Different letters indicate statistically significant differences (Duncan, n = 12, α = 0.05). (c) OsTOR signaling antagonizes basal expression of SA and JA marker genes. Fourth leaves of 3-wk-old TOR RNAi, Raptor RNAi and TOR OX plants were harvested and analyzed by qRT-PCR. Data shown are expressed relative to the expression in azygous controls. Bars depict average expression level ± SD of three technical replicates. Asterisks indicate statistically significant differences (T-test, α = 0.05). (d) Quantification of endogenous JA and SA levels in TOR OX and Raptor RNAi plants. Data are means ± SD from three biological replicates, each representing a pooled sample from at least 10 individual plants. Different letters indicate statistically significant differences (Duncan, α = 0.05). (e, f) Silencing OsRaptor1 primes benzothiadiazole (BTH)- and MeJA-inducible gene expression. Wild-type Kitaake, null transgene controls and Raptor RNAi and TOR OX lines were treated with 500 μM BTH or 100 μM MeJA and sampled 8 h post treatment. Data are normalized to control-treated samples. Bars depict average expression level ± SD of two technical and two biological replicates. Asterisks indicate statistically significant differences compared to similarly treated Kitaake (T-test, α = 0.05).
Discussion
To survive, plants must adapt to changing environmental conditions while optimizing their growth rate and reproductive success. When plants sense the presence of pathogens, they activate a vast repertoire of energetically costly defense programs, resulting in a temporary reduction of investments made in growth. Although it has been often hypothesized that reallocation of energy and limited carbon pools underpins growth-defense trade-offs (Huot et al., 2014), little is known about the molecular mechanisms orchestrating this regulation.
Recently, crosstalk between hormone signaling networks has emerged as a crucial factor in partitioning growth-defense trade-offs with most reports focusing on the reciprocal antagonism between the central defense hormones SA and JA on the one hand, and the ‘growth hormones’ auxin, cytokinins, brassinosteroids and gibberellins, on the other (De Bruyne et al., 2014; Huot et al., 2014; Naseem et al., 2014; Lozano-Duran & Zipfel, 2015; Bjornson et al., 2016; De Vleesschauwer et al., 2016). New molecular insights also highlight the cooperative function of specific signaling components in balancing growth and defense, including the brassinosteroid receptor BRI1, the bacterial flagellin receptor FLS2, the photoreceptor phytochrome B, JA ZIM-domain (JAZ) proteins, and the transcriptional cascade formed by the transcription factors BZR1 and HBI1 (Belkhadirl et al., 2014; Lozano-Duran & Zipfel, 2015; Campos et al., 2016). In adding to this list, our findings uncover the master growth regulator TOR as a negative regulator of plant immunity and support a scenario whereby TOR antagonizes plant defenses by interfering either directly or indirectly with the classic defense hormones SA and JA. Considering the well-established role of TOR in driving eukaryotic cell proliferation and growth, these findings posit TOR at the nexus of growth and defense signaling and establish TOR as a central command element configuring the plant’s growth and immune programs.
Our results demonstrate that TOR activation can condition susceptibility in multiple plant species and against different pathogens, with examples of the latter exhibiting various parasitic habits (e.g. biotrophy, hemibiotrophy and necrotrophy). Although these results are compatible with a conserved role for TOR in balancing growth and defense, it does not follow that active TOR signaling triggers susceptibility to all pathogens. Indeed, we did not see increased susceptibility to B. cinerea in the Arabidopsis TOR OX plants. Moreover, contrary to our data, Meteignier et al. (2017) failed to detect enhanced susceptibility to P. syringae pv. tomato in their TOR OX lines, while Popa et al. (2016) recently found Arabidopsis TOR RNAi plants to be more susceptible to the bacterial wilt pathogen Ralstonia solanacearum. These differences seen between different studies and pathosystems suggest that the emerging new role of TOR in modulating disease resistance depends on specialized features of each interaction (e.g. strength of TOR signaling output), rather than on the overall infection biology and lifestyle of the invading pathogen. Thus, complex nuanced mechanisms seem to underlie TOR modulation of plant biotic stresses.
In the absence of the adaptive immunity displayed by animals, plants rely solely on innate immunity, which involves two tiers of functionally interlinked immune responses termed PTI and ETI (Jones & Dangl, 2006). In addition to recent findings linking ETI to translational repression of TOR (Meteignier et al., 2017), our results hint at a potential role for TOR in intercepting PTI responses. This notion was born out by the stimulatory effect of rapamycin co-treatment on MAMP-induced expression of PTI marker genes and the observation that TOR overexpression disables chitin-induced pathogen resistance. Interestingly, rapamycin had no effect on early PTI events such as medium alkalinization, which raises the possibility that TOR dampens PTI by interfering with downstream signaling. Although these results should be interpreted with caution because it is not known at this stage whether rapamycin merely has a TOR-inhibiting effect or might contribute more broadly by altering the metabolic state of the cell, the involvement of TOR in PTI seems apparent. Clearly, unraveling the molecular intricacies of TOR-PTI signal interactions is an important challenge ahead.
Consistent with recent findings in A. thaliana (Dong et al., 2015; Song et al., 2017), we found that pharmacological and genetic disruption of TOR strongly boosts SA and JA responses in rice. Given the paramount importance of both hormones in orchestrating myriad defense reactions, these findings support the notion that TOR restricts plant immunity at least in part by antagonizing SA and JA. Importantly, such antagonistic interconnection between TOR and SA/JA not only offers a mechanistic framework for how TOR conditions broad-spectrum disease susceptibility, but also reinforces the emerging paradigm that growth-defense antagonism is likely not caused by competition for limited metabolic resources but rather by activation of conflicting hormone signaling pathways (Eichmann & Schafer, 2015; Campos et al., 2016; Havko et al., 2016; Kliebenstein, 2016).
Theoretically, cross talk between signaling pathways may occur at the level of biosynthesis regulation, signal transduction, and/or gene expression. Although the elevated JA titers found in our Raptor RNAi lines could be explained by the rapamycin-induced activation of several JA biosynthetic genes (see Table S4), further work will be required to unveil the exact mechanism(s) by which TOR impinges on SA and JA pathways. To date, few plant TOR substrates and phosphorylation targets have been identified and none of them are known to function in the SA or JA pathways (Xiong & Sheen, 2015). Yet, the effect of TOR on SA and JA responses may also be explained through its crosstalk with the sucrose non-fermenting-related kinase 1 (SnRK1). SnRK1 is the plant ortholog of the budding yeast SNF1 and mammalian AMP-activated protein kinase (AMPK) (Hulsmans et al., 2016). In contrast to TOR, these evolutionarily conserved kinases safeguard cellular energy homeostasis and promote tolerance to adverse conditions, partly through an induction of catabolic processes and a general repression of anabolism. In animals, AMPK and TOR interact in a mutually antagonistic fashion (Hindupur et al., 2015). Aspirin, which is rapidly broken down to SA in vivo, directly activates AMPK and inhibits mTOR signaling in colorectal cancer cells (Din et al., 2012; Hawley et al., 2012). Moreover, JA activates the expression of various SnRK1 interaction partners in Arabidopsis leaves (Nietzsche et al., 2014). Following these data and given the rapidly emerging role of SnRK1 in steering multiple growth- and defense-related processes in plants (Robaglia et al., 2012; Hulsmans et al., 2016), it is tempting to speculate that the TOR-SnRK1 regulatory module acts as an integrative rheostat on which multiple signaling pathways impinge, enabling effective fine-tuning of the balance between growth and defense in a timely and cost-efficient manner.
During infection, pathogens engage in a battle to accommodate their metabolic needs and often exploit host machinery that controls cellular metabolic processes. Considering the vital role of TOR in coordinating and integrating a wide array of basic cellular processes, it is not inconceivable that microbes attempt to subvert host metabolism by targeting TOR. Supporting this hypothesis, it has been reported that TOR is strongly activated by the Cauliflower Mosaic Virus (CaMV) TAV protein, with TAV-TOR binding being critical for viral fitness (Schepetilnikov et al., 2011). Moreover, TAV reduces accumulation of SA and, consequently, lowers basal defense to P. syringae pv. tomato in a TOR-dependent manner (Zvereva et al., 2016). Alternatively, or in parallel, pathogens may usurp plant growth-promoting hormones to subvert host TOR signaling and induce a state of susceptibility. Xoo and many other pathogens produce and secrete auxin themselves and also increase plant auxin biosynthesis and/or signaling on infection (Fu et al., 2011; De Vleesschauwer et al., 2013). Recent evidence showed that auxin triggers TOR activation followed by S6K1 phosphorylation and dissociation and efficient translation reinitiation of polycistronic mRNAs (Schepetilnikov et al., 2011, 2017). Other microbe-produced growth hormones such as cytokinins, gibberellins and brassinosteroids are also thought to activate TOR signaling, potentially explaining their immune-suppressive effects (Deprost et al., 2007; Schepetilnikov et al., 2013; Dong et al., 2015; Zhang et al., 2016).
Despite the agricultural and economic importance of growth-defense balance, past breeding efforts have mostly focused on optimizing and maximizing growth-related traits, tipping the energetic balance to favor growth over defense (Strange & Scott, 2005). In this context, understanding the molecular processes governing plant prioritization and diversion of resources towards growth or defense can provide powerful tools to genetically tailor plants that optimize this balance to maximize fitness under diverse agricultural settings. Based on the results presented in this study, timely alterations of TOR activity distinctly in specific cell types to limit pathogen replication and augmenting protective immune responses may be one approach to circumvent growth-defense trade-offs. Likewise, targeted overexpression of SnRK1 under control of a tightly regulated and stress-inducible promoter may break the link between enhanced stress tolerance and growth restriction. Considering the vital importance of TOR and SnRK1 in orchestrating myriad cellular and metabolic activities that drive cellular growth and immune function, further elucidating the action mechanisms of TOR and unraveling the molecular intricacies of TOR-SnRK1 interplay will create new avenues for developing crops that can maximize biomass accumulation while efficiently defending against stress.
Supplementary Material
Table S1 Primers used for gene cloning and qRT-PCR.
Table S2 List of upregulated genes in rice suspension cells treated for 24 h with 100 μM rapamycin.
Table S3 List of downregulated genes in rice suspension cells treated for 24 h with 100 μM rapamycin.
Table S5 GO terms associated with genes differentially upregulated by rapamycin.
Table S6 GO terms associated with genes differentially downregulated by rapamycin.
Table S7 T1 segregation analysis and characterization of TOR and Raptor transgenic lines used in this study.
Table S8 Query genes used for RiceNet analysis.
Table S9 Top 100 OsTOR interacting proteins as predicted by RiceNet.
Table S10 GO terms associated with the top 100 OsTOR interactors as predicted by RiceNet.
Table S11 RiceCyc metabolic pathways associated with the top100 OsTOR interactors as predicted by RiceNet.
Table S12 Protein domains significantly enriched among the top 100 OsTOR interactors as predicted by RiceNet.
Table S4 Mapman analysis of differentially expressed genes in rice suspension cells treated for 24 h with 100 μM rapamycin.
Fig. S1 Gene structure, protein domain architecture and phylogeny of OsTOR.
Fig. S2 Gene structure, protein domain architecture and phylogeny of the rice Raptor homologs OsRaptor1 and OsRaptor2.
Fig. S3 Influence of OsTOR on cell elongation, senescence and vegetative plant growth.
Fig. S4 Rice growth and resistance to Xanthomonas oryzae pv. oryzae (Xoo) are inversely correlated.
Fig. S5 TOR suppresses immunity in different dicot pathosystems.
Fig. S6 Chemical disruption of TOR signaling by rapamycin and Torin2 has no significant effect on the alkalinization response of PAMP-treated rice cells.
Fig. S7 Effect of rapamycin treatment on the expression of the pathogen associated molecular pattern triggered immunity marker genes OsPAL2 and OsMLO in rice suspension cells treated with insoluble crab shell chitin, Pseudomonas aeruginosa-derived lipopolysaccharides, or fungal xylanase.
Methods S1 Plant materials and growth conditions.
Methods S2 Arabidopsis pathogenicity assays.
Methods S3 Lettuce–Bremia lactucae infection assay.
Acknowledgments
We thank Dr Christian Meyer for sharing seeds of Arabidopsis TOR mutants and Nathalie Van Hese and Silke Deketelaere for their help with the Bremia and Verticillium disease assays. This work was supported by the Special Research Fund of Ghent University (GOA 01GB3013), Research Foundation Flanders (FWO G.0833.12N, FWO postdoctoral fellowship given to D.D.V.), the National Institute of Health (NIH, GM55962) and the National Science Foundation (NSF, IOS-081773) to PCR. The authors also acknowledge the financial support from the Hercules Foundation of the Flemish Government (AUGE/11/016) for the UHPLC-Q Exactive™ mass spectrometry equipment.
Footnotes
MR OSVALDO FILIPE (Orcid ID : 0000-0002-8591-2468)
PROF. MONICA HÖFTE (Orcid ID : 0000-0002-0850-3249)
Author contributions
D.D.V., H.S.S., K.D., P.R. and M.H. conceived and designed the experiments. D.D.V., O.F., G.H., H.S.S., A.H., P.C. and E.D.W. performed the experiments. D.D.V., O.F., H.S.S., J.V.B. and M.H. analyzed the data. P.R. contributed reagents, materials, analysis tools. D.D.V., O.F. and M.H. wrote the paper.
Supporting Information
Additional Supporting Information may be found online in the Supporting Information tab for this article:
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
References
- Bakshi A, Moin M, Kumar MU, Reddy AB, Ren M, Datla R, Siddiq EA, Kirti PB. Ectopic expression of Arabidopsis Target of Rapamycin (AtTOR) improves water-use efficiency and yield potential in rice. Sci Rep. 2017;7:42835. doi: 10.1038/srep42835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadirl Y, Yang L, Hetzel J, Dangl JL, Chory J. The growth-defense pivot: crisis management in plants mediated by LRR-RK surface receptors. Trends in Biochemical Sciences. 2014;39(10):447–456. doi: 10.1016/j.tibs.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson M, Dandekar AM, Chory J, Dehesh K. Brassinosteroid’s multi-modular interaction with the general stress network customizes stimulus-specific responses in Arabidopsis. Plant Science. 2016;250:165–177. doi: 10.1016/j.plantsci.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Brauc S, De Vooght E, Claeys M, Hofte M, Angenon G. Influence of over-expression of cytosolic aspartate aminotransferase on amino acid metabolism and defence responses against Botrytis cinerea infection in Arabidopsis thaliana. Journal of Plant Physiology. 2011;168(15):1813–1819. doi: 10.1016/j.jplph.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Caldana C, Li Y, Leisse A, Zhang Y, Bartholomaeus L, Fernie AR, Willmitzer L, Giavalisco P. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant Journal. 2013;73(6):897–909. doi: 10.1111/tpj.12080. [DOI] [PubMed] [Google Scholar]
- Campos ML, Yoshida Y, Major IT, Ferreira DD, Weraduwage SM, Froehlich JE, Johnson BF, Kramer DM, Jander G, Sharkey TD, et al. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nature Communications. 2016;7:12570. doi: 10.1038/ncomms12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern M, Fitzgerald HA, Canlas PE, Navarre DA, Ronald PC. Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Molecular Plant–Microbe Interactions. 2005;18(6):511–520. doi: 10.1094/MPMI-18-0511. [DOI] [PubMed] [Google Scholar]
- Curvers K, Seifi H, Mouille G, de Rycke R, Asselbergh B, Van Hecke A, Vanderschaeghe D, Hofte H, Callewaert N, Van Breusegem F, et al. Abscisic acid deficiency causes changes in cuticle permeability and pectin composition that influence tomato resistance to Botrytis cinerea. Plant Physiology. 2010;154(2):847–860. doi: 10.1104/pp.110.158972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bruyne L, Hofte M, De Vleesschauwer D. Connecting growth and defense: The emerging roles of brassinosteroids and gibberellins in plant innate immunity. Molecular Plant. 2014;7(6):943–959. doi: 10.1093/mp/ssu050. [DOI] [PubMed] [Google Scholar]
- De Storme N, Geelen D. The Arabidopsis mutant jason produces unreduced first division restitution male gametes through a parallel/fused spindle mechanism in meiosis II. Plant Physiology. 2011;155(3):1403–1415. doi: 10.1104/pp.110.170415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D, Gheysen G, Hofte M. Hormone defense networking in rice: tales from a different world. Trends in Plant Science. 2013;18(10):555–565. doi: 10.1016/j.tplants.2013.07.002. [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D, Seifi HS, Filipe O, Haeck A, Huu SN, Demeestere K, Hofte M. The DELLA protein SLR1 integrates and amplifies salicylic acid- and jasmonic acid-dependent innate immunity in rice. Plant Physiology. 2016;170(3):1831–1847. doi: 10.1104/pp.15.01515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D, Xu J, Hofte M. Making sense of hormone-mediated defense networking: from rice to Arabidopsis. Frontiers in Plant Science. 2014;5:611. doi: 10.3389/fpls.2014.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D, Yang YN, Cruz CV, Hofte M. Abscisic acid-induced resistance against the brown spot pathogen Cochliobolus miyabeanus in rice involves MAP kinase-mediated repression of ethylene signaling. Plant Physiology. 2010;152(4):2036–2052. doi: 10.1104/pp.109.152702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debode J, Declercq B, Hofte M. Identification of cauliflower cultivars that differ in susceptibility to Verticillium longisporum using different inoculation methods. Journal of Phytopathology. 2005;153(5):257–263. [Google Scholar]
- Deng KX, Yu LH, Zheng XZ, Zhang K, Wang WJ, Dong P, Zhang JK, Ren MZ. Target of rapamycin is a key player for auxin signaling transduction in Arabidopsis. Frontiers in Plant Science. 2016;7:291. doi: 10.3389/fpls.2016.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolai M, Bedu M, Robaglia C, Meyer C. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Reports. 2007;8(9):864–870. doi: 10.1038/sj.embor.7401043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deravel J, Lemiere S, Coutte F, Krier F, Van Hese N, Bechet M, Sourdeau N, Hofte M, Lepretre A, Jacques P. Mycosubtilin and surfactin are efficient, low ecotoxicity molecules for the biocontrol of lettuce downy mildew. Applied Microbiology and Biotechnology. 2014;98(14):6255–6264. doi: 10.1007/s00253-014-5663-1. [DOI] [PubMed] [Google Scholar]
- Din FVN, Valanciute A, Houde VP, Zibrova D, Green KA, Sakamoto K, Alessi DR, Dunlop MG. Aspirin inhibits mTOP signaling, activates AMP-activated protein kinase, and induces autophagy in colorectal cancer cells. Gastroenterology. 2012;142(7):1504–1515. doi: 10.1053/j.gastro.2012.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel T, Caldana C, Hanson J, Robaglia C, Vincentz M, Veit B, Meyer C. TOR signaling and nutrient sensing. Annual Review of Plant Biology. 2016;67:261–285. doi: 10.1146/annurev-arplant-043014-114648. [DOI] [PubMed] [Google Scholar]
- Dobrenel T, Marchive C, Sormani R, Moreau M, Mozzo M, Montane MH, Menand B, Robaglia C, Meyer C. Regulation of plant growth and metabolism by the TOR kinase. Biochemical Society Transactions. 2011;39:477–481. doi: 10.1042/BST0390477. [DOI] [PubMed] [Google Scholar]
- Dong P, Xiong FJ, Que YM, Wang K, Yu LH, Li ZG, Ren MZ. Expression profiling and functional analysis reveals that TOR is a key player in regulating photosynthesis and phytohormone signaling pathways in Arabidopsis. Frontiers in Plant Science. 2015;6:677. doi: 10.3389/fpls.2015.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichmann R, Schafer P. Growth versus immunity – a redirection of the cell cycle? Current Opinion in Plant Biology. 2015;26:106–112. doi: 10.1016/j.pbi.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nature Reviews Drug Discovery. 2014;13(2):140–156. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Liu HB, Li Y, Yu HH, Li XH, Xiao JH, Wang SP. Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiology. 2011;155(1):589–602. doi: 10.1104/pp.110.163774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havko NE, Major IT, Jewell JB, Attaran E, Browse J, Howe GA. Control of carbon assimilation and partitioning by jasmonate: an accounting of growth-defense tradeoffs. Plants. 2016;5(1):7. doi: 10.3390/plants5010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Fullerton MD, Ross FA, Schertzer JD, Chevtzoff C, Walker KJ, Peggie MW, Zibrova D, Green KA, Mustard KJ, et al. The ancient drug salicylate directly activates AMP-activated protein kinase. Science. 2012;336(6083):918–922. doi: 10.1126/science.1215327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. Targets for cell-cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253(5022):905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- Henriques R, Bogre L, Horvath B, Magyar Z. Balancing act: matching growth with environment by the TOR signalling pathway. Journal of Experimental Botany. 2014;65(10):2691–2701. doi: 10.1093/jxb/eru049. [DOI] [PubMed] [Google Scholar]
- Herms DA, Mattson WJ. Plant growth and defense. Trends in Ecology & Evolution. 1994;9(12):488–488. doi: 10.1016/0169-5347(94)90319-0. [DOI] [PubMed] [Google Scholar]
- Hindupur SK, Gonzalez A, Hall MN. The opposing actions of target of rapamycin and AMP-activated protein kinase in cell growth control. Cold Spring Harbor Perspectives in Biology. 2015;7:a019141. doi: 10.1101/cshperspect.a019141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsmans S, Rodriguez M, De Coninck B, Rolland F. The SnRK1 energy sensor in plant biotic interactions. Trends in Plant Science. 2016;21(8):648–661. doi: 10.1016/j.tplants.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, He SY. Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Molecular Plant. 2014;7(8):1267–1287. doi: 10.1093/mp/ssu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Current Biology. 2004;14(10):885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ. False idolatry of the mythical growth versus immunity tradeoff in molecular systems plant pathology. Physiological and Molecular Plant Pathology. 2016;95:55–59. [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Seo YS, Coltrane D, Hwang S, Oh T, Marcotte EM, Ronald PC. Genetic dissection of the biotic stress response using a genome-scale gene network for rice. Proceedings of the National Academy of Sciences, USA. 2011;108(45):18548–18553. doi: 10.1073/pnas.1110384108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Oh T, Yang S, Shin J, Hwang S, Kim CY, Kim H, Shim H, Shim JE, Ronald PC, et al. RiceNet v2: an improved network prioritization server for rice genes. Nucleic Acids Research. 2015;43(W1):W122–W127. doi: 10.1093/nar/gkv253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Duran R, Zipfel C. Trade-off between growth and immunity: role of brassinosteroids. Trends in Plant Science. 2015;20(1):12–19. doi: 10.1016/j.tplants.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Maegawa K, Takii R, Ushimaru T, Kozaki A. Evolutionary conservation of TORC1 components, TOR, Raptor, and LST8, between rice and yeast. Molecular Genetics and Genomics. 2015;290(5):2019–2030. doi: 10.1007/s00438-015-1056-0. [DOI] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proceedings of the National Academy of Sciences, USA. 2002;99(9):6422–6427. doi: 10.1073/pnas.092141899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meteignier LV, El Oirdi M, Cohen M, Barff T, Matteau D, Lucier JF, Rodrigue S, Jacques PE, Yoshioka K, Moffett P. Translatome analysis of an NB-LRR immune response identifies important contributors to plant immunity in Arabidopsis. Journal of Experimental Botany. 2017;68(9):2333–2344. doi: 10.1093/jxb/erx078. [DOI] [PubMed] [Google Scholar]
- Miki D, Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant and Cell Physiology. 2004;45(4):490–495. doi: 10.1093/pcp/pch048. [DOI] [PubMed] [Google Scholar]
- Naseem M, Wolfling M, Dandekar T. Cytokinins for immunity beyond growth, galls and green islands. Trends in Plant Science. 2014;19(8):481–484. doi: 10.1016/j.tplants.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Nietzsche M, Schiessl I, Bornke F. The complex becomes more complex: protein-protein interactions of SnRK1 with DUF581 family proteins provide a framework for cell and stimulus type-specific SnRK1 signaling in plants. Frontiers in Plant Science. 2014;5:54. doi: 10.3389/fpls.2014.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Park CJ, Ronald PC. Quantitative measurements of Xanthomonas oryzae pv. oryzae distribution in rice using fluorescent-labeling. Journal of Plant Biology. 2011;54(4):269–274. [Google Scholar]
- Pieterse CMJ, Leon-Reyes A, Van der Ent S, Van Wees SCM. Networking by small-molecule hormones in plant immunity. Nature Chemical Biology. 2009;5(5):308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- Popa C, Li L, Gil S, Tatjer L, Hashii K, Tabuchi M, Coll NS, Arino J, Valls M. The effector AWR5 from the plant pathogen Ralstonia solanacearum is an inhibitor of the TOR signalling pahtway. Science Reports. 2016;6:27058. doi: 10.1038/srep27058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz MU, Gifford ML, Schafer P. Hormone activities and the cell cycle machinery in immunity-triggered growth inhibition. Journal of Experimental Botany. 2015;66(8):2187–2197. doi: 10.1093/jxb/erv106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren MZ, Qiu SQ, Venglat P, Xiang DQ, Feng L, Selvaraj G, Datla R. Target of rapamycin regulates development and ribosomal rna expression through kinase domain in Arabidopsis. Plant Physiology. 2011;155(3):1367–1382. doi: 10.1104/pp.110.169045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren MZ, Venglat P, Qiu SQ, Feng L, Cao YG, Wang E, Xiang DQ, Wang JH, Alexander D, Chalivendra S, et al. Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell. 2012;24(12):4850–4874. doi: 10.1105/tpc.112.107144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexin D, Meyer C, Robaglia C, Veit B. TOR signalling in plants. Biochemical Journal. 2015;470:1–14. doi: 10.1042/BJ20150505. [DOI] [PubMed] [Google Scholar]
- Robaglia C, Thomas M, Meyer C. Sensing nutrient and energy status by SnRK1 and TOR kinases. Current Opinion in Plant Biology. 2012;15(3):301–307. doi: 10.1016/j.pbi.2012.01.012. [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JDG. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annual Review of Phytopathology. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann-Haefelin E, Sabatini DM, Blackwell TK. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/Foxo. Cell Metabolism. 2012;15(5):713–724. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh K, Kondoh H, Sasaya T, Shimizu T, Choi IR, Omura T, Kikuchi S. Selective modification of rice (Oryza sativa) gene expression by rice stripe virus infection. Journal of General Virology. 2010;91:294–305. doi: 10.1099/vir.0.015990-0. [DOI] [PubMed] [Google Scholar]
- Schepetilnikov M, Dimitrova M, Mancera-Martinez E, Geldreich A, Keller M, Ryabova LA. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO Journal. 2013;32(8):1087–1102. doi: 10.1038/emboj.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Kobayashi K, Geldreich A, Caranta C, Robaglia C, Keller M, Ryabova LA. Viral factor TAV recruits TOR/S6K1 signalling to activate reinitiation after long ORF translation. EMBO Journal. 2011;30(7):1343–1356. doi: 10.1038/emboj.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Makarian J, Srour O, Geldreich A, Yang Z, Chicher J, Hammann P, Ryabova LA. GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. EMBO Journal. 2017;36(7):886–903. doi: 10.15252/embj.201694816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B, Ronald PC. Plant innate immunity: perception of conserved microbial signatures. Annual Review of Plant Biology. 2012;63:451–482. doi: 10.1146/annurev-arplant-042811-105518. [DOI] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, et al. A receptor kinas-like protein encoded by the rice disease resistance gene, XA21. Science. 1995;270(5243):1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Song Y, Zhao G, Zhang X, Li L, Xiong F, Zhuo F, Zhang C, Yang Z, Datla R, Ren M, et al. The crosstalk between target of rapamycin (TOR) and jasmonic acid (JA) signaling existing in Arabidopsis and cotton. Sci Rep. 2017;7:45830. doi: 10.1038/srep45830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange RN, Scott PR. Plant disease: a threat to global food security. Annual Review of Phytopathology. 2005;43:83–116. doi: 10.1146/annurev.phyto.43.113004.133839. [DOI] [PubMed] [Google Scholar]
- Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M. A guide to using MapMan to visualize and compare omics data in plants: a case study in the crop species, maize. Plant, Cell & Environment. 2009;32(9):1211–1229. doi: 10.1111/j.1365-3040.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- Xiong FJ, Dong P, Liu M, Xie G, Wang K, Zhuo F, Feng L, Yang L, Li Z, Ren MZ. Tomato FK506 binding protein 12KD (FKBP12) mediates the interaction between rapamycin and target of rapamycin (TOR) Frontiers in Plant Science. 2016;7:1746. doi: 10.3389/fpls.2016.01746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang CB, Sheen J. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature. 2013;496(7444):181–+. doi: 10.1038/nature12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. Journal of Biological Chemistry. 2012;287(4):2836–2842. doi: 10.1074/jbc.M111.300749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiology. 2014;164(2):499–512. doi: 10.1104/pp.113.229948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. Novel links in the plant TOR kinase signaling network. Current Opinion in Plant Biology. 2015;28:83–91. doi: 10.1016/j.pbi.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZZ, Zhu JY, Roh J, Marchive C, Kim SK, Meyer C, Sun Y, Wang WF, Wang ZY. TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Current Biology. 2016;26(14):1854–1860. doi: 10.1016/j.cub.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvereva AS, Golyaev V, Turco S, Gubaeva EG, Rajeswaran R, Schepetilnikov MV, Srour O, Ryabova LA, Boller T, Pooggin MM. Viral protein suppresses oxidative burst and salicylic acid-dependent autophagy and facilitates bacterial growth on virus-infected plants. New Phytologist. 2016;211(3):1020–1034. doi: 10.1111/nph.13967. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Primers used for gene cloning and qRT-PCR.
Table S2 List of upregulated genes in rice suspension cells treated for 24 h with 100 μM rapamycin.
Table S3 List of downregulated genes in rice suspension cells treated for 24 h with 100 μM rapamycin.
Table S5 GO terms associated with genes differentially upregulated by rapamycin.
Table S6 GO terms associated with genes differentially downregulated by rapamycin.
Table S7 T1 segregation analysis and characterization of TOR and Raptor transgenic lines used in this study.
Table S8 Query genes used for RiceNet analysis.
Table S9 Top 100 OsTOR interacting proteins as predicted by RiceNet.
Table S10 GO terms associated with the top 100 OsTOR interactors as predicted by RiceNet.
Table S11 RiceCyc metabolic pathways associated with the top100 OsTOR interactors as predicted by RiceNet.
Table S12 Protein domains significantly enriched among the top 100 OsTOR interactors as predicted by RiceNet.
Table S4 Mapman analysis of differentially expressed genes in rice suspension cells treated for 24 h with 100 μM rapamycin.
Fig. S1 Gene structure, protein domain architecture and phylogeny of OsTOR.
Fig. S2 Gene structure, protein domain architecture and phylogeny of the rice Raptor homologs OsRaptor1 and OsRaptor2.
Fig. S3 Influence of OsTOR on cell elongation, senescence and vegetative plant growth.
Fig. S4 Rice growth and resistance to Xanthomonas oryzae pv. oryzae (Xoo) are inversely correlated.
Fig. S5 TOR suppresses immunity in different dicot pathosystems.
Fig. S6 Chemical disruption of TOR signaling by rapamycin and Torin2 has no significant effect on the alkalinization response of PAMP-treated rice cells.
Fig. S7 Effect of rapamycin treatment on the expression of the pathogen associated molecular pattern triggered immunity marker genes OsPAL2 and OsMLO in rice suspension cells treated with insoluble crab shell chitin, Pseudomonas aeruginosa-derived lipopolysaccharides, or fungal xylanase.
Methods S1 Plant materials and growth conditions.
Methods S2 Arabidopsis pathogenicity assays.
Methods S3 Lettuce–Bremia lactucae infection assay.


