Abstract
Background
In recent influenza seasons, the live attenuated influenza vaccine (LAIV) has not demonstrated the same level of vaccine effectiveness as that observed among children who received the inactivated influenza vaccine (IIV). To better understand this difference, this study compared the mRNA sequencing transcription profile (RNA seq) in children who received either IIV or LAIV.
Methods
Children 3–17 years of age receiving quadrivalent influenza vaccine were enrolled. Blood samples were collected on Day 0 prior to vaccination and again on Day 7 (range 6–10 days) following vaccination. Total RNA was isolated from PAXgene tubes and sequenced for a custom panel of 89 transcripts using the TruSeq Targeted RNA Expression method. Fold differences in normalized RNA seq counts from Day 0 to Day 7 were calculated, log2 transformed and compared between the two vaccine groups.
Results
Of 72 children, 46 received IIV and 26 received LAIV. Following IIV vaccination, 7 genes demonstrated significant differential expression at Day 7 (down-regulated). In contrast, following LAIV vaccination, 8 genes demonstrated significant differential expression at Day 7 (5 up-regulated and 3 down-regulated). Only two genes demonstrated similar patterns of regulation in both groups.
Conclusions
Differential regulation of genes was observed between 2015–16 LAIV and IIV recipients. These results help to elucidate the immune response to influenza vaccines and may be related to the difference in vaccine effectiveness observed in recent years between LAIV and IIV.
Keywords: Influenza vaccine, antibodies, RNA sequencing
INTRODUCTION
In recent influenza seasons, the live attenuated influenza vaccine (LAIV) has offered limited protection from influenza to children in the United States (U.S.). Specifically, the vaccine effectiveness (VE) of LAIV against any influenza in children 2–17 years old was 7% (95%CI = −46 to 40%) in 2013–2014 [1] and 26% (95%CI = 3 to 44) in 2014–2015 [2]. LAIV’s VE against influenza A/H3N2 was especially low among children 2–17 years in 2014–2015 (−6% VE; 95%CI = −40% to 20%), while LAIV offered reasonable protection (74% VE (95%CI= 25% to 91%)) against the less commonly occurring influenza B [2]. In the prior (2013–2014) season, VE of LAIV against (A/H1N1) was poor [1], but influenza A/H1N1 did not circulate widely in 2014–15, thus VE was not calculated. The reason for the reported decline in LAIV VE from previous seasons [3, 4] is unclear. Initially believed to be due to heat instability of the A/H1N1 construct in 2014–2015 [5, 6], manufacturer’s modifications of the vaccine did not improve VE in 2015–2016 season [7]. In response to this low VE, the Advisory Committee on Immunization Practices does not currently support the use of LAIV, thereby removing the intranasal delivery mechanism preferred by many children. Studies are needed to help to determine what accounts for the differences in VE between LAIV and inactivated influenza vaccine (IIV).
In a previous study of children ages 3 to 17 years, post 2014–2015 influenza vaccination antibody titers were significantly higher in response to IIV than to LAIV [8]. Moreover, microneutralization antibodies measuring cross-reactive protection against influenza were significantly higher among recipients of IIV than LAIV. It is likely that other immune responses, including cellular or mucosal immune responses, differ between LAIV and IIV recipients.
RNA sequencing is a technique that has been used to examine the cellular transcriptome such as gene expression, and provides a snapshot at the time of blood collection. Changes in the transcriptome before and after vaccination may identify transcription profiles and pathways of response that can be used for future vaccine development.
This study was designed to compare the RNA seq responses to the 2015–2016 IIV versus LAIV in children during the last year when LAIV use was recommended in the U.S. Specifically, selected gene profiles known to be related to viral infection, as well as the innate and adaptive immune responses to influenza, were compared.
METHODS
This study was approved by the University of Pittsburgh Institutional Review Board. Written informed consent was obtained from the participants’ parents.
Subjects, Eligibility and Enrollment
This study was conducted before influenza was circulating in the region [9]. From September through December 2015, a convenience sample of healthy children aged 3 through 17 years who planned to receive influenza vaccine were recruited from 3 primary care offices in the Pittsburgh area. Exclusion criteria included body weight <17 kilograms, having an immunosuppressive disease, taking immunosuppressive medicine or high dose oral steroids, pregnancy, and having a severe allergy to the influenza vaccine or its components. Parents could choose which form of the quadrivalent vaccine they preferred and the children were grouped by the vaccine received. All children received quadrivalent vaccines: IIV was either Fluzone (Sanofi Pasteur) or Fluarix (GlaxoSmithKline) as determined by their health insurance. Quadrivalent LAIV was FluMist (MedImmune). Demographic data were provided by the families and/or extracted from the electronic medical record.
Specimen Collection and Processing
On Day 0, participants provided blood samples prior to receiving influenza vaccine as part of their clinical care; they returned on Day 7 (range 6–10 days) for an additional blood draw. Within 4–6 hours of collection, blood samples collected into PAXgene tubes (Becton Dickinson) were centrifuged at 3200 rpm for 10–15 minutes and stored at −70°Celcius until further analysis.
RNA sequencing
Total RNA was isolated from PAXgene tubes using the PAXgene Blood RNA Kit following the manufacturer’s protocol. The 89 transcripts selected for testing were based on the knowledge of the investigators and local experts of genes known to have a role in influenza and other viral infections and a review of the medical literature for prior similar studies (See Supplemental Tables for a list of all transcripts). Both LAIV and IIV recipients were tested for all 89 transcripts. RNA quality and quantity were assessed by Agilent Tapestation 2200 and Invitrogen Qubit 2.0 fluorometer. Using 50 ng of total RNA, a multiplexed gene expression profiling panel of the selected targets was performed using the TruSeq Targeted RNA Expression Reference Guide, Rev. C[10]. Single-read 50 base pairs (bp) sequencing was performed on an Illumina NextSeq 500 with an average of 2 million reads per sample. Sequencing libraries were normalized to 2nM and then pooled.
Alignment
The banded Smith-Waterman algorithm was used to align clusters from each sample against the amplicon sequences provided in the kits to determine similar regions between 2 sequences, comparing segments of all possible lengths. If the start of a read matched a probe sequence with no more than 1 mismatch, the full length of the read was aligned against the amplicon target for that sequence. Alignments that included more than 3 indels (insertions or deletions) were excluded from alignment results.
Statistical Analyses
Depth of sequencing, variance estimation and calculation of p and q values
Depth of sequencing was used to compare samples for differences between Day 0 and Day 7. For each RNA transcript, the raw counts and the geometric mean of aligned read counts were calculated for all samples. A scaling factor (median of the ratio of raw counts to the geometric mean) was then used to normalize raw counts, dividing raw counts by the scaling factor to enable comparison of non-differentially expressed transcripts. The bias inherent in estimating variance is correlated with the number of biological replicates; thus, variance was calculated using data from all sample IDs rather than calculating for each sample name separately. The squared coefficient of variation was calculated by dividing the raw variance by the square of the mean.
Fold differences in normalized RNA seq counts from Day 0 to Day 7 were calculated and log2 transformed. A negative binomial distribution was used to model the normalized transcript abundance to derive a P-value for the differential expression (up- or down-regulations) of each transcript. Q-values were computed using the Benjamini-Hochberg procedure to control the false discovery rate by correcting for multiple hypothesis testing.
Patient characteristics
Demographic characteristics of recipients, including age groups (3–8 years vs. 9–17 years), sex, race (Black vs. non-Black), health insurance (public vs. private insurance), parent’s educational level (high school or less vs. some college or higher degree), and household smoking status, were compared by vaccine type (LAIV vs. IIV) using Chi-square or Fisher’s exact tests. Statistical significance was set at P < 0.05.
RESULTS
Seventy-two participants provided adequate Day 0 and Day 7 blood samples for analysis. The demographic characteristics of IIV recipients did not differ from LAIV recipients (Table 1). Differentially expressed genes (DEGs) with a p-value < 0.05 were identified and are shown in Table 2. Following IIV vaccination, 7 genes demonstrated significant differential expression at Day 7, all down-regulated; whereas, LAIV vaccination resulted in a total of 8 genes with significant differential expression at Day 7, with 5 up-regulated and 3 down-regulated. Two genes with significant differential expression at Day 7 were common to both vaccine types, both were down-regulated (CCL 7 and CXCR4). Most interesting was the striking difference between the two vaccines, where IIV vaccination resulted in down-regulated 7 genes compared to LAIV vaccination which resulted primarily in up-regulation of 5 different genes. Figure 1, a heat map of the gene expression, shows the differences between RNA seq responses to the two vaccine types.
Table 1.
Demographic Characteristics of Participants Overall and by 2015–2016 Influenza Vaccine Type
| Variable | Total N=72 n (%) |
LAIV* N=26 n (%) |
IIV** N=46 n (%) |
P-value† |
|---|---|---|---|---|
| Age 9–17 years, ref. = 3–8 years | 58 (80.6) | 20 (76.9) | 38 (82.6) | 0.558 |
| Male, ref. = female | 31 (43.1) | 9 (34.6) | 22 (47.8) | 0.277 |
| Black race, ref. = white and others | 62 (86.1) | 22 (84.6) | 40 (87.0) | 0.783 |
| Parent’s education ≥ some college, ref. = high school or less | 40 (55.6) | 16 (61.5) | 24 (52.2) | 0.442 |
| Public health insurance, ref. = private insurance | 64 (88.9) | 24 (92.3) | 40 (87.0) | 0.488 |
| Smoker in household | 31 (43.1) | 8 (30.8) | 23 (50.0) | 0.113 |
Live attenuated influenza vaccine
Inactivated influenza vaccine
By Chi-square test
Table 2.
Differential Gene Expression in Response to Vaccination with Inactivated Influenza Vaccine (IIV) and Live Attenuated Influenza Vaccine (LAIV)
| Inactivated Influenza Vaccine (IIV) | Live Attenuated Influenza Vaccine (LAIV) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Name | Normalized Mean Counts | Fold Change | Log2 Fold Change | P- value | Q-value | Normalized Mean Counts | Fold Change | Log2 Fold Change | P- value | Q- value | ||
| Day 0 | Day 7 | Day 0 | Day 7 | |||||||||
| CCL 2 | 2250.4 | 666.4 | 0.30 | −1.76 | 0.000 | 0.000 | 1222.5 | 982.8 | 0.80 | −0.31 | 0.418 | 0.995 |
|
| ||||||||||||
| CCL 7 | 218.2 | 21.7 | 0.10 | −3.33 | 0.000 | 0.000 | 148.2 | 15.2 | 0.10 | −3.29 | 0.000 | 0.000 |
|
| ||||||||||||
| CRP | 0.0 | 0.3 | 10.8 | 3.44 | 0.469 | 0.964 | 4.0 | 0.0 | 0.01 | −6.77 | 0.009 | 0.127 |
| CXCL 10 | 194.5 | 214.3 | 1.10 | 0.14 | 0.699 | 0.964 | 118.6 | 297.5 | 2.50 | 1.33 | 0.016 | 0.186 |
|
| ||||||||||||
| CXCR 4 | 85358.2 | 65535.9 | 0.77 | −5.06 | 0.005 | 0.090 | 96750.5 | 69100.4 | 0.71 | −0.49 | 0.024 | 0.208 |
|
| ||||||||||||
| IFI 6 | 5583.3 | 5884.0 | 1.10 | 0.08 | 0.891 | 0.964 | 3132.5 | 10545.8 | 3.40 | 1.75 | 0.000 | 0.007 |
| IFIT 3 | 5185.3 | 5437.8 | 1.00 | 0.07 | 0.697 | 0.964 | 3065.8 | 7878.8 | 2.60 | 1.36 | 0.001 | 0.036 |
| IFNA 1 | 13.3 | 2.5 | 0.19 | −2.40 | 0.014 | 0.175 | 11.3 | 5.6 | 0.50 | −1.01 | 0.449 | 0.995 |
| IL 12B | 13.9 | 1.7 | 0.12 | −3.02 | 0.011 | 0.163 | 3.6 | 5.5 | 1.50 | 0.62 | 0.635 | 0.995 |
| IL 4 | 5.3 | 0.2 | 0.03 | −0.38 | 0.001 | 0.038 | 79.5 | 96.3 | 1.20 | 0.28 | 0.597 | 0.995 |
| IL 8 | 33833.4 | 23295.4 | 0.69 | −0.54 | 0.002 | 0.038 | 30873.1 | 26617.8 | 0.86 | −0.21 | 0.309 | 0.995 |
| MX 1 | 6148.1 | 6471.1 | 1.10 | 0.07 | 0.707 | 0.964 | 4289.6 | 8382.4 | 2.00 | 0.97 | 0.022 | 0.208 |
| TNFSF 10 | 29765.0 | 35952.3 | 1.20 | 0.27 | 0.102 | 0.665 | 22680.5 | 39467.1 | 1.70 | 0.80 | 0.025 | 0.208 |
NOTE: The two genes that are common between the two vaccine types are shaded in gray; bolded P-values indicate significance. Normalized mean counts obtained for each gene on Day 0 pre-, or Day 7 post influenza vaccination; fold change refers to the ratio of mean counts on Day 0 and Mean Counts on Day 7.
Figure 1. Heat maps of RNA-seq responses to IIV and LAIV.
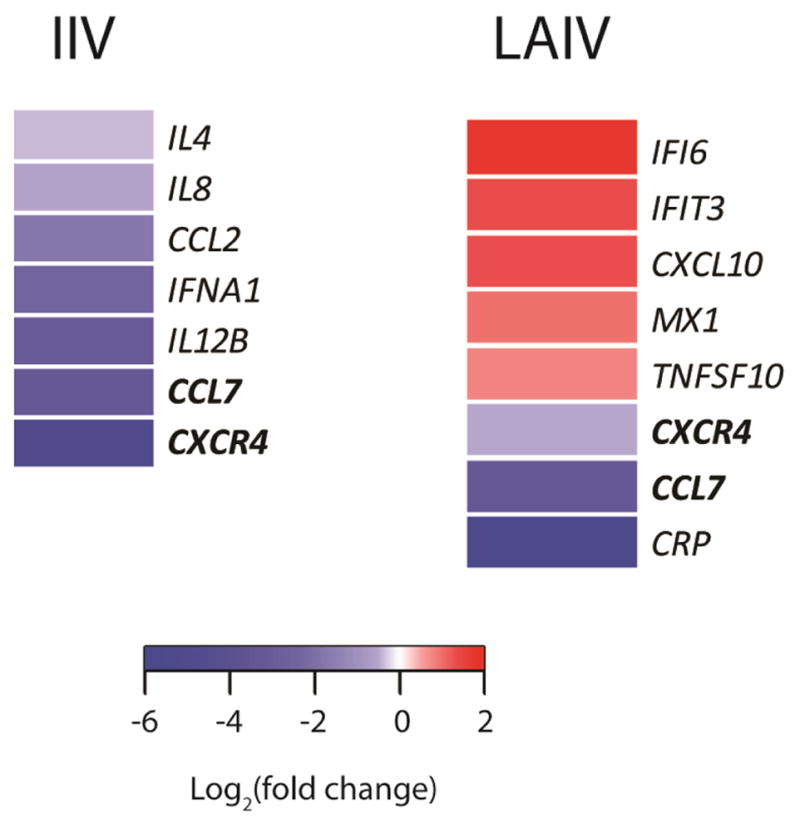
Heat maps representing differential expression of genes between Day 0 (pre) and Day 7 (post) vaccination based on P<0.05. IIV = inactivated influenza vaccine; LAIV = live attenuated influenza vaccine
Discussion
Systems biology approaches have been used in influenza research since 2011. The present study was designed to analyze RNA transcripts following IIV compared to LAIV vaccination during a year when the influenza vaccine was well-matched to the circulating strains. Our results demonstrated a remarkable difference between the two vaccines, where most of the genes expressed following IIV vaccination were down-regulated, while genes expressed after LAIV vaccination were predominantly upregulated. We found differences in transcriptional changes when comparing IIV with LAIV for the 2015-16 season; others have found differences in a previous season's vaccines [11]. The expression of two genes, CCL7 and CXCR4, demonstrated similar changes in magnitude and direction in both IIV and LAIV recipients.
The genes with significant differential expression in response to IIV included the T cell related cytokine interleukin (IL)4, which is a B cell stimulating cytokine produced by activated Type 2 T-helper cells (Th2) and IL12B (IL12p40) a cytokine subunit of IL12 and IL23, which prime Th1 and Th17 responses, respectively. This finding suggests decreased T cell activation 7 days post-vaccination. Interferon alpha 1 (IFN A1), a canonical antiviral gene was also downregulated by vaccination. Finally, monocyte and neutrophil chemokines and receptors were decreased, including CCL2 (MCP-1), CCL7 (MCP-3), and CXCR4 (SDF1 receptor), which mediate monocyte chemotaxis, and IL8, a prominent neutrophil chemoattractant. These data suggest an anti-inflammatory state 7 days post-vaccination.
Unlike the PBMC response to IIV, we found five upregulated DEGs following LAIV that indicates an interferon stimulated gene (ISG) response. This ISG response to LAIV was distinct from that observed with IIV. Among these five genes, two are involved in inducing or regulating apoptosis of infected or abnormal cells: Interferon alpha Inducible Protein 6 (IFI6) and Tumor Necrosis Factor Superfamily member 10 (TFNSF10/TRAIL). Another two upregulated DEGs, Myxovirus Dynamin Like GTPase 1 (MX1) and Interferon Induced Protein with Tetratricopeptide Repeats 3 (IFIT3), are known inhibitors of influenza virus replication and promote interferon signaling. Finally, C-X-C Motif Chemokine Ligand 10 (CXCL10/IP-10) is a type II interferon induced gene that promotes lymphocyte chemotaxis. These upregulated DEGs are largely involved in the canonical interferon signaling pathway that is invoked in response to live virus infection. Why LAIV induces interferon signaling, while IIV did not at this time point, is unknown, but intriguing. The association of LAIV vaccination with upregulation of genes involved in apoptosis may be indications of alternate mechanisms of action [12], and warrant further evaluation of immune responses associated with LAIV vaccination.
The dramatic difference between transcriptomic responses to the two vaccines and previous findings that children who received LAIV generated a less robust IgG antibody response than those who received IIV [8] provide new insights to help explain the lower vaccine effectiveness of LAIV observed during the 2015–16 influenza season [7]. Cao et al., found differential expression in the IIV group of plasma cell-mediated and inflammation genes, but in the LAIV group found IFN and cell-cycle-related transcripts [11]. IIV has been found to induce expression of IFN genes at day 1 post vaccination, while LAIV was not found to be associated with these changes until day 7 [10]. It is possible that IIV induced gene expression occurs earlier than LAIV and this temporal difference in transcriptional regulation may affect protective immunity. The pattern of overexpression of IFN genes has been suggested as a biomarker of vaccination response [13]. Certain IFN-related DEGs were also correlated with antibody response. We add to this important work of Cao et al., by including a larger sample size of children using data from the last season that LAIV was widely used and a season when LAIV did not perform well, as reported by the US Flu VE Network [7].
Identification of predictive signatures of serologic response to vaccination is an emerging field of systems biology. Bucasas et al. were among the first to demonstrate a positive correlation between HA titers and upregulation of interferon-responsive genes early after IIV vaccination [14]. Elevated antibody titers to IIV are correlated with upregulation of B-cell specific transcripts, upregulation of immunoglobulin genes, and proliferation-associated genes. These responses may be predictors of vaccine-specific antibody and plasmablast responses at seven days post vaccination [15], as Kim et al. have shown a similar relationship between antibody titers and circulating plasmablasts following IIV vaccination [16]. More recently, a methodology has been described for isolating and characterizing RNA transcripts in peripheral blood cell subsets compared to whole blood following influenza vaccination [17].
This approach, combining systems biology and predictive modeling to vaccinology, provides a powerful tool for unraveling the molecular mechanisms of vaccine-induced immunity [18, 19]. Systems vaccinology has recently been used to successfully study the immune responses to influenza vaccination in adults [14, 17, 19–23]. The current study adds to the body of systems vaccinology literature for understanding immune response to influenza vaccine among children and highlights the many differences between responses to LAIV and IIV. It is important to note that these differences may not reflect potential differences in response to infection with wild type influenza compared with LAIV [13, 24].
Strengths and Limitations
The primary strength of this study is that it uses one of few data sets available on the transcriptional response to the 2015–2016 influenza vaccine when LAIV had limited effectiveness but was still widely being used. Further, many of the children sampled had been enrolled in the study for two or more years, providing a unique opportunity to evaluate and compare immunity elicited by LAIV to that of IIV and the impact of prior vaccination. Investigators at Stanford confirmed limitations of using current serum antibody assays to monitor vaccine effectiveness, demonstrating that prior vaccination priming was critical for the effective immune response to the current vaccine year as measured by plasmablast and other more in-depth immune responses [25]. Of note, a larger proportion of children received IIV compared to LAIV, as this was a convenience sample of children for whom sufficient specimen volume for both Day 0 and Day 7 were available. A limited set of pre-selected genes was used for testing; however, this selection was based on the knowledge of local experts and review of the medical literature of prior similar studies. It is possible that a larger sample size or examination of other genes would generate additional results and that another mechanism explains the differences in response. Given the limitations in sample size, we report both P and Q values in Table 2, thereby allowing readers to determine which they prefer. Furthermore, these findings should be evaluated in the context of a broad spectrum of humoral antibody responses (IgG and IgA), protein expression and cell mediated responses in the same population. It has recently been suggested that obesity and exposure to cigarette smoke may each be associated with immune dysfunction and may affect response to vaccines [26, 27]. However, Smit et al. did not demonstrate a difference in rates of PCR confirmed influenza in vaccinated obese children compared with vaccinated non-obese children [28]. The current study was not large enough to examine these variables and future studies should further evaluate these relationships. Additional factors that may affect VE, which we could not evaluate in our study, include vaccine match and mutations due to passage of the vaccine virus in eggs during manufacturing [29].
Conclusion
Differential gene expressions were observed between children who received the 2015–16 IIV and those who received the LAIV. These transcriptional profiles help to elucidate the differences in the immune response to influenza vaccines and may help to explain why LAIV has not been as effective as IIV in recent years.
Supplementary Material
Supplemental Table 1. Inactivated Influenza Vaccine (IIV)
Supplemental Table 2. Live Attenuated Influenza Vaccine (LAIV)
Key points – Vaccine RNA seq paper.
Selected RNA sequencing was studied before and after LAIV and IIV administration in children.
After IIV vaccination, 7 genes demonstrated significant differential expression at Day 7 (down-regulated).
In contrast, following LAIV vaccination, 8 genes demonstrated significant differential expression at Day 7 (5 up-regulated and 3 down-regulated).
Only two genes demonstrated similar patterns of regulation in both groups (down-regulated).
Acknowledgments
Funding: This work was supported by the Centers for Disease Control and Prevention (CDC), through cooperative agreements with the University of Pittsburgh [grant U01 IP000467 and IP001035] and by the National Institutes of Health (NIH) [grant UL1 TR001857] to the University of Pittsburgh. The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the CDC or the NIH.
Footnotes
Clinical Trial Registry: N/A
Potential Conflict of Interest: Drs. Zimmerman has research funding from Sanofi Pasteur, Inc. Pfizer, Inc. and Merck & Co., Inc. Dr. Lin has research funding from Sanofi Pasteur, Inc. and Merck & Co, Inc. Dr. Nowalk has research funding from Pfizer, Inc. and Merck & Co., Inc. Dr. Alcorn has research funding from MedImmune, LLC and Merck & Co.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gaglani M, Pruszynski J, Murthy K, Clipper L, Robertson A, Reis M, Chung JR, Piedra PA, Avadhanula V, Nowalk MP, et al. Influenza Vaccine Effectiveness Against 2009 Pandemic Influenza A(H1N1) Virus Differed by Vaccine Type During 2013–2014 in the United States. The Journal of infectious diseases. 2016 doi: 10.1093/infdis/jiv577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimmerman RK, Nowalk MP, Chung J, Jackson ML, Jackson LA, Petrie JG, Monto AS, McLean HQ, Belongia EA, Gaglani M. 2014–2015 influenza vaccine effectiveness in the United States by vaccine type. Clinical Infectious Diseases. 2016;63(12):1564–1573. doi: 10.1093/cid/ciw635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohmit SE, Thompson MG, Petrie JG, Thaker SN, Jackson ML, Belongia EA, Zimmerman RK, Gaglani M, Lamerato L, Spencer SM, et al. Influenza Vaccine Effectiveness in the 2011–2012 Season: Protection Against Each Circulating Virus and the Effect of Prior Vaccination on Estimates. Clinical Infectious Diseases. 2014;58(3):319–327. doi: 10.1093/cid/cit736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osterholm MT, Kelley NS, Sommer A, Belongia EA. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. The Lancet Infectious diseases. 2012;12(1):36–44. doi: 10.1016/S1473-3099(11)70295-X. [DOI] [PubMed] [Google Scholar]
- 5.Cotter CR, Jin H, Chen Z. A single amino acid in the stalk region of the H1N1pdm influenza virus HA protein affects viral fusion, stability and infectivity. PLoS pathogens. 2014;10(1):e1003831. doi: 10.1371/journal.ppat.1003831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Donnell CD, Vogel L, Matsuoka Y, Jin H, Subbarao K. The matrix gene segment destabilizes the acid and thermal stability of the hemagglutinin of pandemic live attenuated influenza virus vaccines. Journal of virology. 2014;88(21):12374–12384. doi: 10.1128/JVI.01107-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson ML, Chung JR, Jackson LA, Phillips CH, Benoit J, Monto AS, Martin ET, Belongia EA, McLean HQ, Gaglani M, et al. Influenza Vaccine Effectiveness in the United States — 2015/16 Season. New England Journal of Medicine. 2017 doi: 10.1056/NEJMoa1700153. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine MZ, Martin JM, Gross FL, Jefferson S, Cole KS, Archibald CA, Nowalk MP, Susick M, Moehling K, Spencer S. Neutralizing Antibody Responses to Antigenically Drifted Influenza A (H3N2) Viruses among Children and Adolescents following 2014–2015 Inactivated and Live Attenuated Influenza Vaccination. Clinical and Vaccine Immunology. 2016;23(10):831–839. doi: 10.1128/CVI.00297-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russell K. Update: Influenza Activity—United States, October 4, 2015–February 6, 2016. MMWR Morbidity and mortality weekly report. 2016:65. doi: 10.15585/mmwr.mm6506a3. [DOI] [PubMed] [Google Scholar]
- 10.TruSeq Targeted RNA Expression Reference Guide Revision C. San Diego CA: 2016. Illumina. [Google Scholar]
- 11.Cao RG, Suarez NM, Obermoser G, Lopez SM, Flano E, Mertz SE, Albrecht RA, García-Sastre A, Mejias A, Xu H. Differences in antibody responses between trivalent inactivated influenza vaccine and live attenuated influenza vaccine correlate with the kinetics and magnitude of interferon signaling in children. Journal of Infectious Diseases. 2014;210(2):224–233. doi: 10.1093/infdis/jiu079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dou Y, Fu B, Sun R, Li W, Hu W, Tian Z, Wei H. Influenza vaccine induces intracellular immune memory of human NK cells. PloS one. 2015;10(3):e0121258. doi: 10.1371/journal.pone.0121258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu W, Higgs BW, Morehouse C, Streicher K, Ambrose CS, Woo J, Kemble GW, Jallal B, Yao Y. A whole genome transcriptional analysis of the early immune response induced by live attenuated and inactivated influenza vaccines in young children. Vaccine. 2010;28(16):2865–2876. doi: 10.1016/j.vaccine.2010.01.060. [DOI] [PubMed] [Google Scholar]
- 14.Bucasas KL, Franco LM, Shaw CA, Bray MS, Wells JM, Niño D, Arden N, Quarles JM, Couch RB, Belmont JW. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. Journal of Infectious Diseases. 2011;203(7):921–929. doi: 10.1093/infdis/jiq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan Y, Tamayo P, Nakaya H, Pulendran B, Mesirov JP, Haining WN. Gene signatures related to B-cell proliferation predict influenza vaccine-induced antibody response. European Journal of Immunology. 2014;44(1):285–295. doi: 10.1002/eji.201343657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JH, Mishina M, Chung JR, Cole KS, Nowalk MP, Martin JM, Spencer S, Flannery B, Zimmerman RK, Sambhara S. Cell-mediated immunity against antigenically drifted influenza A (H3N2) viruses in children during a vaccine mismatch season. Journal of Infectious Diseases. 2016;214(7):1030–1038. doi: 10.1093/infdis/jiw311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoek KL, Samir P, Howard LM, Niu X, Prasad N, Galassie A, Liu Q, Allos TM, Floyd KA, Guo Y. A cell-based systems biology assessment of human blood to monitor immune responses after influenza vaccination. PloS one. 2015;10(2):e0118528. doi: 10.1371/journal.pone.0118528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pulendran B. Systems vaccinology probing humanity’s diverse immunesystems with vaccines. Proceedings of the National Academy of Sciences. 2014;111(34):12300–12306. doi: 10.1073/pnas.1400476111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li G-M. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12(8):786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furman D, Jojic V, Kidd B, Shen-Orr S, Price J, Jarrell J, Tse T, Huang H, Lund P, Maecker HT. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Molecular systems biology. 2013;9(1):659. doi: 10.1038/msb.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, Germain RN, Wang E, Olnes MJ, Narayanan M, Golding H. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157(2):499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haralambieva IH, Ovsyannikova IG, Kennedy RB, Zimmermann MT, Grill DE, Oberg AL, Poland GA. Transcriptional signatures of influenza A/H1N1-specific IgG memory-like B cell response in older individuals. Vaccine. 2016;34(34):3993–4002. doi: 10.1016/j.vaccine.2016.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kennedy RB, Ovsyannikova IG, Haralambieva IH, Oberg AL, Zimmermann MT, Grill DE, Poland GA. Immunosenescence-related Transcriptomic and immunologic changes in Older individuals Following influenza Vaccination. Frontiers in immunology. 2016;7:1–15. doi: 10.3389/fimmu.2016.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer WA, II, Chason KD, Brighton M, Jaspers I. Live attenuated influenza vaccine strains elicit a greater innate immune response than antigenically-matched seasonal influenza viruses during infection of human nasal epithelial cell cultures. Vaccine. 2014;32(15):1761–1767. doi: 10.1016/j.vaccine.2013.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He X-S, Holmes TH, Sanyal M, Albrecht RA, García-Sastre A, Dekker CL, Davis MM, Greenberg HB. Distinct patterns of B-cell activation and priming by natural influenza virus infection versus inactivated influenza vaccination. The Journal of infectious diseases. 2014;211(7):1051–1059. doi: 10.1093/infdis/jiu580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng Y, Kong Y, Barnes PF, Huang F-F, Klucar P, Wang X, Samten B, Sengupta M, Machona B, Donis R. Exposure to cigarette smoke inhibits the pulmonary T-cell response to influenza virus and Mycobacterium tuberculosis. Infect Immun. 2011;79(1):229–237. doi: 10.1128/IAI.00709-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheridan PA, Paich HA, Handy J, Karlsson EA, Hudgens MG, Sammon AB, Holland LA, Weir S, Noah TL, Beck MA. Obesity is associated with impaired immune response to influenza vaccination in humans. International journal of obesity. 2012;36(8):1072–1077. doi: 10.1038/ijo.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smit MA, Wang H-L, Kim E, Barragan N, Aldrovandi GM, El Amin AN, Mascola L, Pannaraj PS. Influenza Vaccine is Protective Against Laboratory-confirmed Influenza in Obese Children. The Pediatric infectious disease journal. 2016;35(4):440–445. doi: 10.1097/INF.0000000000001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie H, Wan X-F, Ye Z, Plant EP, Zhao Y, Xu Y, Li X, Finch C, Zhao N, Kawano T. H3N2 mismatch of 2014–15 northern hemisphere influenza vaccines and head-to-head comparison between human and ferret antisera derived antigenic maps. Scientific reports. 2015:5. doi: 10.1038/srep15279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Inactivated Influenza Vaccine (IIV)
Supplemental Table 2. Live Attenuated Influenza Vaccine (LAIV)


